Abstract
An individual’s health and conditions are associated with a complex interplay between the individual’s genetics and his or her exposures to both internal and external environments. Much attention has been placed on characterizing of the genome in the past; nevertheless, genetics only account for about 10% of an individual’s health conditions, while the remaining appears to be determined by environmental factors and gene-environment interactions. To comprehensively understand the causes of diseases and prevent them, environmental exposures, especially the external exposome, need to be systematically explored. However, the heterogeneity of the external exposome data sources (e.g., same exposure variables using different nomenclature in different data sources, or vice versa, two variables have the same or similar name but measure different exposures in reality) increases the difficulty of analyzing and understanding the associations between environmental exposures and health outcomes. To solve the issue, the development of semantic standards using an ontology-driven approach is inevitable because ontologies can (1) provide a unambiguous and consistent understanding of the variables in heterogeneous data sources, and (2) explicitly express and model the context of the variables and relationships between those variables. We conducted a review of existing ontology for the external exposome and found only four relevant ontologies. Further, the four existing ontologies are limited: they (1) often ignored the spatiotemporal characteristics of external exposome data, and (2) were developed in isolation from other conceptual frameworks (e.g., the socioecological model and the social determinants of health). Moving forward, the combination of multi-domain and multi-scale data (i.e., genome, phenome and exposome at different granularity) and different conceptual frameworks is the basis of health outcomes research in the future.
Keywords: Semantic standard, External exposome, Environmental exposure, Ontology
1. Introduction
In 2005, the concept of the exposome was first proposed by Wild as “encompasses all life-course environmental exposures from the prenatal period onwards, complementing the genome.” (Wild, 2005) It was developed to draw attention to a more comprehensive assessment of environmental exposures, although what they meant by environmental exposure was not very clearly defined in the original paper. Later in 2008, Wild talked about environmental exposures “in a broader sense of all lifestyle, infections, radiation, natural and man-made chemicals and occupational exposures” that are not genetic driven (Wild, 2009). In that sense, these environmental exposures not only refer to the external environment we commonly discussed (e.g., urban–rural environment, climate, and environmental pollutants), but also internal biologic markers of environmental exposures impacted by an individual’s physiology and external environmental exposures such as metabolic factors and inflammation.
In fact, an individual’s phenotypes related to their health and diseases are associated with the complex interplay between the individual’s genetics and their exposures to both internal and external environments (Weatherall, 1999; Raknes, 2018; The National Institute fo, 2020). For different environmental factors, long-term vs. short-term exposures also matter on their impact an individual’s health outcomes, especially for non-communicable diseases. For example, when studying air pollution, short-term exposure to air pollution (i.e., particulate matter 10–2.5, elemental carbon, nitrogen dioxide [NO2], nitrate, and O3) could increase the risk of asthma exacerbation (Mann et al., 2010; Meng et al., 2010), while exposure to certain air pollutants over a long period of time could increase the risk of heart attack, stroke, and atrial fibrillation (DaneshYazdi et al., 2021). It is important to note that individuals’ health determinants are the product of their individual history of exposures (especially those long-term environmental exposures), superimposed on their underlying genetic susceptibilities (Vineis et al., 2020). While much attention has been placed on the characterization of the genome in the past, recent studies suggest that the contributions of environmental factors may play an important role in determining individuals’ health outcomes (The National Institute fo, 2020). For example, genetics has been found to account for only about 10% of diseases, and the remaining causes appear to be determined by environmental factors and gene-environment interactions. Further, a World Health Organization (WHO) report in 2016 estimated that 23% of all deaths and 26% of deaths among children under age 5 worldwide were due to preventable environmental exposures such as indoor and outdoor air pollution, cigarette smoking, and poor diet (World Health Organization, 2016). To comprehensively understand the causes of diseases and prevent them, environmental exposures need to be systematically explored.
1.1. Defining the exposome
The totality of environmental exposures is diverse, dynamic, and may interact with each other. To better study the exposome data, many frameworks have been proposed in the last decade to model and categorize various environmental exposures. Based on a recent scoping review of exposome studies (Haddad et al., 2019), the most commonly used framework, proposed by Wild et al. (Wild, 2012) in 2012, categorizes the exposome into three domains: (1) the internal exposome—exposures that impact the internal environment of the body such as metabolic factors, hormones, and gut microbiota; (2) the general external exposome—social, cultural and ecological contexts in which the person lives their life such as urban-rural environment, traffic, climate factors, and social capital; and (3) the specific external exposome—the specific external agents to which one is exposed such as specific contaminants, poor diet, and lack of exercise. Nevertheless, Wild himself acknowledged “the difficulty in placing a particular exposure in one domain or another; for example, one can debate whether physical activity should be in the internal or specific external domains.” (Wild, 2012) Such difficulty may have arisen from the non-specificity when one describes certain exposures. For example, the exposure to education can either be a specific external exposome factor (i.e., an individual’s own education history) or a general external exposome factor (i.e., the education environment where an individual lives) depending on how the exposure is measured. In fact, some recent studies (Oyana et al., 2015; Loh et al., 2017) as well as the United States Centers for Disease Control and Prevention (CDC) (The National Institute fo, 2020) categorized the exposome into only two domains: the external exposome and the internal exposome, where the external exposome were studied as a whole. Nevertheless, correctly and explicitly defining and representing each domain of the exposome is essential, as it helps us systematically think and structure potential environmental exposures, especially considering the multi-level and cross-domain nature of these exposome factors from heterogenous data sources. Fig. 1 shows a conceptual framework of the exposome integrating the definitions of Wild and CDC.
Fig. 1.
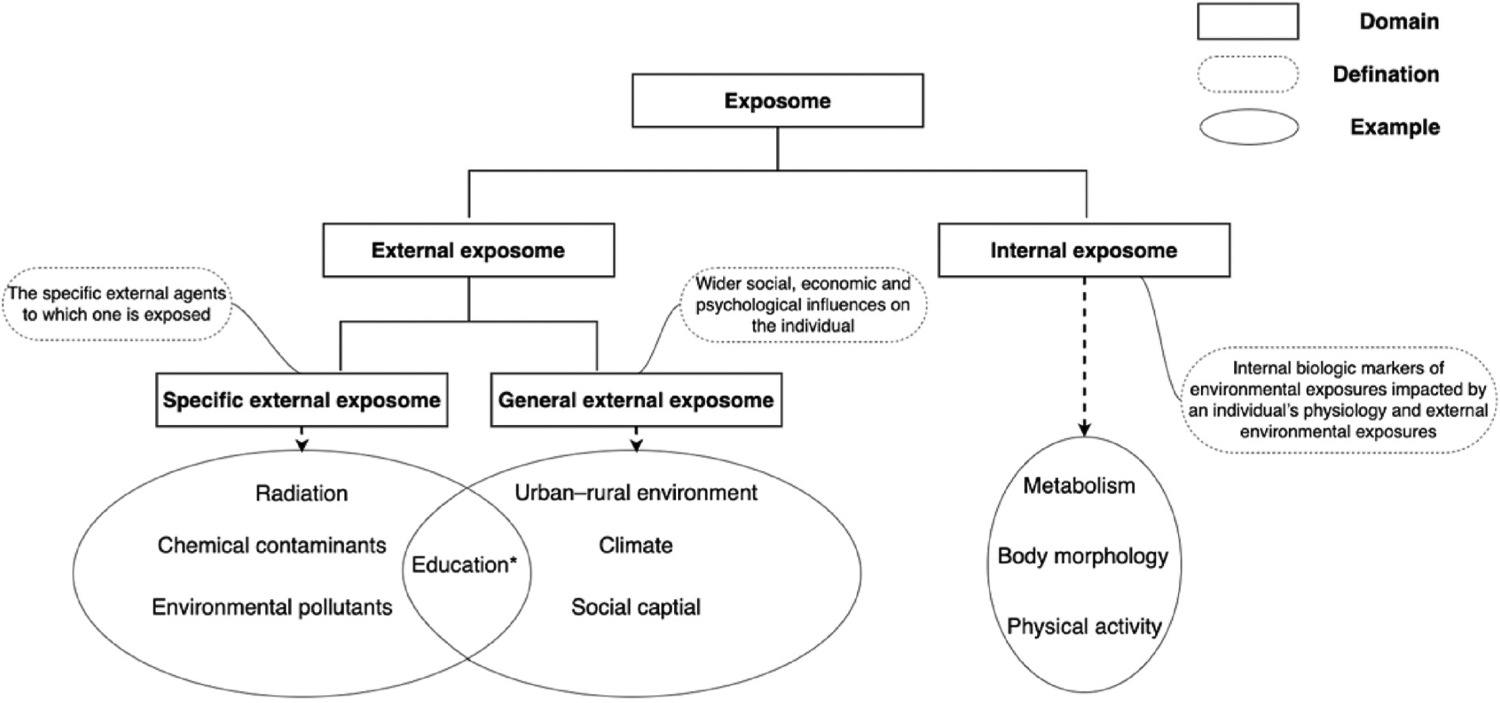
A conceptual framework of the exposome.
*The exposure to Education can either be a specific external exposome (i.e., the individual’s own education history) or general external exposome (i.e., the education environment where the individual lives)
1.2. Opportunities and challenges: the blooming of exposome data and semantic standards
In the past decade, the internal exposome has drawn significant attention, because advancements such as the rise of “-omics” (e.g., transcriptomics, proteomics, and metabolomics) for the development of biomarkers that have already taken place for other purposes (e.g., the precision medicine initiative (White Hourse, 2015)), making measurements of internal exposome factors (e.g., carcinogen metabolism and xenobiotics (Vineis et al., 2017)) more feasible. Meanwhile, advances in technologies (e.g., remote sensing and wearable devices) also allowed us to collect an increasing amount of external exposome data, providing researchers great opportunities to explore the contributions of the external exposome to health outcomes at both individual- and population-levels.
However, the increasing availability of exposome data also comes with challenges. First, measures of environmental exposures usually come from heterogeneous data sources. When accessing, sharing, integrating, managing, processing, reporting on, and interpreting these exposome data, it requires significant effort to overcome unique data challenges, especially the heterogeneous syntax, schema, and semantics of the data from different data domains and sources. Second, when studying health outcomes, it is essential to consider the interplays between the individual/population and their environmental exposures and deal with multi-domain, multi-level, and multi-scale data including the genome (e.g., genomics, and transcriptomics), phenome (e.g., disease phenotype, behavior, and organismal traits), and exposome (e.g., body morphology, environmental pollutants, and social capital), as well as their interactions. To have a comprehensive view of outcomes and factors from these different types of data, priority must be given to enabling integration and reuse of existing data and lowering access barriers as first steps.
To tackle the data discovery, access, and integration barriers, data standards such as common data elements (CDEs) and common data models (CDMs (National Library of Medic, 2020)) play an important role in increasing data interoperability. Nevertheless, although helpful in addressing syntactical and schematic data heterogeneity, CDEs and CDMs are limited in capturing the semantics of the data. On the other hand, biomedical ontologies are widely used as artifacts to represent, manage, and share biomedical knowledge, and to support downstream applications (Schulz and Jansen, 2013; Amith et al., 2018; Ochs et al., 2017). Ontologies have also been widely used in computer systems for metadata representations to facilitate semantic data integrations (SDIs), advancing beyond the traditional data integration approaches that use CDEs and CDMs (Wache et al., 2001). For example, the use of ontologies in the biomedical domain began with the development of Gene Ontology (GO). The original goal of GO was to provide a set of structured vocabularies that can be used to describe gene products in any organism (Gene Ontology Consortium., 2001). More recently, GO has now been widely used to facilitate (1) data annotation and data integration, and (2) sharing of biological knowledge (Blake and Bult, 2006). Another example is the Human Phenotype Ontology (HPO) (Köhler et al., 2017), which was developed to provide a standardized vocabulary of phenotypic abnormalities encountered in human diseases. The HPO has now been widely used for (1) providing a standardized vocabulary for clinical databases, (2) facilitating database searchers for clinical diagnostics, and (3) mapping the phenotypes (rather than the genotypes provided in GO) between human and model organism.
The definition of an ontology often varies depending on one’s viewpoint, however a commonly accepted definition is that an ontology is a formal, computational representation of a domain of knowledge based upon a controlled, standardized vocabulary for describing entities and the semantic relationships between them. Another approach—that of ontological realism—expands upon this definition to emphasize that the terms or classes in an ontology shall represent entities truthfully and unambiguously according to the best available scientific evidence (Smith and Ceusters, 2010). Regardless of the different modeling approaches, the common features of an ontology make it an ideal tool to (1) establish a shared, common understanding of the structure of information, (2) ensure that domain assumptions are explicitly modeled, (3) be adaptive to knowledge changes, and (4) facilitate domain knowledge/data management.
Summarized above, we first introduced the concept of “exposome” to better define and model various environmental exposures. With the blooming of the exposome data, we discussed the opportunities and challenges that came with it, especially those related to the heterogeneity of the exposome data sources, as well as a potential solution (i.e., semantic standards via ontology) for solving those challenges. In the rest of the paper, we will discuss the deficiency of existing efforts on unraveling the heterogeneous syntax, schema, and semantics of the data from different exposome data domains and sources, and compare those efforts with an ontology-based approach. We will then provide an in-depth discussion on the challenges of developing ontologies for external exposome data and future research needs.
2. Existing works on unraveling exposome data
The booming of “-omics” studies came with advances in the development of data standards, including semantic standards, for management and interoperability across studies and systems. The semantic standards, more specifically, benefited internal exposome studies (e.g., Cell Ontology and Gene Ontology) as they established shared controlled vocabularies and made internal exposome data not only understandable across human users but also computable across computer programs. For example, the Gene Ontology (GO) resource aims to “provide a computational representation of our current scientific knowledge about the functions of genes (or, more properly, the protein and non-coding RNA molecules produced by genes) from many different organisms, from humans to bacteria.” (Ashburner et al., 2000) To achieve that goal, it is necessary for the GO to include important representations for classes such as metabolites, hormones, oxidative stress, and inflammation, which are also important for representing the internal exposome.
On the external exposome side, the rising awareness of environmental exposures’ importance has also led to the blooming collections of external exposome data (Martin Sanchez et al., 2014). For example, in our prior work (Hu et al., 2020), we integrated external exposome data from multiple well-validated sources (see Table 1), with over 5500 variables, and spatiotemporally linked them to Florida vital statistics birth records in order to characterize pregnant women’ exposures to their surrounding environment during pregnancy (i.e., climate, air pollution, greenness, walkability, food access, socioeconomics, social capital, housing, and safety). As shown in Table 1, we summarize the characteristics of the reliable and well-validated external exposome data and data sources, grouped into three categories:
Table 1.
Examples of heterogeneous external exposome data and data sources.
| Data Source and Validation Study | Since | Spatial Scale Temporal Scale | #of Vars | Example Measures | |
|---|---|---|---|---|---|
| Natural Environment | |||||
| Air toxicants | EPA National Air Toxics Assessment (NATA) (Logue et al., 2011) | 2002 | Census Tract/1-year | 181 | Acrolein, propylene oxide |
| Meteorology | Environmental Prediction (NCEP) North American Regional Reanalysis (NARR) (North American Regio, 2005) | 1979 | 32km/1-day | 410 | Temperature, humidity, pressure |
| Fine Particulate Matter and Ozone | EPA and CDC’s National Environmental Public Health Tracking Network (irquality dat, 2014) | 2002 | Census Tract/1-day | 6 | PM2.5, O3 |
| Built Environment | |||||
| Walkability | Walk Score (Carr et al., 2010, 2011) | 2009 | 0.0015°/Cross-sectional | 1 | Walk score |
| Food Access | US Department of Agriculture (USDA) Food Access Research Atlas (FARA) (United States Department of Agriculture, 2019) | 2010 | Census Tract/1-year | 42 | Percent of low-access population at 1 mile |
| Green Space | National Aeronautics and Space Administration (NASA)’s Moderate Resolution Imaging Spectroradiometer (MODIS) (Rhew et al., 2011) | 2000 | 250m; 16-day | 3 | Normalized difference vegetation index |
| Social Environment | |||||
| Socio-demographic | American Community Survey (ACS) (Mather et al., 2005) | 2005 | Census Block Group/5-year | 4930 | Migration, education, employment |
| Social Capital | Census Bureau Business Patterns (CBP) (Rupasingha et al., 2006) | 1986 | Zip-code/1-year | 9 | Religious, civic, and social organizations |
| Crime and Safety | Uniform Crime Reporting (UCR) (Lynch and Addington, 2007) | 1974 | County/1-year | 7 | Burglary rate, aggravated assault rate |
| Address Vacancy | US Department of Housing and Urban Development (Garvin et al., 2013) | 2005 | Census Tract/3-month | 289 | Average days addresses vacant |
• Natural Environment
○ Air toxicants data from the National Air Toxics Assessment (NATA) includes estimates for 181 toxicants of the Clean Air Act (Logue et al., 2011).
○ The meteorological data are available from the National Centers for Environmental Prediction (NCEP) North American Regional Reanalysis (NARR) (North American Regio, 2005), which contains extensive meteorology information such as temperature, dew point temperature, specific humidity, relative humidity, pressure, and wind speed.
○ The data on fine particulate matter and ozone are available from the US Environmental Protection Agency (EPA) and CDC’s National Environmental Public Health Tracking Network (ir quality dat, 2014).
• Built Environment
○ Walk Score assess the walkable amenities using data from geographic information systems (GIS) that are publicly available (Carr et al., 2010, 2011).
○ Data on food access are available from the US Department of Agriculture (USDA) Food Access Research Atlas and Food Environment Atlas (United States Department of Agriculture, 2019).
○ The normalized difference vegetation index (NDVI) is derived from the US National Aeronautics and Space Administration (NASA)’s Moderate Resolution Imaging Spectroradiometer (MODIS) on the Terra satellite, which has been validated as a measure of neighborhood greenness and widely used in epidemiological studies (Rhew et al., 2011).
• Social Environment
○ Census block group level socio-demographic status and housing information can be obtained from the American Community Survey (ACS) 5-year estimates (Mather et al., 2005). ACS is the premier source for detailed population and housing information in the US, including a variety of sociodemographic and housing information, such as age, sex, race/ethnicity, origins, disability, migration, education, employment, income and earnings, insurance coverage, acculturation, marital status, poverty, and housing information including count, financial characteristics, occupancy characteristics, and physical characteristics.
○ Social capital data are available from the US Census Bureau Business Patterns (CBP) (Rupasingha et al., 2006), which provides gold-standard economic data by industry. Using the North American Industry Classification System (NACIS) codes, we can identify the number of the establishments at the zip-code level for entities such as religious organizations, civic and social associations, and business associations among others.
○ County-level annual crime measures can be obtained from the Uniform Crime Reporting (UCR) program (Lynch and Addington, 2007), which has the most reliable crime data in the US.
○ Vacant land is an important predictor of health and the gold-standard data can be obtained from the US Department of Housing and Urban Development (HUD), which provides quarterly information on address vacancies at the census tract level since 2005 (Garvin et al., 2013).
Many efforts have been devoted to unraveling external exposome data. Using approaches such as an exposome-wide association study (ExWAS), broadening the concept of environment-wide association study (EWAS) (Hu et al., 2020, 2021; Zheng et al., 2020), we can systematically and efficiently screen the associations between thousands of environmental exposures and health outcomes. Compared with traditional studies which only examine a small subset of environmental exposures, the exposome framework can generate more timely findings as it eliminates the time-consuming process to conduct separate studies on individual exposures. In addition, the agnostic exposome approach leads to much better controls of potential confounding by co-exposures and is less likely to miss potential important predictors of the specific health outcomes. Many of previous projects have implemented ExWAS to explore the external exposome data. For example, the Health and Environment-wide Associations based on Large population Surveys (HEALS) project aims to advance the ExWAS methodology and provide analytical tools in support of European Union-wide environment and health assessments (Health and Environment-wi, 2021). The HEALS project adopted Wild’s definition (Wild, 2005) for categorizing external exposome data, but further developed a conceptual framework to provide an overview of how individual- and population-level external exposome are (1) combined using physiologically based pharmacokinetic models; and (2) linked with omics data to obtain markers of exposure, dose, or health effect and to determine underlying mechanistic pathways between environment and health (Turner et al., 2017).
The Human Early-Life Exposome (HELIX) (Vrijheid et al., 2014) project is another example that aims to characterize children’s exposomes as they progress through early life and explore of the relationships between the early life exposomes with omics markers and health in childhood. The HELIX project plans to measure external environmental exposures for food, water, air pollution, pesticides, noise, and ultraviolet radiation of up to 32,000 mother-child pairs and measure the growth, development, and health of the children, including birth outcomes, postnatal growth and body mass index, asthma and lung function, and neuro-development among others. Agnostic ExWAS approaches were used to study the impact of the early-life exposome on child health. They also reused the definition from Wild’s but further categorized the exposome into internal and external exposome. Similarly, the Children’s Health Exposure Analysis Resource (CHEAR) program (Balshaw et al., 2017), supported by the National Institute for Environmental Health Sciences (NIEHS), aims to support the analysis of the exposome data through the harmonization of studies enabled by a CHEAR ontology for children’s health, exposures and associated data, and metadata standards. Established in 2019, the Human Health Exposure Analysis Resource (HHEAR) program (McGuinness et al., 2019) succeed CHEAR and expanded to promote using the “exposome” concept to characterize of the totality of human environmental exposures.
Other projects such as EXPOsOMICS (Turner et al., 2018) have also aimed to develop approaches to comprehensively integrate both external and internal exposomes at the individual level. Different from ExWAS, EXPOsOMICS took a bottom-up approach that starts with a set of exposures or environmental compartments to determine the pathways or networks by which such exposures lead to disease (Vineis et al., 2017).
3. The needs for semantic standards of external exposome data
It is evident that there is a wealth of external exposome data currently available; nevertheless, these are significantly different from internal exposome data: they are more heterogenous in terms of spatial and temporal scales, formats, and data domains. Without a clear mental model of these data and data sources, it is difficult for researchers to formulate their research questions, especially when these heterogenous external exposome data need to be integrated and explored together. Researchers who are interested in conducting an external ExWAS needs to comprehensively and carefully examine these different external exposome data and data sources, being mindful of the different potential data integration scenarios and asking themselves a number of questions: (1) what are the domains of the measures of interest? (2) what are the spatial and temporal scales of the measures (3) how can we establish the susceptible spatial (e.g., buffer vs. administrative boundary)? and temporal windows (e.g., short-term vs. long-term) to these external exposome measures (4) are there similar measures (e.g., monitored vs. modelled PM2.5)? from different sources? and then decide on (5) which ones to use, (6) whether it make senses to combine these similar measures to a “better” (e.g., more reliable or representative) measure or (7) how to interpret inconsistent results for similar measures from different sources; among many other practical considerations that are needed.
To make conscious decisions when designing an external ExWAS, understanding the heterogeneity of the external exposome across different data sources is critical and semantics standards such as ontologies can facilitate the data integration efforts. With the help of ontologies, researchers can have a consistent understanding of the variables in heterogenous data sources. Further, the context of the variables and relationships between those variables can be explicitly expressed and modeled. For example, if we want to study how the interaction between an individual’s education level with the average education level of the community he lives in - a well-known social determinants of health for disease prevention (Berger et al., 2017) - would affect certain health behavior (e.g. cancer screening), we would like to examine both the individual-level education statuses and the community-level education attainment rates, linked by individuals’ residential addresses, in a single statistical model. When integrating variables (e.g., “education”) with different spatial scales (e.g., individual’s educational attainment vs. the average educational attainment of the community at a census tract level), besides providing a standardized definition for the variable “education”, an ontology can ensure the contextual variables (e.g., “census tract”) are also correctly defined and the relationships among them are unambiguously modeled. For example, as shown in Fig. 2, the individual-level (OPMI_0000111) and census tract-level (NCIT_C171590) educational attainments are connected through the geographic region (GEO_000000372) that a person lives in, and the differences (i.e., level of measures) are clearly modeled. In this way, our integration strategy will be focusing on attaching a community educational attainment rate to each individual person based on the census tract they live in. Having such models is crucial when integrating variables from heterogeneous data sources because the context information (e.g., what is the spatial scale of the educational attainment variable in the source data) will impact the integration strategies and decisions.
Fig. 2.
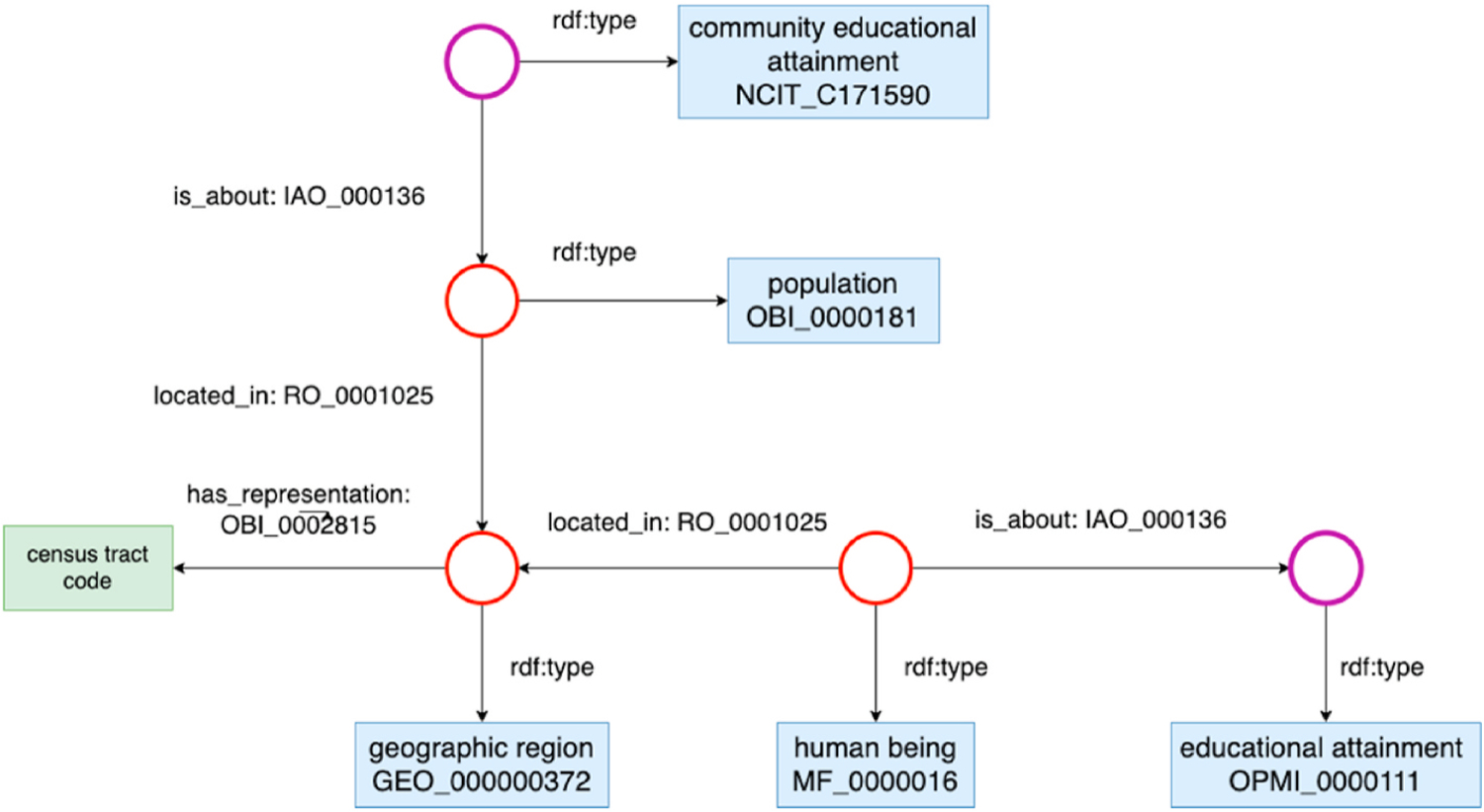
The ontological representation for education attainment at the individual-level and census tract-level. Note that the blue rectangular represents the class, the circle represents the individual of the class, and the green rectangular represents the data values (literals). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Challenges in the development of ontologies for external exposome data
Given the amount of time and effort required to develop and maintain an ontology and the role that they play in enabling semantic interoperability, it is common practice to reuse ontology classes whenever possible. To facilitate this, web repositories, like the National Center for Biomedical Ontology (NCBO) BioPortal (Whetzel et al., 2011), have been developed to support searching and viewing ontologies and their classes. We conducted a systematic search of BioPortal to identify existing ontologies related to external environmental exposures. As shown in Fig. 3, we identified 882 ontologies in BioPortal. During our screening process, we first developed a list of keywords based on the external exposome variables and data sources listed in Table 1 (i.e., natural environment, air toxicants, meteorology, built environment, vacant land, walkability, food access, food environment, green space, social environment, socio-demographic, social capital, crime, safe and hospital bed capacity). We excluded ontologies without keywords hit. We then screened the scope of the ontologies and only included ontologies that focus on the exposome, encompass external environmental exposures, and are actively maintained (e.g., at least a yearly update).
Fig. 3.
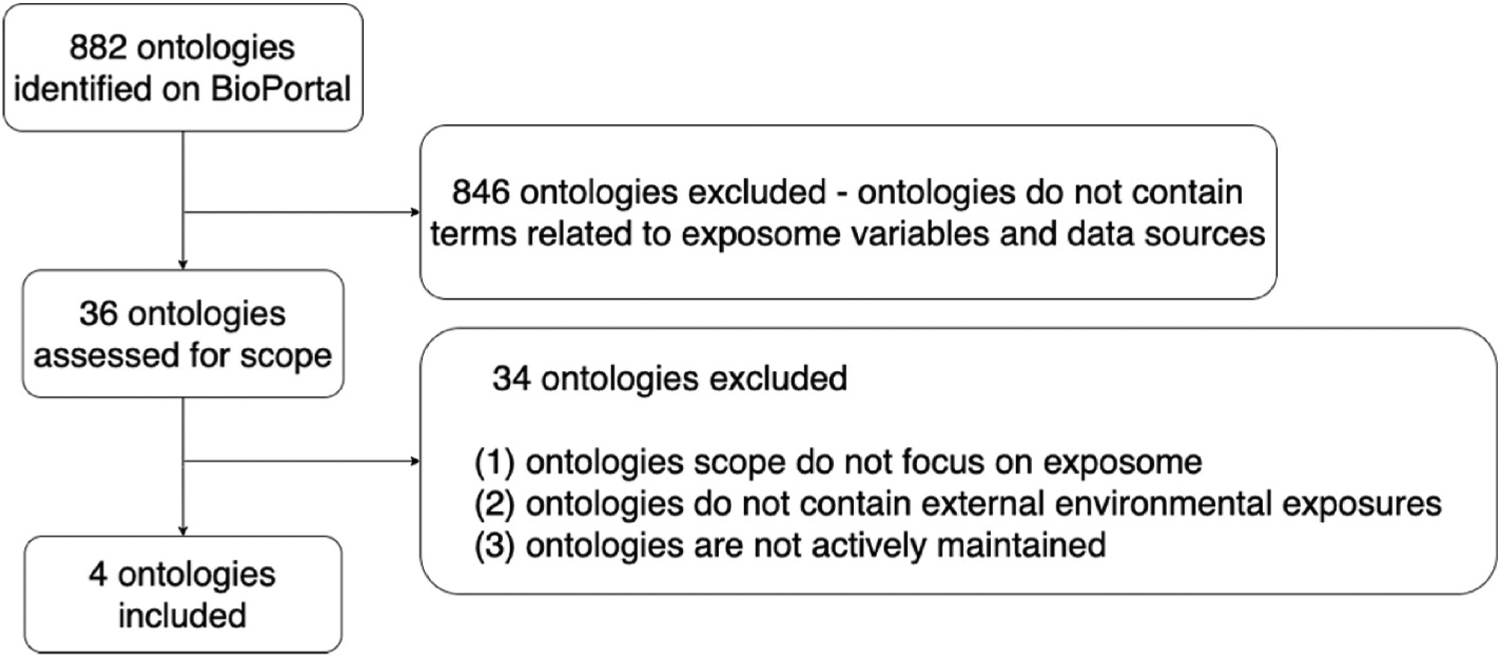
The screening process of ontology designed for the external exposome.
We identified only 4 ontologies that met our inclusion criteria: the Environment Ontology (ENVO), the Human Health Exposure Analysis Resource (HHEAR) ontology, the Child Health Exposure Analysis Resource (CHEAR) ontology, and the Environment Conditions, Treatments, and Exposures Ontology (ECTO). In 2016, Pier et al. proposed ENVO (Buttigieg et al., 2013, 2016) to “promote standardization and interoperability of diverse data sets through the concise, controlled description of environment types across several levels of granularity.” However, as the stated domain of the ontology is not environmental exposures, the development of ENVO is not based on the established exposome framework (Wild, 2005, 2012), where the exposome framework can help ontology developers to structurally and systematically think of all potential external exposures that is important to facilitate the annotation and integration of external exposome data. As such, the coverage of the terms related to external exposome is limited. For example, terms related to the social environment are missing from ENVO. Therefore, when integrating external exposome data containing social environment variables (e.g., crime and safety), ENVO provides little to no support, despite having extensive representation of the built and natural environments. Further, many of the variables defined in ENVO lack the necessary granularity. For example, ENVO reused the classes such as “air pollution” and “soil pollution” from the Chemical Entities of Biological Interest (ChEBI) ontology (Degtyarenko et al., 2008), should have further classified those classes into more granular types of pollutants (e. g., Nitrogen oxide [NO] and Carbon dioxide [CO2]) to meet the needs of the exposome community.
In 2019, McGuinness et al. developed the HHEAR and CHEAR ontologies (Balshaw et al., 2017; McGuinness et al., 2019), which aimed to promote the characterization of the totality of environmental exposures adult humans and children. They followed the exposome framework and both cover a wide range of environmental exposures including chemical, physical, and biological stressors, as well as lifestyle and social environments, from conception through adulthood. However, exposures relevant to the natural environment (e.g., air toxicants and meteorology) and built environment (e.g., food environment and walkability) are still not well captured (listed in Table 1). Additionally, CHEAR and HHEAR make poor use of the is-a hierarchy that lends to the utility of representing hierarchically-structured knowledge in ontologies. For example, the term ‘entity’ (SIO:000000) is classified in both as a subclass of ‘ScienceIndicator’ (ns2:ScienceIndicator), despite having the definition “Every thing [sic] is an entity.” Doing so can not only lead to errors in reasoning (e.g., ‘Cement Floor’ [HHEAR:00269] would inherit the properties of ‘ScienceIndicator’ [ns2:ScienceIndicator]) and contradictions, which creates confusion for human users. The latter is further compounded when the user finds that there are two classes with the label of ‘entity’ (SIO:000000 and BFO:0000001) and a third with the label ‘Entity’ (prov:Entity). Issues like these, along with a lack of documentation or publication of the methods used in the development of both ontologies, fail to address many of the concerns that have plagued efforts for enabling semantic interoperability.
The fourth ontology we reviewed was the Environment Conditions, Treatments, and Exposures Ontology (ECTO) (Environmental conditions, 2021). The stated goal of ECTO, as an Open Biological and Biomedical Ontology (OBO) Foundry (Smith et al., 2007) library ontology, is to represent exposures to experimental treatments and environmental exposures. With nearly 12,000 classes—many of which are imported from ontologies such as the GO (Ashburner et al., 2000), Food Ontology (FOODON) (Dooley et al., 2018), ChEBI (Degtyarenko et al., 2008), and ENVO—it also contains extensive coverage of the external environment. Nevertheless, despite having a wide coverage of different factors belonging to the natural and built environments, there is no representation for exposure to infectious agents. There are also aspects of the social environment that are not represented in ECTO, such as level of education, access to education, exposure to crime, and access to social capital. Something else worth noting is ECTO’s reuse of both OBO Foundry library ontology classes and National Cancer Institute Thesaurus (NCIt) classes in defining its classes for exposure events. Because both sets of classes belong to disjoint parts of the ontology, object properties, like has-exposure-stimulus (RO:0002309), that relate an exposure stimulus to the individual experiencing the exposure are limited or prohibited from making use of Web Ontology Language (OWL) 2 object (W3Working Group., 2012) property range restrictions.
Further, the Exposure Ontology (ExO), is also an OBO Foundry library ontology that was designed for the purpose of supporting the integration of exposure data (Mattingly et al., 2012), but was excluded during our ontology screening process, because of its insufficient coverage of external exposome and has not been actively maintained with spotty updates. To that end, ExO contains 151 classes divided into five categories: “assay” (e.g., “biological marker”, “model”, etc. that have no apparent relationships), “exposure stressor”, “exposure receptor”, “exposure event” (as a subclass of “process”), and “exposure outcome”. Unfortunately, given the expansive nature of the exposome, this small number of classes suggests insufficient coverage of this domain (i.e., external exposome). Also, ExO does not make use of a top-level ontology (e.g., Basic Formal Ontology [BFO] (Arp et al., 2015), an implicit requirement for an OBO Foundry ontology), thus complicating efforts to reuse ExO classes in other ontologies, and vice versa.
In short, there is a great need to develop or further expand these semantic standards and ontologies to alleviate the heterogeneity across different data sources and facilitate data integration efforts. Although there are several ontologies that offer wide coverages of the natural, built, and social environments, there are noticeable gaps in each ontology we reviewed.
Further, external environmental exposures are closely related to spatiotemporal information (e.g., “Ambient concentration of QUINOLINE, during the first two trimesters”, “Soil Temperature at 0 cm subsurface level, during the first trimester”, and “Low food access tract at 1 mile for urban areas and 10 miles for rural areas”). However, none of the 4 included ontologies above contain enough details to represent exposures with the necessary spatiotemporal information. Future work thus should also focus on spatiotemporal constraints to better represent external environmental exposures.
5. Ways forward – ontology-based standardization of external exposome data
In this paper, we organized the definition of the exposome into two categories based on the exposome framework and revealed that no existing ontology currently provides comprehensive representations for the exposome data, especially for the external exposome. To fill this gap, the community needs to (1) develop new ontological resources systematically following the exposome framework to ensure the coverage of external environmental exposures (2) reuse and integrate existing ontologies (e.g., ENVO and ECTO); to ensure interoperability, and (3) create new classes to represent external environmental exposures with finer granularity in order to better model the complexity of the external exposome data, as well as many other best practice in ontology development for environmental health sciences (Mattingly et al., 2016).
Nevertheless, there are also challenges in creating these new ontological resources. For example, as shown in Fig. 1, the concept of “education” can be either a specific or general external exposome, depending on the perspective. As the integrated conceptual framework of the exposome (Fig. 1) looks at the problem mainly from the data collection perspective, when creating new classes (and categorizing the new exposures), other conceptual frameworks (e.g., the level of measure in the traditional socioecological model sense) can help determine the category of the exposome (e.g., individual level vs. community level education measures).
Another important goal of a comprehensive external exposome ontology should be to alleviate the semantic interoperability among the different types of data sources when integrating genome, phenome, and exposome data. The external exposome framework should not be considered in isolation from other conceptual frameworks (e.g., SDoH (Healthy People 2020. Heal, 2010), NIMHD Research Framework (Research Fra, 2019), and the socioecological model (Bronfenbrenner, 1977)). For example, the concept of SDoH has significant overlaps with the general external exposome, which both focus on the wider social, economic, and psychological influences on the individual. From a data perspective, these conceptual frameworks can help us to expand the existing exposome framework (Fig. 1) to better represent the external exposome data. For example, in the NIMHD Research Framework (i.e., an extension to the socioecological model), an individual is embedded within the larger social system and constrained by the physical environment they live in, within which health outcomes are influenced by exposures from different levels (i.e., individual, interpersonal, community, and societal) and domains (i.e., biological, behavioral, physical/built environment, sociocultural environment, and healthcare system). Considering the external environmental exposures in the NIMHD framework is helpful for building semantic standards for the external exposome, especially when the external exposome data need to be integrated with phenome and genome data.
We believe that the combination of multi-domain and multi-scale data (i.e., genome, phenome and exposome at different granularity) and different conceptual frameworks is the basis of health outcomes research in the future. But only with ontology-based semantic standards, these conceptual frameworks can guide researchers to structurally gain a clear mental model of possible exposome factors, facilitate formulation/formalization of research questions, and help identify data integration needs and strategies. Nevertheless, these semantic standards need to have community agreement on the definition and categorization of the classes so that they can be adopted and refined under a community effort.
In conclusion, to fully consider the external exposome data in health outcomes research, we must first overcome the unique data challenges, especially the heterogeneous syntax, schema, and semantics of the data coming from different data domains and sources. Our recommendations are:
Semantic standards for external exposome data should be established, and ontology is the current state-of-the-art for developing such standards in biological and biomedical sciences.
We need (1) community (from environmental sciences to biomedical informatics to ontology development) efforts to develop ontological resources, and (2) standardized ontology evaluation methods and guidelines for quality assurance.
When doing so, besides following the best practice in ontology development (e.g., reuse and integrate existing ontologies), researchers need to create new classes for external environmental exposures with finer granularity to increase coverages of the external exposome ontologies.
It is important to leverage the computational (i.e., make ontology understandable to not just human but also computers) and reasoning power of the ontology for categorizing, querying, and integrating large amounts of exposome data.
Not only exposome data (and their metadata) could be standardized using ontologies; it is also important to clearly document and standardize the data collection and data management processes (potentially using ontologies) (Zhang et al., 2020).
Last but not least, to increase the semantic interpretability when integrating external exposome data with other human health data-sets (e.g., genome and phenome), conceptual frameworks need to be considered for building these semantic standards.
Acknowledgments
This work was supported in part by National Institute of Environmental Health Sciences (NIEHS), United States grant R21ES032762, National Center for Advancing Translational Sciences (NCATS), United States grant UL1TR001427 and the Cancer Informatics Shared Resource (CISR) at the University of Florida Health Cancer Center (UFHCC), United States. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Funding sources
This work was supported in part by NIH grants R21 ES 032762-01 and UL1TR001427 and the Cancer Informatics Shared Resource at the University of Florida Health Cancer Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Amith M, He Z, Bian J, Lossio-Ventura JA, Tao C, 2018. Assessing the practice of biomedical ontology evaluation: gaps and opportunities. J. Biomed. Inf 80, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arp R, Smith B, Spear AD, 2015. Building Ontologies with Basic Formal Ontology. Massachusetts Institute of Technology, Cambridge, Massachusetts. [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. , 2000. Gene Ontology: tool for the unification of biology. Nat. Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balshaw DM, Collman GW, Gray KA, Thompson CL, 2017. The Childrenʼs Health Exposure Analysis Resource: enabling research into the environmental influences on childrenʼs health outcomes. Curr. Opin. Pediatr 29, 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Huang C-C, Rubin CL, 2017. The role of community education in increasing knowledge of breast health and cancer: findings from the Asian breast cancer project in boston, Massachusetts. J. Canc. Educ 32, 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Bult CJ, 2006. Beyond the data deluge: data integration and bio-ontologies. J. Biomed. Inf 39, 314–320. [DOI] [PubMed] [Google Scholar]
- Bronfenbrenner U, 1977. Toward an experimental ecology of human development. Am. Psychol 32, 513–531. [Google Scholar]
- Buttigieg P, Morrison N, Smith B, Mungall CJ, Lewis SE, the ENVO Consortium, 2013. The environment ontology: contextualising biological and biomedical entities. J. Biomed. Semant 4, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttigieg PL, Pafilis E, Lewis SE, Schildhauer MP, Walls RL, Mungall CJ, 2016. The environment ontology in 2016: bridging domains with increased scope, semantic density, and interoperation. J. Biomed. Semant 7, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr LJ, Dunsiger SI, Marcus BH, 2010. Walk Score™ as a global estimate of neighborhood walkability. Am. J. Prev. Med 39, 460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr LJ, Dunsiger SI, Marcus BH, 2011. Validation of Walk Score for estimating access to walkable amenities. Br. J. Sports Med 45, 1144–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh Yazdi M, Wang Y, Di Q, Wei Y, Requia WJ, Shi L, et al. , 2021. Long-term association of air pollution and hospital admissions among medicare participants using a doubly robust additive model. Circulation 120, 050252. CIRCULATIONAHA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, et al. , 2008. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. D344–D350, 36 Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley DM, Griffiths EJ, Gosal GS, Buttigieg PL, Hoehndorf R, Lange MC, et al. , 2018. FoodOn: a harmonized food ontology to increase global food traceability, quality control and data integration. Npj Sci Food 2, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Environmental conditions, treatments and exposures ontology (ECTO). https://github.com/EnvironmentOntology/environmental-exposure-ontology. (Accessed 16 March 2021). [DOI] [PMC free article] [PubMed]
- Garvin E, Branas C, Keddem S, Sellman J, Cannuscio C, 2013. More than just an eyesore: local insights and solutions on vacant land and urban health. J. Urban Health 90, 412–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium, 2001. Creating the gene ontology resource: design and implementation. Genome Res. 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad N, Andrianou XD, Makris KC, 2019. A scoping review on the characteristics of human exposome studies. Curr Pollut Rep 5, 378–393. [Google Scholar]
- Health and environment-wide associations based on large population Surveys. http://www.heals-eu.eu/index.php/welcome-letter/. (Accessed 16 March 2021).
- Healthy People 2020. Healthy People 2020: an Opportunity to Address Societal Determinants of Health in the United States, 2010. https://www.healthypeople.gov/2010/hp2020/advisory/SocietalDeterminantsHealth.htm. (Accessed 13 July 2020).
- Hu H, Zhao J, Savitz DA, Prosperi M, Zheng Y, Pearson TA, 2020. An external exposome-wide association study of hypertensive disorders of pregnancy. Environ. Int 141, 105797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Zheng Y, Wen X, Smith SS, Nizomov J, Fishe J, et al. , 2021. An external exposome-wide association study of COVID-19 mortality in the United States. Sci. Total Environ 768, 144832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA, 2014. Air Quality Data for the Cdc National Environmental Public Health Tracking Network. https://archive.epa.gov/epa/aboutepa/about-national-exposure-research-laboratory-nerl.html. (Accessed 28 October 2020).
- Köhler S, Vasilevsky NA, Engelstad M, Foster E, McMurry J, Aymé S, et al. , 2017. The human phenotype ontology in 2017. Nucleic Acids Res. 45, D865–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue JM, Small MJ, Robinson AL, 2011. Evaluating the national air toxics assessment (NATA): comparison of predicted and measured air toxics concentrations, risks, and sources in Pittsburgh, Pennsylvania. Atmos. Environ 45, 476–484. [Google Scholar]
- Loh M, Sarigiannis D, Gotti A, Karakitsios S, Pronk A, Kuijpers E, et al. , 2017. How sensors might help define the external exposome. Int. J. Environ. Res. Publ. Health 14, 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Addington LA (Eds.), 2007. Understanding Crime Statistics: Revisiting the Divergence of the NCVS and UCR. Cambridge University Press, New York. [Google Scholar]
- Mann JK, Balmes JR, Bruckner TA, Mortimer KM, Margolis HG, Pratt B, et al. , 2010. Short-term effects of air pollution on wheeze in asthmatic children in fresno, California. Environ. Health Perspect 118, 1497–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin Sanchez F, Gray K, Bellazzi R, Lopez-Campos G, 2014. Exposome informatics: considerations for the design of future biomedical research information systems. J. Am. Med. Inf. Assoc 21, 386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather M, Rivers KL, Jacobsen LA, 2005. American Community Survey (ACS). Population Bulletins. [Google Scholar]
- Mattingly CJ, McKone TE, Callahan MA, Blake JA, Hubal EAC, 2012. Providing the missing link: the exposure science ontology ExO. Environ. Sci. Technol 46, 3046–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly CJ, Boyles R, Lawler CP, Haugen AC, Dearry A, Haendel M, 2016. Laying a community-based foundation for data-driven semantic standards in environmental health sciences. Environ. Health Perspect 124, 1136–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness Deborah, Pinheiro Paulo, McCusker James, 2019. Human Health Exposure Analysis Resource. https://bioportal.bioontology.org/ontologies/HHEAR. (Accessed 9 August 2020).
- Meng Y-Y, Rull RP, Wilhelm M, Lombardi C, Balmes J, Ritz B, 2010. Outdoor air pollution and uncontrolled asthma in the San Joaquin Valley, California. J. Epidemiol. Community Health 64, 142–147. [DOI] [PubMed] [Google Scholar]
- National Library of Medicine. NIH CDE Repository. https://cde.nlm.nih.gov/cde/search. (Accessed 25 July 2020).
- NCEP North American Regional Reanalysis (NARR), 2005. Research Data Archive at the National Center for Atmospheric Research, Computational and Information Systems Laboratory, Boulder CO. https://rda.ucar.edu/datasets/ds608.0/.
- Ochs C, Perl Y, Geller J, Arabandi S, Tudorache T, Musen MA, 2017. An empirical analysis of ontology reuse in BioPortal. J. Biomed. Inf 71, 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyana T, Matthews-Juarez P, Cormier S, Xu X, Juarez P, 2015. Using an external exposome framework to examine pregnancy-related morbidities and mortalities: implications for health disparities research. Int. J. Environ. Res. Publ. Health 13, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raknes Solfrid, 2018. Anxious Adolescents: Prevalence, Correlates, and Preventive Cognitive Behavioural Interventions. University of Bergen. [Google Scholar]
- NIMHD., NIMHD. Research framework. https://www.nimhd.nih.gov/about/overview/research-framework.html. (Accessed 28 June 2019).
- Rhew IC, Vander Stoep A, Kearney A, Smith NL, Dunbar MD, 2011. Validation of the normalized difference vegetation index as a measure of neighborhood greenness. Ann. Epidemiol 21, 946–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupasingha A, Goetz SJ, Freshwater D, 2006. The production of social capital in US counties. J Socio-Econ 35, 83–101. [Google Scholar]
- Schulz S, Jansen L, 2013. Formal ontologies in biomedical knowledge representation. Yearb Med Inform 8, 132–146. [PubMed] [Google Scholar]
- Smith B, Ceusters W, 2010. Ontological realism: a methodology for coordinated evolution of scientific ontologies. Appl. Ontol 5, 139–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The OBI Consortium, Smith B, Ashburner M, Rosse C, Bard J, Bug W, et al. , 2007. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol 25, 1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The National Institute for Occupational Safety and Health (NIOSH). Exposome and exposomics. https://www.cdc.gov/niosh/topics/exposome/default.html. (Accessed 21 January 2020).
- Turner MC, Nieuwenhuijsen M, Anderson K, Balshaw D, Cui Y, Dunton G, et al. , 2017. Assessing the exposome with external measures: commentary on the state of the science and research recommendations. Annu. Rev. Publ. Health 38, 215–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- on behalf of the EXPOsOMICS Consortium, Turner MC, Vineis P, Seleiro E, Dijmarescu M, Balshaw D, et al. , 2018. EXPOsOMICS: final policy workshop and stakeholder consultation. BMC Publ. Health 18, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Department of Agriculture, 2019. Food Environment Atlas. https://www.ers.usda.gov/foodatlas/. (Accessed 11 May 2020).
- Vineis P, Chadeau-Hyam M, Gmuender H, Gulliver J, Herceg Z, Kleinjans J, et al. , 2017. The exposome in practice: design of the EXPOsOMICS project. Int. J. Hyg Environ. Health 220, 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vineis P, Robinson O, Chadeau-Hyam M, Dehghan A, Mudway I, Dagnino S, 2020. What is new in the exposome? Environ. Int 143, 105887. [DOI] [PubMed] [Google Scholar]
- Vrijheid M, Slama R, Robinson O, Chatzi L, Coen M, van den Hazel P, et al. , 2014. The human early-life exposome (HELIX): project rationale and design. Environ. Health Perspect 122, 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- W3C OWL Working Group, 2012. OWL 2 Web Ontology Language Document Overview, second ed. https://www.w3.org/TR/2012/REC-owl2-conformance-20121211/. (Accessed 25 March 2021).
- Wache H, Vögele T, Visser U, Stuckenschmidt H, Schuster G, Neumann H, et al. , 2001. Ontology-Based Integration of Information - A Survey of Existing Approaches, pp. 108–117.
- Weatherall D, 1999. From genotype to phenotype: genetics and medical practice in the new millennium. Philos. Trans. R. Soc. Lond. B Biol. Sci 354, 1995–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, et al. , 2011. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 39 (Suppl. l). W541–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White Hourse, The, 2015. THE PRECISION MEDICINE INITIATIVE. https://obamawhitehouse.archives.gov/precision-medicine. (Accessed 10 May 2020).
- Wild CP, 2005. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol 14, 1847–1850. [DOI] [PubMed] [Google Scholar]
- Wild CP, 2009. Environmental exposure measurement in cancer epidemiology. Mutagenesis 24, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild CP, 2012. The exposome: from concept to utility. Int. J. Epidemiol 41, 24–32. [DOI] [PubMed] [Google Scholar]
- World Health Organization, 2016. Preventing Disease through Healthy Environments: a Global Assessment of the Burden of Disease from Environmental Risks. https://www.who.int/quantifying_ehimpacts/publications/preventing-disease/en/. (Accessed 20 January 2020).
- Zhang H, Guo Y, Prosperi M, Bian J, 2020. An ontology-based documentation of data discovery and integration process in cancer outcomes research. BMC Med. Inf. Decis. Making 20, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Chen Z, Pearson T, Zhao J, Hu H, Prosperi M, 2020. Design and methodology challenges of environment-wide association studies: a systematic review. Environ. Res 183, 109275. [DOI] [PMC free article] [PubMed] [Google Scholar]


