Abstract
Almost all small eukaryotic RNAs are processed from transiently stabilized 3′-extended forms. A key question is how and why such intermediates are stabilized and how they can then be processed to the mature RNA. Here we report that yeast U3 is also processed from a 3′-extended precursor. The major 3′-extended forms of U3 (U3-3′I and -II) lack the cap trimethylation present in mature U3 and are not associated with small nucleolar RNP (snoRNP) proteins that bind mature U3, i.e., Nop1p, Nop56p, and Nop58p. Depletion of Nop58p leads to the loss of mature U3 but increases the level of U3-3′I and -II, indicating a requirement for the snoRNP proteins for final maturation. Pre-U3 is cleaved by the endonuclease Rnt1p, but U3-3′I and -II do not extend to the Rnt1p cleavage sites. Rather, they terminate at poly(U) tracts, suggesting that they might be bound by Lhp1p (the yeast homologue of La). Immunoprecipitation of Lhp1p fused to Staphylococcus aureus protein A resulted in coprecipitation of both U3-3′I and -II. Deletion of LHP1, which is nonessential, led to the loss of U3-3′I and -II. We conclude that pre-U3 is cleaved by Rnt1p, followed by exonuclease digestion to U3-3′I and -II. These species are stabilized against continued degradation by binding of Lhp1p. Displacement of Lhp1p by binding of the snoRNP proteins allows final maturation, which involves the exosome complex of 3′→5′ exonucleases.
Eukaryotic cells contain a large number of stable RNA species, nearly all of which are synthesized by posttranscriptional processing from larger precursors. This has long been known for the highly abundant cytoplasmic RNAs, tRNAs, and rRNAs, but more recently it has become clear that is also the case for the small nuclear RNAs (snRNAs), which participate in pre-mRNA splicing, and the small nucleolar RNAs (snoRNAs), which participate in rRNA processing and modification. It is a long-standing mystery why cells use such a strategy, rather than simply terminating transcription at the end of the mature RNA sequence. We will offer a potential explanation for this, at least in the case of the U3 snoRNA.
Analyses of the 3′ end processing of the 5.8S rRNA in Saccharomyces cerevisiae led to the identification of the exosome complex, composed of 11 different 3′→5′ exonucleases (6, 36, 37; E. Petfalski and D. Tollervey, unpublished data). Subsequent work showed that the exosome participates in the 3′ processing of other RNA substrates, including many snRNAs and snoRNAs (5, 55), and also participates in mRNA turnover (9). A homologous complex, designated the PM-Scl complex, is present in human cells and is a target for autoimmune antibodies (6).
In addition to the exosome, normal 3′ processing of the U1, U2, U4, and U5 snRNAs involves cleavage by the endonuclease Rnt1p (1, 5, 14, 45), the yeast homologue of Escherichia coli RNase III (2). Rnt1p cleaves on both sides of extended stem-loop structures with closing AGNN tetraloops (15), and these cleavages are likely to act as entry sites for the exosome complex, with the final trimming performed by the Rex exonucleases and/or the exosome component Rrp6p (5, 54). Rnt1p also acts to separate the individual pre-snoRNAs from polycistronic precursors (15, 16) and processes the 3′ external transcribed spacer of the yeast pre-rRNA (2, 28).
Another 3′ processing factor, the La phosphoprotein, was identified as the target of human autoimmune antibodies and was shown to bind to the poly(U) tracts located at the 3′ ends of all RNA polymerase III transcripts (42, 48). La also binds to 3′ extended precursors to human U1 and the yeast snRNAs (34, 58) and to internal sequences of several viral RNAs, in some cases at sequences that lack poly(U) tracts (4, 23). The yeast homologue of La, Lhp1p (La-homologous protein), is nonessential for viability but is required for normal 3′ processing of tRNAs (56, 59). In the presence of Lhp1p, processing is endonucleolytic, whereas in the absence of Lhp1p this cleavage is inhibited and an alternative, exonucleolytic pathway takes over tRNA 3′ maturation (59). Lhp1p also associates with the newly transcribed U6 snRNA, which is transcribed by RNA polymerase III (39).
Here we show how these factors collaborate in the 3′ processing of the U3 snoRNA.
MATERIALS AND METHODS
Strains.
Growth and handling of S. cerevisiae were by standard techniques. The transformation procedure was as described elsewhere (21). Yeast strains used and constructed in this study are listed in Table 1. Wild-type RNT1 and rnt1-Δ sister strains (15) were used to prepare whole-cell extract. Strain rat1-1 was kindly provided by C. Cole (7). The nonessential gene LHP1 was disrupted and tagged with Staphylococcus aureus protein A (“ProtA” in construct designations) at the carboxy-terminal end of Lhp1p by a PCR strategy (29) in the haploid strain YDL401, using the Kluyveromyces lactis URA3 marker.
TABLE 1.
Yeast strains used in this work
| Strain | Genotype | Reference or source |
|---|---|---|
| YCA12 | MATa ade2-1 his3-Δ200 leu2-3,112 trp1-1 ura3-1 can1-100 RRP6::Kl TRP1 | 6 |
| YDL401 | MATa his3Δ200 leu2Δ1 trp1 ura3-52 gal2 galΔ108 | 29 |
| D150 | MATa ura3-52 leu2-3,112 ade1-100 his4-519 | L. Guarente, personal communication |
| P118 | As YDL401 but GAL10::ProtA-RRP41 | 29 |
| YCA20 | As YDL401 but GAL10::RRP45 | 6 |
| YJK10 | As YDL401 but RNT1::TRP1 | 5 |
| YJK11 | As P118 but RNT1::TRP1 | 5 |
| YJK12 | As YCA20 but RNT1::TRP1 | 5 |
| YJK13 | As YCA12 but RNT1::TRP1 | 5 |
| rat1-1 | MATα his3-Δ200 leu2-Δ1 ura3-52 rat1-1 | 6 |
| RP582 | MATa leu2-3,112 ura3-52 rpb1-1 | 18 |
| ProtA-Nop1 | MATα ade leu trp lys ura3 nop1::URA3 pUN100-ProtA-NOP1 | 26 |
| ProtA-Nop58 | MATa ade2 ade3 leu2 ura3 can1 nop58::HIS3 pRS315-ProtA-NOP58 | 20 |
| ProtA-Nop56 | MATa ade2 ade3 leu2 ura3 nop56::HIS3 pRS315-ProtA-NOP56 | 20 |
| GAL::nop58 | As YDL401 but GAL10::NOP58 | 30 |
| GAL::nop56 | As YDL401 but GAL10::NOP56 | 31 |
| Lhp1p-ProtA | As YDL401 but LHP1::ProtA | This work |
| YCA 35 | MATa his3Δ200 leu2Δ1 trp1 ura3-52 gal2 galΔ108 LHP1::Kl URA | This work |
The oligonucleotides used to construct and test the gene disruption and protein A tagging were 838 (5′ LHP1::URA), 5′-TCTATTTGGTTCTACTGGAACTAAAGTAGCATCTGCAAAGAAGTAGAGAAGTTTGAGAGGGC; 839 (3′ LHP1::URA), 5′-ATATGCTATGATAATGAGATACGAGAACCAGAAGAAACACAAGAACTGGGTAGAAGATCGGTC; 840 (5′ LHP1 test), 5′-ACAGAGTCGCATCTCATCGC; 841 (3′ Kl URA), 5′-GGTAGAAGATCGGTC; 842 (5′ LHP1::ProtA); 5′-GAGGACTCTTCTGCCATTGCCGATGACGATGAGGAGCACAAGGAGGGCGTGGACAACAAATTC; and 843 (3′ LHP1::ProtA), 5′-TCCATTTTAACCAGTAACGGTAATTTTTAATACTAATAAAAAAAGCTGGGTAGAAGATCGGTC.
RNA extraction, Northern hybridization, and primer extension.
For depletion of Rrp41p and Rrp45p, cells were harvested at intervals following the shift from RSG medium (2% galactose, 2% sucrose, 2% raffinose) to medium containing 2% glucose. Otherwise strains were grown in YPD medium. RNA was extracted as described previously (52). Northern hybridization and primer extension were as described previously (12, 51). Standard 6 or 8% acrylamide gels were used to analyze low-molecular-weight RNA species and primer extension reactions. For RNA hybridization and primer extension, the following oligonucleotides were used: 200 (U3), 5′-UUAUGGGACUUGUU; 203 (5′U3), 5′-CUAUAGAAAUGAUCCU; 218 (snR10), 5′-CUIUUAAAUUUICIUU; 230 (anti-U3sub6), 5′-GATTCCTATAGAAACACAG; 250 (scR1), 5′-ATCCCGGCCGCCTCCATCAC; 251 (3′Ex-U3), 5′-GTGGTTAACTTGTCA; 252 (U3ADS), 5′-TTTGTTTTCGCATCCGTCGCTC; 253 (U3DS), 5′-GGAGTCATACTATCAAGAAC; 254 (3′U3), 5′-CCAACTTGTCAGACTGCCATT; 260 (U3 intron), 5′-CAAAAGCTGCTGCAATGG; 261 (U6), 5′-AAAACGAAATAAATTCTTTGTAAAAC; and 310 (tRNATyrGΨA-intron), 5′-AAGATTTCGTAGTGATAA.
Oligonucleotides 200, 203, and 218 are largely composed of 2′-O-methyl RNA.
Expression of the U3 cDNA.
The synthesis of U3A from cDNA constructs was analyzed by expression of the ARS-CEN pU3-wt plasmid carrying an ADE2 marker (11). This U3 intronless construct is under the control of the natural promoter and terminator regions. Expression was analyzed in the GAL::snr17A strain JH84 (24; J. Hughes, personal communication), from which the endogenous U3A was depleted by growth on glucose medium. Alternatively, the pU3 sub6-CBS1 plasmid, which carries the viable mutations U3sub6 and CBS1 (11, 47), was expressed in wild-type yeast strains. U3 synthesized from the cDNA construct was detected by hybridization with a probe specific for the sub6 mutation (47).
In vitro processing reactions.
Synthetic U3-3′ RNAs were obtained by in vitro transcription as described elsewhere (16), using a PCR product as template. The PCR product was generated from genomic DNA using a forward primer carrying a T7 promoter (T7U3DS; 5′-GCGAATTCTAATACGACTCACTATAGGTACTTCTTTTTTGAAGGGAT) and reverse primers 252 (U3ADS) for a longer U3(−60/+177) transcript or 253 (U3DS) for a shorter U3(−60/+139) transcript. Whole-cell extracts were prepared from wild-type and rnt1-Δ sister strains as described previously (16), and recombinant His6-Rnt1p was purified as described previously (5, 16).
In vitro processing of U3-3′ RNA in cell extracts or with recombinant His6-Rnt1p and mapping of the cleavage sites using primer extension were performed as described elsewhere (16). Prior to the reaction, gel-purified RNA substrates (2 nM) were denatured for 2 min at 85°C in Rnt1p buffer (50 mM Tris-HCl [pH 7.6], 200 mM KCl, 0.1 mg of wheat-germ tRNA/ml, 5 mM MgCl2) and cooled to 23°C. The cleavage reaction was performed at 23°C using 100 fmol of recombinant His6-Rnt1 or by incubation in the whole-cell extracts.
RNase A/T1 mapping.
RNase A/T1 mapping was performed as described elsewhere (22). The 32P-labeled antisense probe was transcribed in vitro with T7 polymerase using a PCR template as described above. The PCR product was generated from genomic DNA using forward primer T7antiU3, carrying a T7 promoter (5′-GCGAATTCTAATACGACTCACTATAGGTTTTAAACAATTTAGAAAAGG), and reverse primer 3′antiU3 (5′-GGGCTCTATGGGTGGGTAC). The RNA transcript was gel purified and hybridized to 8 μg of total RNA in 30 μl of piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer (40 mM PIPES) [pH 6.7], 400 mM NaCl, 1 mM EDTA) and 50% formamide. Annealing was performed by heating at 95°C for 2 min followed by incubation at 48°C for several hours. Digestion in RNase buffer (10 mM Tris-HCl [pH 7.5], 300 mM NaCl, 1 mM EDTA) was performed with 5 to 15 U of RNase T2, 0.4 to 2.5 units of RNase T1, and 0.1 to 0.5 μg of RNase A (RNase T2 from GibcoBRL; RNases T1 and A from Boehringer) for 30 min at 25°C. Protected products were recovered by guanidium thiocyanate–phenol-chloroform extraction and separated on an 10% polyacrylamide gel.
RNase H treatment.
Deadenylation was performed essentially as described elsewhere (18). Samples of 30 μg of RNA were annealed with 750 ng of oligo(dT) at 65°C for 1 h and digested with 6 U of RNase H at 30°C for 1 h. The control samples were treated identically except that the oligo(dT) was omitted.
Immunoprecipitation.
For immunoprecipitation of ProtA-Nop1p, ProtA-Nop58p, ProtA-Nop56p, Lhp1p-ProtA, and m32,2,7G-capped RNAs, yeast whole-cell extracts were prepared as described elsewhere (46) except that for immunoprecipitation of m32,2,7G-capped RNAs, cells were resuspended in buffer A (150 mM potassium acetate [KAc; pH 7.5], 20 mM Tris-Ac, 5 mM MgAc) with 1 mM dithiothreitol, 0.5% Triton X-100, and 5 mM phenylmethylsulfonyl fluoride. Immunoprecipitation of ProtA-Nop1p, ProtA-Nop58p, ProtA-Nop56p, and Lhp1p-ProtA with rabbit immunoglobulin G (IgG) agarose beads (Sigma) was performed as previously described (33) at 150 mM salt (KAc) concentration. For immunoprecipitation with m32,2,7G-cap-specific serum (R1131; kindly provided by R. Lührmann), 30 μl of suspension of protein G-Sepharose was washed with phosphate-buffered saline buffer and incubated on a rotating wheel with extract equivalent to 4 units of optical density at 600 nm of cells in 120 μl of buffer A for 2 h at 4°C. After the pellet was washed in buffer A, bound m32,2,7G-capped RNAs were eluted with 10 mM m7G(5′)ppp(5′)G (Pharmacia) in 30 μl of buffer A. The RNAs were extracted with GTC/phenol-chloroform and ethanol precipitated.
RESULTS
Yeast cells contain 3′-extended forms of U3.
Yeast U3 is encoded by two genes, SNR17A, encoding U3A, and SNR17B, encoding U3B (25). U3A is approximately 10-fold more abundant than U3B (25), and all analyses have been performed for U3A. On Northern hybridization, probe 200, to mature U3A, was observed to hybridize to two RNA species of slower gel mobility (U3-3′I and U3-3′II) in total yeast RNA preparations (Fig. 1A, lane 1) that were estimated to be approximately 10 and 20 nucleotides (nt), respectively, longer than the mature U3 (333 nt). A probe complementary to the sequence across the 3′ end of the mature U3A (probe 251), which hybridizes specifically to 3′-extended species, also detected these RNAs as well as a longer species (U3-int 3′) of approximately 470 nt. Both SNR17A and SNR17B contain introns that are excised by the pre-mRNA splicing machinery (38). The size and hybridization pattern of U3-int 3′ indicates that it corresponds to a 3′-extended precursor that retains the intron (Fig. 1D and 6B). It is not clear whether U3-int 3′ has 3′ ends identical to those of U3-3′I and U3-3′II. Synthesis of the U3-3′I and U3-3′II RNAs was not affected by the presence or absence of the intron in the pre-snoRNA, since identical species were observed in strains expressing U3 cDNA constructs (see Materials and Methods) (data not shown).
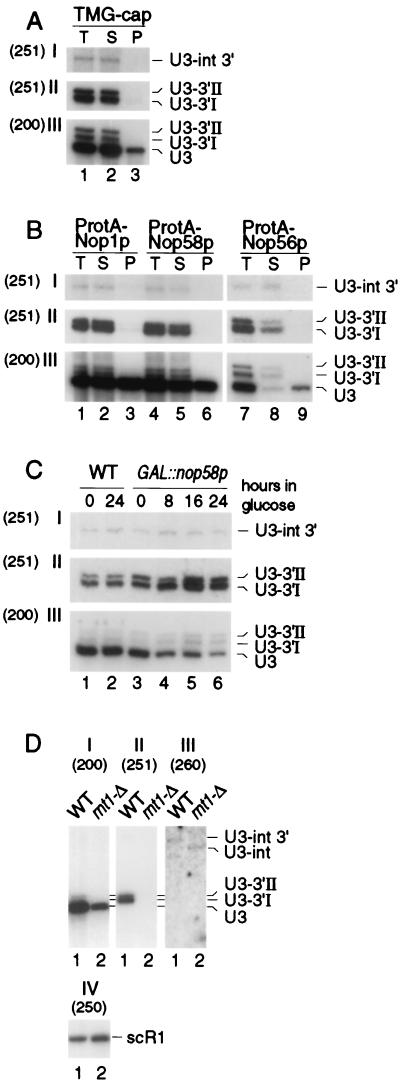
FIG. 1.
Northern analysis of 3′-extended forms of U3 snoRNA. Probes (indicated in parentheses): 251, complementary to the region across the 3′ end of the mature U3A; 200, complementary to mature U3; 260, complementary to the U3A intron; 250, complementary to the scR1 RNA. For panels A and B, input lysates were estimated to contain comparable amounts of U3 snoRNA, and equal fractions of the preparation were loaded for each lane; panels C and D, constant amounts of total RNA were loaded in each lane. (A) Immunoprecipitation with m32,2,7G cap-specific antibody (R1131) on lysates from the wild-type D150 strain. (B) Immunoprecipitation of lysates from strains expressing epitope-tagged fusion proteins ProtA-Nop1p, ProtA-Nop58p, and ProtA-Nop56p. (C) Stability of mature and 3′-extended U3 upon depletion of Nop58p. RNA was extracted from the GAL::nop58 and wild-type (WT) strains following transfer from permissive, galactose medium to repressive, glucose medium for the times indicated. (D) Effects of rnt1-Δ on 3′-extended U3. The level of scR1 RNA is shown as a control for loading. T, total cell lysate; S, immune supernatant; P, immunoprecipitate.
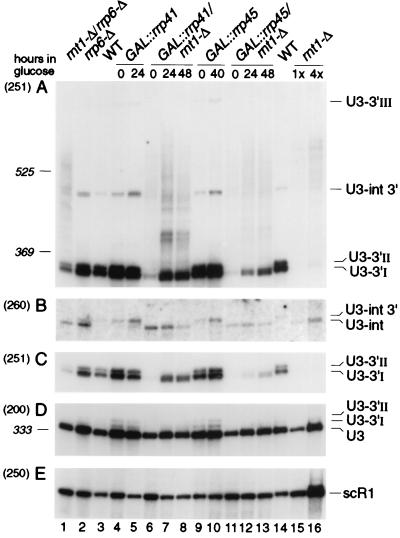
FIG. 6.
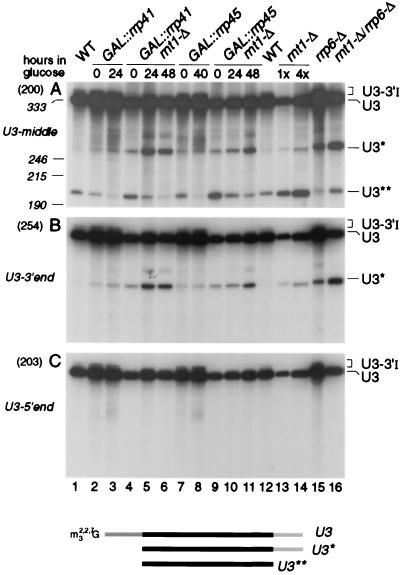
Northern analysis of processing of U3 snoRNA in exosome mutants. RNA was extracted from strains carrying GAL-regulated constructs following transfer from permissive, RSG medium to repressive, glucose medium at 30°C for the times indicated, or from the wild-type (WT), rnt1-Δ, rrp6-Δ, and rnt1-Δ/rrp6-Δ strains grown on glucose medium at 30°C. RNA was separated on an 6% polyacrylamide gel and hybridized with oligonucleotide probes. The panels show successive hybridization of the same filter. Probes are indicated in parentheses on the left and described in Materials and Methods; the positions of RNA species detected are indicated on the right. Panel C presents a weaker exposure of the same gel as panel A. Panels B to E present only relevant regions of the Northern blots. The amount of total RNA loaded in lane 16 is fourfold greater than in lane 15 and other lanes. The positions of migration of scR1 (525 nt) and P (369 nt) RNAs determined by hybridization of the same filter are indicated as size markers. Mature U3 is 333 nt.
The mature U3 carries a 5′ trimethyl guanosine (TMG) cap structure (25) and was precipitated with anti-TMG antibodies (Fig. 1A, lane 3) (generously provided by R. Lührmann, University of Marburg). In contrast, the U3-3′I, U3-3′II, and U3-int 3′ RNAs were not precipitated with anti-TMG and were recovered exclusively in the immune supernatant (Fig. 1A, lane 2). Mature yeast U3, like all box C+D snoRNAs, is associated with Nop1p, Nop56p, and Nop58p (30, 31, 44) and was coprecipitated with protein A-tagged fusion proteins (Fig. 1B, lanes 3, 6, and 9). No association of U3-3′I, U3-3′II, or U3-int 3′ with these proteins was observed, and the RNAs were again recovered exclusively in the immune supernatants (Fig. 1B, lanes 2, 5, and 8).
Genetic depletion of Nop58p leads to the loss of all tested box C+D snoRNAs including U3 (30). The GAL::nop58 strain was pregrown on permissive, galactose medium (0-h sample) and then transferred to glucose to repress synthesis of Nop58p (Fig. 1C). Mature U3 was codepleted with Nop58p, whereas the levels of the U3-3′I and U3-3′II RNAs were increased. The U3-int 3′ species was unaffected.
We conclude that the U3 snoRNA is synthesized from 3′ extended precursors that lack the TMG cap structure. The pre-U3 species are not associated with snoRNP proteins and, unlike the mature snoRNA, do not require Nop58p for stability. Indeed, the accumulation of U3-3′I and U3-3′II in strains depleted of Nop58p indicates that their normal maturation to U3 requires Nop58p binding.
3′ processing of U3 involves cleavage by Rnt1p.
Rnt1p cleaves 3′-extended precursors to the U1, U2, U4, and U5 snRNAs and processes polycistronic pre-snoRNAs. We therefore determined whether it is also involved in the 3′ processing of pre-U3 species. In strains carrying a complete deletion of the RNT1 gene, the level of mature U3 was reduced approximately threefold (Fig. 1D, I; see also Table 2). Strains carrying rnt1-Δ lacked the U3-3′I and U3-3′II RNAs (Fig. 1D, II) and we observed a heterogeneous group of RNAs extending to approximately 600 nt (see Fig. 6A, lane 16, where more RNA is loaded). In addition, the intron-containing precursor was found to be 3′ processed in the rnt1-Δ strain, in contrast to the 3′-extended form seen in the wild type (Fig. 1D, III, lane 2; see also Fig. 6C, lanes 12 to 14). The reduced levels of U3 in the rnt1-Δ strain were initially postulated to be due to impaired splicing (15). However, subsequent work indicated that splicing was not defective in the rnt1-Δ strain (45) and, as shown in Fig. 1D, there is no overall accumulation of intron-containing forms of U3.
TABLE 2.
| Construct | Quantification
|
||||||
|---|---|---|---|---|---|---|---|
| GAL::rrp41 | GAL::rrp41/rnt1-Δ | GAL::rrp45 | GAL::rrp45/rnt1-Δ | rnt1-Δ | rrp6-Δ | rrp6-Δ/rnt1-Δ | |
| U3 | 2.1 | 1.16 | 2.14 | 1 | 0.31 | 2.82 | 0.97 |
| scR1 | 0.83 | 0.87 | 1.66 | 1.85 | 2.27 | 2.16 | 2.34 |
| U3/scR1 | 2.53 | 1.33 | 1.29 | 0.54 | 0.11 | 1.3 | 0.41 |
| U3-3′I + -II | 2.3 | 0.57b | 2.89 | 0.27b | 0.044 | 2.78 | 0.31 |
| U3-3′I + -II/scR1 | 2.77 | 0.65 | 1.74 | 0.15 | 0.019 | 1.29 | 0.13 |
The U3-3′I and -II doublet was quantified as one species. The GAL::rrp41 and GAL::rrp41/rnt1-Δ data are from the 24-h time points; the GAL::rrp45 and GAL::rrp45/rnt1-Δ data are from the 40- and 48-h time points, respectively. Values are relative to the wild-type level, assigned a value of 1.
Species shorter than U3-3′I that appears in the GAL::rrp41/rnt1-Δ and GAL::rrp45/rnt1-Δ strains.
We conclude that 3′ processing of U3 normally involves cleavage by Rnt1p. In the absence of cleavage, long 3′-extended forms are synthesized. The time required for these to be synthesized and then processed may allow assembly of the mature snoRNP proteins, and processing proceeds directly to the 3′ end of the mature snoRNA. This processing is, however, inefficient since mature U3 levels are strongly reduced (Fig. 1D; see also Table 2).
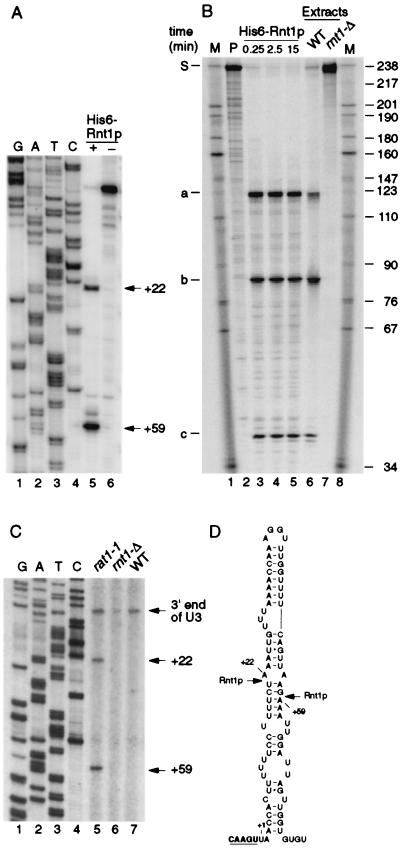
Rnt1p cleaves on both sides of extended stem-loop structures with a closing AGNN tetraloop (15). Inspection of the 3′ flanking sequences revealed the presence of good matches to consensus Rnt1p cleavage sites 3′ to both SNR17A and SNR17B, the genes encoding U3A and U3B, respectively (shown for SNR17A in Fig. 2D). To confirm that these are authentic cleavage sites, the cleavage of the SNR17A site was tested in vitro. The U3(−60/+139) in vitro transcript, which spans the region between positions −60 and +139 with respect to the mature U3 3′ end including the predicted stem-loop structure, was used to map the cleavage site by primer extension (Fig. 2A). Incubation with recombinant His6-Rnt1p (Fig. 2A, lane 5) resulted in the appearance of two primer extension stops that were not detected after incubation in the absence of Rnt1p (Fig. 2A, lane 6). The primer extension stops were at nt +22 and +59, corresponding to cleavage between nt +21/22 and +58/+59, and are in good agreement with the consensus sites of Rnt1p cleavage (Fig. 2D). To demonstrate that in vitro processing is by endonuclease cleavage, a longer transcript was labeled internally; U3(−60/+177) spans the 3′ region of the U3A precursor between positions −60 and +177. Incubation with either recombinant His6-Rnt1p (Fig. 2B, lanes 3 to 5) or an extract from a wild-type (RNT1+) strain of yeast (Fig. 2B, lane 6) led to the appearance of a set of discrete cleavage products that were not observed with the no-enzyme control reaction (Fig. 2B, lane 2) or with an extract from an rnt1-Δ strain of yeast (Fig. 2B, lane 7). The substrate is 237 nt, and comparison to size markers (Fig. 2B, lanes 1 and 8) indicated that the sizes of the three smaller species were in good agreement with the predicted cleavage products: from +59 to the 3′ end of the transcript (predicted size, 119 nt) (band a), from the 5′ end to +21 (predicted size, 81 nt) (band b), and from +22 to +58 (predicted size, 37 nt) (band c).
FIG. 2.
Rnt1p cleaves the 3′ end of the U3 precursor. (A) Mapping of the in vitro Rnt1p cleavage sites. Primer extension was performed with probe 253 on the model U3(−60/+139) RNA incubated with buffer (lane 6) or recombinant His6-Rnt1p (lane 5) as described in Materials and Methods. DNA sequencing reaction on a PCR product encompassing the 3′ end of U3 from positions −60 to +139, using the same primer, was run in parallel (lanes 1 to 4). The primer extension stops at positions +22 and +59 are indicated. (B) In vitro cleavage of an internally labeled model U3(−60/+177) RNA substrate by Rnt1p. 32P-labeled U3(−60/+177) RNA was incubated at 23°C in the following conditions: lane 2, Rnt1p buffer; lanes 3 to 5, Rnt1p buffer with 10 ng of recombinant His6-Rnt1p for the times indicated; lane 6, with whole-cell extract from a wild-type (WT) strain of yeast; lane 7, with whole-cell extract from an rnt1-Δ strain. Lanes 1 and 8, RNA size markers. The positions of DNA size markers are indicated on the right in nucleotides. The obtained cleavage products are labeled a to c on the left, and the predicted origins of these species are as follows: S, substrate (237 nt); a, 3′ end of transcript to position +21/+22 (119 nt); b, 5′ end of transcript to position +58/+59 (81 nt); c, positions +21/+22 to +58/+59 (37 nt). Since in vitro cleavages of U3(−60/+177) are complete (100%), no intermediate cleav- age products are visible. (C) Mapping of the Rnt1p 5′ cleavage site in vitro. Primer extension analysis through the 3′ end of the pre-U3 was performed with primer 252, hybridizing downstream of position +177. RNA was extracted from wild-type (lane 7) and rnt1-Δ (lane 6) strains grown at 30°C and from a rat1-1 strain following transfer to 37°C for 2 h (lane 5). DNA sequencing reactions were run in parallel (lanes 1 to 4). The primer extension stops at positions +59, +22, and +1 (3′ end of U3) are indicated. (D) Computer-predicted RNA structure in the U3 3′ flanking region that contains the Rnt1p cleavage sites. The cleavage sites between nt +21 and +22 and between nt +58 and +59 are indicated by arrows. The 3′ end of mature U3 is underlined.
The 3′ fragments generated by Rnt1p cleavage of the pre-U4 snRNA and the pre-rRNA are strongly stabilized by mutation of the nuclear 5′→3′ exonuclease Rat1p (5, 28), indicating that it normally degrades these regions. The sites of in vivo cleavage of pre-U3 were identified by primer extension using probe 252, which hybridizes in the SNR17A flanking sequence 3′ to the stem-loop structure. In the rat1-1 strain (Fig. 2C, lane 5), primer extension stops were observed at +22 and +59, identical to the in vitro cleavage sites. These were absent from RNA extracted from the rnt1-Δ strain (Fig. 2C, lane 6) but were also not detectable in the wild-type strain (Fig. 2C, lane 7). The stop corresponding to the position of the 3′ end of mature U3 may be a consequence of the stem structure at this position. The level of this stop is unaltered in the rat1-1 strain, suggesting that it is not a cleavage site. We cannot, however, exclude the possibility that a fraction of U3 is processed by endonucleolytic cleavage at the mature 3′ end. RNase MRP was shown not to be involved in this process (data not shown).
We conclude that Rnt1p cleaves the 3′ extended pre-U3 at +21/+22 and +58/+59. Following cleavage the 3′ fragment is degraded by Rat1p. The level of the mature U3 is reduced in strains lacking Rnt1p, indicating that this is normally the major synthesis pathway.
The major 3′-extended forms of U3 do not extend to the Rnt1p cleavage sites.
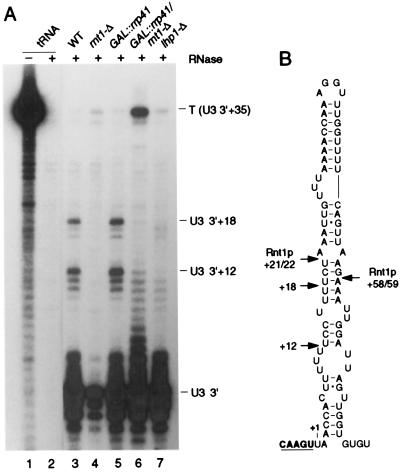
High-resolution Northern hybridization showed that the U3-3′I band was too small to extend to the Rnt1p cleavage sites, and even the larger U3-3′ II species appeared to be slightly smaller than expected. The 3′ ends of these species were therefore determined by RNase protection. For this, the region of SNR17A from 295 to +36 was amplified by PCR using a primer that incorporated a T7 promoter (see Materials and Methods). In addition to the band corresponding to the mature 3′ end of U3, two major protected fragments were detected in RNA from the wild-type strain (Fig. 3A, lane 3) but were absent from the rnt1-Δ strain (Fig. 3A, lane 4). The sizes to these bands correspond to species that extend to U3+12 and U3+18, in good agreement with the gel mobilities of the U3-3′I and U3-3′II RNAs, respectively.
FIG. 3.
Mapping of the 3′-extended forms of U3 by RNase protection. (A) RNA was extracted from wild-type (WT), rnt1-Δ, and lhp1-Δ strains grown at 30°C and from GAL::rrp41 and GAL::rrp41/rnt1-Δ strains following transfer from permissive, RSG medium to repressive, glucose medium at 30°C for 24 and 48 h, respectively. Total E. coli tRNA was used as a control RNA. Positions of the Rnt1p-dependent protected species at +12 and +18 are indicated. (B) Schematic of the U3 3′ flanking region showing the ends of the protected regions and the Rnt1p cleavage sites.
We conclude that following Rnt1p cleavage, the pre-U3 undergoes rapid trimming back to +12 and +18.
The major 3′-extended forms of U3 are stabilized by Lhp1p.
It seemed very likely that some RNA binding factor was responsible for stabilizing the 3′ ends of the U3-3′I and -II species. Inspection of the sequence showed that these RNAs possessed 3′ poly(U) tracts (Fig. 3B). The 3′ poly(U) tracts of RNAs transcribed by RNA polymerase III are bound by the La protein (42, 48), as are the 3′ extended precursors to human U1 (34) and yeast (58) snRNAs. We therefore tested whether the U3-3′I and -II RNAs were being stabilized by binding to Lhp1p, the yeast homologue of La (39, 59).
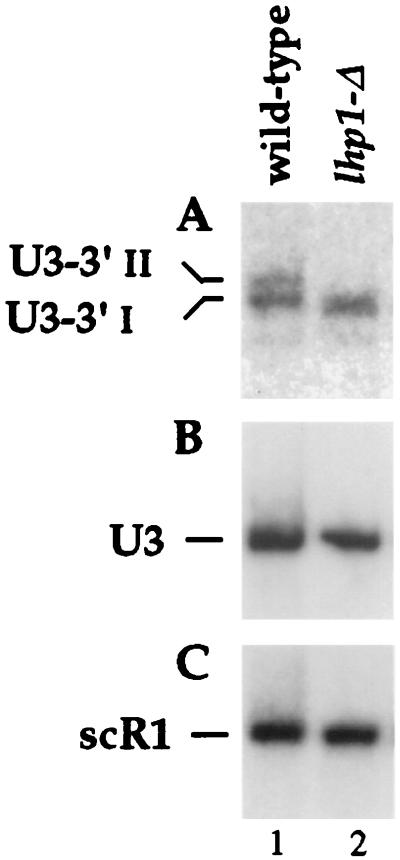
The LHP1 gene is nonessential (59), and a gene disruption was performed by a one-step PCR approach (10) using the K. lactis URA3 marker (see Materials and Methods). RNase protection analysis of RNA from the lhp1-Δ strain showed the loss of the major 3′-extended ends at +18 and +12 and the appearance of shorter, heterogeneous protected fragments corresponding to RNAs from U3+8 to U3+11 (Fig. 3A, lane 7). This result was confirmed by Northern hybridization (Fig. 4). The U3-3′II and U3-3′I species were absent from the lhp1-Δ strain (Fig. 4A), and a species slightly shorter than U3-3′I was detected. The level of mature U3 was unaffected in the lhp1-Δ strain (Figs. 3A and 4B), as were the levels of the truncated U3 degradation intermediates seen in wild-type cells (see Fig. 6; data not shown). These data suggested that both U3-3′I and U3-3′II were stabilized by binding Lhp1p.
FIG. 4.
3′-extended forms of U3 are stabilized by Lhp1p. Lane 1, LHP1 strain; lane 2, lhp1-Δ strain. Total RNA was analyzed by Northern hybridization with probe 251, specific for the 3′-extended U3 (A), probe 200, which hybridizes to the mature U3 (B), and probe 250, which hybridizes to scR1 RNA (C).
To confirm this, a C-terminal fusion between Lhp1p and two copies of the Z domain of S. aureus protein A was constructed and integrated at the chromosomal LHP1 locus by a one-step PCR approach (29) (see Materials and Methods). Western blotting confirmed that the fusion protein was expressed and could be efficiently immunoprecipitated with IgG agarose (data not shown). Immunoprecipitation was performed on two independently isolated Lhp1p-ProtA strains; data are presented for only one strain in Fig. 5. Processing of pre-tRNATyr appeared to be the same in the strain expressing only Lhp1p-ProtA and the wild type (Fig. 5D); however, some accumulation of the shorter 3′-extended pre-U3 species was visible (Fig. 5A), suggesting that the Lhp1p-ProtA fusion protein is underexpressed or otherwise not fully functional.
FIG. 5.
3′-extended forms of U3 are coprecipitated with Lhp1p-ProtA. Lysates from the LHP1+ and LHP1::ProtA strains were immunoprecipitated using IgG agarose. RNA was recovered from the total cell lysate (T), immune supernatant (S), and immunoprecipitate (P) and analyzed by Northern hybridization. Probes are indicated in parentheses and described in Materials and Methods. On prolonged exposure, background precipitation of mature U3 is seen for both the wild-type and Lhp1-ProtA strains (lanes 7 and 8). In panel B, the total and supernatant lanes were heavily overexposed at the exposure needed to visualize the U3-int 3′ and U3-3′III RNAs and were omitted. Approximately fourfold more cell equivalents are loaded for the bound material.
As expected, the tRNATyr primary transcript (Fig. 5D) and the U6 snRNA (Fig. 5E) were immunoprecipitated on IgG agarose from the strain expressing Lhp1p-ProtA (lane 6) but not from the wild type (lane 3). Both U3-3′I and U3-3′II were coprecipitated with Lhp1p-ProtA (Fig. 5A), as were U3-int 3′ and a species of approximately 800 nt designated U3-3′III (Fig. 5B). The species shorter than U3-3′I seen in the Lhp1p-ProtA strain was not coprecipitated and remained in the immune supernatant (Fig. 5A, lane 5). Mature U3 (Fig. 5B and C) and the 3′ processed, intron-containing pre-tRNATyr (Fig. 5D) were recovered at the same low levels in the wild-type and Lhp1-ProtA precipitates. The pre-U3 and pre-tRNA species were more efficiently precipitated than U6, presumably because only the newly synthesized U6 is associated with Lhp1p (35, 39).
We conclude that Lhp1p binds and stabilizes the major 3′-extended forms of U3.
The exosome participates in 3′ processing of U3.
The levels of 3′-extended precursors to other snoRNAs and snRNAs are elevated in strains carrying mutations in the exosome complex (5, 55). To assess the effects of genetic depletion of exosome components on the 3′-extended forms of U3, Rrp41p and Rrp45p were depleted by transfer of GAL::rrp41 and GAL::rrp45 strains (6, 36) from permissive RSG medium (0-h samples) to repressive, glucose medium for the times indicated. A strain deleted for the gene encoding the Rrp6p component of the exosome (6) was also analyzed. In the strains lacking Rrp41p (Fig. 6A and C, lanes 5), Rrp45p (lanes 10), or Rrp6p (lanes 2), the levels of U3-3′I and U3-3′II were higher than in the isogenic wild-type control strains (lanes 3 and 14); these results are quantitated in Table 2. For the GAL::rrp41 strain, this increase was confirmed by RNase protection (Fig. 3A, lane 4), which showed that the accumulated precursors were identical to U3-3′I and -II. Rrp41p is underexpressed in the GAL::rrp41 strain in RSG medium and therefore shows some accumulation of the extended species in the 0-h sample (6, 36). In strains genetically depleted of other exosome components, Rrp4p, Rrp40p, Rrp46p, or Csl4p, increased levels of U3-3′I and -II were also observed (data not shown). In addition, an RNA species that comigrated with the U3-3′III RNA, seen on Lhp1p-ProtA precipitation (Fig. 5), was accumulated in the exosome mutants. On prolonged exposure, this species could also be detected at low levels in wild-type cells. Depletion of the exosome components did not lead to depletion of the mature U3. Indeed, as was previously observed for the U4 and U5 snRNAs, depletion of exosome components led to an increase in the mature U3 snoRNA of approximately twofold (Table 2).
In strains lacking exosome components, the 3′ processed, intron-containing precursor is clearly detected. This is most visible for Rrp6p (Fig. 6B, lane 2) but was also seen for several other exosome mutants (Fig. 6B and data not shown). This species is not detected in the wild type, and we speculate that this processing intermediate is normally a dead-end product that is degraded by the exosome. 3′ processing appears to be dependent on snoRNP protein binding, but assembly with the mature snoRNP proteins may be incompatible with assembly of a functional spliceosome. The exosome also degrades other stalled, intron-containing pre-mRNAs (C. Bousquet-Antonelli, C. Presutti, and D. Tollervey, submitted for publication).
The combination of the deletion of both RNT1 and RRP6 (Fig. 6, lane 1) partially restored synthesis of species with the same gel mobility as the U3-3′I and U3-3′II RNAs. Depletion of Rrp41p or Rrp45p from the strain lacking Rnt1p (Fig. 6A and C, lanes 7, 8, 12, and 13) led to the appearance of heterogeneous RNA species slightly smaller than U3-3′I, similar in size to the species seen in the lhp1-Δ strain (Fig. 4). Consistent with this, RNase protection analysis in the GAL::rrp41/rnt1-Δ strain reveals a ladder of protected RNA fragments extending from mature U3 to position U3+12 (Fig. 3A, lane 6); due to the location of the hybridization probe, only the longer RNAs were detected by Northern hybridization (Fig. 6). A stronger ladder of RNA species extending up to the position of U3-3′III was observed by Northern hybridization (Fig. 6, lanes 7, 8, 12, and 13), which was reflected by the strong protection of the full-length antisense probe (Fig. 3A, lane 6). The combination of each of exosome mutations with rnt1-Δ partially restored the mature U3 levels compared to the rnt1-Δ single mutant strain (Table 2).
We conclude that the exosome complex of 3′→5′ exonucleases participates in the 3′ processing of U3. This processing pathway closely resembles that of the U1, U4, and U5 snRNAs (5, 14, 45, 55). In each case, synthesis of the mature RNA continues in strains depleted of single components of the exosome, indicating either that different components of the complex are partially functionally redundant or that other exonucleases can largely substitute for the exosome.
The level of the mature U3 is elevated in the exosome mutants, indicating competition between the synthetic pathway and degradation of the pre-U3. This was also seen for the U4 and U5 snRNAs (5). Consistent with this model, a truncated U3 species (U3**) was observed in wild-type strains (Fig. 7, lanes 1 and 12) (24, 35). The U3** species was 5′ and 3′ truncated, as shown by its failure to hybridize to probes directed against either the 3′ end of U3 (Fig. 7B) or the 5′ end of U3 (Fig. 7C). In contrast, the U3* species that is accumulated in rrp6-Δ, GAL::rrp41, and GAL::rrp45 strains was truncated only at the 5′ end, indicating that U3 is normally 3′ degraded by the exosome. The level of U3* is further elevated in exosome mutants that also lack Rnt1p, consistent with the model that degradation of pre-U3 is increased in rnt1-Δ strains. The 5′ degradation activity has not been further characterized but is likely due to the 5′→3′ exonuclease Rat1p, which 5′ processes other snoRNAs and degrades pre-rRNA spacer fragments (41).
FIG. 7.
Exosome components participate in the degradation of U3 snoRNA. For Northern analysis of U3 snoRNA in wild-type (WT) and rnt1-Δ and exosome mutant strains. RNA was extracted as described for Fig. 2, separated on an 6% polyacrylamide gel, and hybridized with oligonucleotide probes. The panels show successive hybridization of the same filter. Probes are indicated in parentheses on the left and described in Materials and Methods; the positions of RNA species detected are indicated on the right. The amount of total RNA loaded in lane 14 is fourfold greater than in lane 13 and other lanes. The positions of migration of snRNA190 (190 nt), U5L (215 nt), and snR10 (246 nt) determined by hybridization of the same filter are indicated as size markers. Mature U3 is 333 nt. The locations of the oligonucleotide probes and the predicted structures of the degradation intermediates are shown schematically.
In strains lacking Rnt1p, 3′ extended forms of U1 and U2 snRNAs undergo a low level of polyadenylation (1, 45), and the precursors to several snRNAs and snoRNAs are polyadenylated in exosome mutants (5, 55). To determine whether this was also the case for the 3′-extended U3, RNA was treated in vitro with oligo(dT) and RNase H. Following this deadenylation treatment, the longer 3′-extended species detected in the rnt1-Δ/GAL::rrp41 strain became shorter and more discrete (data not shown), indicating that a low level of polyadenylation had indeed occurred.
DISCUSSION
How is U3 processed?
A model for 3′ processing of the U3A snoRNA is presented in Fig. 8. We postulate that processing is normally initiated by cotranscriptional cleavage by Rnt1p across a stem structure at positions +21 and +58 with respect to the 3′ end of U3. The released 3′ fragment is degraded by the 5′→3′ exonuclease Rat1p, as shown by its accumulation in the rat1-1 strain. The 3′ extended pre-snoRNA is rapidly processed to +12 and +18, since the species extended to +21 is not readily detected in total RNA. The products of Rnt1p cleavage of pre-U4 and pre-U5 are elevated in strains deleted for components of the exosome (5), and we think it probable that the exosome complex also carries out the initial shortening of the pre-U3. We cannot, however, exclude the participation of other exonucleases, such as the Rex1-3p family that carry out the final trimming of several small RNA species (54). The pre-U3 is stabilized against further 3′ degradation by binding of Lhp1p to the 3′ poly(U) tracts at +19 and +13; whether Lhp1p binds to internal poly(U) tracts prior to the start of digestion, or binds to free 3′ poly(U) tracts generated during digestion, cannot be determined at present. The larger U3-3′III species is bound by Lhp1p, suggesting that Lhp1p does bind to internal poly(U) sequences prior to processing, but the endpoints of this have not been mapped and we cannot exclude the possibility that it has a terminal poly(U) tract. It is likely that the poly(U) tracts at +19 and +13 can each bind Lhp1p, although binding may be mutually exclusive.
FIG. 8.
Model for the 3′ processing of the U3A snoRNA. The presence of the poly(U) tracts and stem-loop structure in the 3′ flanking sequence and the intron are indicated. For simplicity, only one poly(U) tract is indicated. In reality, two tracts are present, at +19 and +13, each of which is likely to act as a binding site for Lhp1p. The activity that carries out the initial trimming to +18 and +12 has not been determined but is likely to be the exosome. The endpoints of the U3-int 3′ species have not been determined, but the finding that these species are associated with Lhp1p suggests that they are processed to +18 and +12.
The box C+D snoRNAs, including U3, bind a set of common proteins, Nop1p, Nop56p, and Nop58p (13, 30–32, 40, 44, 53) that probably bind to the box D sequence close to the 3′ end of the snoRNA and the 3′-terminal stem (13, 57). These proteins are not associated with the 3′-extended U3 species, and we propose that their binding displaces Lhp1p from the 3′ flanking sequence. Since the snoRNP proteins bind at the very 3′ end of the snoRNA, this displacement may be steric. Removal of Lhp1p is envisaged to allow the exosome to resume processing, generating the mature snoRNA 3′ end. This is followed by cap trimethylation; in vertebrates this snoRNA modification requires the conserved box C+D snoRNAs (50), probably acting via binding the mature snoRNP proteins. The yeast U3 genes are unusual in that they contain an intron that is excised by the normal pre-mRNA splicing machinery. In wild-type cells this is spliced from the 3′-extended pre-U3, since only the 3′-extended, intron-containing species is detected. The endpoints of the U3-int 3′ species have not been determined, but these species are associated with Lhp1p, suggesting that they may have been largely processed to +18 and +12.
Deletion of Rnt1p strongly reduces synthesis of mature U3 (Table 2). Processing of the long 3′-extended pre-U3 species generated in the absence of Rnt1p cleavage involves the exosome, as shown by their increased levels in rnt1-Δ strains lacking exosome components. We speculate that a processive exosome complex assembles on the long 3′-extended pre-U3, which is able to substantially displace bound Lhp1p and/or the snoRNP proteins and therefore degrades most of the pre-U3 population. Consistent with this model, depletion of exosome components from rnt1-Δ strains restored mature U3 to the wild-type level (Table 2).
In the absence of Lhp1p, the U3 snoRNA was still 3′ processed. The U3-3′I and U3-3′II species were absent but slightly smaller, heterogeneous species were observed, indicating that some other factor(s) can also bind the 3′ poly(U) tract. An obvious candidate is the Lsm complex, which binds to the 3′ poly(U) tract of the U6 snRNA and is required for normal 3′ processing of the RNase P RNA (3, 17, 35, 43). Consistent with this model, mutations in Lsm8p were lethal in combination with the deletion of LHP1 (39).
In otherwise wild-type strains, depletion of exosome components increased the mature U3 level by inhibiting a 3′ degradation pathway that generates the truncated U3** intermediate, indicating competition between the synthetic and degradative pathways during normal U3 synthesis. Similar observations have been reported for the U4 and U5 snRNAs (5).
The processing pathway deduced here for yeast U3 shows similarities to the processing pathways proposed for the U1, U2, U4, and U5 spliceosomal snRNAs. In each case, downstream cleavage by Rnt1p is thought to act as an entry for the exonucleases (1, 5, 14, 45). For U1, U4, and U5, this processing was also shown to involve the exosome complex (5, 55); this has not been addressed for U2. Also in each case, shorter 3′ extended species normally accumulate as transient intermediates, although their 3′ ends have not yet been accurately mapped. In the case of pre-U4 and pre-U5, the Rnt1p cleavage products are 3′ processed by the exosome complex and then trimmed to the mature RNAs by the Rex1-3p family of exonucleases together with the Rrp6p exosome component (54). Other box C+D snoRNAs are 3′ trimmed by Rrp6p (5), but this is not the case for U3.
Inspection of the 3′ flanking sequences reveals that poly(U) tracts are present in the 3′ flanking sequences of the U1, U2, U4, and U5 snRNA genes (Fig. 9). In each case, the Rnt1p cleavage site is adjacent to a poly(U) tract (Fig. 9A). For the U2, U3, and U5L RNAs, the mature RNA regions (uppercase in Fig. 9A) are located relatively close to the Rnt1p cleavage site, with additional poly(U) tracts between the Rnt1p cleavage site and the mature 3′ end. The mature regions of U1, U4, and U5S are more distant, and their 3′ ends are located within a further poly(U) tract (Fig. 9B). Lhp1p is associated with yeast pre-U1, U2, U4, and U5 (58). However, in contrast to the model presented here for U3, Lhp1p is proposed to function as a cofactor for the assembly of the spliceosomal snRNAs with the Sm proteins. The human and plant U3 snoRNAs also have 3′ flanking poly(U) tracts, suggesting that this feature may be conserved throughout eukaryotes (27, 49).
FIG. 9.
Comparison of the 3′ flanking sequence of U3A to those of the U1, U2, U4, and U5 snRNAs. In panel A, the Rnt1p cleavage sites (\) have been aligned. The mature regions of U3, U2, and U5L are in uppercase. For U1, U4, and U5S, the mature regions are further from the Rnt1p cleavage site. These are aligned in the panel B. Poly(U) sequences of four or more residues are underlined.
Why is U3 processed?
The 3′ ends of almost all RNAs from all organisms are generated by 3′ processing rather than transcription termination, but the reasons for this have largely remained obscure. The data presented here provide a possible explanation, at least for U3. The binding sites for the common snoRNP proteins, the box D element and the terminal stem structure, define the 3′ end of the mature U3 snoRNA. Transcription termination at this site would generate an RNA with a monomethylguanosine cap structure and lacking the snoRNP proteins. This could not readily be distinguished from the products of premature termination or failed pre-mRNA splicing. It is likely that these are normally very rapidly degraded by the exosome complex and Rat1p (C. Bousquet-Antonelli, C. Presutti, and D. Tollervey, unpublished data). Delaying or reducing these degradative activities might allow sufficient time for snoRNP assembly and cap trimethylation, but at the expense of allowing greater accumulation of aberrant RNAs. Such a strategy might also allow a greater level of accidental protection of inappropriate RNA species by RNA-binding proteins. Instead, the cell has adopted a mechanism to specifically delay 3′ processing of the snoRNA. Transcription continues beyond the 3′ end of the mature snoRNA, with the transcript normally being cleaved by Rnt1p and protected by binding of Lhp1p. This leaves the mature 3′ end free for binding of the snoRNP proteins. Such a system has the additional advantage of acting as a quality control system. We envisage that the snoRNP proteins, or at least Nop58p, must displace Lhp1p to allow final maturation of the snoRNA. In the absence of Nop58p binding, the 3′ extended pre-U3 accumulates to low levels and is then degraded. Binding of La to pre-tRNAs has also been proposed to function as a quality control system (19), and binding of Lhp1p to the U6 snRNA and pre-tRNAiMet is also likely to antagonize rapid 3′ degradation (8, 39).
We propose that 3′ processing acts as a quality control system in the synthesis of many RNA species and that this underlies its ubiquitous occurrence.
ACKNOWLEDGMENTS
We thank Bertrand Séraphin (EMBL) for the cloned K. lactis URA3 gene and R. Lührmann for the kind gift of R1131 antibodies. J.K. was the recipient of a long-term EMBO fellowship. This work was supported by the Wellcome Trust.
REFERENCES
- 1.About Elela S, Ares M., Jr Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 1998;17:3738–3746. doi: 10.1093/emboj/17.13.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou Elela S, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 3.Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Lührmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′->5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amberg D C, Goldstein A L, Cole C N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 8.Anderson J, Phan L, Cuesta R, Carlson B A, Pak M, Asano K, Bjork G R, Tamame M, Hinnebusch A G. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson J S J, Parker R P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SK12 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beltrame M, Tollervey D. Base-pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caffarelli E, Losito M, Giorgi C, Fatica A, Bozzoni I. In vivo identification of nuclear factors interacting with the conserved elements of box C/D small nucleolar RNAs. Mol Cell Biol. 1998;18:1023–1028. doi: 10.1128/mcb.18.2.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chanfreau G, Elela S A, Ares M, Jr, Guthrie C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chanfreau G, Legrain P, Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J Mol Biol. 1998;284:975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- 16.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper M, Johnston L H, Beggs J D. Identification and characterization of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995;14:2066–2075. doi: 10.1002/j.1460-2075.1995.tb07198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 19.Fan H, Goodier J L, Chamberlain J R, Engelke D R, Maraia R J. 5′ processing of tRNA precursors can be modulated by the human La antigen phosphoprotein. Mol Cell Biol. 1998;18:3201–3211. doi: 10.1128/mcb.18.6.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautier T, Bergès T, Tollervey D, Hurt E. The nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodall G J, Wiebauer K, Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- 23.Heise T, Guidotti L G, Chisari F V. La autoantigen specifically recognizes a predicted stem-loop in hepatitis B virus RNA. J Virol. 1999;73:5767–5776. doi: 10.1128/jvi.73.7.5767-5776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes J M X, Ares M J. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes J M X, Konings D A M, Cesareni G. The yeast homologue of U3 snRNA. EMBO J. 1987;6:2145–2155. doi: 10.1002/j.1460-2075.1987.tb02482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen R, Tollervey D, Hurt E C. A U3 snoRNP protein with homology to splicing factor PRP4 and Gβ domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiss T, Marshallsay C, Filipowicz W. Alteration of the RNA polymerase specificity of U3 snRNA genes during evolution and in vitro. Cell. 1991;65:517–526. doi: 10.1016/0092-8674(91)90469-f. [DOI] [PubMed] [Google Scholar]
- 28.Kufel J, Dichtl B, Tollervey D. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA. 1999;5:909–917. doi: 10.1017/s135583829999026x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lafontaine D, Tollervey D. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 1996;24:3469–3472. doi: 10.1093/nar/24.17.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lafontaine D L J, Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for stability of the snoRNAs. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lafontaine D L J, Tollervey D. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol Cell Biol. 2000;20:2650–2659. doi: 10.1128/mcb.20.8.2650-2659.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lischwe M A, Ochs R L, Reddy R, Cook R G, Yeoman L C, Tan E M, Reichlin M, Busch H. Purification and partial characterization of a nucleolar scleroderma antigen (Mr=34,000; pI, 8.5) rich in NG, NG-dimethylarginine. J Biol Chem. 1985;260:14304–14310. [PubMed] [Google Scholar]
- 33.Lygerou Z, Mitchell P, Petfalski E, Séraphin B, Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- 34.Madore S J, Wieben E D, Pederson T. Eukaryotic small ribonucleoproteins. Anti-La human autoantibodies react with U1 RNA-protein complexes. J Biol Chem. 1984;259:1929–1933. [PubMed] [Google Scholar]
- 35.Mayes A E, Verdone L, Legrain P, Beggs J D. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome; a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonuclease activities. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 37.Mitchell P, Petfalski E, Tollervey D. The 3′-end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 1996;10:502–513. doi: 10.1101/gad.10.4.502. [DOI] [PubMed] [Google Scholar]
- 38.Myslinski E, Ségault V, Branlant C. An intron in the genes for U3 small nucleolar RNAs of the yeast Saccharomyces cerevisiae. Science. 1990;247:1213–1216. doi: 10.1126/science.1690452. [DOI] [PubMed] [Google Scholar]
- 39.Pannone B K, Xue D, Wolin S L. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker K A, Steitz J A. Structural analysis of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol Cell Biol. 1987;7:2899–2913. doi: 10.1128/mcb.7.8.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rinke J, Steitz J A. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1982;29:149–159. doi: 10.1016/0092-8674(82)90099-x. [DOI] [PubMed] [Google Scholar]
- 43.Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Seraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schimmang T, Tollervey D, Kern H, Frank R, Hurt E C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seipelt R L, Zheng B, Asuru A, Rymond B C. U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res. 1999;27:587–595. doi: 10.1093/nar/27.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Séraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 47.Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol Cell Biol. 1999;19:6012–6019. doi: 10.1128/mcb.19.9.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stefano J E. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- 49.Suh D, Busch H, Reddy R. Isolation and characterization of a human U3 small nucleolar RNA gene. Biochem Biophys Res Commun. 1986;137:1133–1140. doi: 10.1016/0006-291x(86)90343-8. [DOI] [PubMed] [Google Scholar]
- 50.Terns M P, Grimm C, Lund E, Dahlberg J E. A common maturation pathway for small nucleolar RNAs. EMBO J. 1995;14:4860–4871. doi: 10.1002/j.1460-2075.1995.tb00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tollervey D, Mattaj I W. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 1987;6:469–476. doi: 10.1002/j.1460-2075.1987.tb04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tyc K, Steitz J A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Hoof A, Lennertz P, Parker R. Three conserved members of the RNAse D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 2000;19:1357–1365. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Horn D J, Yoo C J, Xue D, Shi H, Wolin S L. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA. 1997;3:1434–1443. [PMC free article] [PubMed] [Google Scholar]
- 57.Watkins N J, Newman D R, Kuhn J F, Maxwell E S. In vitro assembly of the mouse U14 snoRNP core complex and identification of a 65-kDa box C/D-binding protein. RNA. 1998;4:582–593. doi: 10.1017/s1355838298980128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue D, Rubinson D, Pannone B K, Yoo C J, Wolin S L. U snRNP assembly in yeast involves the La protein. EMBO J. 2000;19:1650–1660. doi: 10.1093/emboj/19.7.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoo C J, Wolin S L. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]