Abstract
Prime editing is a recent precision genome editing modality whose versatility offers the prospect for a wide range of applications, including the development of targeted genetic therapies. Yet, an outstanding bottleneck for its optimization and use concerns the difficulty in delivering large prime editing complexes into cells. Here, we demonstrate that packaging prime editing constructs in adenoviral capsids overcomes this constrain resulting in robust genome editing in both transformed and non-transformed human cells with up to 90% efficiencies. Using this cell cycle-independent delivery platform, we found a direct correlation between prime editing activity and cellular replication and disclose that the proportions between accurate prime editing events and unwanted byproducts can be influenced by the target-cell context. Hence, adenovector particles permit the efficacious delivery and testing of prime editing reagents in human cells independently of their transformation and replication statuses. The herein integrated gene delivery and gene editing technologies are expected to aid investigating the potential and limitations of prime editing in numerous experimental settings and, eventually, in ex vivo or in vivo therapeutic contexts.
INTRODUCTION
Programmable nucleases based on sequence-tailorable guide RNAs (gRNAs) and CRISPR-associated (Cas) nucleases are powerful genome editing tools (1,2). However, besides off-target mutagenesis (3–9), programmable nucleases often yield complex target allele disruptions and large genomic rearrangements due to double-strand break (DSB) repair by illegitimate recombination processes (10,11). Hence, recent genome editing developments include advancing from DNA cutting to DNA non-cutting technologies based on nicking Cas proteins as such (12–14), or on these RNA-programmable nickases fused to DNA modifying moieties, e.g., base editors and, more recently, prime editors (15,16). Prime editing permits installing any single base-pair substitution in addition to well-defined small insertions or deletions, while requiring neither DSBs nor donor DNA substrates (15). Prime editors consist of an extended gRNA and a Cas9H840A nickase fused to an engineered reverse transcriptase (RT) named, respectively, pegRNA and PE2 (Supplementary Figure S1A). The pegRNA is formed by a gRNA covalently linked at its 3′-end to a RT template encoding the edit-of-interest and a RT primer binding site (PBS). Site-specific genomic DNA nicking yields a 3′-ended DNA flap that, upon PBS annealing, primes RT-mediated DNA synthesis over the RNA template. After DNA copy hybridization to complementary target DNA, the edit is ultimately incorporated in the genome presumably through sequential strand-resolution reactions (Supplementary Figure S1B). Prime editing has two main modalities, i.e. PE2 and PE3. The former system requires the delivery of PE2:pegRNA complexes; the latter relies on the transfer of these complexes together with a conventional gRNA. In the PE3 system, gRNA-directed nicking of the non-edited DNA strand fosters the use of the edited strand as repairing template (Supplementary Figure S1B).
Notwithstanding their enormous potential and versatility, prime editing principles bring to the fore specific shortcomings that will need identification, careful assessment and resolution. The large size of prime editing ribonucleoprotein complexes, composed of a ∼125 nucleotide-long pegRNA and a 238-kDa fusion protein encoded by a 6.3-kb ORF, poses substantial production and delivery issues. Indeed, producing proteins >100 kDa in sufficient quantities is particularly challenging. Moreover, although viral vectors are amongst the most efficient genome-editing tool delivery systems (17), the most commonly used platform, based on ∼15 nm adeno-associated viral (AAV) particles, is unsuitable for transferring full-length prime editing sequences due to its limited packaging capacity (∼4.7-kb) (17).
Fully viral gene-deleted adenoviral vectors (also called high-capacity adenoviral vectors), hereafter named adenovector particles (AdVPs), aggregate a valuable set of characteristics, namely; (i) large packaging capacity (i.e. up to 36-kb), (ii) strict episomal nature, (iii) high genetic stability; (iv) facile cell-tropism modification and (v) efficient transduction of dividing and quiescent cells (17–21). Here, we investigate the feasibility and utility of tailoring these ∼90 nm biological nanoparticles for all-in-one transfer of full-length prime editing components and, as the cellular processes underlying or influencing prime editing outcomes are essentially unknown, exploit the latter characteristic to study the role of cell cycling on this site-specific DNA modifying principle.
MATERIALS AND METHODS
Cells
The human cervix carcinoma (HeLa) cells and the human embryonic kidney 293T (HEK293T) cells (both from American Type Culture Collection) were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific; Cat. No.: 41966-029) containing 5% and 10% fetal bovine serum (FBS; Biowest; Cat. No.: S1860-500), respectively. PEC3.30 cells were maintained in high-glucose DMEM supplemented with 10% FBS, 10 mM MgCl2 and 0.4 μg ml−1 puromycin (Thermo Fisher Scientific; Cat. No.: A11138-03) (22). The generation and characterization of HEK293T.EGFP+ reporter cells harboring a transcriptionally active EGFP allele in the presence of doxycycline, were detailed elsewhere (23). These cells were cultured in high-glucose DMEM containing 10% FBS and 200 ng ml–1 doxycycline. The bone marrow-derived primary human mesenchymal stem cells (hMSCs) were kept in Minimum Essential Medium α (MEM-α) (Thermo Fisher Scientific; Cat. No.: 22561-021) supplemented with 10% FBS, 100 U ml–1 penicillin/streptomycin (Thermo Fisher Scientific; Cat. No.: 15140–122), 1× non-essential amino acids (NEAA; Thermo Fisher Scientific; Cat. No.: 11140–050) and 1× GlutaMax (Thermo Fisher Scientific; Cat. No.: 35050-061) (24,25). The hMSCs were passaged every 3–4 days at low split ratios in culture vessels coated with 0.1% gelatin (Sigma-Aldrich; Cat. No.: G1393) for at least 2 h at 37°C. Collection of human primary cells from bone marrow was carried out from anonymous ‘left-over’ surgery material in accordance with the Best Practices Code of the Dutch Federation of Biomedical Scientific Societies. No informed consent is required for the use of anonymous and non-traceable body materials and the institutional ethics committee of the LUMC waived the need for donor consent. The human myoblasts derived from an healthy donor and the retinal pigment epithelial cells expressing Fucci reporters (RPE-Fucci) (26), have both been described before (27–29). The former cells were cultured in Skeletal Muscle Cell Growth Medium (Ready-to-use, PromoCell; Cat. No.: C-23060) containing 20% FBS, 100 U ml–1 penicillin/streptomycin and 1× GlutaMax; the latter cells were maintained in DMEM/Nutrient Mixture F-12 (DMEM/F-12; Thermo Fisher Scientific; Cat. No.: 31331-028) supplemented with 10% FBS. All cells were kept in a humidified-air 5% CO2 atmosphere and were routinely tested for verifying the absence of mycoplasma.
Recombinant DNA
Standard recombinant DNA techniques were applied for the generation of the various constructs (Supplementary Information). The cloning schemes, annotated maps and nucleotide sequences of BF18_pLV.PURO.pegRNAHBB, BF19_pLV.PURO.pegRNA.gRNAHBB, BF20_pLV.BSD-EGFP.pegRNAEGFP, BF21_pLV.BSD-EGFP.pegRNA.gRNAEGFP, BF46_pLV.PURO.pegRNA.gRNAEGFP, BF22_pUCBM21.U6.gRNAHBB, BF23_pUCBM21.U6.gRNAEGFP, S75_pAdVP.PE2 and S89_pAdVP.PE2.pegRNACTT are available in pages 1–35 of the Supplementary Information. The oligonucleotides used for assembling the various gRNA and pegRNA expression constructs are indicated in Supplementary Table S1.
AdVP production, purification and characterization
The production of AdVP.PE2 and AdVP.PE2.pegRNACTT was done as follows. The bacteriophage P1 Cre recombinase- and adenovirus type 5 E1-expressing PEC3.30 cells were seeded at a density of 1.8 × 106 cells per well of 6-well plates (Greiner Bio-One). Transfection of the AdVP molecular clones was performed 16–18 h later with the aid of 25 kDa linear polyethyleneimine (PEI, Polysciences). In brief, 6.25 μg of MssI-linearized plasmids S75_pAdVP.PE2 and S89_pAdVP.PE2.pegRNACTT were diluted in a total volume of 200 μl of 150 mM NaCl and, after adding 20.6 μl of a 1 mg ml–1 PEI solution (pH 7.4) to each of the DNA transfection reactions, vigorous mixing in a vortex for about 10 s ensued. The DNA-PEI complexes, assembled after a 15-min incubation period at room temperature (RT), were directly added to the medium of the producer cells. Six hours later, transfection media were substituted by fresh medium containing E1-deleted helper AdV vector AdV.SRα.LacZ.1.50 (30) at an MOI of 10 infectious units (IUs) per cell. The helper vector contains its packaging elements flanked by a direct repeat of Cre recombinase loxP recognition sites (floxed.Ψ) and is used for supporting the amplification and selective packaging of AdVP genomes (Supplementary Figure S2). Typically, AdVP producer cells express Cre recombinase and E1 peptides (18–20). In addition to these proteins, PEC3.30 cells also express a thermosensitive form of the adenoviral DNA-binding protein (DBP) that remains inactive during regular culturing at 39°C (30,31). Hence, after helper addition, PEC3.30 cells were transferred from 39°C to 34°C for extra adenoviral protein complementation in the form of properly folded thermosensitive DBP. Upon helper-triggered emergence of complete cytopathic effect (CPE), the producer cells were harvested and subjected to three cycles of freezing and thawing in liquid N2 and 37°C water baths, respectively. Cellular debris were subsequently removed by centrifugation for 10 min at 2000 × g. The vector particles present in the collected supernatants were then amplified via four rounds of propagation in producer cells transduced with helper AdV.SRα.LacZ.1.50 (Supplementary Figure S2). The fourth propagation step involved twenty T175-cm2 culture flasks (Greiner Bio-One) each containing 27 × 106 producer cells. The resulting AdVPs were purified by sequential block and continuous CsCl buoyant density ultracentrifugation steps and were de-salted by ultrafiltration through Amicon Ultra-15 100K MWCO filters (MerckMillipore; Cat. No.: UFC910024) (Supplementary Figure S3). With the exception of the use of a AdVP molecular clone expressing mCherry from the hybrid CAG promoter (Supplementary Figure S4), the production of the reporter vector AdVP.mCherry followed essentially the same methodologies applied for the production of AdVP.PE2 and AdVP.PE2.pegRNACTT. The titers of purified AdVP stocks were determined via previously detailed procedures using the Quant-iT™ PicoGreen™ dsDNA Assay Kit reagents and protocol (Thermo Fisher Scientific; Cat.No.: P11496A) (30,32). Additionally, physical and transducing titers of prime editor-encoding AdVPs were also determined through qPCR assays. Quantification of physical titers was initiated by using the DNeasy Blood & Tissue kit (QIAGEN; Cat. No.: 69506) to isolate vector DNA from purified AdVP stocks. Next, six serial 3-fold dilutions of the extracted vector genomes were prepared for qPCR with iQ™ SYBR® Green Supermix (Bio-Rad; Cat. No.: L010171C) and the primers listed in Supplementary Table S2. Quantification of transducing titers was initiated by plating HeLa cells at a density of 8 × 104 cells per well of 24-well plates (Greinder Bio-One). The next day, the cells were transduced with six serial 3-fold dilutions of each purified AdVP preparation. After approximately 24 h, the transduced cells were harvest for DNA isolation by using the DNeasy Blood & Tissue kit (QIAGEN; Cat. No.: 69506). The resulting DNA was then used for qPCR quantification of transducing vector genome copies. In parallel, a standard curve was generated by using as qPCR template, eight serial 10-fold dilutions of a linearized target DNA-containing plasmid stock containing 1 × 107 GC μl–1. The primers, cycling conditions and components of qPCR mixtures are specified in Supplementary Tables S2 and S3. Data analysis was performed by using Bio-Rad CFX Manager 3.1 software and the titers were calculated based on the Ct values of standard curve and sample dilutions. The AdVP titers obtained via the different titration methods are listed in Supplementary Table S4. The AdVP MOI indicated in this study were based on packaged vector genome copies (GC) determined by the PicoGreen™ titration method (30,32).
The structural integrity of vector genomes packaged in purified adenoviral capsids (Supplementary Figure S3) was assessed essentially as indicated elsewhere (30,32). In brief, 50 μl of purified AdVP stocks were treated with 8 μl of 10 mg ml–1 DNaseI (Sigma-Aldrich; Cat. No.: 10104159001) at 37°C for 30 min. Next, the DNaseI enzyme was inactivated by adding 2.4 μl of 0.5 M ethylenedinitrilotetraacetic acid (EDTA) solution (pH 8.0), 6 μl of 10% (w/v) sodium dodecyl sulphate (SDS) and 1.5 μl of 20 mg ml–1 proteinase K (Thermo Fisher Scientific; Cat. No.: EO0491), and incubating the resulting mixture at 55°C for 1 h. Vector DNA isolation was then done by using the QIAEX II Gel Extraction Kit (QIAGEN; Cat. No.: 20021) following the manufacturer's instructions. Finally, the isolated vector genomes were subjected to restriction enzyme fragment analysis (RFLA) by using the Gel-Doc XR + system and the ImageLab 4.1 software (both from Bio-Rad). Parental plasmids pAdVP.PE2 and pAdVP.PE2.pegRNACTT, digested with the same restriction enzymes applied to vector genomes, served as molecular weight references. The in silico restriction patterns corresponding to intact plasmid and vector DNA were made with the aid of SnapGene (version 5.2.4) software (Supplementary Figure S3).
Transduction experiments
HeLa cells, hMSCs and HEK293T cells (both regular and genetically-modified through lentiviral vector transduction) were seeded at densities of 5 × 104, 1.5 × 105, and 7 × 104 cells per well of 24-well plates (Greinder Bio-One), respectively. After overnight incubations, the cells were either mock-transduced or were transduced with the various AdVPs at the MOIs specified in the figures and respective legends. The cells were harvested at 2 days post-transduction for western blotting and immunofluorescence microscopy analyses and at 3 days post-transduction for prime editing analyses. For the latter analyses, genomic DNA was extracted by using the DNeasy Blood & Tissue kit (QIAGEN; Cat. No.: 69506) following the manufacturer's recommendations. Transduction efficiencies were determined by reporter-directed flow cytometry and direct fluorescence microscopy analyses of parallel cell cultures exposed to AdVP.mCherry.
To compare prime editing activities mediated by AdVP transduction versus plasmid transfection, 1.5 × 105 HEK293T cells were seeded in wells of 24-well plates (Greinder Bio-One). The next day, plasmids encoding prime editing complexes were diluted in 150 mM NaCl to which 3.95 μl of 1 mg ml–1 of 25-kDa linear polyethyleneimine (pH 7.4; Polysciences) were added (Supplementary Table S5). The resulting transfection reactions were vortexed vigorously for about 10 sec and were then incubated for 15 min at RT. Afterwards, the reactions were directly added to the cell media and, after 6 h, the transfection media were substituted by regular culture media. In parallel, HEK293T cells were transduced with AdVPs at the MOIs specified in the Supplementary Table S5. Three days later, genomic DNA from transfected and transduced cells were harvested using the DNeasy Blood & Tissue kit reagents and protocol (QIAGEN; Cat. No.: 69506) for prime-editing activity analysis.
The transduction of dividing myoblasts and non-dividing myotubes was done as follows. Approximately 16–18 h prior to transduction, 5 × 104 human myoblasts were seeded in wells of 24-well plates and, the next day, these cells were exposed to AdVPs at the MOIs indicated in the respective figures. In parallel, 2 × 105 human myoblasts were seeded in the wells of 24-well plates pre-coated with a 0.1% (w/v) gelatin solution. Upon myoblast confluency, differentiation was triggered by switching regular culture medium for mitogen-poor differentiation medium whose composition is indicated below. Two days after differentiation initiation, the cells were treated with AdVPs at the MOIs specified in the corresponding figures. Myoblasts and myotubes were harvested for western blotting and genomic DNA extraction at 2 and 3 days post-transduction, respectively.
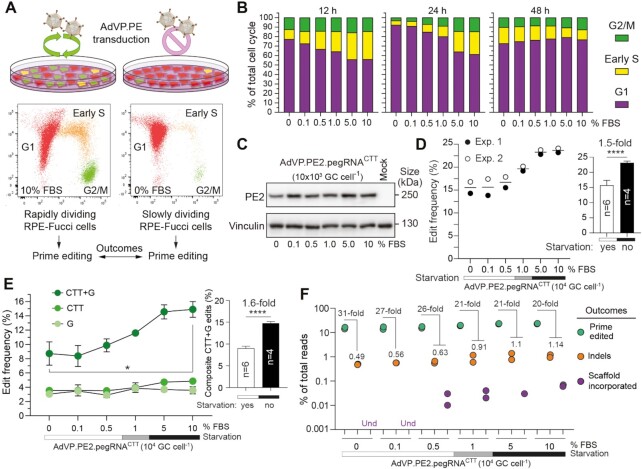
RPE-Fucci cells were seeded at a density of 2 × 105 cells per well of six-well plates. For generating cultures containing different proportions of cycling and non-cycling cells, RPE-Fucci cells were treated at ∼17 h after seeding with DMEM/F-12 medium lacking or containing different FBS concentrations (i.e. 0.1, 0.5, 1, 5 and 10%). Twelve hours later, the RPE-Fucci cells were exposed for an additional 12-h period to 104 GC cell–1 of AdVP.PE2.pegRNACTT in medium containing the various FBS amounts. Finally, the cells were harvested for western blotting and genomic DNA extraction at 60 h post-transduction. At 12, 24 and 48 h after the initiation of the various FBS treatments, the frequencies of RPE-Fucci cells at different phases of the cell cycle were determined by flow cytometry.
The transduction of HEK293T.EGFP+ cells stably expressing EGFP-specific pegRNA and gRNA was done as follows. The cells were seeded at 2 × 105 cells per well in 24-well plates and, after overnight incubation, the cells were exposed to different AdVP.PE2 MOI for 2 days. Quantification of EBFP-positive and EGFP-negative cells was performed at 7 days post-transduction via reporter-directed flow cytometry.
Cell differentiation assays
The capacity of hMSCs to differentiate into osteoblasts and adipocytes and of human myoblasts to differentiate into syncytial myotubes, was assessed as follows. To induce osteogenic differentiation, mock- and AdVP-transduced hMSCs initially seeded at a density of 1 × 104 cells per well of 48-well plates (Greiner Bio-One), were incubated for 2 weeks in osteogenic differentiation medium consisting of MEM-α supplemented with 10% FBS, 100 U ml–1 penicillin/streptomycin, 1× NEAA, 1× GlutaMax, 0.2 mM l-ascorbic acid 2-phosphate (Sigma-Aldrich; Cat. No.: A8960), 10 mM β-glycerophosphate (Sigma-Aldrich; Cat. No.: G6251), 2 μM dexamethasone (Sigma-Aldrich; Cat. No.: D4902) and 100 ng ml–1 of recombinant human bone morphogenetic protein 6 (BMP6; PeproTech; Cat. No.: 120-06). The differentiation medium was replenished every 3–4 days. Alizarin Red S staining was carried out for the detection of calcium deposits. In brief, cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS; pH 7.4) for at least 10 min and were then incubated at RT in the dark for 5 min in a 2% Alizarin Red S (pH 4.25) solution (Sigma-Aldrich; Cat. No.: A5533). After several washes with PBS, the treated cultures were photographed and were subsequently incubated for 30 min at RT, while shaking, in 100 μl of a 10% (v/v) acetic acid solution (Sigma-Aldrich; Cat. No.: 64-19-7) for Alizarin Red S extraction. Next, the samples were heated for 10 min at 85°C and then centrifuged for 15 min at 20 000 × g. The pH of the resulting supernatants was adjusted to 4.1–4.5 with a 32% (w/v) ammonium solution (Merck Millipore; Cat. No.: 105426) and measurements of the absorbance at OD405 nm were done by using a multimode plate reader (PerkinElmer VICTOR™ X3).
To trigger adipogenic differentiation, mock- and AdVP-transduced hMSCs, initially seeded at a density of 2 × 104 cells per well of 24-well plates (Greiner Bio-One), were treated with 100 ng ml–1 of recombinant human bone morphogenetic protein 7 (BMP7; PeproTech; Cat. No.: 120-03) until reaching confluence in 3 days. Next, the cells were incubated for 3 weeks in adipogenic differentiation medium consisting of DMEM supplemented with 10% FBS, 100 U ml–1 penicillin/streptomycin, 50 μM Indomethacin (Sigma-Aldrich; Cat. No.: I7378), 0.25 μM dexamethasone, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX; Sigma-Aldrich; Cat. No.: I5879) and 1.6 μM bovine insulin (Sigma-Aldrich; Cat. No.: I5500). The differentiation medium was replenished every 4 days. Oil Red O staining was performed for the detection of lipid droplets. In brief, the cells were fixed with 4% PFA in PBS (pH 7.4) for ∼1 h and, after two washes with water and a 5-min treatment with 60% 2-propanol, they were incubated for 10 min at RT in a solution of 1.8 mg ml–1 Oil Red O (Sigma-Aldrich; Cat. No.: O-0625) in 60% 2-propanol. After several washes with water, the treated cultures were photographed and were subsequently incubated for 10 min at RT, while shaking, in 2-propanol for Oil Red O extraction. Measurements of the absorbance at OD490nm were carried out with the aid of a multimode plate reader (PerkinElmer VICTOR™ X3).
To induce myogenic differentiation, confluent human myoblasts initially seeded at a density of 2 × 105 cells per well of 24-well plates pre-coated with 0.1% gelatin, were exposed to differentiation medium consisting of phenol red-free DMEM (Thermo Fisher Scientific; Cat. No.: 11880-028) supplemented with 100 U ml–1 penicillin/streptomycin, 100 μg ml–1 human holo-transferrin (Sigma-Aldrich; Cat. No.: T0665) and 10 μg ml−1 human insulin (Sigma-Aldrich; Cat. No.: I9278). Post-mitotic myotubes were detected by immunofluorescence staining with an antibody specific for the late muscle marker, sarcomeric α-actinin (Supplementary Table S6).
Reverse transcription-quantitative PCR (RT-qPCR)
RT-qPCR was applied for quantifying the differentiation abilities of mock- and AdVP-transduced hMSCs. Total RNA from cultures containing undifferentiated and differentiated hMSCs (osteoblasts and adipocytes) was extracted by using the NucleoSpin RNA kit according to the manufacturer's instructions (Macherey Nagel; Cat. No.: 740955). Next, reverse transcription was carried out with the RevertAid RT Reverse Transcription Kit (Thermo Fisher Scientific; Cat. No.: K1691). In brief, 450 ng of RNA was incubated with 0.5 μl of 100 μM random hexamer primers and 0.5 μl of 100 μM Oligo(dT)18 primers in 12-μl reaction volumes at 65°C for 5 min followed by an incubation at 4°C for 2 min. After a brief spinning, the mixtures were immediately chilled on ice and reverse transcription components consisting of 1 μl of 20 U μl–1 RiboLock RNase Inhibitor, 1 μl of 200 U μl–1 RevertAid H Minus M-MuLV Reverse Transcriptase, 2 μl of 10 mM dNTP Mix and 4 μl of 5× Reaction Buffer, were directly added to each sample. Next, the samples were incubated at 25°C for 5 min followed by an incubation at 42°C for 1 h. Finally, the reactions were terminated by heating the samples at 70°C for 5 min. The resulting cDNA templates were then diluted 4-fold in nuclease-free water and 1 μl of diluted cDNA was subjected to qPCR by using the iQ™ SYBR® Green Supermix (Bio-Rad; Cat. No.: L010171C) and the primers listed in Supplementary Table S2. GAPDH transcripts served as RT-qPCR targets for gene expression normalization. The signal outputs were detected by using the CFX Connect Real-Time PCR Detection System (Bio-Rad) and the relative expression of each target gene was analyzed through the 2−ΔΔCt method. The qPCR cycling conditions and mixture components are specified in Supplementary Tables S2 and S3, respectively.
Western blotting
Laemmli buffer consisting of 8.0% glycerol, 3% SDS and 200 mM Tris–HCl (pH 6.8) was applied for lysing cells for 5 min at 100°C. Protein concentrations in the resulting cell lysates were determined by using a DC™ protein assay kit (Bio-Rad; Cat. No.: 5000111) according to the manufacturer's instructions. Proteins were loaded in equal amounts and were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Afterwards, the resolved proteins were transferred onto 45-μm polyvinylidene difluoride (PVDF) membranes (Merck Millipore; Cat. No.: IPVH00010) that were subsequently blocked with 5% (w/v) non-fat dry milk (Campina Elk; Cat. No.: 112349) dissolved in Tris-buffered saline (TBS; 50 mM Tris–HCl pH 7.6; 150 mM NaCl) with 0.1% (v/v) Tween 20 (TBST) at RT for 1 h. After the blocking step, the membranes were incubated overnight at 4°C with primary antibodies raised against S. pyogenes Cas9 (Abcam; Cat. No.: ab191468; 1:1000 dilution), myosin heavy chain (Sigma-Aldrich; Cat. No.: M4276; 1:500 dilution), sarcomeric α-actinin (Sigma-Aldrich; Cat. No.: A7811; 1:1000 dilution), dystrophin (Abcam; Cat. No.:ab15277; 1:500 dilution), ki-67 (Sigma-Aldrich; Cat. No.: AB9260; 1:1000 dilution), α/β-tubulin (Cell signalling; Cat. No.: 2418S; 1:1000 dilution) and vinculin (Sigma-Aldrich; Cat. No.: V9131; 1:1000 dilution) all diluted in TBST supplemented with 5% bovine serum albumin (BSA). Next, the membranes were washed with TBST thrice and the antigens were probed at RT for 2 h with horseradish peroxidase (HRP)-conjugated secondary antibodies raised against mouse IgG (Sigma-Aldrich; Cat. No.: NA931V; 1:5000 dilution) or rabbit IgG (Cell signalling; Cat. No.: 7074S; 1:1000 dilution) diluted in TBST containing 1% (w/v) non-fat dry milk. Target proteins were detected with the Clarity™ Western ECL Substrate (Bio-Rad; Cat. No.: 1705060) and the ChemiDoc Imaging System (Bio-Rad; Cat. No.: 17001402).
Flow cytometry
Flow cytometry was applied for quantifying cell transduction efficiencies and RPE-Fucci cell fractions at different stages of the cell cycle. In brief, cells were washed with PBS (pH 7.4) and, after trypsin treatment and centrifugation, collected cells were resuspended in PBS containing 0.5% BSA and 2 mM EDTA (pH 8.0). Flow cytometry was performed in a BD LSR II flow cytometer (BD Biosciences) using mock-transduced cells as controls for establishing background fluorescence thresholds. At least 10,000 viable single cells were acquired per sample. Data were analysed with the aid of FlowJo 10.5.0 software (Tree Star).
Direct fluorescence microscopy
Transduction of hMSCs and human myoblasts was monitored by direct fluorescence microscopy. The nuclei were stained with 10 μg ml–1 Hoechst 33342 (Thermo Fisher Scientific; Cat. No.: H3570) for 10 min. Subsequently, the cells were washed thrice with PBS, after which, regular culture medium was added to the cell cultures. The mCherry- and Hoechst 33342-specific signals were detected by using an inverted DMI8 fluorescence microscope equipped with a DFC450C camera and acquired images were examined with the LAS X software (Leica Microsystems).
Confocal immunofluorescence microscopy
Cells were fixed with 4% PFA in PBS for 10 min at RT and then washed thrice with PBS before being permeabilized in 0.5% Triton X-100 in TBS (pH 7.6) at RT for 10 min. After three 10-min washes with 0.1% Triton X-100 in TBST, a blocking step was performed by incubating the permeabilized cells in a blocking solution consisting of TBS, 0.1% Triton X-100, 2% BSA and 0.1% sodium azide for 1 h at RT. Next, the cells were incubated overnight at 4°C with the appropriate primary antibodies diluted in blocking solution (Supplementary Table S6). The specimens were subsequently subjected to three 10-min washes with TBST and the target antigens were probed with fluorophore-conjugated secondary antibodies diluted in blocking solution for 1 h in the dark at RT (Supplementary Table S6). Finally, after three 10-min washes with TBST, ProLong Gold Antifade Mounting reagent containing DAPI (Thermo Fisher Scientific; Cat. No.: P36931) was used for mounting the specimens. Immunofluorescence microscopy images were acquired with the aid of an upright Leica SP8 confocal microscope equipped with Leica hybrid detectors HyD and were analysed with the LAS X software (Leica Microsystems).
EdU labeling
The 5-ethynyl-2′-deoxyuridine (EdU) labelling of myoblasts and myotubes was carried out by using the Click-iT® EdU Flow Cytometry Assay Kit (Thermo Fisher Scientific; Cat. No.: C10425). In brief, myoblasts and myotubes were treated with 10 μM EdU solution for 2 h at 37°C and were subsequently fixed with 4% PFA at RT for 15 min. After two washes with 3% BSA in PBS, a permeabilization step was performed by incubating the fixed cells in 0.5% Triton X-100 in TBS for 20 min at RT. Afterwards, the permeabilized cells were washed twice with 3% BSA in PBS and were incubated for 30 min in the dark at RT with 500 μl of Click-iT® reaction cocktail consisting of 427.5 μl of TBS, 20 μl of CuSO4, 2.5 μl of Alexa Fluor® 488 azide and 50 μl of 1× Reaction buffer additive. Next, a blocking TBS solution containing 0.1% Triton X-100, 2% BSA and 0.1% sodium azide was applied for 1 h at RT after which the appropriate primary antibodies diluted in blocking solution were added. After overnight incubation at 4°C in the dark, the specimens were washed thrice for 10 min with TBST and fluorophore-conjugated secondary antibodies diluted in blocking solution were added for 1 h at RT in the dark (Supplementary Table S6). Finally, ProLong Gold Antifade Mounting reagent containing DAPI (Thermo Fisher Scientific; Cat. No.: P36931) was used for mounting the specimens. Immunofluorescence microscopy images were acquired with the aid of an inverted DMI8 fluorescence microscope equipped with a DFC450C camera and with an upright Leica SP8 confocal microscope equipped with Leica hybrid detectors HyD. The resulting micrographs were analysed with LAS X software (Leica Microsystems).
DNA content analysis
Staining with the DNA dye Hoechst 33342 was performed to determine RPE-Fucci cell fractions at different stages of the cell cycle based on their DNA content. In brief, 2 × 105 cells RPE-Fucci cells were seeded in wells of 6-well plates. After overnight incubation, the cells were treated with DMEM/F-12 medium lacking or containing FBS (i.e. 0.1, 0.5, 1, 5 and 10% FBS). At 12 h, 24 h and 48 h after the initiation of the various FBS treatments, the cells were stained with 2 μl of a 10 mg ml–1 Hoechst 33342 solution (Thermo Fisher Scientific; Cat. No.: H3570) for 10 min at 37°C. After several washes with PBS, the DNA content profiles in the various cell populations were determined by using a BD LSR II flow cytometer (BD Biosciences).
Target site genotyping assays
Prime editing activities were assessed by DNA sequencing assays. The hMSCs, human myoblasts and RPE-Fucci cells were exposed to AdVP.PE2.pegRNACTT at the MOIs indicated in the respective figures. At 3 days post-transduction, genomic DNA was extracted via the DNeasy Blood & Tissue Kit reagents and protocol. The 273-bp target-specific PCR product was amplified with Phusion High-Fidelity Polymerase (Thermo Fisher Scientific; Cat. No.: #F-530L) and the primers with adapter tag overhangs listed in Supplementary Table S7. The cycling conditions and PCR mixtures used are specified in Supplementary Tables S8 and S9, respectively. The resulting amplicons were purified with AMPure XP beads (Beckman Coulter; Cat. No.: A63881) and were subsequently subjected to PCR barcoding using Illumina tag-specific primer pairs with unique sequence combinations for demultiplexing and sample identification (Supplementary Table S10). The cycling parameters and PCR mixtures used for the preparation of barcoded amplicons are indicated in Supplementary Tables S8 and S11, respectively. After purification using AMPure XP beads, the concentrations of barcoded amplicons were determined by using the Qubit dsDNA HS assay kit (Thermo Fisher Scientific; Cat. No.: Q32854) with the Qubit2.0 fluorometer. Finally, purified amplicons were pooled together in an equal molar ratio and then were subjected to Illumina MiSeq deep sequencing for obtaining 100 000 paired-end reads.
The analyses of deep sequencing data were carried out with the aid of CRISPResso2 software (33) after demultiplexing and adapter trimming of the paired-end MiSeq raw reads (R1 and R2 fastq files) with Cutadapt version 2.10 (34). CRISPResso2 was run in standard prime editing mode with multiple alleles option for quantifying the frequencies of intended prime-editing events and imprecise byproducts consisting of scaffold-derived sequences and indels. Prime editing activities in heterozygous hMSCs were calculated as: % (number of reads with the desired CTT insertion that do not contain indels)/(number of total aligned reads). The prime-edited reads in homozygous myoblasts, myotubes and RPE-Fucci cells were divided into three sub-types: composite CTT plus G edits, partial CTT edits and partial G edits. The frequencies of each individual outcome was calculated as: % (number of reads with CTT + G insertion that do not contain indels)/(number of total aligned reads); % (number of reads with CTT insertion that do not contain indels)/(number of total aligned reads); and % (number of reads with G mutation that do not contain indels)/(number of total aligned reads), respectively. Indel yields in all the experiments were calculated as: % (number of reads with indels that do not contain scaffold incorporated events)/(number of total aligned reads). The codes applied in each round of the CRISPResso2 analyses are available as Supplementary Information. Prime editing activities were also assessed through the analysis of Sanger sequencing chromatogram peaks using the EditR method (35). In brief, genomic DNA from AdVP-transduced cells was extracted at 3 days post-transduction with the DNeasy Blood & Tissue kit following the manufacturer's recommendations. Next, the target sites were amplified with GoTaq G2 Flexi DNA Polymerase (Promega; Cat. No.: M7805). The primer sequences, PCR mixture compositions and cycling conditions are specified in Supplementary Tables S12 and S13. Afterwards, the resulting amplicons were purified with the QIAEX II Gel Extraction Kit protocol and subjected to automated Sanger sequencing.
Lentiviral vector production and purification
The production of lentiviral vectors encoding pegRNAs and pegRNA/gRNA pairs was essentially carried out as follows. Twenty million HEK293T cells were seeded per 175-cm2 culture flask (Greiner Bio-One). The next day, 30-μg plasmid mixtures were diluted in 150 mM NaCl to a final volume of 1 ml. The plasmid mixtures consisted of a 2:1:1 molar ratio of each lentiviral transfer vector, the packaging construct psPAX2 (Addgene #12260) and the vesicular stomatitis virus glycoprotein-G-pseudotyping construct pLP/VSVG (Invitrogen). The lentiviral transfer vectors used were BF18_pLV.PURO.pegRNAHBB, BF19_pLV.PURO.pegRNA.gRNAHBB, BF20_pLV.BSD-EGFP.pegRNAEGFP, BF21_pLV.BSD-EGFP.pegRNA.gRNAEGFP and BF46_pLV.PURO.pegRNA.gRNAEGFP (Supplementary Information). In parallel, 99 μl of a 1 mg ml−1 PEI solution was diluted in 150 mM NaCl to a final volume of 1 ml. This PEI solution was subsequently added to each of the plasmid mixtures and, after vigorous homogenization in a vortex for approximately 10 s, a 15-min incubation period at RT ensued. Subsequently, the assembled PEI-DNA complexes were directly added to the medium of the HEK293T producer cells. After overnight incubation, the transfection media were replaced by 15 ml of regular culture medium. At 3 days post-transfection, the supernatants containing lentiviral vector particles were collected. Cellular debris were removed by centrifugation and filtration of the supernatants through 0.45-μm pore-sized HT Tuffryn membrane filters (Pall Life Sciences; Cat. No.: PN4184). The resulting clarified supernatants were then gently added onto 5-ml 20% (w/v) sucrose cushions in 35.8-ml polyallomer tubes (Beckman Coulter; Cat. No.: 326823). After ultracentrifugation (30 000 RPM for 2 h at 4°C) in an Optima LE-80K centrifuge (Beckman Coulter) using the SW32Ti rotor, pelleted vector particles were resuspended overnight at 4°C in 400 μl of ice-cold PBS supplemented with 1% BSA. The titers of the purified lentiviral vector stocks were determined by converting 1 ng of p24 antigen to 2500 lentiviral vector transducing units (36,37) after applying the materials and protocol of the RETROTEK HIV-1 p24 antigen ELISA kit (ZeptoMetrix, Cat. No.: 0801111).
Generation of cells stably expressing prime editing RNAs
The generation of HEK293T cells stably expressing HBB-specific and EGFP-specific pegRNAs and gRNAs was initiated by seeding HEK293T cells in regular growth medium at a density of 5 × 104 cells per well of 24-well plates. At ∼16–24 h after seeding, the cells were exposed to medium containing 8 μg ml–1 polybrene and lentiviral vectors at the indicated MOIs. After overnight incubation, the inocula were substituted by fresh culture medium. At 48 h post-transduction, the cells were transferred to 6-well plates containing regular growth medium supplemented with 2 μg ml–1 puromycin (Thermo Fisher Scientific; Cat. No.: A11138-03) or 5 μg ml–1 blasticidin (Thermo Fisher Scientific; Cat. No.: R21001). Parental mock-transduced cells served as negative controls during the drug selection procedure.
Statistical analyses
Statistical analyses were performed with the aid of GraphPad Prism software (version 8.0.1) on datasets derived from independent biological replicates. Statistical significances were calculated with the tests indicated in the various figure legends. P values lower than 0.05 were considered statistically significant.
RESULTS AND DISCUSSION
First- and second-generation adenoviral vectors (AdVs) are rendered replication-defective through the removal of only a few viral ORFs (17,38), which prevents exploiting the aforesaid full 36-kb packaging capacity of adenoviral capsids. Moreover, at high vector doses, so-called ‘leaky’ viral gene expression from the remaining ORFs contributes to cytotoxic effects in vitro and immune responses in vivo (37). Therefore, we focused on assembling third-generation AdVPs whose vector genomes contain exclusively recombinant DNA (Supplementary Figure S2). In particular, recombinant DNA encoding the PE2 fusion protein alone or together with a pegRNA. AdVPs expressing the mCherry reporter were also assembled to monitor transduction efficiencies. Another important aspect to consider concerns the cell tropism of AdVs. Specifically, AdVs with capsids from prototypic adenovirus type-5 enter cells after binding to the Coxsackie B and adenovirus receptor (CAR) (39,40). Yet, scientifically and therapeutically relevant human cell types, e.g., mesenchymal stem cells (hMSCs), muscle progenitor cells (myoblasts); and hematopoietic stem cells, all have a paucity of CAR on their plasmalemmas (41–45). Thus, to test prime editing in both CAR-positive and CAR-negative human cells, recombinant vector constructs were packaged in adenoviral capsids displaying type-50 fibers (Supplementary Figure S2), as these fibers have as primary receptor the ubiquitously expressed type I membrane protein CD46 (46).
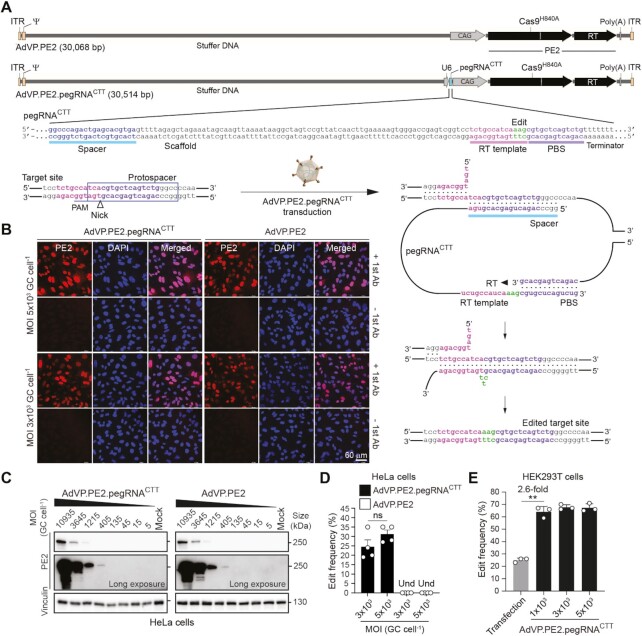
AdVP.PE2 and AdVP.PE2.pegRNACTT, encoding PE2 alone and PE2:pegRNACTT complexes, respectively (Figure 1A), were produced to similar high titers and contained structurally intact DNA with evidence neither for rearranged nor truncated species (Supplementary Figure S3). PE2:pegRNACTT complexes insert a CTT triplet at the long non-coding RNA gene LINC01509, earlier dubbed HEK293 site 3 (HEK3) (15). Transduction of HeLa cells with AdVP.PE2 and AdVP.PE2.pegRNACTT yielded widespread and vector dose-dependent PE2 expression (Figure 1B and C, respectively). Quantification of transduction levels showed that applying AdVPs at a multiplicity of infection (MOI) at or above 3 × 103 vector genome copies per cell (GC cell-1) led to transgene expression in virtually all HeLa cells (Supplementary Figure S4). Importantly, AdVP.PE2.pegRNACTT achieved all-in-one delivery of functional prime editing complexes as demonstrated by robust LINC01509 editing in HeLa and HEK293T cells (Figure 1D and E, respectively). Transfection of the latter cells with plasmids expressing PE2 and pegRNACTT, yielded prime editing frequencies of 25 ± 1.7% (Figure 1E). Transduction of the same easy-to-transfect cells with AdVP.PE2.pegRNACTT led to significantly higher prime editing frequencies (Figure 1E).
Figure 1.
Efficient prime editing in human cells through all-in-one AdVP delivery. (A) Schematics of AdVP genomes encoding PE elements. AdVP.PE2 and AdVP.PE2.pegRNACTT express only PE2 and PE2:pegRNACTT complexes, respectively. The pegRNACTT installs a CTT insertion at the long non-coding RNA gene LINC01509 once coupled to PE2. Hybrid CAG and human U6 promoters drive PE2 and pegRNACTT synthesis, respectively. The pegRNACTT coding sequence is depicted with the respective components highlighted, i.e., sequence-specific spacer, scaffold, primer binding site (PBS) and RT template with embedded edit. A schematics of target site engagement and editing by a PE2:pegRNACTT complex is equally shown. PAM, protospacer adjacent motif (NGG). (B and C) Analyses of PE2 expression in transformed cells. HeLa cells were transduced with AdVP.PE2.pegRNACTT or AdVP.PE2 at the indicated multiplicities-of-infection (MOI). PE2 was detected by immunofluorescence microscopy (panel B) and western blotting (panel C) at 2 days post-transduction. Cas9- and vinculin-specific antibodies served for detecting target (PE2) and loading control proteins, respectively. (D and E) Detection of prime editing in transformed cells. Cervical carcinoma HeLa cells and human embryonic kidney HEK293T cells were transduced with the indicated AdVPs at different MOI (panels D and E, respectively). At 3 days post-transduction, prime editing activities were assessed by target site genotyping assays. HEK293T cells were also co-transfected with plasmids encoding PE2 and pegRNACTT. GC cell–1, vector genome copies per cell. Und, undetected. Graphs in panels D and E present mean ± s.e.m. (n = 4) and mean ± s.d. (n = 3), respectively. Statistical significance between the indicated datasets was assessed with two-tailed Student's t tests; **0.001 < P< 0.01; P > 0.05 was considered non-significant (ns).
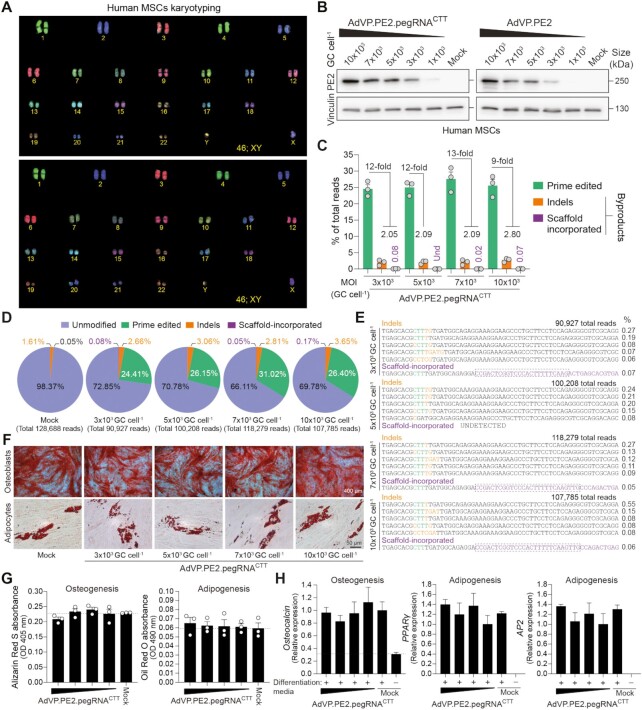
In the foundational study, prime editing frequencies were substantially higher in HEK293T cells than in other cell lines tested (15). Furthermore, the less efficient, yet simpler and less mutagenic PE2 system, was not evaluated in cells other than HEK293T cells (15). Physical and chemical transfection methods permit introducing genome editing reagents into cells in a transient fashion. However, reaching maximum delivery efficiencies without triggering substantial cytotoxic effects is challenging, especially in settings involving non-transformed cells. To start investigating the performance of prime editing in difficult-to-transfect human cells under conditions in which the attendant tools are not limiting, we transduced primary hMSCs (Figure 2A) with AdVP.PE2.pegRNACTT (Figure 1A) and AdVP.mCherry (Supplementary Figure S4A). A vector dose-dependent build-up of PE2 and mCherry was readily detected through western blotting and direct fluorescence microscopy analyses (Figure 2B and Supplementary Figure S5A, respectively). Transduction efficiencies, as determined by flow cytometry, varied from a minimum of 92.4 ± 2.2% to a maximum of 99.2 ± 0.8% (Supplementary Figure S5B). Importantly, AdVP.PE2.pegRNACTT-transduced hMSCs contained the intended edits in up to 31% (27.6 ± 3.8%) of target alleles with limited genomic incorporation of complex small insertions and deletions (indels) and pegRNA scaffold sequences (Figure 2C–E). Indels were found at higher frequencies than scaffold footprints and, together, they reached a combined maximum of 2.05% and 2.80% in hMSCs exposed to the lowest and highest vector concentrations, respectively (Figure 2C). Moreover, hMSCs retained their differentiation capacity regardless of the vector dose applied (Figure 2F), as determined by quantification of osteogenic and adipogenic differentiation using colorimetric and RT-qPCR assays (Figure 2G and H, respectively). Taken together, these data indicate that AdVP-based prime editing achieves efficient and precise genetic modification of target alleles in primary hMSCs.
Figure 2.
Efficient prime editing in primary human mesenchymal stem cells through all-in-one AdVP delivery. (A) Karyotyping of hMSCs. COBRA-FISH analysis was used for confirming the normal diploid status (46;XY) of primary hMSCs. (B) Analyses of PE2 expression in primary hMSCs. hMSCs were transduced with AdVP.PE2.pegRNACTT or AdVP.PE2 at the indicated MOI. At 2 days post-transduction, PE2 expression was assessed by western blotting. Cas9- and vinculin-specific antibodies detected target and loading control proteins, respectively. (C) Relationship between prime edited alleles and byproduct variants in AdVP.PE2.pegRNACTT-transduced hMSCs. hMSCs were transduced with the indicated AdVPs at different MOI. At 3 days post-transduction, prime editing frequencies and unwarranted byproducts (i.e. indels and scaffold-derived insertions), were determined through CRISPResso2 analysis. Byproducts consist of small insertions and deletions (indels; orange bars) plus insertions derived from reverse transcription into the pegRNA scaffold (violet bars). Graph presents mean ± s.e.m. of three biological replicates; Und, undetected. (D) Characterization of prime editing in hMSCs. Pie chart parsing the frequencies of unmodified and modified alleles resulting from a transduction experiment in hMSCs. (E) Characterization of prime editing byproducts in hMSCs. Sequences and frequencies of the most frequent alleles bearing indels and pegRNA scaffold-derived insertions from a transduction experiment in hMSCs are presented. (F) Differentiation of prime-edited hMSCs. Differentiation capacity of mock- and vector-transduced hMSCs was established after their exposure to defined culture conditions. Osteoblasts and adipocytes were identified by Alizarin Red S and Oil Red O staining, respectively. (G) Quantification of osteogenic and adipogenic differentiation through Alizarin Red S and Oil Red O colorimetry, respectively. (H) Quantification of osteogenic and adipogenic differentiation via RT-qPCR targeting the indicated lineage-specific marker transcripts. Data are plotted as mean ± s.e.m. of three technical replicates. In all hMSCs transduction experiments MOIs ranged from 3 × 103 through 10 × 103 genome copies per cell (GC cell–1).
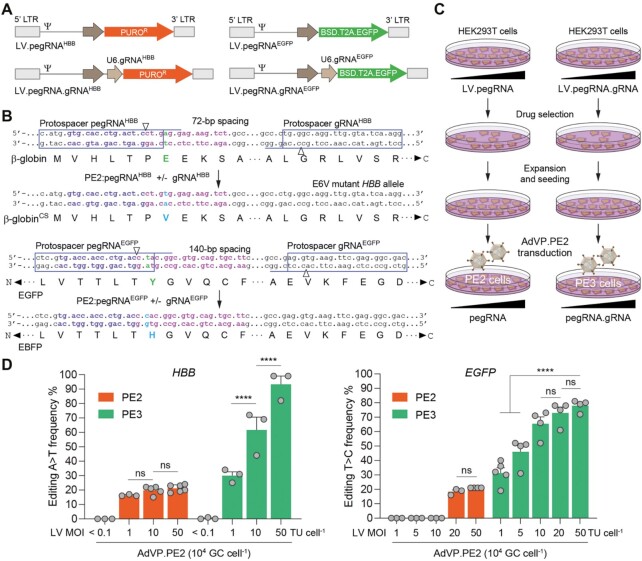
Interestingly, regardless of target cell type, vector dose-dependent accumulation of PE2 components was not accompanied by a measurable increase in prime editing frequencies (Figures 1D, E and 2C). To investigate whether this marked nonlinear dose-response and wide DNA editing plateau is specific to pegRNACTT or to the PE2 system, we compared PE2 and PE3 systems in HEK293T cells containing individual pegRNAs and pegRNA/gRNA pairs, respectively, delivered through lentiviral vector transductions at various MOI (Figure 3A). Cells expressing correspondingly different amounts of these exogenous small RNAs, designed for installing the pathogenic E6V β-globin mutation at HBB (15) or for changing the EGFP fluorophore to that of EBFP (Figure 3B), were subsequently transduced at high MOI with AdVP.PE2 in order to guarantee uniform PE2 protein availability (Figure 3C). A direct correlation between exogenous RNA amounts and prime editing activities was most obvious in cells containing PE3 components with 93% and 78% of HBB and EGFP sequences on average modified, respectively (Figure 3D). These data indicate that, besides being more efficient than the PE2 system on a per dose basis, the PE3 system involving gRNA-directed nicking of the non-edited DNA strand (Supplementary Figure S1B), can readily overcome the maximal activity plateau reached by PE2:pegRNA complexes (Figures 1D, E, 2C and 3D). Moreover, populations of cells containing single- to low-copy numbers of pegRNA expression units had negligible frequencies of edited alleles (Figure 3D). This finding might have implications for genome-wide screens based on single-copy chromosomal integration of pegRNA libraries in test cell populations. Finally, AdVP transduction experiments in EGFP-expressing HEK293T cells followed by reporter-directed flow cytometry confirmed the emergence of cells with EBFP-positive and EGFP-negative phenotypes expected from the activities of the prime editing reagents herein designed for in cellula fluorophore conversion (Supplementary Figure S6).
Figure 3.
Differential prime editing dose-responses of PE2 versus PE3 systems. (A) Lentiviral vectors encoding PE2 and PE3 small RNAs. PE2:pegRNAHBB complexes alone (PE2 system) or together with pairing gRNAHBB (PE3 system) install a A > T transversion at HBB whose β-globin E6V product causes sickle cell (SC) disease. PE2:pegRNAEGFP complexes alone (PE2 system) or together with pairing gRNAEGFP (PE3 system) install a T > C transition, changing the EGFP fluorophore to that of EBFP. HBB-targeting lentiviral vectors code for puromycin N-acetyltransferase and EGFP-targeting lentiviral vectors code for blasticidin S deaminase and EGFP via a BSD.T2A.EGFP expression unit. LTR and Ψ, long terminal repeat and packaging signal elements, respectively. (B) Schematics of HBB and EGFP target sites before and after prime editing. Protospacers and protospacer adjacent motifs of pegRNAs and gRNAs are boxed and underlined, respectively. Genomic sequences with complementarity to pegRNA primer binding sites and reverse transcriptase templates are marked in violet and magenta, respectively. (C) Diagram of the experimental set-up. HEK293T cells stably expressing different amounts of pegRNAs or pegRNA/gRNA pairs were generated by lentiviral vector transduction and puromycin or blasticidin selection. The resulting cell populations were then transduced with AdVP.PE2 at 104 GC cell–1. (D) Comparing PE2 and PE3 editing efficiencies. PE2- and PE3-induced editing frequencies at endogenous HBB and recombinant EGFP alleles were determined at three days after AdVP.PE2 transductions. Data shown represent mean ± s.e.m. of at least three independent biological replicates. Significance between the indicated datasets was calculated with two-way ANOVA followed by Sidak's test for multiple comparisons; ****P < 0.0001; P > 0.05 was considered non-significant (ns).
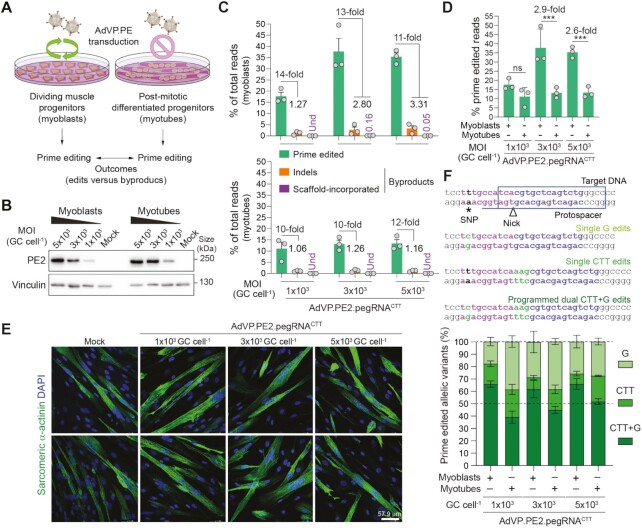
By capitalizing on the capsid-mediated cell nucleus entry mechanisms of adenoviruses (47,48), recombinant vector forms are proficient in transducing dividing and fully post-mitotic cells (17–20,38). Therefore, we next exploited this evolution fine-tuned feature to investigate whether cell replication influences prime editing activities and outcomes. To start addressing these questions, we used human myoblasts and differentiated post-mitotic myotubes as an experimental model (Figure 4A). Irreversible cell cycle withdrawal is a defining feature of the skeletal muscle differentiation program whose regulation involves the MyoD1 family of transcription factors (49). We have confirmed myogenic differentiation of dividing myoblasts into syncytial non-dividing myotubes through combined immunodetection of cell cycling- and late muscle-specific markers (Supplementary Figure S7A and B). In addition, the presence of actively dividing cells in myoblast cultures as well as their absence in post-mitotic myotube cultures was independently ascertained through a proliferating assay comprising the incorporation of the thymidine analogue EdU in newly synthesized DNA chains (Supplementary Figure S7B and C).
Figure 4.
Assessing the impact of cellular replication on prime editing performance. (A) Experimental set-up for characterizing prime editing in dividing versus post-mitotic cells. (B) Analyses of PE2 expression in muscle cells. Muscle progenitor cells were transduced before and after differentiation with AdVP.PE2.pegRNACTT at the specified MOI and, 2 days later, PE2 expression levels were assessed by western blotting. Cas9- and vinculin-specific antibodies detected target (PE2) and loading control proteins, respectively. (C) Characterization of prime editing in dividing versus post-mitotic cells. The frequencies of prime edited and byproduct alleles in myoblasts and myotubes (top and bottom graphs, respectively) exposed to different MOI of AdVP.PE2.pegRNACTT, were determined through CRISPResso2 analysis at 3 days post-transduction. Byproducts consist of small deletions and insertions (indels; orange bars) and pegRNA scaffold-derived insertions (violet bars). Graph presents mean ± s.e.m. of three biological replicates; Und, undetected. (D) Prime editing frequencies in mitotic versus post-mitotic cells. Aggregated prime editing frequencies indicated in panel C highlighting differences in PE2:pgRNACTT activity in myoblasts versus myotubes. Bars and error bars correspond to mean and s.d, respectively. Significance between datasets was calculated with two-way ANOVA followed by Sidak's test for multiple comparisons; ***0.0001 < P < 0.001; P > 0.05 was considered non-significant (ns). (E) Differentiation of AdVP.PE2.pegRNACTT-treated myoblasts. The differentiation capacity of vector-transduced myoblasts was ascertained by immunofluorescence microscopy analysis for the late muscle-specific marker sarcomeric α-actinin after incubating the cells in low-mitogen medium. Nuclei in syncytial myotubes were identified by DAPI staining. Mock-transduced myoblasts served as controls. Two representative micrographs for each experimental condition are shown. (F) Parsing of prime-edited allele variants resulting from a composite prime editing design. Myoblasts and RPE-Fucci cells are homozygous for a SNP in the region complementary to the pegRNACTT RT template permitting assessing composite (CTT + G) versus single (CTT or G) edits instructed by PE2:pegRNACTT complexes. Genomic sequences before and after the delivery of PE2:pegRNACTT complexes into cells containing a SNP in the region complementary to the RT template (magenta nucleotides), are depicted (top panel). Prime editing outcomes instructed by pegRNACTT correspond to alleles containing A > G substitutions, CTT insertions or both modifications. Replicating myoblasts and post-mitotic myotubes were transduced with the all-in-one vector AdVP.PE2.pegRNACTT. Discrimination and quantification of the different target alleles was performed via next-generation deep sequencing analysis on genomic DNA isolated at 3 days post-transduction (bottom panel). Data are plotted as mean ± s.e.m. of three biological replicates.
Similarly to the transduction of HeLa cells and hMSCs (Supplementary Figures S4 and S5, respectively), the transduction of myoblasts with AdVPs was highly efficient (Supplementary Figure S8). In addition, exposing skeletal muscle cells to AdVP.PE2.pegRNACTT at the myoblast and myotube differentiation stages, yielded similar amounts of PE2 protein on a per vector dose basis (Figure 4B). Noticeably, the frequencies of precisely edited alleles were highest (up to 49.6%) in myoblasts (Figure 4C and D), with these muscle progenitors retaining their cell-cycle exit and differentiation capabilities once exposed to poor-mitogen conditions (i.e. no or low serum concentrations) that normally trigger myogenic cell maturation (Figure 4E and Supplementary Figure S9). In comparison with target alleles containing exclusively the programmed edits, those bearing complex indels and pegRNA scaffold sequences were rare in that the frequencies of these variants combined ranged from 1.3% to 3.3% of the total allelic forms found (Figure 4C). In contrast to the results obtained in proliferating myoblasts, in post-mitotic myotubes, alleles exhibiting pegRNA scaffold sequences were not detected (Figure 4C) possibly due to the lower prime editing activity in these cells or their non-replicating status (Supplementary Figure S7).
The ability to instruct different genomic changes at once contributes to the versatility of the prime editing system. In the myoblast-to-myotube cellular differentiation system, an homozygous single nucleotide polymorphism (SNP) present in the region complementary to the pegRNACTT RT template was harnessed to investigate the proportions between edited alleles comprising CTT insertions and A > G substitutions (CTT + G) and those containing only one of the two modifications, i.e., CTT or G (Supplementary Figure S10). A striking difference between prime editing outcomes in myoblasts and myotubes was the prevalence of the instructed CTT + G composite edits in the former, i.e., over 50% of the edited reads (Figure 4F and Supplementary Figure S11). This data suggests that the cellular context has a bearing on the performance of prime editing aiming at simultaneous introduction of different nucleotide changes and extends earlier research showing that the rates of partial edits increase with the distance between the two editing positions and between these positions and the nicking site (50).
To further exploit AdVPs as probes for investigating the relationship between cellular replication and prime editing, we next used retinal pigment epithelial cells (RPE-1) endowed with the Fucci cell-cycle sensors (RPE-Fucci) (Figure 5A) (26,28,29). In contrast to most transformed cells, non-transformed RPE-1 cells have functional G1 and G2 checkpoints (28,29). Most importantly, the Fucci system allowed for flow cytometric quantification of cell fractions in G1, G2/M and early S phases in cultures treated with serum concentrations spanning from mitogen starvation to regular conditions. During the cumulative tracing period, a clear serum concentration-dependent increase in the proportion of cells in S and G2/M was observed (Figure 5B and Supplementary Figure S12A). This observation was independently confirmed by assessing throughout the same cumulative period, the DNA content profiles of RPE-Fucci cells exposed to different serum concentrations (Supplementary Figure S13).
Figure 5.
Characterization of prime editing in rapidly versus slowly dividing cell populations. (A) Experimental set-up for characterizing prime editing in rapidly versus slowly dividing cell populations. RPE-Fucci cell cultures containing varying proportions of cycling and non-cycling cells were established by applying a serum gradient. Fucci reporters in these cells trace cell fractions in G1, early S or G2/M phases, permitting the monitoring of cell division-to-prime editing rates upon AdVP.PE2.pegRNACTT transduction. (B) Tracking cell cycling and prime editing activities in RPE-Fucci cell cultures. Cell cycle analysis was done at the indicated timepoints on RPE-1 cells exposed to various FBS concentrations by flow cytometry. Parallel cultures of RPE-Fucci cells were transduced with AdVP.PE2.pegRNACTT at 12 hours after FBS treatments initiation. (C) Analysis of PE2 expression in RPE-1 cells. PE2 expression levels were determined by western blotting using Cas9- and vinculin-specific antibodies for detecting PE2 and loading control proteins, respectively. (D) Quantification of prime editing outcomes in RPE-1 cells. Frequencies of prime editing and prime editing collateral events were measured by CRISPresso2 analysis at 60 hours post-transduction. Datapoints derive from two independent experiments carried out throughout different days. Bars represent mean ± s.d. of prime editing activities in RPE-Fucci cells exposed to serum starvation (0, 0.1, and 0.5% FBS) or regular culture conditions (5 and 10% FBS). Unpaired two-tailed Student's t test ****P < 0.0001. (E) Parsing of prime-edited allele variants resulting from the composite prime editing design. Statistical significance was assesed by Student's t tests; *0.01 < P < 0.05; ****P < 0.0001. (F) Relationship between prime edited alleles and byproduct variants in AdVP-transduced RPE-Fucci cells. Und, undetected.
AdVP.PE2.pegRNACTT transduction of RPE-Fucci cells treated with these various serum concentrations contained similar amounts of PE2 (Figure 5C). Significantly, prime editing efficiencies were superior in RPE-Fucci populations with the highest frequencies of actively cycling cells (Figure 5D). Interestingly, within the fraction of edited alleles, the frequencies of programmed CTT + G alleles were higher than those corresponding to partially edited CTT or G alleles, especially so in the most actively replicating cell populations (Figure 5E and Supplementary Figure S12B). The direct correlation between RPE-Fucci cell-cycling activities and composite CTT + G allele frequencies, is in line with the data obtained after transduction of myoblasts and post-mitotic myotubes with AdVPs as a higher purity of the programmed CTT + G edits was observed in the proliferating myoblast populations (Figure 4F). Equally consistent with the experiments in muscle cells, complex indels and genomic insertions mapping to pegRNA sequences were rarer in cultures enriched in G1-arrested cells (Figure 5F and Supplementary Figure S12B). In general, the latter prime editing byproducts were the least frequent and were prevalently found in cultures containing high frequencies of mitotically active cells (Figures 2C–E, 4C, 5F and Supplementary Figures S11 and S12B).
Taken together, our data demonstrate that the replication status of target cells influences prime editing activities and outcomes. Edited strand-templated DNA synthesis (Supplementary Figure S14) and/or engagement of cellular factors involved in replication-dependent DNA repair processes might contribute to the herein identified correlation between replication and prime editing activities. Prime editing and non-LTR element retrotransposition share striking similarities, which include genomic DNA nicking followed by target site-primed reverse transcription (51). This fact coupled to the copious abundance of non-LTR retrotransposons in mammalian genomes makes it equally enticing to speculate the participation of conserved retrotransposition-associated processes during prime editing.
Recently, dual AAV strategies involving co-transducing target cells with two split AAV vectors, each encoding N- or C-terminal truncated prime editors, were applied for in situ reconstruction of full-length proteins through vector genome recombination and RNA trans-splicing (52–54) or intein-mediated protein trans-splicing (55,56). These studies yielded important proof-of-concepts for in vivo disease modelling and mutation correction by prime editing. In particular, a dual AAV protein trans-splicing PE3 system corrected 0.6%, 2.3% and 3.1% of defective alleles in livers of alpha-1 antitrypsin deficiency mice at 2, 6 and 10 weeks post-injection, respectively (57). Another study reports that, in the absence of positive selection for gene-corrected cells, dual AAV RNA trans-splicing PE2 and PE3 systems did not lead to detectable correction of defective alleles in livers of hereditary tyrosinemia type 1 mice (58). Notably, however, using the same AAV platform, gene correction levels ranging from 4.1% to 7.4% were measured in RPE65-associated Leber congenital amaurosis mice after sub-retinal injections (58). In another recent study, a dual AAV protein trans-splicing system installed a G > T transversion within Dnmt1 at a frequency of 1.71 ± 1.35% 6 weeks after sub-retinal injections in mice (59). Notwithstanding these important proof-of-concepts, dual AAV designs are complex and relatively inefficient as they operate through only partially controllable intermolecular recombination events for assembling the proper full-length product from the total pool of truncated proteins expressed in co-transduced cells. There is, therefore, a pressing need for additional platforms capable of delivering into different human cell types and animal models emerging RNA-guided gene targeting systems based on large macromolecular complexes, as these systems are starting to offer the prospect for unprecedented genome editing precision in fundamental research, disease modelling and treatment (16).
In this work, besides identifying a direct correlation between prime editing and cell cycling activities, we demonstrate that the AdVP platform is a suitable option for all-in-one delivery of full-length prime editor proteins and cognate RNA molecules into target cells independently of their transformation and replication statuses. Hence this platform is expected to become a valuable addition for investigations on the potential and limitations of prime editing principles and reagents in a broad range of mammalian cells in vitro and in vivo.
DATA AVAILABILITY
All data gathered for and analysed in this study are included in the article and supplementary files. Additional raw datasets that support the findings of this work are available upon request. The flow cytometry datasets were deposited at FlowRepository under accession codes FR-FCM-Z3RU, FR-FCM-Z3RV, FR-FCM-Z4GP, FR-FCM-Z4GQ and FR-FCM-Z4GX. The generated deep sequencing library reads were deposited at the NCBI Sequence Read Archive (SRA) database under BioProject ID PRJNA729964.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Martijn Rabelink and Steve Cramer for performing p24gag ELISA titrations of lentiviral vector particles and assistance with the research logistics, respectively. The authors also thank Karoly Szuhai for the FISH karyotyping, Ignazio Maggio for input on the amplicon deep sequencing procedures, Maarten van Dinther and Midory Thorikay for assistance with the hMSC differentiation assays, Chuannan Fan for his help with the Ki-67 and dystrophin western blot assays (Department of Cell and Chemical Biology, LUMC), and Vincent Mouly from the Center for Research in Myology (Sorbonne University, Paris, France) for providing the human myoblasts. Finally, the authors are grateful to all laboratory members for their support.
Contributor Information
Qian Wang, Department of Cell and Chemical Biology, Leiden University Medical Center, Einthovenweg 20, 2333 ZC Leiden, The Netherlands.
Jin Liu, Department of Cell and Chemical Biology, Leiden University Medical Center, Einthovenweg 20, 2333 ZC Leiden, The Netherlands.
Josephine M Janssen, Department of Cell and Chemical Biology, Leiden University Medical Center, Einthovenweg 20, 2333 ZC Leiden, The Netherlands.
Francesca Tasca, Department of Cell and Chemical Biology, Leiden University Medical Center, Einthovenweg 20, 2333 ZC Leiden, The Netherlands.
Hailiang Mei, Department of Biomedical Data Sciences, Sequencing Analysis Support Core, Leiden University Medical Center, Einthovenweg 20, 2333 ZC Leiden, The Netherlands.
Manuel A F V Gonçalves, Department of Cell and Chemical Biology, Leiden University Medical Center, Einthovenweg 20, 2333 ZC Leiden, The Netherlands.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Prinses Beatrix Spierfonds [W.OR16-13, W.OR21-01]; Dutch Duchenne Parent Project [17.012]; China Scholarship Council–Leiden University Joint Scholarship programme (to Q.W.). This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 765269 (to F.T.). Funding for open access charge: Prinses Beatrix Spierfonds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gasiunas G., Barrangou R., Horvath P., Siksnys V.. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cradick T.J., Fine E.J., Antico C.J., Bao G.. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013; 41:9584–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D.. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013; 31:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O.et al.. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R.. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Bioatechnol. 2013; 31:839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin Y., Cradick T.J., Brown M.T., Deshmukh H., Ranjan P., Sarode N., Wile B.M., Vertino P.M., Stewart F.J., Bao G.. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014; 42:7473–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Q., Liu J., Janssen J.M., Le Bouteiller M., Frock R.L., Gonçalves M.A.F.V. Precise and broad scope genome editing based on high-specificity Cas9 nickases. Nucleic Acids Res. 2021; 49:1173–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Turchiano G., Andrieux G., Klermund J., Blattner G., Pennucci V., El Gaz M., Monaco G., Poddar S., Mussolino C., Cornu T.I.et al.. Quantitative evaluation of chromosomal rearrangements in gene-edited human stem cells by CAST-Seq. Cell Stem Cell. 2021; 28:1136–1147. [DOI] [PubMed] [Google Scholar]
- 10. Frock R.L., Hu J., Meyers R.M., Ho Y.J., Kii E., Alt F.W.. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015; 33:179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kosicki M., Tomberg K., Bradley A.. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018; 36:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen X., Janssen J.M., Liu J., Maggio I., Jong A.E.J., Mikkers H.M.M., Gonçalves M.A.F.V.. In trans paired nicking triggers seamless genome editing without double-stranded DNA cutting. Nat. Commun. 2017; 8:657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakajima K., Zhou Y., Tomita A., Hirade Y., Gurumurthy C.B., Nakada S.. Precise and efficient nucleotide substitution near genomic nick via noncanonical homology-directed repair. Genome Res. 2018; 28:223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hyodo T., Rahman M.L., Karnan S., Ito T., Toyoda A., Ota A., Wahiduzzaman M., Tsuzuki S., Okada Y., Hosokawa Y.et al.. Tandem paired nicking promotes precise genome editing with scarce interference by p53. Cell Rep. 2020; 30:1195–1207. [DOI] [PubMed] [Google Scholar]
- 15. Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A.et al.. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019; 576:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anzalone A.V., Koblan L.W., Liu D.R.. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020; 38:824–844. [DOI] [PubMed] [Google Scholar]
- 17. Chen X., Gonçalves M.A.F.V.. Engineered viruses as genome editing devices. Mol. Ther. 2016; 24:447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jager L., Hausl M.A., Rauschhuber C., Wolf N.M., Kay M.A., Ehrhardt A.. A rapid protocol for construction and production of high-capacity adenoviral vectors. Nat. Protoc. 2009; 4:547–564. [DOI] [PubMed] [Google Scholar]
- 19. Tasca F., Wang Q., Goncalves M. Adenoviral vectors meet gene editing: a rising partnership for the genomic engineering of human stem cells and their progeny. Cells. 2020; 9:953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ricobaraza A., Gonzalez-Aparicio M., Mora-Jimenez L., Lumbreras S., Hernandez-Alcoceba R.. High-capacity adenoviral vectors: expanding the scope of gene therapy. Int. J. Mol. Sci. 2020; 21:3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holkers M., Maggio I., Henriques S.F., Janssen J.M., Cathomen T., Gonçalves M.A.F.V.. Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat. Methods. 2014; 11:1051–1057. [DOI] [PubMed] [Google Scholar]
- 22. Brescia M., Janssen J.M., Liu J., Gonçalves M.. High-capacity adenoviral vectors permit robust and versatile testing of DMD gene repair tools and strategies in human cells. Cells. 2020; 9:869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen X., Rinsma M., Janssen J.M., Liu J., Maggio I., Gonçalves M.A.F.V.. Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res. 2016; 44:6482–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gonçalves M.A.F.V., Swildens J., Holkers M., Narain A., van Nierop G.P., van de Watering M.J., Knaän-Shanzer S., de Vries AA., Gonçalves M.A.et al.. Genetic complementation of human muscle cells via directed stem cell fusion. Mol. Ther. 2008; 16:741–748. [DOI] [PubMed] [Google Scholar]
- 25. Gonçalves M.A.F.V., Janssen J.M., Nguyen Q.G., Athanasopoulos T., Hauschka S.D., Dickson G., de Vries A.A.. Transcription factor rational design improves directed differentiation of human mesenchymal stem cells into skeletal myocytes. Mol. Ther. 2011; 19:1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H.et al.. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008; 132:487–498. [DOI] [PubMed] [Google Scholar]
- 27. Mamchaoui K., Trollet C., Bigot A., Negroni E., Chaouch S., Wolff A., Kandalla P.K., Marie S., Di Santo J., St Guily J.L.. Immortalized pathological human myoblasts: towards a universal tool for the study of neuromuscular disorders. Skelet Muscle. 2011; 1:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krenning L., Feringa F.M., Shaltiel I.A., van den Berg J., Medema R.H.. Transient activation of p53 in G2 phase is sufficient to induce senescence. Mol. Cell. 2014; 55:59–72. [DOI] [PubMed] [Google Scholar]
- 29. Shaltiel I.A., Aprelia M., Saurin A.T., Chowdhury D., Kops G.J., Voest E.E., Medema R.H.. Distinct phosphatases antagonize the p53 response in different phases of the cell cycle. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:7313–7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janssen J.M., Liu J., Skokan J., Goncalves M.A., de Vries A.A.. Development of an AdEasy-based system to produce first- and second-generation adenoviral vectors with tropism for CAR- or CD46-positive cells. J. Gene. Med. 2013; 15:1–11. [DOI] [PubMed] [Google Scholar]
- 31. Havenga M.J., Holterman L., Melis I., Smits S., Kaspers J., Heemskerk E., van der Vlugt R., Koldijk M., Schouten G.J., Hateboer G.et al.. Serum-free transient protein production system based on adenoviral vector and PER.C6 technology: high yield and preserved bioactivity. Biotechnol. Bioeng. 2008; 100:273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Holkers M., Cathomen T., Goncalves M.A.F.V.. Construction and characterization of adenoviral vectors for the delivery of TALENs into human cells. Methods. 2014; 69:179–187. [DOI] [PubMed] [Google Scholar]
- 33. Clement K., Rees H., Canver M.C., Gehrke J.M., Farouni R., Hsu J.Y., Cole M.A., Liu D.R., Joung J.K., Bauer D.E.et al.. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019; 37:224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011; 17:10–12. [Google Scholar]
- 35. Kluesner M.G., Nedveck D.A., Lahr W.S., Garbe J.R., Abrahante J.E., Webber B.R., Moriarity B.S.. EditR: a method to quantify base editing from Sanger sequencing. CRISPR J. 2018; 1:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dautzenberg I.J.C., Rabelink M., Hoeben R.C.. The stability of envelope-pseudotyped lentiviral vectors. Gene Ther. 2021; 28:89–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zufferey R., Dull T., Mandel R.J., Bukovsky A., Quiroz D., Naldini L., Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 1998; 72:9873–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gonçalves M.A.F.V., de Vries A.A.. Adenovirus: from foe to friend. Rev. Med. Virol. 2006; 16:167–186. [DOI] [PubMed] [Google Scholar]
- 39. Bergelson J.M., Cunningham J.A., Droguett G., Kurt-Jones E.A., Krithivas A., Hong J.S., Horwitz M.S., Crowell R.L., Finberg R.W.. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997; 275:1320–1323. [DOI] [PubMed] [Google Scholar]
- 40. Tomko R.P., Xu R., Philipson L.. HCAR and MCAR: The human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:3352–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shayakhmetov D.M., Papayannopoulou T., Stamatoyannopoulos G., Lieber A.. Efficient gene transfer into human CD34(+) cells by a retargeted adenovirus vector. J. Virol. 2000; 74:2567–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Knaän-Shanzer S., van der Velde I., Havenga M.J., Lemckert A.A., de Vries A.A., Valerio D.. Highly efficient targeted transduction of undifferentiated human hematopoietic cells by adenoviral vectors displaying fiber knobs of subgroup B. Hum. Gene Ther. 2001; 12:1989–2005. [DOI] [PubMed] [Google Scholar]
- 43. Knaän-Shanzer S., van de Watering M.J., van der Velde I., Gonçalves M.A., Valerio D., de Vries A.A.. Endowing human adenovirus serotype 5 vectors with fiber domains of species B greatly enhances gene transfer into human mesenchymal stem cells. Stem Cells. 2005; 23:1598–1607. [DOI] [PubMed] [Google Scholar]
- 44. Gonçalves M.A., de Vries A.A., Holkers M., van de Watering M.J., van der Velde I., van Nierop G.P., Valerio D., Knaän-Shanzer S.. Human mesenchymal stem cells ectopically expressing full-length dystrophin can complement Duchenne muscular dystrophy myotubes by cell fusion. Hum. Mol. Genet. 2006; 15:213–221. [DOI] [PubMed] [Google Scholar]
- 45. Gonçalves M.A., Holkers M., Cudré-Mauroux C., van Nierop G.P., Knaän-Shanzer S., van Der Velde I., Valerio D., de Vries A.A.. Transduction of myogenic cells by retargeted dual high-capacity hybrid viral vectors: robust dystrophin synthesis in duchenne muscular dystrophy muscle cells. Mol. Ther. 2006; 13:976–986. [DOI] [PubMed] [Google Scholar]
- 46. Gaggar A., Shayakhmetov D.M., Lieber A.. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 2003; 9:1408–1412. [DOI] [PubMed] [Google Scholar]
- 47. Greber U.F., Flatt J.W.. Adenovirus entry: from infection to immunity. Annu. Rev. Virol. 2019; 6:177–197. [DOI] [PubMed] [Google Scholar]
- 48. Greber U.F., Gomez-Gonzalez A.. Adenovirus - a blueprint for gene delivery. Curr. Opin. Virol. 2021; 48:49–56. [DOI] [PubMed] [Google Scholar]
- 49. Kitzmann M., Fernandez A.. Crosstalk between cell cycle regulators and the myogenic factor MyoD in skeletal myoblasts. Cell. Mol. Life Sci. 2001; 58:571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim H.K., Yu G., Park J., Min S., Lee S., Yoon S., Kim H.H.. Predicting the efficiency of prime editing guide RNAs in human cells. Nat. Biotechnol. 2021; 39:198–206. [DOI] [PubMed] [Google Scholar]
- 51. Beck C.R., Garcia-Perez J.L., Badge R.M., Moran J.V.. LINE-1 elements in structural variation and disease. Annu. Rev. Genomics Hum. Genet. 2011; 12:187–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nakai H., Storm T.A., Kay M.A.. Increasing the size of rAAV-mediated expression cassettes in vivo by intermolecular joining of two complementary vectors. Nat. Biotechnol. 2000; 18:527–532. [DOI] [PubMed] [Google Scholar]
- 53. Sun L., Li J., Xiao X.. Overcoming adeno-associated virus vector size limitation through viral DNA heterodimerization. Nat. Med. 2000; 6:599–602. [DOI] [PubMed] [Google Scholar]
- 54. Yan Z., Zhang Y., Duan D., Engelhardt J.F.. Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6716–6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li J., Sun W., Wang B., Xiao X., Liu X.Q.. Protein trans-splicing as a means for viral vector-mediated in vivo gene therapy. Hum. Gene Ther. 2008; 19:958–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Truong D.J., Kühner K., Kühn R., Werfel S., Engelhardt S., Wurst W., Ortiz O.. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic. Acids. Res. 2015; 43:6450–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu P., Liang S.Q., Zheng C., Mintzer E., Zhao Y.G., Ponnienselvan K., Mir A., Sontheimer E.J., Gao G., Flotte T.R.et al.. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat. Commun. 2021; 9:2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jang H., Jo D.H., Cho C.S., Shin J.H., Seo J.H., Yu G., Gopalappa R., Kim D., Cho S.R., Kim J.H.et al.. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat. Biomed. Eng. 2021; 10.1038/s41551-021-00788-9. [DOI] [PubMed] [Google Scholar]
- 59. Zhi S., Chen Y., Wu G., Wen J., Wu J., Liu Q., Li Y., Kang R., Hu S., Wang J.et al.. Dual-AAV delivering split prime editor system for in vivo genome editing. Mol. Ther. 2021; 10.1016/j.ymthe.2021.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data gathered for and analysed in this study are included in the article and supplementary files. Additional raw datasets that support the findings of this work are available upon request. The flow cytometry datasets were deposited at FlowRepository under accession codes FR-FCM-Z3RU, FR-FCM-Z3RV, FR-FCM-Z4GP, FR-FCM-Z4GQ and FR-FCM-Z4GX. The generated deep sequencing library reads were deposited at the NCBI Sequence Read Archive (SRA) database under BioProject ID PRJNA729964.