Abstract
Nitrogen limitation activates meiosis and meiotic gene expression in yeast, but nitrogen-responsive signal transduction mechanisms that govern meiotic gene expression are poorly understood. We show here that Ume6p, a subunit of the Ume6p-Ime1p meiotic transcriptional activator, undergoes increased phosphorylation in vivo in response to nitrogen limitation. Phosphorylation depends on an N-terminal glycogen synthase kinase 3 (GSK3) target site in which substitutions cause reduced Ume6p-Ime1p interaction and meiotic gene expression, thus arguing that phosphorylation promotes functional Ume6p-Ime1p interaction. Phosphorylation of this site depends on two GSK3 homologs, Rim11p and Mck1p. Prior studies indicate that Rim11p phosphorylates both Ume6p and Ime1p in vitro and is required for Ume6p-Ime1p interaction, but no evidence has linked Mck1p function to Ume6p activity. Here we find that Mck1p-Ume6p interaction is detectable by two-hybrid assays and that meiosis in a partially defective rim11-K68R mutant is completely dependent on Mck1p. These findings argue that nitrogen limitation governs Rim11p/Mck1p-dependent phosphorylation of Ume6p, which in turn is required for Ume6p-Ime1p interaction and meiotic gene activation.
The pathway of meiosis and spore formation of the budding yeast Saccharomyces cerevisiae is initiated in response to nitrogen limitation. Underlying this pathway is a global change in gene expression: within 2 h after starvation, 58 genes are activated over fivefold (8). Many of these genes belong to the set of early meiotic genes, which are required for normal progression through meiosis (20). Expression of early meiotic genes depends on Ume6p and Ime1p, two subunits of a heteromeric transcriptional activator. Ume6p-Ime1p interaction is stimulated by nitrogen limitation (32), but the mechanism through which nitrogen governs interaction is unknown.
Rim11p, a relative of glycogen synthase kinase 3 (GSK3), is required for Ume6p-Ime1p interaction (25, 32). Rim11p phosphorylates both Ime1p and Ume6p in vitro within the interaction region of each protein, and multisite amino acid substitutions that abolish phosphorylation in vitro also disrupt complex formation (4, 24, 25). Thus, regulation of Rim11p-dependent phosphorylation of either substrate would provide a simple mechanism for regulation of Ume6p-Ime1p complex formation. In vitro measurements indicate that Rim11p specific activity is fourfold higher in acetate-grown cells than in glucose-grown cells but have not revealed any response of Rim11p to nitrogen limitation (25).
Rim11p is loosely related to Mck1p (44% identity), a second yeast GSK3 homolog that has diverse functions. Mck1p promotes growth at temperature extremes, diauxic adaptation, centromere segregation, and meiosis (6, 16, 27, 35). In meiosis, Mck1p has two known roles (2, 27). First, it promotes IME1 transcription and is thus required indirectly for Ume6p-Ime1p interaction. Second, it promotes spore maturation, which occurs at the end of the meiotic program. Puziss et al. found that overexpression of Rim11p improves growth of an mck1 mutant at low and high temperatures (30), thus suggesting that Rim11p and Mck1p might have partially overlapping functions. However, mck1 mutant phenotypes are unaffected by a rim11 null mutation, and it has thus seemed equally likely that Rim11p can substitute for Mck1p only when it is overexpressed.
In this study, we have examined the state of Ume6p phosphorylation in vivo. Our findings indicate that Rim11p and Mck1p act together to promote Ume6p phosphorylation and activity. The extent of Ume6p phosphorylation depends on nutrient availability and on the nutritional response protein kinase Rim15p (40), thus indicating that a nutritional control pathway governs phosphorylation by Rim11p and Mck1p. Our results predict a simple genetic interaction between rim11 and mck1 mutations that may provide a generally useful redundancy test for multifunctional protein kinases.
MATERIALS AND METHODS
Strains, genetic methods, and media.
S. cerevisiae strains were derived from the SK-1 genetic background (19) or from crosses between SK-1 strains and strain Y190 (10), as indicated in Table 1. Several mutations have been described previously, including rim11::LEU2 (4), ime1Δ20 (36), PGAL1-IME1::TRP1 (38), ime2Δ4-lacZ (26), ime2-7::HIS3::LEU2 (37), and mck1Δ::TRP1 (27). The mrk1Δ::URA3-Kl mutation as introduced by PCR product-directed gene disruption using oligonucleotides MRK1.-60+kl.URA3 and MRK1.1860.3′+kl.URA3 (Table 2) and plasmid pWJ716 as a template for the Kluyveromyces lactis URA3 gene (11); PCR with outside primers MRK1.-104 and NotI-kl.URA3.1321-3′ confirmed the genotype. Strain construction involved standard methods including mating, meiotic crosses, and transformation (18).
TABLE 1.
Yeast strains used
| Strain | Genotype |
|---|---|
| SK-1 deriveda | |
| AMP107 | a GAL80 |
| AMP1448 | a ime1Δ20 ime2-HIS3::LEU2 arg6 his3Δ |
| AMP1631 | a rim15Δ::TRP1 GAL80 |
| YX230 | a ime1Δ20 ime2-HIS3::LEU2 ume6Δ1::TRP1 arg6 his3Δ |
| YX274 | α ime1Δ20 ime2-4-lacZ::LEU2 ume6Δ1::TRP1 |
| YX276 | a PGAL1-IME1::TRP1 ime2-4-lacZ::LEU2 ume6Δ1::TRP1 his4 met4 |
| YX284 | a ime1Δ20 ume6Δ1::TRP1 his3Δura3::UME6-HA::URA3 |
| YX306 | a/α ume6Δ1::TRP1/ume6Δ1::TRP1 his3Δ/his3Δ |
| YX323 | a ime1Δ20 ume6Δ1::TRP1 his3Δ ura3::UME6-HA::URA3 rim15Δ::TRP1 |
| YX423 | a his3Δ |
| YX424 | α rim11::LEU2 mck1Δ::TRP1 mrk1Δ::URA3-Kl his3Δ |
| YX425 | a rim11::LEU2 his3Δ |
| YX426 | a mck1Δ::TRP1 his3Δ |
| YX427 | a mrk1Δ::URA3-Kl his3Δ |
| YX428 | α rim11::LEU2 mck1ΔTRP1 his3Δ |
| YX429 | a mck1Δ::TRP1 mrk1Δ::URA3-Kl his3Δ |
| YX431 | a rim11::LEU2 mrk1ΔURA3-Kl his3Δ |
| YX446 | α rim15Δ::TRP1 his3Δ |
| YX501 | a/α PGAL1-IME1-17/+ rim11::LEU2/rim11::LEU2 mck1Δ::TRP1/mck1Δ::TRP1 mrk1Δ::URA3-Kl/mrk1Δ::URA3-Kl his3Δ/his3Δ |
| YX502 | a/α PGAL1-IME1-17/+ rim11::LEU2/rim11::LEU2 mck1Δ::TRP1/+ mrk1Δ::URA3-Kl/mrk1Δ::URA3-Kl his3Δ/his3Δ |
| YX503 | a/α PGAL1-IME1-17/+ rim11::LEU2/rim11::LEU2 mck1Δ::TRP1/mck1Δ::TRP1 mrk1Δ::URA3-Kl/+ his3Δ/his3Δ |
| YX504 | a/α PGAL1-IME1-17/+ rim11::LEU2/rim11::LEU2 mck1Δ::TRP1/+ mrk1Δ::URA3-Kl/+ his3Δ/his3Δ |
| Hybrid | |
| AMP1562 | α gal4 gal80 his3 trp1 ade2 ura3 leu2 URA3::GAL1-lacZ lys2 |
| AMP1563 | a gal4 gal80 his3 trp1 ade2 ura3 leu2 URA3::GAL1-lacZ lys2 |
All have the additional markers ura3 leu2 trp1 lys2 ho::LYS2 gal80:LEU2 unless indicated otherwise.
TABLE 2.
Oligonucleotides used
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| UME6-NcoI | CCTTAACTCACCATGGTAGACAAGGCGCG |
| UME6-Asp718 | GCGGTGGTACCGGGGGAAG |
| UME6-T99A | AACAACGCTCCCGTGCACACTCCGTCTGGTTCG |
| UME6-T99A-3′ | GTGCACGGGAGCGTTGTTGGGCGCACATGTCGA |
| UME6-T103A | GTGCACGCTCCGTCTGGTTCGCCGAGTTTG |
| UME6-T103A-3′ | AGACGGAGCGTGCACGGGAGTGTTGTTGGGCGCACA |
| UME6-S107A | GTGCACACTCCGTCTGGTGCACCGAGTTTGAAAGTCC |
| UME6-S107A | CGGTGCACCAGACGGAGTGTGCACGGGAGTGTTGTTGGGCGCACATGTCGA |
| UME6-Ala3 | AACGCTCCCGTGCACGCTCCGTCTGGTGCACCGAGTTTG |
| UME6-Ala3-3′ | CGGTGCACCAGACGGAGCGTGCACGGGAGCGTTGTTGGG |
| UME6-up | GAAGCGCCCACCTTCGCACAGCGCACAGGAACTAGGACACTACCGCACTCAAACCATTTGGCAGATTGTACTGAGAGTGC |
| UME6-dn | GATTTCCTCCAGTTTCATCTGTTTTTTCTTTGGATCAGATACAAAATCTGGTTTGAACGCCGCATCTGTGCGGTATTTCAC |
| MRK1.-60+kl.URA3 | GGGGAAGGGACTTGAAAATAATCTCAAGATTGAAGAGGTGAAAATTGTAAAATAGTCTACTCATGGCAATTCCCGGGGATC |
| MRK1.1860.3′+kl.URA3 | CAAATGCATATTATGTAATAGTAATGATACAATAGACTAAGAAATTTGAAGGATGAGATATAATGGTGGTCAGCTGGAATTC |
| UME6.-100 | GAGACCAGTGATCGTAAG |
| pRS-marker-3′ | CGCATCTGTGCGGTATTTC |
| SmaI+MCK1.1 | CCCGGGAATGTCTACGGAAGAGCAGAA |
| MCK1.1628-3′ | TTATTCAGCAACTTTCGTAG |
| NotI-kl.URA3-1321-3′ | AGCGGCCGCATGGTGGTCAGCTGGAAT T |
| MRK1.-104 | AGAACCCTGCTGCATACAAG |
Strains carrying ume6 mutations were constructed as follows. First, the ume6Δ strain YX230 was constructed by PCR product-directed gene disruption (1, 21) in strain AMP1488, using oligonucleotides UME6-up and UME6-dn (Table 2) and plasmid pRS314 as a TRP1 template. Trp+ His+ transformants (which express the ime2-HIS3 fusion) were purified and tested in PCRs with oligonucleotides UME6.-100 and pRS-marker-3′. Other ume6Δ strains resulted from crosses between YX230 and strains with relevant genotypes. To create strains carrying different ume6 point mutations, integrating URA3-ume6 or HIS3-ume6 plasmids (described below) were linearized within URA3 (with NcoI) or HIS3 (with PstI) and transformed into ura3 ume6Δ or his3 ume6Δ strains.
Yeast and bacterial media, including LB, YPD, YPAc, SC, and sporulation medium, were prepared as described previously (18, 38).
Plasmids. (i) UME6 derivatives.
Plasmid pKB186 carries the hemagglutinin epitope (HA)-tagged UME6-HA allele (25). A 4-kb SpeI-HindIII fragment from pKB186 containing UME6-HA was inserted into SpeI-HindIII-digested vector pRS406 to produce plasmid pYX148, an integrating UME6-HA plasmid. The plasmid carrying untagged Ume6p, pYX147, was created by the same strategy from plasmid pHY14-2 (5). UME6 plasmids with different point mutation were created by PCR-directed sequence alterations. To generate ume6-T99A (pYX285), first-round PCR products were generated with plasmid pYX148 as template and oligonucleotide UME6.-100 paired with UME6-T99A-3′ and oligonucleotide UME6-Asp718 paired with UME6-T99A. Two PCR products were purified and used as template in the second-round PCR with oligonucleotides UME6.-100 and UME6-Asp718. The resulting 0.9-kb DNA fragment was purified, digested with SphI and NheI, and inserted into SphI-NheI-digested plasmid pYX147. An analogous strategy was used to create ume6-T103 (pYX286), ume6-S107A (pYX287), and ume6-Ala3 (pYX281). Presence of desired mutations and absence of secondary mutations were confirmed by sequencing the entire region that had been PCR amplified.
(ii) GBD and GAD fusions.
Fusions of the Gal4p DNA binding domain (GBD) to full-length Rim11p and to Ime1p (residues 294 to 360) have been described elsewhere (25).
To generate fusions of the Gal4 activation domain (GAD) to Ume6p (residues 1 to 232), an NcoI site was introduced at UME6 codon 1 by PCR with oligonucleotides UME6-NcoI and UME6-Asp718; the templates were wild-type or mutant UME6 plasmids. The purified PCR products were digested with Asp718, filled in with Klenow polymerase, then digested with NcoI, and inserted into NcoI-SmaI-digested vector pACTII. Each insert was sequenced to confirm fidelity of the PCR. The resulting GAD-Ume6p fusion plasmids were pYX294 (wild type), pYX282 (Ala3), pYX288 (T99A), pYX289 (T103A), and pYX290 (S107A).
The GAD-Mck1p(1–375) fusion was constructed by introducing an XmaI site at MCK1 bp-1 through a PCR with oligonucleotides SmaI-MCK1.1 and MCK1.1628-3′, cloning the PCR product into vector pGEM(T), and then ligating the XmaI-XhoI-released insert into XmaI-XhoI-digested vector pACTII.
Two-hybrid interaction assays.
Strains AMP1562 and Y190 (10) carrying GBD, GBD-Rim11p, or GBD-Ime1p(294–360) were mated with strain AMP1563 or Y187 (10) carrying GAD or GAD-Ume6p(1–232), and diploids were selected on SC-Trp-Leu plates. For assays, 24-h cultures in SC-Trp-Leu medium were diluted 1/25 into YPAc or SC-Trp-Leu medium and harvested after three doublings. β-Galactosidase assays were conducted on permeabilized cells. Determinations were averages from three independent transformants; the range was less than 20% of the mean value.
Phosphatase treatment.
Whole-cell extracts were prepared as described previously (25) in a mixture containing 20 mM Tris-HCl (pH 7.4), 5% glycerol, 100 mM NaCl, 100 mM KCl, 1 mM EDTA, 0.05% β-mercaptoethanol and protease inhibitors (phenylmethylsulfonyl fluoride [0.15 mg/ml], leupeptin [1 μg/ml], aprotinin [1 μg/ml], and pepstatin [1 μg/ml]). Five milligrams of total protein extract was bound to 5 μg of 12CA5 and 200 μl of 50% protein A-Sepharose beads (Sigma) during a 1-h incubation at 4°C. The beads were washed twice with extraction buffer, washed once with phosphatase buffer (100 mM Tris-HCl [pH 9.6], 2 mM MgCl2, 0.1 mM ZnCl2, protease inhibitors), and suspended in 60 μl of phosphatase buffer; 15-μl aliquots of the suspension were mock treated (with buffer alone), phosphatase treated (with 2 U of calf intestinal phosphatase), or phosphatase treated in the presence of phosphatase inhibitors (5 mM sodium fluoride, 5 mM sodium phosphate, 10 mM sodium pyrophosphate, 1 mM NaVO3, 5 mM EGTA, and 5 mM EDTA). After a 15-min incubation at 37°C, the reaction mixtures were boiled for 5 min with 10 μl of 3× Laemmli buffer.
Immunoblot analysis.
Cells were harvested from mid-log phase in YPD or YPAc or after transfer from YPAc to sporulation medium for 3 h. The cell pellets were suspended at 10 units of optical density at 600 nm per ml of 3× Laemmli buffer and boiled for 5 min. We found that preservation of phosphorylation states was best with 5% sodium dodecyl sulfate (SDS) in the 3× Laemmli buffer. After centrifugation, supernatants were transferred to fresh tubes and loaded on SDS-polyacrylamide gels [6% for full-length Ume6p; 10% for GAD-Ume6p(1–232)] for polyacrylamide gel electrophoresis (PAGE). Protein transfer and detection with anti-HA monoclonal antibody 12CA5 (BabCo), goat anti-mouse antibody conjugated to peroxidase (Boehringer), and enhanced chemiluminescence detection reagents (Amersham) followed standard procedures (25).
RESULTS
Phosphorylation of Ume6p in vivo.
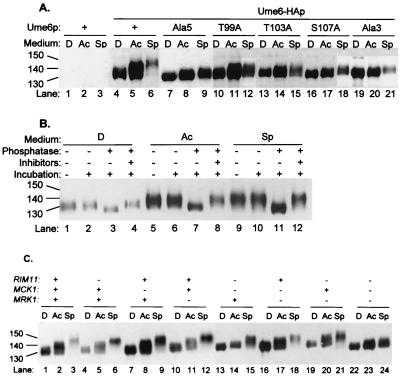
To determine whether Ume6p is phosphorylated in vivo, we examined the electrophoretic mobility of Ume6-HAp. We identified Ume6-HAp through anti-HA immunoblots of UME6 and UME6-HA strains (Fig. 1A, lanes 1 to 3 and 4 to 6, respectively). Ume6-HAp migrated as a disperse protein, and its apparent size was affected by growth conditions: it was 130 kDa in glucose-grown cells (lane 4), 140 kDa in acetate-grown cells (lane 5), and 150 kDa in nitrogen-limited cells (lane 6). Ume6-HAp mobility was unaffected by cell type or IME1 expression (data not shown). The apparent size of Ume6-HAp was reduced to 120 kDa by treatment with phosphatase; absence of phosphatase or presence of phosphatase plus inhibitors had no effect (Fig. 1B). These results indicate that Ume6p is phosphorylated in vivo and that the extent of Ume6p phosphorylation is elevated in response to acetate medium and nitrogen limitation.
FIG. 1.
Phosphorylation of Ume6p in vivo. (A) Mobilities of wild-type and mutant Ume6p derivatives. Strain YX423 carrying integrated UME6 plasmids was grown in rich glucose medium (D; lanes 1, 4, 7, 10, 13, 16, and 19), rich acetate medium (Ac; lanes 2, 5, 8, 11, 14, 17, and 20), or acetate medium lacking nitrogen (sporulation medium [Sp]; lanes 3, 6, 9, 12, 15, 18, and 21). Migration of Ume6-HAp on SDS-PAGE was visualized on an anti-HA immunoblot. The UME6 plasmids specified Ume6p (lanes 1 to 3), wild-type Ume6-HAp (lanes 4 to 6), or mutant Ume6-HAp derivatives with substitutions specified above each set of three lanes (lanes 7 to 21). (B) Phosphatase treatment of Ume6-HAp. Anti-HA immune complexes were prepared from strain YX423 carrying a plasmid specifying Ume6-HAp and grown in rich glucose medium (lanes 1 to 4), rich acetate medium (lanes 5 to 8), or acetate medium lacking nitrogen (lanes 9 to 12). Migration of Ume6-HAp on SDS-PAGE was visualized on an anti-HA immunoblot. Prior to SDS-PAGE, the immune complexes were untreated (lanes 1, 5, and 9) or incubated at 37°C with no addition (lanes 2, 6, and 10), with phosphatase (lanes 3, 7, and 11), or with phosphatase plus phosphatase inhibitors (lanes 4, 8, and 12). (C) Effect of GSK3 homolog defects on Ume6-HAp mobility. SDS-PAGE mobility of Ume6-HAp was examined on an immunoblot of strains YX423 (wild type; lanes 1 to 3), YX425 (rim11; lanes 4 to 6), YX426 (mck1; lanes 7 to 9), YX427 (mrk1; lanes 10 to 12), YX428 (rim11 mck1; lanes 13 to 15), YX429 (mck1 mrk1; lanes 16 to 18), YX431 (rim11 mrk1; lanes 19 to 21), and YX424 (rim11 mck1 mrk1; lanes 22 to 24). Growth conditions and symbols are as in panel A. Sizes are indicated in kilodaltons.
To identify the region of Ume6p required for phosphorylation, we compared electrophoretic mobilities of Ume6-HAp and Ume6-HA–Ala5p, a mutant derivative that fails to undergo phosphorylation by Rim11p in vitro (25). Ume6-HAp was apparently larger than Ume6-HA–Ala5p in both acetate-grown cells and nitrogen-starved cells (Fig. 1A, lanes 4 to 6 and 7 to 9). The Ala5 substitution affects a GSK3 consensus site that includes residues T99, T103, and S107, and we examined effects of alanine substitutions for these residues. Each single substitution and a triple substitution (Ala3) caused a substantial reduction in Ume6-HAp size in nitrogen-starved cells and, to some extent, in acetate-grown cells (Fig. 1A). These results indicate that Ume6p residues T99, T103, and S107 are each required for full levels of phosphorylation promoted by acetate medium and nitrogen limitation.
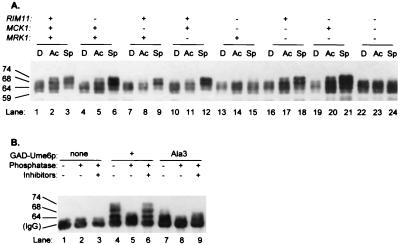
We used a GAD-Ume6p fusion protein that includes the Ume6p N-terminal region (NTR; residues 1 to 232) to confirm that this region is phosphorylated in vivo. GAD-Ume6p(1–232) migrated as 59-, 64-, and 68-kDa forms in glucose-grown cells (Fig. 2A, lane 1). The 68-kDa form accumulated at greater levels in acetate-grown and nitrogen-starved cells, and a 74-kDa form was also detectable in nitrogen-starved cells (Fig. 2A, lanes 2 and 3). A GAD-Ume6p(1–232) mutant derivative carrying the Ala3 triple substitution existed primarily as 59- and 64-kDa forms under all growth conditions (Fig. 2B [lane 7 compared to lane 4] and data not shown). Both the 68- and 74-kDa forms of wild-type GAD-Ume6p(1–232) were sensitive to phosphatase (Fig. 2B, lane 5). These results confirm that the Ume6p NTR is subject to phosphorylation in vivo, that phosphorylation depends on serine and threonine residues in the 99–107 interval, and that Ume6p NTR phosphorylation is stimulated by nitrogen limitation.
FIG. 2.
Phosphorylation of the Ume6p NTR in vivo. (A) Effect of GSK3 homolog defects on mobility of GAD-Ume6p(1–232). Strains carrying a GAD-Ume6p(1–232) plasmid were grown under conditions described in the legend to Fig. 1A, and migration of GAD-Ume6p(1–232) on SDS-PAGE was visualized on an anti-HA immunoblot. The strains had defects in Rim11p, Mck1p, or Mrk1p, as indicated at the top, and are specified in the legend to Fig. 1C. (B) Phosphatase treatment of GAD-Ume6p(1–232). Anti-HA immune complexes were prepared from wild-type strain AMP107 carrying plasmids specifying GAD (lanes 1 to 3), GAD-Ume6p(1 to 232) (lanes 4 to 6), or GAD-Ume6-Ala3p(1–232) (lanes 7 to 9) after growth in acetate medium lacking nitrogen and were visualized after SDS-PAGE on an anti-HA immunoblot. Prior to SDS-PAGE, the samples were incubated at 37°C with no addition (lanes 1, 4, and 7), with phosphatase (lanes 2, 5, and 8), or with phosphatase plus phosphatase inhibitors (lanes 3, 6, and 9). Sizes are indicated in kilodaltons.
Functional activity of Ume6p phosphorylation-negative mutants.
Ume6p functions as both a repressor and an activator. In vegetative cells, it binds to Sin3p and Rpd3p to repress many genes, including early meiotic genes (3, 17, 28, 39). In meiotic cells, it binds to Ime1p to activate early meiotic genes (5, 25, 32). To determine whether these functions are governed by NTR phosphorylation, we assayed repression and activation of the meiotic IME2 promoter in strains expressing wild-type and phosphorylation-defective Ume6p derivatives.
Repression assays were conducted with ime2-HIS3 and ime2-lacZ fusion genes in ime1Δ strains. The ime1Δ mutation ensures that Ume6p functions as a repressor, not an activator, at the IME2 promoter (3). The ime2-HIS3 fusion conferred a His+ phenotype in the absence of Ume6p and a His− phenotype in the presence of Ume6p (Table 3), representing derepressed and repressed states. Presence of each phosphorylation-defective Ume6p derivative also caused a His− phenotype. ime2-lacZ expression measurements indicate that wild-type and mutant Ume6p derivatives all caused 20- to 30-fold repression (Table 3, ime1Δ strain). Northern analysis also indicated that Ume6p and Ume6-Ala5p repress the nonmeiotic gene INO1 equally well (data not shown). Therefore, disruption of Ume6p NTR phosphorylation does not affect repression.
TABLE 3.
Effects of Ume6p phosphorylation-defective mutations on Ume6p function
| Ume6-HAp derivative | ime2-HIS3 expression in ime1Δ straina | Mean ± SE
|
||||
|---|---|---|---|---|---|---|
|
ime2-lacZ expressionb
|
Sporulation (%)c | Two-hybrid interactiond
|
||||
| ime1Δ strain | PGAL1-IME1 strain | GBD-Ime1p(294–300) | GBD | |||
| None | + | 290 ± 12 | 160 ± 1.2 | 0.7 ± 0.6 | 0.4 ± 0.01 | 0.5 ± 0.05 |
| Wild type | − | 15 ± 2.5 | 900 ± 62 | 93 ± 2.4 | 470 ± 62 | 0.4 ± 0.01 |
| T99A | − | 12 ± 0.2 | 8.0 ± 0.4 | 3.0 ± 1.5 | 14 ± 0.5 | 0.2 ± 0.02 |
| Ala3 | − | 12 ± 0.6 | 6.7 ± 0.3 | 3.6 ± 0.9 | 27 ± 2.9 | 0.4 ± 0.03 |
| T103A | − | 12 ± 0.6 | 60 ± 2.5 | 66 ± 14 | 58 ± 4.0 | 0.3 ± 0.03 |
| S107A | − | 13 ± 3.7 | 210 ± 3.0 | 78 ± 5 | 130 ± 13 | 0.2 ± 0.02 |
Growth was measured on SC-His plates of strain YX230 (ume6Δ ime2-HIS3) carrying integrated UME6 mutant plasmids. Growth on SC (containing histidine) was used as a positive control.
β-Galactosidase was measured (in Miller units, average of three independent transformants) in strains YX274 (ime1Δ ime2-lacZ ume6Δ) and YX276 (PGAL1-IME1 ime2-lacZ ume6Δ) carrying integrated UME6 mutant plasmids, after transfer from YPAc medium to sporulation medium for 4 h. A gal80 mutation permits PGAL1-IME1 expression in sporulation medium in the absence of galactose (38).
Determined by microscopic examination of strain YX306 (a/α ume6Δ/ume6Δ) carrying integrated UME6 mutant plasmids after incubation for 20 h in sporulation medium (average of three independent transformants).
Interaction of GAD-Ume6p derivatives with GBD-Ime1p or GBD was determined by β-galactosidase assay of gal1-lacZ expression (Miller units, average of three independent transformants).
Activation assays were conducted with the ime2-lacZ fusion. We compared its expression in PGAL1-IME1 strains and ime1Δ strains to estimate the level of Ime1p-dependent activation. In the absence of Ume6p, Ime1p had little effect on ime2-lacZ expression (Table 3). With wild-type Ume6p, presence of Ime1p caused a 60-fold increase in ime2-lacZ expression. This level of expression is greater than that observed in the absence of Ume6p and thus cannot arise simply from relief of repression. With Ume6-Ala3p and Ume6-T99Ap, Ime1p had no effect on ime2-lacZ expression; with Ume6-T103Ap and Ume6-S107Ap, Ime1p promoted 5- and 16-fold increases in ime2-lacZ expression, respectively. The defects were reflected qualitatively in assays of sporulation and Ume6p-Ime1p interaction (Table 3). These results argue that Ume6p NTR phosphorylation promotes Ime1p interaction and meiotic gene activation. In addition, the three GSK3 site residues differ in functional importance.
Role of the yeast GSK3 family in phosphorylation.
Rim11p immune complexes can phosphorylate the Ume6p NTR in vitro (25). However, we found that a rim11 null mutation had little effect on Ume6-HAp phosphorylation under any growth condition (Fig. 1C, lanes 4 to 6 compared to lanes 1 to 3). Therefore, Rim11p is not required for Ume6p phosphorylation in vivo.
One explanation for this observation is that other protein kinases related to Rim11p promote Ume6p phosphorylation. The closest Rim11p relatives in S. cerevisiae are Mck1p and Mrk1p (4, 14, 27, 30, 35). Single mck1 and mrk1 mutations had little effect on Ume6-HAp phosphorylation (Fig. 1C, lanes 7 to 9 and 10 to 12), as did a rim11 mrk1 double mutation (Fig. 1C, lanes 19 to 21). Double rim11 mck1 and mck1 mrk1 mutations caused some reduction in Ume6-HAp phosphorylation, and a triple rim11 mck1 mrk1 mutation caused a severe defect in Ume6-HAp phosphorylation (Fig. 1C, lanes 13 to 18 and 22 to 24). These defects were particularly evident in nitrogen-starved cells. Therefore, Rim11p, Mck1p, and Mrk1p each contribute to Ume6p phosphorylation in vivo.
To determine the role of each protein kinase in Ume6p NTR phosphorylation, we examined GAD-Ume6p(1–232) electrophoretic mobility in strains lacking the kinases. In mutants lacking any single kinase, as in the wild-type strain, we observed elevated levels of the 68-kDa phosphorylated GAD-Ume6p(1–232) form in nitrogen-starved cells (Fig. 2A, lanes 4 to 12). A similar pattern was observed with most double mutants (lanes 16 to 21), but a mutant lacking both Rim11p and Mck1p had low levels of the 68-kDa GAD-Ume6p(1–232) form during nitrogen limitation (lanes 13 to 15). A mutant lacking all three kinases resembled the mutant lacking Rim11p and Mck1p (lanes 22 to 24). These results argue that Rim11p and Mck1p together account for the bulk of Ume6p NTR phosphorylation in nitrogen-limited cells.
Interaction of Ume6p with Mck1p.
Rim11p and Mck1p may phosphorylate the Ume6p NTR directly and may thus interact with the Ume6p NTR. Rim11p-Ume6p NTR interaction is detectable through two-hybrid assays (25), and so we used this approach to test for Mck1p-Ume6p interaction. Mck1p-Ume6p NTR interaction was detectable and above background levels in quantitative assays (Table 4) and on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates (data not shown). No interaction was detected between Mck1p and the Ime1p C-terminal region (residues 294 to 360), which does interact with Rim11p (4, 24, 32). We also detected no interaction between Mrk1p and the Ume6p NTR (data not shown). These results indicate that both Rim11p and Mck1p are capable of direct or indirect interaction with the Ume6p NTR.
TABLE 4.
Interaction between Ume6p and Mck1p
| GBD construct | Mean gal1-lacZ expression ± SEa
|
|
|---|---|---|
| GAD-Mck1p(1–375) | GAD | |
| GBD-Ume6p(1–232) | 0.10 ± 0.02 | 0.02 ± 0.000 |
| GBD-Ime1p(294–300) | 0.01 ± 0.000 | 0.02 ± 0.005 |
| GBD | 0.01 ± 0.005 | 0.03 ± 0.005 |
Measured by β-galactosidase assays (Miller units, average of three independent transformants).
Shared roles of Rim11p and Mck1p in meiosis.
Our findings indicate that Rim11p and Mck1p are together responsible for Ume6p NTR phosphorylation in nitrogen-limited cells and that this phosphorylation is required for activation of meiotic genes and sporulation. This model predicts that rim11 mck1 double mutants may have a more severe sporulation defect than either single mutant. The assessment is complicated because rim11 null mutants are completely defective in sporulation, as expected from their defect in Ime1p phosphorylation (4, 24). Therefore, we used strains that express a partially defective Rim11p derivative, Rim11-K68Rp, with an ATP binding site alteration. Rim11-K68Rp causes substantially reduced protein kinase activity in vitro, yet strains that express Rim11-K68Rp are capable of efficient sporulation (4). We reasoned that the low level of Rim11-K68Rp kinase activity may be adequate for sporulation only because of the contribution from other protein kinases to Ume6p phosphorylation. We found that Rim11p and Rim11-K68Rp stimulated sporulation to similar levels in strains that express Mck1p (Table 5, strains YX504 and YX502). However, only Rim11p stimulated sporulation in the absence of Mck1p; Rim11-K68Rp did not (strains YX503 and YX501). Sporulation was dependent on Rim11p protein kinase activity in these assays, because the kinase-inactive Rim11-K68Ap did not permit sporulation in any strain. The third GSK3 homolog, Mrk1p, had little effect on sporulation with any Rim11p derivative (strains YX502 and -501 compared to strains YX504 and -503). Therefore, Mck1p can compensate for a reduction in Rim11p activity to promote sporulation. These results support the idea that Rim11p and Mck1p act interchangeably to promote sporulation.
TABLE 5.
Relationship between Rim11p and Mck1p in sporulation
| Strain | Mck1pa | Mrk1pa | Rim11pb | Mean sporulation (%) ± SEMc |
|---|---|---|---|---|
| YX504 | + | + | None | <0.3 ± 0.0 |
| + | 62 ± 2 | |||
| K68R | 55 ± 9 | |||
| K68A | <0.3 ± 0.0 | |||
| YX503 | − | + | None | <0.3 ± 0.0 |
| + | 68 ± 4 | |||
| K68R | <0.3 ± 0.0 | |||
| K68A | <0.3 ± 0.0 | |||
| YX502 | + | − | None | <0.3 ± 0.0 |
| + | 58 ± 17 | |||
| K68R | 33 ± 10 | |||
| K68A | <0.3 ± 0.0 | |||
| YX501 | − | − | None | <0.3 ± 0.0 |
| + | 66 ± 10 | |||
| K68R | <0.3 ± 0.0 | |||
| K68A | <0.3 ± 0.0 |
+, the strain is heterozygous for a mutant allele (MCK1/mck1 or MRK1/mrk1); −, the strain is homozygous for a mutant allele. All strains were also carried a PGAL1-IME1 allele, which is expressed independently of Mck1p and Rim11p (26, 27).
All strains were homozygous rim11/rim11 mutants and expressed the indicated Rim11p derivatives from multicopy plasmids (4).
Average of three independent transformants.
Dependence of Ume6p phosphorylation on Rim15p.
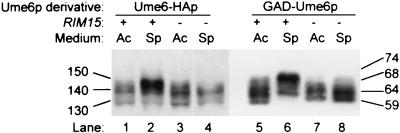
Prior studies indicate that Rim15p stimulates Ume6p-Ime1p interaction (41). To determine whether Rim15p is required for Ume6p phosphorylation, we compared Ume6-HAp forms in wild-type and rim15 mutant strains. The rim15 mutation caused a reduction in accumulation of phosphorylated forms of both Ume6-HAp and GAD-Ume6p(1–232) (Fig. 3). Therefore, Rim15p promotes phosphorylation of the Ume6p NTR.
FIG. 3.
Effect of a rim15Δ mutation on Ume6-HAp and GAD-Ume6p mobility. SDS-PAGE mobility of Ume6-HAp (lanes 1 to 4) and GAD-Ume6p(1–232) (lanes 5 to 8) expressed in strains AMP107 (RIM15; lanes 1, 2, 5, and 6) and AMP1631 (rim15Δ; lanes 3, 4, 7, and 8) was visualized on an immunoblot. Growth conditions and symbols are as for Fig. 1. Sizes are indicated in kilodaltons.
DISCUSSION
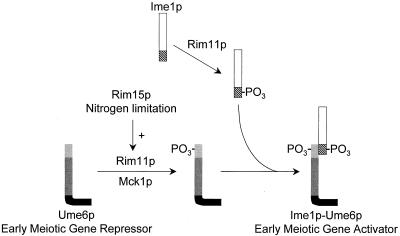
Formation of the Ume6p-Ime1p complex is tightly regulated by nitrogen limitation and by the protein kinase Rim11p (32). Here we have shown that these regulatory signals converge to govern phosphorylation of the Ume6p NTR. Our analysis further indicates that Mck1p and Rim11p both contribute to Ume6p NTR phosphorylation under conditions of nitrogen limitation. The functional significance of Ume6p NTR phosphorylation is supported by the synthetic sporulation defect of the rim11-K68R mck1Δ double mutant and by the sporulation defects caused by Ume6p substitutions that block phosphorylation. Our results are consistent with a model in which nitrogen limitation promotes Ume6p-Ime1p interaction through increased Ume6p NTR phosphorylation (Fig. 4). Growth in the presence of nitrogen results in low levels of Ume6p NTR phosphorylation, thus precluding Ume6p-Ime1p interaction. Limitation for nitrogen causes elevated Ume6p NTR phosphorylation, thus favoring Ume6p-Ime1p interaction, meiotic gene expression, and sporulation.
FIG. 4.
Control of Ume6p NTR phosphorylation. Ume6p exists in one of two states: without NTR phosphorylation or with NTR phosphorylation. NTR phosphorylation is greatest in nitrogen-limited cells and permits formation of an Ume6p-Ime1p complex and activation of early meiotic genes. NTR phosphorylation may be carried out directly by the GSK3 homologs Rim11p and Mck1p; Ime1p phosphorylation is carried out only by Rim11p. NTR phosphorylation also depends on the protein kinase Rim15p, a downstream target of the adenylate cyclase pathway.
Functional roles of Rim11p and Mck1p in meiosis.
The possibility that Rim11p and Mck1p have overlapping functional roles was first suggested by Puziss et al., who found that overexpression of RIM11 can suppress a mitotic mck1 mutant defect (30). However, the lack of a synthetic rim11 mck1 growth defect suggested that overexpression may permit Rim11p to substitute adventitiously for Mck1p. We have presented three findings that argue that Rim11p and Mck1p naturally share a function in nitrogen regulation of meiosis. First, rim11 mck1 double-null mutants are defective in accumulation of phosphorylated GAD-Ume6p after nitrogen limitation. Second, Mck1p interacts with the Ume6p NTR, as we found previously for Rim11p (25). Third, sporulation of the partially defective rim11-K68R mutant is abolished by an mck1Δ mutation. There is no novel sporulation defect of a rim11 mck1 double-null mutant because Rim11p is also required for phosphorylation of Ime1p (4, 24, 32) (Fig. 4). Thus, our results indicate that Rim11p and Mck1p have overlapping roles in promoting Ume6p phosphorylation.
It is likely that the Ume6p NTR is phosphorylated directly by Rim11p and Mck1p, based on three arguments. First, Rim11p immune complexes phosphorylate the Ume6p NTR in vitro (25). Second, both Rim11p (25) and Mck1p are capable of two-hybrid interaction with the Ume6p NTR. Third, Rim11p and Mck1p are both GSK3 family members, and Ume6p phosphorylation depends on GSK3 consensus site residues. Our finding that the putative phosphoacceptor residues differ in functional importance (T99 > T103 > S107) fits with the characterized GSK3 hierarchical phosphorylation mechanism, in which GSK3 prefers S-X-X-X-phosphoserine to S-X-X-X-serine as a substrate (31). According to this view, in Ume6p, phosphorylation of S107 improves phosphorylation of T103, and phosphorylation of T103 improves phosphorylation of T99. Therefore, a simple possibility is that phosphorylation of only T99 is critical for Ume6p to interact with Ime1p, and phosphorylation of T103 and S107 serves to accelerate phosphorylation of T99. The diminished interaction with Ime1p of Ume6-T103Ap and Ume6-S107Ap would then reflect the diminished extent of T99 phosphorylation.
Rim11p and Mck1p have one shared function in meiosis, but each has unique functions as well. This situation—redundancy for one of several functions in a single biological pathway—illustrates a circumstance in which protein kinase missense defects, rather than complete deletions, provide unique insight into functional relationships between protein kinases. Other situations include adventitious substitution of one protein kinase for another (22) and the action of two protein kinases in a linear pathway (12). Rim11p and Mck1p have a lower level of sequence identity (44%) than many other pairs of protein kinases with redundant functions (Tpk1p/Tpk3p, 85%; Cka1p/Cka2p, 61%; Tor1p/Tor2p, 70%; Pkh1p/Pkh2p, 65%; Mkk1p/Mkk2p, 59%), and Rim11p is more homologous to Mrk1p than to Mck1p. It thus seems likely that a systematic analysis of other partially defective protein kinase mutants for synthetic defects may reveal new functional relationships.
The function of the GSK3 homolog Mrk1p remains an enigma. Several studies have failed to detect a mrk1 phenotypic defect, even in backgrounds lacking other GSK3 homologs (13, 14, 42). Our results suggest that Mrk1p contributes to phosphorylation of full-length Ume6p, but its role in Ume6p NTR phosphorylation seems minor, as assessed by GAD-Ume6p mobility and by sporulation assays of rim11-K68R MRK1 and rim11-K68R mrk1 strains. We suggest that Mrk1p may phosphorylate Ume6p at sites outside the NTR and cannot attribute biological significance to the Mrk1p-Ume6p relationship at this time.
Relationship of Rim15p to Rim11p and Mck1p.
Our results argue that Rim15p promotes Ume6p NTR phosphorylation and thus tie Rim15p, Rim11p, and Mck1p to a single biochemical event. Rim15p is inhibited through direct phosphorylation by cyclic AMP (cAMP)-dependent protein kinase, and so our results provide a biochemical connection between the cAMP and meiotic gene expression. However, cAMP levels respond to glucose (40), yet here Rim15p is required for a nitrogen starvation response. Thus, our findings explain one of several mechanisms by which the cAMP pathway governs meiosis but do not explain how nitrogen limitation governs Ume6p NTR phosphorylation.
If Rim11p and Mck1p phosphorylate the Ume6p NTR directly, then Rim15p may affect phosphorylation through an effect on the kinases or Ume6p itself. In vitro assays argue that Rim15p does not govern Rim11p protein kinase activity (41). A second possibility is that Rim15p may serve as a priming kinase (31, 33) that phosphorylates Ume6p S107, thus accelerating phosphorylation of Ume6p T103 by Rim11p and Mck1p. This model predicts that properties of Ume6p-S107Ap in a wild-type strain should mimic properties of Ume6p in a rim15Δ strain. However, we find that the S107A substitution impairs Ume6p-Rim11p interaction, whereas a rim15Δ mutation does not (Y. Xiao and A. P. Mitchell, unpublished data). Also, we have not detected two-hybrid interaction between Rim15p and the Ume6p NTR (Xiao and Mitchell, unpublished). Therefore, we believe that Rim15p acts indirectly, for example, through inhibition of protein phosphatase expression or activity.
A newly defined nitrogen response module.
Nitrogen limitation causes diverse responses in yeast, including expression of nitrogen catabolic genes, changes in permease stability, down-regulation of translational machinery, sporulation, and filamentation. Several individual responses are governed by defined gene products (15, 23, 29), and the TOR signaling cascade may serve as a global regulator of nitrogen response pathways (7, 9, 34, 42). Our finding that Rim11p and Mck1p promote functional, nitrogen-responsive phosphorylation of Ume6p raises two questions. First, does a known nitrogen response pathway govern the activities of Rim11p and Mck1p? And second, do Rim11p and Mck1p govern any responses to nitrogen limitation in addition to activation of meiotic genes? We have observed that rim11 mck1 double mutants are defective in stationary-phase survival (Xiao and Mitchell, unpublished), and so these kinases may together play a role in starvation responses broader than previously thought. Analysis of Rim11p and Mck1p may explain how physiological and differentiation responses are coupled.
ACKNOWLEDGMENTS
We are grateful to past and present members of this lab for many helpful discussions and to Teresa Lamb for comments on the manuscript.
This work was supported by grant GM39531 from the NIH.
REFERENCES
- 1.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benni M L, Neigeborn L. Identification of a new class of negative regulators affecting sporulation-specific gene expression in yeast. Genetics. 1997;147:1351–1366. doi: 10.1093/genetics/147.3.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowdish K S, Mitchell A P. Bipartite structure of an early meiotic upstream activation sequence from Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2172–2181. doi: 10.1128/mcb.13.4.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowdish K S, Yuan H E, Mitchell A P. Analysis of RIM11, a yeast protein kinase that phosphorylates the meiotic activator IME1. Mol Cell Biol. 1994;14:7909–7919. doi: 10.1128/mcb.14.12.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowdish K S, Yuan H E, Mitchell A P. Positive control of yeast meiotic genes by the negative regulator UME6. Mol Cell Biol. 1995;15:2955–2961. doi: 10.1128/mcb.15.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brazill D T, Thorner J, Martin G S. Mck1, a member of the glycogen synthase kinase 3 family of protein kinases, is a negative regulator of pyruvate kinase in the yeast Saccharomyces cerevisiae. J Bacteriol. 1997;179:4415–4418. doi: 10.1128/jb.179.13.4415-4418.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardenas M E, Cutler N S, Lorenz M C, Di Como C J, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 9.Di Como C J, Arndt K T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 10.Durfee T, Becherer K, Chen R, Yeh S H, Yang Y, Killburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with protein the phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 11.Erdeniz N, Mortensen U H, Rothstein R. Cloning-free PCR-based allele replacement methods. Genome Res. 1997;7:1174–1183. doi: 10.1101/gr.7.12.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espinoza F H, Farrell A, Nourse J L, Chamberlin H M, Gileadi O, Morgan D O. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol Cell Biol. 1998;18:6365–6373. doi: 10.1128/mcb.18.11.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajji K, Clotet J, Arino J. Disruption and phenotypic analysis of seven ORFs from the left arm of chromosome XV of Saccharomyces cerevisiae. Yeast. 1999;15:435–441. doi: 10.1002/(SICI)1097-0061(19990330)15:5<435::AID-YEA367>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 14.Hardy T A, Wu D, Roach P J. Novel Saccharomyces cerevisiae gene, MRK1, encoding a putative protein kinase with similarity to mammalian glycogen synthase kinase-3 and Drosophila Zeste-White3/Shaggy. Biochem Biophys Res Commun. 1995;208:728–734. doi: 10.1006/bbrc.1995.1398. [DOI] [PubMed] [Google Scholar]
- 15.Huang H L, Brandriss M C. The regulator of the yeast proline utilization pathway is differentially phosphorylated in response to the quality of the nitrogen source. Mol Cell Biol. 2000;20:892–899. doi: 10.1128/mcb.20.3.892-899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang W, Lim M Y, Yoon H J, Thorner J, Martin G S, Carbon J. Overexpression of the yeast MCK1 protein kinase suppresses conditional mutations in centromere-binding protein genes CBF2 and CBF5. Mol Gen Genet. 1995;246:360–366. doi: 10.1007/BF00288609. [DOI] [PubMed] [Google Scholar]
- 17.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 19.Kane S, Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kupiec M, Byers B, Exposito R E, Mitchell A P. Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 889–1036. [Google Scholar]
- 21.Lorenz M C, Muir R S, Lim E, McElver J, Weber S C, Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- 22.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 23.Magasanik B. Regulation of nitrogen utilization. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 283–317. [Google Scholar]
- 24.Malathi K, Xiao Y, Mitchell A P. Catalytic roles of yeast GSK3beta/Shaggy homolog Rim11p in meiotic activation. Genetics. 1999;153:1145–1152. doi: 10.1093/genetics/153.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malathi K, Xiao Y, Mitchell A P. Interaction of yeast repressor-activator protein Ume6p with glycogen synthase kinase 3 homolog Rim11p. Mol Cell Biol. 1997;17:7230–7236. doi: 10.1128/mcb.17.12.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell A P, Bowdish K S. Selection for early meiotic mutants in yeast. Genetics. 1992;131:65–72. doi: 10.1093/genetics/131.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neigeborn L, Mitchell A P. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes Dev. 1991;5:533–548. doi: 10.1101/gad.5.4.533. [DOI] [PubMed] [Google Scholar]
- 28.Park H D, Luche R M, Cooper T G. The yeast UME6 gene product is required for transcriptional repression mediated by the CAR1 URS1 repressor binding site. Nucleic Acids Res. 1992;20:1909–1915. doi: 10.1093/nar/20.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park H D, Scott S, Rai R, Dorrington R, Cooper T G. Synergistic operation of the CAR2 (ornithine transaminase) promoter elements in Saccharomyces cerevisiae. J Bacteriol. 1999;181:7052–7064. doi: 10.1128/jb.181.22.7052-7064.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puziss J W, Hardy T A, Johnson R B, Roach P J, Hieter P. MDS1, a dosage suppressor of an mck1 mutant, encodes a putative yeast homolog of glycogen synthase kinase 3. Mol Cell Biol. 1994;14:831–839. doi: 10.1128/mcb.14.1.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roach P J. Multisite and hierarchal protein phosphorylation. J Biol Chem. 1991;266:14139–14142. [PubMed] [Google Scholar]
- 32.Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt A, Beck T, Koller A, Kunz J, Hall M N. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shero J H, Hieter P. A suppressor of a centromere DNA mutation encodes a putative protein kinase (MCK1) Genes Dev. 1991;5:549–560. doi: 10.1101/gad.5.4.549. [DOI] [PubMed] [Google Scholar]
- 36.Sia R A, Mitchell A P. Stimulation of later functions of the yeast meiotic protein kinase Ime2p by the IDS2 gene product. Mol Cell Biol. 1995;15:5279–5287. doi: 10.1128/mcb.15.10.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith H E, Driscoll S E, Sia R A, Yuan H E, Mitchell A P. Genetic evidence for transcriptional activation by the yeast IME1 gene product. Genetics. 1993;133:775–784. doi: 10.1093/genetics/133.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith H E, Su S S, Neigeborn L, Driscoll S E, Mitchell A P. Role of IME1 expression in regulation of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6103–6113. doi: 10.1128/mcb.10.12.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strich R, Surosky R T, Steber C, Dubois E, Messenguy F, Esposito R E. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- 40.Thevelein J M, de Winde J H. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- 41.Vidan S, Mitchell A P. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol Cell Biol. 1997;17:2688–2697. doi: 10.1128/mcb.17.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng X F, Schreiber S L. Target of rapamycin proteins and their kinase activities are required for meiosis. Proc Natl Acad Sci USA. 1997;94:3070–3075. doi: 10.1073/pnas.94.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]