Abstract
CRISPR-Cas-guided base editors convert A•T to G•C, or C•G to T•A, in cellular DNA for precision genome editing. To understand the molecular basis for DNA adenosine deamination by adenine base editors (ABEs), we determined a 3.2 Å resolution cryo-EM structure of ABE8e in a substrate-bound state in which the deaminase domain engages DNA exposed within the CRISPR-Cas9 R-loop complex. Kinetic and structural data suggest that ABE8e catalyzes DNA deamination up to ~1,100-fold faster than earlier ABEs due to mutations that stabilize DNA substrates in a constrained tRNA-like conformation. Furthermore, ABE8e’s accelerated DNA deamination suggests a previously unobserved transient DNA melting that may occur during double-stranded DNA surveillance by CRISPR-Cas9. These results explain ABE8e-mediated base editing outcomes and inform the future design of base editors.
ONE SENTENCE SUMMARY
Cryo-EM structure and kinetics of an evolved CRISPR-Cas9 DNA base editor explain rapid Cas9-dependent DNA deamination.
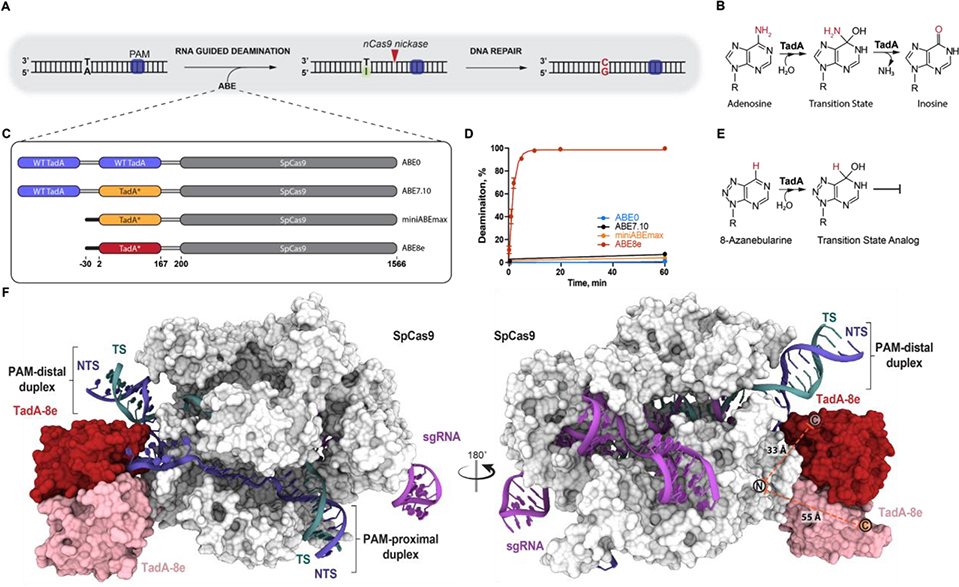
The site-specific conversion of A•T to G•C base pairs and C•G to T•A base pairs in genomic DNA could correct ~60% of all known pathogenic single nucleotide polymorphisms (SNPs) in humans (1). Such conversions can be achieved with CRISPR-Cas9 base editors: RNA-guided Cas proteins fused to a ssDNA deaminase (2, 3). A•T-to-G•C transitions in DNA require the deamination of adenosine to inosine, which is recognized by cellular machinery as guanosine (Fig. 1A–B). With no enzyme capable of deaminating adenine in DNA, E. coli tRNA adenosine deaminase (TadA) was fused to Cas9 and evolved into ABE7.10, which catalyzes the targeted deamination of deoxyadenosine (3–5). ABE7.10 encodes two copies of TadA, an N-terminal wild type (WT) TadA linked to an evolved TadA (TadA-7.10), which is C-terminally linked to a “nickase” (cuts one strand of the dsDNA) version of S. pyogenes Cas9 (nSpCas9) (Fig. 1C). ABE7.10 has since been widely applied as a tool to deaminate genomic DNA in many cell types and organisms (6–12). The ABE7.10 variant was found to catalyze off-target RNA editing in cells, an activity that was reduced by mutations to the TadA-7.10 domain (13) or by removing the N-terminal WT TadA domain generating a truncated version miniABEmax (Fig. 1C) (14). ABE7.10 was further evolved to generate ABE8e which encodes a single TadA domain (TadA-8e) broadly compatible with eight tested Cas effectors (Fig. 1C) (15). In vitro, ABE8e deaminates DNA at a rate 590- and 1,170-fold higher than ABE7.10 and miniABEmax, respectively (15) (Fig. 1D; fig. S1). ABE7.10 and ABE8e contain 14 and 22 substitutions, respectively, within their evolved TadA domains relative to WT TadA (3, 15) (fig. S2A). How these mutations enabled evolved TadA to catalyze deamination of DNA and the molecular basis for the different catalytic rates of ABE7.10 and ABE8e remain unknown.
Fig. 1. Cryo-EM structure of ABE8e in a substrate-bound state.
A) Schematic representation of RNA guided DNA adenosine deamination. B) Mechanism of E. coli TadA catalysed tRNA adenosine deamination. C) Domain architecture of four generations of ABEs encoding SpCas9 N-terminally linked to either wild-type (WT) TadA, evolved TadA7.10 (TadA*, yellow) or evolved TadA8e (TadA*, red). D) Single-turnover kinetics of targeting ABE RNPs measured using dsDNA containing a single adenine. The fraction of deaminated dsDNA is shown plotted as a function of time and fitted to a single phase exponential equation. The extracted apparent deamination rates of ABE7.10 (black), miniABEmax (orange) and ABE8e (red) are 0.0010 ± 3×10−4 min−1, 0.0005 ± 1×10−4 min−1, and 0.585 ± 0.034 min−1, respectively. Data are represented as the mean ± SD from three independent experiments. The deamination data of ABE8e and ABE7.10 are reproduced and were originally published in ref. 15. E) Mechanism of inhibition of adenosine deamination by 8-azanebularine that mimics adenosine deamination reaction intermediate. F) 3.2 Å resolution cryo-EM structure of the SpCas9-ABE8e complex. Subunits are colored: SpCas9 (white), single-guide RNA (sgRNA, purple), target-strand DNA (TS, teal), non-target strand DNA (NTS, blue) and the TadA-8e dimer (red and pink). The theoretical connectivity of the linker region is shown as a dashed orange line between the Cas9 N-terminus and either TadA-8e C-terminus.
To advance our understanding of DNA substrate recognition during SpCas9-guided and TadA-catalyzed DNA adenosine deamination, we used single-particle cryo-electron microscopy (cryo-EM) to determine the structure of a DNA-bound ABE8e complex. The ABE8e complex was trapped in a catalytic conformation using a DNA non-target strand (NTS) containing a transition state analog for adenosine deamination reactions, 8-azanebularine (Fig. 1E) (16). At an overall resolution of 3.2 Å, we resolved the ABE8e9 complex containing Cas9-bound to a single-guide RNA (sgRNA) hybridized to a 20-nucleotide DNA target-strand (TS) within a double-stranded DNA (dsDNA) substrate (Fig. 1F, fig. S3, fig. S4, Table S1). This structure provides the most complete molecular model for a Cas9 R-loop complex, showing Cas9 engaged with the GG dinucleotide protospacer-adjacent motif (PAM) while both PAM-distal and PAM-proximal DNA duplexes are arranged on either side of the complex. The PAM-distal duplex is positioned against SpCas9-Y1016 while the sgRNA-TS heteroduplex is arranged against SpCas9-Y1013, generating a non-coaxially stacked duplex junction separated by SpCas9 (fig. S5, fig. S6A). The ABE8e construct encodes a single TadA-8e domain (Fig. 1A). Cryo-EM density supported unambiguous modelling of a TadA-8e domain secondary structure engaged with the 5′ end of the NTS emerging from SpCas9 (Fig. 1F, fig. S6B). We observed poorly resolved density below the aforementioned TadA-8e domain containing secondary structural features consistent with a second copy of TadA-8e (Fig. 1F, fig. S6B). Further analysis revealed that the second TadA-8e domain was contributed in trans from another ABE8e molecule (fig. S7A); which is unsurprising given that the dimerization interface was unchanged between TadA-WT and TadA-8e (fig. S2). While the linker region was unresolved, measured distances between Cas9 and each TadA-8e domain suggest that the cis-TadA-8e domain is engaged with the NTS (Fig. 1F, Supplementary Text 1).
The cryo-EM structure of the ABE8e complex revealed that the TadA-8e domain engages with the exposed single-stranded region of the PAM distal NTS (Fig. 2A). Consistent with this, in vitro DNA adenosine deamination assays demonstrated that the ABE8e complex rapidly deaminates adenines present in accessible trans-ssDNA but not trans-dsDNA (Fig. 2B). Examining the TadA-8e dimer contacts revealed that one TadA-8e domain contacts the NTS but has no interface with SpCas9 (fig. S8A). In contrast, the second TadA-8e domain does not contact the NTS but forms a non-specific interface with the SpCas9 RuvC domain (fig. S8B). These observations suggested that the presentation of single-stranded NTS by SpCas9 drives accelerated TadA-8e-catalyzed cis-DNA deamination. To explore this idea, we compared the kinetics of adenosine deamination by TadA-8e of a guide-complementary dsDNA (NTS presented as cis-ssDNA) and trans-ssDNA (Fig. 2C and fig. S1E,F S9A). The rate of cis-ssDNA deamination was ~3.7 fold faster relative to trans-ssDNA deamination, confirming that the presentation of the NTS by SpCas9 accelerates the kinetics of TadA-8e. Given that ABE8e was shown to edit more positions within an R-loop than previous iterations of ABEs (15), we hypothesized that the TadA-8e domain of ABE8e is a multiple-turnover enzyme. To test this, we carried out in vitro DNA deamination assays under multiple-turnover conditions (Fig. 2D and fig. S9B). The extracted turnover number for ssDNA in trans was 4.5 ± 0.1 in contrast to 0.97 ± 0.03 for cis-ssDNA. While the observed single-turnover deamination kinetics on cis-DNA reflect the single-turnover kinetics of SpCas9 forming an R-loop (17), the rapid multiple turnover kinetics are consistent with the higher processivity of ABE8e observed in cells relative to previous ABEs (15).
Fig. 2. ABE8e is a multiple-turnover enzyme with a wide editing window.
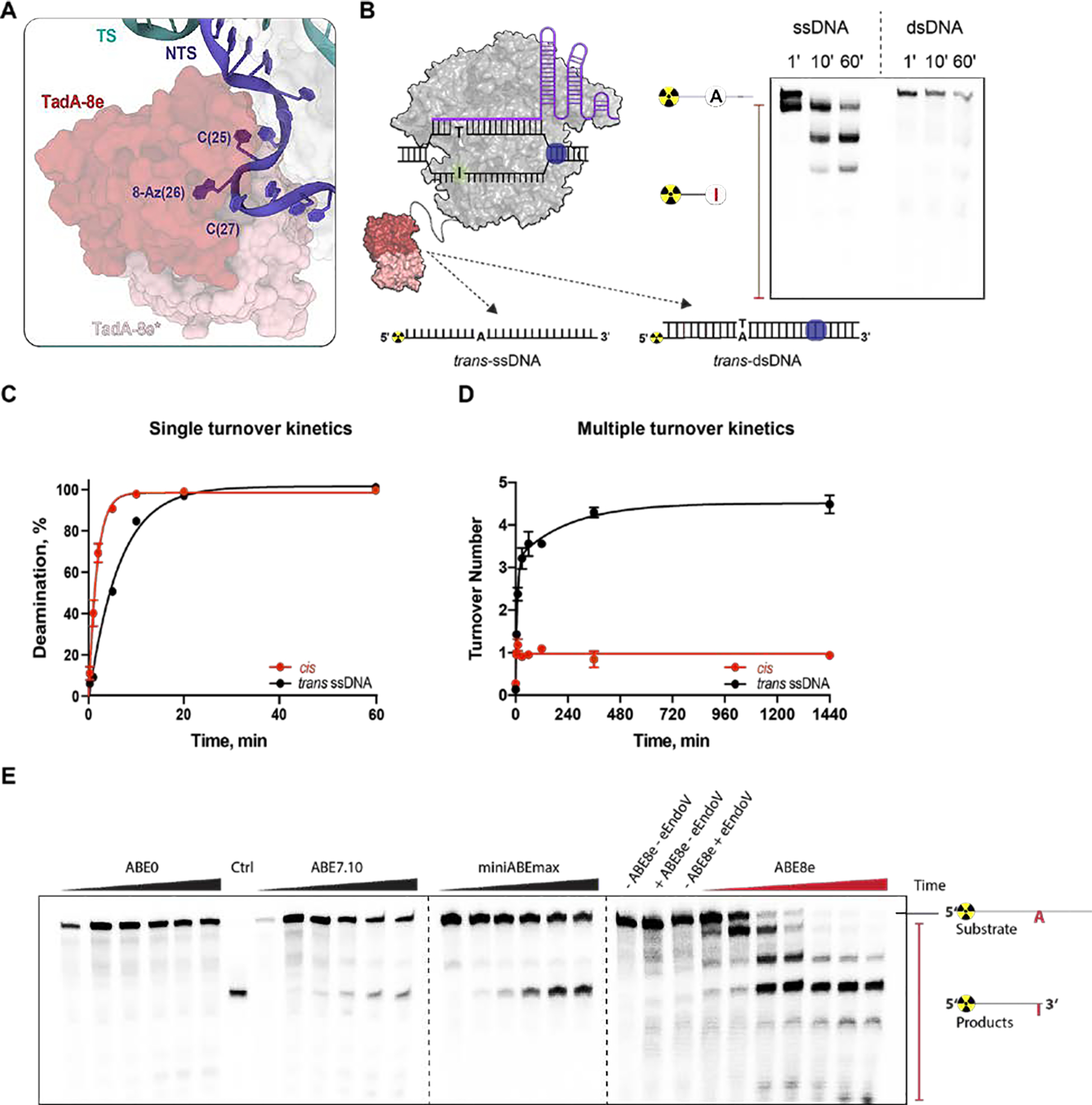
A) Surface exposed and single-stranded topology of the non-target strand (NTS) (cartoon, blue) engaged with the TadA8e domain (surface, red). B) Schematic representation of ABE8e RNP complex acting on radiolabeled trans-ssDNA or trans-dsDNA substrate containing a single adenine. (right) A representative gel showing ABE8e RNP single-turnover deamination of radio-labeled ssDNA and dsDNA over time (min) in trans. C) Single-turnover kinetics of ABE8e RNP measured using either dsDNA (deamination in cis) or ssDNA (deamination in trans) containing single adenine. The fraction of deaminated DNA plotted as a function of time, and fitted to a single exponential equation. Data are represented as the mean ± SD from three independent experiments.The apparent rate of DNA editing in trans is ~3.7 fold lower than the apparent rate of DNA editing in cis (kapp = 0.16 ± 0.01 min−1 vs. kapp = 0.59 ± 0.03 min−1). D) DNA deamination assay in multiple-turnover conditions using ABE8e RNP and either ssDNA (trans) or dsDNA (cis) substrate. Turnover number (a ratio of the deaminated DNA concentration and total concentration of ABE8e) was plotted as a function of time. The extracted turnover number for ssDNA deamination in trans is 4.5 ± 0.1 while for DNA in cis it is 0.97 ± 0.03. E) Gel representing single-turnover kinetics of ABE0, ABE7.10, miniABEmax, and ABE8e measured using dsDNA (cis) containing multiple adenines. Assays were performed in three independent replicates, and time points for ABE0, ABE7.10 and miniABEmax assays were taken at 0, 1, 3, 8, 24 and 32 hours, while for ABE8e at 0, 1, 5, 10, 20, 60 and 180 min. Concentrations of ABE RNPs were 1 μM for single-turnover assays and 25 nM for multiple-turnover assays. Concentrations of DNA were 2.5 nM for single-turnover assays and 250 nM for multiple-turnover assays.
The rapid multiple-turnover trans-ssDNA deamination kinetics of ABE8e suggested that any adenosine in ssDNA, including those transiently exposed to solvent by Cas9, might be deaminated by TadA-8e. To explore this idea, we compared the in vitro editing characteristics of ABE7.10, miniABEmax, and ABE8e using dsDNA where the NTS contained multiple adenines (Fig. 2E). While ABE7.10 and miniABEmax predominantly deaminated a single PAM-distal adenine, ABE8e deaminated multiple adenines, including those present within double-stranded regions in the R-loop (Fig. 2E, fig. S10A, B). Moreover, adenines were deaminated sequentially from the 3′ end of the NTS (Fig. 2E, fig. S10A, B), reminiscent of the biased 3′ to 5′ diffusion of SpCas9 during search of a PAM along dsDNA (18). Thus, we hypothesized that the directional DNA adenosine deamination might be related to the SpCas9 RNP target search mechanism.
Binding to an sgRNA remodels SpCas9 to allow target interrogation through a biased directional search along dsDNA (Fig. 3A) (17, 18). Stable interaction with a PAM initiates probing for complementarity and stable R-loop formation on matched substrates to activate Cas9 for DNA cleavage (Fig. 3B) (17, 19). With this model in mind, we tested whether directional adenosine deamination by ABE8e might be related to the Cas9 target search mechanism. Deamination assays were conducted using either radio-labeled ssDNA or dsDNA substrates and ABE8e in three different states: apo-ABE8e, ABE8e complexed with a targeting sgRNA, and ABE8e complexed with a non-targeting sgRNA (Fig. 3C). Apo-ABE8e was able to deaminate only ssDNA, while ABE8e RNP was able to modify adenines in ssDNA and dsDNA, in contrast with the absence of trans-dsDNA editing by ABE8e engaged in an R-loop complex (Fig. 2B). Moreover, the dsDNA deamination is sgRNA sequence independent as a non-targeting ABE8e RNP also deaminates dsDNA adenines (Fig. 2B) suggesting that stable R-loop formation is not required. To test whether Cas9’s interaction with the PAM affects the observed dsDNA editing by ABE8e, we performed in vitro DNA deamination assay with radiolabeled dsDNA devoid of consensus PAMs (Fig. 3D). To our surprise, ABE8e RNPs containing both the targeting and non-targeting sgRNA deaminated adenine in dsDNA lacking PAMs in the NTS (Fig. 3A). We speculate that during PAM search SpCas9 could transiently melt DNA providing a ssDNA substrate for rapid deamination by the highly active TadA-8e domain. We note that under conditions tested in mammalian cells, ABE8e activity remains strongly guide RNA-dependent with no detectable deamination of adenines flanking target protospacers (15), potentially due to differences in protein or DNA concentration or accessibility between in vitro and cellular conditions (Supplementary Text 2). Future biophysical and single-cell whole genome sequencing studies are necessary to explore the proposed mechanism and identify potential unanticipated ABE8e activity.
Fig. 3. Fast ABE8e kinetics reports on a new step in the SpCas9 target search pathway.
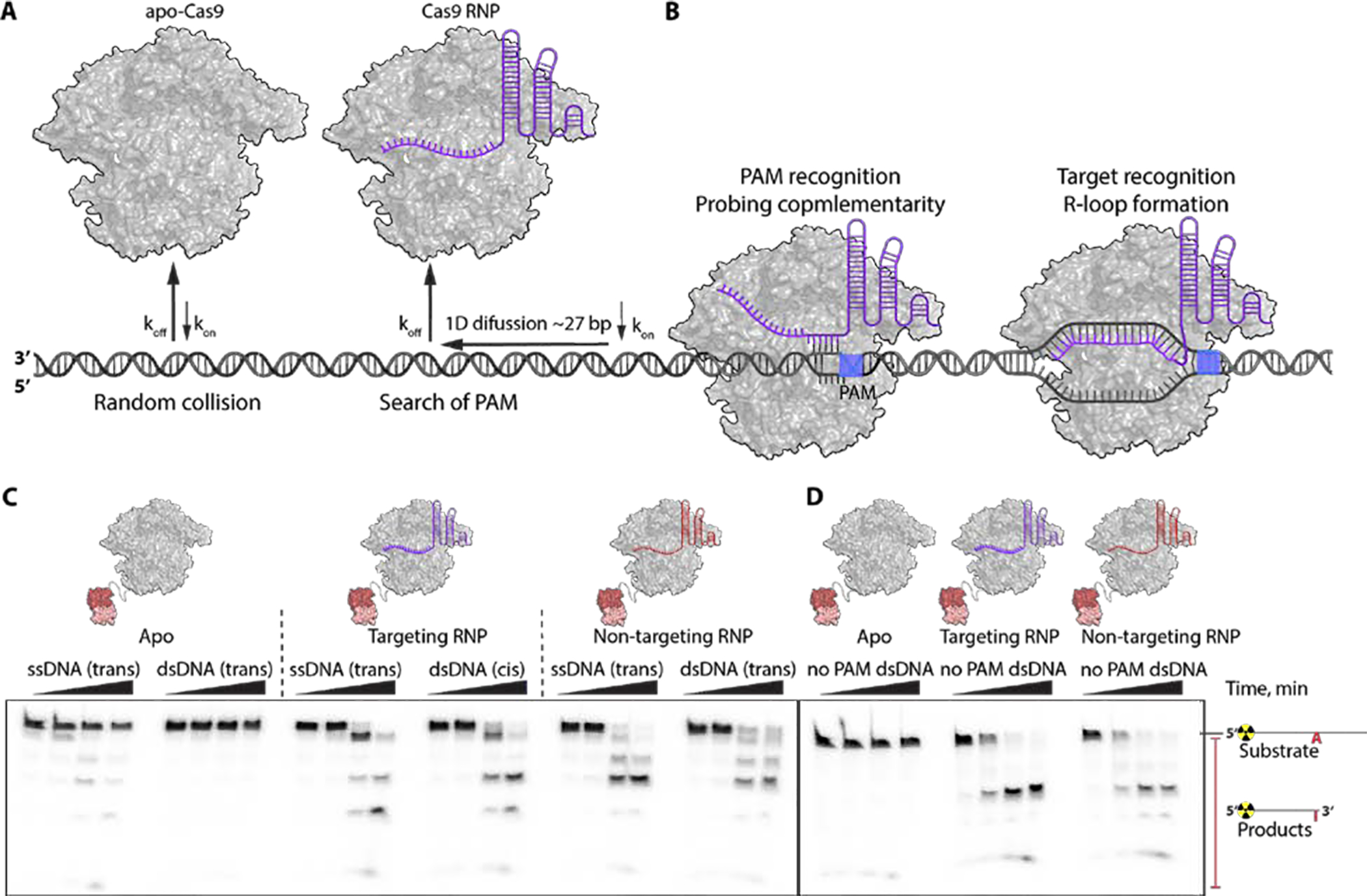
Model for SpCas9 target search mechanism: (A) apo-Cas9 – dsDNA interaction is a random collision; Cas9 bound to gRNA gets primed for a target search which initiates via random interaction with the DNA, followed by unidirectional (NTS-3’ to 5’) Cas9 RNP diffusion along dsDNA (for ~27 bp) in search of PAM and rapid dissociation from DNA unless PAM is encountered; (B) once Cas9 RNP comes across PAM its residence time on DNA increases and it unwinds first few nucleotides adjacent to PAM to probe for guide RNA complementarity; Cas9 RNP at correct target sites initiates an R-loop formation via sequential unwinding (9,10). C) Gel representing single-turnover kinetics of apo-ABE8e acting on either ssDNA (trans) or dsDNA (trans), of ABE8e RNP programmed with targeting gRNA deaminating ssDNA (trans) or dsDNA (cis), and of ABE8e RNP programmed with non-targeting gRNA deaminating ssDNA (trans) or dsDNA (trans). Time points were taken at 0, 1, 10 and 60 min. D) Gel representing single-turnover kinetics of apo-ABE8e, ABE8e targeting RNP and ABE8e non-targeting RNP acting on dsDNA that lacks PAM sequence. Time points were taken at 0, 1, 10 and 60 min. Concentrations of ABE RNPs were 1 μM and concentrations of DNA were 1 nM.
Examining the overall topology of the TadA-8e homodimer relative to WT TadA-8e revealed a striking difference in the conformation of the C-terminal α-helix, α5 (Fig. 4A, fig. S2B). The α5-helix of TadA-8e undergoes a sharp 180° turn at P152 (Fig. 4A), a mutation introduced early during ABE evolution in addition to five other substitutions along the α5-helix of TadA-7.10 (fig. S2). To investigate the importance of the P152 substitution and the cumulative changes within the α5-helix of TadA-8e, we purified the single P152R variant and a stacked variant REIK (P152R, V155E, F156I, N157K), both of which dispalyed significantly reduced rates of cis-DNA deamination relative to wild-type (Fig. 4B, fig. S11). These structural and biochemical observations demonstrate that cumulative substitutions within the α5-helix likely enable effective access to the NTS presented by SpCas9 within an R-loop.
Fig. 4. Mechanism of ABE8e substrate specificity.
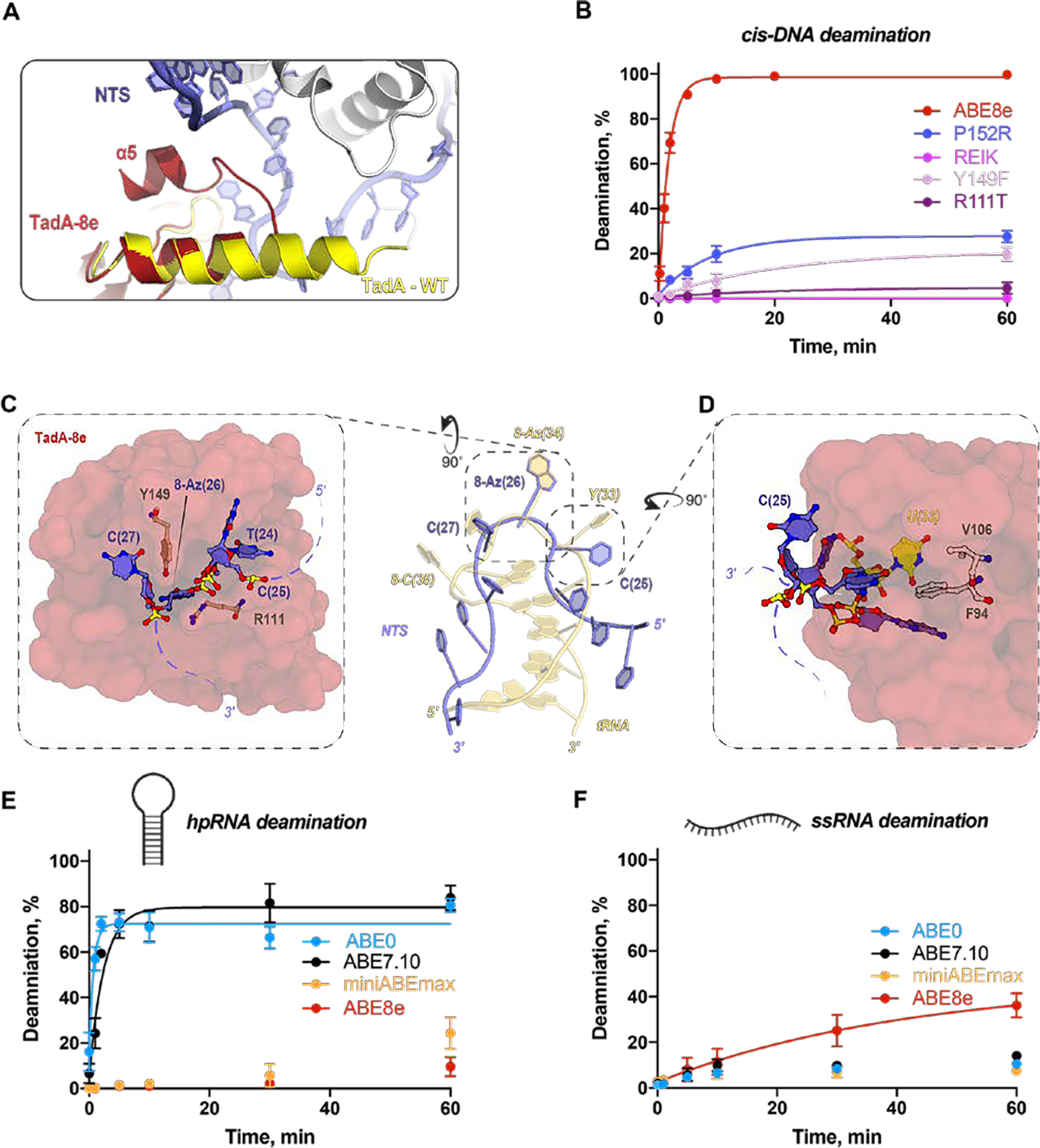
A) Superposition of TadA-8e (cartoon, red) and TadA-WT (cartoon, yellow) showing the altered disposition of ɑ5-helix. B) Single-turnover kinetics of ABE8e and four ABE8e variants using dsDNA (containing a single adenosine in the NTS). The apparent deamination rates are: ABE8e 0.59 ± 0.04 min−1; ABE8e-P152R 0.11 ± 0.02 min−1; ABE8e-Y149F 0.05 ± 0.02 min−1; ABE8e-R111T 0.05 ± 0.04 min−1. and N.D. for ABE8e-REIK. Concentrations of ABE RNPs were 1 μM and concentrations of DNA were 2.5 nM. C) TadA-8e active site showing the non-target strand (NTS; sticks, blue) entering the TadA-8e substrate binding pocket (surface, red) with evolved residue R111 and Y149 shown as sticks (brown). D) TadA-8e active-site proximal pocket (surface, red) with evolved residues F84 and V106 shown as sticks (white). The E. coli tRNA U(33) is shown in yellow. E) Single-turnover RNA deamination kinetics of all four ABEs programmed with targeting sgRNA (Table S2) using hpRNA as a substrate which contains single adenine. The fraction of deaminated hpRNA plotted as a function of time and fitted to a single exponential equation. The extracted apparent deamination rates of ABE0 (blue) and ABE7.10 (black) are 1.38 ± 0.28 min−1 and 0.41 ± 0.08 min−1, respectively. The kinetics of miniABEmax and ABE8e were much slower, and non-exponential. Data are represented as the mean ± SD from three independent experiments. Concentrations of ABE RNPs were 1 μM and concentrations of RNA were 1 nM. F) Single-turnover kinetics of all four ABEs programmed with targeting sgRNA (Table S2) using ssRNA as a substrate which contains single adenine. The fraction of deaminated hpRNA plotted as a function of time and fitted to a single exponential equation. The extracted apparent deamination rate of ABE8e (red) is 0.02 ± 0.01 min−1. The kinetics of ABE0, ABE7.10, and miniABEmax were much slower, and non-exponential. Data are represented as the mean ± SD from three independent experiments. Concentrations of ABE RNPs were 1 μM and concentrations of RNA were 1 nM.
Directed evolution of TadA-7.10 yielded the TadA-8e enzyme capable of rapid adenosine deamination in ssDNA. The ABE8e structure shows 8-azanebularine (8Az) buried deep within the active-site pocket of TadA-8e where it is oriented by the zinc containing active site, consistent with previous reports for E. coli TadA and ADAR2 (20, 21) (Fig. 4C). The nucleotides flanking the editing site, C(25) and C(27), are splayed against TadA-8e generating a sharp U-turn geometry in the NTS, a conformation not unlike that observed in the native tRNA anticodon (Fig. 4C). Throughout ABE8 evolution two residues were particularly conserved, R111 and Y149, both of which are oriented at the entrance to the active-site cleft and near the distorted NTS backbone flanking the editing site (Fig. 4C). To explore the role of R111 and Y149 in TadA-8e catalyzed DNA deamination, we reverted these positions back to ABE7.10 (R111T, Y149F). Under single-turnover conditions, we observed that Y149F had a 7-fold decrease in the apparent rate of cis-DNA deamination while the rate of the R111T mutant resembled that of ABE7.10 (Fig. 4B, fig. S11). Collectively, our structural and biochemical data support a role for R111 and Y149 in ssDNA substrate capture through stabilization of the U-turn conformation and induced fit into the deaminase active center.
WT TadA catalyzes site-specific deamination of tRNA by recognizing both the tRNA structure and anticodon YAC motif (Fig. 4C) (20, 21). A pair of substitutions introduced into ABE7.10 and preserved in ABE8e, A106V and L84F, provide steric bulk within the E. coli tRNA U(33) binding pocket (Fig. 4D). Coupled with the loss of the α5-helix that inserts into the tRNA major-groove (20), the steric clash at E. coli tRNA U(33) could prevent tRNA deamination by ABE8e. Consistent with this structural observation, the kinetics of in vitro hairpin RNA (hpRNA) adenosine deamination revealed that miniABEmax (TadA-7.10) had a dramatically reduced rate of hpRNA deamination relative to ABE0 (WT TadA - WT TadA) and ABE7.10 (WT TadA - TadA-7.10) (Fig. 4E and fig. S12A,D). The kinetics of single-stranded RNA (ssRNA) deamination demonstrated that ABE7.10 and miniABEmax are as slow and inefficient as WT TadA (Fig. 4F and fig. S12A, E), consistent with data showing that miniABEmax has significantly reduced RNA off-target editing in cells (14). In contrast, the additional eight substitutions in ABE8e (relative to miniABEmax) do not affect hpRNA editing but induce a two-fold faster ssRNA deamination rate (Fig. 4E, F and fig. S12D, E). These data suggest that the active site of TadA-8e has evolved to capture single-stranded nucleic acids of any sequence and to exclude more rigid tRNA substrates.
Our results demonstrate how the laboratory-evolved base editor ABE8e achieves rapid deamination on DNA. Due to substitutions near the active center, TadA-8e effectively remodels and induces fit of structurally flexible ssDNA into a narrow substrate binding pocket. The structure of ABE8e also demonstrated that the TadA-8e domain (and likely that of Tad-7.10) does not specifically interact with SpCas9, suggesting that the broader Cas-effector compatibility of TadA-8e stems from the rapid deamination kinetics. Furthermore, ABE8e’s accelerated DNA deamination suggested a previously unobserved transient melting of DNA that may occur during dsDNA surveillance by SpCas9. Our data suggest that site-specific base editors are possible through constraining the deaminase domain within the Cas9 R-loop structure to enable transmission of Cas9 conformational changes upon interaction with on-target DNA.
Supplementary Material
Acknowledgments:
We thank N. Orlova, J. Cofsky, J-J. Liu, K. Soczek, S. Eitzinger, C. Huang, J. Grunewald, K. J. Joung, and members of the Doudna laboratory for comments and discussions. We thank A. Iavarone from QB3/Chemistry Mass Spectrometry Facility (NIH 1S10OD020062–01) at UC Berkeley for native mass spectrometry of ABE8e. EM data were collected at the Bay Area Cryo-EM (BACEM) facility located at UC Berkeley. We thank D. B. Toso for expert electron microscopy assistance, and P. Tobias for computational support.
Funding:
JAD acknowledges support from US NIH RM1HG009490 and U01AI142817–02, the William M. Keck Foundation, the National Multiple Sclerosis Society, NSF 1817593, the Paul Allen Frontiers Group and the Howard Hughes Medical Institute. GJK is supported by an NHMRC Investigator Grant (EL1, 1175568) and previously an American Australian Association Fellowship. PAB acknowledges support from the National Institutes of Health grant R01GM061115. CMP was supported by the National Institutes of Health predoctoral training grant T32GM113770. DRL acknowledges support from US NIH U01 AI142756, RM1 HG009490, R01 EB022376, and R35 GM118062, St. Jude Collaborative Research Consortium, the Bill and Melinda Gates Foundation, and the Howard Hughes Medical Institute. MFR was supported by an HHMI Hanna Gray Fellowship. KTZ was supported by Harvard Chemical Biology Training Grant (T32 GM095450).
Footnotes
Competing interests: The Regents of the University of California have patents issued and pending for CRISPR technologies on which JAD and GJK are inventors. JAD is a co-founder of Caribou Biosciences, Editas Medicine, Scribe Therapeutics, and Mammoth Biosciences. JAD is a scientific advisory board member of Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Mammoth Biosciences, Synthego, and Inari. JAD is a Director at Johnson & Johnson and has research projects sponsored by Biogen and Pfizer. PAB is a consultant with stock options in Beam Therapeutics. DRL is a consultant and cofounder of Editas Medicine, Pairwise Plants, Beam Therapeutics, and Prime Medicine, companies that use genome editing. The authors have filed patent applications on evolved ABEs.
Data availability:
The cryo-EM map of the ABE8e-sgRNA-dsDNA complex at 3.2 Å resolution and model coordinates have been deposited to the EMDB and PDB with accession codes EMD-21308 and 6VPC. All other data generated or analyzed during this study are included in the manuscript or supporting files. Plasmids are available from Addgene.
REFERENCES
- 1.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K, Ovetsky M, Riley G, Sethi A, Tully R, Villamarin-Salomon R, Rubinstein W, Maglott DR, ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Research. 44 (2016), pp. D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR, Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR, Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 551 (2017), pp. 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolf J, tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. The EMBO Journal. 21 (2002), pp. 3841–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J, Malashkevich V, Roday S, Lisbin M, Schramm VL, Almo SC, Structural and Kinetic Characterization ofEscherichia coliTadA, the Wobble-Specific tRNA Deaminase†. Biochemistry. 45 (2006), pp. 6407–6416. [DOI] [PubMed] [Google Scholar]
- 6.Ryu S-M, Koo T, Kim K, Lim K, Baek G, Kim S-T, Kim HS, Kim D-E, Lee H, Chung E, Kim J-S, Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol 36, 536–539 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Qin W, Lu X, Liu Y, Bai H, Li S, Lin S, Precise A•T to G•C base editing in the zebrafish genome. BMC Biol. 16, 139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang P, Sun H, Zhang X, Xie X, Zhang J, Bai Y, Ouyang X, Zhi S, Xiong Y, Ma W, Liu D, Huang J, Songyang Z, Effective and precise adenine base editing in mouse zygotes. Protein Cell. 9, 808–813 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Y, Yu L, Zhang X, Xin C, Huang S, Bai L, Chen W, Gao R, Li J, Pan S, Qi X, Huang X, Zhang L, Highly efficient and precise base editing by engineered dCas9-guide tRNA adenosine deaminase in rats. Cell Discov. 4, 39 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, Zhang X, Wang L, Yin S, Zhu B, Xie L, Duan Q, Hu H, Zheng R, Wei Y, Peng L, Han H, Zhang J, Qiu W, Geng H, Siwko S, Zhang X, Liu M, Li D, Correction to: Increasing targeting scope of adenosine base editors in mouse and rat embryos through fusion of TadA deaminase with Cas9 variants. Protein Cell. 10, 700 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song C-Q, Jiang T, Richter M, Rhym LH, Koblan LW, Zafra MP, Schatoff EM, Doman JL, Cao Y, Dow LE, Zhu LJ, Anderson DG, Liu DR, Yin H, Xue W, Adenine base editing in an adult mouse model of tyrosinaemia. Nature Biomedical Engineering. 4 (2020), pp. 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levy JM, Yeh W-H, Pendse N, Davis JR, Hennessey E, Butcher R, Koblan LW, Comander J, Liu Q, Liu DR, Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat Biomed Eng. 4, 97–110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rees HA, Wilson C, Doman JL, Liu DR, Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci Adv. 5, eaax5717 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grünewald J, Zhou R, Iyer S, Lareau CA, Garcia SP, Aryee MJ, Keith Joung J, CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nature Biotechnology. 37 (2019), pp. 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richter MF, Zhao KT, Eton E, Lapinaite A, Newby GA, Thuronyi BW, W. C., Koblan LW, Zeng J, Bauer DE, Doudna JA, Liu DR, Phage-Assisted Evolution of an Adenine Base Editor with Enhanced Cas Domain Compatibility and Activity. Nat. Biotechnol. In Press (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haudenschild BL, Maydanovych O, Véliz EA, Macbeth MR, Bass BL, Beal PA, A Transition State Analogue for an RNA-Editing Reaction. Journal of the American Chemical Society. 126 (2004), pp. 11213–11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA, DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 507 (2014), pp. 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang M, Zhang L, Sun R, Zhong W, Yang Y, Chen C, Streptococcus pyogenes Cas9 displays biased one-dimensional diffusion on dsDNA to search for a target, doi: 10.1101/754564. [DOI] [Google Scholar]
- 19.Sternberg SH, LaFrance B, Kaplan M, Doudna JA, Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 527, 110–113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Losey HC, Ruthenburg AJ, Verdine GL, Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nature Structural & Molecular Biology. 13 (2006), pp. 153–159. [DOI] [PubMed] [Google Scholar]
- 21.Matthews MM, Thomas JM, Zheng Y, Tran K, Phelps KJ, Scott AI, Havel J, Fisher AJ, Beal PA, Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat. Struct. Mol. Biol 23, 426–433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vik ES, Nawaz MS, Strøm Andersen P, Fladeby C, Bjørås M, Dalhus B, Alseth I, Endonuclease V cleaves at inosines in RNA. Nat. Commun 4, 2271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao M, Hatahet Z, Melamede RJ, Kow YW, Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3’ endonuclease, from Escherichia coli. J. Biol. Chem 269, 16260–16268 (1994). [PubMed] [Google Scholar]
- 25.Dalhus B, Arvai AS, Rosnes I, Olsen ØE, Backe PH, Alseth I, Gao H, Cao W, Tainer JA, Bjørås M, Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair. Nat. Struct. Mol. Biol 16, 138–143 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang P, Xie X, Zhi S, Sun H, Zhang X, Chen Y, Chen Y, Xiong Y, Ma W, Liu D, Huang J, Songyang Z, Genome-wide profiling of adenine base editor specificity by EndoV-seq. Nat. Commun 10, 67 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Véliz EA, Easterwood LM, Beal PA, Substrate analogues for an RNA-editing adenosine deaminase: mechanistic investigation and inhibitor design. J. Am. Chem. Soc 125, 10867–10876 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Scheres SHW, RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA, MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Anders C, Niewoehner O, Duerst A, Jinek M, Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 513, 569–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD, Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD, Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution. International Tables for Crystallography (2012), pp. 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA, Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 351, 867–871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J, Malashkevich V, Roday S, Lisbin M, Schramm VL, Almo SC, Structural and kinetic characterization of Escherichia coli TadA, the wobble-specific tRNA deaminase. Biochemistry. 45, 6407–6416 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Losey HC, Ruthenburg AJ, Verdine GL, Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol 13, 153–159 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R, The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bond CS, Schüttelkopf AW, ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. D Biol. Crystallogr 65, 510–512 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM map of the ABE8e-sgRNA-dsDNA complex at 3.2 Å resolution and model coordinates have been deposited to the EMDB and PDB with accession codes EMD-21308 and 6VPC. All other data generated or analyzed during this study are included in the manuscript or supporting files. Plasmids are available from Addgene.