Abstract
Background
Although vaccines effectively prevent coronavirus disease 2019 (COVID-19) in healthy individuals, they appear to be less immunogenic in individuals with chronic inflammatory disease (CID) or receiving chronic immunosuppression therapy.
Methods
Here we assessed a cohort of 77 individuals with CID treated as monotherapy with chronic immunosuppressive drugs for antibody responses in serum against historical and variant severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viruses after immunization with the BNT162b2 mRNA vaccine.
Findings
Longitudinal analysis showed the greatest reductions in neutralizing antibodies and Fc effector function capacity in individuals treated with tumor necrosis factor alpha (TNF-α) inhibitors (TNFi), and this pattern appeared to be worse against the B.1.617.2 delta virus. Within 5 months of vaccination, serum neutralizing titers of all TNFi-treated individuals tested fell below the presumed threshold correlate for antibody-mediated protection. However, TNFi-treated individuals receiving a third mRNA vaccine dose boosted their serum neutralizing antibody titers by more than 16-fold.
Conclusions
Vaccine boosting or administration of long-acting prophylaxis (e.g., monoclonal antibodies) will likely be required to prevent SARS-CoV-2 infection in this susceptible population.
Funding
This study was supported by grants and contracts from the NIH (R01 AI157155, R01AI151178, and HHSN75N93019C00074; NIAID Centers of Excellence for Influenza Research and Response (CEIRR) contracts HHSN272201400008C and 75N93021C00014; and Collaborative Influenza Vaccine Innovation Centers [CIVIC] contract 75N93019C00051).
Keywords: SARS-CoV-2, mRNA vaccine, immunosuppression, antibody, neutralization, Fc effector functions, variants of concern, TNF inhibitors
Graphical abstract
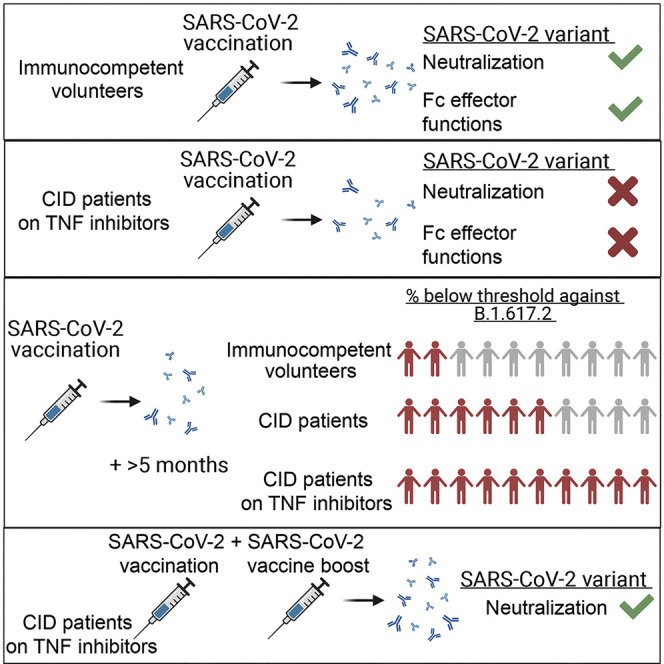
Context and significance
In most individuals, mRNA vaccines effectively prevent severe disease following SARS-CoV-2 infection. However, the protective immunity induced by mRNA vaccines is diminished in immunocompromised individuals, and the effect of variant strains is unexplored. Here we evaluated serum antibody responses in individuals with chronic inflammatory disease after immunization with the Pfizer BNT162b2 mRNA vaccine. The lowest neutralizing antibody titers were observed in individuals treated with TNF-α inhibitors, and this pattern appeared to be worse against the delta virus, with the antibody levels falling below the presumed threshold correlate of protection. Administration of a third vaccine dose substantially boosted serum neutralizing titers. Our data suggest that vaccine boosting is needed to prevent SARS-CoV-2 infection in some immunocompromised populations, especially those receiving TNF-α inhibitor therapies.
Chen et al. assess serum antibodies from BNT162b2 mRNA-vaccinated individuals with chronic inflammatory disease receiving single immunosuppressive drug therapies. Individuals receiving TNF-α inhibitors (TNFi) had reduced antibody neutralizing and Fc effector function activity against the B.1.351 and B.1.617.2 variants. A third vaccine dose markedly boosted neutralizing titers in TNFi recipients.
Introduction
In December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged and the coronavirus disease 2019 (COVID-19) pandemic began. Since then, many antibody-based therapeutic agents and vaccines have been developed,1 , 2 with some given emergency use authorization (EUA) or US Food and Drug Administration approval (e.g., the BNT162b2 mRNA vaccine) in hopes of preventing infection and severe disease. Although several of these countermeasures show efficacy against historical (2019–early 2020) SARS-CoV-2 strains, the emergence of variants of concern (VOC) has prompted questions regarding whether they will retain efficacy.
The SARS-CoV-2 spike protein engages the cell surface receptor angiotensin-converting enzyme 2 (ACE2) for attachment and entry into human cells.3 The S1 component of the spike protein contains the N-terminal domain (NTD) and receptor binding domain (RBD), the latter being the primary target of neutralizing antibodies.4, 5, 6, 7 However, recent studies have shown that therapeutic monoclonal and vaccine-elicited polyclonal antibodies have reduced neutralizing activity against VOC, likely because these strains contain mutations within the RBD and the receptor binding motif (RBM).8, 9, 10, 11, 12 This observation is concerning because serum neutralizing antibody titers are believed to be an in vitro correlate of in vivo protection.13, 14, 15
Most studies on the immunogenicity and efficacy of SARS-CoV-2 vaccines have focused on immunocompetent animals and humans. Because SARS-CoV-2 has been documented to mutate and evolve in immunocompromised hosts,16 , 17 it is important to understand whether vaccine-elicited responses are protective and durable in this population. Just recently, the Centers for Disease Control and Prevention recommended an additional BNT162b2 mRNA dose for immunocompromised individuals, notwithstanding the relatively limited study of effects of immunosuppression on the immunogenicity and protection following SARS-CoV-2 vaccination. Here we examine a cohort of individuals with chronic inflammatory disease (CID) from the COVID-19 Vaccine Responses in Patients with Autoimmune Disease (COVaRiPAD) study18 who received different treatment regimens for functional antibody responses against SARS-CoV-2 after completion of a BNT162b2 mRNA vaccination regimen.
Results
Although the potency of neutralization by monoclonal antibodies (mAbs) can be affected greatly by small numbers of mutations in emerging SARS-CoV-2 strains,8 , 11 , 12 , 19 , 20 it is less clear how polyclonal antibodies derived after vaccination that target multiple epitopes perform. While the efficacy of mRNA vaccine protection in immunocompetent volunteers has been high (>90%) against historical and some emerging SARS-CoV-2 strains (e.g., B.1.1.7 [alpha]),21 , 22 there are concerns regarding breakthrough infections in immunosuppressed individuals or those receiving immunomodulatory drugs because of blunted immune responses, waning immunity, or evasion by variants, including B.1.351 (beta) and B.1.617.2 (delta). We assessed the neutralizing activity of serum from individuals immunized with the BNT162b2 mRNA vaccine against fully infectious SARS-CoV-2 strains, including recombinant WA1/2020 viruses encoding D614G (WA1/2020 D614G) and the B.1.351 (beta) spike (Wash-B.1.351) and a clinical isolate of B.1.617.2 (delta). All viruses were passaged in Vero-TMPRSS2 cells to minimize adventitious generation of furin cleavage site mutations23 and confirmed by deep sequencing (Table S1).
We obtained sera from a cohort of individuals with CID 3 months after BNT162b2 mRNA vaccination. This group included individuals with Crohn’s disease, ulcerative colitis, asthma, multiple sclerosis, ankylosing spondylitis, systemic lupus erythematosus, Sjögren’s syndrome, Hashimoto’s disease, psoriasis, rheumatoid arthritis and undifferentiated inflammatory arthritis, type 1 diabetes, combined variable immune deficiency, alopecia areata, uveitis, vasculitis, scleroderma, psoriatic arthritis, anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis, and microscopic colitis (Table 1 ). Because of disease severity, some individuals received multiple drug interventions. For our analysis, we focused on subjects (n = 77) treated with a single immunomodulatory agent so we could correlate treatment interventions with immunological outcomes. This included individuals receiving chronic treatment with tumor necrosis factor alpha (TNF-α) inhibitors (TNFi) (n = 14), antimetabolites (n = 12), antimalarial agents (n = 10), anti-integrin inhibitors (n = 10), non-steroidal anti-inflammatory drugs (NSAIDs) (n = 9), anti-interleukin-23 (IL-23) inhibitor (n = 9), B cell depletion therapy (BCDT) (n = 5), Bruton’s tyrosine kinase inhibitor (BTKi) (n = 1), nuclear factor-erythroid factor 2-related factor 2 (Nrf2) activator (n = 1), sulfasalazine (SSZ) (n = 1), systemic steroids (n = 2), anti-B lymphocyte stimulator (anti-BLyS) (n = 1), and sphingosine 1-phosphate receptor modulator (S1PR mod) (n = 1).
Table 1.
Patient characteristics
| Total (N = 77) | |
|---|---|
| Variable | N (%) |
| Age (median [range]) | 49 (22-82) |
| Sex | |
| Female | 52 (68) |
| Male | 24 (31) |
| N/A | 1 (1) |
| Race | |
| White | 69 (90) |
| Black | 5 (6) |
| Multiple race | 2 (3) |
| Asian | 1 (1) |
| CIDa | |
| Alopecia areata | 1 (1) |
| ANCA-associated vasculitis | 1 (1) |
| Ankylosing spondylitis | 2 (3) |
| Asthma | 6 (8) |
| Combined variable immune deficiency | 2 (3) |
| Crohn’s disease | 31 (40) |
| Hashimoto’s disease | 5 (6) |
| IBD-associated arthritis | 1 (1) |
| Inflammatory arthritis | 1 (1) |
| Multiple sclerosis | 10 (13) |
| Psoriasis | 5 (6) |
| Psoriatic arthritis | 1 (1) |
| Rheumatoid arthritis | 8 (10) |
| Sarcoidosis | 1 (1) |
| Scleroderma | 1 (1) |
| Sjögren’s syndrome | 5 (6) |
| Systemic lupus erythematosus | 5 (6) |
| Type 1 diabetes | 4 (5) |
| Ulcerative colitis | 5 (6) |
| Uveitis | 2 (3) |
Individuals may have more than one disease diagnosis.
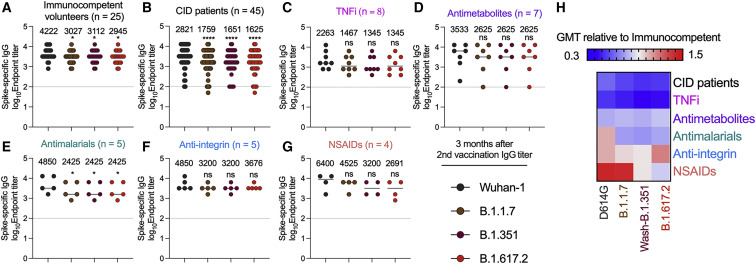
We first used an ELISA to assess serum immunoglobulin G (IgG) binding to spike proteins of Wuhan-1 (historical) or three dominant circulating variants: B.1.1.7, B.1.351, and B.1.617.2. When we analyzed relative binding to the different spike proteins, sera from immunocompetent, healthy volunteers (n = 25)24 and individuals with CID (n = 45) showed slightly reduced (1.5-fold) binding to B.1.1.7, B.1.351, and B.1.617.2 compared with to Wuhan-1 spike protein (Figures 1A and 1B). Serum IgG titers from individuals with CID as a group did not show statistically significant differences against the historical and variant spike proteins compared with those from immunocompetent volunteers (wild type [WT], p = 0.6; B.1.1.7, p = 0.29; B.1.351, p = 0.14; B.1.617.2, p = 0.23) (Figures 1A and 1B). Most drug treatment groups also showed no statistically significant differences in anti-spike IgG binding titers compared with the controls (Figures 1C–1G). As expected, individuals receiving BCDT had substantially lower levels of IgG against the spike proteins (Figure S1).
Figure 1.
Serum IgG titers against SARS-CoV-2 variant spike proteins 3 months after the second vaccination
(A–G) Analyses of spike-specific endpoint IgG serum titers measured by ELISA from humans 3 months after the second vaccination with the BNT162b2 mRNA vaccine. Individuals were grouped as (A) immunocompetent volunteers (n = 25) and (B) individuals with CID (n = 45) or subdivided by immunosuppressive drug class: (C) TNFi (n = 8), (D) antimetabolites (n = 7), (E) antimalarials (n = 5), (F) anti-integrin inhibitors (n = 5), or (G) non-steroidal anti-inflammatory drugs (NSAIDs) (n = 4). Geometric mean titer (GMT) values are shown on the graph. The dotted line represents the limit of detection of the assay. One-way ANOVA with Dunnett’s post-test; ∗p < 0.05, ∗∗∗∗p < 0.0001.
(H) Heatmap of GMT values relative to healthy volunteer GMT values for each SARS-CoV-2 spike protein. Blue, reduction; red, increase.
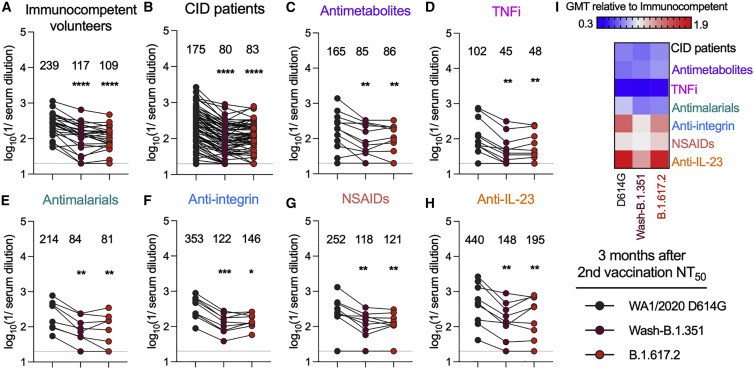
To begin to determine the functional capacity of the antibody response, we interrogated sera obtained from immunocompetent volunteers (n = 25) and individuals with CID (n = 75) 3 months after their second dose of the BNT162b2 mRNA vaccine. As a group, sera from individuals with CID did not show statistically significant differences in neutralizing activity compared with the controls (WA1/2020 D614G (geometric mean titer [GMT]: 239 [immunocompetent] versus 175 [individuals with CID], p = 0.67), Wash-B.1.351 (GMT: 117 [immunocompetent] versus 80 [individuals with CID], p = 0.16), and B.1.617.2 (GMT: 109 [immunocompetent] versus 83 [individuals with CID], p = 0.49) (Figures 2A and 2B; Figure S2A). In the control and CID groups, the B.1.351 and B.1.617.2 variants were neutralized less efficiently than the historical WA1/2020 D614G strain, as reported previously.10 , 12 , 19 , 20 , 25 Among the treatment subgroups, individuals receiving TNFi had lower serum neutralizing titers (GMT: 239 [immunocompetent] versus 102 [TNFi] [WA1/2020 D614G], p = 0.11; 117 versus 45 [Wash-B.1.351], p = 0.01; 109 versus 48 [B.1.617.2], p = 0.048) (Figures 2A and 2D; Figure S2). Other treatment groups (n > 5), including antimalarial, anti-integrin, NSAIDs, and anti-IL-23 inhibitors, had neutralization titers similar to immunocompetent volunteers (Figures 2A and 2E–2H). In addition, sera from all subgroups with more than 5 individuals neutralized Wash-B.1.351 and B.1.617.2 less efficiently than WA1/2020 (Figures 2A–2H). This pattern was also seen in groups with fewer individuals receiving other immunosuppressive agents (Figure S3).
Figure 2.
Serum NT50 against SARS-CoV-2 variant viruses 3 months after the second vaccination
(A–H) Paired analyses of neutralization titers (NT50) in serum measured by focus reduction neutralization test (FRNT) against fully infectious SARS-CoV-2 strains 3 months after the second vaccination with the BNT162b2 mRNA vaccine. Individuals were grouped as (A) immunocompetent volunteers (n = 25) and (B) individuals with CID (n = 75) or subdivided by immunosuppressive drug class: (C) antimetabolites (n = 12), (D) TNFi (n = 12), (E) antimalarials (n = 8), (F) anti-integrin inhibitors (n = 9), (G) NSAIDs (n = 9), or (H) anti-IL-23 inhibitors (n = 9). GMT values are shown on the graph. The dotted line represents the limit of detection of the assay. One-way ANOVA with Dunn’s post-test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(I) Heatmap of GMT values relative to healthy volunteer GMT values for each SARS-CoV-2 spike protein. Blue, reduction; red, increase.
A recent study analyzing the relationship between in vitro neutralization titers and protection against SARS-CoV-2 infection by vaccines estimated a protective serum titer of approximately 1/50 (range, 1/10–1/200, depending on the assay used).26 Given this result, we determined the fraction of individuals in the different groups of our cohort that fell below this cutoff. Three months after completion of the initial series of BNT162b2 mRNA vaccine, only 2 of 25 (8%) immunocompetent volunteers had neutralizing titers (NT50) below 1/50 against B.1.617.2, whereas 27 of 75 (36%) of individuals with CID had titers below this level (Table 2 ). Many of the drug treatment groups also had a higher percentage of individuals below the 1/50 cutoff compared with immunocompetent volunteers, including TNFi (67%), antimetabolites (33%), antimalarial agents (37.5%), NSAIDs (11%), and anti-IL-23 inhibitor (22%) (Table 2).
Table 2.
Numbers (and percentages) of individuals with neutralization titers (NT50) below 1/50
| Class | No. (%) 3 months after second vaccination below NT50 of 1/50 |
No. (%) 5a or 6b months after second vaccination below NT50 of 1/50 |
||||||
|---|---|---|---|---|---|---|---|---|
| Total | WA1/2020 D614G | Wash-B.1.351 | B.1.617.2 | Total | WA1/2020 D614G | Wash-B.1.351 | B.1.617.2 | |
| Immunocompetent volunteers | 25 | 0 (0) | 4 (16) | 2 (8) | 24 | 0 (0) | 5 (22) | 4 (17) |
| Individuals with CID | 75 | 14 (19) | 26 (35) | 27 (36) | 43 | 14 (33) | 24 (56) | 25 (58) |
| TNFi | 12 | 3 (27) | 8 (67) | 8 (67) | 7 | 5 (71) | 6 (86) | 7 (100) |
| Antimetabolites | 12 | 2 (17) | 5 (42) | 4 (33) | 5 | 2 (40) | 2 (40) | 2 (40) |
| Antimalarials | 8 | 0 (0) | 1 (12.5) | 3 (37.5) | 7 | 1 (14) | 4 (57) | 3 (43) |
| Anti-integrin | 9 | 0 (0) | 1 (11) | 0 (0) | 3 | 0 (0) | 0 (0) | 1 (33) |
| NSAIDs | 9 | 1 (11) | 1 (11) | 1 (11) | 5 | 1 (20) | 2 (40) | 2 (40) |
| Anti-IL-23 | 9 | 1 (11) | 2 (22) | 2 (22) | 3 | 0 (0) | 1 (33) | 1 (33) |
Samples from individuals with CID were collected 5 months after completion of vaccination.
Immunocompetent volunteer samples were collected 6 months after completion of vaccination.
We separately grouped the individuals by disease and re-analyzed NT50 values 3 months after the second vaccination. Groups of individuals with certain diseases (e.g., ulcerative colitis, systemic lupus erythematosus, Sjögren’s syndrome, rheumatoid arthritis, and asthma) had NT50 values that were statistically similar to those of immunocompetent volunteers (Figure 2A; Figure S4). Subjects with Crohn’s disease (Table S2) had slightly lower geometric mean NT50 than healthy individuals, although this difference did not attain statistical significance (GMT: 239 [healthy] versus 214 [Crohn’s disease] [WA1/2020 D614G], p = 0.59; 117 versus 83 [Wash-B.1.351], p = 0.27; 109 versus 94 [B.1.617.2], p = 0.40) (Figure 2A; Figure S4). Because TNFi is an important therapeutic target in Crohn’s disease, we analyzed this group by treatment class. Compared with 36% (27 of 75) of all individuals with CID 3 months after the second vaccination who had an NT50 of less than 1/50 against B.1.617.2, 39% (12 of 31) of individuals with Crohn’s disease fell below this threshold (Figure S4). However, 67% (6 of 9) of individuals with Crohn’s disease receiving TNFi had NT50 against B.1.617.2 below the 1/50 cutoff compared with 27% (6 of 22) of those on other treatment regimens (Figure S4). Multivariable regression analysis indicated that the lower NT50 against B.1.617.2 were associated with TNFi treatment and not Crohn’s disease (Table S3).
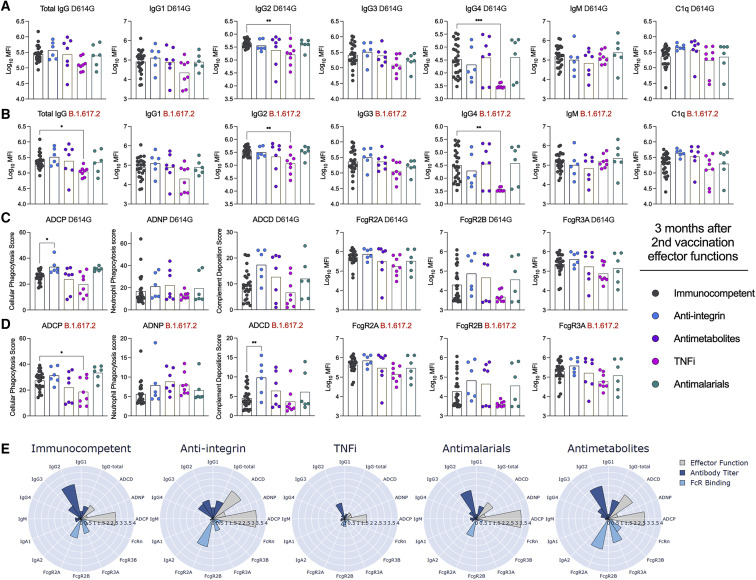
Recent studies have shown that the Fc effector functions of antibodies can contribute to protection against SARS-CoV-2 infection and disease.27, 28, 29, 30, 31, 32 To address whether our immunized individuals with CID had distinct Fc effector function profiles, we analyzed their SARS-CoV-2-specific antibodies in sera for IgG subclass distribution and C1q and Fcγ receptor (FcγR) binding. We focused our analyses on groups with at least 5 individuals (those receiving anti-integrin inhibitors, antimetabolites, TNFi, and antimalarial agents and immunocompetent volunteers) and measured responses against Wuhan-1 D614G, B.1.617.2, and B.1.351 spike proteins (Figure 3 ; Figure S5). There were no differences in levels of IgG1, IgG3, and IgM against SARS-CoV-2 spike proteins and C1q or FcγR (FcγR2A, FcγR2B, or FcγR3A) binding (Figure 3; Figure S5). However, compared with immunocompetent volunteers, individuals treated with TNFi had decreased anti-SARS-CoV-2 total IgG, IgG2, and IgG4 levels against Wuhan-1 D614G (Figure 3A), B.1.617.2 (Figure 3B), and B.1.351 (Figure S5), whereas all other groups had no significant difference. We also assessed for antibody-mediated innate immune effector functions. Although all groups showed similar antibody-dependent neutrophil phagocytosis (ADNP) (Figures 3C and 3D; Figure S5), individuals receiving TNFi had decreased antibody-dependent cellular phagocytosis (ADCP) against B.1.617.2 (Figure 3D) and B.1.351 (Figure S4). For unexplained reasons, individuals receiving anti-integrin inhibitors had enhanced ADCP against Wuhan-1 D614G (Figure 3C) and antibody-dependent complement deposition (ADCD) against B.1.617.2 (Figure 3D) and B.1.351 (Figure S5). When we combined the data, only individuals receiving TNFi showed substantive decreases in antibody effector functions 3 months after the second immunization (Figure 3E).
Figure 3.
Effector functions against SARS-CoV-2 variant viruses 3 months after the second vaccination
(A–D) Serum from humans 3 months after the second vaccination with the BNT162b2 mRNA vaccine were assayed for (A and B) total IgG, IgG subclasses (IgG1, IgG2, IgG3, IgG4, and IgM), and C1q binding or (C and D) antibody-dependent cellular phagocytosis (ADCP), antibody-dependent neutrophil phagocytosis (ADNP), antibody-dependent complement deposition (ADCD), or FcγR (FcγR2A, FcγR2B, or FcγR3A) binding as measured by Luminex. Reponses were measured against (A and C) Wuhan-1 D614G or (B and D) B.1.617.2. Individuals were grouped as immunocompetent volunteers (n = 25) or subdivided by immunosuppressive drug class: TNFi (n = 8), antimetabolites (n = 7), antimalarials (n = 6), or anti-integrin inhibitors (n = 5). One-way ANOVA with Dunnett’s post-test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(E) Composite polar plots depicting shifted Z score only median of antibody titer, FcγR binding, and antibody function against Wuhan-1 D614G, B.1.351, and B.1.617.2 for each group.
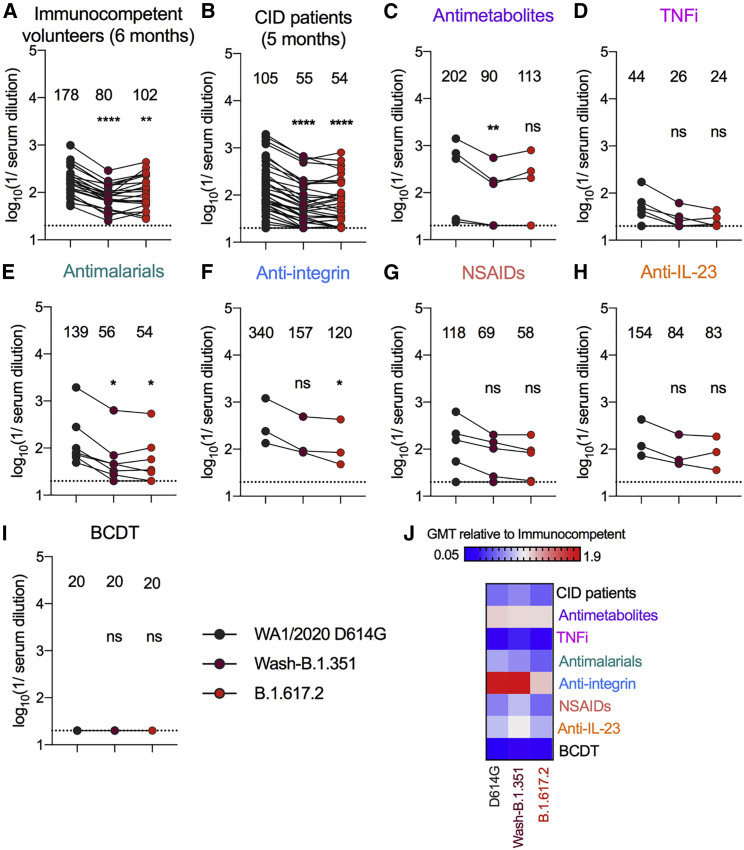
Because serum antibody titers generated by the BNT162b2 vaccine wane over time,33 , 34 we also assessed neutralizing activity 5 months after immunization in individuals with CID (n = 43); for comparison, we used the later, 6-month time point from our immunocompetent cohort (n = 24) because a 5-month time point was not sampled. Sera from individuals with CID showed lower neutralizing activity against all three viruses compared with immunocompetent volunteers (GMT: 178 [immunocompetent] versus 105 [individuals with CID] [WA1/2020 D614G], p = 0.04; 80 versus 55 [Wash-B.1.351], p = 0.02; 102 versus 54 [B.1.617.2], p = 0.008) (Figure S2). Sera from affected individual and immunocompetent subject groups were also less efficient at neutralizing Wash-B.1.351 and B.1.617.2 than WA1/2020 D614G (Figure 4 ). Although there were limited numbers of individuals in each drug treatment group at the 5-month time point, decreased inhibitory activity against all three viruses tested was seen in serum from individuals receiving TNFi (Figure S2B). Moreover, at this time point, all other therapy groups generally had less serum neutralizing activity against Wash-B.1.351 and B.1.617.2 compared with WA1/2020 D614G (Figure 4; Figure S6). Although only 4 of 24 (17%) immunocompetent volunteers had NT50 below 1/50 at the 6-month time point against B.1.617.2, 25 of 43 (58%) of individuals with CID fell below this postulated protective threshold at the 5-month time point (Table 2). Individuals treated with TNFi (100%), antimetabolites (40%), antimalarial agents (43%), anti-integrin inhibitors (33%), NSAIDs (40%), and anti-IL-23 inhibitors (33%) had higher percentages of individuals than healthy volunteers below the 1/50 cutoff against B.1.617.2 (Table 2).
Figure 4.
Serum NT50 of individuals with CID against SARS-CoV-2 variant viruses 5 months after the second vaccination
(A–H) Paired analyses of NT50 in serum measured by FRNT against fully infectious SARS-CoV-2 strains from humans 5 months (individuals with CID) or 6 months (immunocompetent volunteers) after the second vaccination with the BNT162b2 mRNA vaccine; different time points were used based on variability of the separate study designs and availability of samples. Individuals were grouped as (A) immunocompetent volunteers (n = 24) or (B) individuals with CID (n = 43) or subdivided by immunosuppressive drug class: (C) antimetabolites (n = 5), (D) TNFi (n = 7), (E) antimalarials (n = 7), (F) anti-integrin inhibitors (n = 3), (G) NSAIDs (n = 5), (H) anti-IL-23 inhibitors (n = 3), or (I) B cell depletion therapy (BCDT) (n = 3). GMT values are shown on the graph. The dotted line represents the limit of detection of the assay. One-way ANOVA with Dunn’s post-test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(J) Heatmap of GMT values relative to healthy volunteer GMT values for each SARS-CoV-2 spike protein. Blue, reduction; red, increase.
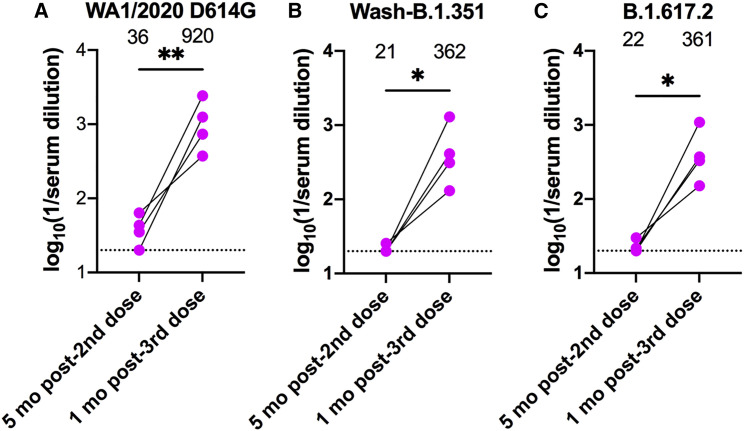
Recently, manufacturers of mRNA and adenovirus-vectored vaccines against SARS-CoV-2 were granted an expansion of the EUA for providing an additional booster dose for at-risk and general adult populations because of waning immunity and breakthrough infections.35 , 36 We examined the effect of a third BNT162b2 mRNA vaccine dose in individuals with CID treated with TNFi. In preliminary studies, the four individuals who received a booster dose showed 16- to 25-fold increases (p < 0.05) in mean neutralizing titers within 1 month against the WA1/2020, Wash-B.1.351, and B.1.617.2 strains compared with their pre-boost titers (Figure 5 ). Notably, none of the four held their TNFi medication at the time of third dose administration. Thus, despite their limited serum neutralizing activity 5 months after completion of the primary mRNA vaccine series, individuals on TNFi show evidence of a robust anamnestic B cell response.
Figure 5.
Serum NT50 of individuals with CID receiving TNFi 1 month after receiving a third dose of the BNT162b2 mRNA vaccine
(A–C) Paired analyses of NT50 in serum measured by FRNT against fully infectious SARS-CoV-2 strains (A) WA1/2020, (B) Wash-B.1.351, and (C) B.1.617.2 from humans 5 months after the second dose or 1 month after the third dose of the BNT162b2 mRNA vaccine. None of the individuals held TNFi medication at the time of third dose administration. GMT values are shown on the graph. The dotted line represents the limit of detection of the assay. Paired t test; ∗p < 0.05, ∗∗p < 0.01.
Discussion
In this study, we evaluated the functional antibody responses after immunization with the Pfizer BNT162b2 mRNA vaccine against historical and emerging SARS-CoV-2 strains in a cohort of adult individuals with CID with a range of diagnoses and treatment interventions. These results were compared with a separate cohort of similarly vaccinated immunocompetent adults24 and showed consistently lower serum antibody neutralizing titers in most individuals with CID after two doses, with a substantial fraction falling below an estimated 1/50 cutoff against B.1.617.2 (delta) that has been proposed as a correlate of protection.26 Subgroup analysis suggested that individuals on TNFi had lower inhibitory titers than other therapeutic groups. Thus, these individuals might be at greatest risk for breakthrough infections, especially with VOC. Similarly, in studies that evaluated Fc effector function of serum antibodies, those receiving TNFi showed a greater decrease in antibody effector functions, providing a second possible mechanism for risk of vaccine failure in these populations.
Our results contrast with a recent report of 84 individuals with psoriasis and 17 healthy controls immunized with the Pfizer BNT162b2 mRNA vaccine.37 In that study, neutralizing activity against SARS-CoV-2 Wuhan-1 was lower in individuals receiving methotrexate than TNFi. The variation in results could be explained by the following differences in study design and analysis: (1) we had only one individual with psoriatic arthritis in our CID cohort; (2) we grouped methotrexate with other antimetabolites because of small numbers; (3) differences in the specific TNFi used; and (4) the neutralization assays used were not the same. In comparison, our data suggesting that TNFi blunts the humoral response to vaccines is consistent with a meta-analysis of 25 observational studies with 5,360 individuals who received Pfizer BNT162b2, Moderna mRNA-1273, or other platforms.38
Our findings corroborate studies that report an association between individuals with CID, including those on TNFi, and reduced antibody responses after vaccination.18 , 39, 40, 41 Poor seroconversion in these individuals has been described after vaccination against the hepatitis A,42 hepatitis B,43 , 44 and influenza45, 46, 47 viruses, which may be potentiated by TNFi and other immunosuppressants.48 Furthermore, a specific effect of TNFi on dampening antibody responses in individuals with inflammatory bowel disease has been observed in those who recovered from SARS-CoV-2 infection41 , 49 or after vaccination.50 Because TNF-α contributes to secondary lymphoid organ (B cell follicles or lymph nodes) development and signaling,51, 52, 53 TNFi may interfere with germinal center reactions and induction of optimal humoral immune responses.
The importance of Fc effector functions for antibody efficacy in vivo against SARS-CoV-2 has been illustrated with several NTD and RBD mAbs in animal models of infection.27 , 28 , 32 , 54 , 55 Consistent with these results, differences in antibody effector functions in serum also are associated with distinct levels of protection in the upper and lower respiratory tract of non-human primates immunized with a recombinant SARS-CoV-2 spike glycoprotein (NVX-CoV2373).56 Moreover, the BNT162b2 and mRNA1273 mRNA vaccines appear to induce different functional profiles in humans with higher RBD- and NTD-specific IgA as well as functional antibodies (ADNP and antibody-dependent natural killer cell activity [ADNK]) seen in mRNA-1273 vaccine recipients.57 Thus, and independent of the lower neutralizing antibody levels observed in the sera of individuals with CID treated with TNFi, diminished Fc effector function profiles could also contribute to a higher frequency of breakthrough infections in this and other immunosuppressed vaccinated populations.58, 59, 60
The lower serological responses to the BNT162b2 vaccine seen in many of the individuals with CID in our cohort appears the be worse against the two viruses with spike genes of B.1.351 and B.1.617.2. This was not unexpected, given that the evolution of more transmissible SARS-CoV-2 VOC with substitutions in the spike protein affects antibody binding. Indeed, neutralization by vaccine-induced sera is diminished against variants expressing mutations in the spike gene at positions L452 and E484 and elsewhere.8 , 12 , 19 , 61, 62, 63 Moreover, several vaccines have shown a reduced ability to prevent symptomatic infection caused by the B.1.351 and B.1.617.2 variants in humans.64, 65, 66, 67 The lower functional (neutralizing and Fc effector functions) antibody responses seen in immunized immunosuppressed individuals with CID combined with inherently less inhibitory activity against emerging variants is consistent with the recent recommendation to administer an additional dose of vaccine to these at-risk populations. In preliminary studies of a small number of TNFi-treated individuals, a third dose of mRNA vaccine resulted in markedly increased serum neutralizing titers against historical and variant SARS-CoV-2 strains, likely suggesting a relatively intact memory B cell response. Longitudinal analysis of these and other boosted individuals receiving TNFi is needed to determine the durability of this augmented serum antibody response.
Limitations of study
We note several limitations of our study. (1) Because of the diversity of immunosuppressants used, the number in each treatment subgroup is relatively small, limiting the power of the statistical analysis. (2) The cohort we used was a subset of a larger CID cohort18 that included vaccinated individuals receiving multiple immunosuppressive treatment modalities. Only those on monotherapy regimens were evaluated in our study to eliminate confounding by use of concomitant immunosuppressive agents. (3) We grouped subjects by treatment intervention but, because of the small numbers, did not account for underlying disease severity or diagnosis, which independently could affect immune responses. (4) Our study focused on subjects immunized with the Pfizer BNT162b2 mRNA vaccine. Separate studies are needed with other vaccine platforms. (5) Our study focused on the effect of immunosuppression on serum antibody responses after vaccination and did not account for T cell and anamnestic responses, which also may confer protection. (6) The small sample size precluded assessment of comorbidities, age, sex, and race as independent variables on humoral responses. There was some sex-skewing between the immunocompetent (64% male) and CID (68% female) cohorts.
From a cohort of 77 individuals with CID on different immunosuppressive therapies, we observed a clear trend toward lower antibody neutralizing and Fc effector function responses after two doses of Pfizer BNT162b2 mRNA vaccination in those receiving TNFi. Although numbers of individuals with CID in the subgroup analysis were small, those few receiving prolonged systemic steroids or BCDT also had reduced serum binding and neutralizing titers. Similar blunted humoral responses after mRNA vaccination were seen in cohorts of individuals treated with systemic glucocorticoids or BCDT.18 , 68, 69, 70 Because the responses we observed are even lower against emerging VOC, boosting and functional monitoring of immunity will be important in these individuals. Future studies are warranted to understand and improve SARS-CoV-2 immunity in immunologically vulnerable populations.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SARS2-2, SARS2-11, SARS2-16, SARS2-31, SARS2-38, SARS2-57, and SARS2-71 | VanBlargan et al.71; Liu et al.72 | N/A |
| HRP conjugated anti-mouse IgG (whole molecule) | Millipore; Southern Biotech; | Sigma 12-349 and #A0293; Southern Biotech Cat #4700-04’ RRID:AB_390192 |
| Anti-human CD66-Pac Blue | Biolegend | Cat# 305112; RRID:AB_2563294 |
| Bacterial and virus strains | ||
| SARS-CoV-2 WA1/2020 D614G | Plante et al.73 | N/A |
| SARS-CoV-2 WA1/2020 B.1.351 spike | Chen et al.12 | N/A |
| SARS-CoV-2 B.1.617.2 | Gift from R. Webby (Memphis, TN) | N/A |
| Biological samples | ||
| COVaRiPAD patient serum | Deepak et al.18 | N/A |
| Immunocompetent serum | Turner et al.24 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 Spike proteins (for ELISA) | Amanat et al.74; Stadlbauer et al.75 | N/A |
| SARS-CoV-2 antigens (for Luminex) | Gift from EO Saphire (La Jolla Institute for Immunology) | N/A |
| o-phenylenediamine dihydrochloride substrate | Sigma-Aldrich | P9187 |
| TrueBlue peroxidase substrate | VWR | KPLI50-78-02 |
| EDC | Thermo Fisher Scientific | A35391 |
| EZ-link Sulfo-NHS-LC-LC-Biotin | Thermo Fisher Scientific | A35358 |
| NeutrAvidin conjugated beads | Thermo Fisher Scientific | F8777 |
| Lyophilized guinea pig complement | Cederlane | CL4051 |
| Gelatin Veronal Buffer with calcium and magnesium | Boston BioProducts | IBB-300 |
| Fluorescein-conjugated IgG fraction to guinea pig complement C3 | MP Bio | Cat# 0855385; RRID:AB_2334913 |
| Sulfo-NHS | Thermo Fisher Scientific | A39269 |
| Experimental models: Cell lines | ||
| Vero-TMPRSS2 | Zang et al.76 | N/A |
| THP-1 | ATCC | Cat# TIB-202; RRID:CVCL_0006 |
| Software and algorithms | ||
| BioSpot | Cellular Technology Limited | N/A |
| IntelliCyt ForeCyt | Intellicyt | V8.1 |
| Python | Sievert, 2020 | V.3.8.5 |
| FlowJo | FlowJo | V10.7.1 |
| GraphPad Prism | GraphPad | V.8.4.3 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Michael S. Diamond (diamond@wusm.wustl.edu).
Materials vailability
All requests for resources and reagents should be directed to the Lead Contact author. This includes mice and viruses. All reagents will be made available on request after completion of a Materials Transfer Agreement.
Experimental model and subject details
Patient samples
CID patient samples were obtained from the COVaRiPAD longitudinal observational study as previously described.18 Immunocompetent volunteer samples were obtained as previously described.24 All individuals were enrolled at Washington University School of Medicine in studies that had received Institutional Review Board approval [202012081 (WU368) and 202012084 (COVaRiPAD)].
Cells
Vero-TMPRSS2 cells76 were cultured at 37°C in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES pH 7.3, 1 mM sodium pyruvate, 1 × non-essential amino acids, and 100 U/ml of penicillin–streptomycin, and 5 μg/mL of blasticidin.
Viruses
The WA1/2020 D614G recombinant strain was obtained from an infectious cDNA clone of the 2019n-CoV/USA_WA1/2020 strain as described previously.73 The beta (B.1.351) variant spike gene was introduced into the WA1/2020 backbone as described previously.12 The B.1.617.2 isolate was obtained a gift from R. Webby (Memphis, TN). All viruses were passaged once in Vero-TMPRSS2 cells and subjected to next-generation sequencing after RNA extraction to confirm the introduction and stability of substitutions (Table S1). All virus experiments were performed in an approved Biosafety level 3 (BSL-3) facility.
Method details
Enzyme-linked immunosorbent assay (ELISA)
Serum antibodies against SARS-CoV-2 spike (S) were measured as previously described.74 , 75 Briefly, polystyrene 96-well plates (Immulon 4HBX; Thermo Fisher Scientific) were coated with 50 μL/well of PBS (pH 7.4) (GIBCO) containing recombinant spike proteins (2 μg/mL) and incubated at 4°C overnight. On the next day, plates were washed with PBS-0.1% Tween 20 (PBS-T) using an automated plate washer (AquaMax 2000; Molecular Devices). Plates were blocked with 220 μL/well of PBS-T, 3% nonfat dry milk (AmericanBio) for 1 h. For serum and secondary antibody dilutions, a solution of PBS-T, 1% nonfat dry milk (AmericanBio) was used. Sera were serially diluted (3-fold) starting at a 1:100 dilution. Dilutions were added to the plates (100 μL/well) for 2 h at room temperature (RT). Plates were washed, and the secondary antibody IgG (whole molecule)-peroxidase antibody was added for 1 h at room temperature. Plates were washed, and the substrate o-phenylenediamine dihydrochloride (SigmaFast OPD; Sigma-Aldrich) was added (100 μL/well) and incubated for 10 min. The reaction was stopped by addition of 50 μL/well of a 3 M HCl solution (Thermo Fisher Scientific). Optical density (OD) was measured (490 nm) using a microplate reader (Synergy H1; BioTek). Analysis was performed using Prism 7 software (GraphPad), and values were reported as area under the curve (AUC).
Focus reduction neutralization test
Serial dilutions of mAbs or sera were incubated with 102 focus-forming units (FFU) of different strains or variants of SARS-CoV-2 for 1 h at 37°C. Antibody-virus complexes were added to Vero-TMPRSS2 cell monolayers in 96-well plates and incubated at 37°C for 1 h. Subsequently, cells were overlaid with 1% (w/v) methylcellulose in MEM supplemented with 2% FBS. Plates were harvested 24 h later by removing overlays and fixed with 4% PFA in PBS for 20 min at room temperature. Plates were washed and sequentially incubated with an oligoclonal pool of SARS2-2, SARS2-11, SARS2-16, SARS2-31, SARS2-38, SARS2-57, and SARS2-7172 , 71 anti-spike antibodies and HRP-conjugated goat anti-mouse IgG (Sigma, 12-349) in PBS supplemented with 0.1% saponin and 0.1% bovine serum albumin. SARS-CoV-2-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot microanalyzer (Cellular Technologies).
Effector function antigens
Antigens used for Luminex based assays: SARS-CoV-2 D614G, beta B.1.351, and delta B.1.617.2 spike antigens all were kindly provided by Erica Ollmann Saphire (La Jolla Institute for Immunology).
Luminex profiling
Serum samples were analyzed by customized Luminex assay to quantify the relative concentration of antigen-specific antibody isotypes, subclasses, and Fcγ-receptor (FcγR) binding profiles, as previously described.77 , 78 Briefly, SARS-CoV-2 antigens were used to profile specific humoral immune responses. Antigens were coupled to magnetic Luminex beads (Luminex Corp) by carbodiimide-NHS ester-coupling (Thermo Fisher). Antigen-coupled microspheres were washed and incubated with plasma or serum samples at an appropriate sample dilution (1:5000 for IgG1 and all low affinity FcγR, and 1:200 for all other readouts) for 2 h at 37°C in 384-well plates (Greiner Bio-One). Unbound antibodies were washed away, and antigen-bound antibodies were detected by using a PE-coupled detection antibody for each subclass and isotype (IgG1, IgG3, IgA1, and IgM; Southern Biotech), and FcγR were fluorescently labeled with PE before addition to immune complexes (FcγR2a, FcγR3a; Duke Protein Production facility). After one hour of incubation, plates were washed, and flow cytometry was performed with an iQue (Intellicyt) and analyzed using IntelliCyt ForeCyt (v8.1). PE median fluorescent intensity (MFI) is reported as a readout for antigen-specific antibody titers.
Antibody-dependent complement deposition (ADCD)
Antibody-dependent complement deposition (ADCD) was conducted as previously described.79 Briefly, SARS-CoV-2 antigens were coupled to magnetic Luminex beads (Luminex Corp) by carbodiimide-NHS ester-coupling (Thermo Fisher). Coupled beads were incubated for 2 h at 37°C with serum samples (1:10 dilution) to form immune complexes and then washed to remove unbound immunoglobulins. To measure antibody-dependent deposition of C3, lyophilized guinea pig complement (Cedarlane) was diluted in gelatin veronal buffer with calcium and magnesium (GBV++) (Boston BioProducts) and added to immune complexes. Subsequently, C3 was detected with an anti-C3 fluorescein-conjugated goat IgG fraction detection antibody (Mpbio). Flow cytometry was performed 5 Laser LSR Fortessa Flow Cytometer and analyzed using FlowJo V10.7.1. ADCD was reported as the median of C3 deposition.
Antibody-dependent cellular (ADCP) and neutrophil (ADNP) phagocytosis
Antibody-dependent cellular phagocytosis (ADCP) and antibody-dependent neutrophil phagocytosis (ADNP) were conducted according to the previously described protocols.80 , 81 SARS-CoV-2 antigens were biotinylated using EDC (Thermo Fisher) and Sulfo-NHS-LCLC biotin (Thermo Fisher) and coupled to yellow-green (505/515) fluorescent Neutravidinconjugated beads (Thermo Fisher), respectively. To form immune complexes, antigen-coupled beads were incubated for 2 h at 37°C with 1:100 diluted serum samples and then washed to remove unbound immunoglobulins. For ADCP, the immune complexes were incubated for 16–18 h with THP-1 cells (1.25 × 105 THP-1 cells/mL) and for ADNP for 1 h with RBC-lyzed whole blood. Following the incubation, cells were fixed with 4% PFA. For ADNP, RBC-lyzed whole blood was washed, stained for CD66b+ (Biolegend) to identify neutrophils, and then fixed in 4% PFA. Flow cytometry was performed to identify the percentage of cells that had phagocytosed beads as well as the number of beads that had been phagocytosis (phagocytosis score = % positive cells × Median Fluorescent Intensity of positive cells/10000). Flow cytometry was performed with 5 Laser LSR Fortessa Flow Cytometer and analyzed using FlowJo V10.7.1.
Quantification and statistical analysis
Statistical significance was assigned when P values were < 0.05 using Prism Version 8 (GraphPad). Specific tests (one-way ANOVA with Dunn’s or Dunnett’s post-test for multiple comparisons), number of subjects (n), geometric mean values, and comparison groups are indicated in the Figure legends. All data were graphed and analyzed in GraphPad Prism v8.4.3. The data used to generate Figure 3E was graphed and analyzed using Python version 3.8.5 and the ‘plotly’ package.82 For each feature, data was first standardized by computing the Z-score, scaling values to zero mean and unit variance. The median resulting values are represented on each polar plot. All other data were graphed and analyzed in GraphPad Prism v8.4.3. Tobit linear regression was performed using Stata/MP 13.1, and the effects were refined to account for left-censoring of data below the limit of detection (LoD).
Acknowledgments
This study was supported by grants and contracts from the NIH (R01 AI157155, R01AI151178, and HHSN75N93019C00074; NIAID Centers of Excellence for Influenza Research and Response (CEIRR) contracts HHSN272201400008C and 75N93021C00014, and Collaborative Influenza Vaccine Innovation Centers (CIVIC) contract 75N93019C00051). The Alter laboratory was supported by the Ragon Institute, the Massachusetts Consortium on Pathogen Readiness (MassCPR), the NIH (3R37AI080289-11S1, R01AI146785, U19AI42790, U19AI135995, U19AI42790, 1U01CA260476, and CIVIC75N93019C00052), Gates Foundation Global Health Vaccine Accelerator Platform funding (OPP1146996 and INV-001650), and the Musk Foundation. M.P. was supported by the Scientist Development Award from the Rheumatology Research Foundation. P.D. is supported by a Junior Faculty Development Award from the American College of Gastroenterology and IBD Plexus of the Crohn's & Colitis Foundation. A.H.J.K. is supported by the Rheumatology Research Foundation, NIH/NIAMS P30 AR073752, and PCORI SDM2017C28224. G.F.W. was supported by grant funding from the NIH (R01 NS106289) and the NMSS (RG-1802-30253). The COVaRiPAD study was supported by The Leona M. and Harry B. Helmsley Charitable Trust, the Washington University Digestive Disease Research Core Center (NIDDK P30DK052574), the Washington University Rheumatic Diseases Research Resource-Based Center (NIAMS P30AR073752), the Judy Miniace Research Fund for the Washington University Lupus Clinic, Siteman Cancer Center grant P30CA091842 from the NIH/NCI, and the Washington University Institute of Clinical and Translational Sciences grant UL1TR002345 from the NIH/NCATS. We thank Richard Webby and Pei-Yong Shi for some of the viruses used in this study and Kimberly E. Taylor at University of California, San Francisco for statistical support.
Author contributions
R.E.C. and L.A.V. performed and analyzed neutralization assays. M.J.G., D.Y., and D.Y.Z. performed and analyzed effector function analyses. J.M.C. performed and analyzed ELISA data. L.D. and S.A.H. performed and analyzed next-generation sequencing of viral stocks. P.D., M.P., R.M.P., J.A.O., S.C., G.F.W., A.H.E., and A.H.J.K. designed the clinical studies and provided human samples. S.B., W.K., and J.S.T. processed clinical samples. R.E.C., A.H.J.K., and M.S.D. performed and oversaw the statistical analysis. R.E.C. and M.S.D. had unrestricted access to all of the data in the paper. P.D., D.A.L., F.K., G.A., A.H.E., A.H.J.K., and M.S.D. obtained funding. G.A. and M.S.D. supervised the research. R.E.C. and M.S.D. wrote the initial draft, with the other authors providing editorial comments. All authors agreed to submit the manuscript, read and approved the final draft, and take full responsibility for its content, including the accuracy of the data.
Declaration of interests
M.S.D. is a consultant for Inbios, Vir Biotechnology, Senda Biosciences, and Carnival Corporation and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Vir Biotechnology, Moderna, and Emergent BioSolutions. F.K. is a coinventor on a patent application for serological assays and SARS-CoV-2 vaccines (international application numbers PCT/US2021/31110 and 62/994,252). A.H.J.K. participated in consulting, advisory board, or speaker’s bureau for Alexion Pharmaceuticals; Aurinia Pharmaceuticals; Exagen Diagnostics, Inc.; and GlaxoSmithKline and received unrelated funding support under a sponsored research agreement from GlaxoSmithKline. The Ellebedy laboratory received funding under sponsored research agreements that are unrelated to current study from Emergent BioSolutions and AbbVie. A.H.E. is a consultant for Mubadala Investment Company and the founder of ImmuneBio Consulting LLC. A.H.E., M.S.D., and J.S.T. are recipients of a licensing agreement with Abbvie Inc. for commercial development of a SARS-CoV-2 mAb not described in this study. J.S.T. is a consultant for Gerson Lehrman Group. S.C. received research funding from Biogen and received speaking and/or consulting fees from Biogen, Novartis, Sanofi Genzyme, Genentech, and Bristol Myers Squibb. P.D. has participated in consulting, advisory board, or speaker’s bureau for Janssen, Pfizer, Prometheus Biosciences, Boehringer Ingelheim, AbbVie, and Arena Pharmaceuticals and received funding under an unrelated sponsored research agreement from Takeda Pharmaceutical, Arena Pharmaceuticals, Bristol Myers Squibb-Celgene, and Boehringer Ingelheim. G.F.W. has received honoraria for consulting from Novartis and Genentech, Inc. and research funding from Biogen, EMD Serono, and Roche. F.K. has consulted for Merck, Curevac, and Pfizer in the past and is currently consulting for Pfizer, Seqirus, and Avimex. The Krammer laboratory is collaborating with Pfizer on animal models of SARS-CoV-2. G.A. is the founder of SeromYx Systems Inc. and an equity holder of Leyden Labs.
Published: December 10, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.medj.2021.11.004.
Supplemental information
Data and code availability
-
(a)
Data. All serological results described in this study are available within the body of the paper. All data (including raw data used to generate neutralizing and binding curves) reported in this paper will be shared by the lead contact upon request
-
(b)
Code. This paper does not report original code.
-
(c)
Additional information. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Sempowski G.D., Saunders K.O., Acharya P., Wiehe K.J., Haynes B.F. Pandemic Preparedness: Developing Vaccines and Therapeutic Antibodies For COVID-19. Cell. 2020;181:1458–1463. doi: 10.1016/j.cell.2020.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Case J.B., Winkler E.S., Errico J.M., Diamond M.S. On the road to ending the COVID-19 pandemic: Are we there yet? Virology. 2021;557:70–85. doi: 10.1016/j.virol.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 6.Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 8.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A., Cai H., Sarkar R., Chen W., Cutler M., et al. Neutralizing Activity of BNT162b2-Elicited Serum. N. Engl. J. Med. 2021;384:1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J., Liu Y., Xia H., Zou J., Weaver S.C., Swanson K.A., Cai H., Cutler M., Cooper D., Muik A., et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature. 2021;596:273–275. doi: 10.1038/s41586-021-03693-y. [DOI] [PubMed] [Google Scholar]
- 11.Chen R.E., Winkler E.S., Case J.B., Aziati I.D., Bricker T.L., Joshi A., Darling T.L., Ying B., Errico J.M., Shrihari S., et al. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature. 2021;596:103–108. doi: 10.1038/s41586-021-03720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbett K.S., Nason M.C., Flach B., Gagne M., O’Connell S., Johnston T.S., Shah S.N., Edara V.V., Floyd K., Lai L., et al. Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science. 2021;373:eabj0299. doi: 10.1126/science.abj0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francica J.R., Flynn B.J., Foulds K.E., Noe A.T., Werner A.P., Moore I.N., Gagne M., Johnston T.S., Tucker C., Davis R.L., et al. Protective antibodies elicited by SARS-CoV-2 spike protein vaccination are boosted in the lung after challenge in nonhuman primates. Sci. Transl. Med. 2021;13:eabi4547. doi: 10.1126/scitranslmed.abi4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbett K.S., Werner A.P., Connell S.O., Gagne M., Lai L., Moliva J.I., Flynn B., Choi A., Koch M., Foulds K.E., et al. mRNA-1273 protects against SARS-CoV-2 beta infection in nonhuman primates. Nat. Immunol. 2021;22:1306–1315. doi: 10.1038/s41590-021-01021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.H., Boucau J., Bowman K., et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark S.A., Clark L.E., Pan J., Coscia A., McKay L.G.A., Shankar S., Johnson R.I., Brusic V., Choudhary M.C., Regan J., et al. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell. 2021;184:2605–2617.e18. doi: 10.1016/j.cell.2021.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deepak P., Kim W., Paley M.A., Yang M., Carvidi A.B., Demissie E.G., El-Qunni A.A., Haile A., Huang K., Kinnett B., et al. Effect of Immunosuppression on the Immunogenicity of mRNA Vaccines to SARS-CoV-2 : A Prospective Cohort Study. Ann. Intern. Med. 2021 doi: 10.7326/M21-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., Ho D.D. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29:747–751.e4. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 21.Abu-Raddad L.J., Chemaitelly H., Butt A.A., National Study Group for COVID-19 Vaccination Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Engl. J. Med. 2021;385:187–189. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chemaitelly H., Yassine H.M., Benslimane F.M., Al Khatib H.A., Tang P., Hasan M.R., Malek J.A., Coyle P., Ayoub H.H., Al Kanaani Z., et al. mRNA-1273 COVID-19 vaccine effectiveness against the B.1.1.7 and B.1.351 variants and severe COVID-19 disease in Qatar. Nat. Med. 2021;27:1614–1621. doi: 10.1038/s41591-021-01446-y. [DOI] [PubMed] [Google Scholar]
- 23.Klimstra W.B., Tilston-Lunel N.L., Nambulli S., Boslett J., McMillen C.M., Gilliland T., Dunn M.D., Sun C., Wheeler S.E., Wells A., et al. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J. Gen. Virol. 2020;101:1156–1169. doi: 10.1099/jgv.0.001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turner J.S., O’Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236.e13. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 27.Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R., et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021;218:e20201993. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M.D., Burke J., Yu J., Feldman J., Hauser B.M., et al. Compromised Humoral Functional Evolution Tracks with SARS-CoV-2 Mortality. Cell. 2020;183:1508–1519.e12. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiakolas A.R., Kramer K.J., Wrapp D., Richardson S.I., Schäfer A., Wall S., Wang N., Janowska K., Pilewski K.A., Venkat R., et al. Cross-reactive coronavirus antibodies with diverse epitope specificities and Fc effector functions. Cell Rep Med. 2021;2:100313. doi: 10.1016/j.xcrm.2021.100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan C.E.Z., Seah S.G.K., Chye H., Massey S., Torres M., Lim A.P.C., Wong S.K.K., Neo J.J.Y., Wong P.S., Lim J.H., et al. The Fc-mediated effector functions of a potent SARS-CoV-2 neutralizing antibody, SC31, isolated from an early convalescent COVID-19 patient, are essential for the optimal therapeutic efficacy of the antibody. PLoS ONE. 2021;16:e0253487. doi: 10.1371/journal.pone.0253487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamin R., Jones A.T., Hoffmann H.H., Schäfer A., Kao K.S., Francis R.L., Sheahan T.P., Baric R.S., Rice C.M., Ravetch J.V., Bournazos S. Fc-engineered antibody therapeutics with improved anti-SARS-CoV-2 efficacy. Nature. 2021 doi: 10.1038/s41586-021-04017-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas S.J., Moreira E.D., Jr., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Polack F.P., Zerbini C., et al. C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine through 6 Months. N. Engl. J. Med. 2021;385:1761–1773. doi: 10.1056/NEJMoa2110345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Israel A., Shenhar Y., Green I., Merzon E., Golan-Cohen A., Schäffer A.A., Ruppin E., Vinker S., Magen E. Large-scale study of antibody titer decay following BNT162b2 mRNA vaccine or SARS-CoV-2 infection. medRxiv. 2021 doi: 10.1101/2021.08.19.21262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergwerk M., Gonen T., Lustig Y., Amit S., Lipsitch M., Cohen C., Mandelboim M., Levin E.G., Rubin C., Indenbaum V., et al. Covid-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med. 2021;385:1474–1484. doi: 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kustin T., Harel N., Finkel U., Perchik S., Harari S., Tahor M., Caspi I., Levy R., Leshchinsky M., Ken Dror S., et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2-mRNA-vaccinated individuals. Nat. Med. 2021;27:1379–1384. doi: 10.1038/s41591-021-01413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahil S.K., Bechman K., Raharja A., Domingo-Vila C., Baudry D., Brown M.A., Cope A.P., Dasandi T., Graham C., Lechmere T., et al. The effect of methotrexate and targeted immunosuppression on humoral and cellular immune responses to the COVID-19 vaccine BNT162b2: a cohort study. Lancet Rheumatol. 2021;3:e627–e637. doi: 10.1016/S2665-9913(21)00212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakuraba A., Luna A., Micic D. Serologic response to coronavirus disease 2019 (COVID-19) vaccination in patients with immune-mediated inflammatory diseases: a systematic review and meta-analysis. Gastroenterology. 2021 doi: 10.1053/j.gastro.2021.09.055. S0016-5085(21)03604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiorino G., Peyrin-Biroulet L., Naccarato P., Szabò H., Sociale O.R., Vetrano S., Fries W., Montanelli A., Repici A., Malesci A., Danese S. Effects of immunosuppression on immune response to pneumococcal vaccine in inflammatory bowel disease: a prospective study. Inflamm. Bowel Dis. 2012;18:1042–1047. doi: 10.1002/ibd.21800. [DOI] [PubMed] [Google Scholar]
- 40.Alexander J.L., Moran G.W., Gaya D.R., Raine T., Hart A., Kennedy N.A., Lindsay J.O., MacDonald J., Segal J.P., Sebastian S., et al. Inflammatory Bowel Disease section of the British Society of Gastroenterology and the the Inflammatory Bowel Disease Clinical Research Group SARS-CoV-2 vaccination for patients with inflammatory bowel disease: a British Society of Gastroenterology Inflammatory Bowel Disease section and IBD Clinical Research Group position statement. Lancet Gastroenterol. Hepatol. 2021;6:218–224. doi: 10.1016/S2468-1253(21)00024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy N.A., Goodhand J.R., Bewshea C., Nice R., Chee D., Lin S., Chanchlani N., Butterworth J., Cooney R., Croft N.M., et al. Contributors to the CLARITY IBD study Anti-SARS-CoV-2 antibody responses are attenuated in patients with IBD treated with infliximab. Gut. 2021;70:865–875. doi: 10.1136/gutjnl-2021-324388. [DOI] [PubMed] [Google Scholar]
- 42.Park S.H., Yang S.K., Park S.K., Kim J.W., Yang D.H., Jung K.W., Kim K.J., Ye B.D., Byeon J.S., Myung S.J., Kim J.H. Efficacy of hepatitis A vaccination and factors impacting on seroconversion in patients with inflammatory bowel disease. Inflamm. Bowel Dis. 2014;20:69–74. doi: 10.1097/01.MIB.0000437736.91712.a1. [DOI] [PubMed] [Google Scholar]
- 43.Pratt P.K., Jr., David N., Weber H.C., Little F.F., Kourkoumpetis T., Patts G.J., Weinberg J., Farraye F.A. Antibody Response to Hepatitis B Virus Vaccine is Impaired in Patients With Inflammatory Bowel Disease on Infliximab Therapy. Inflamm. Bowel Dis. 2018;24:380–386. doi: 10.1093/ibd/izx001. [DOI] [PubMed] [Google Scholar]
- 44.Haykir Solay A., Eser F. High dose hepatitis B vaccine is not effective in patients using immunomodulatory drugs: a pilot study. Hum. Vaccin. Immunother. 2019;15:1177–1182. doi: 10.1080/21645515.2019.1574151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cullen G., Bader C., Korzenik J.R., Sands B.E. Serological response to the 2009 H1N1 influenza vaccination in patients with inflammatory bowel disease. Gut. 2012;61:385–391. doi: 10.1136/gutjnl-2011-300256. [DOI] [PubMed] [Google Scholar]
- 46.Shirai S., Hara M., Sakata Y., Tsuruoka N., Yamamoto K., Shimoda R., Gomi Y., Yoshii H., Fujimoto K., Iwakiri R. Immunogenicity of Quadrivalent Influenza Vaccine for Patients with Inflammatory Bowel Disease Undergoing Immunosuppressive Therapy. Inflamm. Bowel Dis. 2018;24:1082–1091. doi: 10.1093/ibd/izx101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hua C., Barnetche T., Combe B., Morel J. Effect of methotrexate, anti-tumor necrosis factor α, and rituximab on the immune response to influenza and pneumococcal vaccines in patients with rheumatoid arthritis: a systematic review and meta-analysis. Arthritis Care Res. (Hoboken) 2014;66:1016–1026. doi: 10.1002/acr.22246. [DOI] [PubMed] [Google Scholar]
- 48.Andrisani G., Frasca D., Romero M., Armuzzi A., Felice C., Marzo M., Pugliese D., Papa A., Mocci G., De Vitis I., et al. Immune response to influenza A/H1N1 vaccine in inflammatory bowel disease patients treated with anti TNF-α agents: effects of combined therapy with immunosuppressants. J. Crohn’s Colitis. 2013;7:301–307. doi: 10.1016/j.crohns.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dailey J., Kozhaya L., Dogan M., Hopkins D., Lapin B., Herbst K., Brimacombe M., Grandonico K., Karabacak F., Schreiber J., et al. Antibody Responses to SARS-CoV-2 after Infection or Vaccination in Children and Young Adults with Inflammatory Bowel Disease. medRxiv. 2021 doi: 10.1093/ibd/izab207. 2021.2006.2012.21258810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edelman-Klapper H., Zittan E., Shitrit A.B.-G., Rabinowitz K.M., Goren I., Avni-Biron I., Ollech J.E., Lichtenstein L., Banai-Eran H., Yanai H., et al. Decreased Immune Response to COVID-19 mRNA Vaccine in Patients with Inflammatory Bowel Diseases Treated with Anti TNFα. medRxiv. 2021 doi: 10.1101/2021.08.22.21262263. [DOI] [Google Scholar]
- 51.Fu Y.X., Chaplin D.D. Development and maturation of secondary lymphoid tissues. Annu. Rev. Immunol. 1999;17:399–433. doi: 10.1146/annurev.immunol.17.1.399. [DOI] [PubMed] [Google Scholar]
- 52.Murphy K., Weaver C. Garland Science; 2016. Janeway’s immunobiology. [Google Scholar]
- 53.Pasparakis M., Alexopoulou L., Grell M., Pfizenmaier K., Bluethmann H., Kollias G. Peyer’s patch organogenesis is intact yet formation of B lymphocyte follicles is defective in peripheral lymphoid organs of mice deficient for tumor necrosis factor and its 55-kDa receptor. Proc. Natl. Acad. Sci. USA. 1997;94:6319–6323. doi: 10.1073/pnas.94.12.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suryadevara N., Shrihari S., Gilchuk P., VanBlargan L.A., Binshtein E., Zost S.J., Nargi R.S., Sutton R.E., Winkler E.S., Chen E.C., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331.e15. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ullah I., Prévost J., Ladinsky M.S., Stone H., Lu M., Anand S.P., Beaudoin-Bussières G., Symmes K., Benlarbi M., Ding S., et al. Live imaging of SARS-CoV-2 infection in mice reveals that neutralizing antibodies require Fc function for optimal efficacy. Immunity. 2021;54:2143–2158.e15. doi: 10.1016/j.immuni.2021.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gorman M.J., Patel N., Guebre-Xabier M., Zhu A.L., Atyeo C., Pullen K.M., Loos C., Goez-Gazi Y., Carrion R., Jr., Tian J.-H., et al. Fab and Fc contribute to maximal protection against SARS-CoV-2 following NVX-CoV2373 subunit vaccine with Matrix-M vaccination. Cell Rep. Med. 2021;2:100405. doi: 10.1016/j.xcrm.2021.100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaplonek P., Cizmeci D., Fischinger S., Collier A.R., Suscovich T., Linde C., Broge T., Mann C., Amanat F., Dayal D., et al. Subtle immunological differences in mRNA-1273 and BNT162b2 COVID-19 vaccine induced Fc-functional profiles. bioRxiv. 2021 doi: 10.1101/2021.08.31.458247. [DOI] [Google Scholar]
- 58.Hall V.G., Ferreira V.H., Ierullo M., Ku T., Marinelli T., Majchrzak-Kita B., Yousuf A., Kulasingam V., Humar A., Kumar D. Humoral and cellular immune response and safety of two-dose SARS-CoV-2 mRNA-1273 vaccine in solid organ transplant recipients. Am. J. Transplant. 2021 doi: 10.1111/ajt.16766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamar N., Abravanel F., Marion O., Couat C., Izopet J., Del Bello A. Three Doses of an mRNA Covid-19 Vaccine in Solid-Organ Transplant Recipients. N. Engl. J. Med. 2021;385:661–662. doi: 10.1056/NEJMc2108861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin C.X., Moore L.W., Anjan S., Rahamimov R., Sifri C.D., Ali N.M., Morales M.K., Tsapepas D.S., Basic-Jukic N., Miller R.A., et al. Risk of Breakthrough SARS-CoV-2 Infections in Adult Transplant Recipients. Transplantation. 2021;105:e265–e266. doi: 10.1097/TP.0000000000003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Lambson B.E., Vermeulen M., van den Berg K., Rossouw T., Boswell M., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv. 2021 doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 62.Tada T., Dcosta B.M., Samanovic-Golden M., Herati R.S., Cornelius A., Mulligan M.J., Landau N.R. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. bioRxiv. 2021 [Google Scholar]
- 63.McCallum M., Bassi J., Marco A., Chen A., Walls A.C., Iulio J.D., Tortorici M.A., Navarro M.J., Silacci-Fregni C., Saliba C., et al. SARS-CoV-2 immune evasion by variant B.1.427/B.1.429. bioRxiv. 2021 doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E., et al. NGS-SA Group. Wits-VIDA COVID Group Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shinde V., Bhikha S., Hoosain Z., Archary M., Bhorat Q., Fairlie L., Lalloo U., Masilela M.S.L., Moodley D., Hanley S., et al. 2019nCoV-501 Study Group Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021;384:1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. ENSEMBLE Study Group Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moor M.B., Suter-Riniker F., Horn M.P., Aeberli D., Amsler J., Möller B., Njue L.M., Medri C., Angelillo-Scherrer A., Borradori L., et al. Humoral and cellular responses to mRNA vaccines against SARS-CoV-2 in patients with a history of CD20 B-cell-depleting therapy (RituxiVac): an investigator-initiated, single-centre, open-label study. Lancet Rheumatol. 2021;3:e789–e797. doi: 10.1016/S2665-9913(21)00251-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ali A., Dwyer D., Wu Q., Wang Q., Dowling C.A., Fox D.A., Khanna D., Poland G.A., Mao-Draayer Y. Characterization of humoral response to COVID mRNA vaccines in multiple sclerosis patients on disease modifying therapies. Vaccine. 2021;39:6111–6116. doi: 10.1016/j.vaccine.2021.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prendecki M., Clarke C., Edwards H., McIntyre S., Mortimer P., Gleeson S., Martin P., Thomson T., Randell P., Shah A., et al. Humoral and T-cell responses to SARS-CoV-2 vaccination in patients receiving immunosuppression. Ann. Rheum. Dis. 2021;80:1322–1329. doi: 10.1136/annrheumdis-2021-220626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.VanBlargan L.A., Adams L.J., Liu Z., Chen R.E., Gilchuk P., Raju S., Smith B.K., Zhao H., Case J.B., Winkler E.S., et al. A potently neutralizing SARS-CoV-2 antibody inhibits variants of concern by utilizing unique binding residues in a highly conserved epitope. Immunity. 2021;54:2399–2416.e6. doi: 10.1016/j.immuni.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488.e4. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A., Tan J., Bhavsar D., Capuano C., Kirkpatrick E., et al. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 2020;57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brown E.P., Licht A.F., Dugast A.-S., Choi I., Bailey-Kellogg C., Alter G., Ackerman M.E. High-throughput, multiplexed IgG subclassing of antigen-specific antibodies from clinical samples. J. Immunol. Methods. 2012;386:117–123. doi: 10.1016/j.jim.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brown E.P., Dowell K.G., Boesch A.W., Normandin E., Mahan A.E., Chu T., Barouch D.H., Bailey-Kellogg C., Alter G., Ackerman M.E. Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J. Immunol. Methods. 2017;443:33–44. doi: 10.1016/j.jim.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fischinger S., Fallon J.K., Michell A.R., Broge T., Suscovich T.J., Streeck H., Alter G. A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J. Immunol. Methods. 2019;473:112630. doi: 10.1016/j.jim.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Butler A.L., Fallon J.K., Alter G. A Sample-Sparing Multiplexed ADCP Assay. Front. Immunol. 2019;10:1851. doi: 10.3389/fimmu.2019.01851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karsten C.B., Mehta N., Shin S.A., Diefenbach T.J., Slein M.D., Karpinski W., Irvine E.B., Broge T., Suscovich T.J., Alter G. A versatile high-throughput assay to characterize antibody-mediated neutrophil phagocytosis. J. Immunol. Methods. 2019;471:46–56. doi: 10.1016/j.jim.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sievert C. Chapman and Hall/CRC; 2020. Interactive Web-Based Data Visualization with R, plotly, and shiny. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
(a)
Data. All serological results described in this study are available within the body of the paper. All data (including raw data used to generate neutralizing and binding curves) reported in this paper will be shared by the lead contact upon request
-
(b)
Code. This paper does not report original code.
-
(c)
Additional information. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.