Abstract
Background & Aims
Notch pathway signaling maintains gastric epithelial cell homeostasis by regulating stem cell proliferation and differentiation. We previously identified NOTCH1 and NOTCH2 as the key Notch receptors controlling gastric stem cell function. Here, we identify the niche cells and critical Notch ligand responsible for regulating stem cell proliferation in the distal mouse stomach.
Methods
Expression of Notch ligands in the gastric antrum was determined by quantitative reverse-transcriptase polymerase chain reaction and cellular localization was determined by in situ hybridization and immunostaining. The contribution of specific Notch ligands to regulate epithelial cell proliferation in adult mice was determined by inducible gene deletion, or by pharmacologic inhibition using antibodies directed against specific Notch ligands. Mouse gastric organoid cultures were used to confirm that Notch ligand signaling was epithelial specific.
Results
Delta-like 1 (DLL1) and Jagged 1 (JAG1) were the most abundantly expressed Notch ligands in the adult mouse stomach, with DLL1 restricted to the antral gland base and JAG1 localized to the upper gland region. Inhibition of DLL1 alone or in combination with other Notch ligands significantly reduced epithelial cell proliferation and the growth of gastric antral organoids, while inhibition of the other Notch ligands, DLL4, JAG1, and JAG2, did not affect proliferation or organoid growth. Similarly, DLL1, and not DLL4, regulated proliferation of LGR5+ antral stem cells, which express the NOTCH1 receptor.
Conclusions
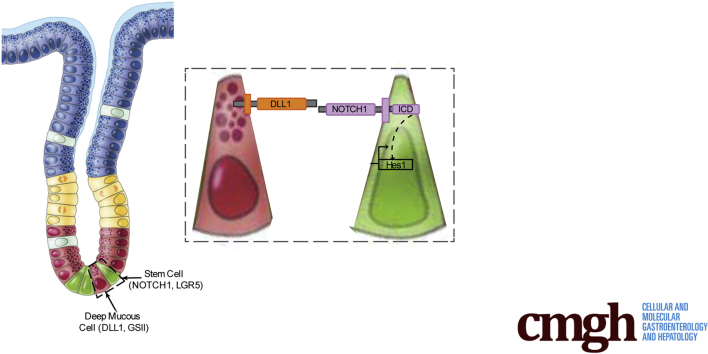
DLL1 is the key Notch ligand regulating epithelial cell proliferation in the gastric antrum. We propose that DLL1-expressing cells at the gland base are Notch niche cells that signal to adjacent LGR5+ antral stem cells to regulate stem cell proliferation and epithelial homeostasis.
Keywords: Notch Signaling, Stem Cell Niche, Gastric Organoids, Notch Inhibitory Antibodies, LGR5 Stem Cells
Abbreviations used in this paper: DLL, Delta-like; fl, floxed; GFP, green fluorescent protein; JAG, Jagged; TX, tamoxifen
Graphical abstract
Summary.
We identify Delta-like 1 as the key Notch ligand that regulates LGR5+ stem cell proliferation in the antral region of the adult mouse stomach. Delta-like 1–expressing cells at the gland base signal to adjacent stem cells to promote proliferation.
The gastric epithelium is renewed by a population of actively cycling stem and progenitor cells that form the differentiated cell lineages that maintain tissue function. Located in the isthmus of gastric corpus glands and at the gland base in the gastric antrum, distinct stem cell populations in these 2 regions of the stomach fuel epithelial cell renewal throughout lifespan.1 The local signaling environment, or “niche,” regulates stem cell function to control the balance of cellular proliferation and differentiation to maintain cellular homeostasis. In the distal, antral region of the stomach, stem cells marked by the R-spondin receptor LGR5 generate all of the differentiated lineages, including deep mucous cells, which lie adjacent to the stem cells at the gland base, surface mucous cells, and endocrine cells.2 Key niche signaling pathways that have been shown to regulate gastric stem cell function include Wnt2,3 and Notch.4, 5, 6, 7 However, the specific signaling ligands regulating stem cell function, and the identification of the niche cells that express these ligands are not understood.
Notch signaling has been shown to be essential for proliferation and self-renewal of gastric stem cells. Notch inhibition using pharmacologic or genetic approaches in adult mouse, as well as in human organoid models, showed decreased gastric stem cell function, including reduced stem and progenitor cell proliferation.4, 5, 6, 7 Conversely, genetic Notch activation in LGR5+ antral stem cells induced stem cell proliferation, antral gland fission, tissue expansion, and eventual hyperplastic polyp formation.4 Further, both mouse and human gastric tumors exhibit increased Notch signaling associated with increased expression of Notch pathway components, including Notch ligands, receptors, and target genes.8, 9, 10 We recently showed that both human gastric cancer cell lines and cancer-derived organoids are dependent on Notch for enhanced growth.8 Overall, these findings demonstrate the importance of Notch signaling to maintain antral stem cell homeostasis and suggest that Notch dysregulation is associated with gastric tumorigenesis. Thus, defining the signaling components and the cellular basis for Notch niche signaling is important to understand stem cell mechanisms driving gastric tissue health and disease.
In mammals there are 4 Notch receptors (NOTCH1–4) and 5 ligands (Delta-like 1 [DLL1], DLL3, DLL4, Jagged 1 [JAG1], and JAG2).11 In contrast to most stem cell niche signals, Notch requires cell-cell interaction for signal transduction, with both Notch ligands and receptors serving as integral membrane proteins. Ligand-receptor engagement on neighboring cells triggers sequential proteolytic cleavage events at the cell membrane to release an intracellular signaling fragment of the Notch receptor, which moves to the nucleus to activate target gene transcription. Previous studies have identified the key Notch receptors regulating LGR5+ antral stem cell function, showing that NOTCH1 and NOTCH2 together regulate antral homeostasis in both mouse and human. Further, NOTCH1 and NOTCH2 have been shown to be activated to signal in gastric stem cells.4,6 However, the Notch ligands and niche cells regulating signaling in gastric stem cells have not been identified.
Here, we identify the key Notch ligand responsible for regulating stem and progenitor cell proliferation in the mouse gastric antrum. We show using both pharmacologic and genetic models of Notch ligand inhibition that DLL1 is the predominant ligand that signals to LGR5+ antral stem cells to support gastric epithelial cell proliferation. Further, we localized DLL1-expressing cells to the antral gland base adjacent to LGR5+ stem cells to define the cellular basis for the Notch niche.
Results
Notch Ligand Expression in the Adult Mouse Stomach: Localization of DLL1 to the Antral Stem Cell Zone
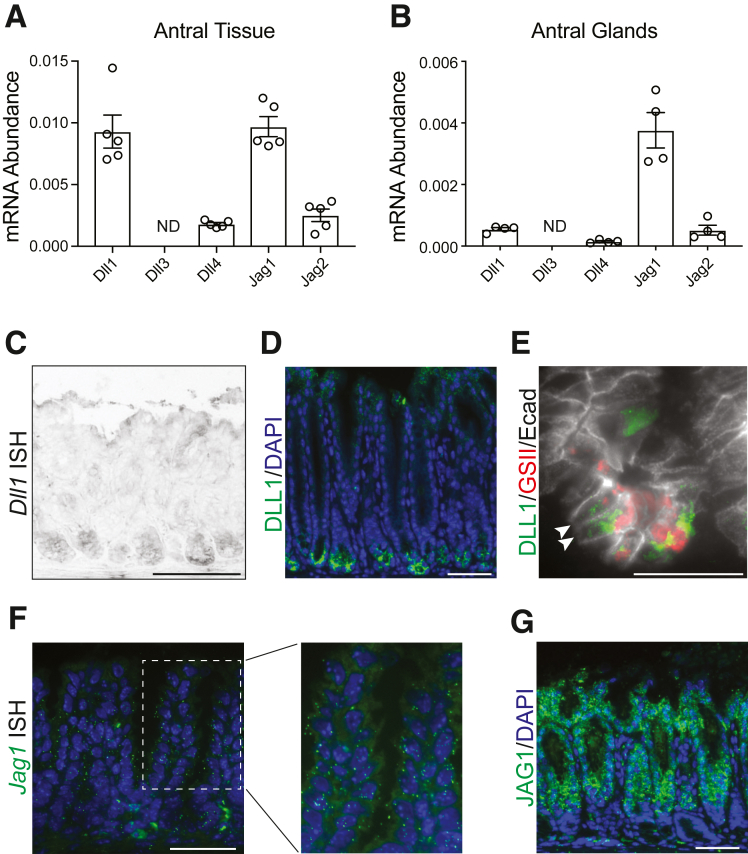
We first characterized expression of the 5 Notch ligands in the adult mouse stomach. Gene expression analysis revealed Dll1 and Jag1 to be the primary Notch ligands expressed in full-thickness gastric antral tissue (Figure 1A), whereas Jag1 was the predominant ligand expressed in isolated antral glands (Figure 1B). We next mapped the cellular localization of these 2 ligands in antral tissue sections. In situ hybridization (Figure 1C) and immunostaining (Figure 1D) revealed that DLL1 is expressed in cells at the base of the antral glands, where LGR5+ stem cells and deep mucous cells are located.2 Visualizing deep mucous cells with GSII lectin staining together with DLL1 and the epithelial membrane marker E-cadherin showed colocalization of GSII and DLL1 in epithelial cells at the gland base (Figure 1E). In contrast, in situ hybridization (Figure 1F) and immunostaining (Figure 1G) showed that JAG1 was absent from the gland base, with expression detected broadly in the mid and upper gland regions.
Figure 1.
Notch ligand expression in the mouse gastric antrum. (A, B) Measurement of the abundance of messenger RNAs (mRNAs) encoding the Notch ligands Dll1, Dll3, Dll4, Jag1, and Jag2 by quantitative reverse-transcriptase polymerase chain reaction in (A) full-thickness antral tissue or (B) isolated antral glands. Data are normalized to the expression of Gapdh and values are presented as mean ± SEM (n = 4–5 mice as indicated). (C, D) Localization of DLL1-expressing cells to the antral gland base by (C) in situ hybridization and (D) immunostaining (green) with DAPI (blue) nuclear counterstain. (E) Immunostaining for DLL1 (green) and the epithelial membrane marker E-cadherin (Ecad) (white), with GSII lectin-labeled deep mucous cells (red). Arrowheads identify epithelial cells at the gland base that coexpress DLL1 and GSII. (F) RNAscope in situ hybridization for Jag1 mRNA. The dashed box is shown at higher magnification in the right panel. (G) Localization of JAG1-expressing cells via immunostaining (green), with DAPI (blue) nuclear stain. Scale bars: (C, D, F, G) 50 μm, (E) 25 μm. ND, not detected.
DLL1 Promotes Gastric Antral Epithelial Proliferation
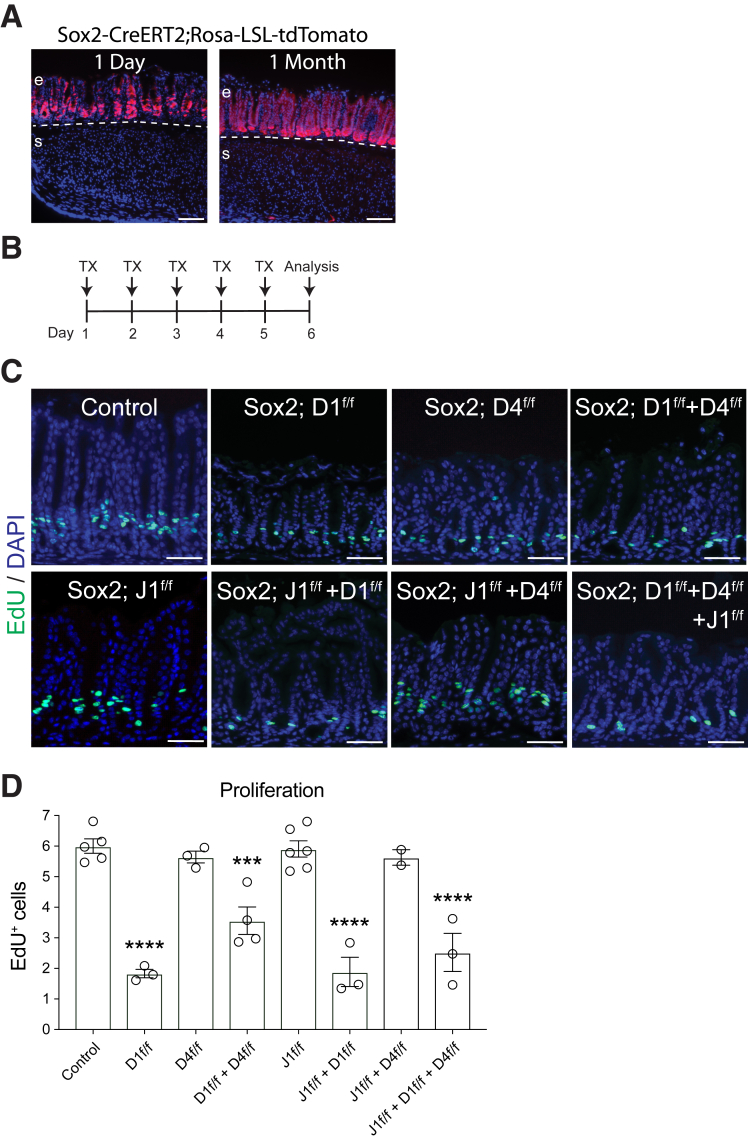
To define the key Notch ligands that regulate epithelial cell proliferation, we examined the effect of inhibition of 1 or more gastric Notch ligands in adult mice. We first used a genetic approach with mice carrying floxed (fl) Dll1, Dll4, and Jag1 alleles crossed to Sox2-CreERT2 mice to delete specific Notch ligands in the gastric epithelium. We showed that Sox2-CreERT2 is broadly expressed in antral epithelial cells, but not in stromal cells, by reporter gene analysis in Sox2-CreERT2; Rosa-LSL-tdTomato mice 1 day post–tamoxifen (TX) treatment (Figure 2A). The reporter analysis also confirmed that Sox2-CreERT2 is expressed in antral stem cells, with full gland lineage tracing observed 1 month post-TX (Figure 2A). Sox2-CreERT2 mice were crossed to Dll1fl/fl, Dll4 fl/fl, and/or Jag1 fl/fl mice. The various genotypes were treated with TX daily for 5 days and analyzed 1 day later to measure the effect on antral cell proliferation (Figure 2B). Importantly, deletion of Dll1 led to a significant reduction in epithelial cell proliferation in comparison with control TX-treated mice (Figure 2C and D). In contrast, deletion of Jag1 or Dll4 did not affect epithelial cell proliferation (Figure 2C and D). Notably, combined deletion of Dll1 with Dll4 and/or Jag1 had a similar effect as single Dll1 deletion (Figure 2C and D). Together, the findings suggest that DLL1 is the sole Notch ligand regulating epithelial cell proliferation in the stomach.
Figure 2.
Genetic deletion of Dll1 affects gastric antral epithelial cell proliferation. (A) Cellular expression pattern of Sox2-CreERT2 in the mouse antrum. Lineage tracing was examined in Sox2-CreERT2; Rosa-LSL-tdTomato mice 1 day or 1 month post–TX treatment as indicated. Dashed line indicates the boundary between epithelium (e) and stroma (s). (B) Schematic of TX treatment to induce Notch ligand deletion in the adult stomachs of Sox2-CreERT2 (Sox2) mice crossed to fl Notch ligand mice (Dll1f/f, Dll4f/f, Jag1f/f). The control group includes TX-treated mice containing fl Notch ligand alleles, but without Sox2-CreERT2. (C) Proliferation was assessed in the gastric antrum in the various TX-treated strains by EdU incorporation (green), with DAPI (blue) nuclear stain. (D) Morphometric quantification of EdU+ cells in control and ligand-deleted mice. The number of EdU+ cells per epithelial area (μm2) × 104 is presented as mean ± SEM (n = 2–6 mice/group as indicated). ∗∗∗P < .001, ∗∗∗∗P < .0001 vs control using 1-way analysis of variance with Dunnett’s posttest. Scale bars: (A) 100 μm, (C) 50 μm.
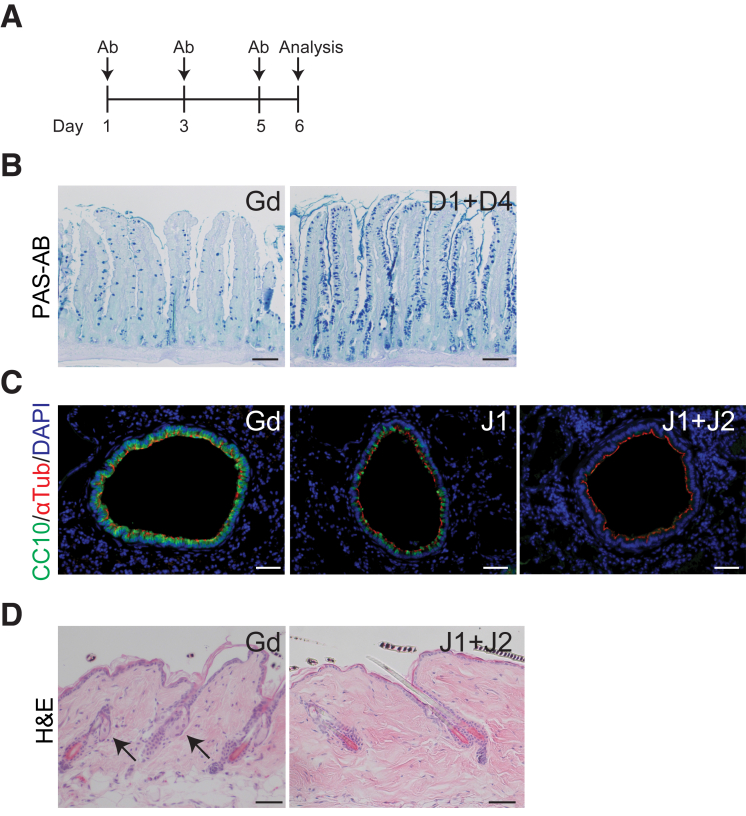
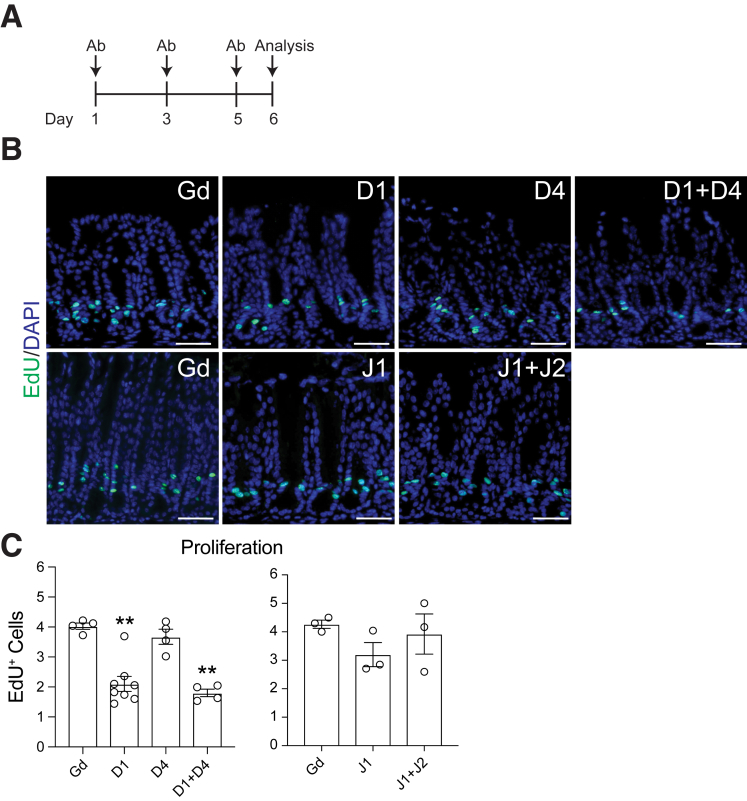
The Sox2-CreERT2 driver is known to be patchy, with only a portion of the antral glands showing recombination.12 Furthermore, TX treatment is known to induce gastric toxicity in the more proximal, corpus region of the stomach.13,14 Thus, to solidify the conclusions of our genetic studies, we used a pharmacologic approach to block Notch ligand signaling with neutralizing antibodies against DLL1, DLL4, JAG1, or JAG2.15, 16, 17 Furthermore, this approach also allowed us to test the role of JAG2. Use of these inhibitory antibodies generates a more complete inhibition of Notch ligand signaling across the tissue, while also avoiding the potential confounding effects of gastric TX toxicity. Mice were treated with inhibitory antibodies every other day for 5 days, and antral proliferation was analyzed on day 6 (Figures 3A and 4A), a time frame similar to our genetic Notch inhibition studies.
Figure 3.
Confirmation of efficacy of inhibition of Notch ligand signaling using neutralizing antibodies. (A) Schematic of antibody treatment with inhibitory antibodies (Ab) directed against Notch ligands DLL1 (D1), DLL4 (D4), JAG1 (J1), and JAG2 (J2) or control Gd antibody. Antibody dosing was as described in the Materials and Methods. Note that the Gd control for D1 and D4 inhibition experiments used a dose of 10 mg/kg, while the Gd dose for J1 and J2 inhibition was 15 mg/kg. (B) The effective block of D1 and D4 signaling was confirmed in small intestine by observation of goblet cell hyperplasia in duodenal sections stained with periodic acid Schiff-Alcian blue (PAS-AB). (C) The effective block of J1 and J2 signaling was demonstrated by observation of club-cell-to-ciliated-cell transformation in lung airway epithelium shown by co-immunostaining for the club cell marker CC10 (green) and the ciliated cell marker acetylated α-tubulin (αTub) (red) with DAPI (blue) nuclear stain. (D) Further effectiveness of J1 and J2 antibody treatment was demonstrated in hematoxylin and eosin (H&E)–stained skin tissue by the loss of mature sebocytes (arrows) in hair follicles. Scale bars: (B) 100 μm or (C, D) 50 μm.
Figure 4.
Inhibitory antibody treatment shows DLL1 signaling promotes gastric antral epithelial cell proliferation. (A) Schematic of Notch ligand inhibitory antibody treatment as described in Figure 3. (B) Proliferation was assessed in Notch inhibitory antibody-treated gastric antral sections by staining for EdU incorporation (green), with DAPI (blue) nuclear stain. (C) Morphometric quantification of EdU+ cells in control and antibody-treated mice. The number of EdU+ cells per epithelial area (μm2) × 104 is presented as mean ± SEM (n = 3–8 mice/group as indicated). ∗∗P < .01 vs Gd using 1-way analysis of variance with Dunnett’s posttest. Scale bars: 50 μm.
We first confirmed effective DLL1 and DLL4 inhibition by analyzing goblet cell differentiation in the intestinal epithelium, observing the expected goblet cell hyperplasia after treatment with combined anti-DLL1 and anti-DLL4 antibodies (Figure 3B).18 To confirm effective inhibition of JAG1 and JAG2 signaling, we analyzed both lung and skin epithelial tissues. Histological analysis showed that treatment with anti-JAG1 + anti-JAG2 antibodies produced the expected loss of differentiated club cells in the lung epithelium, as demonstrated by staining for the club cell marker CC10, and increased differentiation of ciliated cells marked by acetylated α-tubulin (Figure 3C).15 Further validation was demonstrated by loss of mature sebocytes in the skin epithelium after treatment with anti-JAG1 + anti-JAG2 antibodies (Figure 3D).19,20
Having confirmed the efficacy of the antibody treatments, we next analyzed gastric epithelial cell proliferation in Notch ligand inhibitory antibody-treated animals. This analysis confirmed the results of our genetic studies. We observed a significant reduction in proliferation in anti-DLL1–treated animals alone or in combination with anti-DLL4 (Figure 4B and C). Treatment with anti-DLL4 alone had no effect on proliferation. Similarly, treatment with anti-JAG1 alone or combined with anti-JAG2 did not affect epithelial cell proliferation (Figure 4B and C). Taken together, our genetic and pharmacologic inhibition studies showed a prominent role for DLL1 to support epithelial cell proliferation in the antrum.
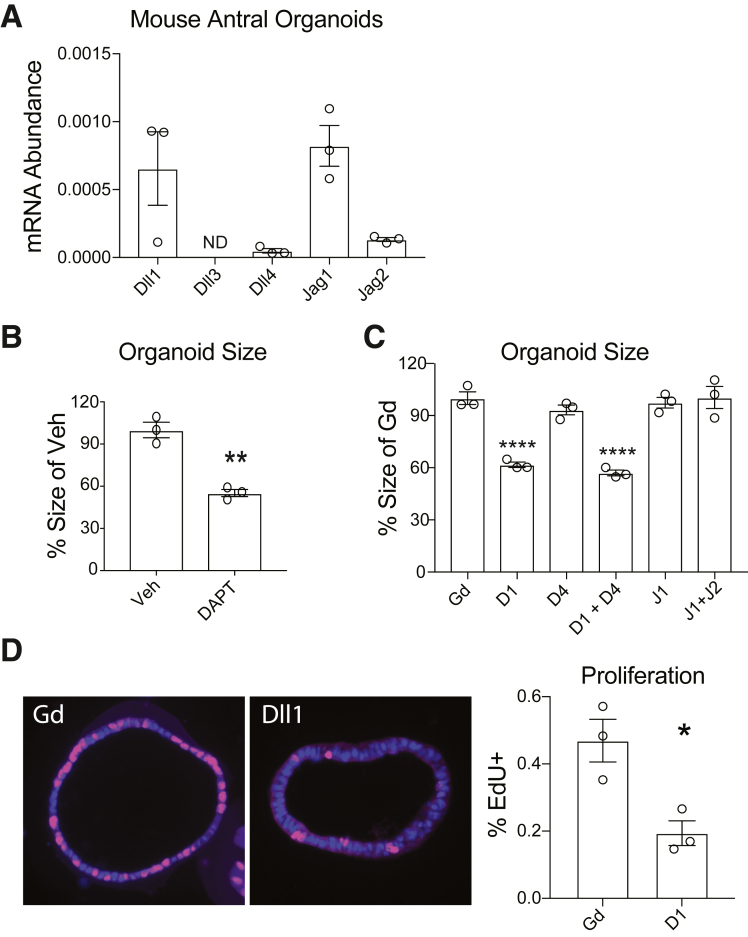
DLL1 Promotes Antral Organoid Growth
Notch signaling occurs in numerous cells in the stomach, including epithelial cells as well as stromal cell types,4,6 thus complicating the identification of Notch niche cells regulating gastric stem cells. We tested whether the DLL1 effect on proliferation was epithelial cell specific using the gastric organoid culture system. Antral organoids have been demonstrated to establish intrinsic Notch pathway activity without addition of Notch ligands to the culture system.6 Thus, this model system allowed us to study gastric epithelial Notch function. We first analyzed messenger RNA expression of Notch ligands in antral organoids. This analysis revealed that Dll1 and Jag1 were the most abundantly expressed Notch ligands (Figure 5A), which is consistent with our findings from analysis of gastric antral full-thickness tissue and isolated antral glands (Figure 1). Addition of the pan-Notch inhibitor DAPT to the culture medium reduced organoid growth (Figure 5B), consistent with previous reports demonstrating the requirement of Notch signaling to support gastric stem cell activity in both mouse and human organoids.4, 5, 6 To define the specific ligands responsible for the Notch effect, we added Notch ligand inhibitory antibodies to the culture media and measured organoid growth. This analysis revealed a significant reduction in organoid growth after anti-DLL1 treatment, either alone or in combination with anti-DLL4 (Figure 5C). Furthermore, treatment with anti-DLL4 alone, or anti-JAG1 or anti-JAG2, did not affect organoid growth (Figure 5C). To confirm that the DLL1 effect on organoid growth was due to reduced proliferation, we measured EdU incorporation in control (Gd) or DLL1 antibody-treated cultures. Quantification of EdU+ cells/total cells showed a significant reduction in proliferation with DLL1 inhibition (Figure 5D). Together, these findings are consistent with our in vivo analysis of Notch ligand function, suggesting that DLL1 expression from an epithelial Notch niche cell promotes antral epithelial cell proliferation.
Figure 5.
DLL1 regulates mouse gastric antral organoid growth. (A) Measurement of the abundance of mRNAs encoding the Notch ligands Dll1, Dll3, Dll4, Jag1, and Jag2 by quantitative reverse-transcriptase polymerase chain reaction in mouse antral organoids. Data are normalized to the expression of Gapdh and values are presented as mean ± SEM (n = 3 technical replicates from 1 organoid line). (B) Organoid size after treatment with the global Notch inhibitor DAPT or vehicle (Veh) was measured by morphometric analysis, and values are presented as mean ± SEM. ∗∗P < .01 vs Veh control using Student’s t test (n = 3 technical replicates from 1 organoid line). (C) Morphometric analysis of organoid growth following Notch ligand inhibitory antibody treatment or Gd control, as indicated. Data are presented as mean ± SEM. ∗∗∗∗P < .0001 vs Gd using 1-way analysis of variance with Dunnett’s posttest (n = 3 technical replicates from 1 organoid line). (D) Analysis of organoid proliferation by measurement of EdU incorporation following DLL1 inhibitory antibody treatment or Gd control. Left panels show representative images of organoid sections stained for EdU (red) and DAPI nuclear stain (blue). The percentage of EdU-positive nuclei are presented as mean ± SEM. ∗P < .05 vs Gd control by Student’s t test (n = 3 independent organoid lines from different mice). ND, not detected.
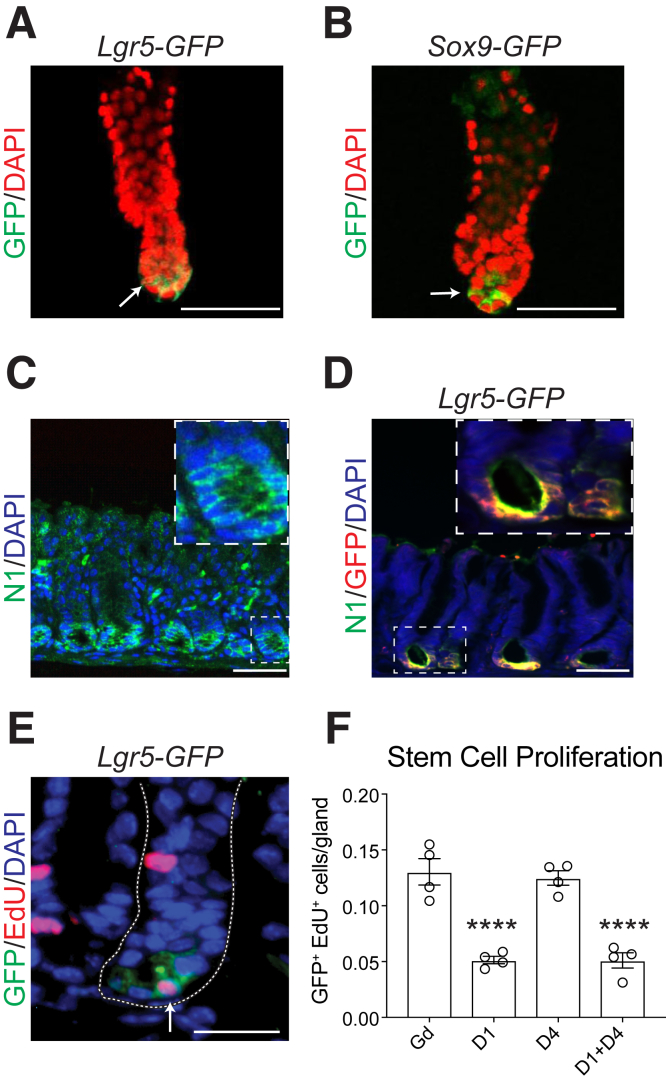
DLL1 Supports LGR5+ Antral Stem Cells
We examined the distribution pattern of antral LGR5+ stem cells by imaging green fluorescent protein (GFP)–labeled stem cells in antral glands prepared from Lgr5-GFP-CreERT2 (Lgr5-GFP) mice. This analysis showed GFP-expressing stem cells interspersed between large cells at the gland base (Figure 6A). A similar pattern of GFP-marked stem cells interspersed with large cells at the gland base was also observed in Sox9-GFP reporter mice (Figure 6B), suggesting that Sox9 marks antral stem cells, as had been previously demonstrated for intestinal stem cells.21 Based on localization of DLL1 to the antral gland base (Figure 1), we hypothesized that DLL1-expressing epithelial cells at the gland base signal directly to neighboring LGR5+ stem cells to regulate their function. Consistent with this signaling axis, immunostaining analysis revealed that the NOTCH1 receptor was expressed at the antral gland base (Figure 6C). Furthermore, costaining for NOTCH1 and GFP in Lgr5-GFP mice revealed that LGR5+ antral stem cells express the NOTCH1 receptor (Figure 6D). This finding is consistent with previous reports demonstrating that NOTCH1 and NOTCH2 receptors regulate LGR5+ antral stem cell function.6
Figure 6.
DLL1 signaling promotes antral LGR5-stem cell proliferation. (A, B) Whole-mount immunostaining for GFP (green) in gastric antral glands isolated from (A) Lgr5-GFP-CreERT2 (Lgr5-GFP) or (B) Sox9-GFP mice. Arrows indicate GFP+ stem cells. DAPI (pseudo-colored red) nuclear stain. (C) Immunostaining for the NOTCH1 receptor (green) in the adult mouse antrum, with DAPI (blue) nuclear stain. The inset shows DLL1 membrane staining at the antral gland base in the gland marked with the dashed line. (D) Co-immunostaining for the NOTCH1 receptor (green) and GFP (red) in Lgr5-GFP mouse antrum, with DAPI (blue) nuclear stain. The marked area is magnified in inset to illustrate NOTCH1/GFP coexpression. (E) Immunostaining for GFP (green) and the proliferation marker EdU (red) in antral sections from Gd- or inhibitory antibody–treated Lgr5-GFP mice. The arrow indicates an EdU+/GFP+ proliferating LGR5+ antral stem cell, and the white dotted line outlines an individual antral gland. DAPI (blue) nuclear stain. (F) Morphometric analysis of EdU+/GFP+ cells after antibody treatment. Data are presented as mean ± SEM (n = 4 mice/group, 32–74 glands/mouse). ∗∗∗∗P < .0001, vs Gd using 1-way analysis of variance with Dunnett’s posttest. Scale bars: (A-D) 50 μm, (E) 25 μm.
Finally, we tested whether DLL1 inhibition affected proliferation of LGR5+ antral stem cells in Lgr5-GFP mice treated with inhibitory antibodies. Costaining for GFP and the proliferation marker EdU showed that the number of actively proliferating LGR5+ stem cells (Figure 6E, arrow) was significantly reduced in mice treated with anti-DLL1, either alone or in combination with anti-DLL4 (Figure 6F). Consistent with our previous findings of overall epithelial cell proliferation (Figures 4 and 5), mice treated with anti-DLL4 alone did not exhibit changes to LGR5+ stem cell proliferation status (Figure 6F). Further, combined block of DLL1 and DLL4 had a similar effect as anti-DLL1 treatment alone. Taken together, these data suggest that DLL1-expressing cells at the gland base signal to neighboring NOTCH1-expressing stem cells to regulate antral stem cell function.
Discussion
Notch signaling has been shown to be essential for gastrointestinal epithelial cell homeostasis, regulating both gastric4, 5, 6, 7 and intestinal22, 23, 24, 25, 26 stem cell proliferation and self-renewal to maintain the active stem cell pool. In this study, we aimed to define the key ligands and cells that make up the Notch stem cell niche in the mouse gastric antrum. Both genetic and pharmacologic approaches were used to block ligand signaling by targeted gene deletion in the gastric epithelium of adult mice or by treatment with inhibitory antibodies against specific Notch ligands DLL1, DLL4, JAG1, and/or JAG2. These studies investigated the consequences of targeting single ligands as well as combinations of ligands on epithelial and stem cell proliferation. Consistent results were obtained from both genetic and pharmacologic approaches to identify DLL1 as the sole Notch ligand regulating epithelial and stem cell proliferation in the adult mouse antrum.
Although we found all Notch ligands except for Dll3 to be expressed in the gastric antrum, effects on epithelial and stem cell proliferation were only observed with DLL1 inhibition, either alone or when combined with DLL4, JAG1, or JAG2. Importantly, combined ligand inhibition did not enhance the DLL1 effect, supporting the conclusion that DLL1 alone is the sole Notch ligand regulating antral epithelial proliferation. Interestingly, JAG1 was the most highly expressed Notch ligand in antral glands. However, JAG1 expression was restricted to differentiated epithelial cells in the mid and upper gland, away from the stem cell compartment at the gland base. In contrast, using gene expression and histological analysis, we mapped DLL1 expression to cells at the base of the antral glands. We also identified GSII-expressing antral deep mucous cells as the potential cellular source of DLL1, as co-immunostaining analyses revealed GSII+/DLL1+ cells at the base of antral glands.
An epithelial cell source for the Notch ligand would be predicted based on the canonical signaling mechanism, with ligand and receptor expressed on the cell membranes of neighboring cells.11 Our functional studies identified an epithelial cell source for DLL1 in the gastric stem cell niche, as gastric antral organoids treated with anti-DLL1 alone or in combination with anti-DLL4 exhibited reduced growth and proliferation. These findings follow previous reports showing that the gastric antral epithelium exhibits Notch activity and that epithelial organoids establish intrinsic Notch signaling to support stem cell growth in culture.4,6 Further, inhibition of JAG1 or JAG2 did not affect organoid growth, supporting our conclusion that DLL1 is the sole Notch ligand regulating gastric antral stem cells.
Lgr5-expressing cells at the antral gland base have been shown to function as active stem cells that maintain the epithelium.2 We visualized antral stem cells marked by Lgr5-GFP and Sox9-GFP in isolated glands, confirming localization to the gland base, and demonstrating that antral stem cells are intercalated between large, differentiated cells. This cellular pattern supports a view that DLL1-expressing cells at the gland base signal directly to adjacent LGR5+/SOX9+ stem cells to regulate stem cell function and antral tissue homeostasis. Previous analysis of mouse reporter strains showed that antral Lgr5-GFP marked stem cells are NOTCH-signaling cells.6 In the present study, we demonstrated by immunostaining NOTCH1 receptor expression in cells at the antral gland base, which agrees with previous expression profiling analysis of isolated gastric Lgr5-GFP stem cells,27 as well as functional studies that identified NOTCH1 and NOTCH2 as the key receptors controlling antral stem cell function.6 Together, our current report and past studies suggest that DLL1-expressing deep mucous cells at the antral gland base function as Notch niche cells to support LGR5+ antral stem cell homeostasis.
Our findings parallel previous reports showing an essential role for Notch to regulate intestinal stem cells.22, 23, 24, 25, 26,28 Similar to the gastric antrum, LGR5-expressing intestinal stem cells lie at the base of the intestinal crypt, intercalated between large, differentiated cells termed Paneth cells.29 Further, intestinal stem cells are also known to be Notch signaling cells, with NOTCH1 as the dominant receptor regulating stem cell Notch function.22 The neighboring Paneth cells are multifunctional cells that secrete antimicrobial proteins and also express stem cell niche factors, such as EGF and Wnt3, as well as Notch ligands DLL1 and DLL4.28, 29, 30 However, in contrast to our finding that DLL1 alone serves as the critical Notch ligand supporting gastric antral stem cell proliferation, both DLL1 and DLL4 together are required to promote stem cell proliferation and tissue homeostasis in the intestine.18 This highlights not only similarities, but also differences in the mechanisms of signaling pathway regulation of stem cell niche function in these 2 related gastrointestinal tissues. Future studies will be needed to determine if antral deep mucous cells have additional stem cell niche function, similar to what has been proposed for intestinal Paneth cells.28,29,31,32
Materials And Methods
Mice
Mice of both sexes 2–3 months of age were used for experiments. The following mouse strains were used: Lgr5-GFP-CreERT2 (Lgr5-GFP) (JAX #008875; The Jackson Laboratory, Bar Harbor, ME),33 Sox9-GFP (from Scott Magness),34,35 Sox2-CreERT2 (JAX #017593),12 Rosa-LSL-tdTomato (JAX #007908; The Jackson Laboratory), Dll4fl/fl (from Freddy Radtke),36 and Dll1fl/fl and Jag1fl/fl (from Julian Lewis).37,38 All mice were maintained on a C57BL/6 background, except for Sox9-GFP mice, which were on a CD-1 background. Mice were housed under specific pathogen–free conditions in automated watering and ventilated cages on a 12-hour light–dark cycle. All experimental procedures were approved by the Institutional Animal Care and Use Committee at the University of Michigan.
Notch Pathway Inhibition
To test Notch ligand inhibition in genetic mouse models, mice were treated by oral gavage with TX (Sigma-Aldrich, St Louis, MO; 100 mg/kg in 100% corn oil) once per day for 5 days, with tissue collection on day 6. Experimental mice were Sox2-CreERT2 crossed to Dll1fl/fl, Dll4fl/fl, or Jag1fl/fl mice. Controls were TX-treated mice with fl Notch ligand alleles, but without Sox2-CreERT2. To test global Notch ligand inhibition, Lgr5-GFP mice were injected intraperitoneally with Notch ligand inhibitory antibodies anti-JAG115 (15 mg/kg), anti-JAG2 (15 mg/kg),15 anti-DLL1 (10 mg/kg),16 or anti-DLL4 (10 mg/kg),16 or an irrelevant control IgG1 antibody targeting herpes simplex virus gD protein (Gd control; 10 mg/kg or 15 mg/kg).39 Antibodies were injected every other day for 5 days, with tissue collection on day 6.
To test effects of Notch ligand inhibition in vitro, mouse gastric antral organoid cultures were treated with Notch ligand inhibitory antibodies (20 μg/mL in culture media) either alone or in combination, and compared with global Notch pathway inhibition using the gamma-secretase inhibitor DAPT (Sigma, St. Louis, MO) (1 μM in culture media). Inhibitors were added directly to organoid culture media and renewed every other day for 5 days, with growth or proliferation analyzed on day 6.
Tissue Collection and Histological Analysis
Mice were fasted overnight with free access to water before tissue collection. In some experiments, mice were injected intraperitoneally with EdU (Life Technologies, Carlsbad, CA; 25 mg/kg) 2 hours prior to tissue collection. Stomachs and intestines were processed for frozen and paraffin tissue sections as previously described.4 Dorsal skin biopsies and lung tissue were collected, fixed in 4% paraformaldehyde in phosphate-buffered saline overnight at 4°C, and embedded in paraffin. Immunostaining of paraffin or frozen tissue sections was done as previously described,40 using antibodies or lectin listed in Table 1. Tissue sections were incubated with primary antibodies followed by appropriate secondary antibodies (1:400; Invitrogen, Waltham, MA), and mounted with ProLong Gold containing DAPI (Invitrogen).
Table 1.
List of Primary Antibodies/Lectin
| Antibody | Dilution | Host Species | Source |
|---|---|---|---|
| Acetylated α-tubulin | 1:200 | Mouse monoclonal | Santa Cruz #sc-23950 |
| CC10 | 1:200 | Goat polyclonal | Santa Cruz #sc-9772 |
| DLL1 | 1:100 | Rabbit polyclonal | Santa Cruz #sc-9102 |
| GFP | 1:200 | Rabbit polyclonal conjugated to Alexa488 | Invitrogen #A-21311 |
| GFP | 1:100 | Chicken polyclonal | Abcam #ab13970 |
| Griffonia simplicifolia (GSII) | 1:500 | Lectin conjugated to Alexa594 | Invitrogen #L-21416 |
| JAG1 | 1:500 | Goat polyclonal | Santa Cruz #sc-6011 |
| NOTCH1 | 1:500 | Rabbit monoclonal | Cell Signaling #3608 |
| E-cadherin | 1:1000 | Rat monoclonal | Invitrogen #13-1900 |
DLL, Delta-like; GFP, green fluorescent protein; JAG, Jagged.
For analysis of proliferation, EdU incorporation was visualized with the Click-iT EdU Alexa Fluor 488, 594, or 647 Imaging Kits (Life Technologies) according to manufacturer’s instructions. To analyze proliferating LGR5+ antral stem cells, tissue sections were costained for GFP and EdU as previously described.4
Lineage Tracing
To define the cellular pattern of Sox2-CreERT2 expression, Sox2-CreERT2; Rosa-LSL-tdTomato mice were injected intraperitoneally with TX (100 mg/kg in 95% corn oil, 5% EtOH) and tdTomato expression was examined in gastric tissue 1 day after a single TX treatment, or 1 month after 5 daily TX injections. Tissues were processed for cryopreservation, and sections were mounted with ProLong Gold containing DAPI nuclear stain and imaged by digital microscopy as described.4
Gene Expression Analysis
Total RNA was isolated from full-thickness antral tissue, isolated antral glands, or gastric antral organoids as described,4 followed by DNase I treatment and column purification using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Complementary DNA was prepared from 500 ng total RNA, and quantitative reverse-transcriptase polymerase chain reaction was performed as described,40 using primers listed in Table 2. Quantitative reverse-transcriptase polymerase chain reaction data are expressed as messenger RNA abundance normalized to Gapdh.
Table 2.
List of Quantitative Reverse-Transcriptase Polymerase Chain Reaction Primers
| Gene | Forward (5′ to 3′) | Reverse (5′ to 3′) | Amplicon Size (bp) |
|---|---|---|---|
| Dll1 | CTGAGGTGTAAGATGGAAGCG | CAACTGTCCATAGTGCAATGG | 245 |
| Dll3 | CTGAGACTATTTCCCCATCCTG | CCCGAAAGAATGAATTTGTAGC | 143 |
| Dll4 | TCGTCGTCAGGGACAAGAATAGC | CTCGTCTGTTCGCCAAATCTTACC | 213 |
| Jag1 | CAGAATGACGCTTCCTGTCG | TGCAGCTGTCAATCACTTCG | 361 |
| Jag2 | TATGACAGCGGCGACACCTTC | CAACACAGATGCCTCCGTTATAGC | 235 |
| Gapdh | TCAAGAAGGTGGTGAAGCAGG | TATTATGGGGGTCTGGGATGG | 350 |
In Situ Hybridization
Dll1 complementary DNA plasmid was linearized and labeled, with probe purification and tissue hybridization performed as previously described.22 Jag1 in situ hybridization was performed as described41 using RNAscope Jag1 probe (ACD 412831; ACDBio, Newark, CA)) and the Fluorescent Multiplex Detection kit (ACD 320851; ACDBio).
Gastric Organoid Culture
Mouse gastric antral organoid culture was carried out as previously described.4,6 L-WRN–conditioned media containing Wnt3a, R-spondin3, and Noggin were generated as described.42 Studies were performed in technical triplicates in an established mouse gastric antral organoid line that had been passaged at least 3 times before analysis. To measure proliferating cells in organoids, EdU (10 μM) was added to cultures 30 minutes before collection. After collection, organoids were fixed for 30 minutes in 2% paraformaldehyde on ice, washed in 70% ethanol solution, briefly stained with eosin to facilitate tracking, and embedded in a 2% agar plug followed by paraffin processing. Sections were stained with the Click-iT EdU Alexa Fluor 647 Imaging Kit (Thermo Fisher Scientific, Waltham, MA).
Gastric Gland Isolation, Immunostaining, and Imaging
Antral tissue was minced, glands were dissociated, and GFP immunostaining was performed in suspension culture using a conjugated GFP antibody (Table 1) as previously described.4 Whole-mount gland imaging used a Zeiss LSM510 inverted confocal microscope (Zeiss, Jena, Germany).
Morphometrics
Morphometric analysis was performed using ImageJ software (1.46r, Wayne Rasband; National Institutes of Health, Bethesda, MD). For EdU incorporation in tissue, the entire length of the antrum for each animal was imaged (n = 2–8 animals per group) and cell counts were normalized to epithelial area (μm2). For measurement of LGR5+ antral stem cell proliferation, the number of GFP/EdU double-positive cells per gastric antral gland was counted in Gd control (n = 253 glands), anti-DLL1 (n = 195 glands), anti-DLL4 (n = 214 glands), and anti-DLL1 + anti-DLL4 (n = 223 glands) (n = 4 mice/group). To measure organoid growth, the area of at least 120 organoids per treatment group was measured. Organoid EdU incorporation was measured on paraffin sections stained for EdU, with the number of EdU-positive nuclei counted divided by the total DAPI-positive nuclei from 54 organoids measured from each treatment group.
Statistical Analysis
GraphPad Prism software (GraphPad Software Version 9, San Diego, CA) was used for statistical analysis. Quantitative data are presented as mean ± SEM and analyzed using Student’s t test or 1-way analysis of variance with Dunnett’s post hoc test. P < .05 was considered statistically significant.
All authors had access to the study data and approved the final manuscript.
Acknowledgments
The authors thank Erin Collin and Lindsay Griffin for mouse colony maintenance, Dr. Sunny Wong for technical help with skin tissue analysis, and Dr. Deborah Gumucio for assistance with Dll1 in situ hybridization. The authors also thank Drs. Scott Magness, Freddy Radtke, Katsuto Hozumi, and Julian Lewis for sharing valuable mouse models via institutional colleagues Dr. Jason Spence, Dr. Ivan Maillard, and Dr. Matthew Schaller.
CRediT Authorship Contributions
Nobukatsu Horita (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Investigation: Lead; Writing – review & editing: Supporting)
Theresa M Keeley, MS (Data curation: Equal; Formal analysis: Equal; Investigation: Lead; Methodology: Lead; Writing – review & editing: Equal)
Elise S Hibdon, PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Equal; Writing – original draft: Lead; Writing – review & editing: Equal)
Elizabeth Delgado (Investigation: Supporting; Writing – review & editing: Supporting)
Daniel Lafkas (Conceptualization: Supporting; Resources: Equal; Writing – review & editing: Supporting)
Christian W Siebel (Conceptualization: Supporting; Resources: Lead; Writing – review & editing: Supporting)
Linda C Samuelson, PhD (Conceptualization: Lead; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Lead; Investigation: Equal; Methodology: Equal; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest These authors disclose the following: Christian W. Siebel and Daniel Lafkas are employees of Genentech, Inc, and own shares of Roche. The remaining authors disclose no conflicts.
Funding Elise S. Hibdon was supported by National Institutes of Health K01-DK111710. The research was funded by National Institutes of Health R01-DK118023 and P01-DK06041 project awards to Linda C. Samuelson and core support from the Michigan Gastrointestinal Research Center Grant (National Institutes of Health P30-DK34933).
References
- 1.Demitrack E.S., Samuelson L.C. Notch as a driver of gastric epithelial cell proliferation. Cell Mol Gastroenterol Hepatol. 2017;3:323–330. doi: 10.1016/j.jcmgh.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker N., Huch M., Kujala P., van de Wetering M., Snippert H.J., van Es J.H., Sato T., Stange D.E., Begthel H., van den Born M., Danenberg E., van den Brink S., Korving J., Abo A., Peters P.J., Wright N., Poulsom R., Clevers H. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 3.Sigal M., Logan C.Y., Kapalczynska M., Mollenkopf H.J., Berger H., Wiedenmann B., Nusse R., Amieva M.R., Meyer T.F. Stromal R-spondin orchestrates gastric epithelial stem cells and gland homeostasis. Nature. 2017;548:451–455. doi: 10.1038/nature23642. [DOI] [PubMed] [Google Scholar]
- 4.Demitrack E.S., Gifford G.B., Keeley T.M., Carulli A.J., VanDussen K.L., Thomas D., Giordano T.J., Liu Z., Kopan R., Samuelson L.C. Notch signaling regulates gastric antral LGR5 stem cell function. EMBO J. 2015;34:2522–2536. doi: 10.15252/embj.201490583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demitrack E.S., Gifford G.B., Keeley T.M., Horita N., Todisco A., Turgeon D.K., Siebel C.W., Samuelson L.C. NOTCH1 and NOTCH2 regulate epithelial cell proliferation in mouse and human gastric corpus. Am J Physiol Gastrointest Liver Physiol. 2017;312:G133–G144. doi: 10.1152/ajpgi.00325.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gifford G.B., Demitrack E.S., Keeley T.M., Tam A., La Cunza N., Dedhia P.H., Spence J.R., Simeone D.M., Saotome I., Louvi A., Siebel C.W., Samuelson L.C. Notch1 and Notch2 receptors regulate mouse and human gastric antral epithelial cell homoeostasis. Gut. 2017;66:1001–1011. doi: 10.1136/gutjnl-2015-310811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim T.H., Shivdasani R.A. Notch signaling in stomach epithelial stem cell homeostasis. J Exp Med. 2011;208:677–688. doi: 10.1084/jem.20101737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hibdon E.S., Razumilava N., Keeley T.M., Wong G., Solanki S., Shah Y.M., Samuelson L.C. Notch and mTOR signaling pathways promote human gastric cancer cell proliferation. Neoplasia. 2019;21:702–712. doi: 10.1016/j.neo.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y., Gao X., Liu J., Kong Q.Y., Wang X.W., Chen X.Y., Wang Q., Cheng Y.F., Qu X.X., Li H. Differential Notch1 and Notch2 expression and frequent activation of Notch signaling in gastric cancers. Arch Pathol Lab Med. 2011;135:451–458. doi: 10.5858/2009-0665-OA.1. [DOI] [PubMed] [Google Scholar]
- 10.Yeh T.S., Wu C.W., Hsu K.W., Liao W.J., Yang M.C., Li A.F., Wang A.M., Kuo M.L., Chi C.W. The activated Notch1 signal pathway is associated with gastric cancer progression through cyclooxygenase-2. Cancer Res. 2009;69:5039–5048. doi: 10.1158/0008-5472.CAN-08-4021. [DOI] [PubMed] [Google Scholar]
- 11.Kopan R., Ilagan M.X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnold K., Sarkar A., Yram M.A., Polo J.M., Bronson R., Sengupta S., Seandel M., Geijsen N., Hochedlinger K. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keeley T.M., Horita N., Samuelson L.C. Tamoxifen-induced gastric injury: effects of dose and method of administration. Cell Mol Gastroenterol Hepatol. 2019;8:365–367. doi: 10.1016/j.jcmgh.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huh W.J., Khurana S.S., Geahlen J.H., Kohli K., Waller R.A., Mills J.C. Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology. 2012;142:21–24. doi: 10.1053/j.gastro.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lafkas D., Shelton A., Chiu C., de Leon Boenig G., Chen Y., Stawicki S.S., Siltanen C., Reichelt M., Zhou M., Wu X., Eastham-Anderson J., Moore H., Roose-Girma M., Chinn Y., Hang J.Q., Warming S., Egen J., Lee W.P., Austin C., Wu Y., Payandeh J., Lowe J.B., Siebel C.W. Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature. 2015;528:127–131. doi: 10.1038/nature15715. [DOI] [PubMed] [Google Scholar]
- 16.Ridgway J., Zhang G., Wu Y., Stawicki S., Liang W.C., Chanthery Y., Kowalski J., Watts R.J., Callahan C., Kasman I., Singh M., Chien M., Tan C., Hongo J.A., de Sauvage F., Plowman G., Yan M. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 17.Tran I.T., Sandy A.R., Carulli A.J., Ebens C., Chung J., Shan G.T., Radojcic V., Friedman A., Gridley T., Shelton A., Reddy P., Samuelson L.C., Yan M., Siebel C.W., Maillard I. Blockade of individual Notch ligands and receptors controls graft-versus-host disease. J Clin Invest. 2013;123:1590–1604. doi: 10.1172/JCI65477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellegrinet L., Rodilla V., Liu Z., Chen S., Koch U., Espinosa L., Kaestner K.H., Kopan R., Lewis J., Radtke F. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. 2011;140:1230–1240. doi: 10.1053/j.gastro.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demehri S., Kopan R. Notch signaling in bulge stem cells is not required for selection of hair follicle fate. Development. 2009;136:891–896. doi: 10.1242/dev.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veniaminova N.A., Grachtchouk M., Doane O.J., Peterson J.K., Quigley D.A., Lull M.V., Pyrozhenko D.V., Nair R.R., Patrick M.T., Balmain A., Dlugosz A.A., Tsoi L.C., Wong S.Y. Niche-specific factors dynamically regulate sebaceous gland stem cells in the skin. Dev Cell. 2019;51:326–340. doi: 10.1016/j.devcel.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Formeister E.J., Sionas A.L., Lorance D.K., Barkley C.L., Lee G.H., Magness S.T. Distinct Sox9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestinal epithelium. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1108–G1118. doi: 10.1152/ajpgi.00004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carulli A.J., Keeley T.M., Demitrack E.S., Chung J., Maillard I., Samuelson L.C. Notch receptor regulation of intestinal stem cell homeostasis and crypt regeneration. Dev Biol. 2015;402:98–108. doi: 10.1016/j.ydbio.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fre S., Huyghe M., Mourikis P., Robine S., Louvard D., Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 24.Riccio O., van Gijn M.E., Bezdek A.C., Pellegrinet L., van Es J.H., Zimber-Strobl U., Strobl L.J., Honjo T., Clevers H., Radtke F. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–383. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Es J.H., van Gijn M.E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D.J., Radtke F., Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 26.VanDussen K.L., Carulli A.J., Keeley T.M., Patel S.R., Puthoff B.J., Magness S.T., Tran I.T., Maillard I., Siebel C., Kolterud A., Grosse A.S., Gumucio D.L., Ernst S.A., Tsai Y.H., Dempsey P.J., Samuelson L.C. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez Vallone V., Leprovots M., Strollo S., Vasile G., Lefort A., Libert F., Vassart G., Garcia M.I. Trop2 marks transient gastric fetal epithelium and adult regenerating cells after epithelial damage. Development. 2016;143:1452–1463. doi: 10.1242/dev.131490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bohin N., Keeley T.M., Carulli A.J., Walker E.M., Carlson E.A., Gao J., Aifantis I., Siebel C.W., Rajala M.W., Myers M.G., Jr., Jones J.C., Brindley C.D., Dempsey P.J., Samuelson L.C. Rapid crypt cell remodeling regenerates the intestinal stem cell niche after notch inhibition. Stem Cell Reports. 2020;15:156–170. doi: 10.1016/j.stemcr.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki N., Sachs N., Wiebrands K., Ellenbroek S.I., Fumagalli A., Lyubimova A., Begthel H., van den Born M., van Es J.H., Karthaus W.R., Li V.S., Lopez-Iglesias C., Peters P.J., van Rheenen J., van Oudenaarden A., Clevers H. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc Natl Acad Sci U S A. 2016;113:E5399–E5407. doi: 10.1073/pnas.1607327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez-Colman M.J., Schewe M., Meerlo M., Stigter E., Gerrits J., Pras-Raves M., Sacchetti A., Hornsveld M., Oost K.C., Snippert H.J., Verhoeven-Duif N., Fodde R., Burgering B.M. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature. 2017;543:424–427. doi: 10.1038/nature21673. [DOI] [PubMed] [Google Scholar]
- 32.Farin H.F., Van Es J.H., Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology. 2012;143:1518–1529. doi: 10.1053/j.gastro.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 33.Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 34.Gong S., Zheng C., Doughty M.L., Losos K., Didkovsky N., Schambra U.B., Nowak N.J., Joyner A., Leblanc G., Hatten M.E., Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 35.Gracz A.D., Ramalingam S., Magness S.T. Sox9 expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol Gastrointest Liver Physiol. 2010;298:G590–G600. doi: 10.1152/ajpgi.00470.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch U., Fiorini E., Benedito R., Besseyrias V., Schuster-Gossler K., Pierres M., Manley N.R., Duarte A., Macdonald H.R., Radtke F. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205:2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hozumi K., Negishi N., Suzuki D., Abe N., Sotomaru Y., Tamaoki N., Mailhos C., Ish-Horowicz D., Habu S., Owen M.J. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol. 2004;5:638–644. doi: 10.1038/ni1075. [DOI] [PubMed] [Google Scholar]
- 38.Brooker R., Hozumi K., Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- 39.Wu Y., Cain-Hom C., Choy L., Hagenbeek T.J., de Leon G.P., Chen Y., Finkle D., Venook R., Wu X., Ridgway J., Schahin-Reed D., Dow G.J., Shelton A., Stawicki S., Watts R.J., Zhang J., Choy R., Howard P., Kadyk L., Yan M., Zha J., Callahan C.A., Hymowitz S.G., Siebel C.W. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 40.Keeley T.M., Samuelson L.C. Cytodifferentiation of the postnatal mouse stomach in normal and Huntingtin-interacting protein 1-related-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1241–G1251. doi: 10.1152/ajpgi.00239.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bohin N., McGowan K.P., Keeley T.M., Carlson E.A., Yan K.S., Samuelson L.C. Insulin-like growth factor-1 and mTORC1 signaling promote the intestinal regenerative response after irradiation injury. Cell Mol Gastroenterol Hepatol. 2020;10:797–810. doi: 10.1016/j.jcmgh.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyoshi H., Stappenbeck T.S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]