The mucus cocoon of terrestrialized African lungfish is a living tissue with potent antimicrobial functions.
Abstract
Terrestrialization is an extreme physiological adaptation by which African lungfish survive dry seasons. For months and up to several years, lungfish live inside a dry mucus cocoon that protects them from desiccation. Light and electron microscopy reveal that the lungfish cocoon is a living tissue that traps bacteria. Transcriptomic analyses identify a global state of inflammation in the terrestrialized lungfish skin characterized by granulocyte recruitment. Recruited granulocytes transmigrate into the cocoon where they release extracellular traps. In vivo DNase I surface spraying during terrestrialization results in dysbiosis, septicemia, skin wounds, and hemorrhages. Thus, lungfish have evolved unique immunological adaptations to protect their bodies from infection for extended periods of time while living on land. Trapping bacteria outside their bodies may benefit estivating vertebrates that undergo metabolic torpor.
INTRODUCTION
Transitioning from water to land was a critical step in vertebrate evolution. African lungfish (Protopterus sp.), the extant relative to all tetrapods, have a dual mode of living, surviving in both aquatic and terrestrial environments (1, 2). Every year, the dry seasons in Africa limit the access to food and water to animals such as lungfish. Unfavorable environmental cues signal lungfish to begin the estivation process by which they curl up, reduce their metabolic activity, and produce copious amounts of mucus that will ultimately harden and form the cocoon (3). While dormancy allows survival during unfavorable periods, it drastically increases vulnerability to predators and pathogens. Estivation is a physiological adaptation that requires extensive tissue remodeling at mucosal barriers (4, 5), yet the involvement of the immune system in this process remains essentially unknown.
Almost a century ago, the investigation of the African lungfish immune system revealed unusually large deposits of granulocytes located in the gut, gonads, and kidneys of these animals (6). It was then proposed that these granulocytes may play a role in estivation (6), but these original observations have not been substantiated by molecular or functional studies. Here, we asked whether the lungfish immune system plays a role in estivation, protecting the skin from pathogen invasion during the vulnerable dormant state. Our results answer the long-standing question of why lungfish invest in maintaining large granulocyte reservoirs during free-swimming periods and reveal unique immunological innovations that allow lungfish to retain a dual mode of living in water and land.
RESULTS AND DISCUSSION
Tissue reservoirs supply granulocytes to the integument during terrestrialization
The presence of very large deposits of granulocytes associated with the gut, kidneys, and gonads of free-swimming lungfish was noted in the 1930s and hypothesized to play a role in terrestrialization (6). Although lungfish granulocytes are eosinophilic on the basis of traditional hematoxylin and eosin (H&E) stain, they are thought to be the functional equivalent of mammalian neutrophils because they are the most abundant myeloid cell in lungfish (6). Granulocyte migration to epithelial tissues is a hallmark of mucosal inflammation in mammals (7), and recruited granulocytes can transmigrate across epithelia and reach the lumen (7–9). To understand the role of lungfish granulocyte reservoirs during terrestrialization, we performed histological analysis on freshwater and lungfish gut and kidney that were terrestrialized for 2 weeks. Hematoxylin and eosin staining confirmed that the gut wall and the kidney are granulocyte-rich structures in freshwater lungfish (Fig. 1, A and C). In terrestrialized animals, reservoir tissues contained lower granulocyte numbers, increased pigment deposition thought to be granulocyte debris (6), and increased number of lymphatic micropumps (Fig. 1, B and D). Lymphatic micropumps are a key structure of the Dipnoi lymphatic system (10), and the observed changes during estivation may be due to changes in cardiovascular homeostasis or inflammation among other factors. We also noted that granulocytes entered circulation as a result of terrestrialization, as evidenced in the Giemsa-stained blood smears (Fig. 1E and fig. S1, A and B).
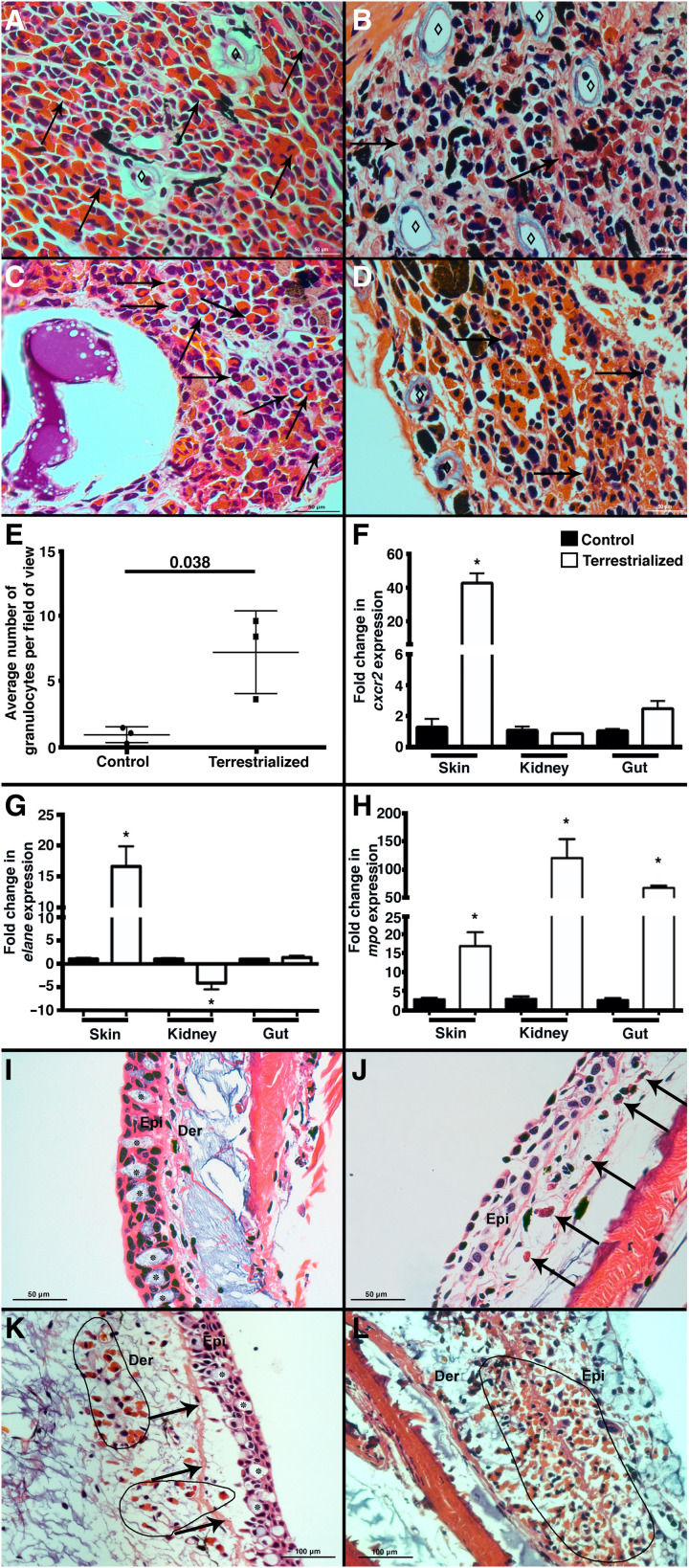
Fig. 1. Lungfish terrestrialization results in the mobilization of granulocytes from reservoir tissues into the integument.
H&E staining of (A) control gut, (B) terrestrialized gut, (C) control kidney, and (D) terrestrialized kidney tissues of African lungfish (n = 3 per group). Black arrows denote examples of granulocytes, and ⋄ denotes lymphatic micropumps. Note the increased abundance of brown and black pigment deposits in terrestrialized samples possibly corresponding to granulocyte debris and senescent granulocytes according to (6). (E) Quantification of granulocyte counts in control and terrestrialized lungfish blood smears (n = 3 animals per group, 10 fields were scored by two independent researchers). Quantification of (F) cxcr2, (G) elane, and (H) mpo mRNA levels by real-time quantitative polymerase chain reaction (RT-qPCR) in the skin, kidney, and gut of control (black bars) and terrestrialized lungfish (white bars) (n = 3). (I) H&E stain of control free-swimming lungfish skin. Asterisks indicate goblet cells. Note the columnar epithelial cells and intact basal membrane. (J) H&E stain of terrestrialized lungfish skin 2 weeks after terrestrialization showing complete terrestrialized features including absence of goblet cells, fully flattened epithelial cells, and a few granulocytes (black arrows). (K) H&E stain of terrestrialized lungfish skin 2 weeks after terrestrialization in the initial stages of tissue remodeling. Note the presence of goblet cells, the detachment of the epidermis from the basal membrane (black arrows), epithelial cells that have not yet flattened, and abundant granulocytes infiltrating the dermis (black circled area). (L) H&E stain of terrestrialized lungfish skin 2 weeks after terrestrialization in advanced stages of terrestrialization showing massive infiltration of granulocytes (black circled area) and severe edema. Epi, epidermis; Der, dermis (n = 3). Data were analyzed by unpaired Student’s t test. *P < 0.05.
We previously reported abundant granulocytes in the epidermis and dermis of terrestrialized animals compared to free-swimming controls (5). Here, granulocyte recruitment to the skin corresponded with increased mRNA expression of three granulocyte markers including C-X-C motif chemokine receptor 2 (cxcr2), myeloperoxidase (mpo), and neutrophil elastase (elane) in the terrestrialized skin (Fig. 1, F to H) (11–16). The cxcr2 gene encodes for the chemokine receptor CXCR2, a marker of neutrophils in mammals and whose increased expression matches neutrophil recruitment in infected and inflamed tissues (11, 12). The mpo product, myeloperoxidase (MPO), is a heme-containing peroxidase mainly expressed in neutrophils and critical for reactive oxygen species formation (13, 14). elane encodes for neutrophil elastase (ELANE), a protease that is released by granulocytes during degranulation and extracellular trap (ET) formation and activates proinflammatory cytokines (15, 16). Expression patterns of these three gene markers were very different in lungfish reservoir tissues. While cxcr2 expression was not significantly modified in kidney or gut, elane expression was significantly down-regulated (4-fold) in the kidney, and mpo expression was highly up-regulated in both gut and kidney (68-fold and 120-fold, respectively) of terrestrialized lungfish compared to free-swimming controls (Fig. 1H).
As previously reported (5), terrestrialization was accompanied by drastic remodeling of the integument of the lungfish. However, close examination of multiple histological sections from six different terrestrialized animals revealed heterogenous stages of skin remodeling (Fig. 1, I to L). Whereas some regions of the skin showed complete flattening of the epidermis and goblet cell exhaustion as we had previously documented (Fig. 1J), others were only partially terrestrialized, which gave us a view of the stepwise remodeling process that may occur in lungfish skin during terrestrialization. There were skin areas where the epidermis was only beginning to show some remodeling, including detachment of the basal membrane and invasion of few granulocytes into an otherwise intact epidermal layer (Fig. 1K). In other areas, the epidermis was in a much more advanced stage of inflammation, with granulocytes flooding both the dermis and the epidermis. Combined, these data indicate that free-swimming lungfish invest in large deposits of granulocytes that are mobilized to barrier tissues such as the skin upon terrestrialization. Whereas our histological observations suggest a stepwise process of skin remodeling starting by granulocyte recruitment to the dermis, transmigration to the epidermis, detachment of the basal membrane, and inflammation of the epidermis [modeled in Fig. 2 (A and B)], further studies are required to ascertain this sequence of events.
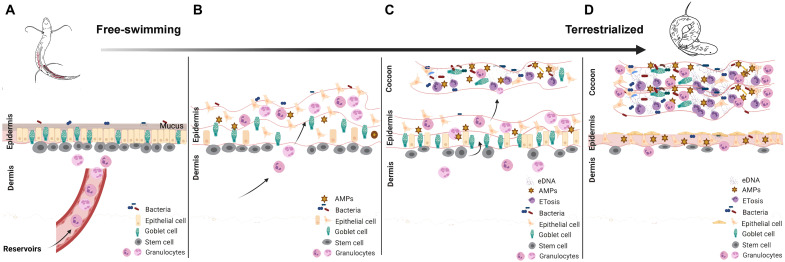
Fig. 2. Graphic drawing showing a proposed model for cocoon formation in African lungfish.
Free-swimming lungfish skin is characterized by a columnar mucosal epithelium. (A) Large numbers of multipotent stem cells with alkaline phosphatase activity (fig. S1) can be observed at the interphase between the epidermis and the dermis. Granulocyte deposits in the tissue reservoirs of free-swimming lungfish become mobilized to the skin via peripheral circulation when lungfish begin to sense lack of food and water. (B) Skin remodeling begins, with increasing numbers of granulocytes infiltrating the dermal and epidermal layers resulting in loosening of the basal membrane and inflammation. (C) The cocoon then starts to form by detachment and shedding of the inflamed epidermis. Many granulocytes are part of the cocoon, and they produce ETs in response to the high microbial load. Epithelial cells, goblet cells, and antimicrobial peptides (AMPs) are also present in the cocoon. The pool of stem cells starts to regenerate the epidermis, while granulocytes continue to arrive from reservoirs maintaining an inflammatory state. (D) In the next stages of estivation (end of the induction phase), the lungfish skin shows complete flattening of the epidermis and goblet cell exhaustion. The cocoon has several layers derived from multiple rounds of epidermal shedding and regeneration, and stem cell numbers are severely reduced. Granulocytes in the cocoon continue to undergo ETosis and are still elevated in the epidermis and dermis compared to free-swimming controls. It is unknown whether the cocoon continues to thicken beyond 2 weeks after terrestrialization. This illustration was created in BioRender. eDNA, extracellular DNA.
The mucus cocoon of terrestrialized lungfish is a living tissue that prevents bacteria invasion
Estivating animals such as lungfish and amphibians form cocoon structures to reduce evaporative water loss in the dry season (17–19). African lungfish live inside their mucus cocoon for months or even years (20). Previous studies described the cocoon of anuran and urodele amphibians as shed epithelial layers, whereas the lungfish cocoon was described as a translucent brownish dried mucus structure (17, 19, 20–23). Estivating lungfish are likely more vulnerable to pathogen attack since they are immobile and continuous skin mucus secretion stops once the cocoon is formed. Thus, we hypothesized that the cocoon may have important immunological functions thus far undescribed. Our histological observations revealed that the lungfish cocoon is not a dead, dry mucus layer but, instead, is a living tissue with a well-defined cellular structure. The thickness of the lungfish cocoon was approximately 350 to 900 μm, although this may vary from body site to body site, and the distance between the epidermis and the cocoon ranged from 15 to 173 μm (Fig. 3A). The cocoon was composed of different cell types including goblet cells, epithelial cells, endothelial cells, and immune cells (Fig. 3B). Transmission electron microscopy confirmed the presence of goblet cells with many secretory mucus granules within the cocoon (fig. S3D). Moreover, abundant nests of cells were observed surrounded by long channels lined by double-layered squamous cells. A series of monolayered vessels running parallel and perpendicular to the main channel were identified. These nests of cells contained endothelium-lined vessels that could potentially serve to transport cells into and within the cocoon (Fig. 3B). Channels were not filled with mucosal secretions based on periodic acid–Schiff staining (Fig. 3C). The cocoon contained abundant granulocytic cells (fig. S2A) that were MPO+, indicating transmigration of granulocytes into the cocoon [fig. S2B; modeled in Fig. 2 (C and D)]. Scanning electron microscopy (SEM) showed that during terrestrialization, the flattened smooth epidermis of freshwater fish (Fig. 3E) became a disrupted surface covered by the cocoon where lungfish cells and groups of bacterial cells were observed (Fig. 3F and fig. S2C).
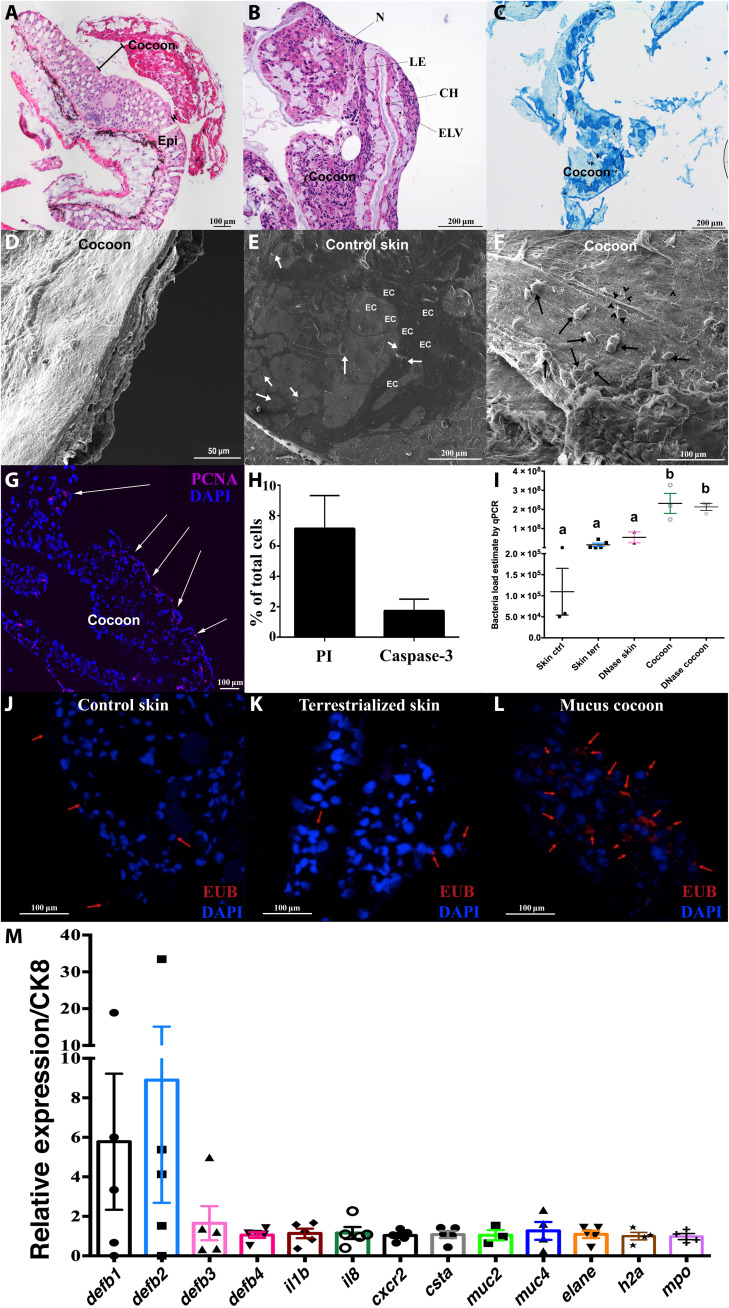
Fig. 3. The lungfish mucus cocoon is a living cellular structure that traps bacteria.
(A) H&E staining of lungfish cocoon (large image composite). (B) Magnified view of an H&E-stained cocoon. N, nest of cells; CH, channels; IC, intercommunicating channels; LE, loss of eosinophilia of nest cells; ELV, endothelium lined vessels. (C) Periodic acid–Schiff staining of lungfish cocoon showing goblet cells (dark blue) and mucus but no positive staining in the channels (n = 3). (D) SEM image of lungfish cocoon cross section showing uneven cellular layers. (E) Coronal SEM image of control lungfish skin showing polygonal epithelial cell (EC) and goblet cell openings (white arrows). (F) Coronal SEM image of cocoon showing lungfish cells (black arrows) and bacteria (black arrow heads). (G) PCNA staining (magenta) of a cocoon cryosection showing proliferating cells at the edge (white arrows). Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; blue). (H) Percent PI+ and activated caspase-3+ cells in cocoon single-cell suspensions (n = 3). (I) Relative quantification of bacterial loads by qPCR in control skin, terrestrialized skin, and mucus cocoon (n = 3 to 4). Different letters indicate statistically significant differences [one-way analysis of variance (ANOVA) and Tukey’s multiple comparison test]. (J to L) Maximum projection of confocal fluorescence images of fluorescence in situ hybridization (FISH) using EUB338 oligoprobe (red) showing bacteria (red arrows) in control skin, terrestrialized skin, and mucus cocoon (n = 3). (M) Gene expression of antimicrobial peptide genes (defb1 to defb4), proinflammatory cytokines (il1b and il8), cystatin A (csta), mucins (muc2 and muc4), granulocyte markers (cxcr2, mpo, and elane), and h2a in the lungfish cocoon (n = 5) by RT-qPCR. Cytokeratin 8 (ck8) was used as house-keeping gene.
Amphibian cocoons form by successive shedding of epithelial layers, resulting in a tightly packed, lamellated structure made up by several dozen layers of cornified cells (17, 21–23). Cornification is a terminal differentiation process and a form of programmed cell death (24) that occurs in amphibian but not in fish skin (25). Uneven cellular layers were observed by SEM on lungfish cocoon cross sections (Fig. 3D), supporting the idea that lungfish form their cocoon by successive shedding of inflamed epidermal layers that regenerate multiple times. In support of this idea, staining of lungfish skin cryosections for alkaline phosphatase activity, a marker of multipotent stem cells (26), showed a large pool of stem cells in the dermis of freshwater lungfish, which was significantly reduced in terrestrialized animals [fig. S1, C to E; modeled in Fig. 2 (A to D)]. Multipotent stem cells with embryonic characteristics have been identified in the mammalian dermis before (27) but, to our knowledge, have not been documented in lungfish. Furthermore, staining with anti–proliferating cell nuclear antigen (PCNA) antibody revealed active proliferation of cells with flattened morphology located in the outer edge of the cocoon (Fig. 3G). Single-cell suspensions of mucus cocoon samples stained with propidium iodide (PI) or activated caspase-3 confirmed that the cocoon is composed of viable cells with only 7% cells being PI+ and 2% being activated caspase-3+ (Fig. 3H). Combined, these data indicate that the lungfish cocoon is a granulocyte-rich, living structure and that lungfish cocoons are sharply different from amphibian cocoons.
Previous studies on amphibian cocoons noted the presence of bacteria both on the outer surface and the deeper layers of the cocoon (21). Our SEM and transmission electron microscopy observations led us to hypothesize that the lungfish cocoon serves as a protective barrier against bacteria during estivation. Relative quantification of bacterial loads by quantitative polymerase chain reaction (qPCR) showed that the cocoon contained approximately three orders of magnitude higher bacterial loads than the control and terrestrialized skin samples (Fig. 3I). In support, fluorescence in situ hybridization (FISH) using universal eubacterial oligoprobes (EUB338) revealed few bacterial cells in the skin of freshwater lungfish or in the skin of terrestrialized animals (Fig. 3, J and K). In contrast, large clumps of bacteria were present in the cocoon (Fig. 3L and fig. S2E). Higher bacterial loads in the cocoon may be the result of skin commensals accumulating during the successive shedding of epidermal layers in the estivation process or of colonizing bacteria from the external environment.
In support of the idea that the cocoon is a living tissue, in all cocoon samples examined, we found active transcription of epithelial cell markers genes [ck8, used as house-keeping gene, which encodes cytokeratin 8 (CK8)], antimicrobial peptide genes (defb1 to defb4, encoding β-defensins 1 to 4), proinflammatory cytokines (il1b and il8, which encode for interleukin 1β and interleukin 8), goblet cell markers (muc2 and muc4 encoding for mucin 2 and mucin 4, respectively, and csta encoding for cystatin A), and granulocyte gene markers [cxcr2, elane, h2a, and mpo; protein products CXCR2, ELANE, histone 2A (H2A), and MPO] (Fig. 3M). Expression levels of the antimicrobial peptide genes defb1 and defb2 were the highest (sixfold and ninefold, respectively, compared to ck8) of all genes examined in the cocoon. Combined, these results provide evidence that lungfish terrestrialization involves the production of a living cellular cocoon that traps bacteria and actively transcribes immune genes to provide prolonged antimicrobial protection.
Terrestrialization induces a proinflammatory environment in the lungfish skin
The drastic tissue remodeling observed during physiological terrestrialization suggested profound changes in the skin transcriptional program of lungfish. Transcriptomic analysis of the skin of freshwater and terrestrialized fish (n = 4) showed unique transcriptional profiles in each group, with a total of 1560 genes differentially expressed in the skin of terrestrialized lungfish compared to freshwater animals using DESeq and 713 genes using EdgeR analyses (Fig. 4, A and B). Gene ontology analysis identified 13 significantly regulated biological processes in lungfish skin due to terrestrialization using both DESeq and EdgeR pipelines. The identified biological processes included immune response, response to hypoxia, signal transduction, and negative regulation of apoptotic process (Fig. 4C). Kyoto Encyclopedia of Genes and Genomes pathway analysis further confirmed that up-regulated genes detected in the skin of terrestrialized lungfish had predicted roles in inflammation and infection, as well as metabolism and cell signaling processes (fig. S3).
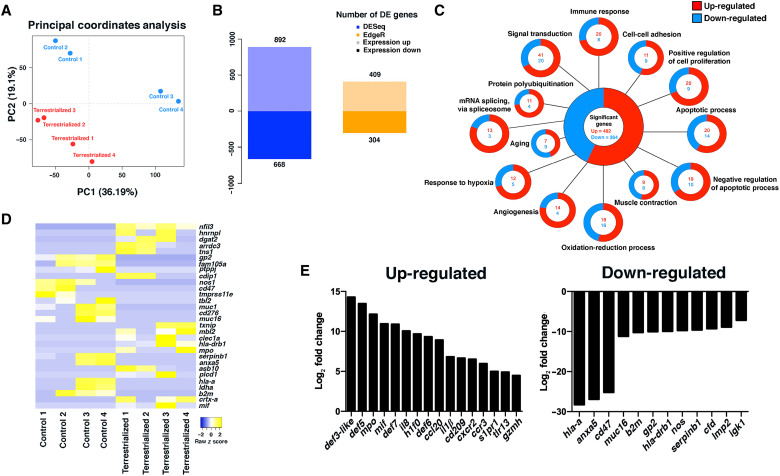
Fig. 4. Terrestrialization results in a global proinflammatory state in the lungfish skin.
(A) Principal components analysis of control and terrestrialized lungfish skin samples used in RNA sequencing (RNA-seq) analyses. (B) Number of differentially up- and down-regulated genes in control and terrestrialized lungfish skin using DESeq and EdgeR. DE, differentially expressed. (C) Gene ontology analyses of RNA-seq results showing the top 13 significantly up- and down-regulated biological processes in lungfish skin due to terrestrialization using both DESeq and EdgeR pipelines. (D) Heatmap of the top 30 significantly regulated genes in terrestrialized lungfish skin compared to controls. (E) Changes in gene expression (log2 fold) of the top 16 up-regulated and 12 down-regulated immune genes in terrestrialized lungfish skin compared to control free-swimming skin.
Terrestrialization resulted in high up-regulation of canonical proinflammatory signature characterized by il8, il1b, mif, mpo, ccl20, and cd209 expression and an antimicrobial signature characterized by up-regulated expression of defb5 in the skin. Conversely, terrestrialization resulted in down-regulation of the ELANE inhibitor serpinb1, mucin 16 (muc16), a marker for goblet cells, anticoagulant molecule annexin A5 (anxa5), and genes related to antigen presentation such as b2m, hla-drb1, hla-a, gp2, and immunoglobulin K light chain (igk) in the lungfish skin (Fig. 4, D and E). This proinflammatory environment may prolong lungfish granulocyte life span, as it has been documented for mammalian eosinophils (28, 29), a question that deserves further investigation. Collectively, our results show that terrestrialization leads to marked tissue remodeling and creates a proinflammatory environment in the lungfish skin. Furthermore, inflammation observed in terrestrialized lungfish skin shows many of the hallmarks of mammalian wounded skin such as neutrophil recruitment and elevated cytokine, chemokine, and antimicrobial peptide expression.
Lungfish granulocytes produce ETs during estivation
From plants to mammals, ET formation is one of the most ancient and conserved mechanisms of innate immunity (30–33). ETs are released from cells and are composed of decondensed chromatin decorated by histones and up to 30 different globular proteins (34–38). ETs play critical roles in infection, inflammation, injury, tissue remodeling, and autoimmunity (39–42). In humans, the presence of ETs in the skin has been described in several pathological states including delayed wound healing in diabetes (43), psoriasis (44–46), and systemic and cutaneous lupus erythematosus (47, 48).
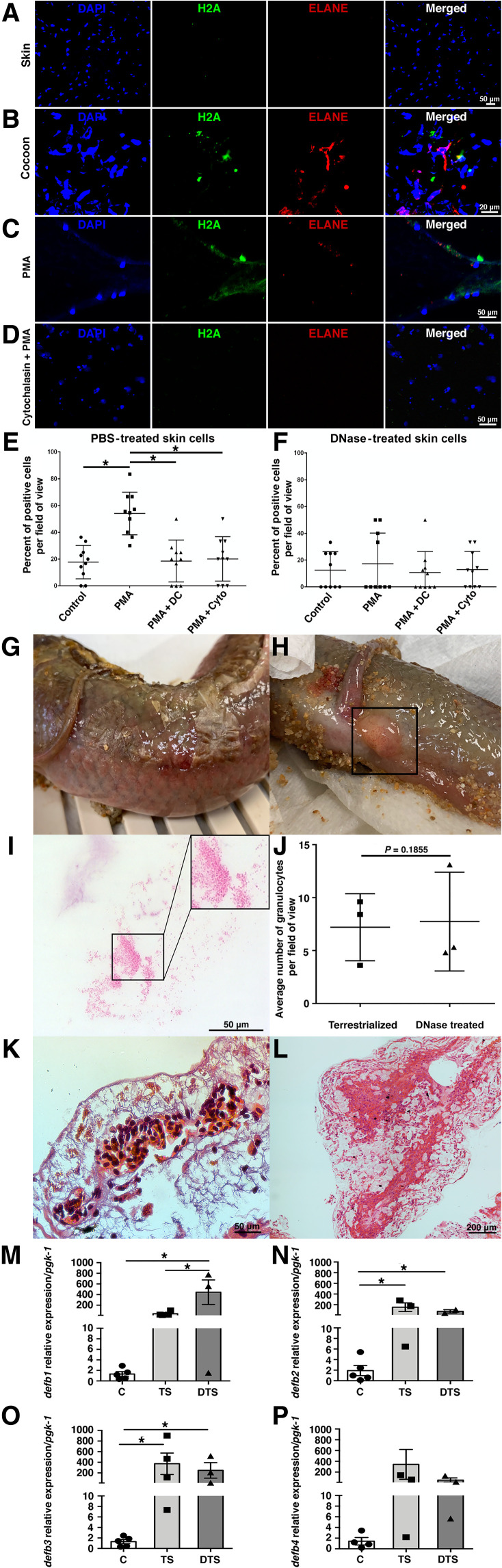
Using immunofluorescence staining with anti-ELANE and anti-H2A antibodies, we found that the skin of terrestrialized lungfish did not contain granulocytes undergoing ETosis in vivo (Fig. 5A). In contrast, the presence of cells undergoing ETosis was found in all cocoon samples examined [Fig. 5B; modeled in Fig. 2 (C and D)]. Skin cell suspensions from terrestrialized animals that were stimulated ex vivo with phorbol 12-myristate 13-acetate (PMA) contained 40 to 60% of the cells undergoing ETosis (Fig. 5, C and E). Lungfish ETs in skin cell suspensions stimulated with PMA had cloud-like or spike-like morphologies. Treatment with the inhibitors diphenyleneiodium chloride (DC) or cytochalasin A (Cyto) before PMA stimulation resulted in similar percentages of cells undergoing ETosis as in the unstimulated controls (Fig. 5, D and E). Cocoon single-cell suspensions treated with PMA did not trigger ET formation. As controls, we used PMA-stimulated gut cell suspensions from control freshwater lungfish and found that ETosis could be inhibited upon treatment with the ETosis inhibitors DC or Cyto (fig. S4, C to E). Collectively, these experiments indicate that during terrestrialization, granulocytes provide lungfish with two different layers of protection: the outer layer found in the microbial-rich cocoon where transmigrating granulocytes actively undergo ETosis and a second immunological layer located in the inflamed skin where infiltrating granulocytes can undergo ETosis upon stimulation.
Fig. 5. Skin ETosis is essential for preventing infection during terrestrialization in lungfish.
Terrestrialized lungfish (A) skin and (B) cocoon stained with DNA stain DAPI (blue), anti-H2A antibody (green), anti-human ELANE antibody (red), and merged. Cell suspensions (50,000 cells per treatment) were either not stimulated (control), stimulated with PMA, or stimulated with PMA and treated with the ETosis inhibitors diphenyleneiodium chloride (PMA + DC) or Cyto (PMA + Cyto). Cells were stained with DNA stain DAPI (blue), anti-H2A (green), anti-human ELANE antibody (red), and merged. (C) Representative confocal microscopy images of terrestrialized lungfish skin stimulated with PMA. (D) Representative confocal microscopy images of terrestrialized lungfish skin stimulated with PMA + Cyto. (E) Quantification of the percentage of cells undergoing ETosis in control terrestrialized lungfish skin (n = 2). PBS, phosphate-buffered saline. (F) Quantification of the percentage of cells undergoing ETosis in in vivo deoxyribonuclease I (DNase I)–treated terrestrialized lungfish skin (n = 2 fish). Counts were performed in n = 10 fields per fish and treatment. (G) Skin lesion and (H) prolapsed anus in DNase I–treated terrestrialized lungfish. (I) Giemsa-stained blood smear of terrestrialized lungfish treated with DNase I showing bacteria in circulation. (J) Average number of granulocytes in blood smears of control terrestrialized and DNase I–treated terrestrialized lungfish (n = 3). (K) Representative H&E stain of DNase I–treated lungfish skin from a wound area showing hemorrhages. (L) DNase I–treated mucus cocoon of terrestrialized lungfish (n = 3). (M to P) Quantification of gene expression levels of antimicrobial peptide genes (defb1 to defb4) in control free-swimming lungfish skin (C), terrestrialized lungfish skin (TS), and DNase I–treated terrestrialized lungfish skin (DTS) (n = 3 to 5). Gene expression levels were normalized to the house-keeping gene pgk-1. Data were analyzed by unpaired Student’s t test. *P < 0.05.
Extracellular DNA is essential for preventing infection during lungfish estivation
In plants, roots exposed to a root-rotting fungal pathogen and treated with deoxyribonuclease I (DNase I) suffer from severe root necrosis (49). To determine the impact of skin ETosis on lungfish terrestrialization, DNase I treatment was performed in vivo by surface spraying the lungfish once a day to deplete extracellular DNA. Spraying treatment effectively eliminated ETs in the skin and cocoon (fig. S4, A and B) and the ability of terrestrialized lungfish skin cell suspensions to form ETs following PMA stimulation (Fig. 5, C to F).
DNase I treatment during terrestrialization resulted in deleterious effects on lungfish including skin lesions, hemorrhages, head edema, and a prolapsed anus (Fig. 5, G, H, and K). Upon closer investigation of blood smears, we found that DNase I treatment was associated with the presence of large numbers of bacterial cocci in circulation, a sign of septicemia (Fig. 5I). These pathologies occurred despite the fact that granulocytes of DNase I–treated terrestrialized animals still entered systemic circulation (Fig. 5J) and that the cocoon structure appeared intact (Fig. 5L). EUB338 FISH staining of DNase I–treated skin and cocoon cryosections showed large numbers of bacteria that were able to penetrate through the cocoon and reach the skin (fig. S4, G and H).
Terrestrialization causes a significant increase in expression of antimicrobial peptides, granulocyte markers, and proinflammatory marker genes in the lungfish skin based on our RNA sequencing (RNA-seq) results. To understand whether DNase I treatment affected this transcriptional pattern, we checked expression levels of relevant gene markers in the skin of DNase I–treated lungfish. In vivo DNase I treatment did not significantly affect the expression of il1b, il8, elane, and defb2 to defb4 in lungfish skin but resulted in increased mRNA expression levels of defb1 and h2a (Fig. 5, M to P, and fig. S4, H to L). Antimicrobial peptides such as LL-37 and α-defensin play active roles in NETosis formation, stabilization, and function (45, 50–52). Whether β-defensins are part of the ET complex in lungfish requires further investigation, but our data point toward a potential compensatory role of defb1 in lungfish skin defense when extracellular DNA is eliminated. Collectively, these results show that extracellular DNA in lungfish skin is essential for microbial control during terrestrialization.
Microbiome profiling of terrestrialized integument provides evidence of extracellular DNA protection during estivation
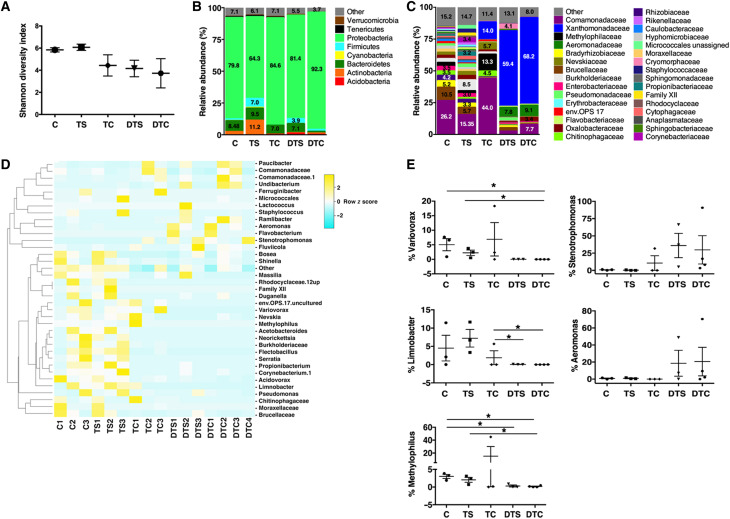
ETs trap and inactivate bacteria (34). We hypothesized that confining bacteria to the outer cocoon layer may be beneficial to estivating lungfish to conserve energy reserves. Microbiome analysis showed that the mucus cocoon has a distinct microbial community compared to that of control and terrestrialized skin. Shannon diversity index indicated that the control and terrestrialized skin have a higher microbial diversity than the rest of the samples including the cocoon, DNase I–treated skin, and DNase I–treated cocoon (Fig. 6A and fig. S5A). At the phylum level, the microbial communities of all were largely dominated by Proteobacteria, which accounted for 75.4 and 65.5% of all diversity in the control and terrestrialized skin and 82.5% of the total diversity in the cocoon (Fig. 6B). No significant changes at the phylum level were detected between the freshwater skin and the terrestrialized skin bacterial communities (Fig. 6B), suggesting that the cocoon and the active ETosis that occurs in the cocoon define the composition and spatial localization of bacterial communities in African lungfish, confining bacteria outside the lungfish body during terrestrialization. In support, the cocoon microbial community was enriched in members of the families Comamonadaceae, Methylophilaceae, and Xanthomonadaceae compared to the skin samples (Fig. 6C). A total of 37 different operational taxonomic units (OTUs) were identified to be significantly different among all treatment groups (Fig. 6D). DNase I treatment resulted in profound changes in the microbial community composition of the cocoon, particularly in the phyla Actinobacteria, Bacteroidetes, and Firmicutes. Specifically, significant increases in abundance of Actinobacteria (P = 0.021) and Firmicutes (P = 0.046) were observed in the DNase I–treated cocoon compared to the untreated cocoon. The abundance of Tenericutes decreased from 1% in the untreated cocoon to 0% in the DNase I–treated cocoon (P = 0.007), whereas Bacteroidetes abundance decreased from 7% of the untreated cocoon to 1.2% in the DNase I–treated cocoon (P = 0.231).
Fig. 6. The lungfish cocoon harbors a unique microbial community and removal of extracellular DNA results in dysbiosis.
(A) Shannon diversity index of the microbial communities of control free-swimming lungfish skin (C), terrestrialized lungfish skin (TS), terrestrialized cocoon (TC), DNase I–treated terrestrialized lungfish skin (DTS), and DNase I–treated terrestrialized lungfish cocoon (DTC) (n = 3). (B) Microbial community composition at the phylum level of the microbial communities of control free-swimming lungfish skin (C), terrestrialized lungfish skin, terrestrialized cocoon, DNase I–treated terrestrialized lungfish skin, and DNase I–treated terrestrialized lungfish cocoon. (C) Microbial community composition at the family level of the microbial communities of control free-swimming lungfish skin, terrestrialized lungfish skin, terrestrialized cocoon, DNase I–treated terrestrialized lungfish skin, and DNase I–treated terrestrialized lungfish cocoon. (D) Heatmap of the top 36 bacterial genera with differential abundances in the bacterial communities of control free-swimming lungfish skin, terrestrialized lungfish skin, terrestrialized cocoon, DNase I–treated terrestrialized lungfish skin, and DNase I–treated terrestrialized lungfish cocoon (P < 0.05). The heatmap was generated using the online free tool Heatmapper using average linkage as a clustering method followed by the Spearman rank correlation for distance measurement. (E) Mean relative abundance at the genus level of Variovorax sp., Stenotrophomonas sp., Limnobacter sp.; Aeromonas sp., and Methylophilus sp. in the microbial communities of control free-swimming lungfish skin, terrestrialized lungfish skin, terrestrialized cocoon, DNase I–treated terrestrialized lungfish skin, and DNase I–treated terrestrialized lungfish cocoon. Data were analyzed in Quantitative Insights Into Microbial Ecology (QIIME) 1.8. Differential abundances were determined by unpaired Student’s t test. *P < 0.05.
In terrestrialized skin, DNase I treatment resulted in several changes that did not reach significance because of high interindividual variability. This may be due to the fact that each lungfish was maintained in an individual tank to avoid physical attack. However, a trend to increased Proteobacteria abundance (65.5% in control terrestrialized skin versus 88.9% in DNase I–treated terrestrialized skin, P = 0.43), decreased Actinobacteria abundance (10.5% in control terrestrialized animals versus 1.3% in DNase I–treated animals, P = 0.08), and Firmicutes abundances (7% in terrestrialized skin to 3.9% in the DNase I–treated terrestrialized skin; P = 0.78) was observed. The dysbiosis caused by DNase I treatment was evident at the family and genus levels (Fig. 6, C to E, and fig. S5, B and C). Increased abundances of pathogenic taxa such as Stenotrophomonas sp. and Aeromonas sp. served in the skin and cocoon treated with DNase I (Fig. 6E). The relative abundances of Variovorax sp., Methylophilus sp., and Limnobacter sp. overall decreased as a result of DNase I treatment in both the skin and the cocoon (Fig. 6E). These results indicate that extracellular DNA and ETosis are critical for the homeostasis of the lungfish cocoon and skin microbial communities during terrestrialization.
Estivating vertebrates form cocoons to prevent water loss during drought periods. Our findings extend the function of the cocoon to not only a physical barrier but also an immunological one. In many ways, the tissue remodeling and inflammation that takes place in the lungfish skin recapitulate mucosal inflammatory disorders and dermatological diseases described in mammals characterized by granulocyte infiltration of epithelia, granulocyte transmigration, and production of ETs (7, 47). However, lungfishes are able to physiologically induce this inflammatory process every dry season, their mucosal barriers restoring once water returns. Thus, future investigations in lungfish may reveal new mechanisms by which barrier tissues are remodeled and restored following inflammatory damage. The lungfish immune system has long been considered enigmatic because of its unusually large deposits of eosinophilic granulocytes. We unravel notable adaptations of the lungfish immune system to support the physiological process of terrestrialization. Our study reveals a remarkable new form of granulocyte-driven antimicrobial defense consisting of an outer living tissue that wraps the lungfish body and traps bacteria. Since estivation can last several months and up to years and our study was limited to the first 2 weeks of estivation, we cannot conclude for how long this outer layer effectively protects lungfish against infection. It appears, however, that this extracorporeal bacterial trapping device is likely advantageous in estivating animals during metabolic torpor.
MATERIALS AND METHODS
Animals
Juvenile Protopterus dolloi (slender lungfish) (300 to 800 g) were obtained from ExoticFishShop.com (https://exoticfishshop.net/) and maintained in individual 10-gallon aquarium tanks with dechlorinated water and a mixture of sand/gravel substrate, at a temperature of 27° to 29°C. Fish were acclimated to laboratory conditions for 4 weeks before being used in experiments. During this acclimation, fish were fed one earthworm every third day, with feeding terminated 48 hours before the start of terrestrialization experiments. All animals used in this study were sampled between 8 a.m. and 11 a.m. All animal studies were reviewed and approved by the Office of Animal Care Compliance at the University of New Mexico (protocol number 11-100744-MCC).
Terrestrialization
After acclimating to laboratory settings, feeding was stopped, and the water level in tanks were lowered to 20 cm and allowed to naturally evaporate (3). With the cessation of food and the lowering of water, the fish entered the induction phase of terrestrialization and began to hyperventilate and profusely secrete mucus from their gills. Secreted mucus combined with the substrate in the tank formed a mucus cocoon that hardened 10 days after the start of the induction phase. Because of the extreme dry climate of New Mexico, the protocol was modified and the fish tanks were sprayed with 1 to 2 ml of water every third day to avoid severe dehydration.
DNase I treatment
To eliminate extracellular DNA, 1 U of DNase I in phosphate-buffered saline (PBS; Worthington, Lakewood, NJ) was administered to the terrestrializing fish (n = 3) by spraying 1 to 2 ml per day, starting at the beginning of the induction phase until sampling 10 days later. Control terrestrialized fish were sprayed once a day with 1 to 2 ml of PBS.
Tissue sampling
Lungfish mucus cocoon was peeled off with sterile forceps and divided into six pieces. One piece was fresh-frozen. The second piece was embedded in Tissue-Tek OCT compound (Sakura Finetek). A third piece was fixed in 4% paraformaldehyde (PFA) for histology. A fourth piece was placed in 1 ml of sucrose lysis buffer (SLB) for microbiome analysis; the fifth piece was preserved in RNAlater (Thermo Scientific) for molecular analysis, and the last piece was subject to cell isolation in Dispase I as explained below. Once the mucus cocoon was removed, the animals were euthanized with blunt force to their second cervical vertebra and then bled through their caudal vein for blood smears. Skin, gut, kidneys, and gonads were dissected and fixed in 4% PFA for histology. A piece of each organ was also placed in 1 ml of RNAlater for gene expression analyses. In addition, a 2 cm–by–2 cm piece of the skin and gut was used for cell isolation, and a final 0.5 cm–by–0.5 cm piece was placed in SLB for DNA extraction and microbiome sequencing.
Cell isolation
Cells from the skin and mucus cocoon were isolated by enzymatic treatment as explained elsewhere (53). Briefly, samples were excised in small pieces with sterile scissors; placed in 10 ml of Dulbecco’s minimum essential medium (DMEM) containing 5% fetal bovine serum (FBS; Peak Serum, Wellington, CO), penicillin/streptomycin (P/S; Gibco), and Dispase I (5 mg/ml; Worthington Biochemical Corporation); and incubated at 4°C on a shaker for 3 hours. Supernatants were collected, filtered through a 100-μm nylon cell strainer into a new 50-ml Falcon tube, and kept on ice. Ten milliliters of fresh Dispase I solution was then added to the tubes containing the tissue pieces and incubated for another 3 hours at 4°C on a shaker. Supernatants were filtered and combined, and the remaining tissue pieces were grounded through the cell strainer with a sterile syringe plunger. Cells were spun down at 400g at 4°C for 10 min; supernatants were discarded, and the cell pellets were washed twice in DMEM containing 5% FBS and P/S. For isolation of gut wall granulocytes from freshwater lungfish, a 1-cm-long piece of the gut was dissected, placed in DMEM containing 5% FBS and P/S, minced into small pieces, and filtered through 100-μm nylon cell strainer while mashing the tissue with a sterile syringe plunger. Cells were washed twice in DMEM containing 5% FBS and P/S. Cells were stained with trypan blue, counted under a hematocytometer, and adjusted to 106 cells/ml. Cell suspensions were then used for cell viability and active caspase-3 staining by flow cytometry or seeded onto slides for ETosis assays.
Flow cytometry
The viability of the cells isolated from either skin or cocoon samples was quantified by staining 105 freshly isolated cells with a 1:500 dilution of PI solution (1 mg/ml; Sigma-Aldrich). Positive controls for cell death consisted of permeabilized cells fixed with 4% PFA for 10 min at room temperature before PI staining. A total of 30,000 events per sample were recorded in an Attune NxT Flow Cytometer (Life Technologies). Cell death was quantified as the percentage of PI+ cells. To determine apoptosis in skin and cocoon cell suspensions, 105 cells of each cell suspension were fixed in 2% PFA for 15 min at room temperature, then permeabilized, and blocked using the Perm/Wash Buffer (Bio-Rad). Next, cells were stained with rabbit anti-human caspase-3 antibody (1:200 dilution; ab13847, Abcam), washed three times in Perm/Wash Buffer, and labeled with secondary antibody fluorescein isothiocyanate donkey anti-rabbit IgG (1:200 dilution; 711-095-152, Jackson ImmunoResearch). After three washes, cells were resuspended in Perm/Wash, and 30,000 events were recorded in the Attune NxT Flow Cytometer.
ETosis assays
Cell suspensions (50,000 cells per slide) isolated from gut, skin, and mucus cocoon were seeded onto slides for 30 min at 27°C in 5% CO2. Positive controls consisted of cells treated with PMA (0.1 μM) for 3 hours at 27°C in 5% CO2 without inhibitors. In addition, cells were treated with PMA, and either the inhibitor Cyto (10 μM) or DC (2 μM) diluted in culture medium (DMEM + 5% FBS + P/S). Negative controls received culture medium only. All slides were fixed in 4% PFA for 30 min at room temperature and stained for ETosis marker visualization as explained below.
Histology and light microscopy
Skin, gonads, and gut samples from free-swimming and terrestrialized fish (n = 3) were fixed in 4% PFA overnight, transferred to 70% ethanol, and embedded in paraffin. Sections were stained using hematoxylin and eosin or periodic acid–Schiff stain for general morphological analyses. Serial coronal sections of one control cocoon and one DNase I–treated cocoon were performed to estimate thickness of the cocoon. Large-field images for cocoon samples stained for hematoxylin and eosin were acquired on a brightfield. Images were captured on a Keyence BZ-X700 digital microscope and Nikon Eclipse Ti-S inverted microscope. Multiple images were taken across the entire cross section using a 20× objective and compiled into a single image using proprietary stitching software from the manufacturer. Blood smears were fixed in 100% methanol for 60 s and stained in 1:10 dilution of Giemsa solution (Sigma-Aldrich) in tap water for 50 min. Quantification of granulocytes in Giemsa-stained blood smears was performed in 10 fields of view per animal with three animals per treatment by two different persons in a blind fashion. Images were acquired and analyzed with a Nikon Eclipse Ti-S inverted microscope and NIS-Elements Advanced Research Software (version 4.20.02).
Immunofluorescence microscopy
Cryoblocks were sectioned (5 μm thick) and stored at −80°C until processing. Cryosections were postfixed with 4% PFA for 3 min followed by 5-min rinse in tap water. Slides were blocked in StartingBlock T20 buffer (Pierce) for 15 min at room temperature and incubated in each of the corresponding primary antibodies diluted in PBT (PBS containing 0.5% Triton X-100 and 0.1% bovine serum albumin) overnight at 4°C. For detection of ETs, slides containing treated and untreated cell suspensions were stained with anti-H2A antibody (1:100; Proteintech, 16441-1-AP) labeled with the Mix-n-Stain CF 555 Antibody Labeling Kit (Sigma-Aldrich), anti-MPO antibody (1:200; Boster Bio, PA1054), or anti-ELANE (1:200; Abcam, ab68672). After three washes in PBS, slides were incubated with secondary antibody Alexa Fluor 647–conjugated donkey anti-rabbit IgG (Abcam, ab150075; 1:200) for 2 hours at room temperature in the dark. Slides were washed twice in PBS and stained with DAPI (4,6-diamidino-2-phenylindole; 1 μg/ml in water; Invitrogen) for 60 s. For the detection of ETs in the cocoon, slides were stained with anti-ELANE (1:200; Abcam, ab68672) overnight, and after two washes in PBS, slides were incubated with secondary antibody Cy3 AffiniPure Goat Anti-Rabbit IgG (H + L) (1:200; Jackson ImmunoResearch, 111-165-144) for 2 hours at room temperature in a humidified chamber. After two washes in PBS, slides were incubated with anti-H2A antibody (1:50) labeled with the Mix-n-Stain CF 647 Antibody Labeling Kit (Sigma-Aldrich) for 2 hours at room temperature in a humidified chamber. Slides were then washed twice in PBS and counterstained with DAPI (0.5 μg/ml in water; Invitrogen) for 10 min. After rinsing in water, slides were mounted in KPL fluorescent mounting media (SeraCare) and observed under a Zeiss LSM 780 laser scanning confocal microscope. Quantification of cells undergoing ETosis was performed as previously described (54) using Photoshop.
Bacterial 16S FISH
Cryosections (10 sections per animal, n = 3) were fixed in 10% formaldehyde for 20 min, washed twice in PBS, and permeabilized in 70% ethanol overnight at 4°C. Slides were hybridized with 5′-end Cy5-labeled EUB338 (anti-sense probe) and 5′-end Cy5-labeled NONEUB (control sense probe complementary to EUB338) oligonucleotide probes (Eurofins Genomics). Hybridizations were performed at 37°C for 8 hours in hybridization buffer (2× SSC/10% formamide) containing labeled probes (1 μg/ml). Slides were then washed with hybridization buffer without probes followed by two more washes in washing buffer (2× SSC) and two washes in PBS at 37°C. Nuclei were stained with DAPI (2.5 μg/ml; Invitrogen) for 30 min at 37°C, washed twice in PBS at 37°C, and mounted in KPL fluorescent mounting media (SeraCare). Sections were observed under a Zeiss LSM 780 laser scanning confocal microscope. Images are shown as maximum projections of the Z-stacks (five stacks of 1 μm each).
Scanning and transmission electron microscopy
Skin from control and skin and cocoon samples from terrestrialized fish were fixed in formaldehyde/glutaraldehyde, 2.5% in 0.1 M sodium cacodylate buffer (pH 7.4) (Electron Microscopy Sciences, Hatfield PA), overnight at 4°C. Samples were then washed three times in sodium cacodylate buffer, postfixed using 1% osmium tetroxide for 1 hour, and washed in sodium cacodylate buffer. Samples were then dehydrated through a graded ethanol series for 1 hour and coated in gold-palladium. Samples were analyzed on a JSM-IT100 InTouchScope scanning electron microscope. For transmission electron microscopy, control and infected skin P. dolloi (n = 2) were fixed overnight at 4°C in 2.5% (v/v) glutaraldehyde in PBS and then transferred to 1% osmium tetroxide (w/v) in PBS for 2 hours at 4°C. Samples were washed in PBS three times for 10 min and then dehydrated in a graded series of ethanol (10 to 100%) through changes of propylene oxide. Samples were then embedded in EPON resin, sectioned, and stained with uranyl acetate and lead citrate before being examined in a PHILIPS TECNAI 12 transmission electron microscope.
Gene expression analysis by real-time qPCR
Tissue from free-swimming and terrestrialized lungfish (n = 3 to 4) was collected using sterile dissecting tools and preserved in 1 ml of RNAlater. Total RNA was extracted from each sample using TRIzol (Invitrogen) and following the manufacturer’s instruction, and 1 μg of RNA was synthesized into cDNA as described in (55). The final cDNA was stored at −20°C. Expression levels of granulocyte markers h2a, csta, cxcr2, and mpo; proinflammatory cytokines il1b and il8; mucins muc2 and muc4; and antimicrobial peptides defb1 to defb4 were measured by real-time qPCR (RT-qPCR) using primers shown in table S1. β-Actin, phosphoglycerate kinase 1 (pgk-1), and ck8 were used as the house-keeping genes.
RNA-seq and assembly
Extracted RNA was cleaned of genomic DNA contamination using TURBO DNase (Invitrogen, Thermo Fisher Scientific, Waltham, MA). Illumina libraries were constructed using Kapa mRNA HyperPrep kits (Roche Sequencing, Pleasanton, CA) and sequenced on an Illumina NextSeq 500 at the University of New Mexico Biology Molecular Core Facility. Quality of FASTQ files were assessed using FastQC (56), and poor quality reads were trimmed by Trimmomatic using default parameters (57). The trimmed ends were assembled into de novo transcriptomes using Trinity (58). The success of the assembly was determined by realigning the raw FASTQ reads to the corresponding assembled transcriptome using Burrows-Wheeler transform (59) and samtools (60). Differential gene analysis was assessed through DESeq and EdgeR in R software. Gene ontology analysis was performed using the bioinformatic database DAVID (61), and scatterplots were made in R software.
Microbiome sequencing and analysis
Whole genomic DNA was extracted using the cetyltrimethylammonium bromide method previously described by Mitchell and Takacs-Vesbach (62), and DNA concentration and purity were assessed using a NanoDrop ND-1000 (Thermo Scientific). Skin and cocoon microbial communities were determined via PCR, where bacterial DNA was amplified using 5PRIME HotMasterMix (Quantabio, Beverly, MA, USA) and primers 28f (5′-GAGTTTGATCNTGGCTCAG-3′) and 519r (5′-GTNTTACNGCGGCKGCTG-3′), targeting the variable V1-V3 regions of the prokaryotic 16S ribosomal DNA gene. All DNA samples were amplified with three independent reactions and pooled before the cleanup.
Amplicons were purified using an Axygen AxyPrep Mag PCR cleanup kit (Thermo Scientific) and eluted in nuclease-free water to a final volume of 30 μl. Unique barcodes were ligated to the Illumina adapters via PCR using the Nextera XT Index Kit v2 Set A (Illumina). All DNA sample concentrations were quantified using the dsDNA HS Assay Kit and Qubit fluorometer (Invitrogen); then, DNA was normalized to 200 ng/μl for DNA library pooling. Pooled samples were cleaned again using an Axygen PCR cleanup kit. Sequencing was performed on the Illumina MiSeq platform using the MiSeq Reagent Kit v3 (600 cycles) at the Clinical and Translational Sciences Center at the University of New Mexico Health Sciences Center. Sequence data were analyzed using Quantitative Insights Into Microbial Ecology (QIIME 1.9) pipeline (63) within the web-based platform Galaxy at the University of New Mexico. OTUs were selected by open reference picking using the sumaclust method. OTUs were aligned in the SILVA 16S/18S database with a 97% identity. Rarefaction analysis was performed in QIIME using several alpha-diversity metrics (PD_whole_tree, chao1, and observed_otus). Core diversity analysis was run with a normalized sampling depth of 10,525 sequences. Alpha-diversity metrics, nonphylogenetic, and phylogenetic beta-diversity analyses were performed in QIIME using the Bray-Curtis metric or the unweighted and weighted UniFrac, respectively. Principal coordinate analysis and taxonomic summaries were produced in QIIME to compare the bacterial communities in all tissue samples per groups.
Bacterial load quantification
The extracted genomic DNA from control skin, terrestrialized skin, and cocoon samples was also used to quantify the total bacterial load via RT-qPCR, using the primers 341F and 805R that amplify the bacterial 16S hypervariable region V3-V4 (table S1). RT-qPCR was performed using the SsoAdvanced Universal SYBR Green Supermix (Roche, USA). Briefly, all DNA samples were normalized to 50 ng/μl, and 2 μl of each sample was used as template into a 20-μl total reaction volume. Ct values and concentrations were calculated on the basis of a standard curve generated with a serial dilution of a purified bacterial DNA with known colony-forming units (CFU) per milliliter. The relative bacterial load in each sample is represented as the reaction mix of mean CFU per milliliter.
Statistical analysis
Data were expressed as means ± SE. Statistical analysis was performed by unpaired Student’s t test or one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests. Differences were considered statistically significant when P < 0.05. Statistical analyses were performed in Prism GraphPad version 6. For RT-qPCR assays, relative changes in gene expression were determined using the Pfaffl method (64).
Acknowledgments
We thank L. Bu and the UNM Center for Advanced Research Computing (CARC) for bioinformatics support, K. Hassan for assistance with preparation and interpretation of histological sections, and M. Spidle for SEM. We thank A. Zychlinsky for insightful suggestions on DNase I treatments. The authors thank Drs. J. Allen and E.S Loker for invaluable feedback on this manuscript.
Funding: This work was funded by the NSF award nos. 1456940 and 1938816 to I.S.
Competing interests: The authors declare that they have no competing interests.
Author contributions: Conceptualization: I.S. Methodology: R.D.H., E.C., O.B., P.M., C.T.A., and I.S. Analysis: R.D.H., E.C., O.B., P.M., and I.S. Writing (original draft preparation): R.D.H. and I.S. Writing (review and editing): R.D.H., E.C., O.B., P.M., C.T.A., and I.S. Funding acquisition: I.S.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data and Microbiome sequencing data are available under BioProject accession PRJNA600941 (www.ncbi.nlm.nih.gov/bioproject/PRJNA600941).
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Table S1
REFERENCES AND NOTES
- 1.Amemiya C. T., Alföldi J., Lee A. P., Fan S., Philippe H., Callum I. M., Braasch I., Manousaki T., Schneider I., Rohner N., Organ C., Chalopin D., Smith J. J., Robinson M., Dorrington R. A., Gerdol M., Aken B., Biscotti M. A., Barucca M., Baurain D., Berlin A. M., Blatch G. L., Buonocore F., Burmester T., Campbell M. S., Canapa A., Cannon J. P., Christoffels A., De Moro G., Edkins A. L., Fan L., Fausto A. M., Feiner N., Forconi M., Gamieldien J., Gnerre S., Gnirke A., Goldstone J. V., Haerty W., Hahn M. E., Hesse U., Hoffmann S., Johnson J., Karchner S. I., Kuraku S., Lara M., Levin J. Z., Litman G. W., Mauceli E., Miyake T., Mueller M. G., Nelson D. R., Nitsche A., Olmo E., Ota T., Pallavicini A., Panji S., Picone B., Ponting C. P., Prohaska S. J., Przybylski D., Saha N. R., Ravi V., Ribeiro F. J., Sauka-Spengler T., Scapigliati G., Searle S. M. J., Sharpe T., Simakov O., Stadler P. F., Stegeman J. J., Sumiyama K., Tabbaa D., Tafer H., Turner-Maier J., van Heusden P., White S., Williams L., Yandell M., Brinkmann H., Volff J.-N., Tabin C. J., Shubin N., Schartl M., Jaffe D. B., Postlethwait J. H., Venkatesh B., Palma F. D., Lander E. S., Meyer A., Lindblad-Toh K., The African coelacanth genome provides insights into tetrapod evolution. Nature 496, 311–316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garofalo F., Amelio D., Icardo J. M., Chew S. F., Tota B., Cerra M. C., Ip Y. K., Signal molecule changes in the gills and lungs of the African lungfish Protopterus annectens, during the maintenance and arousal phases of aestivation. Nitric Oxide 44, 71–80 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Strula M., Paola P., Carlo G., Anegla M. M., Maria U. B., Effects of induced aestivation in Protopterus annectens: A histomorphological study. J. Exp. Zool. 292, 26–31 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Icardo J. M., Loong A. M., Colvee E., Wong W. P., Ip Y. K., The alimentary canal of the African lungfish Protopterus annectens during aestivation and after arousal. Anat. Rec. 295, 60–72 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Heimroth R. D., Casadei E., Salinas I., Effects of experimental terrestrialization on the skin mucus proteome of African lungfish (Protopterus dolloi). Front. Immunol. 9, 1259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordan H. E., Speidel C. C., Blood formation in the African lungfish, under normal conditions and under conditions of prolonged estivation and recovery. J. Morphol. 51, 319–371 (1931). [Google Scholar]
- 7.Sumagin R., Robin A. Z., Nusrat A., Parkos C. A., Transmigrated neutrophils in the intestinal lumen engage ICAM-1 to regulate the epithelial barrier and neutrophil recruitment. Mucosal Immunol. 7, 905–915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkos C. A., Molecular events in neutrophil transepithelial migration. BioEssays 19, 865–873 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Chin A. C., Parkos C. A., Neutrophil transepithelial migration and epithelial barrier function in IBD: Potential targets for inhibiting neutrophil trafficking. Ann. N. Y. Acad. Sci. 1072, 276–287 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Vogel W. O. P., Mattheus U., Lymphatic vessels in lungfishes (Dipnoi): I. The lymphatic vessel system in Lepidosireniformes. Zoomorphology 117, 199–212 (1998). [Google Scholar]
- 11.Baugher P. J., Richmond A., The carboxyl-terminal PDZ ligand motif of chemokine receptor CXCR2 modulates post-endocytic sorting and cellular chemotaxis. J. Biol. Chem. 283, 30868–30878 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Londhe V., Belperio J., Keane M., Burdick M. D., Xue Y. Y., Strieter R. M., CXCR2/CXCR2 ligand biological axis impairs alveologenesis during dsRNA-induced lung inflammation in mice. Pediatr. Res. 58, 919–926 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Kinkade J. M., Pember S. O., Barnes K. C., Shapira R., Spitznagel J. K., Martin L. E., Differential distribution of distinct forms of myeloperoxidase in different azurophilic granule subpopulations from human neutrophils. Biochem. Biophys. Res. Commun. 114, 296–303 (1983). [DOI] [PubMed] [Google Scholar]
- 14.Aratani Y., Myeloperoxidase: Its role for host defense, inflammation, and neutrophil function. Arch. Biochem. Biophys. 640, 47–52 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Korkmaz B., Horwitz M. S., Jenne D. E., Gauthier F., Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol. Rev. 62, 726–759 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor S., Dirir O., Zamanian R. T., Rabinovitch M., Thompson A. A. R., The role of neutrophils and neutrophil elastase in pulmonary arterial hypertension. Front. Med. 5, 217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Withers P. C., Cocoon formation and structure in the estivating Australian desertfrogs, Neobatrachus and Cyclorana. Aust. J. Zool. 43, 429–441 (1995). [Google Scholar]
- 18.Withers P. C., Evaporative water loss and the role of cocoon formation in Australian frogs. Aust. J. Zool. 41, 405–418 (1998). [Google Scholar]
- 19.Withers P. C., Thompson G. G., Cocoon formation and metabolic depression by the aestivating hylid frogs Cyclorana australis and Cyclorana cultripes (Amphibia: Hylidae). J. R. Soc. West. Aust. 83, 39–40 (2000). [Google Scholar]
- 20.Smith H. W., Metabolism of the lung-fish, Protopterus aethiopicus. J. Biol. Chem. 88, 97–130 (1930). [Google Scholar]
- 21.McClanahan L. L. Jr., Shoemaker V. H., Ruibal R., Structure and function of the cocoon of a ceratophryd frog. Copeia 1976, 179–185 (1976). [Google Scholar]
- 22.Ruibal R., Hillman S., Cocoon structure and function in the burrowing hylid frog, Pternohyla fodiens. J. Herpetol. 15, 403–408 (1981). [Google Scholar]
- 23.Christian K., Parry D., Reduced rates of water loss and chemical properties of skin secretions of the frogs Litoria caerulea and Cyclorana australis. Aust. J. Zool. 45, 13–20 (1997). [Google Scholar]
- 24.Eckhart L., Lippens S., Tschachler E., Declercq W., Cell death by cornification. Biochim. Biophys. Acta 1833, 3471–3480 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Spearman R. I. C., Epidermal keratinization in the salamander and a comparison with other amphibia. J. Morphol. 125, 129–143 (1968). [DOI] [PubMed] [Google Scholar]
- 26.Štefková K., Procházková J., Pacherník J., Alkaline phosphatase in stem cells. Stem Cells Int. 2015, 628368 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandes K. J., McKenzie I. A., Mill P., Smith K. M., Akhavan M., Barnabé-Heider F., Biernaskie J., Junek A., Kobayashi N. R., Toma J. G., Kaplan D. R., Labosky P. A., Rafuse V., Hui C. C., Miller F. D., A dermal niche for multipotent adult skin-derived precursor cells. Nat. Cell Biol. 6, 1082–1093 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Levi-Schaffer F., Temkin V., Malamud V., Feld S., Zilberman Y., Mast cells enhance eosinophil survival in vitro: Role of TNF-alpha and granulocyte-macrophage colony-stimulating factor. J. Immunol. 160, 5554–5562 (1998). [PubMed] [Google Scholar]
- 29.Park Y. M., Bochner B. S., Eosinophil survival and apoptosis in health and disease. Allergy Asthma Immunol. Res. 2, 87–101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abi Abdallah D. S., Denkers E. Y., Neutrophils cast extracellular traps in response to protozoan parasites. Front. Immunol. 3, 382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guimarães-Costa A. B., Nascimento M. T. C., Wardini A. B., Pinto-da-Silva L. H., Saraiva E. M., ETosis: A microbicidal mechanism beyond cell death. J. Parasitol. Res. 2012, 929743 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biermann M. H. C., Podolska M. J., Knopf J., Reinwald C., Weidner D., Maueröder C., Hahn J., Kienhöfer D., Barras A., Boukerroub R., Szunerits S., Bilyy R., Hoffmann M., Zhao Y., Schett G., Herrmann M., Munoz L. E., Oxidative burst-dependent NETosis is implicated in the resolution of necrosis-associated sterile inflammation. Front. Immunol. 7, 557 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lázaro-Díez M., Chapartegui-González I., Redondo-Salvo S., Leigh C., Merino D., Segundo D. S., Fernández A., Navas J., Icardo J. M., Acosta F., Ocampo-Sosa A., Martínez-Martínez L., Ramos-Vivas J., Human neutrophils phagocytose and kill Acinetobacter baumannii and A. pittii. Sci. Rep. 7, 4571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D. S., Weinrauch Y., Zychlinsky A., Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Clark S. R., Ma A. C., Tavener S. A., McDonald B., Goodarzi Z., Kelly M. M., Patel K. D., Chakrabarti S., McAvoy E., Sinclair G. D., Keys E. M., Allen-Vercoe E., DeVinney R., Doig C. J., Green F. H. Y., Kubes P., Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat. Med. 13, 463–469 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Brinkmann V., Zychlinsky A., Neutrophil extracellular traps: Is immunity the second function of chromatin? J. Cell Biol. 198, 773–783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delgado-Rizo V., Martínez-Guzmán M. A., Iñiguez-Gutierrez L., García-Orozco A., Alvarado-Navarro A., Fafutis-Morris M., Neutrophil extracellular traps and its implications in inflammation: An overview. Front. Immunol. 8, 81 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Bont C. M., Boelens W. C., Pruijn G. J. M., NETosis, complement, and coagulation: A triangular relationship. Cell. Mol. Immunol. 16, 19–27 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saitoh T., Komano J., Saitoh Y., Misawa T., Takahama M., Kozaki T., Uehata T., Iwasaki H., Omori H., Yamaoka S., Yamamoto N., Akira S., Neutrophil extracellular traps mediate a host defense response to human immunodeficiency virus-1. Cell Host Microbe 12, 109–116 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Kruger P., Saffarzadeh M., Weber A. N., Rieber N., Radsak M., von Bernuth H., Benarafa C., Roos D., Skokowa J., Hartl D., Neutrophils: Between host defence, immune modulation, and tissue injury. PLOS Pathog. 11, e1004651 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daniel C., Leppkes M., Muñoz L. E., Schley G., Schett G., Herrmann M., Extracellular DNA traps in inflammation, injury and healing. Nat. Rev. Nephrol. 15, 559–575 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Binet F., Cagnone G., Crespo-Garcia S., Hata M., Neault M., Dejda A., Wilson A. M., Buscarlet M., Mawambo G. T., Howard J. P., Diaz-Marin R., Parinot C., Guber V., Pilon F., Juneau R., Laflamme R., Sawchyn C., Boulay K., Leclerc S., Abu-Thuraia A., Côté J. F., Andelfinger G., Rezende F. A., Sennlaub F., Joyal J. S., Mallette F. A., Sapieha P., Neutrophil extracellular traps target senescent vasculature for tissue remodeling in retinopathy. Science 369, eaay5356 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Wong S., Demers M., Martinod K., Gallant M., Wang Y., Goldfine A. B., Kahn C. R., Wagner D. D., Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat. Med. 21, 815–819 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin A. M., Rubin C. J., Khandpur R., Wang J. Y., Riblett M., Yalavarthi S., Villanueva E. C., Shah P., Kaplan M. J., Bruce A. T., Mast cells and neutrophils release IL-17 through extracellular trap formation in psoriasis. J. Immunol. 187, 490–500 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu S. C., Yu H. S., Yen F. L., Lin C. L., Chen G. S., Lan C. C., Neutrophil extracellular trap formation is increased in psoriasis and induces human β-defensin-2 production in epidermal keratinocytes. Sci. Rep. 6, 31119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shao S., Fang H., Dang E., Xue K., Zhang J., Li B., Qiao H., Cao T., Zhuang Y., Shen S., Zhang T., Qiao P., Li C., Gudjonsson J. E., Wang G., Neutrophil extracellular traps promote inflammatory responses in psoriasis via activating epidermal TLR4/IL-36R crosstalk. Front. Immunol. 10, 746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffmann J. H., Enk A. H., Neutrophil extracellular traps in dermatology: Caught in the NET. J. Dermatol. Sci. 84, 3–10 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Safi R., Al-Hage J., Abbas O., Kibbi A.-G., Nassar D., Investigating the presence of neutrophil extracellular traps in cutaneous lesions of different subtypes of lupus erythematosus. Exp. Dermatol. 28, 1348–1352 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Wen F., White G. J., Van Etten H. D., Xiong Z., Hawes M. C., Extracellular DNA is required for root tip resistance to fungal infection. Plant Physiol. 151, 820–829 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neumann A., Völlger L., Berends E. T. M., Molhoek E. M., Stapels D. A. C., Midon M., Friães A., Pinguod A., Rooijakkers S. H. M., Gallo R. L., Mörgelin M., Nizet V., Naim H. Y., von Köckritz-Blickwede M., Novel role of the antimicrobial peptide LL-37 in the protection of neutrophil extracellular traps against degradation by bacterial nucleases. J. Innate Immun. 6, 860–868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neumann A., Berends E. T. M., Nerlich A., Molhoek E. M., Gallo R. L., Meerloo T., Nizet V., Naim H. Y., von Köckritz-Blickwede M., The antimicrobial peptide LL-37 facilitates the formation of neutrophil extracellular traps. Biochem. J. 464, 3–11 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Hosoda H., Nakamura K., Hu Z., Tamura H., Reich J., Kuwahara-Arai K., Iba T., Tabe Y., Nagaoaka I., Antimicrobial cathelicidin peptide LL-37 induces NET formation and suppresses the inflammatory response in a mouse septic model. Mol. Med. Rep. 16, 5618–5626 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Xu Z., Parra D., Gómez D., Salinas I., Zhang Y., von Gersdorff Jørgensen L., Heinecke R. D., Buchmann K., LaPatra S., Sunyer J. O., Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc. Natl. Acad. Sci. U.S.A. 110, 13097–13102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carmona-Rivera C., Kaplan M. J., Induction and quantification of NETosis. Curr. Protoc. Immunol. 115, 14.41.1–14.41.14 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Tacchi L., Larragoite E., Salinas I., Discovery of J chain in African lungfish (Protopterus dolloi, Sarcopterygii) using high throughput transcriptome sequencing: Implications in mucosal immunity. PLOS ONE 8, e70650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Andrews, FastQC: A quality control tool for high throughput sequence data (2010); www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 57.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B. W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A., Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang D. W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Mitchell K. R., Takacs-Vesbach C. D., A comparison of methods for total community DNA preservation and extraction from various thermal environments. J. Ind. Microbiol. Biotechnol. 35, 1139–1147 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J., Knight R., QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pfaffl M. W., A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Table S1