Abstract
PKR is a cellular serine/threonine kinase that phosphorylates eukaryotic translation initiation factor 2α (eIF2α) to regulate protein synthesis. PKR also plays a role in the regulation of transcription, programmed cell death and the cell cycle, processes which likely involve other substrates. In a yeast two-hybrid screen, we isolated human protein phosphatase 2A (PP2A) regulatory subunit B56α as a PKR-interacting protein. The interaction between B56α and PKR was confirmed by in vitro binding assays as well as by in vivo coimmunoprecipitation, and this interaction is dependent on the catalytic activity of PKR. Moreover, recombinant B56α was efficiently phosphorylated by PKR in vitro and an isoelectric point shift in B56α was detected in extracts from cells induced with the PKR activator pIC. An in vitro dephosphorylation assay showed that when B56α was phosphorylated by PKR, the activity of PP2A trimeric holoenzyme was increased. A functional interaction between B56α and PKR was observed in cotransfection assays, where a B56α-mediated increase in luciferase expression was inhibited by cotransfection with wild-type PKR. This is likely due to a decreased level of eIF4E phosphorylation caused by an increase in PP2A activity following PKR phosphorylation of B56α. Taken together, our data indicate that PKR can modulate PP2A activity by phosphorylating B56α to regulate cellular activities.
Protein phosphorylation is a critical regulatory mechanism utilized by the cell to regulate a myriad of different enzyme reactions and signaling pathways. The steady-state phosphorylation status of a protein is regulated through the combined activities of kinases and phosphatases. Protein phosphatase 2A (PP2A) (40, 46, 54) is the major cellular serine/threonine phosphoprotein phosphatase and plays important roles in regulating the cell cycle (32, 48), apoptosis (16), transcription (1), translation (6), and signal transduction (19). PP2A consists of three subunits: a 36-kDa catalytic C subunit, a 60-kDa regulatory A subunit, and a regulatory B subunit. PP2A can exist in the form of either AC core dimer (PP2Ac) or heterotrimeric ABC holoenzyme. Free C subunit is not found in the cell. Generally, PP2A is believed to be a negative regulator of cell growth and possibly a tumor suppressor, since inactivation of the regulatory A subunit due to gene mutation is tumorigenic (53). The regulation of PP2A activity can occur at several levels. Structurally, association of different B regulatory subunits with the AC core dimer can result in altered substrate specificity, catalytic activity, and subcellular localization. There are three structurally unrelated B families, B(B55), B′(B56), and B", each having several closely related proteins and isoforms with tissue-specific expression. PP2A activity is also subject to regulation by posttranslational modification. For example, the catalytic subunit of PP2A can be phosphorylated in vitro by tyrosine kinases, including p60v-src, p56tck, epidermal growth factor, and insulin receptor (9). Many of the regulatory B proteins are phosphoproteins (36), but until this study, there was no evidence for a role of B protein phosphorylation in the regulation of PP2A activity.
PKR has long been known to mediate the antiviral activity of interferons. PKR is activated following infection with different viruses and acts to suppress viral replication (for reviews, see references 13, 56, and 57). The antiviral activity of PKR is attributed to its inhibitory phosphorylation of eukaryotic translation initiation factor 2α (eIF2α). The kinase activity of PKR is dependent on but not limited to double-stranded RNA (dsRNA) binding (18). PKR has a bipartite structure consisting of (i) an N-terminal dsRNA binding domain which contains two dsRNA binding motifs responsible for dsRNA binding and (ii) an C-terminal catalytic domain (38).
PKR functions in other cellular activities, including growth regulation, transcription, the cell cycle, and cell death. The expression of functional PKR in yeast results in growth inhibition which can be rescued by coexpressing S51A mutant eIF2α, which is not phosphorylated by PKR (11). Likewise, overexpression of PKR in mammalian cells arrests cell growth and promotes apoptosis (3). Conversely, overexpression of mutant PKR protein causes the transformation of NIH 3T3 cells into cells having a tumorigenic phenotype (4, 28, 33, 39). However, the mechanisms underlying the effects of PKR on mammalian cell growth remain unclear and likely involve regulatory pathways in addition to eIF2α phosphorylation. For example, PKR plays a role in NF-κB activation by dsRNA, tumor necrosis factor alpha, and gamma interferon (56). Mouse embryonic fibroblasts derived from PKR knockout mice show deficient or reduced activation of NF-κB or IRF1 by dsRNA or tumor necrosis factor alpha (29, 59). PKR has also been found to physically interact with Stat1 (58), Stat3 (A. Deb and B. R. G. Williams, unpublished data) and p53 (14, 15), and it may regulate the activities of these transcription factors. PKR has also been implicated in cell cycle regulation (61), controlling the induction of apoptosis (3, 17) and regulating stress-activated cell signaling (12, 20, 23).
Clearly, PKR plays an important role in different pathways of cellular activities. However, with the exception of eIF2α, substrates mediating the activities of PKR remain to be convincingly identified in vivo. Accordingly, to search for novel substrates for PKR, we performed a yeast two-hybrid screen (34) using a mutant of PKR (L362Q) with reduced kinase activity as bait. One interacting clone encoded a regulatory subunit of PP2A, B56α (37), suggesting an interplay between PKR and PP2A. Here, we identify B56α as a novel substrate of PKR and show that by phosphorylating B56α, PKR may modulate PP2A activity, resulting in a potential novel pathway of protein synthesis inhibition.
MATERIALS AND METHODS
Cell culture.
Human glioblastoma T98G, fibrosarcoma 2fTGH, uterus carcinoma HeLa S3, monkey kidney COS-1, and murine 3T3-like pkr+/+ or pkr−/− fibroblasts (C57/BL6 background) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and grown at 37°C with 5% CO2.
Yeast two-hybrid screen of human HeLa S3 cDNA library.
The bait plasmid pGBT9-PKR M3 was constructed by cloning mutant human PKR (M3, L362Q) cDNA (7) into pGBT9 (Clontech) fused to the Gal4 binding domain (Gal4BD). Screening of a HeLa S3 cDNA Gal4 activation domain (Gal4AD) library was performed according to the instructions of the vendor (Clontech).
Construction of the full-length B56α cDNA.
The B56α cDNA clone pGAD GH B56αΔN97 isolated from the HeLa S3 Gal4AD cDNA library has a truncation of about 300 bp encoding the N-terminal 97 amino acids of B56α. This portion of cDNA was prepared by reverse transcription of total mRNA from HeLa S3 cells by using Superscriptase II (Gibco BRL) with an antisense primer (ZX4-dw [5′TAA TAA CAT ATG TCA GGG CTC TCC AAA AAT CTC AAG]) specific to human B56α cDNA followed by a PCR with primer ZX4-up (5′TAA TAA CAT ATG AGC GTC AGG GCC GCG GAG ATG T) and ZX4-dw. This cDNA product was cloned into pBS II KS via BamHI and EcoRI sites to create pBS-B56αN97. A 1.2-kb EcoRI fragment containing the B56α C-terminal cDNA obtained from the original B56α truncation clone pGAD GH B56αΔN97 was then inserted into pBS-B56αN97. The resulting construct, pBS-B56α, contains a cDNA encoding full-length B56α protein. Recombinant B56α expression plasmid pET28c-B56α was constructed by digesting pBS-B56α with BamHI and XhoI and ligating this BamHI- and XhoI-digested B56α fragment to pET28c (Novagen). B56α protein expressed from this vector has a six-histidine epitope tag followed by a T7 tag in the N terminus. To construct the mammalian expression plasmid pZeoSV-B56α, pBS-B56α was digested with BamHI and XhoI and ligated into pZeoSV (Invitrogen).
Recombinant protein preparation and purification.
Both glutathione S-transferase–PKR (6) and histidine-tagged recombinant human PKR protein (8) were produced as described previously. Both recombinant B56α and eIF2α were expressed as six-histidine-tagged proteins and purified by affinity chromatography using His-Bind metal chelation resin according to the instructions of the manufacturer (Novagen). B56α was expressed from the pET28c-B56α expression construct in Escherichia coli BL21(DE3)pLysS cells (Novagen), which were grown at 37°C in 500 ml of Luria-Bertani medium containing 30 μg of kanamycin per ml and 34 μg of chloramphenicol per ml and induced by 1.0 mM IPTG (iospropyl-β-d-thiogalactopyranoside) for 45 min, and purified under natural conditions. Human eIF2α was expressed from pQE-eIF2α (7) in E. coli M15(pREP4) host cells (Qiagen) grown in 4 liters of Luria-Bertani culture and induced with 1 mM IPTG for 4 h and was purified under denaturing conditions. Further purification of eIF2α was performed using a Q Sepharose Fast-Flow anion-exchange column (Pharmacia) with a starting buffer of 50 mM Tris-HCl (pH 7.9) and 50 mM NaCl. After washing of the column with 250 mM NaCl–Tris buffer, eIF2α was eluted by increasing the NaCl concentration to 500 mM. eIF2α protein was concentrated and stored at −80°C after addition of glycerol to 10%.
In vitro phosphorylation assay. (i) Phosphorylation of MBP by PKCα.
Myelin basic protein (MBP) (100 μg) was phosphorylated with 25 ng of protein kinase (PKC) (Upstate Biotechnology) in 40 μl of assay dilution buffer (20 mM MOPS [morpholinepropanesulfonic acid] [pH 7.2], 25 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM dithiothreitol, 1 mM CaCl2), 10 μl of PKC activator (0.5 mg of phosphatidylserine per ml and 0.5 mg of diglycerides per ml in assay dilution buffer), and 10 μl of ATP mixture (75 mM MgCl2, 50 μM ATP, 150 μCi of [γ-32P]ATP). The reaction was carried out at 30°C for 20 min.
(ii) Phosphorylation of B56α or eIF2α by PKR.
Recombinant B56α or eIF2α protein solution (about 200 ng) was mixed with 30 μl of DBGA buffer (10 mM Tris-HCl [pH 7.6], 50 mM KCl, 2 mM Mg acetate, 7 mM 2-mercaptoethanol, 20% glycerol), 20 μl of DBGB buffer (2.5 mM MnCl2 in DBGA), 5 μl of ATP mixture (10 μM ATP and 1.5 μCi of [γ-32P]ATP per ml in DBGA), and 5 μl of poly(rI:rC) (pIC) (12 ng/μl in DBGA). The kinase reaction was carried out at 30°C for 20 min after addition of 100 ng of recombinant PKR to the mixture.
Site-directed mutagenesis of B56α.
Site-directed mutagenesis of B56α was carried out with a reaction kit from Clontech according to the manufacturer's instructions. To mutate serine 28 of B56α to alanine, mutation primer MP-S28A (5′P-CACCCGGAAAGCGGTCCGCAAG-3′) and selection primer B56α-pSelect (5′P-GGGGCCCGGTTCCCAGCTTTTG-3′ with a mutated KpnI site) were annealed with denatured pBS-FLAG-B56α plasmid DNA. T4 DNA polymerase was added, and following DNA synthesis, the gaps were ligated with T4 DNA ligase. The product was digested with KpnI and transformed into mutS E. coli. Second-round selection was performed by isolating plasmid DNA, digesting with KpnI, and transforming into DH5α. Plasmid DNA isolated from individual colonies was analyzed by restriction digestion and sequencing to confirm the success of mutation. Quadruple mutation of B56α (S18A, S28A, S323A, and S436A) was achieved by annealing pBS-FLAG-B56α/S28A with mutation primers MP-S18A (5′P-CCA TCT CGG CCG CGG AGA AAG TG-3′), MP-S323A (5′P-GGC CAA AAA CCT GCG CTC AGA AAG AGG TGA TG-3′), and MP-436A (5′P-GAC CTT ACT AGC GCA TAC AAA GCT G-3′) and selection primer B56α-pSelect2 (5′P-GAT ACC GTC GAG CTC GAG GGG G-3′). B56α mutant fragments were then recloned into the pET28C expression vector, and proteins were expressed and purified as described above.
PAA assay and tryptic phosphopeptide mapping of B56α phosphorylation sites.
Two-dimensional separation of phosphoamino acids (PAA) and phosphopeptides of B56α using thin-layer cellulose (TLC) plates was performed as described previously (5). Recombinant B56α was phosphorylated by PKR and labeled in vitro with [γ-32P]ATP as described above, and the proteins were separated on a sodium dodecyl sulfate (SDS)–8% polyacrylamide gel, excised from the dried gel, eluted out of the gel slice with 50 mM NH4HCO3, and precipitated with 20% cold trichloroacetic acid. For PAA assay, the sample was resuspended in 100 μl of 5.7 M HCl and boiled at 110°C for 1 h. After hydrolysis, the sample was dried in a Speed-Vac (Savant) and resuspended in pH 1.9 buffer (2.2% formic acid and 7.8% acetic acid), which contains 15 parts of buffer to 1 part of cold PAA standards (1.0 mg [each] of phosphoserine, phosphothreonine, and phosphotyrosine per ml) (Sigma). The sample was applied to a TLC plate (C.B.S. Scientific Company, Inc.), and two-dimensional electrophoresis was carried out using the Hunter thin-layer peptide mapping electrophoresis system (model no. HTLE-7000; C.B.S.). The first-dimension separation was done with pH 1.9 buffer at a constant 1,500 V for 30 min, and the second-dimension separation was done in pH 3.5 buffer (5% acetic acid and 2.5% pyridine) at a constant 1,300 V for 25 min. PAA standard colors were developed by spraying 0.25% ninhydrin in acetone and baking the plate at 65°C for 30 min.
For peptide mapping, the sample was dissolved after trichloroacetic acid precipitation in 100 μl of cold perfomic acid, oxidized at 0°C for 60 min, and lyophilized in a Speed-Vac. The oxidized protein pellet was resuspended in 50 μl of 50 mM ammonium bicarbonate (pH 8.0 to 8.3), and TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone)-trypsin (10 μg) was added to cleave the protein at 37°C overnight. After digestion and lyophilization, the peptides were separated on TLC plates in the first dimension by electrophoresis using the Hunter thin-layer peptide mapping electrophoresis system in pH 1.9 buffer at 1,600 V for 1 h. The second-dimension separation was performed by chromatography using an organic solvent buffer (37.5% n-butanol, 25% pyridine, and 7.5% acetic acid) in a chromatography tank. After overnight separation, the plate was dried and exposed to an X-ray film.
In vitro phosphatase assay.
Approximately 2 μg of B56α was phosphorylated by PKR as described above. As an unphosphorylated B56α control, the B56α was incubated in the same phosphorylation reaction mixture except that no PKR was added. The reaction mixtures were concentrated and washed with phosphatase reaction buffer (20 mM MOPS [pH 7.2], 25 mM 2-mercaptoethanol, 10 mM MgCl2, 100 μg of bovine serum albumin per ml) without bovine serum albumin, using a 0.5-ml ultrafree centrifugal filter device spin tube (Biomax 30K NMWL membrane; Millipore), and finally concentrated to 20 μl. For the dephosphorylation of MBP by PP2A AC dimer (PP2Ac), 15 μl of MBP phosphorylation mixture (25 μg of MBP) was diluted to a final volume of 200 μl with phosphatase assay buffer. Dephosphorylation was initiated at 30°C with the addition of 150 ng of PP2Ac (Upstate Biotechnologies). For dephosphorylation of MBP by PP2A-B56α, 15 μl of MBP substrate mix was diluted to a final volume of 180 μl with phosphatase assay buffer, and then 20 μl of B56α, either phosphorylated or not, was quickly mixed with 150 ng of PP2Ac and added to the reaction mixture. At each time point (1, 2, 5, 10, and 20 min), 40 μl was sampled and 5 μl of 10× PP2A stop solution (50 mM EDTA, 1 M NaF, 20 mM NaPPi) was added. All samples were analyzed on SDS-polyacrylamide gels and exposed for PhosphorImager (Molecular Dynamics) analysis. Protein phosphorylation was quantitated by ImageQuant version 1.1 software after scanning the screen using a Storm PhosphorImager (Molecular Dynamics). For the eIF2α dephosphorylation assay, recombinant eIF2α (10 μg) was phosphorylated by PKR in vitro, and the phosphorylation reaction product was washed with phosphatase reaction buffer and concentrated to 40 μl. For each phosphatase assay, 12 μl of eIF2α (3 μg) was used. The rest of the procedure was as for the MBP dephosphorylation assay described above.
Coimmunoprecipitation of human PKR with B56α.
Human glioblastoma T98G cells were lysed with immunoprecipitation lysis buffer (50 mM HEPES [pH 7.5], 0.5% NP-40, 150 mM NaCl, 2 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 1 mM phenyl methyl sulfonyl fluoride, 5 μg of leupeptin per ml, 2 μg of aprotinin per ml) on ice for 30 min. The crude lysate was cleared by centrifugation at 14,000 × g for 20 min, and the supernatant was collected. To assay the endogenous association between PKR and B56α, 1 to 3 mg of protein extract was used for immunoprecipitation with an antibody against human PKR (61). After addition of antibody, the lysate was incubated on ice for 30 min and mixed with 40 μl of protein G-conjugated Sepharose resin (Pharmacia), and the mixture was agitated at 4°C overnight then washed with immunoprecipitation lysis buffer. The proteins were separated with SDS–8% polyacrylamide gel and transferred to an Immobilon-P membrane (Millipore). Western blotting was performed using a polyclonal antibody against human B56α (Santa Cruz). To assay kinase activation-dependent association of PKR with recombinant B56α, T98G cell extract (200 μg) was immunoprecipitated with a monoclonal antibody against human PKR as described above. After washing with immunoprecipitation lysis buffer, the immunoprecipitates were washed three times with DBGA buffer. Recombinant histidine-tagged B56α (1 μg) in 60 μl of kinase reaction buffer (10 mM Tris-HCl [pH 7.6], 50 mM KCl, 2 mM Mg acetate, 7 mM 2-mercaptoethanol, 0.83 mM MnCl2, 100 μM ATP, 20% glycerol) either with or without pIC (1 ng/μl) was added to the immunoprecipitates, and the mixture was incubated at 30°C for 20 min. Following washing with DBGA buffer, the proteins were separated with an SDS–8% polyacrylamide gel and analyzed by Western blot assay using a polyclonal antibody against human B56α. The membrane was stripped and reprobed with a polyclonal antibody against human PKR. T98G and 2fTGH cells (2 × 106) were also transfected with 4 μg of FLAG-B56α expression plasmid DNA (pZeoSV-FLAG-B56α) using Lipofectamine (Gibco BRL). After 24 h of transfection, the cells were treated with 100 μg of pIC per ml in the presence of 100 nM okadaic acid for 1 h. The cell lysates were prepared with cell lysis buffer, and 2-mg cell lysates were used for immunoprecipitation of FLAG-B56α with anti-FLAG M2 affinity gel (Sigma). The gel was washed with cell lysis buffer after agitation at cold room overnight, and the association of PKR was analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting using a polyclonal antibody.
Preparation of endogenous PP2A.
Cell extracts was prepared from HeLa S3 cells as described above. Anti-human PP2A C subunit polyclonal antibody (4 μg) (Upstate Biotechnologies) was added to 200 μg of cell extract and incubated on ice for 30 min. Protein G-Sepharose beads were mixed with the extract and agitated at 4°C overnight, and the immunoprecipitates were washed with immunoprecipitation lysis buffer and used in the kinase assay described above.
IEF–SDS-PAGE two-dimensional analysis of protein phosphorylation.
Human 2fTGH cells and murine 3T3-like pkr+/+ or pkr−/− fibroblasts (2 × 106) were transfected with 4 μg of FLAG-B56α expression plasmid DNA pZeoSV-FLAG-B56α using Lipofectamine reagent. After 24 h of transfection, the cells were treated with 100 μg of pIC per ml in the presence of 100 nM okadaic acid for 1 h. The cells were lysed with cell lysis buffer, and 2 mg of cell lysate was used for immunoprecipitation of FLAG-B56α with anti-FLAG M2 affinity gel. FLAG-B56α was eluted from the gel by adding 125 μl of rehydration buffer {8 M urea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 10 mM dithiothreitol, 0.2% Bio-Lytes 3/10}. IPG strips (7 cm, pH 5 to 8; Bio-Rad) were rehydrated with rehydration buffer containing FLAG-B56α for 12 h before running isoelectric focusing (IEF). The IEF electrophoresis was performed with a Protean IEF Cell (Bio-Rad) according to the instruction manual. Focusing was carried out with 250 V for 15 min with a linear increase from 250 to 4,000 V for 2 h and 4,000 V for 5 h. After focusing, the strips were laid on top of SDS-polyacrylamide gels for second-dimension separation. B56α was examined by Western blotting using a polyclonal antibody against B56α as mentioned above. eIF4E phosphorylation was assayed from 2fTGH cells which were lysed directly with rehydration buffer. About 300 μg of cell lysate was reabsorbed into 17-cm IPG strips (pH 3 to 10), and IEF-PAGE separation was performed as described above. Western blotting of eIF4E was performed using a monoclonal antibody against rabbit eIF4E (Transduction Laboratories, Lexington, Ky.).
Transfection assays of exogenous luciferase expression.
COS-1 cells were plated in six-well plates at 3 × 105 cells per well. The following day six duplicated plates were transfected with 200 ng of luciferase reporter construct pGL2p (Promega), 1.5 μg of pZeoSV-B56α or pZeoSV vector, and 1.0 μg of either pRC-PKR, pRC-PKR(K296R), or pRC vector. The cells were transfected for 3 h using Lipofectamine Plus (Gibco BRL) according to manufacturer's instructions. After 24 h, the cells were lysed with 300 μl of 1× reporter lysis buffer (Promega), and 40 μl of cell extract was assayed for luciferase activity. To examine the effect of okadaic acid on protein expression, cells were transfected with pGL2p (200 ng) in the presence or absence of okadaic acid (100 nM). After 3 h, the medium was replaced with complete Dulbecco modified Eagle medium with or without 100 nM okadaic acid and incubated for a further 4 h. Cell extracts were prepared and luciferase activity was measured as described above. Transfection treatment of murine 3T3-like fibroblasts followed same protocol with the amount of DNA described in the legend to Fig. 7. Okadaic acid treatment of fibroblasts lasted overnight, as they are more resistant to okadaic acid-induced apoptosis than COS-1 cells.
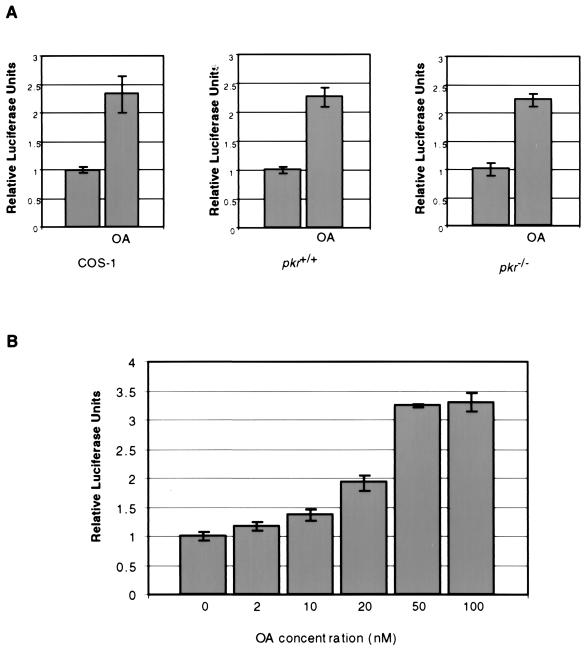
FIG. 7.
Stimulation of protein synthesis by B56α is inhibited by PKR. (A) The PP2A inhibitor okadaic acid (OA) stimulates exogenous luciferase expression in cells. COS-1 (left panel), pkr+/+ (middle panel), or pkr−/− (right panel) cells were transfected with luciferase expression vector pGL2p (200 ng in COS-1 cells and 500 ng in 3T3-like cells) and treated with 100 nM OA (right bars) or left untreated (left bars). Six duplicate samples were prepared for COS-1 cells, and four duplicate samples were prepared for fibroblasts. (B) OA sensitivity assay of exogenous luciferase expression in pkr+/+ cells. pkr+/+ cells were transfected with 200 ng of luciferase expression vector pGL2p and treated with different concentrations of OA. Luciferase units were measured after 24 h of transfection and plotted as relative units against an untreated control. Triplicate samples were assayed. (C) A luciferase reporter, pGL2p (200 ng), was cotransfected into COS-1 cells (left panel) with B56α expression construct pZeoSV-B56α (bars 2, 4, and 6) (1.5 μg), wild-type PKR expression construct pRC-PKR(wt) (bars 3 and 4) (1 μg), or mutant PKR construct pRC-PKR(K296R) (bars 5 and 6) (1 μg). pGL2p (200 ng) was cotransfected into pkr+/+ cells (right panel) with B56α expression construct pZeoSV-B56α (bars 2, 4, and 6) (0.5 μg), wild-type PKR expression construct pRC-PKR(wt) (bars 3 and 4) (0.5 μg), or mutant PKR construct pRC-PKR(K296R) (bars 5 and 6) (0.5 μg). Cell extracts were prepared 24 h after transfection and assayed for luciferase activity, and the values from the means of six (COS-1) or three (pkr+/+) duplicate samples were plotted. (D) Increased expression of exogenous luciferase in pkr−/− cells transfected with B56α. pkr-null cells and wild-type cells were transfected with 200 ng of pGL2p. The cells were also cotransfected with 1 μg of either pZeoSV-β-gal or pZeoSV-B56α. Luciferase units were measured in 24 h after the start of transfection. The mean value from four duplicate samples each was plotted. Error bars indicate standard deviations.
RT-PCR detection of luciferase mRNA.
3T3-like pkr+/+ cells were cotransfected with 500 ng of pGL2p and 1 μg of pZeoSV-B56α or pZeoSV as described earlier. The cells which were transfected with vector pZeoSV either were left untreated or were treated with 50 nM okadaic acid for 24 h. DNA-free mRNA was isolated from the cells by using a High Pure RNA Isolation Kit (Boehringer Mannheim). Reverse transcription (RT) was performed with Superscript II from 1 μg of RNA sample according to the instructions of the manufacturers (Gibco BRL). PCR of luciferase or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was carried out with 1/10 of the RT sample, using 30 cycles of denaturation at 94°C for 20 s, renaturation at 55°C for 20 s, and extension at 72°C for 30 s. Real-time SYBR Green PCR was performed according to the instructions of the manufacturer (Perkin-Elmer Applied Biosystems).
RESULTS
Identification of B56α as a PKR-interacting protein.
To identify PKR-interacting proteins, we performed a yeast two-hybrid screen of a human HeLa S3 Gal4AD fusion cDNA library using mutant PKR M3 (L362Q) Gal4BD fusion protein as bait. Since wild-type PKR inhibits yeast cell growth (11), previous two-hybrid screens have used catalytically inactive PKR (42). However, we reasoned that kinase activity may be important for PKR substrate interactions and therefore selected an L362Q mutant PKR which exhibits low residual autophosphorylation activity but does not inhibit yeast growth (7). A primary screen of the library identified 30 histidine-positive clones, 10 of which were positive for β-galactosidase expression. The cDNAs from these clones were sequenced, and a BLAST search against nonredundant human nucleotide sequences in the National Center for Biotechnology Information database identified two known proteins, while the remainder represented novel genes. The identified proteins are PP2A regulatory subunit B56α (37) and P76, also known as NF90 (26) or M-phase phosphoprotein 4 (35). P76 was also identified as a dsRNA-interacting protein and shown to be phosphorylated by PKR (43). Since B56α is a regulatory subunit of PP2A, we hypothesized that the interaction between PKR and B56α may represent a mechanism by which PKR regulates PP2A activity.
B56α is a substrate for PKR in vitro.
The B56α cDNA obtained from the initial library screen encoded a B56α protein with a truncation of 97 amino acids from the amino terminus (37). A full-length B56α cDNA was constructed by ligation with a cDNA fragment encoding the corresponding amino-terminal peptide (see Materials and Methods), and the recombinant protein was tested for dsRNA-dependent phosphorylation by recombinant human PKR. In the absence of the dsRNA analog pIC, recombinant PKR exhibited only a low level of kinase activity which was insufficient to phosphorylate B56α. However, when PKR was activated by dsRNA, B56α was phosphorylated to a high level (Fig. 1A). Phosphorylation of B56α can be also achieved by using glutathione S-transferase fusion human PKR (data not shown).
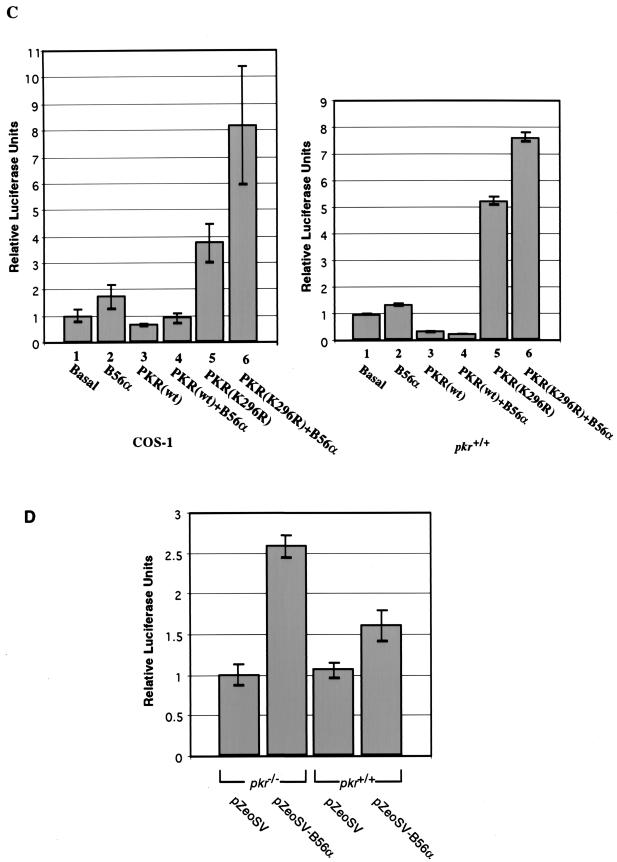
FIG. 1.
Assay of substrate phosphorylation of B56α and eIF2α by PKR. The proteins were separated by SDS-PAGE and visualized by autoradiography. (A) Phosphorylation of B56α by recombinant His-PKR in vitro. Recombinant B56α (200 ng) was mixed with 100 ng of recombinant human PKR in kinase reaction buffer either in the presence (lane 3) or in the absence (lane 2) of pIC. PKR autophosphorylation is shown in lane 1. (B) Comparison of the substrate phosphorylation of eIF2α and B56α by PKR. About 200 ng of recombinant B56α (lanes 3 and 4) or eIF2α (lanes 1 and 2) was reacted with 100 ng of recombinant PKR in kinase reaction buffer either in the presence (lanes 1 and 3) or in the absence (lanes 2 and 4) of pIC. The weaker autophosphorylation of PKR in the presence of eIF2α was due to impurity in the eIF2α preparation (data not shown).
The relative efficiency of phosphorylation of B56α by PKR was compared to that of the well-characterized substrate eIF2α (Fig. 1B). While the basal level of kinase activity of PKR was unable to phosphorylate B56α, eIF2α exhibited a low level of phosphorylation. However, activated PKR phosphorylated both substrates efficiently and to comparable levels (Fig. 1B). A correlation between autophosphorylation and substrate phosphorylation activity of PKR has long been established (22, 44, 51). Although it may hold true for eIF2α phosphorylation, the lack of basal phosphorylation of B56α in this assay may reflect a unique property of the interaction between PKR and B56α.
B56α is multiple phosphorylated on serine and threonine by PKR.
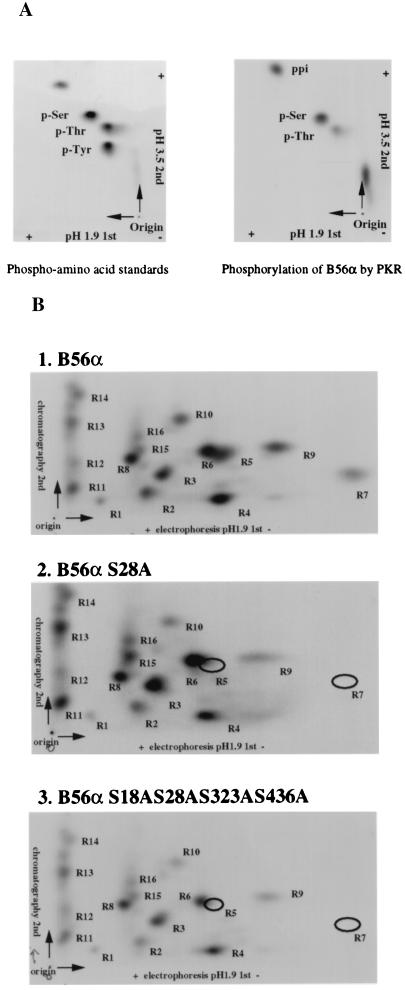
PKR has been characterized as a Ser/Thr kinase, but no consensus phosphorylation site has been described. To determine if B56α was phosphorylated on Ser/Thr by PKR, a PAA assay was performed on phosphorylated protein. Recombinant B56α was phosphorylated by PKR in vitro and hydrolyzed by boiling HCl. The PAA were analyzed by two-dimensional thin-layer electrophoresis. Compared with PAA standards (Fig. 2A, left panel), PKR phosphorylates B56α mainly on serine residues and to a lesser extent also on threonine (Fig. 2A, right panel).
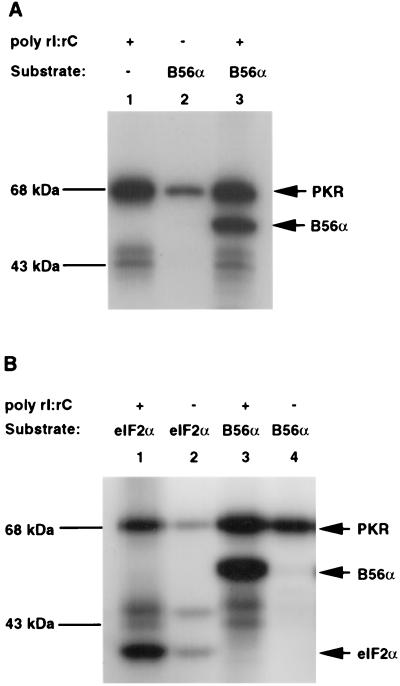
FIG. 2.
PAA assay (A) and peptide mapping (B) of B56α phosphorylation sites by PKR. (A) B56α was radioactively phosphorylated by PKR in vitro and acid hydrolyzed. Amino acids were separated on a TLC plate. PAA standards (p-Ser, p-Thr, and p-Tyr) were also loaded and visualized by color reaction with 0.25% ninhydrin in acetone (left panel). PAA in B56α phosphorylation by PKR were visualized by autoradiography (right panel). ppi, free phosphate. (B) Wild-type (panel 1), single point mutant (S28A) (panel 2), or quadruple point mutant (S18AS28AS323AS436A) (panel 3) B56α was radioactively phosphorylated by PKR in vitro and digested with TPCK-trypsin. Peptides were separated on TLC plates with electrophoresis at the first dimension and chromatography in the second dimension. The positions of phosphopeptides were visualized by autoradiography. The individual peptides were labeled as shown.
To examine whether S18, S28, S323, and S436 are possible candidates for PKR phosphorylation sites, mutant B56α proteins with a single point mutation (S28A) or a quadruple point mutation (S18AS28AS323AS436A) were prepared. Wild-type or mutant B56α was phosphorylated by PKR in vitro and cleaved by TPCK-trypsin. Phosphopeptides were separated on TLC plates with electrophoresis in the first dimension and chromatography in the second dimension (Fig. 2B). PKR-phosphorylated wild-type B56α generated more than 10 tryptic phosphopeptides (Fig. 2B, panel 1), indicating that B56α is phosphorylated at multiple sites. Mutant B56α S28A generated most of the same phosphopeptides as the wild type, except that peptides R5 and R7 disappeared, suggesting that peptides R5 and R7 were phosphorylated at S28 (Fig. 2B, panel 2). Since quadruple mutant B56α showed the same peptide map as single mutant B56α S28A, the residues S18, S323, and S436 are not phosphorylated by PKR (Fig. 2B, panel 3).
Physical association between PKR and B56α requires kinase activity of PKR.
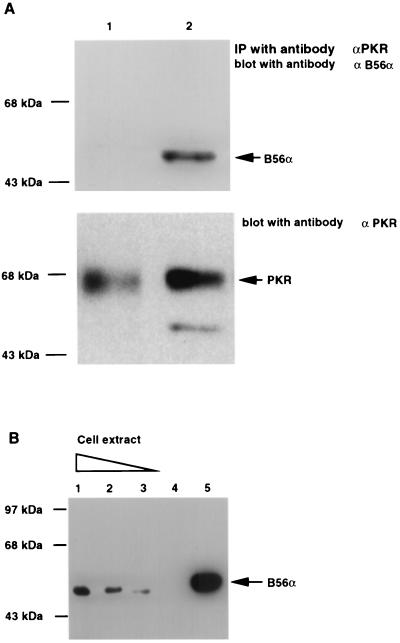
Although recombinant B56α is an efficient substrate for activated PKR, there is an absence of basal phosphorylation of B56α by latent recombinant PKR (Fig. 1). To determine whether activation of PKR was necessary to allow interaction with B56α, we immunoprecipitated endogenous PKR from T98G cell extracts, activated this with dsRNA, and added recombinant B56α. Physical association between PKR and B56α can be detected only under conditions where PKR is activated (Fig. 3A).
FIG. 3.
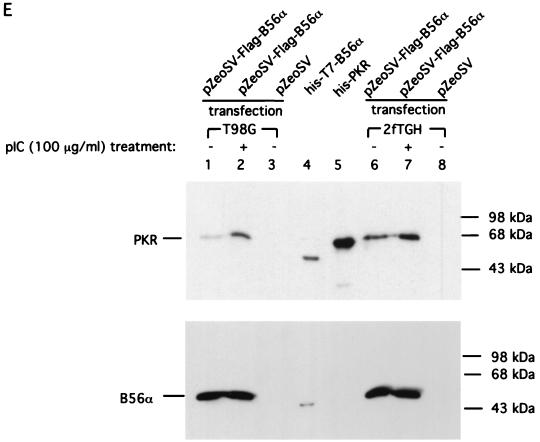
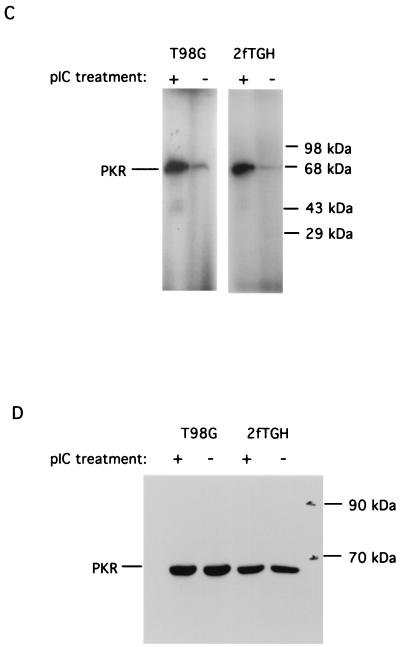
Physical interaction between PKR and B56α. (A) PKR was immunoprecipitated (IP) with monoclonal antibody from 200 μg of T98G cell extract, either activated (lane 2) or not activated (lane 1) by pIC. Recombinant B56α was added and the immunoprecipitates were washed, followed by SDS-PAGE and Western blotting assay with a polyclonal antibody against B56α (top panel). The blot was stripped and reprobed with a polyclonal antibody against human PKR (bottom panel). (B) In vivo association between PKR and B56α. PKR was immunoprecipitated with monoclonal antibody from 3 (lane 1), 2 (lane 2), and 1 (lane 3) mg of T98G cell extract, respectively. The proteins were separated by SDS-PAGE, transferred to a membrane, and immunoblotted with a polyclonal antibody against human B56α. Cell extract (3 mg) without addition of anti-PKR monoclonal antibody was used as a negative control (lane 4). Recombinant His-T7 B56α was used as a positive control (lane 5). The lack of endogenous B56α in panel A is likely due to the less amount of cell lysate. (C) Autophosphorylation of PKR in human T98G and 2fTGH cells in response to pIC treatment. Human T98G and 2fTGH cells were treated with pIC (100 μg/ml) for 1 h. PKR was immunoprecipitated from 150 μg of cell lysate with monoclonal antibody. The kinase activity of PKR was assayed in an in vitro autophosphorylation reaction in the presence of [γ-32P]ATP. (D) Western blot assay of PKR protein levels in cells either treated or not treated with pIC. T98G or 2fTGH cells were either treated or not treated with 100 μg of pIC per ml as described above, and 150 μg of total cell lysate was assayed for PKR protein level by Western blotting using a polyclonal antibody against human PKR. (E) Activation of PKR in cells by pIC increases its affinity with B56α. Human T98G or 2fTGH cells were transfected with FLAG-B56α expression plasmid DNA (pZeoSV-FLAG-B56α) (lanes 1, 2, 6, and 7) or vector DNA (pZeoSV) (lanes 3 and 8). Cells were treated with 100 μg of pIC per ml for 1 h (lanes 2 and 7). Immunoprecipitation of FLAG-B56α was performed with anti-FLAG M2 affinity gel, and the association of PKR was detected by Western blotting using an anti-human PKR polyclonal antibody (top panel). Recombinant His-T7-B56α (lane 4) and His-PKR (lane 5) were also included as positive controls. The polyclonal antibody against human PKR has cross-reaction with His-T7-B56α because His-PKR was used as a source to generate the polyclonal antibody. The membrane was also stripped and reprobed with anti-human B56α polyclonal antibody to examine B56α protein levels (bottom panel).
To examine whether there is an association between PKR and B56α in vivo, we performed a coimmunoprecipitation with a monoclonal antibody against PKR (30) on T98G cell extracts followed by a Western blot assay using a polyclonal antibody against B56α. Since use of a moderate amount of cell extract (200 μg) for coimmunoprecipitation failed to detect the association of PKR with the endogenous B56α (Fig. 3A), we reasoned that endogenous PKR possesses low basal level kinase activity and consequently associates with a smaller amount of B56α. Accordingly, by using an increased amount of cell extract, B56α could be detected by coimmunoprecipitation with PKR (Fig. 3B), suggesting an endogenous interaction between PKR and B56α.
Since the interaction between PKR and B56α requires active PKR (Fig. 3A), the basal level of cellular PKR must be sufficient to allow the endogenous constitutive association between PKR and B56α. If the kinase activity of PKR is increased, we should also be able to see an increase in the association between PKR and B56α. Treatment of human T98G glioblastoma cells and 2fTGH fibroblasts with pIC resulted in the activation of PKR as demonstrated by an autophosphorylation assay (Fig. 3C). This treatment did not cause an obvious increase in the PKR protein level (Fig. 3D). In order to see the association between PKR and B56α in response to pIC activation of PKR, we transfected both T98G and 2fTGH cells with FLAG epitope B56α expression plasmid DNA to enhance the B56α protein level and activated PKR in these cells with pIC. The interaction between PKR and B56α was examined by immunoprecipitation of B56α with anti-FLAG M2 affinity gel followed by western blotting against human PKR. The results (Fig. 3E) demonstrated that there is an increase in the association between PKR and B56α when PKR is activated in cells treated with pIC.
Activation of PKR alters the isoelectric point of B56α.
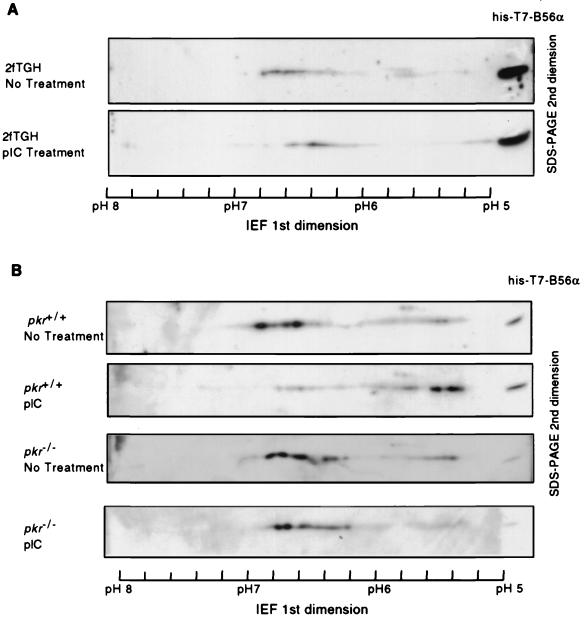
B56α is a substrate for PKR in vitro and shows increased association with PKR upon PKR activation in cells. This suggests that B56α should be a physiological substrate for PKR in vivo. Accordingly, we investigated whether the activation of PKR in the cell resulted in a change in the phosphorylation level of B56α. Human 2fTGH fibroblasts were transfected with FLAG-tagged B56α expression plasmid DNA and treated with pIC to activate PKR protein. B56α was immunoprecipitated from cell lysates and analyzed by two-dimensional IEF and SDS-PAGE to determine the phosphorylation-induced change in its isoelectric point (pI). The pI of B56α from untreated cells spans a range from 6.2 to 6.8, suggesting that B56α is phosphorylated at multiple sites, consistent with the results of B56α phosphopeptide maps in vitro. Treatment of cells with the PKR activator pIC caused a shift in the pI of B56α toward a lower pH value, indicating an increase in acidic forms of B56α, likely due to phosphorylation (Fig. 4A). To assess the possibility that the pIC-induced pI shift in B56α is dependent on PKR, we transfected pkr+/+ or pkr−/− fibroblasts with FLAG epitope B56α expression plasmid DNA and examined the pI status of FLAG-B56α after treatment of the cells with pIC (Fig. 4B). The pI of B56α in murine fibroblasts ranges from 5.2 to 6.8. Treatment of pkr+/+ cells with pIC caused a pI shift in B56α, with an increase its acidic forms compared with untreated cells. This change is not observed in pkr−/− cells. These observations indicate that the pIC-induced pI shift of B56α toward acidic forms in cells results from phosphorylation of B56α by pIC-activated PKR.
FIG. 4.
Mobility shift of B56α in IEF in response to pIC treatment of the cell. (A) Human 2fTGH cells were transfected with FLAG-B56α expression plasmid DNA pZeoSV-FLAG-B56α. The cells were either treated (bottom panel) or not treated (top panel) with 100 μg of pIC per ml to activate PKR. B56α was immunoprecipitated from 2 mg of cell lysate and separated in the first dimension with IEF in a ready-strip IPG strip (pH range, 5 to 8; Bio-Rad) and in the second dimension with an SDS-polyacrylamide gel. Western blotting with a polyclonal antibody against human B56α was performed to detect B56α. Recombinant His-T7-B56α was loaded at the rightmost side and used as reference for alignment. B56α from 2fTGH cells showed pI values varying from 6.2 to 6.8, but it is very likely that more acidic forms of B56α failed to be detected because of a lower expression level of B56α in 2fTGH cells compared with murine fibroblasts in panel B. (B) pI of FLAG-B56α from pkr+/+ or pkr−/− cells treated or not treated with pIC. The experimental procedure is same as for 2fTGH cells in panel A. The pI values range from 5.3 to 6.8.
Phosphorylation of B56α by PKR modulates PP2A activity.
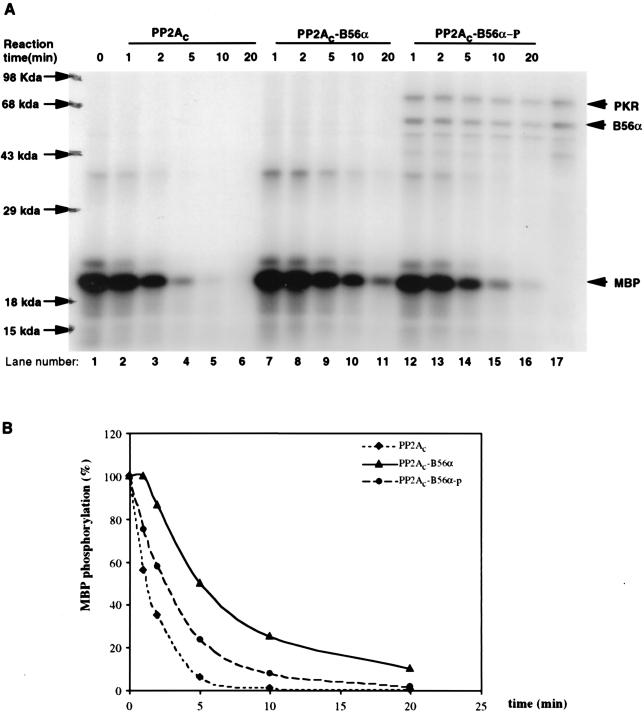
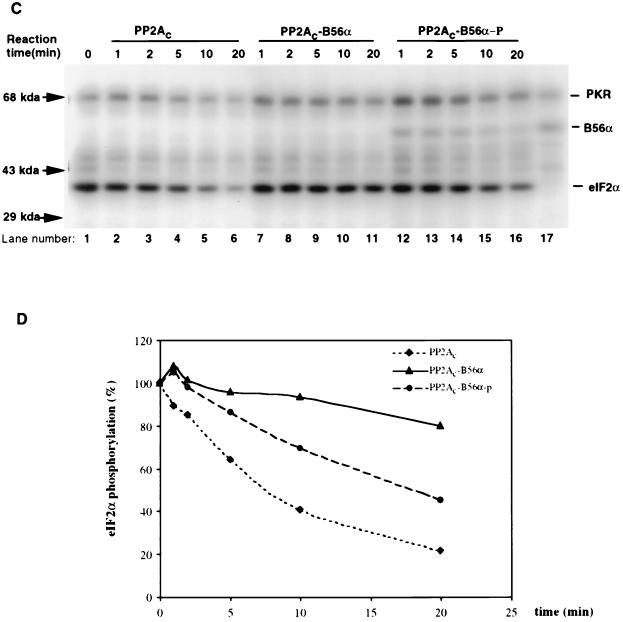
B56α is a regulatory subunit of PP2A and has been found to tightly associate with PP2Ac (37). Although it is also a phosphoprotein in vivo, it is not clear whether this phosphorylation plays a role in regulating PP2A activity. To determine if B56α phosphorylation by PKR could regulate PP2A activity in vitro, a phosphatase activity assay was performed using MBP phosphorylated by PKC as the substrate for PP2A (Fig. 5A). The phosphatase activities of three forms of PP2A were compared. PP2Ac was commercially obtained. PP2Ac-B56α, which is a trimeric holoenzyme with unphosphorylated B56α as the regulatory subunit, was prepared by mixing recombinant B56α with PP2Ac. PP2Ac-B56α-p, which is a trimeric holoenzyme with phosphorylated B56α as the regulatory subunit, was prepared by mixing PKR-phosphorylated B56α with PP2Ac. Excess B56α was added to ensure that PP2A existed as the form of AC-B56α trimeric holoenzyme. The results (Fig. 5A and B) show that whereas unphosphorylated B56α decreased PP2Ac activity on MBP, phosphorylation of B56α by PKR blocked this effect and increased the phosphatase activity of PP2Ac-B56α holoenzyme. Thus, PP2Ac-B56α holoenzyme was less active than PP2Ac, but when B56α was phosphorylated by PKR, phosphatase activity on MBP was enhanced. Similar results were obtained when recombinant eIF2α protein phosphorylated by PKR was used as a substrate for PP2A (Fig. 5C and D), although the recombinant eIF2α is a less efficient substrate for PP2A than MBP.
FIG. 5.
Modulation of PP2A activity on MBP or eIF2α via phosphorylation of B56α by PKR. (A) In vitro PP2A activity on PKC-phosphorylated MBP. MBP was phosphorylated by PKCα with radioactive ATP ([γ-32P]ATP) and used as a substrate for PP2A (either PP2Ac dimer, PP2Ac-B56α, or phosphorylated PP2Ac-B56α). B56α either phosphorylated or not by PKR was quickly premixed with PP2Ac to generate the PP2Ac-B56α or PP2Ac-B56α-p form of PP2A. Dephosphorylation was carried out at 30°C, and equal amounts of reaction mixture were taken out after 1, 2, 5, 10, and 20 min. The reaction was stopped by adding phosphatase stop solution and separated by SDS–12% PAGE. Lane 1, starting level of MBP phosphorylation; lanes 2 to 16, time courses of dephosphorylation of MBP; lane 17, input of PKR and B56α in PP2Ac-B56α-p reaction. (B) Quantitation of the data in panel A. MBP phosphorylation levels were quantitated by ImageQuant version 1.1 analysis following scanning of the gel with a Storm PhosphorImager. (C) In vitro PP2A activity on PKR-phosphorylated eIF2α. Recombinant eIF2α was phosphorylated by recombinant PKR with radioactive ATP ([γ-32P]ATP) and used as a substrate for PP2A (either AC dimer PP2Ac, PP2Ac-B56α, or PP2Ac-B56α-p). Dephosphorylation of eIF2α was carried out in the same way as for MBP dephosphorylation. Lane 1, starting level of eIF2α phosphorylation; lanes 2 to 6, time course of dephosphorylation of eIF2α by PP2Ac core dimer; lanes 7 to 11, time course of dephosphorylation of eIF2α by PP2Ac-B56α; lanes 12 to 16, time course of dephosphorylation of eIF2α by PP2Ac-B56α-p; lane 17, Input of PKR and B56α in PP2Ac-B56α-p reaction. (D) eIF2α phosphorylation levels were quantitated by ImageQuant version 1.1 analysis following scanning of the gel with a Storm PhosphorImager. The abnormal eIF2α phosphorylation percentage with PP2Ac-B56α and PP2Ac-B56α-p at the 5-min time point is likely due to the undermeasurement of the initial eIF2α phosphorylation.
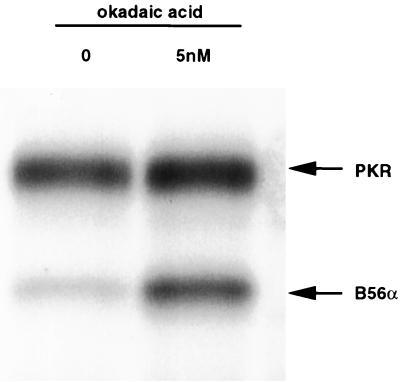
B56α is a target for dephosphorylation by PP2A.
In the in vitro phosphatase activity assay, we observed that the phosphorylation level of B56α was progressively reduced over time, suggesting that B56α itself can be dephosphorylated by PP2A (Fig. 5). This observation was confirmed by an in vitro B56α dephosphorylation assay using immunoprecipitated PP2A (Fig. 6). When okadaic acid was included to inhibit PP2A activity, PKR was able to phosphorylate B56α to a maximal level as determined by labeling with [γ-32P]ATP, whereas in the absence of okadaic acid, B56α phosphorylation was greatly reduced. The finding that B56α is a target for PP2A suggests that there is an autoregulatory mechanism for PP2A activity.
FIG. 6.
Phospho-B56α is dephosphorylated by PP2A. Endogenous PP2A was immunoprecipitated from HeLa S3 cells and mixed with recombinant B56α phosphorylated by PKR in the presence or absence of 5 nM okadaic acid at 30°C for 20 min. The proteins were separated on an SDS-polyacrylamide gel, and an autoradiograph of the dried gel is shown.
PKR inhibits the stimulatory effect of B56α on protein expression.
The identification of B56α as a novel PKR substrate that physically associates with PKR begs the question of the physiological significance of this interaction. Although B56α may direct PP2A to regulate the phosphorylation status and the activity of PKR, the activity of PP2A on PKR is weak (Fig. 6). Since in vitro phosphatase activity assays showed that phosphorylation of B56α by PKR could modulate PP2A activity, we hypothesized that PKR can alter PP2A activity by phosphorylating B56α and affect downstream cellular activities. However, a direct assay of PP2A phosphatase activity immunoprecipitated from the human cells treated with pIC did not show much difference from that from untreated cells (data not shown). This is not unexpected, since B56α is also likely a target for dephosphorylation by PP2A itself (Fig. 6). Therefore, we sought an alternate way to examine the alteration of PP2A activity in vivo resulting from the interaction between PKR and B56α. We found that treatment of cells (COS-1, pkr+/+, or pkr−/− fibroblasts) with the PP2A inhibitor okadaic acid resulted in enhanced expression from a transfected luciferase reporter plasmid, pGL2p, whose luciferase expression is driven by a constitutive simian virus 40 promoter (Fig. 7A). This effect can be observed with a concentration of okadaic acid of less than 10 nM (Fig. 7B). Since PP2A is much more sensitive to okadaic acid than PP1 is, these results support a role for PP2A in regulating protein expression. To examine if overexpression of B56α could influence PP2A activity in this assay and if this activity could be modulated by coexpression with PKR, we cotransfected COS-1 cells with pGL2p and a construct expressing B56α (pZeoSV-B56α), wild-type human PKR (pRC-PKR), or dominant-negative mutant PKR K296R (pRC-K296R) (Fig. 7C, left panel). Since translational control by PKR mediated through eIF2α phosphorylation is a well-established PKR function, luciferase expression in transfected COS-1 cells will be altered by expression of PKR alone (7, 60). In accord with previous reports, wild-type PKR reduces luciferase expression in this assay whereas cotransfection with catalytically inactive PKR results in enhanced expression from the reporter (Fig. 7C, bars 3 and 5). Transfection with the B56α expression construct alone resulted in enhanced luciferase expression, which could be abrogated by coexpression of wild-type PKR (Fig. 7C, bars 2 and 4). Cotransfection of B56α with mutant PKR K296R resulted in striking enhancement of protein synthesis (Fig. 7C, bar 6). These results suggest that overexpression of B56α in COS-1 cells is able to modulate PP2A activity to enhance protein synthesis. This effect can be inhibited by PKR, most likely via a functional interaction with and phosphorylation of B56α by PKR. This observation was confirmed when the experiment was repeated with a murine fibroblast cell line in which the effect of B56α is completely suppressed when cotransfection is with wild-type PKR (Fig. 7C, right panel). The PKR-dependent modulation of PP2A activity was supported by comparing the luciferase expression in pkr-null cells transfected with B56α with that in wild-type cells transfected with B56α. The results show that B56α is more active in the absence of PKR (Fig. 7D).
PKR may modulate PP2A activity to regulate eIF4E phosphorylation.
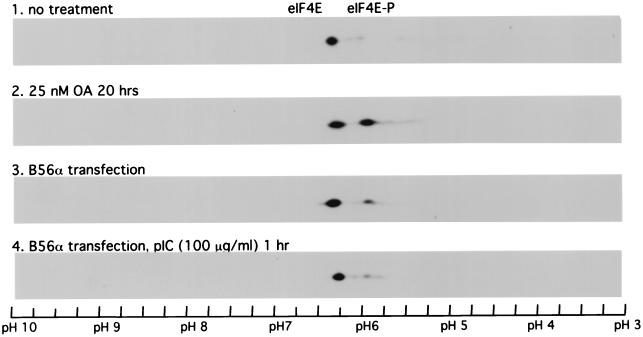
The phosphorylation of eIF2α to inhibit protein translation is a well-established mechanism for PKR-dependent regulation of protein translation. However, the luciferase transfection assay results (Fig. 7) indicate that there is likely a novel mechanism involving PKR upregulation of PP2A activity. When the luciferase mRNA levels in pkr+/+ cells transfected with pGL2p were compared with those in transfected cells treated with okadaic acid or cells which were also cotransfected with B56α by RT-PCR (Fig. 8) or by SYBR Green assay (data not shown), no obvious changes in luciferase mRNA levels were seen, indicating that PP2A is acting at the translational level. Since phosphorylation of eIF4E at serine 209 (25, 31) plays an important role in protein translation initiation, we examined whether expression of B56α or activation of PKR can regulate eIF4E dephosphorylation. Untreated cell extracts analyzed by IEF-PAGE revealed a high percentage of unphosphorylated eIF4E (Fig. 9, panel 1). Treatment with okadaic acid caused an increased level of eIF4E phosphorylation (Fig. 9, panel 2), consistent with enhanced protein translation (Fig. 7). Transfection of 2fTGH cells with B56α expression DNA also resulted in an increased level of eIF4E phosphorylation, likely due to decreased PP2A activity (Fig. 9, panel 3). Interestingly, when 2fTGH cells transfected with a B56α expression plasmid were treated with pIC to activate PKR, the eIF4E phosphorylation level was decreased compared with that in untreated cells (Fig. 9, panel 4). These data suggest that PKR can phosphorylate B56α to increase PP2A activity, resulting in decreased eIF4E phosphorylation.
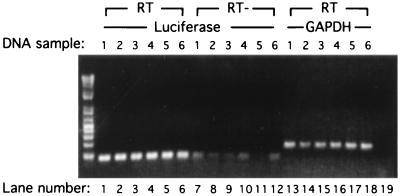
FIG. 8.
RT-PCR assay of luciferase mRNA. pkr+/+ cells were cotransfected with 500 ng of pGL2p and 1 μg of pZeoSV (DNA samples 1, 2, 3, and 4 [lanes 1, 2, 3, 4, 7, 8, 9, 10, 13, 14, 15, and 16]) or pZeoSV-B56α (DNA samples 5 and 6 [lanes 5, 6, 11, 12, 17, and 18]) and either treated with 50 nM okadaic acid overnight (DNA samples 3 and 4 [lanes 3, 4, 9, 10, 15, and 16]) or left untreated (DNA samples 1 and 2 [lanes 1, 2, 7, 8, 13, and 14]). After 24 h of transfection, total cell RNA was prepared (see Materials and Methods) and RT was performed with 1 μg of total RNA primed with oligo(dT) by SuperScriptase II (lanes 1 to 6 and 13 to 18). PCR was carried out with two primers derived from luciferase (lanes 1 to 12) or GAPDH as a control for RNA input (lanes 13 to 18). A PCR without template DNA was also included (lane 19). RNA samples that were not reverse transcribed were also used as templates for luciferase PCR to check the level of plasmid contamination (lanes 7 to 12).
FIG. 9.
IEF–SDS-PAGE analysis of eIF4E phosphorylation in 2fTGH cells. 2fTGH cells were either left untreated (panel 1), treated with 25 nM okadaic acid (OA) for 20 h (panel 2), transfected with B56α expression plasmid DNA pZeoSV-B56α (panel 3), or transfected with pZeoSV-B56α and then treated with 100 μg of pIC per ml for 1 h (panel 4). Cells were lysed, and proteins from 300 μg of cell extract were separated with IEF–SDS-PAGE two-dimensional electrophoresis. eIF4E protein was detected by Western blotting using a monoclonal antibody against rabbit eIF-4E. Phosphorylated eIF4E has a lower isoelectric point and migrates to a lower pH position than unphosphorylated eIF4E.
DISCUSSION
PKR is an established eIF2α kinase and plays an important role in translational control. In fact, thus far, eIF2α is the only well-known physiological substrate established in vivo for PKR. However, there is an increasing amount of evidence supporting a role for PKR in different cellular activities, including transcription, apoptosis, and tumorigenesis. Therefore, it is likely that there are other physiologically important substrates for PKR. Identifying such substrates has become a primary obstacle in understanding PKR function, but yeast two-hybrid screening has provided a powerful tool to obtain interacting proteins (34). However, obtaining proteins which interact with active PKR by using yeast two-hybrid screening has been precluded because of the growth-inhibitory characteristics of PKR. Recently we described a PKR mutant with reduced kinase activity that retained wild-type affinity for eIF2α (7). Using this mutant in a two-hybrid screening has allowed us to identify two novel substrates for PKR, DRBP76 (43) and B56α.
B56α is a cytosolic member of B′ (B56) family and is expressed ubiquitously in many cell types (37). Compared to eIF2α, B56α exhibits different features in its phosphorylation by PKR in vitro. B56α does not interact with or become phosphorylated efficiently by PKR unless PKR is activated by dsRNA (Fig. 1). Nevertheless, some constitutive association between PKR and B56α was observed in vivo (Fig. 3B and E), suggesting interaction likely reflecting basal PKR activity. The isolation of B56α by two-hybrid screening using the PKR M3 mutant with residual kinase activity supports this. The PKR-interacting domain on B56α may be distinct from the phosphorylation sites, since the PKR monoclonal antibody used in this study interferes with B56α phosphorylation by PKR (data not shown). In contrast, the phosphorylation of eIF2 is not affected by the monoclonal antibody (30).
B56α harbors multiple phosphorylation sites, and this is indicated from both in vitro B56α tryptic phosphopeptide mapping and in vivo B56α IEF–SDS-PAGE two-dimensional analysis. Although B56α is phosphorylated by PKR at S28 in vitro (Fig. 2), whether it is a physiological phosphoacceptor in vivo is not known. Since S28A mutant B56α retains the ability to be phosphorylated by PKR efficiently, the significance of phosphorylation of B56α by PKR at site S28 remains to be established. In vivo, B56α exhibits a wide range of isoelectric point values (pI 5.3 to 6.8 in murine fibroblasts [Fig. 4]), which may correspond to various phosphorylated states. Certainly PKR is not the only kinase that regulates the phosphorylation of B56α, because in pkr-null cells B56α still exhibits differentially phosphorylated forms. Other kinases capable of phosphorylating B56α likely include PKC, PKA, and casein kinase II, since B56α has consensus sequences for these kinases. We confirmed that B56α is phosphorylated by PKC in vitro (data not shown). The regulation of B56α at multiple phosphoacceptor sites by different kinases suggests a previously unidentified mechanism by which kinases interact with PP2A.
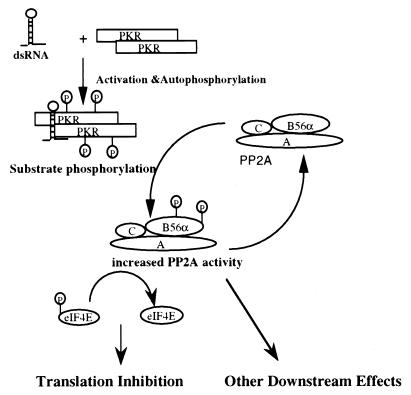
There are two possible physiological consequences resulting from the interaction between PKR and B56α. B56α may direct PP2A to dephosphorylate PKR, resulting in downregulation of PKR activity. PKR phosphorylation of B56α could cause either direct or indirect downregulation of PKR activity by activating dephosphorylation of PKR by PP2A. However, PKR is a poor substrate for PP2A (Fig. 6), and the major PKR phosphatase is likely PP1 (50). PKR dephosphorylation may be PP2A mediated in vivo, but cells transiently expressing B56α and treated with pIC showed no difference in the time course of PKR activation compared to untransfected cells (data not shown). Therefore, we concluded that it is unlikely that B56α directs PP2A to dephosphorylate PKR. PKR may phosphorylate B56α to regulate PP2A activity in vivo. The in vitro phosphatase assay demonstrated that phosphorylation of B56α by PKR could modulate PP2A activity, which was reflected by an increase in phosphatase activity on MBP and recombinant eIF2α (Fig. 5). However, direct evidence for PKR phosphorylation of B56α and modulation of PP2A activity is difficult to obtain due to PP2A dephosphorylating B56α (Fig. 6). Nevertheless, the alteration of PP2A activity by PKR shown indirectly by transfection assays in COS-1 cells and murine fibroblasts (Fig. 7) suggests an in vivo mechanism. Transiently expressed B56α can form trimeric PP2A either by interacting with free AC core dimer or by replacing other B subunits in the PP2A trimeric complex. This change in PP2A oligomeric forms could alter PP2A activity or localization. Overexpressing B56α causes an upregulation of protein synthesis. PKR inhibits this effect, probably by phosphorylating B56α. A model can be proposed from these results which is based on PKR phosphorylation of B56α causing an increase in PP2A activity (Fig. 10). Increased PP2A activity likely affects different cellular activities including translation, transcription, apoptosis, and cell cycle regulation, processes in which PKR has also been implicated (reviewed in references 13 and 57).
FIG. 10.
Schematic model representing the modulation of PP2A activity by phosphorylation of B56α by PKR. PKR is activated by dsRNA and phosphorylates B56α. Phosphorylation of B56α likely enhances enzymatic activity of the PP2Ac-B56α trimeric complex and alters cell activities by modulating the phosphorylation levels of downstream target proteins.
Although the detailed mechanisms of how PP2A activity causes an altered level of protein expression in transfection assays in COS-1 and murine fibroblasts remains to be elucidated, our data indicate that this regulation occurs at a translational level, likely involving the regulation of eIF4E phosphorylation. PP2A has been reported previously to regulate the dephosphorylation of different proteins involved in translation control, including eIF4E (27), eIF2α (10), and EF2 (6). Phosphorylation of eIF2α or EF2 inhibits translation initiation or retards translation elongation, respectively. Phosphorylation of eIF4E increases the efficiency of eIF4E binding to mRNA cap, thus increasing translation initiation (27). PP2A can also dephosphorylate eIF4E-inhibitory protein PHAS-I to facilitate its inhibitory association with eIF4E (31). Thus, alteration of PP2A activity may have different outcomes for protein translation regulation. Reduced PP2A activity would cause an increase in eIF4E and PHAS-I phosphorylation, stimulating translation initiation, but would also result in an increase in the phosphorylation of eIF2α and EF2, leading to translational inhibition. Consistent with this notion, treatment of cells with okadaic acid or transfection with B56α (Fig. 7) results in a reduction of PP2A activity and increases protein translation, likely via an increase in eIF4E phosphorylation (Fig. 9). Interestingly, activation of PKR by pIC also results in a decrease in eIF4E phosphorylation. We would argue that PKR can phosphorylate B56α to increase PP2A activity, leading to eIF4E dephosphorylation. Although increased PP2A activity may also cause an increase in eIF2α dephosphorylation, thus promoting protein translation, the eIF2α phosphatase is PP2Ac whereas the trimeric PP2A is inefficient in eIF2α dephosphorylation (10). Furthermore, active PKR can readily phosphorylate eIF2α, and this must override the dephosphorylation of eIF2α by PP2A. Therefore, we propose that PKR can regulate protein translation by phosphorylating not only eIF2α but also B56α, thus increasing PP2A activity and consequently dephosphorylation of eIF4E.
Because PP2A has a range of actions on cellular activities, phosphorylation of B56α by PKR to modulate PP2A activity may also have a broad spectrum of impact. This could mediate other biological effects of PKR besides translation control, for example, functions in transcription (29, 59) and apoptosis (17), where the activity of PP2A is also involved (45, 49, 52, 62). Other functions of PKR, contributing to cell cycle regulation (61), interleukin-3 signaling (24), and PDGF signaling (41), may also have a link to the regulation of PP2A activity. Additionally, investigating the targets of B56α may implicate PKR in novel signaling pathways or cellular activities. For example, in Wnt signaling and others, B56α subunits associate with the adenomatous polyposis coli (APC) protein to direct PP2A to dephosphorylate specific components of the APC-dependent signaling complex, reducing the abundance of transcription factor beta-catenin and consequently inhibiting the transcription of beta-catenin target genes in mammalian cells and Xenopus embryo explants (47). It would be interesting to determine whether PKR has an impact in this signaling pathway via its phosphorylation of B56α.
B56α is also dephosphorylated by PP2A (Fig. 6), indicating that this enzyme is subject to autoregulation. Previously it has been shown that there is autodephosphorylation on Tyr 307 in the PP2A catalytic C subunit (9). The expression of the C subunit is also subjected to an autoregulating mechanism ensuring that an appropriate level of C subunit protein is maintained (2). Autodephosphorylation of B56α is therefore likely an important mechanism by which PP2A adjusts its activity within a critical range, allowing tight control over cellular activities.
The phosphorylation of B56α by PKR also represents a new paradigm for a kinase to modulate PP2A activity. This is different from the direct interaction mechanisms between kinase and phosphatase described previously (21, 55). Most recently, Westphal et al. (54) reported formation of a stable complex between the trimeric PP2A holoenzyme and Ca2+ calmodulin-dependent kinase IV in which PP2A serves to negatively control kinase activity. PP2A has also been shown to interact with casein kinase 2α in mitogen-starved cells (21). We have demonstrated here that the activity of PP2A can be modulated through phosphorylation of its regulatory subunit B56α by PKR. However, it is unlikely that PKR forms a stable complex with trimeric PP2A holoenzyme, as we failed to detect an association between PKR and the catalytic C subunit by coimmunoprecipitation (data not shown). Since B56α belongs to a family consisting of several phosphoproteins sharing very high homology in amino acid sequence, it is also possible that PKR can phosphorylate other B56 regulatory subunits of PP2A.
ACKNOWLEDGMENTS
We thank Bruce Carpick for preparing the recombinant human wild-type PKR proteins, Ruorong Cai for the mutant PKR L362Q construct, and Kee-Chuan Goh for help with RNA assays.
This study is made possible by a grant from the National Institutes of Health (AI34039-02).
REFERENCES
- 1.Alberts A S, Deng T, Lin A, Meinkoth J L, Schonthal A, Mumby M C, Karin M, Feramisco J R. Protein phosphatase 2A potentiates activity of promoters containing AP-1-binding elements. Mol Cell Biol. 1993;13:2104–2112. doi: 10.1128/mcb.13.4.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baharians Z, Schonthal A H. Autoregulation of protein phosphatase type 2A expression. J Biol Chem. 1998;273:19019–19024. doi: 10.1074/jbc.273.30.19019. [DOI] [PubMed] [Google Scholar]
- 3.Balachandran S, Kim C N, Yeh W C, Mak T W, Bhalla K, Barber G N. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 1998;17:6888–6902. doi: 10.1093/emboj/17.23.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber G N, Wambach M, Thompson S, Jagus R, Katze M G. Mutants of the RNA-dependent protein kinase (PKR) lacking double-stranded RNA binding domain I can act as transdominant inhibitors and induce malignant transformation. Mol Cell Biol. 1995;15:3138–3146. doi: 10.1128/mcb.15.6.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 6.Buss W C, Stepanek J, Queen S A. Association of tissue-specific changes in translation elongation after cyclosporin with changes in elongation factor 2 phosphorylation. Biochem Pharmacol. 1994;48:1459–1469. doi: 10.1016/0006-2952(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 7.Cai R, Williams B R G. Mutations in the double-stranded RNA-activated protein kinase insert region that uncouple catalysis from eIF2alpha binding. J Biol Chem. 1998;273:11274–11280. doi: 10.1074/jbc.273.18.11274. [DOI] [PubMed] [Google Scholar]
- 8.Carpick B W, Graziano V, Schneider D, Maitra R K, Lee X, Williams B R G. Characterization of the solution complex between the interferon-induced, double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J Biol Chem. 1997;272:9510–9516. doi: 10.1074/jbc.272.14.9510. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Martin B L, Brautigan D L. Regulation of protein serine-threonine phosphatase type-2A by tyrosine phosphorylation. Science. 1992;257:1261–1264. doi: 10.1126/science.1325671. [DOI] [PubMed] [Google Scholar]
- 10.Chen S C, Kramer G, Hardesty B. Isolation and partial characterization of an Mr 60,000 subunit of a type 2A phosphatase from rabbit reticulocytes. J Biol Chem. 1989;264:7267–7275. [PubMed] [Google Scholar]
- 11.Chong K L, Feng L, Schappert K, Meurs E, Donahue T F, Friesen J D, Hovanessian A G, Williams B R G. Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. EMBO J. 1992;11:1553–1562. doi: 10.1002/j.1460-2075.1992.tb05200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu W M, Ostertag D, Li Z W, Chang L, Chen Y, Hu Y, Williams B, Perrault J, Karin M. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- 13.Clemens M J, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 14.Cuddihy A R, Li S, Tam N W, Wong A H, Taya Y, Abraham N, Bell J C, Koromilas A E. Double-stranded-RNA-activated protein kinase PKR enhances transcriptional activation by tumor suppressor p53. Mol Cell Biol. 1999;19:2475–2484. doi: 10.1128/mcb.19.4.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuddihy A R, Wong A H, Tam N W, Li S, Koromilas A E. The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 protein and phosphorylates human p53 on serine 392 in vitro. Oncogene. 1999;18:2690–2702. doi: 10.1038/sj.onc.1202620. [DOI] [PubMed] [Google Scholar]
- 16.Deng X, Ito T, Carr B, Mumby M, May W S., Jr Reversible phosphorylation of bcl2 following interleukin 3 or bryostatin 1 is mediated by direct interaction with protein phosphatase 2A. J Biol Chem. 1998;273:34157–34163. doi: 10.1074/jbc.273.51.34157. [DOI] [PubMed] [Google Scholar]
- 17.Der S D, Yang Y L, Weissmann C, Williams B R G. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galabru J, Katze M G, Robert N, Hovanessian A G. The binding of double-stranded RNA and adenovirus VAI RNA to the interferon-induced protein kinase. Eur J Biochem. 1989;178:581–589. doi: 10.1111/j.1432-1033.1989.tb14485.x. [DOI] [PubMed] [Google Scholar]
- 19.Goedert M, Cohen E S, Jakes R, Cohen P. p42 MAP kinase phosphorylation sites in microtubule-associated protein tau are dephosphorylated by protein phosphatase 2A1. Implications for Alzheimer's disease. FEBS Lett. 1992;312:95–99. doi: 10.1016/0014-5793(92)81418-l. [DOI] [PubMed] [Google Scholar]
- 20.Goh K C, Haque S J, Williams B R G. p38 MAP kinase is required for STAT1 serine phosphorylation and transcriptional activation induced by interferons. EMBO J. 1999;18:5601–5608. doi: 10.1093/emboj/18.20.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heriche J K, Lebrin F, Rabilloud T, Leroy D, Chambaz E M, Goldberg Y. Regulation of protein phosphatase 2A by direct interaction with casein kinase 2alpha. Science. 1997;276:952–955. doi: 10.1126/science.276.5314.952. [DOI] [PubMed] [Google Scholar]
- 22.Hovanessian A G. Double-stranded RNA dependent protein kinase(s) in rabbit reticulocyte lysates analogous to kinase from interferon-treated cells. Biochimie. 1980;62:775–778. doi: 10.1016/s0300-9084(80)80132-5. [DOI] [PubMed] [Google Scholar]
- 23.Iordanov M S, Paranjape J M, Zhou A, Wong J, Williams B R G, Meurs E F, Silverman R H, Magun B E. Activation of p38 mitogen-activated protein kinase and c-Jun NH(2)-terminal kinase by double-stranded RNA and encephalomyocarditis virus: involvement of RNase L, protein kinase R, and alternative pathways. Mol Cell Biol. 2000;20:617–627. doi: 10.1128/mcb.20.2.617-627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito T, Jagus R, May W S. Interleukin 3 stimulates protein synthesis by regulating double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci USA. 1994;91:7455–7459. doi: 10.1073/pnas.91.16.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi B, Cai A L, Kaper B D, Minich W B, Mandez R, Beach C M, Stepinski J, Darzynkiewicz E, Rhoads R M. Phosphorylation of eukaryotic protein synthesis factor 4E at ser-209. J Biol Chem. 1995;270:14597–14603. doi: 10.1074/jbc.270.24.14597. [DOI] [PubMed] [Google Scholar]
- 26.Kao P N, Chen L, Brock G, Ng J, Kenny J, Smith A J, Corthesy B. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J Biol Chem. 1994;269:20691–20699. [PubMed] [Google Scholar]
- 27.Kleijn M, Vrins C L, Voorma H O, Thomas A A. Phosphorylation state of the cap-binding protein eIF4E during viral infection. Virology. 1996;217:486–494. doi: 10.1006/viro.1996.0143. [DOI] [PubMed] [Google Scholar]
- 28.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 29.Kumar A, Yang Y L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R G. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laurent A G, Krust B, Galabru J, Svab J, Hovanessian A G. Monoclonal antibodies to an interferon-induced Mr 68,000 protein and their use for the detection of double-stranded RNA-dependent protein kinase in human cells. Proc Natl Acad Sci USA. 1985;82:4341–4345. doi: 10.1073/pnas.82.13.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence J C, Jr, Fadden P, Haystead T A, Lin T A. PHAS proteins as mediators of the actions of insulin, growth factors and cAMP on protein synthesis and cell proliferation. Adv Enzyme Regul. 1997;37:239–267. doi: 10.1016/s0065-2571(96)00016-7. [DOI] [PubMed] [Google Scholar]
- 32.Lee T H, Solomon M J, Mumby M C, Kirschner M W. INH, a negative regulator of MPF, is a form of protein phosphatase 2A. Cell. 1991;64:415–423. doi: 10.1016/0092-8674(91)90649-j. [DOI] [PubMed] [Google Scholar]
- 33.Lengyel P. Tumor-suppressor genes: news about the interferon connection. Proc Natl Acad Sci USA. 1993;90:5893–5895. doi: 10.1073/pnas.90.13.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luban J, Goff S P. The yeast two-hybrid system for studying protein-protein interactions. Curr Opin Biotechnol. 1995;6:59–64. doi: 10.1016/0958-1669(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto-Taniura N, Pirollet F, Monroe R, Gerace L, Westendorf J M. Identification of novel M phase phosphoproteins by expression cloning. Mol Biol Cell. 1996;7:1455–1469. doi: 10.1091/mbc.7.9.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCright B, Rivers A M, Audlin S, Virshup D M. The B56 family of protein phosphatase 2A (PP2A) regulatory subunits encodes differentiation-induced phosphoproteins that target PP2A to both nucleus and cytoplasm. J Biol Chem. 1996;271:22081–22089. doi: 10.1074/jbc.271.36.22081. [DOI] [PubMed] [Google Scholar]
- 37.McCright B, Virshup D M. Identification of a new family of protein phosphatase 2A regulatory subunits. J Biol Chem. 1995;270:26123–26128. doi: 10.1074/jbc.270.44.26123. [DOI] [PubMed] [Google Scholar]
- 38.Meurs E, Chong K, Galabru J, Thomas N S, Kerr I M, Williams B R G, Hovanessian A G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 39.Meurs E F, Galabru J, Barber G N, Katze M G, Hovanessian A G. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mumby M C, Walter G. Protein serine/threonine phosphatases: structure, regulation, and functions in cell growth. Physiol Rev. 1993;73:673–699. doi: 10.1152/physrev.1993.73.4.673. [DOI] [PubMed] [Google Scholar]
- 41.Mundschau L J, Faller D V. Platelet-derived growth factor signal transduction through the interferon-inducible kinase PKR. Immediate early gene induction. J Biol Chem. 1995;270:3100–3106. doi: 10.1074/jbc.270.7.3100. [DOI] [PubMed] [Google Scholar]
- 42.Patel R C, Sen G C. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel R C, Vestal D J, Xu Z, Bandyopadhyay S, Guo W, Erme S M, Williams B R G, Sen G C. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J Biol Chem. 1999;274:20432–20437. doi: 10.1074/jbc.274.29.20432. [DOI] [PubMed] [Google Scholar]
- 44.Romano P R, Garcia-Barrio M T, Zhang X, Wang Q, Taylor D R, Zhang F, Herring C, Mathews M B, Qin J, Hinnebusch A G. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2α kinases PKR and GCN2. Mol Cell Biol. 1998;18:2282–2297. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santoro M F, Annand R R, Robertson M M, Peng Y W, Brady M J, Mankovich J A, Hackett M C, Ghayur T, Walter G, Wong W W, Giegel D A. Regulation of protein phosphatase 2A activity by caspase-3 during apoptosis. J Biol Chem. 1998;273:13119–13128. doi: 10.1074/jbc.273.21.13119. [DOI] [PubMed] [Google Scholar]
- 46.Schonthal A H. Role of PP2A in intracellular signal transduction pathways. Front Biosci. 1998;3:D1262–D1273. doi: 10.2741/A361. [DOI] [PubMed] [Google Scholar]
- 47.Seeling J M, Miller J R, Gil R, Moon R T, White R, Virshup D M. Regulation of beta-catenin signaling by the B56 subunit of protein phosphatase 2A. Science. 1999;283:2089–2091. doi: 10.1126/science.283.5410.2089. [DOI] [PubMed] [Google Scholar]
- 48.Sontag E, Nunbhakdi-Craig V, Bloom G S, Mumby M C. A novel pool of protein phosphatase 2A is associated with microtubules and is regulated during the cell cycle. J Cell Biol. 1995;128:1131–1144. doi: 10.1083/jcb.128.6.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sontag E, Sontag J M, Garcia A. Protein phosphatase 2A is a critical regulator of protein kinase C zeta signaling targeted by SV40 small t to promote cell growth and NF-kappaB activation. EMBO J. 1997;16:5662–5671. doi: 10.1093/emboj/16.18.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szyszka R, Kudlicki W, Kramer G, Hardesty B, Galabru J, Hovanessian A. A type 1 phosphoprotein phosphatase active with phosphorylated Mr = 68,000 initiation factor 2 kinase. J Biol Chem. 1989;264:3827–3831. [PubMed] [Google Scholar]
- 51.Taylor D R, Lee S B, Romano P R, Marshak D R, Hinnebusch A G, Esteban M, Mathews M B. Autophosphorylation sites participate in the activation of the double-stranded-RNA-activated protein kinase PKR. Mol Cell Biol. 1996;16:6295–6302. doi: 10.1128/mcb.16.11.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vlach J, Garcia A, Jacque J M, Rodriguez M S, Michelson S, Virelizier J L. Induction of Sp1 phosphorylation and NF-kappa B-independent HIV promoter domain activity in T lymphocytes stimulated by okadaic acid. Virology. 1995;208:753–761. doi: 10.1006/viro.1995.1207. [DOI] [PubMed] [Google Scholar]
- 53.Wang S S, Esplin E D, Li J L, Huang L, Gazdar A, Minna J, Evans G A. Alterations of the PPP2R1B gene in human lung and colon cancer. Science. 1998;282:284–287. doi: 10.1126/science.282.5387.284. [DOI] [PubMed] [Google Scholar]
- 54.Wera S, Hemmings B A. Serine/threonine protein phosphatases. Biochem J. 1995;311:17–29. doi: 10.1042/bj3110017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Westphal R S, Anderson K A, Means A R, Wadzinski B E. A signaling complex of Ca2+-calmodulin-dependent protein kinase IV and protein phosphatase 2A. Science. 1998;280:1258–1261. doi: 10.1126/science.280.5367.1258. [DOI] [PubMed] [Google Scholar]
- 56.Williams B R G. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 57.Williams B R G. Role of the double-stranded RNA-activated protein kinase (PKR) in cell regulation. Biochem Soc Trans. 1997;25:509–513. doi: 10.1042/bst0250509. [DOI] [PubMed] [Google Scholar]
- 58.Wong A H, Tam N W, Yang Y L, Cuddihy A R, Li S, Kirchhoff S, Hauser H, Decker T, Koromilas A E. Physical association between STAT1 and the interferon-inducible protein kinase PKR and implications for interferon and double-stranded RNA signaling pathways. EMBO J. 1997;16:1291–1304. doi: 10.1093/emboj/16.6.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Y L, Reis L F, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams B R G, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yue Z, Shatkin A J. Double-stranded RNA-dependent protein kinase (PKR) is regulated by reovirus structural proteins. Virology. 1997;234:364–371. doi: 10.1006/viro.1997.8664. [DOI] [PubMed] [Google Scholar]
- 61.Zamanian-Daryoush M, Der S D, Williams B R G. Cell cycle regulation of the double stranded RNA activated protein kinase, PKR. Oncogene. 1999;18:315–326. doi: 10.1038/sj.onc.1202293. [DOI] [PubMed] [Google Scholar]
- 62.Zandi E, Chen Y, Karin M. Direct phosphorylation of IkappaB by IKKalpha and IKKbeta: discrimination between free and NF-kappaB-bound substrate. Science. 1998;281:1360–1363. doi: 10.1126/science.281.5381.1360. [DOI] [PubMed] [Google Scholar]