Abstract
We have previously shown that poly(A) polymerase (PAP) is negatively regulated by cyclin B-cdc2 kinase hyperphosphorylation in the M phase of the cell cycle. Here we show that cyclin B1 binds PAP directly, and we demonstrate further that this interaction is mediated by a stretch of amino acids in PAP with homology to the cyclin recognition motif (CRM), a sequence previously shown in several cell cycle regulators to target specifically G1-phase-type cyclins. We find that PAP interacts with not only G1- but also G2-type cyclins via the CRM and is a substrate for phosphorylation by both types of cyclin-cdk pairs. PAP's CRM shows novel, concentration-dependent effects when introduced as an 8-mer peptide into binding and kinase assays. While higher concentrations of PAP's CRM block PAP-cyclin binding and phosphorylation, lower concentrations induce dramatic stimulation of both activities. Our data not only support the notion that PAP is directly regulated by cyclin-dependent kinases throughout the cell cycle but also introduce a novel type of CRM that functionally interacts with both G1- and G2-type cyclins in an unexpected way.
Almost all eukaryotic mRNAs contain a string of adenylate residues at their 3′ ends. This structure, known as the poly(A) tail, has been implicated in mRNA localization, stability, and translation (reviewed in references 41, 52, and 59). The polyadenylation reaction affects, and is affected by, other steps in mRNA synthesis, such as transcription, splicing, and capping (e.g., references 11, 12, 29, 44, and 62). Therefore, polyadenylation could constitute a significant point of regulation utilized by the cell to control gene expression (reviewed in reference 4). A growing body of evidence suggests this to be the case in early development (59), in cell differentiation (e.g., references 19 and 56) and in the M phase of the cell cycle (9).
mRNA 3′-end formation is achieved in a two-step reaction: endonucleolytic cleavage of the pre-mRNA followed by synthesis of the poly(A) tail. Components of the basal polyadenylation machinery, which constitute a complex array of protein factors, work together to catalyze and tightly couple these two reactions (reviewed in references 8, 34, and 58). Poly(A) polymerase (PAP) is a single subunit enzyme responsible for adding the adenylate residues onto the cleaved mRNA, and it is also required in many cases for the cleavage reaction in vitro. Multiple additional, multi-subunit proteins are involved in 3′ end formation: cleavage-polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factors I and II, and RNA polymerase II. CPSF is required for both steps of the reaction and is responsible for recognizing the polyadenylation signal AAUAAA. CPSF binds very efficiently AAUAAA when complexed with CstF, which is itself required for efficient cleavage in vitro. CstF also binds RNA specifically to the GU-rich element found in many polyadenylation sites. The complexes most likely to be directly involved in the endonucleolytic cleavage of the pre-mRNA are CFI and CFII. The newest known essential component of 3′ processing is RNA polymerase II, specifically the C-terminal domain of its largest subunit (CTD). The CTD was shown to be required for the cleavage reaction in vitro (29), and interactions between the CTD and CstF and CPSF have been observed (44).
Our laboratory and others have collected data supporting the regulation of polyadenylation via control of PAP activity. The U1 snRNP A protein (U1A) is able to repress PAP's polymerase activity via a direct interaction between U1A bound to sequences in the U1A pre-mRNA 3′ untranslated region and the C terminus of PAP, thereby negatively autoregulating its own synthesis (22–24). We and others have been studying the effect of phosphorylation of PAP on its activity in in vitro and in vivo assays (e.g., references 3, 9, and 64). All known vertebrate PAPs contain a C-terminal Ser-Thr-rich domain with multiple cyclin-dependent kinase (cdk) sites. These sites are phosphorylated in vitro and in vivo by cyclin B-p34cdc2 (10). In M-phase cells, where cyclin B-p34cdc2 is most active, PAP is hyperphosphorylated and its activity is repressed (9). Chicken B cells expressing a PAP with two consensus cdk sites mutated show growth defects compared to cells expressing similar levels of the wild-type enzyme (64).
Cyclin B-p34cdc2 is a member of the cdk family, with all members being heterodimers containing a kinase subunit (the cdk) and a regulatory subunit (the cyclin). These kinases are important players in regulating the entry into and progression of the eukaryotic cell cycle (reviewed in references 32 and 46). As such their activities are tightly controlled to ensure a proper cell cycle. One of the most well-studied mechanisms of cdk regulation is the requirement of the cyclin binding to the catalytic subunit for its activation (e.g., references 31, 33, and 40). Binding of the cyclin imparts upon the kinase a structure conducive to catalysis (33).
In addition to influencing the structure of the catalytic subunit, the cyclin also apparently imparts upon the cdk much of its substrate specificity (e.g., references 14, 31, 35, 47, and 48). Initially through analysis of the crystal structure of cyclin A-cdk2 complexed with the cdk-inhibitory protein p27 (51), a motif in p27 was derived that was suggested to be responsible for the interaction of the inhibitor with the cyclin. This sequence, the cyclin recognition motif (CRM), is now known to be shared by inhibitors (p21, p27, and p57) and substrates (e.g., E2F-1, p130, p107, and pRB) alike (1, 2, 6, 15, 38, 42, 43, 45, 49, 53, 65). The CRM's interaction with the cyclin relies on contact with residues in a hydrophobic patch (50, 51, 54), which are strongly conserved in cyclins A, B, D1, and E, although CRM-dependent interactions have only been detected with the G1-specific cyclins, A, D1, and E not with cyclin B1. A cyclin A mutated in its hydrophobic patch was unable to interact with CRM-containing proteins and lost its ability to drive cells out of G1 (54), underlining the importance of CRM-mediated interactions for the cdk in controlling the progression of the cell cycle. When the CRM's interaction with the cyclin is disrupted, decreased phosphorylation of the substrate is frequently observed (1, 2, 38, 43, 54). Not all cdk substrates contain a CRM, and why some do and others do not is not known.
Here we further investigate PAP's regulation by cdks. We extend our previous findings that PAP is a target for cdk phosphorylation by showing that PAP binds directly to cyclins, both in vivo and in vitro. PAP interacts with both G1- and G2-type cyclins and is a substrate for phosphorylation by both types of cyclin-cdk pairs. Cyclin binding is mediated by a stretch of amino acids with similarity to the consensus CRM. An 8-mer peptide spanning PAP's CRM has novel concentration-dependent effects on binding and phosphorylation of PAP by cdks. Unexpectedly, lower concentrations of the 8-mer peptide actually stimulate PAP binding and phosphorylation by cyclin B-cdc2. Higher concentrations abolish the PAP-cyclin interaction and, in in vitro kinase reactions, specifically inhibit both PAP's and pRB's phosphorylation by G2- as well as G1 cdks. With its ability to interact with both G1- and G2-type cyclins, PAP's CRM allows for the possible regulation of polyadenylation throughout the cell cycle, and the novel mechanism by which it interacts with cyclins provides possible insight into the mechanism of substrate targeting by cyclins.
MATERIALS AND METHODS
Coimmunoprecipitation.
Sf9 insect cells were infected with 1 PFU of PAP-expressing per cell and/or 3 PFU of cyclin B1-expressing recombinant baculoviruses. After 40 h at 27°C, cells were harvested and lysed in 50 mM Tris (pH 8.0)–150 mM NaCl–0.1% aprotinin–10 mM benzamidine–30 μg of leupeptin per ml–1 mg of bacitracin per ml–10 mg of α2-macroglobulin per ml–0.35 mM phenylmethylsulfonyl fluoride for 15 min on ice. Lysates were spun at 37,000 × g for 15 min at 4°C, and supernatants were collected. Protein G-Sepharose beads (Pharmacia), anti-PAP polyclonal antisera, and supernatants were rocked for 3 h at 4°C. After an extensive washing with 50 mM Tris (pH 7.2)–200 mM NaCl–0.1% NP-40, samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 10% polyacrylamide gel and Western blot analysis. Filters were probed with a monoclonal cyclin B1 antibody (Santa Cruz).
Far-Western assays.
The modified far-Western assay was carried out as previously described (37). One microgram of protein was used for each strip. After renaturation and blocking, 100 ng of purified cyclin B1-cdc2 was incubated with the strips for 12 h at 4°C. After extensive washing, strips were probed with the anti-cyclin B1 monoclonal antibody.
GST binding assays.
Glutathione S-transferase (GST)-cyclin fusion proteins were expressed in recombinant baculovirus-infected cells. Infection and lysis were carried out as described earlier (9). GST was expressed in Escherichia coli (JM101), induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 3 h. Proteins were affinity purified using glutathione-Sepharose beads (Amersham Pharmacia Biotech AB). After an extensive washing with NETN (20 mM Tris, pH 8.0; 100 mM NaCl; 0.5% NP-40; 1 mM EDTA), proteins were eluted with 120 mM reduced glutathione (Sigma) and dialyzed against 10 mM HEPES (pH 7.5)–5 mM NaCl–0.1 mM EDTA–1 mM dithiothreitol (DTT)–25% glycerol. Two micrograms of each GST protein was rebound to glutathione-Sepharose beads. Unbound proteins were washed away, and in vitro-translated 35S-labeled PAPs (2 μl), purified bacterial PAP (100 ng), or purified hemagglutinin (HA)-cdk2 (100 ng) were incubated with beads for 2 h at 24°C in a total volume of 40 μl. For assays with peptides, 45 ng of purified GST-B1 bound to glutathione-Sepharose beads, various amounts of peptides, and 1 μl of in vitro-translated 35S-labeled PAP were incubated for 2 h at 24°C in a total volume of 200 μl. In vitro-translated proteins were produced using TNT rabbit reticulocyte lysate (Promega). Bovine PAP I (species II in Fig. 2B) was produced from bovine PAP I cDNA subcloned into a pET-14b plasmid containing a T7 promoter. The C-terminal truncated PAP (amino acids [aa] 1 to 434) was in vitro transcribed and translated with the above-mentioned PAP I-pET14b plasmid linearized with DraIII. One N-terminal truncated PAP (309 to 689 aa) was produced from a template constructed by blunt-end ligation of PAP I-pET14b, cut with KpnI and NcoI. The second, N-terminal truncated PAP (539 to 689 aa) was also produced from a template constructed by blunt-end ligation of PAP I-pET14b but cut with SpeI and NdeI.
FIG. 2.
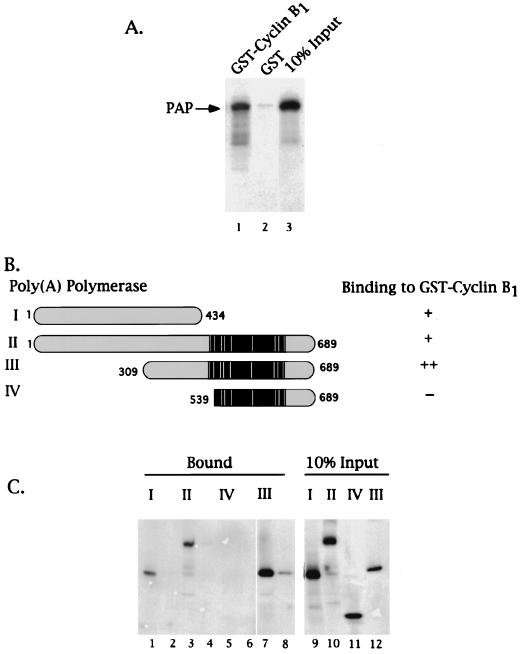
PAP binds cyclin B1 via residues N-terminal of its Ser-Thr-rich regulatory region. GST-cyclin B1 pull-down assays were performed using purified GST or GST fusion proteins bound to a glutathione matrix and in vitro-translated 35S-labeled PAPs (2 μl). (A) Autoradiogram of the eluates of either GST-cyclin B1 (lane 1) or GST (lane 2) glutathione matrices and 10% of the input PAP I (lane 3). An arrow on the left indicates the position of PAP I. (B) Schematic representation of PAP species used in the assay depicted in panel C and summary of results. The black region indicates the Ser-Thr-rich region, and the white bars indicate the sites for cdk phosphorylation. The bipartite nuclear localization signal sequences are boxed in gray. A plus sign indicates observed binding, two plus signs indicate strongest binding, and a minus sign indicates no binding was observed. (C) Autoradiogram of 35S-labeled PAPs bound to either GST-cyclin B1 (lanes 1, 3, 5, and 7) or GST (lanes 2, 4, 6, and 8). Lanes 9 to 12 are 10% of the input PAPs. The roman numerals indicate the PAP species used as graphically represented in panel C.
Protein phosphorylation.
A 200-ng portion of purified pRB, PAP or histone H1 was incubated with 80 ng of purified cdk for 20 min at 30°C in kinase buffer (25 mM HEPES, pH 7.5; 5 mM MgCl2; 100 mM ATP; 0.5 μCi of [γ-32P]ATP; 0.1 mM DTT) in a total volume of 30 μl. Olomoucine (Calbiochem) and roscovitine (Calbiochem) were added where indicated at the concentrations shown in the figure legends. Where indicated, peptides were added at the concentrations shown. Peptides were added to reaction mixtures prior to substrates. The cyclin B1-cdc2 preparations used in the experiments shown in Fig. 7 and in Fig. 8B varied slightly in their specific activities, resulting in small differences in phosphorylation at lower concentrations.
FIG. 7.
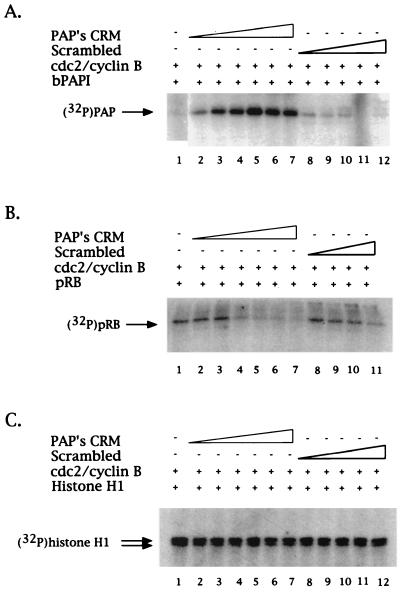
Phosphorylation of PAP by cyclin B1-cdc2 is CRM enhanced. (A) Lower concentrations of CRM stimulate cyclin B1-cdc2 phosphorylation of PAP. Purified PAP and cyclin B1-cdc2 were incubated under kinase conditions in the presence of [γ-32P]ATP and increasing concentrations (4.3, 5.2, 6.1, 7, 7.8, and 8.7 μM) of either PAP's CRM (lanes 2 to 7) or an 8-mer peptide of scrambled sequence (lanes 8 to 12) or else they were incubated with no peptide (lane 1). Phosphorylated proteins were resolved by SDS-PAGE and detected by autoradiography. An arrow on the left indicates the position of PAP. (B) Equivalent concentrations of CRM have either no effect or an inhibitory effect on pRB's phosphorylation by cyclin B1-cdc2. Kinase reactions were carried out as described above. Lanes 2 to 7 contained increasing amounts (4.3, 5.2, 6.1, 7, 7.8, and 8.7 μM) of PAP's CRM, and lanes 8 to 11 contained increasing amounts (4.3, 6.1, 7.8, and 8.7 μM) of the scrambled sequence peptide. An arrow on the left indicates the position of pRB. (C) No CRM peptide effect is observed on phosphorylation of histone H1 by cyclin B1-cdc2. As above, the same concentrations of either the CRM 8mer (lanes 2 to 7) or the scrambled sequence 8-mer (lanes 8 to 12) were added to the kinase assays. An arrow on the left indicates the position of histone H1.
FIG. 8.
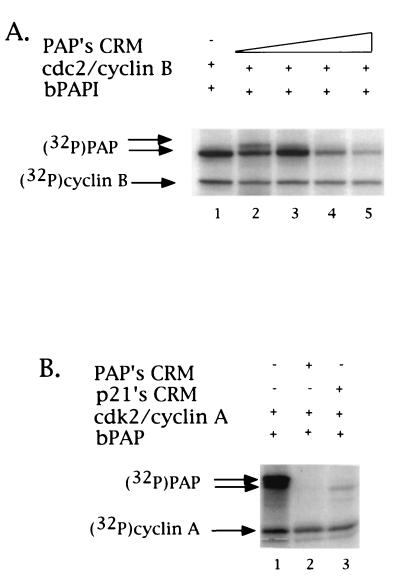
Phosphorylation of PAP by cdks is CRM dependent. (A) Higher concentrations of CRM specifically inhibit cyclin B1-cdc2 phosphorylation of PAP. Kinase reactions were carried out as described in Fig. 7. Samples in lanes 1 to 5 contained increasing amounts of CRM peptide (0, 5.8, 11.6, 17.3, and 22.5 μM). Arrows to the left indicate the positions of PAP and cyclin B1. (B) Phosphorylation of PAP by cyclin A-cdk2 is CRM dependent. Inhibition of cdk activity toward PAP by either by PAP's CRM or p21's CRM was tested. Purified PAP and cyclin A-cdk2 were incubated under kinase conditions in the presence of [γ-32P]ATP (lane 1). Kinase reactions contained 23 μM concentrations of either PAP's CRM (lane 2), p21's CRM (lane 3), or no peptide (lane 1). Arrows to the left indicate the positions of PAP and cyclin A.
RESULTS
PAP interacts both in vivo and in vitro with cyclin B1.
We have been studying how PAP is regulated by cyclin B-p34cdc2 phosphorylation, which involves multiple cdk consensus (S/TPXK/R) and nonconsensus (S/TP) sites and the inhibition of its catalytic activity upon hyperphosphorylation (9, 10). Given that some cdk substrates appear to be targeted by a direct interaction with the cyclin subunit, it seemed that PAP, with its multiple phosphorylation sites, would be a good candidate for such an association. To investigate this possibility, we first sought to test whether PAP can interact with a cyclin in vivo. To this end, Sf9 insect cells were coinfected with recombinant baculoviruses expressing bovine PAP I and human cyclin B1. (PAP I and II arise from alternatively spliced mRNAs [63]. They behave indistinguishably in their interaction with cdk-cyclins and have been used interchangeably in the experiments described here.) Total cell extracts were prepared and subjected to immunoprecipitation with a rabbit polyclonal anti-PAP antibody, and the immunocomplex was analyzed by Western blotting using an anti-cyclin B1 monoclonal antibody (see Materials and Methods). Figure 1A shows that cyclin B was present in the anti-PAP immunocomplex (lane 1) and that this was dependent on coexpression of PAP (lane 2). Lanes 3 and 4 show a Western blot with anti-cyclin B1 antibodies of the lysates prior to immunoprecipitation, which indicates that a significant fraction of the cyclin was associated with PAP. We have been unable to coimmunoprecipitate PAP I and cyclin B1 from uninfected cells. However, this is, to our knowledge, consistent with all other studies examining cyclin-substrate associations and suggests that the interactions are relatively weak and/or transient.
FIG. 1.
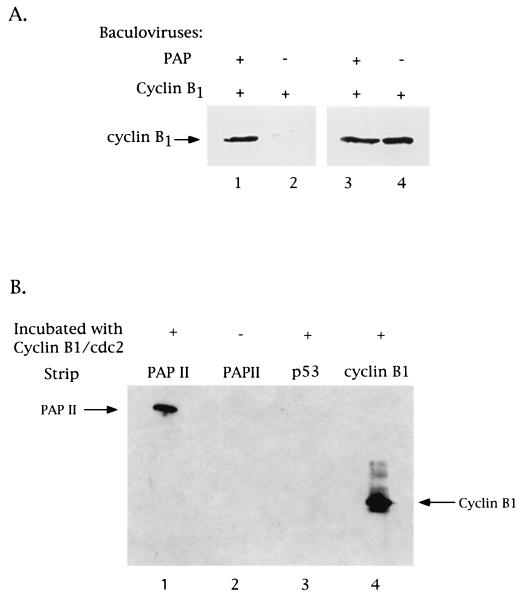
PAP binds cyclin B1 in vivo and in vitro. (A) PAP associates with cyclin B1 in vivo. Coimmunoprecipitation and Western blot analysis of Sf9 insect cell extracts made from cells infected with recombinant baculoviruses expressing either PAP plus cyclin B1 or cyclin B1 alone. Lysates were immunoprecipitated with a polyclonal antibody raised against PAP. The immunoprecipitates (lanes 1 and 2) and 10% of the cell extracts used (lanes 3 and 4) were subjected to SDS-PAGE and subsequent Western blot analysis using a monoclonal antibody against human cyclin B1. The position of cyclin B1 is indicated. (B) PAP binds cyclin B1-cdc2 directly. Purified PAP II (1 μg; strips 1 and 2), purified p53 (1 μg; strip 3), and bacterial whole-cell extract expressing human cyclin B1 (strip 4) were first immobilized on nitrocellulose. Following renaturation by serial dilution with guanidine-HCl, strips 1, 3, and 4 were incubated with purified cyclin B1-cdc2 (100 ng). After washing, cyclin B1 was detected by immunoreactivity to the anti-cyclin B1 antibody. An arrow on the left indicates the position of PAP II. An arrow on the right indicates the position of cyclin B1.
To characterize the PAP-cyclin B1 interaction further, baculovirus-produced and purified histidine-tagged PAP II and human cyclin B1-flu epitope-tagged p34cdc2 proteins were used in a modified far-Western protein-protein interaction assay (Fig. 1B). Purified PAP II (1 μg) was immobilized on nitrocellulose by Western blotting and renatured by serial dilution with guanidine-HCl, and strips were incubated with purified cyclin B1-p34cdc2 (100 ng). After extensive washing (see Materials and Methods), the presence of cyclin B1 bound to PAP II was determined by its immunoreactivity to the anti-cyclin B1 antibody (strip 1). The absence of cross-reactivity of PAP II with the anti-cyclin B1 antibody was established by incubating a strip of PAP II under the above conditions except for excluding incubation with cyclin B1 (strip 2). A strip containing p53 instead of PAP was used to demonstrate the specificity of the interaction (strip 3). Strip 4 contained cyclin B1. The results of this experiment indicate the existence of a direct interaction between PAP and cyclin B1-p34cdc2.
PAP interacts with cyclin B1 via sequences N-terminal of the Ser-Thr-rich regulatory region.
To address the role of p34cdc2, if any, in the interaction of PAP with cyclin B1 and to extend the above results to soluble proteins, a GST pull-down assay was employed. A GST-B1 fusion protein was purified from recombinant baculovirus-infected Sf9 cells, rebound to a glutathione-agarose matrix, and incubated with in vitro-translated 35S-labeled PAP I. After extensive washing and elution, the elute was subjected to SDS-PAGE and the presence of PAP I was determined by autoradiography. In Fig. 2A, PAP I was detected in the eluate of the cyclin B1 matrix (lane 1) but not in that of a GST control (lane 2), confirming the interaction of cyclin B1 with PAP.
The same assay was next used to determine whether the Ser-Thr-rich C terminus of PAP, which contains the seven known sites for cyclin B1-p34cdc2 phosphorylation (10), also contains the residues responsible for associating with cyclin B1. (Although, perhaps arguing against this, we have observed no effect of the phosphorylation status of PAP on the enzyme's ability to bind cyclin B1; results not shown). For this experiment, full-length in vitro-translated [35S]methionine-labeled PAP I was again incubated with GST-cyclin B1 bound to glutathione agarose beads, but this time alongside of both N-terminal and C-terminal truncated PAPs (Fig. 2B). Both the wild-type and N-terminal truncated species contain the Ser-Thr-rich region, the last comprising only this region (species II, III, and IV in panel B), but the C-terminal truncated PAP (species I in panel B) contains only residues N terminal of the regulatory region. As seen in Fig. 2C, lanes 1, 3, 5, and 7, only those species with residues N terminal to the regulatory region retained the ability to bind cyclin B1, proving that the Ser-Thr-rich region is neither necessary nor sufficient for PAP's association with cyclin B1, whereas sequences N terminal to this region, between residues 309 and 434, are sufficient. Preliminary data (not shown) suggest the possibility of a weak, secondary cyclin binding site N-terminal of residue 309, although this has not been studied further.
PAP contains a novel cyclin recognition motif.
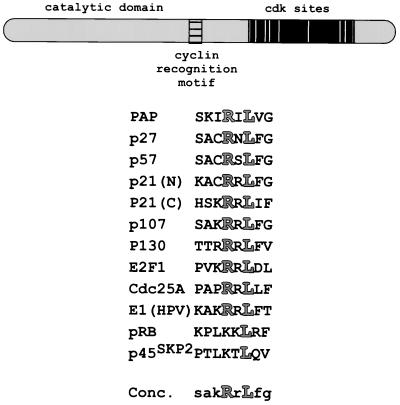
Inspection of the PAP sequence revealed that it contains a stretch of conserved amino acids with similarity to the consensus CRM, situated just N terminal of the Ser-Thr-rich regulatory region (Fig. 3). Although other CRM-containing cdk substrates analyzed to date do not appear to interact with cyclin B1, B-type cyclins do contain the residues in other cyclins necessary to contact the CRM (2, 50, 54). The interaction between PAP and GST-cyclin B1 is also resistant to high salt concentrations (data not shown), which could suggest a hydrophobic association, another trait of a CRM-mediated interaction (50, 54).
FIG. 3.
PAP contains a CRM. A schematic representation of PAP is shown at the top. The cyclin recognition motif is boxed and striped, and the Ser-Thr-rich regulatory region is boxed in black, with white bars representing the cdk sites and gray bars representing the nuclear localization sequences. An alignment of CRMs with the highly conserved arginine and leucine residues highlighted is shown below. The CRM consensus contains the nearly invariant arginine and leucine residues and, in lowercase, the residues found most frequently at the other positions.
To test the functional significance of the putative PAP CRM, a series of experiments was carried out using a synthetic peptide spanning these eight residues of PAP (Fig. 3). These experiments were based on studies of the p21 family of cdk inhibitors and of cdk substrates, including the transcription factor E2F-1, the retinoblastoma protein (pRB), and the related protein p107 (1, 2). We first took advantage of the finding that phosphorylation of pRB by cyclin A-cdk2, cyclin E-cdk2, and cyclin D1-cdk4 can be inhibited by the addition of increasing amounts of CRM-containing peptides (from p21, E2F-1, or pRB) to in vitro kinase assays and tested whether PAP's potential CRM-containing peptide could also inhibit pRB phosphorylation. Baculovirus-produced and purified flu-tagged pRB and human cyclin A-flu-tagged cdk2 were incubated under kinase conditions in the presence of [γ-32P]ATP and analyzed by SDS-PAGE and subsequent autoradiography. As seen in Fig. 4A, PAP's CRM effectively inhibited pRB phosphorylation by cyclin A-cdk2 (lanes 2 and 3). The fact that pRB but not cyclin A phosphorylation (which appears to be a CRM-independent substrate [54]) was inhibited in the same reaction mixture provided an internal control for the specificity of the inhibition. In order to address the sequence specificity of this effect, a peptide with PAP's CRM scrambled was also tested, and it showed no effect on pRB phosphorylation (lanes 4 and 5). Similar results were obtained with cyclin E-cdk2 (data not shown). These results suggest that eight residues of PAP can act as a CRM, functionally interacting with cyclins A and E.
FIG. 4.
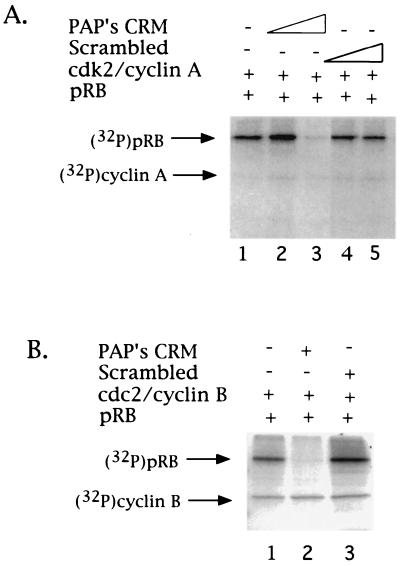
PAP contains a functional CRM. Inhibition of cdk phosphorylation of pRB by an 8-mer PAP-derived peptide. (A) Purified pRB and cyclin A-cdk2 were incubated under kinase conditions in the presence of [γ-32P]ATP and two concentrations (9 and 18 μM) of either an 8-mer PAP CRM-derived peptide (SKIRILVG) (lanes 2 and 3), an 8-mer peptide of scrambled sequence (LRSGIKVI) (lanes 4 and 5), or no peptide (lane 1). Phosphorylated proteins were resolved by SDS-PAGE and detected by autoradiography. Arrows on the left indicate the positions of pRB and cyclin A. (B) Purified pRB and cyclin B1-cdc2 were incubated under kinase conditions in the presence of [γ-32P]ATP and the 8-mer PAP CRM-derived peptide (18 μM) (lane 2), the 8-mer peptide of scrambled sequence (18 μM) (lane 3), or no peptide (lane 1). Arrows on the left indicate the positions of pRB and cyclin B1.
Our data has shown that PAP interacts directly with cyclin B1. We therefore tested the ability of PAP's CRM to interact functionally with cyclin B1 by testing the effects of the 8-mer PAP peptide in the pRB phosphorylation assay. Strikingly, given the inactivity of other CRM's on cyclin B1-cdc2 phosphorylation (2), PAP's CRM also strongly and specifically inhibited phosphorylation of pRB by cyclin B1-cdc2 (Fig. 4B, compare lanes 1 and 3 with lane 2). As observed above with cyclin A, cyclin B1 autophosphorylation was not affected. Together, these results suggest that PAP contains a novel CRM-like sequence capable of functionally interacting with both G1 and G2 cyclins.
PAP interacts with and is phosphorylated by cyclin A-cdk2.
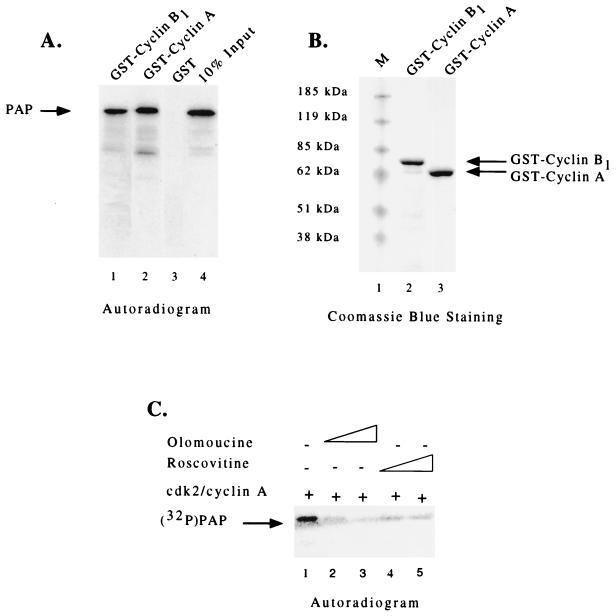
In order to determine whether the inhibition of cyclin A-cdk2 phosphorylation by the PAP peptide reflected an interaction between cyclin A and PAP, we investigated the ability of full-length PAP to bind cyclin A. To this end, a GST binding assay similar to that used in Fig. 2 was employed. In vitro-translated [35S]methionine-labeled PAP I was incubated with glutathione-agarose beads containing GST-cyclin B1, GST-cyclin A, or GST alone. As seen on the autoradiogram depicted in Fig. 5A, similar amounts of PAP I were present in the eluates of both cyclin B1 (lane 1) and cyclin A (lane 2) matrices but not in that of the GST control (lane 3), providing evidence for an interaction between cyclin A and PAP. (Figure 5B shows a Coomassie blue-stained gel of the purified GST-cyclin fusion proteins used.)
FIG. 5.
PAP binds cyclin A and is phosphorylated by cyclin A-cdk2. (A) Autoradiogram of in vitro-translated 35S-labeled PAP bound to GST-cyclin B1 (lane 1), GST-cyclin A (lane 2), or GST (lane 3) glutathione matrices and 10% of the input PAP I (lane 4). An arrow on the left indicates the position of PAP. (B) Coomassie blue-stained SDS-PAGE of purified GST-cyclin A (lane 2) and GST-cyclin B1 (lane 3) fusion proteins used in the binding reactions. (C) Autoradiogram of phosphorylated PAP after incubation with cyclin A-cdk2. Purified PAP and cyclin A-cdk2 were incubated under kinase conditions in the presence of [γ-32P]ATP (lane 1). Specific inhibitors of cdks, olomoucine (14 and 70 μM) (lanes 2 and 3) and roscovitine (7 and 14 μM) (lanes 4 and 5), were added to establish the specificity of the reaction. Phosphorylated proteins were resolved by SDS-PAGE and detected by autoradiography. An arrow on the left indicates the position of PAP.
We next tested whether PAP could bind cyclin A, as well as serve as a substrate for cyclin A-cdk2 phosphorylation. Phosphorylation was examined in an in vitro kinase assay using baculovirus-produced and purified human cyclin A-flu-tagged cdk2 and bacterium-produced and purified His-tagged PAP I. After incubation under kinase conditions in the presence of [γ-32P]ATP, the reaction mixture was analyzed by SDS-PAGE and subsequent autoradiography. As seen in Fig. 5C, lane 1, 32P was efficiently incorporated into PAP. The specificity of cdk phosphorylation was controlled for by the addition of two specific cdk inhibitors, olomoucine and roscovitine, into the kinase reactions. As seen in lanes 2 to 5, incorporation of 32P into PAP was inhibited by both compounds. Taken together, these results demonstrate that PAP can both bind cyclin A and serve as a substrate for phosphorylation by cyclin A-cdk2. This is consistent with the fact that PAP is phosphorylated throughout the cell cycle, not only in the M phase (9).
Novel, concentration-dependent effects of PAP's CRM.
The data presented above show that PAP can interact with both cyclin B1 and cyclin A and, at least in the case of cyclin B1, that these interactions are dependent on residues N terminal of the Ser-Thr-rich PAP regulatory region which encompass the PAP CRM. We next wished to examine the CRM dependence of PAP's interactions with these cdks, and we therefore undertook a series of binding and kinase assays using PAP as a substrate.
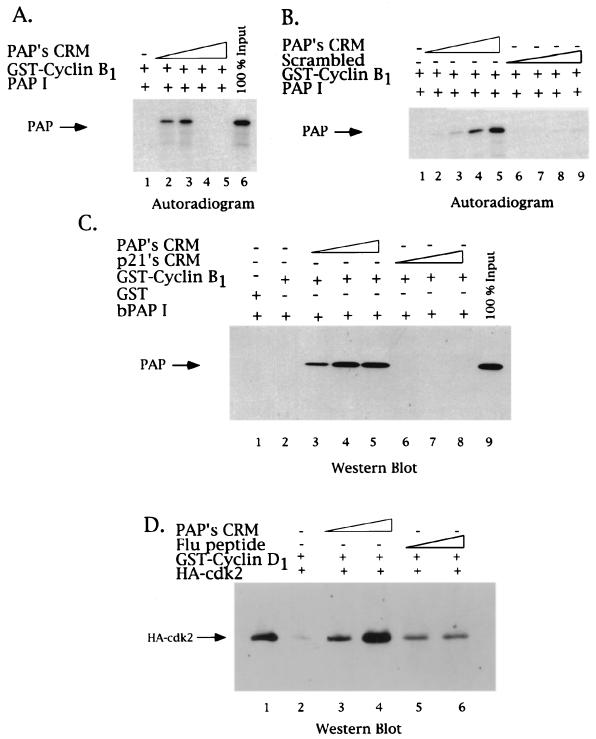
Figure 6A and B show autoradiograms of GST pull-down assays using GST-cyclin B1 and in vitro-translated 35S-labeled PAP I. These experiments were carried out like those in Fig. 2 and 5, except that increasing amounts of the 8-mer PAP CRM peptide were added to the glutathione-GST-cyclin B1 matrix prior to PAP I (see Materials and Methods). Binding studies examining other CRM-dependent interactions with cyclins have demonstrated that addition of increasing amounts of CRM-containing peptides disrupts binding of the CRM-containing protein and the cyclin (e.g., references 1 and 2). Our experiments with PAP's CRM also illustrate a disruption of the interaction between PAP and cyclin B1 (at peptide concentrations of 36 and 72 μM; compare lanes 4 and 5 with lanes 2 and 3 of Fig. 6A). Unexpectedly, however, at lower concentrations of peptide (9 and 18 μM), we observed a dramatic stimulation of binding. Lanes 2 and 3, compared to lane 1, illustrate the striking enhancement of PAP I's binding to cyclin B1: up to 50 to 100% bound at the lower concentrations of peptide (compare to lane 6, which displays 100% of the amount of PAP I used for each reaction). Figure 6B illustrates more thoroughly the dose-dependent enhancement of PAP I-GST-cyclin B1 binding by the CRM (2.25, 4.5, 9, and 18 μM; compare lane 1 with lanes 2 to 5). As a control for sequence specificity, an 8-mer peptide of scrambled CRM sequence was used (lanes 6 to 9). (Note that these experiments were done with a low concentration of GST-cyclin B1, which does not allow significant PAP-cyclin B1 interaction without addition of the lower concentrations of CRM peptide [Fig. 6A to C, lanes 1].) These results together suggest a unique, CRM-dependent interaction of PAP with cyclin B1.
FIG. 6.
PAP's CRM both activates and represses PAP binding to cyclin B1. The effect of the 8-mer PAP CRM-derived peptide in GST-cyclin-PAP pull-down assays was tested. (A) GST-cyclin B1 glutathione matrices were incubated with in vitro-translated 35S-labeled PAP (1 μl) in the absence (lane 1) or presence of increasing amounts of PAP's CRM peptide (9, 18, 36, and 72 μM; lanes 2, 3, 4, and 5, respectively). Samples were washed, and bound proteins were analyzed by SDS–10% PAGE and autoradiography. An arrow on the left indicates the position of PAP. A total of 100% of the 35S-labeled PAP used in each reaction is found in lane 6. (B) Concentration dependence of the PAP CRM stimulatory effect. Lower concentrations of PAP's CRM peptide were used in binding assays similar to those in panel A (2.25, 4.5, 9, and 18 μM; lanes 2, 3, 4, and 5, respectively), as well as identical amounts of the 8-mer peptide of scrambled sequence (lanes 6, 7, 8, and 9). (C) CRM enhancement of PAP-cyclin B1 binding reflects a direct interaction. GST-cyclin B1 glutathione matrices were incubated with purified bacterial PAP in the presence of increasing amounts (2, 8.6, and 17 μM) of either PAP's CRM (lanes 3, 4, and 5), p21's CRM (lanes 6, 7, and 8), or no peptide (lane 1). Samples were subjected to SDS-PAGE and subsequent Western blot analysis using a PAP polyclonal antibody. (D) PAP's CRM can enhance cyclin-cdk association. GST-cyclin D1 glutathione matrices were incubated with purified HA-tagged cdk2 in the presence of increasing amounts (9 and 18 μM) of either PAP's CRM (lanes 3 and 4), a control peptide (lanes 5 and 6), or no peptide (lane 2). The presence of cdk2 in the eluates was detected by Western blot analysis using a monoclonal antibody against the HA epitope. Lane 1 contains the amount of HA-tagged cdk2 used.
To determine whether the CRM-mediated enhancement of the PAP-cyclin association reflects a direct interaction between these two molecules, we changed the sources of PAP I. Bacterium-produced, purified His-tagged PAP I (100 ng) was incubated with the GST-cyclin B1 (1 μg) glutathione matrix in the presence of the lower concentrations of PAP's CRM-containing peptide that stimulated the binding seen in Fig. 6A. After extensive washing and elution, the eluate was subjected to SDS-PAGE and Western blotting with an anti-PAP polyclonal antibody. As seen in Fig. 6C, lanes 3 to 5, compared to lane 2, addition of the peptide strongly stimulated the association of PAP with cyclin B1 (2, 8.6, and 17 μM). Again, nearly 100% of the input PAP bound GST-cyclin B1 at the highest concentration. As a control for specificity, an 8-mer peptide spanning p21's CRM was used. This peptide was chosen because it has been previously reported not to functionally associate with cyclin B1 (2). As seen in lanes 6 to 8, p21's CRM had no effect on PAP-cyclin B1 binding. (The apparent absence of PAP in lane 2 is due to the exposure time of the blot, which was designed to highlight the dose-dependent stimulation of PAP-cyclin binding by the peptide.) Together, these results support a mechanism by which the CRM peptide directly stimulates the PAP-cyclin B1 interaction at low peptide concentrations but subsequently inhibits the interaction at higher peptide concentrations. (Note that the abrupt switch from stimulation to inhibition [e.g., Fig. 6A, lanes 3 and 4] is highly reproducible.) A possible explanation for these results is discussed below.
Although there have been no previous reports suggesting that a CRM peptide could enhance cyclin-substrate interactions, Adams et al. (2) reported that CRM peptides from p21 or E2F increased cyclin-cdk association. To determine if PAP's CRM could also increase cyclin-cdk association, we used a binding assay with GST-cyclin D1 and baculovirus-produced and purified human flu-tagged cdk2. Cyclin D1's binding to cdk2 was assayed for because of its documented weak affinity (39). Binding reactions were performed as described above, and the eluates were analyzed for the presence of cdk2 by SDS-PAGE and subsequent Western blotting using an anti-flu antibody. As shown in Fig. 6C, the prior addition of PAP's CRM peptide (9 and 18 μM) into the binding reaction significantly increased the presence of cdk2 in the eluate (compare lanes 2 to 4), while addition of a control peptide was without significant effect (lanes 5 and 6). These results establish another similarity between PAP's CRM and other characterized CRMs, namely, the ability to stimulate the association of a cyclin and a cdk. Whether these other CRMs might also enhance cyclin-substrate binding, under appropriate conditions, is unknown.
PAP phosphorylation is also first enhanced and then inhibited by the CRM peptide.
We lastly wished to test the CRM dependence of PAP phosphorylation, not only by cyclin B1-cdc2 but also by a G1-specific cyclin-cdk, cyclin A-cdk2. We utilized kinase assays similar to those shown in Fig. 4 but with bacterium-produced and purified His-tagged PAP I instead of pRB. PAP I was first incubated with baculovirus-produced and purified cyclin B1-flu-tagged cdc2 under kinase conditions in the presence of [γ-32P]ATP and analyzed by SDS-PAGE and subsequent autoradiography. As seen in Fig. 7A, the dramatic stimulation of PAP-cyclin B1 binding seen upon addition of low concentrations of CRM is mirrored in a similar stimulation of PAP I phosphorylation: prior addition of PAP's CRM-containing peptide (4.3 to 8.7 μM) led to a dose-dependent increase of phosphorylation by cyclin B1-cdc2 (compare lane 1 with lanes 2 to 7), while addition of the scrambled sequence peptide had no significant effect (lanes 8 to 12). This CRM stimulation of cyclin B1-cdc2 phosphorylation seems to be specific for PAP, since parallel experiments using either the CRM-containing substrate pRB (Fig. 7B) or the CRM-independent histone H1 (Fig. 7C) showed either only inhibition or no effect, respectively. Consistent with the observed inhibition of cyclin B1-PAP binding, addition of higher concentrations of PAP's CRM-containing peptide (5.8 to 22.5 μM) led to a dose-dependent inhibition of PAP phosphorylation by cyclin B1-cdc2, after an initial stimulation (Fig. 8A, compare lane 1 with lanes 2 to 5). (Note that in lane 2 stimulation of PAP's phosphorylation upon addition of 5.8 μM CRM was observed not only by incorporation of 32P but by the shift in mobility, as indicated by the arrows to the left in Fig. 8A.)
To test the CRM dependence of PAP phosphorylation by cyclin A-cdk2, a concentration of the PAP (Fig. 8B, lane 2) or p21 (lane 3) CRM-containing peptide that inhibited PAP phosphorylation by cyclin B1-cdc2 (25 μM) was added to kinase reactions containing PAP and baculovirus-produced and purified cyclin A-flu-tagged cdk2. As shown in Fig. 8B, lanes 1 and 2, PAP phosphorylation was strongly inhibited. In keeping with the known response of cyclin A-cdk2 to p21, the p21 CRM also strongly inhibited PAP phosphorylation by this cyclin-cdk pair (Fig. 8B, lane 3). These results establish the CRM-dependent nature of PAP phosphorylation by both cyclin B1-cdc2 and cyclin A-cdk2 and provide further evidence that PAP's CRM is exceptional in its ability to interact with cyclin B1.
DISCUSSION
One role of cyclin recognition motifs is to target CRM-containing proteins for cdk phosphorylation (e.g., reference 54). The data in this report support a mechanism whereby the complex phosphorylation status of PAP is, at least in part, CRM mediated. We previously reported that PAP is phosphorylated on multiple sites (9). In bovine PAP, these sites consist of three consensus sites (T/SPXK/R) and at least four nonconsensus sites (S/TP), and phosphorylation on all sites appears to be required for inactivation of PAP activity. Complete phosphorylation of the nonconsensus sites in vitro requires a 10-fold-higher concentration of kinase than is required to phosphorylate the consensus sites (10). By binding to an active cyclin B1-cdc2, the CRM can help create a high local concentration of kinase, leading to hyperphosphorylation and subsequent inactivation of PAP in late M phase. The CRM also likely helps to maintain normal levels of PAP phosphorylation throughout the cell cycle.
As mentioned in the introduction, polyadenylation seems to be regulated not only during M phase but also as cells enter S phase. Cyclin B1-p34cdc2 is most active in M phase of the cell cycle, whereas cdk-cyclin pairs such as cyclin A-cdk2 and cyclin E-cdk2 are more active during the transition into, and in, S phase (e.g., references 32 and 46). If the cell cycle machinery also regulates polyadenylation in S phase via cdk phosphorylation, utilization of PAP's CRM likely contributes to targeting the G1-phase cyclin-cdks. As the data above illustrate, PAP's CRM functions to facilitate not only the association of PAP with G1 and G2 cyclins but also the phosphorylation by both types of cdks. This suggests the potential for regulation of PAP and polyadenylation by cdks in the G1, S, and G2 phases of the cell cycle, possibly contributing to the well-documented increase of polyadenylation activity observed upon entry into S phase (5, 7, 20, 28, 30). PAP is phosphorylated on only a subset of cdk sites throughout the cell cycle, and it is not yet known how or if this affects activity.
Much of what we know about CRMs and how they interact with cyclins has come from the crystal structure of a fragment of p27kip1 bound to a fragment of cyclin A-cdk2 (51). This structure shows p27's CRM to be part of a rigid alpha-helical coil, tucked into a groove formed by cyclin A's cyclin box. The major contacts are with residues conserved in the A-, B-, D-, and E-type cyclins (the so-called MRAILVDW motif; reviewed in reference 50). While many examples of A-, E-, and D-type cyclins interacting with CRMs have been reported (e.g., references 1, 2, 6, 15, 38, 42, 43, 45, 49, 53, and 65), there has been no evidence of cyclin B-CRM interactions, and it has in fact been shown in one case that cyclin B does not bind to the E2F-1 CRM (2). Here we have provided evidence that B-type cyclins can indeed interact with a CRM. Moreover, since the PAP CRM can interact with both B- and G1-type cyclins, we suggest that PAP CRM typifies a novel, universal CRM.
When comparing the core residues of multiple CRMs (Fig. 3), the conservation of the +4 arginine and +6 leucine residues is striking. These residues are responsible for multiple contacts with the cyclin, based on the above-mentioned crystal structure (51). The surrounding residues, however, do not form as obvious a pattern. When comparing PAP's CRM to those listed in Fig. 3, it is apparent that the PAP CRM contains more hydrophobic residues. The groove in cyclin A shown to contact the CRM is part of a hydrophobic patch present in all cyclins, containing the above-mentioned MRAILVDW motif (50, 51, 54). When several hydrophobic residues in cyclin A were mutated to alanines, interactions with CRMs were disrupted (54). We therefore propose that the greater hydrophobicity of PAP's CRM contributes to its ability to interact with both G1- and G2-type cyclins.
By studying the structure of the cyclin box, it has been proposed that two CRMs can bind one cyclin molecule (50). In fact, multiple molecules of p21 and p57 (both CRM-containing proteins) have been shown to complex with one cyclin-cdk heterodimer (27, 39, 61). The strong initial stimulation of PAP binding to cyclin B1 by the CRM peptide can be explained as cooperative binding of two CRM-containing molecules, whereby binding of the first (the CRM peptide alone) leads to enhanced binding of the second (PAP). In the case of p21, multiple molecules binding to a cyclin-cdk complex has been implicated in the ability of p21 to function as a cdk inhibitor (25, 26, 61). In the case of PAP, we propose that this property would provide for a rapid response mechanism to sense the end of M phase.
M phase is characterized by the stimulation, rise, and subsequent loss of cyclin B1-cdc2 kinase activity, which is mirrored in the rise and fall of the cyclin B1 subunit (reviewed in reference 36). As mentioned above, the inactivation of PAP through hyperphosphorylation is restricted to the late M phase (10). Cooperative binding of two PAP molecules to cyclin B1, via their CRM's, whereby binding of the first strongly stimulates binding of a second, could allow for rapid regulation by hyperphosphorylation of PAP's activity in late M phase. PAP would be extremely sensitive to a rise in cyclin B1-cdc2 levels, allowing for maintenance of an active PAP in M phase until a threshold level of cyclin B1-cdc2 is reached. A rapid association with cyclin B1-cdc2 would then occur, driving subsequent hyperphosphorylation and inactivation of PAP in late M phase. An extension of this model is that the PAP-cyclin B1-cdc2 complex is activated for binding and phosphorylating other substrates containing a PAP-like CRM. A cooperative interaction, involving an activated CRM-cyclin B1-cdc2 complex, also offers an explanation for the observed precipitous inhibition of PAP binding at elevated CRM peptide concentrations, such that the excess peptide is efficiently bound to the second site, outcompeting the limiting concentration of PAP.
As mentioned in the introduction, the most well-studied mechanism of cdk regulation is cyclin-cdk binding. A number of mechanisms have been shown to influence this interaction. For example, threonine phosphorylation of the cdk subunit has been shown in some cases to stabilize cyclin-cdk binding (13). Cyclin H-cdk7 utilizes the assembly factor MAT-1 to promote an active cyclin-cdk complex (17, 57). Although MAT-1 does not contain a recognizable CRM, it behaves in a manner similar to PAP's CRM. The C terminus of p27 can also serve as a stabilizing factor for the cyclin B1-cdc2 interaction (18). Interestingly, in the past few years data have been collected on the ability of CRM-containing proteins to serve as assembly and stabilization factors for the association of multiple cyclins and their respective cdks (e.g., references 27, 39, 48, 60, and 61). In fact, Adams et al. (2) were able to stimulate cyclin A's-cdk2 binding with the addition of just the CRM from p21 (as well as from E2F-1), thus providing evidence that p21's ability to function as a cyclin-cdk assembly factor lies in its CRM. As to the mechanism, it was speculated that CRM binding to cyclin A induces an allosteric change, thereby promoting complex assembly (2). In this report we provide evidence that PAP's CRM can also stimulate cyclin-cdk association, thereby supporting the generality of this phenomena, and extending it to G2 cyclin-cdk complexes. We propose that CRM-containing proteins provide another regulatory mechanism for cdk activity, ensuring cdk activity at the proper substrates and subcellular localization to allow for proper cell cycle progression.
Early studies of polyadenylation activity during the mammalian cell cycle revealed an increase as cells entered S phase (5, 7, 20, 28, 30) and a decrease during M phase (16, 55). The hyperphosphorylation and inactivation of PAP in M-phase cells (9, 10) coincides with the observed decrease in both polyadenylation and general gene expression during M phase (21). The data presented here both provide a mechanistic underpinning for the regulation of PAP by cyclin B-p34cdc2, in which the PAP CRM plays an important, and complex, role in promoting phosphorylation, and also opens up the possibility of regulation of polyadenylation during other phases of the cell cycle, when cdks other than cyclin B-p34cdc2 are active. Interestingly, all CRM-containing proteins examined to date play active roles in regulating the cell cycle. It is tempting to speculate that PAP also plays such a role and that CRM-mediated phosphorylation of PAP helps modulate polyadenylation, and hence gene expression, throughout the cell cycle.
ACKNOWLEDGMENTS
We thank B. Dynlacht for the bacluloviruses expressing the GST-cyclins, W. Zhao for the vector expressing PAP I, D. F. Colgan for the baculovirus expressing PAP I, K. G. K. Murthy for the bacterial-purified PAP I, and E. Freulich for expert help with baculoviruses. We thank C. Cain, D. F. Colgan, E. Freulich, C. Gaiddon, F. Kleiman, K. G. K. Murthy, L. Ko, Y. Takagaki, and N. Baptiste for helpful discussions.
This work was supported by grants from the NIH to C.P. and J.L.M.
REFERENCES
- 1.Adams P D, Li X, Sellers W R, Baker K B, Leng X, Harper J W, Taya Y, Kaelin W G., Jr Retinoblastoma protein contains a C-terminal motif that targets it for phosphorylation by cyclin-cdk complexes. Mol Cell Biol. 1999;19:1068–1080. doi: 10.1128/mcb.19.2.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams P D, Sellers W R, Sharma S K, Wu A D, Nalin C M, Kaelin W G., Jr Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballantyne S, Bilger A, Astrom J, Virtanen A, Wickens M. Poly(A) polymerases in the nucleus and cytoplasm of frog oocytes: dynamic changes during oocyte maturation and early development. RNA. 1995;1:64–78. [PMC free article] [PubMed] [Google Scholar]
- 4.Barabino S M, Keller W. Last but not least: regulated poly(A) tail formation. Cell. 1999;99:9–11. doi: 10.1016/s0092-8674(00)80057-4. [DOI] [PubMed] [Google Scholar]
- 5.Benz E W, Jr, Getz M J, Wells D J, Moses H L. Nuclear RNA polymerase activities and poly(A)-containing mRNA accumulation in cultured AKR mouse embryo cells stimulated to proliferate. Exp Cell Res. 1977;108:157–165. [PubMed] [Google Scholar]
- 6.Chen I T, Akamatsu M, Smith M L, Lung F D, Duba D, Roller P P, Fornace A J, Jr, O'Connor P M. Characterization of p21Cip1/Waf1 peptide domains required for cyclin E/Cdk2 and PCNA interaction. Oncogene. 1996;12:595–607. [PubMed] [Google Scholar]
- 7.Coleman M S, Hutton J J, Bollum F J. Terminal riboadenylate transferase in human lymphocytes. Nature. 1974;248:407–409. doi: 10.1038/248407a0. [DOI] [PubMed] [Google Scholar]
- 8.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 9.Colgan D F, Murthy K G, Prives C, Manley J L. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- 10.Colgan D F, Murthy K G, Zhao W, Prives C, Manley J L. Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of multiple consensus and non-consensus sites. EMBO J. 1998;17:1053–1062. doi: 10.1093/emboj/17.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooke C, Alwine J C. The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol Cell Biol. 1996;16:2579–2584. doi: 10.1128/mcb.16.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dantonel J C, Murthy K G, Manley J L, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 13.Desai D, Wessling H C, Fisher R P, Morgan D O. Effects of phosphorylation by CAK on cyclin binding by CDC2 and CDK2. Mol Cell Biol. 1995;15:345–350. doi: 10.1128/mcb.15.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dynlacht B D, Flores O, Lees J A, Harlow E. Differential regulation of E2F transactivation by cyclin/cdk2 complexes. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 15.Dynlacht B D, Moberg K, Lees J A, Harlow E, Zhu L. Specific regulation of E2F family members by cyclin-dependent kinases. Mol Cell Biol. 1997;17:3867–3875. doi: 10.1128/mcb.17.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan H, Penman S. Regulation of protein synthesis in mammalian cells. II. Inhibition of protein synthesis at the level of initiation during mitosis. J Mol Biol. 1970;50:655–670. doi: 10.1016/0022-2836(70)90091-4. [DOI] [PubMed] [Google Scholar]
- 17.Fisher R P, Jin P, Chamberlin H M, Morgan D O. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell. 1995;83:47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- 18.Font de Mora J, Uren A, Heidaran M, Santos E. Biological activity of p27kip1 and its amino- and carboxy-terminal domains in G2/M transition of Xenopus oocytes. Oncogene. 1997;15:2541–51. doi: 10.1038/sj.onc.1201420. [DOI] [PubMed] [Google Scholar]
- 19.Foulkes N S, Schlotter F, Pevet P, Sassone-Corsi P. Pituitary hormone FSH directs the CREM functional switch during spermatogenesis. Nature. 1993;362:264–267. doi: 10.1038/362264a0. [DOI] [PubMed] [Google Scholar]
- 20.Getz M J, Elder P K, Benz E W, Jr, Stephens R E, Moses H L. Effect of cell proliferation on levels and diversity of poly(A)-containing mRNA. Cell. 1976;7:255–265. doi: 10.1016/0092-8674(76)90025-8. [DOI] [PubMed] [Google Scholar]
- 21.Gottesfeld J M, Forbes D J. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 22.Gunderson S I, Beyer K, Martin G, Keller W, Boelens W C, Mattaj L W. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994;76:531–541. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 23.Gunderson S I, Polycarpou-Schwarz M, Mattaj I W. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 24.Gunderson S I, Vagner S, Polycarpou-Schwarz M, Mattaj I W. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev. 1997;11:761–773. doi: 10.1101/gad.11.6.761. [DOI] [PubMed] [Google Scholar]
- 25.Harper J W, Elledge S J. Cdk inhibitors in development and cancer. Curr Opin Genet Dev. 1996;6:56–64. doi: 10.1016/s0959-437x(96)90011-8. [DOI] [PubMed] [Google Scholar]
- 26.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tsai L H, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, et al. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tsai L H, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, Fox M P, Wei N. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hauser H, Knippers R, Schafer K P. Increased rate of RNA-polyadenylation. An early response in concanavalin A activated lymphocytes. Exp Cell Res. 1978;111:175–184. doi: 10.1016/0014-4827(78)90247-1. [DOI] [PubMed] [Google Scholar]
- 29.Hirose Y, Manley J L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 30.Hirsch M, Penman S. The messenger-like properties of the poly(A)+ RNA in mammalian mitochondria. Cell. 1974;3:335–339. doi: 10.1016/0092-8674(74)90047-6. [DOI] [PubMed] [Google Scholar]
- 31.Horton L E, Templeton D J. The cyclin box and C-terminus of cyclins A and E specify CDK activation and substrate specificity. Oncogene. 1997;14:491–498. doi: 10.1038/sj.onc.1200851. [DOI] [PubMed] [Google Scholar]
- 32.Hunter T, Pines J. Cyclins and cancer. II. cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 33.Jeffrey P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N P. Mechanism of CDK activation revealed by the structure of a cyclin A-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 34.Keller W, Minvielle-Sebastia L. A comparison of mammalian and yeast pre-mRNA 3′-end processing. Curr Opin Cell Biol. 1997;9:329–336. doi: 10.1016/s0955-0674(97)80004-x. [DOI] [PubMed] [Google Scholar]
- 35.Kelly B L, Wolfe K G, Roberts J M. Identification of a substrate-targeting domain in cyclin E necessary for phosphorylation of the retinoblastoma protein. Proc Natl Acad Sci USA. 1998;95:2535–2540. doi: 10.1073/pnas.95.5.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King R W, Jackson P K, Kirschner M W. Mitosis in transition. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 37.Kohtz J D, Jamison S F, Will C L, Zuo P, Luhrmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 38.Krek W, Ewen M E, Shirodkar S, Arany Z, Kaelin W G, Jr, Livingston D M. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 39.LaBaer J, Garrett M D, Stevenson L F, Slingerland J M, Sandhu C, Chou H S, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 40.Lees E M, Harlow E. Sequences within the conserved cyclin box of human cyclin A are sufficient for binding to and activation of cdc2 kinase. Mol Cell Biol. 1993;13:1194–1201. doi: 10.1128/mcb.13.2.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis J D, Gunderson S I, Mattaj I W. The influence of 5′ and 3′ end structures on pre-mRNA metabolism. J Cell Sci Suppl. 1995;19:13–19. doi: 10.1242/jcs.1995.supplement_19.2. [DOI] [PubMed] [Google Scholar]
- 42.Lin J, Reichner C, Wu X, Levine A J. Analysis of wild-type and mutant p21WAF-1 gene activities. Mol Cell Biol. 1996;16:1786–1793. doi: 10.1128/mcb.16.4.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma T, Zou N, Lin B Y, Chow L T, Harper J W. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc Natl Acad Sci USA. 1999;96:382–387. doi: 10.1073/pnas.96.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 45.Morris M C, Divita G. Characterization of the interactions between human cdc25C, cdks, cyclins and cdk-cyclin complexes. J Mol Biol. 1999;286:475–487. doi: 10.1006/jmbi.1998.2475. [DOI] [PubMed] [Google Scholar]
- 46.Nigg E A. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 47.Pan Z Q, Amin A, Hurwitz J. Characterization of the in vitro reconstituted cyclin A or B1-dependent cdk2 and cdc2 kinase activities. J Biol Chem. 1993;268:20443–20451. [PubMed] [Google Scholar]
- 48.Peeper D S, Parker L L, Ewen M E, Toebes M, Hall F L, Xu M, Zantema A, van der Eb A J, Piwnica-Worms H. A- and B-type cyclins differentially modulate substrate specificity of cyclin-cdk complexes. EMBO J. 1993;12:1947–1954. doi: 10.1002/j.1460-2075.1993.tb05844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petersen B O, Lukas J, r. CS S, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pines J. Cyclin-dependent kinase inhibitors: the age of crystals. Biochim Biophys Acta. 1997;1332:M39–M42. doi: 10.1016/s0304-419x(96)00042-x. [DOI] [PubMed] [Google Scholar]
- 51.Russo A A, Jeffrey P D, Patten A K, Massague J, Pavletich N P. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 52.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 53.Saha P, Eichbaum Q, Silberman E D, Mayer B J, Dutta A. p21CIP1 and Cdc25A: competition between an inhibitor and an activator of cyclin-dependent kinases. Mol Cell Biol. 1997;17:4338–4345. doi: 10.1128/mcb.17.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulman B A, Lindstrom D L, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci USA. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steward D L, Shaeffer J R, Humphrey R M. Breakdown and assembly of polyribosomes in synchronized Chinese hamster cells. Science. 1968;161:791–793. doi: 10.1126/science.161.3843.791. [DOI] [PubMed] [Google Scholar]
- 56.Takagaki Y, Seipelt R L, Peterson M L, Manley J L. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 57.Tassan J P, Jaquenoud M, Fry A M, Frutiger S, Hughes G J, Nigg E A. In vitro assembly of a functional human CDK7-cyclin H complex requires MAT1, a novel 36 kDa RING finger protein. EMBO J. 1995;14:5608–5617. doi: 10.1002/j.1460-2075.1995.tb00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wahle E, Kuhn U. The mechanism of 3′ cleavage and polyadenylation of eukaryotic pre-mRNA. Prog Nucleic Acid Res Mol Biol. 1997;57:41–71. doi: 10.1016/s0079-6603(08)60277-9. [DOI] [PubMed] [Google Scholar]
- 59.Wickens M, Anderson P, Jackson R J. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 60.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 61.Zhang H, Hannon G J, Beach D. p21-containing cyclin kinases exist in both active and inactive states. Genes Dev. 1994;8:1750–1758. doi: 10.1101/gad.8.15.1750. [DOI] [PubMed] [Google Scholar]
- 62.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao W, Manley J L. Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol Cell Biol. 1996;16:2378–2386. doi: 10.1128/mcb.16.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao W, Manley J L. Deregulation of poly(A) polymerase interferes with cell growth. Mol Cell Biol. 1998;18:5010–5020. doi: 10.1128/mcb.18.9.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu L, Harlow E, Dynlacht B D. p107 uses a p21CIP1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes Dev. 1995;9:1740–1752. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]