Abstract
The nuclear receptor peroxisome proliferator-activated receptor γ regulates adipose differentiation and systemic insulin signaling via ligand-dependent transcriptional activation of target genes. However, the identities of the biologically relevant target genes are largely unknown. Here we describe the isolation and characterization of a novel target gene induced by PPARγ ligands, termed PGAR (for PPARγ angiopoietin related), which encodes a novel member of the angiopoietin family of secreted proteins. The transcriptional induction of PGAR follows a rapid time course typical of immediate-early genes and occurs in the absence of protein synthesis. The expression of PGAR is predominantly localized to adipose tissues and placenta and is consistently elevated in genetic models of obesity. Hormone-dependent adipocyte differentiation coincides with a dramatic early induction of the PGAR transcript. Alterations in nutrition and leptin administration are found to modulate the PGAR expression in vivo. Taken together, these data suggest a possible role for PGAR in the regulation of systemic lipid metabolism or glucose homeostasis.
The past several years have witnessed an increasing recognition of the adipocyte as a remarkably dynamic entity that uses diverse signaling pathways to interact with other tissues. A compelling body of evidence now implicates adipocyte-derived proteins such as leptin (25a, 31) and tumor necrosis factor α (10) as endocrine and/or autocrine modulators of distant and local targets. This enables the adipose tissue to exercise feedback regulation over systemic energy homeostasis and metabolism.
In parallel with the improved functional understanding of the adipocyte, there has been a burgeoning interest in studying the molecular control of adipose differentiation. Research efforts directed at the transcriptional regulation of adipogenesis have led to the elucidation of several transcription factors that play key roles in this process, including the peroxisome proliferator-activated receptor γ (PPARγ) (26) and the CCAAT/enhancer binding protein (6) family members. PPARγ, a member of the PPAR subfamily of nuclear hormone receptors, is a ligand-dependent transcription factor expressed in a tissue-selective manner, with the highest levels in the adipose tissue. Much evidence from gain- and loss-of-function studies indicates that PPARγ is a central regulator of adipogenesis and systemic insulin action (reviewed in references 15 and 24), providing a crucial link between these two major aspects of adipocyte biology. PPARγ has been demonstrated to stimulate adipose conversion in a variety of fibroblastic cell lines ectopically expressing PPARγ, as well as in preadipocyte and mesenchymal precursor cell lines (26). Committed myoblasts stably infected with PPARγ and CCAAT/enhancer binding protein α can be induced to undergo transdifferentiation into adipocytes upon PPARγ activation (11). More recently, loss-of-function experiments using PPARγ−/− mice or embryonic stem cells have confirmed the requirement of PPARγ for adipogenesis in vivo and in vitro (2, 13, 20). PPARγ has been shown to heterodimerize with retinoid X receptor and to direct gene expression through response elements in the promoters of the adipocyte fatty acid binding protein (aP2), lipoprotein lipase (LPL), and phosphoenolpyruvate carboxykinase, highlighting its role as a mediator of terminal adipose differentiation (22, 27).
Naturally occurring PPARγ ligands reported to date, such as 15-d-prostaglandin J2 and other prostaglandin J derivatives, several mono- and polyunsaturated fatty acids, oxidized lipids, and fibrates, all bind PPARγ at relatively low affinities (9, 12). In addition, certain members of the thiazolidinedione (TZD) class of synthetic antidiabetic drugs are high-affinity, isoform-selective ligands for PPARγ, and within this group, there exists a strong correlation between the affinity for PPARγ and the potency of antidiabetic action (9, 14, 29). Two heterozygous mutations in the ligand-binding domain of PPARγ have been reported in three human subjects with severe insulin resistance and type 2 diabetes mellitus (3), suggesting that reduced PPARγ function can impair the control of insulin sensitivity.
The systemic nature of the therapeutic response to the TZDs may be explained by direct actions through the relatively low but significant levels of PPARγ present in other insulin-sensitive tissues, such as muscle or liver, or by the existence of heretofore unidentified fat-produced signaling molecules that impinge upon other tissues. The underlying molecular mechanism for the insulin sensitizing action of the TZDs is not understood at present but could involve a higher level expression of certain adipocyte genes, e.g., the insulin-sensitive glucose transporter Glut4, reduction of lipolysis and consequent lowering of circulating free fatty acids, functional antagonism of tumor necrosis factor α action, and/or additional targets yet to be identified (24). Likewise, the transcriptional target genes of PPARγ responsible for setting the adipogenic program in motion have not been fully determined.
With the ultimate goal of enhancing the current understanding of the biological functions of PPARγ, we have initiated studies aimed at identifying additional downstream target genes of PPARγ, based on a subtractive cloning strategy. We report here the primary structure of a novel PPARγ target gene thus isolated, termed PGAR, and describe the expression pattern and regulation data. As a novel, secreted protein produced by fat and a bona fide target of PPARγ, PGAR is a potential contributor to systemic metabolic processes.
MATERIALS AND METHODS
Cell culture.
NIH 3T3 fibroblasts were grown in 10% bovine calf serum (Hyclone)–Dulbecco's modified Eagle medium (DMEM) (Life Technologies) and were stably infected with retroviruses carrying either pBabe-PPARγ or an empty pBabe vector as previously described (26). Briefly, BOSC23 cells were transiently transfected with pBabe-derived expression vectors and the resulting supernatants containing recombinant virus were transferred to the NIH 3T3 fibroblasts, which were subsequently replated and selected in puromycin.
Subtractive hybridization.
These two stable cell lines were cultured to confluence in 10% Cosmic calf serum (Hyclone)–DMEM and treated with pioglitazone (10 μg/ml; Upjohn) dissolved in dimethyl sulfoxide for a period of 3 h. Total RNA was isolated from cultured cells by the acid guanidium thiocyanate method, and the poly(A)+ RNA was prepared from total RNA with PolyATract mRNA isolation system (Promega). The double-stranded cDNA synthesized from 3 μg of poly(A)+ RNA was then digested with RsaI and used for multiple rounds of subtractive hybridization and PCR amplification according to PCR-Select cDNA subtraction protocol (Clontech).
Differential screening and Northern analysis.
The enriched cDNAs were cloned into pCR2.1 vector (Invitrogen) and screened by slot blot analysis for differentially regulated genes. Plasmid DNA from three hundred random clones were spotted onto nylon membranes using a vacuum manifold apparatus (Schleicher & Schuell) and hybridized with α-32P-labeled cDNA probes prepared by reverse transcription from the two poly(A)+ RNA samples used above. Northern blotting was then performed on those clones judged to be differentially regulated on the slot blots. Twenty micrograms of total RNA prepared for the subtraction experiment was fractionated in a 1% agarose-formaldehyde gel, transferred onto nylon, and hybridized with the EcoRI-excised inserts labeled by random-primed labeling (Boehringer Mannheim) with [α-32P]dCTP as described (26). DNA sequence analysis and GenBank database searches using the BLAST program were subsequently carried out on the clones of interest.
Cloning of PGAR cDNA and genomic DNA.
Full-length clones of PGAR were obtained from a 3T3-F442A adipocyte λZAPII cDNA library using a partial clone isolated from the differential screen. The library screening, subcloning, and sequence analysis were done as described (1). The human PGAR gene was independently identified by single-pass sequencing of a plasmid cDNA library constructed from human aortic endothelial cells, and the full-length clone was isolated by screening a lambda phage cDNA library. Mouse genomic clones were isolated by screening a mouse 129SV BAC library (Research Genetics) by PCR. The primers used for screening were as follows: forward primer, 5′-CTCTTAGCCTCCTCACTGGAG-3′; reverse primer, 5′-CACAGTTAGCACCTGTGCATC-3′. Amplification conditions were an initial denaturation at 95°C for 2 min; 35 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 10 min. The positive clone address was 313H16. The PGAR BAC clone was purified by a DNA Megaprep kit (Qiagen) and sequenced with PGAR-specific primers.
Genetic mapping.
The PGAR gene was localized to human chromosome 19 using the GENEBRIDGE4 human-hamster radiation hybrid panel and the MapManager software. The human mapping primers used for PCR amplification were the forward primer 5′-GATCGAGGCTGCAGGATATGC-3′ and the reverse primer 5′-GTCAGTCAATGTGACTGAGTCC-3′. PCRs were performed with an annealing temperature of 52°C and extension times of 50 s at 72°C for 35 cycles with a final extension of 5 min on an MJ Research Peltier PCT-225 thermal cycler. Amplification products were run on a 2% agarose gel, stained postelectrophoresis with SYBR Gold and scanned on a Molecular Dynamics Fluorimager (model 595). PCRs were also performed with mouse primers described above.
Expression of epitope-tagged PGAR and Western analysis.
The coding region of PGAR was subcloned into pcDNA3 (Invitrogen) using oligonucleotides 5′-AATTAACCCTCACTAAAGGG-3′ and 5′-TACGCGTCGACCTAAGCGTAGTCTGGGACGTCGTATGGGTAAGAGGCTGCTGTAGCCTCCAT-3′, thus enabling the attachment of a hemagglutinin antigen (HA) epitope to the carboxy terminus of PGAR protein. COS7 cells were grown to 80% confluence in 10% Cosmic calf serum–DMEM and were transiently transfected with pcDNA or pcDNA-PGAR-HA using Superfect (Qiagen). After 24 h, cells were washed with DMEM and fresh serum-free medium was added. The conditioned medium was collected 48 h posttranfection, filtered (0.22-μm-pore-size filter; Costar), and concentrated 10-fold by centrifugation in a 10-kDa-cutoff spin column (Micron Separations). Cell extracts were prepared with radioimmunoprecipitation assay lysis buffer as described (1). Proteins were separated in a sodium dodecyl sulfate-polyacrylamide gel, transferred to polyvinylidene difluoride membranes, and incubated with a monoclonal antibody against the HA antigen (1:1000; Babco), washed, and then incubated with anti-mouse immunoglobulin G conjugated with horseradish peroxidase (1:3,000; Amersham), followed by detection with enhanced chemiluminescence reagents (Amersham).
RNA extraction from tissues and Northern analysis.
Total RNA was prepared from tissues with Trizol reagent (Life Technologies) according to the manufacturer's instructions and analyzed by Northern blotting as described previously (1).
In situ hybridization.
Fresh frozen sections (8 μm thick) of C57BL/6 mouse embryos and adult mouse tissues were prepared and treated for in situ analysis as previously described (5). A 548-bp fragment of the PGAR gene was PCR amplified using the primers 5′-GGAGCGGCACAGTGGACTTT-3′ and 5′-TACCCTTTTTACGCTCCTGC-3′ and used as the template for the synthesis of 35S-labeled sense and antisense cRNA probes. Labeling and hybridization of probes were performed as described previously (5). White and brown adipose tissues from adult mouse were fixed in 10% formalin, paraffin embedded, and sectioned at 4 μm onto slides. Sections were deparaffinized in xylene, hydrated through a series of graded ethanols, and fixed in 4% formaldehyde–phosphate-buffered saline (PBS) for 10 min and rinsed twice in diethyl pyrocarbonate-PBS. After a 15-min digestion with proteinase K (20 μg/ml; Sigma), sections were immersed in 4% ethanol for 5 min and 0.2 N HCl for 10 min and then rinsed in diethyl pyrocarbonate PBS followed by a rinse in 0.1 M triethanolamine-HCl (pH 8.0). Subsequent treatments, including dehydration through alcohols and hybridization to radiolabeled probes, were similar to those of cryostat sections (5). Slides were dipped in nuclear tack emulsion (NTB-2; Eastman Kodak) and exposed for 60 days. After developing and fixation, slides were stained with hematoxylin and eosin-Y and coverslips were placed on top.
Animal studies with leptin administration.
Eight-week-old female C57BL/6J mice were individually caged 1 week prior to the experiment. Mice received intraperitoneal injections twice daily with PBS (control and pair-fed groups) or leptin (20 mg/kg of body weight/injection), and daily body mass and food intake were monitored. Pair-fed animals were staggered 1 day behind the other groups and fed an amount equal to the average amount of food consumed by the leptin group the previous day. After 1 or 5 days, the animals were sacrificed by cervical dislocation, and total RNA was isolated from pooled peri-uterine white adipose tissue. Leptin treatment or diet restriction led to reduced leptin mRNA as expected (23). The statistical significance in body mass and food intake between groups was determined using an unequal variance Student's t test. At day 5, the average daily food intake per animal was 4.32 g for the control group and 2.58 g for the leptin-treated and pair-fed groups (P < 0.001 between PBS and leptin-treated groups).
RESULTS
PPARγ activation induces a novel angiopoietin-related protein.
To identify direct transcriptional targets of PPARγ, we designed a subtractive cloning strategy that takes advantage of the availability of cellular model systems and the specificity provided by the synthetic PPARγ ligands. Because fibroblastic cells that ectopically express PPARγ display a basal level of spontaneous adipose conversion, we compared two NIH 3T3 cell lines—one infected with a retrovirus containing an empty vector and the other expressing PPARγ. Both cell lines were then treated with a TZD ligand for 3 h. The RNA from these cells was used to create a subtracted library. Subsequent analysis on Northern blots identified and confirmed 12 gene products as being expressed in a ligand-dependent way. After accounting for redundancy, these were found to correspond to known adipocyte-selective genes, including aP2, LPL, and adipocyte differentiation-related protein, and one novel sequence.
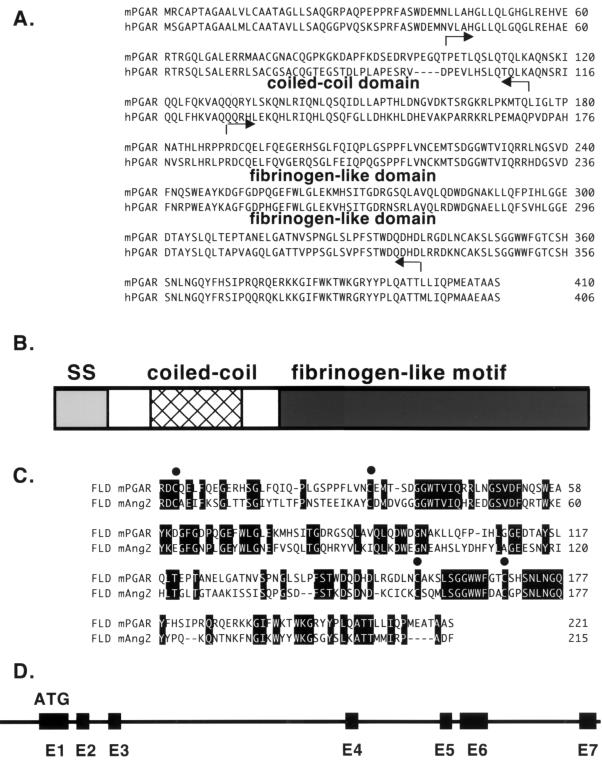
A partial clone of the novel gene, designated PGAR, was used to screen a 3T3-F442A adipocyte library to obtain the full-length clone. An open reading frame encoding 410 amino acid residues was identified, resulting in a predicted 45-kDa protein (Fig. 1A). Hydrophobic sequences were found to be present in the N-terminal regions of PGAR, consistent with secretory signal peptides. Sequence homology searches revealed that the C-terminal half of PGAR bears strong similarity to a family of proteins sharing the so-called fibrinogen-like motif (Fig. 1B). The highest similarity is with angiopoietin-2 (Fig. 1C), followed by angiopoietin-1 and ficolin (8, 16, 25). The N-terminal half, on the other hand, displays little homology to known proteins. The residues 50 to 150 are likely to form a coiled-coil quaternary structure, in common with most other members of the fibrinogen-like protein family, according to the results of a computer algorithm-based analysis of the primary structure. The presence of this motif suggests that PGAR may form multimeric structures or other higher-order structures (18). The PGAR protein also contains three potential N-glycosylation sites and four cysteines that may be available for intramolecular disulfide bonding. The human version of PGAR was also isolated; the inferred sequence is 406 amino acids long and also has a signal peptide in the N terminus. Human and mouse PGAR are 75% identical at the amino acid level (Fig. 1A).
FIG. 1.
Sequence analysis of PGAR. (A) Deduced amino acid sequences of mouse PGAR (mPGAR) and human PGAR (hPGAR). The arrows indicate the limits of the coiled-coil and fibrinogen-like domains. (B) Schematic diagram of the predicted PGAR protein structure. (C) Alignment of the fibrinogen-like domains (FLD) of PGAR and angiopoietin-2 (Ang2). Conserved cysteines are indicated by solid circles. SS, signal sequence. (D) Genomic structure of PGAR. Exons are denoted by black boxes, and introns are denoted by a solid line.
By radiation hybrid mapping, the human PGAR gene was localized to chromosome 19p13.3, 7.8 cR distal of marker AFMA135XB9, and 2.4 cR proximal of marker RP_S28_1. This region is close to the ATHS (atherosclerosis susceptibility) and ML4 (sialolipidosis) loci and syntenic to mouse chromosome 10. A murine genomic clone was subsequently isolated by screening a BAC library by PCR (Fig. 1D). The mouse PGAR gene spans approximately 6 kb. The coding region is contained in seven exons, the last four of which encode the fibrinogen-like domain.
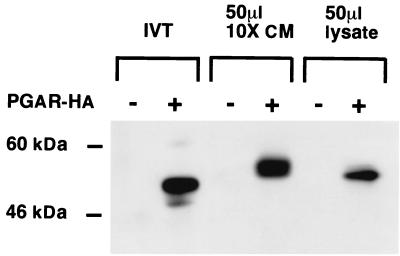
To determine if PGAR protein is secreted, an epitope-tagged PGAR construct was introduced into COS7 cells by transient transfection; the protein was detected in the conditioned medium by immunoblotting. Western analysis showed the processed protein migrating with an apparent molecular mass of 60 kDa (Fig. 2), substantially larger than would be predicted from the amino acid sequence. This may be due to posttranslational modifications such as glycosylation. Consistent with this possibility, the product of an in vitro transcription and translation reaction with the PGAR cDNA migrated with a slightly lower molecular mass.
FIG. 2.
Detection of PGAR in secreted form. Shown is a Western blot of conditioned media and cell lysates of COS7 cells transiently transfected with HA-tagged PGAR construct (+) or an empty vector (−). IVT, 10 μl of in vitro transcription and translation products of pcDNA and pcDNA-PGAR-HA, respectively, was used with the TNT in vitro translation system (Promega); CM, 50 μl of 10-fold-concentrated supernatant collected from pcDNA and pcDNA-PGAR-HA transfected cells was used; lysate, 10 μl of cell lysates from the transfected cells was used. See Materials and Methods for additional details.
PGAR is expressed selectively in fat and placenta.
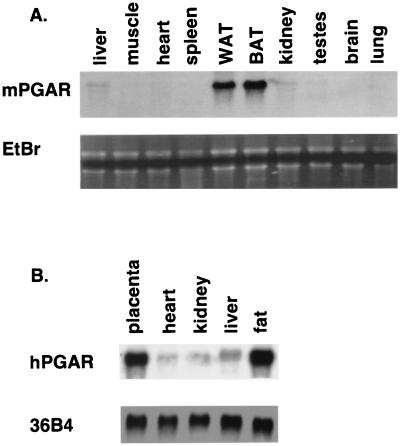
The expression of mouse PGAR was examined at the RNA level in various mouse and human tissues. The tissue distribution of the PGAR transcript in mouse was found to be highly adipose tissue selective, with at least 10- to 20-fold higher expression in white and brown fat over other tissues analyzed (Fig. 3A). Liver and kidney showed the next highest expression, albeit at much lower levels compared to fat. Some of the weaker expression discernible in other tissues may partly be due to fat contamination. A Northern analysis of human tissues (Fig. 3B) likewise showed a single 2.0-kb transcript highly enriched in white fat and placenta.
FIG. 3.
Tissue expression of PGAR. (A) RNA blot analysis of mouse PGAR (mPGAR) in adult mouse tissues. Ten micrograms of total RNA from tissues of 9-week-old male C57BL KS/J mice was fractionated on an agarose gel and transferred to nylon membranes. WAT, white adipose tissue; BAT, brown adipose tissue; EtBr, ethidium bromide staining. (B) Expression of human PGAR (hPGAR) RNA. Each lane contains 10 μg of total RNA prepared from surgical specimens (Brigham and Women's Hospital, Boston, Mass.).
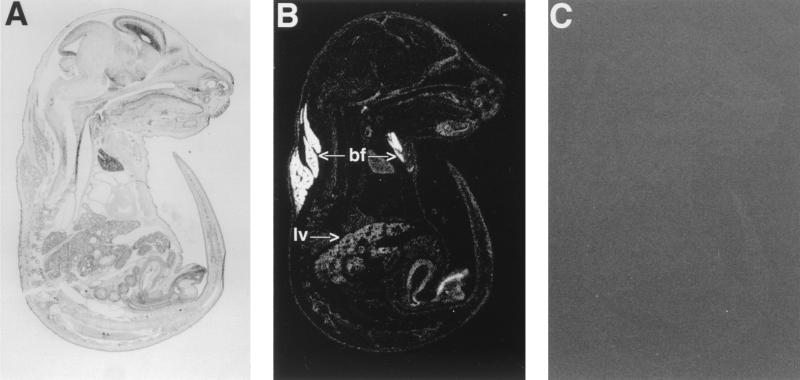
We also investigated the expression of PGAR during embryogenesis by in situ hybridization. Analysis of expression on day E13.5 revealed a low-level signal distributed widely in most organs and connective tissue with the noticeable exception of the central nervous system and the heart (data not shown). By E15.5, however, a strong expression emerged in brown fat, which remained by far the highest-expressing tissue until E18.5 (Fig. 4), the last time of examination before birth. Liver had the next highest signal at this stage.
FIG. 4.
In situ analysis of PGAR in mouse. Shown are parasagittal sections (8 μm thick) of E18.5 mouse embryo hybridized to antisense (B) or sense (C) PGAR RNA probe. Arrows point to subscapular brown fat (bf) and liver (lv). Also shown is a bright-field image of a hematoxylin-and-eosin-stained section (A).
TZDs produce a rapid transcriptional induction of PGAR without requiring protein synthesis.
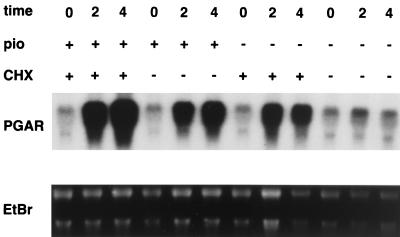
In order to examine the time course by which PGAR is regulated by PPARγ, we determined the PGAR mRNA levels in PPARγ-expressing NIH 3T3 cells after treatment with pioglitazone for 0, 2, and 4 h. Northern blot analysis performed on the cellular RNA showed that PGAR transcript is barely detectable in untreated cells but undergoes at least a 10-fold increase within 2 h after pioglitazone administration (Fig. 5). To determine if protein synthesis is required for this, we also examined the effect of cycloheximide. Interestingly, cycloheximide itself caused a rise in the PGAR mRNA, a phenomenon often associated with immediate-early genes in the literature and generally attributed to enhanced mRNA stability (7). Pretreatment with cycloheximide (5 μg/ml) did not block PPARγ ligand-produced induction but rather caused an additive increase. NIH 3T3 parental cells did not express significant levels of PGAR before or after similar treatments with pioglitazone or cycloheximide (data not shown). The persistence of TZD-mediated induction in the absence of protein synthesis, even after accounting for the effect of cycloheximide alone, is consistent with the notion of PGAR being a direct transcriptional target of PPARγ ligands and is also in agreement with the rapid time course of its induction.
FIG. 5.
Regulation of PGAR mRNA in PPARγ-expressing cells. NIH 3T3 fibroblasts were stably infected with retroviruses carrying pBabe-PPARγ and were cultured in 10% fetal bovine serum–DMEM. Cells were pretreated with cycloheximide (CHX) (5 μg/ml) (lanes 1 to 3 and 7 to 9) or vehicle (lanes 4 to 6 and 10 to 12) for 15 min and subsequently incubated further with pioglitazone (pio) (10 μg/ml) (lanes 1 to 6) or vehicle (lanes 7 to 12). Total RNA was isolated 0, 2, or 4 h after initiation of pioglitazone treatment and analyzed by Northern blotting (20 μg/lane). EtBr, ethidium bromide staining.
Regulation of PGAR expression in adipose differentiation.
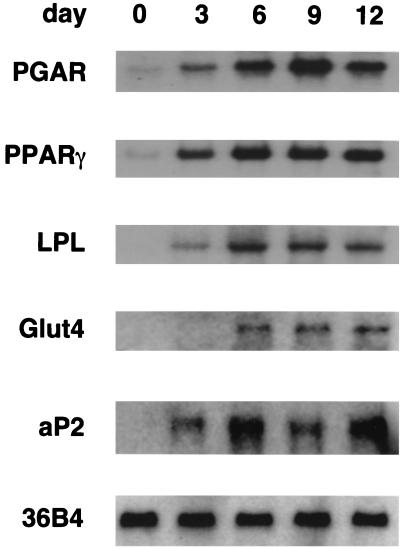
Using the 3T3-L1 preadipocyte culture model of adipogenesis, the expression of PGAR was examined at various time points during hormone-induced differentiation. PGAR levels were found to increase dramatically in early differentiation, preceding well-known markers such as LPL while lagging slightly behind PPARγ (Fig. 6). The latter observation is compatible with PGAR being a target of PPARγ.
FIG. 6.
Induction of PGAR expression during adipocyte differentiation. 3T3-L1 preadipocytes were cultured to confluence and induced to differentiate using established protocols, i.e., 3 days of dexamethasone-isobutylmethylxanthine-insulin treatment followed by insulin treatment. Northern analysis was performed on the total RNA isolated 3, 6, 9, and 12 days after induction (10 μg/lane).
Alterations of PGAR with obesity and changes in nutritional states.
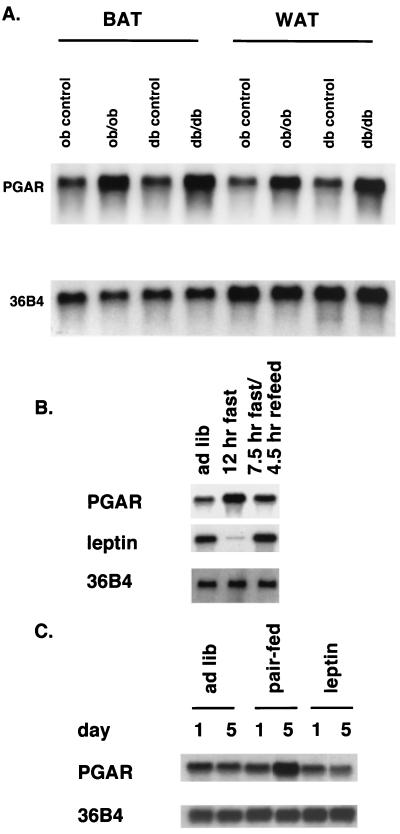
Modulation of gene expression in states of obesity may provide valuable clues on the functional relevance of the protein of interest in metabolic disease states. We observed a uniform three- to fourfold elevation of the PGAR mRNA in obese (ob/ob) and diabetic (db/db) mice relative to their respective congenic lean controls, both in white and brown fat (Fig. 7A). This may reflect a correlation of increased PGAR expression with obese states in general, raising the possibility that PGAR may contribute to some of the pathophysiological features of such states. Alternatively, PGAR mRNA levels may be regulated directly or indirectly by the genetic defects specific for these two models, i.e., leptin and the leptin receptor.
FIG. 7.
Regulation of PGAR in mouse models of obesity and by nutritional changes and leptin administration. (A) Northern analysis of PGAR (10 μg RNA/lane) in white (WAT) and brown (BAT) fat tissues from 9-week-old obese (ob/ob) and diabetic (db/db) mice and their congenic controls. (B) PGAR expression in white adipose tissue of mice subjected to dietary restrictions. Eleven-week-old C57BL KS/J mice were divided into three experimental groups and were either allowed free access to food (lane 1) or subjected to 12 h of fast (lane 2) or 7.5 h of fast followed by refeeding (lane 3). The animals were sacrificed at the end of 12 h, and 10 μg of total RNA extracted from pooled white fat (n = 3 per group) was loaded on each lane. (C) PGAR expression in leptin-treated versus diet-restricted animals. Eight-week-old female C57BL/6J mice were divided into three groups (n = 5). The first two groups received intraperitoneal injections twice daily with either PBS or leptin. The third group, the pair-fed group, was diet-restricted to match the amount of food intake by the leptin-injected group and also received injections with PBS. The animals were sacrificed after 1 or 5 days, and 10 μg of total RNA from pooled white fat was analyzed by Northern blotting. See Materials and Methods for additional details.
Possible effects of changes in nutritional states on PGAR expression were investigated. Northern blotting revealed a 2.5-fold increase in PGAR mRNA levels in white adipose tissue of mouse after a short-term fast (12 h) which was reversed by refeeding (Fig. 7B). This suggests that PGAR levels are responsive to acute metabolic changes in vivo. In a second study, PGAR expression in mice receiving intraperitoneal administration of leptin, with consequent decrease in dietary intake, was compared to that in mice diet-restricted to match the amount of food intake by the former group. PGAR mRNA levels were seen to go up threefold with restrictions of caloric intake, consistent with the findings in the above study, but this rise was not seen in leptin-treated animals (Fig. 7C). That may reflect a fundamental difference in leptin-induced dietary restriction versus externally imposed restriction or may be consistent with a further downstream effect, e.g., leptin-mediated suppression of PGAR upregulation that would ordinarily result from reduced food intake.
DISCUSSION
Activation of the nuclear receptor PPARγ has been demonstrated to elicit an extraordinarily diverse spectrum of responses in different biological settings (3, 4, 11a, 17, 19, 21, 26, 30), with adipogenesis and insulin sensitization constituting two conspicuous examples. While it is expected that multiple effector genes participate in the execution of the specific phenotypical aspects of PPARγ biology, the actual identities of such PPARγ-regulated gene products underlying the insulin sensitizing or differentiation-promoting action of TZDs have remained elusive. In an attempt to address this issue directly, we have focused on identifying the transcriptional targets of PPARγ in a cultured cell system. We report here that TZDs directly stimulate the expression of PGAR, a novel secreted protein with selective tissue distribution.
A number of observations made in this study suggest that PGAR may serve as an intercellular or intertissue signaling molecule. The primary structure of PGAR and its sequence similarity to other secreted proteins of the fibrinogen-like family raise the possibility of a common functional basis of action. Especially noteworthy is the homology to the angiopoietins, which are known to serve as signaling molecules in vascular development (8, 16, 25). The selective localization of PGAR to highly vascular tissues such as fat and placenta could be consistent with a possible function as a modulator of angiogenesis. As an early component of the PPARγ-initiated differentiation program, PGAR is a good candidate for linking adipocyte differentiation to stimulation of angiogenesis, a process known to occur early in adipose differentiation in vivo (28). Placental expression is of additional interest because placenta is a tissue known to undergo extensive vascular remodeling in the adult and is involved in transport of metabolic substrates for the fetus. In this vein, it is noteworthy that PPARγ gene inactivation results in embryonic lethality secondary to defective placental vascularization (2, 13).
The present data are also compatible with a role for PGAR in adipocyte development, for example, by acting as an autocrine factor to potentiate the drive toward terminal differentiation triggered by PPARγ. The early upregulation of PGAR in adipocyte differentiation renders further credence to that notion. PGAR regulation in genetic models of obesity and by nutrition and leptin treatment also provides circumstantial evidence pointing to its postulated role as a physiological signaling molecule relating to metabolism. Notably, the finding that PGAR levels are modulated on an acute basis by dietary deprivation and intake could be suggestive of its function as a component of a blood-borne homeostatic mechanism. The output from such regulatory systems may pertain to the pathways of lipolysis, lipogenesis, appetite, and energy expenditure, among others. Our identification of PGAR as a target of PPARγ places it in a potentially significant biological context, particularly in light of the systemic nature of the activity manifested by the latter molecule in spite of its apparent tissue selectivity. It is possible that PGAR may act upon distal target tissues such as muscle or liver to ultimately influence systemic insulin sensitivity and glucose metabolism. Ectopic expression of PGAR through transgenesis or introduction of purified PGAR protein into animals should shed further light on these questions.
ACKNOWLEDGMENTS
We thank C. Walkey for providing tissue samples from animals subjected to fasting-refeeding protocols. We are also grateful to the members of the Spiegelman laboratory for helpful discussions, P. Puigserver for critical comments on the manuscript, and A. Troy for technical assistance.
This work was funded by a grant from the National Institutes of Health (DK31405 to B.M.S.). J.C.Y. is an NIH predoctoral trainee.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1991. [Google Scholar]
- 2.Barak Y, Nelson M C, Ong E S, Jones Y Z, Ruiz-Lozano P, Chien K R, Koder A, Evans R M. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 3.Barroso I, Gurnell M, Crowley V E F, Agnosti M, Schwabe J W, Soos M A, Maslen G L, Williams T D M, Lewis H, Schafer A J, Chatterjee V K K, O'Rahilly S. Dominant negative mutations in human PPARγ associated with severe insulin resistance, diabetes mellitus, and hypertension. Nature. 1999;402:880–883. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- 4.Bishop-Bailey D, Hla T. Endothelial cell apoptosis induced by the peroxisome proliferator-activated receptor (PPAR) ligand 15-deoxy-delta 12, 14-prostaglandin J2. J Biol Chem. 1999;274:17042–17048. doi: 10.1074/jbc.274.24.17042. [DOI] [PubMed] [Google Scholar]
- 5.Busfield S J, Michnick D A, Chickering T W, Revett T L, Ma J, Woolf E A, Comrack C A, Dussault B J, Woolf J, Goodearl A D J, Gearing D P. Characterization of a neuregulin-related gene, Don-1, that is highly expressed in restricted regions of the cerebellum and hippocampus. Mol Cell Biol. 1997;17:4007–4014. doi: 10.1128/mcb.17.7.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christy R J, Yang V W, Ntambi J M, Geiman D E, Landschulz W M, Friedman A D, Nakbeppu U, Kelly T J, Lane M D. Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev. 1989;3:1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- 7.Cochran B H, Reffel A C, Stiles C D. Molecular cloning of genes regulated by platelet-derived growth factor. Cell. 1983;33:939–947. doi: 10.1016/0092-8674(83)90037-5. [DOI] [PubMed] [Google Scholar]
- 8.Davis S, Aldrich T H, Jones P F, Acheson A, Compton D L, Jain V, Ryan T E, Bruno J, Radziejewski C, Maisonpierre P C, Yancopoulos G D. Isolation of angiopoietin-1, a ligand for the TIE2 receptor, by secretion trap expression cloning. Cell. 1996;87:1161–1169. doi: 10.1016/s0092-8674(00)81812-7. [DOI] [PubMed] [Google Scholar]
- 9.Forman B M, Tontonoz P, Chen J, Brun R P, Spiegelman B M, Evans R M. 15-Deoxy-Δ12,14-prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 10.Hotamisligil G S, Shargill N S, Spiegelman B M. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 11.Hu E, Tontonoz P, Spiegelman B M. Transdifferentiation of myoblasts by the adipogenic transcription factors PPARγ and C/EBPα. Proc Natl Acad Sci USA. 1995;92:9856–9860. doi: 10.1073/pnas.92.21.9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Jiang C, Ting A T, Seed B. PPAR-γ agonists inhibit production of monocyte inflammatory cytokines. Nature. 1998;391:82–86. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- 12.Kliewer S A, Sunddseth S S, Jones S A, Brown P J, Wisely G B, Koble C S, Devchand P, Wahli W, Willson T M, Lenhard J M, Lehmann J M. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc Natl Acad Sci USA. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubota N, Terauchi Y, Miki H, Tamamoto H, Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, Eto K, Tsubamoto Y, Okuno A, Murakami K, Sekihara H, Hasegawa G, Naito M, Toyoshima Y, Tanaka S, Shiota K, Kitamura T, Fujita T, Ezaki O, Aizawa S, Nagai R, Tobe K, Kimura S, Kadowaki T. PPARγ mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- 14.Lehmann J M, Moore L B, Smith-Olivier T A, Wilkison W O, Willson T M, Kliewer S A. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ. J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 15.Lowell B B. PPARγ: an essential regulator of adipogenesis and modulator of fat cell function. Cell. 1999;99:239–242. doi: 10.1016/s0092-8674(00)81654-2. [DOI] [PubMed] [Google Scholar]
- 16.Maisonpierre P C, Suri C, Jones P F, Bartunkova S, Wiegand S J, Radziejewski C, Compton D, McCain J, Aldrich T H, Padopoulos N, Daly T J, Davis S, Sato T N, Yancopoulos G D. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 1997;277:55–60. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- 17.Mueller E, Sarraf P, Tontonoz P, Evans R M, Martin K J, Zhang M, Fletcher C, Singer S, Spiegelman B M. Terminal differentiation of human breast cancer through PPARγ. Mol Cell. 1998;1:465–470. doi: 10.1016/s1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- 18.Procopio W N, Pelavin P I, Lee W M F, Yeilding N M. Angiopoietin-1 and -2 coiled coil domains mediate distinct homo-oligomerization patterns, but fibrinogen-like domains mediate ligand activity. J Biol Chem. 1999;274:30196–30201. doi: 10.1074/jbc.274.42.30196. [DOI] [PubMed] [Google Scholar]
- 19.Ricote M, Li A C, Willson A M, Kelly C J, Glass C K. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 20.Rosen, E. D., P. Sarraf, A. E. Troy, G. Bradwin, K. Moore, D. S. Milstone, B. M. Spiegelman, and R. M. Mortensen. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 4:611–617. [DOI] [PubMed]
- 21.Sarraf P, Mueller E, Jones D, King F, DeAngelo D, Partridge S, Holden S, Chen L, Singer S, Fletcher C, Spiegelman B. Differentiation and reversal of malignant changes in colon cancer through PPARγ. Nat Med. 1998;4:1046–1052. doi: 10.1038/2030. [DOI] [PubMed] [Google Scholar]
- 22.Schoonjans K, Peinado-Onsurbe J, Lefebvre A M, Heyman R A, Briggs M, Deeb S, Staels B, Auwerx J. PPARα and PPARγ activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 1996;15:5336–5348. [PMC free article] [PubMed] [Google Scholar]
- 23.Slieker L J, Sloop K W, Surface P L, Kriauciunas A, LaQuier F, Manetta J, Bue-Valleskey J, Stevens T W. Regulation of expression of ob mRNA and protein by glucocorticoids and cAMP. J Biol Chem. 1996;271:5301–5304. doi: 10.1074/jbc.271.10.5301. [DOI] [PubMed] [Google Scholar]
- 24.Spiegelman B M. PPARγ: adipogenic regulator and thiazolidinedione receptor. Diabetes. 1998;47:507–514. doi: 10.2337/diabetes.47.4.507. [DOI] [PubMed] [Google Scholar]
- 25.Suri C, Jones P F, Patan S, Bartunkova S, Maisonpierre P C, Davis S, Sato T N, Yancopoulos G D. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell. 1996;87:1171–1180. doi: 10.1016/s0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 25a.Tartaglia L A, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards G J, Campfield L A, Clark F T, Deeds J. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 26.Tontonoz P, Hu E, Spiegelman B M. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 27.Tontonoz P, Hu E, Devine J, Beale E G, Spiegelman B M. PPARγ2 regulates adipose expression of the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1994;15:351–357. doi: 10.1128/mcb.15.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wasserman F. The development of adipose tissue. In: Renold A E, Cahill G F, editors. Handbook of physiology. Vol. 5. Washington, D.C.: American Physiology Society; 1965. pp. 87–100. [Google Scholar]
- 29.Wilson T M, Cobb J E, Cowan D J, Wiethe R W, Correa I D, Prakash S R, Beck K D, Moore L B, Kliewer S A, Lehmann J M. The structure-activity relationship between peroxisome-proliferator-activated receptor gamma agonism and the antihyperglycemic activity of the thiazolidinediones. J Med Chem. 1996;39:665–668. doi: 10.1021/jm950395a. [DOI] [PubMed] [Google Scholar]
- 30.Xin X, Yang S, Kowalski J, Gerritsen M E. Peroxisome proliferator-activated receptor gamma ligands are potent inhibitors of angiogenesis in vitro and in vivo. J Biol Chem. 1999;274:9116–9121. doi: 10.1074/jbc.274.13.9116. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman J. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]