Integrative multiomics analysis of myeloma identifies 12 disease subtypes defined by specific patterns of genetic alterations.
Abstract
The remarkable genetic heterogeneity of multiple myeloma poses a substantial challenge for proper prognostication and clinical management of patients. Here, we introduce MM-PSN, the first multiomics patient similarity network of myeloma. MM-PSN enabled accurate dissection of the genetic and molecular landscape of the disease and determined 12 distinct subgroups defined by five data types generated from genomic and transcriptomic profiling of 655 patients. MM-PSN identified patient subgroups not previously described defined by specific patterns of alterations, enriched for specific gene vulnerabilities, and associated with potential therapeutic options. Our analysis revealed that co-occurrence of t(4;14) and 1q gain identified patients at significantly higher risk of relapse and shorter survival as compared to t(4;14) as a single lesion. Furthermore, our results show that 1q gain is the most important single lesion conferring high risk of relapse and that it can improve on the current International Staging Systems (ISS and R-ISS).
INTRODUCTION
Multiple myeloma (MM) is a mostly incurable malignancy of bone marrow terminally differentiated plasma cells, affecting more than 30,000 patients each year in the United States, with a median survival of approximately 6 years (1). While most patients initially respond to standard-of-care treatment, most relapse and become refractory as they undergo multiple lines of therapy. In particular, about 15% of patients fall in the high-risk category and typically relapse within 2 years from diagnosis (2). MM is characterized by remarkable clinical and genomic heterogeneity (3). Recent studies based on next-generation sequencing have revealed complex patterns of primary and secondary genetic alterations across patients (4, 5), and novel precision medicine approaches, where treatment is guided by the genomic profile of the individual patient, are being tested in trials (6, 7). Accurate classification of patients with MM into biologically homogeneous classes is thus essential for diagnosis, prognosis, and clinical management.
Several classifications of MM based on gene expression have been proposed in the past two decades. The TC (translocation/cyclin D) classification included eight groups characterized by different chromosomal translocations and the up-regulation of the cyclin D genes CCND1 and CCND3 (8). The UAMS (University of Arkansas for Medical Sciences) classification, based on unsupervised clustering of gene expression data, proposed seven clusters in part overlapping the TC classes and enriched for clinically relevant features and differential response to therapy (9). A further refinement was proposed by the HOVON Dutch-Belgium Hemato-Oncology Cooperative Group study group, which consisted of six of the UAMS classes and four novel classes enriched for activation of specific genes such as NFκB (nuclear factor κB) and PRL3 (protein-tyrosine phosphatase of regenerating liver 3) and a myeloid signature (10).
Our recent network model of newly diagnosed MM based on gene coexpression, MMNet, revealed a clear molecular separation between patients with immunoglobulin (Ig) translocations and hyperdiploidy and identified three novel subtypes characterized by cytokine signaling (CK), immune signatures (IMM), and MYC translocations (MYC) (11). Another study investigated novel MM subtypes based on a targeted DNA panel (12). The analysis revealed a large cluster comprising most hyperdiploidy (HD) and IgH-translocated patients and two smaller clusters, one enriched for IgH translocations and high number of copy number alterations (CNAs) and one mostly composed of patients with HD, with the fewest CNAs and mutations. Other recent works have described novel approaches to classification of patients with MM based on DNA alterations (13, 14). However, currently, no classification of MM accounts for both genomic and transcriptomic abnormalities, and the different results obtained from different data types, e.g., DNA versus RNA, suggest that a more holistic approach including different omics might further improve patient classification and reveal biologically and clinically informative subtypes of the disease.
Recently, patient similarity networks (PSNs) have emerged as a powerful tool to capture and structure the complexity and diversity of clinical, genetic, and molecular information across a patient population (15). In a PSN, patients are represented as nodes, much like in a social network, and connected with one another based on how similar their genomic and transcriptomic profiles are. The network structure enables effective identification of communities of highly similar patients, allowing a more comprehensive classification than other approaches based on a single measurement. PSNs have been successfully used to dissect the genomic and molecular complexity of several cancers, including medulloblastoma, glioblastoma multiforme, pancreatic ductal adenocarcinoma, and metastatic colorectal cancer (16–19).
In this study, we generated MM-PSN, the first PSN of newly diagnosed MM based on multiomics data from the MMRF (Multiple Myeloma Research Foundation) CoMMpass study (20). Clustering of MM-PSN identified 12 subgroups, revealing novel insights into the co-occurrence of primary translocation events such as t(4;14)-MMSET (multiple myeloma SET domain) and secondary adverse lesions such as gain of 1q and whole-arm deletions of 16q and 17p, which harbors the tumor suppressor TP53.
RESULTS
Multiomics PSN of newly diagnosed MM reveals greater genetic and molecular heterogeneity than current classifications
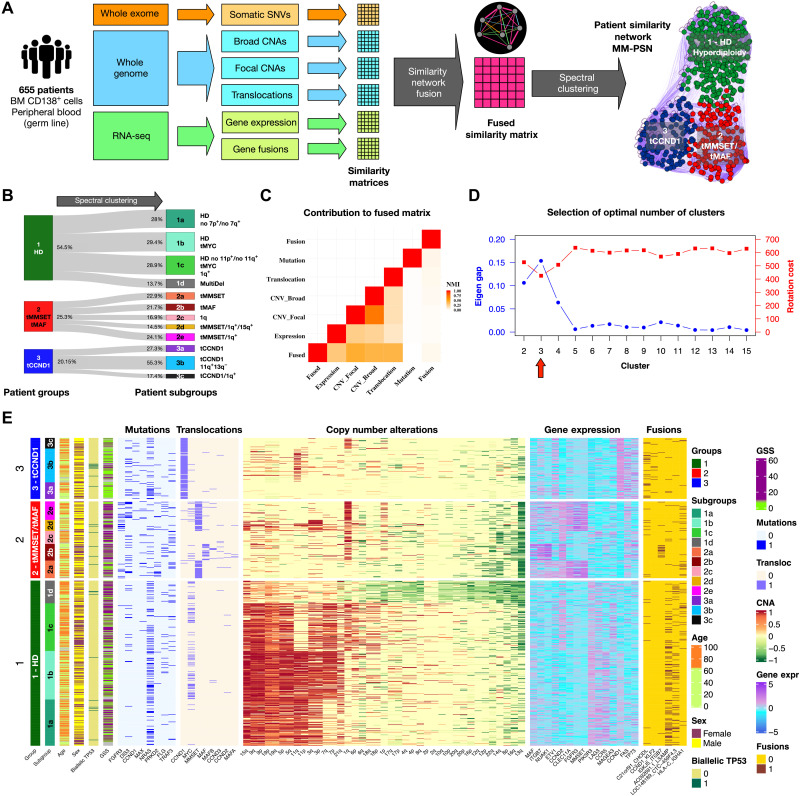
We generated MM-PSN, a multiomics PSN based on whole-exome sequencing (WES), whole-genome sequencing (WGS), and RNA sequencing (RNA-seq) data from 655 tumor samples from newly diagnosed patients with MM enrolled in the MMRF CoMMpass study, using the similarity network fusion (SNF) method (see Table 1 for summary patient characteristics) (17). In MM-PSN, each node represents a patient, and connecting edges represent similarity on the basis of multiple data types. In particular, for each sample, we used (i) gene expression and (ii) gene fusion data from RNA-seq, (iii) somatic single-nucleotide variations (SNVs) from WES, (iv) CNAs (focal and broad), and (v) translocation calls from WGS (Fig. 1, A and B). Translocations and CNAs provided the strongest contribution to MM-PSN, followed by gene expression, gene fusions, and SNVs (Fig. 1C). We then applied spectral clustering to determine groups of highly similar patients sharing features across the five data types. Our evaluation of the network using eigengap and rotation cost suggested 3 as the optimal number of clusters (Fig. 1D; see Methods for further details). Differential feature analysis revealed that the three clusters were enriched for (i) HD and the t(8;14) translocation of MYC (tMYC), (ii) translocations t(4;14) of MMSET/FGFR3 (tMMSET) and t(14;16) of MAF (tMAF), and (iii) translocation t(11;14) of CCND1 (tCCND1), respectively (Fig. 1, A and E). We labeled each group on the basis of these features. Group 1 (HD) included n = 357 patients (54.5%) and was further enriched for mutations in NRAS and an LSAMP:RPL18 gene fusion. Group 2 (tMMSET/tMAF) included n = 166 patients (25.3%) and was enriched for mutations in FGFR3, DIS3, and MAX. Group 3 (tCCND1) included n = 132 patients (20.15%) and was enriched for mutations in CCND1 and NRAS (see Methods and Supplementary Materials for details and statistics). To further dissect intragroup heterogeneity, we reapplied spectral clustering within each group, determining a total of 12 subgroups (Fig.1, B and E). Table 2 provides a summary of the most relevant features enriched in each group and subgroups. The complete data are given in tables S1 to S22.
Table 1. Patient cohort description and demographics.
N/A, Not Available.
| Characteristic | Value |
| No. of patients | 655 |
| Age | 63 (31–93) |
| Gender | |
| M | 382 (50.1%) |
| F | 218 (33.3%) |
| N/A | 109 (16.6%) |
| Ethnicity | |
| White | 420 (64.1%) |
| Black or African American | 83 (12.7%) |
| Asian | 12 (1.8%) |
| Filipino | 1 (0.2%) |
| Honduras | 1 (0.2%) |
| Middle Eastern | 1 (0.2%) |
| N/A | 137 (20.9%) |
| Disease stage (ISS) | |
| I | 128 (19.5%) |
| II | 339 (51.8%) |
| III | 63 (9.6%) |
| N/A | 125 (19.1%) |
| Translocations | |
| MMSET | 88 (13.4%) |
| CCND3 | 9 (1.4%) |
| MYC | 97 (14.8%) |
| MAFA | 4 (0.6%) |
| CCND1 | 126 (19.2%) |
| CCND2 | 6 (0.9%) |
| MAF | 27 (4.1%) |
| MAFB | 10 (1.5%) |
| None | 307 (46.9%) |
| Multiple* | 19 (2.9%) |
| Disease status | |
| PFS < 1 year | 225 (34.4%) |
| PFS < 2 years | 365 (55.7%) |
*CCND1 + MYC: 5; CCND2 + MYC: 2; MAF + MYC: 5; MAFB + MYC: 2; MMSET + MAF: 1; MMSET + MYC: 4.
Fig. 1. Network generation and identification of groups and subgroups.
(A) Somatic genetic variants and transcriptomic features from WES, WGS, and RNA-seq data from 655 patients in the MMRF CoMMpass study was used to generate an MM-PSN using the SNF approach. Edges connecting patients in the network represent similarity based on one or more feature type (e.g., orange edges in the sample network represent similarity based on SNVs and magenta edges represent similarity based on all the types of features). Spectral clustering was used to identify patient groups and then reapplied to identify subgroups enriched for specific features. (B) Representation of MM-PSN where nodes (patients) are colored according to the three main groups identified by spectral clustering. (C) The plot shows the contribution of the different data types to the fused matrix, in terms of normalized mutual information (NMI). (D) Eigen gap (maximum) and rotation cost (minimum) were used to determine 3 as the optimal number of clusters. (E) Overview of MM-PSN patient groups and subgroups. The heatmap shows characterization of the three main groups and 12 subgroups of MM-PSN based on their enrichment for the different genomic and transcriptomic features. GSS, genomic scar score.
Table 2. Summary of the MM-PSN subgroups.
DE, differentially expressed. The symbols ↑ and ↓ indicate positive and negative enrichment in column 3 (Selected enriching features) and up- and down-regulated genes in column 5 (Selected DE genes), respectively. AKTi, AKT inhibitor.
| Group/Subgroup | Size (n) |
Selected enriching
features |
Median
PFS; OS (days) |
Selected DE
genes |
Selected enriched
pathways |
Selected drug
candidates |
Selected
essential genes |
| 1: HD | 357 | HD; mutations in NRAS; AC092691.1:LSAMP, CEP164P1:RPS19, and TPM4:SIK1 fusions |
1094; N/A | - | Inflammation and immune evasion |
- | - |
| 1a: HD/-7− | 100 | HD with no gain of chromosome 7; TPM4:SIK1 fusion; mutational signatures: SBS1 (clock-like) ↑; low clonal complexity |
1082; N/A |
TNF↑, H19↑, KIT↑, FRZB↑, ASS1↓, ETV4↓ |
Interferon signaling, CD40 signaling, IL12 and IL18 signaling, JAK2 targets, DNA repair, cell cycle |
Carboplatin + paclitaxel + sorafenib, palbociclib, NOTCH1 antibody, vandetanib + everolimus, dinaciclib + AKTi |
APOC1, EBF1, MYT1L, PAX5 |
| 1b: HD/tMYC | 105 | HD with MYC translocation; mutations in NRAS; mutational signatures: SBS1 (clock-like)↑; SBS2 and SBS13 (APOBEC)↓ |
1297; N/A |
FAS↓, PIK3R3↑, TIMP1↓, AHR↓, CCR5↑, IL4R↓ |
Unfolded protein response, DNA repair, MET signaling, PI3K-AKT activation by MET, MYC targets, ATR signaling, TRAIL signaling |
Onartuzumab, crizotinib, palbociclib, cetuximab, pictilisib, alpelisib, taselisib, NOTCH1 antibody, Nutlin-3, vandetabib + everolimus, sorafenib |
CCDC78, HOXC4, TMEM121, MYC |
| 1c: HD/tMYC/1q+ | 103 | HD with MYC translocation and 1q gain age > 65 years old; higher number of female patients |
677; N/A |
GRM3↑, GDA↑, FLT3↓, CKB↑ |
IL27 signaling, MYC targets, MET signaling, MAPK1 activation, IRF4 targets |
NOTCH1 antibody, onartuzumab, crizotinib, dinaciclib + AKTI, vandetanib + everolimus |
CCND2, CALCB, CST6, MYT1L |
| 1d: MultiDel | 49 | Multiple chromosome deletions including 1p, 17, 4p, 4q, 8q, 16p, 16q, 13q, and 14q |
1240; 1777 |
EREG↑, LAMC3↑, CXCR5↓, EPCAM↑, MYCN↑, CCND2↑ |
Down-regulation of interferon response, integrin signaling, IL1, IL2 and IL15 signaling, MTOR, FOXO, WNT signaling |
Irinotecan, pictilisib |
CCND2, KRT7 |
| 2: tMMSET/tMAF | 166 | Translocations of MMSET and MAF; gain of 1q; deletion of 13q and 14q; mutations in FGFR3, PRKD2, and DIS3 |
751*; N/A | - | Proliferative signaling | - | - |
| 2a: tMMSET | 38 | Translocation of MMSET; deletion of 12p, 13q, 14q, and 22q; mutations in PRKD2; mutational signatures: SBS5 (clock-like)↑; SBS9 (polymerase ƞ and somatic hypermutation)↑ |
917 (N/A) | MMSET↑, AR↑, TIMP1↑, FLT4↑ |
Cancer testis antigens, FGFR3 signaling |
Tazemetostat, selumetinib, abemaciclib |
CCND2, OVOL2, PFKP, FGFR3 |
| 2b: tMAF | 36 | Translocation of MAF/ MAFA/MAFB; mutations in FLG; ITGB7 fusions; gain of 1q; deletion of 13q, 16q, and 22q; higher number of female patients; high GSS; mutational signatures: SBS2 and SBS13 (APOBEC)↑; SBS5 (clock-like)↓; SBS6 (MMR)↓; SBS10b (POLE-Exo)↑ |
903; 1500 | MAF↑, MAFA↑, NTRKK2↑, CDK6↑, CDKN2B↑, IL4R↑, AR↓, IL6↓, DKK1↓ |
Hypoxia, IL3 and IL4 signaling, Hedgehog, IGF1, PTEN, VEGF, IL2-STAT5 and HES signaling, activation of NTRK2, O-linked mucin glycosylation |
Trametinib, abemaciclib, tenovin-6, anti-TACI |
CCND2, MAF, CDK6, EEF2KMT, NFIA, RHPN1 |
| 2c: 1q | 28 | Gain of 1q; deletion of 13q, 14q, and 16q; mutations in TRAF3; mutational signatures: SBS1 (clock-like)↓ |
610; N/A |
CD4↑, ERBB3↑, IL32↑, ITGB1↑ |
ERK signaling, down-regulation of CTLA4 inhibitory signal, B cell survival |
Trametinib, abemaciclib |
CCND2, AQP7, GIA3, ZNF676 |
| 2d: tMMSET/1q+/15q+ | 24 | Translocation of MMSET; mutations in FGFR3; gain of 1q, 3p, 3q, and 15q; deletion of 13q |
1031; N/A |
ALB↑, MAGEC2↑, CXCL12↓, IL18↓, CCND2↑, MAF↑ |
B-Arrestins, FGFR3 signaling |
Infigratinib, erdafitinib |
CCND2, MAF, IRS2, FGFR3, BAMBI |
| 2e: tMMSET/1q+ | 40 | Translocation of MMSET; mutations in FGFR3 and DIS3; gain of 1q (highest number of copies); deletion of 13q; mutational signatures: SBS1 (clock-like)↓; SBS2 (APOBEC)↑; SBS10b (POLE-Exo) ↑; high clonal complexity |
624; 1033 |
IGF1R↑, MME↑, PAX9↑, MIAT↑, FGFR3↑, CCND2↑ |
FAS, FGFR3, KRAS, and IL2-STAT5 signaling |
Olaparib, trametinib, abermaciclib, erdafitinib, IKKB inhibitor, anti–NY-ESO-1 |
CCND2, MAF, FGFR3, BAMBI, IGF1R |
| 3: tCCND1 | 132 | Translocation of CCND1; gain of 11q; CCND1;KLF2. FOSB:KLF6 and C21orf91:CHODL fusions |
1130; NA | - | Replicative immortality and evasion of growth suppression |
- | - |
| 3a: tCCND1 | 36 | Translocation of CCND1; low GSS; low clonal complexity |
1176; N/A | CCND1↑, MMP9↑, ERG↑, KDR↑, IL18↑, IRF8↑ |
Inflammasomes, complement cascade, apoptosis, Waldenström macroglobulinemia, amyloid fiber formation, MDM2 targets |
Omipalisib, RAFi HG6-64-1, abemaciclib, trametinib |
CCND1, MT2A, MT1G, APOC1 |
| 3b: tCCND1/11q+/13q+ | 73 | Translocation of CCND1; gain of 11q and 13q; focal gain of 13q12.11; biallegic inactivation of TP53; CCND1:KLF2 fusion; low GSS; mutational signatures: SBS1 (clock-like)↓; SBS2 (APOBEC)↓; SBS6(MMR)↑ |
1236; N/A |
CCND1↑, SLC7A11↑, SALL4↑, TP73↑ |
Apoptosis, MDM2 targets, IL2-STAT5 signaling |
Venetoclax, trametinib, RAFi HG6-64-1, abermaciclib |
RUNX1T1, DMKN, BATF3 |
| 3c: tCCND1/1q+ | 23 | Translocation of CCND1 gain of 1q; deletion of 16q |
832; 1590 | CCND1↑, MS4A1↑, KREMEN2↑ |
MDS high-risk signature, APOBEC, MDM2 targets |
RAFi HG6-64-1, abermaciclib, trametinib |
CCND1, ZNF676 |
Group 1 was composed of four subgroups. Subgroups 1a (n = 100; 28%), 1b (n = 105; 29.4%), and 1c (n = 103; 28.9%) were mostly characterized by HD with differences in gain of chromosomes 7 and 11, which were almost not detected in subgroups 1a and 1c, respectively. Subgroups 1b, 1c, and 1d were enriched for tMYC, which was virtually absent in subgroup 1a. In addition, subgroup 1c was significantly enriched for the fusion HLA-C:IGHA1, gain(1q) and del(13q), and had a significantly higher number of female and older patients [chi-square adjusted P (Padj) < 0.001]. Subgroup 1d (n = 49; 13.7%) had a weaker HD signal and was significantly enriched for multiple chromosome deletions instead, including del(1p), del(8p), del(17p), del(14q), del(16q), and del(13q). Genes overexpressed in group 1 included FRZB, KIT, and TNF in subgroup 1a; PIK3R3 and MYC in subgroup 1b; GRM3, MAGEA3, and GDA in subgroup 1c; and EREG and LAMC3 in subgroup 1d.
Group 2 was composed of five subgroups. Subgroups 2a (n = 38; 22.9%), 2d (n = 24; 14.5%), and 2e (n = 40; 24.1%) were enriched for the tMMSET, while subgroup 2b (n = 36; 21.7%) was enriched for translocations of MAF (tMAF, tMAF-A, and tMAF-B). Subgroup 2a was also enriched for mutations in PRKD2 (protein kinase D2), subgroup 2b had a significantly higher number of female patients (chi-square Padj < 0.1), along with subgroup 1c, and was enriched for the IGKJ5:ITGB7 gene fusion and del(16q), and subgroups 2d and 2e were enriched for mutations in FGFR3 (fibroblast growth factor receptor 3). Del(13q) was observed in all five subgroups, while gain(1q) was significantly enriched in all subgroups with the exception of 2a. Of note, subgroup 2d was also enriched for gain of 15q and showed additional HD features, such as gain of chromosomes 3 and 19. Subgroup 2c (n = 28; 16.9%) was the only subgroup in group 2 without any translocation and was mainly characterized by gain(1q), del(14q), and mutations in tumor necrosis factor (TNF) receptor–associated factor 3 (TRAF3). Genes overexpressed in group 2 included CCND2 overall; MMSET across subgroups with t(4;14), 2a, 2d, and 2e; MAF in subgroups 2b, 2d, and 2e; NTRK, CDK6, and IL4R in subgroup 2b; MAGEA3, ERBB3, IL32, and ITGB1 in subgroup 2c; ALB, MAGEC2, and CXCL12 in subgroup 2d; and IGF1R, MME, and MIAT in subgroup 2e.
Group 3 was composed of three subgroups, all enriched for tCCND1. Subgroup 3a (n = 36; 27.3%) had virtually no CNAs, while subgroup 3b (n = 73; 55.3%) was additionally enriched for del(13q), specifically focused at 13q12.11, TP53 biallelic inactivation (mutation + deletion), gain(11q), and other sparse CNAs. Subgroup 3c (n = 23; 17.4%) was enriched for gain(1q). The co-occurrence of gain(1q) and del(16q) was identified in three different subgroups (1c, 2b, and 2c), indicating that it is a recurring whole-arm secondary aberration. Genes overexpressed in group 3 included CCND1 overall; MMP9, KDR, and IL18 in subgroup 3a; SLC7A11 and SALL4 in subgroup 3b; and KREMEN2 in subgroup 3c. To enable classification of patients with MM using MM-PSN, we have developed a freely available multiomics classifier on the basis of a support vector machine (SVM) approach (see Supplementary Methods, Data and materials availability statement, table S38, and fig. S13).
To assess the biological robustness of MM-PSN and the added value of using multiomics data, we performed a comparison with previous classifications based on gene expression, such as the UAMS and our previous MMNet classifications (figs. S1 and S2) (9, 11). To carry out the comparison with UAMS classes, we devised a gene expression classifier trained on the 12 MM-PSN subgroups, using an approach based on SVM (table S39; see Data and materials availability statement). Then, we predicted MM-PSN subgroup membership for 559 newly diagnosed patients before TT2 (total therapy) and TT3 treatments that had been previously assigned to UAMS classes. The results showed substantial agreement among the three systems, where previous classes defined by HD or specific translocations matched the corresponding enriched subgroups in MM-PSN. However, while gene expression–based systems identified broad disease subtypes driven by differential expression reflecting major genetic alterations, MM-PSN revealed a more granular stratification with important biological and clinical implications. Both UAMS and MMNet clustered patients with tMMSET in a unique class, while MM-PSN revealed the existence of three separate subgroups of patients carrying tMMSET, 2a, 2d, and 2e, each associated with different genetic and molecular alterations. Similarly, while HD was previously considered as a homogeneous class of patients, MM-PSN revealed three distinct HD subgroups, 1a, 1b, and 1c. The UAMS HY (hyperdiploid) class had significant overlap with MM-PSN subgroups 1a and 1b but not 1c, which had significant overlap with the low bone disease (LB) UAMS class instead. This provides the genetic basis for the LB class, whose hyperdiploid features were not apparent by simple analysis of its gene expression profile. The CCND1 groups in UAMS (CD1/CD2) and MMNet (CCND1) were represented in the three different MM-PSN subgroups 3a, 3b, and 3c, which revealed significant genetic differences in terms of co-occurring CNAs among patients affected by tCCND1. The UAMS PR (proliferative) class mapped onto multiple MM-PSN classes, and prominently to subgroup 1c. This provides evidence that PR is a genetically heterogeneous class of patients whose transcriptomes may result from different combinations of genomic alterations. Last, subgroup 1d, characterized by prominent multiple deletions, and subgroup 2c, enriched for gain(1q) and del(14q), were poorly represented in both the UAMS and MMNet classifications and identified novel classes whose genetic features were not apparent by gene expression analysis alone.
MM-PSN subgroups correlate with markers of genomic instability, mutational signatures, and subclonal complexity
To estimate genomic instability between groups, we additionally calculated a genomic scar score (GSS) measuring the levels of homologous recombination deficiency in each patient, based on telomeric allelic imbalance (TAI), loss of heterozygosity (LOH), and number of large-scale transitions (LSTs) (fig. S3 and table S23) (21). High GSS has been shown to correlate with poor prognosis in other cancers, while low GSS was associated with superior outcome in a recent study on patients with MM (22). Consistently, with these findings, the lowest GSS levels across all subgroups were found in patients with tCCND1, specifically those in subgroups 3a (tCCND1) (Padj = 1.3 × 10−8) and 3b (tCCND1/11q+/13q−) (Padj = 5 × 10−3), but not 3c (tCCND1/1q+), while patients in 2b (tMAF) had the highest GSS (Padj = 0.04) (Fig. 1E, fig. S3, and table S23). Of note, a subset of patients in subgroup 3b with biallelic loss of TP53 had significantly higher GSS than the rest of the subgroup (P = 0.001). Within group 2 (tMMSET/tMAF), subgroups 2b (tMAF), 2c (1q+/14q−), and 2e (tMMSET/1q+) had higher GSS [Padj(2b) = 0.01; Padj(2c) = 0.03; Padj(2e) = 0.03]. In particular, while all three subgroups had increased LOH levels, GSS was specifically driven by increased TAI levels in subgroups 2b and 2c and by increased LST in subgroup 2e. In group 1 (HD), subgroup 1d (MultiDel) had a significantly high GSS (Padj = 0.02), particularly driven by increased TAI levels.
To further characterize the genetic abnormalities enriched in each subgroup, we extracted mutational signatures from the samples in the cohort, decomposed them into the mutational signatures described in the COSMIC (Catalogue Of Somatic Mutations In Cancer) catalog (v3.2), estimated the contribution of each signature in each sample, and performed enrichment at the subgroup level. Our analysis identified eight single-base substitution (SBS) signatures largely overlapping those detected in a previous work in MM (fig. S3B and table S24) (23). Seven MM-PSN subgroups were significantly enriched for at least one signature (fig. S3C). The clock-like signatures SBS1 and SBS5, indicating a mutational process correlated with age and whose activity is constant over time, were detected in every patient. However, SBS1 activity, which is initiated by spontaneous or enzymatic deamination of 5-methylcytosine to thymine, was significantly higher in HD subgroups 1a (HD/-7−) and 1b (HD/tMYC) and significantly lower in subgroups 2c (1q+/14q−), 2e (tMMSET/1q+), and 3b (tCCND1/11q+/13q−), while SBS5 activity was higher in subgroup 2a (tMMSET) and lower in subgroup 2b (tMAF). Other enriched signatures were the APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like)-related signatures SBS2, which was significantly higher in subgroup 2b (tMAF); SBS13, which was higher in subgroups 2b (tMAF) and 2e (tMMSET/1q+); SBS6, attributable to defective DNA mismatch repair (MMR), which was higher in subgroup 3b (tCCND1/11q+/13q−) and lower in subgroup 2b (tMAF); SBS9, associated with replication by polymerase eta as part of somatic hypermutation in lymphoid cells, which was higher in subgroup 2a (tMMSET); and SBS10b, related to polymerase epsilon exonuclease (POLE-Exo) domain mutations, which was higher in subgroups 2b (tMAF) and 2e (tMMSET/1q+).
Last, we inferred the clonal landscape of tumor cells using SNVs and CNAs and evaluated differences across the MM-PSN subgroups in terms of number of subclones and complexity of the tumor structure, as measured by the number of branches in the phylogenetic tumor trees. The analysis revealed significantly less subclonal tumor populations in subgroups 1a (HD/-7−) and 3a (tCCND1), lower complexity in subgroup 1a (HD/-7−), and a significantly higher complexity in terms of both the number of subclones and branches in subgroup 2e (tMMSET/1q+) (fig. S3D and tables S25 and S26).
MM-PSN reveals prognostic heterogeneity in hyperdiploid and MMSET-translocated patients and improves on current risk classification systems
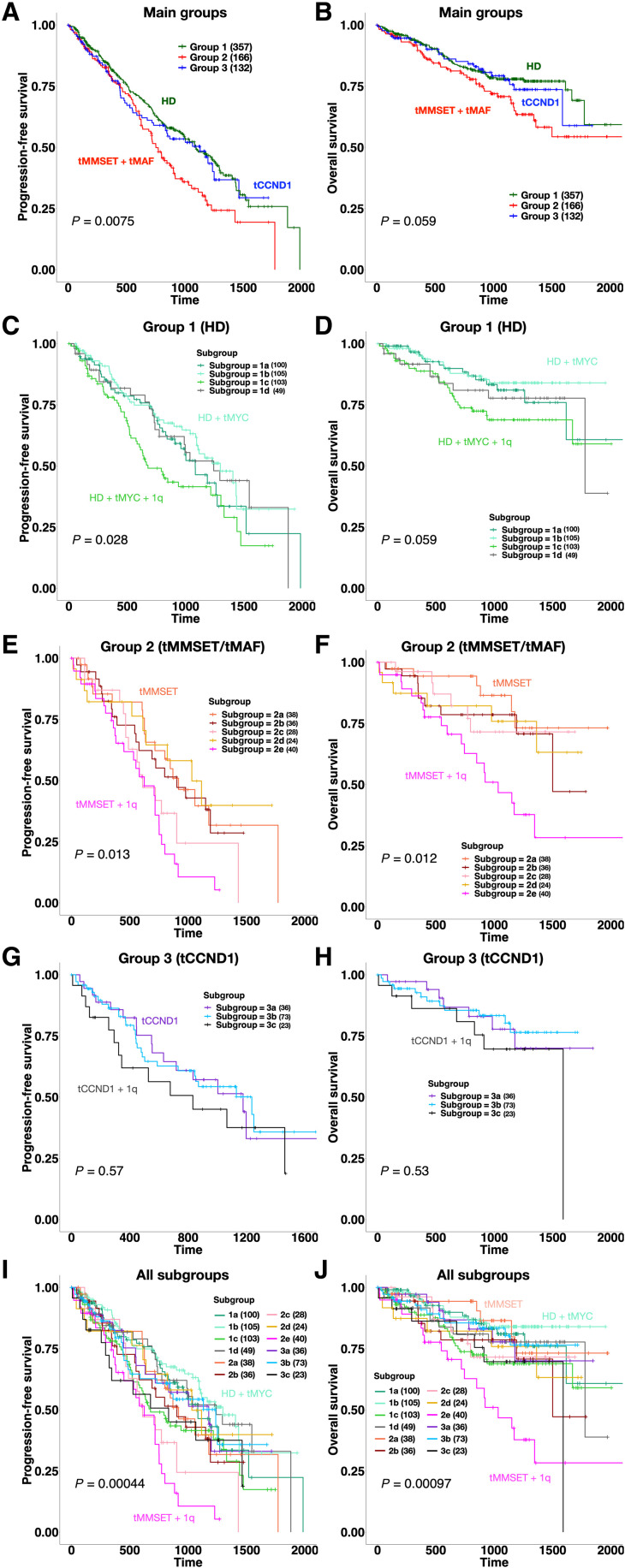
Survival analysis of the three main groups showed that patients in group 2 (tMMSET/tMAF) had shorter progression-free survival (PFS) compared to patients in groups 1 (HD) and 3 (tCCND1) (P < 0.01) (Fig. 2, A and B) and shorter overall survival (OS) compared to patients in group 1 (P = 0.05). While HD has been shown to confer a more favorable prognosis, concurrent gain(1q) in 1c (HD/tMYC/1q+) was associated with higher risk of relapse within group 1 [hazard ratio (HR) = 1.6, P = 0.04] and significantly shorter OS than patients in subgroup 1b (HD/tMYC) (HR = 2.26, P = 0.01) (Fig. 2, C and D). Stratification of the whole cohort based on the co-occurrence of HD and gain(1q) revealed significantly shorter PFS and OS in patients carrying both aberrations, as compared to patients with HD and no 1q alterations (PFS: HR = 1.56, Padj = 0.03; OS: HR = 2.04, Padj = 0.007) (Fig. 3, A and B).
Fig. 2. Survival analysis of MM-PSN identifies high-risk subgroups.
(A and B) PFS and OS plots for the three main patient groups identified by MM-PSN, showing significant poorer outcome for the tMMSET + tMAF group. (C and D) Survival plots for subgroups of group 1 show shorter PFS in patients from subgroup 1C characterized by HD, tMYC, and 1q gain, compared to patients in subgroup 1B, which do not have 1q gain. (E and F) Survival plots for subgroups of group 2 show shorter PFS and OS in patients from subgroup 2E, enriched for tMMSET and 1q gain. (G and H) Survival plots for subgroups of group 3 do not show significant differences in either PFS or OS. (I and J) Survival plots for all 12 subgroups of MM-PSN, indicating poorer outcome of patients in subgroup 2e. P values were calculated using the log-rank test.
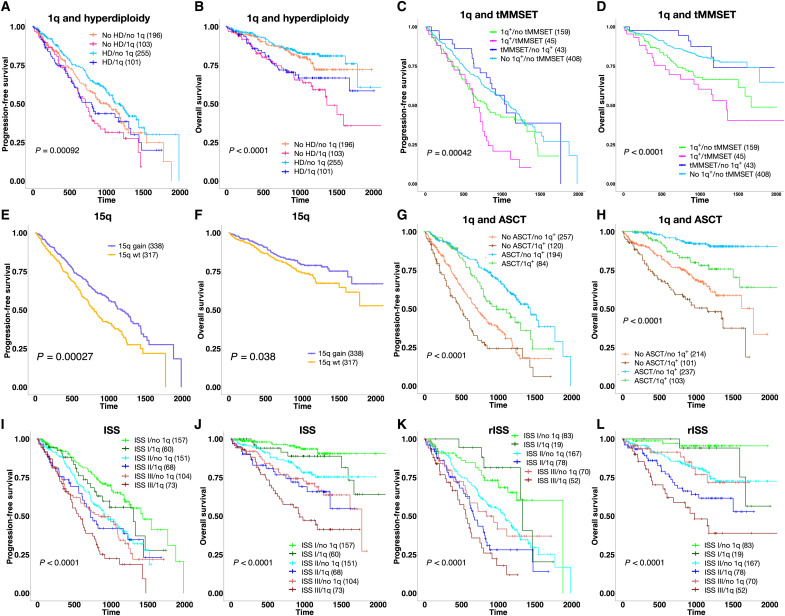
Fig. 3. Prognostic implications of gain(1q), tMMSET, and gain(15q).
(A and B) Survival plots show that 1q gain identifies a subset of HD patients with significantly shorter PFS and OS. (C and D) Survival plots show that patients with 1q gain with or without tMMSET have poorer outcome than patients with tMMSET alone. (E and F) Gain of 15q is associated with better PFS and OS. (G and H) Survival plots show that patients with 1q gain that received autologous stem cell transplant (ASCT) have significantly better PFS and OS compared to patients that did not receive ASCT. (I and J) Gain of 1q significantly stratifies risk for relapse and mortality in patients in International Staging System (ISS) classes I and III and risk of mortality in patients in ISS class II. (K and L) Gain of 1q significantly stratifies risk for relapse and mortality in patients in revised ISS (rISS) class II and risk of mortality in patients in rISS class III. P values were calculated using the log-rank test.
tMMSET is currently considered to confer poor prognosis and identifies high risk in the revised International Staging System (R-ISS) (24). MM-PSN revealed the existence of three separate subgroups of patients carrying tMMSET, each associated with different genetic and molecular alterations and with significantly different risk profiles. While subgroup 2e (tMMSET/1q+) had significantly worse PFS (HR = 2.04, P = 0.002) and OS (HR = 2.71, P = 5 × 10−4) within group 2 and overall, patients in subgroup 2a (tMMSET) had a significantly better prognosis, comparable to that of patients with HD (HR = 2.35, P = 0.005) (Fig. 2, E, F, I, and J). Of note, subgroup 2d, which was enriched for both tMMSET and gain(1q) and had additional HD features such as gain(15q) and gain(3p/3q), had better prognosis than subgroup 2e (HR = 2.59, Padj = 0.04). Conversely, subgroup 2c (1q+/14q−), which was the only subgroup in group 2 without any translocations, had a PFS profile similar to subgroup 2e.
While no significant differences in survival were detected among subgroups of the tCCND1-enriched group 3, a slightly poorer PFS in patients in subgroup 3c enriched for gain of 1q (tCCND1/1q+) was observed (HR = 1.3, P = 0.3) (Fig. 2, G and H). These findings indicate a significant overall role of gain(1q) in driving prognosis.
Multivariate Cox regression analysis confirmed overall shorter PFS in all subgroups enriched for gain(1q) and shorter OS in subgroups 1c (HD/tMYC/1q+) and 2e (tMMSET/1q+), even after adjusting for sex, age, race, and the therapies received (see figs. S4 and S5 for details). Further dissection of the major alterations enriched in the subgroups showed that gain(1q) alone and co-occurring with tMMSET were both significantly associated with worse PFS and OS (PFS: HR = 2.08, P < 0.001, OS: HR = 3.49, P < 0.001), while tMMSET alone was not (figs. S6 and S7). Overall, patients with both tMMSET and gain(1q) (n = 45) had much shorter PFS and OS, compared to patients with tMMSET alone (n = 43) (PFS: HR = 2.3, Padj = 0.01; OS: HR = 3.47, Padj = 0.02) (Fig. 3, C and D). Because the number of 1q copies has been previously reported as being prognostically relevant, we evaluated it across all the subgroups (25, 26). Subgroup 2e had the highest number of copies (P = 0.0001) (fig. S8A). Stratification extended to all patients in CoMMpass with CNA data available (n = 870) confirmed that patients carrying four copies of 1q had worse PFS and OS than patients carrying three copies (PFS: HR = 1.4, Padj = 0.06; OS: HR = 1.6, Padj = 0.04) (fig. S8, B and C). The finding of amp(1q) in group 2e indicates that this subgroup identifies the subclonal progression of 1q copy number. We have previously shown concomitant multiple whole-arm deletions in patients with gain(1q), including 16q and 17p (27, 28). Our analysis revealed significant enrichment for del(16q) in subgroup 2b (tMAF) and significant co-occurrence of gain(1q) with del(16q) in subgroups 1c (HD/tMYC/1q+), 2b (tMAF), and 2c (1q+/14q−), and of gain(1q) with del(17p) in subgroup 2d (tMMSET/1q+/15q−).
Biallelic inactivation of TP53 (mutation and deletion), which was enriched in subgroup 3b (tCCND1/11q+/13q−), was also significantly associated with worse prognosis (PFS: HR = 2.02, Padj = 0.01; OS: HR = 3.08, Padj = 6 × 10−4), although it was overall present in a small fraction of the patient population (n = 22, 3.3%) (figs. S6, S7, and S9). Of note, the translocation of MAF, which has been considered a high-risk alteration in previous studies, was not significantly associated with worse outcome (29). The analysis also showed that male patients, independent of ancestry, had also worse OS (HR = 1.50, P = 0.041). In contrast, gain(15q), which was a widespread alteration in main group 1 (HD) and significantly enriched in subgroup 2d (tMMSET/1q+/15q−), was associated with better PFS and OS (PFS: HR = 0.66, P = 3 × 10−4; OS: HR = 0.7, P = 0.04) (Fig. 3, E and F). This might, in part, explain the better prognosis observed in subgroup 2d compared to subgroup 2e, despite the presence of tMMSET and gain(1q).
The analysis also revealed that neither induction therapy (e.g., bortezomib/carfilzomib with lenalidomide) nor autologous stem cell transplant (ASCT) could overcome the negative prognostic impact of gain(1q). While patients with gain(1q) who received ASCT (n = 108) had better prognosis compared to patients with gain(1q) who did not receive it (n = 217), the outcome was still significantly poorer than in patients without gain(1q) who received ASCT (n = 194) (Fig. 3, G and H, and figs. S6 and S7). Stratification of patients based on co-occurrence of tMMSET and gain(1q) and ASCT showed that ASCT was significantly associated with better PFS in patients with tMMSET and tMMSET + 1q but did not correspond to better OS in patients with tMMSET alone (fig. S10). In the whole cohort (n = 655), we observed better PFS in patients treated with carfilzomib-based therapies (HR = 0.46, P = 0.05), and ASCT was the only treatment that was overall significantly associated with both better PFS and OS (PFS: HR = 0.26, P = 0.001; OS: HR = 0.26, P = 0.001) (figs. S6 and S7).
Given these prognostic findings, we stratified patients on the basis of their risk classes according to the ISS and the presence/absence of gain(1q). ISS is the most used risk score in the clinical setting and is based on serum b2 microglobulin and albumin (30). The rISS additionally includes lactate dehydrogenase as well as the high-risk cytogenetic markers tMMSET, tMAF, and del(17p), but not gain(1q) (24). Our analysis revealed that gain(1q) could identify patients at higher risk of relapse in ISS class III (Padj = 0.07) and rISS classes II and III (rISS II: Padj = 0.01; rISS III: Padj = 0.03) (Fig. 3, I and J). Gain(1q) also identified patients with significantly shorter OS in ISS and rISS classes II and III (ISS II: Padj = 0.04; ISS III: Padj = 0.07; rISS II: Padj = 0.02; rISS III: Padj = 6 × 10−3) (Fig. 3, K and L).
Prognostic findings from MM-PSN replicate in an independent dataset of 559 newly diagnosed patients
To validate our MM-PSN prognostic model based on co-occurrence of tMMSET and gain(1q), we performed survival analysis on the 559 newly diagnosed patients before TT2 and TT3 treatments from the UAMS cohort that were assigned to MM-PSN groups and subgroups with our gene expression classifier. At the group level, we were able to replicate the prognostic findings from MM-PSN, with group 2 having significantly shorter PFS (HR = 2.25, Padj = 6.6 × 10−7) and OS (HR = 1.8, Padj = 1 × 10−4) than groups 1 and 3 (fig. S11, A and B). Furthermore, we validated subgroup 2e as having significantly shorter PFS (HR = 3.08, Padj = 0.02) and OS (HR = 3.64, Padj = 0.01) than subgroup 2a (fig. S11, C and D), thus supporting the existence of two tMMSET-associated disease subtypes with different risk profiles and, specifically, the negative prognostic impact of co-occurrence of tMMSET and gain(1q).
Multiomics enrichment analysis reveals activation of specific oncogenic pathways in MM-PSN subgroups
To characterize the 12 MM-PSN subgroups functionally, we performed a multiomics gene set and pathway activation analysis using the data types contributing the most to the classification: gene expression, CNAs, and translocations. The analysis revealed concordance between different types of alterations and allowed the identification of CNAs and translocations with corresponding gene expression changes, thus more likely to be functional. Table 2 and Fig. 4 provide a summary and a graphical representation of the most representative results, respectively (full data are given in tables S27 and S28). The analysis was performed on a set of 6229 curated canonical pathways and 50 hallmark gene sets from MSigDB (31–33). The results were adjusted for multiple testing and considered significant if the corrected P values were below the 5% cutoff.
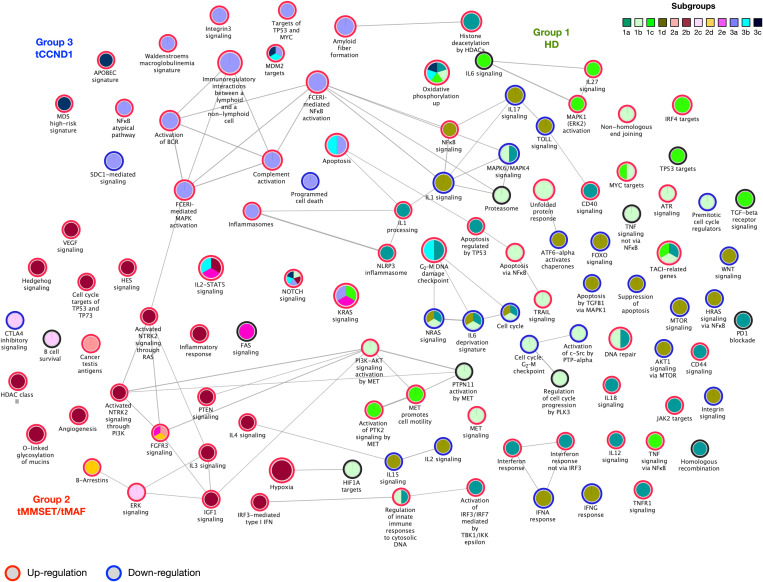
Fig. 4. Pathway activation in MM-PSN subgroups.
Enrichment map for selected pathways that are significantly activated in MM-PSN subgroups. Each circle indicates a pathway, and the colors represent the subgroups with significant up- or down-regulation of the pathway (indicated in red and blue, respectively). Edges connect pathways that share genes, and edge thickness is proportional to the number of shared genes.
Group 1 (HD) was overall enriched for inflammation and immune evasion pathways. In particular, subgroup 1a (HD/-7−) was characterized by significant activation of interferon-related pathways and CD40 signaling, up-regulation of Janus kinase 2 (JAK2) targets, and up-regulation of genes in the interleukin-1 (IL-1)–processing pathway [IL-18, RELA (RELA proto-oncogene, NF-KB subunit), and CASP1 (caspase 1)] as a consequence of copy number gain of their loci on chromosome 11. Subgroup 1b (HD/tMYC) was enriched for apoptosis, (TNF)-related apoptosis-inducing ligand (TRAIL) signaling, proteasome, and unfolded protein response. Furthermore, we observed activation of MET signaling in subgroups 1b and 1c (HD/tMYC/1q+), and of ATR signaling in subgroup 1b, as a consequence of copy number gain and corresponding up-regulation of several genes in these pathways. This may be clinically relevant because both MET and ATR are considered potential therapeutic targets in MM. Subgroup 1c (HD/tMYC/1q+) also showed activation of mitogen-activated protein kinase 1/extracellular signal–regulated kinase 2 (MAPK1/ERK2) and IL27 signaling and up-regulation of interferon regulatory factor 4 (IRF4) targets. In subgroup 1d (MultiDel), we identified several pathways affected by the observed multiple chromosomal deletions, including fas cell surface death receptor (FAS), mechanistic target of rapamycin kinase (MTOR), forkhead box O (FOXO), TOLL, and several IL pathways such as IL-1, IL-2, IL-15, and IL-17.
Group 2 (tMMSET/tMAF) was overall enriched for proliferative pathways. Subgroups 2a (tMMSET), 2d (tMMSET/15q+), and 2e (tMMSET/1q+) were characterized by the activation of the FGF signaling pathway as a result of the t(4;14) translocation involving FGFR3/MMSET and additional activating mutations in the gene. In addition, subgroup 2c was characterized by the activation of ERK signaling and the down-regulation of genes involved in cytotoxic T-lymphocyte-associated protein 4 (CTLA4) inhibitory signaling, while subgroup 2e showed activation of FOS signaling. Subgroup 2b (tMAF) was instead characterized by up-regulation of neurotrophic receptor tyrosine kinase 2 (NTRK2), activation of the hypoxia pathway, and activation of insulin-like growth factor 1 (IGF1), IL, and hedgehog signaling.
Group 3 (tCCND1) was overall enriched for pathways associated with replicative immortality and evasion of growth suppression. All three subgroups exhibited up-regulation of targets of MDM2, which is a critical negative regulator of TP53. Furthermore, subgroup 3b (tCCND1/11q+/13q−) was characterized by higher expression of B-cell lymphoma 2 (BCL2) and low expression of myeloid cell leukemia 1 (MCL1), while subgroup 3a (tCCND1) was enriched for activation of inflammasomes and complement cascade and down-regulation of proapoptotic genes such as BCL2 Associated X (BAX). Subgroup 3a was also enriched for up-regulation of genes involved in Waldenström macroglobulinemia and amyloid fiber formation, suggesting that this group of patients may be at higher risk of developing amyloidogenic plasma cells.
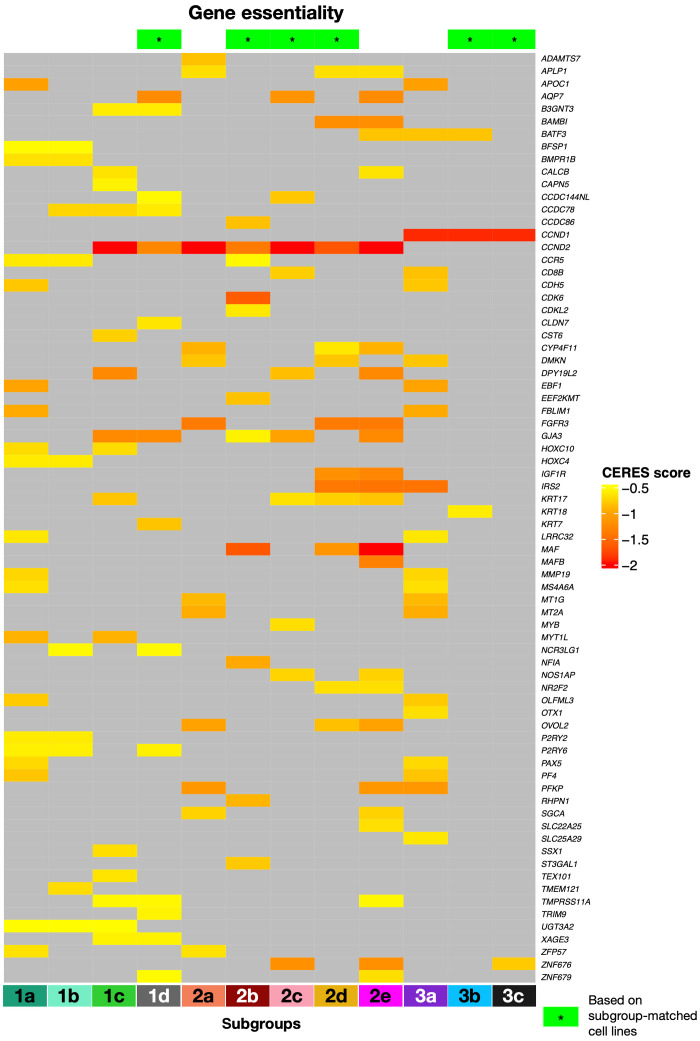
Gene essentiality screenings and multiomics drug repurposing identify potential subgroup-specific vulnerabilities and small-molecule and immuno-oncology candidates for future trials
To investigate the therapeutic implications of the MM-PSN classification, we matched the up-regulated genes in each subgroup with genes essential for cell survival as determined by CRISPR-Cas9 screens [Cancer Dependency Map (DepMap)] in MM cell lines (CERES score < −0.5; see Methods for details) (34, 35). First, we used the MM-PSN classifier to assign 60 MM cell lines to subgroups on the basis of their DNA and RNA profiles. These data enable in vitro studies using cell lines to model the MM-PSN subgroups and are provided as supplementary data (tables S29 and S38). We were able to find at least one representative cell line for 9 out of 12 subgroups and identified essential up-regulated genes in 6 of the 9 subgroups (1d, 2b, 2c, 2d, 3b, and 3c) using the corresponding CRISPR-Cas9 data. We additionally determined essential genes in the remaining subgroups using data from all available MM cell lines. The analysis revealed a total of 213 essential genes, some of which were potential actionable targets (Fig. 5 and table S30). The overexpression of CCND2 was determined to be the major vulnerability across all group 2 (tMMSET/tMAF) and subgroups 1c (HD/1q+) and 1d (MultiDel). CCND2 is a transcriptional target of MAF (musculoaponeurotic fibrosarcoma); thus, its overexpression in subgroup 2b (tMAF) is a direct consequence of tMAF. We also determined that up-regulation of MAF and CCND2 were vulnerabilities in subgroups 2d (tMMSET/1q+/15q+) and 2e (tMMSET/1q+), consistent with recent studies that have reported MAF activation also in tMMSET cell lines and primary samples (36). However, MAF was not identified as essential in subgroup 2a (tMMSET), revealing that not all patients with tMMSET have MAF activation. Because the MAPK kinase (MEK)–ERK pathway has been demonstrated to regulate MAF transcription, patients in MAF-expressing subgroups could be potential candidates for treatment with MEK inhibitors, currently being tested in clinical trials (7, 37). Moreover, a recent study demonstrated that CCND2 down-regulation is achievable using the signal transducer and activator of transcription 3 (STAT3) inhibitor SC99, which has potent anti-MM activity both in vitro and in vivo (38). CDK6 was another major vulnerability identified in the 2b (tMAF), supporting clinical evaluation of CDK4/CDK6 inhibitors, such as palbociclib and abemaciclib, in this class of patients. Another actionable vulnerability identified by our screening was the overexpression of FGFR3 in the tMMSET subgroups 2a, 2d, and 2e (39). Sensitivity to FGFR inhibitors in patients overexpressing FGFR3 is currently being tested in basket trials (7). Among the essential genes up-regulated in the high-risk subgroup 2e (tMMSET/1q+), we identified IGF1R, which has been previously reported as aberrantly expressed in aggressive MM (40, 41) and represents a potential vulnerability that could be exploited therapeutically in this specific class of patients. We also determined the type I melanoma antigen gene MAGEA3 as a potential vulnerability in gain(1q)-enriched subgroups 1c (HD/tMYC/1q+) and 2c (1q+/14q−). We recently showed that MAGEA3 inhibits apoptosis and promotes proliferation in MM, and its overexpression is associated with shorter OS after melphalan chemotherapy and ASCT (42). MAGEA-targeted immunotherapies are currently being evaluated in different cancers, including MM (43–45). In group 3, besides consistent up-regulation of CCND1 across all three subgroups, we identified FLT3 as essential in subgroup 3a (tCCND1). A recent study demonstrated poor prognostic significance of FLT3 overexpression in MM and in vitro anti-MM activity on FLT3-positive cell lines and primary cells, thus supporting our finding of a specific subgroup of patients who could potentially benefit from this treatment (46).
Fig. 5. Gene essentiality screening identifies potential vulnerabilities in MM-PSN subgroups.
Selected genes that are considered essential for cell survival according to the CRISPR-Cas9 screening data retrieved from DepMap. Lower CERES scores indicate higher essentiality. A green star symbol in the heatmap header indicates that the gene was identified based on cell lines that were also specifically matched to the corresponding subgroups. For the other subgroups, genes were identified using all the available MM cell lines in DepMap.
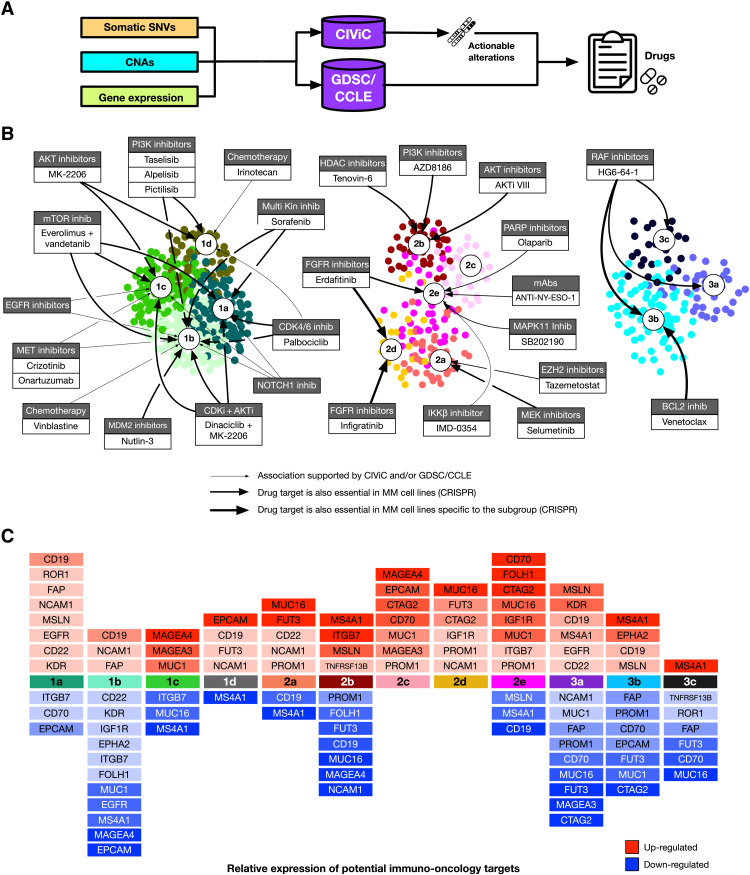
We further investigated the therapeutic implications of MM-PSN by applying our multiomics precision medicine approach previously described to the patients in the network and performing subgroup enrichment (6). The analysis identified general, group-, and subgroup-specific therapeutic options on the basis of actionable DNA and RNA alterations represented in the database CIViC (Clinical Interpretation of Variants in Cancer) (47) as well as from cell line profiles from the Cancer Cell Line Encyclopedia (CCLE) and the Genomics of Drug Sensitivity in Cancer (GDSC) databases (Fig. 6, A and B, tables S31 to S37) (48–50). We further annotated the group/drug associations with gene essentiality from CRISPR screens.
Fig. 6. Multiomics drug repurposing identifies candidate therapeutic options and potential immuno-oncology targets in MM-PSN subgroups.
(A) Schema of the drug repurposing analysis. Somatic SNVs, CNAs, and gene expression profiles are annotated with the pan-cancer database CIViC to determine actionable alterations and the associated drugs. Additional drugs are identified through a machine learning approach matching patient profiles with sensitive cell lines in the databases GDSC and CCLE. (B) Drugs associated with each subgroup. The thickness of the edges represents the strength of the association, as defined by the essentiality of drug targets in MM cell lines according to CRISPR screenings (DepMap). (C) Targets of immuno-oncology therapies up- and down-regulated in each subgroup.
Group 1 (HD) was enriched for several drugs mostly matched to actionable genes in the chromosomes affected by trisomies. For example, the MET inhibitors onartuzumab and crizotinib were top-ranking options in subgroups 1b (HD/tMYC) and 1c (HD/tMYC/1q+) because of gain of MET (chromosome 7) and its consequent up-regulation. This was consistent with the results of our pathway analysis, demonstrating specific activation of MET/HGF in subgroup 1b (HD/tMYC). This is clinically relevant because MET dysregulation has been reported to be a hallmark of aggressive disease in MM, and previous studies have suggested it as a potential therapeutic target in MM. Therefore, subgroups 1b and 1c identify a population of patients with HD MM who may be sensitive to MET inhibition. Other options in group 1 included the CDK4/CDK6 inhibitor palbociclib associated with CCND1 gain in subgroups 1a, 1b, and 1d; the NOTCH1 antibody PF-06293622 associated with gain of NOTCH1 in subgroups 1a, 1b, and 1c; and the phosphatidylinositol 3-kinase (PI3K) inhibitors alpelisib and taselisib associated with gain of phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA) and pictlisib associated with gain of PIK3CA or Erb-B2 receptor tyrosine kinase 2 (ERBB2) and mutations in BRAF in subgroups 1b and 1d. Some of these associations were further supported by matched cell line data from CCLE and GDSC (Fig. 6, A and B). In addition, subgroup 1d was associated with irinotecan because of the amplification of DNA topoisomerase I (TOP1).
In group 2 (tMMSET/tMAF), tazemetostat and selumetinib were significant top-ranking options in subgroup 2a (tMMSET) because of deletion and/or underexpression of SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 (SMARCB1) and deletion of moesin-ezrin-radixin like (MERLIN) tumor suppressor (NF2), respectively. The pan-FGFR inhibitors erdafitinib and infigratinib and the cisplatin/gemcitabine combination were significantly recommended for subgroup 2d (tMMSET/1q+/15q+) because of the enrichment for FGFR3 mutations (7). This was further supported by patient profile matching with GDSC and CCLE drug screenings and by the essentiality of FGFR3 observed in cell lines representative of subgroup 2d. Potential options in subgroup 2e (tMMSET/1q+) included the MAPK11 inhibitor SB202190 because of the underexpression of STK11, the poly(adenosine diphosphate–ribose) polymerase (PARP) inhibitor olaparib because of the overexpression of PARP and/or underexpression of ATM serine/threonine kinase (ATM), and the FGFR inhibitor erdafitinib because of mutations in FGFR3. The use of PARP inhibitors for patients in subgroup 2e is also supported by their increased genomic instability, which is a marker for this therapeutic class as suggested by previous studies in other cancers (51).
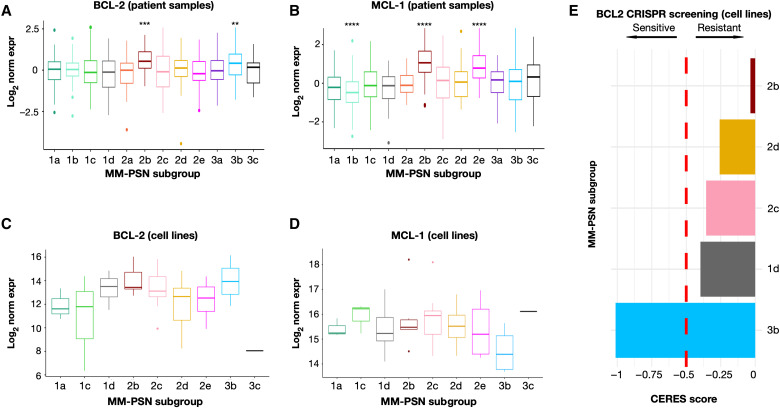
In group 3, we identified 3b (tCCND1/11q+/13q−) as a subgroup of patients with higher sensitivity to BCL2 inhibition. While t(11;14) has been reported as a marker of sensitivity to the BCL2 inhibitor venetoclax, because of the increased dependency on BCL2 for survival of the altered cells, higher expression of BCL2 itself is associated with better response, as determined in our study and in other studies (6, 52, 53). Furthermore, it has been shown that higher expression of the BCL2 family member MCL1 is associated with resistance to venetoclax (53). BCL2 and MCL1 expression profiles across MM-PSN subgroups showed significant up-regulation of BCL2 in subgroups 3b (tCCND1/11q+/13q−) and 2b (tMAF) (Fig. 7, A and B). However, subgroup 2b also showed significant up-regulation of MCL1, indicating resistance (Fig. 7, C and D). Cell line expression exhibited a similar trend, and data from CRISPR screenings confirmed vulnerability to BCL2 inhibition in cell lines associated with subgroup 3b (e.g., SKMM2 and KMS27) but not in cell lines associated with subgroup 2b (e.g., OCIMY5) (Fig. 7E). These findings suggest that MM-PSN subgroup 3b may identify patients that are sensitive to venetoclax. Moreover, the expression pattern of BCL2 and MCL1 in subgroup 2b also indicates a possible benefit for patient in this class from dual targeting of BCL2 and MCL1 (54, 55).
Fig. 7. BCL2 and MCL1 expression in patients and cell lines identifies subgroups at increased sensitivity and resistance to venetoclax.
(A to D) Gene expression profile of BCL2 and MCL1 across MM-PSN subgroups in patients and cell lines suggests sensitivity to venetoclax in subgroup 3b (high BCL2 and low MCL1) and resistance in subgroup 2b (high BCL2 and high MCL1). (E) CRISPR screening indicates increased essentiality of BCL2 in cell lines associated with subgroup 3b and decreased essentiality in subgroup 2b, supporting sensitivity and resistance to venetoclax, respectively. CERES score < −0.5 indicates essentiality. P values were calculated using the Wilcoxon rank sum test. **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Last, we matched up- and down-regulated genes in each subgroup with current chimeric antigen receptor (CAR) T cells and immuno-oncology treatments targets. While transcript expression of such genes does not necessarily correlate with the corresponding surface protein levels, it may still be an important indication of potential actionable targets specific to each subgroup. Of particular relevance is the expression of genes encoding proteins that were recently identified as most enriched on MM cell surface, the MM surfaceome, when compared to B cell malignancy models (56), such as TNFRSF13B, encoding the protein transmembrane activator and calcium modulating ligand (CAML) interactor (TACI), and integrin subunit beta 7 (ITGB7) (Fig. 6C). The mucin gene MUC1, which is encoded in chromosome 1q22, was a recurrent target identified in the gain(1q)-enriched subgroups 1c (HD/tMYC/1q+), 2c (1q+/14q−), and 2e (tMMSET/1q+), while MUC16 was found up-regulated in subgroups 2a (tMMSET), 2d (tMMSET/1q+/15q+), and 2e (tMMSET/1q+). Other relevant targets included IGF1R, which was also identified as an essential gene in subgroup 2e; CTAG2, up-regulated in subgroups 2d and 2e; ITGB7, up-regulated in subgroups 2b (tMAF) and 2e; and TNFRSF13B, up-regulated in subgroup 2b. The latter encodes the TACI protein, which is a member of the TNFR receptor superfamily and is a critical regulator of B cell maturation and differentiation into plasma cells, together with the B cell maturation agent (BCMA) protein (57). BCMA is a suitable therapeutic target for CAR T, bispecific T cell engagers (BiTE), and antibody-drug conjugates, such as belantamab mafodotin, and TACI is now being investigated as a potential additional target for immunotherapies in MM (58, 59). Up-regulation of its transcript in subgroup 2b may indicate greater response in these patients.
In group 3 (tCCND1), the gene MS4A1, encoding the B cell surface protein CD20, was identified as a potential target in subgroups 3b (tCCND1/11q+/13q−) and 3c (tCCND1/1q+). Subgroup 3c was instead characterized by down-regulation of TNFRSF13B, which may correspond to reduced response to TACI targeted therapy.
Last, subgroup 1d (MultiDel) was enriched for the deletion of 16p, which harbors TNFRSF17, the gene encoding BCMA. Patients in this subgroup are at higher risk of biallelic loss of BCMA, which has been shown to trigger resistance to anti-BCMA CAR T therapy in MM (60). Overall, MM-PSN may help inform patient selection in future clinical trials.
DISCUSSION
In this study, we have generated a PSN of newly diagnosed patients with MM, MM-PSN, using five different data types derived from WGS, WES, and RNA-seq and have determined a novel classification of MM consisting of three main groups and 12 subgroups. While several studies have focused on specific genetic lesions to determine their prognostic implications, our analysis aimed at dissecting the intertumor heterogeneity of MM and investigating the co-occurrence of multiple alterations in an integrated fashion.
Overall, our MM-PSN model organized previous knowledge on genetic markers into broad patient groups defined by translocations or HD and then further refined and harmonized such knowledge cohesively with other genetic and molecular markers to determine subgroups of highly similar patients. Our model identified two novel subgroups, one (1d) characterized by multiple chromosome deletions and one (2c) identified by co-occurrence of gain(1q) with del(14q). The MM-PSN classification uncovered novel associations between distinct MM hallmarks with powerful prognostic implications and enabled further refinement of risk stratification.
Overall, the significant representation of specific patterns of genomic and transcriptomic dysregulation in MM-PSN subgroups, as shown in Fig. 1, indicates coherent biological entities and outlines their genetic and molecular features, as well as their prognostic and potential for therapeutic implications. Close inspection of the MM-PSN groups identified a small number of outlier patients who lacked some of the defining features of the groups they were assigned to (fig. S12).
The most significant findings of our study involve the gain of the long arm of chromosome 1, a high-risk feature observed in ~40% of MM cases and in several other types of cancer, including breast cancer, hepatocellular carcinoma, and myeloproliferative neoplasms (61–64). Patients affected by monoclonal gammopathy of undetermined significance, a precursor of MM, and smoldering MM who carry gain(1q) have higher risk of progression to MM, with a median time to progression of 2 years (65). Other studies with newly diagnosed and relapsed/refractory MM patients have confirmed the negative prognostic impact of gain(1q) in different therapeutic regimens (25, 66). MM-PSN identified six subgroups (1c, 2b, 2c, 2d, 2e, and 3c) enriched for gain(1q), all of them associated with other recurrent lesions and having the shortest median time to relapse and death compared to the other subgroups. We have previously described 1q12 pericentromeric instability of satellite DNA as a major cause of many of the secondary subclonal karyotypic events in MM (67). In this regard, a recent study of double-refractory MM found that karyotypic events influenced clustering of patients more than treatment or mutations and that amp(1q) was the only high-risk feature predicting survival (68). These authors suggest that chromosome instability enables subclones to enter different evolutionary trajectories and adapt to selective pressure of therapies and underlying treatment failures. The types of secondary karyotypic associations found in MM-PSN add significantly to the genomic heterogeneity of the disease and are most likely, at least partially, responsible for the resistance to therapy. The subgroup of patients having the poorest outcome in terms of both PFS and OS had co-occurrence of gain(1q) with the t(4;14) translocation involving MMSET. We further confirmed this prognostic implication in an independent dataset. Notably, while the presence of gain(1q) alone was still a significantly deleterious event, the presence of tMMSET alone was not. This is an important result, as tMMSET has always been considered a high-risk feature regardless of other co-occurring lesions (24, 69).
In contrast, and consistent with findings from the Myeloma XI and Myeloma IX trials, gain(1q) appeared to confer poorer prognosis also to patients usually considered at lower risk, such as those carrying HD (26). Hyperdiploid patients with concurrent gain(1q) had significantly shorter PFS and OS than hyperdiploid patients with no gain(1q). A similar trend, although not statistically significant, was also observed in patients with tCCND1 and concurrent gain(1q), who had a median time to progression close to patients with tMMSET and gain(1q).
Our multivariate analysis also revealed a protective effect conferred by gain(15q), whose presence determined a significantly longer PFS and OS. This might also explain the better outcome observed in patients with tMMSET, gain(1q), and gain(15q) (subgroup 2d). Further investigations aimed at dissecting this potential protective effect are currently underway.
Despite increasing evidence supporting the prognostic relevance of gain(1q), current staging systems such as the ISS and its revised version, rISS, do not include it (24, 70). Our results show that gain(1q) could significantly stratify patients in almost all risk classes into high- versus low-risk subclasses, in terms of both PFS and OS. Our results independently confirm findings by Walker et al. (71) and suggest that gain(1q) should be incorporated into staging systems and used in the clinic to determine patient risk.
Functional characterization of the MM-PSN subgroups through pathway activation analysis and drug repurposing based on both DNA and RNA alterations have further revealed meaningful insights with important biological and clinical implications. These findings have immediate implication for precision medicine and clinical trials, as different subgroups of patients may respond to different targeted and immuno-oncology therapies based on their genomic and transcriptomic profiles. A clear example of potential therapeutic implications of MM-PSN is provided by the significant up-regulation of BCL2 in the tCCND1 subgroup 3b (tCCND1/11q/13q) and in the tMAF subgroup 2b (tMAF). In MM, tCCND1 and high BCL2 expression are considered markers of sensitivity to venetoclax, while the up-regulation of MCL1, which is often a consequence of copy number gain of its locus at 1q21, is associated with resistance. Our analysis suggested higher sensitivity to venetoclax in subgroup 3b and resistance in subgroup 2b because of concurrent up-regulation of MCL1. Other examples of applications of MM-PSN in patient selection for specific therapies include the activation of MET signaling in subgroups 1b (HD/tMYC) and 1c (HD/tMYC/1q+), which suggest the use of MET inhibitors for patients in these classes, or the deletion of one copy of the gene encoding the CAR T target BCMA, TNFRSF17, in patients in subgroup 1d, which may also indicate higher risk of failure in these patients, as they are more likely to acquire additional alterations leading to biallelic loss of BCMA.
Our study confirms the advantages of using multiple features to dissect cancer heterogeneity and the ability of PSNs to handle multiple data types to generate clear and interpretable disease models. MM-PSN structures and harmonizes the complexity of MM by associating patients with highly similar genomic and transcriptomic profiles to form more granular and homogeneous classes than achieved by previous classifications. The MM-PSN classification is a valuable and accessible resource that can be used in most clinical settings, because the features defining high risk can be easily detected by fluorescence in situ hybridization/cytogenetics.
While the prognostic impact of gain(1q) has been previously investigated and established in numerous studies, our network model and analysis have revealed a much higher significance and centrality of this genetic lesion in risk assessment of treatment-naive patients with MM. Ongoing research is now focused on a deeper characterization of the MM-PSN subgroups, and, in particular, those enriched for gain(1q), to gain novel insights into the molecular mechanisms and pathways that drive each disease subtype. These studies are fundamental to advance our understanding of MM pathology and paves the way for future research into drug repurposing approaches aimed at novel therapies tailored to specific patient subgroups.
METHODS
Dataset acquisition and primary data generation
WGS, WES, and RNA-seq data were generated from bone marrow aspirates (tumor CD138+ cells) and peripheral blood (control) of 655 treatment-naive newly diagnosed patients with MM enrolled in the MMRF CoMMpass study. Data were provided by MMRF and are available on the database of genotypes and phenotypes (dbGaP) database (www.ncbi.nlm.nih.gov/gap) under accession number “phs000748.”
Raw sequencing data were processed at Translational Genomics Research Institute (TGen), and primary data were generated as described in Supplementary Methods (SM1). Briefly, somatic single-nucleotide variants (SNVs) and small insertions-deletions (INDELs) were identified from matched tumor-normal WES data, CNAs, and structural variants that were identified from matched tumor-normal WGS data. RNA-seq data were used to obtain gene expression estimates and identify gene fusion transcripts.
PSN generation, visualization, and analysis
For the generation of MM-PSN, we applied the SNF method as it did not require a priori feature selection and was shown to outperform methods on the basis of single data types as well as other multiomics approaches such as iCluster (17, 19, 72). We additionally tested the methods iClusterBayes (73), which was discarded because it required feature selection and thus may significantly introduce bias in the results, and ANF (Affinity Network Fusion) (74), whose results were almost identical to those obtained by SNF. MM-PSN was generated using the SNF method implemented in the R package SNFtool (v2.3.0) as described in Supplementary Methods (section S2) (17). Briefly, SNFtool was run on 655 MM tumor samples using gene expression (50,495 genes), SNV (57,736 mutations), gene fusion (13,682 fusions), focal CNA (93 features), and broad CNA data (39 features), and the obtained fused matrix was clustered using the spectral clustering method implemented in the SNFtool package. We selected 3 as the optimal number of clusters, which maximized the eigen gap and minimized the rotation cost, as suggested by the authors of SNF (Fig. 1).
To visualize the network, we retrieved all the patient pairs from the fused similarity matrix W returned by SNF and retained only those with similarity greater than the third quartile values of all possible pairs for improved visualization. These filtered pairs were then imported in Cytoscape (v3.7.2) (75). We used the edge-weighted spring-embedded layout for visualizing the edges shown in Fig. 1A.
Significant features across MM-PSN groups and subgroups were identified by Kruskal-Wallis test (numerical features) and chi-square test (categorical features). Subgroup enrichment for significant features was determined by Wilcoxon rank sum test (numerical features) and chi-square test (categorical features). CNA data were considered numerical [Genomic Identification of Significant Targets in Cancer (GISTIC) scores], while SNV and fusion data were considered binary (presence or absence). Clinical data were split into numerical (e.g., blood markers) and categorical (e.g., sex) variables and analyzed separately. We adjusted the P values for multiple testing using the Bonferroni correction. We considered significant the features with adjusted P values <0.1. Differential gene expression (DE) analysis was performed using the R package edgeR (76), comparing each subgroup of patients with all the other patients.
We calculated subgroup-specific pathway activation by applying a multiomics enrichment approach to DE genes, CNAs, and translocations significantly enriched in each subgroup, using 6229 curated canonical pathways and 50 hallmark gene sets from MSigDB (32, 33, 77). P values were corrected for multiple testing using the Benjamini-Hochberg procedure, and 0.05 was set as the cutoff value for significance. The enrichment map was created using the plugin enhancedGraphics in Cytoscape (75, 78). Additional details on DE analysis, pathway enrichment, and the analysis of mutational signatures, genomic instability, and clonal landscape are provided in Supplementary Methods (section S3).
Gene essentiality screening and drug repurposing analysis
To identify essential genes in MM-PSN subgroups, we first applied the MM-PSN classifier to sequencing data from 60 MM cell lines and assigned them to nine different subgroups. No cell lines were matched with subgroups 1b, 2a, and 3a. The results are given in table S2. We then retrieved the Achilles dataset, which contains the results of genome-scale CRISPR knockout screens for 18,119 genes in 793 cell lines, from the DepMap portal. We identified MM cell line–specific essential genes as those with a CERES score < −0.5, as suggested in DepMap (34, 35). We matched the essential genes with significantly up-regulated genes in each subgroup, which were identified by DE analysis performed with the R package edgeR (76). Ig genes were excluded. Genes with logFC ≥ 1.5 and false discovery rate (FDR) < 0.05 were considered up-regulated. Specifically, up-regulated genes in each subgroup were matched with genes with a CERES score < −0.5 in at least one of the cell lines representative of the subgroup. For subgroups with no representative cell lines, we considered an average CERES score of all MM cell line data available. Overall, we identified 213 essential genes (table S3).
Drug repurposing analysis was performed using an updated version of our multiomics precision medicine platform previously described. For each patient in MM-PSN, we matched SNVs, CNAs, and differentially expressed genes with the actionable alterations reported in the database CIViC (47). Patient-specific differentially expressed genes were identified by calculating z scores from log2 variance stabilizing transformation (VST)-normalized gene expression data and selecting genes with |z scores| ≥ 2. Actionable alterations in CIViC are associated with drugs and/or drug combinations curated from the literature and clinical trials in MM and other cancers. We only considered associations with evidence level of A (validated), B (clinical evidence), C (case study), or D (preclinical). Inferential associations (level E) were discarded. In addition, we implemented a classifier to learn features predicting response to drugs leveraging large cell line collections from the CCLE and the GDSC (48–50). We then applied the classifier to predict potential drugs for the patients with MM. Subgroup enrichment for drugs and drug combinations was assessed by applying Fisher’s exact test to drugs and drug combinations associated with patients in the subgroup (table S4). P values were adjusted for multiple testing with the Benjamini-Hochberg procedure.
CAR T and immuno-oncology targets were curated from the literature and sources in our MM clinic (56, 79). Subgroup-specific targets were identified by matching this gene list with significantly up-regulated genes in each subgroup according to the analysis described above.
Survival analysis
PFS and OS were analyzed by the Kaplan-Meier method, and P values were calculated using the log-rank test. When multiple tests were performed, the P values were corrected for multiple testing using the Benjamini-Hochberg procedure. The HR was calculated using the Cox proportional hazards method (coxph from the R package “survival”).
MM-PSN multiomics and gene expression classifiers
For the generation of the MM-PSN multiomics classifier, we divided the 655 samples into training and validation sets in the ratio of 70:30. The classifier was built by combining the most informative three types of features, i.e., copy number of focal chromosome bands, translocation calls, and gene expression. We applied different strategies for feature filtering and selection to identify the top 50 CNA features that maximized the performance in terms of weighted recall and the top 100 genes with the highest NMI (normalized mutual information) for each of the three main groups and their subgroups. All the features were then further one-Hot–encoded to be fed into a stacking classifier with random forest, XGBoost, and linear SVC as the base learners. The performance on training data was calculated using threefold cross-validation. The final model was developed using full training data and tested on 30% validation dataset separated upfront from the full dataset. Figure S13 (A and B) shows the precision versus recall curves for the training and test sets, respectively. The features included in the classifier are given in tables S34 and S35.
To validate MM-PSN groups and subgroups on an independent gene expression dataset, we trained classification models on the 655 samples using gene expression data only. The 12 subgroups obtained by spectral clustering on the SNF-fused similarity matrix were taken as the ground truth for label assignment. The classifier was developed using a two-step process. First, an SVM classifier was developed to classify the three main groups using the z-scored expression of 134 genes. The 134 gene expression features were selected using the SVC-L1 feature selection method implemented in the Python package Scikit-learn. The samples predicted to be in group 2 were further sent through another SVM classifier based on 1200 features that were selected using the univariate feature selection method f_classif in Scikit-learn to predict membership to the five subgroups of group 2 (2a, 2b, 2c, 2d, and 2e). The best models were selected using 10-fold cross-validation at the group level and 5-fold cross-validation at the subgroup level on the training data. The validation data were generated with Affymetrix GeneChip U133 plus 2.0 arrays from Zhan et al. (9) and were retrieved from the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) (GSE2658). Additional details on the generation and validation of the classifiers are provided in Supplementary Methods (sections S5 and S6).
Acknowledgments
We thank all the patients enrolled in the MMRF CoMMpass study.
Funding: This work was supported by NIH-NCI (R21-CA209875-01A1 and R01-1R01CA244899-01A1), Tisch Cancer Institute (TCI) (NCI support grant: P30 CA196521), and the Multiple Myeloma philanthropic fund. This work was also supported, in part, through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Author contributions: Contribution: S.B., A.L., and S.P. conceived and designed the study. S.B. led the study and performed data analysis. D.T.M., A.A., and V.L. generated genomic metrics and performed data analysis. S.B., P.R., D.T.M., A.A., V.L., and A.L. created the figures. J.K. provided sequencing data. D.M., J.R., S.R., A.C., H.J.C., S.J., and S.P. contributed patient samples and clinical data. K.O., J.R.S., and J.T.D. provided scientific expertise. S.B., A.L., J.R.S., and S.P. wrote the manuscript. All authors revised and approved the final version of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are available on the dbGaP database (www.ncbi.nlm.nih.gov/gap) under accession number phs000748 and in the Supplementary Materials. The MM-PSN classifier is freely available at https://github.com/laganalab/MM-PSN. The dataset used for validation was retrieved from NCBI GEO (accession number GSE2658).
Supplementary Materials
This PDF file includes:
Supplementary Methods
Figs. S1 to S13
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S39
REFERENCES AND NOTES
- 1.Kumar S. K., Rajkumar V., Kyle R. A., van Duin M., Sonneveld P., Mateos M.-V., Gay F., Anderson K. C., Multiple myeloma. Nat. Rev. Dis. Primers. 3, 17046 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Bazarbachi A. H., Al Hamed R., Malard F., Harousseau J.-L., Mohty M., Relapsed refractory multiple myeloma: A comprehensive overview. Leukemia 33, 2343–2357 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Morgan G. J., Walker B. A., Davies F. E., The genetic architecture of multiple myeloma. Nat. Rev. Cancer 12, 335–348 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Lohr J. G., Stojanov P., Carter S. L., Cruz-Gordillo P., Lawrence M. S., Auclair D., Sougnez C., Knoechel B., Gould J., Saksena G., Cibulskis K., McKenna A., Chapman M. A., Straussman R., Levy J., Perkins L. M., Keats J. J., Schumacher S. E., Rosenberg M.; Multiple Myeloma Research Consortium, Getz G., Golub T. R., Widespread genetic heterogeneity in multiple myeloma: Implications for targeted therapy. Cancer Cell 25, 91–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolli N., Avet-Loiseau H., Wedge D. C., Van Loo P., Alexandrov L. B., Martincorena I., Dawson K. J., Iorio F., Nik-Zainal S., Bignell G. R., Hinton J. W., Li Y., Tubio J. M. C., McLaren S., O’Meara S., Butler A. P., Teague J. W., Mudie L., Anderson E., Rashid N., Tai Y.-T., Shammas M. A., Sperling A. S., Fulciniti M., Richardson P. G., Parmigiani G., Magrangeas F., Minvielle S., Moreau P., Attal M., Facon T., Futreal P. A., Anderson K. C., Campbell P. J., Munshi N. C., Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 5, 2997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laganà A., Beno I., Melnekoff D., Leshchenko V., Madduri D., Ramdas D., Sanchez L., Niglio S., Perumal D., Kidd B. A., Miotto R., Shaknovich R., Chari A., Cho H. J., Barlogie B., Jagannath S., Dudley J. T., Parekh S., Precision medicine for relapsed multiple myeloma on the basis of an integrative multiomics approach. JCO Precis. Oncol. 2018, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Study tests targeted drugs for multiple myeloma. Cancer Discov. 9, 459 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Bergsagel P. L., Kuehl W. M., Molecular pathogenesis and a consequent classification of multiple myeloma. J. Clin. Oncol. 23, 6333–6338 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Zhan F., Huang Y., Colla S., Stewart J. P., Hanamura I., Gupta S., Epstein J., Yaccoby S., Sawyer J., Burington B., Anaissie E., Hollmig K., Pineda-Roman M., Tricot G., van Rhee F., Walker R., Zangari M., Crowley J., Barlogie B., Shaughnessy J. D. Jr., The molecular classification of multiple myeloma. Blood 108, 2020–2028 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broyl A., Hose D., Lokhorst H., de Knegt Y., Peeters J., Jauch A., Bertsch U., Buijs A., Stevens-Kroef M., Beverloo H. B., Vellenga E., Zweegman S., Kersten M.-J., van der Holt B., el Jarari L., Mulligan G., Goldschmidt H., van Duin M., Sonneveld P., Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood 116, 2543–2553 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Laganà A., Perumal D., Melnekoff D., Readhead B., Kidd B. A., Leshchenko V., Kuo P.-Y., Keats J., DeRome M., Yesil J., Auclair D., Lonial S., Chari A., Cho H. J., Barlogie B., Jagannath S., Dudley J. T., Parekh S., Integrative network analysis identifies novel drivers of pathogenesis and progression in newly diagnosed multiple myeloma. Leukemia 32, 120–130 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Bolli N., Biancon G., Moarii M., Gimondi S., Li Y., de Philippis C., Maura F., Sathiaseelan V., Tai Y.-T., Mudie L., O’Meara S., Raine K., Teague J. W., Butler A. P., Carniti C., Gerstung M., Bagratuni T., Kastritis E., Dimopoulos M., Corradini P., Anderson K., Moreau P., Minvielle S., Campbell P. J., Papaemmanuil E., Avet-Loiseau H., Munshi N. C., Analysis of the genomic landscape of multiple myeloma highlights novel prognostic markers and disease subgroups. Leukemia 32, 2604–2616 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker B. A., Mavrommatis K., Wardell C. P., Ashby T. C., Bauer M., Davies F. E., Rosenthal A., Wang H., Qu P., Hoering A., Samur M., Towfic F., Ortiz M., Flynt E., Yu Z., Yang Z., Rozelle D., Obenauer J., Trotter M., Auclair D., Keats J., Bolli N., Fulciniti M., Szalat R., Moreau P., Durie B., Stewart A. K., Goldschmidt H., Raab M. S., Einsele H., Sonneveld P., San Miguel J., Lonial S., Jackson G. H., Anderson K. C., Avet-Loiseau H., Munshi N., Thakurta A., Morgan G. J., Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 132, 587–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maura F., Bolli N., Angelopoulos N., Dawson K. J., Leongamornlert D., Martincorena I., Mitchell T. J., Fullam A., Gonzalez S., Szalat R., Abascal F., Rodriguez-Martin B., Samur M. K., Glodzik D., Roncador M., Fulciniti M., Tai Y. T., Minvielle S., Magrangeas F., Moreau P., Corradini P., Anderson K. C., Tubio J. M. C., Wedge D. C., Gerstung M., Avet-Loiseau H., Munshi N., Campbell P. J., Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat. Commun. 10, 3835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pai S., Bader G. D., Patient similarity networks for precision medicine. J. Mol. Biol. 430, 2924–2938 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavalli F. M. G., Remke M., Rampasek L., Peacock J., Shih D. J. H., Luu B., Garzia L., Torchia J., Nor C., Morrissy A. S., Agnihotri S., Thompson Y. Y., Kuzan-Fischer C. M., Farooq H., Isaev K., Daniels C., Cho B.-K., Kim S.-K., Wang K.-C., Lee J. Y., Grajkowska W. A., Perek-Polnik M., Vasiljevic A., Faure-Conter C., Jouvet A., Giannini C., Rao A. A. N., Li K. K. W., Ng H.-K., Eberhart C. G., Pollack I. F., Hamilton R. L., Gillespie G. Y., Olson J. M., Leary S., Weiss W. A., Lach B., Chambless L. B., Thompson R. C., Cooper M. K., Vibhakar R., Hauser P., van Veelen M.-L. C., Kros J. M., French P. J., Ra Y. S., Kumabe T., López-Aguilar E., Zitterbart K., Sterba J., Finocchiaro G., Massimino M., Van Meir E. G., Osuka S., Shofuda T., Klekner A., Zollo M., Leonard J. R., Rubin J. B., Jabado N., Albrecht S., Mora J., Van Meter T. E., Jung S., Moore A. S., Hallahan A. R., Chan J. A., Tirapelli D. P. C., Carlotti C. G., Fouladi M., Pimentel J., Faria C. C., Saad A. G., Massimi L., Liau L. M., Wheeler H., Nakamura H., Elbabaa S. K., Perezpeña-Diazconti M., de León F. C. P., Robinson S., Zapotocky M., Lassaletta A., Huang A., Hawkins C. E., Tabori U., Bouffet E., Bartels U., Dirks P. B., Rutka J. T., Bader G. D., Reimand J., Goldenberg A., Ramaswamy V., Taylor M. D., Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31, 737–754.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang B., Mezlini A. M., Demir F., Fiume M., Tu Z., Brudno M., Haibe-Kains B., Goldenberg A., Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 11, 333–337 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research Network , Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 32, 185–203.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitroda S. P., Khodarev N. N., Huang L., Uppal A., Wightman S. C., Ganai S., Joseph N., Pitt J., Brown M., Forde M., Mangold K., Xue L., Weber C., Segal J. P., Kadri S., Stack M. E., Khan S., Paty P., Kaul K., Andrade J., White K. P., Talamonti M., Posner M. C., Hellman S., Weichselbaum R. R., Integrated molecular subtyping defines a curable oligometastatic state in colorectal liver metastasis. Nat. Commun. 9, 1793 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keats J. J., Craig D. W., Liang W., Venkata Y., Kurdoglu A., Aldrich J., Auclair D., Allen K., Harrison B., Jewell S., Kidd P. G., Correll M., Jagannath S., Siegel D. S., Vij R., Orloff G., Zimmerman T. M., Capone W., Carpten J., Lonial S., Interim analysis of the mmrf CoMMpass trial, a longitudinal study in multiple myeloma relating clinical outcomes to genomic and immunophenotypic profiles. Blood 122, 532–532 (2013). [Google Scholar]
- 21.Sztupinszki Z., Diossy M., Krzystanek M., Reiniger L., Csabai I., Favero F., Birkbak N. J., Eklund A. C., Syed A., Szallasi Z., Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. NPJ Breast Cancer 4, 16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samur M. K., Aktas Samur A., Fulciniti M., Szalat R., Han T., Shammas M., Richardson P., Magrangeas F., Minvielle S., Corre J., Moreau P., Thakurta A., Anderson K. C., Parmigiani G., Avet-Loiseau H., Munshi N. C., Genome-wide somatic alterations in multiple myeloma reveal a superior outcome group. J. Clin. Oncol. 38, 3107–3118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rustad E. H., Yellapantula V., Leongamornlert D., Bolli N., Ledergor G., Nadeu F., Angelopoulos N., Dawson K. J., Mitchell T. J., Osborne R. J., Ziccheddu B., Carniti C., Montefusco V., Corradini P., Anderson K. C., Moreau P., Papaemmanuil E., Alexandrov L. B., Puente X. S., Campo E., Siebert R., Avet-Loiseau H., Landgren O., Munshi N., Campbell P. J., Maura F., Timing the initiation of multiple myeloma. Nat. Commun. 11, 1917 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palumbo A., Avet-Loiseau H., Oliva S., Lokhorst H. M., Goldschmidt H., Rosinol L., Richardson P., Caltagirone S., Lahuerta J. J., Facon T., Bringhen S., Gay F., Attal M., Passera R., Spencer A., Offidani M., Kumar S., Musto P., Lonial S., Petrucci M. T., Orlowski R. Z., Zamagni E., Morgan G., Dimopoulos M. A., Durie B. G. M., Anderson K. C., Sonneveld P., Miguel J. S., Cavo M., Rajkumar S. V., Moreau P., Revised international staging system for multiple myeloma: A report from International Myeloma Working Group. J. Clin. Oncol. 33, 2863–2869 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt T. M., Barwick B. G., Joseph N., Heffner L. T., Hofmeister C. C., Bernal L., Dhodapkar M. V., Gupta V. A., Jaye D. L., Wu J., Goyal S., Chen Z., Boise L. H., Lonial S., Nooka A. K., Kaufman J. L., Gain of Chromosome 1q is associated with early progression in multiple myeloma patients treated with lenalidomide, bortezomib, and dexamethasone. Blood Cancer J. 9, 94 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah V., Sherborne A. L., Walker B. A., Johnson D. C., Boyle E. M., Ellis S., Begum D. B., Proszek P. Z., Jones J. R., Pawlyn C., Savola S., Jenner M. W., Drayson M. T., Owen R. G., Houlston R. S., Cairns D. A., Gregory W. M., Cook G., Davies F. E., Jackson G. H., Morgan G. J., Kaiser M. F., Prediction of outcome in newly diagnosed myeloma: A meta-analysis of the molecular profiles of 1905 trial patients. Leukemia 32, 102–110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawyer J. R., Tian E., Heuck C. J., Epstein J., Johann D. J., Swanson C. M., Lukacs J. L., Johnson M., Binz R., Boast A., Sammartino G., Usmani S., Zangari M., Waheed S., van Rhee F., Barlogie B., Jumping translocations of 1q12 in multiple myeloma: A novel mechanism for deletion of 17p in cytogenetically defined high-risk disease. Blood 123, 2504–2512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawyer J. R., Tian E., Walker B. A., Wardell C., Lukacs J. L., Sammartino G., Bailey C., Schinke C. D., Thanendrarajan S., Davies F. E., Morgan G. J., Barlogie B., Zangari M., van Rhee F., An acquired high-risk chromosome instability phenotype in multiple myeloma: Jumping 1q Syndrome. Blood Cancer J. 9, 62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajkumar S. V., Vincent Rajkumar S., Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. Am. J. Hematol. 95, 548–567 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Greipp P. R., Miguel J. S., Durie B. G. M., Crowley J. J., Barlogie B., Bladé J., Boccadoro M., Child J. A., Avet-Loiseau H., Kyle R. A., Lahuerta J. J., Ludwig H., Morgan G., Powles R., Shimizu K., Shustik C., Sonneveld P., Tosi P., Turesson I., Westin J., International staging system for multiple myeloma. J. Clin. Oncol. 23, 3412–3420 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J. P., Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., Tamayo P., The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsherniak A., Vazquez F., Montgomery P. G., Weir B. A., Kryukov G., Cowley G. S., Gill S., Harrington W. F., Pantel S., Krill-Burger J. M., Meyers R. M., Ali L., Goodale A., Lee Y., Jiang G., Hsiao J., Gerath W. F. J., Howell S., Merkel E., Ghandi M., Garraway L. A., Root D. E., Golub T. R., Boehm J. S., Hahn W. C., Defining a cancer dependency map. Cell 170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyers R. M., Bryan J. G., McFarland J. M., Weir B. A., Sizemore A. E., Xu H., Dharia N. V., Montgomery P. G., Cowley G. S., Pantel S., Goodale A., Lee Y., Ali L. D., Jiang G., Lubonja R., Harrington W. F., Strickland M., Wu T., Hawes D. C., Zhivich V. A., Wyatt M. R., Kalani Z., Chang J. J., Okamoto M., Stegmaier K., Golub T. R., Boehm J. S., Vazquez F., Root D. E., Hahn W. C., Tsherniak A., Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurt E. M., Wiestner A., Rosenwald A., Shaffer A. L., Campo E., Grogan T., Bergsagel P. L., Kuehl W. M., Staudt L. M., Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell 5, 191–199 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Annunziata C. M., Hernandez L., Davis R. E., Zingone A., Lamy L., Lam L. T., Hurt E. M., Shaffer A. L., Kuehl W. M., Staudt L. M., A mechanistic rationale for MEK inhibitor therapy in myeloma based on blockade of MAF oncogene expression. Blood 117, 2396–2404 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z., Mao H., Du X., Zhu J., Xu Y., Wang S., Xu X., Ji P., Yu Y., Cao B., Han K., Hou T., Xu Z., Kong Y., Jiang G., Tang X., Qiao C., Mao X., A novel small molecule agent displays potent anti-myeloma activity by inhibiting the JAK2-STAT3 signaling pathway. Oncotarget 7, 9296–9308 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu F.-T., Li N.-G., Zhang Y.-M., Xie W.-C., Yang S.-P., Lu T., Shi Z.-H., Recent advance in the development of novel, selective and potent FGFR inhibitors. Eur. J. Med. Chem. 186, 111884 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Chng W. J., Gualberto A., Fonseca R., IGF-1R is overexpressed in poor-prognostic subtypes of multiple myeloma. Leukemia 20, 174–176 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Bataille R., Robillard N., Avet-Loiseau H., Harousseau J.-L., Moreau P., CD221 (IGF-1R) is aberrantly expressed in multiple myeloma, in relation to disease severity. Haematologica 90, 706–707 (2005). [PubMed] [Google Scholar]
- 42.Mei A. H.-C., Tung K., Han J., Perumal D., Laganà A., Keats J., Auclair D., Chari A., Jagannath S., Parekh S., Cho H. J., MAGE-A inhibit apoptosis and promote proliferation in multiple myeloma through regulation of BIM and p21Cip1. Oncotarget 11, 727–739 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schooten E., Di Maggio A., van Bergen En Henegouwen P. M. P., Kijanka M. M., MAGE-A antigens as targets for cancer immunotherapy. Cancer Treat. Rev. 67, 54–62 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Zajac P., Schultz-Thater E., Tornillo L., Sadowski C., Trella E., Mengus C., Iezzi G., Spagnoli G. C., MAGE-A antigens and cancer immunotherapy. Front. Med. 4, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Packiriswamy N., Upreti D., Zhou Y., Khan R., Miller A., Diaz R. M., Rooney C. M., Dispenzieri A., Peng K.-W., Russell S. J., Oncolytic measles virus therapy enhances tumor antigen-specific T-cell responses in patients with multiple myeloma. Leukemia 34, 3310–3322 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steiner N., Jöhrer K., Plewan S., Brunner-Véber A., Göbel G., Nachbaur D., Wolf D., Gunsilius E., Untergasser G., The FMS like tyrosine kinase 3 (FLT3) is overexpressed in a subgroup of multiple myeloma patients with inferior prognosis. Cancers (Basel). 12, 2341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Griffith M., Spies N. C., Krysiak K., McMichael J. F., Coffman A. C., Danos A. M., Ainscough B. J., Ramirez C. A., Rieke D. T., Kujan L., Barnell E. K., Wagner A. H., Skidmore Z. L., Wollam A., Liu C. J., Jones M. R., Bilski R. L., Lesurf R., Feng Y.-Y., Shah N. M., Bonakdar M., Trani L., Matlock M., Ramu A., Campbell K. M., Spies G. C., Graubert A. P., Gangavarapu K., Eldred J. M., Larson D. E., Walker J. R., Good B. M., Wu C., Su A. I., Dienstmann R., Margolin A. A., Tamborero D., Lopez-Bigas N., Jones S. J. M., Bose R., Spencer D. H., Wartman L. D., Wilson R. K., Mardis E. R., Griffith O. L., CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 49, 170–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A. A., Kim S., Wilson C. J., Lehár J., Kryukov G. V., Sonkin D., Reddy A., Liu M., Murray L., Berger M. F., Monahan J. E., Morais P., Meltzer J., Korejwa A., Jané-Valbuena J., Mapa F. A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I. H., Cheng J., Yu G. K., Yu J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M. D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R. C., Liefeld T., MacConaill L., Winckler W., Reich M., Li N., Mesirov J. P., Gabriel S. B., Getz G., Ardlie K., Chan V., Myer V. E., Weber B. L., Porter J., Warmuth M., Finan P., Harris J. L., Meyerson M., Golub T. R., Morrissey M. P., Sellers W. R., Schlegel R., Garraway L. A., The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghandi M., Huang F. W., Jané-Valbuena J., Kryukov G. V., Lo C. C., McDonald E. R. III, Barretina J., Gelfand E. T., Bielski C. M., Li H., Hu K., Andreev-Drakhlin A. Y., Kim J., Hess J. M., Haas B. J., Aguet F., Weir B. A., Rothberg M. V., Paolella B. R., Lawrence M. S., Akbani R., Lu Y., Tiv H. L., Gokhale P. C., de Weck A., Mansour A. A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J. M., Porter D. A., Jones M. D., Golji J., Caponigro G., Taylor J. E., Dunning C. M., Creech A. L., Warren A. C., McFarland J. M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y. E., Cherniack A. D., Tsherniak A., Vazquez F., Jaffe J. D., Lane A. A., Weinstock D. M., Johannessen C. M., Morrissey M. P., Stegmeier F., Schlegel R., Hahn W. C., Getz G., Mills G. B., Boehm J. S., Golub T. R., Garraway L. A., Sellers W. R., Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang W., Soares J., Greninger P., Edelman E. J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J. A., Thompson I. R., Ramaswamy S., Futreal P. A., Haber D. A., Stratton M. R., Benes C., McDermott U., Garnett M. J., Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–D961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao E. Y., Shen Y., Pleasance E., Kasaian K., Leelakumari S., Jones M., Bose P., Ch’ng C., Reisle C., Eirew P., Corbett R., Mungall K. L., Thiessen N., Ma Y., Schein J. E., Mungall A. J., Zhao Y., Moore R. A., Den Brok W., Wilson S., Villa D., Shenkier T., Lohrisch C., Chia S., Yip S., Gelmon K., Lim H., Renouf D., Sun S., Schrader K. A., Young S., Bosdet I., Karsan A., Laskin J., Marra M. A., Jones S. J. M., Homologous recombination deficiency and platinum-based therapy outcomes in advanced breast cancer. Clin. Cancer Res. 23, 7521–7530 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Kumar S., Kaufman J. L., Gasparetto C., Mikhael J., Vij R., Pegourie B., Benboubker L., Facon T., Amiot M., Moreau P., Punnoose E. A., Alzate S., Dunbar M., Xu T., Agarwal S. K., Enschede S. H., Leverson J. D., Ross J. A., Maciag P. C., Verdugo M., Touzeau C., Efficacy of venetoclax as targeted therapy for relapsed/refractory t(11;14) multiple myeloma. Blood 130, 2401–2409 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Punnoose E. A., Leverson J. D., Peale F., Boghaert E. R., Belmont L. D., Tan N., Young A., Mitten M., Ingalla E., Darbonne W. C., Oleksijew A., Tapang P., Yue P., Oeh J., Lee L., Maiga S., Fairbrother W. J., Amiot M., Souers A. J., Sampath D., Expression profile of BCL-2, BCL-XL, and MCL-1 predicts pharmacological response to the BCL-2 selective antagonist venetoclax in multiple myeloma models. Mol. Cancer Ther. 15, 1132–1144 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Seiller C., Maiga S., Touzeau C., Bellanger C., Kervoëlen C., Descamps G., Maillet L., Moreau P., Pellat-Deceunynck C., Gomez-Bougie P., Amiot M., Dual targeting of BCL2 and MCL1 rescues myeloma cells resistant to BCL2 and MCL1 inhibitors associated with the formation of BAX/BAK hetero-complexes. Cell Death Dis. 11, 316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Algarín E. M., Díaz-Tejedor A., Mogollón P., Hernández-García S., Corchete L. A., San-Segundo L., Martín-Sánchez M., González-Méndez L., Schoumacher M., Banquet S., Kraus-Berthier L., Kloos I., Derreal A., Halilovic E., Maacke H., Gutiérrez N. C., Mateos M.-V., Paíno T., Garayoa M., Ocio E. M., Preclinical evaluation of the simultaneous inhibition of MCL-1 and BCL-2 with the combination of S63845 and venetoclax in multiple myeloma. Haematologica 105, e116–e120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.I. D. Ferguson, B. P. Escobar, S. T. Tuomivaara, Y.-H. T. Lin, M. A. Nix, K. K. Leung, M. Hale, P. Choudhry, A. Lopez-Girona, E. Ramos, S. W. Wong, J. L. Wolf, T. G. Martin III, N. Shah, S. Vandenberg, S. Prakash, L. Besse, C. Driessen, J. A. Wells, A. P. Wiita, Defining the cell surface proteomic landscape of multiple myeloma reveals immunotherapeutic strategies and biomarkers of drug resistance. bioRxiv 10.1101/2021.01.17.427038 [Preprint]. 19 January 2021; 10.1101/2021.01.17.427038. [DOI]
- 57.Marsters S. A., Yan M., Pitti R. M., Haas P. E., Dixit V. M., Ashkenazi A., Interaction of the TNF homologues BLyS and APRIL with the TNF receptor homologues BCMA and TACI. Curr. Biol. 10, 785–788 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Xu S., Lam K.-P., Transmembrane activator and CAML interactor (TACI): Another potential target for immunotherapy of multiple myeloma? Cancers (Basel). 12, 1045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee L., Draper B., Chaplin N., Philip B., Chin M., Galas-Filipowicz D., Onuoha S., Thomas S., Baldan V., Bughda R., Maciocia P., Kokalaki E., Neves M. P., Patel D., Rodriguez-Justo M., Francis J., Yong K., Pule M., An APRIL-based chimeric antigen receptor for dual targeting of BCMA and TACI in multiple myeloma. Blood 131, 746–758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Samur M. K., Fulciniti M., Aktas Samur A., Bazarbachi A. H., Tai Y.-T., Prabhala R., Alonso A., Sperling A. S., Campbell T., Petrocca F., Hege K., Kaiser S., Loiseau H. A., Anderson K. C., Munshi N. C., Biallelic loss of BCMA as a resistance mechanism to CAR T cell therapy in a patient with multiple myeloma. Nat. Commun. 12, 868 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muthuswami M., Ramesh V., Banerjee S., Viveka Thangaraj S., Periasamy J., Bhaskar Rao D., Barnabas G. D., Raghavan S., Ganesan K., Breast tumors with elevated expression of 1q candidate genes confer poor clinical outcome and sensitivity to Ras/PI3K inhibition. PLOS ONE. 8, e77553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen L., Chan T. H. M., Guan X.-Y., Chromosome 1q21 amplification and oncogenes in hepatocellular carcinoma. Acta Pharmacol. Sin. 31, 1165–1171 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Najfeld V., Tripodi J., Scalise A., Silverman L. R., Silver R. T., Fruchtman S., Hoffman R., Jumping translocations of the long arms of chromosome 1 in myeloid malignancies is associated with a high risk of transformation to acute myeloid leukaemia. Br. J. Haematol. 151, 288–291 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Marcellino B. K., Hoffman R., Tripodi J., Lu M., Kosiorek H., Mascarenhas J., Rampal R. K., Dueck A., Najfeld V., Advanced forms of MPNs are accompanied by chromosomal abnormalities that lead to dysregulation of TP53. Blood Adv. 2, 3581–3589 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rajkumar S. V., Landgren O., Mateos M.-V., Smoldering multiple myeloma. Blood 125, 3069–3075 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mohan M., Weinhold N., Schinke C., Thanedrarajan S., Rasche L., Sawyer J. R., Tian E., Rhee F., Zangari M., Daratumumab in high-risk relapsed/refractory multiple myeloma patients: Adverse effect of chromosome 1q21 gain/amplification and GEP70 status on outcome. Br. J. Haematol. 189, 67–71 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Sawyer J. R., Tricot G., Mattox S., Jagannath S., Barlogie B., Jumping translocations of chromosome 1q in multiple myeloma: Evidence for a mechanism involving decondensation of pericentromeric heterochromatin. Blood 91, 1732–1741 (1998). [PubMed] [Google Scholar]
- 68.Ziccheddu B., Biancon G., Bagnoli F., De Philippis C., Maura F., Rustad E. H., Dugo M., Devecchi A., De Cecco L., Sensi M., Terragna C., Martello M., Bagratuni T., Kastritis E., Dimopoulos M. A., Cavo M., Carniti C., Montefusco V., Corradini P., Bolli N., Integrative analysis of the genomic and transcriptomic landscape of double-refractory multiple myeloma. Blood Adv. 4, 830–844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chng W. J.; International Myeloma Working Group, Dispenzieri A., Chim C.-S., Fonseca R., Goldschmidt H., Lentzsch S., Munshi N., Palumbo A., Miguel J. S., Sonneveld P., Cavo M., Usmani S., Durie B. G. M., Avet-Loiseau H., IMWG consensus on risk stratification in multiple myeloma. Leukemia 28, 269–277 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Abdallah N., Greipp P., Kapoor P., Gertz M. A., Dispenzieri A., Baughn L. B., Lacy M. Q., Hayman S. R., Buadi F. K., Dingli D., Go R. S., Hwa Y. L., Fonder A., Hobbs M., Lin Y., Leung N., Kourelis T., Warsame R., Siddiqui M., Lust J., Kyle R. A., Bergsagel L., Ketterling R., Rajkumar S. V., Kumar S. K., Clinical characteristics and treatment outcomes of newly diagnosed multiple myeloma with chromosome 1q abnormalities. Blood Adv. 4, 3509–3519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walker B. A., Boyle E. M., Wardell C. P., Murison A., Begum D. B., Dahir N. M., Proszek P. Z., Johnson D. C., Kaiser M. F., Melchor L., Aronson L. I., Scales M., Pawlyn C., Mirabella F., Jones J. R., Brioli A., Mikulasova A., Cairns D. A., Gregory W. M., Quartilho A., Drayson M. T., Russell N., Cook G., Jackson G. H., Leleu X., Davies F. E., Morgan G. J., Mutational spectrum, copy number changes, and outcome: Results of a sequencing study of patients with newly diagnosed myeloma. J. Clin. Oncol. 33, 3911–3920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen R., Mo Q., Schultz N., Seshan V. E., Olshen A. B., Huse J., Ladanyi M., Sander C., Integrative subtype discovery in glioblastoma using iCluster. PLOS ONE 7, e35236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mo Q., Shen R., Guo C., Vannucci M., Chan K. S., Hilsenbeck S. G., A fully Bayesian latent variable model for integrative clustering analysis of multi-type omics data. Biostatistics 19, 71–86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.T. Ma, A. Zhang, in 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (2017), pp. 398–403. [Google Scholar]
- 75.Su G., Morris J. H., Demchak B., Bader G. D., Biological network exploration with Cytoscape 3, Curr. Protoc. Bioinformatics 8.13.1–8.13.24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paczkowska M., Barenboim J., Sintupisut N., Fox N. S., Zhu H., Abd-Rabbo D., Mee M. W., Boutros P. C.; PCAWG Drivers and Functional Interpretation Working Group, Reimand J.; PCAWG Consortium , Integrative pathway enrichment analysis of multivariate omics data. Nat. Commun. 11, 735 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morris J. H., Kuchinsky A., Ferrin T. E., Pico A. R., enhancedGraphics: A Cytoscape app for enhanced node graphics. F1000Res. 3, 147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chu F., Cao J., Neelalpu S. S., Versatile CAR T-cells for cancer immunotherapy. Contemp. Oncol. (Pozn.). 2018, 73–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Figs. S1 to S13
Tables S1 to S39