Abstract
Exosomes with diameters of 30–150 nm are small membrane-bound vesicles secreted by a variety of cells. They play an important role in many biological processes, such as tumor-related immune response and intercellular signal transduction. Exosomes have been considered as emerging and noninvasive biomarkers for cancer diagnosis. Recently, a large number of optical and electrochemical biosensors have been proposed for sensitive detection of exosomes. To meet the increasing demands for ultrasensitive detection, nanomaterials have been integrated with various techniques as powerful components. Because of their intrinsic merits of biological compatibility, excellent physicochemical features and unique catalytic ability, nanomaterials have significantly improved the analytical performances of exosome biosensors. In this review, we summarized the recent progress in nanomaterials-based biosensors for the detection of cancer-derived exosomes, including fluorescence, colorimetry, surface plasmon resonance spectroscopy, surface enhanced Raman scattering spectroscopy, electrochemistry, electrochemiluminescence and so on.
Keywords: exosomes, nanomaterials, circulating tumor biomarkers, electrochemical biosensor, optical biosensor
Introduction
Cancer is the main cause of death, and its occurrence and development is a gradual and complicated process. Early diagnosis and treatment of cancer can enormously improve the survival chance of cancer patients. Extracellular vesicles (EVs) are secreted by various cell types and circulate in different body fluids. They were first discovered as “cell junks” about 40 years ago.1,2 The lack of specific and reliable markers makes the clear assignation of EV subtypes extraordinarily difficult. According to operational terms for EV subtypes proposed by the International Society for Extracellular Vesicles (ISEV), EVs can be classified into three groups based on their size: exosomes, small EVs (sEVs) (< 200 nm), medium EVs (mEVs), and large EVs (lEVs).3 Exosomes are nanosized extracellular vesicles (EVs) (30–200 nm). For the convenience of the reader, this review used a relatively broad term of “exosomes” to refer to a heterogeneous mixture of sEVs less than 200 nm in size because it is increasingly utilized in bioassays.4 Compared to circulating tumor cells and circulating DNA, exosomes exist in body fluids (such as serum, urine and ascites) with higher abundance and stability.5 Emerging evidences suggest that most types of cancer secret large numbers of exosomes that carry abundant molecular information stemming from parent tumor cells, including nucleic acids, proteins, bioactive lipids and metabolites.6,7 Exosomes can act as cellular messengers to deliver information between cells via endocytosis.8,9 They also play a key role in cancer metastasis and progression in the tumor microenvironment.10,11 Moreover, various unique cargos in exosomes represent meaningful physiological and pathological states of diseases.12,13 Therefore, exosomes have been recognized as the most reliable non-invasive biomarkers for the early diagnosis and cure of cancers.14–17
Various exosomes-containing complex biological fluids have a plenty of nonvesicular macromolecules, such as proteins, proteases, nuclease and RNA complexes, which may interfere with the analysis of exosomes. For example, proteases and nuclease can digest the biorecognition elements such as antibodies and aptamers. Thus, the growing interests in exosomes and their potential applications in cancer detection have pushed researchers to develop various isolation and detection techniques. Normally, conventional isolation techniques, including ultracentrifugation, density gradient centrifugation and ultrafiltration, are mainly based on the physical properties of exosomes (eg size and density).5,18 However, these techniques confront of some problems, such as low purity, tedious procedures and requirement of expensive instruments. Traditional detection techniques utilized to quantify the isolated exosomes include nanoparticle tracking analysis (NTA), flow cytometry, Western blot, dynamic light scattering, scanning electron microscope (SEM), and transmission electron microscope (TEM).2,19–22 Although these methods have been popularly used, the problems of low reproducibility, time consuming, large sample demand and low sensitivity limit their further applications.
Over the past years, many types of proteins, such as tetraspanins (eg CD9, CD63, CD81 and CD82), adhesion molecules (eg integrins and lactadherin) and lipid rafts (eg cholesterol, phosphatidylserine and ceramide), have been found on the membrane of all types of exosomes.23,24 These biomolecules can be utilized as the targets for the detection of total exosomes. However, exosomes derived from various tumor cells exhibit different cancers-associated antigens on the surface. These distinct antigens have been used as the biomarkers for the determination of certain cancer-derived exosomes.25 For example, MCF-7-secreted exosomes with highly expressed MUC1 on the surface.26 Protein tyrosine kinase 7 and CD147 are over-expressed on the surface of human leukemic lymphoblasts (CCRF-CEM) and colorectal cancer cellular exosomes, respectively.27,28 These proteins are the promising markers for the isolation and detection of exosomes with the aid of biorecognition elements.29,30 By modifying magnetic beads (MBs), chips or microfluidics with antibodies toward exosome membrane markers, immunoaffinity-capture-based techniques are proposed for the isolation and enrichment of exosomes with high selectivity and specificity as well as intact biological activity.23,31–44 However, the expensive cost and instability of antibodies limit their practical applications. Aptamers and peptides are screened to act as biorecognition elements for the isolation and detection of exosomes by the virtue of low cost, small size and relatively excellent stability.45–47 Besides, exosomal glycans provide a valuable route to label exosomes through the interaction of lectin and glycan.48,49 Lipophilic cholesterol anchors can penetrate into the lipid bilayers for membrane modification, which have been regarded as the promising candidates for labeling of exosomes.50,51 In contrast to physical property-based isolation techniques, affinity-based isolation techniques exhibit much higher enrichment efficiency, facilitating the sensitive detection of exosomes.
In recent years, numerous novel biosensors with high sensitivity and efficiency have been established for the determination of exosomes by specific recognition, including colorimetry, fluorescence, surface plasmon resonance (SPR), surface enhanced Raman scattering (SERS) spectroscopy, mass spectrometry and electrochemical, electrochemiluminescent (ECL) and photoelectrochemical (PEC) assays.4,52–59 Moreover, many works based on microfluidic devices implemented with optical or electrochemical techniques have been reported for the comprehensive assays of exosomes.60–64
With the growing development of nanotechnology, myriad nanomaterials and nanostructures have made a great impact on biosensing. Significant advances have made it possible to controllably prepare nanomaterials with various chemical composition, morphology and physicochemical characteristics. For example, nanomaterials with wonderful luminescence properties have been an important alternative to traditional dyes in optical assays, because of their excellent merits of adjustable emission wavelength, high luminescence quantum yield and good photostability. Thanks to the interesting size and shape-dependent localized surface plasmon resonance phenomenon, noble metal nanoparticles (NPs), especially gold and silver, have been widely used to enhance the signal intensity in SPR and SERS assays. Carbon-based nanomaterials (eg, carbon nanotubes and graphene oxide) with a high surface-to-volume ratio and high electrical conductivity are always employed for electrode modification to accelerate the electron transfer and increase the electrode surface area. Moreover, recently, the photothermal and enzyme-mimic properties of nanomaterials have gained considerable interest toward the development of portable bioassays. Various nanomaterials-based signal amplification strategies, coupled with different detection techniques, have been developed for the ultrasensitive detection of biomolecules, including DNA/RNA, proteins, exosomes and cells. In exosomes detection, there are two mainly objectives, including improving the capture of exosomes and enhancing the performance of detection methods. For the former, magnetic beads (MBs) as the classical materials have been increasingly used to selectively capture exosomes from clinical samples, when being decorated with antibodies or aptamers.
In this review, the recent progress in nanomaterials-based biosensors for exosome detection was comprehensively summarized. The detection techniques mainly cover optical and electrochemical assays (Tables 1 and 2). Nanomaterials are involved in semiconductor quantum dots (QDs), metal NPs, metal oxides and sulfides, and carbon-based nanomaterials. The intent of this review is to impart insights into the versatile roles of nanomaterials in assays, and illustrate their potential benefits in further applications.
Table 1.
An Overview on Nanomaterials-Based Optical Methods for Exosome Detection
| Methods | Type of Nanomaterials | Functions | Cancer Types | Target Biomarkers | Response Time (min) | Detection Limit (Particles/μL) | Linear Range (Particles/μL) | Ref. |
|---|---|---|---|---|---|---|---|---|
| Fluorescence | Biorecognition elements-modified MNPs | Capturing exosomes and transforming the detection of exosomes into the detection of DNA with DNA-based signal amplification | Liver cancer | CD63 | Not reported | 0.1 | 1–1 × 104 | [83] |
| Not reported | CD63 and lipid bilayer | 160 | 1.29 × 103 | 5.5 × 103–1.1 × 107 | [84] | |||
| Lung cancer | CD63 | 65 | 1 × 105 | 1 × 107–1 × 109 | [85] | |||
| Lung cancer | CD63 | 95 | 0.1 | 0.3–1 × 104 | [86] | |||
| Liver cancer | CD63 | 255 | 1.16 × 103 | 1.75 × 103–7 × 106 | [90] | |||
| Cervical cancer | CD63 and lipid bilayer | 245 | 480 | 1.66 × 103–1.66 × 106 | [91] | |||
| Prostate cancer | PSMA | 210 | 1 × 102 | 1 × 103–1 × 108 | [92] | |||
| QDs | Fluorescence labels | Lung cancer | EpCAM | 60 | 0.013 | 5 × 102–5 ×109 | [94] | |
| Oral squamous Cell carcinoma | CD63 | 30 | 5 × 102 | 5.0×102 – 1.0×105 | [95] | |||
| Pancreatic cancer | GPC-1 | 90 | Not reported | 1.0×104 – 1.0×106 | [96] | |||
| Human tongue squamous cell carcinoma | Lipid bilayer | 65 | 2 × 103 | 4 × 103–2 × 105 | [97] | |||
| CuNPs | Liver cancer | CD63 and lipid bilayer | 120 | 4.8 × 104 | 7.5 × 104–1.5 × 107 | [98] | ||
| AuNPs | Nanoquenchers | Leukemia cell | CD63 and nucleolin | 180 | 1 × 102 | 1 × 103–1 × 105 | [88] | |
| GO | Breast cancer | PSMA | 40 | 1.6 × 102 | 1.6 × 102–1.6 × 105 | [101] | ||
| Colorectal cancer | CD63 and EpCAM | 40 | 2.1 × 104 | 3 × 104–6 × 105 | [102] | |||
| Breast cancer | GPC-1 | 128 | 6.56 × 104 | 7.8 × 104–3.9 × 109 | [103] | |||
| MOS2-MWCNTs | Breast cancer | CD63 | 70 | 1.48 × 103 | 0–1.11 × 104 | [104] | ||
| Ti3C2 MXenes | Nanoquenchers and luminescent emitters | Breast cancer | CD63 | 70 | 1.4 | 10–1 × 106 | [105] | |
| AuNRs UCNPs |
Quenching the luminescence of UCNPs by AuNRs | Breast cancer | CD63 | 250 | 8.2 | 20–2.0 × 106 | [99] | |
| Colorimetry | AuNPs | Nanocarriers for enzymes | Colorectal cancer | A33 and EpCAM | 220 | 6.7 × 103 | 9.75 × 103–1.95 × 106 | [114] |
| AuNPs | Color indicators | Nasopharyngeal carcinoma | CD63, CD9, CD81, LMP1 and EGFR | 120 | 0.1 | 0.1–1 × 105 | [122] | |
| AuNRs | Breast cancer | CD63 and lipid bilayer | Not reported | 1.6 × 102 | 1.4 × 103–2.8×105 | [127] | ||
| Au NBP@ MnO2 NSs |
Breast cancer | CD63 | 155 | 1.35 × 102 | 8.5 × 102–8.5 × 104 | [128] | ||
| C-IONPs | Nanozymes | Ovarian cancer | CD63 | 1.25 × 103 | 6.25 × 102–1.0 × 104 | [129] | ||
| g-C3N4 nanosheets | Peroxidase-mimetic enzyme | Breast cancer | CD63 | 30 | 1.352 × 106 | 1.9 × 106–3.38 × 107 | [132] | |
| SWCNTs | Breast cancer | CD63 | 40 | 5.2 × 105 | 1.84 × 106–2.21 × 107 | [133] | ||
| CuCo2O4 nanorods | Breast cancer | CD63 | 160 | 4.5 × 103 | 5.6 × 104–8.9 × 105 | [134] | ||
| SPR | GO | Chip substrate | Lung cancer | PD-L1 | 50 | 0.02 | 10–1.0 × 105 | [145] |
| MNPs | Nanolabels | Lung cancer | CD63, EGFR and EpCAM | 55 | 2.37 × 104 | 3.125 × 104–100 × 104 | [148] | |
| AuNPs | Breast cancer | CD63 | 155 | 5 | Not reported | [151] | ||
| SERS | AuNPs | Raman reporters with LSPR | Pancreatic cancer | CD63, GPC-1, EpCAM and CD44V6 | 150 | Not reported | Not reported | [164] |
| Polydopamine-encapsulated Au@Ag NPs | Pancreatic cancer | CD63, GPC-1, EGFR, EpCAM, and MIF | 150 | Not reported | 0.544–27.2 | [161] | ||
| AuNS@4-MBA@Au | Hepatocellular Carcinoma | CD9 and lipid bilayer | 50 | 27 | 40–4.0 × 107 | [162] | ||
| Gold-silver-silver core shell-shell nanotrepangs | Hepatocellular Carcinoma | PSMA, Her2 and AFP | 120 | 35 | 1–1 × 107 | [168] | ||
| TP-DNA@AuNPs | Electromagnetic hot spots to enhance Raman signals | Breast cancer | EpCAM | 150 | 110 | 1 × 103–1 × 107 | [169] |
Abbreviations: QDs, quantum dots; CuNPs, copper nanoparticles; AuNPs, gold nanoparticles; GO, graphene oxide; MWCNTs, multiwall carbon nanotubes; AuNRs, gold nanorods; UCNPs, upconversion nanoparticles; Au NBP@MnO2 NSs, gold nanobipyramid@MnO2 nanosheet nanostructures; C-IONPs, carboxyl group-functionalized iron oxide nanoparticles; SWCNTs, single-walled carbon nanotubes; Au@PDA NPs, polydopamine-functionalized gold nanoparticle; TP-DNA, triangular pyramid DNA; AuNS@4-MBA@Au, gold nanostar@4-mercaptobenzoic acid@nanoshell structures; PSMA, prostate-specific membrane antigen; GPC-1, glypican-1; EpCAM, epithelial cell adhesion molecule; LMP1, latent membrane protein 1; EGFR, epidermal growth factor receptor; PD-L1, programmed death ligand-1; CD44V6, CD44 variant isoform 6; MIF, migration inhibitory factor.
Table 2.
An Overview on Nanomaterials-Based Electrochemical Methods for Exosome Detection
| Methods | Type of Nanomaterials | Functions | Cancer Types | Target Biomarkers | Response Time (min) | Detection Limit (Particles/μL) | Linear Range (Particles/μL) | Ref. |
|---|---|---|---|---|---|---|---|---|
| Electrochemistry (direct detection) | AuNPs | Improving the large surface area, accelerating electrical conductivity and facilitating the immobilization of biorecognition elements | Breast cancer | CD63 | 180 | 4.82 × 102 | 5 × 102–5 × 103 | [181] |
| BPNSs and MOFs | Breast cancer | CD63 | Not reported | 1 × 102 | 1.3 × 102–2.6 × 105 | [182] | ||
| Electrochemistry (sandwich detection) | AuNPs | Nanocarriers for enzymes, electroactive molecules | Liver cancer | CD63 | 255 | 16.6 | 21.6–7.5 × 104 | [189] |
| Zr-MOF | Glioblastoma | EGFR, EGFRvIII, and phosphate groups outside of exosomes | 150 | 7.83 × 103 | 9.5 × 103–1.9 × 107 | [190] | ||
| Zr-MOF and ZIF-8 | Breast cancer | CD63 and phosphate groups outside of exosomes | 190 | 0.3 | 1–1 × 105 | [191] | ||
| ZIF-8 | Breast cancer | CD63 and PD-L1 | 245 | 0.334 | 1–1 × 107 | [192] | ||
| COFs@Au NPs | Colorectal cancer | CD63 and amino acid residues outside of exosomes | 170 | 1.6 × 102 | 5 × 102–5 × 107 | [193] | ||
| Au-NPFe2O3NC | Nanozymes to catalyze the redox reaction | Placental choriocarcinoma | CD63 and PLAP | 90 | 1 | 1–1 × 104 | [194] | |
| Ti3C2 MXene@PB | Showing electroactive properties or consisting of electroactive ions | Ovarian cancer | CD63 | 120 | 2.29 × 102 | 5 × 102–5.0 × 105 | [195] | |
| CdSe QDs | Colorectal adenocarcinoma | CD63, HER-2 and FAM134B | 130 | 1 × 102 | 1 × 102–1 × 107 | [196] | ||
| Electrochemiluminescence | Ru(II) markers-loaded SiNPs | Nanomaterials acts as nanocarriers for molecules | Hepatocellular carcinoma | CD63 | 120 | 60 | 2 × 102–7.5 ×104 | [212] |
| CdS:Eu QDs | Electrochemiluminescence emitter | Breast cancer | CD63 | 240 | 74.1 | 3.4 × 102–1.7 × 104 | [210] | |
| CdS QDs@CaCO3 | Breast cancer | EpCAM | 180 | 21 | 50–1 × 105 | [212] | ||
| g-C3N4@Galinstan-PDA | Cervical cancer | GPC-1 | 120 | 31 | 50–1 × 105 | [211] | ||
| BPQDs@ Ti3C2 MXene | Coreactants | Breast cancer | CD63 and EpCAM | 100 | 37 | 1.1 × 102–1.1 × 107 | [214] | |
| AuNPs@ Ti3C2 MXene | Nanocatalysts | Cervical cancer | CD63 | 240 | 30 | 1 × 102–1 × 105 | [215] | |
| Ti3C2 MXene | Breast cancer | EpCAM | 240 | 1.25 × 102 | 5 × 102–5 × 106 | [213] | ||
| AuNPs@ g-C3N4 | Ovarian cancer | phosphatidylserine | 120 | 39 | 1 × 102–1 × 107 | [216] |
Abbreviations: BPNSs, black phosphorus nanosheets; AuNPs, gold nanoparticles; MOFs, metal organic frameworks; COFs, covalent organic frameworks; Au-NPFe2O3NC, gold-loaded ferric oxide nanocubes; PB, Prussian blue; QDs, quantum dots; PDA, polydopamine; BPQDs, black phosphorus quantum dots; EGFR, epidermal growth factor receptor; EGFRvIII, epidermal growth factor receptor variant (v) III mutation; PD-L1, programmed death ligand-1; PLAP, placental alkaline phosphatase; EpCAM, epithelial cell adhesion molecule; GPC-1, glypican-1.
Nanomaterials-Based Optical Biosensors for Exosome Detection
Fluorescence Biosensors
Fluorescence biosensors have great advantages of simple operation, comparable sensitivity, and multiplex target detection capability. Exosome can be labeled with dyes or dye-modified biorecognition elements through various targeting strategies and then determined by fluorescence imaging or spectroscopy.50,65–72 However, the low fluorescence intensities of dyes always limits the sensitivity for exosome detection.73 Thus, several signal amplification strategies have been proposed to improve the detection sensitivity.74,75 For example, β-galactosidase-labeled antibody was used to label the captured exosomes in which β-galactosidase acted as the reporter enzyme to catalyze the decomposition of fluorescein-di-β-D-galactopyranoside, generating a strong fluorescence signal.41,76 Combined with nano-interface-based microfluidic platforms, exosomes were effectively enriched and sensitively detected.
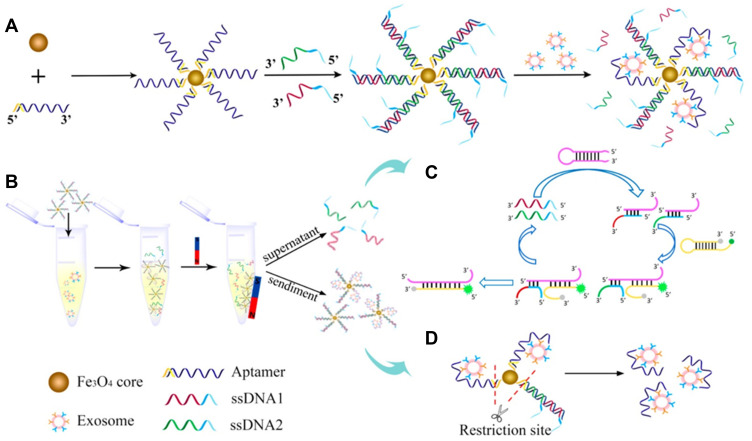
For homogeneous fluorescence detection, it is one of the most powerful strategies to convert the detection of exosomes into the detection of DNA or others, generally producing a “one-to-many” amplification effect.77–81 Owing to the flexible structure, the corresponding DNA can be ultrasensitively and homogeneously detected by previously reported DNA-based signal amplification methods, such as terminal deoxynucleotidyl transferase (TdT)-mediated polymerization and hybridization chain reaction (HCR).82–89 For example, Gao et al reported a dual signal amplification method for indirect detection of exosomes based on the catalytic hairpin DNA cascade reaction (HDCR) and the self-assembly of DNA dendrimer on the surface of gold nanoparticles (Au NPs).90 In this work, streptavidin (SA)-modified MBs were labeled with biotin-modified CD63 aptamer and then bound with probe S through the hybridization. After the addition of exosomes, aptamers bound with CD63 on exosomes and the probe S was released to trigger the HDCR on the nanoparticle surface. Then, the opened metastable hairpin (HP) DNA probes captured the fluorescently-labeled DNA dendrimers. After several rounds of Y-shaped DNA assembly, the complexes of AuNPs, HP and DNA dendrimers were separated. With the aid of β-mercaptoethanol, the fluorescently-labeled dendrimers were released and the fluorescence signal was recorded for the determination of exosomes. This method showed an increased signal-to-noise ratio and had a linear detection range of 1.75×103 – 7.0×106 particles/μL. Pan et al reported a steric hindrance-controlled signal-amplified fluorescent strategy for exosome detection.91 In the absence of exosome, cholesterol-conjugated DNA 1 could hybridize with SA-modified DNA 2 into dsDNA with blunt ends. The formed dsDNA could not been recognized and extended by TdT enzyme because of the absence of single-stranded initiator (more than three deoxynucleotide residues). However, when exosomes were added, DNA 1 was inserted into the lipid membrane through the hydrophobic interaction between cholesterol and lipid bilayer. The huge steric hindrance of exosomes strongly inhibited the hybridization of DNA 1 and SA-modified DNA 2. Therefore, SA-modified DNA 2 could be extended by TdT enzyme to generate abundant G-quadruplex structures, increasing the fluorescence intensity. The “signal-on” method maintained high sensitivity and excellent selectivity for the assay of complex samples. Li et al developed a reversible nanoplatform for fluorescent detection of urinary exosomes by using a superparamagnetic conjunction and molecular beacon.92 As shown in Figure 1, prostate specific membrane antigen (PSMA) aptamer was modified on the surface of superparamagnetic NPs and then hybridized with two ssDNA strands with the decreased hybridization energy. Exosome bound to the aptamer with high affinity resulted in the release of double amounts of ssDNA to initiate the amplification cycle with two hairpin DNA strands. Molecular beacon HP2 was opened and the fluorescence of probe was recovered. The two released ssDNA sequences with low concentration initiated the amplification cycle with two hairpin DNA strands. Numerous molecular beacon HP2 were opened and the fluorescence of probe was recovered. The detection limit of this method was 100 particles/μL for the assay of urine samples.
Figure 1.
Schematic of SMC-MB platform. (A) Construction of SMC-MB platform. (B) Procedure of SMC-MB platform in exosomes analysis. (C) Principle of nonenzymatic amplification cycle. (D) Exosomes purification by restriction enzyme. Reprinted with permission from Li P, Yu X, Han W, et al. Ultrasensitive and reversible nanoplatform of urinary exosomes for prostate cancer diagnosis. ACS Sens. 2019;4:1433–1441. Copyright 2019 American Chemical Society.92
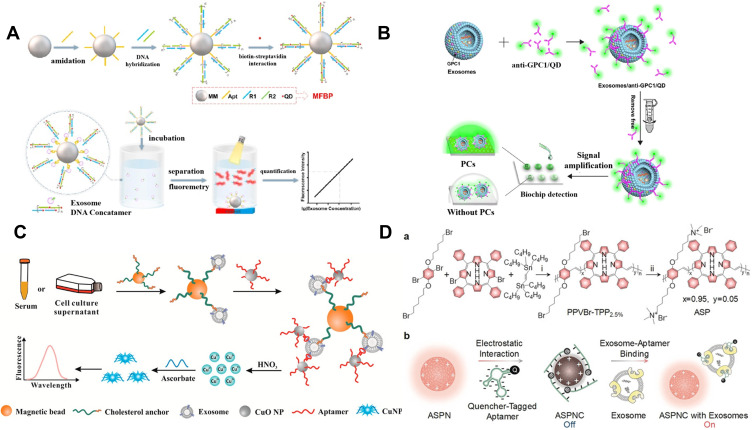
In contrast to traditional fluorescence dyes, QDs possess size/component-tunable luminescence and excellent resistance against photobleaching, which have been widely applied in bioimaging, luminescent biolabels and light-emitting devices.93,94 Wu et al proposed a “one-step” strategy for the detection of exosomes using aptamers as the biorecognition elements and QDs as the signal-amplified reporters (Figure 2A).95 CD63 aptamer was anchored on the surface of magnetic microspheres (MMs) and tethered to the self-assembled DNA concatemers. Then, streptavidin (SA)-conjugated QDs were used to label the biotin-modified DNA concatemers. Exosome preferentially bound to the aptamer and induced the release of QDs-labeled DNA concatemers. After the magnetic separation, the fluorescence signal in the supernatant was monitored for exosome detection. Zhang et al prepared a biomimetic periodic nanostructure-based diagnostic biochip for exosome detection using QDs.96 As displayed in Figure 2B, glypican-1 (GPC1) antibody-modified QDs were utilized to label exosomes. When the solution was dropped on the photonic crystals-coated biochip, the fluorescence was significantly amplified. In addition, QDs-embedded silica NPs were also used to extracellular vesicles via membrane biotinylation strategy in lateral flow assay (LFA).97
Figure 2.
(A) Schematic of magnetic and fluorescent bio-probes (MFBP) constructing process and sensing principle of MFBP-based quantification of exosomes. Reprinted with permission from Wu M, Chen Z, Xie Q, et al. One-step quantification of salivary exosomes based on combined aptamer recognition and quantum dot signal amplification. Biosens Bioelectron. 2021;171:112733–112742. Copyright 2021 Elsevier B.V.95 (B) Schematic of photonic crystals-assisted signal amplification for measurement of tumor-derived exosomes. Reprinted with permission from Zhang J, Zhu Y, Shi J, et al. Sensitive signal amplifying a diagnostic biochip based on a biomimetic periodic nanostructure for detecting cancer exosomes. ACS Appl Mater Interfaces. 2020;12:33473–33482. Copyright 2020 American Chemical Society.96 (C) Schematic of the proposed method for exosome detection based on a copper-mediated signal amplification strategy. Reprinted with permission from He F, Wang J, Yin BC, Ye BC. Quantification of exosome based on a copper-mediated signal amplification strategy. Anal Chem. 2018;90:8072–8079. Copyright 2018 American Chemical Society.98 (D) Schematic of design and sensing mechanism of ASPNC. (a) Synthetic route of ASP. Reagents and conditions: i) tris(dibenzylideneacetone)dipalladium(0) [Pd2(dba)3], tri(p-tolyl)phosphine (TPTP), chlorobenzene, 100°C, 24 h; ii) trimethylamine, tetrahydrofuran (THF), methanol, 24 h. (b) Illustration of the formation of ASPNC and the afterglow detection of exosomes. Reprinted with permission from Lyu Y, Cui D, Huang J, Fan W, Miao Y, Pu K. Near-infrared afterglow semiconducting nano-polycomplexes for the multiplex differentiation of cancer exosomes. Angew Chem Int Ed. 2019;58:4983–4987. Copyright 2019 Wiley-VCH.69
Metal nanoclusters (such as Au, Ag and Cu) with ultrasmall size possess excellent fluorescent properties. Ye’s group proposed a copper-mediated signal-amplified method to quantify exosomes (Figure 2C).98 In this work, cholesterol-modified MBs were used to capture exosomes via the hydrophobic interaction between cholesterol group and lipid membrane. CD63 aptamer-modified CuO NPs were utilized to label exosome and Cu2+ ions were released under acidolysis after the magnetic separation. Numerous released Cu2+ ions could be reduced into fluorescent Cu nanoclusters with poly(thymine) as the template. Furthermore, Lyu et al constructed a luminescent nanosensor for exosome detection by using an afterglow semiconducting polyelectrolyte nanocomplex (ASPNC).69 As displayed in Figure 2D, the backbone of poly(phenylenevinylene) (PPV) modified with cationic quaternary ammonium groups was conjugated with tetraphenylporphyrin (TPP) for the red-shift of the emission and the enhancement of the afterglow signal. The positively charged ASPNC could adsorbed the quencher the black hole quencher 2 (BHQ-2)-labeled aptamer through the electrostatic interaction and the afterglow and fluorescence signal were quenched through the efficient electron transfer between the PPV backbone and BHQ-2. In the presence of exosomes, the specific and strong affinity between exosomal protein and BHQ-2-labeled aptamer resulted in the desorption of aptamer from the ASPNC. Consequently, the afterglow and fluorescence signal were restored.
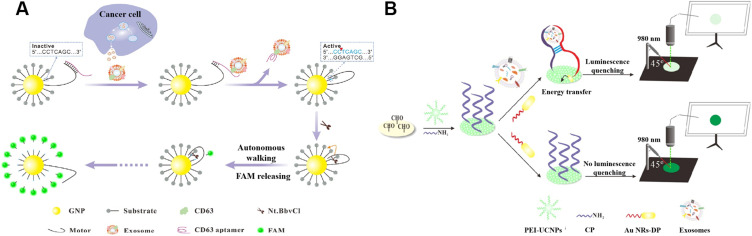
Nanomaterials with excellent fluorescent quenching ability are attractive to develop “on-off” biosensors. Zhang’s reported a dual-signal amplification platform for the analysis of leukemia-derived exosomes based on rolling circle amplification (RCA) and nicking endonuclease-assisted target recycling.88 In addition, Yu et al proposed a 3D DNA motor-based platform for the detection of exosomes by using AuNPs as the tracks.99 As shown in Figure 3A, AuNPs were modified with fluorescein-labeled substrate strands and aptamer-locked motor strands. In the presence of exosomes, the aptamer bound to the target protein and the motor strand was unlocked to trigger the DNA motor process. Powered by restriction endonuclease, the motor strands autonomously walked along the track, leading to the release of many fluorescent molecules. Gold nanorods (AuNRs) with tunable aspect ratio-dependent plasmonic extinction band can also be utilized to quench the luminescence. For instance, Chen’s group reported a simple paper-supported biosensor with AuNRs to quench the luminescence of upconversion nanoparticles (UCNPs) through luminescence resonance energy transfer (LRET).100 As illustrated in Figure 3B, the sequence of aptamer toward CD63 protein is divided into two flexible ssDNA pieces with different sequence (CP and DP). Branched polyethylenimine (PEI)-modified UCNPs and CP were immobilized on the surface of filter paper by the formation of Schiff base. When exosomes were added, CD63 on the surface of exosomes facilitated the combination of DP and CP together into the intact aptamer tertiary, resulting in the close of AuNRs and UCNPs. The distance between AuNRs and UCNPs was shortened to allow for the occurrence of LRET. However, with the CD63, there was no interaction between two fragments and the LRET could not occur.
Figure 3.
(A) Schematic of the exosome-triggered enzyme-powered DNA motors for exosome detection. Reprinted with permission from Yu Y, Zhang WS, Guo Y, Peng H, Zhu M, Miao D, Su G. Engineering of exosome-triggered enzyme-powered DNA motors for highly sensitive fluorescence detection of tumor-derived exosomes. Biosens Bioelectron. 2020;167:112482–112490. Copyright 2020 Elsevier B.V.99 (B) Schematic of paper-supported aptasensor based on the LRET between UCNPs and AuNRs for the determination of exosomes. Reprinted with permission from Chen X, Lan J, Liu Y, et al. A paper-supported aptasensor based on upconversion luminescence resonance energy transfer for the accessible determination of exosomes. Biosens Bioelectron. 2018;102:582–588. Copyright 2018 Elsevier B.V.100
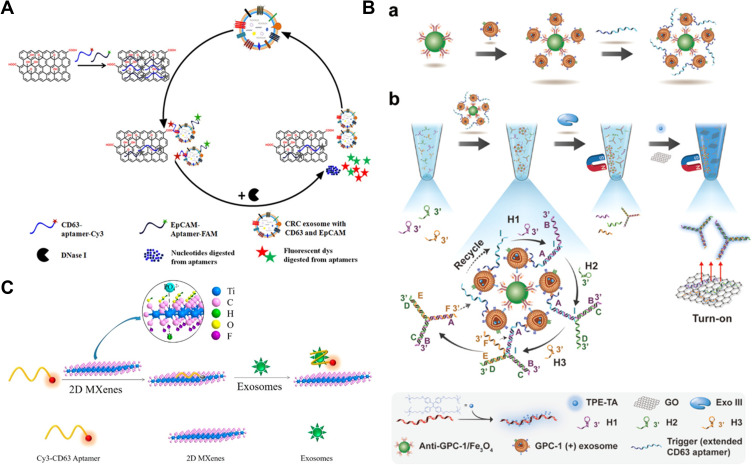
Graphene oxide (GO) can interact with DNA or RNA through π-π stacking interactions and thus quench the fluorescence of dye-labeled DNA/RNA probe through FRET. It is a fascinating nanomaterial to develop DNA-based “signal on/off” fluorescent biosensors for exosome detection.101 Wang et al designed a DNase I enzyme-aided signal amplification strategy for fluorescence analysis of colorectal cancer (CRC) exosomes based on the interaction between GO and aptamer.102 As illustrated in Figure 4A, the fluorescence of two aptamers (CD63 and epithelial cell adhesion molecule or EpCAM) labeled with different fluorophores was quenched by GO. In the presence of exosomes, two aptamers were bound to the target proteins of CD63 and EpCAM on the surface of CRC exosomes and then released from the surface of GO. DNase I promoted the digestion of the aptamers and induced the release of exosomes to liberate more aptamers, thus achieving a signal amplification. Few exosomes resulted in the release of numerous dyes and the restoration of fluorescence. Li et al developed a homogeneous magneto-fluorescent nanosensor for exosome analysis using GO as the quencher to reduce the background signal.103 As shown in Figure 4B, after exosomes were isolated by GPC-1 antibody-coated MBs, an extended CD63 aptamer was used to label the exosomes and the extended terminus served as a toehold to initiate the strand displacement, resulting in the formation of a large number of DNA three-way junctions (TWJ). After the magnetic separation, DNA TWJ in the supernatant could adsorb numerous positively charged derivatives of tetraphenylethene (TPE) aggregation-induced emission luminogens (AIEgens) through the electrostatic interaction. As a result, an enhanced fluorescence signal was observed. Meanwhile, GO was added to quench the fluorescence of AIEgens-stained ssDNA. The novel method achieved a wide linear detection range and the detection limit was calculated to be 6.56×104 particles/μL. In addition, MoS2–multiwall carbon nanotubes nanocomposites were employed to quench the fluorescence of dye-labeled CD63 antibody, which could be restored after the immunoreaction between exosome and antibody.104
Figure 4.
(A) Schematic of enzyme-aided fluorescence amplification based on GO-DNA aptamer interactions for exosome detection. Reprinted with permission from Wang H, Chen H, Huang Z, Li T, Deng A, Kong J. DNase I enzyme-aided fluorescence signal amplification based on graphene oxide-DNA aptamer interactions for colorectal cancer exosome detection. Talanta. 2018;184:219–226. Copyright 2018 Elsevier B.V.102 (B) Schematic of homogeneous magneto-fluorescent nanosensor for tumor-derived exosome isolation and analysis. (a) Tumor-derived exosomes are specifically captured by GPC-1 antibody coated magnetic beads and subsequently bind with extended CD63 aptamers, forming a bead−exosome−aptamer complexes. (b) Captured exosomes are detected in a homogeneous solution by aptamer-triggered DNA TWJs cyclic assembly strategy along with TPE-TA and the GO-based “turn-on” fluorescent system. Reprinted with permission from Li B, Pan W, Liu C, et al. Homogenous magneto-fluorescent nanosensor for tumor-derived exosome isolation and analysis. ACS Sens. 2020;5:2052–2060. Copyright 2020 American Chemical Society.103 (C) Schematic of Cy3-CD63 aptamer was mixed with MXenes aqueous solution and then added exosomes. Reprinted with permission from Zhang Q, Wang F, Zhang H, Zhang Y, Liu M, Liu Y. Universal Ti3C2 MXenes based self-standard ratiometric fluorescence resonance energy transfer platform for highly sensitive detection of exosomes. Anal Chem. 2018;90:12737–12744. Copyright 2018 American Chemical Society.105
As one subclass of 2D transition-metal carbides and carbonitrides materials, ultrathin MXenes have attracted much attention in biomedical applications due to their superior properties similar to those of GO. Based on their outstanding quenching efficiency, MXenes have been intensively utilized to construct fluorescent biosensors for the detection of targets, including DNA, RNA and proteins. Recently, Liu and co-workers reported a Ti3C2 MXenes-based self-standard ratiometric FRET platform for the detection of exosomes (Figure 4C).105 In the work, Cy3-labeled CD63 (Cy3-CD63) aptamers were adsorbed on the surface of MXenes by hydrogen-bond and metal-chelate interactions. The fluorescence of Cy3-CD63 aptamer was quenched by FRET and the intrinsic fluorescence of MXenes showed little change as a standard reference. Exosomes could specifically bind to the aptamers and induce their release from the surface of MXenes, thus leading to the recovery of fluorescence signal.
Colorimetric Biosensors
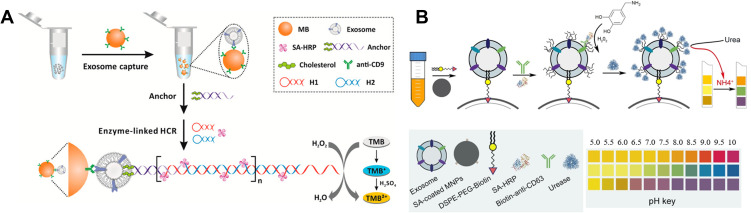
Colorimetric biosensors have attracted extensive attention because of their low cost and convenient readout. The results can be quickly observed with naked eyes. Thus, colorimetric assay is of great importance for point-of-care testing in facility-limited settings. Normally, enzymes are required to catalyze the chromogenic reaction in colorimetric assays.106,107 In traditional ELISA for the detection of exosomes, horseradish peroxidase (HRP) linked with detection antibody was always used to catalyze the reaction between H2O2 and colorimetric substrate 3,3′,5,5′-tetramethylbenzidine (TMB).108,109 Then, the color of solution changes from colorless to blue. However, they face the problems of low reproducibility and sensitivity (with a minimum amount of 3 μg of purified samples).110 Hemin/G-quadruplex with HRP-mimicking catalytic activity has also been widely used in bioassays for signal amplification.111,112 To improve the sensitivity, several strategies have been proposed for signal amplification, such as using immune-magnetic nanoparticles (MNPs) to enrich exosomes and using NPs to enhance the amount of enzymes for signal output.113–115 For example, He et al reported the direct quantification of exosomes based on HCR- and HRP-mediated signal amplification.116 As shown in Figure 5A, after exosomes were captured by CD9 antibody-coated MBs, the bivalent-cholesterol-labeled DNA probes were added to recognize the lipid membrane of exosomes via the hydrophobic cholesterol moiety. The DNA probe triggered the HCR and then numerous SA-HRP conjugates were captured by the DNA polymer for catalyzing the chromogenic reaction. The proposed assay showed a detection limit of 2.2×103 particles/μL. In addition, DNA nanoflowers were also employed to encapsulate HRP, thus improving the loading number of enzyme in signal output.117 Yang et al proposed a pH-responsive paper-based bioassay for the detection of exosomes.118 As shown in Figure 5B, after the SA-coated MNPs-based capture of exosomes, HRP conjugated with CD63 antibody catalyzed the formation of polydopamine film on the exosome surface, thus allowing for the binding of ureases.119 The captured ureases could hydrolyze urea into ammonia and carbon dioxide, resulting in the change of solution pH value from 5 to 10 and the color change of commercially available pH test paper. However, the utilization of natural enzyme is confronted of severe disadvantages of low stability, high-cost and complicated preparation process.
Figure 5.
(A) Schematic of the colorimetric method for exosome detection by a combination of immunoaffinity separation and cholesterol-based signal amplification. Reprinted with permission from He F, Liu H, Guo X, Yin BC, Ye BC. Direct exosome quantification via bivalent-cholesterol-labeled DNA anchor for signal amplification. Anal Chem. 2017;89:12968–12975. Copyright 2017 American Chemical Society.116 (B) Schematic of magnetic capture of exosomes, HRP-mediated PDA engineering of exosomes and urease immobilization for point-of-care testing. Reprinted with permission from Yang Y, Li C, Shi H, Chen T, Wang Z, Li G. A pH-responsive bioassay for paper-based diagnosis of exosomes via mussel-inspired surface chemistry. Talanta. 2019;192:325–330. Copyright 2019 Elsevier B.V.118
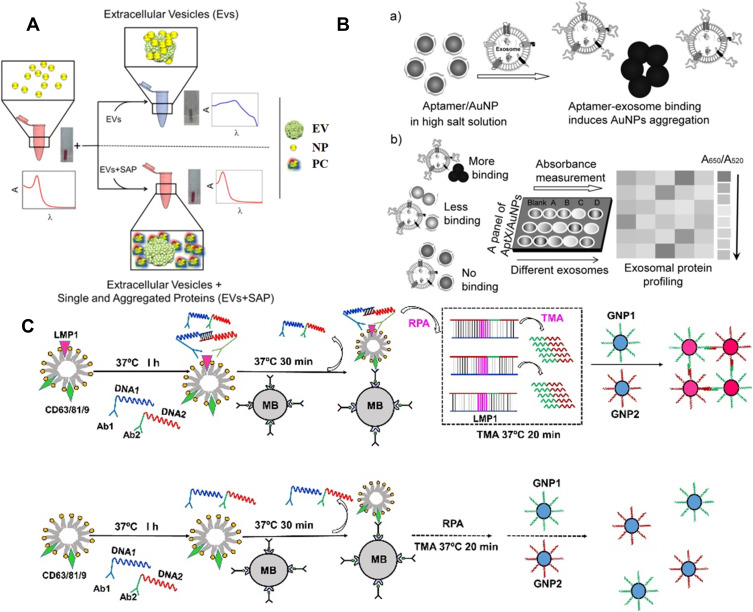
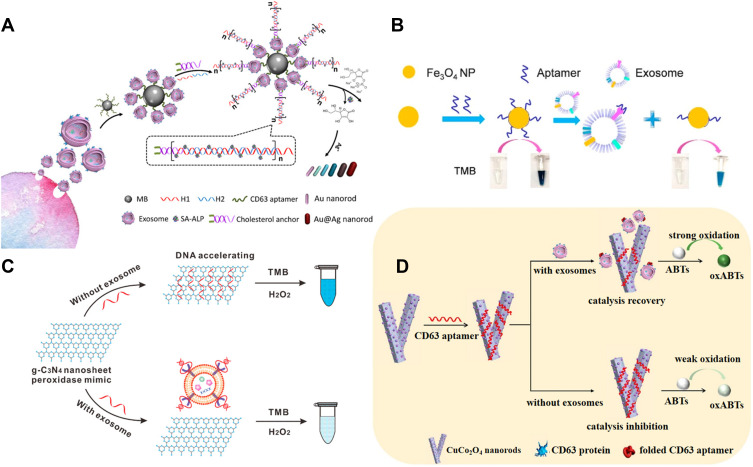
Au and Ag NPs with local surface plasmon resonance (LSPR) characteristics exhibit higher extinction coefficient than the organic chromogens. Such NPs have been widely used as the alternative substrates to develop plasmonic colorimetric methods for bioassays. The detection principles of NPs-based colorimetric strategies can be divided into two subclasses: aggregation/disaggregation and etching/growth. Typically, Maiolo et al presented a simple plasmonic colorimetric strategy for the determination of exosomes.120 As shown in Figure 6A, AuNPs could aggregate at the lipid membrane of exosomes, resulting in the shift and broaden of LSPR absorption spectrum and the change of solution color from red to blue. However, in the presence of exosome and protein contaminants, the formation of protein corona around AuNPs prevented the aggregation, and the LSPR absorption kept unchanged. Tan and co-workers developed a colorimetric aptasensor for profiling of exosomal proteins (Figure 6B).121 In this work, conjugation of aptamer with AuNPs prevented NPs from aggregation in high salt solution. However, the specific interaction between aptamer and exosome made aptamer leave the surface of AuNPs and resulted in the aggregation of AuNPs with the color change from red to blue. Liu et al reported a rapid and convenient colorimetric method for the detection of exosomes at ultralow concentrations by integrating target-induced proximity ligation assay (PLA) with recombinase polymerase amplification (RPA) and transcription-mediated amplification (TMA).122 As illustrated in Figure 6C, after two PLA probes bound to the protein LMP1 on the surface of exosomes, two DNA probes hybridized with each other. Under the RPA and TMA amplification, multiple copies of RNA transcripts were generated, which could induce the aggregation of the DNA-modified AuNPs and the change of solution color. Furthermore, the principle of AuNPs-based color assay have been introduced into LFA for rapid and sensitive analysis of exosomes.123–126 In addition, because AuNR is more sensitive to the change of local media environment, and the AuNR-based color change is more realistic, AuNR-based colorimetric assays exhibit better performances. Zhang et al reported the multicolor visual assay of exosome by the enzyme-catalyzed metallization of AuNR and HCR amplification.127 As depicted in Figure 7A, after exosomes were captured and labeled with cholesterol-modified DNA probes, the terminal DNA probes initiated the HCR assembly. Large numbers of alkaline phosphatase (ALP) molecules were loaded on the exosome surface to catalyze the production of ascorbic acid (AA) and the in-situ formation of silver shells on AuNRs, alongside with a vivid change of solution color. Moreover, Au nanobipyramid@MnO2 nanosheet was also used as the substrate for colorimetric detection of exosomes.128 During the exosome-induced competitive reaction, a large amount of ALP molecules were released into the solution by few exosomes and the free ALP catalyzed the generation of AA to etch Au nanobipyramid@MnO2 nanosheet, accompanied with multicolor change.
Figure 6.
(A) Schematic of nanoplasmonic assay for probing by eye protein contaminants (single and aggregated exogenous proteins, SAP) in EV preparations. Reprinted with permission from Maiolo D, Paolini L, Di Noto G, et al. Colorimetric nanoplasmonic assay to determine purity and titrate extracellular vesicles. Anal Chem. 2015;87:4168–4176. Copyright 2015 American Chemical Society.120 (B) Schematic of the aptamer/AuNP complex for molecular profiling of exosomal proteins. (a) Schematic of the displacement of aptamers from gold nanoparticles by binding with exosome surface protein and the concomitant aggregation of gold nanoparticles. (b) Profiling of different exosome surface proteins with the aptamer/AuNP complex. Reprinted with permission from Jiang Y, Shi M, Liu Y, et al. Aptamer/AuNP biosensor for colorimetric profiling of exosomal proteins. Angew Chem. 2017;129:12078–12082. Copyright 2017 Wiley-VCH.121 (C) Schematic of the PLA-RPA-TMA assay. Reprinted with permission from Liu W, Li J, Wu Y, et al. Target-induced proximity ligation triggers recombinase polymerase amplification and transcription-mediated amplification to detect tumor-derived exosomes in nasopharyngeal carcinoma with high sensitivity. Biosens Bioelectron. 2018;102:204–210. Copyright 2018 Elsevier B.V.122
Figure 7.
(A) Schematic of the mechanism for multicolor visual detection of exosomes based on HCR and enzyme-catalyzed metallization of Au NRs. Reprinted with permission from Zhang Y, Wang D, Yue S, et al. Sensitive multicolor visual detection of exosomes via dual signal amplification strategy of enzyme-catalyzed metallization of Au nanorods and hybridization chain reaction. ACS Sens. 2019;4:3210–3218. Copyright 2019 American Chemical Society.127 (B) Schematic of the visible detection of exosomes based on ssDNA-enhanced Fe3O4 NPs nanozyme activity. Reprinted with permission from Chen J, Xu Y, Lu Y, Xing W. Isolation and visible detection of tumor-derived exosomes from plasma. Anal Chem. 2018;90:14207–14215. Copyright 2018 American Chemical Society.131 (C) Schematic of DNA aptamer accelerating the intrinsic peroxidase-like activity of g-C3N4 NSs for the detection of exosomes. Reprinted with permission from Wang YM, Liu JW, Adkins GB, et al. Enhancement of the intrinsic peroxidase-like activity of graphitic carbon nitride nanosheets by ssDNAs and its application for detection of exosomes. Anal Chem. 2017;89:12327–12333. Copyright 2017 American Chemical Society.132 (D) Schematic of the detection mechanism for the visible detection of exosomes based on ssDNA-enhanced Fe3O4 NPs nanozyme activity. Reprinted with permission from Zhang Y, Su Q, Song D, Fan J, Xu Z. Label-free detection of exosomes based on ssDNA-modulated oxidase-mimicking activity of CuCo2O4 nanorods. Anal Chim Acta. 2021;1145:9–16. Copyright 2021 Elsevier B.V.134
Since Fe3O4NPs were reported to show peroxidase-like activity, more and more nanomaterials called nanozymes have proven to show catalytic ability and have been integrated into colorimetric bioassays.129 Compared to the natural enzymes, the nanozymes show attractive advantages of improved stability, low cost and ease of storage. Surface charge and composition are two crucial roles to regulate the catalytic activity of nanozymes. ssDNA can improve the peroxidase-mimicking activity of nanozymes.130 Chen et al reported the colorimetric assay of exosomes through ssDNA aptamer-enhanced peroxidase activity of Fe3O4NPs (Figure 7B).131 They found that aptamers attached on the surface of Fe3O4NPs could increase the affinity between NPs and TMB, thus leading to the enhancement of peroxidase activity. In the work, anion exchange method was first designed to extract exosomes from plasma. Then, the captured exosomes bound to aptamers from NPs and led to the decrease in the catalytic activity. Moreover, Wang et al developed the ssDNA-enhanced nanozyme-based colorimetric method for exosome detection (Figure 7C).132 They demonstrated that ssDNA could accelerate the intrinsic peroxidase-mimicking activity of graphitic carbon nitride nanosheets (g-C3N4 NSs) through the electrostatic and aromatic stacking interactions between ssDNA and TMB. However, CD63 on the surface of exosomes could competitively bind with ssDNA aptamer and reduce the enhancement of the peroxidase-mimicking activity. The method is sensitive and could determine exosomes in the range from 1.9×106 to 3.38×107 particles/μL. Xia et al employed ssDNA-modified single-wall carbon nanotubes for the detection of exosomes by the same principle.133 On the contrary, Zhang et al reported the label-free colorimetric assay of exosomes based on ssDNA-inhibited oxidase-like activity of CuCo2O4 nanorods (NRs).134 As shown in Figure 7D, CuCo2O4 NRs could catalyze the oxidation of ABTS with O2 as the electron acceptor, instead of volatile H2O2. The negatively charged CD63 aptamers were adsorbed on the CuCo2O4 NRs through the electrostatic interaction and the oxidase-like activity of NRs was inhibited by hindering the electron transfer between NRs and substrates. However, in the presence of exosomes, aptamers were released from the CuCo2O4 NRs and the oxidase-like activity was restored.
Surface Plasmon Resonance Biosensors
Surface plasmon resonance (SPR) is a label-free, real-time sensing technique to study and quantify biomolecular interaction.135 It can monitor the change of refractive index in close proximity to the gold surface (within 200 nm), resulting from binding event-induced increase of thickness. Moreover, it possesses the merits of high signal-to-noise, good compatibility with microfluidic technique and advanced surface modifications. Therefore, exosomes with around 100 nm size and large mass can be detected by SPR, whose size fits within the surface plasmon wave depth. To date, a series of label-free SPR biosensors for exosome detection have been developed by modifying the sensor surface with antibodies specific to the membrane proteins on exosome.136–145 To overcome the slow diffusion-limited mass transfer, magnetic nanoparticles can be utilized to pre-concentrate exosomes on the sensor surface under an external magnetic field gradient.146 However, the poor sensitivity of the methods limits their further applications for analyzing trace targets in complex samples.
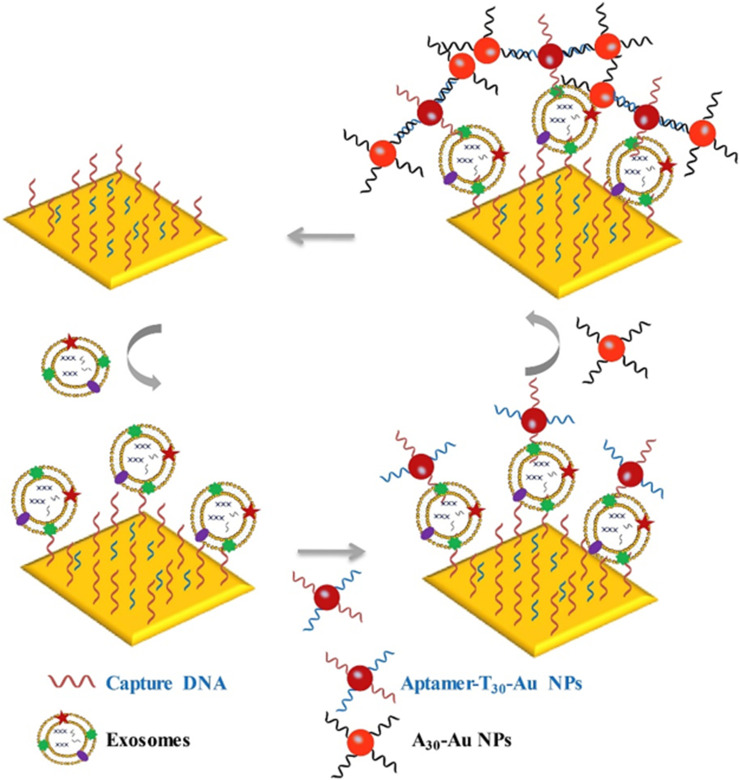
AuNPs can enhance the SPR signal through plasmonic coupling.147–150 Wang et al proposed a SPR aptasensor for quantification of cancerous exosomes with dual AuNPs-assisted signal amplification.151 As illustrated in Figure 8, the gold chip was functionalized with aptamers to capture exosomes. Aptamer/T30-modified AuNPs were further used to label exosomes on the Au film. The sensitivity of the biosensor is very high because of the electronic coupling between the gold film and AuNPs. For increasing the specificity, they further developed a dual-aptamers-based SPR strategy for the determination of human hepatic carcinoma (SMMC-7721) exosomes.152 In addition, combining with DNA-based signal amplification, Ding’s group reported a hydrogel-AuNP supramolecular sphere (H-Au)-based SPR biosensor for analysis of prostate cancer-derived exosomes.153 The H-Au network was prepared via the self-assembly of DNA chains and DNA-modified AuNPs. The PSMA-specific aptamer-functionalized MBs were used to transduce target exosomes into triple DNAs, which could initiate the terminal transferase-catalyzed generation of polyA tails than can bind to the H-Au network on the SPR surface. This biosensor had a wide linear range from 1.00×102 to 1.00×104 particles/μL.
Figure 8.
Schematic of dual AuNP-assisted signal amplification for SPR determination of exosomes. Reprinted with permission from Wang Q, Zou L, Yang X, et al. Direct quantification of cancerous exosomes via surface plasmon resonance with dual gold nanoparticle-assisted signal amplification. Biosens Bioelectron. 2019;135:129–136. Copyright 2019 Elsevier B.V.151
Surface Enhanced Raman Scattering Biosensors
Raman spectroscopy can provide a characteristic fingerprint spectrum. However, the signal intensity is always too weak to be distinguished. Since SERS effect was discovered, a significant interest in SERS study was aroused. Generally, metal nanostructures or nanomaterials can be employed to amplify the signal through chemical and electromagnetic field enhancement. SERS spectroscopy has been used for the design of biosensors by label-free analysis and SERS-tag-based methods.154
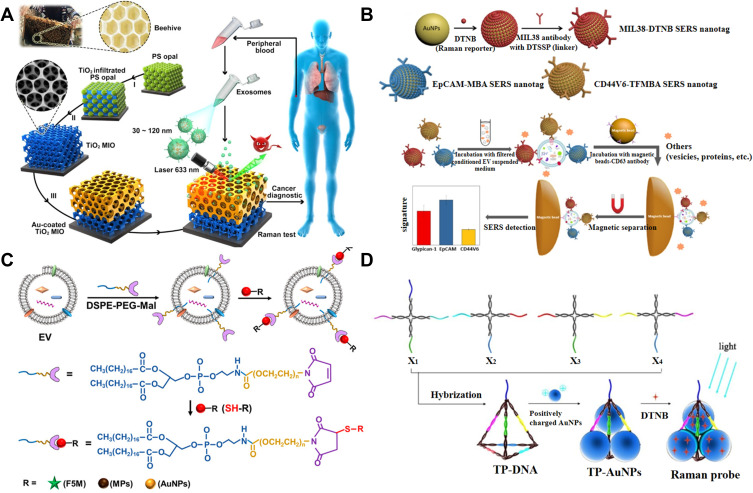
Label-free SERS analysis is mainly based on the use of roughened or nanosized SERS substrate to enhance the weak Raman vibration signal of exosomal biomolecules with fingerprint characters. For example, Avella-Oliver et al reported the label-free SERS analysis of exosomes with large-scale substrates from recordable compact disk by coating it with silver.155 This cost-effective technology provided an alternative solution to perform SERS bioassays in non-specialized environments. Inspired by the concept of beehives, Dong et al suggested that the Au-coated TiO2 macroporous inverse opal (MIO) structures could be used as the SERS substrates for label-free detection of exosomes.156 As shown in Figure 9A, different from traditional SERS substrates, the MIO structure could capture exosomes by its interconnected nanoscale pore networks, exhibiting a prominent “slow light effect” and enhancing the Raman signal of exosomes by the SERS effect of Au layer. The SERS intensity of 1087 cm−1 from the P-O bond within phosphoproteins on the surface of exosomes was used as the detection criterion. Because of the heterogeneity, the Raman spectra of exosomes show complex and inconsistent data, which are difficult to be classified. For this view, principal component analysis was employed to monitor the Raman signal, and a meaningful pattern for exosome analysis was obtained.157–159
Figure 9.
(A) Schematic representation of detection process and design inspiration of the Au-coated TiO2 MIO SERS probe. Reprinted with permission from Dong S, Wang Y, Liu Z, et al. Beehive-inspired macroporous SERS probe for cancer detection through capturing and analyzing exosomes in plasma. ACS Appl Mater Interfaces. 2020;12:5136–5146. Copyright 2020 American Chemical Society.156 (B) Schematic representation of preparation of three types of SERS nanotags and molecular phenotype profiling of exosomes using SERS nanotags and CD63 antibody-functionalized MBs. Reprinted with permission from Zhang W, Jiang L, Diefenbach RJ, et al. Enabling sensitive phenotypic profiling of cancer-derived small extracellular vesicles using surface-enhanced Raman spectroscopy nanotags. ACS Sens. 2020;5:764–771. Copyright 2020 American Chemical Society.164 (C) Schematic of exosomes engineering based on a hydrophobic insertion strategy with DSPE-PEG-Mal. Reprinted with permission from Di H, Zeng E, Zhang P, et al. General approach to engineering extracellular vesicles for biomedical analysis. Anal Chem. 2019;91:12752–12759. Copyright 2019 American Chemical Society.165 (D) Schematic representation of assembling AuNPs in triangular pyramid DNA. Reprinted with permission from Zhang X, Liu C, Pei Y, Song W, Zhang S. Preparation of a novel Raman probe and its application in the detection of circulating tumor cells and exosomes. ACS Appl Mater Interfaces. 2019;11:28671–28680. Copyright 2019 American Chemical Society.169
Au and Ag nanomaterials with LSPR have been utilized as the SERS-active nanotags to enhance the signal intensity of Raman dyes. With immunomagnetic beads and for the capture of exosomes, several aptamer or antibody-modified SERS nanotags have been developed for exosome detection via the formation of antibody-exosome-aptamer sandwich-type immunocomplexes.160–163 Microfluidic Raman biochips were also fabricated to isolate and determine exosomes in situ.43 Typically, Wang’s group reported an effective approach for the detection of exosomes by simultaneously profiling multiple protein biomarkers on the surface.164 As shown in Figure 9B, three specific nanotags for antibody modification were prepared and labeled in the filtered conditional exosome-suspension medium. Then, the antibody-modified CD63-conjugated MBs were added for the sandwich-like immunoassays. The heterogeneous antigens expressed on diverse exosomes limited the applications of the methods based on the antigen-antibody/aptamer interaction. Liu’s group proposed a general, facile, and robust strategy to label exosomes with maleimide (Mal) tags by the hydrophobic insertion.165 As shown in Figure 9C, maleimide-terminated DSPE-PEG (DSPE-PEG-Mal) as labeling probe was inserted into the lipid membranes. Mal group could be conjugated with the thiol-containing species (1,6-hexanedithiol) via the click chemistry and further bound to bare AuNPs for SERS analysis. Wang’s group developed a SERS biosensor for multiple assays of exosomes with gold layer-coated MBs as SERS probes which were modified with three different types of Raman reporters and aptamers.166 Besides, Kwizera et al presented a method for exosome detection by using cationic AuNRs as SERS tags to label exosomes through the electrostatic attraction.167 The “hot spots” generated in the AuNP-AuNP junctions due to the plasmon coupling effect can intensify the Raman signal of SERS molecules. For this view, Ning et al reported the multiple SERS assays of exosomes using gold-silver bimetallic nanotrepangs, in which different Raman reporter molecules were confined in the interfaces of gold core and silver shell.168 Zhang et al designed a novel Raman probe for the assays of exosomes by assembling AuNPs in triangular pyramid DNA (TP-DNA).169 As illustrated in Figure 9D, TP-DNA was prepared through the hybridization of four X-shaped DNA sequences and then the positively charged AuNPs and Raman reporter molecules were entrapped through the electrostatic interactions. Besides, laser-tweezers Raman spectroscopy has also been employed to determine exosomes with individual nanoparticle for signal enhancement.170
Nanomaterials-Based Electrochemical Biosensors for Exosome Detection
Electrochemical Biosensors
Electrochemical biosensor has been recognized as an excellent platform for biological sample analysis due to its advantages of high sensitivity, low cost, rapid response and low sample volume.171,172 Several classic electrochemical techniques are frequently utilized in bioassays, including amperometry, voltammetry, impedimetry and field effect transistor. Nanomaterials mainly play two vital roles in these techniques: as the electrode substrate to improve the electron transfer and as the functional nanotags for signal amplification.173
Direct Detection
Direct electrochemical detection is achieved by monitoring the change of electrical conductivity of electrode by the target-induced electrical signal change.174–177 The method can quantify exosomes without labeling step, thus shortening the response time. For example, Tan and co-workers presented an electrochemical aptasensor for direct determination of cancerous exosomes by using DNA nanotetrahedra to immobilize aptamer on the electrode surface to improve the accessibility of exosomes.178 Davis’s group reported an immunosensor for the analysis of exosomes by electrochemical impedance spectroscopy.179 Vaidyanathan developed a multiplexed device to detect exosomes by alternating current electrohydrodynamic induced nanoshearing.180
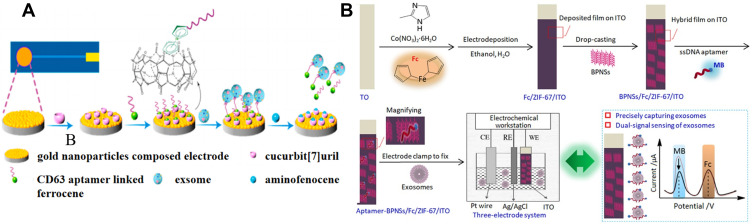
AuNPs with good electrical conductivity and easy of functionalization have been widely utilized to modify the sensor electrode. Cucurbit[7]uril with excellent supramolecular recognition ability toward ferrocene (Fc) has been widely used as the receptor in electrochemical analysis. Liu et al reported a label-free electrochemical aptasensor for exosome detection based on the host-guest interaction between cucurbit[7]uril and Fc.181 As illustrated in Figure 10A, cucurbit[7]uril was immobilized on the AuNPs-modified electrode for the capture of Fc-labeled CD63 aptamer. The aptamer bound to the target exosome with high affinity can be released from the electrode surface, thus resulting in the decrease of electrochemical signal. Sun et al developed a dual-signal and intrinsic self-calibration aptasensor for direct detection of exosomes.182 As shown in Figure 10B, ITO slice was modified with Fc-doped metal-organic frameworks (ZIF-67) by electrodeposition and black phosphorus nanosheets (BPNSs). Then, the methylene blue-labeled ssDNA aptamers were adsorbed on the electrode surface. The platform exhibited dual redox-signal responses from methylene blue and Fc. In the presence of exosomes, the aptamers were desorbed from the electrode surface, leading to the decrease of redox current of methylene blue. In this process, no significant change was observed for the current of Fc. The intrinsic self-calibration aptasensor showed a detection limit down to 0.1 particles/μL.
Figure 10.
(A) Schematic representation of the electrochemical aptasensor for exosomes capture and release based on specific host-guest interactions between cucurbit[7]uril and Fc. Reprinted with permission from Liu Q, Yue X, Li Y, et al. A novel electrochemical aptasensor for exosomes determination and release based on specific host-guest interactions between cucurbit [7]uril and ferrocene. Talanta. 2021;232:122451–122458. Copyright 2021 Elsevier B.V.181 (B) Schematic representation of the construction process and application of a dual-signal and intrinsic self-calibration aptasensor of exosomes based on a functional hybrid thin-film sensing platform aptamer-BPNSs/Fc/ZIF-67/ITO. Reprinted with permission from Sun Y, Jin H, Jiang X, Gui R. Assembly of black phosphorus nanosheets and MOF to form functional hybrid thin-film for precise protein capture, dual-signal and intrinsic self-calibration sensing of specific cancer-derived exosomes. Anal Chem. 2020;92:2866–2875. Copyright 2020 American Chemical Society.182
Field effect transistor (FET) biosensor is a promising label-free detection tool, which can monitor the microelectrical signal caused by the interaction between target and recognition element on the sensing interface.183 Yu et al designed a reduced graphene oxide (rGO)-based FET biosensor for electrical and label-free quantification of exosomes.184 In this paper, 1-pyrenebutanoic acid succinimidyl ester was modified on the rGO surface through the π-π stacking interaction between pyrene and graphene. Then, CD63 antibody was covalently immobilized on the FET surface. The net carrier density changed with the introduction of negatively charged exosomes, thus leading to the left shift of the Dirac point.
Sandwich-Like Methods
Although the label-free electrochemical method is simple, it shows poor sensitivity and selectivity. Therefore, different types of sandwich-like methods were developed for bioassays. Generally, the electrode was modified with a biorecognition element to capture exosome, and the another biorecognition element modified with a signal reporter was added to recognize the captured exosome and produce an electrical signal. Enzymes and electroactive molecules are usually used as the signal reporters for signal amplification.185 For example, Doldan et al reported an electrochemical immunosensor for the determination of exosomes using HRP-conjugated antibody.186 An et al designed an electrochemical aptasensor for the detection of tumor exosomes by the HCR assembly to linking numerous HRP molecules for catalytic redox reactions.187 He and co-workers reported an electrochemical aptasensor for the analysis of exosomes by hemin/G-quadruplex-assisted rolling circle amplification.188 Generally, nanomaterials can act as nanocarriers, nanoelectrocatalysts and electroactive tags for signal amplification in sandwich-like electrochemical assays.
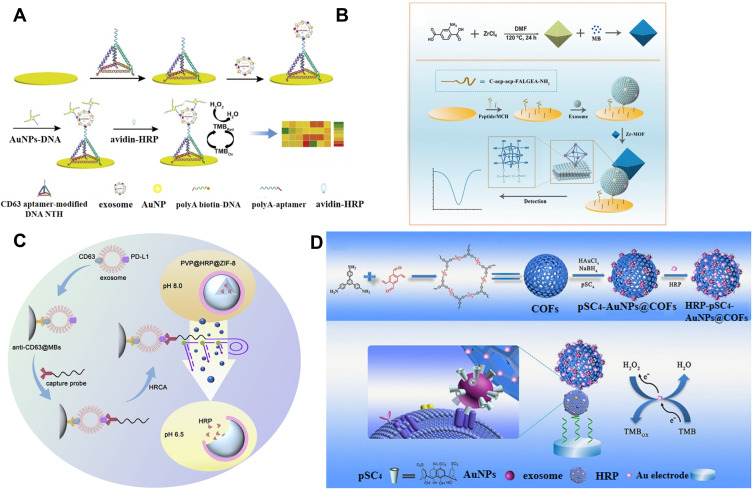
Because of their excellent biocompatibility and large surface area, AuNPs have been widely used to carry various biomolecules (such as proteins, DNA and RNA) for different biological applications. Jiang et al reported an electrochemical aptasensor for exosome detection using AuNPs and enzyme for signal amplification.189 As showed in Figure 11A, aptamer-modified DNA nanotetrahedron (NTH) was employed to modify the electrode, avoiding the problem of the entanglement of aptamer and the spatial hindrance effect. Aptamer and biotin were modified on AuNPs through the interaction between polyA10 and AuNPs. After the formation of sandwich-like complexes, numerous avidin-HRP conjugates were immobilized on the biotin-labeled AuNPs to catalyze the reduction of TMB and H2O2.
Figure 11.
(A) Schematic of the electrochemical aptasensor for exosomes detection by using AuNPs and enzyme for amplification. Reprinted with permission from Jiang J, Yu Y, Zhang H, Cai C. Electrochemical aptasensor for exosomal proteins profiling based on DNA nanotetrahedron coupled with enzymatic signal amplification. Anal Chim Acta. 2020;1130:1–9. Copyright 2020 Elsevier B.V.189 (B) Schematic of the fabrication process of MB@UiO-66-based nanoprobe and the electrochemical biosensor for the detection of GBM-derived exosomes. Reprinted with permission from Sun Z, Wang L, Wu S, et al. An electrochemical biosensor designed by using Zr-based metal-organic frameworks for the detection of glioblastoma-derived exosomes with practical application. Anal Chem. 2020;92:3819–3826. Copyright 2020 American Chemical Society.190 (C) Schematic of identification of PD-L1+ exosomes based on HRCA-responsive PVP@HRP@ZIF-8. Reprinted with permission from Cao Y, Wang Y, Yu X, Jiang X, Li G, Zhao J. Identification of programmed death ligand-1 positive exosomes in breast cancer based on DNA amplification-responsive metal-organic frameworks. Biosens Bioelectron. 2020;166:112452–112460. Copyright 2020 Elsevier B.V.192 (D) Schematic of the fabrication process of COFs-based nanoprobes and the mechanism of the EC biosensor for exosomes detection. Reprinted with permission from Wang M, Pan Y, Wu S, et al. Detection of colorectal cancer-derived exosomes based on covalent organic frameworks. Biosens Bioelectron. 2020;169:112638–112645. Copyright 2020 Elsevier B.V.193
Due to their high specific surface area, flexible porosity, and tunable framework structure, metal−organic frameworks (MOFs) have attracted wide attention in comprehensive applications, including catalysis, sensors and energy conversion and storage. The porosity endows MOFs with the ability to carry plenty of enzyme or electroactive molecules. Sun et al reported a label-free and enzyme-free electrochemical biosensor for the detection of glioblastoma-derived exosomes using Zr-based MOFs.190 As displayed in Figure 11B, Zr-MOFs (UiO-66) prepared from metal ions and organic ligands through a hydrothermal method were utilized to load numerous electroactive methylene blue molecules. After the capture of exosomes by the peptide-modified electrode, methylene blue-loaded Zr-MOFs were anchored by exosomes. Zr-MOFs interacted with phosphate groups in the phospholipid bilayer of exosomes with high affinity via the formation of Zr-O-P bonds. The concentration of exosomes could be determined by measuring the electrochemical signal of methylene blue inside of MOFs. Recently, Gu et al presented a biofuel cells-based self-powered biosensor for exosome detection, in which two types of MOFs (ZIF-8 and UiO-66-NH2) were utilized to load glucose dehydrogenase and electroactive molecules (K3[Fe(CN)6]), respectively.191 Taking the advantage of exogenous stimulus-responsive property, Cao et al reported an electrochemical biosensor for the detection of programmed death ligand-1 positive (PD-L1+) exosomes based on DNA amplification-responsive MOFs.192 As illustrated in Figure 11C, HRP-encapsulated ZIF-8 was prepared by biomineralization-facilitated method and then coated with PVP, which remained intact at weak alkaline pH and would be broken at acidic pH. After the capture of PD-L1+ exosomes by immune-MBs, the anti-PD-L1-linked DNA strands were added to label exosomes and the DNA parts initiated the hyperbranched rolling circle amplification (HRCA). The released H+ ions caused the change of the environment pH to weak acidity, thus leading to the disassembly of MOFs. HRP molecules were released to increase the electrochemical response.
As an emerging kind of porous crystalline materials, covalent organic frameworks (COFs) exhibit great application potential in bioassays. Li’s group reported a COFs-based aptasensor for the analysis of CRC exosomes.193 As shown in Figure 11D, spherical COFs with high porosity were utilized to load para-sulfocalix[4]arene hydrate (pSC4)-modified AuNPs and a large amount of HRP molecules to form HRP-pSC4-AuNPs@COFs. pSC4 could specifically bind with various amino acid residues on the exosome surface. AuNPs could accelerate the charge transfer of carriers. CD63 aptamer was anchored on the surface of Au electrode to capture exosomes. Then, HRP-pSC4-AuNPs@COFs were added to recognize the captured exosomes and HRP catalyzed the oxidation of TMB by H2O2, generating a high electrochemical signal.
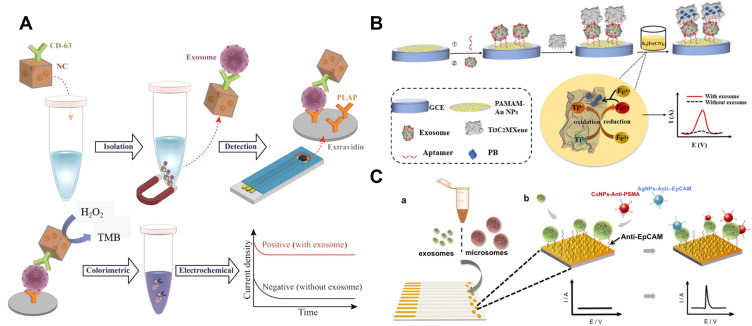
Nanozymes can catalyze the redox reaction between H2O2 and substrate in colorimetric assays, which can be converted into electrochemical assays with improving sensitivity. For example, Boriachek et al reported the direct isolation and subsequent detection of exosomes using gold-loaded ferric oxide nanocubes (Au-NPFe2O3NC).194 As shown in Figure 12A, the Au-NPFe2O3NC was modified with CD63 antibody for the capture of exosomes. After magnetic separation, the complex of Au-NPFe2O3NC and exosomes was attached on the placental alkaline phosphatase (PLAP) antibody-functionalized screen-printed electrode. The signal was measured by the Au-NPFe2O3NC-catalyzed reaction between TMB and H2O2.
Figure 12.
(A) Schematic of the assay for direct exosome isolation and detection from cell culture media based on Au-NPFe2O3NC. Reprinted with permission from Boriachek K, Masud MK, Palma C, et al. Avoiding pre-isolation step in exosome analysis: Direct isolation and sensitive detection of exosomes using gold-loaded nanoporous ferric oxide nanozymes. Anal Chem. 2019;91:3827–3834. Copyright 2019 American Chemical Society.194 (B) Schematic of the electrochemical biosensor for exosomes activity detection signal amplification strategy by using in situ generation of Prussian blue. Reprinted with permission from Zhang H, Wang Z, Wang F, Zhang Y, Wang H, Liu Y. Ti3C2 MXene mediated Prussian blue in situ hybridization and electrochemical signal amplification for the detection of exosomes. Talanta. 2021;224:121879–121886. Copyright 2021 Elsevier B.V.195 (C) Schematic representation of the two-step isolation and analysis of exosomes and microsomes: (a) capture step where vesicles are immobilized on aptamer-modifi ed sensors, (b) electrochemical detection of the captured exosomes/microsomes with Cu and AgNPs. Reprinted with permission from Zhou YG, Mohamadi RM, Poudineh M, et al. Interrogating circulating microsomes and exosomes using metal nanoparticles. Small. 2016;12:727–732. Copyright 2016 Wiley-VCH.197
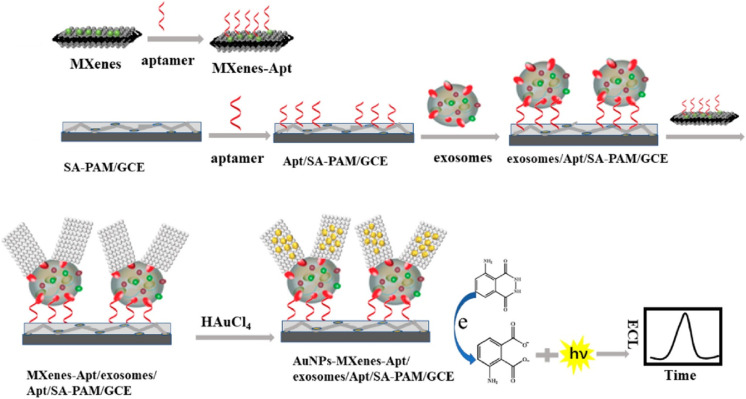
Wang and co-workers developed a sensitive electrochemical biosensor for exosome detection through the in-situ generation of electroactive Prussian blue (Fe[Fe(CN)6]) on the surface of MXenes.195 As illustrated in Figure 12B, the CD63 aptamer-modified poly(amidoamine)-AuNP electrode was used to capture exosomes. Then, aptamer-conjugated MXene was used to recognize the captured exosomes. MXene on the exosome surface served as a reducing carrier to induce in-situ generation of Prussian blue, simplifying the synthesis process. Prussian Blue could produce electrochemical signal at a low potential without the interference of electroactive species. The detection limit of this method was 229 particles/μL. QDs with a large amount of metal ions can also be utilized as the signal transduction labels for the assays of exosomes.196 After acid-assisted dissolution, numerous Cd2+ ions were released, which could be quantified by anodic stripping voltammetry (ASV). Moreover, metal NPs such as Ag and Cu can be utilized as the signal reporters because they can be directly electrochemically oxidized to produce a typical electrochemical peak. Kelley’s group reported the electrochemical detection of exosomes/microsomes with anti-EpCAM aptamers-modified AgNPs and anti-PSMA aptamers-modified CuNPs (Figure 12C).197 After exosomes and microsomes were captured from VCaP cells through a simple centrifugation procedure, aptamers-functionalized NPs were added to label the captured exosomes. Then, linear sweep voltammetry (LSV) was used for the direct electrochemical oxidation of AgNPs or CuNPs.
Magneto-Electrochemical Detection
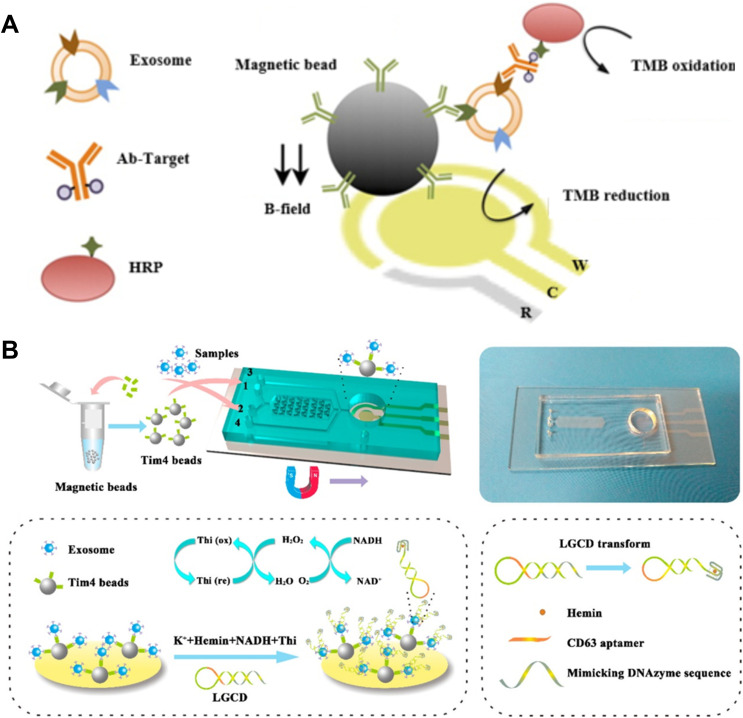
The immobilization of recognition probes on the electrode may suppress the effective recognition between exosome and probe and thus decrease the sensitivity. MBs have been widely used for the capture of exosomes and can be integrated into electrochemical biosensors. MBs can not only simplify the detection procedures, but also concentrate the captured exosomes on the electrode. MBs modified with antibody (immune-MBs) have been used to isolate and enrich exosomes. MBs-based electrochemical techniques for immobilization-free detection of exosomes have been developed.198,199 Lee’s group designed an integrated immuno-magneto-electrochemical sensor for exosome detection (Figure 13A),200 in which MBs were modified with CD63 antibodies to directly capture exosomes in plasma. Next, the captured exosomes were recognized by the HRP-labeled detection antibodies. HRP could catalyze the reaction between TMB and H2O2, thus generating a strong electrochemical signal. To meet the need of portability and sensitivity, Ye’s group reported a two-stage magnetic-based microfluidic platform for on-chip isolation and detection of exosomes.201 As illustrated in Figure 13B, a staggered Y-shaped micropillar mixing pattern was applied to create an anisotropic flow for improving the capture efficiency. Tumor-derived exosomes captured by Tim4-coated MBs were immobilized on the ITO electrode. The ssDNA in a hairpin structure consisted of aptamer and mimicking DNAzyme sequence was employed to label exosomes. After the recognition, hairpin was opened and a G-quadruplex formed with hemin was utilized as NADH oxidase and HRP-mimicking DNAzyme simultaneously. In addition, CdSe QDs were utilized as the signal labels instead of unstable enzymes for exosome detection by an anodic stripping voltammetry.196
Figure 13.
(A) Schematic of the integrated immuno-magneto- electrochemical sensor for exosomes detection. Reprinted with permission from Jeong S, Park J, Pathania D, et al. Integrated magneto-electrochemical sensor for exosome analysis. ACS Nano. 2016;10:1802–1809. Copyright 2016 American Chemical Society.200 (B) Schematic of the Exo PCD-chip and the electrochemical Sensor on the Surface of ITO Electrode. Reprinted with permission from Xu H, Liao C, Zuo P, Liu Z, Ye BC. Magnetic-based microfluidic device for on-chip isolation and detection of tumor-derived exosomes. Anal Chem. 2018;90:13451–13458. Copyright 2018 American Chemical Society.201
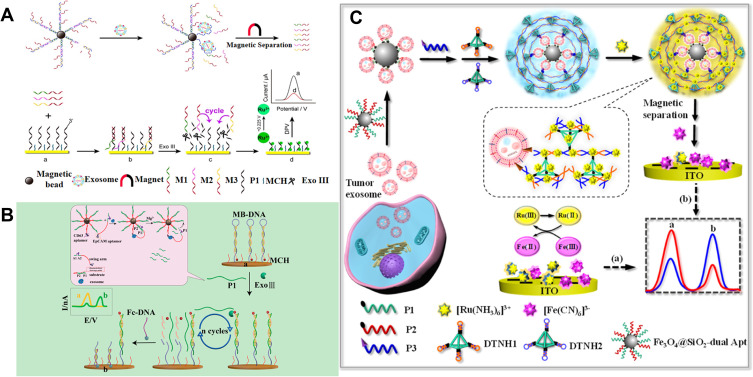
Owing to the flexible structure, aptamer can hybridize with other DNA sequences that can initiate the DNA-based signal amplification or DNA nanomachines.202,203 Therefore, exosome detection can be converted into the analysis of DNA, whose amount is proportional to the number of exosomes. This strategy avoids the direct detection of exosomes on the electrode and many methods can be developed to sensitively determine DNA.204 For instance, Dong et al reported a highly sensitive electrochemical biosensor for exosome detection based on aptamer recognition-induced release of multi-DNA and cyclic enzymatic amplification.205 As shown in Figure 14A, aptamer-messenger DNA (mDNA) complexes were first modified on the MBs. LNCaPcell-derived exosomes bound to aptamer with high affinity, resulting in the release of three kinds of mDNA sequences. After the magnetic separation, the released mDNA in the supernatant initiated the Exo III-assisted cyclic enzymatic amplification reaction, leading to the sharp decrease in the amount of Ru(NH3)63+ on the surface of Au electrode. Zhao et al reported a ratiometric electrochemical biosensor for the detection of exosomes by target-triggered 3D DNA walker and Exonuclease III-assisted cyclic enzymatic amplification.206 As displayed in Figure 14B, MBs were modified with high-density of DNA as 3D DNA walker scaffold. The DNA sequence consisted of CD63 aptamer and DNAzyme substrate. In the presence of exosomes, the recognition of CD63 aptamer on the MBs and EpCAM aptamer on the swing arm simultaneously bound to different target proteins on the same exosome, leading to the close proximity effect between DNAzyme and substrate. After the hybridization, DNA walkers were initiated and a large number of oligonucleotide fragments were released, which could be sensitively detected by the Exonuclease III-assisted cyclic enzymatic amplification.
Figure 14.
(A) Schematic of the highly sensitive electrochemical biosensor for exosomes detection based on aptamer recognition-induced multi-DNA release and cyclic enzymatic amplification. Reprinted with permission from Dong H, Chen H, Jiang J, Zhang H, Cai C, Shen Q. Highly sensitive electrochemical detection of tumor exosomes based on aptamer recognition-induced multi-DNA release and cyclic enzymatic amplification. Anal Chem. 2018;90:4507–4513. Copyright 2018 American Chemical Society.205 (B) Schematic of the ratiometric electrochemical biosensor for the detection of exosomes by target-triggered 3D DNA walker and Exo III-assisted cyclic enzymatic amplification. Reprinted with permission from Zhao L, Sun R, He P, Zhang X. Ultrasensitive detection of exosomes by target-triggered three-dimensional DNA walking machine and exonuclease III-assisted electrochemical ratiometric biosensing. Anal Chem. 2019;91:14773–14779. Copyright 2019 American Chemical Society.206 (C) Schematic of the ratiometric immobilization-free electrochemical sensing system for tumor exosome detection in the absence (a) and in the presence (b) of the tumor exosomes. Reprinted with permission from Yang L, Yin X, An B, Li F. Precise capture and direct quantification of tumor exosomes via a highly efficient dual-aptamer recognition-assisted ratiometric immobilization-free electrochemical strategy. Anal Chem. 2021;93:1709–1716. Copyright 2021 American Chemical Society.207
To improve the capture efficiency, dual aptamers-modified MBs were employed to efficiently capture exosomes.207 As illustrated in Figure 14C, after the capture of tumor exosomes, the cholesterol-modified DNA probe was anchored on the exosome membrane through the hydrophobic interaction, triggering DNA tetrahedron-based hyperbranched HCR to generate sandwich complexes. The complexes could sequestrate a large amount of Ru(NH3)63+ by electrostatic interactions, thus reducing the amount of Ru(NH3)63+ in the solution after the magnetic separation. This caused the change of current ratio of [Fe(CN)6]3− to Ru(NH3)63+.
Electrochemiluminescent Biosensors
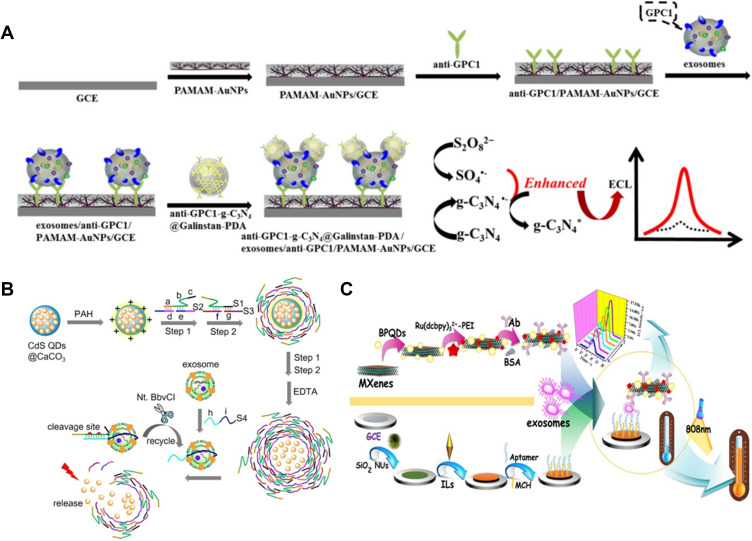
As a powerful technique, ECL has been applied in various bioanalysis due to the remarkable advantages of low background signal, high sensitivity and wide detection range. To meet the need of ultrahigh sensitivity, loading of numerous luminophores into nanomaterials can boost the ECL efficiency and improve the analysis sensitivity.208 Feng et al designed an aptamer-binding DNA walking machine for sensitive ECL detection of tumor exosomes by employing Ru(bpy)32+-loaded silica NPs as the signal reporters.209 To date, several nanomaterials have been utilized as the ECL emitters for diverse biosensing. For example, Sheng and co-workers developed an ECL aptasensor for the analysis of exosomes by the G-quadruplex/hemin DNAzymes-induced quenching of RCL signal of Eu3+-doped CdSQDs.210 Zhang et al designed a sensitive g-C3N4-coated liquid metal nanoprobe-based ECL sensing strategy for exosome detection on the multivalency interface (Figure 15A).211 In the nanoprobe, Galinstan NPs accelerated the electrode transfer and suppressed the g-C3N4 passivation during the electrochemical reduction processes, thus enhancing the ECL signal. Moreover, the antibody-modified PAMAM-Au NPs were used to modify the electrode for multivalent recognition of exosomes with high capture efficiency. This method achieved a detection limit of 31 particles/μL for the determination of HeLa cell-derived exosomes. To simplify the operation procedure and reduce the contamination, Guo et al reported the QDs-based homogeneous ECL sensing of exosomes with stimuli-responsive DNA microcapsules and target recycling system.212 As illustrated in Figure 15B, CdS QDs-loaded CaCO3 microcapsules were sealed by DNA-assembled stimuli-responsive shell layers. Then, the core CaCO3 was removed by treatment with EDTA and the DNA shell-coated CdS QDs microcapsules were formed. The presence of exosomes initiated the nicking endonuclease (Nt.BbvCI)-assisted target recycling and the crosslinked DNA shells were disintegrated. The released QDs were determined by ECL technique. The established method with dual-amplification exhibited a wide detection range of 5×104 to 1×108 particles/μL.
Figure 15.
(A) Schematic of the ECL biosensor for exosomes based on multivalent recognition and signal amplification strategy by anti-GPC1-g-C3N4@Galinstan-PDA nanoprobe. Reprinted with permission from Zhang Y, Wang F, Zhang H, Wang H, Liu Y. Multivalency interface and g-C3N4 coated liquid metal nanoprobe signal amplification for sensitive electrogenerated chemiluminescence detection of exosomes and their surface proteins. Anal Chem. 2019;91:12100–12107. Copyright 2021 Elsevier B.V.211 (B) Schematic of the prepared process of CdS QDs-loaded DNA microcapsules integrated with target recycling amplification for homogeneous ECL detection of tumor exosomes. Reprinted with permission from Guo Y, Cao Q, Zhao C, Feng Q. Stimuli-responsive DNA microcapsules for homogeneous electrochemiluminescence sensing of tumor exosomes. Sens Actuat B Chem. 2021;329:129136–129142. Copyright 2019 American Chemical Society.212 (C) Schematic of the dual-mode biosensor for exosomes detection based on MXenes and black phosphorus quantum dots. Reprinted with permission from Fang D, Zhao D, Zhang S, Huang Y, Dai H, Lin Y. Black phosphorus quantum dots functionalized MXenes as the enhanced dual-mode probe for exosomes sensing. Sens Actuat B Chem. 2020;305:127544–127552. Copyright 2020 Elsevier B.V.214
Nanomaterials with excellent catalytic properties have been increasingly utilized in the development of ECL biosensors. For example, MXenes have been concerned in the areas of catalysis, biosensor and supercapacitors, owing to their excellent electron transfer ability and unique catalytic ability. Wang and co-workers reported a MXenes-catalyzed ECL biosensor for the assay of exosomes.213 Fang et al proposed a self-enhanced ECL and photothermal dual-mode biosensor for exosome detection.214 As shown in Figure 15C, MXenes were employed to carry black phosphorus (BP) quantum dots (BPQDs), Ru(dcbpy)32+ and CD63 antibody. In the nanocomposite, BPQDs not only catalyzed the oxidization of Ru(dcbpy)32+, but also acted as the coreactants. The integration of BPQDs and Ru(dcbpy)32+ into MXenes could shorten the distance and amplify the ECL signal. Moreover, both MXenes and BPQDs with good photothermal property can be used as the photothermal probes. The dual-mode biosensor exhibited a linear range of 1.1×102 ~ 1.1×107 particles/μL. However, the stabilizers for regulating the structure and morphology and biomolecules for the specific recognition may block the active sites and hinder the electron transfer. Thus, Zhang et al reported an ECL biosensor for exosome detection based on the in-situ formation of AuNPs on the aptamer-modified MXenes (Figure 16).215 MXenes were modified with CD63 aptamer to identify exosomes. After the capture of exosomes, aptamer-conjugated MXenes were adsorbed on the surface of exosomes. MXenes with large surface area and strong reduction ability could induce the in-situ formation of AuNPs on their surface without the addition of extra reducing agents. AuNPs with catalytic surface greatly enhanced the ECL signal of luminol. The proposed biosensor showed a detection limit down to 30 particles/μL. In recent years, g-C3N4 was used not only as the carrier of luminol, but also as a catalyst to promote the reaction of luminol with H2O2, thus amplifying the ECL signal for exosome detection.216
Figure 16.
Schematic of the ECL biosensor for exosomes detection based on in situ formation of AuNPs decorated MXenes nanoprobes Reprinted with permission from Zhang H, Wang Z, Wang F, Zhang Y, Wang H, Liu Y. In situ formation of gold nanoparticles decorated Ti3C2 MXenes nanoprobe for highly sensitive electrogenerated chemiluminescence detection of exosomes and their surface proteins. Anal Chem. 2020;92:5546–5553. Copyright 2020 American Chemical Society.215
Photoelectrochemical Methods
A photoelectrochemical (PEC) process involves in the photocurrent as the detection signal generated by photoelectrochemically active materials under light illustration. The separation of the excitation source (light) from the detection signal (electric current) endows PEC biosensors with low background signal, high signal-to-noise ratio and excellent stability. Photosensitive materials play a vital role in the fabrication of PEC biosensors. Typically, Li’s group designed a cathodic PEC aptasensor for the detection of exosomes using p-typeNiO/BiOI/AuNP composites sensitized by CdSe QDs.217 In this work, BiOI with a narrowbandgap (1.8 eV) was immobilized on the NiO-modified ITO to sensitize the wide band gap of NiO (3.6–4.0 eV). Then, AuNPs were deposited on the surface to conjugate EpCAM aptamers through the formation of Au-S bonds. DNA2-modified QDs were bound to the electrode surface through the hybridization, which sensitized the nanocomposite and improved the photocurrent intensity. In the presence of exosomes, aptamer bound to the EpCAM protein on the surface of exosomes, resulting in the release of QDs and the decrease of the photocurrent. Meanwhile, exosomes on the electrode surface hindered the electron transfer between electrode and electron acceptor, leading to the further attenuation of the photocurrent signal.
Other Methods
Chemiluminescence is generated from the exergonic chemical reaction, in which an intermediate molecule in singlet excited state undergoes radiative decay. Chemiluminescence biosensors can sensitively detect targets of interest in the dark without any extra input energy.218 Zhong’s group reported a chemiluminescence strategy for rapid isolation and quantification of exosomes based on CuS-enclosed microgels.219 As displayed in Figure 17, CuS NPs were in-situ generated in porous microgel, which further modified with streptavidin and antibody. After interaction with exosomes, microgels promoted the isolation of exosomes through membrane filtration. A large number of Cu2+ ions were liberated and catalyzed the reaction between H2O2 and luminol derivate, producing strong a chemiluminescence signal. Moreover, Wang et al reported a chemiluminescence immunoassay for exosome detection by using antibody-conjugated superparamagnetic iron oxide particles (SIOPs) and acridinium ester as the signal label.220
Figure 17.
Schematic of CuS-microgel synthesis and the CuS-microgel-based assay for exosome quantification. Reprinted with permission from Jiang Q, Liu Y, Wang L, Adkins GB, Zhong W. Rapid enrichment and detection of extracellular vesicles enabled by cus-enclosed microgels. Anal Chem. 2019;91:15951–15958. Copyright 2019 American Chemical Society.219
Mass spectrometer (MS) is an effective tool to characterize the content of biomolecules at high throughput.221 However, the low sensitivity limits its utilization in detect low concentration of exosomes at the early stage of cancers. The element-labeling strategy endow MS with the advantages of high selectivity and signal amplification. In this detection principle, nanomaterials not only act as matric materials for exosomes capture, but also can amplify MS signal due to the large amount of elements in nanomaterials.222–227 Recently, Zhang et al reported an ultrasensitive inductively coupled plasma−mass spectrometry (ICP-MS) method for the detection of exosomes by using UCNPs as element labels.228 As shown in Figure 18, three different elements (Y, Eu, and Tb) doped UCNPs modified with three different aptamers were linked to AuNPs, forming three statellite-like nanoassemblies. Exosomes with surface proteins could trigger the release of the corresponding aptamers conjugated with UCNPs from AuNPs. Then, the released UCNPs in solution were detected by ICP-MS. Finally, this sensitive and high-throughput method could distinguish exosomes from seven different cell lines.
Figure 18.
Schematic of ultrasensitive inductively coupled plasma−mass spectrometry method for the detection of exosomes by using UCNPs as element labels. Reprinted with permission from Zhang XW, Liu MX, He MQ, Chen S, Yu YL, Wang JH. Integral multielement signals by DNA-programmed UCNP-AuNP nanosatellite assemblies for ultrasensitive ICP-MS detection of exosomal proteins and cancer identification. Anal Chem. 2021;93:6437–6445. Copyright 2021 American Chemical Society.228
Conclusions
In this review, we have systematically reviewed various nanomaterials-based techniques for exosome detection. The unique characteristics of nanomaterials make them fascinating materials for signal transduction and biosensor development. Moreover, the sensitivity and selectivity of biosensors for exosome detection have greatly improved by integration with various elaborate DNA or enzyme-based signal amplification strategies. In spite of significant progress in exosome detection, there are still some challenges to be addressed. For example, stability of nanomaterials and selectivity for complex samples should be improved by appropriate modification with biocompatibility polymers or others. Biorecognition elements used in bioassays may suffer from the digestion of enzymes in real samples. For transforming research in laboratory into clinical practice, it is the key obstacle to develop standardized technical specifications, including collection of biological samples, isolation of exosomes from liposy and detection operation. It is beneficial to enhance the comparability of results and establish a reliable and comprehensive dataset for future study on exosomes. The cost-effective and high-throughput isolation and detection of exosomes is helpful to simultaneous identify a large amount of clinical samples. We believe that nanomaterials-based detection of exosomes will make greater progress with the collective and unremitting efforts of different fields such as chemistry, materials science and clinical diagnosis.
Funding Statement
This research was funded by the Program for Innovative Research Team of Science and Technology in the University of Henan Province (21IRTSTHN005), the National Natural Science Foundation of China (21804085), and the Research Funds for the Henan Key Laboratory of Biomolecular Recognition and Sensing (HKLBRSK1902).
Disclosure
The authors declare no conflict of interest.
References
- 1.Harding C, Stahl P. Transferrin recycling in reticulocytes: pH and iron are important determinants of ligand binding and processing. Biochem Biophys Res Commun. 1983;113:650–658. doi: 10.1016/0006-291X(83)91776-X [DOI] [PubMed] [Google Scholar]
- 2.Wang H, Chen K, Yang Z, et al. Diagnosis of invasive nonfunctional pituitary adenomas by serum extracellular vesicles. Anal Chem. 2019;91:9580–9589. doi: 10.1021/acs.analchem.9b00914 [DOI] [PubMed] [Google Scholar]
- 3.Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (misev2018): a position statement of the international society for extracellular vesicles and update of the misev2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan H, Li Y, Cheng S, Zeng Y. Advances in analytical technologies for extracellular vesicles. Anal Chem. 2021;93:4739–4774. doi: 10.1021/acs.analchem.1c00693 [DOI] [PubMed] [Google Scholar]
- 5.Hyun KA, Kim J, Gwak H, Jung HI. Isolation and enrichment of circulating biomarkers for cancer screening, detection, and diagnostics. Analyst. 2016;141:382–392. doi: 10.1039/C5AN01762A [DOI] [PubMed] [Google Scholar]
- 6.Jeppesen DK, Fenix AM, Franklin JL, et al. Reassessment of exosome composition. Cell. 2019;177:428–445. doi: 10.1016/j.cell.2019.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurunathan S, Kang MH, Kim JH. A comprehensive review on factors influences biogenesis, functions, therapeutic and clinical implications of exosomes. Int J Nanomed. 2021;16:1281–1312. doi: 10.2147/IJN.S291956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, El Andaloussi S, Wood MJ, et al. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. 2012;21:R125–R134. doi: 10.1093/hmg/dds317 [DOI] [PubMed] [Google Scholar]
- 9.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:6697–6712. doi: 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Nedawi K, Meehan B, Micallef J, et al. Intercellular transfer of the oncogenic receptor egfrviii by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725 [DOI] [PubMed] [Google Scholar]
- 11.Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C, Skog J, Hsu CH, et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip. 2010;10:505–511. doi: 10.1039/B916199F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ale Ebrahim S, Ashtari A, Zamani Pedram M, Ale Ebrahim N, Sanati-Nezhad A. Publication trends in exosomes nanoparticles for cancer detection. Int J Nanomed. 2020;15:4453–4470. doi: 10.2147/IJN.S247210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeBleu VS, Kalluri R. Exosomes as a multicomponent biomarker platform in cancer. Trends Cancer. 2020;6:767–774. doi: 10.1016/j.trecan.2020.03.007 [DOI] [PubMed] [Google Scholar]
- 15.Hoshino A, Kim HS, Bojmar L, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182:1044–1061. doi: 10.1016/j.cell.2020.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhat EA, Sajjad N, Thokar FM. Current advancement of exosomes as biomarkers for cancer diagnosis and forecasting. Cancer Treat Res Commun. 2021;28:100417. doi: 10.1016/j.ctarc.2021.100417 [DOI] [PubMed] [Google Scholar]
- 17.Kok VC, Yu CC. Cancer-derived exosomes: their role in cancer biology and biomarker development. Int J Nanomed. 2020;15:8019–8036. doi: 10.2147/IJN.S272378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo HK, Sunkara V, Park J, et al. Exodisc for rapid, size-selective, and efficient isolation and analysis of nanoscale extracellular vesicles from biological samples. ACS Nano. 2017;11:1360–1370. doi: 10.1021/acsnano.6b06131 [DOI] [PubMed] [Google Scholar]
- 19.Clayton A, Court J, Navabi H, et al. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J Immunol Methods. 2001;247:163–174. doi: 10.1016/S0022-1759(00)00321-5 [DOI] [PubMed] [Google Scholar]
- 20.Busatto S, Giacomini A, Montis C, Ronca R, Bergese P. Uptake profiles of human serum exosomes by murine and human tumor cells through combined use of colloidal nanoplasmonics and flow cytofluorimetric analysis. Anal Chem. 2018;90:7855–7861. doi: 10.1021/acs.analchem.7b04374 [DOI] [PubMed] [Google Scholar]
- 21.Zhou H, Yuen PS, Pisitkun T, et al. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006;69:1471–1476. doi: 10.1038/sj.ki.5000273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Pol E, Hoekstra AG, Sturk A, Otto C, van Leeuwen TG, Nieuwland R. Optical and non-optical methods for detection and characterization of microparticles and exosomes. J Thromb Haemost. 2010;8:2596–2607. doi: 10.1111/j.1538-7836.2010.04074.x [DOI] [PubMed] [Google Scholar]
- 23.Sun Y, Haglund TA, Rogers AJ, Ghanim AF, Sethu P. Review: microfluidics technologies for blood-based cancer liquid biopsies. Anal Chim Acta. 2018;1012:10–29. doi: 10.1016/j.aca.2017.12.050 [DOI] [PubMed] [Google Scholar]
- 24.Salomon C, Kobayashi M, Ashman K, Sobrevia L, Mitchell MD, Rice GE. Hypoxia-induced changes in the bioactivity of cytotrophoblast-derived exosomes. PLoS One. 2013;8:79636–79650. doi: 10.1371/journal.pone.0079636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Woo HK, Lee C, et al. EV-Ident: identifying tumor-specific extracellular vesicles by size fractionation and single-vesicle analysis. Anal Chem. 2020;92:6010–6018. doi: 10.1021/acs.analchem.0c00285 [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Shi J, Liu W, et al. A simple, specific and “on-off” type MUC1 fluorescence aptasensor based on exosomes for detection of breast cancer. Sens Actuat B Chem. 2018;276:552–559. doi: 10.1016/j.snb.2018.08.056 [DOI] [Google Scholar]
- 27.Zhao W, Cui CH, Bose S, et al. Bioinspired multivalent DNA network for capture and release of cells. Proc Natl Acad Sci U S A. 2012;109:19626–19631. doi: 10.1073/pnas.1211234109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao D, Ohlendorf J, Chen Y, et al. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS One. 2012;7:46874–46889. doi: 10.1371/journal.pone.0046874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akbarinejad A, Hisey CL, Brewster D, et al. Novel electrochemically switchable, flexible, microporous cloth that selectively captures, releases, and concentrates intact extracellular vesicles. ACS Appl Mater Interfaces. 2020;12:39005–39013. doi: 10.1021/acsami.0c11908 [DOI] [PubMed] [Google Scholar]
- 30.Liu C, Qie Y, Qin W, et al. Emerging immunoassay technologies for the rapid detection of exosomes. Sens Actuat B Chem. 2021;345:130336–130351. doi: 10.1016/j.snb.2021.130336 [DOI] [Google Scholar]
- 31.Ziaei P, Berkman CE, Norton MG. Review: isolation and detection of tumor-derived extracellular vesicles. ACS Appl Nano Mater. 2018;1:2004–2020. doi: 10.1021/acsanm.8b00267 [DOI] [Google Scholar]
- 32.Dong J, Zhang RY, Sun N, et al. Bio-inspired nanovilli chips for enhanced capture of tumor-derived extracellular vesicles: toward non-invasive detection of gene alterations in non-small cell lung cancer. ACS Appl Mater Interfaces. 2019;11:13973–13983. doi: 10.1021/acsami.9b01406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Kannisto E, Yu G, Reid ME, Patnaik SK, Wu Y. An immuno-biochip selectively captures tumor-derived exosomes and detects exosomal RNAs for cancer diagnosis. ACS Appl Mater Interfaces. 2018;10:43375–43386. doi: 10.1021/acsami.8b13971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibsen SD, Wright J, Lewis JM, et al. Rapid isolation and detection of exosomes and associated biomarkers from plasma. ACS Nano. 2017;11:6641–6651. doi: 10.1021/acsnano.7b00549 [DOI] [PubMed] [Google Scholar]
- 35.Zhao Z, Yang Y, Zeng Y, He M. A microfluidic exosearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip. 2016;16:489–496. doi: 10.1039/C5LC01117E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin S, Yu Z, Chen D, et al. Progress in microfluidics-based exosome separation and detection technologies for diagnostic applications. Small. 2020;16:1903916. doi: 10.1002/smll.201903916 [DOI] [PubMed] [Google Scholar]
- 37.Kang YT, Hadlock T, Jolly S, Nagrath S. Extracellular vesicles on demand (EVOD) chip for screening and quantification of cancer-associated extracellular vesicles. Biosens Bioelectron. 2020;168:112535. doi: 10.1016/j.bios.2020.112535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang YT, Purcell E, Palacios-Rolston C, et al. Isolation and profiling of circulating tumor-associated exosomes using extracellular vesicular lipid-protein binding affinity based microfluidic device. Small. 2019;15:e1903600. doi: 10.1002/smll.201903600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding L, Yang X, Gao Z, et al. A holistic review of the state-of-the-art microfluidics for exosome separation: an overview of the current status, existing obstacles, and future outlook. Small. 2021;17:e2007174. doi: 10.1002/smll.202007174 [DOI] [PubMed] [Google Scholar]
- 40.Rana A, Zhang Y, Esfandiari L. Advancements in microfluidic technologies for isolation and early detection of circulating cancer-related biomarkers. Analyst. 2018;143:2971–2991. doi: 10.1039/C7AN01965C [DOI] [PubMed] [Google Scholar]
- 41.Zhang P, Zhou X, He M, et al. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat Biomed Eng. 2019;3:438–451. doi: 10.1038/s41551-019-0356-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su W, Li H, Chen W, Qin J. Microfluidic strategies for label-free exosomes isolation and analysis. TrAC Trends Anal Chem. 2019;118:686–698. doi: 10.1016/j.trac.2019.06.037 [DOI] [Google Scholar]
- 43.Wang Y, Li Q, Shi H, et al. Microfluidic Raman biochip detection of exosomes: a promising tool for prostate cancer diagnosis. Lab Chip. 2020;20:4632–4637. doi: 10.1039/D0LC00677G [DOI] [PubMed] [Google Scholar]
- 44.Yang Q, Cheng L, Hu L, et al. An integrative microfluidic device for isolation and ultrasensitive detection of lung cancer-specific exosomes from patient urine. Biosens Bioelectron. 2020;163:112290. doi: 10.1016/j.bios.2020.112290 [DOI] [PubMed] [Google Scholar]
- 45.Zhu C, Li L, Wang Z, Irfan M, Qu F. Recent advances of aptasensors for exosomes detection. Biosens Bioelectron. 2020;160:112213. doi: 10.1016/j.bios.2020.112213 [DOI] [PubMed] [Google Scholar]
- 46.Zhang K, Yue Y, Wu S, Liu W, Shi J, Zhang Z. Rapid capture and nondestructive release of extracellular vesicles using aptamer-based magnetic isolation. ACS Sens. 2019;4:1245–1251. doi: 10.1021/acssensors.9b00060 [DOI] [PubMed] [Google Scholar]
- 47.Suwatthanarak T, Thiodorus IA, Tanaka M, et al. Microfluidic-based capture and release of cancer-derived exosomes via peptide-nanowire hybrid interface. Lab Chip. 2021;21:597–607. doi: 10.1039/D0LC00899K [DOI] [PubMed] [Google Scholar]
- 48.Feng Y, Guo Y, Li Y, et al. Lectin-mediated in situ rolling circle amplification on exosomes for probing cancer-related glycan pattern. Anal Chim Acta. 2018;1039:108–115. doi: 10.1016/j.aca.2018.07.040 [DOI] [PubMed] [Google Scholar]
- 49.Choi Y, Park U, Koo HJ, et al. Exosome-mediated diagnosis of pancreatic cancer using lectin-conjugated nanoparticles bound to selective glycans. Biosens Bioelectron. 2021;177:112980–112990. doi: 10.1016/j.bios.2021.112980 [DOI] [PubMed] [Google Scholar]
- 50.Zhang P, Dong B, Zeng E, et al. In vivo tracking of multiple tumor exosomes labeled by phospholipid-based bioorthogonal conjugation. Anal Chem. 2018;90:11273–11279. doi: 10.1021/acs.analchem.8b01506 [DOI] [PubMed] [Google Scholar]
- 51.Zhang W, Yu ZL, Wu M, et al. Magnetic and folate functionalization enables rapid isolation and enhanced tumor-targeting of cell-derived microvesicles. ACS Nano. 2017;11:277–290. doi: 10.1021/acsnano.6b05630 [DOI] [PubMed] [Google Scholar]
- 52.Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New technologies for analysis of extracellular vesicles. Chem Rev. 2018;118:1917–1950. doi: 10.1021/acs.chemrev.7b00534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng N, Du D, Wang X, et al. Recent advances in biosensors for detecting cancer-derived exosomes. Trends Biotechnol. 2019;37:1236–1254. doi: 10.1016/j.tibtech.2019.04.008 [DOI] [PubMed] [Google Scholar]
- 54.Kholafazad Kordasht H, Hasanzadeh M. Biomedical analysis of exosomes using biosensing methods: recent progress. Anal Methods. 2020;12:2795–2811. doi: 10.1039/D0AY00722F [DOI] [PubMed] [Google Scholar]
- 55.Xu H, Ye B-C. Advances in biosensing technologies for analysis of cancer-derived exosomes. TrAC Trend Anal Chem. 2020;123:115773–115784. doi: 10.1016/j.trac.2019.115773 [DOI] [Google Scholar]
- 56.Shao B, Xiao Z. Recent achievements in exosomal biomarkers detection by nanomaterials-based optical biosensors - a review. Anal Chim Acta. 2020;1114:74–84. doi: 10.1016/j.aca.2020.02.041 [DOI] [PubMed] [Google Scholar]
- 57.Yu X, Sha L, Dong L, Cao Y, Zhao J. Recent advances in bio-sensing methods for the detection of tumor exosomes. Crit Rev Anal Chem. 2020;1–19. doi: 10.1080/10408347.2020.18022201-19 [DOI] [PubMed] [Google Scholar]
- 58.Zhang L, Gu C, Wen J, Liu G, Liu H, Li L. Recent advances in nanomaterial-based biosensors for the detection of exosomes. Anal Bioanal Chem. 2021;413:83–102. doi: 10.1007/s00216-020-03000-0 [DOI] [PubMed] [Google Scholar]
- 59.Xiong H, Huang Z, Yang Z, et al. Recent progress in detection and profiling of cancer cell-derived exosomes. Small. 2021;17(35):2007971. doi: 10.1002/smll.202007971 [DOI] [PubMed] [Google Scholar]
- 60.Liu C, Xu X, Li B, et al. Single-exosome-counting immunoassays for cancer diagnostics. Nano Lett. 2018;18:4226–4232. doi: 10.1021/acs.nanolett.8b01184 [DOI] [PubMed] [Google Scholar]
- 61.Kashefi-Kheyrabadi L, Kim J, Chakravarty S, et al. Detachable microfluidic device implemented with electrochemical aptasensor (DeMEA) for sequential analysis of cancerous exosomes. Biosens Bioelectron. 2020;169:112622–112631. doi: 10.1016/j.bios.2020.112622 [DOI] [PubMed] [Google Scholar]
- 62.Tayebi M, Zhou Y, Tripathi P, Chandramohanadas R, Ai Y. Exosome purification and analysis using a facile microfluidic hydrodynamic trapping device. Anal Chem. 2020;92:10733–10742. doi: 10.1021/acs.analchem.0c02006 [DOI] [PubMed] [Google Scholar]
- 63.Zhou S, Hu T, Zhang F, et al. Integrated microfluidic device for accurate extracellular vesicle quantification and protein markers analysis directly from human whole blood. Anal Chem. 2020;92:1574–1581. [DOI] [PubMed] [Google Scholar]
- 64.Mohammadi M, Zargartalebi H, Salahandish R, Aburashed R, Wey Yong K, Sanati-Nezhad A. Emerging technologies and commercial products in exosome-based cancer diagnosis and prognosis. Biosens Bioelectron. 2021;183:113176–113203. [DOI] [PubMed] [Google Scholar]
- 65.Liu C, Zhao J, Tian F, Chang J, Zhang W, Sun J. Lambda-DNA- and aptamer-mediated sorting and analysis of extracellular vesicles. J Am Chem Soc. 2019;141:3817–3821. doi: 10.1021/jacs.9b00007 [DOI] [PubMed] [Google Scholar]
- 66.Li Q, Wang Y, Ling L, et al. Rapid and specific detection nanoplatform of serum exosomes for prostate cancer diagnosis. Microchim Acta. 2021;188:283–293. doi: 10.1007/s00604-021-04934-7 [DOI] [PubMed] [Google Scholar]
- 67.Zhand S, Xiao K, Razavi Bazaz S, et al. Improving capture efficiency of human cancer cell derived exosomes with nanostructured metal organic framework functionalized beads. Appl Mater Today. 2021;23:100994–101007. doi: 10.1016/j.apmt.2021.100994 [DOI] [Google Scholar]
- 68.Thorsteinsson K, Olsen E, Schmidt E, Pace H, Bally M. FRET-based assay for the quantification of extracellular vesicles and other vesicles of complex composition. Anal Chem. 2020;92:15336–15343. doi: 10.1021/acs.analchem.0c02271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lyu Y, Cui D, Huang J, Fan W, Miao Y, Pu K. Near-infrared afterglow semiconducting nano-polycomplexes for the multiplex differentiation of cancer exosomes. Angew Chem Int Ed. 2019;58:4983–4987. doi: 10.1002/anie.201900092 [DOI] [PubMed] [Google Scholar]
- 70.Li N, Huang Z, Zhang X, Song X, Xiao Y. Reflecting size differences of exosomes by using the combination of membrane-targeting viscosity probe and fluorescence lifetime imaging microscopy. Anal Chem. 2019;91:15308–15316. doi: 10.1021/acs.analchem.9b04587 [DOI] [PubMed] [Google Scholar]
- 71.Wang F, Zhang Y, Chen D, Zhang Z, Li Z. Single microbead-based fluorescent aptasensor (SMFA) for direct isolation and in situ quantification of exosomes from plasma. Analyst. 2021;146:3346–3351. doi: 10.1039/D1AN00463H [DOI] [PubMed] [Google Scholar]
- 72.Xu R, Fitts A, Li X, et al. Quantification of small extracellular vesicles by size exclusion chromatography with fluorescence detection. Anal Chem. 2016;88:10390–10394. doi: 10.1021/acs.analchem.6b03348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lazaro-Ibanez E, Faruqu FN, Saleh AF, et al. Selection of fluorescent, bioluminescent, and radioactive tracers to accurately reflect extracellular vesicle biodistribution in vivo. ACS Nano. 2021;15:3212–3227. doi: 10.1021/acsnano.0c09873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujii T, Kaneta T. Direct counting of exosomes in a cell culture medium using neither isolation nor preconcentration. Anal Chim Acta. 2020;1119:35–40. doi: 10.1016/j.aca.2020.04.052 [DOI] [PubMed] [Google Scholar]
- 75.Wang L, Yang Y, Liu Y, Ning L, Xiang Y, Li G. Bridging exosome and liposome through zirconium-phosphate coordination chemistry: a new method for exosome detection. Chem Commun. 2019;55:2708–2711. doi: 10.1039/C9CC00220K [DOI] [PubMed] [Google Scholar]
- 76.Zhang P, He M, Zeng Y. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab Chip. 2016;16:3033–3042. doi: 10.1039/C6LC00279J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wan S, Zhang L, Wang S, et al. Molecular recognition-based DNA nanoassemblies on the surfaces of nanosized exosomes. J Am Chem Soc. 2017;139:5289–5292. doi: 10.1021/jacs.7b00319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He D, Ho SL, Chan HN, et al. Molecular-recognition-based DNA nanodevices for enhancing the direct visualization and quantification of single vesicles of tumor exosomes in plasma microsamples. Anal Chem. 2019;91:2768–2775. doi: 10.1021/acs.analchem.8b04509 [DOI] [PubMed] [Google Scholar]
- 79.Tian Q, He C, Liu G, et al. Nanoparticle counting by microscopic digital detection: selective quantitative analysis of exosomes via surface-anchored nucleic acid amplification. Anal Chem. 2018;90:6556–6562. [DOI] [PubMed] [Google Scholar]
- 80.Zhao X, Zeng L, Mei Q, Luo Y. Allosteric probe-initiated wash-free method for sensitive extracellular vesicle detection through dual cycle-assisted CRISPR-Cas12a. ACS Sens. 2020;5:2239–2246. doi: 10.1021/acssensors.0c00944 [DOI] [PubMed] [Google Scholar]
- 81.Chen J, Meng HM, An Y, et al. Structure-switching aptamer triggering hybridization displacement reaction for label-free detection of exosomes. Talanta. 2020;209:120510–120517. doi: 10.1016/j.talanta.2019.120510 [DOI] [PubMed] [Google Scholar]
- 82.Wang L, Pan Y, Liu Y, et al. Fabrication of an aptamer-coated liposome complex for the detection and profiling of exosomes based on terminal deoxynucleotidyl transferase-mediated signal amplification. ACS Appl Mater Interfaces. 2020;12:322–329. doi: 10.1021/acsami.9b18869 [DOI] [PubMed] [Google Scholar]
- 83.Shi L, Ba L, Xiong Y, Peng G. A hybridization chain reaction based assay for fluorometric determination of exosomes using magnetic nanoparticles and both aptamers and antibody as recognition elements. Microchim Acta. 2019;186:796–802. doi: 10.1007/s00604-019-3823-9 [DOI] [PubMed] [Google Scholar]
- 84.Wang X, Shang H, Ma C, Chen L. A fluorescence assay for exosome detection based on bivalent cholesterol anchor triggered target conversion and enzyme-free signal amplification. Anal Chem. 2021;93:8493–8500. doi: 10.1021/acs.analchem.1c00796 [DOI] [PubMed] [Google Scholar]
- 85.Yu X, He L, Pentok M, et al. An aptamer-based new method for competitive fluorescence detection of exosomes. Nanoscale. 2019;11:15589–15595. doi: 10.1039/C9NR04050A [DOI] [PubMed] [Google Scholar]
- 86.Zhang H, Zhou Y, Luo D, et al. Immunoassay-aptasensor for the determination of tumor-derived exosomes based on the combination of magnetic nanoparticles and hybridization chain reaction. RSC Adv. 2021;11:4983–4990. doi: 10.1039/D0RA10159A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang H, Wan K, Zhou Y, et al. A three-dimensional multipedal DNA walker for the ultrasensitive detection of tumor exosomes. Chem Commun. 2020;56:12949–12952. doi: 10.1039/D0CC04360E [DOI] [PubMed] [Google Scholar]
- 88.Huang L, Wang DB, Singh N, Yang F, Gu N, Zhang XE. A dual-signal amplification platform for sensitive fluorescence biosensing of leukemia-derived exosomes. Nanoscale. 2018;10:20289–20295. doi: 10.1039/C8NR07720G [DOI] [PubMed] [Google Scholar]
- 89.Huang Z, Lin Q, Yang B, et al. Cascade signal amplification for sensitive detection of exosomes by integrating tyramide and surface-initiated enzymatic polymerization. Chem Commun. 2020;56:12793–12796. doi: 10.1039/D0CC04881J [DOI] [PubMed] [Google Scholar]
- 90.Gao ML, He F, Yin BC, Ye BC. A dual signal amplification method for exosome detection based on DNA dendrimer self-assembly. Analyst. 2019;144:1995–2002. doi: 10.1039/C8AN02383B [DOI] [PubMed] [Google Scholar]
- 91.Pan Y, Wang L, Deng Y, et al. A simple and sensitive method for exosome detection based on steric hindrance-controlled signal amplification. Chem Commun. 2020;56:13768–13771. doi: 10.1039/D0CC06113A [DOI] [PubMed] [Google Scholar]
- 92.Li P, Yu X, Han W, et al. Ultrasensitive and reversible nanoplatform of urinary exosomes for prostate cancer diagnosis. ACS Sens. 2019;4:1433–1441. doi: 10.1021/acssensors.9b00621 [DOI] [PubMed] [Google Scholar]
- 93.Dobhal G, Ayupova D, Laufersky G, Ayed Z, Nann T, Goreham RV. Cadmium-free quantum dots as fluorescent labels for exosomes. Sensors. 2018;18:3308–3320. doi: 10.3390/s18103308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu N, Li G, Zhou J, et al. A light-up fluorescence resonance energy transfer magnetic aptamer-sensor for ultra-sensitive lung cancer exosome detection. J Mater Chem B. 2021;9:2483–2493. doi: 10.1039/D1TB00046B [DOI] [PubMed] [Google Scholar]
- 95.Wu M, Chen Z, Xie Q, et al. One-step quantification of salivary exosomes based on combined aptamer recognition and quantum dot signal amplification. Biosens Bioelectron. 2021;171:112733–112742. doi: 10.1016/j.bios.2020.112733 [DOI] [PubMed] [Google Scholar]
- 96.Zhang J, Zhu Y, Shi J, Zhang K, Zhang Z, Zhang H. Sensitive signal amplifying a diagnostic biochip based on a biomimetic periodic nanostructure for detecting cancer exosomes. ACS Appl Mater Interfaces. 2020;12:33473–33482. doi: 10.1021/acsami.0c06785 [DOI] [PubMed] [Google Scholar]
- 97.Dong D, Zhu L, Hu J, Pang DW, Zhang ZL. Simple and rapid extracellular vesicles quantification via membrane biotinylation strategy coupled with fluorescent nanospheres-based lateral flow assay. Talanta. 2019;200:408–414. doi: 10.1016/j.talanta.2019.03.069 [DOI] [PubMed] [Google Scholar]
- 98.He F, Wang J, Yin BC, Ye BC. Quantification of exosome based on a copper-mediated signal amplification strategy. Anal Chem. 2018;90:8072–8079. doi: 10.1021/acs.analchem.8b01187 [DOI] [PubMed] [Google Scholar]
- 99.Yu Y, Zhang WS, Guo Y, et al. Engineering of exosome-triggered enzyme-powered DNA motors for highly sensitive fluorescence detection of tumor-derived exosomes. Biosens Bioelectron. 2020;167:112482–112490. doi: 10.1016/j.bios.2020.112482 [DOI] [PubMed] [Google Scholar]
- 100.Chen X, Lan J, Liu Y, et al. A paper-supported aptasensor based on upconversion luminescence resonance energy transfer for the accessible determination of exosomes. Biosens Bioelectron. 2018;102:582–588. doi: 10.1016/j.bios.2017.12.012 [DOI] [PubMed] [Google Scholar]
- 101.Jin D, Yang F, Zhang Y, et al. Exoapp: exosome-oriented, aptamer nanoprobe-enabled surface proteins profiling and detection. Anal Chem. 2018;90:14402–14411. doi: 10.1021/acs.analchem.8b03959 [DOI] [PubMed] [Google Scholar]
- 102.Wang H, Chen H, Huang Z, Li T, Deng A, Kong J. DNase I enzyme-aided fluorescence signal amplification based on graphene oxide-DNA aptamer interactions for colorectal cancer exosome detection. Talanta. 2018;184:219–226. doi: 10.1016/j.talanta.2018.02.083 [DOI] [PubMed] [Google Scholar]
- 103.Li B, Pan W, Liu C, et al. Homogenous magneto-fluorescent nanosensor for tumor-derived exosome isolation and analysis. ACS Sens. 2020;5:2052–2060. doi: 10.1021/acssensors.0c00513 [DOI] [PubMed] [Google Scholar]
- 104.Tayebi M, Tavakkoli Yaraki M, Yang HY, Ai Y. A mos2–mwcnt based fluorometric nanosensor for exosome detection and quantification. Nanoscale Adv. 2019;1:2866–2872. doi: 10.1039/C9NA00248K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang Q, Wang F, Zhang H, Zhang Y, Liu M, Liu Y. Universal Ti3C2 MXenes based self-standard ratiometric fluorescence resonance energy transfer platform for highly sensitive detection of exosomes. Anal Chem. 2018;90:12737–12744. doi: 10.1021/acs.analchem.8b03083 [DOI] [PubMed] [Google Scholar]
- 106.Lee J, Kim H, Heo Y, et al. Enhanced paper-based ELISA for simultaneous EVs/exosome isolation and detection using streptavidin agarose-based immobilization. Analyst. 2019;145:157–164. doi: 10.1039/C9AN01140D [DOI] [PubMed] [Google Scholar]
- 107.Xia Y, Chen T, Chen G, et al. A nature-inspired colorimetric and fluorescent dual-modal biosensor for exosomes detection. Talanta. 2020;214:120851–120861. doi: 10.1016/j.talanta.2020.120851 [DOI] [PubMed] [Google Scholar]
- 108.Hartman ZC, Wei J, Glass OK, et al. Increasing vaccine potency through exosome antigen targeting. Vaccine. 2011;29:9361–9367. doi: 10.1016/j.vaccine.2011.09.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Khodashenas S, Khalili S, Forouzandeh Moghadam M. A cell ELISA based method for exosome detection in diagnostic and therapeutic applications. Biotechnol Lett. 2019;41:523–531. doi: 10.1007/s10529-019-02667-5 [DOI] [PubMed] [Google Scholar]
- 110.Logozzi M, De Milito A, Lugini L, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4:5219–5229. doi: 10.1371/journal.pone.0005219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou Y, Xu H, Wang H, Ye BC. Detection of breast cancer-derived exosomes using the horseradish peroxidase-mimicking dnazyme as an aptasensor. Analyst. 2019;145:107–114. doi: 10.1039/C9AN01653H [DOI] [PubMed] [Google Scholar]
- 112.Xu H, Zheng L, Zhou Y, Ye B-C. An artificial enzyme cascade amplification strategy for highly sensitive and specific detection of breast cancer-derived exosomes. Analyst. 2021;146(18):5542–5549. doi: 10.1039/d1an01071a [DOI] [PubMed] [Google Scholar]
- 113.Chen Z, Cheng SB, Cao P, et al. Detection of exosomes by ZnO nanowires coated three-dimensional scaffold chip device. Biosens Bioelectron. 2018;122:211–216. doi: 10.1016/j.bios.2018.09.033 [DOI] [PubMed] [Google Scholar]
- 114.Huang Z, Lin Q, Ye X, et al. Terminal deoxynucleotidyl transferase based signal amplification for enzyme-linked aptamer-sorbent assay of colorectal cancer exosomes. Talanta. 2020;218:121089–121097. doi: 10.1016/j.talanta.2020.121089 [DOI] [PubMed] [Google Scholar]
- 115.Moura SL, Martin CG, Marti M, Pividori MI. Multiplex detection and characterization of breast cancer exosomes by magneto-actuated immunoassay. Talanta. 2020;211:120657–120666. doi: 10.1016/j.talanta.2019.120657 [DOI] [PubMed] [Google Scholar]
- 116.He F, Liu H, Guo X, Yin BC, Ye BC. Direct exosome quantification via bivalent-cholesterol-labeled DNA anchor for signal amplification. Anal Chem. 2017;89:12968–12975. doi: 10.1021/acs.analchem.7b03919 [DOI] [PubMed] [Google Scholar]
- 117.Zeng R, Wang J, Wang Q, Tang D, Lin Y. Horseradish peroxidase-encapsulated DNA nanoflowers: an innovative signal-generation tag for colorimetric biosensor. Talanta. 2021;221:121600–121607. doi: 10.1016/j.talanta.2020.121600 [DOI] [PubMed] [Google Scholar]
- 118.Yang Y, Li C, Shi H, Chen T, Wang Z, Li G. A pH-responsive bioassay for paper-based diagnosis of exosomes via mussel-inspired surface chemistry. Talanta. 2019;192:325–330. doi: 10.1016/j.talanta.2018.09.067 [DOI] [PubMed] [Google Scholar]
- 119.Xu L, Chopdat R, Li D, Al-Jamal KT. Development of a simple, sensitive and selective colorimetric aptasensor for the detection of cancer-derived exosomes. Biosens Bioelectron. 2020;169:112576–112586. doi: 10.1016/j.bios.2020.112576 [DOI] [PubMed] [Google Scholar]
- 120.Maiolo D, Paolini L, Di Noto G, et al. Colorimetric nanoplasmonic assay to determine purity and titrate extracellular vesicles. Anal Chem. 2015;87:4168–4176. doi: 10.1021/ac504861d [DOI] [PubMed] [Google Scholar]
- 121.Jiang Y, Shi M, Liu Y, et al. Aptamer/AuNP biosensor for colorimetric profiling of exosomal proteins. Angew Chem. 2017;129:12078–12082. doi: 10.1002/ange.201703807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu W, Li J, Wu Y, Xing S, Lai Y, Zhang G. Target-induced proximity ligation triggers recombinase polymerase amplification and transcription-mediated amplification to detect tumor-derived exosomes in nasopharyngeal carcinoma with high sensitivity. Biosens Bioelectron. 2018;102:204–210. doi: 10.1016/j.bios.2017.11.033 [DOI] [PubMed] [Google Scholar]
- 123.Lopez-Cobo S, Campos-Silva C, Moyano A, et al. Immunoassays for scarce tumour-antigens in exosomes: detection of the human NKG2D-ligand, MICA, in tetraspanin-containing nanovesicles from melanoma. J Nanobiotechnol. 2018;16:47–59. doi: 10.1186/s12951-018-0372-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Oliveira-Rodriguez M, Lopez-Cobo S, Reyburn HT, et al. Development of a rapid lateral flow immunoassay test for detection of exosomes previously enriched from cell culture medium and body fluids. J Extracell Vesicles. 2016;5:31803–31814. doi: 10.3402/jev.v5.31803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Oliveira-Rodriguez M, Serrano-Pertierra E, Garcia AC, et al. Point-of-care detection of extracellular vesicles: sensitivity optimization and multiple-target detection. Biosens Bioelectron. 2017;87:38–45. doi: 10.1016/j.bios.2016.08.001 [DOI] [PubMed] [Google Scholar]
- 126.Wu T, Yang Y, Cao Y, et al. Enhanced lateral flow assay with double conjugates for the detection of exosomes. Sci China Chem. 2018;61:1423–1429. doi: 10.1007/s11426-018-9305-6 [DOI] [Google Scholar]
- 127.Zhang Y, Wang D, Yue S, et al. Sensitive multicolor visual detection of exosomes via dual signal amplification strategy of enzyme-catalyzed metallization of Au nanorods and hybridization chain reaction. ACS Sens. 2019;4:3210–3218. doi: 10.1021/acssensors.9b01644 [DOI] [PubMed] [Google Scholar]
- 128.Zhang Y, Jiao J, Wei Y, Wang D, Yang C, Xu Z. Plasmonic colorimetric biosensor for sensitive exosome detection via enzyme-induced etching of gold nanobipyramid@MnO2 nanosheet nanostructures. Anal Chem. 2020;92:15244–15252. doi: 10.1021/acs.analchem.0c04136 [DOI] [PubMed] [Google Scholar]
- 129.Farhana FZ, Umer M, Saeed A, et al. Isolation and detection of exosomes using Fe2O3 nanoparticles. ACS Appl Nano Mater. 2021;4:1175–1186. doi: 10.1021/acsanm.0c02807 [DOI] [Google Scholar]
- 130.Liu B, Liu J. Accelerating peroxidase mimicking nanozymes using DNA. Nanoscale. 2015;7:13831–13835. doi: 10.1039/C5NR04176G [DOI] [PubMed] [Google Scholar]
- 131.Chen J, Xu Y, Lu Y, Xing W. Isolation and visible detection of tumor-derived exosomes from plasma. Anal Chem. 2018;90:14207–14215. doi: 10.1021/acs.analchem.8b03031 [DOI] [PubMed] [Google Scholar]
- 132.Wang YM, Liu JW, Adkins GB, et al. Enhancement of the intrinsic peroxidase-like activity of graphitic carbon nitride nanosheets by ssDNAs and its application for detection of exosomes. Anal Chem. 2017;89:12327–12333. doi: 10.1021/acs.analchem.7b03335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xia Y, Liu M, Wang L, et al. A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens Bioelectron. 2017;92:8–15. doi: 10.1016/j.bios.2017.01.063 [DOI] [PubMed] [Google Scholar]
- 134.Zhang Y, Su Q, Song D, Fan J, Xu Z. Label-free detection of exosomes based on ssDNA-modulated oxidase-mimicking activity of CuCo2O4 nanorods. Anal Chim Acta. 2021;1145:9–16. doi: 10.1016/j.aca.2020.12.018 [DOI] [PubMed] [Google Scholar]
- 135.Shpacovitch V, Hergenroder R. Optical and surface plasmonic approaches to characterize extracellular vesicles. A review. Anal Chim Acta. 2018;1005:1–15. doi: 10.1016/j.aca.2017.11.066 [DOI] [PubMed] [Google Scholar]
- 136.Jackman JA, Yorulmaz Avsar S, Ferhan AR, et al. Quantitative profiling of nanoscale liposome deformation by a localized surface plasmon resonance sensor. Anal Chem. 2017;89:1102–1109. doi: 10.1021/acs.analchem.6b02532 [DOI] [PubMed] [Google Scholar]
- 137.Liu C, Zeng X, An Z, et al. Sensitive detection of exosomal proteins via a compact surface plasmon resonance biosensor for cancer diagnosis. ACS Sens. 2018;3:1471–1479. doi: 10.1021/acssensors.8b00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Picciolini S, Gualerzi A, Vanna R, et al. Detection and characterization of different brain-derived subpopulations of plasma exosomes by surface plasmon resonance imaging. Anal Chem. 2018;90:8873–8880. doi: 10.1021/acs.analchem.8b00941 [DOI] [PubMed] [Google Scholar]
- 139.Rupert DL, Lasser C, Eldh M, et al. Determination of exosome concentration in solution using surface plasmon resonance spectroscopy. Anal Chem. 2014;86:5929–5936. doi: 10.1021/ac500931f [DOI] [PubMed] [Google Scholar]
- 140.Rupert DLM, Shelke GV, Emilsson G, et al. Dual-wavelength surface plasmon resonance for determining the size and concentration of sub-populations of extracellular vesicles. Anal Chem. 2016;88:9980–9988. doi: 10.1021/acs.analchem.6b01860 [DOI] [PubMed] [Google Scholar]
- 141.Thakur A, Qiu G, Ng SP, et al. Direct detection of two different tumor-derived extracellular vesicles by SAM-AuNIs LSPR biosensor. Biosens Bioelectron. 2017;94:400–407. doi: 10.1016/j.bios.2017.03.036 [DOI] [PubMed] [Google Scholar]
- 142.Yang Y, Zhai C, Zeng Q, Khan AL, Yu H. Multifunctional detection of extracellular vesicles with surface plasmon resonance microscopy. Anal Chem. 2020;92:4884–4890. doi: 10.1021/acs.analchem.9b04622 [DOI] [PubMed] [Google Scholar]
- 143.Zhu L, Wang K, Cui J, et al. Label-free quantitative detection of tumor-derived exosomes through surface plasmon resonance imaging. Anal Chem. 2014;86:8857–8864. doi: 10.1021/ac5023056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Im H, Shao H, Park YI, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol. 2014;32:490–495. doi: 10.1038/nbt.2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mao Z, Zhao J, Chen J, Hu X, Koh K, Chen H. A simple and direct spr platform combining three-in-one multifunctional peptides for ultra-sensitive detection of PD-L1 exosomes. Sens Actuat B Chem. 2021;346:130496–130505. doi: 10.1016/j.snb.2021.130496 [DOI] [Google Scholar]
- 146.Reiner AT, Ferrer NG, Venugopalan P, Lai RC, Lim SK, Dostalek J. Magnetic nanoparticle-enhanced surface plasmon resonance biosensor for extracellular vesicle analysis. Analyst. 2017;142:3913–3921. doi: 10.1039/C7AN00469A [DOI] [PubMed] [Google Scholar]
- 147.Di Noto G, Bugatti A, Zendrini A, et al. Merging colloidal nanoplasmonics and surface plasmon resonance spectroscopy for enhanced profiling of multiple myeloma-derived exosomes. Biosens Bioelectron. 2016;77:518–524. doi: 10.1016/j.bios.2015.09.061 [DOI] [PubMed] [Google Scholar]
- 148.Fan Y, Duan X, Zhao M, et al. High-sensitive and multiplex biosensing assay of NSCLC-derived exosomes via different recognition sites based on SPRi array. Biosens Bioelectron. 2020;154:112066–112073. doi: 10.1016/j.bios.2020.112066 [DOI] [PubMed] [Google Scholar]
- 149.Park J, Im H, Hong S, Castro CM, Weissleder R, Lee H. Analyses of intravesicular exosomal proteins using a nano-plasmonic system. ACS Photonics. 2018;5:487–494. doi: 10.1021/acsphotonics.7b00992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang C, Wang C, Jin D, et al. AuNP-amplified surface acoustic wave sensor for the quantification of exosomes. ACS Sens. 2020;5:362–369. doi: 10.1021/acssensors.9b01869 [DOI] [PubMed] [Google Scholar]
- 151.Wang Q, Zou L, Yang X, et al. Direct quantification of cancerous exosomes via surface plasmon resonance with dual gold nanoparticle-assisted signal amplification. Biosens Bioelectron. 2019;135:129–136. doi: 10.1016/j.bios.2019.04.013 [DOI] [PubMed] [Google Scholar]
- 152.Liao G, Liu X, Yang X, et al. Surface plasmon resonance assay for exosomes based on aptamer recognition and polydopamine-functionalized gold nanoparticles for signal amplification. Microchim Acta. 2020;187:251–260. doi: 10.1007/s00604-020-4183-1 [DOI] [PubMed] [Google Scholar]
- 153.Chen W, Li J, Wei X, et al. Surface plasmon resonance biosensor using hydrogel-AuNP supramolecular spheres for determination of prostate cancer-derived exosomes. Microchim Acta. 2020;187:590–600. doi: 10.1007/s00604-020-04573-4 [DOI] [PubMed] [Google Scholar]
- 154.Shin H, Oh S, Hong S, et al. Early-stage lung cancer diagnosis by deep learning-based spectroscopic analysis of circulating exosomes. ACS Nano. 2020;14:5435–5444. doi: 10.1021/acsnano.9b09119 [DOI] [PubMed] [Google Scholar]
- 155.Avella-Oliver M, Puchades R, Wachsmann-Hogiu S, Maquieira A. Label-free SERS analysis of proteins and exosomes with large-scale substrates from recordable compact disks. Sens Actuat B Chem. 2017;252:657–662. doi: 10.1016/j.snb.2017.06.058 [DOI] [Google Scholar]
- 156.Dong S, Wang Y, Liu Z, et al. Beehive-inspired macroporous SERS probe for cancer detection through capturing and analyzing exosomes in plasma. ACS Appl Mater Interfaces. 2020;12:5136–5146. doi: 10.1021/acsami.9b21333 [DOI] [PubMed] [Google Scholar]
- 157.Lee W, Nanou A, Rikkert L, et al. Label-free prostate cancer detection by characterization of extracellular vesicles using Raman spectroscopy. Anal Chem. 2018;90:11290–11296. doi: 10.1021/acs.analchem.8b01831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Park J, Hwang M, Choi B, et al. Exosome classification by pattern analysis of surface-enhanced Raman spectroscopy data for lung cancer diagnosis. Anal Chem. 2017;89:6695–6701. doi: 10.1021/acs.analchem.7b00911 [DOI] [PubMed] [Google Scholar]
- 159.Yan Z, Dutta S, Liu Z, et al. A label-free platform for identification of exosomes from different sources. ACS Sens. 2019;4:488–497. doi: 10.1021/acssensors.8b01564 [DOI] [PubMed] [Google Scholar]
- 160.Hou M, He D, Bu H, et al. A sandwich-type surface-enhanced Raman scattering sensor using dual aptamers and gold nanoparticles for the detection of tumor extracellular vesicles. Analyst. 2020;145:6232–6236. doi: 10.1039/D0AN01385D [DOI] [PubMed] [Google Scholar]
- 161.Li TD, Zhang R, Chen H, et al. An ultrasensitive polydopamine bi-functionalized SERS immunoassay for exosome-based diagnosis and classification of pancreatic cancer. Chem Sci. 2018;9:5372–5382. doi: 10.1039/C8SC01611A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tian YF, Ning CF, He F, Yin BC, Ye BC. Highly sensitive detection of exosomes by SERS using gold nanostar@Raman reporter@nanoshell structures modified with a bivalent cholesterol-labeled DNA anchor. Analyst. 2018;143:4915–4922. doi: 10.1039/C8AN01041B [DOI] [PubMed] [Google Scholar]
- 163.Zong S, Wang L, Chen C, et al. Facile detection of tumor-derived exosomes using magnetic nanobeads and SERS nanoprobes. Anal Methods. 2016;8:5001–5008. doi: 10.1039/C6AY00406G [DOI] [Google Scholar]
- 164.Zhang W, Jiang L, Diefenbach RJ, et al. Enabling sensitive phenotypic profiling of cancer-derived small extracellular vesicles using surface-enhanced Raman spectroscopy nanotags. ACS Sens. 2020;5:764–771. doi: 10.1021/acssensors.9b02377 [DOI] [PubMed] [Google Scholar]
- 165.Di H, Zeng E, Zhang P, et al. General approach to engineering extracellular vesicles for biomedical analysis. Anal Chem. 2019;91:12752–12759. doi: 10.1021/acs.analchem.9b02268 [DOI] [PubMed] [Google Scholar]
- 166.Wang Z, Zong S, Wang Y, et al. Screening and multiple detection of cancer exosomes using an SERS-based method. Nanoscale. 2018;10:9053–9062. doi: 10.1039/C7NR09162A [DOI] [PubMed] [Google Scholar]
- 167.Kwizera EA, O’Connor R, Vinduska V, et al. Molecular detection and analysis of exosomes using surface-enhanced Raman scattering gold nanorods and a miniaturized device. Theranostics. 2018;8:2722–2738. doi: 10.7150/thno.21358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ning CF, Wang L, Tian YF, Yin BC, Ye BC. Multiple and sensitive SERS detection of cancer-related exosomes based on gold-silver bimetallic nanotrepangs. Analyst. 2020;145:2795–2804. doi: 10.1039/C9AN02180A [DOI] [PubMed] [Google Scholar]
- 169.Zhang X, Liu C, Pei Y, Song W, Zhang S. Preparation of a novel Raman probe and its application in the detection of circulating tumor cells and exosomes. ACS Appl Mater Interfaces. 2019;11:28671–28680. doi: 10.1021/acsami.9b09465 [DOI] [PubMed] [Google Scholar]
- 170.Dai Y, Bai S, Hu C, Chu K, Shen B, Smith ZJ. Combined morpho-chemical profiling of individual extracellular vesicles and functional nanoparticles without labels. Anal Chem. 2020;92:5585–5594. doi: 10.1021/acs.analchem.0c00607 [DOI] [PubMed] [Google Scholar]
- 171.Bagheri Hashkavayi A, Cha BS, Lee ES, Kim S, Park KS. Advances in exosome analysis methods with an emphasis on electrochemistry. Anal Chem. 2020;92:12733–12740. doi: 10.1021/acs.analchem.0c02745 [DOI] [PubMed] [Google Scholar]
- 172.Ma X. Recent progress in electrochemical detection of tumor-derived exosomes. Int J Electrochem Sci. 2020;15:7954–7967. doi: 10.20964/2020.08.101 [DOI] [Google Scholar]
- 173.Chen A, Chatterjee S. Nanomaterials based electrochemical sensors for biomedical applications. Chem Soc Rev. 2013;42:5425–5438. doi: 10.1039/c3cs35518g [DOI] [PubMed] [Google Scholar]
- 174.Cavallaro S, Horak J, Hååg P, et al. Label-free surface protein profiling of extracellular vesicles by an electrokinetic sensor. ACS Sens. 2019;4:1399–1408. doi: 10.1021/acssensors.9b00418 [DOI] [PubMed] [Google Scholar]
- 175.Kilic T, Valinhas ATDS, Wall I, Renaud P, Carrara S. Label-free detection of hypoxia-induced extracellular vesicle secretion from MCF-7 cells. Sci Rep. 2018;8:9402–9411. doi: 10.1038/s41598-018-27203-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Suthar J, Parsons ES, Hoogenboom BW, Williams GR, Guldin S. Acoustic immunosensing of exosomes using a quartz crystal microbalance with dissipation monitoring. Anal Chem. 2020;92:4082–4093. doi: 10.1021/acs.analchem.9b05736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yadav S, Boriachek K, Islam MN, et al. An electrochemical method for the detection of disease-specific exosomes. ChemElectroChem. 2017;4:967–971. doi: 10.1002/celc.201600391 [DOI] [Google Scholar]
- 178.Wang S, Zhang L, Wan S, et al. Aptasensor with expanded nucleotide using DNA nanotetrahedra for electrochemical detection of cancerous exosomes. ACS Nano. 2017;11:3943–3949. doi: 10.1021/acsnano.7b00373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li Q, Tofaris GK, Davis JJ. Concentration-normalized electroanalytical assaying of exosomal markers. Anal Chem. 2017;89:3184–3190. doi: 10.1021/acs.analchem.6b05037 [DOI] [PubMed] [Google Scholar]
- 180.Vaidyanathan R, Naghibosadat M, Rauf S, et al. Detecting exosomes specifically: a multiplexed device based on alternating current electrohydrodynamic induced nanoshearing. Anal Chem. 2014;86:11125–11132. doi: 10.1021/ac502082b [DOI] [PubMed] [Google Scholar]
- 181.Liu Q, Yue X, Li Y, et al. A novel electrochemical aptasensor for exosomes determination and release based on specific host-guest interactions between cucurbit [7]uril and ferrocene. Talanta. 2021;232:122451–122458. doi: 10.1016/j.talanta.2021.122451 [DOI] [PubMed] [Google Scholar]
- 182.Sun Y, Jin H, Jiang X, Gui R. Assembly of black phosphorus nanosheets and MOF to form functional hybrid thin-film for precise protein capture, dual-signal and intrinsic self-calibration sensing of specific cancer-derived exosomes. Anal Chem. 2020;92:2866–2875. doi: 10.1021/acs.analchem.9b05583 [DOI] [PubMed] [Google Scholar]
- 183.Ramadan S, Lobo R, Zhang Y, et al. Carbon-dot-enhanced graphene field-effect transistors for ultrasensitive detection of exosomes. ACS Appl Mater Interfaces. 2021;13:7854–7864. doi: 10.1021/acsami.0c18293 [DOI] [PubMed] [Google Scholar]
- 184.Yu Y, Li YT, Jin D, et al. Electrical and label-free quantification of exosomes with a reduced graphene oxide field effect transistor biosensor. Anal Chem. 2019;91:10679–10686. doi: 10.1021/acs.analchem.9b01950 [DOI] [PubMed] [Google Scholar]
- 185.Wang L, Zeng L, Wang Y, et al. Electrochemical aptasensor based on multidirectional hybridization chain reaction for detection of tumorous exosomes. Sens Actuat B Chem. 2021;332:129471–129480. doi: 10.1016/j.snb.2021.129471 [DOI] [Google Scholar]
- 186.Doldán X, Fagúndez P, Cayota A, Laíz J, Tosar JP. Electrochemical sandwich immunosensor for determination of exosomes based on surface marker-mediated signal amplification. Anal Chem. 2016;88:10466–10473. doi: 10.1021/acs.analchem.6b02421 [DOI] [PubMed] [Google Scholar]
- 187.An Y, Jin T, Zhu Y, Zhang F, He P. An ultrasensitive electrochemical aptasensor for the determination of tumor exosomes based on click chemistry. Biosens Bioelectron. 2019;142:111503–111510. doi: 10.1016/j.bios.2019.111503 [DOI] [PubMed] [Google Scholar]
- 188.Huang R, He L, Xia Y, et al. A sensitive aptasensor based on a hemin/g-quadruplex-assisted signal amplification strategy for electrochemical detection of gastric cancer exosomes. Small. 2019;15:1900735–1900742. doi: 10.1002/smll.201900735 [DOI] [PubMed] [Google Scholar]
- 189.Jiang J, Yu Y, Zhang H, Cai C. Electrochemical aptasensor for exosomal proteins profiling based on DNA nanotetrahedron coupled with enzymatic signal amplification. Anal Chim Acta. 2020;1130:1–9. doi: 10.1016/j.aca.2020.07.012 [DOI] [PubMed] [Google Scholar]
- 190.Sun Z, Wang L, Wu S, et al. An electrochemical biosensor designed by using Zr-based metal-organic frameworks for the detection of glioblastoma-derived exosomes with practical application. Anal Chem. 2020;92:3819–3826. doi: 10.1021/acs.analchem.9b05241 [DOI] [PubMed] [Google Scholar]
- 191.Gu C, Bai L, Pu L, Gai P, Li F. Highly sensitive and stable self-powered biosensing for exosomes based on dual metal-organic frameworks nanocarriers. Biosens Bioelectron. 2021;176:112907–112913. doi: 10.1016/j.bios.2020.112907 [DOI] [PubMed] [Google Scholar]
- 192.Cao Y, Wang Y, Yu X, Jiang X, Li G, Zhao J. Identification of programmed death ligand-1 positive exosomes in breast cancer based on DNA amplification-responsive metal-organic frameworks. Biosens Bioelectron. 2020;166:112452–112460. doi: 10.1016/j.bios.2020.112452 [DOI] [PubMed] [Google Scholar]
- 193.Wang M, Pan Y, Wu S, et al. Detection of colorectal cancer-derived exosomes based on covalent organic frameworks. Biosens Bioelectron. 2020;169:112638–112645. doi: 10.1016/j.bios.2020.112638 [DOI] [PubMed] [Google Scholar]
- 194.Boriachek K, Masud MK, Palma C, et al. Avoiding pre-isolation step in exosome analysis: direct isolation and sensitive detection of exosomes using gold-loaded nanoporous ferric oxide nanozymes. Anal Chem. 2019;91:3827–3834. doi: 10.1021/acs.analchem.8b03619 [DOI] [PubMed] [Google Scholar]
- 195.Zhang H, Wang Z, Wang F, Zhang Y, Wang H, Liu Y. Ti3C2 MXene mediated Prussian blue in situ hybridization and electrochemical signal amplification for the detection of exosomes. Talanta. 2021;224:121879–121886. doi: 10.1016/j.talanta.2020.121879 [DOI] [PubMed] [Google Scholar]
- 196.Boriachek K, Islam MN, Gopalan V, Lam AK, Nguyen NT, Shiddiky MJA. Quantum dot-based sensitive detection of disease specific exosome in serum. Analyst. 2017;142:2211–2219. doi: 10.1039/C7AN00672A [DOI] [PubMed] [Google Scholar]
- 197.Zhou YG, Mohamadi RM, Poudineh M, et al. Interrogating circulating microsomes and exosomes using metal nanoparticles. Small. 2016;12:727–732. doi: 10.1002/smll.201502365 [DOI] [PubMed] [Google Scholar]
- 198.Moura SL, Martin CG, Marti M, Pividori MI. Electrochemical immunosensing of nanovesicles as biomarkers for breast cancer. Biosens Bioelectron. 2020;150:111882–111889. doi: 10.1016/j.bios.2019.111882 [DOI] [PubMed] [Google Scholar]
- 199.Wei F, Yang J, Wong DT. Detection of exosomal biomarker by electric field-induced release and measurement (EFIRM). Biosens Bioelectron. 2013;44:115–121. doi: 10.1016/j.bios.2012.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jeong S, Park J, Pathania D, Castro CM, Weissleder R, Lee H. Integrated magneto-electrochemical sensor for exosome analysis. ACS Nano. 2016;10:1802–1809. doi: 10.1021/acsnano.5b07584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xu H, Liao C, Zuo P, Liu Z, Ye BC. Magnetic-based microfluidic device for on-chip isolation and detection of tumor-derived exosomes. Anal Chem. 2018;90:13451–13458. doi: 10.1021/acs.analchem.8b03272 [DOI] [PubMed] [Google Scholar]
- 202.Cao Y, Li L, Han B, Wang Y, Dai Y, Zhao J. A catalytic molecule machine-driven biosensing method for amplified electrochemical detection of exosomes. Biosens Bioelectron. 2019;141:111397–111403. doi: 10.1016/j.bios.2019.111397 [DOI] [PubMed] [Google Scholar]
- 203.Miao P, Tang Y. A multipedal DNA walker for amplified detection of tumor exosomes. Chem Commun. 2020;56:4982–4985. doi: 10.1039/D0CC01817A [DOI] [PubMed] [Google Scholar]
- 204.Guo Y, Cao Q, Feng Q. Catalytic hairpin assembly-triggered DNA walker for electrochemical sensing of tumor exosomes sensitized with Ag@C core-shell nanocomposites. Anal Chim Acta. 2020;1135:55–63. doi: 10.1016/j.aca.2020.08.036 [DOI] [PubMed] [Google Scholar]
- 205.Dong H, Chen H, Jiang J, Zhang H, Cai C, Shen Q. Highly sensitive electrochemical detection of tumor exosomes based on aptamer recognition-induced multi-DNA release and cyclic enzymatic amplification. Anal Chem. 2018;90:4507–4513. doi: 10.1021/acs.analchem.7b04863 [DOI] [PubMed] [Google Scholar]
- 206.Zhao L, Sun R, He P, Zhang X. Ultrasensitive detection of exosomes by target-triggered three-dimensional DNA walking machine and exonuclease III-assisted electrochemical ratiometric biosensing. Anal Chem. 2019;91:14773–14779. doi: 10.1021/acs.analchem.9b04282 [DOI] [PubMed] [Google Scholar]
- 207.Yang L, Yin X, An B, Li F. Precise capture and direct quantification of tumor exosomes via a highly efficient dual-aptamer recognition-assisted ratiometric immobilization-free electrochemical strategy. Anal Chem. 2021;93:1709–1716. doi: 10.1021/acs.analchem.0c04308 [DOI] [PubMed] [Google Scholar]
- 208.Li R, An Y, Jin T, Zhang F, He P. Detection of MUC1 protein on tumor cells and their derived exosomes for breast cancer surveillance with an electrochemiluminescence aptasensor. J Electroanal Chem. 2021;882:115011–115020. doi: 10.1016/j.jelechem.2021.115011 [DOI] [Google Scholar]
- 209.Feng Q, Ma P, Cao QH, Guo YH, Xu JJ. An aptamer-binding DNA walking machine for sensitive electrochemiluminescence detection of tumor exosomes. Chem Commun. 2019;56:269–272. doi: 10.1039/C9CC08051A [DOI] [PubMed] [Google Scholar]
- 210.Qiao B, Guo Q, Jiang J, et al. An electrochemiluminescent aptasensor for amplified detection of exosomes from breast tumor cells (MCF-7 cells) based on G-quadruplex/hemin dnazymes. Analyst. 2019;144:3668–3675. doi: 10.1039/C9AN00181F [DOI] [PubMed] [Google Scholar]
- 211.Zhang Y, Wang F, Zhang H, Wang H, Liu Y. Multivalency interface and g-C3N4 coated liquid metal nanoprobe signal amplification for sensitive electrogenerated chemiluminescence detection of exosomes and their surface proteins. Anal Chem. 2019;91:12100–12107. doi: 10.1021/acs.analchem.9b03427 [DOI] [PubMed] [Google Scholar]
- 212.Guo Y, Cao Q, Zhao C, Feng Q. Stimuli-responsive DNA microcapsules for homogeneous electrochemiluminescence sensing of tumor exosomes. Sens Actuat B Chem. 2021;329:129136–129142. doi: 10.1016/j.snb.2020.129136 [DOI] [Google Scholar]
- 213.Zhang H, Wang Z, Zhang Q, Wang F, Liu Y. Ti3C2 MXenes nanosheets catalyzed highly efficient electrogenerated chemiluminescence biosensor for the detection of exosomes. Biosens Bioelectron. 2019;124–125:184–190. doi: 10.1016/j.bios.2018.10.016 [DOI] [PubMed] [Google Scholar]
- 214.Fang D, Zhao D, Zhang S, Huang Y, Dai H, Lin Y. Black phosphorus quantum dots functionalized MXenes as the enhanced dual-mode probe for exosomes sensing. Sens Actuat B Chem. 2020;305:127544–127552. doi: 10.1016/j.snb.2019.127544 [DOI] [Google Scholar]
- 215.Zhang H, Wang Z, Wang F, Zhang Y, Wang H, Liu Y. In situ formation of gold nanoparticles decorated Ti3C2 MXenes nanoprobe for highly sensitive electrogenerated chemiluminescence detection of exosomes and their surface proteins. Anal Chem. 2020;92:5546–5553. doi: 10.1021/acs.analchem.0c00469 [DOI] [PubMed] [Google Scholar]
- 216.Liu X, Wang Q, Chen J, Chen X, Yang W. Ultrasensitive electrochemiluminescence biosensor for the detection of tumor exosomes based on peptide recognition and luminol-AuNPs@g-C3N4 nanoprobe signal amplification. Talanta. 2021;221:121379–121386. doi: 10.1016/j.talanta.2020.121379 [DOI] [PubMed] [Google Scholar]
- 217.Pei Y, Ge Y, Zhang X, Li Y. Cathodic photoelectrochemical aptasensor based on NiO/BiOI/Au NP composite sensitized with cdse for determination of exosomes. Microchim Acta. 2021;188:51–61. doi: 10.1007/s00604-021-04716-1 [DOI] [PubMed] [Google Scholar]
- 218.Roda A, Mirasoli M, Michelini E, et al. Progress in chemical luminescence-based biosensors: a critical review. Biosens Bioelectron. 2016;76:164–179. doi: 10.1016/j.bios.2015.06.017 [DOI] [PubMed] [Google Scholar]
- 219.Jiang Q, Liu Y, Wang L, Adkins GB, Zhong W. Rapid enrichment and detection of extracellular vesicles enabled by cus-enclosed microgels. Anal Chem. 2019;91:15951–15958. doi: 10.1021/acs.analchem.9b04485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wang Y, Liu Z, Wang X, et al. Rapid and quantitative analysis of exosomes by a chemiluminescence immunoassay using superparamagnetic iron oxide particles. J Biomed Nanotechnol. 2019;15:1792–1800. doi: 10.1166/jbn.2019.2809 [DOI] [PubMed] [Google Scholar]
- 221.Yang JS, Lee JC, Byeon SK, Rha KH, Moon MH. Size dependent lipidomic analysis of urinary exosomes from patients with prostate cancer by flow field-flow fractionation and nanoflow liquid chromatography-tandem mass spectrometry. Anal Chem. 2017;89:2488–2496. doi: 10.1021/acs.analchem.6b04634 [DOI] [PubMed] [Google Scholar]
- 222.Zhang L, Wang H, Zhao G, et al. Anti-Tim4 grafting strongly hydrophilic metal-organic frameworks immunoaffinity flake for high-efficiency capture and separation of exosomes. Anal Chem. 2021;93:6534–6543. doi: 10.1021/acs.analchem.1c00528 [DOI] [PubMed] [Google Scholar]
- 223.Chang M, Wang Q, Qin W, Shi X, Xu G. Rational synthesis of aptamer-functionalized polyethylenimine-modified magnetic graphene oxide composites for highly efficient enrichment and comprehensive metabolomics analysis of exosomes. Anal Chem. 2020;92:15497–15505. doi: 10.1021/acs.analchem.0c03374 [DOI] [PubMed] [Google Scholar]
- 224.Zhang N, Sun N, Deng C. Rapid isolation and proteome analysis of urinary exosome based on double interactions of Fe3O4@TiO2-DNA aptamer. Talanta. 2021;221:121571–121579. doi: 10.1016/j.talanta.2020.121571 [DOI] [PubMed] [Google Scholar]
- 225.Brown BA, Zeng X, Todd AR, et al. Charge detection mass spectrometry measurements of exosomes and other extracellular particles enriched from bovine milk. Anal Chem. 2020;92:3285–3292. doi: 10.1021/acs.analchem.9b05173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Fang X, Duan Y, Adkins GB, et al. Highly efficient exosome isolation and protein analysis by an integrated nanomaterial-based platform. Anal Chem. 2018;90:2787–2795. doi: 10.1021/acs.analchem.7b04861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang Y, Zhang K, Huang X, Qiao L, Liu B. Mass spectrometry imaging of mass tag immunoassay enables the quantitative profiling of biomarkers from dozens of exosomes. Anal Chem. 2021;93:709–714. doi: 10.1021/acs.analchem.0c03904 [DOI] [PubMed] [Google Scholar]
- 228.Zhang XW, Liu MX, He MQ, Chen S, Yu YL, Wang JH. Integral multielement signals by DNA-programmed UCNP-AuNP nanosatellite assemblies for ultrasensitive ICP-MS detection of exosomal proteins and cancer identification. Anal Chem. 2021;93:6437–6445. doi: 10.1021/acs.analchem.1c00152 [DOI] [PubMed] [Google Scholar]