Abstract
While an E3 ubiquitin ligase, RNF8, was initially reported to be required for histone-to-protamine exchange in spermiogenesis, we subsequently demonstrated that RNF8 is not involved in this process. Nevertheless, reflecting a lingering misunderstanding in the field, a growing number of studies have continued to postulate a requirement for RNF8 in the histone-to-protamine exchange. For example, a recent study claimed that a mouse PIWI protein, MIWI, controls RNF8-mediated histone-to-protamine exchange. Here, confirming our earlier conclusions, we show that RNF8 is required neither for the establishment of histone H4K16 acetylation, which is an initial step in histone removal during spermiogenesis, nor for the incorporation of two protamine proteins, PRM1 and PRM2. Thus, whereas RNF8 mediates ubiquitination of H2A on the sex chromosomes in meiosis, during the prior stage of spermatogenesis, our genetic evidence underscores that RNF8 is not involved in histone-to-protamine exchange.
Keywords: sperm, spermiogenesis, histone-to-protamine exchange, MIWI, RNF8
Given controversy in the field, we rigorously re-examined the function of RNF8 and conclude that RNF8 is required for ubiquitination of the sex chromosomes during meiosis but not for histone-to-protamine exchange during spermiogenesis.
Introduction
To prepare fertilization, male germ cells undergo a massive nuclear transformation in which nuclear histones are largely replaced with highly basic proteins, called protamines, to produce condensed sperm nuclei. The mechanism underlying histone-to-protamine exchange remains largely unknown, although several key aspects have been identified. Genome-wide histone acetylation represents an initial signal for histone-to-protamine exchange [1]. Following acetylation, the histone variant H2A.L.2 works with transition proteins, TNP1 and TNP2, for the loading of protamines [2]. A histone-ubiquitin ligase RNF8 was reported to be required for genome-wide histone acetylation and subsequent histone-to-protamine exchange based on the absence, genome-wide, of histone acetylation, and of two protamine proteins, PRM1 and PRM2 in Rnf8-deficient (KO) mice [3]. However, using the same line of Rnf8-KO mice, we showed that genome-wide accumulation of histone H4K16 acetylation and incorporation of PRM1 took place in Rnf8-KO elongating spermatids; the effects of RNF8 on PRM2 deposition in elongating spermatids was not examined at that time [4]. In contrast, we did confirm a role for RNF8 in mediating genome-wide ubiquitination of H2A in elongating spermatids. Thus, our previous study demonstrated that RNF8-mediated ubiquitinated H2A is associated neither with H4K16 acetylation nor with the incorporation of PRM1 in elongating spermatids.
Nevertheless, later studies have continued to postulate a requirement for RNF8 in the histone-to-protamine exchange. Among these, a recent study claimed that a mouse PIWI protein, MIWI, controls RNF8-mediated histone-to-protamine exchange through the direct regulation of RNF8 by MIWI in spermiogenesis in mice [5]. PIWI proteins are evolutionarily conserved proteins that are expressed in germ cells and are associated with Piwi-interacting RNA (piRNA) for suppression of retrotransposons [6, 7]. MIWI is one of the two PIWI proteins that is expressed during the pachytene stage in mice [8] and loss of its function leads to spermiogenesis arrest [9]. Based on the finding that MIWI regulates RNF8, Gou et al. (2017) concluded that MIWI has a piRNA-independent function, but instead has an RNF8-dependent role in histone-to-protamine exchange. Furthermore, an additional study suggested that RNF8 is required for histone-to-protamine exchange based on the presence of testis-specific histone H2B in sperm nuclei of Rnf8-deficient mice [10]. These studies have spawned a lingering view in the field that RNF8 is required for histone-to-protamine exchange [11, 12].
In the present study, to clarify the function of RNF8 in histone-to-protamine exchange, we rigorously examined whether RNF8 is involved in this process in two independent laboratories at different institutions (the Namekawa and Li laboratories). Here, we show that histone-to-protamine exchange takes place in Rnf8-deficient mice, demonstrating that RNF8 is not required for histone-to-protamine exchange in spermiogenesis. Instead, RNF8 has a nuclear function in the ubiquitination of the sex chromosomes during meiosis and in the activation of sex chromosome-linked genes in postmeiotic spermatids [4, 13, 14]. Therefore, we refute the findings of Lu et al. (2010) that RNF8 is required for histone-protamine replacement. Furthermore, as a consequence, the infertility phenotype of MIWI mutant mice reported in Gou et al. (2017) cannot be explained by an RNF8-mediated mechanism in spermiogenesis, challenging the main mechanistic conclusion of that article.
Results and discussion
To confirm whether RNF8 is required for histone-to-protamine exchange, we (the Namekawa and Li laboratories) have independently examined Rnf8-deficient (KO) mice [15]. The results were consistent between our two labs, and here we present representative data. This Rnf8-KO mouse line was generated from an embryonic stem cell line, RRR260, in which the Rnf8 gene was disrupted by a gene trap mutation, and the absence of RNF8 protein was previously confirmed [16]. This same line of Rnf8-KO mice was widely used in previous studies of the function of RNF8 in spermatogenesis [3, 4, 10, 13, 14, 16–19], including the initial study by Lu et al. (2010), which suggested a requirement for RNF8 in mediating histone-to-protamine exchange.
Since Lu et al. reported that RNF8-dependent histone ubiquitination induces H4K16 acetylation as an initial step of histone removal [3], we first revisited whether H4K16 acetylation is RNF8 dependent or not. In accordance with our previous conclusion [4], we confirmed that the distribution of H4K16 acetylation was comparable between testicular sections of Rnf8-KO and control littermate wild-type (WT) testes (Figure 1A). This result was independently confirmed by comparable immunostaining signals of pan-acetylation on histone H4 between Rnf8-KO testes and WT controls (Figure 1B). However, in contrast to H4/H4K16 acetylation, RNF8 is required for H2A ubiquitination on entire nuclei of elongating spermatids [4]. Thus, we conclude that RNF8-dependent histone ubiquitination does not induce genome-wide H4K16 acetylation in elongating spermatids.
Figure 1 .
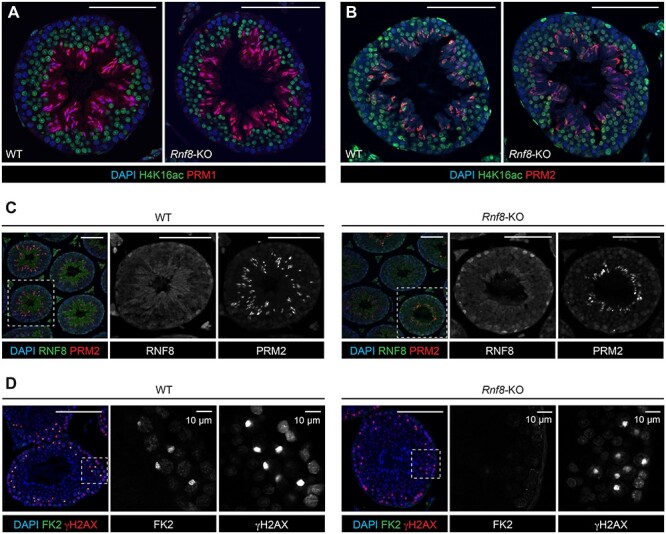
RNF8 is not required for H4K16 acetylation or for histone-to-protamine exchange in spermiogenesis. (A–D) Testis sections from WT and Rnf8-KO mice immunostained with antibodies shown in the panels (H4K16ac: H4K16 acetylation; H4ac: pan-acetyl H4; FK2: ubiquitination). Dashed squares are magnified in panels to the right. Scale bars: 100 μm unless specified.
Next, we examined the distribution of two distinct protamines (PRM1 and PRM2) on testicular sections of Rnf8-KO mice. We found that both PRM1 and PRM2 were present, and the distributions were comparable between Rnf8-KO testes and WT controls (Figure 1A-C). These results demonstrate that incorporation of two kinds of protamines (PRM1 and PRM2) takes place in spermiogenesis in Rnf8-KO testes. Also, consistent with previous studies [3, 4, 13, 14, 16, 19], we confirmed the loss-of-function of RNF8 in Rnf8-KO testes using the FK2 antibody that recognizes ubiquitinated proteins; in Rnf8-KO testes, FK2 signal (ubiquitination) was absent on the sex chromosomes, which were identified using γH2AX, a marker of the DNA damage response that regulates meiotic sex chromosome inactivation [18, 20] (Figure 1D).
To further confirm RNF8-independent protamine incorporation, we prepared smear slides from epididymal sperm of Rnf8-KO mice. Rnf8-KO epididymis contained sperm [3], suggesting that spermiogenic progression was not abrogated in Rnf8-KO testes. Furthermore, we found that PRM1 was present in Rnf8-KO epididymal sperm, and the distribution of PRM1 was comparable between Rnf8-KO and WT sperm (Figure 2A). PRM2 was also present in Rnf8-KO epididymal sperm, and the distribution of PRM2 was comparable between Rnf8-KO and WT sperm (Figure 2B). Importantly, PRM1 and PRM2 were observed in all sperm examined (PRM1 was present in 100% of sperm: n = 163 for WT and n = 202 for the Rnf8-KO; PRM2 also was present in 100% of sperm: n = 140 for WT and n = 135 for the Rnf8-KO). Together, these results confirm that RNF8 is not required for the histone-to-protamine exchange.
Figure 2 .
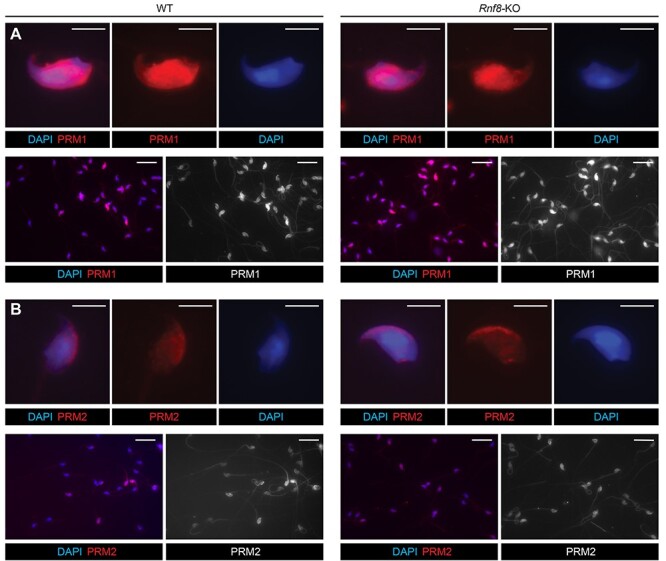
RNF8-independent incorporation of PRM1 and PRM2 in epididymal sperm. (A and B) WT and Rnf8-KO epididymal sperm immunostained with antibodies against PRM1 or PRM2. Upper panels: photos of representative sperm; Scale bars: 5 μm. Bottom panels: photos of multiple sperm; Scale bars: 25 μm.
A possible explanation for why the initial study by Lu et al. did not observe protamine incorporation into sperm nuclei [3] is the inability of Rnf8-KO spermatogenesis to reach the stage of protamine incorporation in certain specific genetic backgrounds or environments. In support of this possibility, Lu et al. showed that the late stages of spermiogenesis were abrogated in the Rnf8-KO testis [3]. On the other hand, Rnf8-KO mice in our colony did not show abrogation of spermiogenic progression, enabling us to confirm that RNF8 is not required for histone-to-protamine exchange. Curiously, the same Rnf8-KO mice were reported to be subfertile in another study [10]. While that study claimed that RNF8 is required for histone-protamine exchange, fertile Rnf8-KO sperm must have underwent normal histone-protamine exchange to enable fertilization, supporting our conclusion that RNF8 is not required for the histone-to-protamine exchange. These studies together suggest that the phenotypic outcomes (spermiogenic progression) may have been different among the various studies due to distinct genetic backgrounds or environments, thereby leading to conflicting conclusions about the role of RNF8 in the histone-to-protamine exchange. Specifically, some factor(s) associated with a particular genetic background and/or environment may have acted as a modifier of spermiogenic progression in Rnf8 KO mice.
In spite of the variable spermiogenic progression among studies, previous studies clearly agree that RNF8 has a function in male meiosis prior to spermiogenesis. RNF8 has a nuclear function in the regulation of sex chromosomes, and Rnf8-KO mice consistently show abrogation of histone ubiquitination on the sex chromosome during meiosis, as also observed by Lu et al. (2010) [3, 4, 13, 14, 16, 19] (Figure 1D). Notably, this phenotype was observed in 100% of the meiotic cells we have examined. This complete phenotype may be due to a requirement for the DDR pathway, which includes RNF8, in the regulation of unsynapsed sex chromosomes during meiosis [18, 20], and DDR-dependent coordination of meiotic progression [21]. Downstream of RNF8-mediated ubiquitination on the sex chromosomes, active epigenetic modifications are established on meiotic sex chromosomes, and a portion of sex chromosome-linked genes are activated in postmeiotic spermatids [4, 13, 14, 17]. Variable abnormal gene expression of sex chromosome-linked genes in Rnf8-KO testes could modulate phenotypic outcomes (i.e. spermiogenic progression) in distinct genetic backgrounds or environments among different studies [3, 4, 10]. Additionally, a chromatin-remodeling nuclear protein, CHD5 has been reported as another factor that controls hyperacetylation of H4 at K5/8/16, histone-to-protamine exchange and chromatin condensation in sperm heads [22, 23]. Notably, the Chd5-KO phenotype shows substantial similarity with the Rnf8-KO phenotype reported by Lu et al. (2010). This is an example of a factor that could potentially be affected by differences in genetic background or environment and thereby account for variable phenotypes of the Rnf8-KO in particular studies.
In the studies by Gou et al. (2017), which claimed that MIWI-dependent regulation of RNF8 is critical for histone-to-protamine exchange, the authors did not directly examine Rnf8-KO mice. However, the authors may have miscited the literature about the function of RNF8. We already described that RNF8 is not required for histone-to-protamine exchange [4], but in Gou et al. (2017), RNF8 was described to be “responsible for histone-to-protamine exchange [3], but not critical for protamine expression [4].” Therefore, the study by Gou et al., appeared to lack the literature-based foundation that RNF8 is responsible for histone-to-protamine exchange. Gou et al. described the phenotype of a novel MIWI mutation of the N-terminal D-box (heterozygous germline-specific knockin of the R218A/L221A mutations on a single allele of the Miwi gene, referred to as Miwi+/DB) [5]. The late stage of spermiogenesis was abrogated in the testes of Miwi+/DB mice [5]. Although the similarity between Miwi+/DB spermiogenesis and Rnf8-KO mice in the Lu et al. (2010) study was described [5], the most straightforward interpretation is that Miwi+/DB spermiogenesis did not reach the stage of histone-to-protamine exchange, and there is no direct evidence that the phenotype of Miwi+/DB spermiogenesis is due to the loss-of-function of RNF8. Instead, the case is built upon the demonstration of an interaction between MIWI and RNF8, and the fact that modulation of MIWI levels has an effect on RNF8 localization to nuclei.
Further, Gou et al. examined RNF8 using a commercial anti-RNF8 antibody. However, the antibody does not specifically recognize RNF8. We detected non-specific signals in sections of Rnf8-KO testes via immunohistochemistry and western blotting, respectively (Figure 1C). In fact, there was no evidence of a specific RNF8 signal on Western blots of lysates of Rnf8-KO testes either (Figure 3). This is a key point because as noted above, the evidence for a MIWI-dependent role of RNF8 in histone-to-protamine exchange was indirect and rested in significant part on a purported role for MIWI in controlling nuclear localization of RNF8; potential utilization of a non-specific antibody for that experiment clearly calls the result and conclusion into question. Taken together, the infertile phenotype of Miwi+/DB mice reported in Gou et al. (2017) cannot be explained by an RNF8-mediated mechanism in spermiogenesis, challenging the main mechanistic conclusion made by Gou et al. (2017).
Figure 3 .
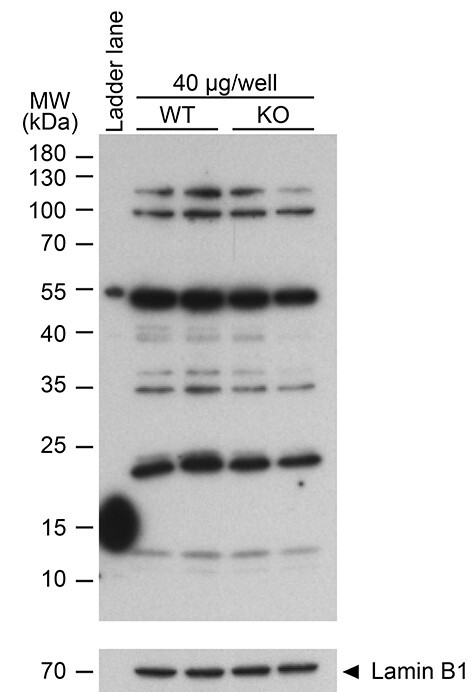
Failure of an anti-RNF8 antibody to specifically recognize RNF8. Western blot of WT and Rnf8-KO lysates from whole mouse testes probed with an anti-RNF8 antibody. 40 μg protein of sample was loaded in each lane. Two independent samples for WT and Rnf8-KO testes are shown. Loading control: Lamin B1. The expected molecular weight of RNF8 is 56–60 kDa. Of critical importance, the band most proximal to the expected size of RNF8 was equally present in extracts of WT and Rnf8-KO testes, and, additionally, no other prominent band displayed a clear difference between these genotypes.
Methods
Detailed information about antibodies used in this study is described in Supplementary Table S1.
Mouse lines
The Rnf8-KO mouse line we utilized was generated previously from the embryonic stem cell line RRR260 (Bay Genomics) [15] and is on a C57BL/6 background. We obtained the Rnf8-KO mouse line from Andre Nussenzweig’s laboratory, which used this line previously [16]. We maintained this same mouse line by intercrossing it for more than 10 years. All subsequent experimental work was performed under protocol no. IACUC2018-0040 approved by the Institutional Animal Care and Use Committee of Cincinnati Children’s Hospital Medical Center.
Immunohistochemistry
For the preparation of testis paraffin blocks, excised testes were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) containing 0.1% Triton X-100 at 4°C overnight. Testes were dehydrated and embedded in paraffin. For histological analyses, 6-μm-thick paraffin sections were deparaffinized. The sections were autoclaved in Target Retrieval Solution, Citrate pH 6.1 (DAKO, S-1700) or antigen unmasking solution, citric acid based (Vector, H-3300), 121°C, 100 kPa (15 psi) for 10 min. The sections were blocked with Blocking One Histo (Nacalai USA, 06349–64) at room temperature (RT) for 10 min; then, the sections were incubated with primary antibodies diluted in PBS at 4°C overnight. The following antibodies were used at the indicated dilutions [format: host anti-protein (source or company with product/catalog number), dilution, Research Resource Identifier (RRID)]: rabbit anti-H4K14ac (Millipore, 06–762), 1/200, AB_2310270; rabbit anti-H4ac (Millipore, 06–866), 1/200, N/A; mouse anti-protamine 1 (Briar Patch Biosciences, Hup1N), 1/200, N/A; goat anti-protamine 2 (Santa Cruz, sc-23,104), 1/200, AB_2284440; mouse anti-ubiquitinated protein antibody FK2 (Enzo Life Sciences, BML-PW8810), 1/200, AB_10541840; rabbit anti-γH2AX antibody (Cell Signaling Technology, 9718), 1/2000, AB_2118009; rabbit anti-RNF8 antibody (Proteintech, 14,112–1-AP), 1/200, N/A. The resulting signals were detected with appropriate secondary antibodies conjugated to Alexa 488 or 555 (ThermoFisher Scientific), diluted 1/1000 in PBS and incubated at RT for 1 h. Sections were counterstained for 5 min with the DNA-binding chemical 4′,6-diamidino-2-phenylindole (DAPI; Sigma, D9542-5MG) diluted to 1 μg/mL in water, then mounted with ProLong™ Gold Antifade Mountant (Thermo Fisher Scientific, P36930). Images were obtained with an A1RSi Inverted Confocal Microscope (Nikon) and processed using NIS-Elements Basic Research (Nikon), Photoshop (Adobe) softwares, and Illustrator (Adobe) softwares.
Smear preparations
Sperm were obtained from cauda epididymides of WT and Rnf8-KO mice at 11 weeks of age. To make sperm suspensions, first, a drop of 400 μL of M2 medium (Sigma, M7167) was prepared on a 35 mm plastic dish and it was fully covered with heavy paraffin oil (Sigma, PX0046–1); then, the dish was kept on a hot plate at 37°C until use. The obtained cauda epididymides were placed in the paraffin oil, and a drop of sperm was squeezed out from the cauda epididymides using a forcep and a needle. The sperm drop was then transferred into the drop of M2 medium. After incubation at 37°C, 5% CO2 for 30 min, sperm swimming up in M2 medium were collected using pipettes. After washing two times with PBS followed by centrifugation at 3000 g for 3 min at RT, sperm pellets were resuspended into 200 μL of PBS and sperm numbers were counted. An aliquot containing 1 × 105 sperm was used to make smears and slides were stored at −80°C until staining was performed.
Immunostaining of sperm
We followed a previously described protocol [24]. Mouse anti-Prm1 antibody (1:500 dilution; Briar Patch Biosciences, Livermore, CA, USA; Hup1N) and mouse anti-Prm2 antibody (1:200 dilution; Briar Patch Biosciences, Livermore, CA, USA; Hup2B) were used. Secondary antibodies conjugated with either Alexa Fluor 488 or 594 (Molecular Probes, Eugene, OR, USA) were used at a dilution of 1:500.
Western blotting
Western blot experiments were replicated twice; for each experiment, all samples were run on the same gel. Detunicated testis pieces obtained from mature adult WT and Rnf8-KO mice were homogenized in RIPA buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate) containing cOmplete Protease Inhibitor Cocktail (Roche, 11 697 498 001); then, the homogenate was incubated on ice for 30 min. After DNA fragmentation by sonication and subsequent centrifugation at 10 000 × g at 4°C, the supernatant was transferred to a new tube before total protein concentration was quantified via Bradford assays. Volumes of lysates containing 40 μg of proteins were separated by electrophoresis on 10% SDS–PAGE gels. Then, the proteins were transferred onto a PVDF membrane (EMD Millipore, IPVH00010). The membranes were blocked with StartingBlockTM T20 (TBS) Blocking Buffer (ThermoFisher Scientific, 37543) at RT for 30 min before incubation with rabbit anti-RNF8 antibody ((Proteintech, 14,112–1-AP), 1/2000, N/A) diluted in Tris-buffered saline containing 0.1% Tween 20 detergent (TBST), at 4°C overnight. On the next day, after washing three times in TBST, 5 min per wash, the blot was incubated with VeriBlot for IP Detection Reagent (HRP) ((Abcam, ab131366), 1/5000, N/A) diluted in TBST at RT, for 1 h. The blot was washed three times in TBST, 5 min per wash, before incubation in Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore, WBKLS0500) at RT for 1 min; then, the blot was imaged using Super RX-N x-ray film (Fujifilm) and a FluorChemQ MultiImage III instrument (Alpha Innotech). To blot loading controls, the initial blot was stripped with Restore Western Blot Stripping Buffer (ThermoFisher Scientific, 21059) at RT for 20 min; then, the stripped blot was washed two times in TBST, 5 min per wash followed by blocking with StartingBlockTM T20 (TBS) Blocking Buffer at RT for 30 min, prior to incubation 4°C overnight with anti-Lamin B1 antibody ((Abcam, ab16048), 1/2000, AB_443298) diluted in TBST. The next day, the blot was washed three times in TBST, 5 min per wash, then incubated with VeriBlot for IP Detection Reagent (HRP), diluted 1/5000 in TBST, at RT for 1 h, and bands were visualized through the procedures described above.
Supplementary Material
Acknowledgements
We thank X. Z. Li for contribution to this project, R. Balhorn for information on Protamine antibodies and usage, and all members of the Namekawa and Li laboratories for discussion and helpful comments.
Footnotes
† Grant Support: This work was supported by the NIH Grants R01GM134731 to P.R.A., and R01GM098605 to S.H.N.
Author Contributions. H.A. and S.H.N. designed the research. H.A. and R.M. and Z.L. performed the experiments. H.A., P.R.A. and S.H.N. wrote the manuscript. S.H.N. supervised the investigation. All authors reviewed the manuscript.
Contributor Information
Hironori Abe, Division of Reproductive Sciences, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA; Department of Microbiology & Molecular Genetics, University of California, California, USA.
Rajyalakshmi Meduri, Department of Biochemistry and Biophysics, Department of Urology, University of Rochester Medical Center, New York, USA.
Ziwei Li, Department of Biochemistry and Biophysics, Department of Urology, University of Rochester Medical Center, New York, USA.
Paul R Andreassen, Division of Experimental Hematology & Cancer Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA.
Satoshi H Namekawa, Division of Reproductive Sciences, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA; Department of Microbiology & Molecular Genetics, University of California, California, USA.
Conflict of interest
The authors declare no competing interests.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Goudarzi A, Shiota H, Rousseaux S, Khochbin S. Genome-scale acetylation-dependent histone eviction during spermatogenesis. J Mol Biol 2014; 426:3342–3349. [DOI] [PubMed] [Google Scholar]
- 2. Barral S, Morozumi Y, Tanaka H, Montellier E, Govin J, de Dieuleveult M, Charbonnier G, Coute Y, Puthier D, Buchou T, Boussouar F, Urahama T et al. Histone variant H2A.L.2 guides transition protein-dependent protamine assembly in male germ cells. Mol Cell 2017; 66:89, e108–101. [DOI] [PubMed] [Google Scholar]
- 3. Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL, Yu X. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell 2010; 18:371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sin HS, Barski A, Zhang F, Kartashov AV, Nussenzweig A, Chen J, Andreassen PR, Namekawa SH. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev 2012; 26:2737–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gou LT, Kang JY, Dai P, Wang X, Li F, Zhao S, Zhang M, Hua MM, Lu Y, Zhu Y, Li Z, Chen H et al. Ubiquitination-deficient mutations in human Piwi cause male infertility by impairing histone-to-protamine exchange during Spermiogenesis. Cell 2017; 169:1090, e1013–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol 2011; 12:246–258. [DOI] [PubMed] [Google Scholar]
- 7. Juliano C, Wang J, Lin H. Uniting germline and stem cells: The function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet 2011; 45:447–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, Zamore PD. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 2013; 50:67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deng W, Lin H. Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2002; 2:819–830. [DOI] [PubMed] [Google Scholar]
- 10. Guo Y, Song Y, Guo Z, Hu M, Liu B, Duan H, Wang L, Yuan T, Wang D. Function of RAD6B and RNF8 in spermatogenesis. Cell Cycle 2018; 17:162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. WE-I-CBOR Y. piRNA-independent PIWI function in spermatogenesis and male fertility. Biol Reprod 2017; 96:1121–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang X, Kang JY, Wei L, Yang X, Sun H, Yang S, Lu L, Yan M, Bai M, Chen Y, Long J, Li N et al. PHF7 is a novel histone H2A E3 ligase prior to histone-to-protamine exchange during spermiogenesis. Development 2019; 146:dev175547. [DOI] [PubMed] [Google Scholar]
- 13. Hasegawa K, Sin HS, Maezawa S, Broering TJ, Kartashov AV, Alavattam KG, Ichijima Y, Zhang F, Bacon WC, Greis KD, Andreassen PR, Barski A et al. SCML2 establishes the male germline epigenome through regulation of histone H2A ubiquitination. Dev Cell 2015; 32:574–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adams SR, Maezawa S, Alavattam KG, Abe H, Sakashita A, Shroder M, Broering TJ, Sroga Rios J, Thomas MA, Lin X, Price CM, Barski A et al. RNF8 and SCML2 cooperate to regulate ubiquitination and H3K27 acetylation for escape gene activation on the sex chromosomes. PLoS Genet 2018; 14:e1007233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Minter-Dykhouse K, Ward I, Huen MS, Chen J, Lou Z. Distinct versus overlapping functions of MDC1 and 53BP1 in DNA damage response and tumorigenesis. J Cell Biol 2008; 181:727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Santos MA, Huen MS, Jankovic M, Chen HT, Lopez-Contreras AJ, Klein IA, Wong N, Barbancho JL, Fernandez-Capetillo O, Nussenzweig MC, Chen J, Nussenzweig A. Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8. J Exp Med 2010; 207:973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sin HS, Kartashov AV, Hasegawa K, Barski A, Namekawa SH. Poised chromatin and bivalent domains facilitate the mitosis-to-meiosis transition in the male germline. BMC Biol 2015; 13:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ichijima Y, Ichijima M, Lou Z, Nussenzweig A, Camerini-Otero RD, Chen J, Andreassen PR, Namekawa SH. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev 2011; 25:959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu LY, Xiong Y, Kuang H, Korakavi G, Yu X. Regulation of the DNA damage response on male meiotic sex chromosomes. Nat Commun 2013; 4:2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ichijima Y, Sin HS, Namekawa SH. Sex chromosome inactivation in germ cells: Emerging roles of DNA damage response pathways. Cell Mol Life Sci 2012; 69:2559–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abe H, Alavattam KG, Hu YC, Pang Q, Andreassen PR, Hegde RS, Namekawa SH. The initiation of meiotic sex chromosome inactivation sequesters DNA damage Signaling from autosomes in mouse spermatogenesis. Curr Biol 2020; 30:408–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li W, Wu J, Kim SY, Zhao M, Hearn SA, Zhang MQ, Meistrich ML, Mills AA. Chd5 orchestrates chromatin remodelling during sperm development. Nat Commun 2014; 5:3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhuang T, Hess RA, Kolla V, Higashi M, Raabe TD, Brodeur GM. CHD5 is required for spermiogenesis and chromatin condensation. Mech Dev 2014; 131:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Balhorn R, Steger K, Bergmann M, Schuppe HC, Neuhauser S, Balhorn MC. New monoclonal antibodies specific for mammalian protamines P1 and P2. Syst Biol Reprod Med 2018; 64:424–447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.


