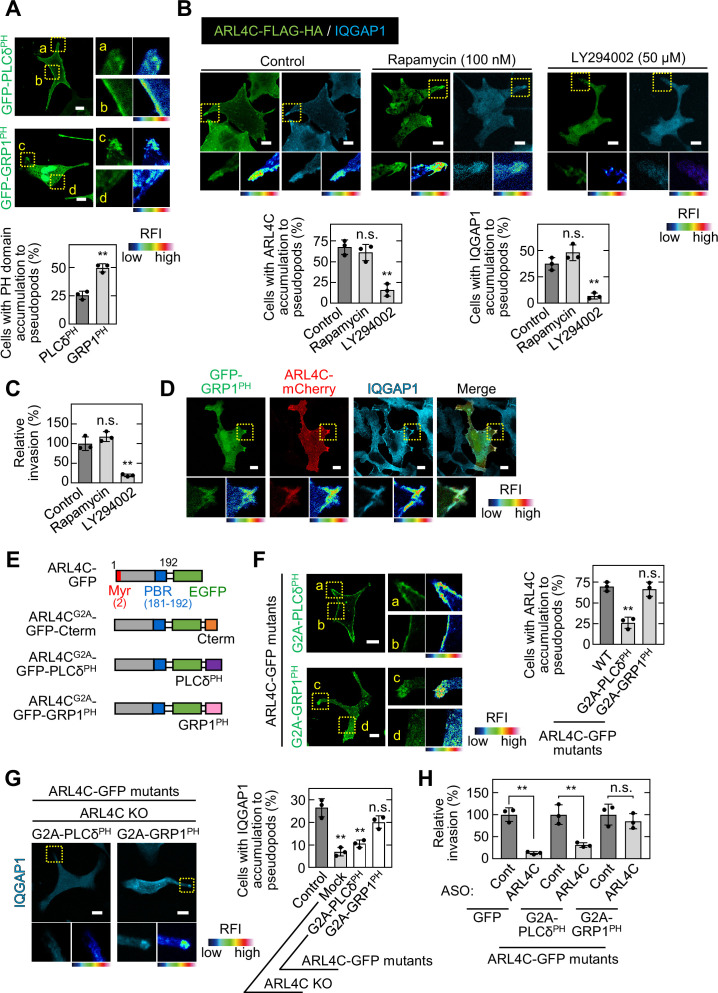
Figure 5. ARL4C recruits IQGAP1 to invasive pseudopods in a PIP3-dependent manner.
(A) S2-CP8 cells were transfected with GFP-PLCδPH or GFP-GRP1PH. The percentages of cells with GFP-PLCδPH or GFP-GRP1PH accumulated at invasive pseudopods compared with the total number of cells were calculated. (B) S2-CP8 cells expressing FRB-CFP, mRFP-FKBP-5-ptase domain, and ARL4C-FLAG-HA were treated with or without rapamycin or LY294002 and stained with anti-HA and anti-IQGAP1 antibodies. The percentages of cells with IQGAP1 or ARL4C-FLAG-HA accumulated at invasive pseudopods compared with the total number of cells were calculated. (C) S2-CP8 cells expressing FRB-CFP and mRFP-FKBP-5-ptase domain were treated with or without rapamycin or LY294002 and subjected to an invasion assay. Invasive abilities are expressed as the percentage of control cells. (D) S2-CP8 cells expressing ARL4C-mCherry and GFP-GRP1PH were stained with anti-IQGAP1 antibody. Images of GFP-GRP1PH, ARL4C-mCherry, and IQGAP1 were merged. (E) A schematic representation of ARL4C-GFP mutants is shown. (F) S2-CP8 cells were transfected with the indicated mutants of ARL4C-GFP. The percentages of cells with ARL4C-GFP mutant accumulated at invasive pseudopods compared with the total number of cells were calculated. (G) ARL4C KO cells expressing control or the indicated mutants of ARL4C-GFP were stained with anti-IQGAP1 antibody. Quantification was performed as in (B). (H) S2-CP8 cells stably expressing GFP or the indicated mutants of ARL4C-GFP were transfected with control or ARL4C ASO and subjected to an invasion assay. Invasive abilities are expressed as the percentage of the same cells transfected with control ASO. (A,F) Enlarged images of the regions in the yellow dashed squares and a false color representation of fluorescence intensity are shown on the right. (a) and (c) show the pseudopods, and (b) and (d) show the cell body. (B,D,G) The regions in the yellow dashed squares are shown enlarged in the left bottom images. The right bottom images are shown in a false color representation of fluorescence intensity. (A–C,F–H) Data are shown as the mean ± s.d. of three biological replicates. p Values were calculated using a two-tailed Student’s t-test (A,H) or one-way ANOVA followed by Bonferroni post hoc test (B,C,F,G). (A,B,D,F,G) False color representations were color-coded on the spectrum. Scale bars in (A,B,D,F,G) 10 μm. KO, knockout. RFI, relative fluorescence intensity. n.s., not significant. **, p < 0.01. See Figure 5—source data 1.