Abstract
Background
We aimed to evaluate a testing program to facilitate control of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission at a large university and measure spread in the university community using viral genome sequencing.
Methods
Our prospective longitudinal study used remote contactless enrollment, daily mobile symptom and exposure tracking, and self-swab sample collection. Individuals were tested if the participant was exposed to a known SARS-CoV-2-infected person, developed new symptoms, or reported high-risk behavior (such as attending an indoor gathering without masking or social distancing), if a member of a group experiencing an outbreak, or at enrollment. Study participants included students, staff, and faculty at an urban public university during the Autumn quarter of 2020.
Results
We enrolled 16 476 individuals, performed 29 783 SARS-CoV-2 tests, and detected 236 infections. Seventy-five percent of positive cases reported at least 1 of the following: symptoms (60.8%), exposure (34.7%), or high-risk behaviors (21.5%). Greek community affiliation was the strongest risk factor for testing positive, and molecular epidemiology results suggest that specific large gatherings were responsible for several outbreaks.
Conclusions
A testing program focused on individuals with symptoms and unvaccinated persons who participate in large campus gatherings may be effective as part of a comprehensive university-wide mitigation strategy to control the spread of SARS-CoV-2.
Keywords: COVID-19 testing, genome sequencing, outbreak, SARS-CoV-2, university
Universities are characterized by congregate living, in-person learning, and active social environments, all of which may contribute to rapid spread of infectious diseases. Between May and August 2020, persons aged 20–29 years accounted for >20% of confirmed coronavirus disease 2019 (COVID-19) cases nationwide, and an even higher proportion in Washington State [1, 2]. Numerous outbreaks on university campuses were observed early in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic [3]. Surveillance strategies ranged from pooled testing of asymptomatic persons to wastewater analysis, with substantial heterogeneity across universities and no clear federal guidance [3–5]. Nationwide shortages of testing supplies and reagents have prevented many universities, including ours, from adopting a strategy of testing all members of the university community on a regular basis. Given these constraints, we sought to identify an approach that would permit early identification of cases and facilitate containment of spread [6, 7].
METHODS
Setting
This study was conducted at a large public university in Seattle, Washington, composed of approximately 60 000 students and 30 000 faculty and staff [8, 9]. In Autumn 2020, the university opened with hybrid in-person classes, and the majority of courses were held fully remotely. Residence halls were populated at a limited capacity [10]. The study population included students, staff, and faculty affiliated with the main campus and 2 smaller campuses located 15 and 35 miles from the main campus.
Study Enrollment
Enrollment began September 24, 2020, during student resident move-in, and continued during the study period. Eligibility criteria included living on-campus or within the main or satellite campus geographic area (~100-mile radius), a valid university identification number, the ability to consent in English, and university-related class or work at least once per month (in-person or remotely). Exclusion criteria were living outside of the geographic area (ie, working remotely but living in another state) and age <13 years. Participants completed informed consent and an online questionnaire that included baseline risk behaviors and demographic information. At enrollment, participants indicated a preference for either email or text communication. Data were managed using REDCap software [11, 12]. Individuals were stratified into risk tiers based on time spent on campus, number of individuals in their household, and university affiliation (Supplementary Table 1).
Attestations and Invitations to Test
Testing was offered for 4 reasons: (1) attestation positivity, (2) outbreaks, (3) baseline surveillance, and (4) holidays. Due to limited testing resources, “attestation positive” study participants were prioritized, followed by outbreak invitations, and finally, baseline and holiday invitations.
A daily attestation survey was used to determine testing eligibility. If “yes” was reported to any of the following, a participant was classified as “attestation positive”: In the last 24 hours, have you (1) experienced new symptoms (“symptoms”), (2) been exposed to a COVID-19-positive individual (“exposure”), or (3) attended a high-risk gathering (“gathering,” defined as attending an indoor gathering of >10 people without social distancing or mask-wearing)? Testing could be offered based on the daily attestation up to every 3 days, and repeat testing was offered immediately if new symptoms were reported. If a “gathering” or “exposure” attestation was reported, the testing invite was delayed 48 hours to account for the incubation period [13], and a second test was offered 72 hours after the first test was completed. During outbreak testing, testing was offered every 3 days to all individuals in the affected group. During outbreak testing, such as in the Greek outbreak, our objective was to identify cases for the purpose of implementing measures such as quarantine to reduce disease transmission. Holiday testing was offered before and after the Thanksgiving break during November 17–December 6, 2020.
Testing Mechanisms
Testing was conducted at in-person kiosks or via mail-in swab kits (Supplementary Methods). Participants affiliated with the main campus who received a testing invitation were offered in-person appointments of their choosing within 72 hours. Samples were collected by observed anterior nasal self-swab at on-campus kiosks. For participants at satellite campuses or who indicated mobility restrictions precluding attendance to an on-campus kiosk, a self-testing kit was sent to and picked up from their residence using rapid courier services [14, 15].
Laboratory Methods and Results Reporting
Samples collected at kiosks were transported to the Northwest Genomics Center at the University of Washington and tested for SARS-CoV-2 using a quantitative reverse transcription polymerase chain reaction (RT-qPCR) laboratory-developed test (LDT). The RT-qPCR consists of assays for 2 SARS-CoV-2 targets in duplicate and the human marker RNase P across 4 multiplexed reactions (Supplementary Data) [16]. A sample was considered positive if 3 or 4 replicates for RNase P had a cycle threshold (Ct) value <36 and SARS-CoV-2 had a value <40 (Supplementary Data). If only 2 SARS-CoV-2 replicate reactions were positive, the result was defined as inconclusive. Inconclusive results were regarded as low-positive results, and participants were counseled identically to participants with positive results [17]. If SARS-CoV-2 was not detected or detected in only 1 replicate, the test was considered negative. Samples were defined as failed and considered “never tested” if RNase P was undetected in 2 or more reactions, or if there was a laboratory or operator error. Midstudy (November 18, 2020), we implemented an extraction-free testing method that yielded similar results, but Ct values from the 2 methods were not directly comparable [16]. Results were provided to participants through a research report hosted on a secure online portal that was accessed using a unique barcode identifier and date of birth. Cases were contacted by university, county, or state public health staff, and contact tracing was initiated. Viral genome sequencing was attempted on all positive samples with Ct values of ≤30 using a hybrid capture enrichment method [18] or a COVID-seq amplicon method (Illumina). Raw sequencing reads were processed using the Seattle Flu Study Assembly Pipeline (GitHub [19]). Viral sequences were aligned and phylogenetic trees constructed using Nextstrain augur software [20]. Trees were visualized using Nextstrain auspice. All assembled genomes were publicly deposited to the Global Initiative on Sharing All Influenza Data (gisaid.org [21]) and Genbank immediately after data generation.
University-Wide Prevention and Mitigation Strategy
Before and during the Autumn quarter, the university deployed a communications campaign focused on masking, social distancing, handwashing, and disinfection of surfaces. Contact tracing was conducted by university public health officials for all students, staff, and faculty, except off-campus Greek community cases, which were handled by county public health. Campus isolation and quarantine housing was provided for infected students who lived in campus housing, consistent with Centers for Disease Control and Prevention recommendations. Students and employees in private residences were given instructions for isolation/quarantine, testing, and precautions. University members also had access to free municipal SARS-CoV-2 testing outside of this research study. Data pertaining to social gatherings of university community members were not collected.
Statistical Methods
Positive and inconclusive results were counted as cases. Ninety-five percent CIs were calculated, and P values were considered significant at an alpha level of .05. Statistical testing for the comparison of averages was completed using Welch’s 2-sample t test. For multivariate regression, the reference group for race was White, and for ethnicity it was non-Latinx/Hispanic.
Persons testing positive for SARS-CoV-2 were categorized as symptomatic, presymptomatic, asymptomatic, and possible asymptomatic. A case who tested positive/inconclusive was defined as symptomatic if they reported symptoms on their daily attestation survey within the 7 days before testing or the day of testing. Presymptomatic was defined as those who only reported symptoms in the week after testing on their daily attestation or follow-up survey. If no symptoms were reported before or after testing on the daily attestation or follow-up survey, a participant was classified as asymptomatic. Participants who did not complete their daily attestations or the follow-up survey and who reported no symptoms at the time of testing were classified as “possible asymptomatic” cases.
A generalized estimating equation with a logit link, robust variance, and independent working correlation matrix was used to analyze risk factors for testing positive, allowing for dependence within individuals longitudinally. Odds ratios (ORs) and their 95% CIs were calculated adjusting for race, Latinx/Hispanic ethnicity, university or Greek affiliation, number of household members, attesting positive, mask wearing behavior, social distancing behavior, and on-campus frequency. Analyses were performed in R, version 3.6.1.
Human Subjects
The University of Washington Institutional Review Board approved this study. All participants (or their guardians) provided written informed consent.
RESULTS
Between September 24 and December 18, 2020, 16 476 individuals enrolled in the study, and 29 783 samples from 11 644 unique individuals were collected and tested for SARS-CoV-2 (Supplementary Figure 1). Twenty-five point five percent (15 930/62 591) of matriculated students during the Autumn quarter were enrolled in at least 1 course with in-person instruction, and 19.9% (8204/41 296) of all matriculated undergraduates and 18.4% (2719/14 765) of graduate students participated in the study [22–24]. Due to remote instruction, many students were not living in the surrounding area and were not eligible for the study. More female (61.4% in the study vs 54% in the student body) and White students (62.6% in the study vs 40.8% in the student body (Table 1; Supplementary Figure 2) were enrolled. Of an estimated 4100 eligible Greek community students, 2672 (65.2%) were enrolled, and these students were more likely to be White than non-Greek students (Supplementary Table 2).
Table 1.
Sociodemographic Characteristics of Study Participants
| Students (n = 11 027), No. (%) or Median [Min, Max] | Staff (n = 3898), No. (%) or Median [Min, Max] | Faculty (n = 1426), No. (%) or Median [Min, Max] | Othera (n = 125), No. (%) or Median [Min, Max] | Overall (n = 16 476), No. (%) or Median [Min, Max] | |
|---|---|---|---|---|---|
| Age, y | 20 [16, 78] | 40 [20, 81] | 45 [21, 83] | 31 [19, 77] | 23 [16, 83] |
| Age group | |||||
| 16–17 y | 52 (0.5) | 0 (0) | 0 (0) | 0 (0) | 52 (0.3) |
| 18–49 y | 10 921 (99.0) | 2709 (69.5) | 870 (61.0) | 105 (84.0) | 14 605 (88.6) |
| 50–64 y | 38 (0.3) | 1051 (27.0) | 387 (27.1) | 12 (9.6) | 1488 (9.0) |
| 65+ | 16 (0.1) | 136 (3.5) | 168 (11.8) | 8 (6.4) | 328 (2.0) |
| Sex assigned at birthb | |||||
| Female | 6769 (61.4) | 2473 (63.4) | 765 (53.6) | 75 (60.0) | 10 082 (61.2) |
| Male | 4211 (38.2) | 1411 (36.2) | 658 (46.1) | 50 (40.0) | 6330 (38.4) |
| Racec | |||||
| American Indian or Alaska Native | 45 (0.4) | 17 (0.4) | 6 (0.4) | 0 (0) | 68 (0.4) |
| Asian | 2736 (24.8) | 486 (12.5) | 189 (13.3) | 21 (16.8) | 3432 (20.8) |
| Black or African American | 220 (2.0) | 127 (3.3) | 34 (2.4) | 2 (1.6) | 383 (2.3) |
| Multiple races | 1099 (10.0) | 229 (5.9) | 44 (3.1) | 8 (6.4) | 1380 (8.4) |
| Native Hawaiian or other Pacific Islander | 30 (0.3) | 26 (0.7) | 1 (0.1) | 0 (0) | 57 (0.3) |
| Other | 333 (3.0) | 109 (2.8) | 38 (2.7) | 3 (2.4) | 483 (2.9) |
| White | 6353 (57.6) | 2805 (72.0) | 1073 (75.2) | 83 (66.4) | 10 314 (62.6) |
| Missing | 211 (1.9) | 99 (2.5) | 41 (2.9) | 8 (6.4) | 359 (2.2) |
| Hispanic/Latinx ethnicity | 896 (8.1) | 265 (6.8) | 79 (5.5) | 12 (9.6) | 1252 (7.6) |
| UW Greek member | 2672 (24.2) | ||||
| Housingd | |||||
| Greek (chapter house and live out) | 1763 (16.0) | 0 (0) | 1 (0.1) | 1 (0.8) | 1765 (10.7) |
| Off-campus housing | 7116 (64.5) | 3863 (99.1) | 1423 (99.8) | 124 (99.2) | 12 526 (76.0) |
| On-campus housing | 2101 (19.1) | 11 (0.3) | 0 (0) | 0 (0) | 2112 (12.8) |
| Shelter, transitional housing, or other | 47 (0.4) | 24 (0.6) | 2 (0.1) | 0 (0) | 73 (0.4) |
| On-campus frequency this quartere | |||||
| Do not come to campus | 3345 (30.3) | 916 (23.5) | 252 (17.7) | 17 (13.6) | 4530 (27.5) |
| ≤1 d/wk | 3301 (29.9) | 1161 (29.8) | 583 (40.9) | 41 (32.8) | 5086 (30.9) |
| ≥2 d/wk | 4381 (39.7) | 1820 (46.7) | 591 (41.4) | 67 (53.6) | 6859 (41.6) |
| Registered for in-person classes | 1712 (15.5) | ||||
| Tested for SARS-CoV-2 before enrollment | 6556 (59.5) | 1625 (41.7) | 632 (44.3) | 67 (53.6) | 8880 (53.9) |
| Campus | |||||
| Main campus | 10 539 (95.6) | 3650 (93.6) | 1285 (90.1) | 124 (99.2) | 15 598 (94.7) |
| Satellite campus A | 191 (1.7) | 95 (2.4) | 60 (4.2) | 0 (0) | 346 (2.1) |
| Satellite campus B | 297 (2.7) | 153 (3.9) | 81 (5.7) | 1 (0.8) | 532 (3.2) |
| In the past 7 d, how often did you wear a face mask in public to protect others from getting sick?f | |||||
| Always | 10 719 (97.2) | 3800 (97.5) | 1373 (96.3) | 122 (97.6) | 16 014 (97.2) |
| Sometimes | 297 (2.7) | 95 (2.4) | 50 (3.5) | 3 (2.4) | 445 (2.7) |
| Never | 2 (0.0) | 0 (0) | 0 (0) | 0 (0) | 2 (0.0) |
| In the past 7 d, how often did you try to stay 6 ft away from people who don’t live with you?g | |||||
| Always | 7552 (68.5) | 3161 (81.1) | 1211 (84.9) | 98 (78.4) | 12 022 (73.0) |
| Sometimes | 3418 (31.0) | 728 (18.7) | 214 (15.0) | 27 (21.6) | 4387 (26.6) |
| Never | 42 (0.4) | 5 (0.1) | 1 (0.1) | 0 (0) | 48 (0.3) |
Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
aThose in the “other” affiliation category included postdoctoral fellows, externs, volunteers, and other university affiliates.
bFour participants reported “other” for sex, and 60 had missing sex.
cThree hundred fifty-nine participants had missing race, and 145 participants chose “prefer not to say” for Hispanic/Latinx ethnicity.
dA Greek “live out” is a private house or apartment that is shared by members of the same Greek chapter. On-campus housing was defined as dormitories, apartments, and family units.
eOne participant had missing “frequency on-campus.”
fFifteen participants did not answer the following question: “In the past 7 days, how often did you wear a face mask in public to protect others from getting sick?”
gNineteen participants did not answer the following question: “In the past 7 days, how often did you try to stay 6 feet away from people who don’t live with you?”
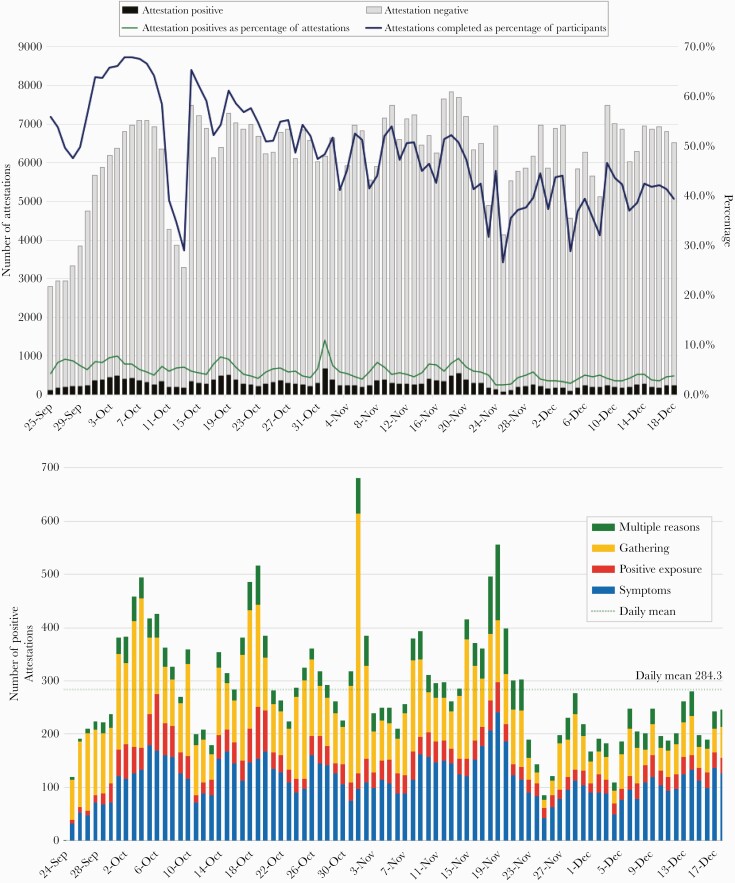
The daily attestation survey was completed by a mean of 47.7% (6203) participants per day (Figure 1A, B). Among participants attesting positive over the study period, 40.4% reported symptoms, 12.1% reported recent exposure to a SARS-CoV-2-positive individual, 36.4% reported attending an indoor gathering with >10 people without distancing or mask wearing, and 11.1% reported multiple reasons. During the study, 409 (2.5%) participants opted to stop receiving attestation alerts and were given the option of completing attestations on the study website. Results on participant preventative behaviors are shown in Supplementary Table 3.
Figure 1.
Daily attestation survey engagement over time. A, Number of daily attestations completed during the study period. Between October 11 and 13, 2020, we experienced an outage of the text messaging service used to send daily attestation survey invites, and this resulted in a reduced response rate. B, Positive daily attestations stratified by reason for positive attestation. A marked increase in positive attestations was observed the day after Halloween, when 487 reported gathering, a 4.7-fold increase from the mean daily gathering attestation positive of 105.
SARS-CoV-2 Testing Results
A total of 11 633 (70.6%) participants were tested at least once (Supplementary Table 4). Tests were resulted and available for participants to view online the day of sample collection (26.3%) or the following day (62.6%), and a minority of students received their results >24 hours after testing. Two hundred sixty-five out of 29 783 samples (0.80%) tested positive or inconclusive for SARS-CoV-2 (Table 2). Among the 265 cases, there were 60.8% (61) symptomatic, 19.6% (52) presymptomatic, 3.4% (9) asymptomatic, and 16.2% (43) possible asymptomatic. Based on the total 29 723 samples tested, 1.4% of participants reporting symptoms at the time of testing were positive (161/11 116) and participants not reporting symptoms at the time of testing had a low likelihood of positivity (0.56%, 104/18 607) (Table 2). The symptoms associated with the highest percent positivity were loss of taste/smell (19/382, 5.0%), fever (52/1518, 3.4%), and chills (36/1365, 2.6%) (Supplementary Table 5). Ninety-two of 256 (34.7%) participants testing positive reported exposure to a known positive case. By group, the Greek community had 1.5% test positivity (1796/12 045), on-campus dorm residents had 1.2% positivity (43/3507), and staff and faculty had 0.4% (19/4417) and 0.3% positivity (4/1467), respectively. Test positivity by affiliation and race is shown in Supplementary Table 6. Overall, during the Autumn quarter, the university was aware of 745 SARS-CoV-2-positive individuals, of whom 31.7% (236/745) were detected as part of this study [25].
Table 2.
Characteristics of Testing Instances by SARS-CoV-2 Result
| Positive | Overalla | Percent Positivity (95% CIb), % | |
|---|---|---|---|
| (n = 265) | (n = 29 783) | ||
| Any symptoms | 161 | 11 116 | 1.4 (1.2–1.7) |
| Attended large indoor gathering | 57 | 6996 | 0.8 (0.6–1.1) |
| Close contact with known positive | 92 | 3877 | 2.4 (1.9–2.9) |
| Baseline test | 35 | 5700 | 0.6 (0.4–0.9) |
| Outbreak test | 83 | 5257 | 1.6 (1.3–2) |
| Holiday test | 3 | 1244 | 0.2 (0–0.7) |
| Walk-in (no invite) | 18 | 1788 | 1.0 (0.6–1.6) |
| Risk tier | |||
| Tier 1 | 220 | 19 239 | 1.1 (1–1.3) |
| Tier 2 | 12 | 4291 | 0.3 (0.1–0.5) |
| Tier 3 | 33 | 6253 | 0.5 (0.4–0.7) |
| Affiliationc | |||
| Student | 240 | 23 802 | 1.0 (0.9–1.1) |
| Staff | 19 | 4417 | 0.4 (0.3–0.7) |
| Faculty | 4 | 1467 | 0.3 (0.1–0.7) |
| Age group | |||
| 13–17 y | 2 | 153 | 1.3 (0.2–4.6) |
| 18–49 y | 254 | 27 783 | 0.9 (0.8–1) |
| 50–64 y | 8 | 1548 | 0.5 (0.2–1) |
| 65+ y | 1 | 299 | 0.3 (0–1.8) |
| Sex assigned at birth | |||
| Female | 180 | 19 234 | 0.9 (0.8–1.1) |
| Male | 84 | 10 474 | 0.8 (0.6–1) |
| House members | |||
| Lives alone | 16 | 2708 | 0.6 (0.3–1) |
| 2 people | 63 | 8051 | 0.8 (0.6–1) |
| 3–5 people | 34 | 7468 | 0.5 (0.3–0.6) |
| 6+ people | 152 | 11 556 | 1.3 (1.1–1.5) |
| UW Greek member | 176 | 12 045 | 1.5 (1.3–1.7) |
| Race | |||
| American Indian or Alaska Native | 0 | 144 | 0.0 (0–2.5) |
| Asian | 32 | 4672 | 0.7 (0.5–1) |
| Black or African American | 2 | 460 | 0.4 (0.1–1.6) |
| Native Hawaiian or other Pacific Islander | 0 | 59 | 0.0 (0–6.1) |
| White | 191 | 20 464 | 0.9 (0.8–1.1) |
| Multiple races | 26 | 2808 | 0.9 (0.6–1.4) |
| Other | 10 | 707 | 1.4 (0.7–2.6) |
| Latinx/Hispanic | 35 | 2000 | 1.8 (1.2–2.4) |
| Housing type | |||
| Apartment (off campus) | 32 | 7403 | 0.4 (0.3–0.6) |
| Dormitory/residence hall (on campus) | 43 | 3507 | 1.2 (0.9–1.6) |
| House/condo/townhouse (off campus) | 62 | 10 433 | 0.6 (0.5–0.8) |
| Permanent supportive/transitional housing (off campus) | 0 | 6 | 0.0 (0–45.9) |
| Homeless shelter (off campus) | 0 | 5 | 0.0 (0–52.2) |
| UW Greek chapter house (off campus) | 104 | 6145 | 1.7 (1.4–2) |
| UW Greek live out (off campus) | 24 | 2168 | 1.1 (0.7–1.6) |
| Other | 0 | 116 | 0.0 (0–3.1) |
| Overall | 265 | 29 783 | 0.9 (0.8–1) |
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; UW, University of Washington.
aOverall includes 60 samples that were never tested.
bConfidence intervals were generated using the Clopper-Pearson exact method.
cEighty-six “other” and 15 “volunteer” affiliations.
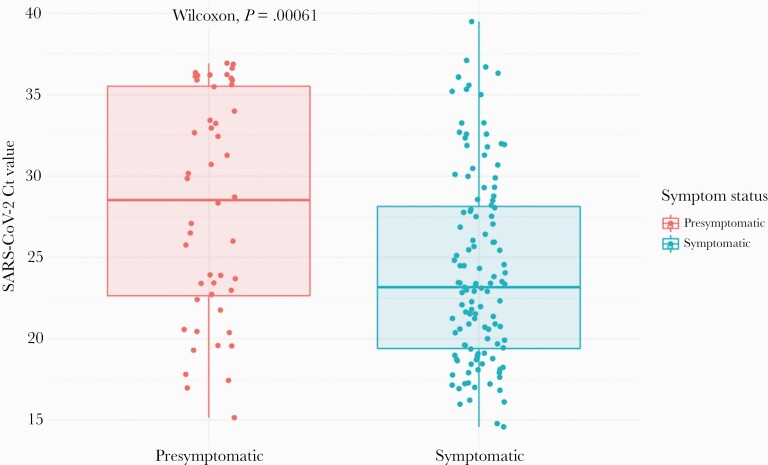
For comparison of Ct values, we restricted comparisons to the time period before the change in testing methods, as methods used before and after November 18, 2020, were not comparable. Among samples collected from September 24 to November 18, 2020, mean Ct values were higher in presymptomatic compared with symptomatic cases (28.7 vs 24.2; P = .001), corresponding to a lower viral load (Figure 2; Supplementary Figure 3).
Figure 2.
Comparison of viral loads in symptomatic (n = 124) vs presymptomatic (n = 48) positive and inconclusive samples. Cycle thresholds for samples (each represented by 1 dot) tested using our protocol with nucleic acid extraction (before November 18) are shown here. Complete data are shown in Supplementary Figure 6. Box plots show the median values and 25th and 75th percentiles, with vertical lines demonstrating the range of values. Abbreviations: Ct, cycle threshold; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Risk Factors for SARS-CoV-2 Infection
On multivariable analysis, Greek affiliation had the strongest association with test positivity (OR, 2.71; 95% CI, 1.84–4.00; P < .001). Latinx/Hispanic ethnicity (OR, 2.12; 95% CI, 1.28–2.18; P = .002) and positive attestations (OR, 1.86; 95% CI, 1.43–2.41; P < .001) were also risk factors for positivity (Supplementary Table 7). Reported frequency of hand washing, mask wearing, and social distancing were not associated with positivity.
Greek Community Outbreak
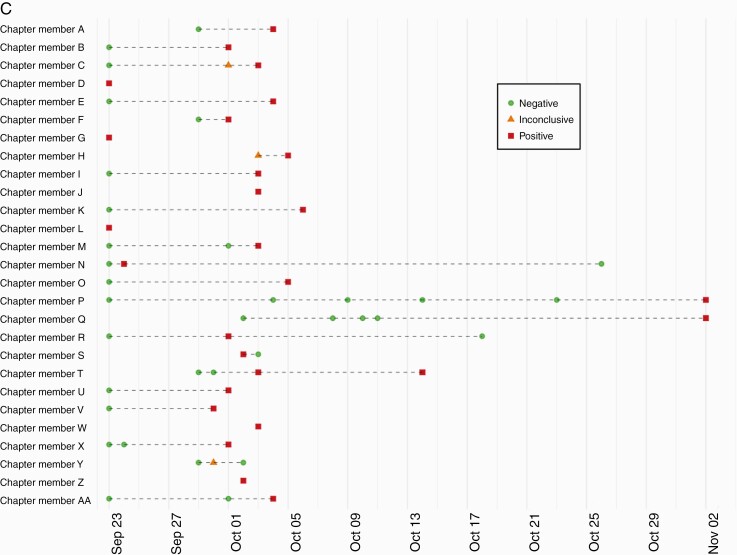
Thirty cases were identified in the Greek community during the first 10 days of the study, which prompted outbreak testing. Test positivity trends in the Greek community demonstrated a unique epidemiologic curve compared with the non-Greek students, employees, and the county (Figure 3A) [26]. Outbreaks within Greek houses were concurrent, but with unique individual timelines, and involved both fraternities and sororities (Figure 3B). Sixty-three point eight percent of Greek members reported sharing a living space with ≥6 people, compared with only 14.0% of non-Greek students. During 37 days of outbreak testing in the Greek community, serial testing frequently identified individuals who tested negative several times before testing positive (Figure 3C).
Figure 3.
Dynamics of a Greek community outbreak. A, Percent positivity over time of university groups and the surrounding county. The employee category includes university staff and faculty, and the Greek-affiliated students category includes all Greek-affiliated students including those living in Greek houses, Greek off-campus housing, and Greek-affiliated dorm residents. B, Greek chapter-level SARS-CoV-2 outbreak dynamics; counts of cases identified by chapter during the Autumn quarter. Sororities are shown in blue and fraternities in red. Chapters with no infections detected (n = 20) or ≤2 infections detected (n = 5) are not shown. C, Example of chapter-level individual SARS-CoV-2 outbreak dynamics within 1 Greek chapter (ending on November 2). Lines represent individual study participants tested multiple times, and dots signify a single test. Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
SARS-CoV-2 Molecular Epidemiology
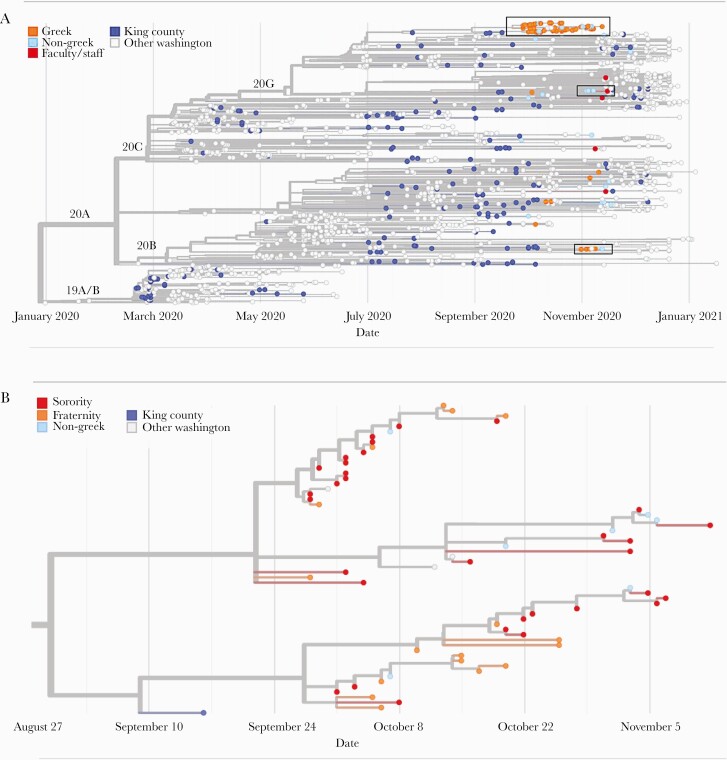
Genome sequences were generated from 88 SARS-CoV-2 samples collected from unique individuals between September 27 and November 28, 2020. Fifty-nine samples were from Greek-affiliated students, 24 from non-Greek-affiliated students, and 5 from faculty/staff. In a phylogenetic tree of 1700 SARS-CoV-2 genomes collected statewide, including the 88 from this study, samples included viruses from each of the 4 major clades (20A, 20B, 20C, 20G) circulating in the county and state during this time frame (Figure 4A).
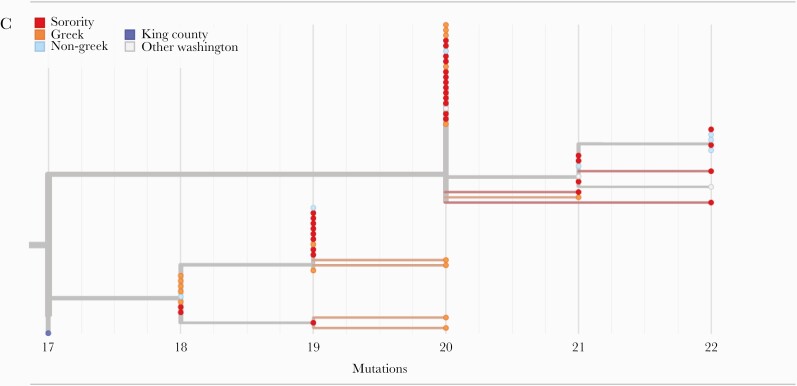
Figure 4.
Molecular epidemiology of a university outbreak. A, Phylogenetic tree of SARS-CoV-2 samples from Washington, including 88 samples from this study. Included here are all SARS-CoV-2 genomes from Washington collected on or after September 25, 2020, a random subsample of 1000 Washington samples collected before September 25, and the Wuhan/Hu-1 reference genome. Samples are positioned on the x-axis by date of collection. B, Detail of a cluster of university genomes organized horizontally by collection date and (C) by divergence, or the number of genetic changes relative to the SARS-CoV-2 reference genome. Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Most viral genomes from this study (56/88, 63.6%) grouped into 1 large cluster that included genomes from 49 Greek-affiliated and 7 non-Greek-affiliated students (large black box in Figure 4A, detailed in Figure 4B, C; Supplementary Figure 4). This cluster also included genomes from 4 samples collected outside of our study. Samples in this cluster were collected between September 27 and November 28, 2020, and all samples in this cluster collected before October 7 originated from Greek-affiliated students.
Closer inspection of this cluster (Figures 4B, C) demonstrates 2 subclusters (branch support values are 0.94 for the larger subcluster and 0.78 for the smaller subcluster) containing both sorority and fraternity members. Samples within clusters are closely related to a maximum pairwise distance between any 2 samples in the same subcluster of 4 single nucleotide changes. Molecular clock estimates place the common ancestor of the larger cluster on September 22, 2020 (95% CI, September 1–29), and the smaller cluster on September 27, 2020 (95% CI, September 20–October 4). These 2 dates are just before the sharp increase in cases observed in our study among Greek students, which peaked on ~October 7–8, 2020 (Figure 3A). The last sample mapping to either of these subclusters was collected on November 12, 2020. This date roughly coincides with the end of the Greek outbreak as measured by the percent positivity rate (Figure 3A). Among the hundreds of genomes sequenced state-wide November 12, 2020–March 17, 2021, none are descendants of the viruses responsible for the Greek community outbreak.
Two smaller clusters of viral genomes from this study are shown in Figure 4A (2 small boxes) and Supplementary Figures 5 and 6. One cluster contains 4 genomes from Greek-affiliated students and 1 from a non-Greek student. This cluster has a most recent common ancestor dating to October 31, 2020 (95% CI, October 17–November 1). The second contains 3 genomes from non-Greek-affiliated students and 1 faculty/staff member, with a most recent common ancestor dating to November 3, 2020 (95% CI, October 11–November 16). Viral genomes from Greek-affiliated students were most likely to cluster with other study samples; 88.1% of genomes from Greek-affiliated students were genetically identical to at least 1 other study SARS-CoV-2 genome, and 45.8% of genomes from non-Greek students and 0% of genomes from faculty/staff were identical to another study genome.
DISCUSSION
We report a large-scale COVID-19 longitudinal study of students, faculty, and staff at a university campus that allocated testing based on self-reported risk of infection, rather than mass surveillance. Most cases were identified through daily attestation surveys, and participants reporting a recent exposure to a case had the highest positivity rate of 2.4%, followed by 1.4% for participants reporting symptoms. Baseline testing had a much lower positivity rate of 0.56%, identifying only 15% of cases. Phylogenetic analysis of SARS-CoV-2 genomes from this study suggests differences in transmission patterns among Greek community members and non-Greek members. SARS-CoV-2 genomes from Greek-affiliated students primarily fell into closely related clusters, suggesting transmission related to Greek-associated housing or activities, while samples from non-Greek students and faculty/staff were more genetically diverse. Most viral genomes from Greek-affiliated students were members of a large cluster that may have resulted from a single SARS-CoV-2 transmission event. Eighty-four percent of the samples in this cluster represented just 5 unique viral genotypes, suggesting rapid spread. Identical viruses were commonly collected from members of several different sororities/fraternities, and this supports the theory that social behaviors, rather than housing arrangements, drove this outbreak.
Outbreak testing was an essential part of our testing strategy. In our study, Greek affiliation was the most important risk factor for testing positive, and more than two-thirds of cases detected in this study were in Greek-affiliated students, similar to findings at other universities [27]. We observed cases in most of our university’s Greek chapters, and the genomic analysis and outbreak dynamics indicate that rapid transmission occurred within and among fraternities and sororities. However, later in the quarter, the decreased test positivity rate among this group became comparable to rates observed among the non-Greek participants. This decline may have been driven in part by our aggressive testing strategy in this group, and effective contact tracing, quarantine, and other risk mitigation strategies. This analysis suggests that policies that limit large gatherings and provide regular testing to high-risk populations can mitigate outbreaks.
Consistent with our findings, random testing of asymptomatic people at the University of Pittsburgh yielded a positivity rate of only 0.4% before widespread vaccination [4]. Among partially vaccinated communities, 1 report indicates that surveillance testing yielded even fewer case detections [28]. Campus outbreaks are known risk factors for broader community transmission [29, 30]. In our study, SARS-CoV-2 test positivity rates in the university community were substantially lower than in the surrounding area. This may have been due to increased test availability, particularly for asymptomatic persons. However, it is also possible that focused testing of high-risk groups prevented and mitigated campus-wide outbreaks. In addition to members of the Greek community, participants who identified as Latinx or Hispanic experienced increased positivity rates. This is consistent with COVID-19 incidence rates in Washington State, where Hispanic individuals have been shown to represent 13% of state residents but 33% of COVID-19 cases [26]. This highlights the need for more targeted and equitable distribution of testing and contact tracing resources in this population.
Testing and behavioral interventions are cost-effective to control outbreaks on college campuses; therefore, prioritization of testing is critical, because laboratory supplies were scarce during our study [31]. We employed several mechanisms to conserve testing resources. First, we used the daily attestation surveys to screen for symptoms predictive of SARS-CoV-2 infection [32] to prioritize tests for those at highest risk. Second, we used mass-produced swabs that are shipped dry and eluted in PCR-friendly buffer to allow an extraction-free RT-qPCR method. By avoiding the need for limiting reagents such as transport media and RNA extraction kits, we avoided supply chain challenges, reduced the price per test, and increased the speed of our testing pipeline, enabling us to maintain a 24-hour turnaround time on average while increasing scale.
As of March 2021, there are only 4 viruses collected statewide that fall into the Greek outbreak cluster, and the last was collected on October 14, 2020. This demonstrates that sustained spread in the surrounding community did not occur in the subset of samples sequenced from the surrounding county/state. This does not fully rule out “spillover” of virus from the Greek outbreak into the outside community, given that a limited subset of samples were sequenced.
Other limitations of this study include that our online format of consent, enrollment, and daily attestation surveys increased participant engagement but was a barrier to participation for individuals with limited technological literacy or access. Additionally, study materials were available in English only. Data about participant exposures, including the daily attestation, were based on self-report, and questions about risk-taking behaviors were asked only at enrollment. After report of a positive exposure or attendance at a high-risk gathering, participants were offered testing twice, and positive cases could have been missed due to infrequent testing during the incubation period. While on-campus testing through this program was often the most convenient testing option, the data included here are not comprehensive in describing on-campus cases as other testing mechanisms were also available. Race and ethnicity representation in the study population was not representative of the university community, potentially due to differences in willingness to participate in studies, and this was likely further skewed by a university campaign to enroll and test members of the Greek community. Because our study included fewer underrepresented minorities than the overall university community, our report may represent an underestimate of the total number of infections. Our ability to speculate on the patterns related to Greek community transmission dynamics are impacted by the availability of other testing mechanisms and inability to sequence all study genomes.
We report here a strategy for SARS-CoV-2 testing on a large university campus using contactless, rapid enrollment, and self-administered testing during Autumn quarter 2020. Most infections were detected in the Greek community, and this group experienced distinct genomic and epidemiologic dynamics compared with other university communities and the surrounding area. This evidence suggests that testing those engaged in high-risk activities, in combination with testing people experiencing symptoms, may be critical steps in stopping on-campus transmission and potentially preventing community spread in a setting of limited resources. Our data also suggest that interventions to reduce large gatherings and promote mask wearing indoors are likely to reduce campus outbreaks.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We would like to thank the study participants. We also thank the UW Environmental Health & Safety COVID-19 Prevention & Response team (Sheryl Schwartz, Megan Gourley, David Coomes, Casey Adams), UW administration and Incident Command leadership group (Margaret Shepherd, Derek Fulwiler, David Hotz, Josh Gana, Erik Johnson, Pamela Schreiber), Chu Lab and BBI HCT team (Jody Sicuro, Dylan McDonald, Devon McDonald, Madison Contor, Cooper Marshall, Lincoln Pothan, Taylor Wilson, Zack Acker, and Evan Sarantinos), Dr. Janet Englund, Dr. Timothy Uyeki, Dr. Michael Jackson, and Dr. Roy Burstein. We gratefully acknowledge the authors and the originating and submitting laboratories of the sequences from GISAID’s EpiFlu Database, on which this research is based.
Financial support. This work was supported by the United States Senate and House of Representative Bill 748, Coronavirus Aid, Relief, and Economic Security Act.
Potential conflicts of interest. H.Y.C. reports consulting with Ellume, Pfizer, The Bill and Melinda Gates Foundation, Glaxo Smith Kline, and Merck. She has received research funding from Gates Ventures, Sanofi Pasteur, and support and reagents from Ellume and Cepheid outside of the submitted work. G.S.G. has received research grants and research support from the NIH, UW, the Bill & Melinda Gates Foundation, Gilead Sciences, Alere Technologies, Merck & Co, Janssen Pharmaceutica, Cerus Corporation, ViiV Healthcare, Bristol-Myers Squibb, Roche Molecular Systems, Abbott Molecular Diagnostics, and TheraTechnologies/TaiMed Biologics, Inc. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Malmgren J, Guo B, Kaplan HG. Continued age shift of confirmed positive COVID-19 incidence over time to children and young adults: Washington State March–August 2020. PLoS One 2020; 16:e0243042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boehmer TK, DeVies J, Caruso E, et al. Changing age distribution of the COVID-19 pandemic - United States, May-August 2020. MMWR Morb Mortal Wkly Rep 2020; 69:1404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hubler S, Hartocollis A. How colleges became the new Covid hot spots. New York Times. 11 September 2020. Available at: https://www.nytimes.com/2020/09/11/us/college-campus-outbreak-covid.html. Accessed 28 January 2021. [Google Scholar]
- 4. O’Donnell C, Brownlee K, Martin E, et al. SARS-CoV-2 control on a large urban college campus without mass testing. medRxiv 2021.01.21.21249825 [Preprint]. 25 January 2021. Available at: 10.1101/2021.01.21.21249825. Accessed 8 October 2021. [DOI] [PubMed] [Google Scholar]
- 5. Christensen K. Tracking and Treating COVID-19 in Human Waste. Michigan Technological University; 2020. [Google Scholar]
- 6. Booeshaghi AS, Tan F, Renton B, et al. Markedly heterogeneous COVID-19 testing plans among US colleges and universities. medRxiv 2020.08.09.20171223 [Preprint]. 11 August 2020. Available at: 10.1101/2020.08.09.20171223. Accessed 8 October 2021. [DOI] [Google Scholar]
- 7. CDC. Communities, Schools, Workplaces, & Events. Centers for Disease Control and Prevention; 2020. [Google Scholar]
- 8. Discover the University of Washington. About the UW blog. Available at: https://www.washington.edu/about/. Accessed 8 October 2021.
- 9. University of Washington. 2020 University of Washington financial report. 2020. Available at: https://finance.uw.edu/financial-report-archive. Accessed 8 October 2021.
- 10.Novel Coronavirus Information Blog. How the pandemic will affect autumn quarter learning (Message to students from the United States). Available at: https://www.washington.edu/coronavirus/2020/08/06/autumn-quarter-learning-message-to-students-from-the-united-states/. Accessed 8 October 2021.
- 11. Harris PA, Taylor R, Minor BL, et al. ; REDCap Consortium. The REDCap Consortium: building an international community of software platform partners. J Biomed Inform 2019; 95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris PA, Taylor R, Thielke R, et al. Research Electronic Data Capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McAloon C, Collins Á, Hunt K, et al. Incubation period of COVID-19: a rapid systematic review and meta-analysis of observational research. BMJ Open 2020; 10:e039652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McCulloch DJ, Kim AE, Wilcox NC, et al. Comparison of unsupervised home self-collected midnasal swabs with clinician-collected nasopharyngeal swabs for detection of SARS-CoV-2 infection. JAMA Netw Open 2020; 3:e2016382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chu HY, Boeckh M, Englund JA, et al. The Seattle Flu Study: a multiarm community-based prospective study protocol for assessing influenza prevalence, transmission and genomic epidemiology. BMJ Open 2020; 10:e037295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Srivatsan S, Heidl S, Pfau B, et al. SwabExpress: An end-to-end protocol for extraction-free covid-19 testing. Clin Chem. 2021:hvab132. doi: 10.1093/clinchem/hvab132. PMID: 34286830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weil AA, Newman KL, Ong TD, et al. Cross-sectional prevalence of SARS-CoV-2 among skilled nursing facility employees and residents across facilities in Seattle. J Gen Intern Med. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bedford T, Greninger AL, Roychoudhury P, et al. ; Seattle Flu Study Investigators. Cryptic transmission of SARS-CoV-2 in Washington State. Science 2020; 370:571–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. seattleflu/assembly. Seattle Flu Study. 2021. Available at: https://github.com/seattleflu/assembly. Accessed 1 February 2021.
- 20. Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018; 34:4121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shu Y, McCauley J. GISAID: Global Initiative on Sharing All Influenza Data – from vision to reality. Euro Surveill 2017; 22:30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Office of Academic Data Management. Quick Stats: Bothell Enrollment Autumn 2020. University of Washington; 2020. [Google Scholar]
- 23.Office of Academic Data Management. Quick Stats: Tacoma Enrollment Autumn 2020. University of Washington; 2020. [Google Scholar]
- 24.Office of Academic Data Management. Quick Stats: Seattle Enrollment Autumn 2020. University of Washington; 2020. [Google Scholar]
- 25.University of Washington. University of Washington COVID-19 case tracking dashboard. Available at: https://www.washington.edu/coronavirus/testing-results/. Accessed 8 October 2021. [Google Scholar]
- 26.Washington Department of Health. COVID-19 data dashboard. Available at: https://www.doh.wa.gov/Emergencies/COVID19/DataDashboard#dashboard. Accessed 8 October 2021. [Google Scholar]
- 27. Vang KE. Participation in fraternity and sorority activities and the spread of COVID-19 among residential university communities — Arkansas, August 21–September 5, 2020. MMWR Morb Mortal Wkly Rep 2021; 70:20–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rubin D, Eisen M, Collins S, et al. SARS-CoV-2 infection in public school district employees following a district-wide vaccination program - Philadelphia County, Pennsylvania, March 21-April 23, 2021. MMWR Morb Mortal Wkly Rep 2021; 70:1040–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pray IW, Kocharian A, Mason J, Westergaard R, Meiman J. Trends in outbreak-associated cases of COVID-19 — Wisconsin, March–November 2020. MMWR Morb Mortal Wkly Rep 2021; 70:114–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Richmond CS, Sabin AP, Jobe DA, et al. SARS-CoV-2 sequencing reveals rapid transmission from college student clusters resulting in morbidity and deaths in vulnerable populations. medRxiv 2020.10.12.20210294 [Preprint]. 14 October 2020. Available at: 10.1101/2020.10.12.20210294. Accessed 8 October 2021. [DOI] [Google Scholar]
- 31. Losina E, Leifer V, Millham L, et al. College campuses and COVID-19 mitigation: clinical and economic value. Ann Intern Med 2021; 174:472–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greater Seattle Coronavirus Assessment Network. SCAN data results and technical report #3. 2020. Available at: https://publichealthinsider.com/wp-content/uploads/2020/08/8.3-SCAN-Technical-Report-3_FINAL_CLEAN.pdf. Accessed 14 February 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.