Abstract
The impact of high levels of RNA polymerase II transcription on mitotic recombination was examined using lys2 recombination substrates positioned on nonhomologous chromosomes. Substrates were used that could produce Lys+ recombinants by either a simple (noncrossover) gene conversion event or a crossover-associated recombination event, by only a simple gene conversion event, or by only a crossover event. Transcription of the lys2 substrates was regulated by the highly inducible GAL1-10 promoter or the low-level LYS2 promoter, with GAL1-10 promoter activity being controlled by the presence or absence of the Gal80p negative regulatory protein. Transcription was found to stimulate recombination in all assays used, but the level of stimulation varied depending on whether only one or both substrates were highly transcribed. In addition, there was an asymmetry in the types of recombination events observed when one substrate versus the other was highly transcribed. Finally, the lys2 substrates were positioned as direct repeats on the same chromosome and were found to exhibit a different recombinational response to high levels of transcription from that exhibited by the repeats on nonhomologous chromosomes. The relevance of these results to the mechanisms of transcription-associated recombination are discussed.
Eukaryotic chromatin is a dynamic package of DNA and protein components, and the structure of chromatin can exert a profound influence on DNA metabolic processes. Chromatin remodeling, for example, accompanies transcription and involves the repositioning of nucleosomes as well as covalent histone modifications which alter DNA-histone interactions (50). In addition, regions of the genome can be maintained in a genetically silent state by protein-mediated compaction of DNA into heterochromatin (15). The role of chromatin structure in maintaining the integrity of DNA has been documented in relation to UV-induced damage, where nucleosome positioning influences the pattern of DNA damage as well as the efficiencies of DNA repair processes (41). Finally, the double-strand breaks that initiate meiotic recombination in yeast occur preferentially in nuclease-sensitive regions of chromatin, which generally correspond to promoters (13).
Although DNA metabolic processes are usually considered in isolation, these processes often occur at the same time and can, in principle, exert either inhibitory or stimulatory effects on each other. Such effects can occur through alterations in chromatin structure, direct coupling between metabolic processes, or steric interference of one process with another. Studies of the repair of UV-induced DNA damage via the nucleotide excision repair pathway were the first to demonstrate that the repair response is related to the transcriptional state of the damaged region (37). Specifically, the transcribed strand of an active gene is targeted for repair, which is initiated by the stalling of the RNA polymerase complex at transcription-blocking lesions. Transcription-coupled repair leads to the enhanced repair of transcribed genes relative to nontranscribed genes, and such preferential DNA repair is evolutionarily conserved from bacteria to humans. More recently, it has been demonstrated that the repair of oxidative damage, which generally involves the base excision repair pathway, also can be transcription coupled (6). Not only can transcription facilitate high-fidelity DNA repair processes, but also high levels of transcription have been associated with increased mutation rates in yeast (7, 27) and bacteria (2, 51). This latter phenomenon could be due to enhanced damage of transcriptionally active DNA or could reflect an interference of high levels of transcription with relatively error-free repair pathways. Finally, transcription has stimulatory effects on recombinational DNA repair in Saccharomyces cerevisiae (20, 28, 42), in Schizosaccharomyces pombe (14), and in mammalian cells (29, 30). Transcription-associated recombination has been best characterized in S. cerevisiae and will be the focus of the studies reported here.
Transcription-associated recombination in yeast was first documented through the identification of the HOT1 mitotic-specific recombination hot spot, which corresponds to the promoter elements for rDNA transcription and hence promotes high levels of transcription by RNA polymerase I (20). HOT1 stimulates both intrachromosomal and interchromosomal recombination, and transcription through the recombination substrates is necessary for the stimulatory effect (45). Mutagenesis of HOT1 has indicated a direct link between transcription and mitotic recombination, with mutations that decrease the transcriptional activity of HOT1 also decreasing the associated recombination (17, 40). Although the HOT1 studies clearly have demonstrated that RNA polymerase I transcription can stimulate recombination, the relevance of these studies to mitotic recombination between non-rDNA sequences, which normally are transcribed by RNA polymerase II, is unclear.
To address the relationship of RNA polymerase II transcription to mitotic recombination in yeast, investigators have used the GAL1-10 promoter, which can be highly induced by galactose or can be controlled genetically by the presence or absence of the positive Gal4p and negative Gal80p regulatory proteins (18). Thomas and Rothstein thus used gal10 alleles oriented as direct repeats in isogenic gal4 and gal80 strains to demonstrate an association between high levels of RNA polymerase II transcription and increased levels of mitotic recombination (42). Two features of the gal10 recombination results indicate that the observed effects might be specific to substrates oriented as direct repeats and therefore may not be generally applicable to other types of recombination events. First, elevated levels of recombination were correlated with transcription through plasmid sequences located between the directly repeated gal10 substrates rather than with transcription through the substrates themselves. Second, high levels of transcription stimulated only plasmid loss events and did not elevate simple gene conversion events. One interpretation of these results is that high levels of transcription through the nonyeast, plasmid sequences lead to the formation of recombination-initiating lesions within the plasmid sequences, which then can be repaired using the single-strand annealing (SSA) mechanism of recombination. In SSA, the ends of a double-strand break are processed by a 5′-to-3′ exonuclease to yield single-stranded regions on both sides of the break. Annealing of complementary single-stranded regions (the direct repeats) and removal of the single-stranded tails results in loss of the sequences between the direct repeats (10).
Nevo-Caspi and Kupiec also have used the GAL1-10 promoter to examine the effect of transcription on recombination between Ty retrotransposons (28). In this system, induction of transcription by galactose increases the loss of a selectable marker inserted into the transcribed Ty, with marker loss occurring either by gene conversion with an unmarked Ty or by recombination involving the flanking δ direct repeats at the ends of the Ty. This latter event is analogous to the plasmid loss events observed with the gal10 direct repeats. Although RNA polymerase II transcription stimulated recombination in the Ty system, it should be noted that aspects of Ty-Ty recombination appear to be unique to these elements (23, 32). In addition, it was not possible to examine the effect of transcription on crossover events. Finally, it has been reported that mitotic recombination between his4 heteroalleles is positively correlated with the activity of the endogenous HIS4 promoter (49).
In addition to the yeast studies documenting an association between high levels of RNA polymerase I or II transcription and elevated mitotic recombination, there are two notable types of yeast mutants that exhibit elevated mitotic recombination rates and may provide information relevant to the mechanism(s) of transcription-associated recombination. First, rDNA recombination is elevated in yeast topoisomerase-deficient top1 and top2 mutants (5, 21), while recombination between the direct repeats that flank the yeast Ty retrotransposon is elevated in a top3 mutant (48). It has been speculated that the increased supercoiling of DNA in toposiomerase-deficient mutants generates recombination-initiating strand breaks, and this basic idea has been extended to explain transcription-associated recombination (see reference 12 for a discussion). In the case of transcription-associated recombination, the supercoiling changes and torsional stress that accompany high levels of transcription (25) similarly could lead to recombination-initiating lesions. The second type of mutant that may provide information relevant to transcription-associated recombination is exemplified by yeast hpr1 mutants, which exhibit a mitotic hyperrecombination phenotype (1). The HPR1 gene product is important in transcript elongation (4), and it has been speculated that elongation problems generate recombination-initiating lesions. It has been further suggested that artificially high transcription of yeast genes that are normally expressed at a low level could lead to elongation problems, even in HPR1 strains (33). An hpr1-like mechanism could therefore account for the HOT1 and GAL1-10 examples of transcription-associated recombination.
To examine the generality of RNA polymerase II transcription-stimulated recombination and to determine whether gene conversion and reciprocal crossover events are similarly affected by transcription, we placed lys2 recombination substrates under the control of either the highly inducible GAL1-10 promoter or the low-level LYS2 promoter. A distinct feature of this system is that transcription of the recombination substrates can be coregulated by fusion to the same promoter or that very different levels of transcription can be achieved by fusion of the substrates to different promoters. Our results obtained with the lys2 substrates extend those obtained in previous studies and provide novel insights into the mechanism(s) of transcription-associated recombination in yeast.
MATERIALS AND METHODS
Growth conditions.
YEP medium (1% yeast extract, 2% Bacto Peptone) was used for all nonselective growth. Synthetic complete medium (39) deficient in lysine (SC − Lys) or uracil (SC − Ura) was used to select Lys+ and Ura+ prototrophs, respectively. Media were supplemented with 2% glucose (YEPD and SCD), with 2% glycerol–2% ethanol (YEPGE and SCGE), or with 2% galactose–2% glycerol–2% ethanol (YEPGGE and SCGGE) for use as carbon sources. Ura− segregants were identified on synthetic minimal medium supplemented with 5-fluoroorotic acid (5-FOA) (3). All yeast strains were propagated at 30°C.
Plasmid constructions.
All plasmid manipulations were done using standard molecular biological techniques (36). Plasmid pSR244 contains the gal80::HIS3 disruption allele and was constructed by replacing a BglII fragment within the GAL80 coding sequence of plasmid pSR243 (a 3.1-kb GAL80 HindIII fragment in pGEM3) with a 1.7-kb HIS3 BamHI fragment. pSR91 contains a 5.5-kb genomic BamHI fragment carrying URA3 in the BamHI site of pUC7 (note that the BamHI sites in the pUC7 polylinker are flanked by EcoRI sites). pSR88 contains the pBM150 (19) BamHI-EcoRI 0.7-kb GAL1-10 promoter (pGAL) fragment with added PstI linkers cloned into the PstI site of pUC7. All lys2 recombination substrates were derived from plasmids pDP4 and pDP6, which are pUC9 derivatives containing the entire LYS2 gene (11). pSR309 was derived from pDP6 by inserting a 1.2-kb EcoRI URA3-containing fragment (from pSR66, which contains the 1.2-kb HindIII genomic URA3 fragment in the HincII site of pUC7) into the polylinker region downstream of the LYS2 sequences.
Plasmid pSR140 (for allele replacement) contains the lys2-Nhe allele and the selectable URA3 marker and was made as follows. First, a 3.7-kb EcoRV LYS2 fragment (missing pLYS and the C-terminal end of Lys2p) from pDP4 was inserted into the HincII site of pUC9 to make pSR135. Next, pSR135 was linearized by partial digestion with NheI and the promoter-proximal NheI site was filled in with the Klenow fragment of DNA polymerase I, an operation that inserts 4 bp and regenerates the NheI site. The resulting lys2-Nhe plasmid (pSR137) was further modified by inserting a 1.2-kb URA3-containing EcoRI fragment (from pSR66) into the pUC9-derived EcoRI site at the 3′ end of lys2-Nhe, creating pSR140.
Plasmid pSR477 (for allele replacement) contains the lys2ΔXho allele and the selectable URA3 marker and was made in two steps. First, pSR445 was made by filling in the XhoI site of pDP6 with Klenow fragment. Second, the 3.7-kb EcoRV lys2ΔXho fragment (missing pLYS and the C-terminal end of Lys2p) from pSR445 was inserted into the filled-in (with Klenow fragment) HindIII site of pM21 (pUC9 containing a 1.2-kb URA3 fragment) to create pSR477.
Plasmids pSR517 and pSR518 (for allele replacement) contain the pLYS-lys2Δ5′ and pGAL-lys2Δ5′ alleles, respectively, plus URA3 as a selectable marker. pSR500 contains a promoterless lys2Δ5′ allele and was constructed by inserting a 3-kb NciI-HindIII fragment (treated with Klenow to fill in the enzyme-generated ends) from pDP6 into the HincII site of pUC9. NciI cuts at position 1773 (relative to the XbaI site upstream of LYS2) and so eliminates the N-terminal one-third of the LYS2 coding sequence; HindIII cuts downstream of the LYS2 coding sequence. pSR513 (pLYS2-lys2Δ5′) was constructed by inserting a HindIII-EcoRV pLYS-containing fragment from pDP6 into pSR500 that had been digested with PstI, treated with T4 polymerase to remove the overhangs, and then digested with HindIII. pSR514 (pGAL-lys2Δ5′) was constructed by inserting a PstI pGAL-containing fragment from pSR88 into pSR500 that had been digested with PstI. pSR517 and pSR518 were constructed by inserting a 5.5-kb URA3-containing EcoRI fragment (from pSR91) into EcoRI-digested pSR513 and pSR514, respectively.
pSR183 contains the pLYS-lys2Δ3′ allele and was constructed by inserting a 5.5-kb URA3-containing EcoRI fragment from pSR91 into the EcoRI site of pDP6ΔB15. The URA3 fragment is just downstream of the lys2 sequences and is transcribed in the same direction. The lys2Δ3′ allele of pDP6ΔB15 was derived by treating SstII-SmaI-digested pDP6 with exonuclease III and mung bean nuclease and is truncated at position 3621 (relative to the XbaI site upstream of LYS2). The lys2Δ3′ allele thus is missing the C-terminal 15% of the LYS2 coding sequence.
pSR234 contains the pGAL-lys2Δ3′ allele and was constructed as follows. The pLYS promotor of pDP6ΔB15 was deleted by digesting the plasmid with HindIII and EcoRV, treating the digest with Klenow fragment, and then ligating in the presence of excess PstI linkers. The resulting plasmid (pSR229) was modified by inserting a pGAL-containing PstI fragment (from pSR88) into the unique PstI site, creating pSR231. Finally, a 5.5 kb EcoRI URA3-containing fragment from pSR91 was inserted into EcoRI-digested pSR231 such that URA3 is just downstream of the lys2 sequences and is transcribed in the same direction (pSR234).
pSR485 contains the pGAL-lys2Δ5′Δ3′ allele and was made in three steps. First, an internal 2-kb NdeI LYS2 fragment from pDP6 was Klenow treated and inserted into the HincII site of pUC9 to make pSR444. Next, the PstI pGAL fragment from pSR88 was inserted at the PstI site upstream of the LYS2 sequences of pSR444 to generate pSR452. Lastly, a 5.5-kb URA3-containing EcoRI fragment from pSR91 was inserted into the lys2-distal EcoRI site of pSR452 to make pSR485.
pSR486 contains the pLYS-lys2Δ5′Δ3′ allele and was made by first inserting an EcoRV-HindIII LYS2 promoter-containing fragment from pDP6 into pSR444 that had been digested with PstI, treated with T4 polymerase to remove the overhangs, and then digested with HindIII (pSR482). A 5.5-kb URA3-containing EcoRI fragment from pSR91 then was inserted into the lys2-distal EcoRI site of pSR482 to make pSR486.
Yeast strain constructions.
All strains used in this study were derived from haploid strain SJR195 (MATa ade2-101oc his3Δ200 ura3ΔNco) by lithium acetate transformation (38). All strain genotypes were confirmed by Southern blot analysis and/or appropriate phenotypic tests. Strains were constructed with various lys2 recombination substrates positioned either as repeats on nonhomologous chromosomes (heterochromosomal repeats) or as direct repeats on the same chromosome (intrachromosomal repeats). Each lys2 recombination substrate was under control of either the low-level LYS2 promoter (pLYS) or the highly inducible GAL1-10 promoter (pGAL). pGAL promoter activity was maintained at a low or high level by placing the lys2 recombination substrates in a GAL80 or gal80::HIS3 background, respectively. One-step disruption of GAL80 was accomplished by transforming cells with NcoI-SmaI-digested pSR244 and selecting His+ transformants.
Strains SJR297 (GAL80) and SJR298 (gal80::HIS3) contain the pGAL-lys2ΔBgl allele, while SJR357 (GAL80) and SJR358 (gal80::HIS3) contain the pLYS-lys2ΔBgl allele (see reference 7 for details). The lys2ΔBgl allele in these strains was replaced with the lys2-Nhe allele by two-step allele replacement (35) using plasmid pSR140. Strains SJR377, SJR378, SJR379, and SJR380 were thus derived by transforming SJR297, SJR298, SJR357, and SJR358, respectively, with AflII-digested pSR140. Because pSR140 does not contain homology to chromosomal sequences upstream of pGAL or pLYS, the promoter regions were not altered. The reversion rate of the pGAL-lys2-Nhe allele is 2.1 × 10−9 in a Gal80+ background and 2.6 × 10−8 in a Gal80− background.
SJR545 (GAL80) contains a pGAL-LYS2 allele and was constructed by replacing the lys2ΔBgl allele of SJR297 with wild-type LYS2 sequences. This was accomplished by two-step allele replacement using XhoI-digested pSR309. The lys2ΔBgl allele of SJR298 (gal80::HIS3) was replaced with wild-type LYS2 sequences by transformation with an EcoRV fragment from pDP6, creating SJR371 (pGAL-LYS2). The LYS2 alleles of SJR371 and SJR545 were replaced with the lys2ΔXho allele, yielding strains SJR565 and SJR566, respectively, by two-step allele replacement using BglII-digested pSR477. The pLYS-LYS2 alleles in strains SJR195 (GAL80) and SJR282 (gal80::HIS3) were similarly replaced with the lys2ΔXho allele, yielding pLYS-lys2ΔXho strains SJR562 and SJR564, respectively. The reversion rate of the pGAL-lys2ΔXho allele is 6.9 × 10−10 in a Gal80+ background and 6.0 × 10−9 in a Gal80− background.
The LYS2 allele of strains SJR195, SJR282, SJR371, and SJR545 were replaced with the lys2Δ5′ allele, yielding strains SJR660, SJR661, SJR662, and SJR663, respectively. The pLYS-lys2Δ5′ and pGAL-lys2Δ5′ alleles were introduced by two-step allele replacement using BstXI-digested pSR517 and pSR518, respectively.
To construct strains with a full-length recombination substrate at the LYS2 locus on chromosome II and a 3′-truncated substrate at the URA3 locus on chromosome V (heterochromosomal conversion-crossover substrates [see Fig. 1A]), strains containing the pLYS-lys2-Nhe or pGAL-lys2-Nhe allele were transformed with SmaI-digested pSR183 (pLYS-lys2Δ3′) or pSR234 (pGAL-lys2Δ3′), both of which contain URA3 as a selectable marker. SmaI cuts in the URA3 sequences of pSR183 and pSR234, thus targeting plasmid integration to the URA3 locus on chromosome V. Following the selection of Ura+ transformants, integration of a single copy of pSR183 or pSR234 was confirmed by Southern analysis. Strains SJR398, SJR400, SJR402, and SJR404 were made by transforming SJR377, SJR378, SJR379, and SJR380, respectively, with SmaI-digested pSR183. Similarly, strains SJR399, SJR401, SJR403, and SJR405 were made by transforming SJR377, SJR378, SJR379, and SJR380, respectively, with SmaI-digested pSR234.
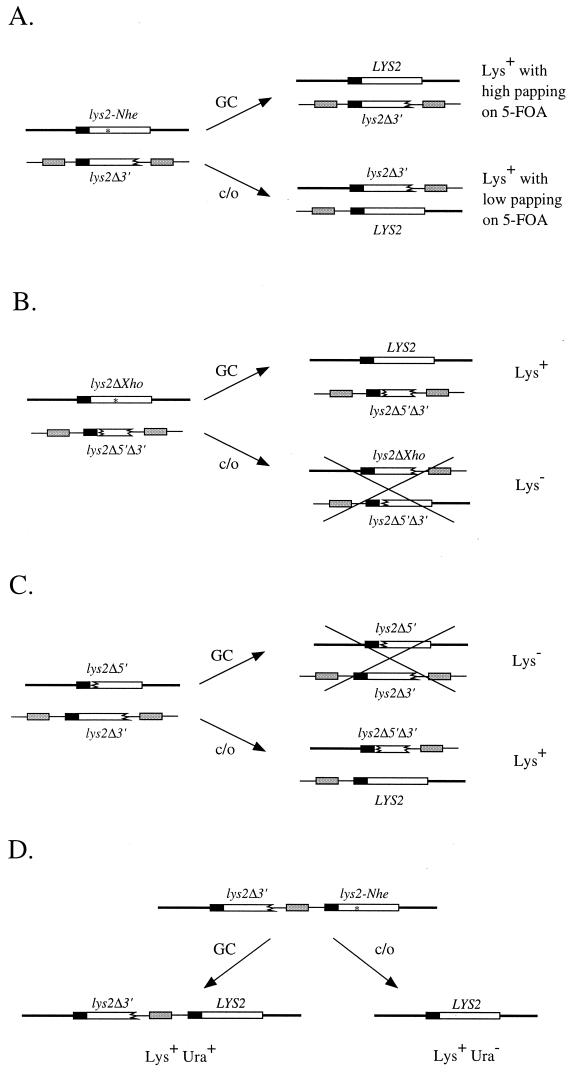
FIG. 1.
Structures of recombination substrates. Open boxes correspond to lys2 sequences (point mutations are indicated with asterisks), and shaded boxes correspond to URA3/ura3 sequences. Thick and thin horizontal lines in panels A to C correspond to chromosomes II and V, respectively. The thick line in panel D corresponds to chromosome V sequences that flank the lys2 direct repeats, and the thin line corresponds to plasmid sequences between the direct repeats. GC, gene conversion; c/o, crossover. (A) Heterochromosomal conversion-crossover substrates; (B) heterochromosomal conversion-only substrates; (C) heterochromosomal crossover-only substrates; (D) intrachromosomal direct-repeat substrates.
To construct strains with a full-length recombination substrate at the LYS2 locus on chromosome II and a 5′- and 3′-truncated substrate at the URA3 locus on chromosome V (heterochromosomal conversion-only substrates [see Fig. 1B]), strains containing the pLYS-lys2ΔXho or pGAL-lys2ΔXho allele were transformed with linearized pSR486 (pLYS-lys2Δ5′Δ3′) or pSR485 (pGAL-lys2Δ5′Δ3′). Ura+ transformants were selected, and the integration of a single copy of pSR485 or pSR486 was confirmed by Southern analysis. SJR578, SJR580, SJR582, and SJR584 were made by transforming SJR563, SJR564, SJR565, and SJR566, respectively, with BglII-digested pSR486. Similarly, SJR577, SJR579, SJR581, and SJR583 were made by transforming SJR563, SJR564, SJR565, and SJR566, respectively, with BglII-digested pSR485.
To construct strains with a 5′-truncated recombination substrate at the LYS2 locus on chromosome II and a 3′-truncated substrate at the URA3 locus on chromosome V (heterochromosomal crossover-only substrates [see Fig. 1C]), strains containing the pLYS-lys2Δ5′ or pGAL-lys2Δ5′ allele were transformed with linearized pSR183 (pLYS-lys2Δ3′) or pSR234 (pGAL-lys2Δ3′). Ura+ transformants were selected, and the integration of a single copy of pSR183 or pSR234 was confirmed by Southern analysis. SJR735 and SJR737 were constructed by transforming SJR662 and SJR663, respectively, with BstXI-digested pSR183. Similarly, SJR733, SJR734, SJR736, and SJR738 were made by transforming SJR660, SJR661, SJR662, and SJR663, respectively, with BstXI-digested pSR234.
To construct strains with lys2 direct repeats at the LYS2 locus (see Fig. 1D), plasmid pSR183 (pLYS-lys2Δ3′) or pSR234 (pGAL-lys2Δ3′) was targeted to integrate at LYS2 by digestion with AflII. SJR432, SJR434, SJR436, and SJR438 were made by transforming SJR377, SJR378, SJR379, and SJR380, respectively, with AflII-digested pSR234. Similarly, SJR431, SJR433, and SJR435 were made by transforming SJR377, SJR378, and SJR379, respectively, with AflII-digested pSR183. Ura+ transformants were selected, and the integration of a single copy of pSR183 or pSR234 was confirmed by Southern analysis. SJR437 was constructed by introducing the gal80::HIS3 allele into SJR435.
Recombination rate analysis.
Recombination rates were determined by the method of the median (24). Briefly, 2-day old colonies were excised from YEPD plates, inoculated into YEPGE liquid medium, and grown for 2 days on a roller drum. Cells were harvested, washed once with sterile H2O, and resuspended in 1 ml of H2O. Aliquots (100 μl) of appropriately diluted cells were plated on SCGGE − Lys medium for prototroph selection and on YEPD to determine the numbers of viable cells. Lys+ colonies were counted on day 3 after selective plating, and this number was used to determine the total number of recombinants per culture. The median number of Lys+ colonies per culture was determined using at least nine independent cultures. The median (or corrected median for the conversion-crossover substrates [see below]) was then used to calculate the recombination rate (number of recombinants per generation). To determine the rates of conversions versus crossovers, the total rate was partitioned using the proportions of gene conversion versus crossover events.
With the heterochromosomal conversion-crossover substrates, a crossover event will yield a reciprocal translocation between chromosomes II and V. Such crossover recombinants will be detected only if both recombinant chromosomes segregate into the same daughter cell. Assuming random chromatid segregation following a G2 recombination event, only 50% of crossovers will be detected. The measured median number of Lys+ recombinants was therefore corrected for undetected exchanges (equivalent to the proportion of detected exchanges assuming random segregation of chromatids) as follows: experimentally determined median × (1 + proportion of observed crossovers). The corrected median was then used to calculate the Lys+ rate. The proportions of conversion versus crossover events determined experimentally with the conversion-crossover substrates also were corrected to account for the inability to detect 50% of the crossover events. If X is the observed number of crossovers out of N total recombinants examined, the corrected proportion of crossovers would be 2X/(N + X). The corrected proportion of gene conversions would then be 1 − (corrected proportion of crossovers). For the corrected values, see Table 1.
TABLE 1.
Effects of transcription on recombination between heterochromosomal conversion-crossover substrates
| Strain | Gal80 |
lys2-Nhe (LYS2 locus)
|
lys2Δ3′ (URA3 locus)
|
Lys+ rate (10−8)a | Conversion (%) | Crossover (%) | ||
|---|---|---|---|---|---|---|---|---|
| Promoter | Transcription | Promoter | Transcription | |||||
| SJR402 | + | LYS | Low | LYS | Low | 4.1 | 38 | 62 |
| SJR404 | − | LYS | Low | LYS | Low | 4.7 (1.1) | 36 | 64 |
| SJR399 | + | GAL | Low | GAL | Low | 4.1 | 47 | 53 |
| SJR401 | − | GAL | High | GAL | High | 15 (3.8) | 19 | 81 |
| SJR398 | + | GAL | Low | LYS | Low | 4.2 | 27 | 73 |
| SJR400 | − | GAL | High | LYS | Low | 37 (8.8) | 43 | 57 |
| SJR403 | + | LYS | Low | GAL | Low | 3.3 | 33 | 67 |
| SJR405 | − | LYS | Low | GAL | High | 45 (14) | 3 | 97 |
Fold increases over the rate in the corresponding Gal80+ strain are given in parentheses.
Partitioning of recombinants.
At least 50 independent Lys+ recombinants from each relevant strain were classified as gene conversion or crossover events. With the heterochromosomal conversion-crossover recombination substrates, the lys2Δ3′ allele on chromosome V is flanked by ura3/URA3 direct repeats (see Fig. 1A). Recombination between the flanking direct repeats occurs at a high level, generating Ura− segregants that can be detected on 5-FOA. A Lys+ recombinant generated by a gene conversion event will maintain the ura3/URA3 direct repeats, whereas a Lys+ recombinant generated by a crossover event will be accompanied by physical separation of the ura3/URA3 direct repeats (16). Thus, a Lys+ recombinant resulting from gene conversion will produce colonies (papillate) at a very high level on 5-FOA while one generated by a crossover event will papillate at a very low level on 5-FOA. Independent Lys+ recombinants were isolated and purified, and three colonies of each were checked for the papillation phenotype on 5-FOA. Correlation between the low-versus high-papillation phenotypes and the occurrence of a crossover versus a gene conversion event was confirmed by Southern blot analysis of random recombinants.
With the intrachromosomal recombination substrates (lys2 direct repeats), the substrates flank a plasmid-carried URA3 gene (see Fig. 1D). Lys+ recombinants due to gene conversion maintain the plasmid sequences, while crossover or SSA events are accompanied by loss of the plasmid sequences and the lys2Δ3′ allele. Gene conversion versus crossover Lys+ recombinants were therefore distinguished by secondary scoring of the Ura phenotype. To select for only gene conversion events, cells were plated on SCGGE medium deficient in both lysine and uracil.
RESULTS
Heterochromosomal conversion-crossover assay system.
The initial assay system used to examine the impact of transcription on mitotic recombination was composed of lys2 heteroalleles positioned on nonhomologous chromosomes in a haploid yeast strain. As shown in Fig. 1A, the LYS2 locus on chromosome II contained the lys2-Nhe frameshift allele, either under control of its normal promoter (pLYS) or fused to the highly inducible GAL1-10 promoter (pGAL). A second copy of lys2 was placed at the URA3 locus on chromosome V by integrating a plasmid containing a lys2 allele truncated at the 3′ end (lys2Δ3′). As with the full-length lys2-Nhe allele, two versions of the lys2Δ3′ allele were constructed, one controlled by pLYS and the other controlled by pGAL. pGAL activity was modulated by constructing pairs of isogenic Gal80+ and Gal80− strains that contained or did not contain, respectively, the negative regulatory protein Gal80p. Although galactose is routinely used to induce high levels of pGAL activity, the use of Gal80+-Gal80− strain pairs allowed promoter activity to be controlled without changing cellular growth conditions. In all the experiments reported here, cells were grown nonselectively in rich medium containing glycerol and ethanol as carbon sources; glucose was not used in order to avoid catabolite repression of pGAL activity. Under these growth conditions, transcription from pGAL, as assayed by β-galactosidase production from a pGAL-lacZ fusion, was approximately 1,000-fold higher in a Gal80− strain than in a Gal80+ strain (data not shown). To identify Lys+ recombinants, nonselectively grown cells were plated on lysine-deficient medium supplemented with galactose as well as glycerol and ethanol. In the presence of galactose, pGAL is highly active even in the presence of Gal80p, thus allowing the identification of pGAL-LYS2 recombinants as well as pLYS-LYS2 recombinants in Gal80+ strains.
There are three unique features of the lys2-Nhe/lys2Δ3′ assay system that are relevant to assessing the effects of transcription on recombination. First, the assay system can detect both noncrossover and crossover events, which are assumed to arise via alternative resolutions of common heteroduplex recombination intermediates. For simplicity, we refer to the noncrossover recombinants simply as gene conversion events and the crossover-associated recombinants as simply crossovers. As illustrated in Fig. 1A, a simple phenotypic test can be used to distinguish gene conversion from crossover events (see Materials and Methods for details). A second important feature of the assay system is that it allows one to identify the donor versus the recipient allele in simple gene conversion events. Because only the full-length lys2-Nhe allele can undergo gene conversion to yield a wild-type LYS2 gene, it must always be the recipient, and the lys2Δ3′ allele the donor, in simple gene conversion events. The third relevant feature of the assay system is that the recombination substrates can be fused either to the same promoter or to different promoters. Strains were therefore constructed that had both lys2 alleles fused to pLYS, both alleles fused to pGAL, or one allele fused to pLYS and the other fused to pGAL. This allows assessment of the effect of transcription on recombination when both alleles are transcribed at the same high or low level and when the two substrates are transcribed at very different levels.
The recombination data obtained with the heterochromosomal conversion-crossover assay are presented in Table 1. The total rate of Lys+ recombinants was measured for each strain, and at least 50 independent recombinants were analyzed to determine the relative proportions of gene conversion versus crossover events. Strains SJR402 and SJR404 are control Gal80+ and Gal80− strains, respectively, that have both lys2 substrates fused to pLYS. These strains exhibited identical recombination rates and identical conversion-crossover distributions of recombinants (approximately 40% conversions and 60% crossovers), thus demonstrating that the presence or absence of Gal80p does not have any general effect on recombination. When both lys2 recombination substrates were fused to pGAL, the rate of recombination was elevated 3.8-fold in the Gal80− high-transcription strain (SJR401) relative to the Gal80+ low-transcription strain (SJR399). In addition, there was shift in the ratio of gene conversion to crossover events, with a greater proportion of crossover events under high-transcription conditions. It should be noted that the recombination rates in the Gal80+ strains (SJR399 and SJR402) are not affected by the presence of pGAL versus pLYS.
Strains SJR398 and SJR400 (Gal80+ and Gal80−, respectively) have the full-length lys2-Nhe allele fused to pGAL and the lys2Δ3′ allele fused to pLYS, while strains SJR403 and SJR405 (Gal80+ and Gal80−, respectively) have the opposite promoter configuration. When only the lys2-Nhe allele was highly transcribed, the total recombination rate was elevated 8.8-fold, while recombination was elevated 14-fold when only the lys2Δ3′ allele was highly transcribed. Thus, with both sets of strains, transcription of only one substrate appears to elevate recombination to a two- to fourfold-higher level than was observed when both substrates were highly transcribed. Although the total rate of recombination was elevated similarly when either the lys2-Nhe or the lys2Δ3′ allele was highly transcribed, there was a striking asymmetry in the distributions of conversion versus crossover events. When only the lys2-Nhe allele was highly transcribed, the ratio of gene conversion to crossover events was similar to that observed under low transcription conditions. When only the lys2Δ3′ allele was highly transcribed, however, the proportion of gene conversion events dropped dramatically, from 33 to 3%. The partitioning of total recombinants into gene conversion versus crossover events thus indicates that gene conversion is elevated only when the lys2-Nhe allele, which acts as the recipient allele in simple gene conversion events, is highly transcribed. High transcription of only the lys2Δ3′ donor allele has little, if any, impact on the conversion rate of the lys2-Nhe recipient allele. Although a trivial explanation for asymmetric effects of transcription on gene conversion is that pGAL-LYS2 recombinants form colonies more efficiently than do pLYS2-LYS2 recombinants on selective media, reconstruction experiments with both types of recombinants do not support this possibility (reference 7 and data not shown). As described below, the striking asymmetry in conversion versus crossover events observed when only one lys2 allele was highly transcribed was confirmed by using additional lys2 substrates that can produce prototrophic recombinants only via a simple gene conversion event.
Heterochromosomal conversion-only assay system.
The initial assay system was modified so that only gene conversion events unassociated with crossing over would be detected. As shown in Fig. 1B, the lys2 allele positioned on chromosome V was truncated at both the 5′ and 3′ ends (lys2Δ5′Δ3′) instead of only at the 3′ end. The lys2 allele at the LYS2 locus on chromosome II was again full length, but the position of the frameshift mutation was changed (lys2ΔXho allele with a filled-in XhoI site) so that the mutation was roughly in the center of the homology region. Recombination between the lys2ΔXho and lys2Δ5′Δ3′ alleles can produce Lys+ recombinants via a gene conversion event, with the full-length allele acting as the recipient of genetic information and the truncated allele acting as the donor. Resolution of the event as a crossover, however, will yield a 5′-truncated allele and a 3′-truncated allele as products, neither of which will produce a Lys+ phenotype. Thus, the system detects only simple gene conversion events that are unassociated with crossing over. As with the conversion/crossover assay system, the lys2 alleles were fused to either the same promoter or different promoters and the promoter activity of pGAL was regulated by the presence or absence of Gal80p.
Recombination rate data obtained with the conversion-only substrates are presented in Table 2 and can be summarized as follows. First, the presence or absence of Gal80p does not affect the rate of gene conversion when both substrates are fused to pLYS (compare SJR578 and SJR580). Second, when Gal80p is present (low-transcription conditions), the conversion rate of the pLYS2-lys2ΔXho allele (SJR578 and SJR577) is the same as that of the pGAL-lys2ΔXho allele (SJR583 and SJR584). Third, under high-transcription conditions where both alleles are highly transcribed, the gene conversion rate is elevated 3.8-fold (compare SJR583 and SJR581). Finally, the effect of highly transcribing the substrates on the rate of gene conversion is asymmetric; transcription of only the lys2ΔXho allele elevates gene conversion 11-fold while transcription of only the lys2Δ5′Δ3′ allele elevates gene conversion only 2.7-fold. As with the conversion/crossover substrates, transcription of both alleles has less impact on recombination than does transcription of only the lys2ΔXho allele.
TABLE 2.
Effects of transcription on recombination between heterochromosomal conversion-only substrates
| Strain | Gal80 |
lys2ΔXho (LYS2 locus)
|
lys2Δ5′Δ3′ (URA3 locus)
|
Lys+ rate (10−8)a | ||
|---|---|---|---|---|---|---|
| Promoter | Transcription | Promoter | Transcription | |||
| SJR578 | + | LYS | Low | LYS | Low | 1.3 |
| SJR580 | − | LYS | Low | LYS | Low | 1.2 (0.9) |
| SJR583 | + | GAL | Low | GAL | Low | 0.89 |
| SJR581 | − | GAL | High | GAL | High | 3.4 (3.8) |
| SJR584 | + | GAL | Low | LYS | Low | 1.3 |
| SJR582 | − | GAL | High | LYS | Low | 14 (11) |
| SJR577 | + | LYS | Low | GAL | Low | 0.98 |
| SJR579 | − | LYS | Low | GAL | High | 2.6 (2.7) |
Fold increases over the rate in the corresponding Gal80+ strain are given in parentheses.
Heterochromosomal crossover-only assay system.
To detect only recombination events that are resolved as crossovers, the lys2 allele at the LYS2 locus on chromosome II was truncated at the 5′ end (lys2Δ5′) and used in combination with the standard lys2Δ3′ allele at the URA3 locus on chromosome V. As illustrated in Fig. 1C, only a crossover event will generate a full-length LYS2 allele. With the exception of the 5′ or 3′ truncation, the alleles are wild type in sequence, and so repair processes will not influence the detection of recombinants. The substrates were both fused to pGAL, or one was fused to pGAL and one was fused to pLYS, and recombination rates were examined in Gal80+ and Gal80− strains. It should be noted that when both substrates are fused to pGAL, it is formally possible to restore the 5′ end of the lys2Δ5′ by a noncrossover (simple gene conversion) mechanism.
The rate data obtained with the crossover assay are presented in Table 3, which shows that high-level transcription of both alleles results in a modest (2.3-fold) elevation in the crossover rate (compare SJR738 and SJR736). In the case where only the lys2Δ3′ allele is highly transcribed, the crossover rate was elevated 12-fold (compare SJR733 and SJR734). In contrast to the strong stimulation of recombination that accompanied transcription of the lys2Δ3′ allele, high-level transcription of only the lys2Δ5′ allele was not associated with an increased crossover rate. In agreement with results obtained using the other heterochromosomal substrates, transcription of both alleles stimulated recombination less than did transcription of only one allele (compare SJR734 and SJR736).
TABLE 3.
Effects of transcription on recombination between heterochromosomal crossover-only substrates
| Strain | Gal80 |
lys2Δ5′ (LYS2 locus)
|
lys2Δ3′ (URA3 locus)
|
Lys+ rate (10−8)a | ||
|---|---|---|---|---|---|---|
| Promoter | Transcription | Promoter | Transcription | |||
| SJR738 | + | GAL | Low | GAL | Low | 2.9 |
| SJR736 | − | GAL | High | GAL | High | 6.8 (2.3) |
| SJR737 | + | GAL | Low | LYS | Low | 2.7 |
| SJR735 | − | GAL | High | LYS | Low | 4.0 (1.5) |
| SJR733 | + | LYS | Low | GAL | Low | 1.6 |
| SJR734 | − | LYS | Low | GAL | High | 19 (12) |
Fold increases over the rate in the corresponding Gal80+ strain are given in parentheses.
Intrachromosomal direct-repeat assay system.
The heterochromosomal conversion-crossover assay system was derived by transforming a lys2-Nhe strain with an integrating plasmid containing URA3 and the lys2Δ3′ allele, with the plasmid being targeted to integrate at the URA3 locus by appropriate enzyme digestion. To directly compare transcriptional effects on heterochromosomal versus intrachromosomal recombination, the plasmid also was targeted to integrate at the LYS2 locus by digestion within the lys2Δ3′ sequences. As shown in Fig. 1D, this generates lys2Δ3′/lys2-Nhe direct repeats, with the direct repeats flanking the remainder of the plasmid sequences, including the selectable URA3 marker. Lys+ recombinants that retain the URA3 marker correspond to simple gene conversion events unassociated with crossing over, while loss of the URA3 marker indicates the loss of plasmid sequences. Plasmid sequences can be lost either via a crossover event between the lys2 direct repeats or by an SSA mechanism. As with the heterochromosomal repeats, the direct repeats were fused to either the same promoter (pLYS or pGAL) or different promoters and recombination rates were then measured in Gal80+ versus Gal80− strains.
Recombination data for the lys2 direct repeats are presented in Table 4. In the absence of high levels of transcription (Gal80+ strains), all direct-repeat substrates produced Lys+ recombinants at comparable rates and the proportions of recombinants classified as simple gene conversion events were similar (22 to 48%). The greatest transcription-associated stimulation of recombination was seen when both lys2 substrates were fused to pGAL. Relative to the Gal80+ strain, recombination in a Gal80− strain was elevated 36-fold (compare SJR432 and SJR434), with 97% of the events corresponding to plasmid loss events. When only the lys2-Nhe allele was fused to pGAL, transcription stimulated recombination 14-fold (compare SJR436 and SJR438), with similar increases in both gene conversion and plasmid loss events. Finally, when only the lys2Δ3′ allele was fused to pGAL, recombination was stimulated 5.7-fold by high levels of transcription (compare SJR431 and SJR433). In the latter case, all of the recombinants analyzed correspond to plasmid loss events.
TABLE 4.
Effects of transcription on recombination between direct-repeat substrates
| Strain | Gal80 |
lys2-Nhe
|
lys2Δ3′
|
Lys+ rate (10−5)a | Conversion (%) | Plasmid loss (%) | ||
|---|---|---|---|---|---|---|---|---|
| Promoter | Transcription | Promoter | Transcription | |||||
| SJR435 | + | LYS | Low | LYS | Low | 1.9 | 48 | 52 |
| SJR437 | − | LYS | Low | LYS | Low | 2.3 (1.2) | 48 | 52 |
| SJR432 | + | GAL | Low | GAL | Low | 2.0 | 41 | 59 |
| SJR434 | − | GAL | High | GAL | High | 71 (36) | 3 | 97 |
| SJR436 | + | GAL | Low | LYS | Low | 1.6 | 22 | 78 |
| SJR438 | − | GAL | High | LYS | Low | 23 (14) | 14 | 86 |
| SJR431 | + | LYS | Low | GAL | Low | 2.1 | 33 | 67 |
| SJR433 | − | LYS | Low | GAL | High | 12 (5.7) | 0 | 100 |
Fold increases over the rate in the corresponding Gal80+ strain are given in parentheses.
In addition to measuring the total recombination rate and then partitioning events into conversions versus crossovers, the rate of simple gene conversion (noncrossover) events can be measured directly by selecting for Lys+ recombinants that remain Ura+. This was done for the Gal80− strains that produced very few gene conversion events (SJR433 and SJR434), as well as for the isogenic Gal80+ control strains (SJR431 and SJR432). The directly measured conversion rates for SJR431 and SJR432 were 3.0 × 10−6 and 4.1 × 10−6, respectively, which are in good agreement with estimates made based on the data presented in Table 4. With the Gal80− strains, the conversion rates were 5.3 × 10−7 for SJR433 and 4.3 × 10−6 for SJR434. Thus, when both direct repeats are fused to pGAL (SJR432 and SJR434), high levels of transcription stimulate only plasmid loss events, with gene conversion events being unaffected. When only the lys2Δ3′ donor allele is fused to pGAL (SJR431 and SJR433), high levels of transcription stimulate plasmid loss events while reducing the rate of gene conversion approximately sixfold.
DISCUSSION
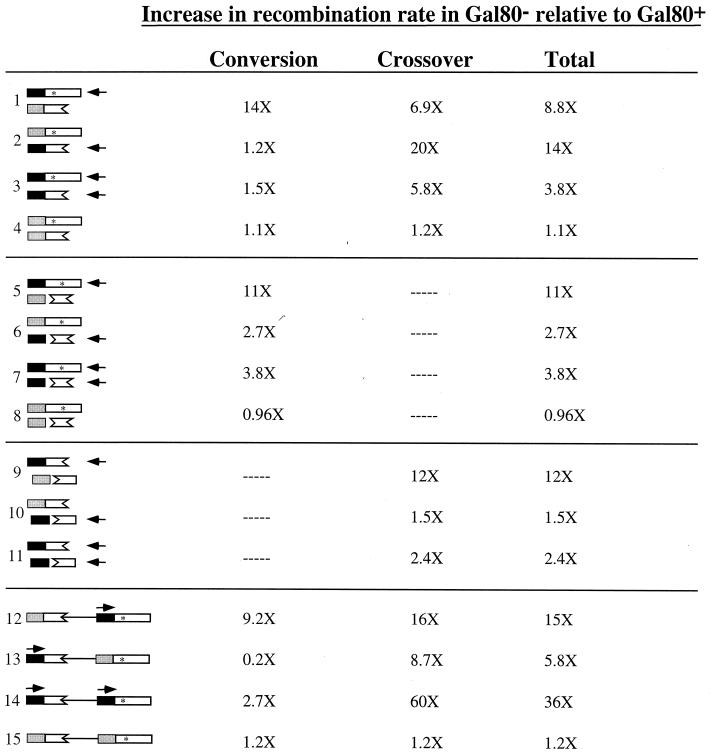
The impact of RNA polymerase II transcription on mitotic recombination in yeast was examined using lys2 alleles under control of the low-level LYS2 promoter (pLYS) and/or the highly inducible GAL1-10 promoter (pGAL). In most assays, the lys2 alleles were positioned on nonhomologous chromosomes (heterochromosomal substrates) to avoid some of the mechanistic ambiguities associated with repeats positioned on the same chromosome (intrachromosomal substrates). Three types of heterochromosomal assays were used: an assay that detects both crossover- and noncrossover-associated gene conversion events, an assay that detects only gene conversion events that are unassociated with crossing over, and an assay that detects only crossover events (Fig. 1A to C, respectively). In addition to the heterochromosomal assays, an intrachromosomal direct-repeat assay (Fig. 1D) was established to compare and contrast the impact of transcription on recombination between direct repeats with that for repeats on nonhomologous chromosomes. The data obtained using these assays are summarized in Fig. 2. In the discussion that follows, the transcription-associated recombination observed in our system is compared to that observed in HOT1 experiments (RNA polymerase I transcription) and in experiments that utilized gal10 direct repeats (RNA polymerase II transcription). In addition, possible models for transcription-associated recombination are considered in relation to the results reported here.
FIG. 2.
Summary of recombination rate data. Substrate sequences are as follows: open boxes correspond to lys2 sequences (asterisks indicate point mutations), solid boxes correspond to pGAL, and shaded boxes correspond to pLYS. Horizontal arrows indicate substrates that are highly transcribed in gal80 strains. Substrates 1 to 4 are heterochromosomal conversion-crossover substrates, substrates 5 to 8 are heterochromosomal conversion-only substrates, substrates 9 to 11 are heterochromosomal crossover-only substrates, and substrates 12 to 15 are intrachromosomal direct-repeat substrates.
The results obtained using the heterochromosomal lys2 conversion-crossover assay demonstrate that high levels of RNA polymerase II transcription can stimulate both gene conversion and crossover events (substrates 1 to 4 in Fig. 2). Our observation that RNA polymerase II-promoted transcription stimulates both gene conversion and crossover events agrees with RNA polymerase I-promoted (HOT1) recombination results obtained using allelic sequences (20). With the HOT1 system, however, the examination of conversion tract lengths led to the suggestion that crossovers might correspond to break-induced recombination (BIR) events, in which a broken end is used to prime DNA replication that extends all the way to the end of the unbroken, template chromosome (47). Because BIR is a nonreciprocal process that replaces all the information distal to the break with information from the invaded chromosome, only one of the exchange products normally associated with a true reciprocal crossover event is produced. In our heterochromosomal assay system, BIR would yield only half of a reciprocal translocation, resulting in what is presumably a lethal aneuploidy. Southern blot analysis of representative Lys+ recombinants classified as crossovers based on phenotypic analysis confirmed the presence of both translocation products (data not shown). The data reported here thus demonstrate that transcription-stimulated recombination is indeed associated with the production of reciprocal crossover products.
In addition to demonstrating transcriptional stimulation of both gene conversion and crossover events, experiments with the conversion-crossover lys2 substrates showed that there is a transcription-associated asymmetry in simple gene conversion events. This asymmetry was evident when only one of the substrates was highly transcribed (compare substrates 1 and 2 in Fig. 2). Specifically, transcription of only the full-length lys2-Nhe allele, which acts as the recipient in gene conversion events, stimulated gene conversion 14-fold, whereas transcription of only the truncated lys2Δ3′ allele, which acts as a donor in gene conversion events, did not significantly stimulate gene conversion. This asymmetry also was evident in strains containing substrates that produce Lys+ recombinants only via a gene conversion mechanism (substrates 5 and 6 in Fig. 2). When the recipient allele (lys2ΔXho) was highly transcribed, gene conversion was stimulated 11-fold; when the donor allele (lys2Δ5′Δ3′) was highly transcribed, gene conversion was stimulated only 2.6-fold. It is generally assumed that mitotic recombination is limited by the occurrence of initiating DNA lesions and that the allele that suffers the initiating lesion will be the recipient in gene conversion events (31). Our data thus suggest that one effect of high levels of RNA polymerase II transcription is to increase the occurrence of recombination-initiating lesions. We note that a similar gene conversion asymmetry was seen in HOT1 experiments when only one allele was highly transcribed by RNA polymerase I and that a link between transcription and initiating lesions was suggested (46). An alternative explanation for the pattern of transcription-associated gene conversion observed here invokes a strong transcription-associated bias in recognition and/or repair of mismatches present in recombination intermediates. Although this is a formal possibility, it cannot explain the stimulatory effects of transcription on recombination when the substrates contain no potential mismatches (i.e., the crossover-only substrates).
In addition to using lys2 substrates that produce prototrophs only via a simple gene conversion mechanism, we constructed substrates that can produce Lys+ recombinants only via a crossover event (substrates 9 to 11 in Fig. 2). As with the gene conversion-only assay, the crossover-only assay revealed an interesting asymmetry when only one of the substrates was highly transcribed. Specifically, high transcription of only the lys2Δ3′ allele stimulated crossovers 12-fold whereas high transcription of only the lys2Δ5′ allele stimulated crossovers only 1.5-fold. We suggest that this asymmetry is related to the issue of recombination-initiating lesions (see above) and reflects the distance of the substrate overlap region from pGAL. In the case where recombination is stimulated, the overlap is separated from pGAL by a nonhomologous region; in the case where recombination is not stimulated appreciably, pGAL directly abuts the substrate overlap region. Thus, the further the overlap region is from pGAL, the higher is the probability that a recombination-initiating lesion will occur. Voelkel-Meiman and Roeder observed a similar gradient in HOT1 gene conversion experiments and suggested that the conversion gradient could be related to breaks associated with transcription-induced torsional stress (46). A similar argument could account for the asymmetry observed with our crossover-only substrates.
Although there are clear similarities between the recombination results we obtained with pGAL-promoted transcription and those obtained with HOT1-promoted transcription, there is one notable exception. With all of the heterochromosomal substrates used here, high-level transcription of only one substrate from pGAL had a greater stimulatory effect on recombination than did high-level transcription of both substrates. Although this seems counterintuitive, it suggests that high transcription of a donor sequence may actually inhibit strand invasion. This could be tested by using the HO endonuclease to introduce recombination-initiating double-strand breaks in the recipient substrate and then assaying repair efficiency using donor substrates that are subject to high versus low transcription. In contrast to our results, there appeared to be an additive effect of highly transcribing both substrates in the HOT1 experiments (46). The difference in HOT1-versus pGAL-associated recombination could reflect an inherent difference in RNA polymerase I and RNA polymerase II transcription or a difference in the assay systems used.
The results obtained with the heterochromosomal lys2 substrates contrast with those obtained by the Rothstein laboratory using gal10 direct repeats. With the gal10 direct repeats, high levels of RNA polymerase II transcription stimulated only events involving loss of sequences between the direct repeats (plasmid loss events) and failed to stimulate simple gene conversion events (42). With direct repeats, plasmid loss can be due to a true reciprocal crossover event involving Holliday junction resolution or can result from the alternative, nonconservative mechanism of SSA (see reference 3 for a description of recombination mechanisms). The absence of transcription-stimulated gene conversion events in the gal10 direct-repeat system suggests that most of the transcription-associated recombination events observed in this system corresponded to SSA events. The overlapping RAD52/RAD1 genetic requirements of the plasmid loss events (43), as well as the observation that transcription between the direct repeats rather than through the repeats was correlated with enhanced recombination (42), is consistent with this interpretation. As discussed in more detail below, the necessity of transcribing sequences between the direct repeats probably is related to the direct-repeat hyperrecombination phenotype observed in hpr1 mutants (33).
To more directly compare heterochromosomal and intrachromsomal transcription-stimulated recombination events, the same lys2 substrates were positioned on nonhomologous chromosomes (substrates 1 to 4 in Fig. 2) or as direct repeats on the same chromosome (substrates 12 to 15 in Fig. 2). In considering results obtained with the direct-repeat substrates, it should be borne in mind that pGAL is a bidirectional promoter. Fusion of pGAL to the downstream, full-length lys2-Nhe allele thus not only results in high-level transcription of lys2-Nhe but also probably results in high-level transcription of the upstream plasmid sequences that are between the lys2 substrates (substrates 12 and 14 in Fig. 2). Furthermore, when both lys2 alleles are fused to pGAL (substrate 14 in Fig. 2), the transcription complexes may converge in the region between the substrates. As observed with the heterochromosomal lys2 repeats, high levels of transcription stimulated both simple gene conversion events and plasmid loss events in the lys2 direct-repeat assay. This is in contrast to lack of simple gene conversion stimulation with gal10 direct repeats (42). In addition, an asymmetry in gene conversion events was evident with the lys2 direct repeats, which was similar to that described above for the heterochromosomal repeats. That is, simple gene conversion events were stimulated only when the recipient, full-length allele was highly transcribed (compare direct-repeat substrates 12 and 13 in Fig. 2). The partitioning of substrate 13 recombinants into conversions versus plasmid loss events indicated that the rate of simple conversion events actually decreased when the donor allele was highly transcribed. This was confirmed by direct measurement of the conversion rates for substrates 12 and 13. Although the reason for this behavior is unclear, it suggests that transcription not only increases the frequency of recombination-initiating lesions but also may alter the recombinational repair of lesions generated by other mechanisms. In the case of the lys2 direct repeats, transcription of the region between the substrates may channel lesions into the SSA pathway. In contrast to the results obtained with the heterochromosomal lys2 repeats, high-level transcription of both substrates in the direct-repeat assay stimulated the total rate of recombination to a greater extent than did high-level transcription of just one substrate. This increased stimulation, however, pertained only to plasmid loss events and may be a consequence of convergent transcription complexes. The different behavior of the heterochromosomal and intrachromosomal lys2 repeats under high-transcription conditions is consistent with SSA being a unique and important recombination mechanism for the intrachromosomal direct repeats.
There are two general models that can account for transcription-stimulated recombination. In the first model, transcription alters the repair of the preexisting DNA lesions by specifically channeling them into the recombination repair pathway or by facilitating the process of recombination. For example, there could exist a recombination subpathway that is analogous to transcription-coupled nucleotide excision repair. Although there is no evidence for transcription-coupled recombination, it is intriguing that at least one recombination protein is associated with a eukaryotic transcription complex (26). Alternatively, high levels of transcription might interfere with competing DNA repair pathways and thereby favor recombination as a DNA repair mechanism. Finally, the open chromatin structure associated with transcription could directly facilitate recombination by increasing the efficiency with which a duplex molecule is invaded by a single strand of DNA. In in vitro recombination studies involving interaction between a single-stranded DNA and a duplex DNA molecule, the assembly of nucleosomes on the duplex molecule was found to inhibit the strand exchange reaction (22, 34). The inhibition was relieved, however, if the chromatin template was transcribed (22). As noted above, transcription of the recipient lys2 allele affected gene conversion more than did transcription of the donor allele (11-fold and 2.6-fold, respectively) in our in vivo conversion-only assay. If the primary effect of transcription in our system were to facilitate the invasion of a duplex molecule, one would expect the greatest elevation in gene conversion rate to accompany transcription of the donor sequence. The relatively small stimulation in gene conversion that accompanied high-level transcription of the donor molecule may indeed reflect more efficient invasion of transcriptionally active DNA, with high levels of transcription perhaps replacing the need for one or more recombination proteins. This possibility could be addressed by examining transcription-associated recombination in appropriate yeast mutant strains. In relation to the role of transcription-associated chromatin remodeling in facilitating strand invasion, we note that transcription of a target sequence in mammalian cells has been reported to increase the efficiency of gene targeting (44).
The second general model that has been proposed to explain transcription-stimulated recombination invokes transcription-associated DNA damage. First, through alterations in chromatin structure and nucleosome positioning, high levels of transcription may increase the accessibility of DNA to endogenous damaging agents. In addition, the transient regions of single-stranded DNA in transcription bubbles may be more susceptible to damage than is duplex DNA (2). Enhanced DNA damage indeed has been invoked to explain the elevated mutation rates that accompany high levels of transcription in yeast (27). Second, the process of transcription generates torsional stress on the DNA template, with regions of positive and negative supercoiling accumulating ahead of and behind, respectively, the transcription machinery (25). Torsional stress has been invoked to explain the increase in rDNA recombination that accompanies the loss of topoisomerase I or II activity in yeast (5, 21). In addition, recombination between the long terminal repeats (δ) of the yeast Ty retrotransposon is increased in top3 mutants (48). The polarity we observed with the lys2 crossover-only substrates and that observed with HOT1-stimulated gene conversion (46) would be consistent with a torsional-stress model. The role of torsional stress in transcription-stimulated recombination could be addressed by examining recombination between lys2 substrates in appropriate topoisomerase-deficient yeast mutants.
A third source of RNA polymerase II transcription-associated DNA damage relates to observations made with yeast hpr1 mutants, which were initially identified as hyperrecombination mutants in a direct-repeat assay (1). Chavez and Aguilera have correlated the increase in direct repeat recombination in hpr1 mutants with a failure to efficiently elongate transcription through the plasmid sequences that separate the direct repeats (4). They thus have hypothesized that a stalled transcription complex either directly induces recombination-initiating strand breaks or impedes the DNA replication machinery, thereby stimulating recombinational bypass. Piruat and Aguilera have noted that all examples of transcription-associated recombination in yeast involve high-level transcription of sequences that normally are expressed at a low level in yeast or are foreign to the yeast genome and thus may be mechanistically similar to hpr1-associated recombination (33). Specifically, in the HOT1 system, his4 alleles were fused to the rDNA promoter (20); in the gal10 direct-repeat system, transcription of plasmid sequences (pBR322) between the repeats was correlated with elevated recombination (42); and in our system, lys2 alleles were fused to pGAL. If transcription elongation problems are indeed triggering recombination in these systems, one would predict that transcription-associated recombination should be further elevated in an hpr1 mutant background. In the gal10 direct-repeat system, eliminating Hpr1p and inducing transcription have synergistic effects on recombination (9), suggesting that elongation problems are responsible for much of the transcription-associated recombination observed in this system. With the heterochromosomal repeats, we can eliminate the transcription of nonyeast plasmid sequences as the causative factor, because we saw elevated recombination rates when no plasmid sequences were linked to the highly transcribed lys2 allele (e.g., substrate 1 in Fig. 2). Elimination of Hpr1p in our strains should indicate whether the transcription-stimulated recombination is related to potential elongation problems through the lys2 recombination substrates. Finally, it has been demonstrated that convergence of RNA polymerase III transcription and DNA replication impedes replication fork movement (8). Although it is not known whether pausing of replication forks triggers recombination in yeast, we note that a similar pausing associated with high levels of RNA polymerase II transcription potentially could trigger the transcription-associated recombination observed in our lys2 system.
In summary, the results reported here demonstrate that high levels of RNA polymerase II transcription can stimulate both mitotic gene conversion and reciprocal crossover events. The asymmetry observed in simple (noncrossover) gene conversion events suggests that the primary effect of high-level transcription is to increase the frequency of recombination-initiating lesions, although the data also indicate that transcription may facilitate the invasion of a duplex chromatin template. Therefore, there may be multiple, independent mechanisms that are important for the elevated recombination associated with high levels of transcription in yeast. The assay system developed for this study should be useful for genetically determining the contributions of these mechanisms to transcription-associated recombination.
ACKNOWLEDGMENTS
We acknowledge the contributions of Merrilyn Michelitch, Tamara Murphy, Anna Speke, and Miyono Hendrix in constructing some of the substrates and strains used in this analysis. We thank Jennifer Freedman and Takura Nakagawa for helpful comments on the manuscript and members of the laboratory for fruitful discussions.
This work was supported by National Institutes of Health grant GM38464 to S.J.-R. D.S. and A.D. were partially supported by the Emory University Graduate Division of Biological and Biomedical Sciences.
REFERENCES
- 1.Aguilera A, Klein H L. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beletskii A, Bhagwat A S. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 4.Chavez S, Aguilera A. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 1997;11:3459–3470. doi: 10.1101/gad.11.24.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christman M F, Dietrich F S, Fink G R. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988;55:413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- 6.Cooper P K, Nouspikel T, Clarkson S G, Leadon S A. Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- 7.Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- 8.Deshpande A M, Newlon C S. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- 9.Fan H-Y, Cheng K K, Klein H L. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1Δ of Saccharomyces cerevisiae. Genetics. 1996;142:749–759. doi: 10.1093/genetics/142.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fishman-Lobell J, Rudin N, Haber J E. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol. 1992;12:1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleig U N, Pridmore R D, Philippsen P. Construction of LYS2 cartridges for use in genetic manipulations of Saccharomyces cerevisiae. Gene. 1986;46:237–245. doi: 10.1016/0378-1119(86)90408-7. [DOI] [PubMed] [Google Scholar]
- 12.Gangloff S, Lieber M R, Rothstein R. Transcription, topoisomerases and recombination. Experientia. 1994;50:261–269. doi: 10.1007/BF01924009. [DOI] [PubMed] [Google Scholar]
- 13.Goldman A S H, Lichten M. The efficiency of meiotic recombination between dispersed sequences in Saccharomyces cerevisiae depends upon their chromosomal location. Genetics. 1996;144:43–55. doi: 10.1093/genetics/144.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimm C, Schaer P, Munz P, Kohli J. The strong ADH1 promoter stimulates mitotic and meiotic recombination at the ADE6 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1991;11:289–298. doi: 10.1128/mcb.11.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 16.Harris S, Rudnicki K S, Haber J E. Gene conversions and crossing over during homologous and homeologous ectopic recombination in Saccharomyces cerevisiae. Genetics. 1993;135:5–16. doi: 10.1093/genetics/135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang G S, Keil R L. Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple cis-acting sequences. Genetics. 1995;141:845–855. doi: 10.1093/genetics/141.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones E W, Pringle J, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 193–281. [Google Scholar]
- 19.Johnston M, Davis R W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keil R L, Roeder G S. cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 21.Kim R A, Wang J A. A subthreshold level of DNA topoisomerase leads to the excision of yeast rDNA as extrachromosomal rings. Cell. 1989;57:975–985. doi: 10.1016/0092-8674(89)90336-x. [DOI] [PubMed] [Google Scholar]
- 22.Kotani H, Kmiec E B. Transcription activates RecA-promoted homologous pairing of nucleosomal DNA. Mol Cell Biol. 1994;14:1949–1955. doi: 10.1128/mcb.14.3.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kupiec M, Petes T D. Meiotic recombination between repeated transposable elements in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2942–2954. doi: 10.1128/mcb.8.7.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lea D E, Coulson C A. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 25.Lui L F, Wang J C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Pickert P, Lees E, Anderson C W, Linn S, et al. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 27.Morey N J, Greene C N, Jinks-Robertson S. Genetic analysis of transcription-associated mutation in Saccharomyces cerevisiae. Genetics. 2000;154:109–120. doi: 10.1093/genetics/154.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nevo-Caspi Y, Kupiec M. Transcriptional induction of Ty recombination in yeast. Proc Natl Acad Sci USA. 1994;91:12711–12715. doi: 10.1073/pnas.91.26.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nickoloff J A. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol Cell Biol. 1992;12:5311–5318. doi: 10.1128/mcb.12.12.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nickoloff J A, Reynolds R J. Transcription stimulates homologous recombination in mammalian cells. Mol Cell Biol. 1990;10:4837–4845. doi: 10.1128/mcb.10.9.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paques F, Haber J E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parket A, Kupiec M. Ectopic recombination between Ty elements in Saccharomyces cerevisiae is not induced by DNA damage. Mol Cell Biol. 1992;12:4441–4448. doi: 10.1128/mcb.12.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piruat J I, Aguilera A. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 1998;17:4859–4872. doi: 10.1093/emboj/17.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramdas J, Mythili E, Muniyappa K. Nucleosomes on linear duplex DNA allow homologous pairing but prevent strand exchange promoted by RecA protein. Proc Natl Acad Sci USA. 1991;88:1344–1348. doi: 10.1073/pnas.88.4.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 38.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 39.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–20. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 40.Steward S E, Roeder G S. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3464–3472. doi: 10.1128/mcb.9.8.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoma F. Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J. 1999;18:6585–6598. doi: 10.1093/emboj/18.23.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 43.Thomas B J, Rothstein R. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thyagarajan B, Johnson B L, Campbell C. The effect of target site transcription on gene targeting in human cells in vitro. Nucleic Acids Res. 1995;23:2784–2790. doi: 10.1093/nar/23.14.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voelkel-Meiman K, Keil R L, Roeder G S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- 46.Voelkel-Meiman K, Roeder G S. A chromosome containing HOT1 preferentially receives information during mitotic interchromosomal gene conversion. Genetics. 1990;124:561–572. doi: 10.1093/genetics/124.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voelkel-Meiman K, Roeder G S. Gene conversion tracts stimulated by HOT1-promoted transcription are long and continuous. Genetics. 1990;126:851–867. doi: 10.1093/genetics/126.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallis J W, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 49.White M A, Detloff P, Strand M, Petes T D. A promoter deletion reduces the rate of mitotic, but not meiotic, recombination at the HIS4 locus in yeast. Curr Genet. 1992;21:109–116. doi: 10.1007/BF00318468. [DOI] [PubMed] [Google Scholar]
- 50.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcription regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 51.Wright B E, Longacre A, Reimers J M. Hypermutation in derepressed operons of Escherichia coli K12. Proc Natl Acad Sci USA. 1999;96:5089–5094. doi: 10.1073/pnas.96.9.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]