Abstract
The green rice leafhopper, Nephotettix cincticeps (Uhler), is an important rice pest and a vector of the rice dwarf virus in Asia. Here, we produced a high-quality chromosome-level genome assembly of 753.23 Mb using PacBio (∼110×) and Hi-C data (∼94×). It contained 163 scaffolds and 950 contigs, whose scaffold/contig N50 lengths reached 85.36/2.57 Mb. And 731.19 Mb (97.07%) of the assembly was anchored into eight pseudochromosomes. Genome completeness was attained to 97.0% according to the insect reference Benchmarking Universal Single-Copy Orthologs (BUSCO) gene set (n = 1,367). We masked 347.10 Mb (46.08%) of the genome as repetitive elements. Nine hundred sixty-two noncoding RNAs were identified and 14,337 protein-coding genes were predicted. We also assigned GO term and KEGG pathway annotations for 10,049 and 9,251 genes, respectively. Significantly expanded gene families were primarily involved in immunity, cuticle, digestion, detoxification, and embryonic development. This study provided a crucial genomic resource for better understanding on the biology and evolution in family Cicadellidae.
Keywords: Chiasmini, green rice leafhopper, insect genomics, comparative genomics, genome annotation, gene family evolution
Significance
Most species of leafhoppers (Hemiptera: Cicadellidae) are important pests of food crops. A high-quality genome could play a key role in appreciating pest biology, evolution, and devising pest control strategies. Right now, only two cicadellid genomes are currently available, including Homalodisca vitripennis and Empoasca onukii. In this study, the genome of the green rice leafhopper, Nephotettix cincticeps, was sequenced and analyzed. The chromosome-level genome of Nephotettix cincticeps provides a valuable resource for studying the phylogeny and biology of leafhoppers.
Introduction
Leafhoppers (Auchenorrhyncha: Cicadellidae) are currently the largest family of sap-sucking herbivores and comprise the most abundant known vectors of plant pathogens of any insect family in the Hemiptera (Dietrich 2013). Many species in Cicadellidae feed on economically significant plants and are thus considered as important pests, mostly because of injuries incurred to plants from direct feeding injury. And some species can transmit plant pathogens (Weintraub and Beanland 2006). The green rice leafhopper (GRLH), Nephotettix cincticeps (Uhler) (Cicadellidae: Deltocephalinae), is a potent pest of rice. It is widely distributed among rice-producing areas in Asia, and able to transmit rice viral pathogens, notably the rice dwarf virus, in a persistent-propagative manner during the sap-sucking process. The facilitated infection can severely damage host rice plants, manifesting as stunted growth, white chlorotic spots on leaves, and delayed panicle development. In this way, outbreaks of Nephotettix cincticeps have caused serious losses in rice yield and quality in China, Japan, Korea, the Philippines, and Nepal (Ruan et al. 1981; Zheng et al. 1997; Zhu et al. 2005; Honda et al. 2007; Wei and Li 2016; Jia et al. 2021). Accordingly, access to a high-quality GRLH genome could play a fundamental role in studies of pest biology, evolution, and control. To date, only two cicadellid genomes, Homalodisca vitripennis (Germar) (Cicadellidae: Deltocephalinae) and Empoasca onukii Matsuda (Cicadellidae: Deltocephalinae), are available on NCBI GenBank (accessed July 10, 2021), whose genome sizes are 1.45 Gb and 599.26 Mb, respectively. Furthermore, although a chromosome-level genome assembly of Empoasca onukii was uploaded (GCA_018831715.1), the annotations are unavailable to the public. On the other hand, the scaffold N50 length of the Homalodisca vitripennis assembly (GCA_000696855.2) was smaller than 50 kb, indicating the low-quality assembly contiguity. In this study, we provided a de novo chromosome-level genome assembly of Nephotettix cincticeps using PacBio long reads and Hi-C sequencing. We annotated its protein-coding genes, as well as repetitive elements and noncoding RNAs (ncRNAs). Gene family evolution was analyzed across the main hemipteran groups. Furthermore, chromosomal syntenic correspondence was investigated between Nephotettix cincticeps and the brown planthopper Nilaparvata lugens (Stål) (Delphacidae: Delphacinae) to reveal their chromosomal evolution.
Results and Discussion
Genome Assembly
Sequencing platforms generated: 93.62 Gb (143×) of Illumina short reads, 85.76 Gb (110×) of PacBio long reads, 70.80 Gb (94×) of Hi-C data for the genome assembly, and 10.87 Gb of transcriptome data used for annotations. After implementing the quality control, 97.82 Gb of Illumina reads were retained for the analyses of genome survey and genome polishing. The genome survey indicated that the sequenced strain had an approximate genome size of 720 Mb (718.12‒724.33) and showed a very high heterozygosity (1.30‒1.38%).
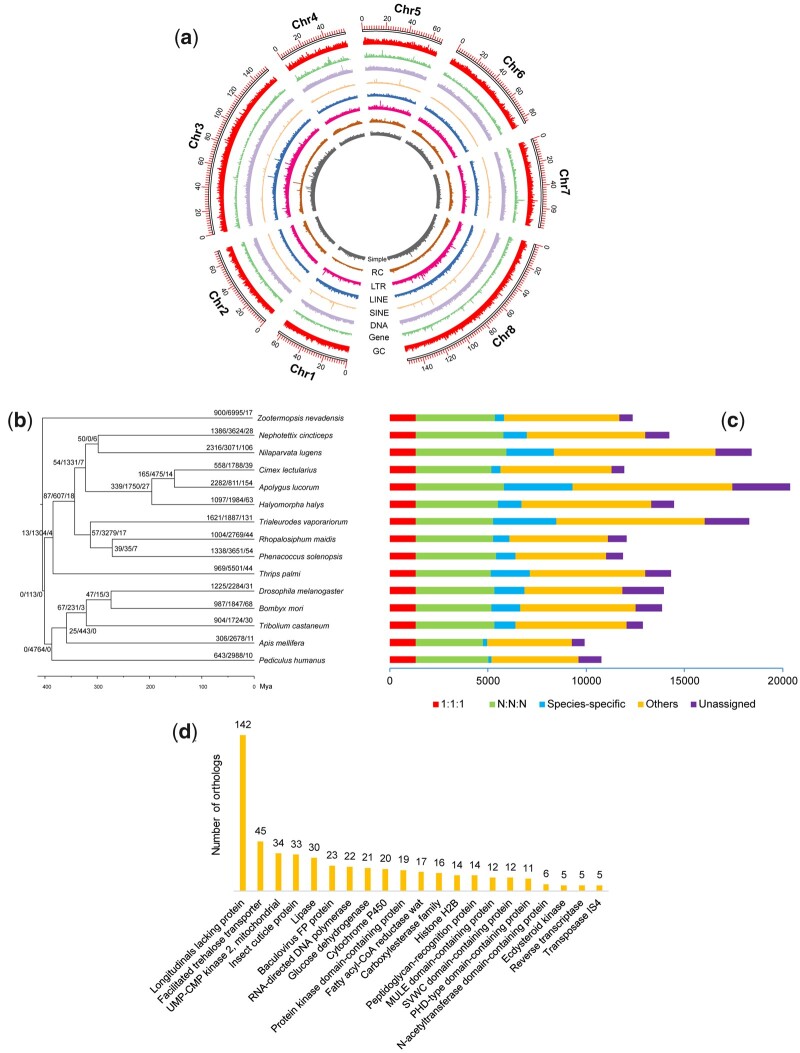
The 85.76 Gb (110×) of PacBio long reads had a N50 of 25.17 kb and a mean length of 14.39 kb. After self-correcting, 56.61 Gb (75×) of Raw PacBio long reads were generated. These corrected reads had a N50 and a mean length of 26.84 and 22.00 kb, respectively. After initial Raven assembly, polishing, redundancy removal, Hi-C scaffolding, and contaminant detection, we generated a high-quality contiguous chromosome-level genome assembly (table 1).The final assembly had a length of 753.23 Mb, comprising 163 scaffolds and 950 contigs, with a scaffold/contig N50 length of 85.36/2.57 Mb, a GC content of 34.48%, and a gap ratio of 0.01%. Among them, 703 contigs (97.07%, 731.19 Mb) were anchored into eight pseudochromosomes (fig. 1). Benchmarking Universal Single-Copy Orthologs (BUSCO) completeness reached 97.0% (1.8% complete and duplicated, 2.0% fragmented, 1.0% missing). High mapping rates for both Illumina (95.65%) and PacBio (97.87%) reads also confirmed the integrity of our assembly. A clean Hi-C contact heatmap (supplementary fig. S1, Supplementary Material online) and low ratio of BUSCO duplicates (1.8%) indicated that no obvious redundant regions were observed within the assembly. When compared with the two available Cicadellidae genomes, the genome size of Nephotettix cincticeps (Deltocephalinae) was slightly larger than that of Empoasca onukii (Typhlocybinae, 599.26 Mb) but much smaller than that of Homalodisca vitripennis (Cicadellinae, 1.45 Gb). This indicated that cicadellid genome sizes can vary greatly among subfamilies of leafhoppers.
Table 1.
Genome Assembly and Annotation Statistics for Nephotettix cincticeps
| Content | Nephotettix cincticeps |
|---|---|
| Genome assembly | |
| Assembly size (Mb) | 753.23 |
| Number of scaffolds/contigs | 163/950 |
| Longest scaffold/contig (Mb) | 154.23/17.07 |
| N50 scaffold/contig length (Mb) | 85.36/2.57 |
| GC content (%) | 34.48 |
| Gaps (%) | 0.01 |
| BUSCO completeness (%) | 97.0 |
| Single copy (%) | 95.2 |
| Duplicated (%) | 1.8 |
| Fragmented (%) | 2.0 |
| Missing (%) | 1.0 |
| Protein-coding genes | |
| Number | 14,337 |
| Mean gene length (bp) | 17,386.5 |
| Gene ratio (%) | 33.42 |
| Exons/introns/CDS per gene | 9.5/8.2/9.2 |
| Exon/intron/CDS ratio (%) | 4.49/28.92/3.29 |
| Mean exon/intron/CDS length (bp) | 245.3/1,827.5/185.3 |
| Genes with GO/KEGG pathway annotations | 10,049/9,251 |
| Repetitive elements | 347.10 Mb (46.08%) |
| Number of ncRNAs | 962 |
Fig. 1.
Genome characteristics of Nephotettix cincticeps. (a) Circos tracks showing element distributions in 100-kb sliding windows from outer to inner: chromosome length, GC content, density of protein-coding genes, DNA transposons, SINE/LINE/LTR retrotransposons, rolling-circle, and simple repeats. (b) Gene family evolution and statistics of orthologs. Node values represent the number of expanded, contracted, and rapidly evolving families; “1:1:1” denoted the shared single-copy genes, “N:N:N” indicates multicopy genes shared by all species, “Others” are the unclassified orthologs, “Unassigned” are orthologs that cannot be assigned to any orthogroups. (c) Significantly expanded families. Families lacking functional annotations are not shown. Functional enrichment of GO (d) and KEGG (e) categories for significantly expanded gene families.
Genome Annotation
Among the genome, 347.10 Mb (46.08%) was masked as repetitive elements. The dominant repeat categories were unclassified (21.10%), retrotransposons (10.70%), DNA transposons (7.63%), rolling-circles (5.26%), and simple repeats (1.21%) (fig. 1a and supplementary table S1, Supplementary Material online). Within the retrotransposons, a large portion were LINE (6.66%) and LTR (3.61%) elements, particularly the families L2 (3.21%) and Gypsy (2.49%). Notably, the retrotransposon L2 elements act as a source of functional micro-RNAs (miRNAs) and target sites (Piriyapongsa et al. 2007; Spengler et al. 2014). The families TcMar-Tc1 (3.44%), TcMar-Mariner (1.27%), and TcMar-Tigger (0.89%) accounted for a major part of the DNA transposons. Both TcMar-Tc1 and TcMar-Mariner, often called “parasitic” mobile elements (Capy et al. 2000), can contribute to the evolution of more complex genomes (Lynch and Conery 2003) such as more mobile elements, larger genome sizes (Liu et al. 2016), or horizontal transmission (Lohe et al. 1995; Lampe et al. 2003). These transposons may play important roles in the adaptations of insect taxa to a wide range of environmental conditions.
Overall, we identified 962 ncRNAs: 60 ribosomal RNAs (rRNAs), 60 miRNAs, 130 small nuclear RNAs (snRNAs), three long noncoding RNAs (lncRNAs), 408 tRNAs (21 isotypes), five riboswitches, eight ribozymes, and 288 other ncRNAs (supplementary table S2, Supplementary Material online). The snRNAs were classified into 96 spliceosomal RNAs in six groups (U1, U2, U4, U5, U6, and U11), five minor spliceosomal RNAs in three groups (U4atac, U6atac, and U12), 25 C/D box small nucleolar RNAs (snoRNAs), three H/ACA box snoRNAs, and one other snoRNA.
We predicted 14,337 protein-coding gene models from 16,817 sequences (isoform included) with a mean length of 17,386.5 bp, for which the gene content accounted for 33.42% of the genome (table 1). The average number of exons/introns/CDS per gene was 9.5/8.2/9.2 and their mean length was 245.3/1827.5/185.3 bp. The BUSCO completeness assessment (in protein mode) for protein sequences reached 96.0%, indicating the high quality of these predictions of protein-coding genes. Of the 14,337 genes, 13,183 (91.95%), 11,823 (82.46%), and 12,337 (86.05%) genes respectively matched the UniprotKB, InterProScan, and eggNOG records. After integrating the above annotation results, 10,049, 9,251, 10,386, 2,827, and 11,663 genes were assigned to gene ontology (GO) terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, Reactome pathways, Enzyme Codes, and COG categories, respectively.
Gene Family Evolution
We selected 15 Pterygota (Insecta: Dicondylia) species including eight hemipterans used for gene family inference, and obtained 15,591 orthogroups (gene families) comprising 189,907 (90.47%) genes. Among them, 4,172 orthogroups and 20,685 genes were assigned as being species-specific; another 4,035 were orthogroups present in all species and 1,328 consisted of single-copy genes (fig. 1c). For the Nephotettix cincticeps, 13,125 (91.54%) genes were assigned to 8,856 orthogroups, of which 235 orthogroups and 1,204 genes were species-specific genes.
The reconstructed phylogenetic tree of all 15 species based on 747 482 amino acid sites was similar with that reported by Misof et al. (2014) (fig. 1b). Thysanoptera was a sister to Hemiptera but Psocodea was clustered with Holometabola insects. Eight hemipteran species were separated into three clades (Sternorrhyncha + (Auchenorrhyncha + Heteroptera)). The Hemiptera was diverged at the middle Mississippian period (339.5–346.8 Ma). Two Auchenorrhyncha taxa species (Fulgoroidea and Membracoidea) were later separated in the early Permian period (296.3–299.6 Ma).
Gene family evolution analyses revealed that 1,386 and 3,624 gene families underwent expansions and contractions, respectively. Of them, 28 gene families were significantly expanded without incurring any significantly contracted ones (fig. 1b). These significantly expanded families were primarily associated with immunity, cuticle, digestion, detoxification, and embryonic development (fig. 1d). Immunity related families constitute the main components of insect antimicrobial defenses, such as Peptidoglycan-recognition protein (14) and SVWC domain-containing protein (12) (Royet and Dziarski 2007; Chen et al. 2011; Royet et al. 2011). Detoxification-related families included cytochrome P450 (20), carboxylesterase (16), and ecdysteroid kinase (5). Interestingly, the fatty acyl-CoA reductase wat genes are essential for gas filling, wax ester synthesis and hydrophobic tracheal coating (Jaspers et al. 2014). Those may be linked to the production of brochosomes in leafhoppers, a process similar to the production of epidermal wax blooms in other sap-sucking hemipterans, which can protect them from entrapment by their exudates (Rakitov and Gorb 2013). Functional enrichment for those significantly expanded gene families also reflected strong representation in immunity and cuticle constituent, particularly in terms of those GO/KEGG items numbering higher than 20 (supplementary figs. S2 and S3, Supplementary Material online).
Materials and Methods
Sample Collection and Sequencing
The specimens Nephotettix cincticeps used for sequencing were collected from rice plants in the Chunhua Modern Agriculture Demonstration Garden, Jiangning district, Nanjing, China, on September 12, 2020. Female adult individuals were washed using ddH2O for DNA and RNA sequencing: seven for Illumina and PacBio whole-genome sequencing, two for transcriptome sequencing, and one for Hi-C sequencing, respectively. Genomic DNA was extracted using the Qiagen Blood & Cell Culture DNA Mini Kit. Libraries consisting of 350-bp and 40-kb insert sizes were constructed using the TruSeq DNA PCR-Free LT Library Preparation Kit and the SMRTbell DNA Template Prep Kit 2.0, and then sequenced on the HiSeq NovaSeq 6000 and PacBio Sequel II platforms. RNA was extracted using the TRIzol Reagent and a library was constructed using the TruSeq RNA v2 Kit. For the Hi-C sequencing, DNA preparation (crosslinking, digesting using the restriction enzyme MboI, ligation, etc.), library construction, and sequencing, as well as other library preparations and sequencing tasks were performed at Berry Genomics (Beijing, China).
Genome Assembly
Quality control of the obtained Illumina clean reads was performed using BBTools suite v38.82 (Bushnell 2014): the script “clumpify.sh” removed any duplicates; “bbduk.sh” carried out the quality trimming (>Q20), length filtering (>15 bp), polymer trimming (>10 bp), and the corrected any overlapping paired reads. To assess the genome characteristics, we surveyed the genome using GenomeScope v2.0 (Ranallo-Benavidez et al. 2020). The k-mer frequency distributions were estimated with 21-mers, based short reads, using the script “khist.sh” (BBTools). The maximum k-mer coverage cutoff was set as 10,000.
Raw PacBio long reads which were longer than 10 kb were self-corrected, using NextDenovo v2.3.1 (https://github.com/Nextomics/NextDenovo), and assembled in Raven v1.3.1 software under its default parameters (Vaser and Šikić 2021). The resulting assembly was then polished with two rounds of Illumina reads, by using NextPolish v1.3.1 (Hu et al. 2020), after which redundant haplotypic duplication was removed by Purge_Dups v1.0.1 (Guan et al. 2020), using a minimum alignment score of 60 (-a 60). Minimap2 v2.17 (Li 2018) was used as a sequence aligner in the polishing and purging steps.
For the Hi-C data set, its alignment to the genome, removal of duplicates, and mining of Hi-C contacts were all conducted in Juicer v1.6.2 (Durand et al. 2016). To generate pseudochromosomes, we used the 3D-DNA v180922 pipeline (Dudchenko et al. 2017) to correct misassemblies present, and to anchor, order, and orient the contigs. Possible assembly errors produced in the first 3D-DNA round were manually corrected, using the Assembly Tools module within Juicebox (Durand et al. 2016), and the pseudochromosomes refined further in a second 3D-DNA round.
We removed potential contaminants through BlastN-like MMseqs2 v11-e1a1c (Steinegger and Söding 2017) searches against the UniVec and NCBI nucleotide (nt) databases. Besides the assembly contiguity indicators, we also evaluated assembly quality in terms of genome completeness and read mapping rate. The BUSCO v5.0.0 pipeline (Manni et al. 2021) was implemented to assess genome completeness against the insect gene set (insecta_odb10, n = 1,367). Finally, all raw PacBio and Illumina reads were mapped to the genome using Minimap2 and the corresponding mapping rate estimated by Samtools v1.9 (Danecek et al. 2021).
Genome Annotations
Three essential genomic elements, namely protein-coding genes, repetitive elements, and ncRNAs, were annotated for the GRLH genome. We used RepeatMasker v4.1.0 (Smit et al. 2013–2015) to mask repetitive elements based on a custom repeat library, which included a de novo library and the Dfam 3.1 (Hubley et al. 2016) and RepBase-20181026 databases (Bao et al. 2015). That de novo repeat library was built using RepeatModeler v2.0.1 (Flynn et al. 2020), with additional LTR discovery pipeline activated (-LTRStruct). Next, we identified the ncRNAs using Infernal v1.1.3 (Nawrocki and Eddy 2013) and tRNAscan-SE v2.0.7 (Chan and Lowe 2019). To reduce potential errors (e.g., pseudogenes), we retained only high-confidence tRNAs by using the tRNAscan-SE built-in script: “EukHighConfidenceFilter.”
Protein-coding gene models were predicted via the MAKER v3.01.03 pipeline (Holt and Yandell 2011), by integrating ab initio, transcriptome, and protein homology-based evidence. Ab initio gene models were predicted using BRAKER v2.1.5 (Brůna et al. 2021), which integrated two ab initio predictor tools, Augustus v3.3.4 (Stanke et al. 2004) and GeneMark-ES/ET/EP 4.59_lic (Brůna et al. 2020), thereby simultaneously incorporating transcriptome and protein homology evidence. To align the transcriptome data to the genome, we used HISAT2 v2.2.0 (Kim et al. 2019) to generate BAM alignments, with arthropod reference proteins retrieved from the OrthoDB10 v1 database (Kriventseva et al. 2019). Transcripts fed into MAKER were assembled using the genome-guided assembler StringTie v2.1.4 (Kovaka et al. 2019). The protein sequences passed on to MAKER as evidence of protein homology were downloaded from NCBI for five species: Apis mellifera, Drosophila melanogaster, Thrips palmi, Nilaparvata lugens, and Rhopalosiphum maidis. Gene functions were assigned by searching the UniProtKB database, using Diamond v0.9.24 (Buchfink et al. 2015) in its more sensitive mode and an e-value of 1e-5. Protein domains, GO terms, and pathways (KEGG, Reactome) were assigned by applying eggNOG-mapper v2.0.1 (Huerta-Cepas et al. 2017) against the eggNOG v5.0 database (Huerta-Cepas et al. 2019) and likewise InterProScan 5.47–82.0 (Finn et al. 2017) against five databases: Pfam (El-Gebali et al. 2019), Superfamily (Wilson et al. 2009), Gene3D (Lewis et al. 2018), SMART (Letunic and Bork 2018), and CDD (Marchler-Bauer et al. 2017) databases.
Gene Family Evolution
Besides Nephotettix cincticeps, to carry out gene family and evolution analyses, we downloaded high-quality nonredundant protein sequences from the NCBI of 14 insect species: one Polyneoptera member (Zootermopsis nevadensis), four belonging to Endopterygota (Apis mellifera, Bombyx mori, Drosophila melanogaster, Tribolium castaneum), one Psocodea member (Pediculus humanus), one Thysanoptera member (Thrips palmi), and eight belonging to Hemiptera (Apolygus lucorum, Cimex lectularius, Halyomorpha halys, Nephotettix cincticeps, Nilaparvata lugens, Phenacoccus solenopsis, Rhopalosiphum maidis, Trialeurodes vaporariorum). Sequence orthology were inferred using OrthoFinder v2.3.8 (Emms and Kelly 2019) for which Diamond served as the sequence aligner.
Single-copy orthologs inferred from OrthoFinder were used to reconstruct the insect phylogeny. For each ortholog, the protein sequences were aligned by MAFFT v7.450 (Katoh and Standley 2013) in the L-INS-I mode. Unreliable homologous regions within the alignment were stringently trimmed, using BMGE v1.12 (Criscuolo and Gribaldo 2010) (-m BLOSUM90 -h 0.4). The phylogeny of 15 species was then reconstructed using IQ-TREE v2.0.7 (Minh et al. 2020), with the following set of parameters: “-m MFP –mset LG –msub nuclear –rclusterf 10 -B 1,000 –alrt 1,000 –symtest-remove-bad –symtest-pval 0.10.” The ensuing tree was fed into MCMCTree within the PAML v4.9j package (Yang 2007) as the guide tree, to estimate the divergence times. We selected six fossils from the PBDB database (https://www.paleobiodb.org/navigator/) for conducting the stem node calibration: root (Pterygota <443.4 Ma), Holometabola (>382.7 Ma), Coleoptera (311.4‒323.2 Ma), Hemiptera (314.6‒323.2 Ma), Aphidomorpha (279.3‒298.9 Ma), and Cicadomorpha (298.9‒307.0 Ma).
Gene family evolution (expansions and contractions) was inferred using CAFÉ v4.2.1 (Han et al. 2013), setting the significance level to 0.01 for the model of single birth–death parameter “lambda.” We next performed GO and KEGG functional enrichment analyses for those significantly expanded families, this done using the R package “clusterProfiler” v3.14.3 (Yu et al. 2012). For the enrichment scores, their significance level was set to 0.01 (P value) with a false discovery rate control cut-off of 0.05 (q value).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to Dr Feng Zhang (Nanjing Agriculture University, China) and Dr Haoxi Li (Guizhou University, China) for revising the manuscript. This work was supported by the Science and Technology Project of Guangxi Zhuang Autonomous Region Branch Company of China Tobacco Company (Grant No. 2021–02 and Contract No. 202145000024006) and the Program of Excellent Innovation Talents, Guizhou Province (Grant No. (2016) –4022).
Data Availability
Genome assembly and raw-sequencing data have been deposited at NCBI under the accession numbers JAGISJ000000000 and SRR14340695‒SRR14340698, respectively. Genome annotations are available on FigShare at this weblink: https://doi.org/10.6084/m9.figshare.15044001.v1.
Literature Cited
- Bao WD, Kojima KK, Kohany O.. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 6(11):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brůna T, et al. 2021. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a Protein database. NAR Genomics Bioinform. 3(1):lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brůna T, Lomsadze A, Borodovsky M.. 2020. GeneMark-EP+: eukaryotic gene prediction with self-training in the space of genes and proteins. NAR Genomics Bioinform. 2(2):lqaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60. [DOI] [PubMed] [Google Scholar]
- Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner (No. LBNL-7065E). Berkeley (CA): Lawrence Berkeley National Lab (LBNL). Available from: https://sourceforge.net/projects/bbmap/. Accessed June 22, 2020.
- Capy P, Gasperi G, Biémont C, Bazin C.. 2000. Stress and transposable elements: co-evolution or useful parasites? Heredity 85(2):101–106. [DOI] [PubMed] [Google Scholar]
- Chan PP, Lowe TM.. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, et al. 2011. Identification and functional characterization of Dicer2 and five single VWCdomain proteins of Litopenaeus vannamei. Dev Comp Immunol. 35(6):661–671. [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S.. 2010. BMGE (Block Mapping and Gathering with Entropy): selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. 2021. Twelve years of SAMtools and BCFtools. GigaScience 10(2):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich CH. 2013. Overview of the phylogeny, taxonomy and diversity of the leafhopper (Hemiptera: Auchenorrhyncha: Cicadomorpha: Membracoidea: Cicadellidae) vectors of plant pathogens. Proceedings of the 2013 International Symposium on Insect Vectors and Insect-Borne Diseases. Taiwan Agricultural Research Institute Special Publication No. 173, Taichung.
- Dudchenko O, et al. 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356(6333):92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, et al. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3(1):95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, et al. 2019. The Pfam protein families database in 2019. Nucleic Acids Res. 47(D1):D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2017. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 45(D1):D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J, et al. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 117(17):9451–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, et al. 2020. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics 36(9):2896–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Thomas G, Lugo-Martinez J, Hah MW.. 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 30(8):1987–1997. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12(1):491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, et al. 2007. Retention of rice dwarf virus by descendants of pairs of viruliferous vector insects after rearing for 6 years. Phytopathology 97(6):712–716. [DOI] [PubMed] [Google Scholar]
- Hu J, et al. 2020. NextPolish: a fast and efficient genome polishing tool for long read assembly. Bioinformatics 36(7):2253–2255. [DOI] [PubMed] [Google Scholar]
- Hubley R, et al. 2016. The Dfam database of repetitive DNA families. Nucleic Acids Res. 44(D1):D81–D89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 34(8):2115–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47(D1):D309–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers MH, et al. 2014. The fatty acyl-CoA reductase Waterproof mediates airway clearance in Drosophila. Dev Biol. 385(1):23–31. [DOI] [PubMed] [Google Scholar]
- Jia WJ, et al. 2021. Identification and characterization of a novel rhabdovirus in green rice leafhopper, Nephotettix cincticeps. Virus Res. 296:198281. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, et al. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37(8):907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S, et al. 2019. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20(1):278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, et al. 2019. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 47(D1):D807–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe DJ, Witherspoon DJ, Soto-Adames FN, Robertson HM.. 2003. Recent horizontal transfer of mellifera subfamily mariner transposons into insect lineages representing four different orders shows that selection acts only during horizontal transfer. Mol Biol Evol. 20(4):554–562. [DOI] [PubMed] [Google Scholar]
- Letunic L, Bork P.. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46(D1):D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis T, et al. 2018. Gene3D: extensive prediction of globular domains in proteins. Nucleic Acids Res. 46(D1):D435–D439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, et al. 2016. The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nat Commun. 7:11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe AR, Moriyama EN, Lidholm DA, Hartl DL.. 1995. Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol Biol Evol. 12(1):62–72. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2003. The origins of genome complexity. Science 302(5649):1401–1404. [DOI] [PubMed] [Google Scholar]
- Manni M, et al. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 38(10):4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45(D1):D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, et al. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346(6210):763–767. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR.. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29(22):2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J, Mariño-Ramírez L, Jordan IK.. 2007. Origin and evolution of human microRNAs from transposable elements. Genetics 176(2):1323–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakitov R, Gorb SN.. 2013. Brochosomes protect leafhoppers (Insecta, Hemiptera, Cicadellidae) from sticky exudates. J R Soc Interface. 10(87):20130445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo-Benavidez TR, Jaron KS, Schatz MC.. 2020. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 11(1):1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royet J, Dziarski R.. 2007. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 5(4):264–277. [DOI] [PubMed] [Google Scholar]
- Royet J, Gupta D, Dziarski R.. 2011. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat Rev Immunol. 11(12):837–851. [DOI] [PubMed] [Google Scholar]
- Ruan YL, et al. 1981. Studies on rice dwarf virus disease: I. The history, symptoms and transmission. Acta Phytophyla Sin. 8(1):27–34. [Google Scholar]
- Smit AFA, Hubley R, Green P.. 2013. –2015. RepeatMasker Open-4.0. Available from: http://www.repeatmasker.org. Accessed September 17, 2020.
- Spengler RM, Oakley CK, Davidson BL.. 2014. Functional microRNAs and target sites are created by lineage-specific transposition. Hum Mol Genet. 23(7):1783–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Steinkamp R, Waack S, Morgenstern B.. 2004. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 32(Web Server Issue):W309–W312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinegger M, Söding J.. 2017. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol. 35(11):1026–1023. [DOI] [PubMed] [Google Scholar]
- Vaser R, Šikić M.. 2021. Time- and memory-efficient genome assembly with Raven. Nat Comput Sci. 1(5):332–336. [DOI] [PubMed] [Google Scholar]
- Wei T, Li Y.. 2016. Rice reoviruses in insect vectors. Annu Rev Phytopathol. 54:99–102. [DOI] [PubMed] [Google Scholar]
- Weintraub PG, Beanland L.. 2006. Insect vectors of phytoplasmas. Annu Rev Entomol. 51(1):91–111. [DOI] [PubMed] [Google Scholar]
- Wilson D, et al. 2009. SUPERFAMILY—sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 37(Database Issue):D380–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng HH, et al. 1997. Recovery of transgenic rice plans expressing the rice dwarf virus outer coat protein (S8). Theor Appl Genet. 94(3):522–527. [Google Scholar]
- Zhu SF, et al. 2005. The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiol. 139(4):1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome assembly and raw-sequencing data have been deposited at NCBI under the accession numbers JAGISJ000000000 and SRR14340695‒SRR14340698, respectively. Genome annotations are available on FigShare at this weblink: https://doi.org/10.6084/m9.figshare.15044001.v1.