Abstract
Background
Our goal was to identify genetic risk factors for severe otitis media (OM) in Aboriginal Australians.
Methods
Illumina® Omni2.5 BeadChip and imputed data were compared between 21 children with severe OM (multiple episodes chronic suppurative OM and/or perforations or tympanic sclerosis) and 370 individuals without this phenotype, followed by FUnctional Mapping and Annotation (FUMA). Exome data filtered for common (EXaC_all ≥ 0.1) putative deleterious variants influencing protein coding (CADD-scaled scores ≥15] were used to compare 15 severe OM cases with 9 mild cases (single episode of acute OM recorded over ≥3 consecutive years). Rare (ExAC_all ≤ 0.01) such variants were filtered for those present only in severe OM. Enrichr was used to determine enrichment of genes contributing to pathways/processes relevant to OM.
Results
FUMA analysis identified 2 plausible genetic risk loci for severe OM: NR3C1 (Pimputed_1000G = 3.62 × 10−6) encoding the glucocorticoid receptor, and NREP (Pimputed_1000G = 3.67 × 10−6) encoding neuronal regeneration-related protein. Exome analysis showed: (i) association of severe OM with variants influencing protein coding (CADD-scaled ≥ 15) in a gene-set (GRXCR1, CDH23, LRP2, FAT4, ARSA, EYA4) enriched for Mammalian Phenotype Level 4 abnormal hair cell stereociliary bundle morphology and related phenotypes; (ii) rare variants influencing protein coding only seen in severe OM provided gene-sets enriched for “abnormal ear” (LMNA, CDH23, LRP2, MYO7A, FGFR1), integrin interactions, transforming growth factor signaling, and cell projection phenotypes including hair cell stereociliary bundles and cilium assembly.
Conclusions
This study highlights interacting genes and pathways related to cilium structure and function that may contribute to extreme susceptibility to OM in Aboriginal Australian children.
Keywords: otitis media, genetic susceptibility, NR3C1 glucocorticoid receptor, NREP neuronal regeneration related protein, stereociliary bundles, cilium assembly
Genome-wide analysis identifies common noncoding putative regulatory, as well as common and rare potentially deleterious variants directly influencing protein coding, in genes and pathways related to cilium structure and function that may contribute to severe otitis media in Aboriginal Australians.
Aboriginal Australian children have high rates of conductive hearing loss associated with otitis media (OM), which commonly occurs within 3 months of birth and progresses to severe chronic disease including chronic suppurative otitis media (CSOM) in 60% of children [1]. Otitis media involves interaction between environmental [2] and genetic [3] risk factors. Caucasians show high heritability for susceptibility to OM, increasing from 49%–71% from ages 2–4 [4]. Candidate gene and genome-wide association studies (GWAS) (reviewed [3]) identified genes/gene regions contributing to susceptibility. However, hypothesis-free genome-wide studies have not been employed for OM in Aboriginal Australians.
GWAS typically employ common variants that influence, or are in linkage disequilibrium (LD) with, single nucleotide variants (SNVs) enriched for expression quantitative trait loci (eQTLs [5]). Exome sequencing identifies putative deleterious variants influencing protein coding, as used to demonstrate that a rare variant within the middle ear-specific gene A2ML1 (α2-macroglobulin-like 1) co-segregated with early-onset OM in an indigenous Filipino pedigree [6]. The variant lay within a founder haplotype shared by 3 unrelated otitis-prone European-American and Hispanic-American children, but was absent in non-otitis-prone children and in >60 000 exomes including the Exome Aggregation Consortium (ExAC) database [7]. De novo mutations are also thought to contribute to genetic risk as, for example, in both rare and common forms of neurodevelopmental diseases [8]. A novel mutation of large effect in an important gene can contribute to a complex disease phenotype [8].
Here we present a community-based study that uses both GWAS analysis of common variants, as well as exome analysis of common and rare variants, to identify SNVs that could contribute to genetic risk for severe OM in a Western Australian Aboriginal population.
METHODS
Ethical Considerations and Study Population
Study participants were from an Aboriginal community of Martu ancestry [9] in Western Australia. A memorandum of understanding with the community included permission to access hard-copy and electronic clinical records. Ethical approval was obtained from the Western Australian Aboriginal Health Ethics Committee (Reference 227 12/12). Participants (or the parent/guardian if <18 years old) signed informed consent forms to take part in the study, for access to clinical records, and to provide a DNA sample. Post-quality control (QC) variant data are lodged in the European Genome-phenome Archive (accession number EGAS00001001004). Following feedback to community, permission to publish was provided by the Board of the local Aboriginal Health Service comprising elders representing extended families residing in the area.
Defining OM Phenotypes
Clinical records provided entries dating back >20 years. Every incidence of OM was noted for all consenting individuals, including records designated by clinical staff as acute OM (AOM), OM with effusion (OME), CSOM, perforations/healed perforations, tympanic sclerosis, and details of myringoplasty. Individuals were classified as: (i) severe OM (N = 21), based on repeated episodes including multiple entries for CSOM and/or perforations or tympanic sclerosis monitored over ≥3 consecutive years (N = 17), or minimally 3 diagnoses of CSOM or perforations in infants <2 years old (N = 4); (ii) intermediate OM, based on at least one diagnosis of CSOM or perforations monitored over ≥3 consecutive years (N = 21); (iii) mild OM, based on a single or maximally 3 episodes of AOM (but never CSOM or perforations) monitored over ≥3 years (N = 28); and (iv) no history of any form of OM over a minimal period of 3 contiguous years (N = 41). There were 280 individuals of unknown phenotype. Supplementary Table 1 summarizes records for individuals with a definitive OM phenotype; Supplementary Table 2 provides data on age and sex. The overall OM study design is presented in Supplementary Figure 1.
GWAS for Common Regulatory Variants
The study examined both adult metabolic diseases [10] and childhood OM. DNAs from saliva (Oragene; DNA Genotek, Ontario, Canada) were genotyped using the Illumina® Omni2.5 BeadChip (Centre for Applied Genomics, Toronto, Ontario, Canada) [10]. Imputation against 1000 Genomes (1000G) haplotypes [Phase I integrated variant set release (v3)], QC procedures, and analysis of population substructure are reported elsewhere [10]. Post-QC data were available for 1 075 436 genotyped and 6 724 284 imputed SNVs for 391 individuals. This included the 21 individuals (9 males, 12 females; mean ± SD 7.29 ± 3.81 years) with a definitive severe OM phenotype. For GWAS analysis this severe OM group was compared with 370 individuals with less severe forms or no record of OM, or phenotype unknown. A case-control analysis was performed using Fisher’s Exact Test to take account of small sample size, with a genomic control to correct for relatedness. Specifically, after comparing the genotype distribution in cases and controls at each SNV using Fisher’s Exact Test, the resulting P-values were converted to equivalent chi-squared test statistics, the genomic control inflation factor λ was calculated as the median of the chi-squared test statistics obtained from the genotyped SNVs divided by 0.456 [11], and the raw chi-squared test statistic at each SNV was divided by λ to give the final corrected chi-squared test statistic and resulting P-value for that SNV. Manhattan plots were generated in R using mhtplot() in the genetic analysis package “gap.” Regional association plots were created using LocusZoom [12].
Post-GWAS Annotation in FUMA
The FUnctional Mapping and Annotation (FUMA) Package [13] was used to characterize regions of association based on positional, eQTL, and chromatin interaction mapping. GWAS summary statistics were loaded into FUMA. SNP2GENE mapping (GWAS P < 10−5) identified independent significant SNVs based on 1000G multi-ethnic LD data. Independent significant SNVs and those in LD were annotated for gene function using ANNOVAR, regulatory functions (Regulome DB score), and 15-core chromatin state predicted by ChromHMM for 127 tissue/cell types. Effects of SNVs on gene expression were determined using eQTLs from multiple tissue/cell types from databases: eQTLgen (44 tissue types); BIOSQTL (BIO_eQTL_gene level, whole peripheral blood, 2116 healthy donors); DICE (B and T cells, monocytes, neutrophils, NK cells); and GTEx v8 (whole blood; cultured fibroblasts).
Exome Analysis
Exome sequences were available for 72 unrelated individuals (35 pure Martu) from the post-QC GWAS sample [14]. SNV data was filtered for variants with Combined Annotation Dependent Depletion (CADD)-scaled scores ≥15 predicted to have medium (missense; splice region) to high (splice-acceptor/splice-donor; stop-gain/stop-loss; start loss) impact on protein function. Exome data included 15 unrelated children with severe OM (mean ± SD 7.86 ± 2.93 years) and 9 unrelated children (mean ± SD 13.63 ± 8.12 years) with mild OM (single episode of acute OM over ≥3 consecutive years). Extreme phenotype case-control analysis compared these 2 groups using Fisher’s Exact Test under allele-wise or dominant models and an allele-wise Trend Test for common CADD-scaled ≥ 15 variants (defined as ExAC_all ≥ 0.1). Genes with variant associations at P < .05 were analyzed in Enrichr [15] for enrichment of gene sets previously associated with phenotypes relevant to OM. Exome data were also filtered for rare (defined as ExAC_all ≤ 0.01) CADD-scaled ≥ 15 high/medium impact variants only ever seen in severe OM, and never in mild OM or in 48 phenotype unknown individuals. This gene list was also analyzed in Enrichr.
RESULTS
Characteristics of the Study Population
The 391 post-QC GWAS individuals belonged to a small number of inter-related extended pedigrees [10] (Supplementary Figure 2A). Principal component analysis (Supplementary Figure 2B) demonstrated introgression of predominantly Caucasian origin, with a tight cluster of 195 individuals of pure Martu Aboriginal ancestry.
GWAS and Integrative post-GWAS Analyses
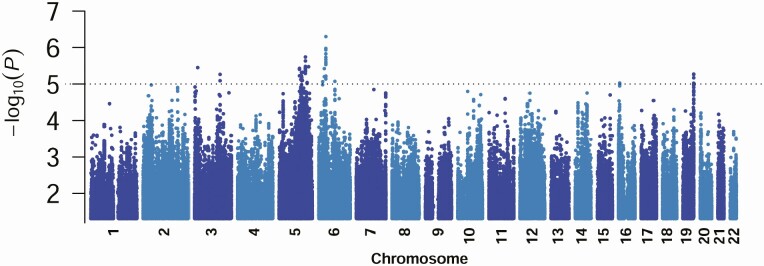
The GWAS Manhattan plot for imputed data comparing 21 severe OM cases with 370 controls (Figure 1) showed no systematic bias (λs 0.998; Supplementary Figure 3). No hits were observed at P < 5 × 10−8. SNP2GENE identified 17 genomic loci associated with severe OM (Table 1; Supplementary Table 3). Positionally mapped SNVs mostly localized to noncoding sequence, 44% intronic, 36% intergenic, 17% intronic in noncoding RNA genes, and 3% other (including 1% exonic).
Figure 1.
Manhattan plot of results from analysis for the 6.72M high-quality 1000G imputed SNV variants. For clarity only variants showing GWAS P-values < .05 are plotted. Data are for the case-control GWAS analysis undertaken using Fisher’s Exact Test with a genomic control correction to look for associations between SNVs and severe OM. The Y-axis indicates –log10P values for association, the X axis indicates the positions across each chromosome. The dotted line indicates the P = 1 × 10−5 cut-off used in the post-GWAS FUMA analysis. Abbreviations: FUMA, FUnctional Mapping and Annotation; GWAS, genome-wide association studies; SNV, single nucleotide variants.
Table 1.
Summary of SNP2GENE Results for Lead Independent Significant GWAS SNPs and Associated Gene and eQTL Information
| Genomic Locus | a Lead IndSigSNP Chr:bp:alleles | rsID | GWAS P-value | Nearest Gene(s) | Type of Gene | Distance Nearest Gene bp | Functional Location | b N Pos Mapped SNVs | N eQTL SNPs | eQTL Database | c eQTL Type/genes influenced |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3:16868616: A:T | rs1866862 | 3.49E-06 | PLCL2 | Protein coding | 0 | intronic | 41 | 63 | BIOSQTL BIOSQTL eQTLGen GTEx/v8 eQTLCat eQTLCat eQTLCat eQTLCat eQTLCat | 63 gene-level PLCL2 25 gene-level PLCL2:PLCL2-AS1 41 cis_eQTLs PLCL2 34 whole blood PLCL2 6 monocyte PLCL2 6 neutrophil PLCL2 5 neutrophil_CD15 PLCL2 5 blood PLCL2 6 fat PLCL2 |
| 2 | 3:138244928: C:G | rs9855074 | 5.38E-06 | CEP70 | Protein coding | 0 | intronic | 8 | 7 | eQTLGen BIOSQTL eQTLCat eQTLCat DICE | 7 cis-EQTL FAIM 2 gene level FAIM 2 monocyte CEP70 7 LCLd CEP70: FAIM 3 B cell naïve CEP70 |
| 3 | 5:111067185: C:CTAA | rs147647553 | 3.67E-06 | STARD4-AS1 NREP | Antisense Protein coding | 0 | ncRNA_ intronic intronic | 33 | 0 | nil | nil |
| 4 | 5:118132897: A:G | rs421765 | 7.48E-06 | CTC-448D22.1 | LincRNA | 11742 | intergenic | 63 | 64 | eQTLGen eQTLGen eQTLCat | 15 cis_eQTL DMXL1 49 cis_eQTL DTWD2 3 iPSC TNFAIP8 |
| 5 | 5:122006325: A:G | rs13180735 | 4.81E-06 | RP11-166A12.1 ARGFXP1 | LincRNA | 0 | ncRNA_ intronic | 6 | 2 | eQTLCat eQTLCat eQTLgen | 2 CEDAR_platelet SNX2 2 LCLd PPIC 2 cis-EQTL CTB-36H16.2 |
| 6 | 5:123136698: C:T | rs257141 | 6.61E-06 | KRT18P16 | Pseudogene | 163600 | intergenic | 10 | 0 | nil | nil |
| 7 | 5:124041059: C:G | rs6860814 | 8.10E-06 | ZNF608 | Protein coding | 0 | intronic | 3 | 0 | nil | nil |
| 8 | 5:126502084: A:G | rs10076402 | 4.71E-06 | CTB-88F18.2 | Pseudogene | 4142 | intergenic | 9 | 9 | eQTLGen BIOSQTL eQTLCat eQTLGen | 1 cis_eQTL C5orf63:MARCH3 3 gene level C5orf63:MARCH3 2 T cell C5orf63 3 cis_eQTL FBN2 |
| 9 | 5:142676404: G:T | rs258814 | 3.62E-06 | NR3C1 | Protein coding | 0 | intronic | 29 | 24 | BIOSQTL eQTLCat | 24 gene level NR3C1 18 blood NR3C1 |
| 10 | 5:143288470: C:G | rs13170657 | 1.83E-06 | CTB-57H20.1 | Sense overlapping | 80132 | intergenic | 62 | 0 | nil | nil |
| 11 | 5:152466290: C:T | rs11744443 | 9.06E-06 | AC091969.1 | LincRNA | 0 | ncRNA_ intronic | 20 | 0 | nil | nil |
| 12 | 5:160326888: A:G | rs10780120 | 3.31E-06 | RP11-109J4.1 | LincRNA | 31896 | intergenic | 57 | 57 | eQTLCat | 57 brain ATP10B |
| 13 | 6:21464051: C:G | rs9460636 | 8.59E-06 | RP11-204E9.1 | LincRNA | 58655 | intergenic | 7 | 0 | nil | nil |
| 14 | 6:38637538: A:G | rs62396381 | 4.95E-07 | GLO1:BTBD9 | Protein coding | 6162 | intergenic | 153 | 130 | eQTLGen eQTLGen eQTLGen eQTLGen eQTLGen eQTLGen BIOSQTL eQTLCat DICE | 61 cis_eQTL GLO1 30 cis_eQTL_GLO1:ZFAND3 7 cis_eQTL GLO1:MDGA1 17 cis_eQTL MDGA1:ZFAND3 6 cis_eQTL GLO1:MDGA1:ZFAND3 1 cis_eQTL MDGA1:ZFAND3:CCDC167 3 gene level GLO1 1 monocyte naïve GLO1 4 B cell naïve RP1–153P14.8 |
| 15 | 6:87510924: C:T | rs4291075 | 8.42E-06 | RP1-253B10.2 | Pseudogene | 32273 | intergenic | 8 | 0 | nil | nil |
| 16 | 16:6614413: A:G | rs2192643 | 9.33E-06 | RP11-420N3.2 RBFOX1 | Transcript Protein coding | 0 | ncRNA_ intronic | 16 | 0 | nil | nil |
| 17 | 19:59028585: A:G | rs11545185 | 5.30E-06 | ZBTB45 | Protein coding | 0 | exonic | 44 | 43 | eQTLCat eQTLCat eQTLCat eQTLCat eQTLCat | e 38 blood MZF1 43 monocytes CD14 MZF1 42 T cell CD4 MZF1 28 T cell CD8 MZF1 3 monocyte LPS MZF1 |
Note: More details of genomic loci are provided in Supplementary Table 3.
Abbreviations: a Lead IndSigSNP = GWAS top SNV; b Pos, positionally mapped SNVs for the locus from SNP2GENE analysis; c eQTL type/genes whose expression is associated with these eQTLs (note BIOSQTL = blood). d LCL, T cell leukemia cell line; e Many eQTLs associated with expression of numerous genes: ZNF584, ZNF132, CTD-2619J13.17, MZF1, AC016629.3, UBE2M, SLC27A5, CTD-2619J13.14, ZNF324, ZNF446, TRIM28, RP55, CHMP2A, ZNF544, A1BG-AS1, ZNF837, ZNF329, A1BG, MIR4754, AC010642.1, AC016629.8, CTD-2619J13.9, AC020915.1 (see Supplementary Figure 10), only those eQTLs associated with MZF1 gene expression are shown as the most plausible candidate in relation to cells relevant to OM.
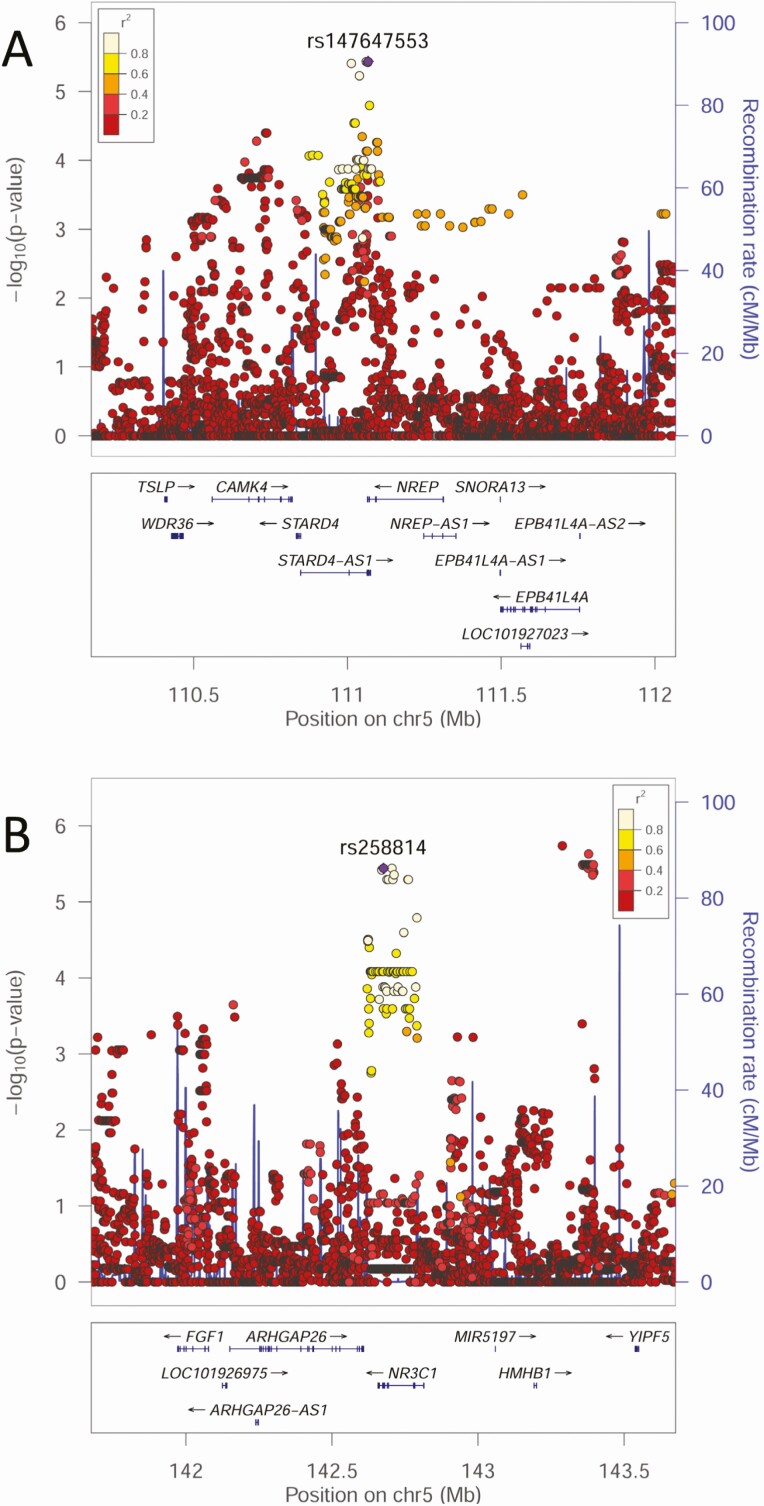
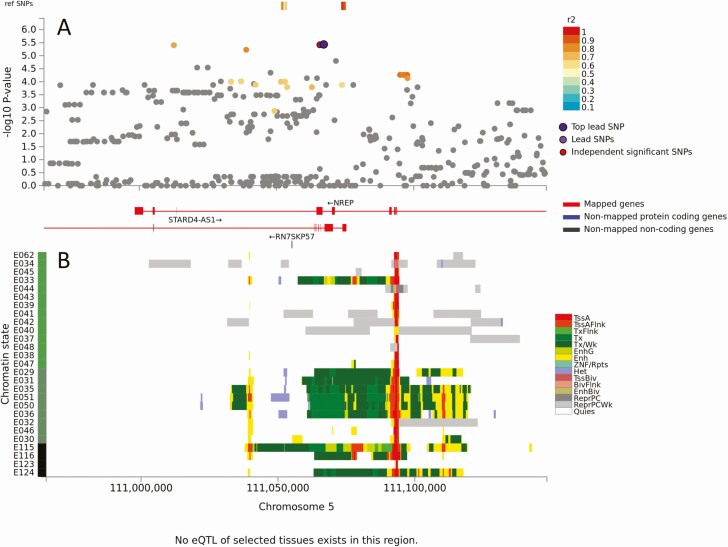
No eQTLS mapped to genomic loci 3, 6, 7, 10, 11, 13, 15 and 16 (ie, there were no expression-related data to support candidacy of genes at these loci). Top SNVs at 4 of these loci (6, 10, 13, 15) were intergenic >30kb from the nearest annotated gene (Table 1). Those at loci 7, 11 and 16, were not located within genes of functional significance for OM (Supplementary Table 4). These 7 loci were not considered further. One gene with no eQTL support was NREP at locus 3 (GWAS P = 3.67 × 10−6, Figure 2A), despite multiple associated SNVs lying in a region of strong transcriptional activity in hematopoetic stem cells and blood (Figure 3). NREP encodes neuronal regeneration-related protein, variants at which associate with esophageal microbiomes [16]. Given OM association with nasopharyngeal microbiomes [17], this could represent a candidate where expression might not relate to eQTL in public domain databases.
Figure 2.
LocusZoom plots for GWAS associations identified as plausible genetic risk factors for severe OM following post-GWAS annotation: (A) NREP and (B) NR3C1. The –log10P values (left y-axis) are shown in the top section of each plot. Dots representing individual SNVs are color coded (see key) based on their population-specific LD r2 with the top SNV (annotated by rs ID) in the region. The right Y-axis is for recombination rate (blue line), based on HapMap data. The bottom section of each plot shows the positions of genes across the region. Abbreviations: GWAS, genome-wide association studies; LD, linkage disequilibrium; OM, otitis media; SNV, single nucleotide variants.
Figure 3.
Results of positional, chromatin interaction, and eQTL activity mapping in FUMA for NREP. (A) Maps the top lead SNV, and SNVs in LD with it according to the r2 color-coded key, across the region of genomic locus 3. There was one additional independent significant SNV located adjacent to the top lead GWAS SNV. (B) Chromatin-15 states color coded for transcriptional/enhancer activity as shown in the key. Y-axis color coding relates to cell/tissue types in which chromatin interaction was mapped. There were no eQTLs mapping to genomic locus 3. Full explanation of keys provided as preamble to supplementary figures. Abbreviations: eQTL, expression quantitative trait loci; FUMA, Functional Mapping and Annotation; GWAS, genome-wide association studies; LD, linkage disequilibrium; SNV, single nucleotide variants.
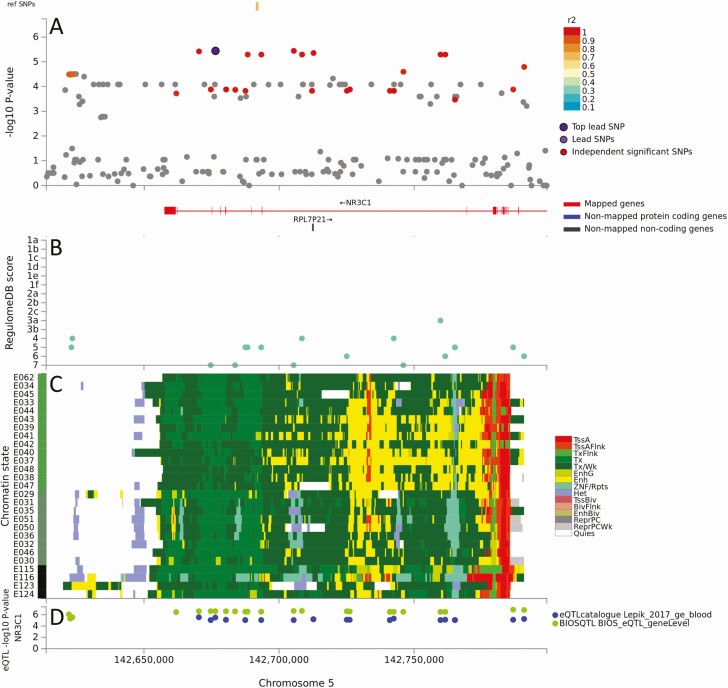
To determine which genes in 9 loci (1, 2, 4, 5, 8, 9, 12, 14, 17) with mapped eQTLs might be risk factors for severe OM, we considered: (i) BIOSQTL gene-level (=blood) and eQTLGen cis-eQTL (=multiple tissues/cell types) support for regulation of specific genes within each locus (Table 1); (ii) eQTL-catalogue/GTEx v8/DICE support for regulation of specific genes in tissues (whole blood; brain) and cell types (immune cells) relevant to OM (Table 1); and (iii) public domain information on gene function consistent with OM pathogenesis (Supplementary Table 4). Table 1 lists eQTLs, noting that these are not always associated with expression of the gene nearest the top GWAS SNV. Of these 9 loci, we noted OM-relevant eQTL cell/tissue expression data for: (i) PLCL2/PLCL2-AS1 in whole blood, monocytes, neutrophils at locus 1; (ii) FAIM in lymphoid cells, CEP70 in monocytes and naive B cells, at locus 2; (iii) cis-eQTL for DMXL1/DTND2 at locus 4, but no relevant cell/tissue specificity; (iv) SNX2 in platelets, PPIC in lymphoid cells, at locus 5; (v) MARCH3 in blood, C5orf63 in blood/T cells, cis-eQTL for FBN2 but no cell/tissue specificity, at locus 8; (vi) NR3C1 in whole blood at locus 9; (vii) ATP10B in brain at locus 12; (viii) cis-eQTL for multiple genes at locus 14, including GLO1 in monocytes but no eQTL for BTBD9, at locus 14; and (ix) strong eQTL in immune cells for many genes, including MZF1 (myeloid zinc finger protein), SLC27A5 and TRIM28, at locus 17. Figure 4 and Supplementary Figures 4 to 11 provide detailed graphical outputs for SNP2GENE mapping. Figure 2B and Supplementary Figure 12 show parallel LocusZoom plots of GWAS mapping. Although we cannot discount roles for PLCL2/PLCL2-AS1, FAIM, DMXL1, DTND2, SNX2, PPIC, MARCH3, FBN2, ATP10B, GLO1, MZF1, SCL27A5, and TRIM28 in OM, filtering on gene function related to pathogenesis of OM disease (Supplementary Table 4) identified NR3C1 and CEP70 as the most plausible genetic risk factors. NR3C1 encodes the glucocorticoid receptor with a clear peak of association (GWAS P = 3.62 × 10−6, Figure 2B) and all eQTLs (Figure 4) chromatin-mapped to regions of strong transcriptional activity in lymphoid and myeloid cells and regulating expression (best Regulome score 3b, indicating transcription factor binding and matched transcription factor motif) only of this gene. CEP70 shares eQTL with FAIM (Table 1; Supplementary Figure 5) but the lead GWAS SNV (GWAS P = 5.38 × 10−6) lies within CEP70 (Supplementary Figures 5 and 12B) which is important in ciliogenesis (discussed below).
Figure 4.
Results of positional, chromatin interaction, and eQTL activity mapping in FUMA for NR3C1. (A) Maps the top lead SNV, and SNVs in LD with it according to the r2 color-coded key, across genomic locus 9. There were no additional independent significant SNV. (B) RegulomeDB scores (Y-axis) for eQTL mapping across the locus (X-axis). (C) Chromatin-15 states color coded for transcriptional/enhancer activity as shown in the key. Y-axis color coding relates to cell/tissue types in which chromatin interaction was mapped. (D) eQTL activity for NR3C1 (Y-axis) in different cells/tissues from public domain databases as shown in the key. Full explanation of keys provided as preamble to supplementary figures. Abbreviations: eQTL, expression quantitative trait loci; FUMA, FUnctional Mapping and Association; LD, linkage disequilibrium; SNV, single nucleotide variants.
Exome Variants Associated with Severe CSOM
A total of 59 987 medium to high impact variants in 72 exomes were identified. Of these, 11 816 had CADD-scaled scores ≥ 15, subsets of which were used as follows. First, we filtered for 1838 common variants (ExAC_all ≥ 0.1) and carried out extreme phenotype analysis comparing 15 severe with 9 mild OM children. Associations at P ≤ .05 were observed for 63 variants in 61 genes (Supplementary Table 5), which were taken forward for gene set enrichment analysis (Table 2). Comparison with MGI Mammalian Phenotype Level 4, GO Biological Process 2018, Jensen Compartments and Jensen Tissues tables showed significant enrichment for genes involved in ear function (GRXCR1, CDH23, LRP2, FAT4, ARSA, EYA4, SLC52A3, CTBP2), notably with stereocilium and stereociliary bundles. Second, we filtered for rare variants (ExAC_all ≤ 0.01) only ever observed in severe OM (N = 15) and never in mild OM (N = 9) or in 48 OM phenotype unknown exomes. This filter identified 1094 variants in 1011 genes with medium (N = 1035 missense; N = 7 splice-region) to high (N = 37 stop-gain; N = 9 splice-donor; N = 4 splice-acceptor; N = 1 stop-lost; N = 1 start-lost) impact (Supplementary Table 6). The 15 severe OM exomes each carried unique variants at mean ± SD 80 ± 37 of the 1011 genes. Analysis of these 1011 genes in Enrichr showed significant (Table 3) enrichment for genes expressed in NK cells, adult lung cells, and monocytes, and for genes associated with MGI Mammalian Phenotype Level 4 “abnormal ear physiology,” GO Molecular Function 2018 ATPase activity and actin binding, NCI Nature 2016 β1- and α6β4-integrin ligand interactions, and Syndacan-1-mediated signaling, CheA 2016 tables including SMAD4 (TGFβ signaling), HNF4A (hypoxia), and CEBPD (inflammation) ChipSeq data, and with Jensen Compartments including cell projections, lamellipodium, actin-based cell projection, ciliary membrane, α7β1-integrin, cilium, and stereocilium. Supplementary Table 7 provides a complete listing of each gene-set, including genes involved in the ciliary membrane, stereociliary bundles, actin-based cell projections, and the basal body (Figure 5). The STRING diagram in Supplementary Figure 13 shows extensive interactions between the 44 genes with putative deleterious variants that overlap significantly (P = 1.16 × 10−4; Padjusted = 0.011) with the 482 genes associated with “cilium” in the Jenson Compartment database.
Table 2.
Results of Enrichr Analysis for Gene Sets Identified from 63 Putative Pathogenic Variants (CADD-scaled ≥ 15; ExAC_all ≥ 0.1) from 61 Genes Associated with Extreme OM Phenotype
| Table and Term | Overlap | P-value | Odds Ratio | Combined Score | Genes |
|---|---|---|---|---|---|
| MGI Mammalian Phenotype Level 4 | |||||
| MP:0004515 abnormal vestibular hair cell stereociliary bundle morphology | 2 of 10 | 4.05E-04 | 65.57 | 512.18 | GRXCR1; CDH23 |
| MP:0003878 abnormal ear physiology | 2 of 11 | 4.94E-04 | 59.61 | 453.78 | CDH23; LRP2 |
| MP:0002856 abnormal vestibular ganglion morphology | 2 of 12 | 5.92E-04 | 54.64 | 406.11 | ARSA; GRXCR1 |
| MP:0004522 abnormal orientation of cochlear hair cell stereociliary bundles | 2 of 14 | 8.13E-04 | 46.84 | 333.23 | CDH23; FAT4 |
| MP:0004363 stria vascularis degeneration | 2 of 15 | 9.37E-04 | 43.72 | 304.85 | GRXCR1; CDH23 |
| MP:0002857 cochlear ganglion degeneration | 3 of 64 | 9.85E-04 | 15.37 | 106.39 | ARSA; GRXCR1; CDH23 |
| MP:0004742 abnormal vestibular system physiology | 2 of 20 | 0.002 | 32.79 | 209.51 | GRXCR1; CDH23 |
| MP:0004532 abnormal inner hair cell stereociliary bundle morphology | 2 of 22 | 0.002 | 29.81 | 184.76 | GRXCR1; CDH23 |
| MP:0004491 abnormal orientation of outer hair cell stereociliary bundles | 2 of 23 | 0.002 | 28.51 | 174.19 | CDH23; FAT4 |
| MP:0004748 increased susceptibility to age-related hearing loss | 2 of 25 | 0.003 | 26.23 | 155.89 | CDH23; LRP2 |
| MP:0004521 abnormal cochlear hair cell stereociliary bundle morphology | 2 of 28 | 0.003 | 23.42 | 133.91 | GRXCR1; CDH23 |
| MP:0011967 increased or absent threshold for auditory brainstem response | 5 of 354 | 0.004 | 4.63 | 25.06 | ARSA; GRXCR1; EYA4; CDH23; LRP2 |
| MP:0004527 abnormal outer hair cell stereociliary bundle morphology | 2 of 34 | 0.005 | 19.29 | 102.89 | GRXCR1; CDH23 |
| MP:0004736 abnormal distortion product otoacoustic emission | 2 of 41 | 0.007 | 15.99 | 79.47 | EYA4; CDH23 |
| MP:0001967 deafness | 3 of 129 | 0.007 | 7.62 | 37.63 | ARSA; GRXCR1; CDH23 |
| MP:0004738 abnormal auditory brainstem response | 4 of 44 | 0.008 | 14.90 | 72.01 | EYA4; CDH23 |
| MP:0006325 impaired hearing | 3 of 140 | 0.009 | 7.03 | 33.11 | GRXCR1; CDH23; LRP2 |
| MP:0004362 cochlear hair cell degeneration | 2 of 50 | 0.010 | 13.11 | 60.13 | GRXCR1; CDH23 |
| MP:0000031 abnormal cochlea morphology | 2 of 59 | 0.014 | 11.11 | 47.44 | CDH23; FAT4 |
| Go Biological Process 2018 | |||||
| Sensory perception of sound (GO:0007605) | 4 of 81 | 1.09E-04 | 16.19 | 147.68 | GRXCR1; SLC52A3; CDH23; LRP2 |
| Jensen Compartments | |||||
| Stereocilium | 2 of 37 | 0.006 | 17.72 | 91.61 | GRXCR1; CDH23 |
| Stereocilium bundle | 2 of 44 | 0.008 | 14.90 | 72.01 | GRXCR1; CDH23 |
| Cluster of actin-based cell projections | 3 of 154 | 0.008 | 7.34 | 35.47 | GRXCR1; CDH23; LRP2 |
| Jensen Tissues | |||||
| Hair cell | 3 of 91 | 0.003 | 10.81 | 63.89 | GRXCR1; CTBP2; CDH23 |
| Vestibular hair cell | 2 of 27 | 0.003 | 24.29 | 140.62 | GRXCR1; CDH23 |
| Inner hair cell | 2 of 37 | 0.006 | 17.72 | 91.61 | CTBP2; CDH23 |
Table 3.
Results of Enrichr Analysis for Gene Sets Identified from 1094 Putative Pathogenic Variants (CADD-scaled ≥ 15) ExAC_all ≤ 0.01) from 1011 Genes Found Only With Extreme OM Phenotype
| Table and Term | Overlapa | P-value | Adjusted P-value | Odds Ratio | Combined Score |
|---|---|---|---|---|---|
| MGI Mammalian Phenotype Level 4 | |||||
| MP:0003878 abnormal ear physiology | 6 of 11 | 6.10E-06 | .016 | 10.79 | 129.56 |
| GO Molecular Function 2018 | |||||
| ATPase activity (GO:0016887) | 29 of 203 | 4.22E-07 | 4.86E-04 | 2.83 | 41.48 |
| Actin binding (GO:0003779) | 28 of 254 | 9.27E-05 | .053 | 2.18 | 20.25 |
| Jenson Compartments | |||||
| Cell projection | 150 of 1774 | 1.52E-10 | 3.46E-07 | 1.67 | 37.82 |
| Lamellipodium | 26 of 168 | 3.44E-07 | 2.62E-04 | 3.06 | 45.57 |
| Actin-based cell projection | 25 of 172 | 1.89E-06 | 8.62E-04 | 2.88 | 37.90 |
| Cell projection membrane | 34 0f 284 | 2.88E-06 | 8.22E-04 | 2.37 | 30.22 |
| Cell projection part | 78 of 913 | 3.78E-06 | 9.59E-04 | 1.69 | 21.10 |
| Ciliary membrane | 15 of 79 | 8.35E-06 | .002 | 3.76 | 43.92 |
| Basement membrane | 15 od 91 | 4.82E-05 | .006 | 3.26 | 32.41 |
| Integrin alpha7-beta1 complex | 9 of 38 | 8.98E-05 | .009 | 4.69 | 43.66 |
| Cilium | 44 of 482 | 1.16E-04 | .011 | 1.81 | 16.37 |
| Filopodium | 14 of 90 | 1.62E-04 | .012 | 3.08 | 26.86 |
| Cluster of actin-based cell projections | 17 of 134 | 4.29E-04 | .026 | 2.51 | 19.46 |
| Stereocilium | 8 of 37 | 4.32E-04 | .025 | 4.28 | 33.14 |
| Ciliary part | 31 of 332 | 7.76E-04 | .039 | 1.85 | 13.23 |
| Microtubule associated complex | 17 of 142 | 8.39E-04 | .040 | 2.37 | 16.78 |
| NCI Nature 2016 | |||||
| Beta1 integrin cell surface interactions Homo sapiens | 12 of 66 | 1.02E-04 | .021 | 3.60 | 33.04 |
| Syndecan-1-mediated signaling events Homo sapiens | 9 of 46 | 4.22E-04 | .044 | 3.87 | 30.07 |
| Alpha6 beta4 integrin-ligand interactions Homo sapiens | 4 of 11 | 0.002 | .067 | 7.19 | 46.27 |
| Tissue Protein Expression From Human Proteome | |||||
| nk cells | 30 of 301 | 3.18E-04 | .010 | 1.97 | 15.88 |
| adult lung | 29 of 301 | 6.89E-04 | .010 | 1.91 | 13.87 |
| monocytes | 26 of 301 | 0.005619 | .056 | 1.71 | 8.85 |
| CheA 2016 | |||||
| SMAD4 21799915 ChIP-Seq A2780 Human | 175 of 2464 | 1.38E-06 | 8.92E-04 | 1.40 | 18.96 |
| FOXA2 19822575 ChIP-Seq HepG2 Human | 203 of 2968 | 2.27E-06 | 7.34E-04 | 1.35 | 17.58 |
| SMARCD1 25818293 ChIP-Seq ESCs Mouse | 150 of 2119 | 1.08E-05 | .002 | 1.40 | 16.01 |
| DROSHA 22980978 ChIP-Seq HELA Human | 45 of 456 | 1.48E-05 | .002 | 1.95 | 21.71 |
| HNF4A 19822575 ChIP-Seq HepG2 Human | 364 of 6083 | 5.31E-05 | .007 | 1.18 | 11.65 |
| CEBPD 23245923 ChIP-Seq MEFs Mouse | 45 of 504 | 1.61E-04 | .017 | 1.77 | 15.42 |
| CTNNB1 20460455 ChIP-Seq HCT116 Human | 76 of 988 | 1.69E-04 | .016 | 1.52 | 13.21 |
| SOX9 24532713 ChIP-Seq HFSC Mouse | 99 of 1384 | 2.70E-04 | .022 | 1.42 | 11.63 |
| AR 19668381 ChIP-Seq PC3 Human | 219 of 3519 | 3.87E-04 | .028 | 1.23 | 9.67 |
| TCF4 23295773 ChIP-Seq U87 Human | 235 of 3812 | 3.88E-04 | .025 | 1.22 | 9.58 |
| RUNX2 22187159 ChIP-Seq PCA Human | 213 of 3423 | 4.79E-04 | .028 | 1.23 | 9.41 |
a Full details of the genes contributing to each gene set are provided in Supplementary Table 7.
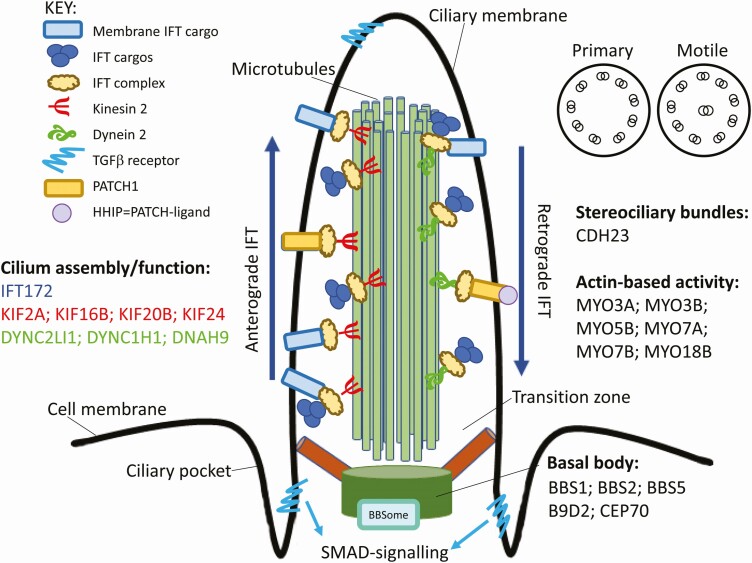
Figure 5.
Model of cilium assembly and function highlighting some of the genes that carry putative deleterious variants (CADD-scaled score ≥15) associated with severe OM and/or where such variants were only ever observed in individuals with severe OM. Details of the full lists of genes carrying such variants are provided in Table 2 and in Supplementary Table 7. Abbreviation: OM, otitis media.
Discussion
Here we examine association between severe OM and common and rare variants. Noncoding regulatory variants were evaluated using post-GWAS annotation in FUMA. Putative deleterious variants influencing protein coding were identified from exome data using a CADD-scaled score cut-off ≥15. CADD scores correlate with pathogenicity [18] but there is no hard cut-off to identify pathogenic variants. CADD authors suggest a cut-off at 15, the median value for all possible canonical splice site changes and nonsynonymous variants [18]. We use CADD-scaled ≥ 15 as an indicator, not a definitive measure of variant pathogenicity. We hypothesize that carriage of multiple such variants influencing genes in functional pathways relevant to OM could contribute to genetic risk.
Whilst we acknowledge the limitations of our small cohort and that the use of probands only in the exome analysis precluded assignment of likely pathogenic variants as de novo versus inherited, 2 interesting themes emerge where functional links are plausible: (i) GWAS associations with NR3C1 and NREP that suggest gene-by-environment interactions; and (ii) exome-based associations with gene-sets enriched for mammalian phenotypes/processes including abnormal ear, stereociliary bundles, cilium assembly, integrin interactions, and syndecan-1 signaling. We focus our discussion on these themes.
Glucocorticoid hormones bind to NR3C1 and regulate gene expression through transrepression of proinflammatory or transactivation of anti-inflammatory pathways [19]. The lead NR3C1 SNV and numerous eQTLs in LD mapped to regions of strong transcriptional activity in lymphoid and myeloid cells. NR3C1 could thus act as a risk factor for severe OM through perturbation of endogenous regulation of inflammation, as seen in childhood asthma [20]. Environmental stresses also act through NR3C1-mediated endogenous corticosteroid responses. In Africa, significant correlations were observed between newborn birth weight/NR3C1 promoter methylation and culturally relevant measures of maternal prenatal stress [21]. Adverse childhood experiences also lead to novel methylation of NR3C1 [22]. Genetic variants modifying epigenetic signatures at NR3C1 could contribute to effects of maternal perinatal and early childhood stress on OM, an interesting hypothesis given the early onset of severe OM in Aboriginal Australians.
Association between severe OM and NREP could also relate to gene-by-environment interactions. NREP (= P311) binds eukaryotic translation initiation factor 3 to promote translation of isoforms 1–3 of transforming growth factor β (TGFβ1–3) [23]. NREP promotes lung [24] and renal [25] fibrosis through stimulation of TGFβ1–3 and SMADs (homologues of “mothers against decapentaplegic, drosophila”). Murine [26] and human [27] studies demonstrate the importance of TGFβ pathways in OM. Also, distinct community types of esophageal microbiomes defined by respiratory bacteria Streptococcus and Prevotella are influenced by SNVs at NREP [16]. Timing of nasopharyngeal colonization with respiratory bacteria (Moraxella catarrhalis, Haemophilus influenzae, Streptococcus pneumoniae) predicts the onset of persistent OM in Aboriginal infants [28]. Nasopharyngeal microbial composition differs between OM-prone and nonprone children [17, 29]. Future studies could examine association between NREP and microbiomes of CSOM-prone Aboriginal children, complementing knowledge of rare variants at A2ML1 and SPINK5 and middle ear microbiomes in Filipino OM patients [30, 31].
Common regulatory variants at CEP70 were associated with severe OM. CEP70 is a centrosomal/basal body protein that regulates ciliogenesis during zebrafish embryogenesis [32], gene depletion causing dysfunctional shortened cilia that affect ear development. Exomes from children with severe vs mild OM highlighted common (EXaC_all > 0.1) variants influencing protein coding in genes (GRXCR1, CDH23, ARSA, FAT4, CTBP2) previously associated with abnormal hair cell stereociliary bundles. Similarly, analysis of rare variants influencing protein coding only seen in severe OM showed significant enrichment for genes in pathways/processes involved in “abnormal ear” function (LMNA, CDH23, LRP2, MYO7A, FGFR1), abnormal stereociliary bundles, and cilium assembly (Figure 5). This included genes that affect assembly/function of primary and motile cilia. In addition to developmental anomalies, human primary cilia dyskinesis is associated with persisting middle ear secretion retention, suppurative infection, and chronic OM [33]. The ciliated mucosa close to the Eustachian tube contains goblet cells which secrete mucins that prevent pathogen adherence (reviewed [34]). Thus, in addition to any contribution to hearing loss (reviewed [35]) that variants in cilium assembly might have, it is likely that they contribute to increased access of bacterial pathogens to the middle ear.
Analysis of rare variants influencing protein coding also identified gene-sets previously associated with Syndecan-1 signaling (COL16A1, COL4A4, COL6A2, COL12A1, COL4A3, CASK, COL6A3, HPSE, MET) and β1- and/or α6β4-integrin cell surface interactions (FGB, LAMA2, LAMA1, COL4A4, COL4A3, LAMA3, F13A1, LAMB1, THBS2). Syndecan-1 is a cell surface proteoglycan that mediates microbial attachment/entry and elicits inflammatory responses [36]. Integrins play a role during tympanic membrane damage and repair [37], and act as cell surface receptors for TGFβ [38]. TGFβ signaling is regulated by clathrin-dependent endocytosis at the base and proximal part of cilia [39]. TGFβ receptors localize to the ciliary tip and endocytic vesicles at the ciliary base where TGFβ stimulation increases SMAD activation. Stunted primary cilia show reduced TGFβ signaling [39]. These observations provide interesting hypotheses for ways in which variants influencing TGFβ pathways and cilium assembly/function may interact to influence severe OM.
Overall, our study highlights interacting genes and pathways that may contribute to extreme susceptibility to OM in Aboriginal Australian children. While common regulatory variants identified through the GWAS appeared biased towards immune-related mechanisms, analysis of common and rare protein-coding variants were mechanistically related to cilium assembly and function.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Glossary
Nonstandard Abbreviations
- AHS
Aboriginal Health Service
- AOM
acute otitis media
- CADD
Combined Annotation Dependent Depletion
- CSOM
chronic suppurative otitis media
- eQTL
expression quantitative trait loci
- ExAC
Exome Aggregation Consortium
- FUMA
FUnctional Mapping and Annotation
- GWAS
genome-wide association studies
- LD
linkage disequilibrium
- NREP
neuronal regeneration-related protein
- OM
otitis media
- OME
OM with effusion
- QC
quality control
- SNV
single nucleotide variants
- TGFβ
transforming growth factor β
Notes
Acknowledgments. We gratefully acknowledge the tremendous contribution made by the Aboriginal community, the board, and the staff of the local Aboriginal Health Service (AHS) where our study was based, and the support of local schools in the area. Without this support the study would not have been possible. We also acknowledge the generous in-kind support provided by the AHS for travel and accommodation to allow the field collection of samples used in the study, and the generosity of the Board of the AHS in allowing access to clinical records through our Memorandum of Understanding and for granting us permission to publish our findings.
Financial support. The study was supported by the Australian National Health and Medical Research Council (APP634301) and the Perron Foundation. H. J. C.’s contribution to the study was supported by 2 Wellcome Trust Senior Fellowships (087436/Z/08/Z and 102858/Z/13/Z). M. F. was supported by a Raine Visiting Fellowship to the University of Western Australia. S. E. J. held a BrightSpark Foundation Research Fellowship.
Potential conflicts of interest. No authors reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Disclaimer. The study sponsor had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all study data and had final responsibility for the decision to submit for publication.
References
- 1. Morris PS, Leach AJ, Halpin S, et al. An overview of acute otitis media in Australian Aboriginal children living in remote communities. Vaccine 2007; 25:2389–93. [DOI] [PubMed] [Google Scholar]
- 2. Uhari M, Mäntysaari K, Niemelä M. A meta-analytic review of the risk factors for acute otitis media. Clin Infect Dis 1996; 22:1079–83. [DOI] [PubMed] [Google Scholar]
- 3. Geng R, Wang Q, Chen E, Zheng QY. Current understanding of host genetics of otitis media. Front Genet 2019; 10:1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rovers M, Haggard M, Gannon M, Koeppen-Schomerus G, Plomin R. Heritability of symptom domains in otitis media: a longitudinal study of 1373 twin pairs. Am J Epidemiol 2002; 155:958–64. [DOI] [PubMed] [Google Scholar]
- 5. Croteau-Chonka DC, Rogers AJ, Raj T, et al. Expression quantitative trait loci information improves predictive modeling of disease relevance of non-coding genetic variation. PLoS One 2015; 10:e0140758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Santos-Cortez RL, Chiong CM, Reyes-Quintos MR, et al. ; University of Washington Center for Mendelian Genomics . Rare A2ML1 variants confer susceptibility to otitis media. Nat Genet 2015; 47:917–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lek M, Karczewski KJ, Minikel EV, et al. ; Exome Aggregation Consortium . Analysis of protein-coding genetic variation in 60 706 humans. Nature 2016; 536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet 2012; 13:565–75. [DOI] [PubMed] [Google Scholar]
- 9. Tonkinson R. The Mardu Aborigines, living the dream in Australia’s desert. New York: Holt, Reinhart & Winston, 1991. [Google Scholar]
- 10. Anderson D, Cordell HJ, Fakiola M, et al. First genome-wide association study in an Australian aboriginal population provides insights into genetic risk factors for body mass index and type 2 diabetes. PLoS One 2015; 10:e0119333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Devlin B, Roeder K. Genomic control for association studies. Biometrics 1999; 55:997–1004. [DOI] [PubMed] [Google Scholar]
- 12. Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010; 26:2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017; 8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang D, Anderson D, Francis RW, et al. Reference genotype and exome data from an Australian Aboriginal population for health-based research. Sci Data 2016; 3:160023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016; 44:W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deshpande NP, Riordan SM, Castaño-Rodríguez N, Wilkins MR, Kaakoush NO. Signatures within the esophageal microbiome are associated with host genetics, age, and disease. Microbiome 2018; 6:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lappan R, Imbrogno K, Sikazwe C, et al. A microbiome case-control study of recurrent acute otitis media identified potentially protective bacterial genera. BMC Microbiol 2018; 18:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46:310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barnes PJ. Corticosteroid effects on cell signalling. Eur Respir J 2006; 27:413–26. [DOI] [PubMed] [Google Scholar]
- 20. Akurugu WA, Van Heerden CJ, Mulder N, Zollner EW. Hypothalamic-pituitary-adrenal axis suppression in asthma: A glucocorticoid receptor polymorphism may protect. Pediatr Allergy Immunol 2020. doi: 10.1111/pai.13379. [DOI] [PubMed] [Google Scholar]
- 21. Mulligan CJ, D’Errico NC, Stees J, Hughes DA. Methylation changes at NR3C1 in newborns associate with maternal prenatal stress exposure and newborn birth weight. Epigenetics 2012; 7:853–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lewis CR, Breitenstein RS, Henderson A, et al. Harsh parenting predicts novel HPA receptor gene methylation and NR3C1 methylation predicts cortisol daily slope in middle childhood. Cell Mol Neurobiol 2020. doi: 10.1007/s10571-020-00885-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yue MM, Lv K, Meredith SC, Martindale JL, Gorospe M, Schuger L. Novel RNA-binding protein P311 binds eukaryotic translation initiation factor 3 subunit b (eIF3b) to promote translation of transforming growth factor β1-3 (TGF-β1-3). J Biol Chem 2014; 289:33971–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duan FF, Barron G, Meliton A, Mutlu GM, Dulin NO, Schuger L. P311 promotes lung fibrosis via stimulation of transforming growth factor-β1, -β2, and -β3 translation. Am J Respir Cell Mol Biol 2019; 60:221–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yao Z, Yang S, He W, et al. P311 promotes renal fibrosis via TGFβ1/Smad signaling. Sci Rep 2015; 5:17032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tateossian H, Morse S, Parker A, et al. Otitis media in the Tgif knockout mouse implicates TGFβ signalling in chronic middle ear inflammatory disease. Hum Mol Genet 2013; 22:2553–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rye MS, Wiertsema SP, Scaman ES, et al. FBXO11, a regulator of the TGFβ pathway, is associated with severe otitis media in Western Australian children. Genes Immun 2011; 12:352–9. [DOI] [PubMed] [Google Scholar]
- 28. Leach AJ, Boswell JB, Asche V, Nienhuys TG, Mathews JD. Bacterial colonization of the nasopharynx predicts very early onset and persistence of otitis media in Australian aboriginal infants. Pediatr Infect Dis J 1994; 13:983–9. [DOI] [PubMed] [Google Scholar]
- 29. Walker RE, Walker CG, Camargo CA Jr, et al. Nasal microbial composition and chronic otitis media with effusion: A case-control study. PLoS One 2019; 14:e0212473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frank DN, Giese APJ, Hafren L, et al. Otitis media susceptibility and shifts in the head and neck microbiome due to SPINK5 variants. J Med Genet 2020. doi: 10.1136/jmedgenet-2020-106844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Santos-Cortez RL, Hutchinson DS, Ajami NJ, et al. Middle ear microbiome differences in indigenous Filipinos with chronic otitis media due to a duplication in the A2ML1 gene. Infect Dis Poverty 2016; 5:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilkinson CJ, Carl M, Harris WA. Cep70 and Cep131 contribute to ciliogenesis in zebrafish embryos. BMC Cell Biol 2009; 10:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morgan LC, Birman CS. The impact of primary ciliary dyskinesia on the upper respiratory tract. Paediatr Respir Rev 2016; 18:33–8. [DOI] [PubMed] [Google Scholar]
- 34. Fuchs JC, Tucker AS. Development and integration of the ear. Curr Top Dev Biol 2015; 115:213–32. [DOI] [PubMed] [Google Scholar]
- 35. Schwander M, Kachar B, Müller U. Review series: The cell biology of hearing. J Cell Biol 2010; 190:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Teng YH, Aquino RS, Park PW. Molecular functions of syndecan-1 in disease. Matrix Biol 2012; 31:3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim SW, Kim J, Seonwoo H, et al. Latent progenitor cells as potential regulators for tympanic membrane regeneration. Sci Rep 2015; 5:11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nishimura SL. Integrin-mediated transforming growth factor-beta activation, a potential therapeutic target in fibrogenic disorders. Am J Pathol 2009; 175:1362–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clement CA, Ajbro KD, Koefoed K, et al. TGF-β signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Rep 2013; 3:1806–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.