Abstract
The TRAP coactivator complex is a large, multisubunit complex of nuclear proteins which associates with nuclear hormone receptors (NRs) in the presence of cognate ligand and stimulates NR-mediated transcription. A single subunit, TRAP220, is thought to target the entire complex to a liganded receptor through a domain containing two of the signature LXXLL motifs shown previously in other types of coactivator proteins to be essential for mediating NR binding. In this work, we demonstrate that each of the two LXXLL-containing regions, termed receptor binding domains 1 and 2 (RBD-1 and RBD-2), is differentially preferred by specific NRs. The retinoid X receptor (RXR) displays a weak yet specific activation function 2 (AF2)-dependent preference for RBD-1, while the thyroid hormone receptor (TR), vitamin D3 receptor (VDR), and peroxisome proliferator-activated receptor all exhibit a strong AF2-dependent preference for RBD-2. Using site-directed mutagenesis, we show that preference for RBD-2 is due to the presence of basic-polar residues on the amino-terminal end of the core LXXLL motif. Furthermore, we show that the presence and proper spacing of both RBD-1 and RBD-2 are required for an optimal association of TRAP220 with RXR-TR or RXR-VDR heterodimers bound to DNA and for TRAP220 coactivator function. On the basis of these results, we suggest that a single molecule of TRAP220 can interact with both subunits of a DNA-bound NR heterodimer.
Nuclear hormone receptors (NRs) make up a family of ligand-activated transcription factors that regulate the expression of target genes involved in development, differentiation, and homeostasis (39, 47, 60). The ligands for NRs, small hydrophobic molecules including steroids, retinoids, thyroid hormone, and vitamin D3, bind to the C-terminal ligand binding domain (LBD) of their cognate NR and induce conformational changes which modulate receptor activity (44). Transcriptional activation by NRs can be mediated by two separable activation functions (AFs): AF1, located at the N terminus (30, 45, 56), and AF2, located in the LBD (3, 11, 15, 52). While AF1 is poorly conserved among NR family members, the AF2 domain is highly conserved and essential for ligand-dependent activation (11, 47). Recent structural studies suggest that ligand binding regulates AF2 activity by changing the stereospecific position of the most C-terminal LBD α helix (helix 12), a motif previously shown to be indispensable for AF2 function (44, 47). The ligand-induced repositioning of helix 12 places it in close proximity to α helices 3, 4, and 5 and is thought to generate a hydrophobic binding surface for transcriptional coactivator proteins (12, 25, 46, 53).
The best-characterized NR coactivators identified thus far are members of the SRC/p160 family of proteins, which include SRC-1/NCoA-1, TIF2/GRIP1, and pCIP/RAC3/AIB-1/ACTR/TRAM-1 (reviewed in references 42 and 58). While the exact mechanism of action of the SRC/p160 proteins is unclear, their ability to associate with histone acetyltransferases (HATs) such as CBP/p300 (6, 7, 23, 29, 42, 57, 61) and pCAF (4) and the presence of intrinsic HAT activity in some family members (7, 54) suggest a role in chromatin remodeling. Each member of this family has a central receptor interaction domain (RID) containing three copies of a consensus leucine-rich motif, LXXLL (also termed NR box), with conserved spacing between the motifs (42, 58). Crystallographic and biochemical studies reveal that the surface of a single LXXLL motif directly contacts the ligand-activated AF2 domain of NRs, thereby providing a molecular basis for NR-coactivator recruitment (12, 46, 53). That distinct LXXLL motifs within one SRC/p160 protein might selectively interact with different NRs is supported by mutagenesis studies showing that specific NR boxes are selectively required for the functional activity of different NRs (13, 38, 41, 57, 61). Studies examining NRs bound to DNA as either homodimers or heterodimers further suggest that a single molecule of SRC/p160 protein might simultaneously contact both AF2 domains of the receptor dimer via multiple LXXLL motifs (28, 46, 62). Amino acid residues immediately flanking the core LXXLL sequence and proper spacing between the motifs have been proposed to modulate the affinity and specificity of distinct NR-SRC/p160 interactions (12, 26, 38, 41).
A different set of NR coactivators termed the TRAP complex was first identified as a large multimeric group of novel proteins that copurify with the thyroid hormone receptor (TR) from HeLa cells cultured in thyroid hormone (triiodothyronine [T3]) (17). The ability of the TRAP complex to markedly stimulate TR-mediated transcription in vitro on naked DNA templates and in the absence of TATA-binding protein-associated factors suggested that TRAPs mediate a novel NR-coactivator pathway or activation step distinct from those mediated by SRC/p160 proteins and CBP/p300 and possibly involving a more direct influence on the basal transcription machinery (19). Several, if not all, of the subunits of the TRAP complex have been identified in other large transcriptional coregulatory complexes, including DRIP (48), NAT (55), SMCC (21), and CRSP (51). As evidenced by its ability to bind TR and other NRs in an avid ligand-dependent fashion, the 220-kDa component of the complex (referred to as TRAP220) has been proposed to target and possibly anchor the entire TRAP complex to a ligand-activated NR (64). Interestingly, and analogous to the SRC/p160 proteins, sequence analysis of TRAP220, also termed TRIP2 (35), RB18A (14), PBP (67), and DRIP205 (49), reveals the presence of two LXXLL motifs in the central region of the protein. Excluding the two NR boxes, TRAP220 displays no other close homology to the SRC/p160 proteins. While the region containing the two LXXLL motifs has been shown to facilitate ligand-dependent interactions with TR (64), the details of the TRAP220-NR interface are poorly understood.
In an effort to define more precisely the specific structural and molecular determinants responsible for TRAP220-NR interactions, we performed extensive mutagenesis of the TRAP220 protein and studied its ability to physically and functionally interact with NRs. We report that the two LXXLL-containing domains of TRAP220, referred to here as receptor binding domains 1 and 2 (RBD-1 and RBD-2), are differentially preferred by various NRs and that specific ligand-dependent interactions with RBD-2 are due in part to the presence of basic amino acid residues flanking the N-terminal side of the LXXLL motif in RBD-2. We also demonstrate that both RBD-1 and RBD-2 are necessary for efficient interaction of TRAP220 with various NR heterodimers bound to DNA and for TRAP220 transcriptional coactivation function in vivo. These results suggest that a single molecule of TRAP220 can functionally interact with dimerized NRs in vivo in a 1:2 stoichiometric ratio.
MATERIALS AND METHODS
Plasmid construction.
The bacterial expression vector for human TRα (hTRα) was generated by subcloning the full-length hTRα NdeI-BamHI fragment of pET11-10His-hTRα (16) into pET3a (Novagen), generating pET3a-hTRα. Construction of full-length FLAG-tagged hTRα and hTRβ expression vectors (pFLAG-hTRα and pFLAG-TRβ) and the three hTRα deletion mutants Δ1 (residues 1 to 123), Δ2 (residues 122 to 211), and Δ3 (residues 213 to 410) have all been described elsewhere (17, 18, 65). Construction of the pET11-10His-hTRβ bacterial expression vector was generated by subcloning the full-length hTRβ NdeI-BamHI fragment of pFLAG-TRβ into the NdeI-BamHI sites of pET11d-10His (16). Construction of hTRα deletion mutant Δ4 (residues 1 to 401) was generated by introducing a stop codon into the open reading frame of pFLAG-hTRα (residue 402) using PCR and the oligonucleotide 5′CGG GAT CCT TAG AAG AGT GGG GGG AAG AGT CC3′. Similarly, the AF2 mutants of hTRα (E-403-K) and hTRβ (E-457-K, E-460-K) were generated by introducing site-directed mutations into the open reading frames of pFLAG-hTRα and pFLAG-TRβ, using PCR and the oligonucleotides 5′CGG GAT CCT TAG ACT TCC TGA TCC TCA AAG ACC TT3′ and 5′CGC CTA GGG ATT AGG AAC TTG TGA AAG TCC TTG3′, respectively.
The human retinoid X receptor alpha (hRXRα) AF-2 mutant (F-450-A, E-453-K, E-456-K) was generated by introducing site-directed mutations into the open reading frame of pFLAG-RXRα (17), using PCR and the antisense oligonucleotide 5′GC GAA TTC CTA AGT CAT TTG GTG CGG CGC CTT CAG CAT CTT CAT AAG GCC GGT G3′. The full-length human TRAP220 (hTRAP220) expression vectors pGEM-HA-TRAP220 and pSG5-HA-TRAP220 were described previously (65). The FLAG-tagged full-length TRAP220 and associated deletion mutants were generated using a modified pRSET vector (Invitrogen) in which the 6-histidine tag leader sequence was replaced with a FLAG tag sequence (10) and an oligonucleotide containing stop codons in all three reading frames, 5′TCG GTG AGT GAG TGA GCG GAG CT3′, was inserted into the SacI site, thus generating the expression vector pT7-FLAG-Tristop. Full-length pFLAG-TRAP220 was generated by subcloning a partially digested NdeI-SacI TRAP220 fragment from pGEM-HA-TRAP220 into pT7-FLAG-Tristop vector. FLAG-tagged N-terminal TRAP220 deletion mutants N1, N2, and N3 were generated by first creating NdeI sites at amino acid residues 275, 557, and 842 within the open reading frame of pGEM-HA-TRAP220 by PCR amplification using the 5′ oligonucleotides 5′GCA CCA TTA CAT ATG GGG TCA CAT CCA G3′, 5′CAG GCA ACA ACC ATA TGA GTG GTA CCA C3′, and 5′AGC TGA TCA TAT GGC AGA TGC TGC TGG AAG, respectively, together with the common 3′ primer 5′CCT GGT TTG CTG TCT AAT CC3′, which spans an internal ApaI site. The PCR fragments were then digested with NdeI and ApaI and subsequently subcloned together with an ApaI-SacI fragment derived from pGEM-HA-TRAP220 containing the 3′ end of TRAP220 into the NdeI/SacI sites of pT7-FLAG-Tristop. The N-terminal TRAP220 deletion mutant N4 was generated by subcloning the smallest NdeI-SacI fragment of pGEM-HA-TRAP220 into pT7-FLAG-Tristop. The C-terminal TRAP220 deletion mutants C1 through C7 were generated by first digesting pGEM-HA-TRAP220 with the following restriction enzymes within the open reading frame: AflII (C1), EcoRI (C2), KpnI (C3), XhoI (C4), SpeI (C5), ApaI (C6), and BamHI (C7). The sticky ends were blunted using Klenow enzyme or T4 DNA polymerase, and the cDNA was redigested with NdeI. The resulting NdeI/blunt-ended fragments were then subcloned into an NdeI-SacI-digested pT7-FLAG-Tristop vector in which the SacI end of the vector had been preblunted. The C-terminal TRAP220 deletion mutant C8 was generated by introducing a stop codon into the open reading frame of pFLAG-TRAP220 (amino acid residue 1423) using PCR and the oligonucleotide 5′GCC ATT TGA GGC CTA AGC CCT TCT CCA CTA C3′.
GST (glutathione S-transferase)-TRAP220-RBD, GST-TRAP220-RBD- 1, and GST-TRAP220-RBD-2 were constructed by first PCR amplifying hTRAP220 amino acids 501 to 738, 501 to 635, and 622 to 701 with primers creating BamHI and EcoRI restriction sites at the 5′ and 3′ ends of the cDNA, respectively. The PCR fragments were then ligated into pGEX-2TK (Pharmacia Biotech, Piscataway, N.J.) predigested with BamHI and EcoRI, thus generating the in-frame GST fusion protein expression vectors. The expression vector pBK-RSV-f:RXRα was generated by subcloning the BglII-EcoRI fragment of pFLAG-RXRα into the BamHI-EcoRI sites of pBK-RSV (Stratagene). The pSG5-hVDR construct and p4xVDRE-Ld-Luc reporter were generously provided by K. Ozato (National Institute of Child Health and Human Development, National Institutes of Health). The pSG5-mPPARα expression vector was kindly provided by S. Green (Zeneca, Manchester, United Kingdom).
Site-directed TRAP220-RBD mutagenesis.
Point and deletion mutations of GST-TRAP220-RBD, -RBD-1, -RBD-2, and pSQ5-HA-TRAP220 were introduced using a commercial kit as instructed by the manufacturer (GeneEditor mutagenesis system; Promega, Madison, Wis.). Briefly, the DNA templates were alkaline denatured and then hybridized with the appropriate selection and mutagenic oligonucleotides. After the annealing reaction, mutant strand synthesis and ligation was obtained by adding T4 DNA polymerase and T4 DNA ligase. All mutations were verified by sequencing (University of Maryland School of Medicine Biopolymer Facility). The mutagenic oligonucleotides were as follows. Primer 95 (5′pACC CAA TTC TTA CCA GTG CGG CGC AAA TCA CAG GGA ACG G3′ [LL residues 607 and 608 to AA]) was used to generate constructs GST-RBD/M95, GST-RBD-1/M95, and pSG5-HA-TRAP220/M95. Primer 96 (5′pACC CGA TGC TCA TGA ACG CTG CTA AAG ATA ATC CTG CCC AG3′ [LL residues 648 and 649 to AA]) was used for GST-RBD/M96, GST-RBD-2/M96, and pSG5-HA-TRAP220/M96. Both primers were used to generate GST-RBD/M95/96. Primer 97 (5′pTTG CAA ATC ACA GGG AAC GGG GGG TCT ACC GCC GGC AAC ACC AAG AAC CAC CCG ATG CTC3′ [deletion of residues 617 to 635 {Δ617–635}]) was used for GST-RBD/M97. Primer 98 (5′pCC CCT CCT CAT CAC ACG CCG CCA CCT GTC CCG ATG CTC ATG AAC CTT CTT AAA GAT AAT C3′ [Δ633–642]) was used for GST-RBD/M98 and GST-RBD-2/M98. Primer 106 (5′pCAC CCG ATG CTC ATG AAC CTT CTT AAA GAT GGA AGC AGC CCT TTA GAA AGG CAG AAC TCC3′ [Δ652–661]) was used for GST-RBD-2/M106. Primer 107 (5′pTCG ATG GCC GGC AAC ACC GCG GCC GCC CCG ATG CTC ATG AAC CTT C3′ [KNH residues 640 to 642 to AAA]) was used for GST-RBD/M107, GST-RBD-2/M107, and pSG5-HA-TRAP220/M107. Primer 108 (5′pATG CTC ATG AAC CTT CTT GCA GCT GCT CCT GCC CAG GAT TTC TC3′ [KDN residues 650 to 652 to AAA]) was used for GST-RBD-2/M108. Primer 109 (5′pCTT AAA GAT AAT CCT GCC GCG GCT GCC TCA ACC CTT TAT GGA AGC3′ [QDF residues 655 to 657 to AAA]) was used for GST-RBD-2/M109. Primer 110 (5′pAAC ACC AAG AAC CAC GCG ATG CTC ATG AAC CTT C3′ [P residue 643 to A]) was used for GST-RBD-2/M110. Primer 111 (5′pAAC ACC AAG AAC CAC CCG GCG CTC ATG AAC CTT C3′ [M residue 644 to A]) was used for GST-RBD-2/M111. Primer 112 (5′pAAG AAC CAC CCG ATG CTC GCG GCC CTT CTT AAA GAT AAT C3′ [MN residues 646 to 647 to AA]) was used for GST-RBD-2/M112.
Expression and purification of GST- and His-tagged proteins.
Escherichia coli BL21(DE3)pLysS cells harboring the pGEX-2TK fusion constructs were grown in Luria-Bertani medium containing ampicillin (100 μg/ml) and subsequently induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 30°C. Bacteria were then harvested by centrifugation, resuspended in lysis buffer (1 M NaCl, 20 mM Tris-Cl [pH 7.3], 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 3 mM β-mercaptoethanol [β-ME], 0.03% NP-40), briefly sonicated, and then centrifuged at 10,000 rpm for 10 min at 4°C. To purify the GST fusion proteins, lysate from 50 ml of culture was mixed with 100 μl (packed resin) of glutathione-Sepharose 4B (Pharmacia Biotech) for 3 to 5 h at 4°C, washed three times with lysis buffer, and then washed twice with BC100/NP-40 (20 mM Tris-Cl [pH 7.9 at 4°C], 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM PMSF, 3 mM β-ME, 0.05% NP-40). The resin was finally resuspended in a 50% slurry of BC100 (without NP-40) as a working stock solution for the GST pull-down assays. To produce the purified fusion protein for the avidin-biotin DNA complex assay, supernatant from 500 ml of induced culture was mixed with 0.5 ml (packed resin) of glutathione-Sepharose 4B, and the fusion proteins were purified as described above. To elute the fusion proteins from the glutathione-Sepharose, the resin was incubated in 0.8 ml of elution buffer (100 mM Tris-Cl [pH 8.0], 100 mM KCl, 10% glycerol, 10 mM reduced glutathione, 1 mM dithiothreitol) at 4°C overnight. The supernatant was then dialyzed in BC100 for 8 h at 4°C. Expression of His10-hTRβ in E. coli and purification by Ni2+-nitrilotriacetic acid (Qiagen) column chromatography were essentially as described elsewhere (16).
GST pull-down assay.
In general, 0.5 to 1 μg of GST fusion protein was added to 250 μl of binding buffer (20 mM HEPES [pH 7.9], 100 mM KCl, 0.5 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 0.05% NP-40, 0.5% powdered milk) together with 1 to 5 μl of in vitro-translated [35S]methionine-labeled NRs (from a 50-μl labeling reaction) (TNT; Promega Corp.) which were generated from the pFLAG-hTRα, pFLAG-RXRα, pSG5-mPPARα, and pSG5-hVDR expression vectors. The reaction mixtures were incubated for 1 h at 4°C on a rocker. Protein complexes were isolated by pelleting the beads and washing three times in binding buffer followed by resuspension in sodium dodecyl sulfate (SDS)-sample loading buffer. After SDS-polyacrylamide gel electrophoresis (PAGE) fractionation, bound 35S-labeled NRs were visualized by autoradiography. The ligands T3 (1 μM, final concentration; Sigma), 9-cis retinoic acid (RA) (1 μM, final concentration; Sigma), 1,25-dihydroxyvitamin D3 [1,25-(OH)2D3; 0.5 μM, final concentration; BioMol], and WY-14643 (100 μM, final concentration; BioMol) were added to reaction mixtures as indicated.
Immunoprecipitation.
One microliter of in vitro-translated 35S-labeled TR or mutant derivatives (either FLAG tagged or untagged, depending on the experiment) (TNT; Promega) was incubated in 20 μl of BC100/NP-40 containing 1 μM T3 for 30 min at 4°C; 1 μl of 35S-labeled TRAP220 (either FLAG or hemagglutinin epitope [HA] tagged) was then added, and the reaction was allowed to proceed for an additional 30 min at 4°C on a rocker. Then 2 μl (packed) of anti-FLAG antibodies coupled to agarose beads (M2 affinity resin; Sigma) was added, and the reaction mixture was rocked for another hour at 4°C. The reaction volume was then increased to 400 μl with BC100/NP-40 and incubated an additional hour at 4°C with rocking. The beads were then pelleted by gentle centrifugation and washed three times with 0.5 ml of BC300/NP-40 (equivalent to BC100/NP-40 but with the KCl concentration increased to 300 mM). After the final wash, all the supernatant was carefully removed by aspiration using a 27.5 gauge needle. The beads were then suspended in 20 μl of sample loading buffer, boiled for 3 min, and fractionated by SDS-PAGE. Immunoprecipitated 35S-labeled proteins were then visualized by autoradiography. Experiments examining TR interaction with N- and C-terminal FLAG-TRAP220 deletion mutants (Fig. 2C) were performed as described above except that the in vitro-translated FLAG-TRAP220 mutants (N1 to N4 and C1 to C8) were not radiolabeled.
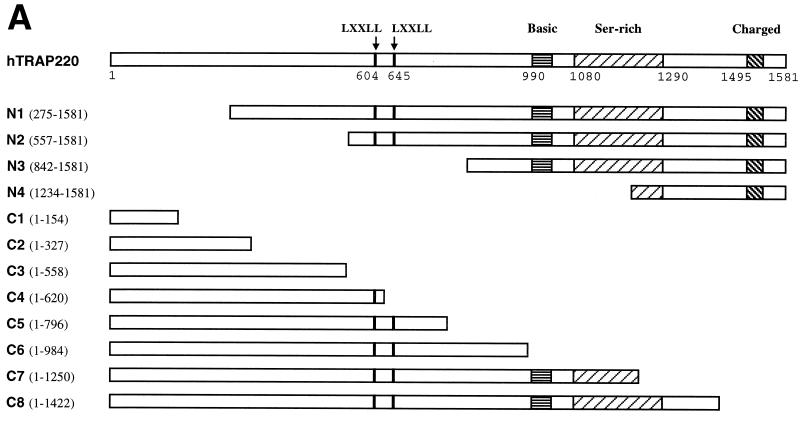
FIG. 2.
Delineation of a minimal TRAP220 RBD. (A) Schematic representation of the hTRAP220 protein including the N- and C-terminal deletion mutants used in this study. Locations of the two LXXLL motifs (black bars) and regions rich in basic, serine, and charged amino acid residues are indicated. (B) In vitro-translated 35S-labeled TRAP220 and various deletion mutants fractionated by SDS-PAGE. The deletion mutant C1 (approximately 17 kDa) was too small to be resolved on this gel. (C) TR binds only TRAP220 deletion mutants containing one or both of the LXXLL motifs. 35S-labeled hTRα was incubated with in vitro-translated unlabeled FLAG-TRAP220 or FLAG-tagged TRAP220 deletion mutants in the presence or absence of T3 as indicated. Protein complexes were coprecipitated with anti-FLAG M2 affinity resin.
SPR analysis.
Surface plasmon resonance (SPR) analysis was performed with a BIACORE 3000 system (Biacore, Inc.). Anti-GST antibody was immobilized on research-grade CM5 sensor chips using the amine coupling kit and the GST kit provided by the manufacturer (Biacore). The immobilization procedure was as follows: flow rate, 5 μl/min, 30 μl of N-hydroxysuccinimide–1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride mix injected to activate the surface, 35 μl of anti-GST antibody (diluted to 30 μg/ml in 10 mM acetate [pH 5.0]) injected, followed by 35-μl injection of ethanolamine to block unreacted groups on the surface. This procedure resulted in approximately 15,000 resonance units of anti-GST immobilized on the surface. The anti-GST surface was preconditioned by multiple cycles of binding (recombinant GST) and regeneration with 10 mM glycine (pH 2.2). The GST–RBD-2 ligands (wild type or mutants) were captured on the anti-GST surface to a level of 1,000 resonance units by using the manual injection mode. The His10-hTRβ analyte was diluted to a final concentration of 20 ng/ml, or 363 nM, with or without T3 (3 μM, final concentration) into HBS-EP (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% polysorbate-20 [pH 7.4]) and injected for 10 min. Overlay plots of the SPR response signals were prepared using BIAevaluation 3.0.2.
TRAP220-NR complex formation on DNA.
Double-stranded oligonucleotides containing 5′-BamHI overhangs on either end for DR4 (5′gatc TCA GGT CAC AGG AGG TCA GC3′), TREpal (5′gatc TCA GGT CAT GAC CTG A3′), the osteopontin/Spp-1 gene promoter vitamin D response element (VDRE; 5′gatc CAC AAG GTT CAC GAG GTT CAC GTC CG3′), and a nonspecific control element (5′gatc TCA TTT CAT GAA ATG A3′) were filled in using biotinylated dUTP (Boehringer Mannheim), dATP, and dCTP (GIBCO-BRL Life Technologies) and purified by ethanol precipitation. GST-RBD or the various GST-RBD mutant proteins were 32P-labeled with heart muscle protein kinase (Sigma product no. P-2645) and purified by NICK columns (Amersham Pharmacia Biotech) in BC100 at a concentration of 1 ng/μl. Typical binding reaction mixtures contained 150 ng of biotinylated oligonucleotide, 7.5 ng of 32P-labeled GST-RBD protein, and 5 μl of unlabeled in vitro-translated hRXRα together with 5 μl of either unlabeled hTRα or (from a 50-μl translation reaction) (TNT; Promega) in a total of 250 μl of IPA buffer (20 mM Tris-Cl [pH 7.9 at 4°C], 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM PMSF, 3 mM β-ME, 0.05% NP-40, 0.5% milk) containing either T3 (1 μM, final concentration), 9-cis RA (1 μM, final concentration), or 1,25-(OH)2D3 (0.5 μM, final concentration) as indicated. Protein-DNA complexes were allowed to assemble for 1 h at 4°C and then captured by adding 12.5 μl (packed resin) of streptavidin-agarose beads (GIBCO-BRL Life Technologies catalog no. 15942-014). Protein-DNA complexes were isolated by gently pelleting the beads, washing four times in IPA buffer, and fractionation by SDS-PAGE. Bound GST-RBD proteins were later visualized by autoradiography.
Transient transfection.
NIH 3T3 cells were routinely maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS); 24 h before transfection, cells were seeded in 12-well plates at a density of 8 × 104 cells per well in DMEM containing 10% charcoal–dextran-stripped FBS (HyClone Laboratories, Inc., Logan, Utah). A DNA mixture containing 0.66 μg of p4xVDRE-Ld-Luc, 0.33 μg of pSG5-hVDR, 0.16 μg of the internal control plasmid pSV-β-gal (Promega), 2 μg of herring sperm DNA, and 0.33 μg of either the empty pSG5 vector, pSG5-HA-TRAP220, pSG5-HA-TRAP220/M95, pSG5-HA-TRAP220/M96, or pSG5-HA-TRAP220/M107 was added to each well by the calcium phosphate transfection method. The precipitate from each set of transfections was removed after 16 h and replaced with fresh DMEM containing either 10% charcoal–dextran-stripped FBS with vehicle alone or vehicle plus 1,25-(OH)2D3 (2.5 × 10−8 M, final concentration) as stated in the legend to Fig. 7. After 48 h, transfected cells in each well were harvested with a cell lysis buffer supplied in a kit (Promega luciferase assay system). Luciferase activity was determined by first adding a commercial assay solution to the lysate as instructed by the manufacturer (Promega) and then measuring in a Lumat LB 9507 luminometer (EG&G Wallac, Inc., Gaithersburg, Md.). The β-galactosidase activity of the lysed transfected cells (as above) was determined using a kit (Promega) β-galactosidase enzyme assay system) according to the manufacturer's instructions. Luciferase activity was normalized to β-galactosidase activity and expressed as relative luciferase light units.
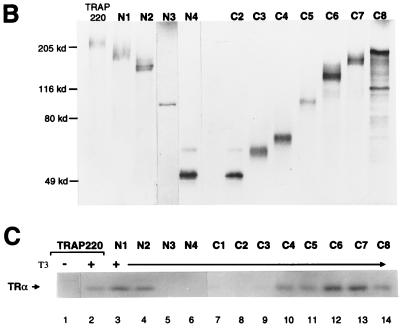
FIG. 7.
Functional role of RBD-1 and RBD-2 in TRAP220 transcriptional coactivation. (A) Schematic diagram of TRAP220 expression constructs. The wild-type sequence (TRAP220) is compared to the three site-directed mutant sequences (95, 96, and 107). Site-directed mutations introduced into the full-length open reading frame of the TRAP220 cDNA are indicated below the constructs in bold. (B) Both RBD-1 and RBD-2 are essential for optimal TRAP220 coactivation of VDR-mediated transcription. NIH 3T3 cells were transiently transfected with 0.66 μg of p4xVDRE-Ld-Luc, 0.33 μg of pSG5-hVDR, 0.16 μg of the internal control plasmid, and 0.33 μg of either empty pSG5 vector, pSG5-TRAP220, pSG5-TRAP220/M95, or pSG5-TRAP220/M96 in the presence (+) or absence (−) of 1,25-(OH)2-D3 (2.5 × 10−8 M, final concentration). Relative luciferase activities were determined from three independent transfections, although similar results were obtained from multiple transfections. (C) Basic/polar residues N terminal to the core LXXLL motif of TRAP220 RBD-2 are necessary for optimal TRAP220 coactivation function. Transient transfections were carried out as for panel B, including the expression vector pSG5-TRAP220/M107. The data are presented as the mean plus or minus the standard deviation of triplicate results.
RESULTS
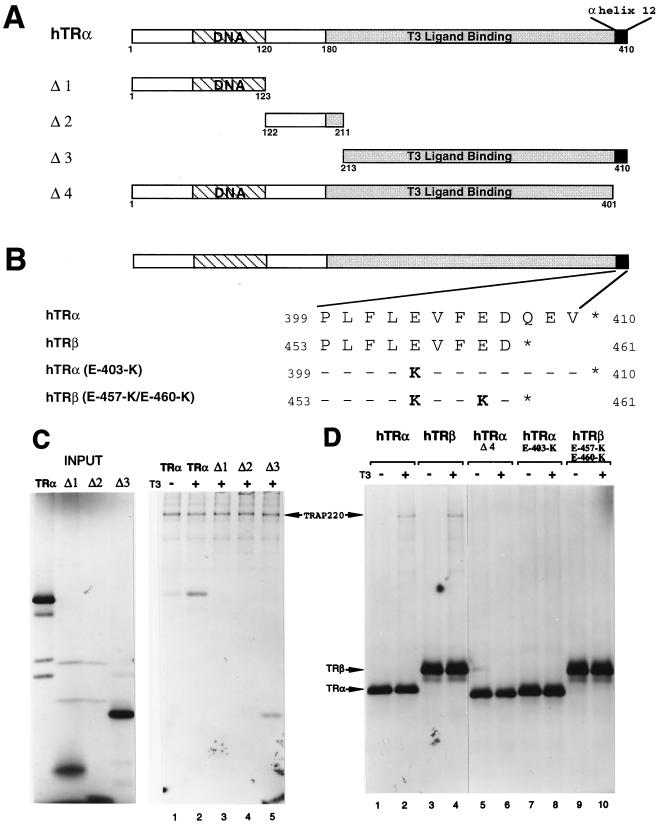
TR interaction with TRAP220 is AF2 dependent.
TRAP220 was first identified by its ability to associate and copurify with TR from T3-treated cells (17). However, whether the association was dependent on TR's AF2 domain was never firmly established. To address this question, we first tested TRα deletion mutants for the ability to immunoprecipitate with TRAP220 in the presence of T3 (Fig. 1A). Consistent with an AF2-dependent interaction, only the TRα deletion mutant containing the entire LBD was capable of associating with TRAP220 (Fig. 1C, lane 5). To more specifically examine the role of the TR's AF2 domain in TRAP220 binding, and given the essential role of the C-terminal helix 12 in mediating AF2 activity (44), we introduced point mutations into the C-terminal helix 12 of both TRα and TRβ (Fig. 1B) and created a deletion mutation of the C-terminal 12 amino acids of TRα (Fig. 1A). The TR AF2 mutants were then tested for binding to TRAP220 in the presence of T3 (Fig. 1D). The specific AF2 point mutations used here were chosen because they drastically impair T3-dependent transcriptional activation by TR but have no effect on T3 or DNA binding affinity (2, 3, 63). As shown in Fig. 1D (lanes 6, 8, and 10), all three TR AF2 mutants were completely deficient in T3-dependent binding to TRAP220. These findings demonstrate that the AF2 domain of TR is absolutely required for TRAP220 binding and thus, analogous to the SRC/p160 family of proteins, TRAP220 can be defined as an AF2-dependent coregulatory factor.
FIG. 1.
TRAP220 interaction with TR is dependent on the AF2 domain. (A) Schematic representation of the hTRα protein including the deletion mutants used in this study. The regions delineating the DNA binding and ligand binding domains are indicated. (B) The amino acid sequences of the conserved C-terminal helix 12 of hTRα and hTRβ and the various site-directed mutants. Only residues that are changed within the mutants are shown (in bold). (C) TRAP220 interacts with the TRα LBD in a T3-dependent fashion. 35S-labeled FLAG-TRAP220 was incubated with 35S-labeled hTRα or deletion derivatives (A) in the presence or absence of T3 as indicated. Protein complexes were coprecipitated with anti-FLAG antibodies coupled to agarose beads (M2 affinity resin). (D) TRAP220 interaction with TR is dependent on the integrity of helix 12. 35S-labeled HA-TRAP220 was incubated with 35S-labeled FLAG-TRα, FLAG-TRβ, or site-directed helix 12 mutants (B) in the presence or absence of T3 as indicated. As above, protein complexes were coprecipitated with M2 affinity resin.
Delineation of the minimal TRAP220 RBD.
TRAP220 contains two closely spaced LXXLL motifs in the central region of the protein (Fig. 2A, residues 604 to 608 and 645 to 649) and each motif can independently mediate T3-dependent interactions with TR (64). Although numerous other studies have demonstrated the importance of the signature LXXLL motifs in mediating coregulatory protein interactions with NRs (13, 24, 34, 57, 61), more recent findings suggest the presence of additional RIDs in some SRC/p160 family members which act in cis with the LXXLL motifs or function independently (1, 26, 37). To determine whether TRAP220 contains additional NR binding domains separable from the region containing the two LXXLL motifs, we generated a series of N- and C-terminal deletion mutations of TRAP220 (Fig. 2A and B) and tested whether the truncated proteins would bind to TRα in the presence of T3. Only TRAP220 deletion mutants containing one or both of the LXXLL motifs were capable of binding to TRα (Fig. 2C), thus suggesting that the region containing these motifs, which we have termed the RBD, is minimally required for ligand-dependent TR binding. While our findings fail to identify additional independent NR binding domains outside the RBD, we cannot rule out the possibility that polypeptide sequences found in the N- or C-terminal ends of TRAP220 might serve to stabilize or enhance the binding of NRs via association at the RBD.
Differential preference for LXXLL motifs in the TRAP220 RBD by different NRs.
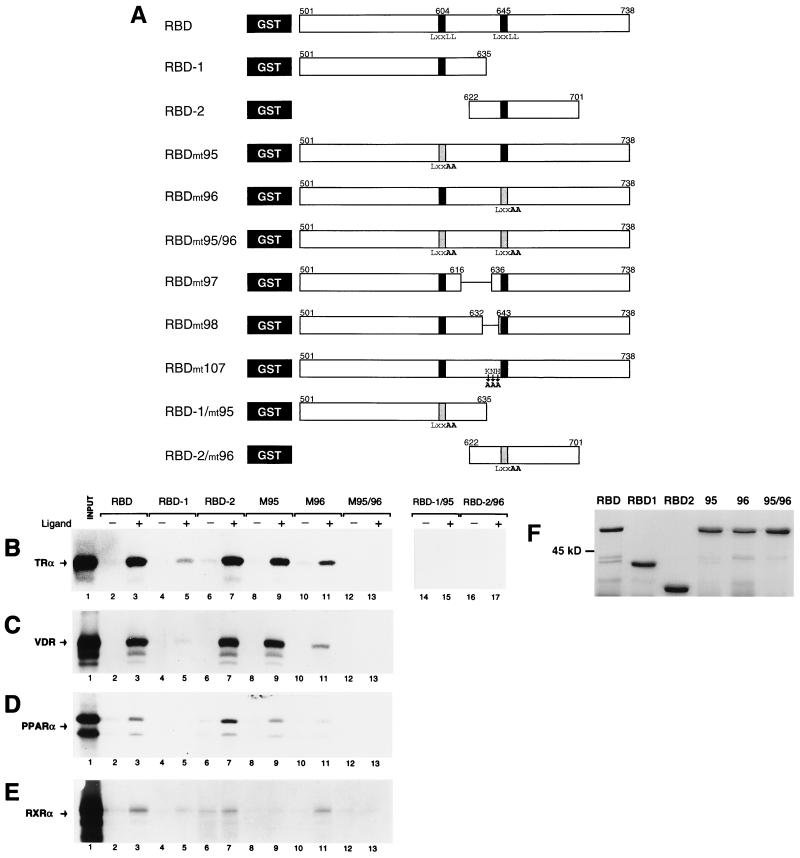
The presence of two LXXLL motifs in the TRAP220 RBD may represent two alternate NR binding sites, each equally potent in contacting a given NR AF2 domain. Conversely, and as evidenced with certain members of the SRC/p160 family, the two motifs might display different affinities for different specific NRs (12, 13, 38, 41, 57, 61). To begin to resolve this issue, each TRAP220 LXXLL core motif plus flanking polypeptide sequence, termed RBD-1 (Fig. 3A, residues 501 to 635) and RBD-2 (residues 622 to 701), was tested independently for ligand-dependent binding to various NRs using a GST pull-down assay (Fig. 3B to E). In agreement with earlier studies (64), TRα showed a significantly stronger T3-dependent binding to RBD-2 than to RBD-1 (Fig. 3B, lanes 4 to 7). Similarly, both VDR and the peroxisome proliferator-activated receptor α (PPARα) displayed a clear preference for binding to RBD-2 in the presence of ligand and showed almost no binding to RBD-1 (Fig. 3C and D, lanes 4 to 7). In contrast to TR, VDR, and PPAR, RXRα displayed a weak yet reproducible, ligand-dependent preference for binding to RBD-1 as well as a considerable amount of non-ligand-dependent association with RBD-2 (Fig. 3E, lanes 4 to 7).
FIG. 3.
Differential NR preference for the two LXXLL motifs in the TRAP220 RBD. (A) Schematic representation of the various GST fusion proteins used in this study. The full-length RBD (residues 501 to 738) including both LXXLL motifs (indicated by back bars) or constructs containing each separate LXXLL motif plus flanking region (RBD-1 and RBD-2; residues 501 to 635 and 622 to 701) were fused to the GST moiety (black box) (see Materials and Methods). Site-directed mutations are indicated below the constructs in bold. Deletions are indicated within the constructs as a horizontal line. Mutations within the core LXXLL motif (LXXLL to LXXAA) are also indicated as a shaded bar. (B to E) Interaction of TRα (B), VDR (C), PPARα (D), and RXRα (E) with the LXXLL-containing regions of TRAP220's RBD using a GST pull-down assay. 35S-labeled NRs were incubated with the indicated GST fusion protein in the presence or absence of cognate ligand (see Materials and Methods). The designation of GST fusion protein usage indicated above panel B is the same for panels C to E. In lane 1 of each panel, 50% of the input was loaded. (F) Purification and size analysis of the mutant GST fusion proteins used for panels B to E. RBD-1/95 and RBD-2/96 (not shown) were expressed and purified at similar concentrations. Proteins used in the binding reactions were normalized to equal concentrations before being added (see Materials and Methods).
An essential role for the conserved leucine residues within a consensus LXXLL motif for the binding of NRs has been firmly established by numerous mutagenesis studies in which substitution of one or more leucines by alanines completely abolishes physical and functional interactions between SRC/p160 proteins and NRs (13, 24, 34, 57, 61). To verify that the core LXXLL motifs within TRAP220 RBD-1 and RBD-2 are critical for the NR binding observed here, we replaced the last two leucine residues of each core motif with alanines (LXXLL to LXXAA) (RBD-1/mt95 and RBD-2/mt96 [Fig. 3A]). As shown in Fig. 3B (lanes 14 to 17), mutation of the core motifs completely abrogated ligand-dependent binding of TRα to both RBD-1 and RBD-2. This result demonstrates that the integrity of the core LXXLL motif in RBD-1 and RBD-2 is crucial for TRAP220-NR binding. Furthermore, in light of studies showing that a core LXXLL motif does not per se constitute an AF2 domain binding surface (12), this result suggests that the regions flanking the core LXXLL motifs in both TRAP220 and the SRC/p160 family of proteins may share common structural and molecular determinants.
As shown here, RBD-1 and RBD-2 display a differential preference for ligand-dependent interactions with different NRs when the two domains are physically separated (Fig. 3B to E, lanes 4 to 7). We sought to more thoroughly confirm this observation by examining LXXLL motif specificity within the context of the entire RBD (Fig. 3A, residues 501 to 738). To this end, we inactivated either the first core LXXLL motif, the second motif, or both motifs within the full-length RBD (Fig. 3A, residues 501 to 738), replacing the last two leucines of each motif with alanines (LXXLL to LXXAA), and then tested the mutant proteins for NR binding. In agreement with our initial results, inactivation of the first motif (RBDmt95) had minimal effect on the ligand-dependent binding of RBD to TRα, VDR, and PPARα (Fig. 3B to D, lanes 3 versus 9), whereas inactivation of the second motif (RBDmt96) significantly decreased the binding efficiency of all three receptors (Fig. 3B to D, lanes 3 and 9 versus 11). By contrast, inactivation of the first motif abolished ligand-dependent binding of RXRα to RBD (Fig. 3E, lanes 3 versus 9), while inactivation of the second motif did not significantly affect the ligand-dependent binding efficiency of RXRα with RBD (Fig. 3E, lanes 3 versus 11). None of the receptors bound to RBD when both motifs were inactivated (RBDmt95/96) (Fig. 3B to E, lanes 12 and 13), again underscoring the fundamental importance of the core LXXLL motif in facilitating the AF2 interactions. Taken together, these data show that RBD-2 is the preferential contact site for AF2-dependent binding of TRα, VDR, and PPARα. Although the interaction is much weaker, RBD-1 appears to be the preferential site for AF2-dependent binding of RXRα.
The role of adjacent residues in mediating RBD-2 specificity.
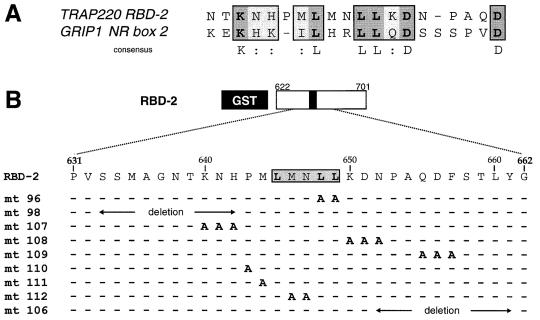
Analogous to the TRAP220 RBD, members of the SRC/p160 family contain multiple copies of the signature LXXLL motif (termed NR boxes) within their RID. Interestingly, biochemical and mutational studies of the p160 coactivator GRIP1 reveal a functional and physical preference of both TR and VDR for interaction with the second LXXLL motif (termed NR box 2) in the RID (12, 13). Amino acid residues flanking the core LXXLL motif have been proposed to determine the specificity and affinity of distinct NRs for individual NR boxes in the SRC/p160 proteins (12, 41). Given the preference of TRα and VDR for TRAP220 RBD-2, we hypothesized that residues adjacent to the core LXXLL motif in RBD-2 might be similar to those found adjacent to NR box 2 of GRIP1. Indeed, alignment of TRAP220 RBD-2 with GRIP1 NR box 2 (Fig. 4A) revealed several conserved amino acids including a cluster of basic residues on the N-terminal side of the core LXXLL motif and two conserved aspartic acid residues on the C-terminal side.
FIG. 4.
Effect of adjacent residues in mediating NR binding to the core LXXLL motif of RBD-2. (A) Comparison of the core LXXLL motif plus immediate flanking residues of TRAP220 RBD-2 with the corresponding region of GRIP1 NR box 2 (13). Conserved residues are boxed; identical residues are indicated in dark shading with bold letters; similar residues (:) are indicated by light shading. (B) Site-directed mutagenesis of the residues within and adjacent to the core LXXLL motif of RBD-2. Only residues that have been changed (to alanine) are shown (in bold). Deletions are indicated by a horizontal line with arrows spanning the deleted region. (C) Basic/polar residues N terminal to the core LXXLL motif are critical for NR binding to RBD-2. 35S-labeled NRs were incubated with the indicated GST fusion protein (above panel C) in the presence or absence of cognate ligand. The designation of GST fusion protein usage indicated above panel C is the same for panels D and E. In lane 1 of each panel, 100% of the input was loaded. (F) Purification and size analysis of the mutant GST fusion proteins used in panels C to E. Mutants 96, 98, and 111 (not shown) were expressed and purified at similar concentrations. Proteins used in the binding reactions were normalized to equal concentrations before being added (see Materials and Methods).
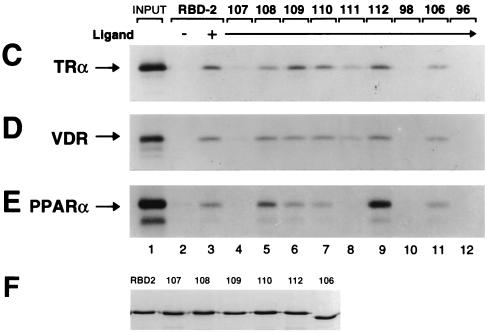
To begin to identify the molecular determinants underlying the specific preference of TRα, VDR, and PPARα for RBD-2, we systematically substituted amino acid residues within and adjacent to the core LXXLL motif of RBD-2 with alanines (Fig. 4B). The mutant proteins were subsequently tested for ligand-dependent binding to NRs (Fig. 4C to E). As expected, substitution of the last two leucines in the core LXXLL motif (residues 648 and 649; RBDmt96) abolished ligand-dependent interaction of RBD-2 with TRα, VDR, and PPARα (Fig. 4C to E, lane 12). Replacement of the hydrophobic methionine found on the immediate N-terminal side of the LXXLL sequence (residue 644; RBDmt111) only slightly reduced RBD-2 binding to TRα and VDR, yet drastically disrupted binding to PPARα (Fig. 4C to E, lane 8). By contrast, replacement of the MN spacer region (residues 646 and 647; RBDmt112) had no effect on RBD-2 binding to TRα and VDR yet significantly enhanced binding to PPARα (Fig. 4C to E, lane 9). These findings suggest that the AF2 domain of PPARα is extremely sensitive to the stereochemical properties of the amino acid side chains found within the core MLMNLL region, whereas the core motif requirements for TRα and VDR binding are less stringent. Replacement of the single proline residue (residue 643; RBDmt110), which presumably interrupts the α-helical structure of RBD-2 on the N-terminal side of the core LXXLL motif, had no significant effect on the binding of TRα, VDR, or PPARα (Fig. 4C to E, lane 7).
Recent structural and biochemical analyses of GRIP1 concluded that the higher affinity of TRα for NR box 2 reflects a favorable interaction between basic residues amino terminal to the core LXXLL motif (Fig. 4A) and acidic residues found in helix 12 of TR's AF2 domain (12). Conversely, mutagenesis experiments with another member of the SRC/p160 family, NCoA-1, demonstrated the importance of specific amino acids carboxy terminal to the core LXXLL in differentially mediating functional interactions with distinct NRs (41). To grossly define amino acid residues adjacent to the LXXLL motif of RBD-2 which may be important for the preferential binding of NRs, we deleted 10 residues on either side of the core LXXLL motif (residues 633 to 642 and 652 to 661, RBDmt98 and -106; Fig. 4B) and subsequently tested the mutants for NR binding. The N-terminal deletion completely abolished ligand-dependent binding of RBD-2 to TRα, VDR, and PPARα (Fig. 4C to E, lane 10), while the C-terminal deletion only barely decreased binding (Fig. 4C to E, lane 11). To more precisely identify the flanking amino acids essential for preferential NR binding, we systematically replaced residues adjacent to the core LXXLL motif of RBD-2 with clusters of alanines (Fig. 4B). Replacement of the regions containing either of the two conserved C-terminal aspartic acid residues (residues 650 to 652 and 655 to 657, RBDmt108 and -109) did not significantly decrease ligand-dependent binding of TRα, VDR, or PPARα to RBD-2 (Fig. 4C to E, lanes 5 and 6). In fact, RBDmt108 modestly enhanced RBD-2 binding to PPARα. By contrast and consistent with the N-terminal deletion mutation, replacement of the three basic/polar residues KNH (residues 640 to 642; RBDmt107) N terminal to the LXXLL motif severely disrupted RBD-2 binding to TRα, VDR, and PPARα (Fig. 4C to E, lane 4).
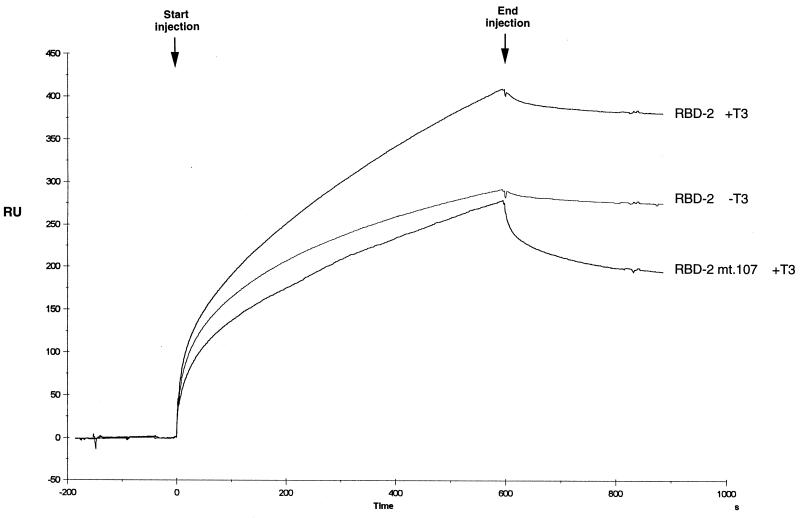
To further characterize the binding of TR to RBD-2, we initiated SPR experiments in which the interaction between two macromolecules can be effectively measured in real time. To facilitate these studies, wild-type RBD-2 or RBD-2/mt107 was immobilized on the surface of a sensor chip and subjected to a constant flow injection of TRβ across the chip surface in either the presence or absence of T3. Analysis of the SPR sensorgram (Fig. 5) revealed a higher affinity of TRβ for RBD-2 in the presence versus the absence of T3, as evidenced by an apparently higher on rate during the period of injection (0 to 600 s). Interestingly, binding affinity of TRβ for RBD-2/mt107 is significantly reduced, as indicated by an apparently lower on rate during the injection period. Moreover, TRβ in association with RBD-2/mt107 exhibited a higher postinjection off rate (600 to 800 s) (Fig. 5), indicating that the TRβ–RBD-2/mt107 complex is much less stable than the TRβ–RBD-2 complex. Taken together with results of the GST pull-down assays above, and in agreement with the previous GRIP1 NR box 2 mutagenesis studies (12), our findings indicate that the specific binding preference of TR, VDR, and PPARα for RBD-2 is due, at least in part, to a cluster of basic/polar amino acid residues amino terminal to the core LXXLL motif. Consistent with this conclusion, the core LXXLL motif of RBD-1 lacks analogous residues immediately flanking its N terminus and likely explains why RBD-1 only weakly associates with TR, VDR, and PPAR (Fig. 3).
FIG. 5.
Mutation of basic/polar residues N terminal to the core LXXLL motif of TRAP220 RBD-2 disrupts ligand-dependent binding of TR as determined by SPR. The sensorgram measures the real-time binding of hTRβ to GST-RBD-2 or GST-RBD-2/mt107 (Fig. 4B) immobilized on the surface of a sensor chip in the presence or absence of T3 as indicated. The arrows indicate the start and the end of a constant-flow injection of the analyte His10-hTRβ over the chip surface. After the 10-min injection was finished, the analyte was replaced by running buffer (HBS-EP [see Materials and Methods]). RU, resonance units.
TRAP220 RBD interactions with NR heterodimers bound to DNA.
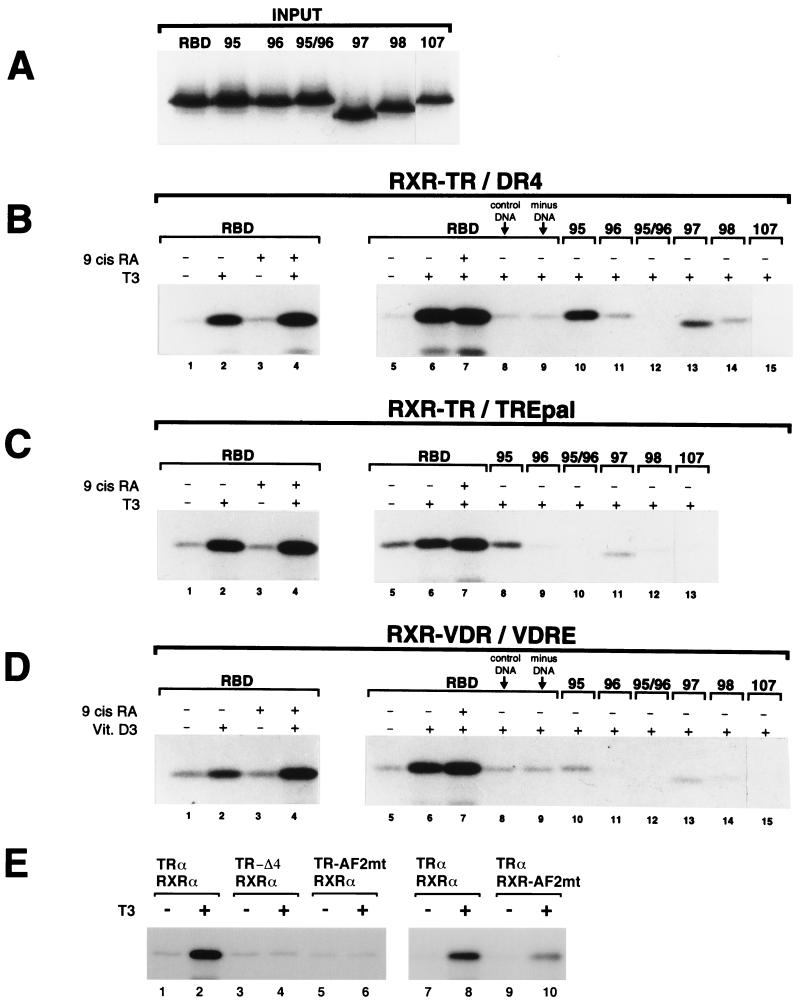
In view of the fact that TR, VDR, and PPAR bind to DNA as heterodimers with RXR (40), the preference of TR, VDR, and PPAR for TRAP220's RBD-2, and weaker preference of RXR for RBD-1, may have implications for TRAP220 binding to DNA-bound NR heterodimers. To begin to investigate this matter, we radiolabeled TRAP220 RBD and various mutant derivatives (Fig. 3A and 6A) and examined their ability to form complexes with either RXR-TR or RXR-VDR heterodimers bound to DNA (Fig. 6B to D). To examine whether each subunit of an RXR-TR heterodimer is capable of recruiting TRAP220 independently, we tested for complex formation in the presence of either T3 or the RXR-specific ligand, 9-cis RA. When RXR-TR heterodimers are bound to a specific T3 response element (DR4 or TREpal), addition of a saturating concentration of T3 (1 μM) induced a robust interaction with TRAP220 RBD, whereas addition of a saturating concentration of 9-cis RA (1 μM) had no effect (Fig. 6B and C, lanes 1 to 3). However, when T3 and 9-cis RA were added together, the overall binding was slightly greater than that observed with T3 alone (Fig. 6B and C, lanes 2 and 4), suggesting that 9-cis RA promotes the overall binding of TRAP220 RBD to the heterodimer. Similar results were obtained using RXR-VDR heterodimers bound to a VDRE (Fig. 6D, lanes 1 to 4). This finding appears to be in contrast to a previous study in which a partial RXR-TR–TRAP220-RBD complex is observed in the presence of 9-cis RA alone (59) and is likely accounted for by our more stringent washing conditions of the complexes (see Materials and Methods). Indeed, the failure of RXR to independently recruit RBD in the presence of 9-cis RA alone may further reflect an allosteric inhibition of RXR's AF2 domain by its heterodimeric partner (20, 33, 62, 66). Consistent with the findings here, such an inhibition might be alleviated only by the ligand-induced binding of an LXXLL motif to the AF2 domain of RXR's partner, after which time RXR's AF2 motif could interact with a second LXXLL motif (62).
FIG. 6.
Interaction of TRAP220 RBD with DNA-bound RXR-TR and RXR-VDR heterodimers. (A) GST-RBD wild-type and GST-RBD mutant proteins (Fig. 3A) were purified and 32P-labeled (see Materials and Methods). Panel A shows 15% of the labeled input used in the experiments described below. (B and C) Both the presence and proper spacing of RBD-1 and RBD-2 are necessary for efficient binding of TRAP220 RBD to an RXR-TR heterodimer bound to a specific TRE. Unlabeled in vitro-translated hRXRα and hTRα were incubated with 32P-labeled GST-RBD wild-type or mutant proteins (panel A and indicated above the lanes) together with a biotinylated DR4 (B) or biotinylated TREpal (C) in the presence or absence of T3 or 9-cis RA as indicated. DNA-bound ternary protein complexes were precipitated by adding streptavidin-agarose beads, and the proteins were subsequently fractionated by SDS-PAGE. The reaction shown in panel B, lane 8, contained a nonspecific biotinylated DNA element (see Materials and Methods) in place of a specific TRE. The reaction shown in Fig. 6B, lane 9, did not contain any DNA. (D) Both the presence and proper spacing of RBD-1 and RBD-2 are necessary for efficient binding of TRAP220 RBD to an RXR-VDR heterodimer bound to a specific VDRE. Unlabeled in vitro-translated hRXRα and hVDR were incubated with 32P-labeled GST-RBD wild-type or mutant proteins (panel A and indicated above the lanes) together with a biotinylated VDRE (derived from the osteopontin gene promoter) in the presence or absence of 1,25-(OH)2-D3 (Vit. D3) or 9-cis RA as indicated. As above, the addition of a nonspecific DNA element or the omission of DNA altogether (lanes 8 and 9) was performed as a control. (E) The AF2 domains from both partners of a DNA-bound RXR-TR heterodimer are required for efficient binding to RBD. Unlabeled wild-type hRXRα and hTRα receptors were dimerized with TR or RXR proteins containing AF2 mutations (as indicated above the lanes) and incubated with wild-type 32P-labeled GST-RBD protein together with a biotinylated DR4 in the presence or absence of T3. Specific AF2 mutants used in this experiment include TRα-AF2mt (hTRα, E-403-K), TRα-Δ4 (C-terminal deletion of helix 12, Δ401), and hRXRα-AF2mt (F-450-A, E-453-K and E-456-K) (see Materials and Methods for details).
The ability of two ligands to modestly enhance the overall binding of TRAP220 RBD to a DNA-bound heterodimer may indicate that two molecules of TRAP220 are recruited into the complex, one TRAP220 protein per heterodimer subunit. Alternatively, RBD-1 and RBD-2 from a single TRAP220 molecule might differentially interact with the AF2 domain of each heterodimer subunit, possibly stabilizing the overall binding of TRAP220 to the heterodimer. Consistent with the second supposition, mutations inactivating the first, the second, or both of the core LXXLL motifs of RBD (RBDmt95, -96 and -95/96, respectively [Fig. 3A]) severely disrupted or completely abolished RBD binding to both RXR-TR and RXR-VDR in the presence of one ligand (T3 and vitamin D3, respectively) (Fig. 6B and D, lanes 10 to 12; Fig. 6C, lanes 8 to 10). When 9-cis RA was added simultaneously with T3 or vitamin D3, similar results were obtained (data not shown). The specific nature of DNA binding element appears to play a role in complex formation since RXR-TR could modestly interact with an RBD containing only one functional LXXLL motif (RBDmt95) when bound to a DR4 but not when bound to a TREpal (Fig. 6B, lane 10, versus 6C, lane 8). This finding indicates that TRAP220 RBD is capable of associating with a DNA-bound heterodimer through a single RBD-2 contact site, although the interaction is weaker than when both RBD-1 and RBD-2 are present. Interestingly, mutations which removed or replaced the conserved cluster of basic residues N terminal to the second LXXLL motif of RBD-2 (RBDmt98 and -107 [Fig. 3A]) also abolished RBD binding to the heterodimers (Fig. 6B and D, lanes 14 and 15; Fig. 6C, lanes 12 and 13), again demonstrating the importance of these residues in mediating RBD-2 function. Finally, we found that mutations affecting the spacing between the two LXXLL motifs of RBD (RBDmt97 and -98 [Fig. 3A]) also severely abrogated RBD interactions with both RXR-TR and RXR-VDR heterodimers (Fig. 6B and D, lanes 13 and 14; Fig. 6C, lanes 11 and 12).
To examine whether the AF2 domains from both partners of a DNA-bound NR heterodimer are required for efficient binding to TRAP220, we tested RBD for binding to RXR-TR heterodimers in which the AF2 motif from either TR or RXR was mutated (Fig. 6E). In agreement with our earlier findings (Fig. 1), deletion or site-directed mutagenesis of the C-terminal helix 12 of TRα abolished RBD binding to an RXR-TR heterodimer (Fig. 6E, lanes 4 and 6). Similarly, and consistent with a role for RXR in promoting the overall binding of TRAP220 to a DNA-bound heterodimer, site-directed mutagenesis of RXRα's AF2 motif also significantly decreased the binding of RBD to RXR-TR (Fig. 6E, lane 10).
In sum, these findings indicate that both RBD-1 and RBD-2, as well as proper spacing between the LXXLL motifs, are required for an optimal interaction between TRAP220 and RXR-TR or RXR-VDR heterodimers bound to DNA. Given the biochemical preference of TR and VDR for RBD-2 and the weaker association of RXR with RBD-1, these results suggest that RBD-1 and RBD-2 from a single TRAP220 molecule may associate with the AF2 domains from each heterodimer partner. Inherent in this hypothesis is that unliganded RXR still associates with RBD-1, possibly stabilizing or facilitating the ligand-dependent interaction between RBD-2 and the other heterodimer subunit. As suggested by the results here, addition of RXR's ligand might further stabilize the RXR–RBD-1 interaction and presumably strengthen the overall association of TRAP220 with the heterodimer.
Role of RBD-1 and RBD-2 in TRAP220 transcriptional function.
Our experiments examining TRAP220 RBD interactions with DNA-bound NR heterodimers in vitro revealed a requirement of both RBD-1 and RBD-2 for optimal binding (Fig. 6). We next examined the functional role of these domains in TRAP220-mediated transcriptional coactivation in vivo. Toward this end, RBD-1 and RBD-2 were selectively inactivated within a full-length TRAP220 mammalian expression vector by replacing the last two leucines of their core LXXLL motifs with alanines (LXXLL to LXXAA) (TRAP220 mt.95 and TRAP220 mt.96 [Fig. 7A]). When wild-type TRAP220 was transiently cotransfected into NIH 3T3 cells with an hVDR expression vector, VDR-dependent transcription in the presence of ligand was enhanced more than threefold (Fig. 7B and C). Consistent with previous findings (64), cotransfection of TRAP220 lacking a functional RBD-2 (mt.96) was devoid of transcriptional coactivation function (Fig. 7B). Interestingly, when TRAP220 lacking a functional RBD-1 (mt.95) was cotransfected, the observed coactivation function was also significantly diminished (Fig. 7), albeit not to the level observed with the RBD-2 mutant (mt.96). These findings closely parallel our in vitro binding studies (Fig. 6) in which RBD mutants lacking RBD-2 are completely devoid of binding to DNA-bound NR heterodimers, while mutants lacking RBD-1 are capable of only modest interactions on specific response elements. Furthermore, our results parallel analogous studies in which both LXXLL motifs of DRIP205 (i.e., TRAP220) were shown to be equally required for the functional interaction with VDR in vivo (49).
To confirm the functional importance of the conserved cluster of basic/polar residues N terminal to the core LXXLL motif of RBD-2, we replaced these residues with alanines in the context of the full-length TRAP220 protein, leaving the two core LXXLL motifs of RBD-1 and RBD-2 intact (TRAP220 mt.107 [Fig. 7A]). In strong agreement with our earlier in vitro binding studies (Fig. 4 to 6), cotransfection of this mutant TRAP220 protein displayed significantly less coactivation function compared to the wild-type protein (Fig. 7C). These findings underscore the importance of the amino acids flanking the core LXXLL motif of RBD-2, not only in determining physical NR binding preference but in mediating transcriptionally functional interactions with hormone-activated NRs. Taken together, the results of Fig. 6 and 7 demonstrate a functional requirement for both RBD-1 and RBD-2 in mediating TRAP220 transcriptional coactivation activities.
DISCUSSION
Hormone-dependent transactivation by NRs involves specific interactions with coactivators via ligand-induced allosteric changes in the conserved AF2 domain (42, 58). Here we show that the transcriptional coregulatory factor TRAP220 associates with NRs in an AF2-dependent fashion through both of its two signature LXXLL motifs found within a minimal RBD. We further demonstrate that the two LXXLL-containing regions, RBD-1 and RBD-2, are differentially preferred by specific NRs and that preference for RBD-2 is due to the presence of basic/polar residues on the amino-terminal end of the core LXXLL motif. Finally, we show that the presence and proper spacing of both RBD-1 and RBD-2 are required for the functional interaction of TRAP220 with DNA-bound NR heterodimers.
Crystallographic studies have established a conserved mechanism for NR-coactivator binding in which the conserved leucines of a signature LXXLL motif pack into a hydrophobic groove formed by conserved residues in helices 3, 4, 5, and 12 of the receptor LBD (12, 46, 53). Given the presence of multiple LXXLL motifs within the SRC/p160 proteins and TRAP220, and the observation of distinct preferences for individual motifs, the existence of a conserved “structural code” flanking the core motifs has been proposed in order to account for their differential usage by NRs (12, 41). Using site-directed mutagenesis, we identified three basic/polar residues, KNH (residues 640 to 642), on the N-terminal side of the core LXXLL motif of RBD-2 which are absolutely required for strong AF2-dependent binding to TR, VDR, and PPAR (Fig. 4 to 6) and for TRAP220 transcriptional coactivation function in vivo (Fig. 7). By contrast, the core LXXLL motif of RBD-1 lacks analogous residues on its N terminus and only weakly associates with TR, VDR, and PPAR (Fig. 3).
Interestingly, three basic amino acids conserved across all members of the SRC/p160 family of proteins are found flanking the N terminus of the second LXXLL motif (NR box 2) (Fig. 4A) (12). Comparable to the case with RBD-2, both TR and VDR display a functional and physical preference for NR box 2 of the p160 coactivator GRIP1 (12, 13), and removal of the N-terminal cluster of basic residues severely compromises interaction with TR (12). Similarly, the estrogen receptor displays a distinct preference for NR box 2 of both TIF2 (the human orthologue of GRIP1) (61) and SRC-1 (13, 28, 38). Replacement or removal of the conserved cluster of basic residues from the NR box 2 of SRC-1 dramatically inhibited ligand-dependent interaction with the estrogen receptor (38). Moreover, introduction of the basic residues at the N terminus of low-affinity LXXLL motifs is sufficient to transform them into high-affinity binding sites for NRs (12, 38).
One possible explanation for the functional importance of these residues comes from recent structural studies examining the TRβ-NR box 2 interface (12). These studies found that the conserved basic residues N terminal to the core LXXLL motif are in close proximity with conserved acidic residues at the C terminus of the TRβ LBD (helix 12 residues E460 and D461). Thus, in addition to a primary hydrophobic interaction between a core LXXLL motif and the AF2 domain, a second electrostatic interaction may occur reflecting a favorable contact between positively charged residues N terminal to the LXXLL motif and negatively charged residues of the NR LBD (12). In view of this hypothesis, it is interesting that lysine residues flanking the core LXXLL motif of NR box 1 in the p160 coactivator ACTR have been proposed to be targets for HAT-mediated transcriptional attenuation in which acetylation of the conserved lysine neutralizes their positive charge and disrupts the electrostatic association with NRs (8).
In addition to providing alternate binding sites for different NRs, the presence of multiple LXXLL motifs in TRAP220, the SRC/p160 proteins, and other NR-coregulatory factors might fulfill other physiologically important functions such as stabilizing the formation of a DNA-bound NR-coactivator complex. Given that NRs typically bind DNA as homo- or heterodimers, multiple LXXLL motifs from a single coactivator molecule might conceivably provide contact sites for each dimer subunit, thereby stabilizing ternary complex formation and possibly enhancing the affinity of the complex for DNA. One line of evidence suggesting that a single molecule of TRAP220 can simultaneously interact with both partners of an RXR-NR heterodimer via its two core LXXLL motifs comes from our experiments examining TRAP220 RBD interactions with NRs bound to DNA (Fig. 6). In general, we found that an efficient interaction between TRAP220 RBD and either RXR-TR or RXR-VDR heterodimers required the presence of both RBD-1 and RBD-2, as well as proper spacing between the two LXXLL motifs (Fig. 6B to D). Reciprocally, we found that the AF2 domains from both receptor partners were also required for an optimal interaction with TRAP220 RBD (Fig. 6E). We did observe modest binding of an RBD mutant (lacking a functional RBD-1; RBDmt95) with RXR-TR bound to a DR4 response element, thus indicating that TRAP220 can form complexes with RXR-TR heterodimers via a single LXXLL contact involving RBD-2. This observation is consistent with previous transient assays in which a TRAP220 protein lacking a functional RBD-1 was still capable of enhancing T3-dependent transcription from a DR4 reporter gene (64). Nonetheless, when RXR-VDR was bound to a high-affinity VDRE from the osteopontin gene promoter, both RBD-1 and RBD-2 were required in order for RBD to effectively associate with the heterodimer (Fig. 6D). Furthermore, we found that both RBD-1 and RBD-2 were functionally required for optimal TRAP220 transcriptional coactivation of VDR-mediated gene expression in vivo (Fig. 7).
In view of these results, the question arises as to the molecular configuration of a TRAP220–RXR-NR complex. Given the clear biochemical preference of TR, VDR, and PPAR for binding to RBD-2, and the much weaker yet specific preference of RXR for binding to RBD-1 (Fig. 3 and 4), our findings suggests a model of ligand-dependent interaction between TRAP220 and a DNA-bound RXR-NR heterodimer in which RXR's partner contacts RBD-2 and RXR simultaneously contacts RBD-1. Addition of cognate ligand for RXR's partner (e.g., TR, VDR, or PPAR) presumably promotes a specific interaction between that NR's AF2 domain and RBD-2. This action may additionally alleviate a putative allosteric inhibition of the RXR AF2 domain (20, 62), thus permitting RXR interactions with RBD-1 in the absence of RXR's ligand. Although we have no direct binding data supporting this step, the interaction is suggested by (i) our findings showing that RBD-1, in addition to RBD-2, is essential for an optimal interaction of RBD with DNA-bound RXR-TR or RXR-VDR in the absence of RXR's ligand (Fig. 6B to D); (ii) our findings showing that disruption of RXR's AF2 domain disrupts the binding of RBD to a DNA-bound RXR-TR heterodimer in the presence of T3 (Fig. 6E); and (iii) our in vivo studies showing that RBD-1 and RBD-2 are both functionally required for optimal TRAP220 coactivation of VDR-mediated gene expression in the absence of RXR's ligand (Fig. 7). Indeed, an intrinsic affinity of unliganded RXR for RBD-1 would presumably be manifest only when RXR's heterodimeric partner is ligand-dependently bound to RBD-2 (20, 62).
Addition of ligand for RXR, in addition to ligand for its heterodimeric partner, might further strengthen the RXR–RBD-1 interaction and presumably stabilize the overall association of TRAP220 with the heterodimer. Consistent with this notion, numerous studies have shown that RXR ligands enhance ligand-dependent transcriptional effects of VDR, PPAR, and the RA receptor (5, 9, 20, 31, 32, 43). Although synergistic effects of RXR ligands and T3 on RXR/TR-mediated transcription have also been reported for specific promoters (27, 50), other studies suggest that RXR ligands may inhibit T3-dependent transcription (20, 22), possibly by promoting the formation of RXR homodimers (36). Thus, in the case of RXR-TR heterodimers, the presence of T3 alone may be sufficient to induce an optimal interaction between TRAP220's RBD-1 and RBD-2 and the AF2 domains of RXR and TR.
ACKNOWLEDGMENT
This work was supported by NIH grant DK54030-02 (to J.D.F.).
REFERENCES
- 1.Alen P, Claessens F, Verhoeven G, Rombauts W, Peeters B. The androgen receptor amino-terminal domain plays a key role in p160 coactivator-stimulated gene transcription. Mol Cell Biol. 1999;19:6085–6097. doi: 10.1128/mcb.19.9.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baniahmad A, Leng X, Burris T P, Tsai S Y, Tsai M J, O'Malley B W. The τ 4 activation domain of the thyroid hormone receptor is required for release of a putative corepressor(s) necessary for transcriptional silencing. Mol Cell Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barettino D, Vivanco Ruiz M M, Stunnenberg H G. Characterization of the ligand-dependent transactivation domain of thyroid hormone receptor. EMBO J. 1994;13:3039–3049. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco J C, Minucci S, Lu J, Yang X J, Walker K K, Chen H, Evans R M, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlberg C, Bendik I, Wyss A, Meier E, Sturzenbecker L J, Grippo J F, Hunziker W. Two nuclear signalling pathways for vitamin D. Nature. 1993;361:657–660. doi: 10.1038/361657a0. [DOI] [PubMed] [Google Scholar]
- 6.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 9.Chen J Y, Clifford J, Zusi C, Starrett J, Tortolani D, Ostrowski J, Reczek P R, Chambon P, Gronemeyer H. Two distinct actions of retinoid-receptor ligands. Nature. 1996;382:819–822. doi: 10.1038/382819a0. [DOI] [PubMed] [Google Scholar]
- 10.Chiang C M, Roeder R G. Expression and purification of general transcription factors by FLAG epitope-tagging and peptide elution. Pept Res. 1993;6:62–64. [PubMed] [Google Scholar]
- 11.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P J, Stallcup M R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 14.Drane P, Barel M, Balbo M, Frade R. Identification of RB18A, a 205 kDa new p53 regulatory protein which shares antigenic and functional properties with p53. Oncogene. 1997;15:3013–3024. doi: 10.1038/sj.onc.1201492. [DOI] [PubMed] [Google Scholar]
- 15.Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fondell J D, Roy A L, Roeder R G. Unliganded thyroid hormone receptor inhibits formation of a functional preinitiation complex: implications for active repression. Genes Dev. 1993;7:1400–1410. doi: 10.1101/gad.7.7b.1400. [DOI] [PubMed] [Google Scholar]
- 17.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fondell J D, Brunel F, Hisatake K, Roeder R G. Unliganded thyroid hormone receptor alpha can target TATA-binding protein for transcriptional repression. Mol Cell Biol. 1996;16:281–287. doi: 10.1128/mcb.16.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fondell J D, Guermah M, Malik S, Roeder R G. Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc Natl Acad Sci USA. 1999;96:1959–1964. doi: 10.1073/pnas.96.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forman B M, Umesono K, Chen J, Evans R M. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 21.Gu W, Malik S, Ito M, Yuan C X, Fondell J D, Zhang X, Martinez E, Qin J, Roeder R G. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 22.Hallenbeck P L, Phyillaier M, Nikodem V M. Divergent effects of 9-cis-retinoic acid receptor on positive and negative thyroid hormone receptor-dependent gene expression. J Biol Chem. 1993;268:3825–3828. [PubMed] [Google Scholar]
- 23.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 25.Henttu P M, Kalkhoven E, Parker M G. AF-2 activity and recruitment of steroid receptor coactivator 1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol Cell Biol. 1997;17:1832–1839. doi: 10.1128/mcb.17.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong H, Darimont B D, Ma H, Yang L, Yamamoto K R, Stallcup M R. An additional region of coactivator GRIP1 required for interaction with the hormone-binding domains of a subset of nuclear receptors. J Biol Chem. 1999;274:3496–3502. doi: 10.1074/jbc.274.6.3496. [DOI] [PubMed] [Google Scholar]
- 27.Kakizawa T, Miyamoto T, Kaneko A, Yajima H, Ichikawa K, Hashizume K. Ligand-dependent heterodimerization of thyroid hormone receptor and retinoid X receptor. J Biol Chem. 1997;272:23799–23804. doi: 10.1074/jbc.272.38.23799. [DOI] [PubMed] [Google Scholar]
- 28.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 30.Kato S, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 31.Keller H, Dreyer C, Medin J, Mahfoudi A, Ozato K, Wahli W. Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator-activated receptor-retinoid X receptor heterodimers. Proc Natl Acad Sci USA. 1993;90:2160–2164. doi: 10.1073/pnas.90.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kliewer S A, Umesono K, Noonan D J, Heyman R A, Evans R M. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld M G, Heyman R A, Glass C K. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature. 1994;371:528–531. doi: 10.1038/371528a0. [DOI] [PubMed] [Google Scholar]
- 34.Le Douarin B, Nielsen A L, Garnier J M, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J W, Choi H S, Gyuris J, Brent R, Moore D D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 36.Lehmann J M, Zhang X K, Graupner G, Lee M O, Hermann T, Hoffmann B, Pfahl M. Formation of retinoic X receptor homodimers leads to repression of T3 response: hormonal cross talk by ligand-induced squelching. Mol Cell Biol. 1993;13:7698–7707. doi: 10.1128/mcb.13.12.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma H, Hong H, Huang S M, Irvine R A, Webb P, Kushner P J, Coetzee G A, Stallcup M R. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol Cell Biol. 1999;19:6164–6173. doi: 10.1128/mcb.19.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mak H Y, Hoare S, Henttu P M, Parker M G. Molecular determinants of the estrogen receptor-coactivator interface. Mol Cell Biol. 1999;19:3895–3903. doi: 10.1128/mcb.19.5.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 41.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna N J, Lanz R B, O'Malley B W. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 43.Minucci S, Leid M, Toyama R, Saint-Jeannet J P, Peterson V J, Horn V, Ishmael J E, Bhattacharyya N, Dey A, Dawid I B, Ozato K. Retinoid X receptor (RXR) within the RXR-retinoic acid receptor heterodimer binds its ligand and enhances retinoid-dependent gene expression. Mol Cell Biol. 1997;17:644–655. doi: 10.1128/mcb.17.2.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 45.Nagpal S, Saunders M, Kastner P, Durand B, Nakshatri H, Chambon P. Promoter context- and response element-dependent specificity of the transcriptional activation and modulating functions of retinoic acid receptors. Cell. 1992;70:1007–1019. doi: 10.1016/0092-8674(92)90250-g. [DOI] [PubMed] [Google Scholar]
- 46.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 47.Parker M G. Steroid and related receptors. Curr Opin Cell Biol. 1993;5:499–504. doi: 10.1016/0955-0674(93)90016-j. [DOI] [PubMed] [Google Scholar]
- 48.Rachez C, Suldan Z, Ward J, Chang C P, Burakov D, Erdjument-Bromage H, Tempst P, Freedman L P. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rachez C, Gamble M, Chang C-P B, Atkins G B, Lazar M A, Freedman L P. The DRIP complex and SRC-1/p160 coactivators share similar nuclear receptor binding determinants but constitute functionally distinct complexes. Mol Cell Biol. 2000;20:2718–2726. doi: 10.1128/mcb.20.8.2718-2726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosen E D, O'Donnell A L, Koenig R J. Ligand-dependent synergy of thyroid hormone and retinoid X receptors. J Biol Chem. 1992;267:22010–22013. [PubMed] [Google Scholar]
- 51.Ryu S, Zhou S, Ladurner A G, Tjian R. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 52.Saatcioglu F, Bartunek P, Deng T, Zenke M, Karin M. A conserved C-terminal sequence that is deleted in v-ErbA is essential for the biological activities of c-ErbA (the thyroid hormone receptor) Mol Cell Biol. 1993;13:3675–3685. doi: 10.1128/mcb.13.6.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 54.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 55.Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 56.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 57.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 58.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 59.Treuter E, Johansson L, Thomsen J S, Warnmark A, Leers J, Pelto-Huikko M, Sjoberg M, Wright A P, Spyrou G, Gustafsson J A. Competition between thyroid hormone receptor-associated protein (TRAP) 220 and transcriptional intermediary factor (TIF) 2 for binding to nuclear receptors. Implications for the recruitment of TRAP and p160 coactivator complexes. J Biol Chem. 1999;274:6667–6677. doi: 10.1074/jbc.274.10.6667. [DOI] [PubMed] [Google Scholar]
- 60.Tsai M J, O'Malley B W. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 61.Voegel J J, Heine M J, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 63.Wong J, Shi Y B, Wolffe A P. Determinants of chromatin disruption and transcriptional regulation instigated by the thyroid hormone receptor: hormone-regulated chromatin disruption is not sufficient for transcriptional activation. EMBO J. 1997;16:3158–3171. doi: 10.1093/emboj/16.11.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan C X, Ito M, Fondell J D, Fu Z Y, Roeder R G. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang J, Fondell J D. Identification of mouse TRAP100: a transcriptional coregulatory factor for thyroid hormone and vitamin D receptors. Mol Endocrinol. 1999;13:1130–1140. doi: 10.1210/mend.13.7.0295. [DOI] [PubMed] [Google Scholar]
- 66.Zhang J, Hu X, Lazar M A. A novel role for helix 12 of retinoid X receptor in regulating repression. Mol Cell Biol. 1999;19:6448–6457. doi: 10.1128/mcb.19.9.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu Y, Qi C, Jain S, Rao M S, Reddy J K. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]