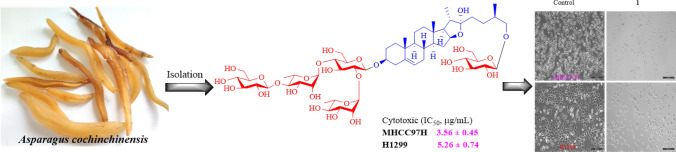
Graphic Abstract
Phytochemical investigation on the roots of Asparagus cochinchinensis led to the isolation of one new furostanol saponin, named 26-O-β-d-glucopyranosyl-22α-hydroxyl-(25R)-Δ5(6)-furost-3β,26-diol-3-O-α-l-rhamnopyranosyl-(1 → 2)-[β-d-glucopyranosyl-(1 → 4)-α-l-rhamnopyranosyl-(1 → 4)]-β-d-glucopyranoside (1), along with three known congeners (2‒4). The structure of new saponin was elucidated via comprehensive inspection of its HRMS and NMR spectral data as well as chemical technology, whereas those of known ones were identified by comparison of their NMR and MS spectral data with those reported in literatures. All isolated saponins were evaluated for their cytotoxic effects on two human liver (MHCC97H) and lung adenocarcinoma (H1299) cancer cells in vitro. Among them, both 1 and 2 showed significant cytotoxicity against above mentioned cell lines. Further studies revealed that these two saponins could significantly inhibit their proliferation of MHCC97H and H1299 cells.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13659-021-00321-0.
Keywords: Steroid saponins, Asparagus cochinchinensis, Cytotoxicity, Structural elucidation
Introduction
Steroid saponins, whose aglycones were usually a spirostanol or its derivatives [1], were commonly found from roots, tubers, leaves, blooms or seeds in more than 100 families of plants [2, 3]. Compared with other glycosides, the strong foam-forming property in aqueous solution of steroidal saponins was their main feature [2, 4]. Previous researches revealed steroidal saponins possessed various pharmacological activities, such as antifungal [5], hypocholesterolemic [6], antimitotic [7] and cAMP phosphodiesterase inhibitory [8] effects. Among them, a large number of publications have revealed steroid saponins shared different cytotoxic properties that promoted their potential as anti-cancer drugs or adjuvants [9, 10].
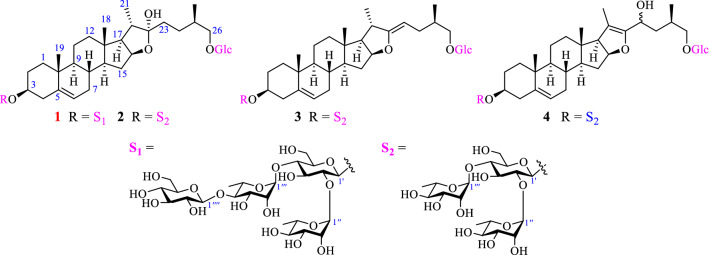
Asparagus cochinchinensis, belonging to the genus Asparagus (Liliaceae), is well-known as “Tianmendong” in China. Its roots have been historically used in Chinese folk medicine for the treatment of cough, acute and chronic bronchitis, chronic pharyngitis, hemorrhoids, and tumors for thousands of years [11]. Apart from steroidal saponins [12], phenolic compounds [13], norlignans [14] and alkaloids [15] have been isolated from this plant as revealed by previous phytochemical studies. However, steroidal saponins obtained from title species were proved to be its major and bioactive components responsible for its cytotoxic [16], anti-inflammatory [17], hepatotoxic and nephrotoxic [18], and anti-neuroinflammatory [11] properties. In continuation of a search for bioactive constituents from plants of the Yunnan province [19], a chemical investigation was performed on the roots of A. cochinchinens. As a result, a total of steroidal saponins (1‒4) were isolated and identified including one new and three previously described furostan-type steroidal saponins. Their cytotoxic effects on two human cancer cells MHCC97H and H1299 were also evaluated (Fig. 1).
Fig. 1.
Structures of 1‒4
Results and Discussion
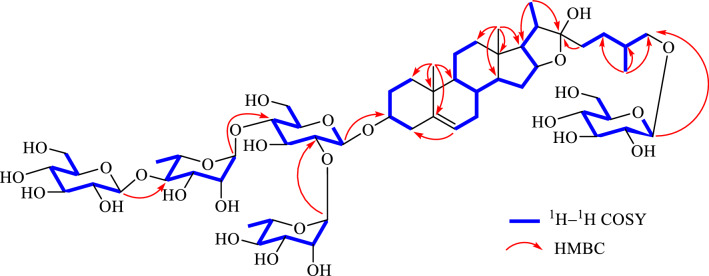
Saponin 1 was obtained as a white amorphous powder. It had a molecular formula of C57H94O27 as determined by the observed (+)-HRESIMS protonated ion peak at m/z 1233.5879 [M + Na]+ (calcd for C57H94O27Na, 1233.5875). It showed a positive reaction to the Ehrlich’s reagent (red color), suggesting a furostanol skeleton [20]. The 13C NMR spectrum (Table 1) displayed 57 carbons, of which 27 were assigned to the aglycone part and the remaining 30 were attributed to five hexose units. With the aid of the HSQC experiment, the 1H and 13C NMR spectrum (Table 1) attributable to the aglycone moiety showed resonances for four characteristic steroidal methyls at δH 0.83 (3H, s, CH3-19), 0.93 (3H, d, J = 6.6 Hz, CH3-27), 1.00 (3H, s, CH3-18), and 1.26 (3H, d, J = 6.7 Hz, CH3-21), together with their corresponding carbons at δC 16.3 (CH3-19), 17.3 (CH3-27), 19.2 (CH3-18), 16.3 (CH3-21); two oxygenated methines at δH 3.82 (1H, m) and 4.88 (1H, m), along with their corresponding carbons at δC 77.8 (CH-3) and 80.8 (CH-16); an olefinic group at δH 5.26 (1H, brs) as well as δC 121.6 (CH-6) and 140.6 (C-5); and a ketal carbon at δC 110.4 (C-22). The abovementioned data indicated that the aglycone of 1 should be a furostanol one as that of protodioscin (2) [21]. Moreover, the aglycone of 1 was further confirmed a by the following diagnostic 1H‒1H COSY, HMBC, and ROESY correlations (Figs. 2 and 3). The 1H‒1H COSY experiment revealed three structural fragments including CH2-1‒CH2-2-CH-3‒CH2-4, CH-6‒CH2-7‒CH-8/(‒CH-9‒CH2-11‒CH2-12)/‒CH-14‒CH2-15‒CH-16‒CH-17‒CH-20‒CH3-21, and CH2-23‒CH2-24‒CH-25/(‒CH3-27)/‒CH2-26. Moreover, the observed HMBC from δH 1.00 (CH3-18) to δC 39.7 (CH2-12), 40.4 (C-13), 56.4 (CH-14), and 63.6 (CH-17), from δH 0.83 (CH3-19) to δC 37.3 (CH2-1), 140.6 (C-5), 50.1 (CH-9), and 36.9 (C-10), and from both δH 1.26 (CH3-21) and δH 2.00 (H-23a) to δC 110.4 (C-22) established the aglycone of 1 to be 22α-hydroxyl-(25R)-furost-Δ5(6)-3β,26-diol. The ROESY correlations of δH 1.00 (Me-18) with 1.51 (H-8)/2.17 (H-20)/1.94 (H-23b) and of δH 0.83 (Me-19) with 1.51 (H-8) and 1.68 (H-1a) verified these protons were placed at the same side, whereas the observed ROESY correlations of δH 0.94 (H-1b) with 3.82 (H-3)/0.86 (H-9), of δH 1.02 (H-14) with 0.86 (H-9)/1.87 (H-17), and of δH 1.87 (H-17) with 4.88 (H-16) indicated these protons were located at the other side. Additionally, the 25R configuration of 1 was assigned according to the small chemical shift difference between Ha-26 and Hb-26 at Δab = 0.34 ppm (Δab > 0.57 ppm for 25S, and Δab < 0.48 ppm for 25R) [22]. In view of aforementioned evidence, the aglycone of 1 was thus elucidated as 22α-hydroxyl-(25R)-furost-Δ5(6)-3β,26-diol.
Table 1.
1H and 13C NMR spectral data of 1 (600 and 150 MHz, pyridine-d5)
| No | Aglycone moiety | No | Sugar moiety | ||
|---|---|---|---|---|---|
| δC | δH (mult., J) | δC | δH (mult., J) | ||
| 1 | 37.3, CH2 |
a 1.68 m b 0.94 m |
3-O-Glc | ||
| 1′ | 100.1, CH | 4.88 d (7.7) | |||
| 2 | 29.9, CH2 |
a 1.98 m b 1.79 m |
2′ | 77.6, CH | 4.10 m |
| 3 | 77.8, CH | 3.82 m | 3′ | 73.8, CH | 4.26 m |
| 4 | 38.7, CH2 |
a 2.71 m b 2.64 m |
4′ | 77.1, CH | 4.31 m |
| 5 | 140.6, C | 5′ | 76.7, CH | 4.30 m | |
| 6 | 121.6, CH | 5.26 br s | 6′ | 61.0, CH2 |
a 4.12 m b 3.98 m |
| 7 | 32.1, CH2 | 1.83 2H m | |||
| 8 | 31.5, CH | 1.51 m | 2′-O-Rha | ||
| 9 | 50.1, CH | 0.86 m | 1″ | 101.6, CH | 6.27 br s |
| 10 | 36.9, C | 2″ | 71.4, CH | 4.76 m | |
| 11 | 20.9, CH2 | 1.38 2H m | 3″ | 72.5, CH | 4.75 m |
| 12 | 39.7, CH2 |
a 1.70 m b 1.06 m b1.06 m |
4″ | 73.8, CH | 4.26 m |
| 13 | 40.4, C | 5″ | 69.3, CH | 4.84 m | |
| 14 | 56.4, CH | 1.02 m | 6″ | 18.4, CH3 | 1.68 3H d (6.0) |
| 15 | 32.2, CH2 | 1.40 2H m | 4′-O-Rha | ||
| 16 | 80.8, CH | 4.88 m | 1‴ | 101.8, CH | 5.74 br s |
| 17 | 63.6, CH | 1.87 m | 2‴ | 71.7, CH | 4.76 m |
| 18 | 19.2, CH3 | 1.00 3H s | 3‴ | 72.2, CH | 4.59 m |
| 19 | 16.3, CH3 | 0.83 3H s | 4‴ | 84.9, CH | 4.35 m |
| 20 | 40.6, CH | 2.17 m | 5‴ | 68.3, CH | 4.93 m |
| 21 | 16.3, CH3 | 1.26 3H d (6.7) | 6‴ | 18.2, CH3 | 1.60 3H d (6.0) |
| 22 | 110.4, C | 4″-O-Glc | |||
| 23 | 36.9, CH2 |
a 2.00 m b 1.94 m b 1.94 m |
1″″ | 106.4, CH | 5.14 d (7.7) |
| 24 | 28.1, CH2 |
a 1.97 m b 1.63 m |
2″″ | 76.7, CH | 3.98 m |
| 25 | 34.0, CH | 1.93 m | 3″″ | 78.2, CH | 3.70 m |
| 26 | 74.9, CH2 |
a 3.55 dd (9.0, 6.1) b 3.88 m |
4″″ | 71.0, CH | 4.12 m |
| 27 | 17.3, CH3 | 0.93 3H d (6.6) | 5″″ | 76.3, CH | 4.00 m |
| 6″″ | 62.2, CH2 |
a 4.45 d (12.4) b 4.28 m |
|||
| 26-O-Glc | |||||
| 1‴″ | 104.6, CH | 4.73 d (7.8) | |||
| 2‴″ | 75.0, CH | 3.82 m | |||
| 3‴″ | 78.2, CH | 3.99 m | |||
| 4‴″ | 71.7, CH | 4.12 m | |||
| 5‴″ | 78.3, CH | 4.10 m | |||
| 6‴″ | 62.5, CH2 |
a 4.45 m b 4.28 m |
|||
Fig. 2.
Key 1H ‒ 1H COSY and HMBC correlations of 1
Fig. 3.
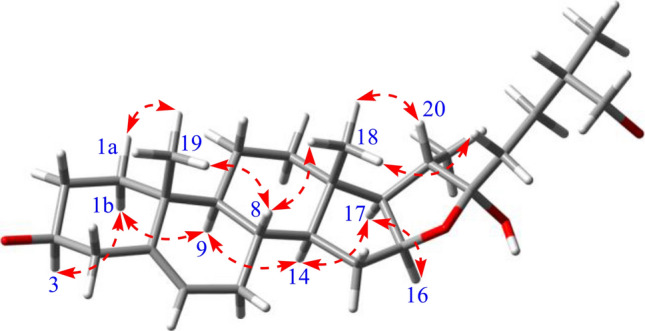
Key ROESY correlations for the aglycone moiety of 1
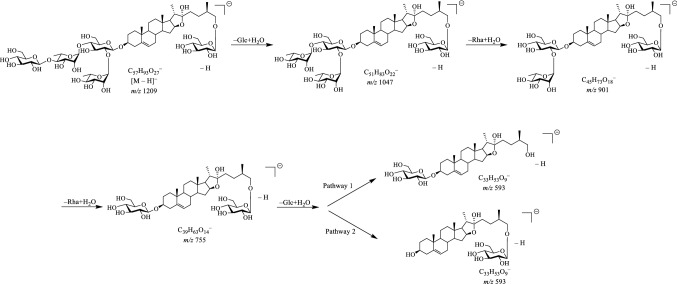
As for the sugar units of 1, its 1H NMR spectrum (Table 1) displayed the presence of five anomeric proton signals at δH 4.73 (1H, d, J = 7.8 Hz, H-1‴″), 4.88 (1H, d, J = 7.7 Hz, H-1′), 5.14 (1H, d, J = 7.7 Hz, H-1″″), 5.74 (1H, brs, H-1‴) and 6.27 (1H, brs, H-1″), which showed correlations in the HSQC spectrum with five anomeric carbons at δC 104.6 (CH-1‴″), 100.1 (CH-1′), 106.4 (CH-1″″), 101.8 (CH-1‴), and 101.6 (CH-1″). With the assistance of MS spectrum, the sugar moiety of 1 was preliminary determined. Specifically, the [M ‒ H]‒ ion (m/z 1209.6) displayed 1 had a molecular weight (MW) of 1210.6 Da in the negative ion mode of ESI-MSn. The observed ions with m/z values of 1047.5, 901.5, and 755.4 indicated the sequential cleavage of two rhamnopyranosyl units followed by the cleavage of a glucopyranosyl moiety from the parent [M ‒ H]‒ ion (m/z 1209.6), respectively. Likewise, the MS2 spectrum also afforded m/z value of 593.4 that was indicative of the loss of one glucopyranosyl group from the C-3 position or the C-26 position (Scheme 1). Also, acid hydrolysis of 1 also gave d-glucoses and l-rhamnoses as the sugar residue, which was confirmed by HPLC analysis of their corresponding PMP derived adducts. All the anomeric protons of d-glucose possessed β-configurations due to their 3JH1, H2 coupling constants (7.8, 7.7, and 7.7 Hz), and both anomeric protons of l-rhamnoses shared α-configurations due to the chemical shifts of C-3 (δC 72.5 and 72.2) and C-5 (δC 69.3 and 68.3), respectively. In the HMBC spectrum, the long-range correlations from δH 4.88 (H-1′) to δC 77.8 (CH-3), from δH 4.73 (H-1‴″) to δC 74.9 (CH2-26), from δH 6.27 (H-1″) to δC 77.6 (CH-2′), from δH 5.74 (H-1‴) to δC 77.1 (CH-4′), and from δH 5.14 (H-1″″) to δC 84.9 (CH-4‴) established the sequence for 3-O-sugar chain as an [α-l-rhamnopyranosyl-(1 → 2)]-[β-d-glucopyranosyl-(1 → 4)-α-l-rhamnopyranosyl-(1 → 4)]-β-d-glucopyranosyl moiety and for 26-O-sugar chain as β-d-glucopyranosyl moiety, respectively. Based on the above information presented, the structure of 1 was thus elucidated to be 26-O-β-d-glucopyranosyl-22α-hydroxyl-(25R)-Δ5(6)-furost-3β,26-diol-3-O-α-l-rhamnopyranosyl-(1 → 2)-[β-d-glucopyranosyl-(1 → 4)-α-l-rhamnopyranosyl-(1 → 4)]-β-d-glucopyranoside.
Scheme 1.
The fragmentation process of 1 in the ESI- MS negative scan
Additionally, three known steroidal glycosides were identified as protodioscin (2) [21], (25R)-26-O-β-d-glucopyranosyl-3β,20α,26-trihydroxyfurostan-5, 22-diene-3-O-α-l-rhamnopyranosyl-(1 → 2)-[α-L-rhamnopyranosyl-(1 → 4)]-O-β-d-glucopyranoside (3) [23], and dioscoreside H (4) [24] by comparison of their spectroscopic data with those reported in the literatures.
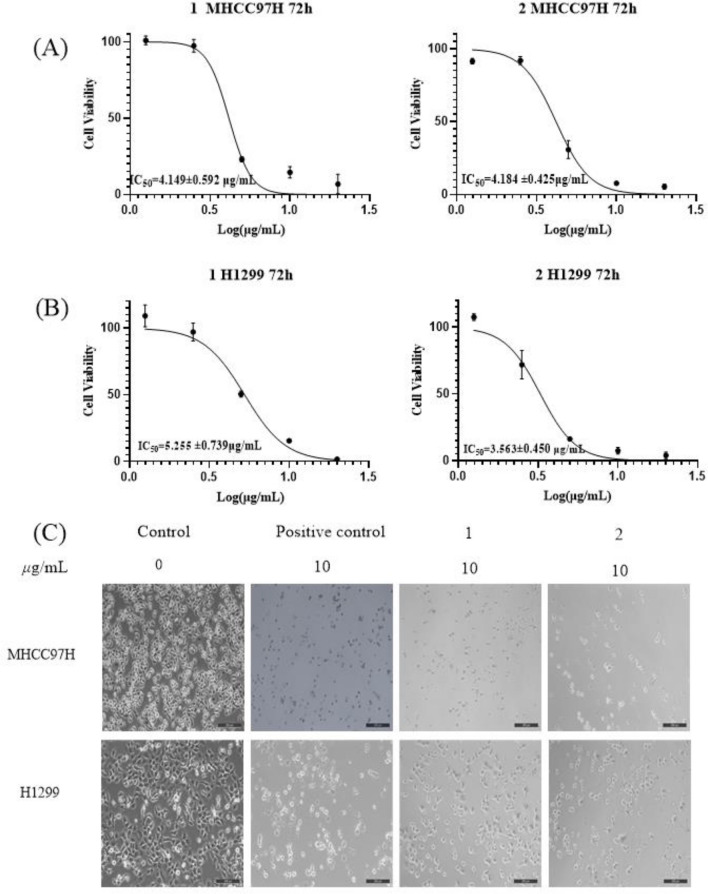
The steroid saponins obtained from species of Liliaceae have shown the potential to significantly inhibit the proliferations of various human tumor cell lines in vitro [25–29]. Therefore, all isolated compounds were evaluated for their cytotoxicity against MHCC97H and H1299 by the MTT method. More specifically, compared with the IC50 values of positive control doxorubicin hydrochloride, and both 1 and 2 displayed strong cytotoxicity against MHCC97H and H1299 cells with IC50 values of 3.56 ± 0.45/4.18 ± 0.43 μg/mL and 5.26 ± 0.74/4.15 ± 0.59 μg/mL, respectively (see Fig. 4). Furthermore, as can be seen from Fig. 4, compared with the positive control doxorubicin hydrochloride, saponins 1 and 2 could significantly inhibit their proliferation (Table 2).
Fig. 4.
Effects of 1 and 2 on MHCC97H and H1299 cells proliferation (n = 3). A The IC50 values of 1 and 2 against MHCC97H; B The IC50 values of 1 and 2 against H1299; C Inhibition effects of MHCC97H and H1299 cells proliferation by 1 and 2 after cultivation for 72 h
Table 2.
Cytotoxicity of saponins 1 and 2 (IC50 ± SD, μg/mL)
| Compound | H1299 | MHCC97H |
|---|---|---|
| 1 | 5.26 ± 0.74 | 3.56 ± 0.45 |
| 2 | 4.15 ± 0.59 | 4.18 ± 0.43 |
| Doxorubicin hydrochloridea | 0.86 ± 0.39 | 0.20 ± 0.08 |
aPositive control
Moreover, all obtained steroid saponins were evaluated for their antimicrobial activity against Escherichia coli (ML-35P), Bacillus cereus (CMCC(B) 63303), Candida albicans (ATCC 2091), Bacillus subtilis (ATCC 6633), Streptococcus hemolyticus (ATCC 19615), Listeria monocytogenes (ATCC 19114), Pseudomonas aeruginosa (PO01), Staphylococcus aureus (ATCC 4330), Salmonella Typhimurium (SL1344) and Staphylococcus epidermidis (CMCC 26069) by the microdilution broth susceptibility assay. The results (see Table 3) revealed that saponins 1‒4 showed moderate antimicrobial activity against C. albicans and B. subtilis, while only saponin 3 showed weak antimicrobial activity against S. aureus (63.30 ± 0.55 μg/mL).
Table 3.
Antimicrobial activity of saponins 1‒4 (IC50 ± SD, μg/mL)
| Compound | C. albicans | B. subtilis | S. aureus |
|---|---|---|---|
| 1 | 55.11 ± 0.32 | 47.93 ± 0.18 | NAa |
| 2 | 72.05 ± 0.49 | 69.30 ± 0.16 | NAa |
| 3 | 52.05 ± 0.31 | 47.19 ± 0.19 | 63.30 ± 0.55 |
| 4 | 52.05 ± 0.31 | 30.07 ± 0.22 | NAa |
| Streptomycin sulfata | 40.88 ± 0.33 | 93.49 ± 0.50 | 22.97 ± 0.24 |
NA no activity (> 100 μg/mL)
aPositive control
Experimental
General Experiment Procedures
Optical rotation was measured on a Autopol VI automatic polarimeter. The IR spectrum were measured on a Thermo Nicolet iS10 infrared spectrophotometer with KBr disk. The NMR spectra were obtained on Bruker DRX-400 and DRX-600 spectrometers. Chemical shifts (δ) were expressed in ppm with reference to the solvent signals. Both ESI and HRESIMS spectra were performed on an UPLC-IT-TOF spectrometer. Semi-preparative HPLC was performed on a Waters 600 with a COSMOSIL C18 (10 × 250 mm, Nacalai Tesque Corporation, Japan) column. Analytical HPLC was performed on a Shimadzu SIL-20A Series HPLC system equipped with a reverse-phase COSMOSIL C18 column (4.6 mm × 250 mm, 5 μm, Nacalai Tesque Corporation, Japan). Column chromatography was carried out using silica gel (100‒200 mesh, Qingdao Haiyang Chemical, Qingdao, Co., Ltd., People’s Republic of China) and macro-porous absorption resin (D101, Donghong Chemical Co., Ltd., People’s Republic of China). The PMP (Chengdu Aikeda Chemical Reagent Co., Ltd., China) was purchased from Beijing 4A Biotech Co., Ltd. (Beijing, China). Fractions were monitored by TLC, and spots were visualized by heating silica gel plates sprayed with Ehrlich’s reagent.
Plant Materials
The roots of A. cochinchinensis was purchased from ‘Luosiwan’ Chinese herbal medicine Market, Kunming, Yunnan Province, in November 2019, identified by Dr. Xu-Jie Qin. A voucher specimen (No. Luo 20191106) has been deposited at State Key Laboratory of Phytochemistry and Plant Resource in West China, Kunming Institute of Botany, Chinese Academy of Sciences.
Extraction and Isolation
The air-dried roots of A. cochinchinensis (5.0 kg) were extracted with 90% aqueous EtOH at 80 ℃ (15 L × 4, each time for 3 h). The solvent was removed under reduced pressure to yield an amber residue (2.5 kg). The residue was subjected to column chromatography over an macroporous resin column eluted first with H2O then successively with 25%, 70%, and 90% EtOH, respectively. The 70% EtOH partition was evaporated under reduced pressure to obtain a total steroidal saponin moiety. The total saponins (153 g) was subjected to a silica gel column eluting with a CHCl3‒MeOH‒H2O gradient (80:20:2 → 65:35:10) to yield five fractions (Fr. A‒Fr. E). Fraction C (105 g) was chromatographed on a silica gel column (CHCl3‒MeOH‒H2O, 9:1:0.1) to give saponin 2 (70 g) and Fr. C1. Fr. C1 (230.5 mg) was further purified by semi-preparative HPLC to afford 1 (29.8 mg; tR = 12 min; MeCN‒H2O, 28:72, 3.0 mL/min). Fraction D (12 g) was separated on a silica gel column (CHCl3‒MeOH‒H2O, 8:2:0.2) and then purified by semi-preparative HPLC to yield saponins 3 (3.4 mg, tR = 20.5 min; MeCN‒H2O, 35:65, 1.0 mL/min) and 4 (2.6 mg, tR = 23.5 min; CH3CN‒H2O, 35:65, 1.0 mL/min).
Spectroscopic Data of 1
26-O-β-d-glucopyranosyl-22α-hydroxyl-(25R)-Δ5(6)-furost-3β,26-diol-3-O-α-l-rhamnopyranosyl-(1 → 2)-[β-d-glucopyranosyl-(1 → 4)-α-l-rhamnopyranosyl-(1 → 4)]-β-d-glucopyranoside (1): white amorphous powder, [α] ‒46.86 (c 0.11, MeOH); IR (νmax): 3417, 2933, 2851, 1635, 1453, 1382, 1045 cm‒1; HRESIMS m/z 1233.5879 [M + Na]+ (calcd for C57H94O27Na, 1233.5875). 1H (pyridine-d5, 600 MHz) and 13C (pyridine-d5, 150 MHz) NMR spectral data, see Table 1.
Acid Hydrolysis of 1
The acid hydrolysis of compound 1 was carried out by a previously reported procedure [19]. Compound 1 (2.0 mg) was refluxed at 120 °C for 2 h with 2 M TFA on an oil bath. The aglycone was removed by the extraction with CHCl3 (5.0 mL) for three times. The reaction residue was filtered after neutralizing with 60.0 μL of NaOH (0.3 M). After removing the solvent under reduced pressure, the residue was refluxed at 75 °C for 1 h with 60.0 μL of PMP (0.5 M in methanol). Moreover, the reaction was quenched with 60.0 μL of HCl (0.3 M) and the reaction mixture was extracted with CHCl3 (5.0 mL, three times). Then, the aqueous layer was analyzed over HPLC (18% acetonitrile: 82% sodium phosphate (pH 6.8; 1.5 mL/min). Likewise, the standard monosaccharides d-glucose (1.0 mg) and l-rhamnose (1.0 mg) were derivatized with PMP by the same way as 1, and HPLC analyses were performed under the same conditions as 1. The sugar units in 1 were identified as d-glucose (tR = 14.5 min) and l-rhamnose (tR = 17.0 min) by comparison of the retention times of the corresponding derivatives.
Cytotoxicity Assay
The cytotoxicity of isolated compounds was determined to use the MTT method with a slight modification [30]. Briefly, two human cancer (MHCC97H and H1299) cell lines were incubated in 96-well plates at a density of 2 × 103 cells/well in DMEM medium supplemented with 10% fetal bovine serum at 37 ℃ with 5% CO2. After overnight incubation, cells were treated with tested compounds at different concentrations (20.00, 10.00, 5.00, 2.50, and 1.25 μg/mL) for 72 h. Subsequently, the culture mediums were exchanged by DMEM medium which contained 10% MTS reagent [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] and then cultured for another 4 h. The absorbance was recorded on a microplate reader at 490 nm.
Antimicrobial Activity Assay
The antimicrobial activity of isolated steroid saponins against 10 strains using the microdilution broth susceptibility assay [31]. The strains frozen in the refrigerator at ‒ 80 ℃ were activated and inoculated on standard tryptone soy broth agar (TSA) plates at 37 ℃ for 8 h to observe the bacterial growth. Subsequently, single colonies were selected and inoculated in tryptone soy broth (TSB) plates. After cultivated at 37 ℃ in shaker (120 rpm) for 8 h, the absorbance of bacterial solution was measured and its concentration was adjusted to 105 CFU/mL. Whereafter, an inoculum of 105 CFU/mL was made to sterile 96-well plate containing tested compounds at different concentrations (100.00, 50.00, 25.00, 12.50, 6.25 and 3.13 μg/mL) at 37 ℃ for 8 h. The wells containing only broth served as growth control. The absorbance of bacterial solution was recorded on a microplate reader at 600 nm.
Conclusion
In summary, a chemical examination of the roots of A. cochinchinensis led to the identification of one new furostanol glycoside 26-O-β-d-glucopyranosyl-22α-hydroxyl-(25R)-Δ5(6)-furost-3β,26-diol-3-O-α-l-rhamnopyranosyl-(1 → 2)-[β-d-glucopyranosyl-(1 → 4)-α-l-rhamnopyranosyl-(1 → 4)]-β-d-glucopyranoside (1) and three known one (2‒4). Meanwhile, compounds 1 and 2 exhibited cytotoxic and anti-proliferative effects on two human (MHCC97H and H1299) cancer cell lines. At the same time, compounds 1‒4 displayed moderate antimicrobial activity against C. albicans and B. subtilis, and compound 3 displayed weak antimicrobial activity against S. aureus.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (Grant Nos. 31770388 and U1802281) and the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (Grant No. 2019QZKK0502).
Declarations
Conflict of interest
The authors declare no competing financial interest.
References
- 1.Mahato SB, Ganguly AN, Sahu NP. Phytochemistry. 1982;21:959–978. doi: 10.1016/S0031-9422(00)82400-0. [DOI] [Google Scholar]
- 2.Man SL, Gao WY, Zhang YJ, Huang LQ, Liu CX. Fitoterapia. 2010;81:703–714. doi: 10.1016/j.fitote.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Kaunda JS, Zhang YJ. Nat. Product. Bioprospect. 2019;9:77–137. doi: 10.1007/s13659-019-0201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vincken JP, Heng L, Groot A, Gruppen H. Phytochemistry. 2007;68:275–297. doi: 10.1016/j.phytochem.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Sautour M, Mitaine-Offer AC, Miyamoto T, Dongmo A, Lacaille-Dubois MA. Planta Med. 2004;70:90–92. doi: 10.1055/s-2004-815467. [DOI] [PubMed] [Google Scholar]
- 6.Sauvaire Y, Ribes G, Baccou JC, Mariani MML. Lipids. 1991;26:191–197. doi: 10.1007/BF02543970. [DOI] [PubMed] [Google Scholar]
- 7.Liu MJ, Wang Z, Ju Y, Zhou JB, Wang Y, Wong RNS. Biol. Pharm. Bull. 2004;27:1059–1065. doi: 10.1248/bpb.27.1059. [DOI] [PubMed] [Google Scholar]
- 8.Nikaidoa T, Ohmotoa T, Kubo S, Mimaki Y, Sashida Y. Phytochemistry. 1992;31:2445–2450. doi: 10.1016/0031-9422(92)83296-B. [DOI] [PubMed] [Google Scholar]
- 9.Podolak I, Galanty A, Sobolewska D. Phytochem. Rev. 2010;9:425–474. doi: 10.1007/s11101-010-9183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachran C, Bachran S, Sutherland M, Bachran D, Fuchs H. Mini-Rev. Med. Chem. 2008;8:575–584. doi: 10.2174/138955708784534445. [DOI] [PubMed] [Google Scholar]
- 11.Jian R, Zeng KW, Li J, Li N, Jiang Y, Tu PF. Fitoterapia. 2013;84:80–84. doi: 10.1016/j.fitote.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Shen Y, Xu CL, Xuan WD, Li HL, Liu RH, Xu XK, Chen HS. Arch. Pharmacal Res. 2011;34:1587–1591. doi: 10.1007/s12272-011-1001-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhang HJ, Sydara K, Tan GT, Ma CY, Southavong B, Soejarto DD, Pezzuto JM, Fong HHS. J. Nat. Prod. 2004;67:194–200. doi: 10.1021/np030370b. [DOI] [PubMed] [Google Scholar]
- 14.Li XN, Chu C, Cheng DP, Tong SQ, Yan JZ. Nat. Prod. Commun. 2012;7:1357–1358. [PubMed] [Google Scholar]
- 15.Li XN, Chu C, Cheng DP, Tong SQ, Yan JZ. Chem. Nat. Compd. 2014;50:326–328. doi: 10.1007/s10600-014-0943-7. [DOI] [Google Scholar]
- 16.Liu B, Li BX, Zhou D, Wen XY, Wang YJ, Chen G, Li N. Bioorg. Chem. 2021;4:105237. doi: 10.1016/j.bioorg.2021.105237. [DOI] [PubMed] [Google Scholar]
- 17.Sung JE, Lee HA, Kim JE, Yun WB, An BS, Yang SY, Kim DS, Lee CY, Lee HS, Lee HS, Bae CJ, Hwang DY. Int. J. Mol. Med. 2017;40:1365–1376. doi: 10.3892/ijmm.2017.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sung JE, Choi JY, Kim JE, Lee HA, Yun WB, Park JJ, Kim HR, Song BR, Kim DS, Lee CY, Lee HS, Lim Y, Hwang DY. Lab. Anim. Res. 2017;33:57–67. doi: 10.5625/lar.2017.33.2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu PF, Cheng GG, Zhao LQ, Khan A, Yang XW, Zhang BY, Li MC, Liu YP, Luo XD. J. Agric. Food Chem. 2021;69:6229–6239. doi: 10.1021/acs.jafc.1c00869. [DOI] [PubMed] [Google Scholar]
- 20.Kiyosawa S, Hutoh M, Komori T, Nohara T, Hosokawa I, Kawasaki T. Chem. Pharm. Bull. 1968;16:1162–1164. doi: 10.1248/cpb.16.1162. [DOI] [PubMed] [Google Scholar]
- 21.Shao Y, Poobrasert O, Kennelly E, Chin CK, Ho CT, Huang MT, Garrison SA, Cordell GA. Planta Med. 1996;63:258–262. doi: 10.1055/s-2006-957667. [DOI] [PubMed] [Google Scholar]
- 22.Agrawal PK. Steroids. 2005;70:715–724. doi: 10.1016/j.steroids.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Shao B, Guo HZ, Cui YJ, Ye M, Han J, Guo D. Phytochemistry. 2007;68:623–630. doi: 10.1016/j.phytochem.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Hu TY, Shen GL, Yang Y, Gu ZB. Zhongchengyao. 2015;37:2682–2686. [Google Scholar]
- 25.Negi JS, Singh P, Joshi GP, Rawat MS, Bisht VK. Phcog. Rev. 2010;4:215–220. doi: 10.4103/0973-7847.70921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song B, Huang WL, Li Y, Zhang YY, Zhang HW, Jiang Y, Deng C, Song XM, Liu JL. Nat. Prod. Res. 2019;35:1478–6419. doi: 10.1080/14786419.2019.1616723. [DOI] [PubMed] [Google Scholar]
- 27.Qin XJ, Zhang LJ, Zhang Y, Ni W, Yang XZ, Yu Q, Yan H, An LK, Liu HY. Bioorg. Chem. 2020;99:103788. doi: 10.1016/j.bioorg.2020.103788. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Li HZ, Zhang YJ, Jacob MR, Khan SI, Li XC, Yang CR. Steroids. 2006;71:712–719. doi: 10.1016/j.steroids.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Qin XJ, Ni W, Chen CX, Liu HY. Nat. Product. Bioprospect. 2018;8:265–278. doi: 10.1007/s13659-018-0179-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buttkea TM, McCubreya JA, Owen TC. J. Immunol. Methods. 1993;157:233–240. doi: 10.1016/0022-1759(93)90092-L. [DOI] [PubMed] [Google Scholar]
- 31.Farooq U, Khan S, Naz S, Khan A, Khan A, Ahmed A, Rauf A, Bukhari SM, Khan SA, Kamil A, Riaz N, Khan AR. China J. Nat. Med. 2017;15:944–949. doi: 10.1016/S1875-5364(18)30011-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.