Abstract
Background
Colistin is classified as the highest priority and critically important antimicrobial for human medicine by WHO as it is the last resort agent for treatment of carbapenem-resistant Enterobacteriaceae in humans. Additional research is necessary to elucidate the genetic structure of mcr-1 resistance genes, commonly found on plasmids, using WGS.
Objectives
To map and compare the genetic characteristics of 35 mcr-1-mediated colistin-resistant Enterobacteriaceae isolated from chicken meat to highlight the genetic variation of the mcr-1-containing plasmids.
Methods
Sequencing was performed using Illumina HiSeq2500, Novaseq6000 and ONT’s GridION. GridION data was locally basecalled and demultiplexed using ONT’s Albacore 2.3.4 followed by Porechop 2.3. Quality filtering was performed using Filtlong 2.0. Hybrid Assembly was performed using Unicycler 4.7. Plasmids were compared with reference sequences in plasmid-RefSeq and pATLAS.
Results
A total of 35 mcr-1 positive Enterobacteriaceae were investigated, which resulted in 34 qualitatively robust hybrid assemblies of 2 Klebsiella pneumoniae and 32 Escherichia coli. mcr-1.1 was present in 33/34 isolates. One isolate contained an mcr-1.1-like resistance gene, due to a deletion of one codon. Two mcr-1.1 genes were located on the chromosome, while the majority of the mcr-1 genes were found on IncX4 type plasmids (n = 19). Almost all plasmids identified in this study were highly similar to plasmids found in human-derived strains.
Conclusions
The mcr-1.1-containing plasmids from retail chicken show high sequence similarity to human mcr-1.1 plasmids, suggesting that this may be a contributor to the presence of colistin resistance in humans.
Introduction
In 2015, a plasmid-mediated colistin resistance gene was reported in China.1 From that moment on, many more mobile colistin resistance (mcr) genes and variants have been detected all over the globe.2 This discovery represents a mechanism for an easy transferable resistance mechanism to colistin, which is seen as a last-resort antibiotic to treat carbapenem-resistant Enterobacteriaceae.3 In Europe, colistin is used to treat infections caused by Enterobacteriaceae in sheep, cows, pigs, goats and chicken.4 Therefore, the detection of mcr-1-harbouring Enterobacteriaceae isolates in chicken meat was self-evident.5,6
In order to understand the molecular epidemiology and resistance mechanism of mcr genes, WGS approaches should be used. Characteristically, high-throughput sequencing platforms (e.g. Illumina) are used in order to sequence the full bacterial genome.7 However, short reads from these high-throughput sequencers can make it challenging to reconstruct plasmids and therefore they are inaccurate for studying antibiotic resistance epidemiology.8 Single-molecule sequencing platforms such as the Oxford Nanopore Technologies (ONT) MinION, GridION and PromethION are able to sequence long fragments of DNA. Subsequently, with the use of a hybrid assembly, increased information content can be generated since the genome completeness is increased and the location of resistance genes in the genome can be determined.9
In this study, short- and long-read sequencing platforms were used in order to study the mcr-1-containing Enterobacteriaceae isolated from retail chicken meat.5,6 We used a hybrid-assembly approach to extract the plasmid sequences that contain mcr-1 and studied the plasmid relationship compared with publicly available mcr-1 plasmid sequences.
Methods
Sample collection
In total, 35 confirmed mcr-1-holding Enterobacteriaceae were subjected to Illumina short read and ONT sequencing. The isolates derived from previous studies,5,6 with the exception of EC-MCR34. All samples derived from three prevalence surveys in Dutch retail chicken meat performed in 2009, 2014 and 2015, which were initially performed to study the presence of ESBL-producing Enterobacteriaceae.10,11 The isolates in this study were genotypical mcr-1 PCR positive and phenotypical colistin resistant.5
Illumina sequencing
The 35 samples were sequenced using paired-end Illumina HiSeq2500.
The library prep for 35 samples was performed using the Nextera XT DNA library prep kit and the Nextera XT Index Kit v2 (Illumina, Eindhoven, The Netherlands), according to the manufacturer’s instructions. Libraries were subsequently purified using Agencourt AMPure XP beads (Beckman Coulter, Woerden, The Netherlands) and quantified using the Quant-iT dsDNA HS-kit (Thermo Fisher, Bleiswijk, The Netherlands) and using a Fragment Analyzer (Agilent, Amstelveen, The Netherlands.) Samples were then loaded on a HiSeq2500 system and run for 251 cycles (PE125) using HiSeq Rapid SBS Kit v2 chemistry.
Due to low quality, EC-MCR10 and EC-MCR21 were re-sequenced using the Illumina NovaSeq 6000 The library prep for these two samples was performed using the Nextera XT DNA library prep kit and the IDT for Illumina Nextera DNA Unique Dual Indexes (Illumina), according to the manufacturer’s instructions. Libraries were subsequently purified using Agencourt AMPure XP beads (Beckman Coulter) and quantified using the Quant-iT dsDNA HS-kit (Thermo Fisher) and using a Fragment Analyzer (Agilent). Samples were then loaded on an S1 flow cell on the NovaSeq6000 system and run for 301 cycles (PE150).
Fastq read sequence files were generated using bcl2fastq2 version 2.18. Initial quality assessment was based on data passing the Illumina Chastity filtering. Subsequently, reads containing PhiX control signal were removed using an in-house filtering protocol. In addition, reads containing (partial) adapters were clipped (up to a minimum read length of 50 bp). The second quality assessment was based on the remaining reads using the FASTQC quality control tool version 0.11.5.
ONT sequencing
All 35 samples were sequenced using the ONT GridION (Oxford Nanopore Technologies, Oxford, UK). Libraries were prepared using shearing by needle shearing (KP-MCR01–02 and EC-MCR03–31) or using the Covaris G-tube (EC-MCR32–35). The library was prepared using the ONT 1D ligation sequencing kit (SQK-LSK109) with the native barcoding kit (EXP-NBD103). Samples KP-MCR01–02 and EC-MCR03–29 were loaded on FLO-MIN107 R9.5.1 flow cells and the remaining on a FLO-MIN106 R9.4.1 flow cell.
Sequence data availability
All data is available from the National Center for Biotechnology Information (NCBI) under BioProject number PRJEB44175. Raw short-read Illumina and long-read ONT sequencing data and metadata for all 35 isolates used in this study are available from the NCBI Sequence Read Archive database under accession numbers ERR5727763 to ERR5727797 (short read) and ERR5726838 to ERR5726872 (long read).
Assembly
GridION data were locally basecalled and demultiplexed using ONT’s Albacore 2.3.4 followed by Porechop 2.3 to demultiplex the unclassified reads. Quality filtering was performed using Filtlong 2.0 using the following settings: (i) maximum size of 500 Mbp; (ii) keep 90% percentage of the best reads of the data; and (iii) minimum size of 1000 bp. The long-read quality was evaluated using FastQC and NanoPlot v1.13.0 and the short-read quality using FastQC. Hybrid assembly was performed using Unicycler 4.7 using default settings and a minimum length of 1000 bp and subsequently assessed using QUAST 5.0.12 Genetic characterization of the hybrid assemblies was performed using the online service of goseqit.com.
The coverage of the ONT sequence reads was calculated by mapping the long reads back to the assembly using minimap2 (v2.13) and SAMtools (v1.9) using the in-house scripts. Sequence annotation was done using Bakta (v1.1).13
Plasmid analysis
The mcr-1 plasmid sequences were manually identified and extracted from the assembly graphs (.gfa files) using Bandage.14 The mcr-1 gene sequence (AKF16168.1) was used to locate the mcr-1 -containing plasmids. mcr-1 gene-containing plasmids from RefSeq plasmid database and pATLAS (accessed April 8, 2020) were retrieved.15 Any duplicates entries were removed prior subsequent analyses. In total 69 publicly available plasmids and mcr-1-containing plasmids from this study were used. Plasmid sequences were clustered using Plasmidsimilarity (v0.3.0, https://github.com/Casperjamin/Plasmidsimilarity). In short, dissimilarity among plasmids was calculated using the Jaccard index, using the complete k-mer composition (all subsequences in a sequence of length k) of each plasmid sequence, using k length of 31 bp. Antimicrobial resistance (AMR) genes, virulence genes and plasmid origin of replications were identified with Abricate (v1.0.1, default settings) using the NCBI, virulence factor database and PlasmidFinder database respectively (retrieved on 10 September 2019).16,17
Results and discussion
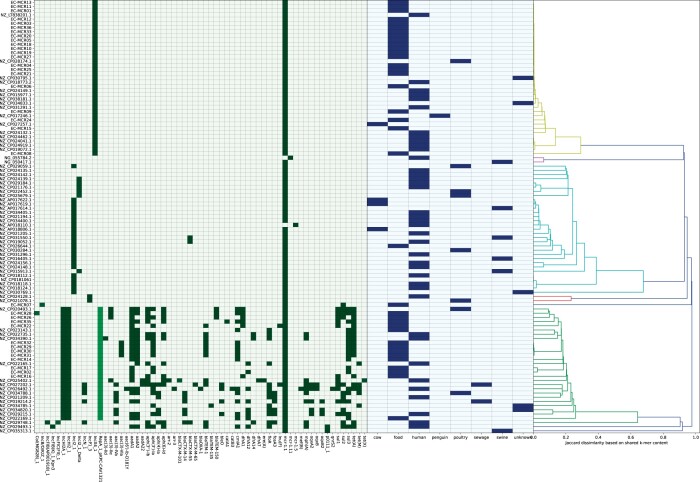
A total of 35 mcr-1-positive Enterobacteriaceae were investigated, which resulted in 34 qualitatively robust hybrid assemblies of 2 Klebsiella pneumoniae and 32 Escherichia coli isolates (Table S1, available as Supplementary data at JAC-AMR Online). The hybrid assembly substantially improved the reconstruction of the microbial genome (data not shown). The mcr-1.1 gene was present in 33/34 isolates (Table 1). The most common STs for E. coli were ST624 (n = 7), ST10 (n = 5) and ST997 (n = 4). The two K. pneumoniae isolates belonged to ST107 and ST1944. One isolate contained an mcr-1.1-like resistance gene, due to a mutation in the start codon, but still remained resistant to colistin.5 The second codon in mcr-1.1 is ATG and will likely replace the first codon as start codon, leading to a truncated but functional gene. Two mcr-1.1 genes were located on the chromosome, while the majority of the mcr-1 genes were found on IncX4 type plasmids (n = 19, Table 1), which is a common plasmid type harbouring mcr-1 found in Europe.18,19 Except for the IncX4, mcr1.1 plasmids, all mcr1.1 genes, plasmid or chromosomal, were flanked by IS30 transposases (Table 1). All the IncX4 mcr-1 plasmids shared, on average, 0.93 (standard deviation 0.08) of their k-mer content and did not contain any additional resistance genes. As a result, these plasmids were highly similar in size (average 33 kb, range 23 kb to 35 kb, Table 1). Additionally, the IncX4 plasmids found in this study were also highly similar to plasmids present in public databases, which originated from clinical isolates (Figure 1). Furthermore, the bacterial hosts of these IncX4 plasmids showed various STs (Table 1), indicating the widespread nature of this plasmid, most likely driven by conjugation. All IncX4 mcr1.1-containing plasmids carried a virB type IV secretion system, required for conjugation (data not shown). The AWGS0007 mcr1.1 plasmid (IncB/O/K/Z) encoded an IncI-1-type conjugal transfer protein TrbA. All other plasmids carried specific incompatibility group-associated conjugation machinery (data not shown). The IncHI2+IncHIA2 plasmids showed high k-mer similarity among each other (mean 0.72, standard deviation 0.12), but less than the IncX4 plasmids. These were generally much larger in size, ranging from 151 kb to 267 kb and additionally encoded a heterogenous set of AMR genes. It should be noted that no plasmids with an IncI origin of replication (ORI) containing mcr-1.1 were encountered in the strains in this study (Table 1, Figure 1) and only one IncI plasmid outside this study (NZ.CP02554.1) was derived from food origin.1
Table 1.
Overview of mcr-1-positive isolates with corresponding Inc type, size and other genetic characteristics
| Sample | Species | ST | Mcr type | Inc type on mcr1.1 plasmid | Other AMR genes | Transposase gene located near mcr1.1 | Contig no. | Contig size | Study reference |
|---|---|---|---|---|---|---|---|---|---|
| KP-MCR01 | K. pneumoniae | ST107 | 1.1 | IncX4 | — | — | 6 | 33 303 | 5 |
| KP-MCR02 | K. pneumoniae | ST1944 | 1.1 | IncHI2, IncHI2Aa | aph(3')-Ia a , sul3, aadA1a, dfrA12 | IS30-like element ISApl1 family transposase | 2 | 211 949 | 5 |
| EC-MCR03 | E. coli | ST10 | 1.1 | IncX4 | — | — | 7 | 33 303 | 5 |
| EC-MCR04 | E. coli | ST8262 | 1.1b | IncX4 | — | — | 5 | 33 310 | 5 |
| EC-MCR05 | E. coli | ST8262 | 1.1 | IncX4 | — | — | 6 | 33 310 | 5 |
| EC-MCR06 | E. coli | ST1564 | 1.1 | IncX4 | — | — | 6 | 33 303 | 5 |
| EC-MCR07 | E. coli | ST752 | 1.1 | IncB/O/K/Z | sul2 | IS30-like element ISApl1 family transposase | 5 | 93 122 | 5 |
| EC-MCR08 | E. coli | ST10 | 1.1 | IncX4 | — | — | 5 | 23 832 | 5 |
| EC-MCR09 | E. coli | ST162 | 1.1 | IncX4 | — | — | 5 | 35 016 | 5 |
| EC-MCR11 | E. coli | ST1842 | 1.1 | IncX4 | — | — | 3 | 33 310 | 5 |
| EC-MCR12 | E. coli | ST10 | 1.1 | IncX4 | — | — | 6 | 33 303 | 5 |
| EC-MCR13 | E. coli | ST641 | 1.1 | IncX4 | — | — | 7 | 33 303 | 5 |
| EC-MCR14 | E. coli | ST155 | 1.1 | IncHI2, IncHI2Aa | aadA2, cmlA1a, aadA1a, sul3 | IS30-like element ISApl1 family transposase | 2 | 243 755 | 5 |
| EC-MCR15 | E. coli | ST10 | 1.1 | IncX4 | — | — | 4 | 34 755 | 5 |
| EC-MCR16 | E. coli | ST997 | 1.1 | IncHI2, IncHI2Aa | tet(A), sul1, aadA1a, dfrA1a, aph(6)-Id, aph(3'')-Iba | IS30-like element ISApl1 family transposase | 2 | 214 156 | 5 |
| EC-MCR17 | E. coli | ST57 | 1.1 | IncHI2, IncHI2Aa | aadA1 a , sul3, aph(3')-Iaa | IS30-like element ISApl1 family transposase | 2 | 211 552 | 5 |
| EC-MCR18 | E. coli | ST997 | 1.1 | IncX4 | — | — | 5 | 33 310 | 5 |
| EC-MCR19 | E. coli | ST997 | 1.1 | IncX4 | — | — | 5 | 33 310 | 5 |
| EC-MCR20 | E. coli | ST624 | 1.1 | IncX4 | — | — | 5 | 33 310 | 5 |
| EC-MCR21 | E. coli | ST624 | 1.1 | IncX4 | — | — | 6 | 33 310 | 5 |
| EC-MCR22 | E. coli | ST10 | 1.1 | IncHI2, IncHI2Aa | bla TEM-1, tet(A), sul1, aadA1a, dfrA1, lnu(F), aph(3')-Ia | IS30-like element ISApl1 family transposase | 2 | 234 218 | 5 |
| EC-MCR23 | E. coli | ST93 | 1.1 | none | — | IS30-like element ISApl1 family transposase | 1 | chromosomal | 5 |
| EC-MCR24 | E. coli | ST48 | 1.1 | IncX4 | — | — | 6 | 34 639 | 5 |
| EC-MCR25 | E. coli | ST624 | 1.1 | IncX4 | — | — | 3 | 33 310 | 5 |
| EC-MCR26 | E. coli | ST997 | 1.1 | IncHI2, IncHI2Aa, IncQI | tet(A), sul1, aadA1a, dfrA1a, aph(6)-Idc, aph(3'')-Iba,c, sul2a, aph(3')-Ia, aac(3)-IIe, blaTEM-150a | IS30-like element ISApl1 family transposase | 2 | 267 214 | 5 |
| EC-MCR27 | E. coli | ST1011 | 1.1 | IncX4 | — | — | 8 | 33 310 | 5 |
| EC-MCR28 | E. coli | ST354 | 1.1 | IncHI2, IncHI2Aa, IncQI, Col(MG828) | tet(A)a, sul1, aadA1a,c, dfrA1a, aph(6)-Idc, aph(3'')-Iba,c, sul2a, aadA2, cmlA1a, sul3, aac(3)-IIe, blaTEM-150a | IS30-like element ISApl1 family transposase | 3 | 252 468 | 5 |
| EC-MCR29 | E. coli | ST624 | 1.1 | IncHI2, IncHI2Aa | cmlA1, aadA1a,c, sul3, aph(3')-Ia, blaTEM-1a, tet(A), aadA2, aac(3)-VIa | IS30-like element ISApl1 family transposase | 2 | 261 285 | 5 |
| EC-MCR30 | E. coli | ST624 | 1.1 | IncHI2, IncHI2Aa | cmlA1, aadA1c, sul3, aph(3')-Ia, blaTEM-1, tet(A), aadA2, aac(3)-VIa | IS30-like element ISApl1 family transposase | 2 | 261 102 | 5 |
| EC-MCR31 | E. coli | ST624 | 1.1 | IncHI2, IncHI2Aa | aadA2, cmlA1, aadA1c, sul3, aph(3')-Ia, blaTEM-1, tet(A), aac(3)-VIa | IS30-like element ISApl1 family transposase | 2 | 260 457 | 5 |
| EC-MCR32 | E. coli | ST624 | 1.1 | IncHI2, IncHI2Aa | cmlA1 a , aadA1a,c, sul3, aph(3')-Ia, blaTEM-1a, tet(A), aadA2, aac(3)-VIa | IS30-like element ISApl1 family transposase | 2 | 261 285 | 5 |
| EC-MCR33 | E. coli | ST1564 | 1.1 | IncX4 | — | — | 4 | 33 303 | 5 |
| EC-MCR34 | E. coli | ST117 | 1.1 | none | — | IS30-like element ISApl1 family transposase | 1 | chromosomal | |
| EC-MCR35 | E. coli | ST2079 | 1.1 | IncHI2, IncHI2Aa | tet(A), sul1, aadA1a,c, dfrA1a, aph(6)-Id, aph(3'')-Iba, sul3, cmlA1a, aadA2, catA1a | IS30-like element ISApl1 family transposase | 2 | 248 481 | 6 |
Identity or alignment length is not 100%.
Substitution in second base pair of first starting codon.
Resistance gene detected twice.
Figure 1.
Heatmap and dendrogram showing all plasmids analysed in this study. The dendrogram represents the similarity among plasmid sequences based on the Jaccard dissimilarity of 31-mers of each plasmid. Coloured cells in the heatmap indicate either the presence of this gene or the origin of replication of this plasmid.
None of the mcr-1 carrying plasmids in our study carried other genes encoding ESBL resistance. Three isolates contained resistance genes (TEM-52c, SHV-12 and CTX-M-1, respectively), however, these resistance genes were not present on the mcr-1.1 plasmid. Additionally, one mcr-1.1 plasmid encoded virulence factors, as it contained five genes of the aerobactin gene cluster (NZ.CP029748.1).
One novel mcr-1-containing plasmid was found (EC-MCR07) with a size of 93 kb, which also encoded the sulphonamide resistance gene sul2. This plasmid shared barely any sequence similarity as, on average, only a fraction of 0.02 (standard deviation 0.069) of all k-mers were shared with the other plasmids. This was the only plasmid with an IncB/O/K/Z ORI.
The two strains with a chromosomal mcr-1.1 gene (EC-MCR23 and EC-MCR34) had no other known resistance genes within the same chromosomal region (within 50 kb, data not shown), indicating the mobilization of colistin resistance as a sole passenger of its mobile genetic element IS30. We observed multiple different isolates from retail meat with similar plasmids, which might be caused by the spread of these plasmids within the farms or by individual introduction since these are common plasmids. In addition, similar isolates with identical plasmids are found, which could indicate a batch effect.
Conclusions
In this study we aimed to elucidate the plasmid backbones from mcr-1-containing plasmids obtained from retail chicken in the Netherlands. In the strains collected here, mcr-1 resided often but not always in various plasmids, indicating the high mobility nature of this gene in E. coli as a host. Most plasmid backbones found in this study were also found in human clinical isolates. This indicates the possibility of retail meat to be a significant contributor to the dissemination of mobile colistin resistance in the Netherlands.
Supplementary Material
Acknowledgements
We thank Carlo Verhulst for technical assistance and Pepijn Huizinga and Marjolein Kluytmans-van den Bergh for sample collection.
Funding
This work was funded by ZonMw for Enabling Technologies Hotels (Funding: 40-43500-98-15). Development of Plasmidsimilarity was made possible by a grant from the Dutch workgroup for molecular diagnostics of infectious diseases (WMDI).
Transparency declarations
None to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC-AMR Online.
References
- 1. Liu YY, Wang Y, Walsh TR. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16: 161–8. [DOI] [PubMed] [Google Scholar]
- 2. Sun J, Zhang H, Liu YH. et al. Towards understanding MCR-like colistin resistance. Trends Microbiol 2018; 26: 794–808. [DOI] [PubMed] [Google Scholar]
- 3. Poulakou G, Bassetti M, Righi E. et al. Current and future treatment options for infections caused by multidrug-resistant Gram-negative pathogens. Future Microbiol 2014; 9: 1053–69. [DOI] [PubMed] [Google Scholar]
- 4. Catry B, Cavaleri M, Baptiste K. et al. Use of colistin-containing products within the European Union and European Economic Area (EU/EEA): development of resistance in animals and possible impact on human and animal health. Int J Antimicrob Agents 2015; 46: 297–306. [DOI] [PubMed] [Google Scholar]
- 5. Schrauwen EJA, Huizinga P, van Spreuwel N. et al. High prevalence of the mcr-1 gene in retail chicken meat in the Netherlands in 2015. Antimicrob Resist Infect Control 2017; 6: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kluytmans-van den Bergh MF, Huizinga P, Bonten MJ. et al. Presence of mcr-1-positive Enterobacteriaceae in retail chicken meat but not in humans in the Netherlands since 2009. Euro Surveill 2016; 21: pii=30149. [DOI] [PubMed] [Google Scholar]
- 7. Perez-Losada M, Arenas M, Castro-Nallar E.. Microbial sequence typing in the genomic era. Infect Genet Evol 2018; 63: 346–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arredondo-Alonso S, Willems RJ, van Schaik W. et al. On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb Genom 2017; 3: e000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. George S, Pankhurst L, Hubbard A. et al. Resolving plasmid structures in Enterobacteriaceae using the MinION nanopore sequencer: assessment of MinION and MinION/Illumina hybrid data assembly approaches. Microb Genom 2017; 3: e000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kluytmans JA, Overdevest IT, Willemsen I. et al. Extended-spectrum β-lactamase-producing Escherichia coli from retail chicken meat and humans: comparison of strains, plasmids, resistance genes, and virulence factors. Clin Infect Dis 2013; 56: 478–87. [DOI] [PubMed] [Google Scholar]
- 11. Overdevest I, Willemsen I, Rijnsburger M. et al. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg Infect Dis 2011; 17: 1216–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wick RR, Judd LM, Gorrie CL. et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017; 13: e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Swengers O, Jelonk L, Dieckmann M. et al. Bakta: rapid & standardized annotation of bacterial genomes via alignment-free sequence identification. bioRxiv 2021; doi:10.1101/2021.09.02.458689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wick RR, Schultz MB, Zobel J. et al. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 2015; 31: 3350–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brooks L, Kaze M, Sistrom M.. A curated, comprehensive database of plasmid sequences. Microbiol Resour Announc 2019; 8: e01325-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carattoli A, Hasman H.. PlasmidFinder and in silico pMLST: identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol Biol 2020; 2075: 285–94. [DOI] [PubMed] [Google Scholar]
- 17. Chen L, Zheng D, Liu B. et al. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res 2016; 44: D694–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matamoros S, van Hattem JM, Arcilla MS. et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep 2017; 7: 15364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schuele L, Fleres G, Strutzberg-Minder K. et al. Detection of a small IncX4 plasmid carrying the mcr-1.1 gene in a pig oral fluid sample by shotgun metagenomic sequencing. J Glob Antimicrob Resist 2021; 24: 205–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.