Abstract
Infections caused by multidrug-resistant (MDR) bacteria are an increasing global healthcare concern. In this study, we developed a dual-ligand-functionalised Au25(SR1)x(SR2)18−x-type gold nanocluster and determined its antibacterial activity against MDR bacterial strains. The pyridinium ligand (SR1) provided bactericidal potency and the zwitterionic ligand (SR2) enhanced the stability and biocompatibility. By optimising the ligand ratio, our gold nanocluster could effectively kill MDR Gram-positive bacteria via multiple antibacterial actions, including inducing bacterial aggregation, disrupting bacterial membrane integrity and potential, and generating reactive oxygen species. Moreover, combining the optimised gold nanocluster with common antibiotics could significantly enhance the antibacterial activity against MDR bacteria both in in vitro and animal models of skin infections. Furthermore, the fluorescence of the gold nanocluster at the second near-infrared (NIR-II) biological window allowed for the monitoring of its biodistribution and body clearance, which confirmed that the gold nanoclusters had good renal clearance and biocompatibility. This study provides a new strategy to combat the MDR challenge using multifunctional gold nanomaterials.
A NIR-II fluorescent Au25 nanocluster capped with optimal dual functional ligands can effectively aggregate and kill Gram-positive drug-resistant bacteria while maintaining good biocompatibility.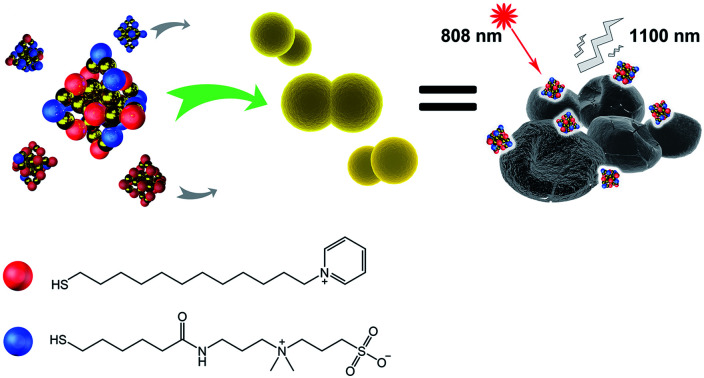
Introduction
Since the discovery of antibiotics in 1928, the unrestrained global use of antibiotics has imposed a highly selective pressure on all bacterial species, which has accelerated the acquisition and accumulation of drug-resistant genes via horizontal transmission.1 Several multidrug-resistant (MDR) pathogens, or the so-called ‘superbugs,’ have emerged over the past 50 years.2,3 Moreover, some drug-resistant bacteria that were previously considered less harmful, like Methicillin-resistant Staphylococcus epidermidis (MRSE), have gained much attention due to their role in the development of drug-resistant strains.4,5 The widespread occurrence of MRSE inevitably increases the risk of infection and promotes intra- and inter-species horizontal transfer of the MDR gene.6 MDR-related infections have imposed a significant burden on the world economy and healthcare systems, yet the development of new antibiotics has largely stalled over the past 20 years.7 Therefore, there is an urgent need to develop new strategies to address this significant global health problem.
Recently, gold nanomaterials, including gold nanoparticles (GNPs),8–10 gold nanorods (GNRs)11 and gold nanoclusters (GNCs),12,13 have emerged as potentially effective antibacterial agents. They can exhibit several antibacterial actions, such as delivering antibiotics, producing reactive oxygen species (ROS) and offering photothermal treatments.14,15 Among them, sub-2 nm GNCs have a distinct advantage of rapid renal clearance, which can greatly reduce the potential long-term toxicity.16 Recent studies have focused on synthesising atomically precise GNCs17 to achieve better quality control, which is important for potential clinical approval. Among all formula-precise GNCs, the Au25(SR)18-type GNC is the most widely studied owing to its high stability, low toxicity, feasible preparation and stable near-infrared (NIR) fluorescence,18 making it a powerful tool for bioimaging, drug delivery and therapy.19,20 However, most Au25(SR)18 GNCs reported so far are capped with a single-type ligand, limiting the ability to tune their biocompatibility and antibacterial potency, as the requirements for the two are often incompatible. Consequently, most antibacterial nanomaterials have only demonstrated effectiveness in vitro but not in vivo.12
Herein, we synthesised a pyridinium–zwitterionic dual-ligand functionalised Au25(SR1)x(SR2)18−x GNC to address MDR bacteria-induced infections, especially MRSE (Fig. 1A). The pyridinium ligand is derived from cetylpyridinium chloride (CPC), a commercial mouth mucosa aseptic additive approved by the US Food and Drug Administration, which affords antibacterial ability although it suffers from poor prospects in in vivo applications.21,22 The zwitterionic (ZW) ligand was introduced due to its biocompatibility, biosafety, and low biofouling properties.23,24 Previously, our group and other researchers showed that ZW ligand-coated nanoparticles are highly stable, biocompatible and strongly resist biofouling.25,26 By adjusting the GNC surface capping pyridinium/ZW ligand ratio, GNCs that displayed low cytotoxicity and excellent antibacterial activity against clinically isolated Gram-positive drug-resistant species were obtained. Besides, we discovered that the combination of our GNCs with conventional antibiotics could significantly enhance the bactericidal potency against MRSE, allowing us to overcome MDR bacterial infections at the cellular level and in vivo skin infection models. Furthermore, we exploited the stable NIR-II fluorescence of our GNC to directly monitor the in vivo biodistribution, revealing that the GNC was efficiently cleared without inducing observable long-term toxic effects. All these results show that our optimised Au25(SR1)x(SR2)18−x GNC is part of a new generation of multifunctional fluorescent nanomaterials for treating MDR bacterial infections induced by Gram-positive strains.
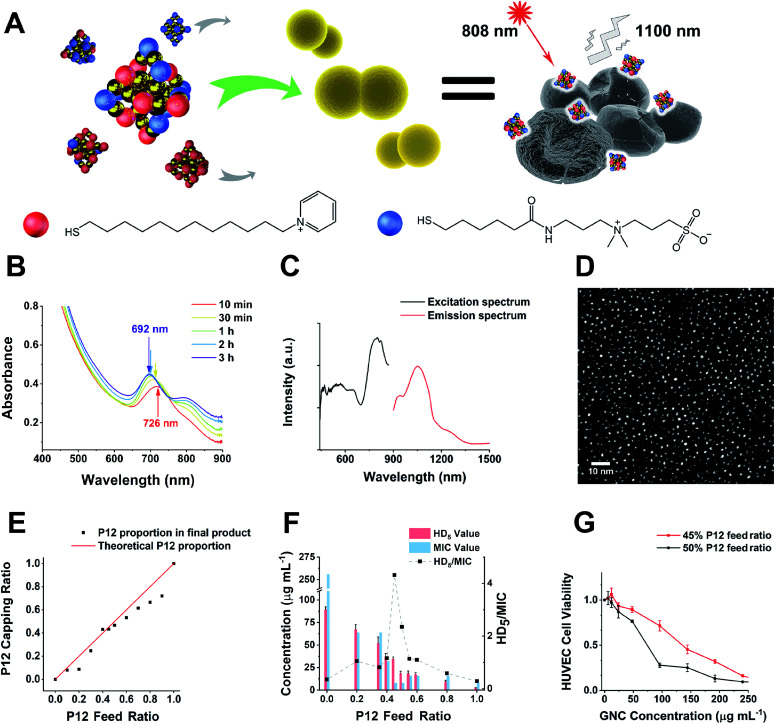
Fig. 1. (A) A schematic diagram of this study. GNCs with a specific P12/C5 composition aggregate planktonic bacteria by interacting with the bacterial cell envelope while maintaining good biocompatibility. Local interactions of GNCs with bacteria cause cell content leakage and structural deformation, which induce cell death. The distribution of GNCs is tracked using NIR fluorescence upon excitation with an 808 nm laser. Golden spheres indicate gold atoms, red spheres indicate the P12 ligands, and blue spheres indicate the C5 ligands. (B) Time-dependent evolution of the UV-vis spectra of the 100% C5-capped GNC (representative species) during the synthesis process. (C) NIR fluorescence excitation (black line, monitored with λEM = 1100 nm) and emission (red line, λEX = 810 nm) spectra of the 100% C5-capped GNC. (D) Double spherical aberration-corrected TEM image (scale bar: 10 nm) of the 100% C5-capped GNC prepared on an ultra-thin carbon film supported copper grid. The UV-vis spectra, NIR fluorescence spectra and TEM images of GNCs with other P12/C5 compositions show no obvious differences from those of this representative species. (E) A plot of the GNC surface P12 capping ratio versus the P12 feed ratio. The red line indicates the theoretical output, assuming no ligand affinity difference, while the black dots represent the actual P12 ratio. (F) HD5/MIC values for dual-ligand GNCs prepared under different P12 feed ratios. The greater the HD5/MIC, the higher the biosafety and antibacterial ability of the material. A sudden change in haemolytic potency occurs as the feed ratio approaches 50% (the MIC value is from anti-S. aureus results). (G) Comparison of the cytotoxicity of the 45%- and 50%-P12 GNCs toward human umbilical vein endothelial cells (HUVECs) with 24 h incubation.
Results and discussion
Synthesis and characterisation of Au25(SR1)x(SR2)18−x-type GNCs
In this study, alkyl-thiolated ZW and pyridinium ligands (abbreviated as C5 and P12, hereafter) were synthesised, and their chemical structures were confirmed by NMR and mass spectrometry (MS), respectively (see ESI Fig. S1–S9†). GNCs with different surface ligand cappings were then prepared by varying the feed ratios between 100% C5 and 100% P12 ligands. The formation of the Au25 NC was characterised by a UV-visible spectrum (UV-vis) absorption peak at ∼690 nm with a shoulder at ∼810 nm.27 The characteristic absorption peak was blue-shifted from ∼730 to ∼690 nm during the first 2 h. Then, the spectrum for the Au25 NC stabilised after 3 h, suggesting that the synthesis of the Au25 NC was complete (Fig. 1B).28 Using this reaction time, we synthesised GNCs with increasing feed ratio of P12 (from 0% to 100%). All mixed P12/C5 groups successfully produced the Au25(P12)x(C5)18−x-type GNCs, indicated by obvious absorption peaks at 690 nm (see ESI Fig. S10†).
Under 808 nm laser excitation, the GNCs showed a strong fluorescence peak at ∼1100 nm with a shoulder at ∼950 nm, regardless of the ligand composition, which is consistent with a previous report29 (Fig. 1C). Such fluorescence characteristics are ideal for sensitive fluorescence imaging in the second near-infrared (NIR-II) biological window, which facilitates enhanced tissue penetration depth and resolution.30 Despite having a moderate absolute fluorescence quantum yield (0.54%) compared with small molecular NIR-II emitting dyes,31,32 our GNCs exhibited higher stability, biocompatibility and anti-photobleaching properties, which are extremely beneficial to bioimaging.33
The GNC size obtained under different ligand feed ratios was measured by double spherical aberration-corrected TEM (DSAC-TEM) analysis, which revealed the GNC granule size to be around 1.2 nm with negligible difference between each different-ligand group (Fig. 1D and Table 1). These results are consistent with the ultra-small size of Au25 NCs reported in the literature.34
Characterisation of Au25(P12)x(C5)18−x GNCs prepared at different P12 feed ratios.
| P12 feed ratio (%) | Zeta potential ± SD (mV) | Size ± SD (nm) | Primary product (P12/C5) | Secondary product(s) (P12/C5) | Deconvoluted MW (Da) |
|---|---|---|---|---|---|
| 0 | −4.7 ± 1.1 | 1.18 ± 0.21 | 0/18 | — | 11 282 |
| 10 | 8.8 ± 0.6 | 1.15 ± 0.23 | 1/17 | 2/16 | 11 176 |
| 20 | 14.1 ± 2.0 | 1.21 ± 0.32 | 1/17 | 2/16 | 11 167 |
| 30 | 24.1 ± 0.9 | 1.11 ± 0.41 | 4/14 | 5/13 | 10 954 |
| 40 | 36.3 ± 2.3 | 1.15 ± 0.22 | 7/11 | 8/10 | 10 710 |
| 45 | 37.5 ± 1.6 | 1.19 ± 0.24 | 8/10 | 7/11 & 9/9 | 10 708 |
| 50 | 43.3 ± 1.5 | 1.16 ± 0.23 | 8/10 | 7/11 & 9/9 | 10 662 |
| 60 | 49.5 ± 3.1 | 1.14 ± 0.28 | 9/9 | 8/10 | 10 575 |
| 70 | 56.9 ± 3.1 | 1.15 ± 0.26 | 11/7 | 10/8 & 12/6 | 10 466 |
| 80 | 62.5 ± 1.9 | 1.17 ± 0.17 | 12/6 | 11/7 | 10 400 |
| 90 | 66.5 ± 1.0 | 1.14 ± 0.27 | 13/5 | 12/6 | 10 327 |
| 100 | 71.4 ± 1.6 | 1.18 ± 0.22 | 18/0 | — | 9956 |
In addition, the zeta potentials of GNCs prepared under different P12 ligand feed ratios were measured and are summarized in Table 1. The zeta potential of GNCs gradually and progressively shifted from around −5 mV to >+70 mV as the P12 ligand feed ratio was increased from 0 to 100%, indicating successful incorporation of the P12 ligand onto the GNC surface.
The GNCs were further characterised by electrospray ionisation MS (ESI-MS), which yielded the general molecular formula of diverse Au25(P12)x(C5)18−x (x = 0–18, depending on the P12/C5 feed ratio, see ESI Fig. S11†). After mass deconvolution, the exact mass of the dominant peaks was matched with the theoretical molecular weight (Table 1 and ESI Table S1†). The relative abundance of each GNC was derived from its corresponding peak in the mass spectrum35 and the relationship between the GNC surface ligand composition and corresponding ligand feed ratio is summarized in Table 1. It is apparent that except for those prepared with 100% P12 or 100% C5, all other GNCs prepared were a mixture of differently formulated Au25(P12)x(C5)18−x GNCs, which could be explained by the putative forming mechanism of GNCs. Upon adding the dual-ligand solution to chloroauric acid, the thiol groups quickly reduced the trivalent Au(iii) ions to Au(i) to form Au(i)–ligand complexes.36 Due to the random reduction and combination of Au(i) with different capping ligands, the complex units would have multiple types. The subsequent addition of sodium borohydride to reduce Au(i) for cluster formation and sodium hydroxide (for controlling ligand etching ability) allowed the precise reduction of some Au(i)–ligand complexes to Au(0), forming a stochastic mixture of dual-ligand capped GNCs with a variety of ligand capping ratios.37 However, controlled mainly by our ligand feed ratio, these synthesis conditions could allow for some specific GNCs to be the main product. The relative abundance of each GNC formulation could be derived from the corresponding MS peak intensity.35
A plot of the average content of P12 in the GNCs against the feed ratio (Fig. 1E) revealed that the product line was mostly below the feed line. Therefore, the C5 ligand appeared to bind more strongly to the gold kernel than P12, possibly because of the lower electrostatic repulsion among the C5 (neutral overall) than P12 (positively charged) ligands. The only exception was observed at a feed ratio of 40–45%, where the product composition (mainly Au25(P12)8(C5)10) matched the feed ratio, suggesting that the products of this ligand ratio were exceptionally stable.
Antibacterial screening
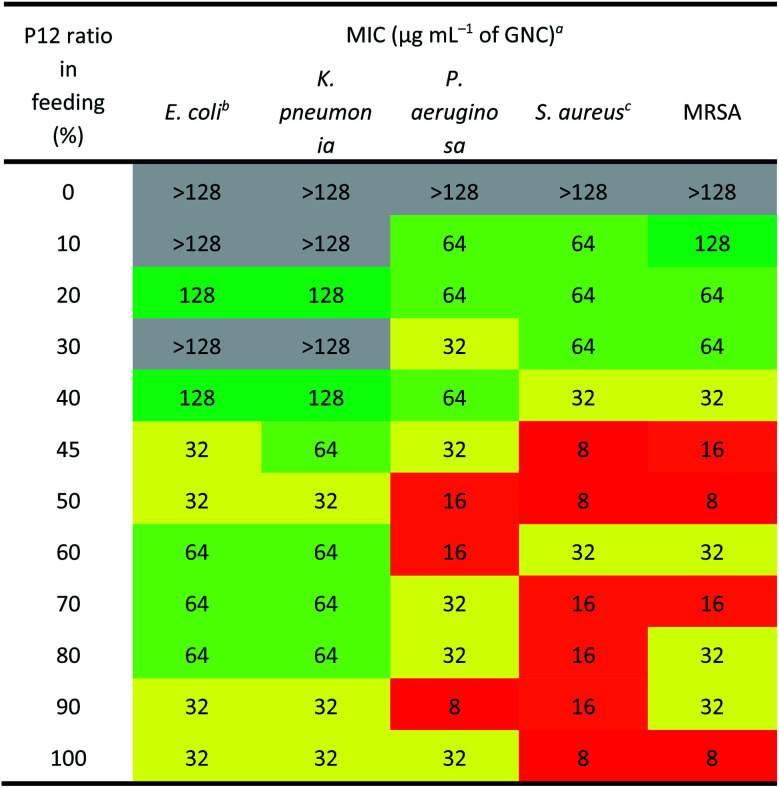
The antibacterial activity of GNCs was assessed using a few common Gram-positive (Staphylococcus aureus and MRSA) and Gram-negative (Escherichia coli, Klebsiella pneumonia and Pseudomonas aeruginosa) species (Table 2 [16 h] and ESI Table S2† [24 h]). The relationship between the antibacterial activity and the P12 ligand feed ratio (i.e. surface positive charge density) of GNCs was interesting: first, the activity increased with increasing P12 ligand feed ratio (up to 50%), then there was a slight decrease (from 50% to 70%), and finally it increased again (from 70% to 100%). This trend was totally unexpected; since the antibacterial properties of GNCs were mainly derived from the incorporated, positively charged P12 ligands, and not the C5 ligands, we had anticipated the antibacterial activity to be positively correlated with the P12 content (GNC surface positive charge density). The fact that the GNC with a 50% P12 feed ratio exhibited comparable antibacterial activity to that with a 100% P12 ligand feed ratio suggests that there must be an optimal window to tune the GNC antibacterial properties and biocompatibility.
Antibacterial activity of different GNCs against five common pathogens (the colour code represents the antibacterial activity: red for high, yellow for medium, green for low and grey for negligible, and the same below).
|
MIC—minimum inhibitory concentration.
Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa are Gram-negative bacteria.
Staphylococcus aureus and Methicillin-resistant Staphylococcus aureus (MRSA) are Gram-positive bacteria.
Table 2 reveals that the GNCs were more potent towards Gram-positive strains than towards Gram-negative ones. This phenomenon can be attributed to the differences in the surface structure between Gram-positive and -negative bacteria. The surface of Gram-positive bacteria is negatively charged;38 thus, they exhibit a strong electrostatic interaction with cationic materials. For example, wall teichoic acid (WTA), a distinct cell wall component in Gram-positive bacteria, consists of repeating poly(glycerol phosphate) units and a phosphodiester terminus. WTA constitutes the polyanionic network, making the cell envelope highly negatively charged and susceptible to the binding of cationic materials.39,40 Besides, anionic lipids, such as phosphatidylglycerol (PG) and cardiolipin, constitute ∼80% of the total lipids in Gram-positive bacterial membranes but only ∼30% in Gram-negative strains.41,42 Moreover, negatively charged phospholipids, such as PGs, are present on both sides of bacterial cell membranes rather than only the inside layer found on mammalian cells. This difference can provide selectivity between mammalian cells and bacteria for some positively charged antibacterial agents.43
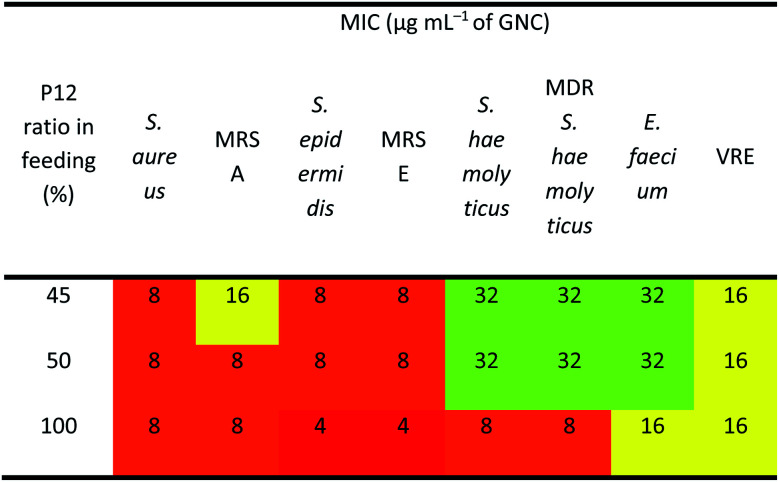
As shown in Table 2, the three best-performing GNCs (45%, 50% and 100% P12 feed ratios) were selected for further tests using more Gram-positive strains, including Staphylococcus epidermidis, Staphylococcus haemolyticus, Enterococcus faecium and their corresponding drug-resistant strains (MRSE, MDR S. haemolyticus and vancomycin-resistant Enterococcus (VRE)). Table 3 reveals that although the GNCs with 100% P12 exhibited the best antibacterial potency among the three groups, especially against MRSE, those with 45% and 50% P12 feed ratios also showed similar activity. These results further confirmed that antibacterial potency is not linearly correlated with the active P12 ligand content. Besides, during antibacterial tests, we observed the aggregation of GNCs only with ≥60% P12 feed ratios in LB culture media, suggesting significant non-specific interactions with negatively charged serum proteins in cell culture media, which could adversely affect their antibacterial potency (ESI Fig. S12†). This result was consistent with their high positive zeta potentials. This represents a key drawback for such strongly positively-charged GNCs and other nanomaterials, which can greatly limit their applications as antibacterial reagents. Therefore, in synthesising mixed-ligand capped GNCs, we must balance the active/inert ligand (P12/C5 here) ratio and determine their actual antibacterial properties in order to find the optimal ligand ratio.
The antibacterial activity toward Gram-positive bacteria of selected GNCs.
|
Biocompatibility and cytotoxicity
To determine the optimal ligand ratio, we performed a haemolytic test (ESI Fig. S13†). The resulting HD5 (the GNC dose that causes a 5% haemolysis value against the P12 ligand feed ratio) was calculated for optimising the P12 feed ratio (Fig. 1F). The GNC haemolytic efficiency was enhanced (HD5 reduced) with increasing P12 ligand content, which also indicated that increasing the ZW ligand ratio could reduce the cytotoxicity of GNCs. The GNC prepared with 45% P12 still maintained a relatively high HD5 of 34 μg mL−1, about twice that of the GNC prepared with 50% P12, while retaining almost the same minimum inhibitory concentration (MIC). Accordingly, GNCs prepared with >45% P12 could be too cytotoxic, and thus unfavourable for in vivo applications. To determine the optimal P12 feed ratio, we plotted the HD5/MIC ratio versus the P12 feed ratio to collectively depict the change of biosafety and antibacterial activity of dual-ligand GNCs. When the feed ratio of P12 approached 45%, a sharp peak appeared, indicating that the GNC prepared with a 45% P12 feed ratio has a high antibacterial ability (low MIC) while maintaining low haemolyticity (high HD5), and hence high biosafety, which is important for biomedical applications. This result also highlighted the importance of incorporating a certain proportion of ZW ligand in order to maintain good overall biocompatibility.
We further evaluated the cytotoxicity of GNCs prepared with 45% and 50% P12 ligand feed ratios toward human umbilical vein endothelial cells (HUVECs). Compared with the 50%-P12 GNC, HUVECs demonstrated a much higher tolerance toward the 45%-P12 GNC (Fig. 1G), indicating a striking shift in cytotoxicity as the P12 feed ratio reaches 50%. This result is consistent with the significantly lower HD5 value for the 50% P12 GNC observed above.
The use of excess cationic P12 ligand on the GNC capping can lead to high cytotoxicity, adversely affecting its potential for in vivo application. However, too low P12 ligand content can lead to low antibacterial activity. The key here is to balance the active/inert ligand ratio to achieve high stability and biocompatibility without compromising antibacterial potency.
The above results comprehensively proved that GNCs prepared with 45% P12 ligand feed ratio displayed excellent antibacterial activity, low cytotoxicity, and good stability. Therefore, the 45% P12 ligand feed ratio was considered optimal for preparing GNCs (denoted as GNC hereafter) for subsequent analyses.
Combinational antibacterial analyses
We considered that, besides the direct killing of bacteria, GNC could sensitise drug-resistant bacteria toward varied antibiotics to restore their antibacterial potency.44 To investigate this potential, we selected MRSE as the model microbe, to which our GNC demonstrated high inhibition. Seven antibiotics used to treat Gram-positive bacterial infections from four main categories: β-lactams (penicillins and carbapenem), glycopeptide, macrolides and tetracycline, were tested. The results demonstrated that, except for vancomycin and tetracycline, all the other antibiotics exhibited low efficacy against this bacterial strain, with MIC values ≥32 μg mL−1 (ESI Table S3†). The test revealed high drug resistance of MRSE to several antibiotics.
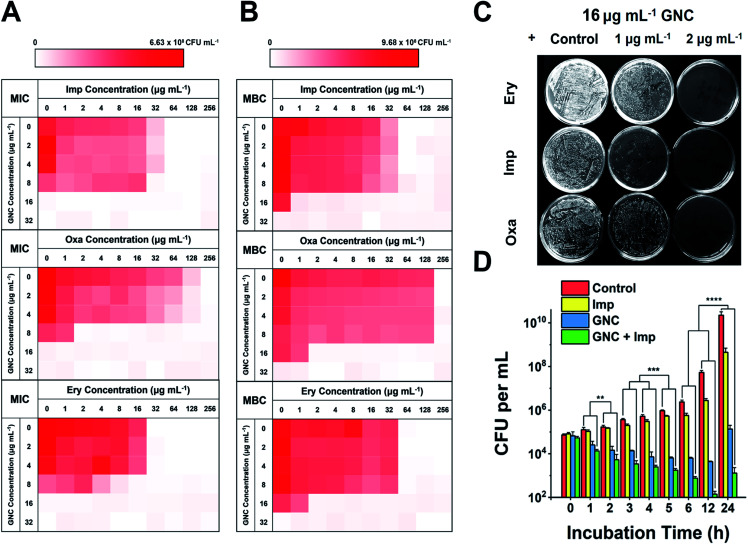
A checkerboard method was further employed to evaluate the antibacterial properties of GNC–antibiotic combinational therapy.45,46 Imipenem (Imp), oxacillin (Oxa) and erythromycin (Ery), representing three classes of antibiotics, were applied with and without the GNC. Then, the fractional inhibitory concentration index (FICI) was calculated after 24 h of incubation. The calculated FICIs show that in joint applications, GNC exhibited additive antibacterial ability with all three antibiotics (Fig. 2A). Particularly, the Oxa group showed an exciting 128-fold decrease in antibiotic dosage needed to inhibit bacterial growth compared to the application of the antibiotic alone.
Fig. 2. Combinational antibacterial assay of GNC with antibiotics. (A) 24 h checkerboard results of the GNC with imipenem (Imp), oxacillin (Oxa), and erythromycin (Ery) (OD600 values are given in the colour scale; a darker colour represents a higher bacterial concentration). The FICI was 1, 0.508, and 0.75 for GNC and Imp, Oxa, and Ery, respectively. (B) 48 h checkerboard results of GNC with Imp, Oxa, and Ery. The FICI was ∼0.5 for all three groups. This experiment revealed the combinational bactericidal effect of GNC with varied types of antibiotic. (C) Undiluted plate-coating results of MRSE colonies after treatment with PBS buffer (control), different concentrations of Ery/Imp/Oxa and 16 μg mL−1 GNC to visually display changes in colonies. (D) Time-dependent killing curves demonstrate a superior antibacterial efficiency of the combination of GNC and Imp compared to their separate applications. PBS buffer (control), Imp only (2 μg mL−1), GNC only (16 μg mL−1), and a combination of GNC (16 μg mL−1) and Imp (2 μg mL−1) (n = 3, ** 0.01 < P < 0.05, ***P < 0.01 and ****P < 0.001).
We further prolonged the incubation to 48 h and re-measured the FICI for each group (Fig. 2B). The FICIs of all three groups were closer to 0.5, indicating that the additive effect of GNC and antibiotics was enhanced. Specifically, adding 16 μg mL−1 GNC and 2 μg mL−1 of any of the three tested antibiotics could prevent the growth of MRSE. We further took out the bacterial mixture and spread it onto agar plates to check the survival of MRSE and the agar plates remained unstained for the test groups compared with innumerable colonies for the PBS control (Fig. 2C). The results indicated that GNC had a remarkable ability to re-sensitise MRSE toward several antibiotics. Owing to the higher bactericidal efficiency, Imp was selected as the model antibiotic for further studies.
The MRSE-killing efficiency of this combination was measured by plotting a time-dependent killing graph (Fig. 2D and ESI Fig. S14†). Compared to the PBS control, adding Imp did not significantly inhibit bacterial growth due to the inherent high drug resistance. However, the GNC alone showed considerable bactericidal properties, giving rise to 105-fold lower MRSE densities at 24 h. The combined group showed the best results, where the population of the MRSE colony was ∼40- and 100-fold smaller than that of the GNC-only group at 12 and 24 h, respectively. Impressively, the bacterial count of the GNC + Imp combination group was more than 107-fold lower than that of the control group at 24 h, demonstrating the excellent additive potency of the combined GNC and Imp in MRSE killing.
Antibacterial mechanisms
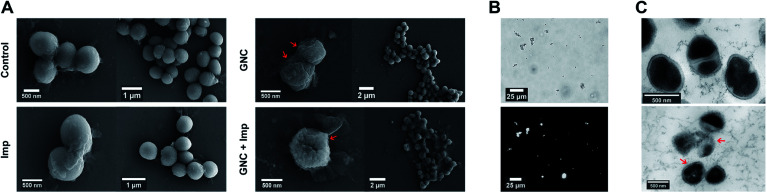
In general, cationic membrane-active molecules can employ several mechanisms to affect bacterial membrane function, for instance, attracting phospholipids to form defects and membrane leakage or perturbing membranes to induce lipid flip-flop.42 The damaged membrane causes detrimental membrane de-polarisation and spillage of cell contents, allowing antibiotics to penetrate the impaired bacterial defence for swift killing.47 The SEM images of MRSE cells revealed significant morphological differences after each different treatment (Fig. 3A). For the negative control, all MRSE cells were spherical without apparent broken parts or wrinkles. Almost identical morphologies were observed for the Imp group, suggesting that the treatment with Imp alone did not cause noticeable damage to MRSE cells. In contrast, the GNC-treated group showed significantly altered cell morphologies and collapsed structures. The treated MRSE cells were tightly clustered together on a larger scale. A closer view revealed that the surface was covered with small grooves and wrinkles, indicating viability loss. This phenomenon was even more evident for the combined group (indicated with red arrows), where the grooves and wrinkles were deeper with a concave surface, indicating that the GNC + Imp combination could effectively agglutinate and deform bacterial cells, resulting in efficient bacterial killing.
Fig. 3. GNC antibacterial mechanism investigation. (A) Typical SEM images of MRSE after a 4 h treatment with PBS buffer (control), 2 μg mL−1 Imp, 16 μg mL−1 GNC, and a combination of Imp (2 μg mL−1) and GNC (16 μg mL−1). (B) Bright-field image (upper) and the corresponding NIR fluorescence image (lower) showing that the GNC + Imp treated MRSE bacteria were extensively aggregated. (C) TEM images of the PBS buffer- (control group, upper) and 16 μg mL−1 GNC + 2 μg mL−1 Imp-treated MRSE (lower).
After treatment with GNC + Imp, the MRSE cells formed irregular clusters which could hinder migration. This phenomenon was further confirmed by the corresponding NIR fluorescence image of the aggregated MRSE cells (Fig. 3B). This result indicated that GNCs covered the envelope of MRSE cells, allowing them to aggregate by neutralising their surface negative charges and form ‘bacterial clusters’ to limit their spread and prolong the interaction.
TEM images (Fig. 3C) further confirmed the interaction of GNC with MRSE cells, which demonstrated the adhesion and penetration of GNCs within the cell envelope. Compared with the intact shape and apparent division septa for bacterial cells in the control group, the GNC + Imp treated group showed an abnormal cell division phenomenon: uneven division and empty cell walls were observed. Moreover, GNCs were mainly distributed within particular spaces on the cell envelope, indicating specific binding (indicated with red arrows).48
The antibacterial mechanism of GNC was further investigated. Although GNC binding-induced bacterial aggregation was already observed, it is still useful to investigate the specific binding target. The high abundance of negatively charged WTA may serve as a binding target for the positively charged GNCs via electrostatic interactions. Isothermal titration calorimetry (ITC) was conducted between GNCs and WTA, which revealed a multiple-binding endothermic interaction (ESI Fig. S15A†), indicating that WTA could initiate bacteria–GNC interactions.49 To further verify this, a fluorescence competition assay with TR-cadaverine was carried out. This dye can form a complex with WTA to quench its fluorescence. If GNC can compete with TR-cadaverine in binding with WTA, adding GNC would result in dye release and fluorescence recovery. Fig. S15B (ESI†) shows that the fluorescence of TR-cadaverine significantly increased with an increase in GNC concentration and was saturated at ∼8 μg mL−1. Further increasing the GNC concentration could quench the dye fluorescence, possibly via dynamic quenching at high concentrations.50 These results confirmed that WTA was a binding target for the GNC.
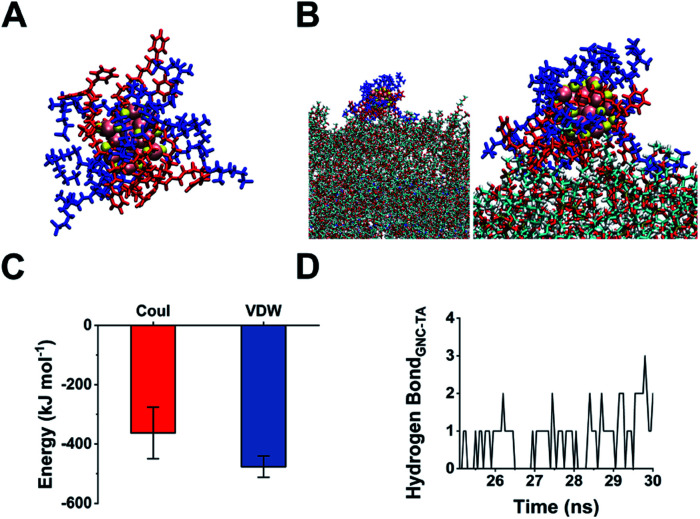
The 45% P12 feed ratio yielded Au25(P12)8(C5)10 as the primary product. Thus, we used this structure to simulate GNC binding with WTA by molecular binding simulation.51 A surface coverage state was established by simulating the GNC configurations (Fig. 4A). Due to the electrostatic attraction between the cationic pyridinium and terminal anionic sulfonate groups, the ligands on the GNC surface are bent rather than pointing outward and form a ‘cage-like’ cap to stabilise the entire structure. This configuration explains the relatively high stability of GNC. Upon mixing with WTA, the electrostatic interactions between the GNC surface pyridinium groups and WTA phosphonate groups appeared to support GNC invasion of the bacterial cell envelope (Fig. 4B). During this process, the cationic ligands interacted with the oppositely charged WTA layer, while the C5 ligands mainly pointed away from the surface. The initial electrostatic interactions further promote extensive contact among the non-charged areas and strengthen their interactions via van der Waals forces, which eventually contribute ∼57% of the overall interaction (Fig. 4C). Moreover, the weak hydrogen bonding between the WTA hydroxyl groups and pyridinium π-ring can further enhance the interaction (Fig. 4D). WTA plays a vital role in drug resistance by providing attaching sites for other proteins that can replace synthases inhibited by penicillin antibiotics.45 Thus, the binding of GNCs to WTA may compromise the bacterial drug resistance and partially explain the sensitisation of MDR strains towards antibiotics when co-treated with GNCs.
Fig. 4. (A) Snapshot of the structure of an obtained Au25(P12)8(C5)10 cluster with 45% P12 feed ratio: blue indicates C5; red indicates P12; yellow indicates thiol groups; pink indicates gold atoms. (B) Left: snapshot of the GNC interaction with WTA; right: scaled-up illustration of the interacting part. (C) Interaction energy between GNC and WTA. Coul represents Coulomb force, while VDW denotes van der Waals force. (D) Number of hydrogen bond interactions between GNC and WTA units during a 5 ns simulation.
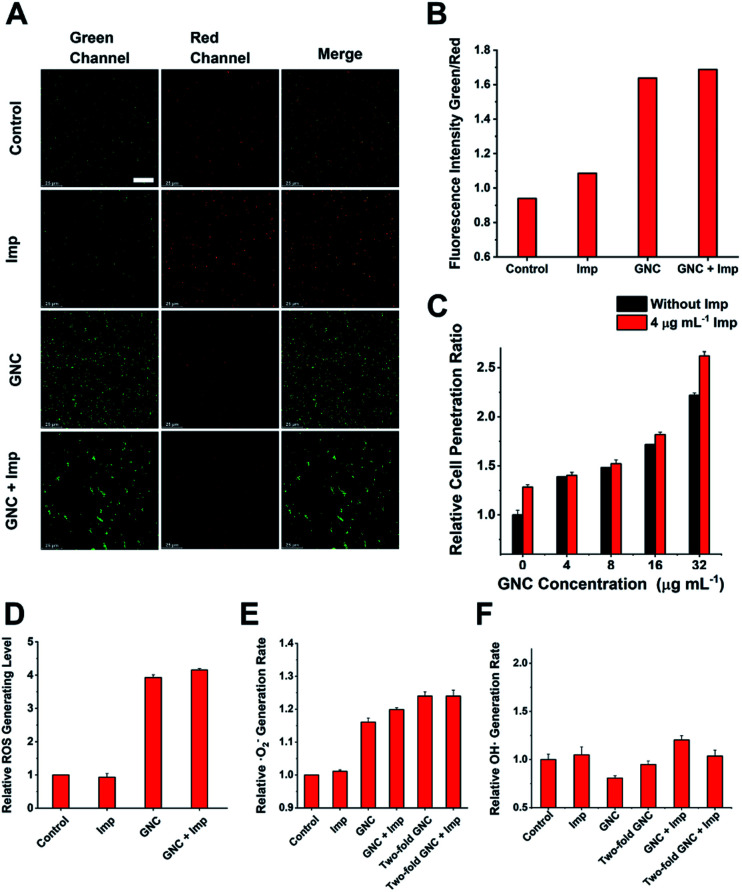
Upon binding to the bacterial cell membrane, the positive charges on the GNC could induce membrane potential dissipation and cause cell death. In this study, the membrane potential was determined by measuring the ratio of green to red fluorescence using a fluorescent kit to stain the bacteria, where the green to red fluorescence ratio is linear with the membrane potential dissipation rate (Fig. 5A and B). Adding only Imp did not cause a significant change of green fluorescence, indicating that Imp had no apparent effect on the bacterial membrane potential. However, a sharp decrease in the red fluorescence and an increase in green fluorescence were observed for the GNC-treated MRSE cells, suggesting significant membrane potential dissipation. Membrane potential dissipation can severely interfere with normal physiological activities and cause cell death; thus, this result was consistent with the SEM and TEM images. Moreover, clustered bacteria were also observed in the GNC groups, indicating that membrane potential dissipation was related to the GNC-induced bacterial agglomeration.52
Fig. 5. Investigating other possible antibacterial properties. (A) Membrane potential of MRSE cells after treatment in PBS buffer (control), 4 μg mL−1 Imp, 16 μg mL−1 GNC, and a combination of Imp (4 μg mL−1) and GNC (16 μg mL−1) (scale bar: 25 μm). (B) Comparison of the green-to-red fluorescence intensity ratios in different groups. (C) Comparison of the cell penetration rate at different concentrations of GNC with/without the addition of 4 μg mL−1 Imp (n = 4). (D) Total ROS generation rate for the combination group compared with the PBS control and individual application (n = 3). (E) Superoxide anion generation rate of the PBS (control), single applications and the combination group (n = 3). (F) ROS colorimetric analysis results with the nitro-blue tetrazolium method, which demonstrates no correlation between the ·OH generation rate and the addition of either GNC or Imp (n = 3).
Inserting a positively charged amphiphilic material has been shown to lyse cell membranes.53,54 The collapse of the MRSE cell structure, as observed in the SEM images, indicated that GNC could affect cell envelope completeness. To investigate whether the GNC treatment affected the integrity of cell membranes, we used a cell-integrity analysis kit, in which the dye can penetrate damaged membranes and bind to intracellular proteins. A positive correlation between the GNC concentration and dye incorporation confirmed that GNC could damage the integrity of the cell membrane (Fig. 5C). The GNC + Imp combination exhibited even higher membrane permeation than the GNC-only treatment, consistent with the superior bactericidal potency. The antibacterial mechanism of Imp is to inhibit the synthesis of cell walls, thereby bursting the bacterial cells. By damaging the bacterial cell membrane, GNC can effectively enhance the ability of antibiotics to traverse bacterial cell membranes, resulting in additive bactericidal potency.
GNCs have been shown to generate ROS, which have bactericidal effects.12 Therefore, we studied ROS generation in all treatments (Fig. 5D). Imp alone did not produce significant ROS, while GNC exhibited a good ability to generate ROS. Further addition of Imp did not increase ROS production. Therefore, only GNC is responsible for ROS production in the GNC + Imp combination. To investigate the exact species of ROS responsible here, we used two test kits to detect common ROS species ·O2− and ·OH (Fig. 5E). There was an increase in the production of ·O2− but not ·OH in the GNC treatments (Fig. 5F). Hence, ·O2− was confirmed to be the ROS species generated by the GNC treatment. Its high reactivity can damage cell membranes and other essential biological macromolecules to exert bactericidal effects. Some ROS-generating antibacterial materials have been found to have higher antibacterial activity against Gram-positive over Gram-negative strains, allowing them to be used even as Gram-selective staining reagents.55–57 The ROS generation ability of our GNC may also account for its higher antibacterial effect towards Gram-positive species like MRSE or S. aureus over Gram-negative ones.
Based on the above experimental results, our GNC exhibits four antibacterial mechanisms. (1) By binding to WTA, the GNC could interact with and aggregate bacterial cells to control their spread and initiate subsequent antibacterial steps. (2) Its positive charges can interrupt the bacterial membrane potential, which may interfere with normal bacterial functions by disrupting the functions of some crucial enzymes.58 (3) Simultaneously, the amphiphilic cationic ligands can disrupt the cell membrane, thus enhancing the permeation of antibiotics and improving their bactericidal effect, which, we believe, is the most important antibacterial mechanism of our GNC.59 (4) It can effectively generate ROS to damage the bacterial membrane and other vital genetic and cellular molecules. Combining all four bactericidal actions, the combination of GNC and antibiotics can effectively eliminate Gram-positive MDR bacteria like MRSE.
Stability and cytotoxicity evaluation
The cytotoxicity of combined therapy on somatic (NIH 3T3 and HUVEC) cell lines was investigated by Calcein-AM/propidium iodide staining, where live/dead cells were stained in green/red, respectively. Confocal fluorescence images (ESI Fig. S16A and B†) reveal that most cells were alive with hardly noticeable numbers of dead cells, suggesting that both cell lines maintained high viability after 24 h incubation with 70 μg mL−1 GNC + 30 μg mL−1 Imp. This was further verified by live-cell counting (ESI Fig. S16C†), where the percentage of live cells was above 80%, comparable to that of the controls.
The stability of nanomaterials during storage is crucial for their potential practical applications. We measured the UV-vis spectrum of freshly prepared GNC solution and that after 3 month storage at 4 °C in a normal refrigerator (ESI Fig. S17†). We did not observe any deformation nor the emergence of new peaks in the UV-vis spectrum. In addition, there were no changes in the physical appearance, aggregation, or precipitation, confirming the good stability of the GNC.
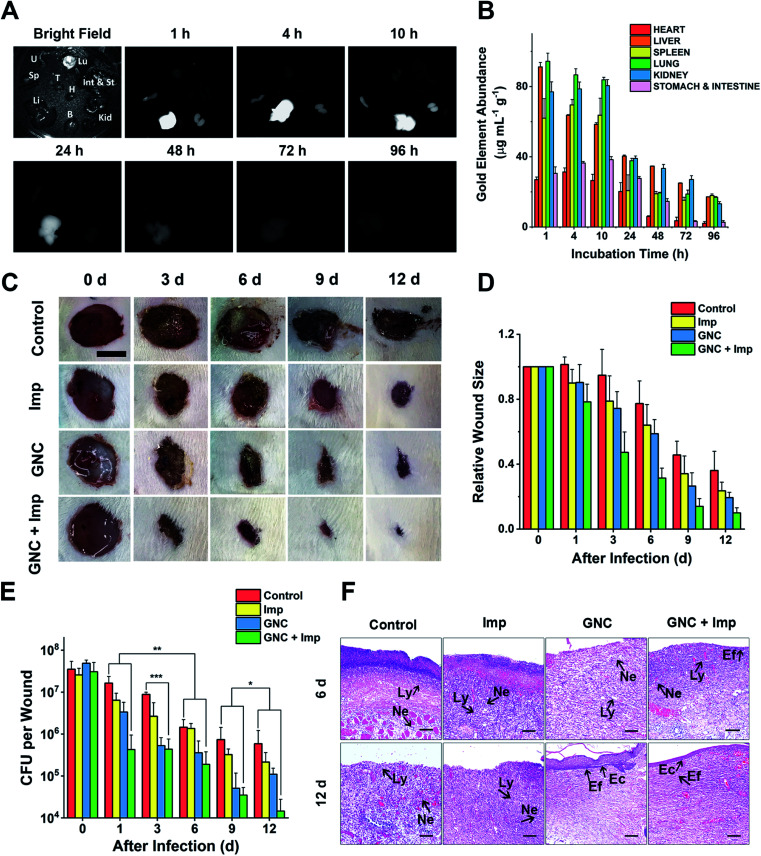
NIR-II fluorescence is an attractive imaging modality that is well suited for in vivo applications. Owing to its attractive NIR fluorescence, GNC can act as a fluorescent probe for organ distribution tracking.60 We evaluated the GNC stability in vitro using simulated biological fluids. The GNC fluorescence (128 μg mL−1) was highly stable and showed no observable changes after 15 days incubation in PBS supplemented with up to 10% human serum albumin (HSA, see ESI Fig. S18†). Thus, its stable NIR fluorescence can be used to directly evaluate the GNC organ distribution with a NIR animal imager using 808 nm laser irradiation (Fig. 6A). Upon intravenous injection of GNC + Imp, strong NIR fluorescence was observed primarily in the liver with weak signals in the spleen and kidneys within the first hour, suggesting preferential accumulation in these organs. The NIR fluorescence in the main organs was maintained for 10 h, then it gradually faded away after 24 h and became almost invisible at 96 h post-injection, indicating that most of the GNCs were cleared from the body. The gold content measured from critical organs harvested at different post-injection times was consistent with the NIR fluorescence results (>80% of the Au content was cleared after 96 h, Fig. 6B). These results indicate that the GNC has an adequate body clearance time and is suitable for intravenous administration. Moreover, the blood routine as well as liver and kidney function indicators further confirmed that a high dosage of GNC + Imp did not induce any notable change when compared with the control group, suggesting minimal in vivo toxicity (ESI Fig. S19A–F†).
Fig. 6. In vivo experiments demonstrating the clearance time and treatment effect. (A) NIR fluorescence of crucial organs of mice harvested at different time points under bright-field and 808 nm laser radiation (B: brain, H: heart, Int & St: intestine and stomach, Kid: kidneys, Li: liver, Lu: lungs, Sp: spleen, T: thymus and U: uterus). (B) Gold content (average weight) in different organs, harvested at different times post-injection (n = 4). (C) Representative images of changes in wound size in different groups within 12 days post-treatment (scale bar: 10 mm). (D) Comparison of the relative wound size vs. time in different groups: PBS (control), 24 μg mL−1 Imp solution, 64 μg mL−1 GNC solution, and the combination of 24 μg mL−1 Imp (∼0.12 mg kg−1, final concentration) and 64 μg mL−1 GNC solution (∼0.32 mg kg−1, final concentration, same below) (n = 3). (E) Plate coating results of the PBS control, single application, and combined therapy groups (n = 3, *P > 0.05, ** 0.01 < P < 0.05 and ***P < 0.01). (F) The hematoxylin–eosin staining graphs in different groups. After 12 days, the epidermis was fully reconstructed in the GNC + Imp group (Ly: lymphocyte; Ne: neutrophil; Ec: epithelial cell and Ef: elongated fiber).
Skin infection model experiments
As a coagulase-negative strain, MRSE can cause soft tissue and skin infections and frequently induces infection in surgical sites;5 thus, we established a skin infection model by creating wounds with removal of epidermis and dermis on the back of rats and infecting them with MRSE, where conventional wound treatment methods find it difficult to sustain antibacterial ability. Different groups were treated with either the PBS control, Imp only, GNC only or GNC + Imp. The effect was monitored by measuring the wound size and bacterial count after plate coating. Compared with the negative control, the wound healing rate for the rats treated with either Imp or GNC was higher. Notably, the wound healing of the GNC + Imp combinational group was the fastest. The size of the wound was significantly smaller than that of other groups, and the normal skin was almost fully reconstructed in 9 days (Fig. 6C and D). Moreover, the scab size was significantly smaller than in other groups, indicating a markedly reduced bacterial infection.
To quantify the clearance of bacteria on the skin wound, we performed plate coating to compare the number of colonies on differently treated wounds (Fig. 6E). The MRSE counts of the negative control and Imp groups were slowly reduced over time due to autoimmunity, and the GNC-treated group showed a more significant decrease. Treatment with GNC + Imp resulted in the most rapid reduction of the bacterial population, and by day 12, the bacterial count was >10-fold lower than that of the GNC-only group and almost two orders of magnitude lower than that of the negative control group. This in vivo result was consistent with the superior in vitro antibacterial potency of GNC + Imp over that of GNC or Imp only. Pathological analyses further confirmed a complete repair of the wound tissue for the GNC + Imp group but not for the negative control and GNC- or Imp-only groups. The reconstruction of well-stratified skin layers and significantly reduced immune cells indicated the successful removal of the lesions in the GNC + Imp group (Fig. 6F).
Conclusion
In this study, we synthesised a series of formula-defined pyridinium–zwitterionic ligand-functionalised Au25(SR1)x(SR2)18−x GNCs as antibacterial nanomaterials. By fine-tuning the ligand feed ratios, we obtained GNCs that exhibited both excellent antibacterial ability and high stability, successfully addressing a major issue in antibacterial gold nanomaterial development. Besides, the GNCs employ multiple antibacterial mechanisms, giving rise to high potency against Gram-positive MDR bacteria. The optimized GNC can significantly reduce the dosage of antibiotics required to treat MDR bacterial infections, thereby greatly enhancing the efficacy of frontline antibiotics. Compared to other nanomaterials without defined chemical formulae, which could cause difficulties in quality control and mechanism research, our Au25(SR1)x(SR2)18−x GNCs are potentially better suited for medical applications. Moreover, biocompatible Au25 GNCs capped with two or more ligands can incorporate more functions, thus widening the scope of their biomedical applications. We envisage that dual-/multi-ligand-functionalised GNCs will find broad applications in chemistry, physics, biology, and biomedical sciences.
Data availability
All other data have already been provided in the ESI.†
Author contributions
D. Z. and Z. P. conceived this study. Z. P. performed all the experiments and analysed the data except those clarified below. Q. L. and J. Q. took and analysed the TEM images. Y. J. and Y. G. contributed some of the bacterial experiments. W. Y. carried out the animal experiments. F. H. contributed MDR bacteria strains. Z. P. wrote the manuscript. D. Z. and X. J. co-supervised the project and obtained funding for this project.
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
We acknowledge the financial support from the Shenzhen Science and Technology Programme (KQTD20190929172743294). This work was partly funded by the UK Biotechnology and Biological Sciences Research Council (grant no: BB/R007829/1) to DZ.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc03056f
Notes and references
- Fernandes P. Nat. Biotechnol. 2006;24:1497–1503. doi: 10.1038/nbt1206-1497. [DOI] [PubMed] [Google Scholar]
- Cassandra W. Nature. 2017;543:15. doi: 10.1038/nature.2017.21550. [DOI] [PubMed] [Google Scholar]
- Wang L. Hu C. Shao L. Int. J. Nanomed. 2017;12:1227–1249. doi: 10.2147/IJN.S121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Nat. Rev. Microbiol. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner-Lastinger L. M. Abner S. Edwards J. R. Kallen A. J. Karlsson M. Magill S. S. Pollock D. See I. Soe M. M. Walters M. S. Dudeck M. A. Infect. Control Hosp. Epidemiol. 2020;41:1–18. doi: 10.1017/ice.2019.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen A. M. Kjeldsen G. Ericson Sollid J. U. Antimicrob. Agents Chemother. 2004;48:285–296. doi: 10.1128/AAC.48.1.285-296.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services Centers Dis. Control Prev. 2019:1–113. [Google Scholar]
- Jiang Z. Sahar A. Li X. Robinson S. M. Rotello V. M. Saha K. Riley M. A. Moyano D. F. Gupta A. ACS Nano. 2014;8:10682–10686. doi: 10.1021/nn5042625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. Mumtaz S. Li C. H. Hussain I. Rotello V. M. Chem. Soc. Rev. 2019;48:415–427. doi: 10.1039/C7CS00748E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong H. T. T. Adnan N. N. M. Barraud N. Basuki J. S. Kutty S. K. Jung K. Kumar N. Davis T. P. Boyer C. J. Mater. Chem. B. 2014;2:5003–5011. doi: 10.1039/C4TB00632A. [DOI] [PubMed] [Google Scholar]
- Ma K. Li Y. Wang Z. Chen Y. Zhang X. Chen C. Yu H. Huang J. Yang Z. Wang X. Wang Z. ACS Appl. Mater. Interfaces. 2019;11:29630–29640. doi: 10.1021/acsami.9b10373. [DOI] [PubMed] [Google Scholar]
- Zheng K. Setyawati M. I. Leong D. T. Xie J. ACS Nano. 2017;11:6904–6910. doi: 10.1021/acsnano.7b02035. [DOI] [PubMed] [Google Scholar]
- Nasaruddin R. R. Chen T. Yan N. Xie J. Coord. Chem. Rev. 2018;368:60–79. doi: 10.1016/j.ccr.2018.04.016. [DOI] [Google Scholar]
- Xie Y. Liu Y. Yang J. Liu Y. Hu F. Zhu K. Jiang X. Angew. Chem., Int. Ed. 2018;57:3958–3962. doi: 10.1002/anie.201712878. [DOI] [PubMed] [Google Scholar]
- Xie Y. Zheng W. Jiang X. ACS Appl. Mater. Interfaces. 2020;12:9041–9049. doi: 10.1021/acsami.9b21777. [DOI] [PubMed] [Google Scholar]
- Yu M. Xu J. Zheng J. Angew. Chem., Int. Ed. 2019;131:4156–4172. doi: 10.1002/ange.201807847. [DOI] [Google Scholar]
- Jin R. Nanoscale. 2015;7:1549–1565. doi: 10.1039/C4NR05794E. [DOI] [PubMed] [Google Scholar]
- Kang X. Chong H. Zhu M. Nanoscale. 2018;10:10758–10834. doi: 10.1039/C8NR02973C. [DOI] [PubMed] [Google Scholar]
- Yu Z. Xiao H. Zhang X. Yang Y. Yu Y. Chen H. Meng X. Ma W. Yu M. Li Z. Li C. Liu H. ACS Nano. 2020;14:13536–13547. doi: 10.1021/acsnano.0c05541. [DOI] [PubMed] [Google Scholar]
- Li D. Liu Q. Qi Q. Shi H. Hsu E. C. Chen W. Yuan W. Wu Y. Lin S. Zeng Y. Xiao Z. Xu L. Zhang Y. Stoyanova T. Jia W. Cheng Z. Small. 2020;16:1–9. doi: 10.1002/smll.202003851. [DOI] [PubMed] [Google Scholar]
- Petkovic M. Seddon K. R. Rebelo L. P. N. Pereira C. S. Chem. Soc. Rev. 2011;40:1383–1403. doi: 10.1039/C004968A. [DOI] [PubMed] [Google Scholar]
- Sreenivasan P. K. Haraszthy V. I. Zambon J. J. Lett. Appl. Microbiol. 2013;56:14–20. doi: 10.1111/lam.12008. [DOI] [PubMed] [Google Scholar]
- Chen S. Li L. Zhao C. Zheng J. Polymer. 2010;51:5283–5293. doi: 10.1016/j.polymer.2010.08.022. [DOI] [Google Scholar]
- Shaoyi J. Zhiqiang C. Adv. Mater. 2009;22:920–932. [Google Scholar]
- Murthy A. K. Stover R. J. Hardin W. G. Schramm R. Nie G. D. Gourisankar S. Truskett T. M. Sokolov K. V. Johnston K. P. J. Am. Chem. Soc. 2013;135:7799–7802. doi: 10.1021/ja400701c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y. Sakonsinsiri C. Nehlmeier I. Fascione M. A. Zhang H. Wang W. Pöhlmann S. Turnbull W. B. Zhou D. Angew. Chem., Int. Ed. 2016;128:4816–4820. doi: 10.1002/ange.201600593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z. Nachammai V. Zhang B. Yan N. Leong D. T. Jiang D. E. Xie J. J. Am. Chem. Soc. 2014;136:10577–10580. doi: 10.1021/ja505429f. [DOI] [PubMed] [Google Scholar]
- Jin R. Qian H. Wu Z. Zhu Y. Zhu M. Mohanty A. Garg N. J. Phys. Chem. Lett. 2010;1:2903–2910. doi: 10.1021/jz100944k. [DOI] [Google Scholar]
- Liu H. Hong G. Luo Z. Chen J. Chang J. Gong M. He H. Yang J. Yuan X. Li L. Mu X. Wang J. Mi W. Luo J. Xie J. Zhang X. D. Adv. Mater. 2019;31:1–9. doi: 10.1002/adma.201901015. [DOI] [PubMed] [Google Scholar]
- Hong G. Antaris A. L. Dai H. Nat. Biomed. Eng. 2017;1:1–22. doi: 10.1038/s41551-016-0001. [DOI] [Google Scholar]
- Antaris A. L. Chen H. Diao S. Ma Z. Zhang Z. Zhu S. Wang J. Lozano A. X. Fan Q. Chew L. Zhu M. Cheng K. Hong X. Dai H. Cheng Z. Nat. Commun. 2017;8:1–11. doi: 10.1038/s41467-016-0009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. Ding M. Zeng X. Xiao Y. Wu H. Zhou H. Ding B. Qu C. Hou W. Er-bu A. G. A. Zhang Y. Cheng Z. Hong X. Chem. Sci. 2017;8:3489–3493. doi: 10.1039/C7SC00251C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldeek F. Muhammed M. A. H. Palui G. Zhan N. Mattoussi H. ACS Nano. 2013;7:2509–2521. doi: 10.1021/nn305856t. [DOI] [PubMed] [Google Scholar]
- Song M. Zhou G. Lu N. Lee J. Nakouzi E. Wang H. Li D. Science. 2020;367:40–45. doi: 10.1126/science.aax6511. [DOI] [PubMed] [Google Scholar]
- Ishida Y. Narita K. Yonezawa T. Whetten R. L. J. Phys. Chem. Lett. 2016;7:3718–3722. doi: 10.1021/acs.jpclett.6b01725. [DOI] [PubMed] [Google Scholar]
- Yuan X. Zhang B. Luo Z. Yao Q. Leong D. T. Yan N. Xie J. Angew. Chem., Int. Ed. 2014;53:4623–4627. doi: 10.1002/anie.201311177. [DOI] [PubMed] [Google Scholar]
- Chen T. Fung V. Yao Q. Luo Z. Jiang D. E. Xie J. J. Am. Chem. Soc. 2018;140:11370–11377. doi: 10.1021/jacs.8b05689. [DOI] [PubMed] [Google Scholar]
- Malanovic N. Lohner K. Biochim. Biophys. Acta, Biomembr. 2016;1858:936–946. doi: 10.1016/j.bbamem.2015.11.004. [DOI] [PubMed] [Google Scholar]
- Pasquina-Lemonche L. Burns J. Turner R. D. Kumar S. Tank R. Mullin N. Wilson J. S. Chakrabarti B. Bullough P. A. Foster S. J. Hobbs J. K. Nature. 2020;582:294–297. doi: 10.1038/s41586-020-2236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z. V. Gunsolus I. L. Qiu T. A. Hurley K. R. Nyberg L. H. Frew H. Johnson K. P. Vartanian A. M. Jacob L. M. Lohse S. E. Torelli M. D. Hamers R. J. Murphy C. J. Haynes C. L. Chem. Sci. 2015;6:5186–5196. doi: 10.1039/C5SC00792E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P. Kizhakkedathu J. N. Straus S. K. Biomolecules. 2018;8:4. doi: 10.3390/biom8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand R. M. R. F. Walker C. Epand R. M. R. F. Magarvey N. A. Biochim. Biophys. Acta, Biomembr. 2016;1858:980–987. doi: 10.1016/j.bbamem.2015.10.018. [DOI] [PubMed] [Google Scholar]
- Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- Zhao Y. Chen Z. Chen Y. Xu J. Li J. Jiang X. J. Am. Chem. Soc. 2013;135:12940–12943. doi: 10.1021/ja4058635. [DOI] [PubMed] [Google Scholar]
- Foxley M. A. Wright S. N. Lam A. K. Friedline A. W. Strange S. J. Xiao M. T. Moen E. L. Rice C. V. ACS Med. Chem. Lett. 2017;8:1083–1088. doi: 10.1021/acsmedchemlett.7b00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. Singh A. Santa Maria J. Younghoon K. Brown S. Swoboda J. G. Mylonakis E. Wilkinson B. J. Walker S. ACS Chem. Biol. 2010;6:106–116. doi: 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N. Ma S. Eur. J. Med. Chem. 2019;184:111743. doi: 10.1016/j.ejmech.2019.111743. [DOI] [PubMed] [Google Scholar]
- Hayden S. C. Zhao G. Saha K. Phillips R. L. Li X. Miranda O. R. Rotello V. M. El-Sayed M. A. Schmidt-Krey I. Bunz U. H. F. J. Am. Chem. Soc. 2012;134:6920–6923. doi: 10.1021/ja301167y. [DOI] [PubMed] [Google Scholar]
- Wang B. Zhang L. Sung C. B. Granick S. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18171–18175. doi: 10.1073/pnas.0807296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubertret B. Calame M. Libchaber A. J. Nat. Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- Caudill E. R. Hernandez R. T. Johnson K. P. O'Rourke J. T. Zhu L. Haynes C. L. Feng Z. V. Pedersen J. A. Chem. Sci. 2020;11:4106–4118. doi: 10.1039/C9SC05436G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. Liu Y. Wang L. Ali A. Yao Q. Jiang X. Gao Y. Biomaterials. 2020;253:120124. doi: 10.1016/j.biomaterials.2020.120124. [DOI] [PubMed] [Google Scholar]
- Pendleton J. N. Gilmore B. F. Int. J. Antimicrob. Agents. 2015;46:131–139. doi: 10.1016/j.ijantimicag.2015.02.016. [DOI] [PubMed] [Google Scholar]
- Chompoosor A. Saha K. Ghosh P. S. Macarthy D. J. Miranda O. R. Zhu Z.-J. Arcaro K. F. Rotello V. M. Small. 2010;6:2246–2249. doi: 10.1002/smll.201000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M. Zhou C. Wu S. Yu B. Zhang Z. Song N. Lee M. M. S. Xu W. Xu F. J. Wang D. Wang L. Tang B. Z. J. Am. Chem. Soc. 2019;141:16781–16789. doi: 10.1021/jacs.9b07162. [DOI] [PubMed] [Google Scholar]
- Li J. Zhou H. Wang J. Wang D. Shen R. Zhang X. Jin P. Liu X. Nanoscale. 2016;8:11907–11923. doi: 10.1039/C6NR02844F. [DOI] [PubMed] [Google Scholar]
- Barnes R. J. Molina R. Xu J. Dobson P. J. Thompson I. P. J. Nanopart. Res. 2013;15:1432. doi: 10.1007/s11051-013-1432-9. [DOI] [Google Scholar]
- Cui Y. Zhao Y. Tian Y. Zhang W. Lü X. Jiang X. Biomaterials. 2012;33:2327–2333. doi: 10.1016/j.biomaterials.2011.11.057. [DOI] [PubMed] [Google Scholar]
- Godoy-Gallardo M. Eckhard U. Delgado L. M. de Roo Puente Y. J. D. Hoyos-Nogués M. Gil F. J. Perez R. A. Bioact. Mater. 2021;6:4470–4490. doi: 10.1016/j.bioactmat.2021.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y. Chen J. Fang H. Heath G. Wo Y. Wang W. Li Y. Guo Y. Evans S. D. Chen S. Zhou D. Chem. Mater. 2016;28:3041–3050. doi: 10.1021/acs.chemmater.6b00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other data have already been provided in the ESI.†