Abstract
A DNA double-strand break (DSB) created by the HO endonuclease in Saccharomyces cerevisiae will stimulate recombination between flanking repeats by the single-strand annealing (SSA) pathway, producing a deletion. Previously the efficiency of SSA, using homologous sequences of different lengths, was measured in competition with that of a larger repeat further from the DSB, which ensured that nearly all cells would survive the DSB if the smaller region was not used (N. Sugawara and J. E. Haber, Mol. Cell. Biol. 12:563–575, 1992). Without competition, the efficiency with which homologous segments of 63 to 205 bp engaged in SSA was significantly increased. A sequence as small as 29 bp was used 0.2% of the time, and homology dependence was approximately linear up to 415 bp, at which size almost all cells survived. A mutant with a deletion of RAD59, a homologue of RAD52, was defective for SSA, especially when the homologous-sequence length was short; however, even with 1.17-kb substrates, SSA was reduced fourfold. DSB-induced gene conversion also showed a partial dependence on Rad59p, again being greatest when the homologous-sequence length was short. We found that Rad59p plays a role in removing nonhomologous sequences from the ends of single-stranded DNA when it invades a homologous DNA template, in a manner similar to that previously seen with srs2 mutants. Δrad59 affected DSB-induced gene conversion differently from msh3 and msh2, which are also defective in removing nonhomologous ends in both DSB-induced gene conversion and SSA. A msh3 rad59 double mutant was more severely defective in SSA than either single mutant.
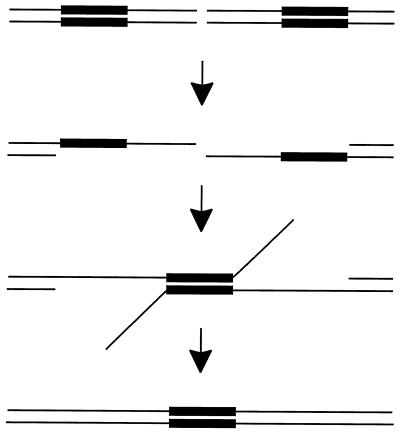
DNA double-strand breaks (DSBs) generated in the chromosomes of a G1 haploid cell are lethal unless repaired. DSBs created within a sequence that is repeated elsewhere in the genome can be repaired by gene conversion (for a review, see reference 37). In contrast, a DSB occurring within a unique sequence cannot be repaired in this manner but can instead be repaired by nonhomologous joining of DNA ends (10, 19, 56). A third pathway exists if the DSB lies in a unique sequence between two repeated sequences, where it can stimulate the formation of a deletion between the repeats by the single-strand annealing (SSA) pathway (14, 22, 23, 27). DNA is resected by a 5′-to-3′ exonuclease on each side of the DSB, leaving 3′-ended single-stranded tails, as diagrammed in Fig. 1. Annealing can take place between the complementary sequences located on opposite sides of the DSB; this is followed by nucleolytic removal of any remaining tails, DNA synthesis to fill in the gaps, and ligation.
FIG. 1.
Single-strand annealing model. When a double-strand break is created in vivo, one strand on each side of the DSB is resected in the 5′-to-3′ direction, leaving a 3′ tail. When complementary sequences on opposite sides are exposed, they can anneal, forming a branched intermediate. The single-stranded tails are removed by a nuclease, the gaps are filled in, and any remaining nicks are ligated, finally resulting in a deletion product.
Certain fundamental properties of SSA have been documented in Saccharomyces cerevisiae by creating a DSB in vivo, using the HO endonuclease under the control of the GAL10 promoter. Analysis of SSA both on plasmids and on the chromosome has confirmed that 5′-to-3′ resectioning occurs, creating 3′ tails (14, 51). When a DSB is situated within one of the two repeats, it can be repaired either by SSA or by gene conversion. Increasing the distance between the two repeats decreases the efficiency of SSA in competition with gene conversion, consistent with a need for more time for 5′-to-3′ resection to expose complementary homologies (14, 16).
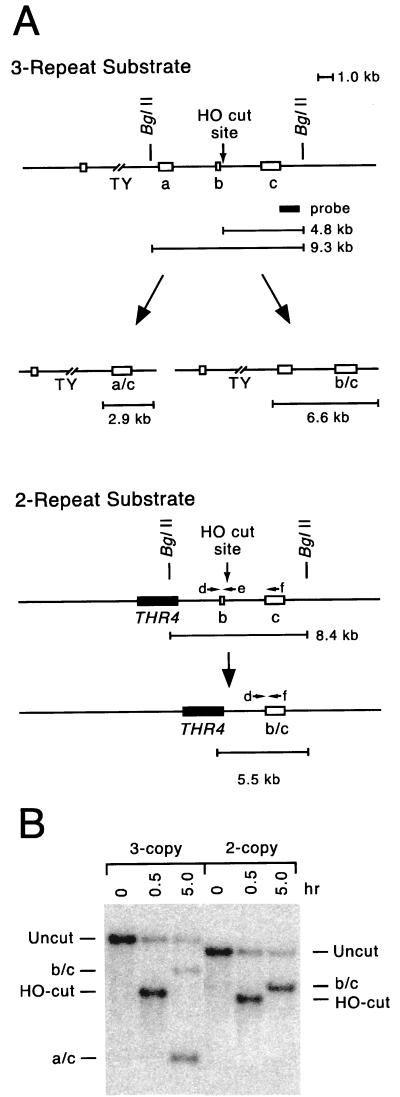
The efficiency of SSA is strongly influenced by the length of the flanking repeats and also by their degree of sequence identity (51, 53). The dependence of SSA on the homologous-sequence length was characterized by using repeats of differing lengths (51). A minimum size of between 63 and 89 bp was determined; however, in these measurements, we utilized substrates containing three repeats of the URA3 sequence, with the more distant pair having 1.17 kb of homologous sequence and the middle repeat differing in length (Fig. 2) (51). The 1.17-kb repeats ensured that cells would survive the induction of a DSB, since recombination between such large flanking repeats could nearly always occur. It was possible, though, that the presence of the 1.17-kb repeat reduced the opportunity for cells to use the smaller sequence as often as would occur in the absence of competition. In this paper, we show that the ability of small sequences to engage in SSA markedly increases when competition is absent. We have therefore reinvestigated the dependence of SSA on the length of homologous sequence and the minimum size requirements when only two repeats are present.
FIG. 2.
Competition between repeated sequences. (A) Strains that have two or three repeated sequences derived from the HindIII restriction fragment containing URA3 were constructed (open boxes). The three-repeat substrate possesses the ura3-52 sequence (a), a 205-bp sequence (b), and the URA3 HindIII restriction fragment (c). Intervening sequences are derived from pUC9, lambda phage, and the 117-bp HO cut site from MATa (51). A DSB at the HO cut site initiates SSA between the repeats and results in two deletion products, a/c and b/c. In the two-repeat substrate, only the b/c product is generated. Primers labeled d, e, and f were used to screen colonies based on structure by PCR analysis (see text). Structures are oriented from centromere distal to centromere proximal (left to right). (B) DNA from tNS1379 (two-repeat strain) and tNS62 (three-repeat strain) was prepared for Southern analysis before and 5 h after HO induction. Locations of the probe and BglII sites used for the Southern blots are shown in panel A. For the two-repeat strain, the Southern blot shows the uncleaved DNA band before induction, the HO-cleaved band (4.8 kb) at 0.5 h, and the product band (5.5 kb) 5 h after induction. The three-repeat strain yielded the uncleaved DNA band (9.3 kb) and two product bands (2.9 and 6.6 kb).
We have also examined genes that influence the dependence of SSA on the length of homologous sequences. Previous studies have shown that the absence of Rad52p, a strand-annealing protein (31, 49), nearly eliminates SSA when the homologous sequences are 1 to 2 kb in length (14, 45, 51). However, when the DSB is induced within much larger arrays, such as the CUP1 or rRNA gene arrays, the events become RAD52 independent (36), since large regions of homology appear to compensate for the lack of Rad52p. We have surveyed other members of the RAD52 epistasis group and found that none of the others is nearly as important as RAD52. Deletions of RAD50 and XRS2 delay but only partially reduce SSA (17, 51); moreover, deletions of RAD51, RAD54, RAD55, and RAD57 did not inhibit SSA between flanking repeated sequences of 1 to 2 kb in length (17). In this study we examined the recA homolog RAD51, using short (205-bp) repeats to show that SSA can truly be carried out independently of Rad51p.
Another pair of proteins whose role exhibits homologous-sequence length dependence during SSA comprises Msh2p and Msh3p (53). These proteins, along with the Rad1p-Rad10p excision endonuclease protein complex (13, 53), are involved in removing nonhomologous 3′-ended DNA tails from annealed intermediates. However, whereas Rad1p and Rad10p are required to clip off the tails regardless of the length of the annealed homologous regions, the requirement for Msh2p and Msh3p decreases dramatically as the annealed regions increase from 205 bp (at which size Msh2p and Msh3p are nearly as essential as Rad52p) to 1.17 kb (at which size SSA is 80 to 90% as efficient as in a wild-type strain). Msh2p and Msh3p are imagined to stabilize the short, annealed heteroduplexes and allow the Rad1p-Rad10p complex to cleave the single-stranded tail at the branched junctions on the ends of the annealed regions (13, 53). As the annealed regions become longer, and therefore more stable, the requirement for Msh2p and Msh3p decreases.
Interestingly, Msh2p and Msh3p are also required for removal of nonhomologous sequences from the ends of invading strands during DSB-induced gene conversion even when the length of homologous sequence shared by the donor and recipient is 2 kb (9, 38, 53). They presumably act in a similar manner by binding to the junctions between single-stranded DNA and double-stranded DNA, promoting the Rad1p-Rad10p-mediated cleavage of the single-stranded nonhomologous tail so that the 3′ end of the invading DNA strand can be used as a primer to initiate new DNA synthesis. The need for Msh2p and Msh3p in gene conversion, even with long homologous regions adjacent to the nonhomologous ends, presumably reflects the difference between the instability of a strand invasion structure (involving an invading single strand and a duplex template) and the more stable intertwinings of two complementary single strands during SSA.
Continuing our survey of RAD genes, we have investigated the role of RAD59 both in SSA and in DSB-induced gene conversion. RAD59 was identified as a mutation that eliminated RAD52-dependent spontaneous recombination in a rad51 background (4). rad59 mutants are defective in several spontaneous and DSB-induced recombination assays, and epistasis analysis has revealed that mutations in RAD59 and RAD51 can act synergistically in recombination assays (3–5). RAD59 is a homolog of RAD52, and overexpression of RAD52 will relieve γ-ray sensitivity in rad59 strains, which suggests a possible overlap of functions (4). This was also suggested by a synergistic defect observed in a rad59 rad52R70K double mutant (3). In this study, we showed that SSA is impaired in a rad59 background nearly as much as in a rad52 background and we further characterize RAD59's role in DSB-induced gene conversion.
MATERIALS AND METHODS
Strains and plasmids.
Yeast strains used in this study are shown in Table 1. Yeast strains used for studying SSA were derived from tNR85 (45) containing the GAL10::HO TRP1 plasmid pFH800 (35). Strains with two ura3 sequences were derived from strains with three copies (Fig. 2A) (51) by deletion of the ura3-52 allele via transplacement of pNSU196 cut with PvuI. pNSU196 contains the THR4 gene as a selectable marker (HindIII fragment isolated from p323 provided by G. Simchen) cloned into the HindIII site of pUC19. The downstream HindIII site was subsequently filled in with the Klenow fragment of DNA polymerase. The sequence upstream of URA3 (1.9-kb SalI-to-HindIII sequence) was cloned between the SalI site and the remaining HindIII site. This upstream sequence was derived from a partial HindIII digestion of pNSU133, which consists of the BglII fragment of URA3-FL100 from pSK180 (M. Rose) cloned into the BglII site of plink 15-5 (26) modified by filling in the HindIII site. PvuI cuts pNSU196 in the pUC19 vector sequence and in the sequence upstream of ura3, which targets the fragment to the homologous sequence in the strains with three repeats of the ura3 sequence.
TABLE 1.
Strains used in this study
| Strain | Genotypea |
|---|---|
| JKM146 | Δho Δhml::ADE1 MATa-inc Δhmr::ADE1 ade1 leu2-3,112 lys5 trp1::hisG ura3-52 ade3::GAL::HO |
| tNR85 | Δho HML matΔ::LEU2 hmrΔ3 leu2-3,112 trp1 thr4 ura3-52 pFH800 |
| tNS709 | tNR85 (THR4 ura3 [29 bp]-cut site-URA3) |
| tNS747 | tNR85 (THR4 ura3 [142 bp]-cut site-URA3) |
| tNS759 | tNR85 (THR4 ura3 [415 bp]-cut site-URA3) |
| tNS1103 | tNR85 (THR4 ura3 [205 bp]-cut site-URA3) |
| tNS1110 | tNR85 (THR4 ura3 [235 bp]-cut site-URA3) |
| tNS1142 | tNR85 (THR4 ura3 [63 bp]-cut site-URA3) |
| tNS1157 | tNR85 (THR4 ura3 [89 bp]-cut site-URA3) |
| tNS1379 | Δho HML matΔ::leu2::hisG hmrΔ3 leu2-3,112 ura3-52 trpl thr4 (ura3 [205 bp]-cut site-URA3) pFH800 |
| tNS1530 | tNR85 (THR4 ura3 [1,170 bp]-cut site-URA3) |
| tNS1551 | tNS759 msh3::KanMX |
| tNS1573 | tNS1379 rad59::KanMX |
| tNS1585 | tNS1379 srs2::KanMX2 |
| tNS1597 | tNS1379 rad51::LEU2 |
| tNS1703 | JKM146 rad59::KanMX2 |
| tNS1706 | JKM146 msh3::LEU2 rad59::KanMX2 |
| tNS1712 | tNS1530 rad59::KanMX2 |
| tNS1785 | tNS759 rad59::KanMX2 |
| tNS1853 | JKM146 srs2::LEU2 rad59::KanMX2 |
| tNS1967 | tNS759 rad59::NatMX4 msh3::KanMX |
| tNS1984 | tNS709 rad59::KanMX2 |
| tNS1990 | tNS709 rad52::KanMX2 |
| YFP218 (tNS1924) | JKM146 srs2::LEU2 |
| YFP259 | JKM146 msh3::LEU2 |
Cut site refers to the 117-bp BglII-HincII MATa HO endonuclease recognition site.
rad59, msh3, and srs2 deletions were constructed by creating rad59::KanMX2, msh3::KanMX2, rad59::NatMX4, or srs2::KanMX2 disruption PCR products in accordance with the procedures of Wach et al. (57) and Goldstein and McCusker (15). pAM50 (6) was used to construct rad51::LEU2 disruptions, pmsh3::LEU2 (46) was used to construct msh3::LEU2 disruptions, pmsh3::TRP1 (34) was used to construct msh3::TRP1 disruptions, and pΔ2R (F. Fabre) was used to create srs2::LEU2 disruptions. These deletions remove all or most of the open reading frames.
To study the homology dependence of DSB-induced gene conversion, pNSU216 (52), pGI9, pGI1, pGI7, and pGI19 were individually transformed into JKM146 or tNS1703. pNSU216 consists of the 3,845-bp EcoRI-to-HindIII sequence of MATα inserted between the EcoRI and HindIII sites of YCp50 and the MATα-inc EcoRI-to-HindIII sequence blunt-end ligated into the SmaI site in an inverted orientation. In pGI9, pGI1, pGI7, and pGI19, the MATα sequence was replaced with shorter MATα sequences derived from PCR-amplified DNA.
Strains used for assaying the removal of nonhomologous DNA ends during DSB-induced gene conversion were constructed by individually transforming plasmids pFP122, pFP121, pFP118, pFP140, and pFP120 (38), which respectively contain 0, 10, 20, and 30 bp on each side and a combination of 308 and 610 bp of nonhomologous sequences adjacent to the DSB, into derivatives of JKM146 containing the mutations msh3::LEU2 (YFP259), rad59::KanMX2 (tNS1703), srs2::LEU2 (YFP218), msh3::LEU2 rad59::KanMX2 (tNS1706), and srs2::LEU2 rad59::KanMX2 (tNS1853).
Media.
Selective and rich media were described by Sherman et al. (48). YP-lactate, YP-glycerol, and YP-galactose were described previously (38, 45). YEPEG consists of 1% (wt/vol) yeast extract, 2% (wt/vol) peptone, 2.6% (wt/vol) glycerol, 2.6% (wt/vol) ethanol, and 1% (wt/vol) succinic acid.
Galactose inductions.
Galactose inductions were carried out as described previously (45). Cultures were grown in liquid YP-lactate medium at 30°C and induced with 2% galactose. Aliquots were taken at appropriate time points, and DNA was extracted by a glass bead-phenol-sodium dodecyl sulfate protocol (45). Southern blotting was carried out in accordance with the procedure of Church and Gilbert (8), and blots were analyzed by using a Molecular Dynamics PhosphorImager. The level of SSA was calculated by dividing the intensity of the product band by the intensity of the 0-h band and normalizing both signals to LEU2 bands that appeared when the blot was reprobed with the HpaI-SalI LEU2 sequence. This value was also divided by the fraction of cells containing pFH800 (GAL::HO) prior to induction to normalize for the fraction of cells able to induce the HO gene.
To determine the frequency with which a 29-bp sequence recombined by SSA, which was too low to measure accurately by densitometry, tNS709, tNS1984, and tNS1990 were individually induced in liquid culture with 2% galactose at 30°C and cells were plated on yeast peptone dextrose (YPD) and synthetic dextrose agar (SD)-Trp media before and after a 3-h induction. Colonies grown on YPD were replica plated to SD-Trp plates to assess retention of pFH800. Colonies grown on SD-Trp selective medium were counted to give an overall frequency of survivors retaining pFH800. To distinguish between colonies with recombined product and those that either had not been cleaved by HO endonuclease or had precisely or imprecisely ligated the cleaved fragments back together, we first induced the colonies again with galactose to determine whether they possessed an intact HO cut site. Colonies were patched onto YEPEG plates, replica plated to YP-galactose plates, and incubated for 6 to 18 h at 30°C. Cells with an intact HO cut site were unable to grow when replica plated to SD-Trp, which scores for retention of the GAL::HO plasmid. The colonies that were insensitive to galactose were further analyzed by PCR using the distal primer d (GCACCATATGCGGTGTG) and the proximal primers e (TGGCCAAATCGATTAGCCGA) and f (TGAGTAGCAGCACGTTCC) (Fig. 2), resulting in a 196-bp product (primers d and e) if an SSA deletion event occurred or a 334-bp product (primers d and f) if the original structure was retained. Colonies that failed to yield a PCR product were analyzed by Southern blotting as described in the legend to Fig. 2.
The dependence of DSB-induced gene conversion on sequence homology was assayed by growing cultures in liquid YP-lactate overnight at 30°C and then plating the cells on YPD and YP-galactose plates. Colonies were scored for retention of plasmids (Ura+), and the frequency of retention was calculated as the fraction of colonies grown on galactose medium that retained the plasmid, on which HO was induced, divided by the fraction of plasmid retention by colonies grown on YPD. The efficiency of removal of nonhomologous sequence from the single-stranded tails during DSB-induced gene conversion was assayed by the same procedure except that YP-glycerol was used in place of YP-lactate as described by Pâques and Haber (38). Normally two genotypes (10 strains) were tested in parallel.
RESULTS
Competition between homologous sequences during SSA.
To characterize the effect of competition in SSA, we constructed pairs of strains with the SSA substrates shown in Fig. 2A. Each construct possessed an HO cut site situated between two homologous ura3 sequences, but the three-repeat construct contained an additional homologous sequence on the centromere-distal side. We compared these two strains to determine how the presence of the 1.17-kb distal sequence influences the frequency with which the more proximal segments participate in SSA. In the example shown in Fig. 2, the proximal sequence was 205 bp in length.
Cultures of each strain were grown in YP-lactate medium and induced for 5 h with galactose to express the galactose-regulated HO endonuclease. Aliquots were obtained prior to induction and at various time points after addition of galactose. DNA samples were extracted and prepared for Southern analysis (Fig. 2B). The Southern blot in Fig. 2B shows the uncut DNA before induction, at 0 h, and the product present in the 5-h lane. The strain with three homologous sequences had two products, resulting from recombination with either the 205-bp region b in Fig. 2A or the 830-bp region a of the ura3-52 locus, proximal to a Ty1 insertion. Previous studies have shown that recombination with the 335 bp distal to the Ty1 insertion in ura3-52 occurs only very rarely (45, 51). What is apparent from examination of this blot is that the quantity of product in the two-repeat strain was much larger than that of the equivalent product, b/c, in the three-repeat strain. Quantitation of the bands in these blots (see Materials and Methods) revealed that there was about fivefold more of this product in the two-repeat strain than in the three-repeat strain (79% vs. 16% product relative to the amount of the original, uncut DNA). In the three-repeat strain, the distal ura3-52 sequence appears to have been in competition with the middle sequence, causing an underestimation of the efficiency with which a 205-bp homologous segment participated in SSA.
Dependence of SSA on the length of homologous sequence in the absence of competition.
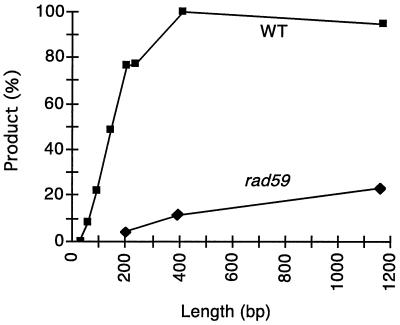
We then examined the dependence of SSA on the length of homologous sequences in the absence of competition, using strains with the 1.17-kb URA3 sequence to the right of the HO cut site and with various lengths of ura3 sequence homology to the left. Each strain was induced as described above, and the effectiveness of SSA was evaluated by densitometry after Southern analysis (Fig. 3). As the homologous-sequence length increased, the amount of product also increased until it reached a plateau at approximately 400 bp, at which point nearly all cells survived by forming a deletion.
FIG. 3.
Homologous-sequence length dependence of SSA. Strains containing the two-repeat substrate (Fig. 2A) were constructed with various amounts of homologous DNA. The sizes tested were 29, 63, 89, 205, 235, 415, and 1,170 bp. The frequency of SSA was determined, as described in detail in Materials and Methods, by quantitating the product band and dividing by the quantity of uncut uninduced band and normalizing to an independent band (after reprobing with leu2) and to the number of cells containing the GAL::HO plasmid. Strain tNS709, with 29-bp repeats, was analyzed separately to exclude NHEJ and religation events as well as uninduced cells (wild type [WT]) as described in the text.
We were also interested in determining the smallest amount of homologous sequence that could be used in SSA. Previously, using the three-repeat substrates, we found that a sequence of between 63 and 89 bp in length was required (51); however, competition may have minimized the recombination potential of the middle repeat. By testing two-repeat strains with 29-, 63-, and 89-bp repeats, we were able to establish that SSA could use homologous sequences as small as 29 bp, although at a very low frequency. Upon HO induction, only a small fraction of cells (0.4%) are capable of forming colonies. In addition to SSA deletion events, several other outcomes can arise by nonhomologous end joining (NHEJ). First, ends at a DSB can be religated to recreate the HO cut site. These sites will be cleaved again by HO endonuclease, but nevertheless, these events will give rise to a small but detectable subpopulation in our experiments. Second, it is possible to recover mutations of the HO cut site arising by NHEJ events such that the HO cut site is no longer cleavable (20, 30). These mutations, including deletions, small base pair insertions, and events that capture small segments of Ty1 elements or mitochondrial DNA, have been documented in previous studies (20, 29, 30, 42, 55, 59). To identify the large-deletion events that arose from SSA, survivors were analyzed for the ability to be recut by HO endonuclease and then analyzed by PCR and/or by Southern hybridizations (see Materials and Methods). With 29-bp repeats, bona fide SSA deletion events occurred at a frequency of 0.17%. Survivors retaining the HO cut site arose at a frequency of 0.31%, and those resulting from other NHEJ events arose at a frequency of 0.04%.
Shen and Huang (47) first showed that as the length of homologous sequence falls below the minimum effective processing segment (MEPS), the homology dependence becomes nonlinear. This could be the result of an alternate pathway acting on substrates too short for the principal pathway. We calculated the MEPS to be 33 bp for SSA by linear regression according to the method described by Shen and Huang, using data points that fell well within the linear range (63 to 235 bp). Given that the 29-bp-repeat substrates that we studied are close to or slightly below the MEPS, we asked whether formation of these deletions still required RAD52, since we had shown that SSA between longer repeats is highly dependent on Rad52p (45, 51). This is also of interest since NHEJ events sometimes appear to join microhomologies of 1 to 5 bp in a manner that is independent of RAD52 (20, 30). Hence, we also wanted to explore whether recombination between 29-bp repeats was RAD52 dependent, as in SSA, or RAD52 independent, as in NHEJ. A similar analysis in a Δrad52 derivative showed that the 29-bp repeats formed the specific deletion expected by SSA at a frequency 26-fold lower than that of the wild type (0.006%), indicating that Rad52p indeed plays an important role in recombination of short deletions of 29 bp, as it does in SSA involving longer regions of homology. The total frequency of survivors in the Δrad52 strain, 0.7%, was comparable to the wild type-frequency (0.5%), consistent with the idea that the non-SSA survivors arose by the NHEJ pathway, which is RAD52 independent.
Role of RAD51 and RAD59 in SSA.
Two RAD52-dependent pathways of spontaneous mitotic recombination have been identified; one depends on Rad51p and the other depends on Rad59p (3–5). Previously we demonstrated that SSA can be carried out independently of Rad51p (17). Those experiments, however, utilized repeats that were greater than 1 kb in length, and SSA deficiencies in some mutants are only observed when the homologous-sequence length is shorter (53). Because of this, we reexamined the effect of rad51 on SSA in a strain with the 205-bp repeats. As much product was made in a rad51 strain as in the wild type, confirming that SSA can proceed independently of the RAD51 pathway (Fig. 4).
FIG. 4.
Effect of rad51 and rad59 mutations on SSA. SSA was initiated in wild-type (WT; tNS1379), rad51 (tNS1597), and rad59 (tNS1573) backgrounds, using the two-repeat substrates with 205-bp repeated ura3 sequences (black boxes). Cultures were induced for 5 h with 2% galactose. DNA samples were digested with BglII and probed with a sequence adjacent to ura3 (cross-hatched box), revealing the uncleaved sequence (8.4 kb), the HO cut sequence (4.8 kb), and the product band (5.5 kb).
In contrast, SSA exhibited a significant dependence on RAD59. For a two-repeat strain with 205-bp repeats on each side of the HO cut site, only 2% of the rad59 mutant cells were capable of giving rise to viable colonies bearing the GAL::HO plasmid (compared with 98% for the wild-type strain). Viable colonies may possess an SSA product, or they may represent uninduced cells or the product of the religation of DNA ends. This reduction was also seen when the process was visualized on a Southern blot (Fig. 4), on which only a very faint product band was present. Densitometric analysis (see Materials and Methods) also showed that the level of product formation fell to approximately 2%. To assess whether the function of RAD59 depends on the amount of homologous sequence, two other strains, differing only in the length of homologous sequence (0.415 or 1.17 kb), were constructed. Increasing the amount of homologous sequence increased the amount of product, but only up to 24% of the level seen in wild-type strains (Fig. 3). We also examined the frequency of deletion events by using the 29-bp repeats as described earlier. We found that deletion events occurred at a frequency of 0.02%, a rate approximately eightfold lower than that of the wild-type strain, indicating that Rad59p is also required for SSA between short repeats.
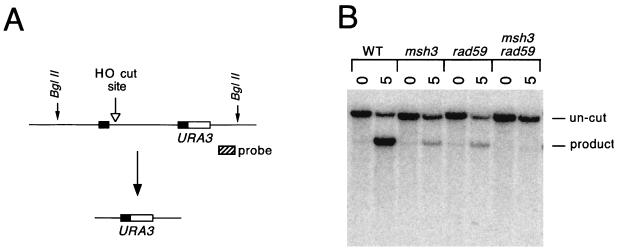
Msh2p and Msh3p play important roles in SSA because they appear to stabilize the junctions between the double- and single-stranded DNA of the annealed intermediate and/or because they target the Rad1p-Rad10p endonuclease to the single-stranded tails (13, 53). To investigate whether Rad59p plays a role in this process, a rad59 msh3 double mutant was constructed. The effect on SSA was measured by using flanking repeats with 0.415 kb of homologous sequence, based on Southern and densitometric analysis, as shown in Fig. 5. Compared with the wild type, which showed little or no impairment of SSA (Fig. 3), msh3 and rad59 reduced SSA product formation to 13 and 15%, respectively, of the wild-type level, while the double mutant yielded approximately 2% of the wild-type product level. Hence, Rad59p's role is distinct from that of Msh2p and Msh3p.
FIG. 5.
SSA in a rad59 msh3 double mutant. (A) HO endonuclease initiates SSA by cleaving between 415-bp sequences (black boxes) of ura3, resulting in a deletion product. (B) Wild-type (WT; tNS759), msh3 (tNS1551), rad59 (tNS1789), and msh3 rad59 (tNS1967) strains were induced for 5 h with galactose. At 0 and 5 h, DNA samples were obtained, digested with BglII, and probed with a sequence adjacent to ura3 (cross-hatched box in panel A), revealing the uncut band (8.6 kb), the HO-cut fragment (4.8 kb), and the product band (5.5 kb).
Role of RAD59 in HO-induced gene conversion.
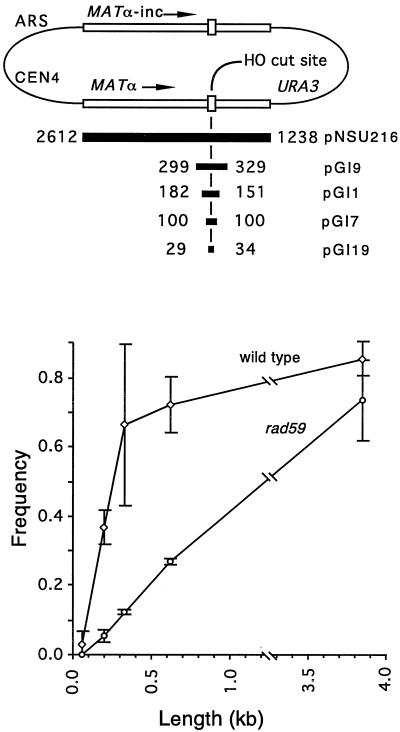
We also investigated whether Rad59p plays a role in DSB-induced gene conversion by using a set of centromeric plasmids containing inverted copies of the MAT locus (Fig. 6). An HO-induced DSB at MATα can be repaired by using a MATα-inc donor sequence, either on the plasmid or on chromosome III, that cannot be cleaved by HO endonuclease. To explore the dependence of DSB-induced gene conversion on homologous-sequence length in wild-type and rad59 backgrounds, a series of such plasmids with various amounts of homologous sequence flanking the HO cut site was constructed. Cells were plated on medium containing either galactose or glucose, and the relative plasmid stability frequencies were determined. At all homologous-sequence lengths examined, DSB-induced gene conversion was reduced in rad59 mutants, with the greatest dependence being seen when the lengths of homologous sequence were smallest. This is reminiscent of what we observed in SSA studies, where the effect of rad59Δ was reduced as the length of homologous sequence increased.
FIG. 6.
Rad59p plays a role in DSB-induced gene conversion. A series of five plasmids containing different lengths of homologous sequences derived from MATα was cleaved in vivo with HO endonuclease; the cut ends used a donor MATα-inc sequence to repair the DSB. Cells that cannot repair the DSB due to limited homology or due to the rad59 mutation will lose the plasmid. Cells were grown in YP-lactate medium and plated on medium containing galactose or glucose, and the colonies were scored for retention of the (Ura+) plasmid. Frequencies (see Materials and Methods) with 1 standard deviation (error bars) were plotted against the total homologous-sequence length.
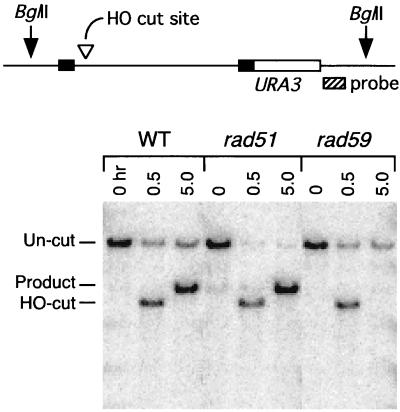
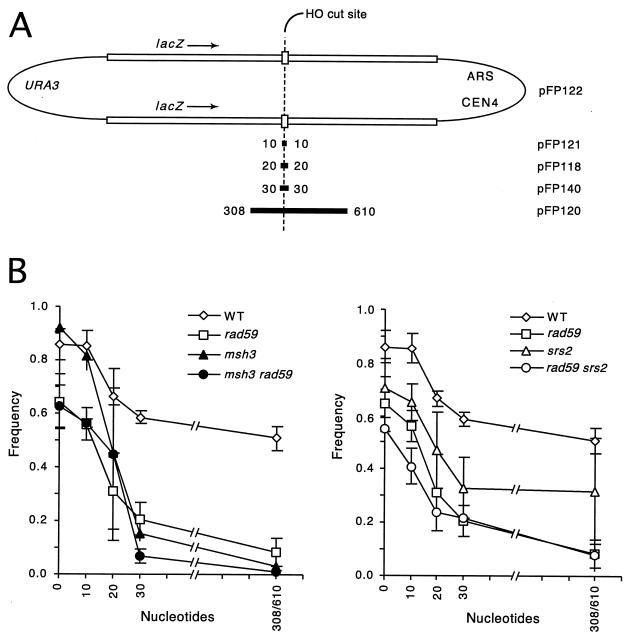
We also asked whether Rad59p is involved in the removal of nonhomologous sequences present at the ends of the invading strands created by a DSB. We investigated this by using a plasmid substrate, containing two copies of the lacZ sequence (2.8 kb), in which gene conversion is initiated by a DSB in one copy that is repaired by using the second copy as a donor template (38). A series of such plasmids with various lengths of nonhomologous tail adjacent to the DSB was constructed (Fig. 7A). The inability to remove the nonhomologous tail results in the loss of the plasmid. Figure 7B shows that wild-type strains were very efficient in repairing DSBs when there was 0 or 10 nucleotides (nt) of nonhomologous sequence present. Consistent with our results obtained while examining DSB-induced gene conversion between MAT sequences, rad59 mutants were defective even when the ends of the DSB were homologous to the donor. However, when longer, nonhomologous tails were used, the rad59 mutation had a greater effect. For example, when the ends of the DSB were completely homologous to the donor sequence, the ratio of successful gene conversions between rad59 and the wild type was 0.74; this decreased to 0.36 with nonhomologous tails on either side of the DSB of 30 nt and to 0.18 when the two lengths of nonhomologous sequence were 308 and 610 nt. This indicates that Rad59p is important for the removal of nonhomologous tails.
FIG. 7.
Effect of rad59 mutation on removal of nonhomologous tails in DSB-induced gene conversion. (A) Plasmid pFP122 contains two copies of the lacZ sequence with embedded 117-bp HO cut sites derived from MATa. The lower HO cut site contains a single point mutation making it uncleavable and allowing it to serve as a donor sequence. pFP121, pFP118, pFP140, and pFP120 were designed to generate various amounts of nonhomologous single-stranded tails on both sides, after DSB induction. Black bars show the extents of deletions of the lacZ and HO cut site sequences (labeled in nucleotides). (B) Cultures containing pFP122, pFP121, pFP118, pFP140, or pFP120 were plated on rich medium (YPD) and galactose medium (YP-galactose), allowed to grow into colonies, and scored for retention of the plasmid (Ura+ colonies). The frequency of plasmid retention was calculated as described in Materials and Methods. WT, wild type.
In view of this involvement with nonhomologous-tail removal, we compared RAD59 with MSH3, which is involved, along with MSH2, RAD1, and RAD10, in the nucleolytic removal of the nonhomologous single-stranded tails in DSB-induced gene conversion. As seen in Fig. 7, rad59 mutants differ from msh3 mutants in having an effect when the nonhomologous tails are 10 nt in length or less. The rad59 msh3 double mutant was also examined; it behaved similarly to rad59 for the 0- and 10-nt substrates, as was expected since msh3 has little effect on these substrates by itself. With 20 nt on either side of the DSB, or with a combination of 308- and 610-nt nonhomologous tails, there is no evidence of an additive effect, and there is only a small effect with the 30-nt tails. Overall, it appears that the rad59 and msh3 mutants behave similarly when nonhomologous tails of 20 nt or more must be removed.
We observed, though, that in this assay the rad59 mutant behaved similarly to a strain deficient in the Srs2p helicase (38). To examine this, we constructed and tested an srs2 mutant as well as a rad59 srs2 double mutant (Fig. 7B). Both single mutants inhibited recombination when nonhomology was absent from the ends next to the DSB, and the double mutant was statistically indistinguishable from either single-deletion mutant. When tails were removed, rad59 and rad59 srs2 mutants exhibited a phenotype slightly more defective than that evident for srs2 alone, shown in Fig. 7, but almost identical to the phenotype for srs2 previously published (38). Our observations of the rad59, srs2, and msh3 mutants suggest that the rad59 mutant exhibits characteristics of both srs2 and msh3 mutants, which we consider further below (see Discussion).
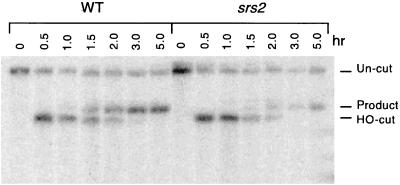
Since the behavior of the rad59 mutant in the nonhomologous tail assay was very similar to results observed for Δsrs2 (38), we also tested the effect of the srs2 deletion on SSA. Southern analysis showed that Δsrs2 also caused an approximately 3-fold reduction in SSA (Fig. 8), but this was not as drastic as the 50-fold effect of the rad59 mutation. Srs2p and Rad59p appear to play different roles in SSA.
FIG. 8.
Effect of the srs2 mutation on SSA. SSA was initiated in the wild type (WT; tNS1379) and the srs2 mutant (tNS1585) by using the two-repeat substrates with 205-bp repeated ura3 sequences as described in the legend to Fig. 4. HO endonuclease gene expression was induced for 5 h with galactose, and DNA samples were subjected to Southern analysis as described in the legend to Fig. 4, revealing the uncleaved sequence (8.4 kb), the HO cut sequence (4.8 kb), and the product band (5.5 kb).
DISCUSSION
An important observation made in this study was that the level of SSA between two repeats can be influenced by the presence of a third homologous repeat. When the third repeat is present, SSA between the other two copies is reduced. This indicates that there is a competition between the two copies on one side of the DSB for the repeat on the other side. We also showed that in the absence of competition, SSA can utilize repeats as small as 29 bp in a manner that is RAD52 and RAD59 dependent.
The fact that the presence of a third, distal competing sequence reduces the use of the proximal homologous segment suggests that SSA does not occur by an irreversible sliding mechanism by which the two single-stranded tails encounter each other at their ends and then slide by each other. If this process were irreversible, such that sliding occurred in only one direction, then there would exist only a single opportunity for the two complementary strands to initiate annealing, and therefore it would not matter if there were a third repeat further down the DNA strand. The opportunity to recognize the middle copy in the three-repeat strain would be the same as the total opportunity in the two-repeat strain, and hence the SSA levels would be similar. Our observations are not consistent with this model (Fig. 2). Instead, we favor the idea that the two copies on the same side of the DSB compete with each other, based on their relative sizes and proximity to the DSB, by a collision mechanism. We suggest that single-stranded DNA tails randomly collide with each other until they find their complementary strands. A collision model would be a straightforward interpretation of observations made by Haber and Leung (16), who found that DSBs on two different chromosomes, each containing the same flanking homologous sequences, gave rise to a reciprocal translocation by SSA as often as they led to a pair of intrachromosomal deletions. All of these results can also be explained by a two-step mechanism in which the DNA strands collide with each other and then engage in a homology search mechanism, such as reversible sliding (7).
This competition may also be influenced by the sequence closer to the DSB being made single stranded before the distal sequence. Hence, when the two sequences are both 1.17 kb, there is a strong preference for the copy closer to the DSB, an observation we previously termed the proximity effect (51). If the closer sequence is short, it may fail to recombine before the larger, more distal sequence becomes single stranded, but once available, this region will be preferred given its greater homology. The 205-bp repeats recombine at a higher rate in the two-repeat strains due to the lack of the competing distal copy.
An alternative view of homology searching in the three-repeat context proposes that the most distal copy acts as a default substrate. Instead of freely competing in recombination, the default copy is utilized only when the copy closest to the DSB is too small to efficiently recombine. The absence of the default copy in the two-repeat strains would force the sequences to repeatedly attempt to recombine, ultimately yielding a higher level of recombination. Multiple attempts make it necessary to postulate that there is a window of opportunity during which recombination must be completed or the cell dies. For example, cells in which the DSB is not repaired could adapt from DSB-induced cell cycle arrest and fatally resume growth (21). It is also possible that 5′-to-3′ resectioning of DNA will interfere with the expression of a short-lived and essential gene product.
Our results can also be explained by the Ty1 element in the ura3-52 allele, in some unexplained manner, causing the ura3-52 allele to preferentially recombine with the wild-type copy. This is unlikely, though, in view of an earlier observation (51). When the middle copy was the full-length 1.17-kb URA3 sequence, it recombined more frequently than the ura3-52 allele, at a ratio of 9:1. When the Ty1 was replaced by a wild-type URA3 sequence, the ratio dropped to 3:1. Hence, the Ty1 element does not increase recombination with the ura3-52 allele but instead decreases it. This may be because the Ty1 element moves part of the ura3 sequence an additional 6.1 kb away from the DSB.
Homologous-sequence length dependence in SSA.
Shen and Huang (47) proposed that a single MEPS possesses the ability to recombine at a characteristic rate. Accumulation of MEPS with larger lengths would result in a proportional increase in recombination rates, with the overall result being a linear relationship between the recombination rate and the length of homologous sequence. Our measurements of SSA show that the relationship of MEPS to successful recombination is initially linear but levels off as the maximum level of SSA is approached. When the SSA rate becomes high, as in this study, then multiple MEPS may initiate recombination simultaneously. This possibility increases with longer homologous sequences, and consequently the curve levels off.
Using a variety of recombination assays, both linear and nonlinear size dependencies have been observed in other organisms, and minimum size requirements range from approximately 23 to 300 bp (1, 2, 18, 24, 25, 44, 47, 50, 58). For the purpose of comparing SSA to spontaneous recombination in S. cerevisiae, we consider a study by Jinks-Robertson et al. (18), who showed that spontaneous recombination exhibited a linear dependence and a MEPS of approximately 250 bp, using either direct or inverted ura3 sequences. SSA is similar to spontaneous recombination in the sense that for sizes greater than its MEPS, it exhibits approximate linearity from 29 to 415 bp. The two processes differ, however, in that SSA can utilize homologous-sequence lengths much smaller than the MEPS of 250 bp. This suggests that spontaneous recombination between direct repeats may occur by a mechanism other than SSA or that there are assay-specific differences involved that lead to differing observations. Two other studies have examined the homology dependence of spontaneous recombination in S. cerevisiae. Using inverted his3 sequences in a plasmid-based gene conversion assay, Ahn et al. (1) documented a logarithmic relationship. In a different study, Datta et al. (11) arrived at an MEPS of 28 bp by introducing mismatches in inverted repeats to study homology dependence. These differing observations may be due to assay-specific properties or to differences in recombination mechanisms.
We showed that SSA can occur with flanking homologous sequences as small as 29 bp. NHEJ events induced by a DSB also yield deletion events that use regions of microhomology as small as 1 to 5 bp (20). Although these events may involve the annealing of strands, it is clear that they arise from a genetic pathway different from SSA since they are RAD52 and RAD1 independent (20, 30). This suggests that there is a lower limit for SSA of between 5 and 29 bp.
Role of Rad51p and Rad59p in SSA.
We have extended our previous studies to show that Rad51p plays no significant role in SSA, even with sequence homologies as small as 205 bp. On the other hand, Rad59p is required for SSA. This is consistent with the observations that Rad59p is a homolog of Rad52p, which itself has the ability to anneal DNA strands in vitro (31, 49), and that overexpression of RAD52 can overcome at least some deficiencies of rad59 mutants (4). These observations led to the conjecture that Rad59p might also possess or promote strand-annealing activity. This is supported by in vitro studies showing that Rad59p can anneal complementary strands (40). There are two aspects of SSA in which Rad59p may participate: in strand annealing itself, and in the stabilization of annealed intermediates to allow the clipping off of nonhomologous tails. Tail removal requires Rad1p-Rad10p regardless of the size of the annealed regions, whereas the role for Msh2p-Msh3p is much larger when the regions are only a few hundred base pairs in length. Rad59p's role in SSA also has a larger impact when the sequences are short. The very severe effect of a Δmsh3 Δrad59 strain with 415-bp flanking homologous sequences suggests that their roles are nonoverlapping. This does not preclude Rad59p playing a role in tail removal, but it would appear from these data that its principal phenotype reflects a defect in annealing per se.
Although there may be some overlap in function, both Rad52p and Rad59p are required for efficient SSA. A contemporaneous study by Bai et al. (3) also concluded that Rad59p plays a role in SSA and further found that a hypomorphic rad52-R70K allele exhibited a similar defect in recombination.
We also have shown that Srs2p plays a relatively minor role in SSA, compared to its more significant involvement in gene conversion. This is consistent with the idea that a helicase, such as Srs2p, is not required to assist complementary single strands in annealing, whereas it seems to be important in the invasion of a single-stranded DNA into a duplex homologous sequence (see below).
Role of Rad59p in DSB-induced gene conversion.
We have also shown that Rad59p is important for efficient DSB-induced gene conversion, both when the DSB ends are completely homologous and, even more so, when the ends are nonhomologous. This seems to point to two distinctive processes that require Rad59p. One process is the Rad1p-Rad10p-, Msh2p-Msh3p-dependent removal of nonhomologous tails during DSB-induced gene conversion. The participation of Rad59p in this step could account for the ninefold-detrimental effect of rad59 in the DSB-induced gene conversion assay reported by Bai and Symington (4), in which tails of 47 and 74 bp must be removed.
The second role of Rad59p is seen even when the ends of DSB are perfectly matched to the donor template. This can be explained by the ability of Rad59p to carry out or promote strand annealing. Annealing of DNA strands is an integral step of several models of DSB-induced gene conversion (41, 54), including the synthesis-dependent strand-annealing model (12, 28, 32, 33, 39). For example, in the latter model, the two newly synthesized DNA strands must dissociate from their respective templates and be annealed. Rad59p may be important at this step for the completion of gene conversion.
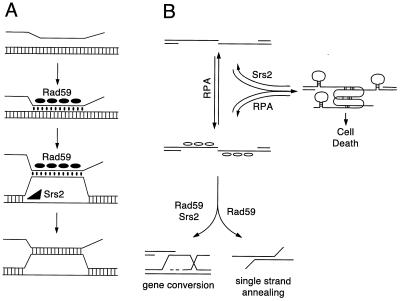
Another way to explain the two roles of Rad59p in DSB-induced gene conversion is in terms of the formation and stabilization of a D loop during strand invasion. The first step in gene conversion is the identification of a duplex DNA homologous to the single-stranded end of the DSB that has been produced by 5′-to-3′ resection. The first encounter, mediated by recombination proteins such as Rad51p, is presumed to result in an unstable paranemic joint which can then be converted into a plectonemic joint (Fig. 9A). When the end of the single-stranded tail is not homologous to the double-stranded DNA, it may be difficult or even impossible for the transition from paranemic to plectonemic joint to occur (43). We suggest that Rad59p may act by catalyzing the formation of a paranemic joint by stabilizing these interactions. This would be important for any strand invasion, even those in which the ends of the DNA are completely homologous to the template, but it would become even more important when there are nonhomologous tails.
FIG. 9.
Models demonstrating how nonhomologous tails might impede DSB-induced gene conversion in rad59 mutants. (A) During DSB-induced gene conversion, a single-stranded DNA intermediate searches duplex DNA for a homologous sequence and initially forms a paranemic joint that is converted to a plectonemic joint. The presence of nonhomologous sequences at the end of the invading strand destabilizes the paranemic joint or impedes the conversion to a plectonemic joint (43). Rad59p (closed ellipses) facilitates the process by annealing the strands and stabilizing the joint. A stable paranemic joint in turn provides the opportunity for the tail to be removed or for a topoisomerase to act. In the absence of Rad59p, longer tails pose a greater impediment to the formation of a plectonemic joint. This is consistent with a previous interpretation of the srs2 mutation, which behaves similarly in the DSB-induced gene conversion assay (38). Srs2p (black triangle) is proposed to stabilize the same structure by unwinding the donor duplex, allowing easier formation of a paranemic joint and, later, a plectonemic joint. (B) When nonhomologous tails become long, they may impede SSA or DSB-induced gene conversion by forming secondary structures in a process mediated by the annealing of microhomologous sequences. The single-stranded DNA binding protein RPA (open ellipses) may reduce secondary structures by forming complexes with single-stranded DNA. These complexes likely lie in equilibrium with the unbound state as the single-stranded tail searches for homologous sequence. As such, the secondary structures remain competitors to successful SSA or gene conversion. A mutation such as rad59 or srs2 that cripples SSA or gene conversion will shift the equilibrium toward the secondary structures, eventually leading to cell death. Substrates with short tails or no tails do not have this competing pathway, and hence it only reveals itself with structures with long tails in a mutant background. Mutations in srs2 decrease SSA to a lesser extent than do rad59 mutations, which is consistent with the likelihood that a helicase is not required for annealing of complementary strands. Srs2p may promote SSA indirectly by unwinding microhomology-based secondary structures or by aiding in the homology search process by unwinding improperly annealed single-stranded tails.
We favor this interpretation in part because rad59 mutants behave in a manner similar to srs2 mutants in the DSB-induced gene conversion assay in the absence of nonhomologous tails. Because Srs2p is a helicase, Pâques and Haber (38) have proposed that Srs2p promotes strand invasion by unwinding the donor duplex to facilitate formation of a longer plectonemic joint (Fig. 9A). Pâques and Haber further have proposed that this will stabilize the junction and facilitate the removal of any nonhomologous tails of the invading strand. Thus, both srs2 and rad59 mutants would be defective in stabilizing the paranemic joint and in the removal of nonhomologous tails.
In addition to this proposed role in strand invasion and strand annealing, Rad59p may also participate directly in the tail removal pathway mediated by Msh2p, Msh3p, and Rad1p-Rad10p, given the phenotypic similarity of rad59 and msh3 mutants in the removal of nonhomologous tails that are 20 nt or longer. With respect to SSA, the rad59 msh3 double mutant may be more severely defective than either single mutant if rad59 and msh3 are each partially defective at two separate steps in the tail removal process.
One other question that intrigues us is why, as they get longer, nonhomologous tails apparently impede gene conversion. As single-stranded tails form, they can become entangled in secondary structures mediated by microhomologies which in turn may prevent longer homologous sequences from finding each other (Fig. 9B). This pathway would compete with the DSB-induced gene conversion pathway, especially if gene conversion were impeded by a mutation in a gene such as rad59 or srs2. When tails are absent or short, this pathway no longer competes with gene conversion, and hence the effect is seen only with longer tails. A similar inhibition of SSA may also occur, and the effect of srs2 on SSA, albeit small compared to that of rad59 or msh3, may reflect its role in this process.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM20056.
We thank members of the Haber lab and three anonymous reviewers for helpful comments regarding this work.
REFERENCES
- 1.Ahn B Y, Dornfeld K J, Fagrelius T J, Livingston D M. Effect of limited homology on gene conversion in a Saccharomyces cerevisiae plasmid recombination system. Mol Cell Biol. 1988;8:2442–2448. doi: 10.1128/mcb.8.6.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayares D, Chekuri L, Song K-Y, Kucherlapati R. Sequence homology requirements for intermolecular recombination in mammalian cells. Proc Natl Acad Sci USA. 1986;83:5199–5203. doi: 10.1073/pnas.83.14.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai Y, Davis A P, Symington L S. A novel allele of RAD52 that causes severe DNA repair and recombination deficiencies only in the absence of RAD51 or RAD59. Genetics. 1999;153:1117–1130. doi: 10.1093/genetics/153.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai Y, Symington L S. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- 5.Bartsch S, Kang L E, Symington L S. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol Cell Biol. 2000;20:1194–1205. doi: 10.1128/mcb.20.4.1194-1205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basile G, Aker M, Mortimer R K. Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol Cell Biol. 1992;12:3235–3246. doi: 10.1128/mcb.12.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg O G, Winter R B, Von Hippel P H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 8.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colaiácovo M P, Pâques F, Haber J E. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics. 1999;151:1409–1423. doi: 10.1093/genetics/151.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Critchlow S E, Jackson S P. DNA end-joining: from yeast to man. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 11.Datta A, Hendrix M, Lipsitch M, Jinks-Robertson S. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci USA. 1997;94:9757–9762. doi: 10.1073/pnas.94.18.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson D O, Holloman W K. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci USA. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 14.Fishman-Lobell J, Rudin N, Haber J E. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol. 1992;12:1291–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein A L, McCusker J H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 16.Haber J E, Leung W Y. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc Natl Acad Sci USA. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov E L, Sugawara N, Fishman-Lobell J, Haber J E. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinks-Robertson S, Michelitch M, Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanaar R, Hoeijmakers J H, van Gent D C. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 20.Kramer K M, Brock J A, Bloom K, Moore J K, Haber J E. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S E, Moore J K, Holmes A, Umezu K, Kolodner R D, Haber J E. Saccharomyces Ku70, Mre11/Rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 22.Lin F-L, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin F-L M, Sperle K, Sternberg N. Repair of double-stranded DNA breaks by homologous DNA fragments during transfer of DNA into mouse L cells. Mol Cell Biol. 1990;10:113–119. doi: 10.1128/mcb.10.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liskay R M, Letsou A, Stachelek J L. Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics. 1987;115:161–168. doi: 10.1093/genetics/115.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liskay R M, Stachelek J L, Letsou A. Homologous recombination between repeated chromosomal sequences in mouse cells. Cold Spring Harbor Symp Quant Biol. 1984;49:183–189. doi: 10.1101/sqb.1984.049.01.021. [DOI] [PubMed] [Google Scholar]
- 26.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. [Google Scholar]
- 27.Maryon E, Carroll D. Characterization of recombination intermediates from DNA injected into Xenopus laevis oocytes: evidence for a nonconservative mechanism of homologous recombination. Mol Cell Biol. 1991;11:3278–3287. doi: 10.1128/mcb.11.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGill C, Shafer B, Strathern J. Coconversion of flanking sequences with homothallic switching. Cell. 1989;57:459–467. doi: 10.1016/0092-8674(89)90921-5. [DOI] [PubMed] [Google Scholar]
- 29.Moore J K, Haber J E. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature. 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 30.Moore J K, Haber J E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortensen U H, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasmyth K A. Molecular genetics of yeast mating type. Annu Rev Genet. 1982;16:439–500. doi: 10.1146/annurev.ge.16.120182.002255. [DOI] [PubMed] [Google Scholar]
- 33.Nassif N, Penney J, Pal S, Engels W R, Gloor G B. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.New L, Liu K, Crouse G F. The yeast gene MSH3 defines a new class of eukaryotic MutS homologues. Mol Gen Genet. 1993;239:97–108. doi: 10.1007/BF00281607. [DOI] [PubMed] [Google Scholar]
- 35.Nickoloff J A, Singer J D, Hoekstra M F, Heffron F. Double-strand breaks stimulate alternative mechanisms of recombination repair. J Mol Biol. 1989;207:527–541. doi: 10.1016/0022-2836(89)90462-2. [DOI] [PubMed] [Google Scholar]
- 36.Ozenberger B A, Roeder G S. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol Cell Biol. 1991;11:1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pâques F, Haber J E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pâques F, Haber J E. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pâques F, Leung W Y, Haber J E. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol Cell Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petukhova G, Stratton S A, Sung P. Single strand DNA binding and annealing activities in the yeast recombination factor Rad59. J Biol Chem. 1999;274:33839–33842. doi: 10.1074/jbc.274.48.33839. [DOI] [PubMed] [Google Scholar]
- 41.Resnick M A. The repair of double strand breaks in DNA: a model involving recombination. J Theor Biol. 1975;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 42.Ricchetti M, Fairhead C, Dujon B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature. 1999;402:96–100. doi: 10.1038/47076. [DOI] [PubMed] [Google Scholar]
- 43.Riddles P W, Lehman I R. The formation of paranemic and plectonemic joints between DNA molecules by the recA and single-stranded DNA-binding proteins of Escherichia coli. J Biol Chem. 1985;260:165–169. [PubMed] [Google Scholar]
- 44.Rubnitz J, Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984;4:2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudin N, Haber J E. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol Cell Biol. 1988;8:3918–3928. doi: 10.1128/mcb.8.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selva E M, New L, Crouse G F, Lahue R S. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen P, Huang H V. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. [Google Scholar]
- 49.Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 50.Singer B S, Gold L, Gauss P, Doherty D H. Determination of the amount of homology required for recombination in bacteriophage T4. Cell. 1982;31:25–33. doi: 10.1016/0092-8674(82)90401-9. [DOI] [PubMed] [Google Scholar]
- 51.Sugawara N, Haber J E. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugawara N, Ivanov E L, Fishman-Lobell J, Ray B L, Wu X, Haber J E. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature. 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- 53.Sugawara N, Pâques F, Colaiácovo M, Haber J E. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 55.Teng S C, Kim B, Gabriel A. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature. 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 56.Tsukamoto Y, Ikeda H. Double-strand break repair mediated by DNA end-joining. Genes Cells. 1998;3:135–144. doi: 10.1046/j.1365-2443.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- 57.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 58.Watt V M, Ingles C J, Urdea M S, Rutter W J. Homology requirements for recombination in Escherichia coli. Proc Natl Acad Sci USA. 1985;82:4768–4772. doi: 10.1073/pnas.82.14.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu X, Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol Cell. 1999;4:873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]