Abstract
Motivation
Quality control (QC) is a critical step in single-cell RNA-seq (scRNA-seq) data analysis. Low-quality cells are removed from the analysis during the QC process to avoid misinterpretation of the data. An important QC metric is the mitochondrial proportion (mtDNA%), which is used as a threshold to filter out low-quality cells. Early publications in the field established a threshold of 5% and since then, it has been used as a default in several software packages for scRNA-seq data analysis, and adopted as a standard in many scRNA-seq studies. However, the validity of using a uniform threshold across different species, single-cell technologies, tissues and cell types has not been adequately assessed.
Results
We systematically analyzed 5 530 106 cells reported in 1349 annotated datasets available in the PanglaoDB database and found that the average mtDNA% in scRNA-seq data across human tissues is significantly higher than in mouse tissues. This difference is not confounded by the platform used to generate the data. Based on this finding, we propose new reference values of the mtDNA% for 121 tissues of mouse and 44 tissues of humans. In general, for mouse tissues, the 5% threshold performs well to distinguish between healthy and low-quality cells. However, for human tissues, the 5% threshold should be reconsidered as it fails to accurately discriminate between healthy and low-quality cells in 29.5% (13 of 44) tissues analyzed. We conclude that omitting the mtDNA% QC filter or adopting a suboptimal mtDNA% threshold may lead to erroneous biological interpretations of scRNA-seq data.
Availabilityand implementation
The code used to download datasets, perform the analyzes and produce the figures is available at https://github.com/dosorio/mtProportion.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Single-cell RNA-seq (scRNA-seq) experiments have improved the resolution of our knowledge about cellular composition and cellular behavior in complex tissues (Sandberg, 2014). A critical step during scRNA-seq data processing is to perform quality control (QC) over the cells sequenced transcriptomes (Hwang et al., 2018). The QC process usually involves applying user-defined thresholds for different metrics computed for each individual cell to filter out doublets and ‘low-quality’ cells (Luecken and Theis, 2019). Commonly used QC metrics include the total transcript counts (also known as the library size), the number of expressed genes and the mitochondrial proportion (mtDNA%, i.e. the ratio of reads mapped to mitochondrial DNA-encoded genes to the total number of reads mapped). Defining the proper thresholds of QC metrics is a complex task that requires a vast knowledge of the cellular diversity in the tissue under study. Thresholds may be uniquely set for each sample, as they are dependent on the cells or tissue being processed (Ji and Sadreyev, 2019).
This study focuses on the systematic determination of a threshold for mtDNA%—the fraction of mitochondrial counts per cell—in scRNA-seq QC. Mitochondrial content is known to interact with the nuclear genome, drive alternative splicing and regulate nuclear gene expression and is also associated with cancer, degenerative diseases and aging (Guantes et al., 2015; Muir et al., 2016). High numbers of mitochondrial transcripts are indicators of cell stress, and therefore mtDNA% is a measurement associated with apoptotic, stressed and low-quality cells (Ilicic et al., 2016; Lun et al., 2016; Zhao et al., 2002). However, mtDNA% threshold depends highly on the tissue type and the questions being investigated (AlJanahi et al., 2018). The mtDNA% threshold is of economic and biological importance. A wrongly defined, very stringent mtDNA% threshold may cause bias in the recovered cellular composition of the tissue under study. This bias may force the researchers to increase the sample size to capture enough cells (which may not have the normal biological behavior of the cell type) under the threshold, and thus increase the cost of the experiment. Inversely, a relaxed threshold of mtDNA% may allow apoptotic, low-quality cells to remain in the analysis, resulting in the identification of wrong biological patterns.
To reduce the bias caused by the use of arbitrary mtDNA% thresholds, Ma et al. (2019) proposed an unsupervised method to optimize the threshold for each given input data. This computationally expensive data-driven procedure, which defines the threshold as a function of the distribution of the data, due to the lack of reference values, is not able to identify bias induced during the library preparation. Without such standard references, the values of mtDNA% thresholds fluctuate with different input datasets. For example, a largely failed experiment may generate a dataset, in which most cells have an inflated mtDNA%. Accordingly, the optimized threshold based on these inflated values may be unreasonably high. Therefore, having a uniform and standardized threshold for scRNA-seq data analysis is essential. It improves the reproducibility of experiments and simplifies the automatization of bioinformatic pipelines (McCarthy et al., 2017).
Through analysis of bulk RNA-seq data produced by the Illumina Body Tissue Atlas, Mercer et al. (2011) reported the mtDNA% for 16 human tissues. They found that the mtDNA% ranges from 5% or less in tissues with low energy requirements up to ∼30% in the heart due to the high energy demand of cardiomyocytes. Based on that study, early publications of scRNA-seq datasets used the 5% threshold reported for tissues with low energy demands (e.g. adrenal, ovary, thyroid, prostate, testes, lung, lymph and white blood cells) as default for data QC (Lukassen et al., 2018). Furthermore, the 5% threshold has been adapted as the default parameter by Seurat—one of the most popular software packages for scRNA-seq data analysis (Satija et al., 2015). These have made a 5% practical standard for scRNA-seq data analyses. Nevertheless, due to the lack of reference values for mtDNA% in different species, technologies, tissues and cell types, the optimal value for a standardized threshold is still an open question in the field.
PanglaoDB is a scRNA-seq database providing uniformly processed, annotated count matrices for thousands of cells from hundreds of scRNA-seq experiments. The data source of PanglaoDB is the sequence read archive (SRA) database of the National Center for Biotechnology Information (NCBI). With the datasets from PanglaoDB, it is possible to systematically evaluate the optimal threshold of mtDNA% for different experimental settings that may vary across platforms, technologies, species, tissues or cell types (Franzen et al., 2019; Svensson et al., 2019). Here, we present a systematic analysis of the mtDNA% in more than 5 million cells reported in over one thousand datasets in PanglaoDB (Franzen et al., 2019). We compared the mtDNA% reported for different technologies, species, tissues and cell types. By analyzing the data provided by hundreds of experiments together, we reach the consensus reference values for more than 40 human tissues and more than 120 mouse tissues. Furthermore, we evaluated the validity of using the 5% threshold in different humans and mice tissues and showed that omitting the mtDNA% as a QC filter led to erroneous biological interpretations of the data.
2 Materials and methods
Datasets in the PanglaoDB database, available at the time of analysis (in January 2020), were downloaded and processed using R 3.6.2 (R Core Team, 2013) through an ‘in-house’ script using the XML (Lang and CRAN Team, 2012) and xml2 (Wickham et al., 2018) packages. The library size (total number of counts), the total number of detected genes and the total number of counts that match with the mitochondrial genes (mitochondrial counts) were estimated for all cells in each of downloaded datasets. The SRA/SRS identifiers, species, protocol, tissue, cell type and barcode of each experiment were obtained and associated with each cell.
Only the cells with more than counts and with the total number of counts greater than two times the average library size in the same sample were retained for analysis. In addition, a polynomic regression of degree (to account for saturation) was applied to establish the confidence intervals of the predicted total number of genes as a function of the library size per cell. Cells with an observed total number of genes below or above expectation limits were removed from the analysis. The same procedure was applied a second time to establish the confidence intervals of the predicted mitochondrial counts as a function of library size. An ordinary least squares (OLS) regression model was used to fit the data and cells with exceptionally high or low mitochondrial counts were removed from the analysis.
Subsequently, the mtDNA% value was computed for each cell as the ratio between the mitochondrial counts and the library size of the cell. The mtDNA% values were then compared between cells from different settings: species, technologies, tissues and cell types. To compare the mtDNA% between humans and mice cells, we used the Welch two-sample t-test and used the Wilcoxon sum-rank test to cross-validate the results. To evaluate the reliability of the threshold, a comparison to evaluate whether the mean was < threshold value was performed using the t-test for each tissue and cell type independently using the data generated by the 10× Genomics Chromium system, after filtering out groups with less than cells.
Example datasets (SRS3703557, SRS3545826 and SRS2397417) were downloaded from the PanglaoDB along with cell clustering results and cell type information. Count matrices were processed using the ‘Seurat’ R package to generate low-dimensional representations. Differential expression analysis was performed using ‘MAST’ (Finak et al., 2015) to compare the transcriptome profile of clusters exhibiting a median mtDNA% higher than 5% against that of other clusters with the same cell type but with a median mtDNA% lower than 5%. Using the sorted list of fold-changes reported by MAST, we performed Gene Set Enrichment Analysis (GSEA) to test the enrichment of the ‘Apoptosis’ pathway from the KEGG database (Kanehisa and Goto, 2000) in the clusters with increased mtDNA%. To run the GSEA analysis efficiently, we used the multilevel function included in the ‘fgsea’ R package (Korotkevich et al., 2019).
3 Results
We downloaded a total of 5 530 106 cells reported in 1349 datasets from the PanglaoDB database. From those, we removed 278 607 cells with a total number of counts smaller than 1000 or above two times the average library size in the sample where it was sequenced. Also, 80 225 cells with no mitochondrial counts were removed. The remaining 5 171 274 cells were used to establish the 95% confidence intervals of the predicted total number of genes as a function of the library size per cell (Supplementary Fig. S1). We found that the relationship between the number of genes and the library size is monotonically positive (), which is consistent with that previously reported (Svensson et al., 2019). We also found that the expected total number of genes reaches saturation at a point close to the library size counts. In this step, we removed 157 960 cells because they have a total number of quantified genes above () or below () the 95% confidence interval limit defined from the prediction.
Next, we accounted for outliers in the mitochondrial counts in relative to the library size. This procedure has been shown to be critical to differentiate apoptotic cells of preapoptotic and healthy cells in a supervised experiment (Ordonez-Rueda et al., 2020). To do so, we used the OLS regression and computed the confidence interval of prediction between the mitochondrial counts and the library size with data from all 5 013 314 cells. We found that the relationship is noisy but positive and linear (Supplementary Fig. S2; ). Following this procedure, we identified 333 712 cells with mitochondrial counts above () or below () the computed confidence interval limits, which were also removed. After this step, 4 679 602 cells were retained for the study.
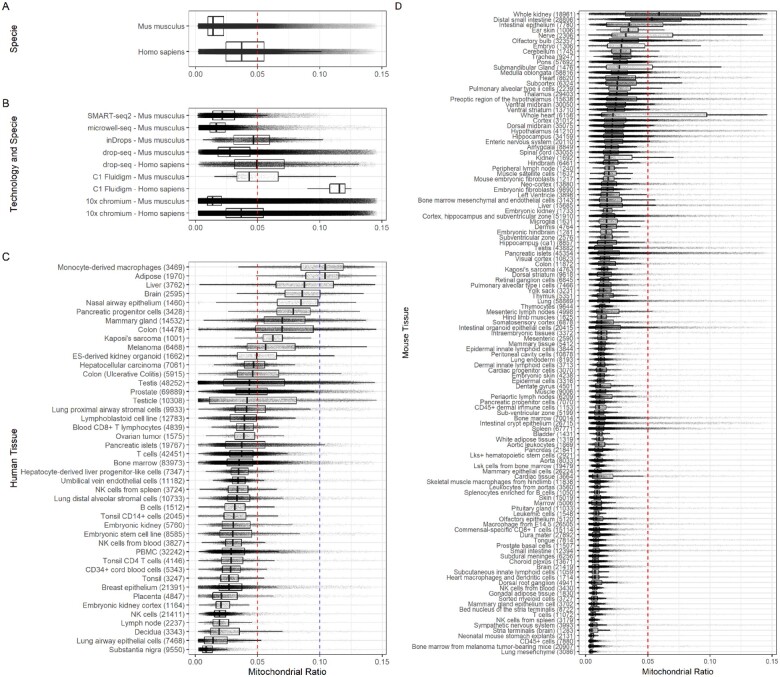
With the cleaned dataset, we estimated that the mtDNA% per cell is distributed between the minimum of 0.17% and the maximum of 14.64%, considerably lower than the upper limit previously reported (up to 30% in heart) using the bulk RNA-seq generated by the Illumina Body Tissue Atlas (Mercer et al., 2011). Next, we performed a comparison to evaluate whether there is a difference in the average mtDNA% cross different species. The PanglaoDB database contains human and mouse datasets; therefore, our comparison was between human and mouse. We performed the Welch two-sample t-test and use the Wilcoxon-sum rank test to validate the results. Both tests converged to the same conclusion, that is, the average mtDNA% in human cells is significative higher than that in mice cells (, in both cases) as is displayed inFigure 1A.
Fig 1.
Boxplots showing the differences in mtDNA% across species, technologies and tissues. Each dot represents a cell; the red line is the early established 5% threshold, and the blue line is the 10% threshold for human cells proposed here. In parenthesis (C and D), the number of cells in the stated tissue. (A) The difference in mtDNA% between human and mice cells. (B) The differences in mtDNA% between human and mice cells by the technology used to generate the data. (C) Boxplots of mtDNA% across 44 human tissues. (D) Boxplots of mtDNA% across 121 mouse tissues
Then, we compared the mitochondrial content between human and mouse data, stratified by the type of scRNA-seq technologies, by which the data are obtained. These technologies include drop-seq, C1 Fluidigm and 10× Genomics. Our results confirm our previous finding. In all cases wherever data allowed, no matter which technology is used, the same pattern was recovered. That is, human cells have significantly larger mtDNA% than mice cells (Fig. 1B). Most importantly, for all cases where mitochondrial content in humans was evaluated, the 75th percentile was located above the threshold, suggesting that the early defined 5% is not appropriate for human cells. Note that 91.3% () of cells analyzed here were processed using the 10× Genomics chromium system. Next, we decided to perform the comparison of the mitochondrial content between tissues and cell types using only the data generated using the 10× Genomics technology.
For humans, we identified 44 tissues, for which more than 1000 cells are available in the database. From those 44 tissues, 13 (29.5%) showed an average mtDNA% significantly higher than 5%. The 13 human tissues are nasal airway epithelium, monocyte-derived macrophages, testicle, colon (ulcerative colitis), liver, colon, melanoma, mammary gland, ES-derived kidney organoid, pancreatic progenitor cells, adipose, Kaposi’s sarcoma and brain. However, as displayed in Figure 1C, 18 of the 44 human tissues (40%) have a portion of the interquartile range over the 5% threshold. Only two of them, monocyte-derived macrophages and adipose, have an average mtDNA% higher than 10%. This result supports our observation that the early defined 5% is not appropriate for human tissues. We conclude that the new standardized threshold for human tissues should be 10% instead. At the cell-type level, we found similar patterns. From 37 different cell types with more than 1000 cells derived from human samples, 13 of them (35.1%) have an average mtDNA% greater than 5%, but none of them have an average mtDNA% greater than 10% (Supplementary Fig. S3). The 13 cell types are hepatocytes, epithelial cells, neutrophils, cholangiocytes, smooth muscle cells, keratinocytes, Langerhans cells, spermatocytes, ductal cells, beta cells, luminal epithelial cells, macrophages and embryonic stem cells. Furthermore, only 4 of them (epithelial cells, Langerhans cells, spermatocytes and macrophages) have a portion of the interquartile range above the 10% threshold. For mice, when the mtDNA% was compared across the cell types with at least 1000 cells reported in the database, 7 of 74 cell types showed an average mtDNA% greater than 5%. The 7 cell types are proximal tubule cells, distal tubule cells, hepatocyte, cardiomyocytes, Leydig cells, intercalated cells and choroid plexus cell (Supplementary Fig. S4). In contrast to the identified 44 human tissues, there are many more mouse tissues (121) with more than 1000 cells reported in the database. Among them, only 3 (2.5%) showed an average mtDNA% significantly higher than 5% (whole kidney, whole heart and distal small intestine). Furthermore, only 6 mouse tissues (whole kidney, intestinal epithelium, whole heart, nerve, distal small intestine and submandibular gland) have a portion of the interquartile range over the 5% threshold (Fig. 1D). These findings indicate that the 5% threshold early proposed in the field is an appropriate standardized threshold for mouse tissues.
To evaluate the effect of mtDNA% in the analysis of single-cell RNA-seq data, we downloaded three datasets from the PanglaoDB database using accessions: SRS3703557, SRS3545826 and SRS2397417. The first dataset contains 9238 cells from the mouse heart, the second 7448 cells from mouse lung and the third 9057 cells from the human umbilical vein.
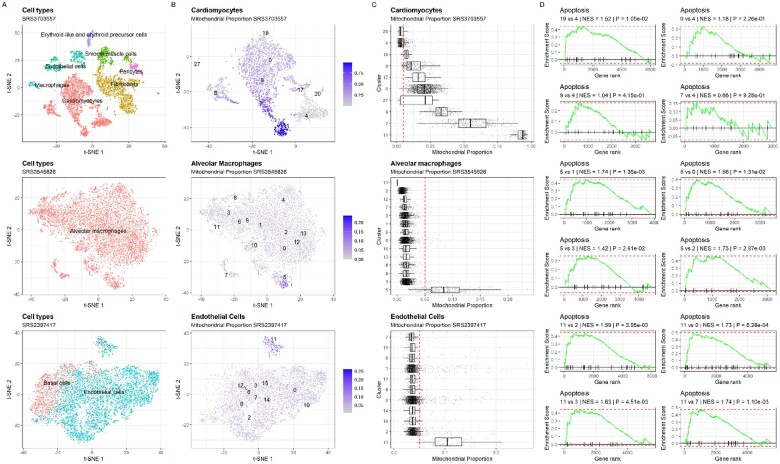
First, we evaluated the effect of genes encoded in the mitochondrial genome (for short, mtGenes) on the cell clustering results. We compared the low-dimensional representations generated by t-SNE using the PCA result as a prior with and without the mtGenes. We found that, in all three examples, the mitochondrial content does not affect significantly the structure of the data in low dimensional representation, allowing to recover clearly in both cases (with and without mtGenes) the clusters reported by the PanglaoDB database (Fig. 2 and Supplementary Fig. S5). These results confirm findings previously reported in an evaluation study of different computational pipelines for scRNA-seq data preprocessing (Germain et al., 2020). We also found that even without considering the mtGenes for the generation of the low-dimensional representation, low-quality cells with high mtDNA% tend to cluster together (Fig. 2B; Clusters 19, 0, 9, 7 and 11 in SRS3703557; cluster 5 in SRS3545826 and cluster 11 in SRS2397417).
Fig 2.
Case examples showing the effect of omitting the mtDNA% QC filter in the analysis of scRNA-seq data. (A) t-SNE representation of all the cell populations included in the dataset generated by excluding the mitochondrial genes from the list of highly variable genes before principal component analysis (PCA). Each dot represents a cell and they are colored by cell type. (B) t-SNE representation of cell type used as an example colored in the function of the mtDNA% in each cell. Clusters reported by the PanglaoDB are labeled. (C) Boxplot showing the distribution of the mtDNA% across clusters. The red line is the early established 5% threshold. (D) GSEA analysis of the Apoptosis pathway between clusters with a high proportion of low-quality cells and others containing high-quality cells
Next, we evaluated the significance of the threshold to identify low-quality cells. For each example, after identifying the clusters containing cells with high mtDNA% of a cell type, we used MAST to compare their expression profiles against other clusters with most of the cells below the threshold. The fold-change values reported by MAST were used as the input of GSEA analysis to test for the significance of the Apoptosis pathway in the KEGG database.
For the first example, we focused on cardiomyocytes, a cell type associated with high mtDNA% with nine clusters reported by the PanglaoDB database (Fig. 2C) for this dataset. We compared cells with a higher mtDNA% level in clusters 19, 0, 9, 7 and 11, which form a larger cluster, against cells with a lower mtDNA% level in cluster 4. We found that the number of genes with detectable expression decreases with the increase of mtDNA% in cells (as shown by the x-axes of the first two rows of subplots in Fig. 2D). This anticorrelation is expected as the increased mtDNA% is likely to be associated with cell breakout events. When a breakout occurs (due to the differences in copy numbers given by a single nucleus and several mitochondria by cell), generate an increase in the abundance of mitochondrial content, and reducing the reads that will be mapped to nuclear genes, resulting in fewer genes’ expression detected. Indeed, we found even a small increase of the mitochondrial content (comparing cluster 19 and cluster 0 versus cluster 4) led to a huge decrease in the number of expressed genes (>6000 genes versus ∼5000 genes, see the first row of subplots in Fig. 2D). The number of genes included in the GSEA analysis, in turn, influences the value of Normalized Enrichment Score (NES), which is used to assess the significance of the apoptosis pathway. Despite this influence, a positive NES value was recovered in all the cases for the tested cardiomyocytes clusters, as well as in the other two examples of mouse alveolar macrophages (cluster 5 against others) and human endothelial cells (cluster 11 against others), suggesting a consistently higher expression (positive log2 fold-change) of apoptotic pathway genes among cells with mtDNA% above the threshold (Fig. 2D).
In summary, we reported a new set of mtDNA% reference values across human and mice tissues and cell types for scRNA-seq QC (Supplementary Tables S1–S4). Based on our analytical results, we suggest a standardized mtDNA% threshold of 10% for scRNA-seq QC of human samples. For mouse samples, we found that the early defined threshold of 5% accurately discriminates between healthy and low-quality cells, bringing to evidence that under a well-performed scRNA-seq QC, clusters containing cells with high mtDNA% exhibiting signatures of apoptosis, like those shown in the example datasets, should be excluded from being used to make biological interpretations. Thus, we suggest that all published mouse studies, in which scRNA-seq QC was based on the mtDNA% value greater than 5%, should be re-evaluated because the use of any mtDNA% higher than 5% is likely to be an overshoot over the threshold, resulting in apoptotic cells being utilized in the subsequent analyses.
Supplementary Material
Acknowledgements
The authors thank Dr. Chapkin and lab members from the Texas A&M nutrition department for their thoughtful questions that motivated this article.
Funding
This work was supported by Texas A&M University 2019 X-grant for J.J.C.
D.O. was supported by the 2020 Award of Texas A&M Institute of Data Science (TAMIDS) Data Resource Development Program.
Conflict of Interest: none declared.
Contributor Information
Daniel Osorio, Department of Veterinary Integrative Biosciences, Texas A&M University, College Station, TX 77843-4458, USA.
James J Cai, Department of Veterinary Integrative Biosciences, Texas A&M University, College Station, TX 77843-4458, USA; Department of Electrical & Computer Engineering, Texas A&M University, College Station, TX 77843-4458, USA; Interdisciplinary Program of Genetics, Texas A&M University, College Station, TX 77843-4458, USA.
References
- AlJanahi A.A. et al. (2018) An introduction to the analysis of single-cell RNA-sequencing data. Mol. Ther. Methods Clin. Dev., 10, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G. et al. (2015) MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol., 16, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen,et O.al. (2019) PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford), 2019, baz046, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain P.-L. et al. (2020) pipeComp, a general framework for the evaluation of computational pipelines, reveals performant single-cell RNA-seq preprocessing tools. bioRxiv 2020.02.02.930578. [DOI] [PMC free article] [PubMed]
- Guantes R. et al. (2015) Global variability in gene expression and alternative splicing is modulated by mitochondrial content. Genome Res., 25, 633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B. et al. (2018) Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med., 50, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilicic T. et al. (2016) Classification of low quality cells from single-cell RNA-seq data. Genome Biol., 17, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji F., Sadreyev R.I. (2019) Single-cell RNA-seq: introduction to bioinformatics analysis. Curr. Protoc. Mol. Biol., 127, e92. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G. et al. (2019) Fast gene set enrichment analysis.bioRxiv 060012.
- Lang D.T. and CRAN Team. (2012) XML: Tools for parsing and generating XML within R and S-Plus. R package version 3.98-1.20. https://CRAN.R-project.org/package=XML
- Luecken M.D., Theis F.J. (2019) Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol., 15, e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S. et al. (2018) Single-cell RNA sequencing of adult mouse testes. Sci. Data, 5, 180192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A.T. et al. (2016) A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res, 5, 2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A. et al. (2019) EnsembleKQC: an unsupervised ensemble learning method for quality control of single cell RNA-seq sequencing data. In: International Conference on Intelligent Computing. 15th International Conference, ICIC 2019, Nanchang, China. Springer, pp. 493–504.
- McCarthy D.J. et al. (2017) Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics, 33, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T.R. et al. (2011) The human mitochondrial transcriptome. Cell, 146, 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir R. et al. (2016) Mitochondrial content is central to nuclear gene expression: profound implications for human health. Bioessays, 38, 150–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez-Rueda D. et al. (2020) Apoptotic cell exclusion and bias-free single-cell selection are important quality control requirements for successful single-cell sequencing applications. Cytometry A, 97, 156–167. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
- Sandberg R. (2014) Entering the era of single-cell transcriptomics in biology and medicine. Nat. Methods, 11, 22–24. [DOI] [PubMed] [Google Scholar]
- Satija R. et al. (2015) Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol., 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson V. et al. (2019) A curated database reveals trends in single-cell transcriptomics.bioRxiv 742304. [DOI] [PMC free article] [PubMed]
- Wickham H. et al. (2018) xml2: parse XML. R Package Version 1.2 .2. https://CRAN.R-project.org/package=xml2 [Google Scholar]
- Zhao Q. et al. (2002) A mitochondrial specific stress response in mammalian cells. EMBO J., 21, 4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.