In Nrf2-activated tumors, G6pd inhibition depletes TCA intermediates and suppresses tumor growth.
Abstract
The KEAP1/NRF2 pathway promotes metabolic rewiring to support redox homeostasis. Activation of NRF2 occurs in many cancers, often due to KEAP1 mutations, and is associated with more aggressive disease and treatment resistance. To identify metabolic dependencies in cancers with NRF2 activation, we performed a metabolism-focused CRISPR screen. Glucose-6-phosphate dehydrogenase (G6PD), which was recently shown to be dispensable in Ras-driven tumors, was a top dependency. G6PD catalyzes the committed step of the oxidative pentose phosphate pathway that produces NADPH and nucleotide precursors, but neither antioxidants nor nucleosides rescued. Instead, G6PD loss triggered tricarboxylic acid (TCA) intermediate depletion because of up-regulation of the alternative NADPH-producing enzymes malic enzyme and isocitrate dehydrogenase. In vivo, G6PD impairment markedly suppressed KEAP1 mutant tumor growth, and this suppression was further augmented by TCA depletion by glutaminase inhibition. Thus, G6PD inhibition–induced TCA depletion is a therapeutic vulnerability of NRF2-activated cancer.
INTRODUCTION
NFE2L2 (hereafter NRF2) acts as a primary line of defense against reactive oxygen species (ROS) to maintain oxidative homeostasis by regulating the expression of a plethora of genes involved in metabolic rewiring and ROS clearance (1). NRF2 transcriptionally induces several major mediators of the redox defense system including enzymes involved in the metabolism of glutathione (GSH) (2–4), reduced form of nicotinamide adenine dinucleotide phosphate (NADPH) (5), redoxin protein family members (6, 7), drug biotransformation and efflux via multidrug resistance–associated proteins (8), and enzymes involved in iron homeostasis (9). Increased production of the reducing cofactor NADPH is essential to support the redox cycling mechanisms of enzymes that detoxify ROS and xenobiotics (10, 11). To maintain the levels of NADPH and other antioxidant substrates, NRF2 diverts glutamine, glucose, and glycolytic intermediates into anabolic pathways such as the pentose phosphate pathway (PPP), glutaminolysis, and de novo serine synthesis (10, 12, 13).
The PPP is thought to play a pivotal role in supporting cancer cell survival and growth by generating both NADPH and ribose phosphate for nucleotide synthesis (14). Multiple enzymes involved in the oxidative PPP (oxPPP) such as glucose-6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (PGD), and in the non-oxPPP such as transaldolase and transketolase (TKT) are transcriptionally activated by NRF2 (10), leading to increased PPP flux (10, 11). G6PD, which catalyzes the first and rate-limiting step of the PPP, accounts for most of the cellular NADPH produced in most cultured cancer cells (15) and can promote tumor initiation (16). However, despite the potential importance of the PPP in cancer metabolism, this pathway has not been successfully therapeutically targeted in cancer (17, 18).
Maintaining oxidative homeostasis through chronic activation of the NRF2 pathway results in a unique set of metabolic requirements and associated targetable metabolic dependencies (10, 13, 19–21). Kelch-like ECH-associated protein 1 (hereafter KEAP1) is the negative regulator of NRF2 and is one of the most frequently mutated genes in lung adenocarcinoma (LUAD) (22, 23). KEAP1 mutation and subsequent NRF2 accumulation lead to metabolic reprogramming to promote the endogenous antioxidant response, which confers proliferative and survival advantages to tumor cells (10, 19, 20). We previously demonstrated that Keap1 loss strongly promotes Kras-driven LUAD progression (20) and metastasis (24). A plethora of emerging clinical data suggest that patients with KEAP1 mutations have poor overall survival and do not respond to the current standard of care, radiation (25), immunotherapy (22, 26), and/or chemotherapy (22, 27, 28). Therefore, there is a great need to identify therapeutic vulnerabilities in this genetic subtype of lung cancer with KEAP1/NRF2 mutations.
Here, we performed a CRISPR/Cas9–based genetic screen of metabolic genes to identify synthetic lethal interactions with Keap1 mutations. We found that G6PD loss of function (LOF) selectively impairs KEAP1 mutant cells. Using both genetic and pharmacological approaches in multiple in vivo models, we demonstrate the selective dependency of KEAP1 mutant tumors on G6PD. Dependence of KEAP1 mutant lung cancer cells on G6PD was not rescued by reducing agents or antioxidants. However, it was reversed by supplementation with tricarboxylic acid (TCA) cycle precursors. Metabolomic analysis of G6PD-depleted KEAP1 mutant cells showed significant depletion of TCA intermediates, especially the substrates of malic enzyme (ME) and isocitrate dehydrogenase (IDH). Furthermore, combined inhibition of G6PD and glutaminase, a major enzyme feeding into the TCA whose inhibition preferentially impairs KEAP1 mutant cells (13, 20, 21, 25, 29), leads to further tumor growth suppression. Thus, oxPPP inhibition, potentially combined with glutaminase inhibition, holds therapeutic potential for KEAP1 mutant cancers.
RESULTS
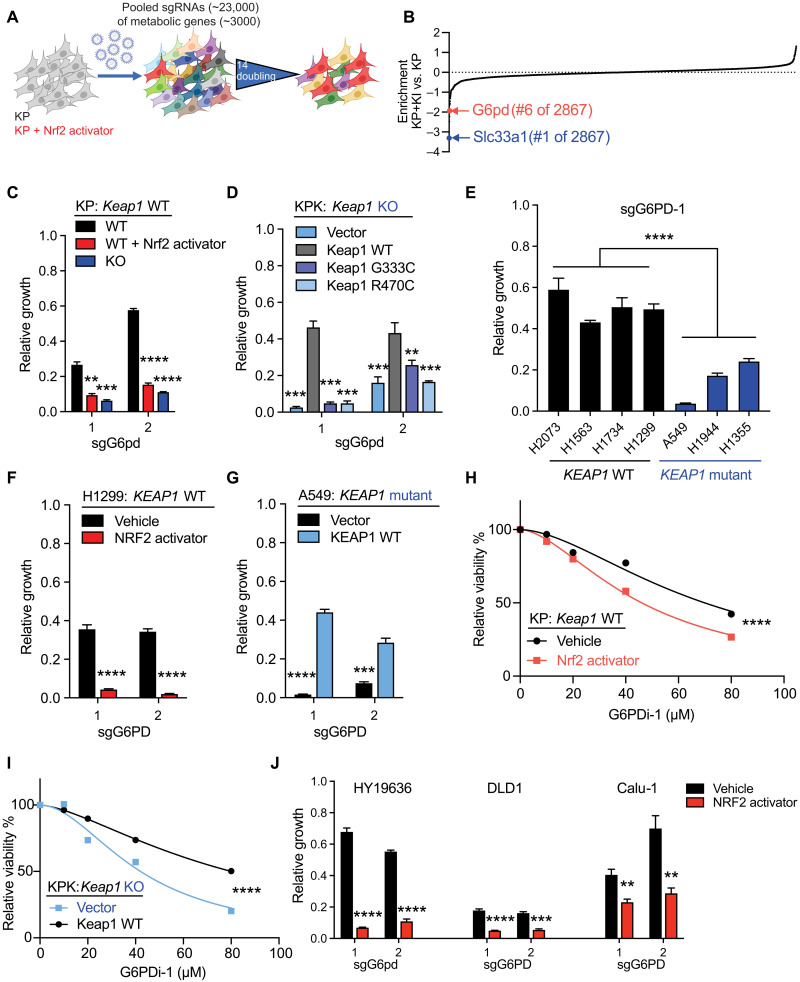
CRISPR/Cas9 metabolic screen identifies dependence on G6pd in Keap1 mutant LUAD
To identify novel metabolic vulnerabilities in Keap1 mutant cells, we performed a focused CRISPR/Cas9–based screen with single-guide RNAs (sgRNAs) targeting 3000 metabolic genes (30) in KrasG12D/+; p53−/− (KP) murine LUAD cell lines in the presence or absence of a pharmacological NRF2 activator, KI696 (KP + KI) (Fig. 1A) (13, 31). To identify the sgRNAs differentially depleted in KP + KI as compared to KP cells, we calculated a relative depletion score. The top-scoring synthetic lethal hit, Slc33a1, was recently independently shown to be synthetically lethal with KEAP1 mutations (32), supporting the rigor of our screen. Among the top most depleted sgRNAs in cells with Nrf2 activation were those targeting G6pd (Fig. 1B; fig. S1, A to C; and table S1). We validated our screen results by demonstrating that G6pd LOF is synthetic lethal in a panel of mouse LUAD cell lines with either genetic loss of Keap1 or pharmacologic activation of Nrf2 with KI696 (Fig. 1C and fig. S1D). In Keap1 null cells, we were able to rescue sensitivity to G6pd loss by reexpression of wild-type (WT) Keap1 but not missense LOF mutants of Keap1 (Fig. 1D). Furthermore, inducible short hairpin RNA (shRNA) knockdown of G6pd led to a similar growth suppression (fig. S1, E and F). Using a panel of KEAP1 WT and mutant human LUAD cell lines, we observed increased sensitivity to G6PD depletion in KEAP1 mutant compared to WT cell lines (Fig. 1E and fig. S1, G and H). Moreover, we could modulate the sensitivity of human cells by NRF2 activation in KEAP1 WT cells (Fig. 1F) or KEAP1 complementation in KEAP1 mutant cells (Fig. 1G). To further validate these genetic studies, we performed pharmacological inhibition of G6PD using a small-molecule inhibitor, G6PDi-1 (17), and observed that either NRF2 pharmacological activation or KEAP1 mutation increased vulnerability to pharmacologic inhibition of G6PD (Fig. 1, H and I). Furthermore, to determine whether genetic loss of the other oxPPP enzymes is synthetic lethal with Nrf2 activation, we performed LOF of 6-phosphogluconolactonase, Pgd, and Tkt. We observed that Nrf2 activation did not lead to selective sensitivity to loss of these downstream oxPPP enzymes (fig. S1, I and J), which is in agreement with the CRISPR screen results (table S1). Cells with Keap1 mutation or pharmacological Nrf2 activation were less sensitive to both genetic loss and pharmacological inhibition of Pgd with 6-aminonicotinamide (6-AN; fig. S1, I to L), suggesting divergent roles of G6pd and Pgd in Keap1 mutant cells, while Tkt deficiency suppressed both Keap1 WT and mutant cell growth (fig. S1, I and J). Furthermore, to determine whether G6pd synthetic lethality with Nrf2 activation occurs in other tumor lineages other than lung, we used PDAC (pancreatic ductal adenocarcinoma), colon, and lung squamous cells and observed that pharmacological activation of Nrf2 sensitized these cells to G6pd loss (Fig. 1J and fig. S1M). Together, our data shows that KEAP1/NRF2 mutation predicts sensitivity to genetic loss or pharmacologic inhibition of G6PD.
Fig. 1. G6PD is synthetic lethal with NRF2 activation and KEAP1 mutations.
(A) Schematic of pooled sgRNA library screen. KP (Keap1 WT) and KP + KI (Keap1 WT + Nrf2 activator) cells infected with sgRNA library virus were passaged for 14 population doublings before collection. (B) Enrichment of genes according to difference between KP + KI versus KP. Negative score signifies increased sensitivity in KP + KI cells. A full table is shown in table S1. (C) Proliferation of KP (Keap1 WT), KP treated with Nrf2 activator, or KPK [Keap1 knockout (KO)] cells with G6pd KO (n = 3). (D) Proliferation of KPK (Keap1 KO) overexpressing vector, Keap1 WT, or Keap1 LOF cDNA (G333C and R470C) cells with G6pd KO (n = 3). (E) Proliferation of KEAP1 WT or KEAP1 mutant human cell lines with sgG6PD-1 KO (n = 3). (F) Proliferation of NCI-H1299 (KEAP1 WT) and H1299 treated with NRF2 activator cells with G6PD KO (n = 3). (G) Proliferation of A549 (KEAP1 mutant) overexpressing KEAP1 WT or vector with G6PD KO (n = 3). (H) Relative viability of KP (Keap1 WT) and KP + KI (Keap1 WT + Nrf2 activator) treated with G6PDi-1 (G6PD inhibitor) for 3 days (n = 4). (I) Relative viability of KPK (Keap1 KO) overexpressing Keap1 WT or Vector treated with G6PDi-1 (G6PD inhibitor) for 3 days (n = 4). (J) Proliferation of HY19636 (Keap1 WT PDAC), DLD1 (KEAP1 WT colon cancer), and Calu-1 (KEAP1 WT lung squamous) treated with NRF2 activator cells or vehicle with G6PD KO (n = 3). **P < 0.01, ***P < 0.001, and ****P < 0.0001.
G6PD dependency leads to defects in central carbon metabolism
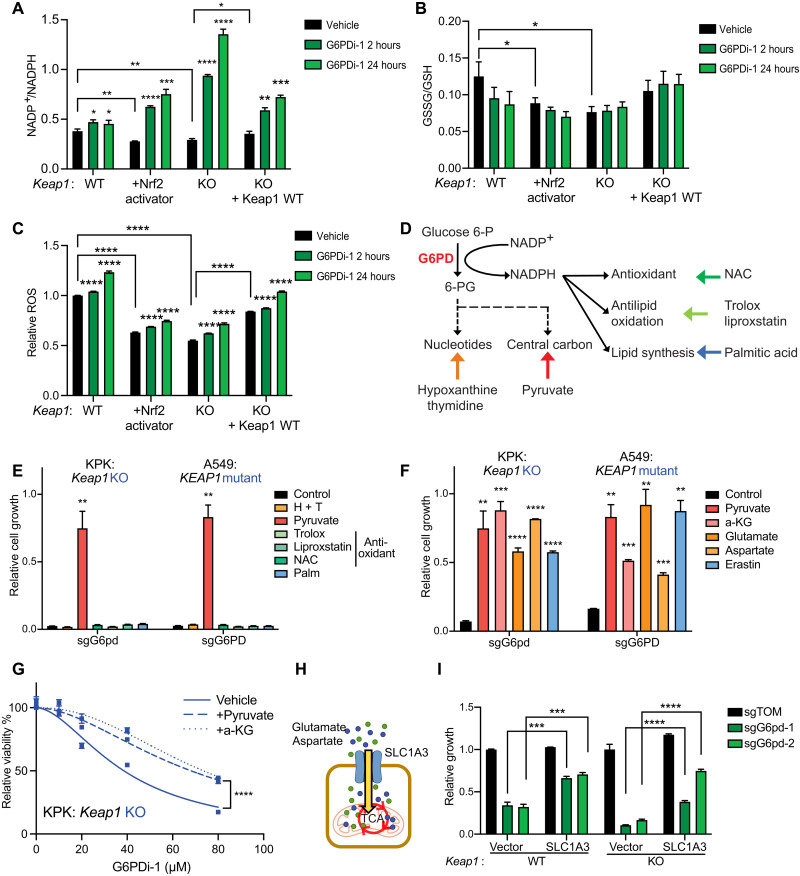
The PPP is the primary source of NADPH (15), an important redox cofactor that is required to reduce oxidized GSH and thioredoxin. To determine the impact of G6pd depletion or pharmacological inhibition on NADPH, we measured the NADP+/NADPH ratio. As expected, based on Nrf2’s role as a stimulator of redox defense, Keap1 mutant cells have a more reduced NADP+/NADPH ratio compared to Keap1 WT cells. Upon G6pd depletion or pharmacological inhibition, we observed a shift toward a more oxidized NADP+/NADPH ratio in both Keap1 mutant and WT cells (Fig. 2A). The change in NADP+/NADPH ratio was greater in cells with pharmacological Nrf2 activation or Keap1 loss, consistent with G6pd being particularly important to NADPH homeostasis in this context. The change in the NADP+/NADPH ratio may result in an elevated GSSG (oxidized gluthathione)/GSH ratio and ROS levels. Keap1 mutant cells have a lower GSSG/GSH ratio and ROS compared to WT cells (13, 20). However, G6pd loss or pharmacological inhibition had a minimal effect on both the GSSG/GSH ratio and ROS levels in both Keap1 mutant and WT cells (Fig. 2, B and C, and fig. S2, A to J), suggesting that other mechanisms might be driving the growth dependency on G6PD.
Fig. 2. G6PD loss or inhibition is rescued by central carbon metabolites but not antioxidants.
(A) NADP+/NADPH ratio of KP, KP + Nrf2 activator, KPK + vector, and KPK + Keap1 WT cells treated with 50 μM G6PDi-1 for 2 or 24 hours (n = 3), data measured by liquid chromatography–mass spectrometry (LC-MS). (B) GSSG/GSH ratio of KP, KP + Nrf2 activator, KPK + vector, and KPK + Keap1 WT cells treated with 50 μM G6PDi-1 for 2 or 24 hours (n = 3), data measured by LC-MS. (C) Relative ROS of KP, KP + Nrf2 activator, KPK + vector, and KPK + Keap1 WT cells treated with 50 μM G6PDi-1 2 or 24 hours (n = 3). (D) Schematic depicting G6PD biosynthesis pathway and corresponding rescue molecules. (E) Proliferation of KPK (Keap1 KO) and A549 (KEAP1 mutant) cells with G6PD KO in media supplemented with 30 mM hypoxanthine + 16 mM thymidine (H + T), 2 mM pyruvate, 50 μM trolox, 100 nM liproxstatin, 0.5 mM N-acetyl-l-cysteine (NAC), and 100 μM palmitic acid (Palm). Data were normalized to KPK sgTOM or A549 sgTOM (n = 3). (F) Proliferation of KPK (Keap1 KO) and A549 (KEAP1 mutant) cells with G6PD KO in media supplemented with 2 mM pyruvate, 2 mM dimethyl-2-oxoglutarate [a-ketoglutarate (a-KG) precursor], 6 mM glutamate, 20 mM aspartate, and 500 nM erastin. Data were normalized to KPK sgTOM or A549 sgTOM (n = 3). (G) Relative viability of KPK cells treated with G6PDi-1 and supplemented with 2 mM pyruvate and 2 mM a-KG for 3 days (n = 4). (H) Schematic depicting SLC1A3 up-taking extracellular glutamate and aspartate. (I) Proliferation of KP and KPK cells expressing SLC1A3 or an empty vector control with G6pd KO. Data were normalized to KP + vector with sgTOM group and KPK + vector with sgTOM group (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To identify which G6PD/PPP-derived metabolites are critical to KEAP1 mutant cell growth, we assessed the ability of downstream metabolites (nucleotides, antioxidants, and central carbon metabolites) to rescue cell growth in the context of G6PD loss (Fig. 2, D and E, and fig. S2, K to N). Unexpectedly, nucleotide precursors or antioxidants as single agents or in combination did not rescue the growth of KEAP1 mutant cells with G6PD LOF. The lack of rescue by any of the three different antioxidants is in agreement with the minimal changes in ROS levels and GSH buffering upon G6PD depletion in KEAP1 mutant cells (Fig. 2, B and C, and fig. S2, A to J). In contrast, supplementation of pyruvate, a metabolite that supports central carbon metabolism, fully rescued the growth of KEAP1 mutant cells with G6PD LOF. Pyruvate can serve as a TCA input via pyruvate dehydrogenase or pyruvate carboxylase. In addition, it can alter cellular redox by increasing the NAD+/NADH ratio. To differentiate between these mechanisms, we tested whether redox-neutral carbon substrates can also rescue the growth inhibition of KEAP1 mutant cells after G6PD loss. We observed that multiple other carbon sources for TCA cycle, including glutamate, a-ketoglutarate (a-KG), and aspartate, also rescued G6PD LOF (Fig. 2F and fig. S2, O to R) or pharmacological inhibition (Fig. 2G and fig. S2S).
NRF2 activation can predispose cells to TCA metabolite depletion by inducing the expression of the system XC− transporter SLC7A11, which exchanges extracellular cystine for intracellular glutamate. The XC− inhibitor erastin blocks the efflux of glutamate and increases intracellular glutamate levels (13). Erastin rescued KEAP1 mutant cells in the context of G6PD LOF (Fig. 2F and fig. S2, O to R). Moreover, expression of SLC1A3, a glutamate and aspartate importer (33) (Fig. 2H), rescued the growth of Keap1 mutant cells in response to G6pd loss (Fig. 2I). Thus, multiple modes of restoring TCA metabolites and associated amino acids, including pyruvate, a-KG, aspartate, and glutamate supplementation, all rescued the synthetic lethality between KEAP1 and G6PD. None of these metabolic supplementations were able to rescue the growth of cells lacking Pgd (fig. S2T), consistent with a distinctive role for G6pd in supporting growth and central carbon metabolism in Keap1 mutant cells.
G6PD deficiency depletes TCA metabolites and represses cell respiration in Keap1 mutants
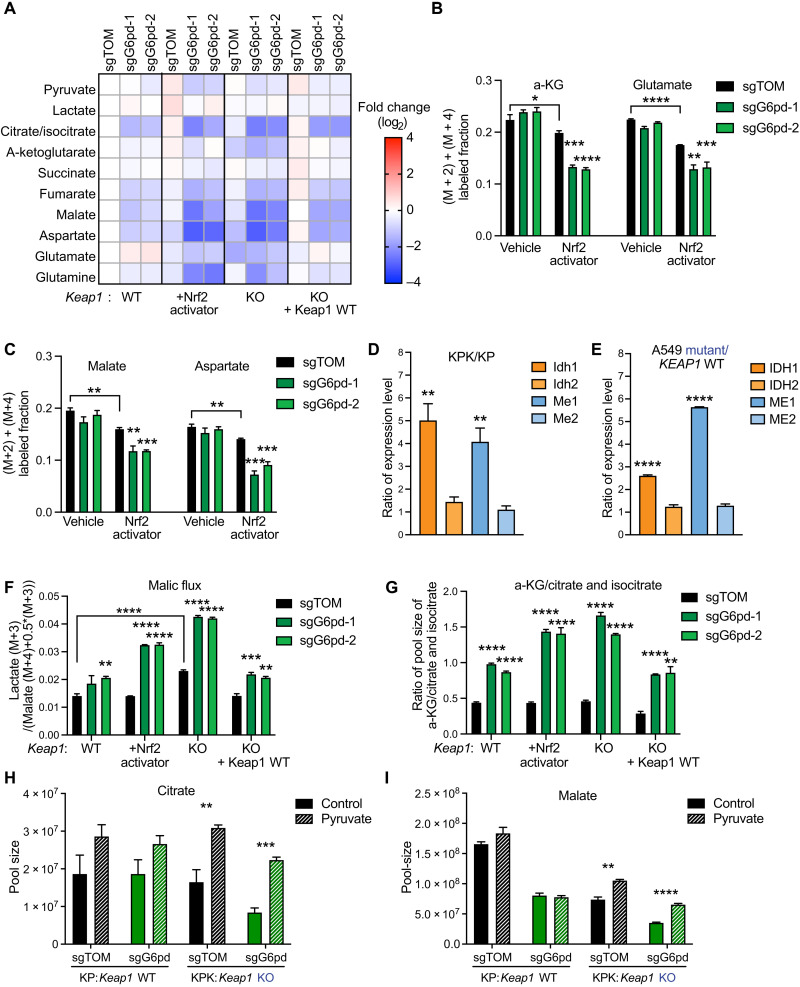
To determine how G6pd inhibition or loss affects central carbon metabolism, we analyzed both TCA metabolite pools and labeling with U-13C-glucose tracer. G6PD inhibition had minimal effect on the pool sizes of glycolytic intermediates and 13C incorporation (fig. S3, A and B). Pharmacological inhibition and genetic loss of G6pd decreased TCA cycle metabolite pools, particularly in the context of Keap1 mutations and Nrf2 activation (Fig. 3A and fig. S3, C and D). This was associated with decreased fractional glucose contribution to TCA intermediates and associated amino acids in the Nrf2-activated cells, which was further accentuated by pharmacological G6pd inhibition (Fig. 3, B and C, and fig. S3E). Consistent with the decreased TCA intermediate levels and labeling, we observed decreased oxygen consumption in response to loss or inhibition of G6PD in both mouse and human LUAD cells (fig. S3, F to H). Hypothetically, glucose flux through G6PD and PPP could be a substantial source of pyruvate/lactate and downstream TCA intermediates. To explore this possibility, we performed [1,2-13C] glucose tracing (fig. S3I) (34) and found that PPP carbon flux into pyruvate/lactate (M + 1) is minimal compared to glycolytic flux [lactate (M + 2)] (fig. S3J), and accordingly, the PPP is not a major route from glucose to the TCA cycle. Instead, as we have previously shown, the decreased glucose contribution to TCA in cells with Nrf2 activation might be explained by exchange of intracellular glutamate for extracellular glutamine (13). Overall, SLC7A11 overexpression and associated glutamate export from cells explains the impact of Nrf2 activation but not of G6pd LOF or inhibition on TCA intermediates.
Fig. 3. G6PD disruption leads to an impairment of central carbon metabolism.
(A) Heatmap of TCA cycle metabolite pool sizes in KP, KP + Nrf2 activator, KPK + Keap1 WT cDNA, and KPK + vector cells with G6pd KO. Data were normalized to KP sgTOM group and calculated as log2(fold change) (n = 3). (B and C) Mass isotopomer analysis of TCA cycle metabolites (malate, aspartate, a-KG, and glutamate) in KP and KP + KI cells cultured for 1.5 hours with U-13C-glucose (n = 3). (D) Quantitative PCR (qPCR) validation of Idh1, Idh2, Me1, and Me2 expression in KP/KPK cells. Data of mRNA expression were normalized to actin and then calculated as the ratio of Keap1 KO/Keap1 WT (n = 4). (E) qPCR validation of IDH1, IDH2, ME1, and ME2 expression in A549 + KEAP1 WT /A549 + vector cells. Data of mRNA expression were normalized to ACTIN and then calculated as the ratio of KEAP1 mutant/KEAP1 WT (n = 4). (F) Malic flux/glycolysis of KP, KP + Nrf2 activator, KPK + vector cells, and KPK + Keap1 WT with G6pd KO. Flux is calculated as (lactate M + 3) / (malate (M + 4) + 0.5*malate (M + 3)) of 3-hour U-13C-glutamine tracing (n = 3). (G) Idh activity of KP, KP + Nrf2 activator, KPK + vector, and KPK + Keap1 WT cells with G6pd KO. Proxy Idh activity was calculated by a-KG/citrate and isocitrate pool-size ratio and then normalized to KP sgTOM (n = 3). (H and I) Pool sizes (TIC) of citrate (H) and malate (I) in KP and KPK sgTOM or sgG6pd cells cultured for 2 days with 2 mM pyruvate (n = 3), data measured by LC-MS. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
On the basis of prior studies, G6PD LOF can lead to compensatory increase of IDH and ME activity to maintain NADPH pools, which could further drain carbons from the TCA cycle (15). In agreement with prior studies, we observed that NRF2 activation induces cytoplasmic IDH1 and ME1 (Fig. 3, D and E, and fig. S3, K and L), while mitochondrial IDH2 and ME2 remain unchanged (10). Therefore, we hypothesized that, together with elevated levels of the substrate NADP+ in the context of G6PD LOF, the higher levels of cytoplasmic IDH1 and ME1 enzymes in KEAP1 mutant cells will consume TCA intermediates to make NADPH. To estimate the ME flux, we used U-13C-glutamine tracer, which generates M + 3 pyruvate from TCA-derived M + 3/M + 4 malate via ME. The fraction of M + 3 pyruvate and lactate normalized to M + 3/M + 4 malate indicates the relative flux of ME compared with glycolysis, without regard to the isozyme, compartment, or cofactor involved (35). We observed an increase in malic flux upon G6pd loss (Fig. 3F).
For IDH, we checked the product/substrate pool size ratio as a proxy readout and observed that G6pd loss or inhibition significantly increased the α-KG/citrate and isocitrate ratio, consistent with higher Idh activity (Fig. 3G and fig. S3, M and N). Moreover, pyruvate rescued the depleted citrate and malate pool size in G6pd depleted Keap1 knockout (KO) cells (Fig. 3, H and I, and fig. S3O), without affecting the NADP+/NADPH status. Together, these data indicate that in the absence of G6PD, KEAP1 mutant cells fail to maintain TCA intermediate homeostasis due to drainage of TCA intermediates by the combined activities of SLC7A11, IDH, and ME.
G6PD is required for KEAP1 mutant tumor growth
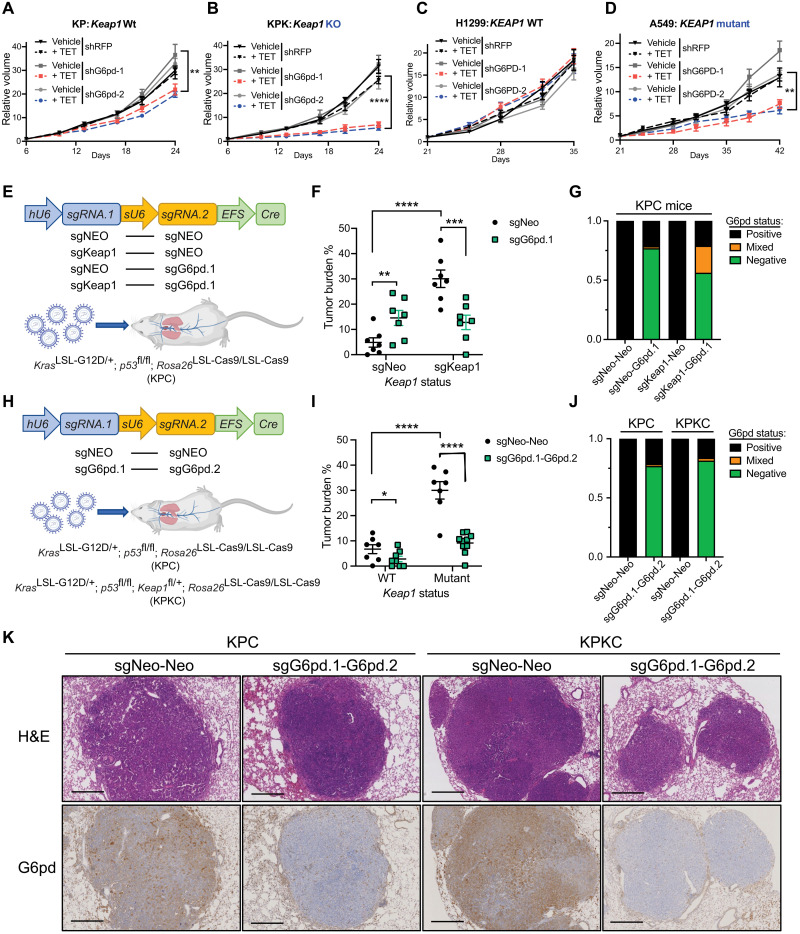
To determine whether G6pd is required for Keap1 mutant tumor growth in vivo, we subcutaneously inoculated Keap1 WT and mutant cells expressing doxycycline-inducible shRNAs against G6pd or control red fluorescent protein (RFP) into C57BL/6J mice. Inducible knockdown of G6pd much more strongly attenuated tumor growth in Keap1 mutant than WT tumors (Fig. 4, A and B, and fig. S4A). Furthermore, inducible knockdown of G6PD repressed A549 (KEAP1 mutant) tumor growth but not H1299 (KEAP1 WT) tumor growth (Fig. 4, C and D, and fig. S4, B to D).
Fig. 4. G6PD is required for KEAP1 mutant LUAD.
(A and B) Relative tumor growth of subcutaneous KP (A) and KPK (B) tumors with inducible shG6pd in animals receiving TET or control diet. TET stands for doxycycline diet activating shRNA expression. Diet was changed at day 6. Data were normalized to first measurement at day 6 (n = 7). (C and D) Relative tumor growth of subcutaneous H1299 (KEAP1 WT) (C) and A549 (KEAP1 mutant) (D) tumors with inducible shG6PD in animals receiving TET or control diet. Diet is changed at day 21. Data were normalized to first measurement at day 21 (n = 8 for H1299 and n = 6 for A549). (E) Schematic figure of KPC mice intratracheally infected with pUSEC lentiviruses containing double sgRNAs targeting G6pd, Keap1, or Neo (Control). (F) Quantification of tumor burden (tumor area/total lung area) in KPC mice after infection with pUSEC lentiviruses (n = 8 for sgNeo-Neo and sgKeap1-Neo and n = 9 for sgNeo-G6pd.1 and sgKeap1-G6pd.1). (G) G6pd status analysis based on IHC staining of lung bearing tumors (n = 4). (H) Schematic figure of KPC or KPKC mice intratracheally infected with pUSEC lentiviruses containing double sgRNAs targeting Neo-Neo and G6pd.1-G6pd.2. (I) Quantification of tumor burden of mice infected with pUSEC lentiviruses (n = 7 for Keap1 WT sgNeo-Neo and sgG6pd.1-G6pd.2, n = 8 for Keap1 mutant sgNeo-Neo, and n = 10 for Keap1 mutant sgG6pd.1-G6pd.2). (J) G6pd status analysis based on IHC staining (n = 3). (K) Representative IHC staining of serial sections from lung tumors of KPC and KPKC mice 16 weeks after infection with pUSEC lentiviruses against Neo-Neo and G6pd.1-G6pd.2. First panels, hematoxylin and eosin (H&E) staining analyses; second panels, G6pd IHC analyses. Note that Keap1 mutant group has higher staining of G6pd, proving G6pd is Keap1/Nrf2 substrate. Scale bars, 500 μm. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Detailed statistics analysis of (A) to (D) is presented in table S3.
To test the importance of G6pd in autochthonous tumorigenesis, we used multiple LUAD GEMMs (genetically engineered mouse models), which better recapitulate the heterogeneity, histological progression, and tumor microenvironment of human LUAD. First, we used animals with conditional Kras, p53, and Cas9 alleles (KrasLSL-G12D/+; p53fl/fl; Rosa26LSL-Cas9/LSL-Cas9; hereafter KPC). KPC mice were intratracheally infected with bifunctional pUSEC (hU6::sgRNA1-sU6::sgRNA2-EFS::Cre) (24) lentiviruses that enable the expression of Cre recombinase and two independent sgRNAs (36) (Fig. 4E). We assessed whether G6pd loss attenuated growth of Keap1 mutant as opposed to WT tumors. In agreement with previous work (18), loss of G6pd in Keap1 WT tumors (sgNeo-sgG6pd.1) did not suppress tumor growth (Fig. 4F and fig. S4, E and F). However, substantial suppression was observed for Keap1 mutant tumors (sgKeap1-sgG6pd) (Fig. 4F and fig. S4, E and F). Immunohistochemical analysis revealed that CRISPR/Cas9–dependent editing of Keap1 was efficient, as 90% of tumors had increased Nqo1, a bona fide Nrf2 target (fig. S4, G and H). However, CRISPR/Cas9–based G6pd editing was largely selected against as we observed that 40% of sgKeap1-sgG6pd tumors retained the G6pd (Fig. 4G and fig. S4G).
To ensure more robust loss of both Keap1 and G6pd, we used KPC GEMMs with conditional loss of Keap1 (KrasLSL-G12D/+; p53fl/fl; Keap1fl/+; Rosa26LSL-Cas9/LSL-Cas9; hereafter KPKC). Animals were intratracheally infected with pUSEC (hU6::sgRNA1-sU6::sgRNA2-EFS::Cre) lentivirus, expressing two independent sgRNAs against G6pd (sgG6pd.1-sgG6pd.2) or controls (sgNeo-sgNeo) (Fig. 4H). Similar to our KPC model, we observed that G6pd loss led to preferential attenuation of tumor growth in Keap1 mutant tumors (Fig. 4I). We observed enhanced efficiency in knocking out G6pd with a second guide targeting G6pd (sgG6pd.1-sgG6pd.2) resulting in G6pd loss in 80% of tumors (Fig. 4, J and K) compared with 60% when using a single guide (Fig. 4G). The robust deletion of G6pd completely abrogated the protumorigenic effect of Keap1 loss as compared to the G6pd WT control group (sgNeo-sgNeo) (Fig. 4I and fig. S4, I and J). Together, these data demonstrate that G6pd is required for Keap1 mutant tumor growth and highlight the potential therapeutic benefits of targeting G6pd in genetically stratified Keap1 mutant tumors.
G6PD inhibition synergizes with glutaminase inhibition in KEAP1 mutant tumors
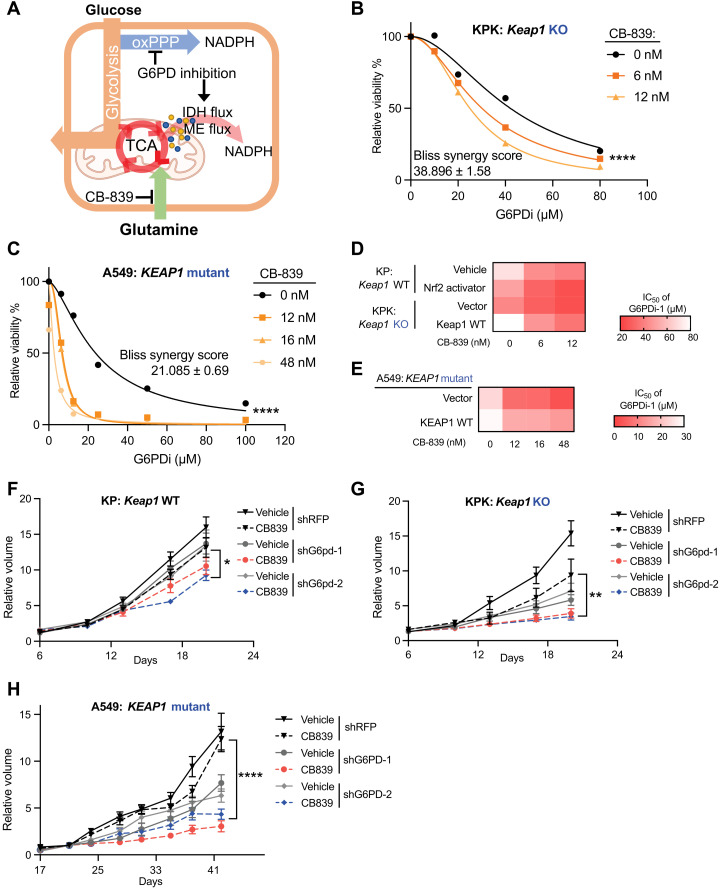
Our studies suggest that G6PD loss or inhibition leads to growth suppression in KEAP1 mutant cells through depletion of TCA intermediates (Figs. 2 and 3). Therefore, we hypothesized that G6PD inhibition might synergize with therapeutic strategies that suppress TCA anaplerosis (Fig. 5A). Consistent with this, Keap1 mutant tumors are sensitive to the glutaminase inhibitor CB-839 (13, 20, 21, 25, 29), which blocks glutamine-driven anaplerosis (13, 37). Keap1 mutant mouse LUAD cells exhibited increased sensitivity to a G6pd inhibitor (G6PDi-1) in the presence of low concentrations of CB-839 that do not affect cell viability (Fig. 5, B and D; fig. S5A; and Table 1). The human LUAD cell line A549 (KEAP1 mutant) also showed the same G6PDi-1-CB-839 synergy (Fig. 5, C and E; fig. S5B; and Table 2), while overexpression of WT KEAP1 cDNA largely rescued this sensitivity. Bliss analysis indicated that the synergy of CB-839 and G6PDi-1 is dependent on NRF2 activation and KEAP1 mutation status (Tables 3 and 4).
Fig. 5. Inhibition of G6PD and glutaminase synergize to attenuate KEAP1 mutant LUAD.
(A) Schematic of G6PD inhibition corporation with glutaminase inhibitor to repress cell glucose metabolism and respiration. (B) Relative viability of KPK (Keap1 KO) cells cultured with 0, 6, and 12 nM CB-839 and treated with G6PDi-1 for 3 days. Data were normalized to CB-839 0 nM and G6PDi-1 vehicle group (n = 4). (C) Relative viability of A549 (KEAP1 mutant) cells cultured with 0, 12, 16, and 48 nM CB-839 and treated with G6PDi-1 for 3 days. Data were normalized to CB-839 0 nM and G6PDi-1 vehicle group (n = 3). (D) Heatmap of KP, KP + Nrf2 activator, KPK + vector, and KPK + Keap1 WT cells median inhibitory concentration (IC50) of G6PDi-1 in different CB-839 treatment. (E) Heatmap of A549 + vector and A549 + KEAP1 WT cells IC50 of G6PDi-1 in different CB-839 treatment. (F) Relative tumor growth of subcutaneous KP (Keap1 WT) tumors with hairpin against G6pd in animals treated with either CB-839 or vehicle. Data were normalized to first measurement at day 6 (n = 6). (G) Relative tumor growth of subcutaneous KPK (Keap1 KO) tumors with hairpin against G6pd in animals treated with either CB-839 or vehicle. Data were normalized to first measurement at day 6 (n = 6 for shRFP and shG6pd-1 and n = 8 for shG6pd-2). (H) Relative tumor growth of subcutaneous A549 (KEAP1 mutant) tumors with hairpin against G6PD in animals treated with either CB-839 or vehicle. Data were normalized to second measurement at day 21 (n = 6). *P < 0.05, **P < 0.01, and ****P < 0.0001. Detailed statistics analysis of (F) to (H) is presented in table S3.
Table 1. IC50 of G6PDi-1 with supplement of CB-839 in mouse cell lines (n = 4).
| G6PDi-1 IC50 (μM) | CB-839 (nM) | ||
| 0 | 6 | 12 | |
| WT | 70 | 44.41 | 40.405 |
| WT + Nrf2 activator | 50.65 | 31.315 | 24.9 |
| KO + vector | 42.08 | 30.09 | 24.98 |
| KO + Keap1 WT | 80.05 | 46.9 | 36.595 |
Table 2. IC50 of G6PDi-1 with supplement of CB-839 in human cell lines (n = 3).
| G6PDi-1 IC50 (μM) | CB-839 (nM) | |||
| 0 | 12 | 16 | 48 | |
| A549 + vector | 23.745 | 6.935 | 6.475 | 2.69 |
| A549 + KEAP1 WT | 29.055 | 16.6 | 16.43 | 14.015 |
Table 3. Bliss synergy score of G6PDi-1 with CB-839 in mouse cell lines (n = 4).
| Bliss synergy score | |
| WT | 6.581 ± 1.49 |
| WT + Nrf2 activator | 13.665 ± 1.45 |
| KO + vector | 38.896 ± 1.58 |
| KO + Keap1 WT | 19.658 ± 1.42 |
Table 4. Bliss synergy score of G6PDi-1 with CB-839 in human cell lines (n = 3).
| Bliss synergy score | |
| A549 + vector | 21.085 ± 0.69 |
| A549 + KEAP1 WT | 3.119 ± 1.04 |
Since currently available G6pd inhibitors are not suitable for in vivo studies, we used an inducible G6pd knockdown tumor model to assess the synergy of G6pd LOF with CB-839 synergy in vivo. We observed that knockdown of G6pd suppressed tumor growth and that addition of CB-839 even further attenuated tumor growth of Keap1 mutant tumors (Fig. 5, F and G, and fig. S5, C and D). CB-839 combined with G6PD knockdown markedly inhibited tumor growth in human KEAP1 mutant LUAD tumors (Fig. 5H and fig. S5E). These data support the potential for combining glutaminase and G6PD inhibition to target KEAP1-mutated LUAD, a major genetic subtype of lung cancer with unmet clinical need.
DISCUSSION
Emerging clinical data highlight that alterations in the KEAP1/NRF2 pathway result in resistance to multiple therapies (22, 25–28). Therefore, identification of new vulnerabilities to target these tumors has been of great interest. Previous preclinical studies from our group and others have identified synthetic lethal interactions in KEAP1/NRF2 mutant LUAD (12, 13, 20, 21, 32). However, it remains to be seen whether any of these metabolic targets alone or in combination with existing therapies will pave the way to viable therapeutic approaches in the clinic.
Although previous studies have suggested that G6PD or the PPP enhances tumor growth by protecting cells from ROS (38) and promoting nucleotide synthesis (16), there is a lack of in vivo studies demonstrating the importance of the PPP in tumorigenesis. Although G6pd was not required for Kras-driven mouse LUAD with WT Keap1 (18), in this study, we demonstrate that G6PD genetic loss or pharmacological inhibition selectively suppresses growth of a genetic subset of LUAD with KEAP1 mutations and NRF2 activation. In our study, we used orthogonal genetic approaches to selectively deplete G6PD in tumors and determine the tumor cell–autonomous dependence on G6PD. In addition, we used the first on-target cellular G6PD inhibitor, G6PDi-1 (17). Our study provides a preclinical platform to test the efficacy of new generation G6PD inhibitors in KEAP1 mutant LUAD. Although the therapeutic index of G6PD inhibitors remains unknown, a large percentage of people are deficient in G6PD and are asymptomatic (39, 40), suggesting that pharmacological inhibition of G6PD may be tolerated.
Tumors with NRF2 activation have high antioxidant capacity and can buffer redox stress by inducing multiple enzymes that generate NADPH in addition to G6PD, including cytosolic IDH1 and ME1 (10, 11). At the same time, NADPH demand may be enhanced by elevated GSH and thioredoxin. How these factors balance was unclear. Here, we show that genetic loss or pharmacological inhibition of G6PD does not affect ROS levels and redox homeostasis, indicating that NADPH production from alternative pathways is sufficient, at least in culture, for redox balance in KEAP1 mutant cells. However, we observed that in the absence of G6PD, there was an impairment in TCA metabolite levels. This can be reversed with pyruvate, TCA intermediates, or TCA-related amino acids. On the basis of prior studies, G6PD depletion induced more cytosolic IDH1 and ME1 flux, which depletes TCA metabolites (15). IDH1 and ME1 are NRF2 targets. We show that G6PD loss increased flux through IDH1 and ME1 in KEAP1 mutant cells. These findings are in line with our previous observations that NRF2 activation rewires cellular metabolism toward increased antioxidant metabolism at the expense of respiration, TCA cycle anaplerosis, and amino acid synthesis (13, 21). This rewiring leaves KEAP1 mutant cells vulnerable to manipulations that deplete TCA intermediates.
Furthermore, we demonstrate that this is not specific to lung cancer and that pancreatic cancer cells with Nrf2 activation are also more dependent on G6pd. These data suggest that tumors of other lineages that acquire Nrf2 activation by genetic (41–46), epigenetic (47–49), or posttranscriptional (50) alterations may also require G6pd for NADPH and TCA cycle homeostasis and therefore be susceptible to G6pd inhibition.
When viewed in typical pathway diagrams, the PPP and TCA are the least closely connected among all central carbon pathways, with the PPP branching from the top of glycolysis and the TCA fed from lower glycolysis, as well as by amino acid and fatty acid catabolism. These diagrams, however, obscure redox connections: cytosolic NADPH can be made by the oxPPP or by ME and IDH. ME, particularly, drains TCA intermediates. Thus, the PPP helps maintain TCA homeostasis not directly by providing carbon but by providing NADPH that satiates other TCA-draining, NADPH-producing enzymes. Here, we show that this previously unrecognized function of G6PD is critical to TCA homeostasis in the context of KEAP1 mutation, which leads to NRF2-dependent increases of multiple NADPH-producing enzymes that contribute to the cataplerosis of TCA cycle intermediates.
Furthermore, we demonstrate that not all oxPPP enzymes are synthetic lethal with KEAP1 mutations. We demonstrate that Keap1 mutant cells are less sensitive to both genetic and pharmacological inhibition of Pgd. Our findings also demonstrate that unlike G6PD, Pgd loss cannot be rescued by TCA cycle metabolites, suggesting that these two oxPPP enzymes sometimes have divergent roles in cellular metabolism. This discrepancy is also noted in T cells, where inhibition of G6PD led to decreased cytokine production (17), while loss of PGD enhanced antitumor function (51). In this context, oxPPP should not be simply viewed as a linear pathway but two steps with distinct biological outcomes. Prior studies have suggested that inhibition of PGD with 6-AN may be a therapeutic target in LUAD (52). However, the tumor cell–autonomous and non-cell–autonomous effects and the metabolic mechanisms underlying 6-AN sensitivity remain unknown. In addition, many studies have demonstrated that 6-AN is not a PGD-specific inhibitor and has off-target activity against glycolysis and NAD+ biosynthesis (53, 54).
Last, the glutaminase inhibitor CB-839 is a well-established means of suppressing KEAP1 mutant cells and tumors in mouse models (13, 20, 21, 25, 29) and is currently being tested in multiple phase 2 clinical trials in patients with KEAP1 or NRF2 mutant LUAD (KEAPSAKE: NCT04265534 and BeGIN: NCT038724270). In this study, we demonstrate that CB-839 synergizes with G6PD inhibition to suppress KEAP1 mutant tumor growth. Implementation of this strategy depends on the development of new G6PD inhibitors for in vivo testing in preclinical models and eventually advancement into clinic use. Notably, mutations that lead to G6PD deficiency are well tolerated, and phase 1 studies with CB-839 showed minimal toxicity (55). Therefore, combined glutaminase and G6PD inhibition could be a promising new therapeutic approach for patients with KEAP1 mutant tumors.
MATERIALS AND METHODS
Mice
All animal studies described were approved by the NYU Langone Medical Center Institutional Animal Care and Use Committee (IACUC). Animals were housed according to IACUC guidelines in ventilated caging in a specific pathogen–free animal facility. For transplantation experiments, 5 × 105 mouse cells were implanted subcutaneously into C57BL/6J female mice (the Jackson Laboratory, 000664), or 1 × 106 human cells were subcutaneously implanted into NSG (NOD scid gamma mouse) mice (the Jackson Laboratory, 005557) approximately 6 to 8 weeks in age. Tumor volume was measured by caliper, and volume was calculated (0.5 × length × width2). After tumor volume reaching 50 to 100 mm3, animals were randomized and assigned to a treatment group. Animals either received CB-839 (200 mg/kg) or vehicle (Calithera) twice daily administered through oral gavage as previously described (56) or doxycycline diet (Bio-Serv, S3888). No animals were excluded from analysis. Tumor growth was tracked for a minimum of six tumors per experimental group.
KrasLSL-G12D/+; p53fl/fl; Rosa26LSL-Cas9/LSL-Cas9 mice is bred as described before (24); KrasLSL-G12D/+; p53fl/fl; Keap1fl/+; Rosa26LSL-Cas9/LSL-Cas9 mice is bred from KrasLSL-G12D/+; p53fl/fl; Rosa26LSL-Cas9/LSL-Cas9 mice and Keap1fl/fl mice (Taconic, 8799). Mice with the appropriate genotype aged 6 to 8 weeks were randomly selected to begin tumor initiation studies with pUSEC virus. Mice were intratracheally infected with lentiviruses as described (57). Total tumor burden and grading analyses were conducted on >6 mice per genotype. No mice were excluded from analyses. The total lung area occupied by each tumor was measured on hematoxylin and eosin (H&E)–stained slides using QuPath software (58). All tumor burden and IHC (immunohistochemistry) analyses were done in a blinded fashion in which the researcher was unaware of which genotype the samples had.
Virus packaging and concentration
Lentivirus was produced by cotransfection of human embryonic kidney–293 (HEK293) cells with viral plasmid with packaging vectors [psPAX2 and VSV-G (vesicular stomatitis virus G)] using PEI Pro (PolyPlus). Retrovirus was produced by transfection of Plat-E cells with viral plasmid using PEI Pro (PolyPlus). Forty-eight hours after transfection, virus is harvested and filtered with 0.45 μM polyvinylidene difluoride (PVDF) filter, and receipt cells are infected with virus in the presence of polybrene (8 μg/ml; Millipore). For large scale of virus production and concentration, virus is harvested and filtered with 0.45 μM PVDF filter, then concentrated by ultracentrifuging at 25,000 rpm for 2 hours, and dissolved in phosphate-buffered saline overnight. Virus tittering is performed as previously reported (59).
Cell lines and culture
Murine KrasG12D/+; p53−/− WT and Keap1 mutant isogenic clonal cell lines and HY19636/HY15549 cell lines were previously established (20, 60). Murine cell lines were originally derived from female mice. Human cell lines were acquired from American Type Culture Collection. All cell lines tested negative for mycoplasma (PlasmoTest, InvivoGen). All cell lines were cultured in a humidified incubator at 37°C and 5% CO2. Cells were maintained in either Dulbecco’s Modified Eagle’s Medium (DMEM) or RPMI 1640 (Cellgro, Corning) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich) and gentamicin (Invitrogen).
Metabolic CRISPR/Cas9 genetic screen
Metabolic library is a gift from K. Birsoy (30). Virus was produced by cotransfection of HEK293 cells with lentiviral library pool with packaging vectors (psPAX2 and VSV-G) using PEI Pro (PolyPlus). The titer of lentiviral supernatants was determined by infecting target cells with several amounts of virus in the presence of polybrene (8 μg/ml; Millipore), counting the number of puromycin-resistant infected cells after 3 days of selection. KP and KP + KI cells were infected at a multiplicity of infection of ~0.3 and selected with puromycin (8 μg/ml) 72 hours after infection. An initial pool of cells was harvested for genomic DNA extraction. Remaining cells were cultured for 14 doublings, after which cells were harvested for genomic DNA extraction. sgRNA inserts were polymerase chain reaction (PCR)–amplified and then purified and sequenced on a MiSeq instrument (Illumina) according to prior studies (30). Sequencing reads were mapped, and the abundance of each sgRNA was tallied. Gene score is defined as the median log2 fold change in abundance between the initial and final populations for all sgRNAs targeting the gene. The differential gene score is the difference between the KP and KP + KI cell gene scores.
Cell proliferation and viability assays
For cell proliferation assays conducted under different drug or media conditions as indicated, cells growing in DMEM were trypsinized, counted (Countess II, Thermo Fisher Scientific), and plated into 12-well plate dishes (BD/Falcon) in 2 ml of RPMI media. For rescue experiments, cells were treated with indicated drugs as legends or 30 mM hypoxanthine (Acros Organics, 122010250), 16 mM thymidine (Sigma-Aldrich, T1895), 50 μM trolox (Acros Organics, AC218940010), 100 nM liproxstatin (Selleck Chemicals, S7699), 0.5 mM N-acetyl-l-cysteine (Acros Organics, 160280250), 100 μM palmitic acid (Cayman Chemical, 21911), 2 mM pyruvate (Thermo Fisher Scientific, 11360070), 2 mM dimethyl-2-oxoglutarate (a-KG precursor, Sigma-Aldrich, 349631), 6 mM glutamate (Sigma-Aldrich, G8415), 20 mM aspartate (Sigma-Aldrich, A7219), and 500 nM erastin (EMD Millipore, 329600), or 1 μM KI-696 [Nrf2 activator; provided by C. Thomas, National Cancer Institute (NCI)]. In experiments where the Nrf2 activator (KI696) was used, cells were pretreated with 1 μM for at least 7 days before the start of the experiment. Proliferation experiments were carried out for 5 days after drug treatment and collected by staining. Mouse cells are plated 2000 per well, and human cells are plated 10,000 per well. Cells were stained with a 0.5% crystal violet (Fisher Scientific) solution in 20% methanol. Plates were then washed and dried, and crystal violet was eluted in 400 μl of 10% acetic acid and measured at 595-nm wavelength by spectrometer (Molecular Devices).
For cell viability assays, cells were plated in a white, opaque 96-well plate (Corning) with clear bottom at a density of 2000 cells per well for mouse cells and 5000 cells per well for human cells in RPMI-1640 media and drug added at the same day. After 3 days, cell viability in the presence of all compounds was assessed by CellTiter-Glo (Promega) and measured by spectrometer (Molecular Devices). All data are represented as relative to KEAP1 WT group vehicle-treated condition. G6PDi-1 was synthesized as previously described (17), and 6-AN was obtained from Cayman Chemical (10009315).
Synergy score
The expected drug combination responses of G6PDi-1 with CB-839 were calculated on the basis of ZIP reference model using SynergyFinder 2.0 (61). Scores higher than 10.0 are regarded as significant synergy.
Quantitative PCR analysis
mRNA was harvested from cells using an RNeasy mini kit (QIAGEN) according to the manufacturer’s protocol. Extracted mRNA was used to synthesize cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosciences, Thermo Fisher Scientific) according to the manufacturer’s protocol. Gene expression was analyzed by quantitative PCR (qPCR) on a QuantStudio 3 (Applied Biosciences, Thermo Fisher Scientific). A list of specific primers used can be found in table S2.
Immunoblotting
Cells were collected and lysed in 200 μl of ice-cold radioimmunoprecipitation assay buffer (Thermo Fisher Scientific) supplemented with 5 μM EDTA and 1× protease and phosphatase inhibitor (Thermo Fisher Scientific) and mixed on a rotator at 4°C for 10 min. Supernatant was collected after centrifugation at 15,000 rpm for 15 min. Protein concentrations were determined using the Bio-Rad DC Protein Assay. Twenty milligrams of total protein was separated on a 4 to 12% bis-tris gradient gel (Invitrogen) and then transferred onto PVDF membrane (Bio-Rad). Goat anti-Rabbit immunoglobulin G (IgG) (H + L) horseradish peroxidase (HRP) and Goat anti-Mouse IgG (H + L) HRP secondary antibody were used as secondary antibodies (Invitrogen, Thermo Fisher Scientific). Antibodies to ACTIN (ab8226) and G6PD (ab993) were obtained from Abcam Inc. Antibody to HSP90 (610418) was obtained from BD Biosciences. Antibody to NQO1 (HPA007308) was obtained from Sigma-Aldrich.
Immunohistochemistry
Mice were euthanized by carbon dioxide asphyxiation. Lung with tumors were dissected, infused, and fixed with 10% formalin overnight; transferred to 70% ethanol; and then embedded in paraffin. Sections (3 mM thick) were cut and stained with H&E. Immunohistochemistry on paraffin-embedded sections was performed using antibodies against Nqo1 (1:100; Sigma-Aldrich, HPA007308) and G6pd (1:5000; Abcam, ab993). Chromogenic IHC was performed on a Leica Bond RX, and stained slides were imaged on a Leica SCN400 F whole-slide scanner. For Nqo1 and G6pd staining, antigen retrieval was performed using antigen retrieval buffer pH 6 (Leica) for 20 min. For detection, Leica Bond Polymer Refine Detection secondary antibody (Leica, no. DS9800) was used according to the manufacturer’s protocol for Nqo1 and G6pd.
ROS and lipid ROS analysis
ROS in cultured cells were measured by incubating cells with 5 mM CM-H2DCFDA (Life Technologies, C6827) for 30 min at 37°C. 2,7-Dichlorofluorescein diacetate fluorescence was acquired on the BD LSRFortessa (BD Biosciences) flow cytometer at fluorescein isothiocyanate (FITC) channel and analyzed using FlowJo software (Tree Star, Ashland, Oregon).
Lipid ROS in cultured cells were measured by incubating cells with 10 μM Image-iT Lipid Peroxidation Sensor (Life Technologies, C10445) for 30 min at 37°C. Fluorescence was acquired on the BD LSRFortessa (BD Biosciences) flow cytometer at FITC and PE-CF594 channel and analyzed using FlowJo software (Tree Star), and the ratio of FITC/PE-CF594 was regarded as lipid ROS level.
Mitochondrial respiration
Oxygen consumption rate experiments were performed using the XFe-96 apparatus from Seahorse Bioscience (Agilent, Santa Clara, CA). Cells were seeded at least five replicates for each condition in RPMI to reach 90% confluence at the day of measurement. Medium was completely replaced with reconstituted Seahorse RPMI with 10 mM glucose and 2 mM glutamine (no sodium bicarbonate) adjusted to pH 7.4 and incubated for 60 min at 37°C in a CO2-free incubator before measurements.
sgRNA and shRNA cloning and cell line generation
KO of target genes was achieved by cloning sgRNAs into lentiCRISPRv2 puro and lentiCRISPRv2 hygro backbones (Addgene plasmid nos. 98290 and 98291, respectively) (62). Briefly, backbones were digested with Bsm BI (New England Biosciences, Ipswich, MA) and purified with a gel extraction kit (QIAGEN). sgRNAs primers were designed according to sgRNA Designer (Broad Institute) and were annealed and phosphorylated using T4 PNK (New England Biosciences). Phosphorylated and annealed oligos were then ligated into the purified digested backbones using Quick Ligase (New England Biosciences). Vectors were transduced into cells and selected with puromycin (8 μg/ml) or hygromycin (600 μg/ml) for 5 days. KO of target gene was subsequently verified by Western blot. Oligos were obtained from Integrated DNA Technologies (Coralville, IA). Double guide sgRNAs were designed according to previous report (36); briefly, sU6 promoter is inserted to 2sgRNA ultramer through PCR and Gibson assembly with pDonor_sU6 (Addgene plasmid no. 69351), then digested with Bbs I, and ligated to Bsm BI–digested lentiCRISPRv2 puro or pUSEC backbone.
Doxycycline-induced knockdown of G6pd was achieved by cloning hairpin-targeting G6pd into the pLKO.1-TETON-Puro vector (Addgene plasmid no. 21915) (63). Briefly, hairpins are designed according to Genetic perturbation platform (Broad Institute) and annealed hairpin is ligated into the Eco RI + Age I–digested backbone with Quick Ligase (New England Biosciences) at a 3:1 insert:vector molar ratio. Vectors were transduced into cells through virus and selected with puromycin (8 μg/ml). Knockdown of G6pd was verified by Western blot and qPCR analysis following 72 hours of treatment with doxycycline (1 μg/ml). Cell lines transduced with the first two efficient shRNAs, along with an anti-RFP shRNA as control, were used in the study. Oligos were obtained from Integrated DNA Technologies (Coralville, IA).
Keap1-complemented cells were generated by cloning mouse Keap1 cDNA into Gibson compatible lentiviral backbone with a hygromycin resistance cassette according to previous paper (64). Keap1 G333C and R470C point mutation were generated by QuikChange II Site-Directed Mutagenesis (Agilent). Cells overexpressing SLC1A3 were generated by cloning human SLC1A3 into the pMXS-blast retroviral backbone. Cells infected were selected with either puromycin (8 μg/ml) or blasticidin (600 μg/ml).
Gas chromatography–mass spectrometry analysis of polar metabolites and stable isotope tracing
For all experiments involving stable isotope tracers, the isotope tracer nutrient was substituted for unlabeled nutrient at the same concentration normally found in the base RPMI media. In addition, dialyzed FBS was used as a supplement in place of FBS.
For analysis of cells, 1 × 105 cells were seeded in 1 ml of RPMI 1640 in 12-well plates. For tracing experiments, media contained 11 mM U-13C–d-glucose or 1,2-13C–d-glucose (Cambridge Isotope Laboratory) with normal glutamine or 2 mM U-13C–d-glutamine with normal glucose. Cells were cultured for different hours and then washed twice in ice-cold 0.9% saline and collected by scraping in 250 μl of 80% (v/v) of ice-cold methanol containing norvaline (1.4 mg/ml; Sigma-Aldrich, N7627). Samples were vortexed for 10 min at 4°C and then centrifuged at 15,000 rpm for 5 min. Supernatant was transferred to fresh tubes and then dried in a speed vac (Thermo Fisher Scientific). Dried metabolite extracts were then derivatized with 20 μl O-methoxyamine-hydrochloride reagent (Sigma-Aldrich, 33045) in pyridine (Sigma-Aldrich) at a concentration of 20 mg/ml for 60 min at 37°C and 30 μl of N-tert-butyldimethylsilyl-N-methyltrifluoracetamide with 1% tert-butyldimethyl chlorosilane (Sigma-Aldrich, 375934) for 30 min at 37°C. After derivatization, samples were analyzed by gas chromatography–mass spectrometry (GC-MS) using an HP-5MS column (Agilent Technologies) in an Agilent Intuvo 9000 gas chromatograph coupled to an Agilent 5977B mass spectrometer. Helium was used as the carrier gas at a flow rate of 1.2 ml/min. One microliter of sample was injected in split mode (split 1:1) at 270°C. After injection, the GC oven was held at 100°C for 1 min and then increased to 300°C at 3.5°C/min. The oven was then ramped to 320°C at 20°C/min and held for 5 min at 320°C. The MS system operated under electron impact ionization at 70 eV; the MS source and quadrupole were held at 230° and 150°C, respectively; the detector was used in scanning mode; and the scanned ion range was 10 to 650 mass/charge ratio (m/z). Mass isotopomer distributions were determined by integrating the appropriate ion fragments for each metabolite (65) using MATLAB (MathWorks) and an algorithm adapted from Fernandez et al. (66) that corrects for natural abundance. For all data, total or relative metabolite pool sizes are normalized to cell counts for each condition.
Isotope tracing and metabolite extraction for liquid chromatography–MS
For extraction of water-soluble metabolites for liquid chromatography–MS (LC-MS) analysis (67), cells were plated and grown to 60 to 90% confluency in six-well plates. At the start of an experiment, the appropriate medium was added to cells, which included isotope tracers and/or chemical inhibitors as described. Cells were incubated at 37°C at 5% CO2 for 3 hours (unless otherwise noted). For all experiments involving small-molecule agents, dimethyl sulfoxide concentrations were <0.2%. After 3 hours, medium was removed by aspiration and metabolome extraction was performed (without any wash steps) by the addition of −20°C extraction solvent (40:40:20 acetonitrile:methanol:water + 0.5% formic acid; 50 to 100 μl of extraction solvent per 1 μl of packed cell volume). After a 1- to 2-min incubation on dry ice, the extract was neutralized by the addition of 15% NH4HCO3 (w/v; 70 μl per 800 μl of extraction solvent). The samples were incubated at −20°C for ~30 min, at which point the wells were scraped and the extract was transferred to 1.5-ml Eppendorf tubes and centrifuged (20 min, 16,000g, 4°C). The resulting supernatant was frozen on dry ice and kept at −80°C until LC-MS analysis.
LC-MS analysis
LC-MS measurement was performed with a quadrupole Orbitrap mass spectrometer (Q Exactive or Q Exactive Plus, Thermo Fisher Scientific) operating in negative ion mode and coupled to hydrophilic interaction chromatography via electrospray ionization (68). Scans were performed from m/z 70 to 1000 at 1 Hz and 140,000 resolution. LC separation was on a XBridge BEH Amide column (2.1 mm by 150 mm by 2.5 mm particle size and 130 Å pore size; Water, Milford, MA) using a gradient of solvent A [20 mM ammonium acetate and 20 mM ammonium hydroxide in 95:5 water:acetonitrile (pH 9.45)] and solvent B (acetonitrile). Flow rate was 150 ml/min. The LC gradient was 0 min, 90% B; 2 min, 90% B; 3 min, 75%; 7 min, 75% B; 8 min, 70%, 9 min, 70% B; 10 min, 50% B; 12 min, 50% B; 13 min, 25% B; 14 min, 25% B; 16 min, 0% B, 20.5 min, 0% B; 21 min, 90% B; and 25 min, 90% B. Autosampler temperature was 5°C, and injection volume was 10 μl.
For improved detection of NAD(P)+ and NAD(P)H (69, 70), the injection volume was increased to 15 μl, and selected ion monitoring scans with m/z window ranging from 640 to 765 were added. The LC gradient was modified as 0 min, 85% B; 2 min, 85% B; 3 min, 60% B; 9 min, 60% B; 9.5 min, 35% B; 13 min, 5% B; 15.5 min, 5% B; 16 min, 85% B; and 20 min, 85% B.
Data were analyzed using the El-MAVEN (Elucidata) software (71), with compounds identified on the basis of exact mass and retention time match to commercial standards. For tracer experiments, isotope labeling was corrected for natural 13C abundance (72).
Quantification and statistical analysis
Values are presented as means ± SEM. n presented in figure legends stands for number of technical repeats. For statistical analysis, we used GraphPad Prism software (GraphPad): two-way analysis of variance (ANOVA) for tumor growth, F test for median inhibitory concentration curve, and one-sided Student’s t test for cell proliferation, metabolite analysis, and gene expression analysis. For synergy score, score higher than 10 is regarded as significant synergy. All experiments were repeated at least twice. All cell proliferation experiments and metabolite analysis contained three biological replicates. Cell viability assays contained at least three biological replicates, and animal experiments and materials generated and analyzed from animal experiments contained a minimum of 6 biological replicates with a maximum of 10.
Acknowledgments
We would like to thank Calithera Biosciences for providing CB-839 for in vivo use and A. Kimmelman and D. Biancur for providing technical assistance with the Seahorse machine and the KP PDAC lines. We would like to thank C. Thomas (NCI/NIH) for providing KI696. We would like to thank C. Jiang (Shenzhen Bay Laboratory) and colleagues for providing G6PDi-1. We thank the NYU Langone Experimental Pathology Laboratory, Flow Cytometry Cores, and the Animal Resources Facility staff for support.
Funding: W.L.W. is supported by a National Institutes of Health training grant (T32GM007308 and 1F30CA247020). R.G.P. is supported by the William Rom fellowship, the Stony Wold-Herbert Fund, and a National Institutes of Health training grant (T32CA009161). K.W. is supported by a National Institutes of Health training grant (T32GM136573). T.P. research was supported by National Institutes of Health grants (R37CA222504 and R01CA227649) and an American Cancer Society Research Scholar Grant (RSG-17-200-01–TBE). J.D.R. research is supported by National Institutes of Health grant (R01CA163591) and Ludwig Cancer Research. Funding for G6PDi-1 synthesis was provided by the Princeton University–PKU Shenzhen collaboration.
Author contributions: Conceptualization and Methodology: H.D., Z.C., J.D.R., and T.P. Investigation: H.D., Z.C., K.W., S.M.H., W.L.W., S.E.L., and R.G.P. Writing (original draft): H.D., S.E.L., J.D.R., and T.P. Visualization: H.D. and Z.C. Supervision: S.E.L., W.L.W., J.D.R., and T.P. Funding acquisition: T.P. Writing (review and editing): All authors.
Competing interests: T.P. has received Honoraria/Consulting fees from Calithera Biosciences and Vividion Therapeutics and research support from Bristol Myers Squibb, Dracen Pharmaceuticals, and Agios Pharmaceuticals. J.D.R. is an advisor and stockholder in Kadmon Pharmaceuticals, Colorado Research Partners, L.E.A.F. Pharmaceuticals, Bantam Pharmaceuticals, Barer Institute, and Rafael Pharmaceuticals; a paid consultant of Pfizer; a founder, director, and stockholder of Farber Partners, Serien Therapeutics, and Sofro Pharmaceuticals; a founder and stockholder in Toran Therapeutics; inventor of patents and patent applications held by Princeton University, including patent application related to G6PD inhibitors; and a director of the Princeton University-PKU Shenzhen collaboration. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Source data for Fig. 1B is provided in table S1.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Legends for tables S1 to S3
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S3
REFERENCES AND NOTES
- 1.Wu W. L., Papagiannakopoulos T., The pleiotropic role of the KEAP1/NRF2 pathway in cancer. Annu. Rev. Cancer Biol. 4, 413–435 (2020). [Google Scholar]
- 2.Moinova H. R., Mulcahy R. T., Up-regulation of the human gamma-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochem. Biophys. Res. Commun. 261, 661–668 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Wild A. C., Moinova H. R., Mulcahy R. T., Regulation of γ-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J. Biol. Chem. 274, 33627–33636 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Chanas S. A., Jiang Q., Mahon M. M., McWalter G. K., McLellan L. I., Elcombe C. R., Henderson C. J., Wolf C. R., Moffat G. J., Itoh K., Yamamoto M., Hayes J. D., Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem. J. 365 (Pt. 2), 405–416 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thimmulappa R. K., Mai K. H., Srisuma S., Kensler T. W., Yamamoto M., Biswal S., Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 62, 5196–5203 (2002). [PubMed] [Google Scholar]
- 6.Sakurai A., Nishimoto M., Himeno S., Imura N., Tsujimoto M., Kunimoto M., Hara S., Transcriptional regulation of thioredoxin reductase 1 expression by cadmium in vascular endothelial cells: Role of NF-E2-related factor-2. J. Cell. Physiol. 203, 529–537 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Wakabayashi N., Dinkova-Kostova A. T., Holtzclaw W. D., Kang M. I., Kobayashi A., Yamamoto M., Kensler T. W., Talalay P., Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. U.S.A. 101, 2040–2045 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi A., Suzuki H., Itoh K., Yamamoto M., Sugiyama Y., Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein 1 in mouse embryo fibroblasts. Biochem. Biophys. Res. Commun. 310, 824–829 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Alam J., Stewart D., Touchard C., Boinapally S., Choi A. M. K., Cook J. L., Nrf2, a Cap’n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 274, 26071–26078 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., Motohashi H., Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22, 66–79 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Singh A., Happel C., Manna S. K., Acquaah-Mensah G., Carrerero J., Kumar S., Nasipuri P., Krausz K. W., Wakabayashi N., Dewi R., Boros L. G., Gonzalez F. J., Gabrielson E., Wong K. K., Girnun G., Biswal S., Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J. Clin. Invest. 123, 2921–2934 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeNicola G. M., Chen P. H., Mullarky E., Sudderth J. A., Hu Z., Wu D., Tang H., Xie Y., Asara J. M., Huffman K. E., Wistuba I. I., Minna J. D., DeBerardinis R. J., Cantley L. C., NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 47, 1475–1481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sayin V. I., LeBoeuf S. E., Singh S. X., Davidson S. M., Biancur D., Guzelhan B. S., Alvarez S. W., Wu W. L., Karakousi T. R., Zavitsanou A. M., Ubriaco J., Muir A., Karagiannis D., Morris P. J., Thomas C. J., Possemato R., Vander Heiden M. G., Papagiannakopoulos T., Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patra K. C., Hay N., The pentose phosphate pathway and cancer. Trends Biochem. Sci. 39, 347–354 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L., Zhang Z., Hoshino A., Zheng H. D., Morley M., Arany Z., Rabinowitz J. D., NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat. Metab. 1, 404–415 (2019). [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Xu Y., Lu W., Ghergurovich J. M., Guo L., Blair I. A., Rabinowitz J. D., Yang X., Upregulation of antioxidant capacity and nucleotide precursor availability suffices for oncogenic transformation. Cell Metab. 33, 94–109.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghergurovich J. M., García-Cañaveras J. C., Wang J., Schmidt E., Zhang Z., TeSlaa T., Patel H., Chen L., Britt E. C., Piqueras-Nebot M., Gomez-Cabrera M. C., Lahoz A., Fan J., Beier U. H., Kim H., Rabinowitz J. D., A small molecule G6PD inhibitor reveals immune dependence on pentose phosphate pathway. Nat. Chem. Biol. 16, 731–739 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghergurovich J. M., Esposito M., Chen Z., Wang J. Z., Bhatt V., Lan T., White E., Kang Y., Guo J. Y., Rabinowitz J. D., Glucose-6-phosphate dehydrogenase is not essential for K-Ras-driven tumor growth or metastasis. Cancer Res. 80, 3820–3829 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeNicola G. M., Karreth F. A., Humpton T. J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K. H., Yeo C. J., Calhoun E. S., Scrimieri F., Winter J. M., Hruban R. H., Iacobuzio-Donahue C., Kern S. E., Blair I. A., Tuveson D. A., Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475, 106–109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero R., Sayin V. I., Davidson S. M., Bauer M. R., Singh S. X., LeBoeuf S. E., Karakousi T. R., Ellis D. C., Bhutkar A., Sánchez-Rivera F. J., Subbaraj L., Martinez B., Bronson R. T., Prigge J. R., Schmidt E. E., Thomas C. J., Goparaju C., Davies A., Dolgalev I., Heguy A., Allaj V., Poirier J. T., Moreira A. L., Rudin C. M., Pass H. I., Vander Heiden M. G., Jacks T., Papagiannakopoulos T., Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 23, 1362–1368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeBoeuf S. E., Wu W. L., Karakousi T. R., Karadal B., RaElle Jackson S., Davidson S. M., Wong K.-K., Koralov S. B., Sayin V. I., Papagiannakopoulos T., Activation of oxidative stress response in cancer generates a druggable dependency on exogenous non-essential amino acids. Cell Metab. 31, 339–350.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arbour K. C., Jordan E., Kim H. R., Dienstag J., Yu H. A., Sanchez-Vega F., Lito P., Berger M., Solit D. B., Hellmann M., Kris M. G., Rudin C. M., Ni A., Arcila M., Ladanyi M., Riely G. J., Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin. Cancer Res. 24, 334–340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Cancer Genome Atlas Research Network , Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lignitto L., Le Boeuf S. E., Homer H., Jiang S., Askenazi M., Karakousi T. R., Pass H. I., Bhutkar A. J., Tsirigos A., Ueberheide B., Sayin V. I., Papagiannakopoulos T., Pagano M., Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of Bach1. Cell 178, 316–329.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Binkley M. S., Jeon Y. J., Nesselbush M., Moding E. J., Nabet B. Y., Almanza D., Kunder C., Stehr H., Yoo C. H., Rhee S., Xiang M., Chabon J. J., Hamilton E., Kurtz D. M., Gojenola L., Owen S. G., Ko R. B., Shin J. H., Maxim P. G., Lui N. S., Backhus L. M., Berry M. F., Shrager J. B., Ramchandran K. J., Padda S. K., Das M., Neal J. W., Wakelee H. A., Alizadeh A. A., Loo B. W. Jr., Diehn M., KEAP1/NFE2L2 mutations predict lung cancer radiation resistance that can be targeted by glutaminase inhibition. Cancer Discov. 10, 1826–1841 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cristescu R., Mogg R., Ayers M., Albright A., Murphy E., Yearley J., Sher X., Liu X. Q., Lu H., Nebozhyn M., Zhang C., Lunceford J. K., Joe A., Cheng J., Webber A. L., Ibrahim N., Plimack E. R., Ott P. A., Seiwert T. Y., Ribas A., McClanahan T. K., Tomassini J. E., Loboda A., Kaufman D., Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, eaar3593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank R., Scheffler M., Merkelbach-Bruse S., Ihle M. A., Kron A., Rauer M., Ueckeroth F., König K., Michels S., Fischer R., Eisert A., Fassunke J., Heydt C., Serke M., Ko Y. D., Gerigk U., Geist T., Kaminsky B., Heukamp L. C., Clement-Ziza M., Büttner R., Wolf J., Clinical and pathological characteristics of KEAP1- and NFE2L2-mutated non-small cell lung carcinoma (NSCLC). Clin. Cancer Res. 24, 3087–3096 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Goeman F., de Nicola F., Scalera S., Sperati F., Gallo E., Ciuffreda L., Pallocca M., Pizzuti L., Krasniqi E., Barchiesi G., Vici P., Barba M., Buglioni S., Casini B., Visca P., Pescarmona E., Mazzotta M., de Maria R., Fanciulli M., Ciliberto G., Maugeri-Saccà M., Mutations in the KEAP1-NFE2L2 pathway define a molecular subset of rapidly progressing lung adenocarcinoma. J. Thorac. Oncol. 14, 1924–1934 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Galan-Cobo A., Sitthideatphaiboon P., Qu X., Poteete A., Pisegna M. A., Tong P., Chen P. H., Boroughs L. K., Rodriguez M. L. M., Zhang W., Parlati F., Wang J., Gandhi V., Skoulidis F., DeBerardinis R. J., Minna J. D., Heymach J. V., LKB1 and KEAP1/NRF2 pathways cooperatively promote metabolic reprogramming with enhanced glutamine dependence in KRAS-mutant lung adenocarcinoma. Cancer Res. 79, 3251–3267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu X. G., Chudnovskiy A., Baudrier L., Prizer B., Liu Y., Ostendorf B. N., Yamaguchi N., Arab A., Tavora B., Timson R., Heissel S., de Stanchina E., Molina H., Victora G. D., Goodarzi H., Birsoy K., Functional genomics in vivo reveal metabolic dependencies of pancreatic cancer cells. Cell Metab. 33, 211–221.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies T. G., Wixted W. E., Coyle J. E., Griffiths-Jones C., Hearn K., McMenamin R., Norton D., Rich S. J., Richardson C., Saxty G., Willems H. M. G., Woolford A. J. A., Cottom J. E., Kou J. P., Yonchuk J. G., Feldser H. G., Sanchez Y., Foley J. P., Bolognese B. J., Logan G., Podolin P. L., Yan H., Callahan J. F., Heightman T. D., Kerns J. K., Monoacidic inhibitors of the Kelch-like ECH-associated protein 1: Nuclear factor erythroid 2-related factor 2 (KEAP1:NRF2) protein-protein interaction with high cell potency identified by fragment-based discovery. J. Med. Chem. 59, 3991–4006 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Romero R., Sánchez-Rivera F. J., Westcott P. M. K., Mercer K. L., Bhutkar A., Muir A., González Robles T. J., Lamboy Rodríguez S., Liao L. Z., Ng S. R., Li L., Colón C. I., Naranjo S., Beytagh M. C., Lewis C. A., Hsu P. P., Bronson R. T., Vander Heiden M. G., Jacks T., Keap1 mutation renders lung adenocarcinomas dependent on Slc33a1. Nat. Cancer 1, 589–602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birsoy K., Wang T., Chen W. W., Freinkman E., Abu-Remaileh M., Sabatini D. M., An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jang C., Chen L., Rabinowitz J. D., Metabolomics and isotope tracing. Cell 173, 822–837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L., Shah S., Fan J., Park J. O., Wellen K. E., Rabinowitz J. D., Malic enzyme tracers reveal hypoxia-induced switch in adipocyte NADPH pathway usage. Nat. Chem. Biol. 12, 345–352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vidigal J. A., Ventura A., Rapid and efficient one-step generation of paired gRNA CRISPR-Cas9 libraries. Nat. Commun. 6, 8083 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altman B. J., Stine Z. E., Dang C. V., From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 619–634 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayes J. D., Dinkova-Kostova A. T., Tew K. D., Oxidative stress in cancer. Cancer Cell 38, 167–197 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frank J. E., Diagnosis and management of G6PD deficiency. Am. Fam. Physician 72, 1277–1282 (2005). [PubMed] [Google Scholar]
- 40.Nkhoma E. T., Poole C., Vannappagari V., Hall S. A., Beutler E., The global prevalence of glucose-6-phosphate dehydrogenase deficiency: A systematic review and meta-analysis. Blood Cells Mol. Dis. 42, 267–278 (2009). [DOI] [PubMed] [Google Scholar]
- 41.The Cancer Genome Atlas Research Network , Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaramillo M. C., Zhang D. D., The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 27, 2179–2191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konstantinopoulos P. A., Spentzos D., Fountzilas E., Francoeur N., Sanisetty S., Grammatikos A. P., Hecht J. L., Cannistra S. A., Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res. 71, 5081–5089 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Shibata T., Kokubu A., Gotoh M., Ojima H., Ohta T., Yamamoto M., Hirohashi S., Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 135, 1358–1368, 1368.e1–4 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Kim Y. R., Oh J. E., Kim M. S., Kang M. R., Park S. W., Han J. Y., Eom H. S., Yoo N. J., Lee S. H., Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J. Pathol. 220, 446–451 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Sato Y., Yoshizato T., Shiraishi Y., Maekawa S., Okuno Y., Kamura T., Shimamura T., Sato-Otsubo A., Nagae G., Suzuki H., Nagata Y., Yoshida K., Kon A., Suzuki Y., Chiba K., Tanaka H., Niida A., Fujimoto A., Tsunoda T., Morikawa T., Maeda D., Kume H., Sugano S., Fukayama M., Aburatani H., Sanada M., Miyano S., Homma Y., Ogawa S., Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 45, 860–867 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Fabrizio F. P., Costantini M., Copetti M., la Torre A., Sparaneo A., Fontana A., Poeta L., Gallucci M., Sentinelli S., Graziano P., Parente P., Pompeo V., Salvo L. D., Simone G., Papalia R., Picardo F., Balsamo T., Flammia G. P., Trombetta D., Pantalone A., Kok K., Paranita F., Muscarella L. A., Fazio V. M., Keap1/Nrf2 pathway in kidney cancer: Frequent methylation of KEAP1 gene promoter in clear renal cell carcinoma. Oncotarget 8, 11187–11198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muscarella L. A., Barbano R., D’Angelo V., Copetti M., Coco M., Balsamo T., la Torre A., Notarangelo A., Troiano M., Parisi S., Icolaro N., Catapano D., Valori V. M., Pellegrini F., Merla G., Carella M., Fazio V. M., Parrella P., Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient’s outcome. Epigenetics 6, 317–325 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanada N., Takahata T., Zhou Q., Ye X., Sun R., Itoh J., Ishiguro A., Kijima H., Mimura J., Itoh K., Fukuda S., Saijo Y., Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer 12, 66 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldstein L. D., Lee J., Gnad F., Klijn C., Schaub A., Reeder J., Daemen A., Bakalarski C. E., Holcomb T., Shames D. S., Hartmaier R. J., Chmielecki J., Seshagiri S., Gentleman R., Stokoe D., Recurrent loss of NFE2L2 exon 2 is a mechanism for Nrf2 pathway activation in human cancers. Cell Rep. 16, 2605–2617 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Daneshmandi S., Cassel T., Lin P., Higashi R. M., Wulf G. M., Boussiotis V. A., Fan T. W. M., Seth P., Blockade of 6-phosphogluconate dehydrogenase generates CD8+ effector T cells with enhanced anti-tumor function. Cell Rep. 34, 108831 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Best S. A., Ding S., Kersbergen A., Dong X., Song J. Y., Xie Y., Reljic B., Li K., Vince J. E., Rathi V., Wright G. M., Ritchie M. E., Sutherland K. D., Distinct initiating events underpin the immune and metabolic heterogeneity of KRAS-mutant lung adenocarcinoma. Nat. Commun. 10, 4190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Street J. C., Alfieri A. A., Koutcher J. A., Quantitation of metabolic and radiobiological effects of 6-aminonicotinamide in RIF-1 tumor cells in vitro. Cancer Res. 57, 3956–3962 (1997). [PubMed] [Google Scholar]
- 54.Belfi C. A., Chatterjee S., Gosky D. M., Berger S. J., Berger N. A., Increased sensitivity of human colon cancer cells to DNA cross-linking agents after GRP78 up-regulation. Biochem. Biophys. Res. Commun. 257, 361–368 (1999). [DOI] [PubMed] [Google Scholar]
- 55.G. Vidal, K. Kalinsky, E Stringer-Reasor, F. Lynce, J. Cole, F Valdes-Albini, H. Soliman, P. Nikolinakos, A. Silber, A De Michele, H. Ali, D. Graham, J. Giguere, A. Brufsky, Y. Liang, S. Holland, G. Fiji, B O’Keeffe, K. Gogineni, Efficacy and safety of CB-839, a small molecule inhibitor of glutaminase, in combination with paclitaxel in patients with advanced triple negative breast cancer (TNBC): Initial findings from a multicenter, open-label phase 2 study, San Antonio Breast Cancer Symposium, San Antonio, TX, 4 December 2018.
- 56.Davidson S. M., Papagiannakopoulos T., Olenchock B. A., Heyman J. E., Keibler M. A., Luengo A., Bauer M. R., Jha A. K., O’Brien J. P., Pierce K. A., Gui D. Y., Sullivan L. B., Wasylenko T. M., Subbaraj L., Chin C. R., Stephanopolous G., Mott B. T., Jacks T., Clish C. B., Vander Heiden M. G., Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metab. 23, 517–528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DuPage M., Dooley A. L., Jacks T., Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 4, 1064–1072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bankhead P., Loughrey M. B., Fernández J. A., Dombrowski Y., McArt D. G., Dunne P. D., McQuaid S., Gray R. T., Murray L. J., Coleman H. G., James J. A., Salto-Tellez M., Hamilton P. W., QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sánchez-Rivera F. J., Papagiannakopoulos T., Romero R., Tammela T., Bauer M. R., Bhutkar A., Joshi N. S., Subbaraj L., Bronson R. T., Xue W., Jacks T., Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bardeesy N., Aguirre A. J., Chu G. C., Cheng K. H., Lopez L. V., Hezel A. F., Feng B., Brennan C., Weissleder R., Mahmood U., Hanahan D., Redston M. S., Chin L., DePinho R. A., Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc. Natl. Acad. Sci. U.S.A. 103, 5947–5952 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ianevski A., Giri A. K., Aittokallio T., SynergyFinder 2.0: Visual analytics of multi-drug combination synergies. Nucleic Acids Res. 48, W488–W493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiederschain D., Susan W., Chen L., Loo A., Yang G., Huang A., Chen Y., Caponigro G., Yao Y. M., Lengauer C., Sellers W. R., Benson J. D., Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 8, 498–504 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Akama-Garren E. H., Joshi N. S., Tammela T., Chang G. P., Wagner B. L., Lee D. Y., Rideout III W. M., Papagiannakopoulos T., Xue W., Jacks T., A modular assembly platform for rapid generation of DNA constructs. Sci. Rep. 6, 16836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lewis C. A., Parker S. J., Fiske B. P., McCloskey D., Gui D. Y., Green C. R., Vokes N. I., Feist A. M., Vander Heiden M. G., Metallo C. M., Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell 55, 253–263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fernandez C. A., Des Rosiers C., Previs S. F., David F., Brunengraber H., Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J. Mass Spectrom. 31, 255–262 (1996). [DOI] [PubMed] [Google Scholar]
- 67.Lu W., Wang L., Chen L., Hui S., Rabinowitz J. D., Extraction and quantitation of nicotinamide adenine dinucleotide redox cofactors. Antioxid. Redox Signal. 28, 167–179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L., Xing X., Chen L., Yang L., Su X., Rabitz H., Lu W., Rabinowitz J. D., Peak annotation and verification engine for untargeted LC–MS metabolomics. Anal. Chem. 91, 1838–1846 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Z., Chen L., Liu L., Su X., Rabinowitz J. D., Chemical basis for deuterium labeling of fat and NADPH. J. Am. Chem. Soc. 139, 14368–14371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang L., Garcia Canaveras J. C., Chen Z., Wang L., Liang L., Jang C., Mayr J. A., Zhang Z., Ghergurovich J. M., Zhan L., Joshi S., Hu Z., McReynolds M. R., Su X., White E., Morscher R. J., Rabinowitz J. D., Serine catabolism feeds NADH when respiration is impaired. Cell Metab. 31, 809–821.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Melamud E., Vastag L., Rabinowitz J. D., Metabolomic analysis and visualization engine for LC−MS data. Anal. Chem. 82, 9818–9826 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Su X., Lu W., Rabinowitz J. D., Metabolite spectral accuracy on Orbitraps. Anal. Chem. 89, 5940–5948 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Legends for tables S1 to S3
Tables S1 to S3