ABSTRACT
Serratia marcescens, a member of the order Enterobacterales, is adept at colonizing health care environments and is an important cause of invasive infections. Antibiotic resistance is a daunting problem in S. marcescens because, in addition to plasmid-mediated mechanisms, most isolates have considerable intrinsic resistance to multiple antibiotic classes. To discover endogenous modifiers of antibiotic susceptibility in S. marcescens, a high-density transposon insertion library was subjected to sub-MICs of two cephalosporins, cefoxitin, and cefepime, as well as the fluoroquinolone ciprofloxacin. Comparisons of transposon insertion abundance before and after antibiotic exposure identified hundreds of potential modifiers of susceptibility to these agents. Using single-gene deletions, we validated several candidate modifiers of cefoxitin susceptibility and chose ydgH, a gene of unknown function, for further characterization. In addition to cefoxitin, deletion of ydgH in S. marcescens resulted in decreased susceptibility to multiple third-generation cephalosporins and, in contrast, to increased susceptibility to both cationic and anionic detergents. YdgH is highly conserved throughout the Enterobacterales, and we observed similar phenotypes in Escherichia coli O157:H7 and Enterobacter cloacae mutants. YdgH is predicted to localize to the periplasm, and we speculate that it may be involved there in cell envelope homeostasis. Collectively, our findings provide insight into chromosomal mediators of antibiotic resistance in S. marcescens and will serve as a resource for further investigations of this important pathogen.
KEYWORDS: AmpC, Serratia marcescens, TIS, TnSeq, YdgH, cefepime, cefoxitin, cephalosporin, ciprofloxacin, fluoroquinolone
INTRODUCTION
Serratia marcescens, a member of the order Enterobacterales, was historically regarded as an environmental bacterium with low inherent pathogenicity (1, 2). However, over the past 50 years, it has been increasingly recognized as an important cause of invasive infections (3). S. marcescens can transiently colonize the gastrointestinal tract and skin and is a frequent cause of sporadic health care-associated pneumonia and urinary tract and bloodstream infections (4–7). Because S. marcescens is commonly isolated from tap water (3) and clinical isolates frequently have high nucleotide identity to environmental isolates, it is thought that many infections result from sporadic exposures (8). However, since many isolates produce tenacious biofilms (9) and can have intrinsic resistance to common biocides (10, 11), S. marcescens also causes hospital outbreaks (12), either through hand hygiene lapses or from a contaminated point source (13, 14). These outbreaks particularly affect vulnerable patients in adult and neonatal intensive care units (15, 16).
Antibiotic resistance is another crucial factor that is thought to facilitate S. marcescens infection of vulnerable hosts; recent receipt of antibiotics is a risk factor for S. marcescens infection (3). Expansion of S. marcescens in the gut under antibiotic pressure (17, 18) might facilitate transmission, either by allowing subsequent colonization of nosocomial point sources or by increasing the likelihood of incidental transmission by health care staff (19). S. marcescens is intrinsically resistant to polymyxins (20, 21) and often has elevated MICs to tetracyclines, macrolides, nitrofurantoin, and fosfomycin (22). Importantly, S. marcescens also encodes a chromosomal Ambler class C beta-lactamase, AmpC, which at basal levels imparts resistance to penicillins and early-generation cephalosporins (e.g., cephalexin, cefuroxime, and cefoxitin) (22). In most instances of infection, the majority of S. marcescens cells initially produce this basal, low level of AmpC. However, a small subpopulation, due to stochastic mutation that leads to high-level AmpC production, can be selected and amplified when patients are treated with late-generation cephalosporins (e.g., moxalactam, ceftriaxone, ceftazidime, and cefepime) (23, 24); in this way, S. marcescens can become resistant to broader-spectrum beta-lactams, because at high levels, AmpC can hydrolyze these antibiotics as well. Disturbingly, there have also been increasing reports of dissemination of S. marcescens clones containing a mobilizable chromosomal genomic island (25) containing the class D beta-lactamase SME, which efficiently hydrolyzes carbapenems (26–29). Infections with isolates that combine high-level expression of both AmpC and SME have been described (30); widespread dissemination of such highly beta-lactam-resistant clones, on a background of fluoroquinolone nonsusceptibility rates as high as 20% (https://sentry-mvp.jmilabs.com/), would leave clinicians to choose among only the most expensive and toxic last-line treatment options.
To comprehensively identify loci that contribute both to basal growth and to antibiotic susceptibility in S. marcescens, here, we use transposon-insertion site sequencing (TIS) mutagenesis, a powerful approach that couples transposon mutagenesis with DNA sequencing. In TIS, a library of mutants is created in which each bacterial cell harbors a transposon randomly inserted into the genome. Transposon insertions typically result in loss-of-function mutants, and in a high-density library, genes for which transposon-insertion mutants are absent or underrepresented in the library are often critical for growth in vitro (sometimes termed “essential” genes). These genes may be potential antibiotic targets.
The library can additionally be subjected to biologically relevant conditions, and analyzing the abundance of mutants before and after exposure can identify genes that contribute to pathogen fitness in said condition. Mutants that are underrepresented after exposure to a condition of interest correspond to loci important for survival under that condition (31). This approach has facilitated rapid genome-scale identification of genes that contribute to phenotypes of interest (32), such as to identify genes that alter fitness in ex vivo and in vivo models of infection (33, 34), to investigate the function of uncharacterized genes (35), and to identify genes that alter antibiotic susceptibility. For example, TIS has identified novel modifiers of antibiotic susceptibility in many pathogens, including Pseudomonas aeruginosa (36), Mycobacterium tuberculosis (37), and Klebsiella pneumoniae (38).
Here, we created a dense transposon library in S. marcescens and used TIS to provide genome-scale insight into genes that contribute to in vitro growth, as well as modifiers of cephalosporin and fluoroquinolone susceptibility. These analyses led to the identification of ydgH, a conserved gene that, when deleted, leads to decreased cephalosporin susceptibility.
RESULTS
Genes contributing to in vitro growth of S. marcescens ATCC 13880.
We began by generating a high-density transposon-insertion mutant library in a spontaneous streptomycin-resistant mutant of S. marcescens ATCC 13880, an environmental, nonclinical isolate that is the type strain for the species. Using a protocol adapted from Escherichia coli (39), we isolated nearly 2 million individual mutant colonies, each containing a genomic insertion of the mariner-based Himar1 transposon TnSC189 (40). Mariner transposons integrate at TA dinucleotides; the resulting pooled library included insertions at 57% (77,082) of possible (134,884) genomic TA sites. To ensure that our library had sufficient complexity to allow subsequent analyses, we determined the percentage of possible insertions achieved per gene. As expected for a high-complexity library (32), a histogram of the resulting percentages revealed a bimodal distribution with a minor peak consisting of genes tolerating relatively few insertions and a major peak of disrupted genes centered around 70% TA site disruption (Fig. 1A). Of the 4,363 S. marcescens genes annotated, 4,138 (94.8%) were isolated with at least one insertion (see Table S1 in the supplemental material). This allowed us to perform a comprehensive analysis of genes involved in in vitro growth of S. marcescens.
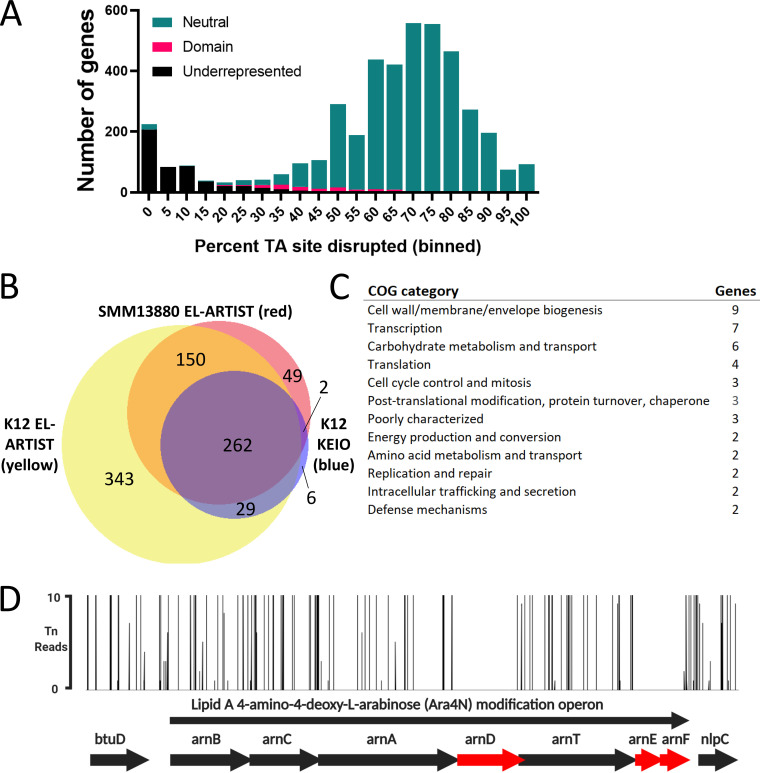
FIG 1.
Serratia marcescens ATCC 13880 transposon-insertion sequencing reveals genes important for in vitro growth. (A) S. marcescens genes binned by percentage of TA sites disrupted. Overlaid is the hidden Markov model-based analysis assignment (EL-ARTIST pipeline) of whether insertions within the gene are underrepresented, differ by domain within the gene (“domain”), or are found to be distributed neutrally within the gene. The left histogram peak contains predominantly underrepresented genes where insertions are tolerated in only a low percentage of sites. (B) Venn diagram illustrating overlap between S. marcescens ATCC 13880 (GenBank accession number CP072199) underrepresented genes (red) with E. coli K-12 underrepresented genes (yellow), with the K-12 KEIO collection as an additional comparator (blue). (C) Genes underrepresented in S. marcescens ATCC 13880 but not in E. coli K-12, either by EL-ARTIST analysis or by the analysis resulting from the KEIO collection. Clusters of orthologous genes (COG) categories of those genes are tabulated. (D) Lipid A 4-amino-4-deoxy-L-arabinose modification operon, demonstrating that transposon insertions are underrepresented (red) in arnD, arnE, and arnF. btuD and nlpC are genes flanking this operon.
We used a previously developed pipeline (ARTIST) that compares actual reads of insertion sites to multinomially resampled simulation and a hidden Markov model-based analysis. We and others have previously shown that this workflow, which has been used in several bacterial species, including Vibrio cholerae strains, V. parahaemolyticus, V. vulnificus, Edwardsiella piscicida, and multiple Escherichia coli strains (39, 41–45), allows accurate categorization of genes into the three categories of “neutral,” “domain,” and “underrepresented” (46, 47). Neutral genes such as entE, which synthesizes the siderophore enterobactin, may be crucial under certain physiologic conditions, but in vitro, in this case when grown in LB, tolerate transposon insertion throughout the span of the gene and are dispensable for growth (Fig. S1A, left). In contrast, underrepresented genes (often referred to as “essential” genes) such as purA, which encodes the adenylosuccinate synthetase involved in purine metabolism, can sustain insertions at few or no sites while still allowing growth (Fig. S1A, middle). Finally, “domain” genes, such as helD, which encodes a DNA helicase involved in unwinding of duplex DNA, can be found with insertions in certain domains or regions of the gene but not in others (Fig. S1A, right).
In this analysis, out of the 4,363 S. marcescens genes annotated, we identified 483 underrepresented for growth in LB, as well as an additional 104 domain genes (Table S2). The remaining 3,771 genes were classified as neutral. Compared to a prior analysis in S. marcescens, we identified fewer genes as essential for in vitro growth, although a strict comparison is difficult since this prior effort used a clinical strain and utilized a lower-density library with 32,000 unique insertion mutants (34, 48). Binning by clusters of orthologous genes (COG) functional category revealed that, as expected, the most frequent categories for underrepresented genes were for core cellular processes such as translation (including many tRNA synthetases and ribosomal proteins), cell envelope biogenesis (including enzymes involved in peptidoglycan and lipopolysaccharide [LPS] synthesis), and coenzyme metabolism (including enzymes involved in central metabolism) (Table S2). These categories are common for essential genes in other organisms (49).
S. marcescens was formerly classified as a member of the Enterobacteriaceae, but modern genome-based phylogenetics have reassigned Serratia species into the sister Yersiniaceae family. We were eager to identify both underrepresented genes shared between S. marcescens and the common model E. coli lab strain, E. coli K-12, as well as those specific to S. marcescens, and so compared those identified here to those previously identified using the same approach in E. coli K-12 (39), as well as to those identified in E. coli K-12 using single-gene knockouts (the KEIO collection described previously [50]). Emphasizing the conserved physiology across the order Enterobacterales, of the 463 underrepresented genes we identified in S. marcescens that were also identified in E. coli (with an E value of 1 × 10−10 and percent identity of >30%) (51), 412 (89%) (Fig. 1B) were also underrepresented in E. coli K-12 by transposon insertion; of the 299 genes in E. coli K-12 identified as essential by single-gene knockout that were also identified in S. marcescens, 264 (88%) were also underrepresented in our analysis (Fig. 1B).
Genes underrepresented in S. marcescens (but not in E. coli K-12) (n = 49; Table S3) were most commonly assigned to COG functional categories that included cell envelope biogenesis (9 genes), transcription (7 genes), and carbohydrate metabolism and transport (6 genes) (Fig. 1C). Additional investigation of these genes may identify divergent biology in S. marcescens that could be targets for novel narrow-spectrum antimicrobials. An intriguing example involves the lipid A 4-amino-4-deoxy-l-arabinose (Ara4N) modification (arn) operon. In Enterobacteriaceae as well as in Pseudomonas aeruginosa, this operon, when upregulated by PhoP/PhoQ, can lead to decreased susceptibility to cationic polypeptides like polymyxins by addition of positively charged Ara4N moieties to lipid A (52). In S. marcescens, which is known to be intrinsically resistant to polymyxins through arn (20), we detected an absence of insertions in arnD, arnE, and arnF (Fig. 1D).
In addition to the arn operon described above, compared to E. coli K-12, we also found that two of the three components of the AcrAB-TolC RND family multidrug efflux pump were underrepresented in S. marcescens ATCC 13880. mlaE, which encodes the inner membrane permease component that facilitates the transport of cell membrane lipids between the inner and outer membranes was underrepresented as well (Table S3) (53). Components of peptidoglycan recycling are also underrepresented in S. marcescens (compared to E. coli K-12); these include nlpD, which serves to activate cell wall amidases that act in daughter cell separation (54), and murQ, encoding a component of the cytoplasmic peptidoglycan recycling machinery (55).
Antibiotic screen in S. marcescens ATCC 13880.
We then subjected our high-density insertion library to antibiotics with the goal of discovering novel loci that modify antibiotic susceptibility. We focused our initial efforts here on cephalosporins and fluoroquinolones, as they are the antibiotic classes with the fastest worldwide growth in consumption (56) and are currently clinically useful against serious S. marcescens infections (23, 57). Within the cephalosporins, we chose cefoxitin, an early-generation cephalosporin that is readily hydrolyzed by the AmpC beta-lactamase and to which S. marcescens is relatively resistant, and cefepime, a late-generation cephalosporin that is negligibly hydrolyzed by AmpC and to which S. marcescens is relatively susceptible (24). We chose ciprofloxacin because, among the fluoroquinolones, its MICs are typically lowest for S. marcescens (https://sentry-mvp.jmilabs.com/) (Fig. 2A). We performed this screen at sub-MICs that over the course of the treatment resulted in a ≤10-fold decrease in CFU (Fig. 2B; see also Fig. S1B); this preserved library diversity so that when we sequenced the resulting libraries, we could identify enough TA insertion mutants per gene to allow us to identify genes that had small as well as large effects on growth and survival (32). Sequencing the resulting libraries allowed us to identify genes that when disrupted by transposon insertion (presumably resulting in null or hypofunction) led to depletion or enrichment of that mutant when exposed to antibiotic (Table S4). Genes with fewer insertion mutants (depleted) represent candidate loci that support resistance, whereas genes with more abundant insertion mutants (enriched) represent genes that may support susceptibility. As expected, under the screening conditions, we measured robust induction of AmpC by cefoxitin (Fig. S1C) (24).
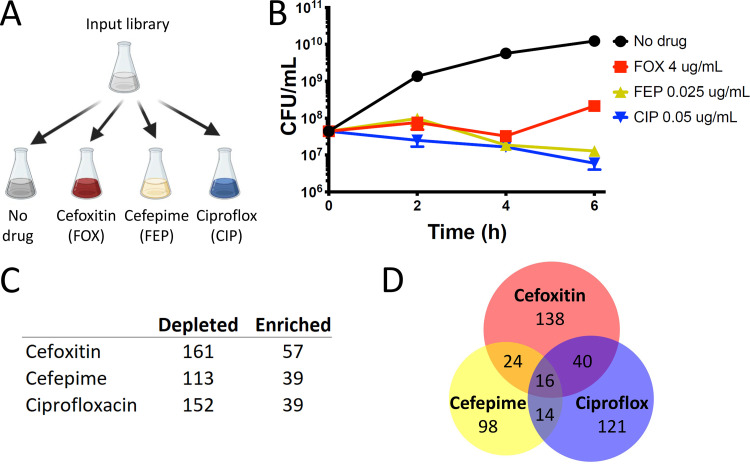
FIG 2.
Antibiotic screen revealing S. marcescens genes important for growth and survival. (A) Screen schematic. An input library (containing about 2 million insertion mutants) is grown to an optical density (OD) of 0.1 and divided between the following 4 conditions for an additional 6 h of growth: LB alone; LB plus cefoxitin 4 μg/ml; LB plus cefepime 0.025 μg/ml; and LB plus ciprofloxacin 0.05 μg/ml. (B) Under the screening conditions, the library without drug selection undergoes a more than 2-log expansion, while those with antibiotic decrease in CFU to a similar extent (about 0.5 log), with the library in cefoxitin undergoing expansion after 4 h, likely due to upregulation of AmpC beta-lactamase. Libraries were harvested for analysis at 6 h. (C) Number of genes with insertion mutants showing a ≥4-fold change at 6 h with a Mann-Whitney U P value of ≤ 0.01, compared to outgrowth in LB alone. All data are tabulated in Table S4 in the supplemental material. (D) Venn diagram illustrating genes showing coordinate enrichment or depletion. Individual genes comprising the depicted sets are tabulated in Table S5.
Compared to outgrowth of the input library in no antibiotic, we found 57, 39, and 39 genes enriched and 161, 113, and 152 genes depleted in cefoxitin, cefepime, and ciprofloxacin, respectively (P ≤ 0.01; ≥4-fold change in abundance) (Fig. 2C and Table S5). Many of the genes depleted in ciprofloxacin act in pathways related to the mechanism of action of fluoroquinolone antibiotics, including those involved in DNA replication (such as holE) and DNA damage repair (such as recD, recG, and xseA), supporting the validity of our approach (Fig. S1D and Table S5) (58, 59). Similarly, we identified many genes depleted in cefepime related to peptidoglycan homeostasis (such as nlpD, pbpG, slt, and dacA) and envelope integrity (such as nlpA) (Fig. S1E and Table S5) (60).
Analysis of coordinately significantly enriched or depleted genes allowed identification of genes that are candidate modifiers of susceptibility to multiple antibiotics (Fig. 2D and Table S5). As we predicted, in all 3 antibiotics, we identified enrichment in insertion mutants in ompF, which encodes an outer membrane porin that facilitates the permeation of cephalosporins and hydrophilic fluoroquinolones like ciprofloxacin (61–63). All 3 also had enrichment of lonP, which encodes a key bacterial serine protease whose deletion has been shown to lead to increased efflux of diverse antibiotics (64, 65). Interestingly, we also found enrichment in cefoxitin, cefepime, and ciprofloxacin in the setting of slyA insertion. SlyA is a member of the MarR family of transcriptional regulators known to be activated by small molecules; it is activated by salicylate, and its best-characterized role seems to be as a countersilencer, alleviating H–NS-mediated repression (66). It has a diverse regulon (67), but it has not previously been reported to be involved in regulation of antibiotic susceptibility. Further study is needed to characterize this and other genes we have identified (Fig. 2D and Table S5) in which insertion leads to coordinate depletion or enrichment in multiple antibiotics.
Modifiers of cefoxitin susceptibility.
To better understand modifiers of intrinsic resistance in S. marcescens, we focused our attention on modifiers of cefoxitin susceptibility, to which basal AmpC levels provide some baseline resistance. Of the 161 genes significantly depleted in cefoxitin (Fig. 3A), we identified many predicted to participate in peptidoglycan homeostasis, such as multiple membrane-bound lytic murein transglycosylases (mltC, mltD, and mltF), as well as penicillin-binding protein 1B (mrcB) and dihydrodipicolinate synthase (dapA) (Fig. 3B). Importantly, mutants in ampC were also depleted, while we saw enrichment in mutants in the N-acetylmuramoyl-l-alanine amidase gene paralog that we previously denoted amiD2 and suggested may be the most important for AmpC derepression in S. marcescens (68).
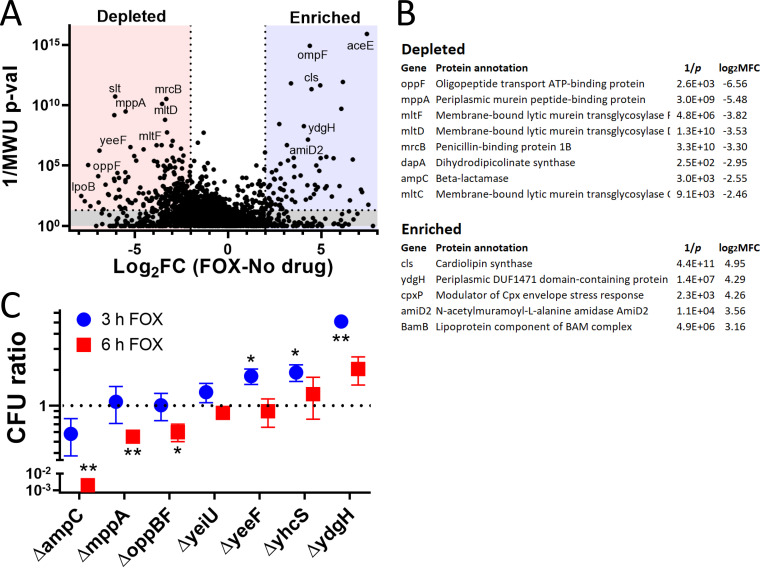
FIG 3.
Whole-genome screen for modifiers of cefoxitin susceptibility. (A) Volcano plot illustrating candidate genes important for cefoxitin susceptibility. On the x axis is the log2 fold change (log2FC) in insertion mutant abundance in cefoxitin (FOX) compared to that in no drug. The y axis is the inverse Mann-Whitney U P value (1/MWU p-val), which roughly measures the concordance between mutants with insertions at individual TA sites across a gene. Genes were depleted (red) if the log2FC was ≤2 and the 1/MWU p-val was ≥100. Genes were enriched (blue) if the log2FC was ≥2 and the 1/MWU p-val was ≥100. (B) Selected genes enriched or depleted in cefoxitin were tabulated. Genes important for envelope integrity and peptidoglycan recycling were depleted. ydgH, an enriched, poorly characterized gene, is also highlighted. (C) Growth of various S. marcescens gene deletion mutants in cefoxitin 4 μg/ml. Growth is depicted as CFUmutant/CFUWt (in cefoxitin) divided by CFUmutant/CFUWt (in LB alone). Although no mutant had large defects in LB alone, due to stochastic errors in dilution to starting CFU, this improved the repeatability of the experiment. *, P ≤ 0.05; **, P ≤ 0.01; unpaired two-tailed t test, unadjusted.
Toward our aim of discovering novel modifiers of cefoxitin susceptibility, we specifically focused on genes in which insertion mutants were predominantly enriched or depleted in cefoxitin but not in cefepime and/or ciprofloxacin as well (Table S5). We constructed in-frame deletions in 5 largely uncharacterized loci identified in our screen that were depleted (mppA, oppBF, yeiU, yeeF, and yhcS) and in one locus (ydgH) that was enriched. We compared the growth of the mutants in cefoxitin to that of wild-type S. marcescens ATCC 13880 in cefoxitin in a separate test tube. We used the ΔampC mutant as a control. The final CFU ratio, expressed as CFUmutant/CFUWt (Wt, wild type) was also corrected for any minor differences in growth that were observed in mutants in LB alone. CFU were determined after both 3 and 6 h in cefoxitin (at the same concentration as that used in the screen).
As expected, we observed a large progressive decrease in the CFU ratio in the ΔampC mutant (Fig. 3C). We also observed less pronounced but significant decreases in CFU ratios at 6 h in the ΔmppA mutant and in a mutant with a deletion of the entire opp operon, the ΔoppBF mutant (oppB, oppC, and oppF were all depleted in the screen). In contrast, mutants with deletions of yeiU, yeeF, and yhcS, in which insertions were depleted in our pooled screen, did not have reduced CFU ratios when these single-gene deletion mutants were tested in isolation (Fig. 3C).
The ΔydgH mutant has decreased susceptibility to cefoxitin.
ydgH, which was enriched in the screen, had a large increase in CFU ratio at 3 h of cefoxitin (Fig. 3C). ydgH was originally identified through proteomic analyses as encoding a possible secreted effector in Salmonella enterica; however, YdgH was only detected in low abundance in samples from mutants deleted for a type III secretion system regulatory protein (and not in wild-type samples) (69). Efforts to identify cognate eukaryotic targets were unsuccessful (70).
In S. marcescens ATCC 13880, ydgH is predicted to encode a 951-amino-acid protein. ydgH is upstream of the ydgI gene; ydgI is predicted, by similarity to P. aeruginosa ArcD, to encode a putative arginine:ornithine antiporter (71). It is unlikely that ydgH and ydgI are part of an operon, as we identified high-confidence σ70 promoters 5′ to both open reading frames, as well as a transcriptional terminator 3′ to ydgH (Fig. S2) (72, 73). Furthermore, in contrast to ydgH, which contained enrichment of insertions throughout the gene in our TIS, there was no enrichment of insertions in ydgI in the presence of cefoxitin (Fig. 4A).
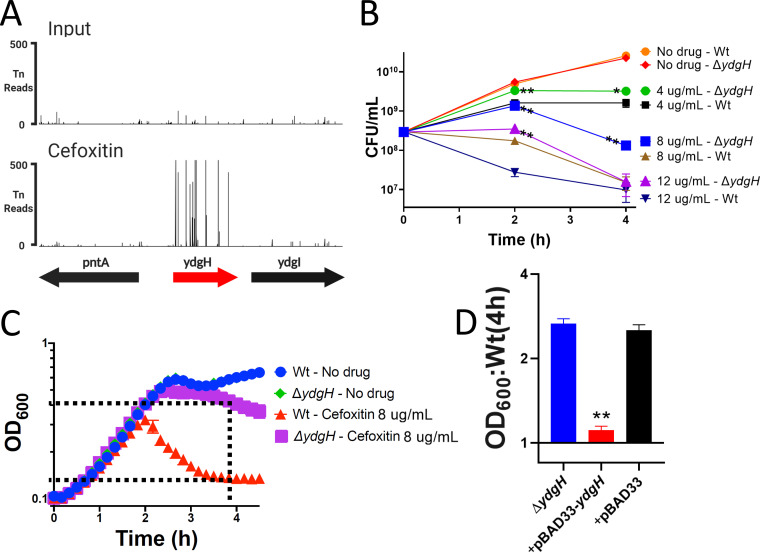
FIG 4.
YdgH contributes to basal S. marcescens cefoxitin susceptibility. (A) The YdgH locus on the y axis with transposon-insertion (Tn) reads on the y axis, demonstrating the large enrichment of YdgH insertion mutants in cefoxitin (below, compared to the input library; above, on the same scale). (B) The ΔydgH mutant had increased growth in cefoxitin compared to that of the wild type (Wt) at multiple drug concentrations and at multiple time points. (C) Schematic illustrating optical density at 600 nm (OD600) ratio used in subsequent figures. The OD600 of the ΔydgH mutant at 4 h is divided by that of the Wt. (D) Exogenous ydgH rescues the cefoxitin phenotype in the ΔydgH mutant. *, P ≤ 0.05; **, P ≤ 0.01; unpaired two-tailed t test, unadjusted.
We observed that the ΔydgH mutant had significantly higher CFU counts than those of the wild type (Wt) across a range of cefoxitin concentrations at both 2 and 4 h, but it grew indistinguishably from the Wt in the absence of cefoxitin (Fig. 4B). We noticed smaller relative differences between the ΔydgH mutant and the Wt at later time points in these experiments, as well as those shown in Fig. 3C, which we speculate results from upregulation of AmpC and eventual hydrolysis of cefoxitin in both the ΔydgH mutant and the Wt.
To enable higher-throughput screening of other compounds by spectrophotometry, for subsequent experiments, we used the optical density at 600 nm (OD600) ratio of the ΔydgH mutant to the Wt (as depicted in Fig. 4C). We first used this approach to confirm that the absence of ydgH was necessary and sufficient to confer enhanced growth in cefoxitin. The ΔydgH mutant was transformed with either empty pBAD33 or pBAD33 containing the ydgH open reading frame; we observed that pBAD33-ydgH rescued the cefoxitin phenotype but that the empty vector did not (Fig. 4D).
The ΔydgH mutant has increased susceptibility to detergents.
To identify if ydgH modifies susceptibility to other antibiotics, and to acquire initial clues to its function, we assayed the susceptibility of the ΔydgH mutant to a range of antibiotics and detergents. For many compounds tested, there was a narrow concentration range that was sufficiently inhibitory to allow us to assay for potential effects. To enhance the accessibility of this large data set, we present the spectrophotometric ratio in the main text figures and the full growth curves from which they are derived in the supplemental figures. The complete data at all concentrations tested are in Table S6.
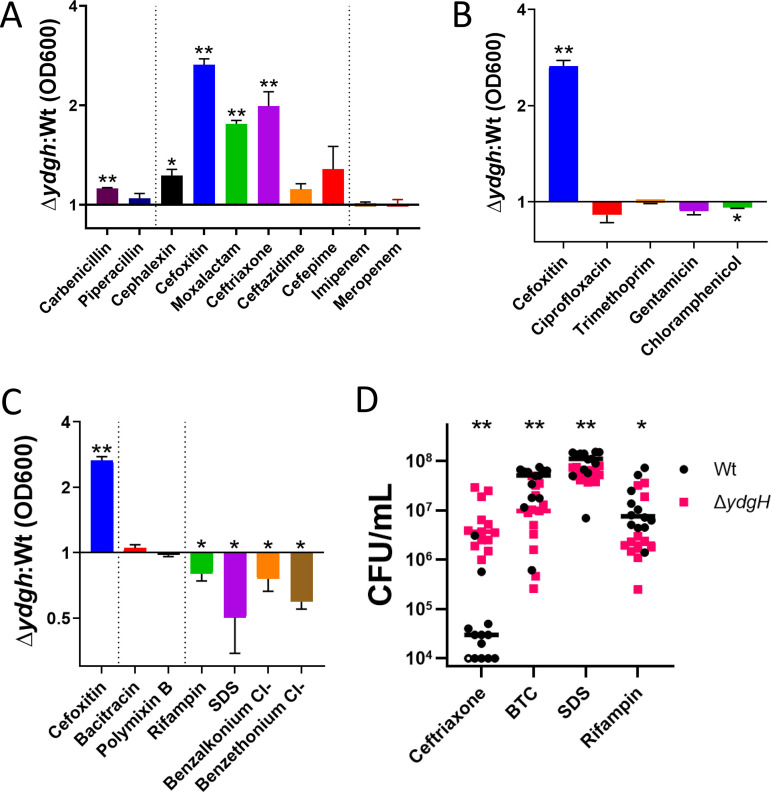
We began with beta-lactams; we found that in addition to the second-generation cephalosporin cefoxitin, the ΔydgH mutant also had significant reductions in susceptibility to the third-generation cephalosporins moxalactam and ceftriaxone but not to the first-generation cephalosporin cephalexin or the antipseudomonal cephalosporins cefepime or ceftazidime (Fig. 5A and Fig. S3). There was no prominent phenotype for the penicillins carbenicillin or piperacillin, nor for the carbapenems imipenem or meropenem (Fig. 5A and Fig. S4). These distinct phenotypes are not attributable to differences in inherent susceptibilities to the assayed antibiotics, as the third-generation cephalosporins, in which the mutant had phenotypes, and the antipseudomonal cephalosporins, in which the mutant did not, were active across similar concentration ranges in S. marcescens ATCC 13880 (Table S6). We hypothesized that the differences seen between the beta-lactams might be attributable to them being differential substrates for AmpC, but the ΔydgH mutant did not have different AmpC activity from that of the Wt (Fig. S4E).
FIG 5.
YdgH deletion leads to decreased cephalosporin susceptibility and increased detergent susceptibility. (A) OD600 ratios demonstrating that the ΔydgH mutant has decreased second- and third-generation cephalosporin susceptibility (including to cefoxitin, moxalactam, and ceftriaxone) but no large differences in susceptibility to first-generation cephalosporins, antipseudomonal cephalosporins, penicillins, or carbapenems. (B) OD600 ratios demonstrating that the ΔydgH mutant has no large differences in non-beta-lactam antibiotic susceptibility. Cefoxitin ratio reproduced for reference. (C) OD600 ratios demonstrating that the ΔydgH mutant has no large differences in bacitracin or polymyxin susceptibility, but has small but significant increases in susceptibility to rifampin, and more broadly to the detergents sodium dodecyl sulfate (SDS), benzethonium chloride, and benzalkonium chloride. Cefoxitin ratio reproduced for reference. (D) Viability experiments of the Wt and the ΔydgH mutant in ceftriaxone, benzethonium chloride (BTC), sodium dodecyl sulfate (SDS), and rifampin confirmed the results of the turbidity experiments. *, P ≤ 0.05; **, P ≤ 0.01; unpaired two-tailed t test (A, B, and C) or Wilcoxon rank-sum test (D), unadjusted.
In contrast to the differential phenotypes observed with different beta-lactam antibiotics, we did not see a strong effect for antibiotics with cytoplasmic targets, such as ciprofloxacin, trimethoprim, gentamicin, and chloramphenicol (Fig. 5B and Fig. S5), nor to antimicrobials to which S. marcescens has high intrinsic resistance, such as bacitracin and polymyxin B (Fig. 5C and Fig. S6). Intriguingly, in contrast to the decreased susceptibility observed with second- and third-generation cephalosporins, we observed small, but significant, increased susceptibility of the ΔydgH mutant (compared to the Wt) to rifampin, and more broadly to detergents, including the anionic detergent sodium dodecyl sulfate (SDS), as well as the cationic detergents benzalkonium chloride and benzethonium chloride (Fig. 5C and Fig. S6).
To confirm these findings from our higher-throughput turbidity screen, we performed viability analyses of Wt S. marcescens and the ΔydgH mutant at 4 h in ceftriaxone, benzethonium chloride, SDS, and rifampin. As predicted, we observed significantly increased viability of the ΔydgH mutant in ceftriaxone and significantly decreased viability in rifampin, SDS, and benzethonium chloride (Fig. 5D).
Conservation within Enterobacterales.
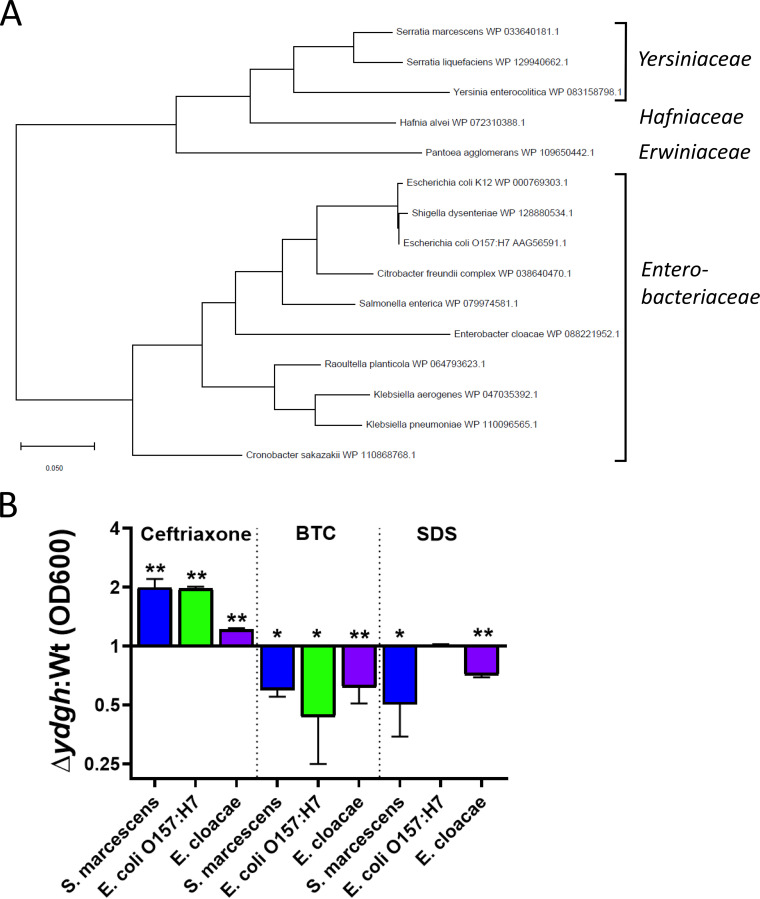
To gain further insight into this relatively uncharacterized gene, we performed a phylogenetic analysis and discovered that ydgH is closely conserved among the Enterobacterales (but not in other Gram-negative bacteria) (Fig. 6A). As expected, S. marcescens YdgH has greatest similarity to homologs identified in other Yersiniaceae species, followed by the closely related sister families Hafniaceae and Erwiniaceae, followed by the more distantly related Enterobacteriaceae. We hypothesized that if the function of YdgH was conserved, we would observe similar phenotypes in distantly related Enterobacterales. To test this idea, we constructed in-frame deletions of ydgH in the pathogens E. coli O157:H7 EDL933 and Enterobacter cloacae ATCC 13047 and assayed the resulting mutants in ceftriaxone, to which the S. marcescens ΔydgH mutant had decreased susceptibility, and in benzethonium chloride and SDS, to which the S. marcescens ΔydgH mutant had increased susceptibility. We used ceftriaxone for these assays because E. cloacae has high-level intrinsic resistance to cefoxitin.
FIG 6.
YdgH deletion results in conserved phenotypes in Escherichia coli O157:H7 EDL933 and Enterobacter cloacae ATCC 13047. (A) The YdgH phylogeny was constructed based on amino acid substitutions by using the maximum-likelihood method and the JTT matrix-based model in MEGA X. The relevant higher-order families are indicated. (B) All 3 mutants have decreased susceptibility to ceftriaxone (E. cloacae has high intrinsic resistance to cefoxitin, so ceftriaxone was chosen), as well as increased susceptibility to benzethonium chloride. Growth of the E. cloacae ΔydgH mutant was decreased in SDS, although growth of E. coli O157:H7 was not. Due to inherent differences in Wt susceptibility, drug concentrations differed considerably between isolates and were as follows: ceftriaxone, 4 μg/ml for E. cloacae, 0.04 μg/ml for E. coli O157:H7, and 0.06 μg/ml for S. marcescens; benzethonium chloride (BTC), 10 μg/ml for E. cloacae, 18 μg/ml for E. coli O157:H7, and 52 μg/ml for S. marcescens); SDS, 0.8% for E. cloacae, 0.13% for E. coli O157:H7, and 5% for S. marcescens. *, P ≤ 0.05; **, P ≤ 0.01; unpaired two-tailed t test, unadjusted.
We observed broadly consistent results, with the E. coli O157:H7 ΔydgH mutant having similar phenotypes in ceftriaxone and benzethonium chloride (although not in SDS), and the E. cloacae ΔydgH mutant having similar phenotypes in benzethonium chloride and SDS (although with only a small difference in ceftriaxone) (Fig. 6B).
DISCUSSION
Here, we present a genome-scale analysis of the essential genome of the type strain of S. marcescens, a medically important nosocomial pathogen. We report a curated resource of genes that alter susceptibility to beta-lactams and fluoroquinolones, arguably the two most useful antibiotic classes for treatment of S. marcescens infections. We also validate and characterize ydgH, a largely uncharacterized gene conserved in the Enterobacterales that, when deleted, leads to decreased susceptibility to second- and third-generation cephalosporins but increased susceptibility to ionic detergents.
A striking feature of the underrepresented genes we identified here in S. marcescens ATCC 13880 (compared to E. coli K-12) is their involvement in envelope biogenesis and homeostasis. These genes are involved in multiple structural and functional compartments, including in LPS modification (arnD, arnE, and arnF) and phospholipid transport (mlaE), as well as peptidoglycan regulation (murQ and nlpD). These results hint at potentially important differences in S. marcescens and E. coli envelope physiology. It is known that in addition to the higher Ara4N content in lipid A, S. marcescens also possesses additional core oligosaccharide substitutions (74, 75). We anticipate that further genetic investigations, including additional TIS to identify underrepresented genes in other relatively neglected Enterobacterales, as well as more detailed bioinformatic analyses using additional comparators, will suggest additional divergence in envelope biology, potentially enabling the development of narrow-spectrum antibiotics or antibiotic adjuvants.
The high-density library we created should be a useful resource for investigation of other clinically relevant phenotypes in S. marcescens. This approach has recently allowed identification of genes that facilitate bacteremia and the production of hemolysins in clinical strains of this pathogen (34, 76). Additional phenotypes crucial for pathogenesis, such as immune system evasion, biofilm formation, and colonization of target organs such as the bladder and lungs, will be of interest. The data set that we have generated here on cephalosporin and fluoroquinolone resistance-modifying genes should also serve as an important resource facilitating future investigations. While they cannot identify mutations in essential genes like classical selection screens and the sequencing of resistant clinical isolates, genome-scale transposon-based antibiotic screens are an important complement to these techniques because they can identify genes, such as ydgH, that regulate antibiotic susceptibility yet may have conditional tradeoffs in fitness, like those we identify here.
Our study has important limitations. We chose the model ATCC strain S. marcescens 13880 in which to conduct our screen because of its wide use and previous characterization, but clinical strains, which often accumulate mutations not only in antibiotic genes, might yield different results. Additionally, the results from our high-throughput screens, while subjected to analysis steps to limit false discoveries (46), nevertheless are susceptible to both statistical and experimental factors that necessitate any initial data be regarded as hypothesis generating. Exemplifying this caution are the results from Fig. 3, where we were able to validate only 50% of the genes that we predicted would modify cefoxitin susceptibility. This pattern is commonly seen in transposon-insertion mutagenesis analyses and may occur because subsequent single-mutant experiments are fundamentally different from the initial, high-throughput pooled experiment (where other mutants may complement for the one of interest) or be due to polar effects resulting from the disruption of downstream genes.
Additionally, inferring the biological function of underrepresented (“essential”) genes such as those identified here can be far from straightforward. Our finding, that arnD, arnE, and arnF are underrepresented in S. marcescens might be a good example. The most obvious explanation, about which we speculate above, is that the Ara4N modification to LPS is necessary in S. marcescens. However, if this is true, why would arnA, arnB, arnC, and arnT not also be underrepresented? There are numerous possibilities. The first is that Ara4N modification is not essential, and instead, arnD, a formylase, and arnEF, the two components of the flippase (77), may have other substrates (that are essential for growth in LB). A second possibility is that arnABCT has unidentified homologues that can substitute, enabling Ara4N modification. A final possibility might be that arnDEF, when mutated, causes the accumulation of toxic intermediates (analogous to recent findings questioning the previously posited involvement of colanic acid biosynthesis in the Rcs response) (78). Additional experimentation (such as with CRISPR interference [CRISPRi] or using single-gene deletion in a strain carrying an inducible plasmid) is needed to confirm the initial findings and ultimately differentiate between these possibilities.
Our initial characterization of ydgH suggests that it does not act directly through the beta-lactamase AmpC; we show here that beta-lactamase activity is not increased in the ΔydgH mutant. The increased susceptibility of ΔydgH to detergents is likely an important clue regarding its function. YdgH is predicted to localize to the periplasm based on bioinformatic analysis and proteomics data sets (79, 80) and may in fact be present there in high concentrations in Enterobacterales (81, 82). One possibility is that in the periplasm, YdgH plays a role in the maturation of outer membrane-bound biomolecules. In support of this speculation, overexpression of micA, a small RNA that is a primary effector of the σE envelope stress response, leads to upregulation of ydgH (83). Alternatively, its absence may lead to envelope stress, leading to activation of σE (60). Since downregulation of outer membrane porins that facilitate the entry of cefoxitin is a prime target of the σE response, this could explain both the increased susceptibility to detergents (and rifampin) and the decreased susceptibility to cefoxitin (and other cephalosporins particularly dependent on shared outer membrane porins for their entry).
Clearly, further mechanistic studies are needed to uncover the function of ydgH, as well as other genes that we identify here as modifiers of antibiotic susceptibility in S. marcescens. Such work will reveal novel bacterial cell biology, as well as potentially tractable new antibiotic targets.
MATERIALS AND METHODS
Transposon-insertion library construction.
S. marcescens ATCC 13880 was obtained from the ATCC, and a spontaneous streptomycin-resistant mutant was isolated on LB plus streptomycin (1,000 μg/ml). This mutant grew indistinguishably from the wild type in the absence of streptomycin. Conjugation was performed to transfer pSC189 (40) from E. coli SM10λpir to a spontaneous streptomycin-resistant mutant of S. marcescens ATCC 13880. Prior to conjugation, the two strains were cultured separately overnight in LB plus chloramphenicol and LB plus streptomycin, respectively, pelleted and washed twice in LB, resuspended 10-fold in LB, and combined 1:1 for a final volume of 50 μl on a 0.45-μm mixed-cellulose ester filter (Millipore) on an LB agar plate and incubated at 37°C for 1 h. The filter was then eluted with 750 μl LB, and the filter washed twice more with 500 μl LB to ensure that all transconjugants were eluted. Four such reaction mixtures were then combined and plated on 245- by 245-mm2 (Corning) LB agar plates containing chloramphenicol and streptomycin. After 16 h of incubation at 37°C, colonies were harvested in LB plus 25% glycerol (vol/vol) and stored at −80°C. This was repeated 3 times, generating aliquots of “A,” “B,” and “C” libraries. The OD of the resulting libraries was measured (after appropriate dilution) prior to freezing and ranged from 56 to 108.
Antibiotic screen.
One aliquot each of the above libraries were thawed and combined (in proportion to its OD) in 50 ml of LB to yield a final OD of 3. One-milliliter aliquots of each were then inoculated in 100 ml LB to roughly yield cultures with an OD of 0.03. Each resulting culture was incubated with continuous agitation in a 500-ml Erlenmeyer flask until an OD of 0.1. Twenty milliliters of the above-described culture were then added to 4- to 125- ml flasks, either an empty control flask or to flasks containing antibiotic, to make final concentrations of cefoxitin 4 μg/ml, cefepime 0.025 μg/ml, or ciprofloxacin 0.05 μg/ml. Preparatory experiments allowed estimation of the resulting CFU, so after 6 h, to enable plating of roughly equal number of colonies, 100 μl of the LB culture (after dilution in LB), 5 ml of the cefoxitin culture (after washing and concentration in LB), and 20 ml of the cefepime and ciprofloxacin cultures (after washing and concentration in LB) were plated on LB agar without antibiotics. Surviving colonies were allowed to grow for 16 h of incubation at 37°C before colonies were harvested and stored as described above. To determine beta-lactamase activity under screening conditions, samples from 2, 4, and 6 h after incubation with antibiotic were taken, essentially as described above, except that a single aliquot of the A library was thawed rather than the 3 libraries combined. At the appropriate time points, a 1-ml aliquot was taken, pelleted at 13,000 relative centrifugal force (RCF) for 5 min at room temperature, resuspended in 1 ml of 50 mM NaPO4 (pH 7) buffer, and flash frozen in liquid nitrogen and stored at −80°C. After thawing, samples were lysed on ice using a Sonic Dismembrator with one pulse of 10 s on setting 8. Samples were clarified at 4°C and 21,000 RCF for 10 min, and the supernatants were transferred to new tubes. A Qubit protein assay (Invitrogen) was used to determine protein concentrations. An aliquot (1 μg) of total protein was added to 7.8 nmol nitrocefin (Sigma) and absorbance at 495 nm measured kinetically for 10 min at room temperature. The slope of the line from the first 5 data points was used to measure beta-lactamase activity (with 1 unit of activity representing hydrolysis of 1 μM of nitrocefin per minute). Activity is expressed per gram of clarified lysate. AmpC is the only beta-lactamase identified in S. marcescens ATCC 13880, and ΔampC mutants have essentially no beta-lactamase activity, so beta-lactamase activity accurately represents AmpC activity. For the assay of AmpC activity in the ΔydgH mutant to allow greater accuracy of a sample with lower relative activity, 2-μg aliquots of similarly obtained and processed mid-logarithmic-phase samples were assayed.
Characterization of transposon-insertion libraries.
Libraries were prepared essentially as described in Warr et al. (39). Prior to sequencing using a MiSeq V3 cartridge, equimolar DNA fragments for the original harvested library, and the resulting libraries after growth in LB alone, cefoxitin, cefepime, and ciprofloxacin, were pooled after addition of barcodes to allow multiplexing. After trimming, reads were mapped, using Bowtie and allowing 1 mismatch, to the S. marcescens ATCC 13880 genome deposited below. Reads that mapped to multiple sites were randomly distributed between them. ARTIST accounts for multiple testing by performing a Benjamini-Hochberg test in the initial sliding-window step (to identify an initial genomic region), as well as stipulating a false discovery rate of 5% in the final Mann-Whitney U P values (below). Briefly, for analysis of underrepresented, domain, and neutral genes, EL-ARTIST was subsequently used. Con-ARTIST was used to analyze conditional enrichment or depletion of transposon-insertion mutants, comparing the original input library to the library obtained after outgrowth in LB alone, cefoxitin, cefepime, and ciprofloxacin; these data are presented in Tables S4 and S5 in the supplemental material. Genes conditionally depleted or enriched after growth in antibiotics were compared to those conditionally depleted or enriched in the library outgrown in LB alone and, if the original P value was ≤0.01 and the adjusted fold change was ≥4, deemed significant. For further details on the ARTIST pipelines, we refer readers to the initial application and the subsequent methods paper, as well as to an explanatory review (32, 46, 47). Artemis was used to generate the TA insertion plots shown in Fig. 1 and 4. BioVenn was used to compare sets of conditionally enriched and depleted genes between antibiotics and between S. marcescens and E. coli K-12 data sets and to illustrate the results (84). Microsoft Excel was used to generate tables. GraphPad Prism was used to depict all remaining data.
Molecular biology.
Allelic exchange using pTOX3 was used to make all in-frame deletions (68) including mppA (GenBank accession number WP_033640165.1), oppB (accession number WP_004932150.1), oppC (accession number WP_004932149.1), oppD (accession number WP_016927496.1), oppF (accession number WP_004932143.1), yeiU (accession number WP_033639836.1), yeeF (accession number WP_004935642.1), yhcS (accession number WP_016929157.1), ydgH (accession number WP_033640181.1), and ampC (accession number WP_033640466.1), all in S. marcescens ATCC 13880, as well as ydgH in E. coli O157:H7 EDL933 (GenBank accession number WP_000769322.1) and Enterobacter cloacae ATCC 13047 (GenBank accession number WP_013096870.1). pTOX3-derivative vectors were constructed essentially as described previously. Primer sequences are given in Table S7. Restriction enzyme-cut pTOX3 was incubated with purified AB and CD PCR products, along with a half-reaction mixture of HiFi DNA assembly master mix (NEB) according to the manufacturer’s directions. Reaction products were routinely desalted using “lily pad dialysis” (the entire reaction volume was placed on a slowly rotating 0.05-μm HA filter [Millipore] floating on the surface of MilliQ water) and subsequently electroporated directly into electrocompetent donor strains E. coli MFDλpir or SM10λpir. Selection and counterselection were performed as described previously (68). Deletion mutants were identified by colony PCR with subsequent Sanger sequencing of the reaction products. Primers used for the construction of pBAD33-ydgH are in Table S7. pBAD33-ydgH or pBAD33 were electroporated into the ΔydgH mutant as described previously (85). To ensure sufficient expression of YdgH, ΔydgH pBAD33-ydgH (and empty vector) were induced in arabinose 1% (vol/vol) for 4 h in the mid-log phase prior to back-dilution and incubation with cefoxitin (or vehicle). To allow for confirmation of expression, a FLAG tag was cloned into the C terminus. The YdgH phylogeny was constructed based on amino acid substitutions by using the maximum-likelihood method and the Jones-Taylor-Thornton (JTT) matrix-based model in MEGA X (86, 87).
ΔydgH mutant chemical screen.
Growth curves to identify phenotypes in the ΔydgH mutant were performed using the indicated concentration of chemical (Sigma) in microplate format (Bioscreen C growth plate reader) using the constant shaking setting at 37°C. Antibiotics were dissolved as recommended by the Clinical and Laboratory Standards Institute (CLSI) and stored in small aliquots at −80°C for up to 3 months.
Viability assays.
To validate candidate genes predicted to affect cefoxitin susceptibility (in Fig. 3), individual overnight cultures of Wt S. marcescens and single mutants were back-diluted and grown to the log phase until all cultures reached an OD of >0.25. They were then diluted again to 0.1 in either LB or LB plus cefoxitin 4 μg/ml. Aliquots were plated for CFU at 3 h and 6 h after cefoxitin. The final CFU ratio was calculated by dividing the CFU ratio of the mutant:Wt in cefoxitin by the CFU ratio of the mutant:Wt in LB alone. To validate the turbidity findings (from Fig. 5), the ΔydgH mutant and the Wt were back-diluted from overnight cultures twice to ensure they were both in the logarithmic phase and to allow initial synchronization of OD, and then when the OD was 0.2 to 0.5, back-diluted a final time to an OD of 0.005 in ceftriaxone 0.1 μg/ml, SDS 4% (wt/vol), benzethonium chloride 0.05 μg/ml, or rifampin (20 μg/ml). After 4 h of incubation at 37°C with intermittent agitation in a SpectraMax ABS Plus instrument (Molecular Devices), samples were plated for CFU. Although these concentrations were chosen to be roughly comparable to the growth curves shown in Fig. S3 and S6, they cannot be directly compared because these cultures were incubated in a different instrument than that for those used to generate the growth curves.
Genome sequencing.
DNA was isolated as directed using the Qiagen DNeasy plant minikit and submitted to the University of Massachusetts Medical School Deep Sequencing Core, which performed de novo assembly using PacBio sequencing and closed and finished using Hierarchical Genome Assembly Process (HGAP). The resulting genome sequence, obtained from the core, was manually polished using Illumina reads and deposited (GenBank accession number CP072199). It is 99.93% identical to GenBank accession number CP041233.1, which was not available at the at the time of sequencing. S. marcescens ATCC 13880 genes were identified using the BASys pipeline (incorporating Glimmer) and manually polished (88). The subsequent gene list was used to generate the look-up table incorporated into the EL-ARTIST and Con-ARTIST pipelines described above.
Bioinformatics and statistical analyses.
To ensure the accuracy of homology identification, for those S. marcescens genes identified as underrepresented, a translated protein BLAST search was used to identify E. coli K-12 homologs and results manually adjudicated. COG categories for appropriate genes of interest were identified in the Clusters of Orthologous Genes database and manually tabulated (89). UniProt and EcoCyc were frequently used to glean initial functional and sequence data about a particular gene product (90, 91). All statistics performed on processed data, depicted in figures were with an unpaired two-tailed t test (except those in Fig. 5D, which due to nonnormality were tested using the Wilcoxon rank-sum test). No correction for multiple comparisons was made. Significance is denoted as follows: *, P ≤ 0.05; **, P ≤ 0.01. The ydgH gene schematic was constructed in Benchling using predicted σ70 promoter regions with a score of >90 in BacPP (72). Rho-independent terminators were identified using ARNold (73).
Data availability.
The genome sequence for the Serratia marcescens subsp. marcescens ATCC 13880 chromosome was deposited in GenBank under accession number CP072199. Reads for all sequenced libraries have been deposited in the GEO database under accession number GSE169651.
ACKNOWLEDGMENTS
We thank members of the Waldor lab, including Brandon Sit, Karthik Hullahalli, Carole Kuehl, and Troy Hubbard for helpful discussion.
J.E.L. is supported by T32 AI-007061, the Harvard Catalyst Medical Research Investigator Training fellowship, and K08 AI-155830. M.K.W. is supported by R01-AI-042347 and by the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Yu VL. 1979. Serratia marcescens: historical perspective and clinical review. N Engl J Med 300:887–893. 10.1056/NEJM197904193001604. [DOI] [PubMed] [Google Scholar]
- 2.Adeolu M, Alnajar S, Naushad S, S Gupta R. 2016. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol 66:5575–5599. 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 3.Mahlen SD. 2011. Serratia infections: from military experiments to current practice. Clin Microbiol Rev 24:755–791. 10.1128/CMR.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sligl W, Taylor G, Brindley PG. 2006. Five years of nosocomial Gram-negative bacteremia in a general intensive care unit: epidemiology, antimicrobial susceptibility patterns, and outcomes. Int J Infect Dis 10:320–325. 10.1016/j.ijid.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 6.Koenig SM, Truwit JD. 2006. Ventilator-associated pneumonia: diagnosis, treatment, and prevention. Clin Microbiol Rev 19:637–657. 10.1128/CMR.00051-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, Sievert DM. 2016. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol 37:1288–1301. 10.1017/ice.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandner-Miranda L, Vinuesa P, Cravioto A, Morales-Espinosa R. 2018. The Genomic basis of intrinsic and acquired antibiotic resistance in the genus Serratia. Front Microbiol 9:828. 10.3389/fmicb.2018.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ray C, Shenoy AT, Orihuela CJ, González-Juarbe N. 2017. Killing of Serratia marcescens biofilms with chloramphenicol. Ann Clin Microbiol Antimicrob 16:19. 10.1186/s12941-017-0192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langsrud S, Møretrø T, Sundheim G. 2003. Characterization of Serratia marcescens surviving in disinfecting footbaths. J Appl Microbiol 95:186–195. 10.1046/j.1365-2672.2003.01968.x. [DOI] [PubMed] [Google Scholar]
- 11.de Frutos M, López-Urrutia L, Domínguez-Gil M, Arias M, Muñoz-Bellido JL, Eiros JM, Ramos C. 2017. Serratia marcescens outbreak due to contaminated 2% aqueous chlorhexidine. Enferm Infecc Microbiol Clin 35:624–629. 10.1016/j.eimc.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 12.Moradigaravand D, Boinett CJ, Martin V, Peacock SJ, Parkhill J. 2016. Recent independent emergence of multiple multidrug-resistant Serratia marcescens clones within the United Kingdom and Ireland. Genome Res 26:1101–1109. 10.1101/gr.205245.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merino JL, Bouarich H, Pita MJ, Martínez P, Bueno B, Caldés S, Corchete E, Jaldo MT, Espejo B, Paraíso V. 2016. Serratia marcescens bacteraemia outbreak in haemodialysis patients with tunnelled catheters due to colonisation of antiseptic solution: experience at 4 hospitals. Nefrologia 36:667–673. 10.1016/j.nefro.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Novosad SA, Lake J, Nguyen D, Soda E, Moulton-Meissner H, Pho MT, Gualandi N, Bepo L, Stanton RA, Daniels JB, Turabelidze G, Van Allen K, Arduino M, Halpin AL, Layden J, Patel PR. 2019. Multicenter outbreak of Gram-negative bloodstream infections in hemodialysis patients. Am J Kidney Dis 74:610–619. 10.1053/j.ajkd.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Regev-Yochay G, Smollan G, Tal I, Pinas Zade N, Haviv Y, Nudelman V, Gal-Mor O, Jaber H, Zimlichman E, Keller N, Rahav G. 2018. Sink traps as the source of transmission of OXA-48-producing Serratia marcescens in an intensive care unit. Infect Control Hosp Epidemiol 39:1307–1315. 10.1017/ice.2018.235. [DOI] [PubMed] [Google Scholar]
- 16.Cristina ML, Sartini M, Spagnolo AM. 2019. Serratia marcescens infections in neonatal intensive care units (NICUs). Int J Environ Res Public Health 16:610. 10.3390/ijerph16040610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moles L, Gómez M, Moroder E, Jiménez E, Escuder D, Bustos G, Melgar A, Villa J, Del Campo R, Chaves F, Rodríguez JM. 2019. Serratia marcescens colonization in preterm neonates during their neonatal intensive care unit stay. Antimicrob Resist Infect Control 8:135. 10.1186/s13756-019-0584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escribano E, Saralegui C, Moles L, Montes MT, Alba C, Alarcón T, Lázaro-Perona F, Rodríguez JM, Sáenz de Pipaón M, Del Campo R. 2019. Influence of a Serratia marcescens outbreak on the gut microbiota establishment process in low-weight preterm neonates. PLoS One 14:e0216581. 10.1371/journal.pone.0216581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montagnani C, Cocchi P, Lega L, Campana S, Biermann KP, Braggion C, Pecile P, Chiappini E, de Martino M, Galli L. 2015. Serratia marcescens outbreak in a neonatal intensive care unit: crucial role of implementing hand hygiene among external consultants. BMC Infect Dis 15:11. 10.1186/s12879-014-0734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin QY, Tsai Y-L, Liu M-C, Lin W-C, Hsueh P-R, Liaw S-J. 2014. Serratia marcescens arn, a PhoP-regulated locus necessary for polymyxin B resistance. Antimicrob Agents Chemother 58:5181–5190. 10.1128/AAC.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merkier AK, Rodríguez MC, Togneri A, Brengi S, Osuna C, Pichel M, Cassini MH, Centrón D, Serratia marcescens Argentinean Collaborative Group. 2013. Outbreak of a cluster with epidemic behavior due to Serratia marcescens after colistin administration in a hospital setting. J Clin Microbiol 51:2295–2302. 10.1128/JCM.03280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stock I, Grueger T, Wiedemann B. 2003. Natural antibiotic susceptibility of strains of Serratia marcescens and the S. liquefaciens complex: S. liquefaciens sensu stricto, S. proteamaculans and S. grimesii. Int J Antimicrob Agents 22:35–47. 10.1016/S0924-8579(02)00163-2. [DOI] [PubMed] [Google Scholar]
- 23.Tamma PD, Doi Y, Bonomo RA, Johnson JK, Simner PJ, Antibacterial Resistance Leadership Group. 2019. A primer on AmpC beta-lactamases: necessary knowledge for an increasingly multidrug-resistant world. Clin Infect Dis 69:1446–1455. 10.1093/cid/ciz173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacoby GA. 2009. AmpC beta-lactamases. Clin Microbiol Rev 22:161–182. 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mataseje LF, Boyd DA, Delport J, Hoang L, Imperial M, Lefebvre B, Kuhn M, Van Caeseele P, Willey BM, Mulvey MR. 2014. Serratia marcescens harbouring SME-type class A carbapenemases in Canada and the presence of blaSME on a novel genomic island, SmarGI1-1. J Antimicrob Chemother 69:1825–1829. 10.1093/jac/dku040. [DOI] [PubMed] [Google Scholar]
- 26.Bush K, Pannell M, Lock JL, Queenan AM, Jorgensen JH, Lee RM, Lewis JS, Jarrett D. 2013. Detection systems for carbapenemase gene identification should include the SME serine carbapenemase. Int J Antimicrob Agents 41:1–4. 10.1016/j.ijantimicag.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Hopkins KL, Findlay J, Meunier D, Cummins M, Curtis S, Kustos I, Mustafa N, Perry C, Pike R, Woodford N. 2017. Serratia marcescens producing SME carbapenemases: an emerging resistance problem in the UK? J Antimicrob Chemother 72:1535–1537. [DOI] [PubMed] [Google Scholar]
- 28.Queenan AM, Torres-Viera C, Gold HS, Carmeli Y, Eliopoulos GM, Moellering RC, Jr, Quinn JP, Hindler J, Medeiros AA, Bush K. 2000. SME-type carbapenem-hydrolyzing class A beta-lactamases from geographically diverse Serratia marcescens strains. Antimicrob Agents Chemother 44:3035–3039. 10.1128/AAC.44.11.3035-3039.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 54:969–976. 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hemarajata P, Amick T, Yang S, Gregson A, Holzmeyer C, Bush K, Humphries RM. 2018. Selection of hyperproduction of AmpC and SME-1 in a carbapenem-resistant Serratia marcescens isolate during antibiotic therapy. J Antimicrob Chemother 73:1256–1262. 10.1093/jac/dky028. [DOI] [PubMed] [Google Scholar]
- 31.Cain AK, Barquist L, Goodman AL, Paulsen IT, Parkhill J, van Opijnen T. 2020. A decade of advances in transposon-insertion sequencing. Nat Rev Genet 21:526–540. 10.1038/s41576-020-0244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chao MC, Abel S, Davis BM, Waldor MK. 2016. The design and analysis of transposon insertion sequencing experiments. Nat Rev Microbiol 14:119–128. 10.1038/nrmicro.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valentino MD, Foulston L, Sadaka A, Kos VN, Villet RA, Santa Maria J, Jr, Lazinski DW, Camilli A, Walker S, Hooper DC, Gilmore MS. 2014. Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. mBio 5:e01729-14–e01714. 10.1128/mBio.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson MT, Mitchell LA, Zhao L, Mobley HLT. 2017. Capsule production and glucose metabolism dictate fitness during Serratia marcescens bacteremia. mBio 8:e00740-17. 10.1128/mBio.00740-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 11:435–442. 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2:e00315-10–e00310. 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellerose MM, Proulx MK, Smith CM, Baker RE, Ioerger TR, Sassetti CM. 2020. Distinct bacterial pathways influence the efficacy of antibiotics against Mycobacterium tuberculosis. mSystems 5:e00396-20. 10.1128/mSystems.00396-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jana B, Cain AK, Doerrler WT, Boinett CJ, Fookes MC, Parkhill J, Guardabassi L. 2017. The secondary resistome of multidrug-resistant Klebsiella pneumoniae. Sci Rep 7:42483. 10.1038/srep42483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warr AR, Hubbard TP, Munera D, Blondel CJ, Abel Zur Wiesch P, Abel S, Wang X, Davis BM, Waldor MK. 2019. Transposon-insertion sequencing screens unveil requirements for EHEC growth and intestinal colonization. PLoS Pathog 15:e1007652. 10.1371/journal.ppat.1007652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang SL, Rubin EJ. 2002. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296:179–185. 10.1016/s0378-1119(02)00856-9. [DOI] [PubMed] [Google Scholar]
- 41.Hubbard TP, Chao MC, Abel S, Blondel CJ, Abel Zur Wiesch P, Zhou X, Davis BM, Waldor MK. 2016. Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proc Natl Acad Sci USA 113:6283–6288. 10.1073/pnas.1601718113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hubbard TP, Billings G, Dörr T, Sit B, Warr AR, Kuehl CJ, Kim M, Delgado F, Mekalanos JJ, Lewnard JA, Waldor MK. 2018. A live vaccine rapidly protects against cholera in an infant rabbit model. Sci Transl Med 10:eaap8423. 10.1126/scitranslmed.aap8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warr AR, Giorgio RT, Waldor MK. 2020. Genetic analysis of the role of the conserved inner membrane protein CvpA in EHEC resistance to deoxycholate. J Bacteriol 203:e00661-20. 10.1128/JB.00661-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin K, Guan Y, Ma R, Wei L, Liu B, Liu X, Zhou X, Ma Y, Zhang Y, Waldor MK, Wang Q. 2018. Critical role for a promoter discriminator in RpoS control of virulence in Edwardsiella piscicida. PLoS Pathog 14:e1007272. 10.1371/journal.ppat.1007272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carda-Diéguez M, Silva-Hernández FX, Hubbard TP, Chao MC, Waldor MK, Amaro C. 2018. Comprehensive identification of Vibrio vulnificus genes required for growth in human serum. Virulence 9:981–993. 10.1080/21505594.2018.1455464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pritchard JR, Chao MC, Abel S, Davis BM, Baranowski C, Zhang YJ, Rubin EJ, Waldor MK. 2014. ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet 10:e1004782. 10.1371/journal.pgen.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chao MC, Pritchard JR, Zhang YJ, Rubin EJ, Livny J, Davis BM, Waldor MK. 2013. High-resolution definition of the Vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nucleic Acids Res 41:9033–9048. 10.1093/nar/gkt654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao L, Anderson MT, Wu W, T Mobley HL, Bachman MA. 2017. TnseqDiff: identification of conditionally essential genes in transposon sequencing studies. BMC Bioinformatics 18:326. 10.1186/s12859-017-1745-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo H, Lin Y, Liu T, Lai F-L, Zhang C-T, Gao F, Zhang R. 2021. DEG 15, an update of the Database of Essential Genes that includes built-in analysis tools. Nucleic Acids Res 49:D677–D686. 10.1093/nar/gkaa917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pearson WR. 2013. An introduction to sequence similarity (“homology”) searching. Curr Protoc Bioinformatics Chapter 3:Unit3.1. 10.1002/0471250953.bi0301s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolard A, Schniederjans M, Haüssler S, Triponney P, Valot B, Plésiat P, Jeannot K. 2019. Production of norspermidine contributes to aminoglycoside resistance in pmrAB mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother 63:e01044-19. 10.1128/AAC.01044-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ekiert DC, Bhabha G, Isom GL, Greenan G, Ovchinnikov S, Henderson IR, Cox JS, Vale RD. 2017. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169:273–285.e17. 10.1016/j.cell.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uehara T, Parzych KR, Dinh T, Bernhardt TG. 2010. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J 29:1412–1422. 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uehara T, Suefuji K, Jaeger T, Mayer C, Park JT. 2006. MurQ Etherase is required by Escherichia coli in order to metabolize anhydro-N-acetylmuramic acid obtained either from the environment or from its own cell wall. J Bacteriol 188:1660–1662. 10.1128/JB.188.4.1660-1662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, Laxminarayan R. 2014. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis 14:742–750. 10.1016/S1473-3099(14)70780-7. [DOI] [PubMed] [Google Scholar]
- 57.Siedner MJ, Galar A, Guzman-Suarez BB, Kubiak DW, Baghdady N, Ferraro MJ, Hooper DC, O’Brien TF, Marty FM. 2014. Cefepime vs other antibacterial agents for the treatment of Enterobacter species bacteremia. Clin Infect Dis 58:1554–1563. 10.1093/cid/ciu182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hooper DC. 1999. Mode of action of fluoroquinolones. Drugs 58(Suppl 2):6–10. 10.2165/00003495-199958002-00002. [DOI] [PubMed] [Google Scholar]
- 59.Correia S, Poeta P, Hébraud M, Capelo JL, Igrejas G. 2017. Mechanisms of quinolone action and resistance: where do we stand? J Med Microbiol 66:551–559. 10.1099/jmm.0.000475. [DOI] [PubMed] [Google Scholar]
- 60.Mitchell AM, Silhavy TJ. 2019. Envelope stress responses: balancing damage repair and toxicity. Nat Rev Microbiol 17:417–428. 10.1038/s41579-019-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cohen SP, McMurry LM, Hooper DC, Wolfson JS, Levy SB. 1989. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother 33:1318–1325. 10.1128/AAC.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lovelle M, Mach T, Mahendran KR, Weingart H, Winterhalter M, Gameiro P. 2011. Interaction of cephalosporins with outer membrane channels of Escherichia coli: revealing binding by fluorescence quenching and ion conductance fluctuations. Phys Chem Chem Phys 13:1521–1530. 10.1039/c0cp00969e. [DOI] [PubMed] [Google Scholar]
- 63.Chenia HY, Pillay B, Pillay D. 2006. Analysis of the mechanisms of fluoroquinolone resistance in urinary tract pathogens. J Antimicrob Chemother 58:1274–1278. 10.1093/jac/dkl404. [DOI] [PubMed] [Google Scholar]
- 64.Griffith KL, Shah IM, Wolf RE, Jr.. 2004. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol Microbiol 51:1801–1816. 10.1046/j.1365-2958.2003.03952.x. [DOI] [PubMed] [Google Scholar]
- 65.Nicoloff H, Andersson DI. 2013. Lon protease inactivation, or translocation of the lon gene, potentiate bacterial evolution to antibiotic resistance. Mol Microbiol 90:1233–1248. 10.1111/mmi.12429. [DOI] [PubMed] [Google Scholar]
- 66.Will WR, Brzovic P, Le Trong I, Stenkamp RE, Lawrenz MB, Karlinsey JE, Navarre WW, Main-Hester K, Miller VL, Libby SJ, Fang FC. 2019. The evolution of SlyA/RovA transcription factors from repressors to countersilencers in Enterobacteriaceae. mBio 10:e00009-19. 10.1128/mBio.00009-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Curran TD, Abacha F, Hibberd SP, Rolfe MD, Lacey MM, Green J. 2017. Identification of new members of the Escherichia coli K-12 MG1655 SlyA regulon. Microbiology (Reading) 163:400–409. 10.1099/mic.0.000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lazarus JE, Warr AR, Kuehl CJ, Giorgio RT, Davis BM, Waldor MK. 2019. A new suite of allelic exchange vectors for the scarless modification of proteobacterial genomes. Appl Environ Microbiol 85:e00990-19. 10.1128/AEM.00990-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niemann GS, Brown RN, Gustin JK, Stufkens A, Shaikh-Kidwai AS, Li J, McDermott JE, Brewer HM, Schepmoes A, Smith RD, Adkins JN, Heffron F. 2011. Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect Immun 79:33–43. 10.1128/IAI.00771-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sontag RL, Nakayasu ES, Brown RN, Niemann GS, Sydor MA, Sanchez O, Ansong C, Lu S-Y, Choi H, Valleau D, Weitz KK, Savchenko A, Cambronne ED, Adkins JN. 2016. Identification of novel host interactors of effectors secreted by Salmonella and Citrobacter. mSystems 1:e00032-15. 10.1128/mSystems.00032-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rudd KE. 2000. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res 28:60–64. 10.1093/nar/28.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Avila e Silva S, Echeverrigaray S, Gerhardt GJL. 2011. BacPP: bacterial promoter prediction—a tool for accurate sigma-factor specific assignment in enterobacteria. J Theor Biol 287:92–99. 10.1016/j.jtbi.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 73.Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. 2001. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res 29:4724–4735. 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yethon JA, Vinogradov E, Perry MB, Whitfield C. 2000. Mutation of the lipopolysaccharide core glycosyltransferase encoded by waaG destabilizes the outer membrane of Escherichia coli by interfering with core phosphorylation. J Bacteriol 182:5620–5623. 10.1128/JB.182.19.5620-5623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vinogradov E, Lindner B, Seltmann G, Radziejewska-Lebrecht J, Holst O. 2006. Lipopolysaccharides from Serratia marcescens possess one or two 4-amino-4-deoxy-l-arabinopyranose 1-phosphate residues in the lipid A and D-glycero-d-talo-oct-2-ulopyranosonic acid in the inner core region. Chemistry 12:6692–6700. 10.1002/chem.200600186. [DOI] [PubMed] [Google Scholar]
- 76.Anderson MT, Mitchell LA, Mobley HLT. 2017. Cysteine biosynthesis controls Serratia marcescens phospholipase activity. J Bacteriol 199:e00159-17. 10.1128/JB.00159-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan A, Guan Z, Raetz CRH. 2007. An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J Biol Chem 282:36077–36089. 10.1074/jbc.M706172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ranjit DK, Young KD. 2016. Colanic acid intermediates prevent de novo shape recovery of Escherichia coli spheroplasts, calling into question biological roles previously attributed to colanic acid. J Bacteriol 198:1230–1240. 10.1128/JB.01034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 80.Sueki A, Stein F, Savitski MM, Selkrig J, Typas A. 2020. Systematic localization of Escherichia coli membrane proteins. mSystems 5:e00808-19. 10.1128/mSystems.00808-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han M-J, Kim JY, Kim JA. 2014. Comparison of the large-scale periplasmic proteomes of the Escherichia coli K-12 and B strains. J Biosci Bioeng 117:437–442. 10.1016/j.jbiosc.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 82.Schmidt A, Kochanowski K, Vedelaar S, Ahrné E, Volkmer B, Callipo L, Knoops K, Bauer M, Aebersold R, Heinemann M. 2016. The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol 34:104–110. 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hammann P, Parmentier D, Cerciat M, Reimegård J, Helfer A-C, Boisset S, Guillier M, Vandenesch F, Wagner EGH, Romby P, Fechter P. 2014. A method to map changes in bacterial surface composition induced by regulatory RNAs in Escherichia coli and Staphylococcus aureus. Biochimie 106:175–179. 10.1016/j.biochi.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 84.Hulsen T, de Vlieg J, Alkema W. 2008. BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9:488. 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tu Q, Yin J, Fu J, Herrmann J, Li Y, Yin Y, Stewart AF, Müller R, Zhang Y. 2016. Room temperature electrocompetent bacterial cells improve DNA transformation and recombineering efficiency. Sci Rep 6:24648. 10.1038/srep24648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 87.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Van Domselaar GH, Stothard P, Shrivastava S, Cruz JA, Guo A, Dong X, Lu P, Szafron D, Greiner R, Wishart DS. 2005. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res 33:W455–W459. 10.1093/nar/gki593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.UniProt Consortium. 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47:D506–D515. 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karp PD, Ong WK, Paley S, Billington R, Caspi R, Fulcher C, Kothari A, Krummenacker M, Latendresse M, Midford PE, Subhraveti P, Gama-Castro S, Muñiz-Rascado L, Bonavides-Martinez C, Santos-Zavaleta A, Mackie A, Collado-Vides J, Keseler IM, Paulsen I. 2018. The EcoCyc database. EcoSal Plus 8:10. 10.1128/ecosalplus.ESP-0006-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Merged supplemental figures with legends. Download aac.00786-21-s0001.pdf, PDF file, 1.8 MB (1.8MB, pdf)
Table S1. Download aac.00786-21-s0002.xlsx, XLSX file, 4.1 MB (4.1MB, xlsx)
Table S2. Download aac.00786-21-s0003.xlsx, XLSX file, 0.2 MB (225.4KB, xlsx)
Table S3. Download aac.00786-21-s0004.xlsx, XLSX file, 0.02 MB (22.8KB, xlsx)
Table S4. Download aac.00786-21-s0005.xlsx, XLSX file, 0.8 MB (856.8KB, xlsx)
Table S5. Download aac.00786-21-s0006.xlsx, XLSX file, 0.10 MB (99.4KB, xlsx)
Table S6. Download aac.00786-21-s0007.xlsx, XLSX file, 0.4 MB (411.7KB, xlsx)
Table S7. Download aac.00786-21-s0008.xlsx, XLSX file, 0.01 MB (12.4KB, xlsx)
Data Availability Statement
The genome sequence for the Serratia marcescens subsp. marcescens ATCC 13880 chromosome was deposited in GenBank under accession number CP072199. Reads for all sequenced libraries have been deposited in the GEO database under accession number GSE169651.