ABSTRACT
Aspergillus fumigatus causes a series of invasive diseases, including the high-mortality invasive aspergillosis, and has been a serious global health threat because of its increased resistance to the first-line clinical triazoles. We analyzed the whole-genome sequence of 15 A. fumigatus strains from China and found that long terminal repeat retrotransposons (LTR-RTs), including Afut1, Afut2, Afut3, and Afut4, are most common and have the largest total nucleotide length among all transposable elements in A. fumigatus. Deleting one of the most enriched Afut4977-sac1 in azole-resistant strains decreased azole resistance and downregulated its nearby gene, sac1, but it did not significantly affect the expression of genes of the ergosterol synthesis pathway. We then discovered that 5'LTR of Afut4977-sac1 had promoter activity and enhanced the adjacent sac1 gene expression. We found that sac1 is important to A. fumigatus, and the upregulated sac1 caused elevated resistance of A. fumigatus to azoles. Finally, we showed that Afut4977-sac1 has an evolution pattern similar to that of the whole genome of azole-resistant strains due to azoles; phylogenetic analysis of both the whole genome and Afut4977-sac1 suggests that the insertion of Afut4977-sac1 might have preceded the emergence of azole-resistant strains. Taking these data together, we found that the Afut4977-sac1 LTR-RT might be involved in the regulation of azole resistance of A. fumigatus by upregulating its nearby sac1 gene.
KEYWORDS: Aspergillus fumigatus, long terminal repeat retrotransposon, Afut4, azole resistance, sac1
INTRODUCTION
Aspergillus fumigatus is one of the most commonly encountered pathogenic fungi in the clinic. Its spores are ubiquitous, with 10 to 200 CFU/m3 in the air, and are small enough (diameter, approximately 2 to 3 μm) to be spread by wind and to easily reach the host alveoli (1). A. fumigatus can cause serious infections and allergies (2), such as allergic bronchopulmonary aspergillosis, chronic pulmonary aspergillosis (CPA), and invasive aspergillosis. Moreover, invasive aspergillosis is considered the deadliest aspergillosis that occurs in the lungs, with a fatality rate of 30% to 95% (3, 4).
Antifungal therapy combined with immunomodulation is considered the most effective treatment option to improve aspergillosis's clinical prognosis (5). Moreover, as first-line drugs in the clinic, triazoles play an essential role in preventing and treating aspergillosis (6). However, since the first emergence of azole-resistant isolates in 1997 (7), the resistance of A. fumigatus to azoles has increased alarmingly and has become a significant public health problem recently (8–10).
Azoles can bind to the cytochrome P450 14α-sterol demethylase (Cyp51) to inhibit the conversion from lanosterol to ergosterol, which is an essential component of the A. fumigatus cell membrane (11, 12). To date, the molecular mechanisms of azole resistance of A. fumigatus are either cyp51 mediated or non-cyp51 mediated. Two paralogs of cyp51 genes, cyp51A and cyp51B, have been reported in A. fumigatus. cyp51A contributes largely to azole resistance through point mutations and overexpression, whereas cyp51B is either functionally redundant or an alternative under particular conditions (13–15). Several point mutations in Cyp51A, such as G448S, G54A, G54W, P216L, and M220V/K/T, have been verified to be related to azole resistance by genetic reconstitution experiments (16, 17). Overexpression of cyp51A is usually caused by the tandem repeats (TR) in the promoter region, including TR34 and TR46 (18). Furthermore, the combinations of a TR with a specific mutation in Cyp51A, such as TR34/L98H and TR46/Y12F/T289A, were frequently found in azole-resistant A. fumigatus clinical samples (19). Alternatively, several non-cyp51-mediated azole resistance mechanisms are related to overexpression of multidrug efflux pumps, interference in ergosterol biosynthesis, stress adaptation, and biofilm formation (15, 20); however, some azole-resistance mechanisms remain to be further investigated in A. fumigatus.
Long terminal repeat retrotransposons (LTR-RTs) are RNA retrotransposons bound at the head and tail ends by a long terminal repeat (LTR). LTR-RTs use RNA as an intermediate in the “copy-paste” manner and have been identified as intracellular viruses (21). LTR-RTs exist throughout eukaryotic genomes, such as those of mammals, plants, and fungi, with scattered distribution and play an essential role in genome evolution (22), genomic stability (23), stress response (24), and gene regulation (25). The research on LTR-RTs of fungi mainly describe their function in response to environmental stresses in yeast, such as low oxygen and high temperature. The best-studied LTR-RT, Ty1 in Saccharomyces cerevisiae, is arguably considered the best model to understand the activation of LTR-RTs by environmental stress and the impact of the activation on adjacent gene expression (26). Similar research has found that the transcription of Tf1 in Schizosaccharomyces pombe was activated, and it was inserted near 32 genes under thermal stimulation, increasing the expression of six of these genes (27). Recently, the involvement of LTR-RTs in antifungal resistance was reported in several filamentous fungi. For example, a 519-bp LTR sequence was inserted into the promoter of the mfs1 gene, leading to the multidrug resistance (MDR) phenotype in Zymoseptoria tritici that causes wheat spot blotch (28). Similarly, LTR-RT-derived fragments induce a rearrangement in the promoter region of the superfamily transporter gene mfsM2 in Botrytis, resulting in the overexpression of mfsM2 and the MDR phenotype (29, 30).
To date, LTR-RTs of A. fumigatus have been found and grouped into Afut1, Afut2, Afut3, and Afut4, all of which belong to the Ty3/Gypsy family (31–33). Notably, the integration of Afut4 was considered to be a recent event due to its integrity (intact open reading frames and two identical 184-bp LTRs) relative to that of Afut1, Afut2, and Afut3 (33). However, the function of LTR-RTs is still poorly understood in A. fumigatus.
In this study, LTR-RTs in genomes of azole-resistant and azole-sensitive A. fumigatus strains were analyzed and characterized by whole-genome sequencing of 15 strains. Further, the function and regulation of LTR-RTs on azole resistance of A. fumigatus, especially Afut4, were investigated.
RESULTS
Loss of Afut4977-sac1 in A. fumigatus STJ0105 reduced its resistance to voriconazole and posaconazole.
Whole-genome sequencing of 15 A. fumigatus strains from China, including 8 azole-resistant and 7 azole-sensitive strains, was performed (Table 1). In particular, all 8 azole-resistant A. fumigatus strains harbor the Cyp51A TR34/L98H mutation (34). Subsequently, the transposable elements (TEs) in all 15 genomes were identified and counted by RepeatModeler and classified into different families. It was shown that LTR-RTs have the greatest length of all kinds of TEs, significantly larger than the other TEs (see Fig. S1A in the supplemental material). It was found that several Afut4 copies, one type of LTR-RTs, were significantly enriched in azole-resistant strains (P < 0.05) (Table 2; Fig. S1B). Among them, an Afut4 was found to be located 977 bp upstream of the sac1 gene (phosphoinositide phosphatase SacI; AFUA_4G08050), and it had the most significant enrichment with azole resistance (P < 0.01); this specific Afut4977-sac1 was subsequently studied. Afut4977-sac1 is located inversely 977 bp upstream of sac1 in chromosome 4. In addition, the sequence between Afut4977-sac1 and sac1 includes the 5′ untranslated region (UTR) of sac1 and a 579-bp fragment that inserts upstream of sac1 together with Afut4977-sac1 (Fig. 1A and B; Fig. S3A). This Afut4977-sac1-flanking 579-bp fragment was found to be somewhat similar to A. fumigatus transposon Taf1, with a lot of gaps and mismatches by the BLAST algorithm optimized for somewhat similar sequences. Afut4977-sac1 and the flanking 579-bp fragment were located together in the genome of all eight azole-resistant strains and one azole-sensitive strain but not in the other azole-sensitive strains, including Af293 and CEA17Δku80 (Table 1).
TABLE 1.
Genome features and MICs of 15 A. fumigatus strains
| Strain | Size (bp) | No. of scaffolds | N50 (bp) | No. of genes | Repeat region (%) | Cyp51A mutation(s) | MIC (mg/liter)a |
Azole resistance | ||
|---|---|---|---|---|---|---|---|---|---|---|
| ITC | VRC | POS | ||||||||
| C94 | 28,816,918 | 94 | 825,047 | 8,824 | 5.05 | TR34/L98H | ≥16 | 2 | 1 | Resistant |
| C96 | 28,676,732 | 45 | 2,607,325 | 8,921 | 2.29 | TR34/L98H/S297T/F495I | ≥16 | 1 | 0.5 | Resistant |
| C116 | 28,867,951 | 43 | 1,501,208 | 8,871 | 5.23 | TR34/L98H | ≥16 | 4 | 0.5 | Resistant |
| XJ138 | 29,142,407 | 46 | 1,853,060 | 8,907 | 5.09 | TR34/L98H | ≥16 | 2 | 0.5 | Resistant |
| E739 | 28,816,696 | 32 | 1,960,044 | 8,928 | 3.53 | TR34/L98H/S297T/F495I | ≥16 | 2 | 0.5 | Resistant |
| C821 | 29,118,210 | 46 | 1,592,331 | 8,922 | 3.91 | TR34/L98H | ≥16 | 4 | 1 | Resistant |
| C1664 | 28994183 | 66 | 1,169,047 | 8,859 | 4.30 | TR34/L98H | ≥16 | 8 | 1 | Resistant |
| STJ0105 | 29,148,970 | 67 | 1,499,649 | 8,874 | 4.53 | TR34/L98H | ≥16 | 8 | 2 | Resistant |
| C79 | 28,522,771 | 65 | 1,040,756 | 8,798 | 4.27 | D262Y | 0.25 | 0.5 | 0.06 | Sensitive |
| C490 | 28,971,844 | 66 | 2,259,216 | 8,976 | 2.82 | N248K | 1 | 0.25–0.5 | 0.125 | Sensitive |
| E509 | 28,966,977 | 63 | 2,394,484 | 8,984 | 2.99 | N248K | 0.5–1 | 0.25 | 0.06–0.125 | Sensitive |
| E631 | 28,676,852 | 62 | 2,159,055 | 8,917 | 1.90 | None | 0.25 | 0.25–0.5 | 0.125 | Sensitive |
| E691 | 28,758,967 | 88 | 1,247,161 | 8,965 | 1.87 | None | 0.25 | 0.5 | 0.25 | Sensitive |
| E1069 | 28,786,365 | 36 | 3,185,189 | 8,917 | 3.24 | A9T | 0.25–0.5 | 0.5 | 0.125 | Sensitive |
| E1109 | 28,073,344 | 37 | 2,455,631 | 8,772 | 3.16 | None | 0.25 | 0.5 | 0.06 | Sensitive |
VRC, voriconazole; POS, posaconazole; ITC, itraconazole.
TABLE 2.
Enrichments of different specified Afut4 copies in azole-resistant strains
The enrichment of each Afut4 insertion was based on Fisher's exact test.
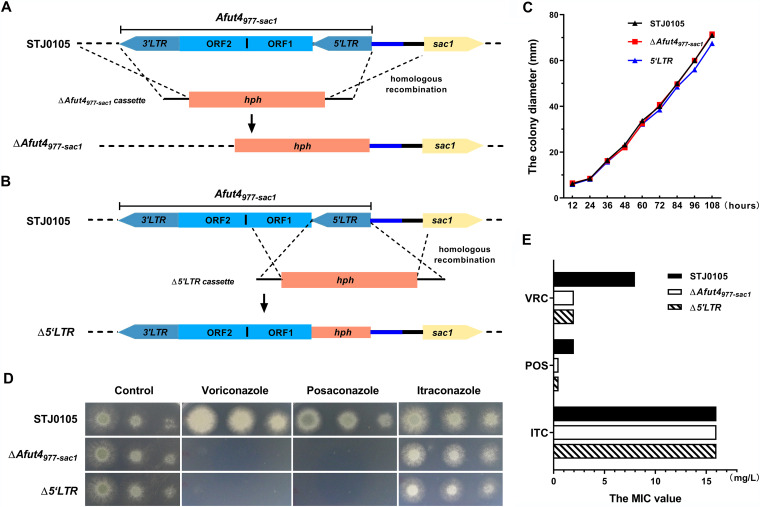
The yellow rectangle represents sac1 (AFUA_4G08050) in chromosome 4 (Chr4). The blue rectangle represents AFUA_6G03860 in chromosome 6 (Chr6). The black rectangle represents AFUA_4G11380 in chromosome 6 (Chr6). The red rectangles represent different Afut4 copies at a different genome locus.
FIG 1.
Azole resistance of STJ0105, ΔAfut4977-sac1, and Δ5'LTR. (A and B) Schematic depiction of ΔAfut4977-sac1 and Δ5'LTR by homologous recombination from STJ0105. Afut4977-sac1 is located inversely 977 bp upstream of sac1 in chromosome 4. The 5'LTR and 3'LTR are similar long repeats at both ends, starting at 5′-TG and ending in CA-3′. Both ORF1 and ORF2 are open reading frames of Afut4. The blue line is a 579-bp sequence that inserted upstream of the sac1 gene with Afut4977-sac1. The black line contains the probable 5′UTR of the sac1 gene. (C) Colony diameters of STJ0105, ΔAfut4977-sac1, and Δ5'LTR. A total of 5 × 105 conidia were inoculated centrally in AMM and cultured at 37°C. The colony diameter was measured every 12 h. (D) Drug plate point assay. Colony growth of STJ0105, ΔAfut4977-sac1, and Δ5'LTR in the presence of VRC (2 mg/liter), POS (0.5 mg/liter), and ITC (8 mg/liter). For the plate point assay, a 5-μl slurry of the indicated spores from the stock suspensions (107, 106, and 105 CFU/ml) was spotted onto AMM. All plates were incubated at 37°C for 2 to 5 days. (E) MIC values for VRC, POS, and ITC, as determined by the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST).
Because STJ0105 had a higher MIC value to voriconazole (VRC) and posaconazole (POS), it was chosen as a research model to explore the function of Afut4977-sac1 insertion and its role in azole resistance (Table 1). Two mutants, ΔAfut4977-sac1 and Δ5'LTR were constructed by homologous recombination. The full-length Afut4977-sac1 was completely knocked out in ΔAfut4977-sac1, while in Δ5'LTR, the 5'LTR adjacent to sac1 was knocked out but 3'LTR and two open reading frames (ORFs) of Afut4977-sac1 remained untouched (Fig. 1A and B; Fig. S2A and S2B). The additional 579-bp flanking fragment between Afut4977-sac1 and sac1 was still kept in both ΔAfut4977-sac1 and Δ5'LTR. The three strains' growth characteristics were not significantly different, and two of the mutants showed radial growth and conidiation similar to their parental strain (Fig. 1C; Fig. S2C). Intriguingly, as depicted in Fig. 1D, compared to their parental strain STJ0105, the resistance of ΔAfut4977-sac1 and Δ5'LTR to VRC and POS decreased dramatically; however, no change was detected in the susceptibility of either ΔAfut4977-sac1 or Δ5'LTR to itraconazole (ITC) in comparison with STJ0105. Furthermore, the MIC value of VRC for ΔAfut4977-sac1 and Δ5'LTR was 2 mg/liter, 4-fold lower than the MIC of 8 mg/liter for STJ0105; similarly, the MIC value of POS for ΔAfut4977-sac1 and Δ5'LTR (0.4 mg/liter) was also dramatically lower than that for STJ0105 (2 mg/liter). In contrast, the MIC value of ITC for ΔAfut4977-sac1 and Δ5'LTR (≥16 mg/liter) was equal to that for STJ0105 (≥16 mg/liter) (Fig. 1E and Table 3). It could be speculated that the MIC of itraconazole for STJ0105 caused by the TR34/L98H mutation is so high (≥16 mg/liter), even saturated, that it might mask the change of susceptibility to itraconazole caused by the deletion of Afut4977-sac1 or its 5'LTR. Collectively, the aforementioned data demonstrated that the lack of Afut4977-sac1 or its 5'LTR might not affect the general growth characteristics of A. fumigatus but reduces the azole resistance of A. fumigatus to VRC and POS.
TABLE 3.
A. fumigatus constructions used in this study
| Strain | Genotype | MIC (mg/liter)a |
Reference or source | ||
|---|---|---|---|---|---|
| ITC | VRC | POS | |||
| ΔAfut4 | STJ0105 ΔAfut4::hph | ≥16 | 2 | 0.5 | This study |
| Δ5′LTR | STJ0105 Δ5′LTR::hph | ≥16 | 2 | 0.5 | This study |
| CEA17Δku80 | Δku80::pyrG+ | 0.5 | 0.5 | 0.125 | da Silva Ferreira et al., 2006 (39) |
| sac1 OE | Δku80 prtA::gpdA(p)::sac1 | 0.25 | 0.25 | 0.0625 | This study |
| sac1teton | Δku80 sac1(p)::ptrA-tetOn::sac1 | 2 | 2 | 0.5 | This study |
VRC, voriconazole; POS, posaconazole; ITC, itraconazole.
Transcriptomic profiling of STJ0105, ΔAfut4977-sac1, and Δ5′LTR.
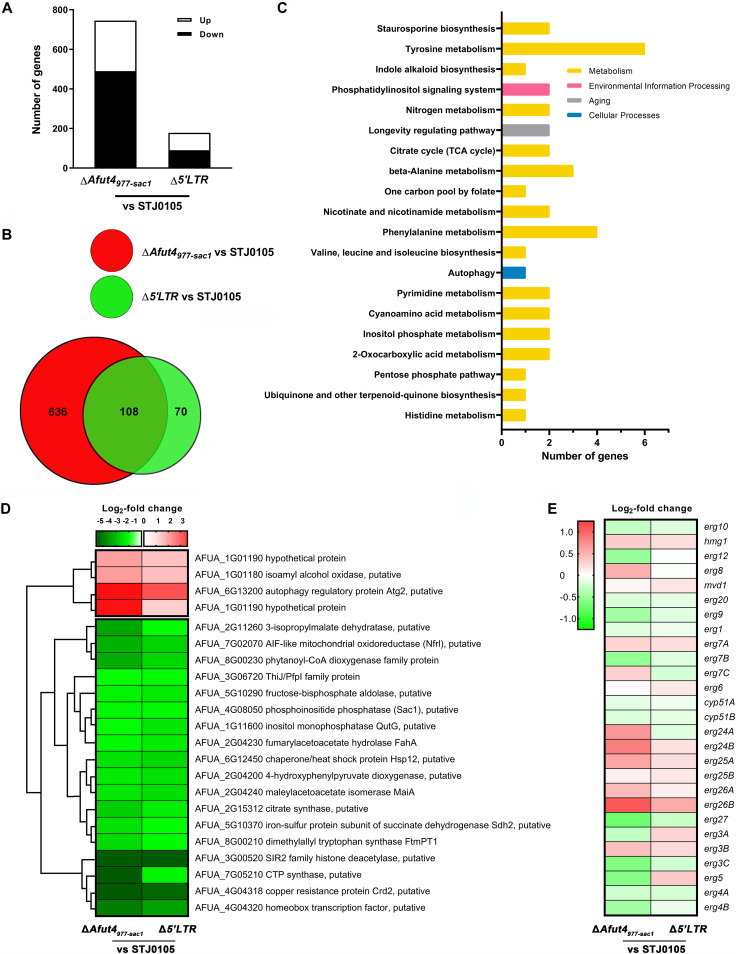
Given the remarkable reduction of azole resistance of ΔAfut4977-sac1 and Δ5'LTR, transcriptomic sequencing analyses (RNA-seq) of STJ0105, ΔAfut4977-sac1, and Δ5'LTR were performed. Compared with parental strain STJ0105, in ΔAfut4977-sac1, 254 genes were upregulated (log2 fold change ≥ 1; Q < 0.05) and 490 genes were downregulated (log2 fold change ≤ −1; Q < 0.05), while 88 genes were upregulated (log2 fold change ≥ 1; Q < 0.05) and 90 genes were downregulated (log2 fold change ≤ −1; Q < 0.05) in Δ5'LTR (Fig. 2A; Table S2). This revealed that the knockout of the shorter 5'LTR instead of full-length Afut4977-sac1 resulted in fewer differentially expressed genes (DEGs). As shown in the Venn diagram (Fig. 2B), 108 DEGs (|log2 fold change| ≥ 1; Q < 0.05) were shared in both ΔAfut4977-sac1 and Δ5'LTR analyzed by KEGG classification and enrichment. Twenty-two genes involved in the 20 most enriched pathways were accordingly selected for further hierarchical cluster analysis to narrow the scope of candidate genes (Fig. 2C). The transcription of the sac1 gene (phosphoinositide phosphatase SacI; AFUA_4G08050) located near ΔAfut4977-sac1 declined significantly in both ΔAfut4977-sac1 and Δ5'LTR relative to STJ0105 (Fig. 2D). We hypothesized that deletion of either Afut4977-sac1 or its 5'LTR might decrease sac1 expression and subsequently play some role in the decline of azole resistance in ΔAfut4977-sac1 and Δ5'LTR, considering that the mutation of SacI might affect the sensitivity of S. cerevisiae to azoles (35).
FIG 2.
RNA sequencing analysis of STJ0105, ΔAfut4977-sac1, and Δ5'LTR. (A) A summary of the differently expressed genes (DEGs) in ΔAfut4977-sac1 relative to STJ0105 (ΔAfut4977-sac1 vs STJ0105) and Δ5'LTR relative to STJ0105 (Δ5'LTR vs STJ0105) (|log2 fold change ≥ 1|; Q < 0.05). (B) Venn diagram showing the differentially and commonly shared DEGs (|log2 fold change ≥ 1|; Q < 0.05) in ΔAfut4977-sac1 versus STJ0105 and Δ5'LTR versus STJ0105. (C) The top 20 enriched pathways of DEGs (|log2 fold change| ≥ 1; Q < 0.05) from the 108 shared DEGs in both ΔAfut4977-sac1 versus STJ0105 and Δ5'LTR versus STJ0105. According to KEGG annotations and classifications, the 58 significant DEGs were classified into different biological pathways. KEGG enrichment analysis was carried out by using the phyper function in R software. The top 20 enriched pathways were screened from all the biological pathways. (D) Hierarchical cluster analysis of the log2 fold change of 22 significant DEGs in the top 20 enriched pathways. (E) Log2 fold change of genes involved in ergosterol biosynthesis in ΔAfut4977-sac1 and Δ5'LTR versus STJ0105.
However, none of these 22 genes were directly linked to ergosterol biosynthesis, which is the main targeted pathway for the azole drug (Fig. 2D). The expression value of genes involved in ergosterol biosynthesis of ΔAfut4977-sac1 and Δ5'LTR was compared with that of parental strain STJ0105 to confirm the role of deletion of either Afut4977-sac1 or its 5'LTR in ergosterol biosynthesis (Fig. 2E). It was shown that most of the genes involved in ergosterol biosynthesis were similarly upregulated or downregulated in both ΔAfut4977-sac1 and Δ5'LTR. However, almost all of the genes showed no significant difference (|log2 fold change|<1; Q < 0.05) among STJ0105, ΔAfut4977-sac1, and Δ5'LTR, except that expression of erg26B in ΔAfut4977-sac1 is more than 2-fold greater than that of STJ0105 (log2 fold change = 1.09; Q < 0.05). However, there is no report that erg26 can affect azole resistance, although its expression is upregulated in response to azole (36). These results indicate that the knockout of either Afut4977-sac1 or its 5'LTR may not significantly change the ergosterol synthesis pathway.
Afut4977-sac1 enhanced transcription of its adjacent gene, sac1.
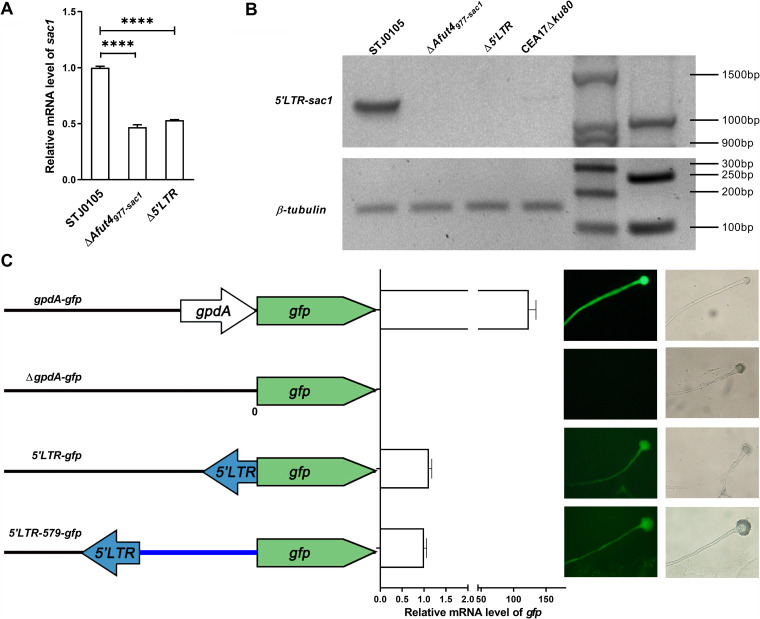
To study the possible regulation by Afut4977-sac1 of sac1 transcription, the mRNA levels of sac1 in STJ0105, ΔAfut4977-sac1, and Δ5'LTR were detected again by quantitative real-time PCR (qPCR). As shown in Fig. 3A, the mRNA level of sac1 in both ΔAfut4977-sac1 and Δ5'LTR was lower than in STJ0105. These data suggested that either full-length Afut4977-sac1 or its 5'LTR upstream of sac1 might upregulate the expression of sac1. As the 5'LTR may harbor transcriptional regulatory elements or regions, we hypothesized that 5'LTR might act as a promoter or enhancer that boosts sac1 transcription. Indeed, a chimeric transcript from 5'LTR to sac1 was found only in STJ0105, not in ΔAfut4977-sac1, Δ5'LTR, or CEA17Δku80, a control strain that has no Afut4977-sac1 insertion upstream of sac1 (Fig. 3B). These data indicated that the 5'LTR might have a cryptic promoter activity toward its nearby gene.
FIG 3.
Elevated transcription of the adjacent sac1 by Afut4977-sac1 in STJ0105. (A) Expression level of sac1 in STJ0105, ΔAfut4997-sac1, and Δ5'LTR. Total RNA was prepared from the culture of each strain. The levels of the indicated mRNAs were determined by qPCR. P values were calculated using unpaired Student’s t tests: ****, P < 0.0001. Error bars represent the standard error of the mean. (B) Reverse transcription PCR detection of 5'LTR-sac1 transcript in STJ0105, ΔAfut4977-sac1, Δ5'LTR, and CEA17Δku80. The 5′ primer 5'LTR-sac1-F was located in 5'LTR of Afut4 and the 3′ primer 5'LTR-sac1-R was located in sac1 (Table S1). (C) Promoter activity assay of 5'LTR. Left, schematic diagram of the plasmid used in the assay. Only one copy of each plasmid was integrated after the histone 2A locus of the transformant genome via a single crossover. Middle, expression level of the reporter gfp gene under the control of 5'LTR or 5'LTR-579 (the 5′LTR of Afut4997-sac1 and the flanking 579-bp sequence) relative to the positive control (gpdA-gfp) and negative control (ΔgpdA-gfp), respectively. Total RNA was prepared from the culture of STJ0105 carrying the corresponding plasmid. Levels of the gfp mRNAs were determined by qPCR. The error bars represent the standard error of the mean. Right, fluorescence images of STJ0105 carrying the corresponding plasmid.
Three plasmids were constructed from backbone plasmid pJW103, which carries a strong promoter, gpdA, followed by a reporter gene encoding a green fluorescent protein (gdpA-gfp) to verify the regulation by 5'LTR of the transcription of its downstream gene. pJW103 is an integrative A. fumigatus expression plasmid that can specifically integrate after the histone 2A locus of the A. fumigatus genome via single crossover (37, 38). All strains carrying only one copy of pJW103 or other derived plasmids were confirmed by Southern blotting (Fig. S4B and S4C). As shown in Fig. 3C, in the 5'LTR-gfp plasmid, the promoter gpdA was replaced with 5'LTR. Similarly, in the 5'LTR-579-gfp plasmid, the promoter was replaced with 5'LTR and its flanking 579-bp fragment. In the ΔgpdA-gfp plasmid, the promoter gpdA was deleted. These plasmids were transformed into STJ0105, the mRNA level was measured, and green fluorescence of green fluorescent protein (GFP) was detected in the strains. As shown in Fig. 3C, STJ0105 transformed with the gpdA-gfp plasmid displayed strong GFP RNA and protein levels in the hyphae and conidiophore, whereas deletion of the gpdA promoter resulted in full deterioration of the GFP fluorescence in the hyphae and conidiophore. The addition of 5'LTR upstream of gfp could significantly promote the transcription of gfp; similar gfp transcription and green fluorescence were also observed in the strain with the 5'LTR-579-gfp plasmid. Multiple transformants carrying the same plasmid show similar corresponding phenotypes. These results demonstrated that 5'LTR of Afut4977-sac1 could upregulate the transcription of its adjacent gene, sac1, in STJ0105.
Overexpression of sac1 promotes the azole resistance of A. fumigatus.
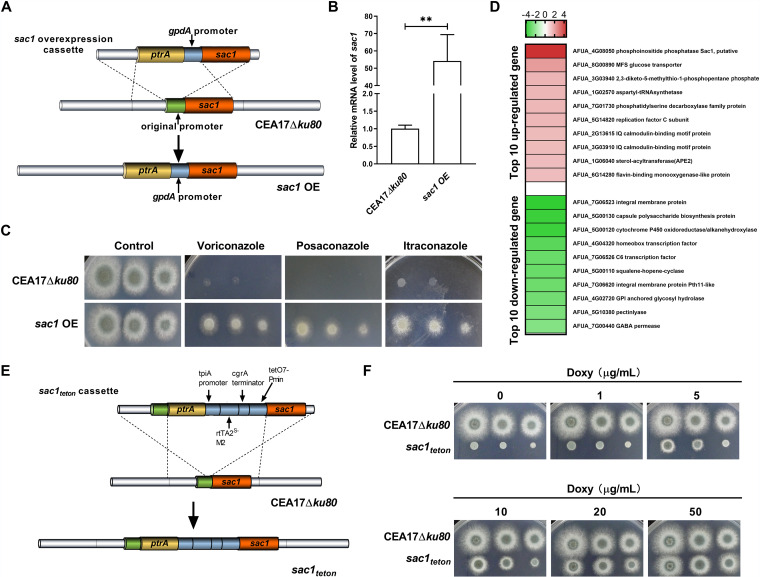
A mutant named sac1 OE with overexpression of sac1 was constructed from CEA17Δku80 by replacing endogenous promoters of sac1 with the strong promoter gpdA to clarify whether higher expression of sac1 is correlated with elevated azole resistance of A. fumigatus (Fig. 4A; Fig. S5A and S5B). CEA17Δku80 is a nonhomologous end-joining-deficient pyrG+ A. fumigatus strain with a high frequency of homologous recombination (Table 3) (39). The mRNA level of sac1 in sac1 OE was significantly higher than in CEA17Δku80 (Fig. 4B). When cultured in solid Aspergillus minimal medium (AMM), sac1 OE displayed a colony and conidiophore morphology similar to that of CEA17Δku80 (Fig. S5C). Furthermore, the sensitivity of sac1 OE to triazoles was tested. As depicted in Fig. 4C, under treatment with 0.5 mg/liter VRC, 0.2 mg/liter POS, and 0.8 mg/liter ITC, sac1 OE showed higher resistance to the triazoles than CEA17Δku80. The MIC values of VRC, POS, and ITC for sac1 OE were significantly higher than those for CEA17Δku80 (2 mg/liter versus 0.5 mg/liter, 0.5 mg/liter versus 0.125 mg/liter, and 2 mg/liter versus 0.5 mg/liter, respectively) (Table 3). Collectively, the above results indicated that overexpression of sac1 could promote the resistance of A. fumigatus to triazoles. Then, transcriptomic (RNA-seq) analysis of sac1 OE and its parental strain, CEA17Δku80, was also performed and results were compared. The top 20 genes with the largest difference, including 10 upregulated and 10 downregulated, are listed in Fig. 4D. Remarkably, both 2.73-fold upregulated AFUA_1G06040 (sterol O-acyltransferase) and 8.12-fold downregulated AFUA_5G00110 (squalene-hopene-cyclase) in sac1 OE relative to CEA17Δku80 are closely involved in steroid biosynthesis according to KEGG annotation. In addition, the ergosterol content of sac1 OE analyzed by liquid chromatography (LC)/mass spectrometry (MS) assays was about 1.22-fold greater than that of the parental strain, CEA17Δku80 (Fig. S5D).
FIG 4.
Azoles resistance of sac1 OE. (A) Schematic depiction of the construction of sac1 OE. The original promoter of sac1 was replaced with the promoter replacement cassette consisting of a 5′ fragment located approximately 1 kb upstream of the start codon, a pyrithiamine resistance cassette, the gpdA promoter, and the 3′ fragment which encompasses the transcribed region beginning with the start codon by homologous recombination. (B) The mRNA level of sac1 in CEA17Δku80 and sac1 OE. Levels of the sac1 mRNAs were determined by qPCR. P values were calculated using unpaired Student’s t tests: **, P < 0.01. Error bars represent the standard error of the mean. (C) Drug plate point assay. Colony growth of CEA17Δku80 and sac1 OE in the presence of VRC (0.5 mg/liter), POS (0.2 mg/liter), and ITC (0.8 mg/liter). For the plate point assay, a 5-μl slurry of the indicated spores from the stock suspensions (107, 106, and 105 CFU/ml) was spotted onto AMM with the indicated drugs. All plates were incubated at 37°C for 2 to 5 days. (D) Top 10 upregulated and downregulated genes in sac1 OE relative to CEA17Δku80 according to the transcriptome data. (E) Schematic depiction of the construction of sac1teton. The original promoter of sac1 was replaced with the promoter replacement cassette consisting of a 5′ fragment located approximately 1 kb upstream of the start codon, a pyrithiamine resistance cassette, the teton promoter, and the 3′ fragment which encompasses the transcribed region beginning with the start codon by homologous recombination. (F) Colonies of sac1teton and CEA17Δku80 under different concentrations of exogenous doxycycline (Doxy).
To further study the function of sac1, a conditional knockout strain named sac1teton was successfully constructed (Fig. 4E; Fig. S6A and S6B), because of failure of the complete knockout of the sac1 gene. The native promoter of sac1 was replaced by a doxycycline-dependent teton promoter by homologous recombination (40). sac1teton showed defective polar growth when sac1 could not be expressed without exogenous doxycycline (Fig. 4F). As the concentration of doxycycline increased, sac1 began to be expressed and the polar growth of sac1teton was restored. However, the function of sac1 and the azole resistance caused by the upregulated sac1 expression need further research.
Afut4977-sac1 evolution pattern converges with whole-genome evolution under azole stress.
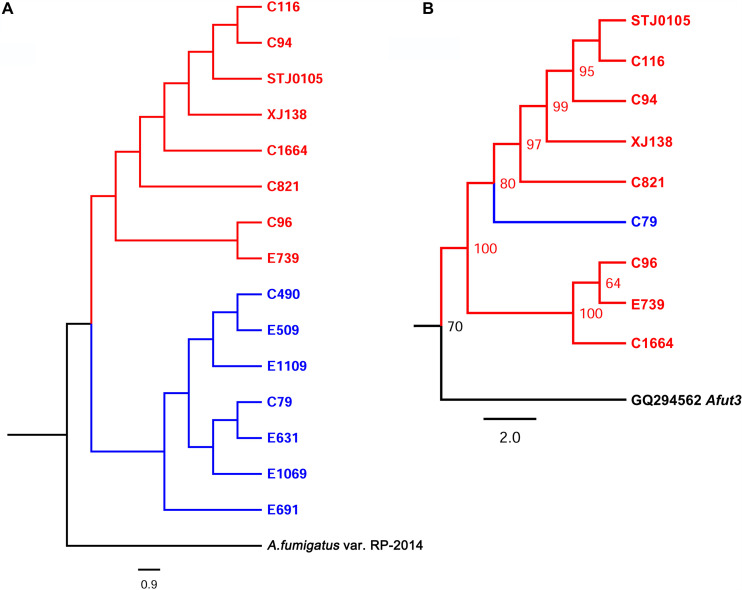
As LTR-RTs can perform autonomous transposition in the whole genome and the eventual prevalence of LTR-RTs in different genomic regions depends on selection processes and “host control,” the phylogenetic relationship between Afut4977-sac1 and the whole genome in A. fumigatus was analyzed. In the whole genome (Fig. 5A), azole-resistant A. fumigatus strains, which all carry the Cyp51A TR34/L98H mutation (34), and azole-sensitive strains were completely divided into two evolutionary branches that might experience different evolutionary processes. The Afut4977-sac1 phylogenetic tree has an evolutionary pattern similar to that of the whole genome. One exception was that an azole-sensitive strain, C79, was grouped into the evolutionary branch of all resistant strains in the phylogenetic tree of Afut4977-sac1 (Fig. 5B), which suggested that the Afut4977-sac1 insertion might happen before the evolutionary divergence of azole-resistant and azole-sensitive strains. Moreover, C1664 was clustered with C96 and E739 in the phylogenetic tree of Afut4977-sac1 instead of with C116, C94, STJ0105, and C821 in the phylogenetic tree of the whole genome. In summary, the Afut4977-sac1 evolution pattern might converge with whole-genome evolution under azole stress in A. fumigatus.
FIG 5.
Phylogenetic analysis of the whole genome and Afut4977-sac1. (A) Whole-genome phylogenetic analysis. A strain of A. fumigatus var. RP-2014 was chosen as the outgroup. (B) Afut4977-sac1 phylogenetic analysis. Sequences of Afut4977-sac1 upstream of sac1 of 15 strains were aligned, and SNP sites were determined for phylogenetic analysis. The nucleotide sequence of Afut3 was chosen as the outgroup.
DISCUSSION
A. fumigatus is a life-threatening pathogenic fungus and causes aspergillosis ranging from disseminated invasive aspergillosis in immunocompromised patients to chronic infections and allergic syndromes (41). Azole resistance has become a thorny incident that hinders clinical treatment and increases the mortality rate of A. fumigatus-infected patients. The cpy51A-related mechanisms of azole resistance have been studied extensively in A. fumigatus. However, it was recently highlighted that around 20% to 50% of clinical azole-resistant isolates have unknown mechanisms of azole resistance (8, 42, 43), and non-cyp51A-mediated mechanisms have been increasingly reported (15, 20).
In this study, we used whole-genome sequencing and bioinformatics analysis to find that LTR-RTs, especially Afut4977-sac1 upstream of sac1, were specifically enriched in azole-resistant strains. Afut4977-sac1 insertion caused a higher resistance to azole drugs, VRC, and POS in an A. fumigatus strain, STJ0105. Deletion of either full-length Afut4977-sac1 (ΔAfut4977-sac1) or the 5'LTR region of Afut4977-sac1 (Δ5'LTR) destroyed the azole-resistance of STJ0105 but did not affect the resistance of STJ0105 to ITC. STJ0105 is a clinical isolate with the TR34/L98H mutation that is a combination of a 34-bp tandem repeat (TR) in the promoter region and a leucine-to-histidine substitution at codon 98 (L98H) in cyp51A (18, 34). In line with the well-known pan-azole resistance characteristics of A. fumigatus strains with the TR34/L98H mutation (44–47), STJ0105 exhibited high resistance to ITC (MIC ≥ 16 mg/liter) and relatively mild resistance to VRC and POS. Thereby, it could be deduced that the decrease in resistance to azole drugs caused by deletion of Afut4977-sac1 might be nondetectable due to the strong resistance to ITC (MIC ≥ 16 mg/liter) caused by the TR34/L98H mutation in STJ0105; in contrast, this decrease was obvious in resistance to VRC and POS by deletion of Afut4977-sac1 in STJ0105. In addition, STJ0105 can even grow at an ITC concentration of 32 mg/liter (data not shown). Any potential change in resistance to itraconazole may be masked by itraconazole saturation. In addition, the enriched Afut4, especially Afut4977-sac1, might function as the additional genetic background that caused a higher resistance to azoles in STJ0105. Certainly, the possibility of differing regulation by Afut4977-sac1 of resistance to ITC, VRC, and POS could not be excluded, and further investigations are necessary.
Another novel finding was that full-length Afut4977-sac1, especially its 5'LTR in STJ0105, conferred a higher expression of its proximal gene, sac1. This finding indicated that Afut4977-sac1, especially its 5'LTR, might function as a promoter. It is known that LTR, as the regulatory element of an LTR-RT, is capable of regulating the transcription of its neighboring genes (26, 48–52). In mammalian cells, LTR is known to function as a promoter in several cases (53–55). For example, LTR-driven transcription was shown in the heritable Opitz syndrome-related gene produce Mid1 (56), endothelin B receptor (57), and insulin-like growth factor INSL4 (58). Likewise, in plants, an LTR-RT Renovator served as a promoter to its downstream rice blast resistance gene, Pit, leading to upregulation of Pit and disease resistance in Nipponbare (59). Similar upregulation of LTR-RTs to its downstream gene was also demonstrated in S. pombe, a fungus, in which six genes were activated by a Tf1 insertion (a type of LTR-RT) (27). Interestingly, in most studies of LTR-RTs, the LTR was usually found to be inserted upstream of a gene in the same ORF orientation to promote its downstream genes. Interestingly, in STJ0105, although the Afut4977-sac1 insertion is in opposite orientation to the nearby sac1 gene, it still greatly enhanced the expression of sac1. Such orientation-opposed regulation by LTR-RTs is rare but has been reported. The solitary LTR of an LTR-RT called human endogenous retrovirus K (HERV-K) could direct transcription in both orientations relative to the downstream reporter gene (60). In S. cerevisiae, the reversed insertion of Ty1 (an active LTR-RT) drives the vicinal reporter gene (61). Furthermore, in Jingxian blood orange, Tcs2, an active LTR-RT, was inserted at just 450 bp upstream of ATG of the Ruby gene in the opposite orientation to the Ruby gene and upregulated the expression of Ruby in the same manner as the regulation by Afut4977-sac1 of sac1 in this study (62).
In this study, a novel azole resistance mechanism was attributable to the sac1 gene, a phosphoinositol metabolism-regulating gene. Overexpression of sac1 not only elevated the resistance of A. fumigatus to VRC (from 0.5 mg/liter to 2 mg/liter) and POS (from 0.125 mg/liter to 0.5 mg/liter) but also raised the resistance to ITC (from 0.5 mg/liter to 2 mg/liter). It was reported that mutation of SacI altered the drug sensitivity of S. cerevisiae to azoles (35). However, the function of SacI in A. fumigatus is still unknown and is predicted to be involved in inositol phosphate metabolism and the phosphatidylinositol signaling system according to KEGG classification. During the study, the mutant with a complete knockout of the sac1 gene in A. fumigatus could not be successfully constructed, so a conditional knockout strain named sac1teton was successfully constructed (see Fig. S6A and S6B in the supplemental material), in which the expression of the sac1 gene is controlled by the teton promoter. In the presence of exogenous doxycycline, the teton promoter can be activated and the targeted gene starts to be expressed; otherwise, it is not expressed. As shown in Fig. 4F, there was a serious growth defect of sac1teton in the absence of doxycycline, which corroborated the unsuccessful screen of the null mutant. With an increase in the doxycycline concentration, growth was restored by the increase of expression of sac1. These results were understandable, since no or little expression of sac1 could result in deterioration of the key conversion from phosphatidylinositol 4-phosphate (PI4P) to phosphatidylinositol (PI), which is critical for trafficking along the early secretory pathway (63), leading to a serious growth defect in A. fumigatus.
To our knowledge, the mutation of SacI protein in S. cerevisiae led to the accumulation of PI4P and delayed endocytosis and vacuolar protein sorting in combination with cold sensitivity and high sensitivity to multiple drugs (64). PI4P, as an important ligand, assists the Osh4 protein in transporting sterols between the Golgi apparatus and the plasma membrane with vesicular trafficking to maintain the normal functions of sterols (65). SacI protein can interact with oxidized sterol binding protein to promote intracellular lipid and sterol transport independent of vesicles (66). SacI protein is also an important factor in efficient ATP uptake into the endoplasmic reticulum (ER) (67). In addition, as a suppressor of actin, SacI protein is also essential for actin organization, hyphal development, cell wall integrity, and pathogenicity (68).
RNA sequencing of sac1 OE and its parental strain, CEA17Δku80, was then performed and the results were compared further to explore the potential role of SacI in azole resistance. Among the top 20 genes with the largest difference, AFUA_1G06040 (sterol O-acyltransferase APE2) was significantly upregulated, while AFUA_5G00110 (squalene-hopene-cyclase) was significantly downregulated in sac1 OE compared with its parent strain. It is well known that azole resistance in fungi is highly related to cyp51A-mediated ergosterol biosynthesis, which is one part of the steroid biosynthesis pathway. These data implied that sac1 might also affect azole resistance by regulating the steroid biosynthesis pathway. Predictably, a 20% higher ergosterol level was detected in sac1 OE. However, this seems to imply that the massive change in azole susceptibility caused by high sac1 expression may also be relevant to the toxic sterol level, the changed ATP uptake, and the cell wall integrity. The exact mechanism needs to be further clarified.
Because of their widespread and abundant insertions, LTR-RTs can cause the rearrangement of genomes and lead to DNA damage (69), directly disrupt gene function or produce harmful mutations, and even endanger the survival of host fungi. Therefore, the wide distribution of LTR-RTs is ultimately controlled, selected, and eventually silent during the host’s long-term evolutionary process. Hence, we hypothesized that the active Afut4977-sac1 in azole-resistant strains might improve the potential adaptation of its host A. fumigatus to azole stress. Afut4977-sac1 was much more prevalent in azole-resistant strains than in sensitive strains in quantity (Fig. 1B), which means that the Afut4977-sac1 in azole-resistant strains was once activated, transposed to different positions in the genome, and left many copies. Several lines of evidence show that LTR-RTs can be activated by environmental stress. Tnt1, the first known plant retrotransposon, could be activated by pathogens, tissue culture, compounds related to plant defense, wounding, freezing, and other abiotic stresses (49). Under hypoxic stress, solo LTRs of Tf2, widely distributed throughout the genome of S. pombe, could be activated to regulate the expression of adjacent coding or noncoding sequences (70). Similarly, the transposition of the transposon impala in A. fumigatus could also be activated by prolonged exposure to low temperatures (71). And the transposon integration upstream of the start codon of the cyp51A gene might also be related to the elevated azole resistance (72).
The phylogenetic tree of Afut4977-sac1 is similar to that of the whole genome of azole-resistant A. fumigatus. In addition to the proven Afut4977-sac1-related azole resistance, it could be deduced that during the process of azole resistance formation, Afut4977-sac1 was activated by azole stress and transposed to leave a large number of copies; some of these copies were retained due to the associated survival advantage of resisting azole stress, resulting in an evolutionary model that is similar to the evolution of the whole genome under azole stress. However, the retrotransposition of LTR-RTs is a very low probability event, usually less than 1%, even under laboratory-induced culture conditions (51). Besides, the insertion site and the flanking sequence brought by Afut4977-sac1 insertion were quite similar among different azole-resistant strains (Fig. S3A). Thereby, it could not be excluded that Afut4977-sac1 insertion might spread in different azole-resistant strains through sexual reproduction (73). The mix of one exceptional azole-sensitive strain, C79, in the Afut4977-sac1-based phylogenetic tree cluster suggested that either an earlier Afut4977-sac1 insertion event occurred before the evolutionary divergence between azole-resistant and azole-sensitive strains or a probable genetic exchange occurred through meiotic recombination between C79 and an Afut4977-sac1-insertion-carried strain. Nevertheless, this hypothesis should be further investigated with more sequencing data and bioinformatics analysis.
This study unraveled a novel azole resistance mechanism in A. fumigatus. LTR-RTs, especially Afut4977-sac1, were enriched in azole-resistant A. fumigatus and might play a role in azole resistance by modulating the expression of its downstream gene, sac1.
MATERIALS AND METHODS
Strains and culture conditions.
The 15 strains of A. fumigatus used for whole-genome sequencing in this work are listed in Table 1, and the constructed strains in this work are listed in Table 3. A. fumigatus strain STJ0105 was the parental strain for Afut4 and 5'LTR deletion and also served as a plasmid transformation strain. The nonhomologous end-joining-deficient A. fumigatus strain CEA17Δku80 (a generous gift from Jean Paul Latgé) served as the parental strain for sac1 overexpression.
The conidia of A. fumigatus were propagated on Aspergillus minimal medium (AMM) for 5 to 8 days at 37°C and were collected with a phosphate buffer solution containing 0.1% Tween 20 (0.1% PBST). The conidia were passed through a filter (40 μm) to remove hyphal fragments and enumerated using a hemocytometer.
DNA, RNA extraction, and cDNA preparation.
Conidia (3 × 108 CFU/ml) were cultured in AMM liquid medium at 37°C, 200 rpm, for 18 h. Mycelia were ground with liquid nitrogen with a mortar and pestle for the next DNA and RNA extractions. DNA extraction followed the instructions of the Biospin fungus genomic DNA extraction kit (BSC14s1; BioFlux). For RNA extraction, total RNA was isolated from the mycelia using TransZol Up (ET111-01; Transgene) according to the manufacturer’s instructions. First-strand cDNA synthesis was performed with an anchored oligo(dT)18 primer using the TransScript one-step genomic DNA (gDNA) removal and cDNA synthesis supermix (AT311; Transgene) according to the manufacturer’s instructions.
Whole-genome sequencing and analysis.
Genomic DNA was extracted using a MagPure plant DNA kit (catalog no. MD5118-05F; Magen, China) according to the manufacturer’s protocol. DNA concentration and purity were determined with a Qubit fluorometer and a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Carlsbad, CA, USA). DNA integrity was assessed by 0.5% agarose gel electrophoresis. Whole-genome sequencing was performed on the MGISEQ-2000 platform at BGI (Shenzhen, China). The raw sequencing data were processed using the following steps: (i) removal of reads containing sequencing adapter; (ii) removal of reads whose low-quality base ratio (base quality ≤ 5) is more than 50%; (iii) removal of reads whose unknown base (“N” base) ratio is more than 10%. Clean data were aligned to the human reference genome using Burrows-Wheeler Aligner (BWA) (68). Picard was used to removing duplicated sequence reads. Realignment was performed with the Genome Analysis Toolkit (GATK) (74). Single-nucleotide polymorphisms (SNPs) were called using HaplotypeCaller of GATK.
Identification and annotation of LTR-RTs.
Identification of LTR-RTs and TEs was performed by RepeatModeler (70) based on the repeat databases (Repbase Update) copyrighted by the Genetic Information Research Institute (GIRI). Classification of Afut1∼Afut4 LTR-RTs was based on BLASTn similarity alignment against known Afut1∼Afut4 sequences from GenBank. The presence or absence of a functional gene was analyzed 3,000 bp upstream or downstream of the intact LTR-RTs. For the intact LTR-RTs inserted near the gene, the statistical difference of insertion frequency between azole-resistant and -sensitive strains was based on Fisher's exact test.
Strain construction.
All primers used in this work are shown in Table 3. For the construction of the Afut4 and 5'LTR deletion cassette as well as sac1 overexpression cassette, fusion PCR was used as described previously (75). Briefly, approximately 1.5 kb of the upstream and downstream flanking sequences of the Afut4 or 5'LTR was amplified from STJ0105 genomic DNA (gDNA) using primers ΔAfut4-up and ΔAfut4-dw and primers Δ5'LTR -up and 5'LTR-dw, respectively. The selection marker hph (approximately 3 kb in length) from plasmid pdht-hph-hdIII-sacI donated by K. J. Kwon-Chung (National Institutes of Health, USA) was amplified with primers ΔAfut4-hph and Δ5'LTR-hph. respectively. Next, the three aforementioned PCR products were combined into the Afut4 or 5'LTR deletion cassette with the primers ΔAfut4-up-F/ΔAfut4-dw-R and Δ5'LTR-up-F/Δ5'LTR-dw-R, respectively. Then, the Afut4 or 5'LTR deletion cassette was transformed into STJ0105. The promoter replacement cassette of sac1 overexpression consisted of a 5′ fragment located approximately 1 kb upstream of the start codon of sac1, a pyrithiamine resistance cassette, the gpdA promoter, and the 3′ fragment which encompasses the transcribed region beginning with the start codon. The 5′ fragment and the 3′ fragment were amplified from CEA17Δku80 gDNA with primers sac1OE-up and sac1OE-dw, respectively. The fragment including the pyrithiamine resistance cassette and gpdA was amplified from pJW103 (76) with primer sac1OE-ptrA. The sac1 overexpression cassette was constructed by fusion PCR and purified for transformation. Similarly, the promoter replacement cassette of sac1teton consisted of a 5′ fragment located approximately 1 kb upstream of the start codon of sac1, a pyrithiamine resistance cassette, the teton promoter, and the 3′ fragment which encompasses the transcribed region beginning with the start codon. The 5′ fragment and the 3′ fragment were amplified from CEA17Δku80 gDNA with primers sac1teton-up and sac1teton-dw, respectively. The fragment including the pyrithiamine resistance cassette and teton promoter was amplified from pCH008 (donated by Johannes Wagener, University of Munich, Germany) with the primer sac1teton-ptrA. The sac1teton cassette was constructed by fusion PCR and purified for transformation. A. fumigatus protoplasts were generated and transformed essentially as described previously (75). The resulting protoplasts were transferred to AMM plates containing 1.2 M sorbitol and 0.1 μg/ml pyrithiamine (P0256; Sigma) or 200 μg/ml hygromycin B (H8080; Solarbio). Trans5α (CD201; Transgene) was used for the construction of plasmids and was propagated in Luria-Bertani (LB) broth at 37°C.
Plasmid construction.
The strong promoter gpdA was removed from pJW103 by digestion with restriction enzymes PstI and PmeI (MssI). The gpdA-deleted linear plasmid pJW103 was linked to the sequence fragments amplified with the primer pair Blank-F/R to form the ΔgpdA-gfp plasmid. The 5'LTR and 5'LTR-579 (5'LTR and the downstream 579-bp sequence) fragment were amplified from the STJ0105 gDNA with primer pairs 5'LTR-up-F/R and 5'LTR-579-up-F/R, respectively. The plasmids 5'LTR-gfp and 5'LTR-579-gfp were constructed by cloning the corresponding PCR products into the gpdA-deleted linear plasmid pJW103 using the pEASY-Uni seamless cloning and assembly kit (CU101; Transgene). All plasmids were transformed into STJ0105 according to the method described above. The strains carrying pJW103 or derived plasmids were checked by PCR (data not shown) and Southern blotting (Fig. S4B and S4C) (37). To visualize the promoter activity of 5'LTR, the strains with the indicated plasmids were cultured and observed. Images were captured using an Olympus BX51 microscope (Olympus, Japan).
Drug spot assay and MIC value test.
To test the sensitivity of A. fumigatus to azoles, POS, VRC, and ITC were supplemented in AMM. For the plate point assay, a 5-μl slurry of the indicated spores from the stock suspensions (107, 106, and 105 CFU/ml) was spotted onto the AMM. All plates were incubated at 37°C for 2 to 5 days. MIC values were determined by the method for the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST) (77).
RNA sequencing and analysis.
A. fumigatus conidia (3 × 108) of strains, including STJ0105, ΔAfut4977-sac1, Δ5'LTR, CEA17Δku80, and sac1 OE, were inoculated in triplicate into AMM liquid medium and cultured at 37°C, 200 rpm, for 18 h. Total RNA was extracted using TransZol Up (ET111-01; Transgene) according to the manufacturer’s instructions. The concentration of the extracted RNA samples was determined using a Nanodrop system (NanoDrop, Madison, WI, USA), and the integrity of the RNA was examined by the RNA integrity number (RIN) using an Agilent 2100 bioanalyzer (Agilent, Santa Clara, USA). The sequencing data were filtered with SOAPnuke (v1.5.2) (78) by removing reads containing sequencing adapter, removing reads whose low-quality base ratio (base quality less than or equal to is more than 20%), and removing reads whose unknown base (“N” base) ratio is more than 5%; afterward, clean reads were obtained and stored in FASTQ format. The clean reads were mapped to the reference genome using HISAT2 (v2.0.4) (79). Bowtie2 (v2.2.5) (80) was applied to align the clean reads to the reference coding gene set, and then the expression level of genes was calculated by RSEM (v1.2.12) (81). The heat map was drawn by pheatmap (v1.0.8) (82) according to the gene expression in different samples. Essentially, differential expression analysis was performed using DESeq2 (v1.4.5) (83) with a Q value of ≤0.05. To gain insight into the change of phenotype, KEGG (https://www.kegg.jp/) enrichment analysis of annotated differentially expressed genes was performed by Phyper based on the hypergeometric test. The significant levels of terms and pathways were corrected by Q value with a rigorous threshold (Q value ≤ 0.05) by Bonferroni.
Quantitative real-time PCR (qPCR).
For quantitative gene expression, a TransStart top green qPCR supermix (AQ131; Transgene) and a Roche LightCycler 96 system were used in accordance with the manufacturers’ instructions. Primer pairs used for Afut1, Afut4, sac1, gfp, and β-tubulin are shown in Table S1. Cycle conditions include two sections according to the manufacturer’s instructions. Relative quantification relates the PCR signal of the target transcript in a sample to a control based on the 2−ΔΔCT method (84). β-tubulin was used as a reference gene for A. fumigatus. Relative expression ratios were calculated by first calculating the cycle threshold (CT) changes in sample and control as ΔCTsample = CT(target) − CT(reference) and ΔCTcontrol = CT(target) − CT(reference), followed by calculating ΔΔCT = ΔCTsample − ΔCTcontrol and relative fold change = 2−ΔΔCT.
Phylogenetic analysis of whole genome and Afut4977-sac1.
For whole-genome phylogenetic analysis, all the SNP sites in the whole genome were determined. Model selection and phylogenetic analysis were performed by IQTree v1.6.8 with 1,000 bootstrap replicates to assess confidence in tree topologies (85). A. fumigatus var. RP-2014 was chosen as the outgroup. For Afut4 phylogenetic analysis, all the Afut4977-sac1 LTR-RTs in different A. fumigatus strains were identified by BLASTN alignment, and the nucleotide sequences were determined according to the coordinates in BLASTN results. All the Afut4977-sac1 LTR-RTs in different A. fumigatus strains were aligned by MAFFT v7.273 (86), and SNP sites were determined for phylogenetic analysis by IQTree with 1,000 bootstrap replicates. The nucleotide sequence of Afut3 from GenBank accession no. GQ294562 was chosen as the outgroup.
Statistical analysis.
Data shown in the figures either are from a representative experiment in triplicate or are presented as the mean ± standard error (SE) of results of three independent experiments. The significance of differences between the two groups was assessed by unpaired Student’s t tests with a 95% confidence interval, using GraphPad Prism software (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
ACKNOWLEDGMENTS
We thank Zongwei Li and Dingchen Li for bioinformatics analysis, Yong Chen for A. fumigatus collection, Jean Paul Latgé for providing the CEA17Δku80 strain, and Johannes Wagener for providing the various plasmids.
We declare no potential conflicts of interest.
This work was supported by the National Natural Science Foundation of China under grant no. 81971914, 8217082306, and 81772163.
The funders had no role in study design, data collection, and interpretation or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
Contributor Information
Fangyan Chen, Email: chenfangyan@163.com.
Li Han, Email: hanlicdc@163.com.
REFERENCES
- 1.Dagenais TR, Keller NP. 2009. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol Rev 22:447–465. 10.1128/CMR.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Latgé JP. 1999. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev 12:310–350. 10.1128/CMR.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szalewski DA, Hinrichs VS, Zinniel DK, Barletta RG. 2018. The pathogenicity of Aspergillus fumigatus, drug resistance, and nanoparticle delivery. Can J Microbiol 64:439–453. 10.1139/cjm-2017-0749. [DOI] [PubMed] [Google Scholar]
- 4.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 5.Lehrnbecher T, Kalkum M, Champer J, Tramsen L, Schmidt S, Klingebiel T. 2013. Immunotherapy in invasive fungal infection–focus on invasive aspergillosis. Curr Pharm Des 19:3689–3712. 10.2174/1381612811319200010. [DOI] [PubMed] [Google Scholar]
- 6.Patterson TF, Thompson GR, 3rd, Denning DW, Fishman JA, Hadley S, Herbrecht R, Kontoyiannis DP, Marr KA, Morrison VA, Nguyen MH, Segal BH, Steinbach WJ, Stevens DA, Walsh TJ, Wingard JR, Young JA, Bennett JE. 2016. Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 63:e1–e60. 10.1093/cid/ciw326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Resendiz Sharpe A, Lagrou K, Meis JF, Chowdhary A, Lockhart SR, Verweij PE, ISHAM/ECMM Aspergillus Resistance Surveillance Working Group. 2018. Triazole resistance surveillance in Aspergillus fumigatus. Med Mycol 56:83–92. 10.1093/mmy/myx144. [DOI] [PubMed] [Google Scholar]
- 8.Meis JF, Chowdhary A, Rhodes JL, Fisher MC, Verweij PE. 2016. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos Trans R Soc Lond B Biol Sci 371:20150460. 10.1098/rstb.2015.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhary A, Sharma C, Meis JF. 2017. Azole-resistant aspergillosis: epidemiology, molecular mechanisms, and treatment. J Infect Dis 216:S436–S444. 10.1093/infdis/jix210. [DOI] [PubMed] [Google Scholar]
- 10.Hagiwara D, Watanabe A, Kamei K, Goldman GH. 2016. Epidemiological and genomic landscape of azole resistance mechanisms in aspergillus fungi. Front Microbiol 7:1382. 10.3389/fmicb.2016.01382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker JE, Warrilow AG, Price CL, Mullins JG, Kelly DE, Kelly SL. 2014. Resistance to antifungals that target CYP51. J Chem Biol 7:143–161. 10.1007/s12154-014-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snelders E, Karawajczyk A, Schaftenaar G, Verweij PE, Melchers WJ. 2010. Azole resistance profile of amino acid changes in Aspergillus fumigatus CYP51A based on protein homology modeling. Antimicrob Agents Chemother 54:2425–2430. 10.1128/AAC.01599-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu W, Sillaots S, Lemieux S, Davison J, Kauffman S, Breton A, Linteau A, Xin C, Bowman J, Becker J, Jiang B, Roemer T. 2007. Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog 3:e24. 10.1371/journal.ppat.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellado E, Garcia-Effron G, Buitrago MJ, Alcazar-Fuoli L, Cuenca-Estrella M, Rodriguez-Tudela JL. 2005. Targeted gene disruption of the 14-alpha sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob Agents Chemother 49:2536–2538. 10.1128/AAC.49.6.2536-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen P, Liu J, Zeng M, Sang H. 2020. Exploring the molecular mechanism of azole resistance in Aspergillus fumigatus. J Mycol Med 30:100915. 10.1016/j.mycmed.2019.100915. [DOI] [PubMed] [Google Scholar]
- 16.Camps SM, van der Linden JW, Li Y, Kuijper EJ, van Dissel JT, Verweij PE, Melchers WJ. 2012. Rapid induction of multiple resistance mechanisms in Aspergillus fumigatus during azole therapy: a case study and review of the literature. Antimicrob Agents Chemother 56:10–16. 10.1128/AAC.05088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnan Natesan S, Wu W, Cutright JL, Chandrasekar PH. 2012. In vitro-in vivo correlation of voriconazole resistance due to G448S mutation (cyp51A gene) in Aspergillus fumigatus. Diagn Microbiol Infect Dis 74:272–277. 10.1016/j.diagmicrobio.2012.06.030. [DOI] [PubMed] [Google Scholar]
- 18.Mellado E, Garcia-Effron G, Alcázar-Fuoli L, Melchers WJ, Verweij PE, Cuenca-Estrella M, Rodríguez-Tudela JL. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob Agents Chemother 51:1897–1904. 10.1128/AAC.01092-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Linden JW, Camps SM, Kampinga GA, Arends JP, Debets-Ossenkopp YJ, Haas PJ, Rijnders BJ, Kuijper EJ, van Tiel FH, Varga J, Karawajczyk A, Zoll J, Melchers WJ, Verweij PE. 2013. Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clin Infect Dis 57:513–520. 10.1093/cid/cit320. [DOI] [PubMed] [Google Scholar]
- 20.Pérez-Cantero A, López-Fernández L, Guarro J, Capilla J. 2020. Azole resistance mechanisms in Aspergillus: update and recent advances. Int J Antimicrob Agents 55:105807. 10.1016/j.ijantimicag.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Benachenhou F, Sperber GO, Bongcam-Rudloff E, Andersson G, Boeke JD, Blomberg J. 2013. Conserved structure and inferred evolutionary history of long terminal repeats (LTRs). Mob DNA 4:5. 10.1186/1759-8753-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao M, Ma J. 2013. Co-evolution of plant LTR-retrotransposons and their host genomes. Protein Cell 4:493–501. 10.1007/s13238-013-3037-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiss RA. 2016. Human endogenous retroviruses: friend or foe? APMIS 124:4–10. 10.1111/apm.12476. [DOI] [PubMed] [Google Scholar]
- 24.Grandbastien MA. 2015. LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochim Biophys Acta 1849:403–416. 10.1016/j.bbagrm.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Elbarbary RA, Lucas BA, Maquat LE. 2016. Retrotransposons as regulators of gene expression. Science 351:aac7247. 10.1126/science.aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curcio MJ, Lutz S, Lesage P. 2015. The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae. Microbiol Spectr 3:MDNA3-0053-2014. 10.1128/microbiolspec.MDNA3-0053-2014. [DOI] [PubMed] [Google Scholar]
- 27.Feng G, Leem YE, Levin HL. 2013. Transposon integration enhances expression of stress response genes. Nucleic Acids Res 41:775–789. 10.1093/nar/gks1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omrane S, Audeon C, Ignace A, Duplaix C, Aouini L, Kema G, Walker AS, Fillinger S. 2017. Plasticity of the MFS1 promoter leads to multidrug resistance in the wheat pathogen Zymoseptoria tritici. mSphere 2:e00393-17. 10.1128/mSphere.00393-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mernke D, Dahm S, Walker AS, Laleve A, Fillinger S, Leroch M, Hahn M. 2011. Two promoter rearrangements in a drug efflux transporter gene are responsible for the appearance and spread of multidrug resistance phenotype MDR2 in Botrytis cinerea isolates in French and German vineyards. Phytopathology 101:1176–1183. 10.1094/PHYTO-02-11-0046. [DOI] [PubMed] [Google Scholar]
- 30.Kretschmer M, Leroch M, Mosbach A, Walker AS, Fillinger S, Mernke D, Schoonbeek HJ, Pradier JM, Leroux P, De Waard MA, Hahn M. 2009. Fungicide-driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea. PLoS Pathog 5:e1000696. 10.1371/journal.ppat.1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neuveglise C, Sarfati J, Latge JP, Paris S. 1996. Afut1, a retrotransposon-like element from Aspergillus fumigatus. Nucleic Acids Res 24:1428–1434. 10.1093/nar/24.8.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paris S, Latge JP. 2001. Afut2, a new family of degenerate gypsy-like retrotransposon from Aspergillus fumigatus. Med Mycol 39:195–198. 10.1080/mmy.39.2.195.198. [DOI] [PubMed] [Google Scholar]
- 33.Novikova OS, Fet V, Blinov AG. 2007. LTR retrotransposons from genomes of Aspergillus fumigatus and A. nidulans. Mol Biol (Mosk) 41:830–838. (In Russian.) [PubMed] [Google Scholar]
- 34.Snelders E, van der Lee HA, Kuijpers J, Rijs AJ, Varga J, Samson RA, Mellado E, Donders AR, Melchers WJ, Verweij PE. 2008. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med 5:e219. 10.1371/journal.pmed.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes WE, Pocklington MJ, Orr E, Paddon CJ. 1999. Mutations in the Saccharomyces cerevisiae gene SAC1 cause multiple drug sensitivity. Yeast 15:1111–1124. . [DOI] [PubMed] [Google Scholar]
- 36.Blosser SJ, Merriman B, Grahl N, Chung D, Cramer RA. 2014. Two C4-sterol methyl oxidases (Erg25) catalyse ergosterol intermediate demethylation and impact environmental stress adaptation in Aspergillus fumigatus. Microbiology (Reading) 160:2492–2506. 10.1099/mic.0.080440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertuzzi M. 2010. Sensory perception in model and pathogenic fungi: engineering misappropriation of response. Ph.D. thesis. Imperial College, London, United Kingdom. [Google Scholar]
- 38.Dichtl K, Helmschrott C, Dirr F, Wagener J. 2012. Deciphering cell wall integrity signalling in Aspergillus fumigatus: identification and functional characterization of cell wall stress sensors and relevant Rho GTPases. Mol Microbiol 83:506–519. 10.1111/j.1365-2958.2011.07946.x. [DOI] [PubMed] [Google Scholar]
- 39.da Silva Ferreira ME, Kress MR, Savoldi M, Goldman MH, Härtl A, Heinekamp T, Brakhage AA, Goldman GH. 2006. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell 5:207–211. 10.1128/EC.5.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helmschrott C, Sasse A, Samantaray S, Krappmann S, Wagener J. 2013. Upgrading fungal gene expression on demand: improved systems for doxycycline-dependent silencing in Aspergillus fumigatus. Appl Environ Microbiol 79:1751–1754. 10.1128/AEM.03626-12. [DOI] [PMC free article] [PubMed]
- 41.Cadena J, Thompson GR, 3rd, Patterson TF. 2016. Invasive aspergillosis: current strategies for diagnosis and management. Infect Dis Clin North Am 30:125–142. 10.1016/j.idc.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 42.Bueid A, Howard SJ, Moore CB, Richardson MD, Harrison E, Bowyer P, Denning DW. 2010. Azole antifungal resistance in Aspergillus fumigatus: 2008 and 2009. J Antimicrob Chemother 65:2116–2118. 10.1093/jac/dkq279. [DOI] [PubMed] [Google Scholar]
- 43.Howard SJ, Cerar D, Anderson MJ, Albarrag A, Fisher MC, Pasqualotto AC, Laverdiere M, Arendrup MC, Perlin DS, Denning DW. 2009. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis 15:1068–1076. 10.3201/eid1507.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiederhold NP, Gil VG, Gutierrez F, Lindner JR, Albataineh MT, McCarthy DI, Sanders C, Fan H, Fothergill AW, Sutton DA. 2016. First detection of TR34 L98H and TR46 Y121F T289A Cyp51 mutations in Aspergillus fumigatus isolates in the United States. J Clin Microbiol 54:168–171. 10.1128/JCM.02478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Ingen J, van der Lee HA, Rijs TA, Zoll J, Leenstra T, Melchers WJ, Verweij PE. 2015. Azole, polyene and echinocandin MIC distributions for wild-type, TR34/L98H and TR46/Y121F/T289A Aspergillus fumigatus isolates in the Netherlands. J Antimicrob Chemother 70:178–181. 10.1093/jac/dku364. [DOI] [PubMed] [Google Scholar]
- 46.Mohammadi F, Hashemi SJ, Zoll J, Melchers WJ, Rafati H, Dehghan P, Rezaie S, Tolooe A, Tamadon Y, van der Lee HA, Verweij PE, Seyedmousavi S. 2016. Quantitative analysis of single-nucleotide polymorphism for rapid detection of TR34/L98H- and TR46/Y121F/T289A-positive Aspergillus fumigatus isolates obtained from patients in Iran from 2010 to 2014. Antimicrob Agents Chemother 60:387–392. 10.1128/AAC.02326-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Astvad KM, Jensen RH, Hassan TM, Mathiasen EG, Thomsen GM, Pedersen UG, Christensen M, Hilberg O, Arendrup MC. 2014. First detection of TR46/Y121F/T289A and TR34/L98H alterations in Aspergillus fumigatus isolates from azole-naive patients in Denmark despite negative findings in the environment. Antimicrob Agents Chemother 58:5096–5101. 10.1128/AAC.02855-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mager DL, Stoye JP. 2015. Mammalian endogenous retroviruses. Microbiol Spectr 3:MDNA3-0009-2014. 10.1128/microbiolspec.MDNA3-0009-2014. [DOI] [PubMed] [Google Scholar]
- 49.Galindo-Gonzalez L, Mhiri C, Deyholos MK, Grandbastien MA. 2017. LTR-retrotransposons in plants: engines of evolution. Gene 626:14–25. 10.1016/j.gene.2017.04.051. [DOI] [PubMed] [Google Scholar]
- 50.Mita P, Boeke JD. 2016. How retrotransposons shape genome regulation. Curr Opin Genet Dev 37:90–100. 10.1016/j.gde.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandmeyer S, Patterson K, Bilanchone V. 2015. Ty3, a position-specific retrotransposon in budding yeast. Microbiol Spectr 3:MDNA3-0057-2014. 10.1128/microbiolspec.MDNA3-0057-2014. [DOI] [PubMed] [Google Scholar]
- 52.Zaratiegui M. 2013. Influence of long terminal repeat retrotransposons in the genomes of fission yeasts. Biochem Soc Trans 41:1629–1633. 10.1042/BST20130207. [DOI] [PubMed] [Google Scholar]
- 53.Beyer U, Moll-Rocek J, Moll UM, Dobbelstein M. 2011. Endogenous retrovirus drives hitherto unknown proapoptotic p63 isoforms in the male germ line of humans and great apes. Proc Natl Acad Sci USA 108:3624–3629. 10.1073/pnas.1016201108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen CJ, Lock WM, Mager DL. 2009. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448:105–114. 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 55.Conley AB, Piriyapongsa J, Jordan IK. 2008. Retroviral promoters in the human genome. Bioinformatics 24:1563–1567. 10.1093/bioinformatics/btn243. [DOI] [PubMed] [Google Scholar]
- 56.Landry JR, Rouhi A, Medstrand P, Mager DL. 2002. The Opitz syndrome gene Mid1 is transcribed from a human endogenous retroviral promoter. Mol Biol Evol 19:1934–1942. 10.1093/oxfordjournals.molbev.a004017. [DOI] [PubMed] [Google Scholar]
- 57.Medstrand P, Landry JR, Mager DL. 2001. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J Biol Chem 276:1896–1903. 10.1074/jbc.M006557200. [DOI] [PubMed] [Google Scholar]
- 58.Bieche I, Laurent A, Laurendeau I, Duret L, Giovangrandi Y, Frendo JL, Olivi M, Fausser JL, Evain-Brion D, Vidaud M. 2003. Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol Reprod 68:1422–1429. 10.1095/biolreprod.102.010322. [DOI] [PubMed] [Google Scholar]
- 59.Hayashi K, Yoshida H. 2009. Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J 57:413–425. 10.1111/j.1365-313X.2008.03694.x. [DOI] [PubMed] [Google Scholar]
- 60.Domansky AN, Kopantzev EP, Snezhkov EV, Lebedev YB, Leib-Mosch C, Sverdlov ED. 2000. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett 472:191–195. 10.1016/s0014-5793(00)01460-5. [DOI] [PubMed] [Google Scholar]
- 61.Servant G, Pennetier C, Lesage P. 2008. Remodeling yeast gene transcription by activating the Ty1 long terminal repeat retrotransposon under severe adenine deficiency. Mol Cell Biol 28:5543–5554. 10.1128/MCB.00416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C. 2012. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24:1242–1255. 10.1105/tpc.111.095232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Foti M, Audhya A, Emr SD. 2001. Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell 12:2396–2411. 10.1091/mbc.12.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tani M, Kuge O. 2014. Involvement of Sac1 phosphoinositide phosphatase in the metabolism of phosphatidylserine in the yeast Saccharomyces cerevisiae. Yeast 31:145–158. 10.1002/yea.3004. [DOI] [PubMed] [Google Scholar]
- 65.Moser von Filseck J, Vanni S, Mesmin B, Antonny B, Drin G. 2015. A phosphatidylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nat Commun 6:6671. 10.1038/ncomms7671. [DOI] [PubMed] [Google Scholar]
- 66.Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F, De Camilli P. 2015. INTRACELLULAR TRANSPORT. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science 349:428–432. 10.1126/science.aab1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kochendorfer KU, Then AR, Kearns BG, Bankaitis VA, Mayinger P. 1999. Sac1p plays a crucial role in microsomal ATP transport, which is distinct from its function in Golgi phospholipid metabolism. EMBO J 18:1506–1515. 10.1093/emboj/18.6.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang B, Yu Q, Jia C, Wang Y, Xiao C, Dong Y, Xu N, Wang L, Li M. 2015. The actin-related protein Sac1 is required for morphogenesis and cell wall integrity in Candida albicans. Fungal Genet Biol 81:261–270. 10.1016/j.fgb.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 69.Burns KH, Boeke JD. 2012. Human transposon tectonics. Cell 149:740–752. 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sehgal A, Lee CY, Espenshade PJ. 2007. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet 3:e131. 10.1371/journal.pgen.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carr PD, Tuckwell D, Hey PM, Simon L, d'Enfert C, Birch M, Oliver JD, Bromley MJ. 2010. The transposon impala is activated by low temperatures: use of a controlled transposition system to identify genes critical for viability of Aspergillus fumigatus. Eukaryot Cell 9:438–448. 10.1128/EC.00324-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Albarrag AM, Anderson MJ, Howard SJ, Robson GD, Warn PA, Sanglard D, Denning DW. 2011. Interrogation of related clinical pan-azole-resistant Aspergillus fumigatus strains: G138C, Y431C, and G434C single nucleotide polymorphisms in cyp51A, upregulation of cyp51A, and integration and activation of transposon Atf1 in the cyp51A promoter. Antimicrob Agents Chemother 55:5113–5121. 10.1128/AAC.00517-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pyrzak W, Miller KY, Miller BL. 2008. Mating type protein Mat1-2 from asexual Aspergillus fumigatus drives sexual reproduction in fertile Aspergillus nidulans. Eukaryot Cell 7:1029–1040. 10.1128/EC.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR, Oakley B. 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc 1:3111–3120. 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 76.Dichtl K, Ebel F, Dirr F, Routier FH, Heesemann J, Wagener J. 2010. Farnesol misplaces tip-localized Rho proteins and inhibits cell wall integrity signalling in Aspergillus fumigatus. Mol Microbiol 76:1191–1204. 10.1111/j.1365-2958.2010.07170.x. [DOI] [PubMed] [Google Scholar]
- 77.Subcommittee on Antifungal Susceptibility Testing of the ESCMID European Committee for Antimicrobial Susceptibility Testing. 2008. EUCAST Technical Note on the method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia-forming moulds. Clin Microbiol Infect 14:982–984. 10.1111/j.1469-0691.2008.02086.x. [DOI] [PubMed] [Google Scholar]
- 78.Li R, Li Y, Kristiansen K, Wang J. 2008. SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714. 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 79.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kolde R. 2015. Package 'pheatmap', v 1.0.8. https://mran.microsoft.com/snapshot/2017-09-01/web/packages/pheatmap/pheatmap.pdf.
- 83.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 85.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aac.00291-21-s0001.pdf, PDF file, 1.4 MB (1.4MB, pdf)
Table S2. Download aac.00291-21-s0002.xlsx, XLSX file, 0.1 MB (117.2KB, xlsx)
Table S3. Download aac.00291-21-s0003.xlsx, XLSX file, 0.04 MB (36KB, xlsx)
Table S4. Download aac.00291-21-s0004.xlsx, XLSX file, 0.1 MB (121.1KB, xlsx)