Ancient human DNA from southern Iberia reveals social and genomic changes during the emergence of the Bronze Age in Europe.
Abstract
The emerging Bronze Age (BA) of southeastern Iberia saw marked social changes. Late Copper Age (CA) settlements were abandoned in favor of hilltop sites, and collective graves were largely replaced by single or double burials with often distinctive grave goods indirectly reflecting a hierarchical social organization, as exemplified by the BA El Argar group. We explored this transition from a genomic viewpoint by tripling the amount of data available for this period. Concomitant with the rise of El Argar starting ~2200 cal BCE, we observe a complete turnover of Y-chromosome lineages along with the arrival of steppe-related ancestry. This pattern is consistent with a founder effect in male lineages, supported by our finding that males shared more relatives at sites than females. However, simple two-source models do not find support in some El Argar groups, suggesting additional genetic contributions from the Mediterranean that could predate the BA.
INTRODUCTION
During the last centuries of the third millennium BCE, the societies of Europe, the Near East, and Egypt experienced large-scale social and political upheavals. Settlement abandonment, depopulation, the disappearance of communication networks, and major political disruptions at the end of the Akkadian empire and the Old Kingdom in Egypt have often been interpreted in the light of a climatic crisis, known as the 4.2k event (1–3). Recently, the possibility of substantial population movements, causing social instability during the third millennium BCE, has been proposed as a further explanation for the changes observed at the end of the Copper Age (CA) in central and western Europe (4–7). Signs of social and economic turnover are particularly marked in the southern half of the Iberian Peninsula (8), where the CA is associated with exceptional demographic growth, a diversity of monumental settlements and funerary structures, widespread copper metallurgy, and a sophisticated, large-scale manufacture and exchange of symbolic goods, among others [e.g., (9, 10)]. Moreover, this period is characterized by a diversity of settlement types, including fortified sites, ditched enclosures, and so-called megasites, some of which exceeded 100 ha in size (e.g., Valencina de la Concepción and Marroquíes Bajos) and all of which were formed at around 3300 to 2800 BCE, therefore predating the Bell Beaker horizon.
This period is also associated with a major increase in interconnectedness and mobility. On the basis of available radiogenic (Sr) isotope studies, the percentage of southern Iberian individuals who were buried in locations other than where they grew up ranges between 8 and 74% (11, 12). Ivory from Africa and the Near East (13–15), amber from Sicily (16), and ostrich eggshells from Africa (17) are indicative of transregional connections. However, evidence of a strong political centralization and economic inequality remains elusive or inconclusive (18–21).
Archaeogenetics has suggested that the remarkable development during the (south) Iberian CA was coupled with a strong population continuity attested since the Neolithic [e.g., (4, 6, 7, 22–27)]. However, the Late CA anthropological and archaeological records from the north and central Iberia show the first individuals carrying “steppe-related ancestry” by ~2400 calibrated (cal) BCE, which are often but not exclusively linked to Bell Beaker–associated artifacts (6, 7). In parallel, African ancestry was also reported in one individual, which suggests discrete movement/mobility of people (7).
The beginning of the Bronze Age [Early Bronze Age (EBA)] in Iberia (2200 to 1550 cal BCE) marks a clear population turnover, suggested by both the omnipresence of steppe-related ancestry in all individuals directly postdating 2200 BCE. An even more notable shift can be observed in the frequency of Y-chromosome haplogroups in males, which are almost exclusively of the R1b-P312 type that was completely absent in Iberia before 2400 BCE (6, 7, 25, 26).
The turn from Late CA to the EBA at the end of the third millennium BCE saw the demise of fortified settlements such as Los Millares and ditched megasites such as Valencina and Perdigões in southern Iberia, while in the southeast it is concomitant with the rise of new hilltop occupations of smaller sizes (<0.5 ha) (8, 28, 29). Substantial and densely built hilltop settlements, distinguished by a specific intramural burial rite and characteristic ceramic and metal types, appear around 2200 BCE in the fertile tertiary basins framed by mountain ranges, running parallel to the coast of southeastern Iberia (8). This area of circa (c.) 3500 km2 is considered to be the core of the El Argar “culture,” which is one of the most outstanding examples of an early complex society in European prehistory with evidence for social stratification (8, 30, 31). The origins of El Argar are still unclear, as there are no hybrid contexts where El Argar elements appear in Late CA settlements or vice versa. Although the early El Argar material record shares some traits with the Bell Beaker complex (30), such as V-perforated buttons, Palmela-type points, or perforated stone plaques so-called “archers’ wristguards,” the characteristic Bell Beaker pottery is absent. Upon the discovery of the monumental fortification in the 5-ha hilltop settlement of La Bastida, dated to around 2200 BCE, a possible eastern Mediterranean contribution was reconsidered (32). The intramural burials in large storage vessels (pithoi), the circulation of silver rings and bracelets, and the characteristic footed Argar cup have also been interpreted as signs of Aegean or Near Eastern contacts (33), although all these features emerged during later phases of El Argar. The genealogy of different characteristic material traits of early El Argar, such as apsidal buildings, intramural burial, metal casting technology, and the halberds as distinguished weapons, is reminiscent of several social developments in southeastern, central, and western Europe, with possible links of still uncertain origin (31).
Between 2000 and 1800/1750 BCE, El Argar expanded across a wider region of southeastern Iberia and entered the central Spanish Meseta. Characteristic El Argar items, such as halberds, are also present beyond this territory. The leading figures within El Argar society seem to have been a class of warriors armed with halberds and daggers. These weapons, occasionally associated with golden arm rings, also became the insignia of political domination in the male elite burials of central Europe around 2000 to 1800 BCE, a time when this specialized weapon had already gone out of use in the rest of Europe (34). At the same time, a growing part of the population, particularly children, was buried in cists, artificial caves, pottery vessels, or pits inside the distinctive settlements of El Argar.
During its final phase (1800/1750 to 1550 BCE), El Argar reached an outstanding level of economic and social development. A vast amount of grinding tools, large workshops, and storage facilities found in the larger hilltop settlements (1 to 6 ha) imply that certain groups managed to control the flow of resources and the workforce of wider regions (35). The establishment of a dominant and hereditary class, as well as increasing social asymmetries, become recognizable in the intramural burials, monumental architecture (31, 36), and the spatial relation between both (31, 36, 37).
Lull and colleagues (30, 38) have argued that social conflict and environmental degradation resulting from deforestation and extensive dry farming could have led to the abandonment or destruction of all El Argar settlements around 1550 BCE. However, the rise of a similar economic organization, architecture, and funerary record in inner Alicante, as highlighted by Cabezo Redondo, suggests that during the peak of El Argar at least some groups managed to establish themselves in a territory that was influenced by the Valencian BA, a neighboring “cultural group” located in southeastern Iberia.
In this study, we aimed to understand the importance of population dynamics in the collapse of the highly dynamic Iberian CA world, the rise and development of El Argar, as well as its relations between neighboring BA groups in western Europe and the Balearic Late BA (LBA). We also explore the genetic makeup of the BA groups in Iberia and the Balearic Islands in relation to the other western and central Mediterranean islands, such as Sardinia and Sicily. In total, we characterize the genomic profiles of 96 BA individuals associated with El Argar and contemporaneous societies, as well as 34 CA and 6 LBA individuals.
RESULTS
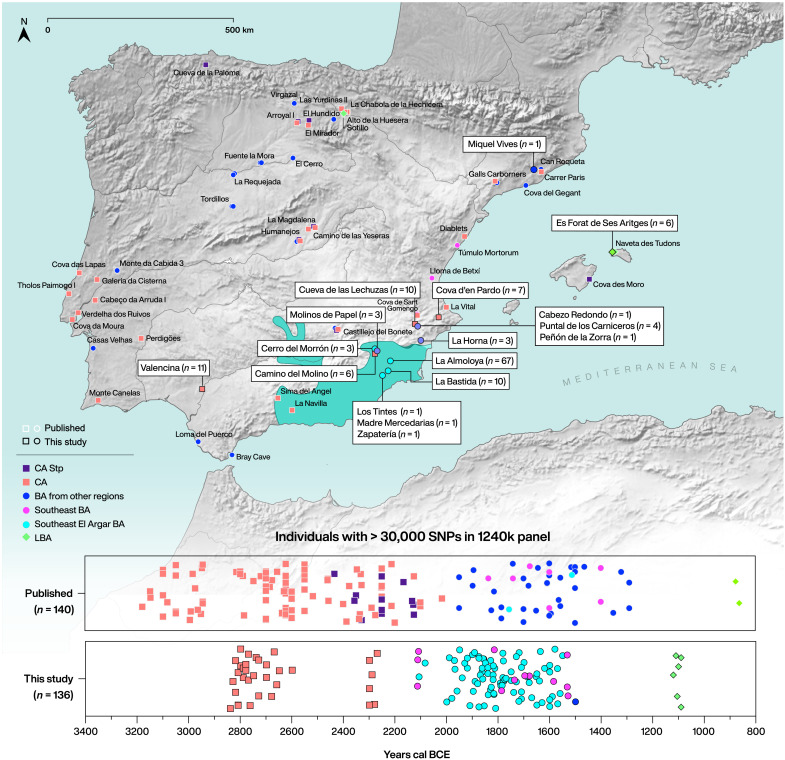
Here, we report genome-wide data in the form of 1.24 million ancestry-informative single-nucleotide polymorphisms (SNPs) for 136 southern Iberian individuals, covering a time span of 2000 years from the Late Neolithic (LN)/CA (3300 cal BCE) to the LBA (1200/1000 cal BCE) (Fig. 1, table S1.1, and text S1). Five of these individuals were also shotgun-sequenced to 1.5× to 5.2× coverage to reconstruct the complete genomes to add to the growing record of ancient western Eurasian genomes for future studies. Our new dataset includes various groups and types of sites such as sepulchral caves from the LN/CA (Cova d’en Pardo, n = 7; Cueva de las Lechuzas, n = 10), simple pit burials of pre–Bell Beaker CA megasites [Valencina (PP4-Montelirio), n = 11], a collective hypogeum with Late CA burials (Camino del Molino, n = 6), and EBA pit graves (Molinos de Papel, n = 3) to provide a diverse comparative dataset for the Argaric societies, which are at the center of our study. For the latter, we include the almost complete and archaeologically well-defined sites of La Almoloya (n = 67) and La Bastida (n = 10), alongside Cerro del Morrón (n = 3) and Lorca town [Madres Mercedarias Church (n = 1), Los Tintes (n = 1), and Zapatería (n = 1) sectors]. We also analyzed individuals of the so-called Valencian BA, including Cabezo Redondo (n = 1), Peñón de la Zorra (n = 1), Puntal de los Carniceros (n = 4), and La Horna (n = 3), individuals of the BA from Catalonia (Miquel Vives, n = 1), as well as LBA individuals from the Balearic Islands of Menorca (Es Forat de ses Aritges, n = 6) (Fig. 1, table S1.1, and text S1).
Fig. 1. Geographic locations as well as cultural and chronological information of the studied sites.
Map of Iberia with sites mentioned in the main text (table S1.1). The area shaded in teal highlights the maximum extent of EBA El Argar. Chronological time scale for published and new individuals analyzed in this study (bottom panel). Directly radiocarbon-dated individuals are plotted according to their mean calibrated date (2-sigma range), and jitter option within their specific time range was applied for individuals that were dated by archaeological context (table S2.1). Random jitter was applied to the Y axis to prevent overplotting.
We initially screened 244 individuals for DNA preservation by shotgun sequencing ~5 million Illumina single end reads per partial uracil-DNA glycosylase (UDG)–treated DNA library (39), followed by assessment of % endogenous human DNA (>0.1%), average read length between 40 and 75 base pairs (bp), and characteristic ancient DNA (aDNA) damage patterns >3% (tables S1.1 and S1.2). Libraries fulfilling these criteria were retained for hybridization capture of 1.24 million informative sites (1240k SNP panel) in the human genome (22). Libraries were independently captured for mitochondrial genomes (tables S1.3 and S1.4 and text S2) [(40) modified following (41)] and mappable regions of the Y chromosome (table S1.1 and text S3) (42). Following standard aDNA processing pipelines (43), we quantified contamination rates at the autosomal (in males) (44) and mitochondrial levels (both genetic sexes) (45), showing low contamination estimates for the individuals in this study (<3% for nuclear and <1% for mitochondrial DNA; see Materials and Methods). By determining the genetic sex (46), we observed two cases of sex chromosome aneuploidies, one XXY (Klinefelter) individual and one XXX (trisomy X) individual (table S1.1, text S4, and fig. S1). We obtained endogenous human aDNA contents ranging from 1.3 to 29.5% on the targeted 1240k SNPs, which correspond to 2004 to 831,086 SNP positions covered (tables S1.1 and S1.2). For population genetic analyses, we only retained individuals with more than 40,000 SNPs on the 1240k panel, excluding 18 low-coverage individuals in downstream analyses. We performed kinship analysis (47–49) for all newly typed individuals (fig. S2; tables S1.5, S1.6, and S1.7; and text S5), the result of which will be featured in a separate study. From each pair of first-degree relatives, we then excluded the individual with lower SNP coverage. In summary, for in-depth population genetic analysis, we merged genome-wide data from 122 newly typed individuals with a comparative set of previously published ancient and modern-day individuals, which are reported by the Reich Lab (https://reich.hms.harvard.edu/datasets; please see the Supplementary Materials for a detailed list of references), as well as genotypes from recently published studies (table S2.1) (50–53).
Genetic substructure in the Iberian CA
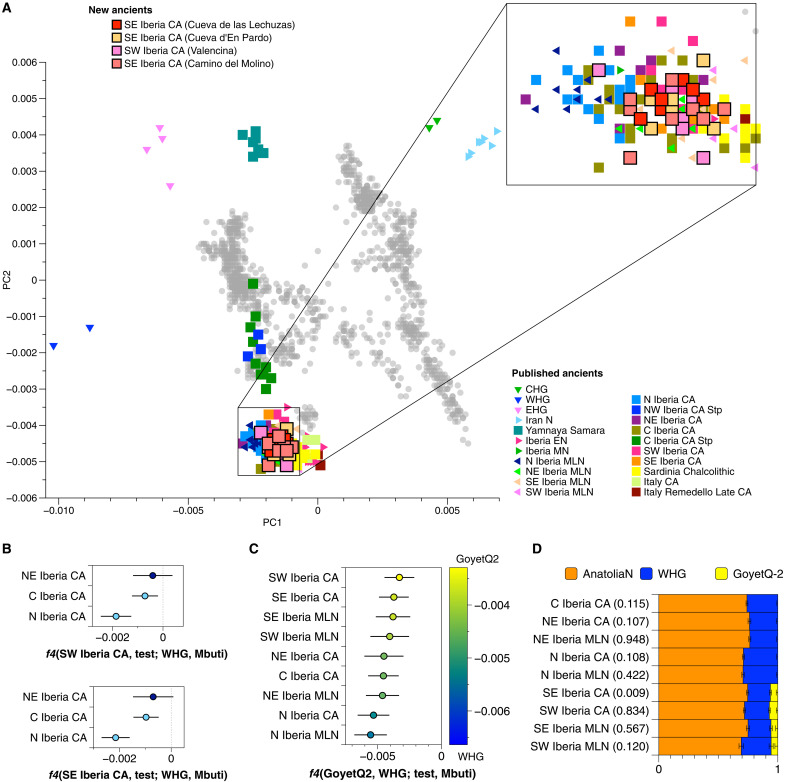
We first explored the genetic affinities of the newly typed CA individuals by performing a principal components analysis (PCA) with relevant ancient individuals projected onto PCs calculated from a set of modern-day West Eurasian populations genotyped with the Human Origins (HO) SNP panel (Fig. 2A) (54). The new CA individuals from southern Iberia fall onto a position that partially overlaps with previous Middle Neolithic (MN), Middle/Late Neolithic (MLN), and CA (non-steppe) groups from Iberia but are slightly positively shifted in their coordinates for PC1 toward previously published Early Neolithic (EN) groups from Iberia and later groups such as Sardinia Chalcolithic, suggesting an equally small hunter-gatherer (HG) ancestry contribution in the CA individuals of southern Iberia. These results run counter to previous suggestions of steppe-related ancestry diffusion into southern Iberia in the early CA based on supposed formal similarities in one element of the Valencina material culture (55). To formally test the differential contribution of the main HG source in European Neolithic farmers, called Western HG (WHG), which represents the predominant form of ancestry in prefarming western Europe (4, 56), we calculated f4-statistics of the form f4(SE/SW_Iberia_CA, N/NE/C_Iberia_CA; WHG, Mbuti) and obtained a significantly negative value for N_Iberia_CA and C_Iberia_CA (Fig. 2B and table S2.2), indicative of higher WHG ancestry in these regions, a signal that is also present in the preceding MLN period, as shown by f4-statistics of the form f4(SE/SW_Iberia_MLN, N_Iberia_MLN; WHG, Mbuti) (table S2.2).
Fig. 2. Key population genetic analyses of CA groups.
(A) PCA of published and newly genotyped CA groups projected onto 2018 modern-day West Eurasians (gray dots). Ancient individuals projected and their correspondent labels are listed in table S2.1. (B) f4-statistics showing significant differences in terms of WHG ancestry in northern and southern CA groups (error bars indicate 3 standard errors) (table S2.2 also includes the results for MLN groups). (C) f4-statistics highlighting the higher affinity of southern CA Iberian groups to GoyetQ2 (error bars indicate 3 standard errors) (table S2.3). (D) Modeling Iberian MLN and CA without steppe-related ancestry with a three-way qpAdm admixture model using distal sources Anatolia_N, WHG, and GoyetQ2. Southern MLN and CA Iberian groups show an extra minor ancestry component represented by GoyetQ2 (error bars indicate 1 standard error; numbers in brackets are P values for qpAdm model) (table S2.4).
Leveraging insights from previous studies about the differential HG legacy in western Europe (7, 57–59), we explored the HG ancestry of geographically diverse CA Iberians in different ways. We first calculated f4-statistics of the form f4(GoyetQ2, WHG; test, Mbuti), where test represents all MLN and CA groups (Fig. 2C and table S2.3). Despite the resulting f4-values being significantly negative for all test groups (indicating WHG admixture), we observed a geographically related gradient with southern groups (MLN and CA) showing lower negative values, indicating a differential relationship with GoyetQ2, who acts as a proxy for Magdalenian-related ancestry (Fig. 2C). We confirmed this subtle signal using qpAdm outgroup-based ancestry modeling with Anatolia_Neolithic, WHG (Loschbour and KO1), EHG [Eastern European HGs; the predominant form of ancestry in pre-agropastoralist eastern Europe; (4)], and GoyetQ2 as distal sources (table S2.4). Here, we found that the CA groups from southern Iberia differ with respect to the overall quantity of HG ancestry, but the latter is also qualitatively different. Estimates of Magdalenian-associated HG ancestry in southern Iberian CA individuals vary from 6.1 ± 1.3% to 7.2 ± 1.3%, which reflects the geographic structure of local HG groups (7, 57) and suggests a certain amount of genetic continuity since the Neolithic (Fig. 2D and table S2.4). In the case of SE_Iberia_CA, adding GoyetQ2 as a third source improves the model fit slightly from P = 2.53 × 10−6 to P = 0.008. Adding Iran_N or Jordan_PPNB as a fourth source (Anatolia, WHG, GoyetQ2, and Iran_N/Jordan_PPNB) further improves the model fit for SE_Iberia_CA by an order of magnitude, i.e., from P = 0.008 to P = 0.014 (with Iran_N) or P = 0.027 (with Jordan_PPNB) (table S2.4, text S7, and fig. S3). In turn, adding other populations as a fourth source does not improve the model fit (text S7). The unknown source contributing to SE_Iberia_CA groups is likely to have carried an excess of Levantine and/or Iran_N-like ancestries compared to the distal source Anatolia_Neolithic, as these together have been found admixed in Anatolian and Levantine CA groups (~6000 to 5000 BCE) (53). This finding suggests a subtle contribution that was spread early along the Mediterranean or, alternatively, different sources of early farmer ancestry during the Neolithic transition with varying proportions of Levantine and/or Iran_N-like components when compared to Anatolia_N used here. Removing Anatolian HG (AHG) from the outgroups also improves the model fit (P = 0.045; table S2.4), indicating that the Neolithic ancestry is not well represented by using Anatolia_N as a distal proxy and might come from another farmer group more similar to AHG than Anatolia_N (text S7). More individuals from the Neolithic and CA across the Mediterranean will be needed to track this contribution more confidently.
The Magdalenian-related ancestry was not detectable in other contemporaneous Mediterranean populations (e.g., Sardinia_Neolithic, Sardinia_Chalcolithic, Sardinia_EBA, Sicily_EBA, and Italy_CA), which renders gene flow in the direction from southern Iberia to the central Mediterranean less plausible. However, applying the same qpAdm model (text S7, fig. S3, and table S2.5), we detect a previously unreported amount of Iran_N-like ancestry in central Mediterranean groups from Sardinia, ranging from 2.8 ± 1.2% in Sardinia_Chalcolithic to 5.8 ± 1% in Sardinia_Nuragic_BA. Adding Iran_N as a source to model Sicily_EBA improves the model fit but without reaching P values ≥0.05, making gene flow from the Italian Peninsula to Sardinia more likely (Italy_CA also shows Iran_N-like ancestry) than from Sicily (Sicily_EBA) (text S7, fig. S3, and table S2.5). Notably, when modeling Sicily_EBA, we obtain P values ≥0.05 by removing AHG from the outgroups in a three-source model (Anatolia_Neolithic, WHG, and Yamnaya_Samara) or by removing Morocco_Iberomaurusian in a four-source model (Anatolia_Neolithic, WHG, Yamnaya_Samara, and Iran_N), which points to genetic substructure in the Mediterranean before the arrival of steppe-related ancestry. However, this finding does not rule out limited genetic input from other Mediterranean populations to southeastern CA Iberians (text S7).
Genetic turnover in the southern Iberian BA and the rise of El Argar
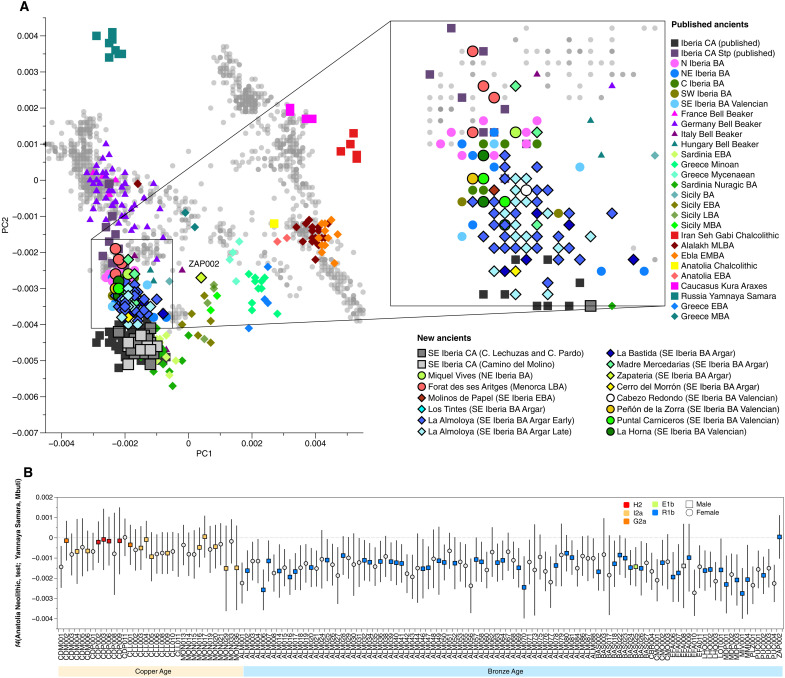
When compared to the preceding CA groups, individuals associated with El Argar BA and SE_Iberia_BA form a dense cloud in PC space that is shifted toward populations with steppe-related ancestry from central Europe and positioned between individuals from the Iberian CA and those of the north Iberian BA (Fig. 3A). A similar genetic shift is noticeable in the ADMIXTURE (60) results (text S8 and fig. S4), where Iberian BA individuals show an additional component when compared to preceding CA individuals, and this is further supported by an f4-test of the form f4 (Anatolia_Neolithic, test; Yamnaya_Samara, Mbuti), in which test iterates through newly reported CA and BA individuals (Fig. 3B and table S2.6). Here, the shift to negative f4-values in EBA individuals indicates shared drift with Yamnaya_Samara individuals, which is absent in earlier CA individuals.
Fig. 3. Evidence for steppe-related ancestry in EBA individuals from southeastern Iberia.
(A) West Eurasian PCA calculated with modern populations (gray dots) (23) on which relevant ancient CA, Bell Beaker and BA groups discussed in the text were projected (correspondent labels are listed in table S2.1).(B) f4-statistics showing the increased affinity to Yamnaya_Samara (absent in CA individuals), which implies the presence of steppe-related ancestry in EBA Iberians. This plot also shows the almost-complete turnover in Y-chromosome lineages in male individuals (color-filled squares) during the southeastern Iberian EBA (error bars indicate 3 standard errors) (table S2.6).
This observation suggests a substantial amount of steppe-related ancestry in El Argar BA individuals, which we tested formally and directly with f4-statistics of the form f4(Argar_Iberia_BA/SE_Iberia_BA, SE_Iberia_CA; Yamnaya_Samara, Mbuti) (fig. S5A and table S2.7). Significantly positive f4-values confirmed the presence of steppe-related ancestry in all BA individuals. We then tested for differences in affinity to steppe-related ancestry by contrasting northern versus southern BA individuals using f4(N/NE/C_Iberia_BA, Argar_Iberia_BA/SE_Iberia_BA; Yamnaya_Samara, Mbuti) (fig. S5B and table S2.8). The resulting f4-values confirmed a smaller amount of steppe-related ancestry in individuals from the Argaric sites La Almoloya and La Bastida compared to the rest of Iberia_BA groups, especially when compared to those from northern Iberia (fig. S5B and table S2.8), despite the complete turnover to lineage R1b-P312 (except for one subadult male in La Bastida) visible in the Y-chromosome record (Fig. 3B, table S2.6, and text S8). However, at the intrasite level, we observe no significant differences with respect to the amount of steppe-related ancestry between the early and late phase of La Almoloya and La Bastida based on PCA and formal f4-statistics (Fig. 3A and fig. S5), which suggests that the contribution is homogenized across the population.
The newly analyzed LBA individuals from the Balearic Islands (Aritges_LBA) showed less steppe-related ancestry than the published Late CA individual from Mallorca (previously named Mallorca_EBA), confirming a decrease of steppe-related ancestry over time in agreement with (51). The amount of steppe-related ancestry in Mallorca_CA_Stp is similar to the first CA groups in Iberia (C_Iberia_CA_Stp and NW_Iberia_CA_Stp) (fig. S5A) and could have arrived at the same time, as there is no clear evidence of human occupation in the archipelago before their arrival (61, 62).
Olalde and colleagues (7) had proposed for the Iberian Peninsula that the first contribution of steppe-related ancestry was diluted during the BA by admixture with descendants of local CA groups, but increased again during the LBA–Iron Age during a second pulse. We do not detect this second pulse in the Balearic Islands, in agreement with the findings of (51), albeit based on one LBA individual from Menorca (fig. S5, A and B).
The situation observed in southern Iberia offers a specific example of the demographic changes occurring during the transition from CA to EBA societies around 2200 BCE. The absence of steppe-related ancestry in our newly typed individuals from the Early CA at Valencina (c. 2900 to 2800 BCE) and the Late CA collective burial cave Camino del Molino, which is contemporaneous with the Beaker Horizon (albeit without Beaker pottery) and the first appearance of steppe-related ancestry in a double tomb with Bell Beaker–associated artifacts at Molinos de Papel, a site only 400 m away from the latter, provides the temporal framework for the arrival of steppe-related ancestry in southern Iberia. Taking the youngest date from Camino del Molino [Beta-261524, 3830 ± 40 years before present (BP)] and the oldest from Molinos de Papel (MAMS-11826, 3780 ± 30 BP) as reference points, we can show that both C14 dates overlap at the 95% level and date the arrival of steppe-related ancestry in southeastern Iberia to ~2200 cal BCE. At a cross-regional scale, none of the south Iberian CA individuals analyzed so far show evidence of any steppe-related ancestry, whereas the oldest individuals from the El Argar sites of La Bastida (BAS024 and BAS025) and La Almoloya (ALM019), directly dated to the 21st century BCE based on the radiocarbon evidence, show clear evidence of such ancestry. Consequently, the population mixture must have occurred in the preceding 150 years at the latest. Estimating the time of admixture of the putative sources using the program DATES (Distribution of Ancestry Tracts of Evolutionary Signals; https://github.com/priyamoorjani/DATES) produced compatible admixture dates in all new BA groups, ranging from ~2550 BCE ± 189 years to ~1642 BCE ± 133 years (table S2.9 and text S8). This could have occurred after the collapse of the CA social system in some aggregation megasites such as Valencina at around 2350 BCE (63) and more or less contemporary in others, such as Los Millares at around 2300/2200 BCE (64). The demise of CA cultures ~2350 to 2200 cal BCE has been linked to the so-called 4.2–thousand year cal BP climatic event, as there is a temporal overlap between this event and the collapse of large megasites (63, 65–68), although direct, conclusive, paleoclimatic evidence for Iberia remains sparse (8, 69). The social and economic changes that lead into the BA, as well as the arrival of the new genetic ancestry, appear to have spread from North to South Iberia (7, 70) and thus might be linked. However, it remains unclear whether this expansion was opportunistic (i.e., followed the consequences of a potential climatic deterioration) or causal to the actual shifts observed in archaeology. Notably, the arrival of new genetic ancestry is not paralleled by the same social changes in all regions of Iberia. In the coastal regions of southeastern Iberia, the shift to single or double burial practices inside settlements occurred ~2200 BCE, coinciding with the beginning of El Argar. However, in inner Iberia, the associated grave goods and settlement pottery are more related to late Bell Beaker CA (leaf-shaped tanged point, V-perforated button, and incised Beaker pottery) than to the material culture of the early El Argar “core” territory (8). In La Mancha, the southern part of the central Spanish plateau, the first individual with a confirmed steppe-ancestry component (7), a male from tomb 4 of Castillejo del Bonete dated to ~2100 BCE, coincides with the foundation of monumental settlements, such as Las Motillas fortifications.
This shows that while the genetic contribution of steppe-related ancestry to Iberia was a long-term process starting around 2400 cal BCE in the northern and central regions (7), from where it spread southward over ~300 years. At a local scale, this change might have occurred faster. A similar situation might have existed in central Portugal, where we still find individuals with no steppe-related ancestry in collective CA burials (Galería da Cisterna and Cova da Moura) around 2300 to 2200 cal BCE. However, after 2100 cal BCE, all individuals from all sites carry steppe-related ancestry, in line with R1b-P312 becoming the predominant Y-chromosomal lineage present not only in El Argar but also in the rest of BA Iberia.
Mediterranean and central European ancestries shaped the genetic profile of southeastern BA groups in Iberia
To explore the genetic turnover and the contribution of the local groups to the newly formed BA genetic profile in Iberia, we systematically tested a series of qpAdm models. We started by using the distal ancestry sources Anatolia_N, WHG, GoyetQ2, Yamnaya_Samara, and Iran_N to model the genetic ancestry components of Iberian BA groups (table S2.10 and fig. S6). We found that the local traces of GoyetQ2, a characteristic but variable component of southern Iberia CA individuals, were no longer detectable, suggesting a dissolution of geographic substructure in BA Iberia with respect to HG ancestry. We explain this by the spread of steppe-related ancestry from North to South (7) that also contributed northern and central Iberian ancestry to the South, diluting the subtle GoyetQ2 signal to a level beyond the limits of detection (text S8). By using the same qpAdm model, we also observed that Almoloya_Argar_Early, Almoloya_Argar_Late, SE_CabezoRedondo_BA, and Bastida_Argar cannot be modeled with Yamnaya_Samara as a single source but find better support with a combination of Iran_N and Yamnaya_Samara, however, without reaching P values ≥0.05 in Almoloya_Argar_Early and Late and SE_CabezoRedondo_BA (table S2.10 and fig. S6). These El Argar groups (Almoloya and Bastida) are also slightly shifted to the right on the PC1 axis, in the direction of Mediterranean BA groups with excess Iran_N-like ancestry, such as “Minoans,” who only carry Iran_N-like ancestry but not steppe-related ancestry, or “Mycenaeans,” who carry a mix of both (71), and that has also been shown for some BA individuals from Sicily_MBA (51) and for Sardinians here (Fig. 3A).
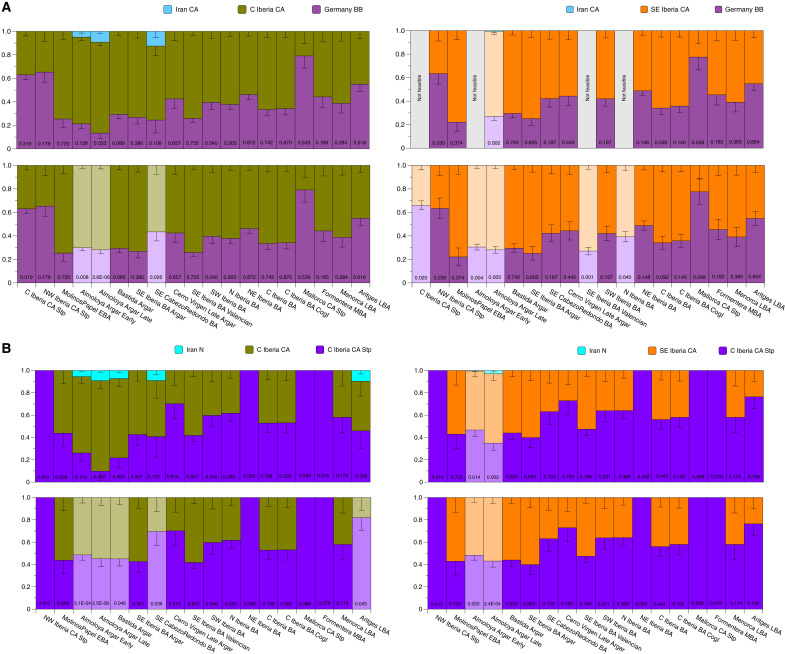
To explore the reasons for the model rejection observed in southeastern BA groups, we tested several qpAdm models (text S8). We used two- and three-way competitive qpAdm models to test whether a third source with Iran_N-like ancestry was needed. In the two-way model (table S2.11), Germany_Bell_Beaker (the largest number of individuals representing BB) was used as a fixed proximal ancestry source together with a local CA source (either N_Iberia_CA, C_Iberia_CA, or SE_Iberia_CA). For target groups in which these two sources were rejected, we iteratively tested central and eastern Mediterranean populations as a third source, as some of them are known to carry Iran_N-like ancestry (Fig. 4A and table S2.11). Using C_Iberia_CA as a local source of ancestry and Germany_Bell_Beaker as a proxy for steppe ancestry, only models with Almoloya_Argar_Early and Late as a target were rejected, suggesting that a third component was required for these Iberian BA groups. We found that adding any Mediterranean population as a third source improves the model fit, albeit without reaching P values >0.05. However, we obtained P values >0.05 when using Iran_Chalcolithic as a third source, which suggests that a higher proportion of Iran_N-like ancestry is needed (text S8 and table S2.11). Notably, when we exchanged C_Iberia_CA with the local SE_Iberia_CA, the model did not find support for a larger number of Iberian BA groups (N_Iberia_BA, SE_Iberia_BA, Almoloya_Argar_Early and Late, and C_Iberia_CA_Stp), and adding a third source in this constellation did not improve the model fit either (Fig. 4A and table S2.11).
Fig. 4. Modeling genetic ancestry in Iberian BA groups.
(A) Modeling Iberian BA individuals with steppe-related ancestry as a two- and three-way qpAdm admixture model using proximal sources C_Iberia_CA/SE_Iberia_CA, Germany_Bell_Beaker, and Iran_C (table S2.11). (B) Modeling Iberian BA individuals with steppe-related ancestry as a two- and three-way qpAdm admixture model using proximal sources C_Iberia_CA_Stp, C_Iberia_CA/SE_Iberia_CA, and Iran_N (table S2.1). P values for each group are given inside each column. Faded colors indicate rejected models when applying a P value cutoff of ≤0.05.
To rule out that the rejection of models involving Germany_Bell_Beaker + C_Iberia_CA in La Almoloya (Early and Late) is due to specific north/central European LN admixture in Germany_Bell_Beaker that was not present in the source of steppe-related ancestry in Iberia, we fixed the source of direct steppe-related ancestry to Yamnaya_Samara, as we expect the same distal steppe source contributing to all Iberian groups, and iterated through the proximal local CA sources, which in these models contribute 100% of the Chalcolithic (i.e., Neolithic farmer) ancestry. We added distal Iran_N (instead of Iran_Chalcolithic, which could contribute other confounding CA ancestries) as a third source only when needed (table S2.12 and fig. S7). By using C_Iberia_CA and Yamnaya_Samara in a two-way model, we could successfully model all Iberian BA groups, but again to the exclusion of the southeastern Almoloya_Argar_Early and Late, and SE_CabezoRedondo_BA groups. Notably, Bastida_Argar also failed for the distal model 2 (table S2.10). However, these three groups returned values ≥0.05 in the proximal local CA substrate model when Iran_N was added as a third source (fig. S7 and table S2.12). In turn, when we exchanged C_Iberia_CA with a local SE_Iberian_CA source, most of the models were rejected, which confirms the genetic substructure in Iberia_CA groups. Using SE_Iberia_CA as substrate only improves the model fit for southeastern Iberian groups, suggesting that the first individuals who brought steppe-related ancestry to Iberia admixed locally with the different CA groups (table S2.12). Notably, SE_Iberia_CA is a sufficiently supported local proxy for Balearic CA_Stp, BA, and LBA groups. We interpret this signal with caution, as Mallorca_CA_Stp and Menorca_LBA are only represented by a single individual each; however, this signal is well supported in the newly typed Aritges_LBA group from Menorca, which consists of six individuals (fig. S7 and table S2.12).
We then tried to further specify the proximal sources without affecting the power of resolution to distinguish between them. We exchanged Yamnaya_Samara by C_Iberia_CA_Stp as the most proximal local source with steppe-related ancestry (6) and combined these with the different Iberian CA sources (table S2.13 and text S8) in two-way models, only adding Iran_N as a third source when needed. Using increasingly more possible proximal sources for steppe-related ancestry, we could replicate the results of the distal models. However, we can show that Almoloya_Argar_Early and Late, Bastida_Argar, SE_CabezoRedondo_BA, and Aritges_LBA still require Iran_N as a third source in all models with C_Iberia_CA and C_Iberia_CA_Stp. In contrast, Iran_N is not required when SE_Iberia_CA is used as a local substrate, although models for Almoloya_Argar_Early and Late are still rejected at P ≤ 0.05. This finding is best explained by either an unresolved distinctive ancestry of La Almoloya individuals or, alternatively, the ability to detect such subtle signals only in groups consisting of a larger number of individuals, as is the case for Almoloya_Argar_Early (n = 36) and Almoloya_Argar_Late (n = 22), which provide the statistical power to reject simpler models. When SE_Iberia_CA is used instead of C_Iberia_CA to model La Bastida, the model is no longer rejected, which suggests a direct contribution of local CA groups to those of the southeastern Iberian BA. Together, the results hint at an additional, albeit subtle, Iran_N-enriched ancestry that is present in SE_Iberia_CA groups and thus potentially predating the El Argar BA.
We also tested whether Almoloya_Argar_Early and Late can be modeled when Iran_N is exchanged by geographically more proximal Iran_N-rich sources involving central and eastern Mediterranean populations (table S2.14 and text S8) and by moving Iran_N to the outgroups. However, this did not result in any supported models (text S8). While we were not able to find a proximal Mediterranean source, we note that adding a central Mediterranean population to the outgroups (Sicily_EBA, Greece_EBA, or Greece_MBA) decreases the model support (P values) for Almoloya_Argar_Early and Almoloya_Argar_Late, indirectly attesting to the importance of central Mediterranean BA. However, we observed no changes in P values by adding other Mediterranean groups to the outgroups (text S8 and table S2.15).
The complete turnover on the Y chromosome to R1b-Z195 (a lineage derived from P312), observed in all males at La Almoloya (29 male individuals tested) and La Bastida (except for one E1b lineage dated around 2134 to 1947 cal BCE; 7 males tested), is another independent source of evidence of gene flow predominantly during the time of the CA-BA transition (table S1.1 and text S3). Notably, Y lineage R1b-Z195, the most common Y lineage in BA Iberia, ultimately derives from a common ancestor R1b-P312 in central Europe but already differs from other derived Bell Beaker R1b variants reported from central Europe and the British Isles. R1b-Z195 has been found in Sicily_CA_Stp (previously assigned to the EBA and thus considered outliers) and Sicily_EBA (51). However, the genealogical and geographic link to other R1b variants still remains unclear. The subtle presence of Iran_N-like ancestry in El Argar and Y haplogroup R1b-Z195 in Sicily opens the possibility of gene flow not only from Iberia to Sicily as previously suggested (51) but also in the opposite direction, implying reciprocal contact with the western and central Mediterranean during the BA, although direct contacts are hardly noticeable in the archaeological record.
Together, these results suggest a dual genetic contribution to the formation of the BA genetic profile of southeastern Iberians in addition to a local CA genetic substrate. The major additional ancestry source resembles central European Bell Beaker groups, which first contributed ancestry to northern Iberia, followed by a southward spread in the form of C_Iberia_CA_Stp. A second minor ancestry component is an Iran_N-rich/central Mediterranean source, which is restricted to individuals from BA El Argar contexts. The timing of the last contributing component remains unclear and points to either a Neolithic legacy that persisted throughout the local CA or a subtle trace of connections to insular central Mediterranean BA groups.
A late Argar genetic outlier makes links to North Africa and the central Mediterranean
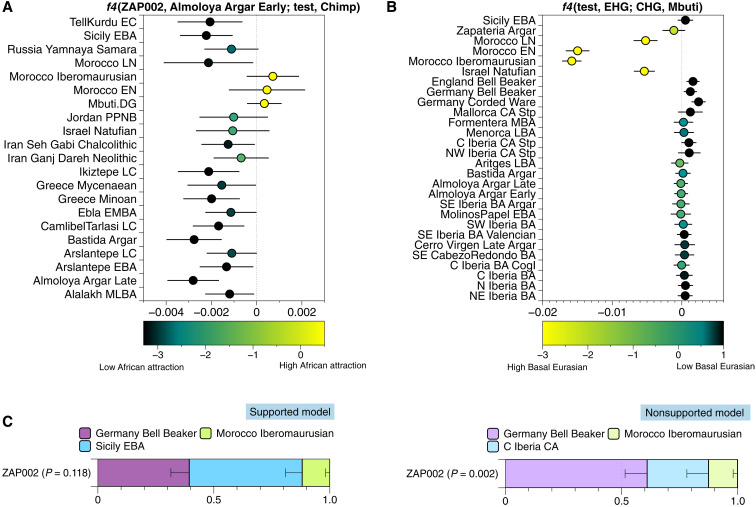
The PCA also revealed a genetic outlier among the newly typed individuals (Fig. 3A). The male individual ZAP002 from the late Argaric site of Lorca-Zapatería falls outside the Iberian_BA cluster on the PCA plot, with a clear shift toward the central Mediterranean. We formally tested for a specific attraction to groups from the eastern Mediterranean, the Near East, and Africa with an f4-admixture test of the form f4(ZAP002, Almoloya_Argar_Early; eastern_Med/Africa, Chimp) (Fig. 5A, table S2.16, and text S9). The resulting f4-values were consistent with zero, showing that ZAP002 and Almoloya_Argar_Early (the Argar group with the largest number of individuals) are symmetrically related to eastern Mediterranean and African groups. However, we noticed a near-significant positive f4-value with a z score = 2.4 for Morocco Iberomaurusian. The attraction to Mbuti in an analogous f4-test suggests either African ancestry or high levels of Basal Eurasian ancestry shared with Morocco Iberomaurusian and Natufians (table S2.16). We also confirmed a negative deviation from zero in f4(test, EHG; CHG, Mbuti), indicative of either African or Basal Eurasian ancestry. Here, we assume that the split of EHG and CHG groups [Caucasus HG; a basal form of HG ancestry south of the Caucasus (72)] is sufficiently deep to result in negative values for any test population. Following this rationale, the finding of negative values suggests a high amount of shared deep ancestry with Mbuti, which also explains the different f4-value for ZAP002 in Fig. 3B when compared to other El Argar individuals. We observe negative values for ZAP002 (z score = −2.14), as well as Morocco Iberomaurusian and Natufian individuals, who provide additional support for this claim (Fig. 5B, table S2.17, and text S9).
Fig. 5. Summary of genetic analyses that explore the genetic outlier status of individual ZAP002.
(A) f4-statistics to detect Basal Eurasian ancestry and/or African ancestry. Cladality test with ZAP002 and individuals from the site La Almoloya (ALM) and a set of test populations including Africans and Near Eastern (NE) groups, and chimpanzee in the outgroup. The results show that only Moroccan, Iberomaurusian, and Mbuti show a shift toward positive f4-values (table S2.16). (B) f4-statistics to detect either high Basal Eurasian or North African ancestry in which negative values imply excess affinity to Mbuti (table S2.17). (C) Supported and nonsupported qpAdm admixture models for outlier individual ZAP002. On the left, C_Iberia_CA is located in the outgroups and the model finds support with Sicily EBA as a source, pointing to a nonlocal origin of individual ZAP002 (table S2.18).
Using qpAdm, we further explored candidate sources of distinct ancestries such as Iberian CA and Germany_Bell_Beaker and a candidate list of central Mediterranean sources from Sardinia and Sicily. Using the same set of outgroups as for the main southeastern Iberian BA groups, we did not find a statistically well-supported model for ZAP002. However, moving Morocco_Iberomaurusian from the outgroups to the sources resulted in a model fit (≥0.05) when a central Mediterranean population was included as a source. We found that ZAP002 does not require a local Iberia_CA source and can be modeled as a three-way mixture of Germany_Bell_Beaker, Morocco_Iberomaurusian, and either Sicily EBA, Sardinia EBA, or Sardinia Nuragic BA (Fig. 5C and table S2.18). These qpAdm results are congruent with the location of ZAP002 in PC space, where he clusters with Sicily EBA individuals (Fig. 3A). Rotating the Iberian CA source to the outgroups in the same model still upholds the model fit (Fig. 5C, table S2.18, and text S9), suggesting an entirely nonlocal origin of individual ZAP002 despite being granted the same burial treatment as other individuals from El Argar (pithos burial and Argaric pottery) (Fig. 5C).
Insights into phenotypic variation, demography, and social correlates of CA and EBA El Argar societies
We explored the phenotypic variants included in the 1240k SNP panel among all well-preserved Iberian CA and BA individuals (>400,000 SNPs covered). In addition to 14 variants associated with the ability to digest lactose and alcohol, adaptation to metabolize fatty acids, predisposition to celiac disease, and resistance to infectious diseases, we also investigated 41 SNPs encoding for skin/hair and eye pigmentation (figs. S8 and S9, tables S2.19 and table S2.20, and text S10). We found no significant increase or decrease in allele frequency of any derived allele between the CA and BA (fig. S9 and table S2.20).
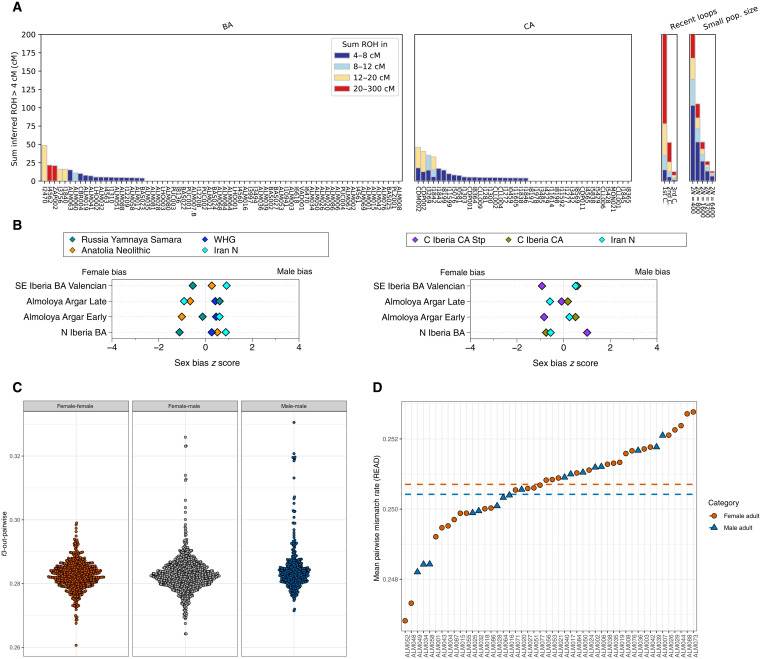
To gain insight into the effective population size during the CA and BA, we applied hapROH to identify runs of homozygosity (73, 74). We found no notable differences in short ROH (<2 cM) patterns between the two chronocultural periods, indicating that all of the individuals under study were drawn from populations with comparably and sufficiently large effective population sizes. We also found no evidence for a decline of CA population size before the emerging BA. In both groups, we did not detect signs of inbreeding, represented by long ROH segments (20 to 300 cM) (Fig. 6A and table S2.21).
Fig. 6. Biological kinship and female-male dynamics at the site La Almoloya.
(A) Sum of all ROH segments measured with hapROH for CA and BA above 400,000 SNPs in 1240k panel (table S2.21). (B) qpAdm z scores between autosomes and the X chromosome showing no signal for male bias related to steppe ancestry in the model with distal sources (right) or proximal sources (left) (table S2.22). (C) f3-outgroup statistics of the form f3(female, female; Mbuti), f3(female, male; Mbuti), and f3(male, male; Mbuti), highlighting a closer relationship among males than among females or closer relationships whenever they involve at least one male individual (first- and second-degree related pairs are excluded from the calculation) (table S2.23). (D) PWMR values of individuals compared to the PWMR average of the site La Almoloya. Dashed lines indicate a lower PWMR average and, thus, higher pairwise relatedness in adult males than adult females (table S2.24).
To assess a potential sex bias in the steppe-related ancestry contribution as postulated previously (7), we applied distal and proximal qpAdm models (text S8) to the X chromosome and autosomes (Fig. 6B and table S2.22). As males contribute, on average, only one-third of the X chromosomes to the next generation (75), a lower proportion of any ancestral component on the X chromosome than on the autosomes would thus be indicative of male bias concerning the respective component (76, 77). In turn, if the ancestral component is statistically higher on the X chromosome, this indicates a female bias. On the basis of this rationale, we do not observe significant male bias in steppe-related ancestry using either distal or proximal sources (Fig. 6B and table S2.22). The fact that the male bias is not detectable could be indicative of an already balanced ancestral component in both sexes, as is reflected in the work of Mittnik et al. (77), where the male bias in the steppe component is only detected in Corded Ware, but no longer in Bell Beaker or BA populations. We also took advantage of the recent extensive excavations and sampled all individuals with suitable morphological preservation (n = 86) from the site La Almoloya, resulting in high-quality genome-wide data for 67 individuals that allowed insight into an El Argar community. We estimated the genetic relatedness using a combination of the three methods: pairwise mismatch rate (PWMR) (48), READ (Relationship Estimation from Ancient DNA) (49), and lcMLkin (47) for pairs of individuals with more than 1000 shared SNPs (see Materials and Methods, fig. S2, and text S5). All reconstructed pedigrees will be published in an accompanying paper, fully integrated into the anthropological and archaeological context. Thus, we report here the main observations of biological relatedness at the metalevel. We found no first-degree relationships between adult women (0 of 30 adult women analyzed). All first-degree relationships among adults involved at least one adult male (three first-degree relationships among adult men and women). We also found no second-degree relationships between adult women. The few second-degree relationships involving adults were all between males (4 of 18 adult males analyzed).
We then formally tested whether the males of La Almoloya had more close relatives at the site than the females. We calculated f3-output statistics after removing first- and second-degree pairs of the form f3(female, female; Mbuti) and f3(male, male; Mbuti) and found that despite a similar average value in female:female and male:male comparisons (Fig. 6C and table S2.23), males have a more skewed f3-distribution than females, which suggests a more close genetic relationships (e.g., at third to fourth degree) among males with other males (P = 1.9 × 10−18) and males with other females (P = 2.2 × 10−4) than for females with other females (see Materials and Methods). We specifically tested for signals that suggest patrilocality using the PWMR extracted from READ (P0_Normalized) (49) and built a matrix to calculate the biological relatedness of each adult individual to the entire group (Fig. 6D and table S2.24). We observe a tendency that males are, on average, more closely related to other individuals from the site than females, despite there also being some adult females with relatives at the site, which renders the formal test for patrilocality nonsignificant (see Materials and Methods). At the intrasite level, this result could also suggest a founder effect from the male line with a higher reproductive rate.
DISCUSSION
Our focused study on the southeastern part of Iberia highlights the potential of genetic analysis on a regional level in the light of broader-scale population dynamics in Europe. Overall, we were able to show that the structure of CA populations shows persistent genetic stability and continuity since the Neolithic. This is coupled with a differential HG ancestry in the South in addition to potential early contact between southeastern Iberian CA and Mediterranean populations, which is also revealed by numerous threads of archaeological evidence (13–17). We provide nuanced evidence for the arrival of steppe-related ancestry in southeastern Iberia by 2200 cal BCE, when widespread social and cultural changes occurred across much of southern Iberia, including the end of the ditched enclosures system of population aggregation and the emergence of BA groups. In the Southeast, all sampled BA groups can be shown to carry steppe-related ancestry, while the more extensively sampled El Argar groups also carry excess Iran_N-like ancestry, which has also been observed in other CA and BA Mediterranean groups. The analysis of one outlier from El Argar reveals a central Mediterranean migrant individual with additional North African ancestry. Overall, we propose that El Argar has likely formed from a mixture of new groups arriving from north-central Iberia, which already carried central European steppe-related ancestry (and the predominant Y-chromosome lineage) and local southeastern Iberian CA groups that differed from other regions in Iberian in that they carried excess Iran_N-like ancestry similar to eastern and/or central Mediterranean groups. Moreover, the large sample sizes of early and late El Argar groups show that the Iran_N-like ancestry contribution in the local CA stratum was not enough to explain this type of ancestry satisfyingly, which, in turn, argues for a continued connection to and genetic influence from the Mediterranean BA at least until the end of the El Argar period.
Last, we were able to shed light onto the population structure of Early Bronze El Argar societies at the intrasite level. In La Almoloya, a closer genetic relationship among males is a strong indicator of patrilineality.
MATERIALS AND METHODS
Experimental design
Archaeological samples
We processed 244 individuals from the Iberian Peninsula and the Balearic Islands dated to a time span from the CA (3000 cal BCE) to the LBA (1200 cal BCE). A detailed description of the archaeological context, sites, and individuals is reported in text S1 and summarized in table S1.1.
Laboratory work
All the laboratory work was performed in the dedicated aDNA facilities of the Department of Archaeogenetics of the Max Planck Institute for the Science of Human History in Jena, Germany.
Sampling
We preferentially sampled petrous bones and teeth (essentially molars). Details of skeletal elements samples are listed in table S1.1. Before sampling, all samples were irradiated with ultraviolet light on either side for 30 min each, after which the surface was cleaned with low-concentration bleach solution (3%). For the petrous bones, we either drilled from the outside, removing the outer layer, or cut the petrous pyramid longitudinally to drill the dense part directly from either side. Molars were cut below the enamel-dentine junction, and tooth powder was drilled from the pulp chamber. We collected between 50 and 100 mg of bone or tooth powder per sample for DNA extractions.
Extraction
DNA was extracted following the modified version of (78) described in www.protocols.io/view/ancient-dna-extraction-from-skeletal-material-baksicwe.
Library preparation
We generated double-indexed double-stranded (79) UDG (“ds UDG-half”) (39) libraries using 25 μl of DNA extract and following established protocols (80). Libraries produced per individual are reported in table S1.1.
Shotgun screening and in-solution enrichment (1240k, MT-capture, and Y-capture)
Libraries were sequenced in-house on an Illumina HiSeq 4000 platform to an average depth of 5 million reads and after demultiplexing processed through EAGER (43). After an initial quality filter based on the presence of aDNA damage and endogenous DNA higher than 0.1%, libraries were subsequently enriched using in-solution capture probes synthetized by Agilent Technologies for ~1240k SNPs along the nuclear genome and independently for the complete mitogenome (40) following (41) and mappable regions of the Y chromosome (42), both in-house protocols. The captured libraries were sequenced for 20 to 40 million reads on average using either a single end (1 × 75-bp reads) or paired end configuration (2 × 50-bp reads). Results are reported in tables S1.2 and S1.3.
aDNA data processing
Read processing and DNA damage
After demultiplexing based on a specific pair of indexes, raw sequencing data were processed with EAGER (1.92.59) (43). First, the adaptor sequences were clipped with Adapter removal (v2.3.1) (81) and then mapped against the Human Reference Genome hs37d5 with BWA (v0.7.12) (82) aln and samse commands (-l 16500, -n0.01, -q30). We removed duplicates (reads with same start and end and same orientation) with DeDup (v0.12.2) (43). Last, we determined the deamination rate pattern in our UDG-half libraries using mapDamage (v.2.0.6) (83). All our libraries showed expected deamination patterns for UDG-half treatment, and according to this, we trimmed 2 bp on each terminal side before genotyping. A summary of quality statistics is given in table S1.2.
Genetic sex determination
We determined genetic sex at BAM file level by calculating the coverage on the X and Y chromosomes and the autosomes using a bed file of the regions captured by the 1240k SNP array. We normalized the X and Y reads by the autosomal coverage resulting in the X ratio and likewise for the Y ratio (table S1.1 and fig. S1). For uncontaminated libraries, we expect an X ratio of 1 and a Y ratio of 0 for females and an X and Y ratio of 0.5 for males (46). Individuals that fall in an intermediate position indicate the presence of DNA contamination from the opposite sex.
Contamination estimation
As first instance, we inspected the scatterplot of X/autosomes ratio versus Y/autosomes ratio from the sex determination to look for outliers that would indicate contamination of the opposite sex. We then estimated exogenous human contamination at nuclear and mitochondrial level. We used method 2 in ANGSD, which estimates heterozygosity at polymorphic sites on the X chromosome in males (44), applying a contamination threshold of 3% in individuals with at least 100 X-SNP positions covered twice. In cases where the number of SNPs was lower and for all other libraries from female individuals, we quantified the heterozygosity on the individual mitochondrial reads using ContamMix 1.0.10 (45) by comparing the consensus mitogenome sequence reconstructed from the MT-capture data with a comparative mitochondrial dataset of 311 global mitogenomes. Only one individual had a rate of contamination of 7.8%, while all others were below 1% (table S1.1). We visually inspected the reads with Geneious (v9.1.8) (84), looking specifically for background contamination at all variable sites but found no consistent pattern of contamination that would affect the haplotype calls.
Genotyping
We used our trimmed BAM files to call genotypes with the tool pilleupCaller (https://github.com/stschiff/sequenceTools), which randomly chooses one allele at every SNP position generating pseudo-haploid genotypes. We generated genotype files using the respective SNP lists of the 1240k panel (22) and the Affymetrix HO panel with the intersecting SNPs (~600,000 SNPs) (23, 54). Both of them were restricted to only autosomal SNPs for population genetics downstream analysis. Numbers of SNPs covered at least once in both HO and 1240k are given in table S1.1.
Mitochondrial haplogroup assignment
Following targeted enrichment, we could reconstruct complete mitochondrial genomes of 144 individuals, with an average coverage of 1.3 to 964.6 X (tables S1.3 and S1.4). To process mitochondrial DNA data, we extracted reads from mito-capture data and complemented with reads from 1240k capture and shotgun sequencing when necessary using SAMtools v1.3.1 (85) and mapping to the mitochondrial reference, exclusively. Then, we remapped reads to the revised Cambridge Reference Sequence (rCRS) using Geneious (v9.1.8) (84). To determine haplogroups from the consensus sequences reliably, we applied a threshold of at least 3000 reads per individual (table S1.4). Upon visual inspection of the assembly, we called and exported the consensus sequences in FASTA format to be used in HaploGrep2 v2.1.1 [available via https://haplogrep.uibk.ac.at/ for an automated mitochondrial haplogroup assignment based on PhyloTree (mtDNA tree build 17, available via www.phylotree.org/)] (86). Using Geneious (v9.1.8), we also visually double-checked whether specific private SNPs were covered and shared in pairs of individuals with an attested degree of relatedness (table S1.4).
Y-haplogroup assignment
By using an in-house protocol for Y capture (42), we were able to assign Y-chromosome haplogroups in male individuals. We genotyped the Y-chromosome reads using a Y-SNP list from the ISOGG (International Society of Genetic Genealogy) dataset included in the 1240k and Y-capture probes and by using an in-house script (A.B.R.) (table S1.1). This procedure allowed us to check in a semi-automatic way which positions were covered to assign an ancestral or derived haplogroup or to make corrections to calls in cases where, for instance, a more derived haplogroup was called because of residual ancient damage (C to T or G to A mismatches) in terminal read positions at diagnostic SNPs.
Kinship estimation
We calculated the PWMR (48, 87) in all pairs of individuals from our pseudo-haploid dataset to double-check for potential duplicate individuals and to determine first- and second-degree relatives. We split our individuals into two groups according to their genetic ancestry profiles (CA and BA individuals), assuming that the median PWMR of the two groups provides a background for unrelated individuals (77). We considered one-half of the unrelated value as baseline for identical samples/twins and the midpoint between the two for first-degree relatives (77). After running PWMR, we found no duplicates and removed one of each pair of first-degree relatives for downstream population genetic analyses (in all cases, the individuals with lower SNP coverage) (table S1.5 and text S5).
For the kinship study of La Almoloya, we compared and combined kinship estimates from several methods. We used READ (49) to determine first- and second-degree relatedness among individuals based on the proportion of nonmatching alleles (P0) in nonoverlapping windows of 1 mega base pairs (Mbps) and calculated standard errors. The normalized value of P0 is expected to be between 0 and 0.625 for identical twins, between 0.625 and 0.8125 for first-degree related individuals, between 0.8125 and 0.90625 for second-degree related individuals, and greater than 0.90625 for all other degrees of relatedness (which are reported as “unrelated”) (table S1.6 and text S5).
We also used the method LcMLkin (47), which uses genotype likelihoods to estimate the three k-coefficients (k0, k1, or k2) defined by (88), which define the probability that two individuals have zero, one, or two alleles identical by descendent (IBD) at a random site in the genome. As the method has to deal with missing data, it uses fractions of sites (haplotypes) that are identical by state (IBS). The length of these analyzed fractions can be modified with the thinning parameter --thin. We performed LcMLkin, exploring different thinning parameters (100,000, 50,000, 10,000, and 5000) to observe the behavior of first-degree estimates and the potential to distinguish between possible parent-offspring or sibling relationships (table S1.7 and text S5). To inform this approach contextually, we combined the genetic data with the archaeological information following this order of relevance: (i) We defined the anthropological age at death to determine sibling relatedness among subadults (before reaching reproductive age) or to inform on the directionality of parent-offspring relatives, as infants cannot be parents. (ii) When two unrelated adult male and female both are first degree related to (an)other individual(s), we identified these as parent-offspring relationship and used these trios to identify potential cases of half siblings. (iii) We used the stratigraphic information to confirm the directionality (Who died first?) to establish the type of first-degree relatedness, e.g., assuming that mothers cannot have passed away long before neonates. (iv) We used uniparentally inherited haplogroups to corroborate the established pedigrees.
Phenotypic and functional variants
We explored a list of 56 SNPs associated with known phenotypic traits. Forty-one of these are linked to physical appearance (skin, eye, or hair color), and we used the HIrisPlex method to calculate the probabilities of phenotype prediction (fig. S8 and table S2.19) (89). The other 15 SNPs are associated with the capacity to digest lactose in adulthood (rs4988235 in LCT), alcohol dehydrogenase (rs1229984 in ADH1B), adaptation to metabolize fatty acids (rs174546 in FADS1), predisposition to celiac disease (rs272872 in SLC22A4 and rs653178 in ATXN2/SH2B3), or resistance against infectious diseases [rs4833103 in the gene cluster TLR1-TLR6-TLR10 and rs2269424 in the major histocompatibility complex (MHC)]. We calculated the genotype likelihood to be homozygous or heterozygous for the ancestral (no effect) or derived (effect) allele based on the number of reads from BAM files after removing duplicates. We restricted the analysis to individuals with more than 400,000 SNPs covered in the 1240k panel, and results are summarized in fig. S9 and table S2.20.
Population genetic analysis
Dataset
We merged our final dataset with previously published datasets of ancient and modern individuals reported by the Reich Lab (https://reich.hms.harvard.edu/datasets; please see table S2.1 for a detailed list of references) as well as genotypes from recently published studies (table S2.1) (50–53).
Grouping and labeling individuals
A detailed description of new labels used in population genetics analysis is described in text S6 and table S2.1. We only included new individuals with more than 40,000 of the 1240k SNPs covered in the downstream population genetics analysis and lowered this threshold to >30,000 SNPs only for individuals forming part of a consistent cluster of individuals. In addition, we excluded groups that have consistently fewer than 60,000 SNPs in qpAdm modeling (table S2.1).
Principal components analysis
We computed PCA using smartpca software from the EIGENSOFT package (v6.0.1) (90) with the lsqproject and SHRINKMODE option YES and an extended list of modern populations from Eurasia and North Africa (23) and the Caucasus (91). The newly typed ancient individuals were projected onto PC1 and PC2.
F-statistics
F-statistics were computed with ADMIXTOOLS (https://github.com/DReichLab). For F4-statistics, we used the qpDstat and with the activated f4-mode. F3-statistics were calculated using qp3Pop. We used the 1240k panel in all our f-statistics to increase the number of SNPs covered by the ancient individuals. Standard errors were calculated with the default block jacknife.
Admixture modeling
We used ADMIXTURE (60) to define the main genetic cluster profiles of CA and BA Iberian individuals. We pruned our data for linkage disequilibrium in PLINK (92) with parameters --indep-pairwise 200 25 0.4 and –maf 0.01, which retained 243,848 autosomal SNPs for the HO dataset. ADMIXTURE was run with default fivefold cross-validation (--cv = 5), varying the number of ancestral populations between K = 2 and K = 18 with different random seeds. Each K was replicated five times.
We used the qpWave and qpAdm programs from the ADMIXTOOLS v3.0 package (https://github.com/DReichLab), with the “allsnps: YES” option to minimally reduce the number of SNPs used and subsequently increase the power to reject models, to model the ancestry in our new reported individuals. With qpWave, we estimated the minimum number of independent contributing ancestral sources of gene flow needed to explain a target population. With qpAdm, we quantified the proportion of genetic ancestry contributed by each source. The ancestry proportions in the target population are inferred on the basis of how the target population is differentially related to a set of reference/outgroups via the source populations.
For all the models applied here, we have used a set of 12 outgroups (Mbuti.DG, Ethiopia_4500BP.SG, Ust_Ishim.DG, Russia_MA1_HG.SG, Italy_Villabruna, Belgium_GoyetQ116_1, Han.DG, Onge.DG, Papuan.DG, AHG, CHG, and Morocco_Iberomaurusian). When we modeled ancestries with proximal sources, we added additional populations or rotated them to the outgroup set. The corresponding rationale is explained in each section of the Supplementary Materials.
Admixture dating
We used DATES v.753 to estimate the date of admixture among different previously fitted ancestral sources (https://github.com/priyamoorjani/DATES). The method works on modeling genotypes of the admixed test population as a linear combination of two population sources calculating the ancestry covariance coefficient. The results are given in generations (28 years, one generation; table S2.9) (93).
Sex bias
To test for potential sex bias during periods of admixture, we ran the distal (model 4; described in text S8) and proximal qpAdm models (models 2 and 4b; described in text S8) for the autosomes and in the X chromosome separately using the parameters allsnps: YES and chrom23: YES. If there was a sex bias related to any of the modeled sources of ancestry, we expect to observe increasing (female bias) or decreasing (male bias) amounts of this ancestry on the X chromosome compared to the autosomes, as the paternal contribution on X chromosome is about one-third when compared to the autosomes (the father only contributes one X chromosome to the female offspring). If the ancestry proportion is significantly higher in the autosomes, it implies that the contribution was male driven. Significance was calculated with z scores following previous studies (76, 77) (table S2.22).
Runs of homozygosity
We used hapROH to calculate the portion of the genome under runs of homozygosity (73), which can be applied to low-coverage genotype data. We included all CA and BA individuals from Iberia, which had more than 400,000 SNPs covered in the 1240k panel (table S2.21).
Testing for skewness in f3-statistics
To test for a significant change in the skewness of the distributions of f3-statistics of the form f3(female, female; Mbuti), f3(male, female; Mbuti), and f3(male, male; Mbuti), we measured the sample skewness for each distribution, denoted bff, bmf, and bmm, respectively. We then looked at the values for the pairwise skewness differences of the form δi, j = (bi − bj), where i, j ∈ (mm, mf, ff). We estimated the standard deviation of the δi, j, denoted by taking 1000 bootstrap samples. We then tested the null hypothesis that the δi, j = 0 by calculating a z score of the form and then calculated a P value using a standard normal distribution. Our P values were ad hoc adjusted via the Bonferroni method to account for multiple hypothesis testing (table S2.23).
Testing for patrilocality via mean PWMR
To test the overall mean relatedness for each individual, we considered the PWMR for each pair of individuals, denoted pij (for individuals i and j). We then calculated the mean PWMR for each individual such that . For the being attributed to groups A and B (male and female), we tested for a significant difference in sample location using a Wilcoxon rank sum test as implemented in the stats library for the R statistical software package (table S2.24).
Acknowledgments
We thank all members of the Archaeogenetics Department of the Max Planck Institute for the Science of Human History, especially the Population Genetics and PALEORIDER groups, for continued support and discussions. We thank G. Brandt, A. Wissgott, S. Clayton, and K. Prüfer for sequencing and handling of the raw data; A. Ghalichi for help with ADMIXTURE; and S. O’Reilly for the design of Fig. 1. We are indebted to the Museo de Lorca, Museo de Alcoy, Museo de Villena, Museo de Sevilla, and Museo de Alicante and all archaeologists and students involved in the excavations.
Funding: This work was supported by the Max Planck Society (V.V.-M. and W.H.); European Research Council (ERC) grant 771234—PALEoRIDER (W.H.); Spanish Ministry of Economy, Industry and Competitiveness project HAR2017-85962-P (C.O., C.R.-H., M.I.F., E.C.B., C.V.-F., V.L., R.M., and R.R.); AGAUR 2017SGR1044 (C.O., C.R.-H., M.I.F., E.C.B., C.V.-F., V.L., R.M., and R.R.); ICREA Academia program (R.R.); John Templeton Foundation grant 61220 (D.R.); and Paul Allen Family Foundation (D.R.). D.R. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: V.V.-M., R.R., and W.H. conceived the study. C.O., C.R.-H., R.M., M.I.F., E.C.B., C.V.-F., K.W.A., D.C.S.-G., G.G.A., M.S.H.P., M.P.d.M.I., A.R., V.B., J.P., A.M., J.L., J.S., A.P.M., J.K., L.G.S., V.L., and R.R. provided archaeological material and advised on the archaeological background and interpretation. F.A., M.H., W.H., C.F., V.V.-M., and C.O. performed the laboratory work. V.V.-M., W.H., A.B.R., and A.C. performed data analysis, with J.K. providing feedback in the analysis and W.H. providing guidance and methodologies. V.V.-M., R.R., W.H., and A.B.R. wrote the manuscript with input from all coauthors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Genomic data (BAM format) are available at the European Nucleotide Archive under accession number PRJEB46907.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S9
Legends for tables S1 and S2
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.Weiss H., Courty M.-A., Wetterstrom W., Guichard F., Senior L., Meadow R., Curnow A., The genesis and collapse of third millennium north Mesopotamian civilization. Science 261, 995–1004 (1993). [DOI] [PubMed] [Google Scholar]
- 2.C. Kuzucuoǧlu, C. Marro, Sociétés Humaines Et Changement Climatique À la Fin Du Troisième Millénaire: Une Crise A-t-elle Eu Lieu en Haute Mésopotamie?: Actes Du Colloque de Lyon, 5–8 Décembre 2005 (Institut français d’études anatolienne Georges-Dumézil, 2007).
- 3.H. Meller, H. W. Arz, R. Jung, R. Risch, 2200 BC—A Climatic Breakdown as a Cause for the Collapse of the Old World? (Tagungen des Landesmuseums für Vorgeschichte Halle, 12. Landesamt für Denkmalpflege und Archäologie Sachsen-Anhalt, Landesmuseum für Vorgeschichte, 2015). [Google Scholar]
- 4.Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K., Fu Q., Mittnik A., Bánffy E., Economou C., Francken M., Friederich S., Pena R. G., Hallgren F., Khartanovich V., Khokhlov A., Kunst M., Kuznetsov P., Meller H., Mochalov O., Moiseyev V., Nicklisch N., Pichler S. L., Risch R., Rojo Guerra M. A., Roth C., Szécsényi-Nagy A., Wahl J., Meyer M., Krause J., Brown D., Anthony D., Cooper A., Alt K. W., Reich D., Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allentoft M. E., Sikora M., Sjögren K.-G., Rasmussen S., Rasmussen M., Stenderup J., Damgaard P. B., Schroeder H., Ahlström T., Vinner L., Malaspinas A.-S., Margaryan A., Higham T., Chivall D., Lynnerup N., Harvig L., Baron J., Casa P. D., Dąbrowski P., Duffy P. R., Ebel A. V., Epimakhov A., Frei K., Furmanek M., Gralak T., Gromov A., Gronkiewicz S., Grupe G., Hajdu T., Jarysz R., Khartanovich V., Khokhlov A., Kiss V., Kolář J., Kriiska A., Lasak I., Longhi C., McGlynn G., Merkevicius A., Merkyte I., Metspalu M., Mkrtchyan R., Moiseyev V., Paja L., Pálfi G., Pokutta D., Pospieszny Ł., Price T. D., Saag L., Sablin M., Shishlina N., Smrčka V., Soenov V. I., Szeverényi V., Tóth G., Trifanova S. V., Varul L., Vicze M., Yepiskoposyan L., Zhitenev V., Orlando L., Sicheritz-Pontén T., Brunak S., Nielsen R., Kristiansen K., Willerslev E., Population genomics of Bronze Age Eurasia. Nature 522, 167–172 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Olalde I., Brace S., Allentoft M. E., Armit I., Kristiansen K., Booth T., Rohland N., Mallick S., Szécsényi-Nagy A., Mittnik A., Altena E., Lipson M., Lazaridis I., Harper T. K., Patterson N., Broomandkhoshbacht N., Diekmann Y., Faltyskova Z., Fernandes D., Ferry M., Harney E., de Knijff P., Michel M., Oppenheimer J., Stewardson K., Barclay A., Alt K. W., Liesau C., Ríos P., Blasco C., Miguel J. V., García R. M., Fernández A. A., Bánffy E., Bernabò-Brea M., Billoin D., Bonsall C., Bonsall L., Allen T., Büster L., Carver S., Navarro L. C., Craig O. E., Cook G. T., Cunliffe B., Denaire A., Dinwiddy K. E., Dodwell N., Ernée M., Evans C., Kuchařík M., Farré J. F., Fowler C., Gazenbeek M., Pena R. G., Haber-Uriarte M., Haduch E., Hey G., Jowett N., Knowles T., Massy K., Pfrengle S., Lefranc P., Lemercier O., Lefebvre A., Martínez C. H., Olmo V. G., Ramírez A. B., Maurandi J. L., Majó T., McKinley J. I., McSweeney K., Mende B. G., Modi A., Kulcsár G., Kiss V., Czene A., Patay R., Endrődi A., Köhler K., Hajdu T., Szeniczey T., Dani J., Bernert Z., Hoole M., Cheronet O., Keating D., Velemínský P., Dobeš M., Candilio F., Brown F., Fernández R. F., Herrero-Corral A.-M., Tusa S., Carnieri E., Lentini L., Valenti A., Zanini A., Waddington C., Delibes G., Guerra-Doce E., Neil B., Brittain M., Luke M., Mortimer R., Desideri J., Besse M., Brücken G., Furmanek M., Hałuszko A., Mackiewicz M., Rapiński A., Leach S., Soriano I., Lillios K. T., Cardoso J. L., Pearson M. P., Włodarczak P., Price T. D., Prieto P., Rey P.-J., Risch R., Rojo Guerra M. A., Schmitt A., Serralongue J., Silva A. M., Smrčka V., Vergnaud L., Zilhão J., Caramelli D., Higham T., Thomas M. G., Kennett D. J., Fokkens H., Heyd V., Sheridan A., Sjögren K.-G., Stockhammer P. W., Krause J., Pinhasi R., Haak W., Barnes I., Lalueza-Fox C., Reich D., The Beaker phenomenon and the genomic transformation of northwest Europe. Nature 555, 190–196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olalde I., Mallick S., Patterson N., Rohland N., Villalba-Mouco V., Silva M., Dulias K., Edwards C. J., Gandini F., Pala M., Soares P., Ferrando-Bernal M., Adamski N., Broomandkhoshbacht N., Cheronet O., Culleton B. J., Fernandes D., Lawson A. M., Mah M., Oppenheimer J., Stewardson K., Zhang Z., Jiménez Arenas J. M., Toro Moyano I. J., Salazar-García D. C., Castanyer P., Santos M., Tremoleda J., Lozano M., García Borja P., Fernández-Eraso J., Mujika-Alustiza J. A., Barroso C., Bermúdez F. J., Viguera Mínguez E., Burch J., Coromina N., Vivó D., Cebrià A., Fullola J. M., García-Puchol O., Morales J. I., Oms F. X., Majó T., Vergès J. M., Díaz-Carvajal A., Ollich-Castanyer I., López-Cachero F. J., Silva A. M., Alonso-Fernández C., Delibes de Castro G., Jiménez Echevarría J., Moreno-Márquez A., Pascual Berlanga G., Ramos-García P., Ramos-Muñoz J., Vijande Vila E., Aguilella Arzo G., Esparza Arroyo Á., Lillios K. T., Mack J., Velasco-Vázquez J., Waterman A., Benítez de Lugo Enrich L., Benito Sánchez M., Agustí B., Codina F., de Prado G., Estalrrich A., Fernández Flores Á., Finlayson C., Finlayson G., Finlayson S., Giles-Guzmán F., Rosas A., Barciela González V., García Atiénzar G., Hernández Pérez M. S., Llanos A., Carrión Marco Y., Collado Beneyto I., López-Serrano D., Sanz Tormo M., Valera A. C., Blasco C., Liesau C., Ríos P., Daura J., de Pedro Michó M. J., Diez-Castillo A. A., Flores Fernández R., Francès Farré J., Garrido-Pena R., Gonçalves V. S., Guerra-Doce E., Herrero-Corral A. M., Juan-Cabanilles J., López-Reyes D., McClure S. B., Merino Pérez M., Oliver Foix A., Sanz Borràs M., Sousa A. C., Vidal Encinas J. M., Kennett D. J., Richards M. B., Werner Alt K., Haak W., Pinhasi R., Lalueza-Fox C., Reich D., Science 363, 1230–1234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.V. Lull, R. Micó, C. Rihuete Herrada, R. Risch, Transition and Conflict at the End of the 3rd Millennium BC in South Iberia, H. Meller, H. W. Arz, R. Jung, R. Risch, Eds. (Landesmuseum für Vorgeschichte, 2015). [Google Scholar]
- 9.R. Chapman, Archaeologies of Complexity (Routledge, 2003). [Google Scholar]
- 10.P. B. Ramírez, J. A. Soler Díaz, Ídolos: Miradas Milenarias (Museo Arqueológico de Alicante, 2020). [Google Scholar]
- 11.Bonilla M. D.-Z., Beck J., Bocherens H., Díaz-del-Río P., Isotopic evidence for mobility at large-scale human aggregations in Copper Age Iberia: The mega-site of Marroquíes. Antiquity 92, 991–1007 (2018). [Google Scholar]
- 12.Valera A. C., Žalaitė I., Maurer A. F., Grimes V., Silva A. M., Ribeiro S., Santos J. F., Barrocas Dias C., Addressing human mobility in Iberian Neolithic and Chalcolithic ditched enclosures: The case of Perdigões (South Portugal). J. Archaeol. Sci. Rep. 30, 102264 (2020). [Google Scholar]
- 13.A. Banerjee, J. A. López Padilla, T. X. Schumacher, Marfil y elefantes en la Península Ibérica y el Mediterráneo occidental, Iberia Archaeologica 16.1. (Zabern, 2012). [Google Scholar]
- 14.Schuhmacher T. X., Frühbronzezeitliche Kontakte im westlichen und zentralen Mittelmeerraum und die Rolle der Iberischen Halbinsel. Madrider Mitteilungen 45, 147–180 (2004). [Google Scholar]
- 15.Schuhmacher T. X., Elfenbeinobjekte des Chalkolithikums und der Frühen Bronzezeit auf der Iberischen Halbinsel. Studien zu Herkunft, Austausch, Verarbeitung und sozialer Bedeutung von Elfenbein. Iberia Archaeologica 16, 610 (2012). [Google Scholar]
- 16.Murillo-Barroso M., Martinón-Torres M., Amber sources and trade in the prehistory of the Iberian peninsula. Eur. J. Archaeol. 15, 187–216 (2012). [Google Scholar]
- 17.R. J. Harrison, A. Gilman, Trade in the second and third millennia BC between the Maghreb and Iberia, in Ancient Europe and the Mediterranean: studies in honour of Hugh Hencken, V. Markotic, Ed. (Aris & Phillips, 1977), pp. 90–104. [Google Scholar]
- 18.V. Hurtado Pérez, L. García Sanjuán, Los inicios de la Jerarquización Social en el Suroeste de la Península Ibérica (c. 2500–1700 ane): Problemas conceptuales y empíricos (1997); http://roderic.uv.es/bitstream/handle/10550/29498/2134.pdf?sequence=1.
- 19.Chapman R., Producing inequalities: Regional sequences in later prehistoric Southern Spain. J. World Prehist. 21, 195–260 (2008). [Google Scholar]
- 20.P. Díaz del Río, Labor in the making of Iberian Copper Age lineages (2011); https://digital.csic.es/handle/10261/35427.
- 21.R. Risch, in Überschuss ohne Staat-Politische Formen in der Vorgeschichte: 10. Mitteldeutscher Archäologentag vom 19. bis 21. Oktober 2017 in Halle (Saale) (Landesmuseum für Vorgeschichte, 2018), pp. 45–65. [Google Scholar]
- 22.Mathieson I., Lazaridis I., Rohland N., Mallick S., Patterson N., Roodenberg S. A., Harney E., Stewardson K., Fernandes D., Novak M., Sirak K., Gamba C., Jones E. R., Llamas B., Dryomov S., Pickrell J., Arsuaga J. L., de Castro J. M. B., Carbonell E., Gerritsen F., Khokhlov A., Kuznetsov P., Lozano M., Meller H., Mochalov O., Moiseyev V., Guerra M. A. R., Roodenberg J., Vergès J. M., Krause J., Cooper A., Alt K. W., Brown D., Anthony D., Lalueza-Fox C., Haak W., Pinhasi R., Reich D., Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazaridis I., Nadel D., Rollefson G., Merrett D. C., Rohland N., Mallick S., Fernandes D., Novak M., Gamarra B., Sirak K., Connell S., Stewardson K., Harney E., Fu Q., Gonzalez-Fortes G., Jones E. R., Roodenberg S. A., Lengyel G., Bocquentin F., Gasparian B., Monge J. M., Gregg M., Eshed V., Mizrahi A.-S., Meiklejohn C., Gerritsen F., Bejenaru L., Blüher M., Campbell A., Cavalleri G., Comas D., Froguel P., Gilbert E., Kerr S. M., Kovacs P., Krause J., McGettigan D., Merrigan M., Merriwether D. A., O’Reilly S., Richards M. B., Semino O., Shamoon-Pour M., Stefanescu G., Stumvoll M., Tönjes A., Torroni A., Wilson J. F., Yengo L., Hovhannisyan N. A., Patterson N., Pinhasi R., Reich D., Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipson M., Szécsényi-Nagy A., Mallick S., Pósa A., Stégmár B., Keerl V., Rohland N., Stewardson K., Ferry M., Michel M., Oppenheimer J., Broomandkhoshbacht N., Harney E., Nordenfelt S., Llamas B., Gusztáv Mende B., Köhler K., Oross K., Bondár M., Marton T., Osztás A., Jakucs J., Paluch T., Horváth F., Csengeri P., Koós J., Sebők K., Anders A., Raczky P., Regenye J., Barna J. P., Fábián S., Serlegi G., Toldi Z., Gyöngyvér Nagy E., Dani J., Molnár E., Pálfi G., Márk L., Melegh B., Bánfai Z., Domboróczki L., Fernández-Eraso J., Antonio Mujika-Alustiza J., Alonso Fernández C., Jiménez Echevarría J., Bollongino R., Orschiedt J., Schierhold K., Meller H., Cooper A., Burger J., Bánffy E., Alt K. W., Lalueza-Fox C., Haak W., Reich D., Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature 551, 368–372 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martiniano R., Cassidy L. M., Ó’Maoldúin R., McLaughlin R., Silva N. M., Manco L., Fidalgo D., Pereira T., Coelho M. J., Serra M., Burger J., Parreira R., Moran E., Valera A. C., Porfirio E., Boaventura R., Silva A. M., Bradley D. G., The population genomics of archaeological transition in west Iberia: Investigation of ancient substructure using imputation and haplotype-based methods. PLOS Genet. 13, e1006852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valdiosera C., Günther T., Vera-Rodríguez J. C., Ureña I., Iriarte E., Rodríguez-Varela R., Simões L. G., Martínez-Sánchez R. M., Svensson E. M., Malmström H., Rodríguez L., Bermúdez de Castro J.-M., Carbonell E., Alday A., Hernández Vera J. A., Götherström A., Carretero J.-M., Arsuaga J. L., Smith C. I., Jakobsson M., Four millennia of Iberian biomolecular prehistory illustrate the impact of prehistoric migrations at the far end of Eurasia. Proc. Natl. Acad. Sci. U.S.A. 115, 3428–3433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brace S., Diekmann Y., Booth T. J., van Dorp L., Faltyskova Z., Rohland N., Mallick S., Olalde I., Ferry M., Michel M., Oppenheimer J., Broomandkhoshbacht N., Stewardson K., Martiniano R., Walsh S., Kayser M., Charlton S., Hellenthal G., Armit I., Schulting R., Craig O. E., Sheridan A., Parker Pearson M., Stringer C., Reich D., Thomas M. G., Barnes I., Ancient genomes indicate population replacement in Early Neolithic Britain. Nat. Ecol. Evol. 3, 765–771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.F. Molina González, J. A. Cámara Serrano, J. Capel Martínez, T. Nájera Colino, L. Sáez Pérez, Los Millares y la periodización de la Prehistoria Reciente del Sudeste, in II - III Simposios de Prehistoria Cueva de Nerja (Fundación Cueva de Nerja, Nerja, 2004), pp. 142–158. [Google Scholar]
- 29.A. C. Valera, Social change in the late 3rd millennium BC in Portugal: The twilight of enclosures, in 2200 BC—A Climatic Breakdown as a Cause for the Collapse of the Old World?, H. Meller, R. Risch, R. Jung, H. W. Arz, Eds. (Landesmuseum für vorgeschichte, 2015), pp. 409–427. [Google Scholar]
- 30.V. Lull, La“ cultura” de El Argar: Un modelo para el estudio de las formaciones económico-sociales prehistóricas (Ediciones Akal, 1983). [Google Scholar]
- 31.V. Lull, R. Micó, C. Rihuete Herrada, R. Risch, in Sozialarchäologische Perspektiven: Gesellschaftlicher Wandel 5000–1500 v. Chr. Zwischen Atlantik und Kaukasus, S. Hansen, J. Müller, Eds. (Archäologie in Eurasien, 2011), vol. 24, pp. 381–414. [Google Scholar]
- 32.Lull V., Micó R., Rihuete-Herrada C., Risch R., The La Bastida fortification: New light and new questions on Early Bronze Age societies in the western Mediterranean. Antiquity 88, 395–410 (2014). [Google Scholar]
- 33.Schubart H., Mediterrane Beziehungen der El Argar-Kultur. Madrider Mitteilungen 14, 41–59 (1973). [Google Scholar]
- 34.V. Lull, R. Micó, C. Rihuete-Herrada, R. Risch, N. Escanilla, in Actas del Congreso de Cronometrías Para la Historia de la Península Ibérica (CEUR Workshop Proceedings 2024), J. A. Barceló, I. Bogdanovic. B. Morell, Eds. (IberCrono, Barcelona, 2017), pp. 143–162. [Google Scholar]
- 35.R. Risch, Recursos naturales, medios de producción y explotación social. Un análisis económico de la industria lítica de Fuente Álamo (Almería), 2250-1400 ANE. (P. von Zabern. Mainz, 2002). [Google Scholar]
- 36.Lull V., Pérez R. M., Herrada C. R., Risch R., Property relations in the Bronze Age of South-western Europe: An archaeological analysis of infant burials from El Argar (Almeria, Spain). Proc. Prehist. Soc. 71, 247–268 (2005). [Google Scholar]
- 37.V. Lull, J. Estévez. Propuesta metodológica para el estudio de las necrópolis argáricas, in Homenaje a Luis Siret (1934–1984), Consejería de Cultura de la Junta de Andalucía (Dirección General de Bellas Artes, 1986), pp. 441–452. [Google Scholar]
- 38.Lull V., Micó R., Rihuete-Herrada C., Risch R., Funerary practices and kinship in an Early Bronze Age society: A Bayesian approach applied to the radiocarbon dating of Argaric double tombs. J. Archaeol. Sci. 40, 4626–4634 (2013). [Google Scholar]
- 39.Rohland N., Harney E., Mallick S., Nordenfelt S., Reich D., Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20130624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maricic T., Whitten M., Pääbo S., Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLOS ONE 5, e14004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu Q., Hajdinjak M., Moldovan O. T., Constantin S., Mallick S., Skoglund P., Patterson N., Rohland N., Lazaridis I., Nickel B., Viola B., Prüfer K., Meyer M., Kelso J., Reich D., Pääbo S., An early modern human from Romania with a recent Neanderthal ancestor. Nature 524, 216–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohrlach A. B., Papac L., Childebayeva A., Rivollat M., Villalba-Mouco V., Neumann G. U., Penske S., Skourtanioti E., van de Loosdrecht M., Akar M., Boyadzhiev K., Boyadzhiev Y., Deguilloux M.-F., Dobeš M., Erdal Y. S., Ernée M., Frangipane M., Furmanek M., Friederich S., Ghesquière E., Haluszko A., Hansen S., Küßner M., Mannino M., Özbal R., Reinhold S., Rottier S., Salazar-García D. C., Soler Diaz J., Stockhammer P. W., Roca de Togores Muñoz C., Aslihan Yener K., Posth C., Krause J., Herbig A., Haak W., Using Y-chromosome capture enrichment to resolve haplogroup H2 shows new evidence for a two-Path Neolithic expansion to Western Europe. Sci. Rep. 11, 15005 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peltzer A., Jäger G., Herbig A., Seitz A., Kniep C., Krause J., Nieselt K., EAGER: Efficient ancient genome reconstruction. Genome Biol. 17, 60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu Q., Mittnik A., Johnson P. L. F., Bos K., Lari M., Bollongino R., Sun C., Giemsch L., Schmitz R., Burger J., Ronchitelli A. M., Martini F., Cremonesi R. G., Svoboda J., Bauer P., Caramelli D., Castellano S., Reich D., Pääbo S., Krause J., A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 23, 553–559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mittnik A., Wang C.-C., Svoboda J., Krause J., A molecular approach to the sexing of the triple burial at the upper paleolithic site of Dolní Věstonice. PLOS ONE 11, e0163019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.M. Lipatov, K. Sanjeev, R. Patro, K. R. Veeramah, Maximum likelihood estimation of biological relatedness from low coverage sequencing data. bioRxiv 023374 [Preprint]. 29 July 2015. 10.1101/023374. [DOI]
- 48.Kennett D. J., Plog S., George R. J., Culleton B. J., Watson A. S., Skoglund P., Rohland N., Mallick S., Stewardson K., Kistler L., LeBlanc S. A., Whiteley P. M., Reich D., Perry G. H., Archaeogenomic evidence reveals prehistoric matrilineal dynasty. Nat. Commun. 8, 14115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monroy Kuhn J. M., Jakobsson M., Günther T., Estimating genetic kin relationships in prehistoric populations. PLOS ONE 13, e0195491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antonio M. L., Gao Z., Moots H. M., Lucci M., Candilio F., Sawyer S., Oberreiter V., Calderon D., Devitofranceschi K., Aikens R. C., Aneli S., Bartoli F., Bedini A., Cheronet O., Cotter D. J., Fernandes D. M., Gasperetti G., Grifoni R., Guidi A., La Pastina F., Loreti E., Manacorda D., Matullo G., Morretta S., Nava A., Fiocchi Nicolai V., Nomi F., Pavolini C., Pentiricci M., Pergola P., Piranomonte M., Schmidt R., Spinola G., Sperduti A., Rubini M., Bondioli L., Coppa A., Pinhasi R., Pritchard J. K., Ancient Rome: A genetic crossroads of Europe and the Mediterranean. Science 366, 708–714 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernandes D. M., Mittnik A., Olalde I., Lazaridis I., Cheronet O., Rohland N., Mallick S., Bernardos R., Broomandkhoshbacht N., Carlsson J., Culleton B. J., Ferry M., Gamarra B., Lari M., Mah M., Michel M., Modi A., Novak M., Oppenheimer J., Sirak K. A., Stewardson K., Mandl K., Schattke C., Özdoğan K. T., Lucci M., Gasperetti G., Candilio F., Salis G., Vai S., Camarós E., Calò C., Catalano G., Cueto M., Forgia V., Lozano M., Marini E., Micheletti M., Miccichè R. M., Palombo M. R., Ramis D., Schimmenti V., Sureda P., Teira L., Teschler-Nicola M., Kennett D. J., Lalueza-Fox C., Patterson N., Sineo L., Coppa A., Caramelli D., Pinhasi R., Reich D., The spread of steppe and Iranian-related ancestry in the islands of the western Mediterranean. Nat. Ecol. Evol. 4, 334–345 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marcus J. H., Posth C., Ringbauer H., Lai L., Skeates R., Sidore C., Beckett J., Furtwängler A., Olivieri A., Chiang C. W. K., Al-Asadi H., Dey K., Joseph T. A., Liu C.-C., Der Sarkissian C., Radzevičiūtė R., Michel M., Gradoli M. G., Marongiu P., Rubino S., Mazzarello V., Rovina D., La Fragola A., Serra R. M., Bandiera P., Bianucci R., Pompianu E., Murgia C., Guirguis M., Orquin R. P., Tuross N., van Dommelen P., Haak W., Reich D., Schlessinger D., Cucca F., Krause J., Novembre J., Genetic history from the Middle Neolithic to present on the Mediterranean island of Sardinia. Nat. Commun. 11, 939 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skourtanioti E., Erdal Y. S., Frangipane M., Balossi Restelli F., Yener K. A., Pinnock F., Matthiae P., Özbal R., Schoop U.-D., Guliyev F., Akhundov T., Lyonnet B., Hammer E. L., Nugent S. E., Burri M., Neumann G. U., Penske S., Ingman T., Akar M., Shafiq R., Palumbi G., Eisenmann S., D’Andrea M., Rohrlach A. B., Warinner C., Jeong C., Stockhammer P. W., Haak W., Krause J., Genomic history of Neolithic to Bronze Age Anatolia, Northern Levant, and Southern Caucasus. Cell 181, 1158–1175.e28 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D., Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heyd V., Kossinna’s smile. Antiquity 91, 348–359 (2017). [Google Scholar]
- 56.Fu Q., Posth C., Hajdinjak M., Petr M., Mallick S., Fernandes D., Furtwängler A., Haak W., Meyer M., Mittnik A., Nickel B., Peltzer A., Rohland N., Slon V., Talamo S., Lazaridis I., Lipson M., Mathieson I., Schiffels S., Skoglund P., Derevianko A. P., Drozdov N., Slavinsky V., Tsybankov A., Cremonesi R. G., Mallegni F., Gély B., Vacca E., Morales M. R. G., Straus L. G., Neugebauer-Maresch C., Teschler-Nicola M., Constantin S., Moldovan O. T., Benazzi S., Peresani M., Coppola D., Lari M., Ricci S., Ronchitelli A., Valentin F., Thevenet C., Wehrberger K., Grigorescu D., Rougier H., Crevecoeur I., Flas D., Semal P., Mannino M. A., Cupillard C., Bocherens H., Conard N. J., Harvati K., Moiseyev V., Drucker D. G., Svoboda J., Richards M. P., Caramelli D., Pinhasi R., Kelso J., Patterson N., Krause J., Pääbo S., Reich D., The genetic history of Ice Age Europe. Nature 534, 200–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Villalba-Mouco V., van de Loosdrecht M. S., Posth C., Mora R., Martínez-Moreno J., Rojo-Guerra M., Salazar-García D. C., Royo-Guillén J. I., Kunst M., Rougier H., Crevecoeur I., Arcusa-Magallón H., Tejedor-Rodríguez C., García-Martínez de Lagrán I., Garrido-Pena R., Alt K. W., Jeong C., Schiffels S., Utrilla P., Krause J., Haak W., Survival of Late Pleistocene hunter-gatherer ancestry in the Iberian peninsula. Curr. Biol. 29, 1169–1177.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Rivollat M., Jeong C., Schiffels S., Küçükkalıpçı İ., Pemonge M.-H., Rohrlach A. B., Alt K. W., Binder D., Friederich S., Ghesquière E., Gronenborn D., Laporte L., Lefranc P., Meller H., Réveillas H., Rosenstock E., Rottier S., Scarre C., Soler L., Wahl J., Krause J., Deguilloux M.-F., Haak W., Ancient genome-wide DNA from France highlights the complexity of interactions between Mesolithic hunter-gatherers and Neolithic farmers. Sci. Adv. 6, eaaz5344 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.M. S. van de Loosdrecht, M. A. Mannino, S. Talamo, V. Villalba-Mouco, C. Posth, F. Aron, G. Brandt, M. Burri, C. Freund, R. Radzeviciute, R. Stahl, A. Wissgott, L. Klausnitzer, S. Nagel, M. Meyer, A. Tagliacozzo, M. Piperno, S. Tusa, C. Collina, V. Schimmenti, R. D. Salvo, K. Prüfer, J.-J. Hublin, S. Schiffels, C. Jeong, W. Haak, J. Krause, Genomic and dietary transitions during the Mesolithic and Early Neolithic in Sicily. bioRxiv 2020.03.11.986158 [Preprint]. 12 March 2020; 10.1101/2020.03.11.986158. [DOI]
- 60.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alcover J. A., The first Mallorcans: Prehistoric colonization in the Western Mediterranean. J. World Prehist. 21, 19–84 (2008). [Google Scholar]
- 62.Micó R., Radiocarbon dating and Balearic prehistory: Reviewing the periodization of the prehistoric sequence. Radiocarbon 48, 421–434 (2006). [Google Scholar]
- 63.García Sanjuán L., Vargas Jiménez J. M., Cáceres Puro L. M., Costa Caramé M. E., Díaz-Guardamino Uribe M., Díaz-Zorita Bonilla M., Fernández Flores Á., Hurtado Pérez V., López Aldana P. M., Méndez Izquierdo E., Pajuelo Pando A., Rodríguez Vidal J., Wheatley D., Bronk Ramsey C., Delgado-Huertas A., Dunbar E., Mora González A., Bayliss A., Beavan N., Hamilton D., Whittle A., Assembling the dead, gathering the living: Radiocarbon dating and Bayesian modelling for Copper Age Valencina de la Concepción (Seville, Spain). J. World Prehist. 31, 179–313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aranda Jiménez G., Díaz-Zorita Bonilla M., Hamilton D., Milesi L., Sánchez Romero M., The radiocarbon chronology and temporality of the megalithic cemetery of Los Millares (Almería, Spain). Archaeol. Anthropol. Sci. 12, 104 (2020). [Google Scholar]
- 65.A. C. Valera, Social change in the late 3rd millennium BC in Portugal: The twilight of enclosures, in 2200 BC: A Climatic Break-down as a Cause for the Collapse of the Old World, H. Meller, H. Arz, R. Jung, R. Risch, Eds. (Landesmuseum für Vorgeschichte, Halle, 2014), pp. 409–428. [Google Scholar]
- 66.Blanco-González A., Lillios K. T., López-Sáez J. A., Drake B. L., Cultural, demographic and environmental dynamics of the Copper and Early Bronze Age in Iberia (3300–1500 BC): Towards an interregional multiproxy comparison at the time of the 4.2 ky BP event. J. World Prehist. 31, 1–79 (2018). [Google Scholar]
- 67.Hinz M., Schirrmacher J., Kneisel J., Rinne C., Weinelt M., The Chalcolithic–Bronze Age transition in southern Iberia under the influence of the 4.2 kyr event? A correlation of climatological and demographic proxies. J. Neolithic Archaeol. 21, 1–26 (2019). [Google Scholar]
- 68.Schirrmacher J., Kneisel J., Knitter D., Hamer W., Hinz M., Schneider R. R., Weinelt M., Spatial patterns of temperature, precipitation, and settlement dynamics on the Iberian Peninsula during the Chalcolithic and the Bronze Age. Quat. Sci. Rev. 233, 106220 (2020). [Google Scholar]
- 69.Lillios K. T., Blanco-González A., Drake B. L., López-Sáez J. A., Mid-late Holocene climate, demography, and cultural dynamics in Iberia: A multi-proxy approach. Quat. Sci. Rev. 135, 138–153 (2016). [Google Scholar]
- 70.V. Lull, R. Micó, C. Rihuete, R. Risch, in The Matter of Prehistory. Papers in Honour of Antonio Gilman Guillén, P. D. del Río, K. Lillios, I. Sastre, Eds. (Biblioteca Praehistorica Hispana, 36—Consejo Superior de Investigaciones Científicas, 2020), pp. 191–209. [Google Scholar]
- 71.Lazaridis I., Mittnik A., Patterson N., Mallick S., Rohland N., Pfrengle S., Furtwängler A., Peltzer A., Posth C., Vasilakis A., McGeorge P. J. P., Konsolaki-Yannopoulou E., Korres G., Martlew H., Michalodimitrakis M., Özsait M., Özsait N., Papathanasiou A., Richards M., Roodenberg S. A., Tzedakis Y., Arnott R., Fernandes D. M., Hughey J. R., Lotakis D. M., Navas P. A., Maniatis Y., Stamatoyannopoulos J. A., Stewardson K., Stockhammer P., Pinhasi R., Reich D., Krause J., Stamatoyannopoulos G., Genetic origins of the Minoans and Mycenaeans. Nature 548, 214–218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones E. R., Gonzalez-Fortes G., Connell S., Siska V., Eriksson A., Martiniano R., McLaughlin R. L., Gallego Llorente M., Cassidy L. M., Gamba C., Meshveliani T., Bar-Yosef O., Müller W., Belfer-Cohen A., Matskevich Z., Jakeli N., Higham T. F. G., Currat M., Lordkipanidze D., Hofreiter M., Manica A., Pinhasi R., Bradley D. G., Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun. 6, 8912 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ringbauer H., Novembre J., Steinrücken M., Parental relatedness through time revealed by runs of homozygosity in ancient DNA. Nat. Commun. 12, 5425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ringbauer H., Steinrücken M., Fehren-Schmitz L., Reich D., Increased rate of close-kin unions in the central Andes in the half millennium before European contact. Curr. Biol. 30, R980–R981 (2020). [DOI] [PubMed] [Google Scholar]
- 75.Goldberg A., Rosenberg N. A., Beyond 2/3 and 1/3: The complex signatures of sex-biased admixture on the X chromosome. Genetics 201, 263–279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mathieson I., Alpaslan-Roodenberg S., Posth C., Szécsényi-Nagy A., Rohland N., Mallick S., Olalde I., Broomandkhoshbacht N., Candilio F., Cheronet O., Fernandes D., Ferry M., Gamarra B., Fortes G. G., Haak W., Harney E., Jones E., Keating D., Krause-Kyora B., Kucukkalipci I., Michel M., Mittnik A., Nägele K., Novak M., Oppenheimer J., Patterson N., Pfrengle S., Sirak K., Stewardson K., Vai S., Alexandrov S., Alt K. W., Andreescu R., Antonović D., Ash A., Atanassova N., Bacvarov K., Gusztáv M. B., Bocherens H., Bolus M., Boroneanţ A., Boyadzhiev Y., Budnik A., Burmaz J., Chohadzhiev S., Conard N. J., Cottiaux R., Čuka M., Cupillard C., Drucker D. G., Elenski N., Francken M., Galabova B., Ganetsovski G., Gély B., Hajdu T., Handzhyiska V., Harvati K., Higham T., Iliev S., Janković I., Karavanić I., Kennett D. J., Komšo D., Kozak A., Labuda D., Lari M., Lazar C., Leppek M., Leshtakov K., Vetro D. L., Los D., Lozanov I., Malina M., Martini F., McSweeney K., Meller H., Menđušić M., Mirea P., Moiseyev V., Petrova V., Price T. D., Simalcsik A., Sineo L., Šlaus M., Slavchev V., Stanev P., Starović A., Szeniczey T., Talamo S., Teschler-Nicola M., Thevenet C., Valchev I., Valentin F., Vasilyev S., Veljanovska F., Venelinova S., Veselovskaya E., Viola B., Virag C., Zaninović J., Zäuner S., Stockhammer P. W., Catalano G., Krauß R., Caramelli D., Zariņa G., Gaydarska B., Lillie M., Nikitin A. G., Potekhina I., Papathanasiou A., Borić D., Bonsall C., Krause J., Pinhasi R., Reich D., The genomic history of southeastern Europe. Nature 555, 197–203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mittnik A., Massy K., Knipper C., Wittenborn F., Friedrich R., Pfrengle S., Burri M., Carlichi-Witjes N., Deeg H., Furtwängler A., Harbeck M., von Heyking K., Kociumaka C., Kucukkalipci I., Lindauer S., Metz S., Staskiewicz A., Thiel A., Wahl J., Haak W., Pernicka E., Schiffels S., Stockhammer P. W., Krause J., Kinship-based social inequality in Bronze Age Europe. Science 366, 731–734 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Dabney J., Knapp M., Glocke I., Gansauge M.-T., Weihmann A., Nickel B., Valdiosera C., García N., Pääbo S., Arsuaga J.-L., Meyer M., Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U.S.A. 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kircher M., Sawyer S., Meyer M., Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meyer M., Kircher M., Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, pdb.prot5448 (2010). [DOI] [PubMed] [Google Scholar]
- 81.Schubert M., Lindgreen S., Orlando L., AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 9, 88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H., Durbin R., Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ginolhac A., Jónsson H., Orlando L., Schubert M., Johnson P. L. F., mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A., Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weissensteiner H., Pacher D., Kloss-Brandstätter A., Forer L., Specht G., Bandelt H.-J., Kronenberg F., Salas A., Schönherr S., HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 44, W58–W63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van de Loosdrecht M., Bouzouggar A., Humphrey L., Posth C., Barton N., Aximu-Petri A., Nickel B., Nagel S., Talbi E. H., El Hajraoui M. A., Amzazi S., Hublin J.-J., Pääbo S., Schiffels S., Meyer M., Haak W., Jeong C., Krause J., Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science 360, 548–552 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Cockerham C. C., Higher order probability functions of identity of allelles by descent. Genetics 69, 235–246 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Walsh S., Liu F., Wollstein A., Kovatsi L., Ralf A., Kosiniak-Kamysz A., Branicki W., Kayser M., The HIrisPlex system for simultaneous prediction of hair and eye colour from DNA. Forensic Sci. Int. Genet. 7, 98–115 (2013). [DOI] [PubMed] [Google Scholar]
- 90.Patterson N., Price A. L., Reich D., Population structure and eigenanalysis. PLOS Genet. 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang C.-C., Reinhold S., Kalmykov A., Wissgott A., Brandt G., Jeong C., Cheronet O., Ferry M., Harney E., Keating D., Mallick S., Rohland N., Stewardson K., Kantorovich A. R., Maslov V. E., Petrenko V. G., Erlikh V. R., Atabiev B. C., Magomedov R. G., Kohl P. L., Alt K. W., Pichler S. L., Gerling C., Meller H., Vardanyan B., Yeganyan L., Rezepkin A. D., Mariaschk D., Berezina N., Gresky J., Fuchs K., Knipper C., Schiffels S., Balanovska E., Balanovsky O., Mathieson I., Higham T., Berezin Y. B., Buzhilova A., Trifonov V., Pinhasi R., Belinskij A. B., Reich D., Hansen S., Krause J., Haak W., Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions. Nat. Commun. 10, 590 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., Bender D., Maller J., Sklar P., de Bakker P. I. W., Daly M. J., Sham P. C., PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moorjani P., Sankararaman S., Fu Q., Przeworski M., Patterson N., Reich D., A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years. Proc. Natl. Acad. Sci. U.S.A. 113, 5652–5657 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.V. Lull, C. Rihuete Herrada, R. Risch, E. Celdrán Beltrán, M. I. Fregeiro Morador, C. Oliart Caravatti, C. Velasco Felipe, La Almoloya (Pliego-Murcia): Murcia: Integral, Sociedad Para el Desarrollo Rural (Integral - Sociedad para el Desarrollo Rural, Bullas, Spain, 2015).
- 95.Lull V., Rihuete Herrada C., Risch R., Bonora B., Celdrán Beltrán E., Fregeiro Morador M. I., Molero C., Moreno C., Oliart Caravatti C., Velasco-Felipe C., Andúgar L., Haak W., Villalba-Mouco V., Micó R., Emblems and spaces of power during the Argaric Bronze Age at La Almoloya, Murcia. Antiquity 95, 329–348 (2021). [Google Scholar]
- 96.V. Lull, R. Micó, C. Rihuete Herrada, R. Risch, La Bastida y Tira del Lienzo (Totana-Murcia): Murcia: Integral, Sociedad Para el Desarrollo Rural (Integral - Sociedad para el Desarrollo Rural, Bullas, Spain, 2015).
- 97.Knipper C., Rihuete-Herrada C., Voltas J., Held P., Lull V., Micó R., Risch R., Alt K. W., Reconstructing Bronze Age diets and farming strategies at the early Bronze Age sites of La Bastida and Gatas (southeast Iberia) using stable isotope analysis. PLOS ONE 15, e0229398 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Szécsényi-Nagy A., Roth C., Brandt G., Rihuete-Herrada C., Tejedor-Rodríguez C., Held P., García-Martínez-de-Lagrán Í., Arcusa Magallón H., Zesch S., Knipper C., Bánffy E., Friederich S., Meller H., Bueno Ramírez P., Barroso Bermejo R., de Balbín Behrmann R., Herrero-Corral A. M., Flores Fernández R., Alonso Fernández C., Jiménez Echevarria J., Rindlisbacher L., Oliart C., Fregeiro M.-I., Soriano I., Vicente O., Micó R., Lull V., Soler Díaz J., López Padilla J. A., Roca de Togores Muñoz C., Hernández Pérez M. S., Jover Maestre F. J., Lomba Maurandi J., Avilés Fernández A., Lillios K. T., Silva A. M., Magalhães Ramalho M., Oosterbeek L. M., Cunha C., Waterman A. J., Roig Buxó J., Martínez A., Ponce Martínez J., Hunt Ortiz M., Mejías-García J. C., Pecero Espín J. C., Cruz-Auñón Briones R., Tomé T., Carmona Ballestero E., Cardoso J. L., Araújo A. C., Liesau von Lettow-Vorbeck C., Blasco Bosqued C., Ríos Mendoza P., Pujante A., Royo-Guillén J. I., Esquembre Beviá M. A., Dos Santos Goncalves V. M., Parreira R., Morán Hernández E., Méndez Izquierdo E., Vega y Miguel J., Menduiña García R., Martínez Calvo V., López Jiménez O., Krause J., Pichler S. L., Garrido-Pena R., Kunst M., Risch R., Rojo-Guerra M. A., Haak W., Alt K. W., The maternal genetic make-up of the Iberian Peninsula between the Neolithic and the Early Bronze Age. Sci. Rep. 7, 15644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.J. A. Soler Díaz, C. Roca de Togores, Ritual funerario en la Cova d’En Pardo ca. 3.350–2.850 CAL ANE: Espacialidad, cronología y territorio cultural, in Cova d’En Pardo: Arqueología en la Memoria: Excavaciones de M. Tarradell, V. Pascual y E. Llobregat (1961–1965), Catálogo de Materiales del Museo de Alcoy y Estudios a Partir de las Campañas del MARQ (1993–2007) en la Cavidad de Planes, Alicante. (Museo Arqueológico de Alicante—MARQ, 2012), pp. 205–248.
- 100.J. A. S. Díaz, C. Roca de Togores Muñoz, Estudio de los restos humanos encontrados en las intervenciones practicadas en los años 1961 y 1965 en la Cova d’En Pardo, Planes, Alicante: análisis antropológico y aproximación a su contexto cultural. Saguntum Extra 2 (1999), pp. 369–377.
- 101.C. Roca de Togores Muñoz, J. A. S. Díaz, Restos humanos en la Cova d’en Pardo (Planes). Problemática y avance de resultados de la investigación antropológica en una cavidad de inhumación múltiple excavada en dos etapas: 1961-1965 y 1993-2007, in Cova d’En Pardo: Arqueología en la Memoria: Excavaciones de M. Tarradell, V. Pascual y E. Llobregat (1961–1965), Catálogo de Materiales del Museo de Alcoy y Estudios a Partir de las Campañas del MARQ (1993–2007) en la Cavidad de Planes, Alicante (Museo Arqueológico de Alicante—MARQ, 2012), pp. 193–204.
- 102.V. Lull, R. Micó, C. Rihuete, R. Risch, La Cova des Càrritx y la Cova des Mussol. Ideología y Sociedad en la Prehistoria de de Menorca (Consell Insular de Menorca, 1999). [Google Scholar]
- 103.M. De Cet, Long-term Social Development on a Mediterranean Island: Menorca Between 1600 BCE and 1900 CE (In Kommission bei Verlag Dr. Rudolf Habelt GmbH, 2017). [Google Scholar]
- 104.Günther T., Valdiosera C., Malmström H., Ureña I., Rodriguez-Varela R., Sverrisdóttir Ó. O., Daskalaki E. A., Skoglund P., Naidoo T., Svensson E. M., Bermúdez de Castro J. M., Carbonell E., Dunn M., Storå J., Iriarte E., Arsuaga J. L., Carretero J.-M., Götherström A., Jakobsson M., Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc. Natl. Acad. Sci. U.S.A. 112, 11917–11922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.de Barros Damgaard P., Marchi N., Rasmussen S., Peyrot M., Renaud G., Korneliussen T., Moreno-Mayar J. V., Pedersen M. W., Goldberg A., Usmanova E., Baimukhanov N., Loman V., Hedeager L., Pedersen A. G., Nielsen K., Afanasiev G., Akmatov K., Aldashev A., Alpaslan A., Baimbetov G., Bazaliiskii V. I., Beisenov A., Boldbaatar B., Boldgiv B., Dorzhu C., Ellingvag S., Erdenebaatar D., Dajani R., Dmitriev E., Evdokimov V., Frei K. M., Gromov A., Goryachev A., Hakonarson H., Hegay T., Khachatryan Z., Khaskhanov R., Kitov E., Kolbina A., Kubatbek T., Kukushkin A., Kukushkin I., Lau N., Margaryan A., Merkyte I., Mertz I. V., Mertz V. K., Mijiddorj E., Moiyesev V., Mukhtarova G., Nurmukhanbetov B., Orozbekova Z., Panyushkina I., Pieta K., Smrčka V., Shevnina I., Logvin A., Sjögren K.-G., Štolcová T., Taravella A. M., Tashbaeva K., Tkachev A., Tulegenov T., Voyakin D., Yepiskoposyan L., Undrakhbold S., Varfolomeev V., Weber A., Sayres M. A. W., Kradin N., Allentoft M. E., Orlando L., Nielsen R., Sikora M., Heyer E., Kristiansen K., Willerslev E., 137 ancient human genomes from across the Eurasian steppes. Nature 557, 369–374 (2018). [DOI] [PubMed] [Google Scholar]
- 106.Mittnik A., Wang C.-C., Pfrengle S., Daubaras M., Zariņa G., Hallgren F., Allmäe R., Khartanovich V., Moiseyev V., Tõrv M., Furtwängler A., Andrades Valtueña A., Feldman M., Economou C., Oinonen M., Vasks A., Balanovska E., Reich D., Jankauskas R., Haak W., Schiffels S., Krause J., The genetic prehistory of the Baltic Sea region. Nat. Commun. 9, 442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rodríguez-Varela R., Günther T., Krzewińska M., Storå J., Gillingwater T. H., MacCallum M., Arsuaga J. L., Dobney K., Valdiosera C., Jakobsson M., Götherström A., Girdland-Flink L., Genomic analyses of pre-European conquest human remains from the Canary Islands reveal close affinity to modern North Africans. Curr. Biol. 28, 1677–1679 (2018). [DOI] [PubMed] [Google Scholar]
- 108.Narasimhan V. M., Patterson N., Moorjani P., Rohland N., Bernardos R., Mallick S., Lazaridis I., Nakatsuka N., Olalde I., Lipson M., Kim A. M., Olivieri L. M., Coppa A., Vidale M., Mallory J., Moiseyev V., Kitov E., Monge J., Adamski N., Alex N., Broomandkhoshbacht N., Candilio F., Callan K., Cheronet O., Culleton B. J., Ferry M., Fernandes D., Freilich S., Gamarra B., Gaudio D., Hajdinjak M., Harney É., Harper T. K., Keating D., Lawson A. M., Mah M., Mandl K., Michel M., Novak M., Oppenheimer J., Rai N., Sirak K., Slon V., Stewardson K., Zalzala F., Zhang Z., Akhatov G., Bagashev A. N., Bagnera A., Baitanayev B., Bendezu-Sarmiento J., Bissembaev A. A., Bonora G. L., Chargynov T. T., Chikisheva T., Dashkovskiy P. K., Derevianko A., Dobeš M., Douka K., Dubova N., Duisengali M. N., Enshin D., Epimakhov A., Fribus A. V., Fuller D., Goryachev A., Gromov A., Grushin S. P., Hanks B., Judd M., Kazizov E., Khokhlov A., Krygin A. P., Kupriyanova E., Kuznetsov P., Luiselli D., Maksudov F., Mamedov A. M., Mamirov T. B., Meiklejohn C., Merrett D. C., Micheli R., Mochalov O., Mustafokulov S., Nayak A., Pettener D., Potts R., Razhev D., Rykun M., Sarno S., Savenkova T. M., Sikhymbaeva K., Slepchenko S. M., Soltobaev O. A., Stepanova N., Svyatko S., Tabaldiev K., Teschler-Nicola M., Tishkin A. A., Tkachev V. V., Vasilyev S., Velemínský P., Voyakin D., Yermolayeva A., Zahir M., Zubkov V. S., Zubova A., Shinde V. S., Lalueza-Fox C., Meyer M., Anthony D., Boivin N., Thangaraj K., Kennett D. J., Frachetti M., Pinhasi R., Reich D., The formation of human populations in South and Central Asia. Science 365, eaat7487 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Klinefelter H. F. Jr., Reifenstein E. C. Jr., Syndrome characterized by gynecomastia, aspermatogenesis without A-Leydigism, and increased excretion of follicle-stimulating hormone. J. Clin. Endocrinol. 2, 615–627 (1942). [Google Scholar]
- 110.Jacobs P., Baikie A. G., MacGregor T. N., Maclean N., Harnden D. G., Evidence for the existence of the human “super female.” Lancet 274, 423–425 (1959). [DOI] [PubMed] [Google Scholar]
- 111.Boada R., Janusz J., Hutaff-Lee C., Tartaglia N., The cognitive phenotype in Klinefelter syndrome: A review of the literature including genetic and hormonal factors. Dev. Disabil. Res. Rev. 15, 284–294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ebenesersdóttir S. S., Sandoval-Velasco M., Gunnarsdóttir E. D., Jagadeesan A., Guðmundsdóttir V. B., Thordardóttir E. L., Einarsdóttir M. S., Moore K. H. S., Sigurðsson Á., Magnúsdóttir D. N., Jónsson H., Snorradóttir S., Hovig E., Møller P., Kockum I., Olsson T., Alfredsson L., Hansen T. F., Werge T., Cavalleri G. L., Gilbert E., Lalueza-Fox C., Walser J. W. III, Kristjánsdóttir S., Gopalakrishnan S., Árnadóttir L., Magnússon Ó. Þ., Gilbert M. T. P., Stefánsson K., Helgason A., Ancient genomes from Iceland reveal the making of a human population. Science 360, 1028–1032 (2018). [DOI] [PubMed] [Google Scholar]
- 113.Otter M., Schrander-Stumpel C. T., Curfs L. M. G., Triple X syndrome: A review of the literature. Eur. J. Hum. Genet. 18, 265–271 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rafique M., AlObaid S., Al-Jaroudi D., 47, XXX syndrome with infertility, premature ovarian insufficiency, and streak ovaries. Clin. Case Rep. 7, 1238–1241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schroeder H., Margaryan A., Szmyt M., Theulot B., Włodarczak P., Rasmussen S., Gopalakrishnan S., Szczepanek A., Konopka T., Jensen T. Z. T., Witkowska B., Wilk S., Przybyła M. M., Pospieszny Ł., Sjögren K.-G., Belka Z., Olsen J., Kristiansen K., Willerslev E., Frei K. M., Sikora M., Johannsen N. N., Allentoft M. E., Unraveling ancestry, kinship, and violence in a Late Neolithic mass grave. Proc. Natl. Acad. Sci. U.S.A. 116, 10705–10710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Walsh S., Chaitanya L., Breslin K., Muralidharan C., Bronikowska A., Pospiech E., Koller J., Kovatsi L., Wollstein A., Branicki W., Liu F., Kayser M., Global skin colour prediction from DNA. Hum. Genet. 136, 847–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chaitanya L., Breslin K., Zuñiga S., Wirken L., Pośpiech E., Kukla-Bartoszek M., Sijen T., de Knijff P., Liu F., Branicki W., Kayser M., Walsh S., The HIrisPlex-S system for eye, hair and skin colour prediction from DNA: Introduction and forensic developmental validation. Forensic Sci. Int. Genet. 35, 123–135 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S9
Legends for tables S1 and S2
References
Tables S1 and S2