Abstract
Reactive oxygen species (ROS) generate oxidized bases and single-strand breaks (SSBs), which are fixed by base excision repair (BER) and SSB repair (SSBR), respectively. Although excision and repair of damaged bases have been extensively studied, the function of the sliding clamp, proliferating cell nuclear antigen (PCNA), including loading/unloading, remains unclear. We report that, in addition to PCNA loading by replication factor complex C (RFC), timely PCNA unloading by the ATPase family AAA domain-containing protein 5 (ATAD5)-RFC–like complex is important for the repair of ROS-induced SSBs. We found that PCNA was loaded at hydrogen peroxide (H2O2)-generated direct SSBs after the 3′-terminus was converted to the hydroxyl moiety by end-processing enzymes. However, PCNA loading rarely occurred during BER of oxidized or alkylated bases. ATAD5-depleted cells were sensitive to acute H2O2 treatment but not methyl methanesulfonate treatment. Unexpectedly, when PCNA remained on DNA as a result of ATAD5 depletion, H2O2-induced repair DNA synthesis increased in cancerous and normal cells. Based on higher H2O2-induced DNA breakage and SSBR protein enrichment by ATAD5 depletion, we propose that extended repair DNA synthesis increases the likelihood of DNA polymerase stalling, shown by increased PCNA monoubiquitination, and consequently, harmful nick structures are more frequent.
INTRODUCTION
DNA synthesis occurs during DNA replication at replication forks and also during repair of various types of DNA damage at the final step, termed repair DNA synthesis. DNA is synthesized by DNA polymerases, which are frequently tethered by a sliding clamp, proliferating cell nuclear antigen (PCNA), homotrimer in eukaryotic cells (1–4). A ring-shaped PCNA homotrimer is opened and loaded on to DNA at the single-strand DNA (ssDNA)/double-strand DNA junction with the 3′-hydroxyl (3′-OH) end by the heteropentameric replication factor complex C (RFC) complex, which is composed of a large RFC1 subunit and four small RFC proteins, RFC 2–5. During DNA replication, the PCNA-loadable primer-template junction is provided by DNA polymerase α/primase. During DNA repair, a 3′-OH end in a nick or a gap, which are formed during incision/excision processes, or a 3′ overhang, which invades sister chromatid DNA during homologous recombination (HR), provides a structure for PCNA loading for DNA synthesis.
After DNA synthesis is complete, PCNA is unloaded from DNA by the ATPase family AAA domain-containing protein 5 (ATAD5)-RFC-like complex (RLC) (enhanced level of genomic instability 1 [Elg1]-RLC in budding yeast) composed of ATAD5 and RFC 2–5 in human cells (5–7). The importance of ATAD5 in maintaining genomic integrity and suppressing tumorigenesis is supported by a high level of tumor onset and severe genomic instability in Atad5 haploinsufficient mice, embryonic lethality in mice caused by Atad5 homozygous deficiency, and the frequent observations of somatic mutations in the ATAD5 gene in human tumors (5,8–11). Accumulating data suggest that a deficiency of ATAD5 (Elg1) causes PCNA to remain persistently on DNA, which can result in defects in DNA replication and repair (6,7,12,13). In human cells, a defect in PCNA unloading reduces the DNA replication rate, inhibits fork regression, which is required for fork restart in replication stress, and causes R-loop formation by increasing collisions between PCNA and transcription machinery (5,14,15). In budding yeast, alleviating PCNA accumulation by either disassembly-prone PCNA mutants or G2/M-specific Elg1 expression rescues genome instability, such as hyper-recombination and telomere lengthening in elg1Δ mutants (16). In addition, it was shown that DNA damage sensitivity and recombination rates correlate with the extent of PCNA accumulation on DNA, using site-specific mutations in ELG1 with different PCNA-unloading activities (17).
Endogenous reactive oxygen species (ROS) generate thousands of oxidative lesions, such as oxidized bases and single-strand breaks (SSBs), per cell on a daily basis, which could alter important genetic information by interfering with replication and transcription. The oxidized bases and SSBs are repaired by base excision repair (BER) and SSB repair (SSBR), respectively (18–22). Oxidized bases are removed by DNA glycosylases, and the resulting abasic sites are processed by apurine/apyrimidine (AP) endonuclease 1 (APE1), which generates a SSB with a 3′-OH moiety on one end and 5′-deoxyribose phosphate (5′-dRP) on the other. DNA polymerase β (pol β) is recruited by APE1 and incorporates a single nucleotide and removes the 5′-dRP, followed by ligation with DNA ligase III/X-ray repair cross-complementing protein 1 (XRCC1). This short-patch BER (SP-BER, also referred as single-nucleotide BER) is the dominant pathway to repair oxidized, as well as alkylated, bases.
Alternatively, when pol β-mediated 5′-dRP removal is not possible, pol δ/ϵ incorporates 2–13 nucleotides, which is referred to as long-patch BER (LP-BER) (23,24). Recently, formation of a 9-nucleotide gap at the 5′ end of the lesion site by RECQ1 DNA helicase and ERCC1-XPF endonuclease, following gap-filling DNA synthesis, has been suggested as a sub-pathway of LP-BER (25). ROS-generated direct SSBs are repaired by components of PCNA-dependent LP-BER in the following manner: strand displacement synthesis by the cooperation of pol δ and flap endonuclease 1 (FEN1) and nick ligation by DNA ligase I. In the SSBR pathway, recognition of SSBs by poly (ADP) ribose polymerase 1 and 2 (PARP1/2) and the recruitment of XRCC1 to the SSB site by PARP1 or its PARylation activity are critical (26). When the 3′-terminus of a SSB, which is produced directly by ROS or indirectly during the BER process, is not suitable for gap filling, several end-processing enzymes bound to the scaffold XRCC1 clean the ends to generate a 3′-OH.
Defects in specific repair pathways, such as double-strand break (DSB)-induced HR and mismatch repair by ATAD5 (Elg1) depletion, were previously reported (11,27,28). Alleviating PCNA accumulation was shown to rescue hypermutation associated with a defect of mismatch repair in elg1Δ mutants (28). In the BER/SSBR process for oxidative DNA damages, PCNA would remain on the DNA after gap-filling DNA synthesis or strand-displacement synthesis was complete in ATAD5-depleted cells. Defects in the regulation of PCNA loading/unloading and the consequences for the BER/SSBR process have not been thoroughly investigated.
ATAD5-depleted cells were reported to be sensitive to various damaging agents (29,30), which suggests that ATAD5 plays a role in DNA damage repair. In budding yeast, loss of elg1 is associated with increased frequency of spontaneous HR (16,27,31). In addition, in human cells, long-term depletion of ATAD5 increases spontaneous HR and sister chromatid exchange (SCE) (11). SSBs are a well-known DNA intermediate that promotes SCE, as exemplified by increased SCE in BER/SSBR-defective cells (32). This suggests a possible role of ATAD5 in BER/SSBR. Here, we report that ATAD5-RLC is important for the repair of the ROS-induced direct SSBs. Starting with the observation that mNeonGreen-ATAD5 signal accumulates on chromatin upon oxidative DNA damage in a PCNA-loading and end-processing-protein-dependent manner in cells at G1 phase, we find that timely unloading of PCNA by ATAD5 is important to properly terminate H2O2-induced repair DNA synthesis. Otherwise, more DNA is synthesized resulting in increased DNA breakage and H2O2 sensitivity. We propose that an increased likelihood of polymerase stalling, which results in increased exposure of nick or flap structures during extended DNA synthesis, due to ATAD5 depletion as a possible mechanism.
MATERIALS AND METHODS
Cell lines, cell culture and cell synchronization
HeLa, U2OS, HeLa-ATAD5mNeonGreen-AID, HeLa-ATAD5AID, U2OS-ATAD5AID, U2OS-ATAD5−/− and control wild-type cells, HEK293AD, and HEK293AD-ATAD5−/− cells were cultured in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum (FBS; GE Healthcare, Little Chalfont, UK), 100 U/ml penicillin G (Life Technologies, Carlsbad, CA, USA) and 100 μg/ml streptomycin (Life Technologies) in a humidified atmosphere of 5% CO2 at 37°C. MRC-5 cells were cultured in Minimum Essential Medium with Earle’s Balanced Salts containing 10% FBS (GE Healthcare), 100 U/ml penicillin G (Life Technologies) and 100 μg/ml streptomycin (Life Technologies) in a humidified atmosphere of 5% CO2 at 37°C. To generate HEK293AD-ATAD5−/−: CLIP vector and HEK293AD-ATAD5−/−: CLIP-ATAD5 cells, HEK293AD-ATAD5−/− cells were infected by lentivirus generated using pLVX-CMV-CLIP or pLVX-CMV-CLIP-ATAD5 DNA and selected with puromycin. To enrich cells at the G1 phase, cells were treated with 1 μM CDK 4/6 inhibitor PD 0332991 for 24 h and 20 μM CDK 7/9 inhibitor PHA-767491 for 1 h before drug treatment. To obtain mitotic cells, cells were treated with 50 ng/ml nocodazole for 5 h, and then cells were shaken off the plate, collected and washed with culture medium. Cells were then incubated in fresh medium for 4 h (G1) before drug treatment.
Generation of HeLa-ATAD5mNeonGreen-AID and HeLa-ATAD5AID cells
HeLa cells expressing osTIR1-9xMyc were first generated by retroviral infection with pBABE-Blast-osTIR1-9xMyc from Andrew Holland (Addgene plasmid # 80073). To generate a pair of CRISPR/Cas9 plasmids targeting the endogenous ATAD5 locus, we used pSpCas9n(BB)-2A-GFP from Feng Zhang (Addgene plasmid #48140) according to the previous protocol (33). Briefly, a pair of complementary single guide-RNA oligomers targeting the region near the ATG start codon of the endogenous ATAD5 locus were annealed and ligated to the BbsI-digested pSpCas9n(BB)-2A-GFP vector. To generate a donor plasmid for tagging endogenous ATAD5 with exogenous DNA, genomic DNA was amplified with homology arms to the target locus (about 250 bp each) using Phusion DNA polymerase (New England Biolabs, Ipswich, MA, USA) and cloned into a pGEM T-easy vector (Promega, Madison, WI, USA). The mNeonGreen and mini auxin-inducible degron (mAID) or mAID cassette was then cloned into the donor plasmid between the ATG codon and the second codon of the ATAD5 gene. HeLa-osTIR1-9xMyc cells were co-transfected with a pair of CRISPR/Cas9 plasmids and donor plasmid by nucleofection (Lonza, Basel, Switzerland) (14). Forty-eight hours after transfection, high-GFP-expressing cells were sorted into 96-well plates using a FACSAria Fusion (BD Biosciences, San Jose, CA, USA). PCR of genomic DNA was used to identify positive clones (Forward primer: GGGTACCGGCGAGAAGAGTGCTTGTCCT, Reverse primer: ACTGGTCGCAGTTAAATGAACATAC). Indole-3-acetic acid (IAA; 500 μM), a natural auxin, was added to the culture medium to induce degradation of AID-tagged ATAD5.
Plasmids, small interfering RNAs (siRNAs) and transfection
The wild-type ATAD5 cDNA or mutant ATAD5 cDNA with an E1173K mutation and mNeonGreen cDNA were cloned into the pcDNA5-FRT-TO vector to express C-terminal mNeonGreen-tagged ATAD5. The following synthetic duplex siRNAs were purchased from Bioneer (Daejeon, Korea): ATAD5 3′ UTR (5′-GUAUAUUUCUCGAUGUACA-3′) (6), ATAD5 exon #1 (5′-GCGCAAUAAUGUAUACUUU-3′), ATAD5 exon #3 (5′-UGAAUGAUGUGCUAGGAAA-3′), RFC1 (Bioneer #SN-5981-1), RFC4 (Bioneer #SN-5984-1), XRCC1 (Bioneer #SN7515-1), PNKP (5′-CCGGAUAUGUCCACGUGAA-3′), APE1 (5′-GUCUGGUACGACUGGAGU-3′), OGG1 (5′-UCCAAGGUGUGCGACUGCUGCGACA-3′) (29), MYH (5′- UCACAUCAAGCUGACAUAUCAAGUA-3′) (29) and control siRNA (Bioneer #SN-1002). Transfection of cells with plasmid DNA was performed using X-tremeGENE™ HP (Roche, Basel, Switzerland) and transfection with siRNAs (20 nM) was performed using RNAiMAX (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Cells were analyzed 48 h after transfection, unless otherwise specified in the text.
Reagents and antibodies
The following drugs were used in this study: hydrogen peroxide (Sigma-Aldrich, St. Louis, MO, USA), hydroxyurea (Sigma-Aldrich), methyl methanesulfonate (Sigma-Aldrich), camptothecin (Sigma-Aldrich), cisplatin (Sigma-Aldrich), CDK4/6 inhibitor (PD 0332991; Selleck Chemicals, Houston, TX, USA), CDC7 inhibitor (PHA-767491; Selleck Chemicals), nocodazole (Sigma-Aldrich), poly (ADP ribose) glycohydrolase (PARG) inhibitor (PDD00017273; Merck) and indole-3-acetic acid (Sigma-Aldrich). The following antibodies were used: anti-mouse-PCNA (PC10, sc-56; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-RFC1 (B-5, sc-271656; Santa Cruz Biotechnology), anti-RFC4 (H-183, sc-20996; Santa Cruz Biotechnology), anti-LAMIN B1 (sc-20682; Santa Cruz Biotechnology), anti-CHK1 (sc-8408; Santa Cruz Biotechnology), anti-FEN1 (sc-28355; Santa Cruz Biotechnology), anti-XRCC1 (sc-56254; Santa Cruz Biotechnology), anti-RPA2 (A300-244A), anti-pRPA2(S4/S8) (A300-245A) (Bethyl Laboratories, Hamburg, Germany), anti-α TUBULIN (T9026; Sigma-Aldrich), anti-rabbit-PCNA (ab18197; Abcam, Cambridge, UK), anti-DNA Polymerase δ (ab10362; Abcam), anti-β tubulin (ab-15568; Abcam), anti-GAPDH (ab181603; Abcam); anti-histone H3 (Thermo Fisher Scientific); anti-ubiquityl-PCNA (Lys164) (D5C7P), anti-phospho-CHK1 (S345) (133D3) (both Cell Signaling Technology, Danvers, MA, USA), anti-PAR (4335-MC-100) (Trevigen, Gaithersburg, MD, USA) and anti-γH2AX (05–636) (Merck Millipore, Burlington, MA, USA) antibodies. The anti-human ATAD5 antibody was raised in rabbits against an N-terminal fragment (1–297 aa) (34).
Click chemistry and unscheduled DNA synthesis (UDS) assay
To distinguish cells in S-phase, cells were labeled with EdU for 30–60 min. For UDS assay, EdU was added during H2O2 treatment or for 3 h after 10 J/m2 254 nm UV irradiation. After EdU labeling, cells were harvested and processed using the Click-iT® EdU Imaging kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. In brief, cells were pre-extracted with cytoskeleton (CSK) buffer (10 mM PIPES, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA and 0.5% Triton X-100™) for 10 min on ice and fixed with 4% paraformaldehyde for 20 min at room temperature before the click reaction.
405-nm UV-laser microirradiation
Cells were pre-incubated with 10 μM Hoechst 33342 for 10 min before microirradiation. UV microirradiation was applied to cells plated on LabTek™ Chambered Coverglasses (Thermo Fisher Scientific) using a Diode 405 nm laser (100% power) projected through a C-Apochromat 40×/1.2W Korr FCS M27 objective, via a bleaching module (5 iterations) on an LSM880 confocal microscope (Carl Zeiss, Oberkochen, Germany) platform.
CLIP-tag labeling
HEK293AD-ATAD5−/−: CLIP vector or HEK293AD-ATAD5−/−: CLIP-ATAD5 cells were labeled with 3 μM CLIP-Cell TMR-Star (New England Biolabs) (benzylcytosine-tetramethylrhodamine) for 1 h. Cells were then washed three times with culture medium and incubated with culture medium for 20 min. H2O2 was added to culture medium during 20 min incubation time.
Immunostaining, image acquisition and image analysis
Cells plated on LabTek™ chamber slides (Thermo Fisher Scientific) were fixed and stained as described previously (6) with slight modifications. Briefly, the cells were pre-extracted with CSK buffer for 10 min on ice and fixed with 4% paraformaldehyde for 20 min at room temperature. For PAR immunostaining, PARG inhibitor was added together with CSK buffer during the pre-extraction step and cells were fixed with 4% paraformaldehyde followed by methanol at –20°C for 10 min. After washing with phosphate-buffered saline (PBS) and incubating in blocking buffer (10% FBS in PBS) for 30 min, cells were incubated with the indicated antibodies diluted in blocking buffer at 4°C overnight. After three washes with 0.05% Triton X-100™ in PBS, Alexa Fluor®-conjugated secondary antibodies (Thermo Fisher Scientific) were added and incubated for 30 min. After washing, cells were mounted using ProLong® Gold antifade reagent (Vector Laboratories, Burlingame, CA, USA). Confocal images were acquired using an LSM880 confocal microscope (Carl Zeiss) with a 40×/1.2 lens objective. Image acquisition and analysis were performed with Zen 2.6 (blue edition) (Carl Zeiss) software. For nuclear intensity analysis, nuclei were first masked, and mean intensity within the masked regions was measured.
Cell survival assay
Cells were seeded in 96-well culture plates. After 24 h, cells were treated with drugs and cell survival was measured by the MTT assay or by using the CellTiter-Glo® 2.0 Assay kit (Promega) according to the manufacturer’s instructions. For the MTT assay, MTT reagent was added together with a different range of H2O2 for 4 h, the purple formazan crystal was dissolved with DMSO, and the colored solution was quantified using a plate reader. For the CellTiter-Glo® 2.0 Assay, in brief, cells were lysed with CellTiter-Glo® reagent and the luminescence signal was read using a plate reader. Percent cell survival was normalized to that of control cells.
Clonogenic survival assay
Control wild-type cells and U2OS ATAD5−/− cells were seeded onto 60-mm plates at a density of 5 × 102 cells/60 mm and 5.5 × 102 cells/60 mm, respectively. Cells were treated with MMS for 1 h and washed with PBS followed by addition of fresh medium and incubation for 14 days. Cells were then washed with PBS and stained with 2% methylene blue in 70% EtOH for 10 min.
Sister chromatid exchanges
Cells were incubated for 4 h with 0.2 μg/ml colcemid, and then metaphase cells were harvested by trypsinization. The cells were swollen in 75 mM KCl for 15 min at 37°C and fixed with methanol:acetic acid (3:1) twice. Cells were dropped on to glass microscope slides and stained with 5% Giemsa stain. Images were acquired using a fluorescence microscope (BX53; Olympus). Images of at least 35 metaphase cells were taken randomly for each condition.
Alkaline COMET assay (single-cell gel electrophoresis)
The COMET assay was performed using a CometAssay® kit (Trevigen) according to the manufacturer’s instructions. In brief, each cell suspension was mixed with COMET LM Agarose at 37°C and the mixture was spread on a COMET slide (Trevigen). After solidification of the agarose, the slide was immersed in a lysis solution (Trevigen) for 1 h at 4°C. Images were acquired with a fluorescence microscope (BX53; Olympus, Tokyo, Japan), and the tail moment was calculated using CometScore software version 2.0.
Flow cytometry
Cells were washed with PBS, fixed with 70% ethanol for 1 h, and then incubated with 0.1 mg/ml RNase A at 37°C for 1 h. DNA was stained with 0.05 mg/ml propidium iodide. Flow cytometry was performed on a FACSVerse™ flow cytometer using BD FACSuite™ software (BD Biosciences). Data analysis was performed using FlowJo software.
Immunoblot analysis
A Triton X-100™-soluble and insoluble fraction were isolated and subjected to immunoblot analysis according to the methods described previously (34) with slight modifications. In brief, the soluble fraction was isolated by incubating cells in buffer A (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES, pH 6.8, 1 mM EGTA, 0.2% Triton X-100™, 0.1 M phenylmethylsulfonyl fluoride, phosphatase inhibitors, and protease inhibitors [Roche]) for 5 min on ice followed by centrifugation. Then, the detergent-insoluble fraction was isolated by resuspending the pellet in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100™, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 0.1 M phenylmethylsulfonyl fluoride, phosphatase inhibitors and protease inhibitors) with Benzonase® nuclease for 40 min on ice followed by sonication and centrifugation. Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane. Blocking of the membranes and blotting with primary antibodies were performed in Tris-buffered saline containing 0.1% Tween® 20 supplemented with 5% skim milk powder. Proteins were visualized using secondary horseradish peroxidase-conjugated antibodies (Enzo Life Sciences, Farmingdale, NY, USA) and enhanced chemiluminescence reagent (Thermo Fisher Scientific). Signals were detected using an automated imaging system (ChemiDoc™; Bio-Rad Laboratories, Hercules, CA, USA).
Reverse transcription quantitative (real-time) PCR (RT-qPCR)
Total RNA was extracted using a TRIzol® Reagent (Invitrogen) protocol. Total RNA (1.5 μg) was used to synthesize cDNA using the SuperScript® IV First-Strand Synthesis System (Invitrogen) according to the manufacturer’s instructions. RT-qPCR was performed using SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) in a QuantStudio 7 Flex system (Applied Biosystems) according to the manufacturer’s instructions. Gene expression was normalized to GAPDH expression. The following primers were used: PNKP-forward (For) (5′-CTGACCCAGGTTACGGACC-3′), PNKP-reverse (Rev) (5′- TCCCGGTAGTTGAGGGGTT-3′), APE1-For (5′-CAATACTGGTCAGCTCCTTCG-3′), APE1-Rev (5′-TGCCGTAAGAAACTTTGAGTGG-3′), OGG1-For (5′-ACCCTGGCTCAACTGTATCACCAC-3′), OGG1-Rev (5′-CCGCTCCACCATGCCAGTGATG), MYH-For (5′-GTATATGGGCTGGCCTTGGAAG-3′), MYH-Rev (5′-CTGTTGGCCCTGATACACACG-3′), GAPDH-For (5′- AGCCACATCGCTCAGACAC-3′), GAPDH-Rev (5′-GCCCAATACGACCAAATCC-3′).
Statistical analysis
Prism 8 software (GraphPad, San Diego, CA, USA) was used to generate graphs and analyze data. For all data, two-tailed paired Student's t-tests were used.
RESULTS
Generation of a cell line endogenously expressing fluorescent-protein-tagged ATAD5
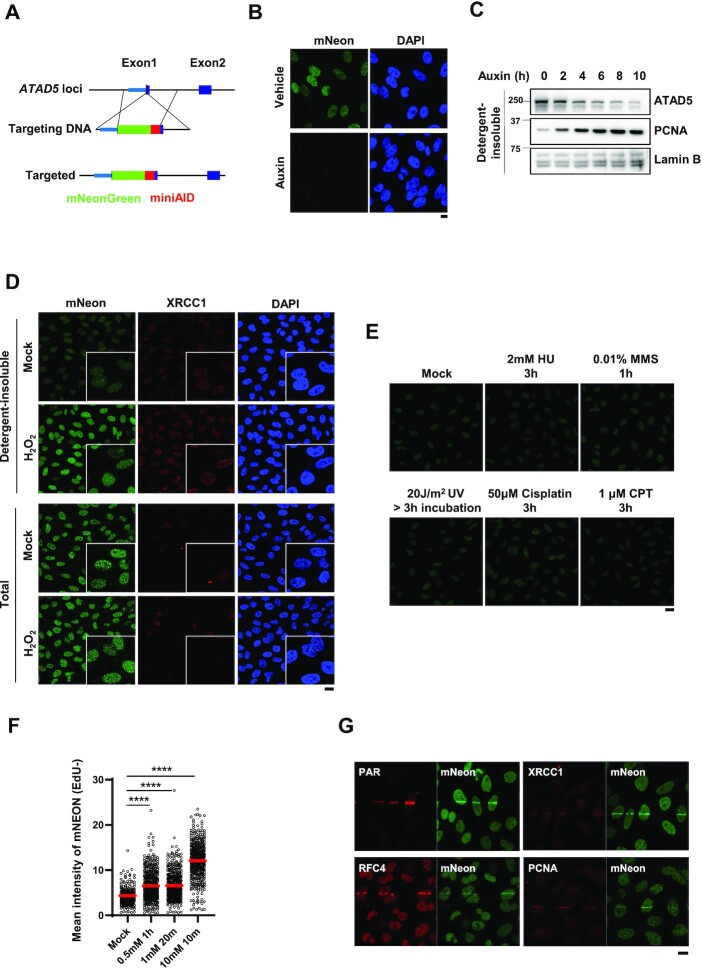
To investigate the effects of DNA-damaging agents on cellular localization and movement of endogenous ATAD5 protein, we generated a cell line endogenously expressing mNeonGreen fluorescent protein-tagged ATAD5 (Figure 1A and Supplementary Figure S1A). For this purpose, we inserted the mNeonGreen and auxin-inducible degron (AID) sequences at the N-terminus of the ATAD5 gene locus immediately after the ATG start codon using a Cas9 nickase mutant with paired guide RNAs in HeLa human cancer cells (35). We used the mini AID sequences to minimize the unwanted effects of the tagged peptide on ATAD5 protein functionality (7). Through genomic DNA PCR and sequencing of the PCR product, we confirmed that the targeting DNA was inserted in the correct position (Supplementary Figure S1B). The insertion occurred in one allele of the ATAD5 locus, whereas deletions and indels before and within the ATG start codon, respectively, occurred in two other alleles (Supplementary Figure S1C and 1D). In the HeLa-ATAD5mNeonGreen-AID cells, protein level of mNeonGreen-AID-tagged ATAD5 was slightly higher than that of endogenous ATAD5 in parental cell (Supplementary Figure S1E). However, cell cycle profile between two cells were comparable (Supplementary Figure S1F). In addition, when the HeLa-ATAD5mNeonGreen-AID cells were treated with auxin, the mNeonGreen signal disappeared and mNeonGreen-AID-tagged ATAD5 was degraded (Figure 1B and C), and the amount of chromatin-bound PCNA and monoubiquitinated PCNA were concomitantly increased (Figure 1C and Supplementary Figure S1G), which verifies that the mNeonGreen-AID-tagged ATAD5 is well expressed and functional, at least for unloading PCNA and facilitating PCNA deubiquitination (6,34). Therefore, we used the microscopic mNeonGreen signal to track the endogenous ATAD5 protein.
Figure 1.
ATAD5 accumulates on chromatin upon oxidative DNA damage. (A) The scheme for generation of a HeLa cell line expressing mNeonGreen-mAID-tagged ATAD5 (HeLa-ATAD5mNeonGreen-AID cell). (B) HeLa-ATAD5mNeonGreen-AID cells were treated with auxin for 6 h and fixed for immunostaining. (C) HeLa-ATAD5mNeonGreen-AID cells were treated with auxin for indicated times, and then detergent-insoluble proteins were fractionated and subjected to immunoblotting. (D and E) HeLa-ATAD5mNeonGreen-AID cells were treated with 10 mM H2O2 for 10 min (D) or with various DNA-damaging agents as indicated (E) and fixed after pre-extraction with CSK buffer (upper panel in D and E) or without pre-extraction (lower panel in D). (D) The fixed cells were subjected to immunostaining with an anti-XRCC1 antibody. Four-fold magnified images are shown in the corner. (F) HeLa-ATAD5mNeonGreen-AID cells were incubated with EdU for 1 h before detergent-pre-extraction. Cells were treated with H2O2 as indicated, detergent-pre-extracted and fixed for EdU-click reactions. mNeonGreen signals were quantified in strong-EdU-signal-negative cells. Red bar indicates mean value. Statistical analysis: two-tailed unpaired Student's t-test; ****P < 0.001. (G) HeLa-ATAD5mNeonGreen-AID cells were subjected to 405-nm UV laser microirradiation. Two minutes after microirradiation, cells were fixed for immunostaining as indicated. (B and G) Scale bar: 10 μm. (D and E) Scale bar: 20 μm.
ATAD5 accumulates on DNA upon oxidative DNA damage
Using the HeLa-ATAD5mNeonGreen-AID cells, we checked effects of various DNA-damaging agents on endogenous ATAD5 protein by examining the mNeonGreen signal in fixed cells where detergent-soluble cytoplasmic and nucleoplasmic proteins had been removed. The remaining detergent-insoluble proteins include chromatin-associated proteins and nuclear matrix proteins and proteins associated with the nuclear matrix. Each treatment generates a different type of DNA damage as follows: H2O2 induces oxidative DNA damage; hydroxyurea (HU) depletes intracellular nucleotide pools, which leads to replication fork stalling or fork breakage; methyl methanesulfonate (MMS) is a DNA alkylating agent; 254-nm-wavelength ultraviolet (UV)-C radiation generates DNA photoadducts; cisplatin induces DNA intra- and inter-strand crosslinks; and camptothecin (CPT), a DNA topoisomerase I inhibitor, traps the topoisomerase I cleavage complex on DNA, which leads to DNA breaks. We found that H2O2 treatment increased detergent-insoluble mNeonGreen and XRCC1 signals in most cells (Figure 1D); however, total mNeonGreen signal was not changed after H2O2 treatment (Figure 1D). Other DNA-damaging agents did not increase the detergent-insoluble mNeonGreen signal under our treatment conditions (Figure 1E). These results suggest that ATAD5 is recruited to sites of oxidative DNA damage, which consists of oxidized bases, SSBs, or double-strand breaks (DSBs). H2O2-generated mNeonGreen signal in most regions of the nucleus suggests that SSBs may be the main cause for accumulation of ATAD5 on chromatin because ROS, such as H2O2, were reported to induce 2000-fold more SSBs than DSBs (36). The alkylated base formed by the MMS treatment is removed by SP-BER generating an intermediate SSB (20). The absence of an increase in detergent-insoluble mNeonGreen signal upon MMS treatment (Figure 1E) suggests that ATAD5 specifically accumulates on DNA with SSBs generated by ROS.
ATAD5 is a replication fork-associated protein whose roles are known to be important in S-phase (6,14). ROS-generated DNA lesions are repaired differently depending on the stage of cell cycle. Therefore, to examine the effect of H2O2 treatment on ATAD5 in phases other than S-phase, cells were incubated with 5-ethynyl-2′-deoxyuridine (EdU), a thymidine homologue, which is incorporated into DNA during DNA replication and thus labels cells in S-phase with a strong EdU signal. We found that H2O2 treatment under three different treatment conditions increased detergent-insoluble mNeonGreen signal in most EdU-negative cells (Figure 1F). The same three different H2O2 treatment conditions did not lead to cell death, determined by acridine orange and propidium iodide staining (Supplementary Figure S1H). This suggests that ATAD5 is recruited to sites of oxidative DNA damage in cells in G1 phase.
We examined H2O2-induced accumulation of ATAD5 on chromatin using a different system, HEK293AD-ATAD5−/− cells stably expressing CLIP-tagged ATAD5. The CLIP-tag is derived from O6-alkylguanine-DNA alkyltransferase and can be covalently labeled in living cells using O2-benzylcytosine derivatives conjugated to chemical dyes (37). CLIP-tagged ATAD5 reduced the amount of chromatin-bound PCNA and monoubiquitinated PCNA that was increased by ATAD5 deficiency (Supplementary Figure S1I), proving the functionality of CLIP-tagged ATAD5. H2O2 treatment increased CLIP-tag-bound tetramethylrhodamine (TMR) signal (Supplementary Figure S1J and K).
We also performed 405-nm UV laser microirradiation experiments. Laser microirradiation generates a mixture of DNA lesions, such as photoadducts, base lesions, SSBs and DSBs (38). We found that ATAD5mNeonGreen-AID signal was increased on the microirradiated strip and co-localized with BER/SSBR proteins such as PAR, XRCC1, RFC4 and PCNA (Figure 1G). Taken together with the H2O2-specific increase in mNeonGreen signal, this result suggests that ATAD5 accumulates around oxidative DNA damage lesions.
ATAD5 accumulation upon oxidative DNA damage depends on RFC1
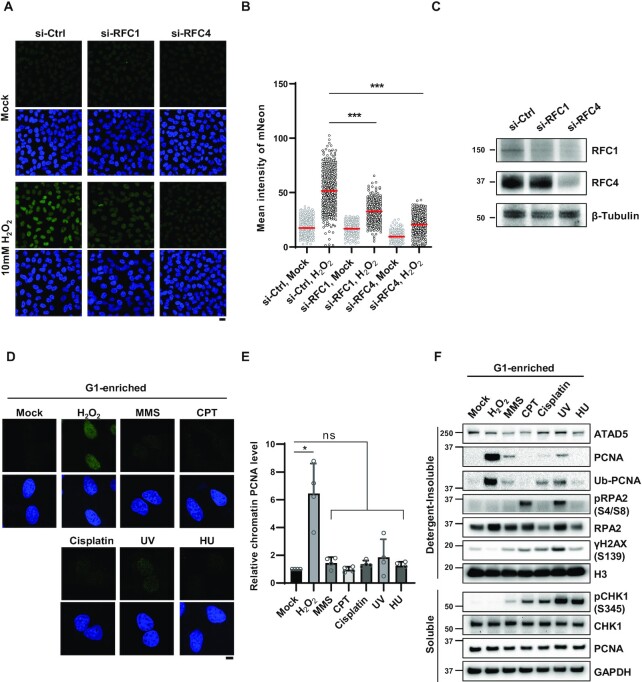
Next, we investigated the mechanism underlying ATAD5 accumulation at sites of oxidative DNA damage. Oxidative DNA damage is repaired by the BER/SSBR pathway; LP-BER and SSBR are completed by repair DNA synthesis following RFC-mediated PCNA loading (18–22). Because the main function of ATAD5 is PCNA unloading, we reasoned that ATAD5-RLC is recruited to oxidative damage sites to unload PCNA that has completed its task. As expected, depletion of RFC1, a large subunit of the RFC complex for PCNA loading, by small interfering RNA (siRNA) significantly decreased mNeonGreen signals induced by treatment (Figure 2A–C). Depletion of RFC4, a small subunit shared by the RFC and ATAD5-RLC, also decreased H2O2-induced mNeonGreen signals. This was also repeated under lower H2O2 treatment conditions (Supplementary Figure S2A and B). This result suggests that ATAD5-RLC moves to sites of oxidative DNA damage in a manner dependent on RFC-mediated PCNA loading.
Figure 2.
ATAD5 accumulation upon oxidative DNA damage depends on RFC1. (A–C) HeLa-ATAD5mNeonGreen-AID cells transfected with small interfering RNAs (siRNAs) as indicated for 48 h. Cells were treated with 10 mM H2O2 for 10 min, detergent-pre-extracted and then fixed for detection of mNeonGreen signal. (A) Representative images are shown; scale bar: 20 μm. (B) Quantification of mNeonGreen signals. Three independent experiments were performed and one representative result is displayed. Red bar indicates mean value. (C) Results of immunoblotting of whole cell extracts prepared 48 h after transfection. (D and E) HeLa cells were treated with nocodazole for 5 h, shaked-off and released into fresh medium followed by incubation for 4 h before treatment with DNA-damaging agents under conditions used in Figure 1D and E. After damage treatment, cells were, pre-extracted with detergent before fixation and immunostained with an anti-PCNA antibody. (D) Representative images are shown; scale bar: 10 μm. (E) The intensity of PCNA signal was quantified. PCNA intensity of four independent experiments are normalized. Error bars represent standard deviation of the mean (n = 4). (F) U2OS-ATAD5AID cells previously reported (14) were enriched at G1 phase by treatment with PD 0332991 and PHA-767491, and treated with DNA-damaging agents under conditions used in Figure 1D and E. After drug treatment, detergent-soluble and -insoluble proteins were fractionated and subjected to immunoblotting. (B and E) Statistical analysis: two-tailed unpaired (B) and paired (E) Student's t-test; ***P < 0.005, *P < 0.05, and ns: not significant.
UV-C-generated DNA photoadducts and CPT-generated DSBs are repaired by nucleotide excision repair (NER) and HR pathways, respectively, using repair DNA synthesis as the final step. However, the detergent-insoluble mNeonGreen-ATAD5 signal was not increased by DNA-damaging agents other than H2O2 (Figure 1E). We speculated that the different amount of PCNA loaded on the DNA by each treatment could be the cause of the difference. Because large amounts of PCNA are already associated with chromatin for DNA replication during S-phase, we investigated this by using cells enriched at G1 phase by releasing from nocodazole arrest for HeLa cells (Figure 2D and E) or by treatment with the cyclin-dependent kinase (CDK) 4/6 inhibitor PD 0332991 for U2OS cells (Supplementary Figure S2C). The firing of new replication origins was also blocked by the CDK7 inhibitor PHA-767491, which excluded cells that escaped from G1 arrest (Figure 2F). Both G1 enrichment procedures greatly reduced the detection of cells with chromatin-bound PCNA signal in immunostained images (Figure 2D and E) and chromatin-bound PCNA level in immunoblots (Figure 2F), suggesting that most cells were in G1 phase. When the G1-enriched cells were treated with DNA-damaging agents under the same conditions, the level of chromatin-bound PCNA was dramatically increased by H2O2 treatment as shown in both immunostaining (Figure 2D and E) and immunoblot assays (Figure 2F). The H2O2-induced increase in chromatin-bound PCNA was observed by different treatment conditions that include 0.25 mM H2O2 for 1h (Supplementary Figure S2D and E). Although a slight increase in chromatin-bound PCNA was observed with MMS treatment and UV-C irradiation, it was much lower than that induced by H2O2 treatment (Figure 2F). These data are consistent with the increase in mNeonGreen-ATAD5 signal only upon H2O2 treatment (Figure 1D and E). We concluded that other DNA-damaging agents used did not lead to detectable PCNA loading, at least under the treatment conditions in our experiments. Contrary to the mNeonGreen signal from the immunostaining data (Figure 1F), the increase in detergent-insoluble ATAD5 protein level with H2O2 treatment was not observed in immunoblots (Figure 2F). This could be the result of the cross-linking step during slide preparation for microscopy, which allows for the detection of a weak association of ATAD5 with detergent-resistant structures.
When a replication fork is stalled by DNA lesions or intrinsic replication blocks, PCNA is monoubiquitinated by the E2-E3 RAD6-RAD18 complex and the resulting monoubiquitinated PCNA recruits translesion synthesis (TLS) polymerases to bypass the DNA lesions (39,40). Consistent with previously reported cell-cycle-independent and RAD18-dependent PCNA monoubiquitination upon H2O2 treatment (41–43), PCNA monoubiquitination was also markedly increased under our H2O2 treatment conditions (Figure 2F and Supplementary Figure S2E).
Accumulation of PCNA and ATAD5 upon oxidative DNA damage depends on 3′- terminal-processing enzymes
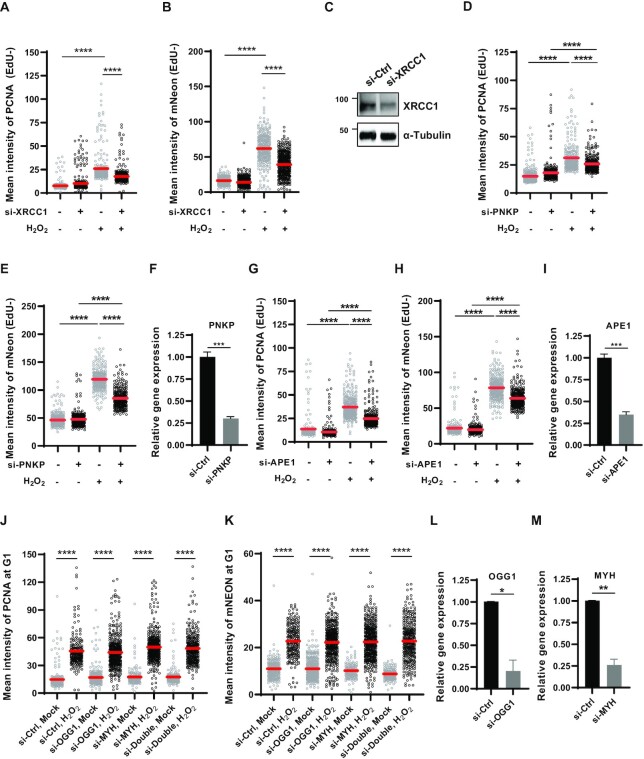
A PCNA homotrimer is loaded at a ssDNA/double-strand DNA junction with a 3′-OH end or even on to single-nicked DNA (2). SSBs generated by ROS are damaged at the 3′-terminus, regardless of whether SSBs are directly generated or formed as intermediates during BER of oxidized bases. The core scaffolding protein XRCC1 engages several end-processing proteins to help generate the 3′-OH that allows PCNA loading by the RFC complex (44). Consistently, XRCC1 depletion reduced both detergent-insoluble PCNA and mNeonGreen signals induced by H2O2 treatment in non-S phase cells (Figure 3A–C). This was also repeated under lower H2O2 treatment conditions (Supplementary Figure S3A and B). The majority of direct SSBs result in 3′-phosphate and 3′-phosphoglycolate ends and are converted to 3′-OH by polynucleotide kinase 3′-phosphatase (PNKP) (45,46) and APE1 (47–50), respectively. XRCC1 has been reported to interact with PNKP and stimulate its enzymatic activity (51). In line with this, PNKP depletion reduced detergent-insoluble PCNA and mNeonGreen signals induced by H2O2 treatment at levels similar to XRCC1 depletion (Figure 3D–F). Although the interactions between XRCC1 and APE1 are still unclear, APE1 depletion also reduced detergent-insoluble PCNA and mNeonGreen signals induced by H2O2 treatment (Figure 3G–I). This was repeated under lower H2O2 treatment conditions (Supplementary Figure S3C and D).
Figure 3.
Accumulation of PCNA and ATAD5 upon oxidative DNA damage depends on 3′-terminal-processing enzymes. (A–M) HeLa-ATAD5mNeonGreen-AID cells were transfected with siRNAs as indicated and incubated for 48 h before 10 mM H2O2 treatment for 10 min. (A,B,D,E,G and H) Cells were incubated with EdU for 30 min before detergent-pre-extraction. (J and K) Before H2O2 treatment, cells were treated with nocodazole for 5 h, shaked-off and released into fresh medium followed by incubation for 4 h. (A, B, D, E, G, H, J and K) After H2O2 treatment, cells were detergent-pre-extracted and fixed for EdU-click reactions (A, B, D, E, G, H) and PCNA immunostaining. PCNA (A, D, G, J) and mNeonGreen (B, E, H, K) signal intensity was quantified. (A, B, D, E, G, H) Quantification was performed in strong-EdU-signal-negative cells. Three independent experiments were performed and one representative result is displayed. Red bar indicates mean value. Statistical analysis: two-tailed unpaired Student's t-test; ****P < 0.001. (C) Forty-eight hours after transfection, protein was extracted and subjected to immunoblotting. (F, I, L, M) Forty-eight hours after transfection, RNA was extracted and subjected to reverse transcription-quantitative PCR (RT-qPCR) with the specific primer set for each gene.
APE1 plays a role in repairing oxidized bases as a core BER protein. 7,8-Dihydro-8-oxo-guanine (8-oxoG) is primarily removed by two DNA glycosylases, 8-oxoguanine DNA glycosylase (OGG1) and MutY homolog DNA glycosylase (MYH). Although bifunctional glycosylase OGG1 has a lyase activity to generate a break at the AP site, APE1 also participates in SSB generation and processing of 3′-termini. Therefore, it is possible that the reduction in H2O2-induced chromatin PCNA signals by APE1 depletion may result from reduced PCNA loading during the BER process (Figure 3G). However, depletion of either OGG1 or MYH and depletion of both proteins did not reduce H2O2-induced detergent-insoluble PCNA and mNeonGreen signals in cells at G1 phase (Figure 3J–M). This suggests that PCNA loading upon H2O2 treatment primarily occurs during repair of direct SSBs but not in the BER process to repair oxidized bases. According to a previous report, an 8-oxoG-containing plasmid was primarily processed by LP-BER in cells (52). It is possible that the effects of LP-BER deficiency can be masked by extensive generation of direct SSBs and followed PCNA loading under our H2O2 treatment conditions (53,54).
Repair of oxidative damage-induced DNA breaks is defective in ATAD5-depleted cells
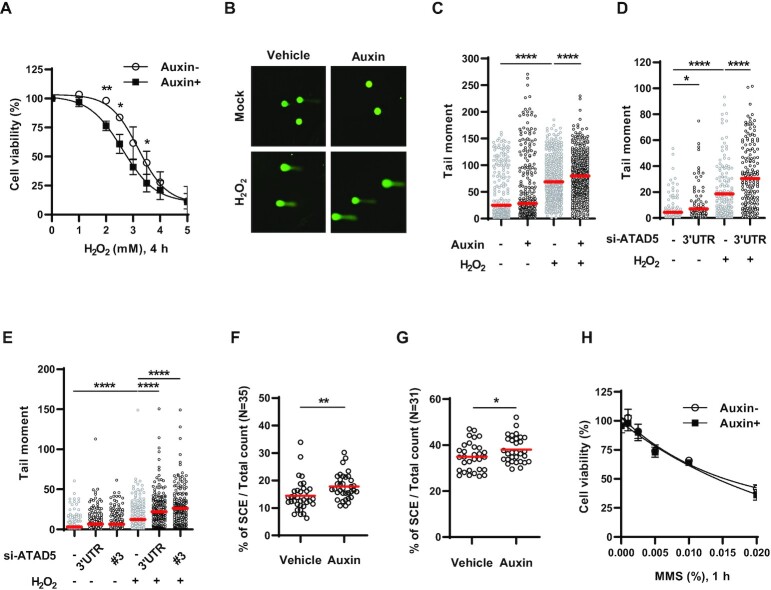
Next, we examined the effects of ATAD5 depletion on H2O2 sensitivity. Because long-term ATAD5 depletion causes many cellular defects (13), we used a U2OS cell line endogenously expressing mini AID-tagged ATAD5 (ATAD5AID) to rapidly deplete endogenous ATAD5 by short auxin treatment (14). We found that cell survival upon acute H2O2 treatment was reduced in ATAD5-depleted cells (Figure 4A). PCNA and ATAD5 increased their association with chromatin upon H2O2 treatment (Figures 1D and 2D–F) and SSBs with 3′-OH ends were used as PCNA-loadable structures (Figure 3). Therefore, we speculated that augmented H2O2 sensitivity induced by ATAD5 depletion results from defects in repair of SSBs generated by oxidative DNA damage. We tested this possibility using the alkaline COMET assay (single-cell gel electrophoresis), which detects DNA strand breaks at the single-cell level (Figure 4B–E). H2O2 treatment increased alkaline tail moment in both U2OS and HeLa cells, as previously reported (53), which was further increased when ATAD5 was depleted by auxin treatment or siRNA. This suggests that SSBs generated by oxidative DNA damage are not properly repaired in ATAD5-depleted cells.
Figure 4.
H2O2 sensitivity and single-strand DNA breakage are increased in ATAD5-depleted cells. (A) U2OS-ATAD5AID cells pre-treated with auxin were treated with H2O2 for 4 h with MTT reagents, and cell survival was measured by quantification of MTT formazan. Error bars represent standard deviation of the mean (n = 3). (B–E) U2OS-ATAD5AID cells treated with auxin for 6 h (B and C) and U2OS cells (D) or HeLa cells (E) transfected with ATAD5 siRNA for 48 h were treated with 0.1 mM H2O2 for 1 h and collected for an alkaline COMET assay. (B) Representative images of an alkaline COMET assay. (C–E) The tail moment was calculated from ∼200 cells and plotted. Three independent experiments were performed and one representative result is displayed. Red bar indicates mean value. Statistical analysis: two-tailed unpaired Student's t-test; ****P < 0.001, *P < 0.05. (F and G) HeLa-ATAD5AID cells (F) and U2OS-ATAD5AID cells (G) were pre-treated with auxin for 24 and 48 h, respectively, and fixed to prepare metaphase spreads. Sister chromatid exchange (SCE) events were counted in metaphase spreads. Statistical analysis: two-tailed unpaired Student's t-test; **P < 0.01, *P < 0.05. (H) U2OS-ATAD5AID cells pre-treated with auxin were treated methyl methanesulfonate (MMS) for 1 h, and cell survival was measured by cellular ATP quantitation. Error bars represent standard deviation of the mean (n = 3). (D and E) 3′UTR: siRNA targeting 3′-untranlsated regions (UTR) of ATAD5 gene, #3: siRNA #3 targeting an exon region of ATAD5 gene.
Long-term depletion of ATAD5 by short hairpin RNA increases spontaneous HR and SCE in human cells (11). In budding yeast, elg1 loss shows increased spontaneous HR frequency (16,27,31). Because SSBs are well known to promote SCE (32), the increased SSBs induced by ATAD5 depletion might be the reason for the increased spontaneous HR and SCE. However, reduced DNA replication and delayed S-phase progression, which is observed in yeast elg1 mutants and in human cells depleted of ATAD5 by siRNA (6,7,12), were also reported to increase SCE frequency (55,56). Therefore, we checked the effects of ATAD5 depletion on SCE under conditions in which the effect of ATAD5 depletion on S-phase progression is minimized by using the U2OS-ATAD5AID cells and newly generated HeLa-ATAD5AID cells (Supplementary Figure S4A and B). Auxin-mediated ATAD5 depletion increased SCE in both HeLa and U2OS cells (Figure 4F and G). This suggests that impaired repair of endogenously generated SSBs by ATAD5 depletion can lead to increased spontaneous HR and SCE.
Alkylated bases formed by MMS exposure are removed by monofunctional glycosylases, and the subsequent coordinated activity of APE1, Pol β, and DNA ligase 3-XRCC1 completes BER (20). Consistent with a relay mode of the BER process (57–59), MMS treatment did not increase detection of detergent-insoluble ATAD5 and PCNA (Figures 1E and 2D–F), although APE1 generates an intermediate SSB with a 3′-OH moiety, which is PCNA-loadable. In line with this result, ATAD5-depleted cells did not show any change in sensitivity to acute MMS treatment (Figure 4H). Previous reports showed higher MMS sensitivity in ATAD5-depleted cells by a clonogenic survival assay (11,29,30). Using ATAD5-knockout U2OS cells (60), we also observed the same result (Supplementary Figure S4C). As previously suggested (29), MMS sensitization of ATAD5-depleted cells, which was observed in the clonogenic survival assay, might reflect HR defects rather than BER defects induced by ATAD5 depletion.
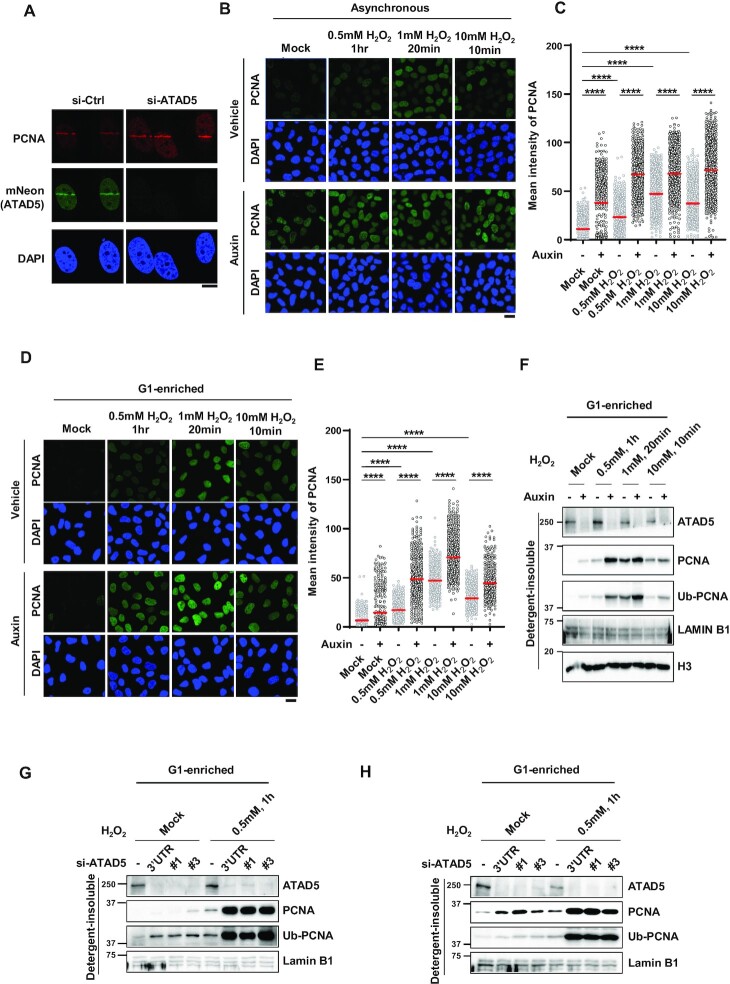
PCNA accumulates on DNA upon oxidative damage in ATAD5-depleted cells
ATAD5 depletion causes PCNA accumulation on DNA during DNA replication (6). Because DNA synthesis is the last step of the SSBR and LP-BER pathway for repairing oxidative DNA damage, we reasoned that PCNA would remain or accumulate on DNA at the damaged sites by ATAD5 depletion due to defects in PCNA unloading. As expected, ATAD5 depletion increased chromatin-bound PCNA signal on 405 nm UV-microirradiated strips (Figure 5A). Next, we examined effects of ATAD5 depletion on PCNA level on DNA in response to H2O2 treatment using U2OS-ATAD5AID cells. We found that chromatin-bound PCNA levels increased under various H2O2 treatment conditions, which was further increased by ATAD5 depletion (Figure 5B and C). We confirmed this result under conditions in which cells undergoing DNA replication are maximally excluded by enriching for cells in G1 phase by treatment with PD 0332991 and PHA-767491 (Figure 5D–F). Under these conditions, chromatin-bound PCNA levels were increased upon H2O2 treatment, and they were further increased by ATAD5 depletion (Figure 5D and E). Immunoblotting analysis showed the same result. Upon H2O2 treatment, PCNA accumulated on chromatin and further increased after ATAD5 depletion by auxin treatment (Figure 5F). When ATAD5 was depleted by siRNA, both U2OS and HeLa cells enriched at G1 phase showed the same results (Figure 5G and H). Taken together, these data indicate that PCNA accumulates on DNA upon oxidative DNA damage in ATAD5-depleted cells. Interestingly, PCNA monoubiquitination was also further increased in ATAD5-depleted cells upon H2O2 treatment (Figure 5F-H), which suggests that DNA polymerase stalling is also increased by ATAD5 depletion.
Figure 5.
PCNA accumulates on DNA upon oxidative damage in ATAD5-depleted cells at G1 phase. (A) HeLa-ATAD5mNeonGreen-AID cells were transfected with ATAD5 siRNA targeting 3′UTR and subjected to 405-nm UV laser microirradiation. Two minutes after microirradiation, cells were fixed for PCNA immunostaining; scale bar: 10 μm. (B–F) Asynchronous U2OS-ATAD5AID cells (B and C) or U2OS-ATAD5AID cells G1-enriched by treatment with PD 0332991 and PHA-767491 (D–F) were treated with auxin for 16 h and then treated with H2O2 as indicated. (B–E) After H2O2 treatment, cells were detergent-pre-extracted, fixed and immunostained with anti-PCNA antibody. (B and D) Representative images of PCNA immunostaining; scale bar: 20 μm. (C and E) Quantification of chromatin-bound PCNA signal intensity. Three independent experiments were performed and one representative result is displayed. Red bar indicates mean value. Statistical analysis: two-tailed unpaired Student’s t-test; ****P < 0.001. (G and H) U2OS cells (G) or HeLa cells (H) transfected with ATAD5 siRNAs were enriched at G1 phase by treatment with PD 0332991 and PHA-767491 (G) or by releasing cells from nocodazole arrest (H), and then treated with 0.5 mM H2O2 for 1 h. #1 and #3: siRNAs targeting an exon region of ATAD5 gene. (F–H) After H2O2 treatment, detergent-insoluble proteins were isolated and subjected to immunoblotting.
Unscheduled DNA synthesis is increased in ATAD5-depleted cells
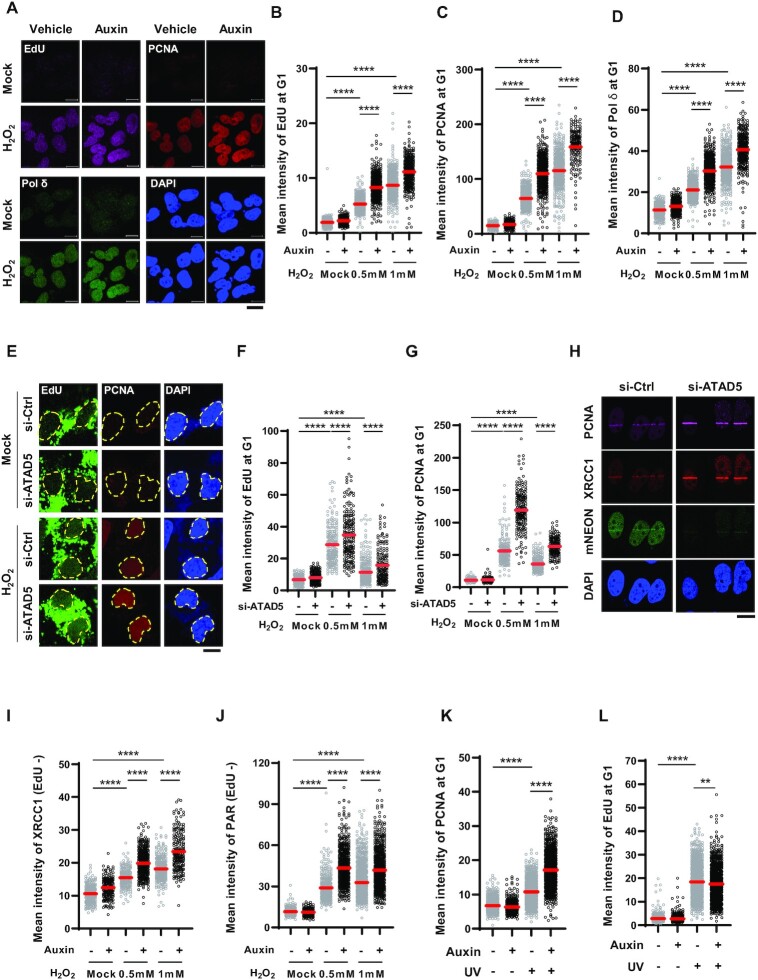
Considering conventional DNA replication process, PCNA unloading would occur after repair DNA synthesis is completed as the last step in the BER/SSBR process. This raises the question of how a defect in PCNA unloading induced by ATAD5 depletion led to the increase in tail moments in the alkaline COMET assay and H2O2 sensitivity (Figure 4). We checked whether unscheduled DNA synthesis (UDS) after H2O2 treatment is affected by ATAD5 depletion by measuring EdU incorporation in cells at G1 phase released from nocodazole arrest. As previously reported, H2O2 treatment increased the EdU signal (61) (Figure 6A and B), and chromatin-bound PCNA and DNA Pol δ signals were also increased upon H2O2 treatment (Figure 6A, C and D). Unexpectedly, the H2O2-induced EdU signal was further increased by ATAD5 depletion (Figure 6A and B), which was accompanied by an increase in PCNA and Pol δ signals in the same cells. When ATAD5 was depleted by siRNA, both U2OS and HeLa cells enriched at G1 phase showed the same results (Supplementary Figure S5A–I). We investigated whether this is cancer-cell-specific by using a human lung fibroblast cell line, MRC-5. Like U2OS cells, H2O2 treatment increased EdU and PCNA signals, and these signals were further increased by ATAD5 depletion in MRC-5 cells (Figure 6E–G). Taken together, these results suggest that repair DNA synthesis induced by oxidative DNA damage is prolonged by ATAD5 depletion and that this may be a common phenotype occurring in ATAD5-depleted human cells.
Figure 6.
Unscheduled DNA synthesis (UDS) is increased in ATAD5-depleted cells. (A–G) U2OS-ATAD5AID cells treated with auxin (A–D) and MRC-5 cells transfected with ATAD5 siRNA targeting 3′UTR (E–G) were enriched at the G1 phase by releasing cells from nocodazole arrest; cells were treated with 0.5 mM H2O2 for 1 h or 1 mM H2O2 for 20 min with EdU incorporation, detergent-pre-extracted, and fixed for EdU-click reaction and immunostaining. UDS was measured based on EdU signal incorporation. (A and E) Representative images of immunostained cells treated with 1 mM H2O2 for 20 min are shown; scale bar: 10 μm. (E) The dotted line represents the edge of the nucleus. (B–D, F, G) The mean signal intensity of EdU (B and F), PCNA (C and G) and Pol δ (D) was quantified. (H) HeLa-ATAD5mNeonGreen-AID cells were transfected with ATAD5 siRNA targeting 3′UTR and then subjected to 405-nm UV laser microirradiation. Two minutes after microirradiation, cells were fixed and immunostained as indicated; scale bar: 10 μm. (I and J) U2OS-ATAD5AID cells enriched at G1 phase by treatment with PD 0332991 and PHA-767491 were treated with auxin for 12 h before H2O2 treatment. Cells were then detergent-pre-extracted and fixed for immunostaining of XRCC1 (I) or PAR (J). Cells were incubated with EdU for 30 min before detergent-pre-extraction. (K and L) U2OS-ATAD5AID cells were treated with auxin for 12 h, irradiated with 10 J/m2 UV-C, incubated with EdU for 3 h, and subjected to PCNA immunostaining (K) and UDS assay (L). Intensity of PCNA (K) and EdU (L) signals was quantified. (B–D, F, G, I–L) Three independent experiments were performed and one representative result is displayed. Red bar indicates mean value. Statistical analysis: two-tailed unpaired Student's t-test; ****P < 0.001, **P < 0.01.
Prolonged repair DNA synthesis would not inhibit completion of the repair process in the BER/SSBR pathways if the repair DNA synthesis is properly followed by flap removal and nick ligation. However, because DNA breaks and H2O2 sensitivity are increased upon H2O2 treatment in ATAD5-depleted cells, we hypothesized that prolonged repair DNA synthesis is associated with defects in the post-DNA-synthesis processes. It has been reported that Okazaki fragments incompletely processed during DNA replication accumulate PARP1 and XRCC1 to repair nicks or gaps (62). We found that detergent-insoluble FEN1 signal increased upon H2O2 treatment in both control and ATAD5-depleted cells (Supplementary Figure S6A and B). This suggests that FEN1 is normally recruited to the damaged sites in ATAD5-depleted cells. However, it cannot be excluded that FEN1 stays at the damaged sites by binding to PCNA remaining or accumulated on DNA and that FEN1 does not act on its substrate in ATAD5-depleted cells. We found that the H2O2-induced XRCC1 signal on the 405-nm UV-microirradiated strip was increased by ATAD5 depletion (Figure 6H). In addition, detergent-insoluble XRCC1 signal induced by H2O2 treatment was further increased by ATAD5 depletion (Figure 6I and Supplementary Figure S6C). In line with the dependence of XRCC1 recruitment on PARP activity, detergent-insoluble PAR signal induced by H2O2 treatment was also further increased by ATAD5 depletion (Figure 6J and Supplementary Figure S6D). These data suggest that nick or flap structures are exposed more frequently due to the extended repair DNA synthesis as a result of ATAD5 depletion, and consequently, more PARP and XRCC1 are recruited.
The NER pathway also utilizes repair DNA synthesis as a final step. Therefore, we examined whether ATAD5 depletion affects repair DNA synthesis upon photoadduct formation. Both chromatin-bound PCNA and EdU signals were increased by UV-C irradiation (Figure 6K and L). Whereas the level of chromatin-bound PCNA was further increased by ATAD5 depletion, the EdU signal was not increased (Figure 6K and L and Supplementary Figure S6E). This suggests that PCNA, which is loaded on DNA during the NER process, is retained on DNA by an ATAD5-depletion like SSBR/LP-BER process, but the PCNA remaining on DNA does not result in prolonged repair DNA synthesis during NER.
PCNA unloading-defective ATAD5 fails to restore defects in ATAD5-depleted cells
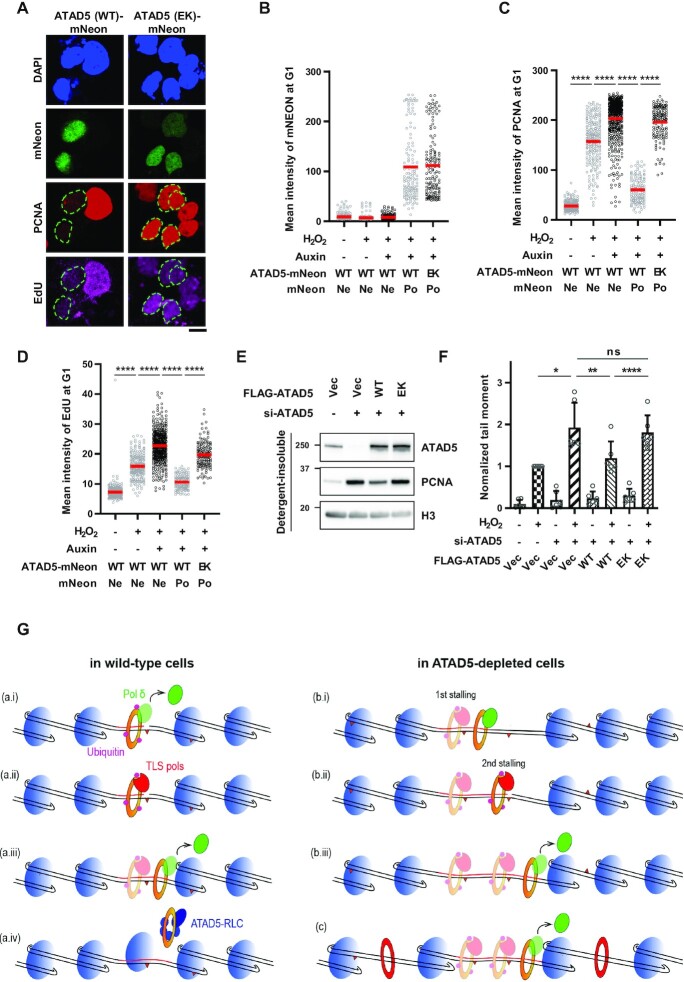
PCNA loaded on DNA upon oxidative DNA damage remains at the damaged sites in ATAD5-depleted cells (Figure 5). We checked whether the PCNA remaining or accumulated on DNA is a cause of defects observed in ATAD5-depleted cells using a recovery experiment. For this purpose, we expressed mNeonGreen-tagged wild-type or PCNA-unloading-defective ATAD5 E1173K ATPase mutant in U2OS-ATAD5AID cells and depleted endogenous ATAD5 by auxin treatment (5). Cells transfected with each DNA showed a similar mean value and distribution of mNeonGreen signal (Figure 7A and B). We found that H2O2 treatment increased chromatin-bound PCNA and EdU signals, and both signals were further increased by ATAD5 depletion in non-transfected mNeonGreen-negative cells (Figure 7C and D). The increased H2O2-induced chromatin-bound PCNA and EdU signals by ATAD5 depletion were reduced by wild-type ATAD5, marked by a positive mNeonGreen signal, but not by the PCNA-unloading-defective ATAD5. In addition, we found that the wild-type ATAD5, but not the PCNA-unloading-defective ATAD5, reduced H2O2-induced tail moment, which was increased by ATAD5 depletion (Figure 7E and F). Taken together, these results suggest that PCNA remaining on DNA as a result of ATAD5 depletion caused prolonged repair DNA synthesis and increased SSBs under conditions of oxidative stress.
Figure 7.
PCNA-unloading-defective ATAD5 fails to restore defects in ATAD5-depleted cells. (A–D) U2OS-ATAD5AID cells were transfected with cDNA expressing mNeonGreen protein-tagged wild-type (WT) ATAD5 or PCNA-unloading-defective ATAD5 E1173K (EK) mutant for 48 h before 1 mM H2O2 treatment for 20 min with EdU incorporation. Auxin was added 12 h before H2O2 treatment. Cells were then detergent-pre-extracted and fixed for immunostaining and UDS assay. (A) Representative images of cells treated with auxin and H2O2. The dotted line represents the edge of the nucleus; scale bar: 10 μm. (B–D) mNeonGreen (B), PCNA (C) and EdU signals (D) were quantified and displayed; Ne: negative, Po: positive. (E and F) U2OS cells were transfected with a combination of WT or E1173K mutant ATAD5 (EK) cDNA and ATAD5 siRNA targeting 3′UTR for 48 h. (E) Detergent-insoluble proteins were fractionated and immunoblotted. (F) Cells were treated with 0.1 mM H2O2 for 1 h and subjected to an alkaline COMET assay. (B–D) Three independent experiments were performed and one representative result is displayed. Red bar indicates mean value. (F) Six independent experiments were normalized. Error bars represent standard deviation of the mean. (B–D, F) Statistical analysis: two-tailed unpaired (B–D) and paired (F) Student's t-test (F); ****P < 0.001, **P < 0.01, *P < 0.05 and ns: not significant. (G) Graphical model for extended DNA synthesis and frequent exposure of nicks in ATAD5-depleted cells. (a) In wild-type cells treated with H2O2, (a, i) when Pol δ encounters DNA lesion on template DNA during repair DNA synthesis, it stalls and leaves from PCNA, and PCNA is monoubiquitinated. (a, ii) TLS polymerases are recruited and bypass the DNA lesion. (a, iii,iv) PCNA-Pol δ then takes over DNA synthesis until unloaded by ATAD5-RLC. After PCNA unloading, nucleosome is assembled. The remaining DNA lesions on template DNA are removed by another round of repair mechanism. (b, c) In ATAD5-depleted cells, repair DNA synthesis is extended due to the less nucleosome compaction occurred locally around clustered DNA damages (b, i) or globally (c) by PCNA remaining on DNA (red), or other yet-clear mechanism, (b, ii and c) which increases the likelihood that Pol δ will encounter DNA lesions and expose nicks. (b, iii and c) Pol δ leaves PCNA and DNA synthesis is terminated.
DISCUSSION
When ATAD5 (Elg1) is depleted in cells, PCNA accumulates and stays longer behind replication forks (6,7), and this is the primary cause of defects in DNA replication/repair and resulting genomic instability (13). Here, we find that timely unloading of PCNA by ATAD5-RLC is also important for the repair of the ROS-induced direct SSBs. Our results strongly suggest that when proper unloading of PCNA does not occur, DNA is synthesized over longer distances, and this may increase the likelihood of DNA polymerase stalling that occurs at the oxidized DNA lesions, leading to more frequent exposure of nick or flap structures (Figure 7G). This can explain the increased DNA breakage and H2O2 sensitivity of ATAD5-depleted cells (Figure 4).
An increase in ROS-induced EdU signal in ATAD5-depleted cells (Figure 6) can represent either an increased number of repair events or extended repair DNA synthesis with the same number of DNA damage sites. BER pathway choice between SP-BER and LP-BER for a given oxidized DNA lesion can affect the events of repair DNA synthesis. Several mechanisms have been suggested to regulate BER pathway choice, including cell cycle stage, type of DNA glycosylase and nucleosomal location of the DNA lesion (63–65). However, these effects can be ruled out because our results were obtained using cells at G1 phase and the choice of DNA glycosylase and location of lesion formation are determined before PCNA participates in the repair process. In addition, the PCNA-dependent pathway does not appear to be meaningfully related to the BER process initiated by two main DNA glycosylases for 8-oxoG under our experimental conditions (Figure 3J and K).
The increased EdU signal induced by ATAD5 depletion may be related to the extended repair DNA synthesis associated with altered nucleosome deposition (Figure 7G, b and c). PCNA-dependent repair DNA synthesis during LP-BER/SSBR is similar in many respects to Okazaki fragment maturation; both utilize strand-displacement synthesis carried out by the co-operation of Pol δ and FEN1, and both processes are coupled to chromatin assembly. Polymerases can invade nucleosomes, but they stall and dissociate when they approach the dyad due to increased resistance (66,67). During Okazaki fragment maturation in budding yeast, chromatin assembly factor 1 (CAF-1)-mediated rapid histone deposition constrains Pol δ extension, and therefore, loss of CAF-1 significantly increases the average length of Okazaki fragments (68). According to two recent reports, PCNA retention on DNA can lead to defects in replication-coupled nucleosome assembly in elg1Δ mutants (69,70). A reduction in nucleosome compaction was also reported in human cells depleted of ATAD5 or a CAF-1 complex subunit (71). This suggests that PCNA remaining on DNA physically inhibits nucleosome assembly, which can lead to extended Okazaki fragment maturation. Rapid recruitment of CAF-1 to SSBs in G1-phase cells also suggests that repair DNA synthesis during LP-BER/SSBR is coordinated with nucleosome assembly (72). Multiple rounds of repair can occur around clustered DNA damages upon oxidative damage, and therefore PCNA signals and EdU signals are composite ones from all repair events (42), which might explain higher PCNA and ATAD5 signals by H2O2 compared to other DNA damage treatment. The multiple rounds of repair may accumulate more PCNAs around DNA lesions with each round of DNA synthesis when ATAD5 is depleted, which may lead to loose nucleosome compaction, consequently contributing to extending repair DNA synthesis (Figure 7G, b). A slight increase in ROS-induced EdU signal by ATAD5 depletion (Figure 6B) suggests that strand-displacement synthesis proceeds slightly further due to reduced nucleosome compaction, but it soon stops due to nucleosomal resistance.
Previous reports showed the requirement of CAF-1 for NER-associated chromatin formation in vitro and colocalization of CAF-1 and NER subunits in UV-induced repair foci of human cells (73,74). However, in contrast to LP-BER/SSBR and Okazaki fragment maturation, NER utilizes PCNA-dependent gap-filling DNA synthesis, and repair patch size is determined by a dual incision process (75,76). Consistently, the UV-C-induced chromatin PCNA signal, but not the EdU signal, was increased in ATAD5-depleted cells (Figure 6K and L).
The PCNA unloading activity of yeast Elg1-RLC can be inhibited by several replisome proteins in vitro (5). This suggests that competition between the PCNA-interacting replisome proteins and ATAD5/Elg1-RLC for PCNA binding suppresses PCNA unloading until DNA replication is completed. However, according to two reports (77,78), ATAD5 mutant protein with a defect in interaction with BET family proteins, the acetyl-histone-binding chromatin reader, displayed reduced chromatin PCNA level and DNA replication rate. This suggests that there might be a mechanism that negatively regulates PCNA unloading by ATAD5-RLC, otherwise, PCNA is prematurely unloaded during DNA replication. According to a previous report, histone acetylation mark is changed upon H2O2 treatment (79). This suggests that the PCNA unloading by ATAD5-RLC can be also regulated via a similar mechanism during repair DNA synthesis.
ROS-induced DNA breakage is increased in ATAD5-depleted cells (Figure 4). We propose that extended repair DNA synthesis can be a cause of the increased breaks and H2O2 sensitivity induced by ATAD5 depletion. H2O2 treatment increased monoubiquitinated PCNA in cells at G1 phase (Figure 2F). This is consistent with cell cycle-independent, but RAD18-dependent, PCNA monoubiquitination and accumulation of TLS Polη in the chromatin upon H2O2 treatment (41–43). This suggests that when Pol δ encounters oxidized DNA lesions, such as 8-oxoG and AP sites, on the template strands during repair DNA synthesis, it stalls and is replaced by Polη, which can bypass 8-oxoG, one of the most abundant oxidized bases, and AP sites (80,81) (Figure 7G, a). DNA lesions that have been bypassed will be repaired in the next rounds of the repair process. Different from budding yeast, human Pol δ was reported to maintain a loose association with PCNA while replicating DNA, and therefore, human Pol δ rapidly dissociates from PCNA upon stalling and leaves PCNA on DNA (82). This strongly suggests that extended strand displacement synthesis may increase the chance of nicks being exposed by dissociation of Pol δ, either spontaneously or upon stalling, in ATAD5-depleted cells (Figure 7G, b and c). Augmented H2O2-induced PCNA monoubiquitination as a result ATAD5 depletion supports increased events of DNA polymerase stalling (Figure 5F). In addition, PAR/XRCC1 signals, increased by ATAD5 depletion upon H2O2 treatment (Figure 6H–J), may represent a situation in which the SSBR proteins bind to the nick. It has been reported that mismatch repair proteins play a replication-independent role against clustered DNA lesions and the role might be a prerequisite for PCNA monoubiquitination (42,83). Therefore, further studies will be needed to elucidate the involvement of the noncanonical mismatch repair process in the increase in H2O2-induced PCNA monoubiquitination by ATAD5 depletion.
Monoubiquitinated PCNA recruits error-prone TLS polymerases to bypass the DNA lesions (39,40). Termination of the potentially mutagenic TLS is performed individually or cooperatively by ubiquitin-specific protease 1 (USP1), USP7 and/or USP10, depending on the cell-cycle stage and the type of damage (41,84,85). USP1-associated factor 1 (UAF1) is required for the optimal activity and protein stability of USP1 (86). Because ATAD5 interacts with UAF1 and this interaction is important for many cellular processes, including USP1-mediated PCNA deubiquitination (14,15,34,60), in ATAD5-depleted human cells, monoubiquitinated PCNA increases without exogenous DNA damage (34). Therefore, it is possible that the augmented H2O2-induced PCNA monoubiquitination induced by ATAD5 depletion represents a defect in USP1-mediated PCNA deubiquitination, and further, this can contribute to defects observed in ATAD5-depleted cells upon H2O2 treatment. However, it has been reported that USP7 primarily contributes to the deubiquitination of H2O2-induced monoubiquitinated PCNA; whereas, USP1 primarily suppresses replication-coupled PCNA monoubiquitination and UV-induced mutagenesis (41). Even in G1 phase, the contribution of USP1 to PCNA deubiquitination under conditions of oxidative stress cannot be completely ruled out and functional interactions between ATAD5 and USP7 for USP7-mediated PCNA deubiquitination remain to be investigated. However, we think that increased DNA-polymerase-stalling events is a more probable explanation for the augmented PCNA monoubiquitination in ATAD5-depleted cells.
Detergent-insoluble PCNA and mNeonGreen-ATAD5 signal were correlatively increased upon H2O2 treatment at the G1 cells. In contrast, detergent-insoluble mNeonGreen-ATAD5 signal was not increased in the S-phase cells where PCNA was abundant in chromatin for DNA replication (Figure 5B). In addition, detergent-insoluble mNeonGreen-ATAD5 signal was detected in all unperturbed cells (Figure 1D). Consistently, endogenous ATAD5 protein was primarily detected in detergent-insoluble protein fraction (Supplementary Figure S1E). This suggests that localization of ATAD5 is differently regulated during DNA repair and replication. Additionally, this suggests that a majority of ATAD5 is associated with chromatin and/or nuclear matrix throughout the cell cycle although further detailed analysis is required. Previous reports showed that replication along each replication domain occurs in a spatially stationary fashion in living cells (87,88). In addition, a study in budding yeast showed that DNA is replicated by passing through a stationary replication factory (89). Based on these findings, DNA replication machinery including ATAD5-RLC probably performs whole genome duplication in a distributive, ordered and stationary manner via a replication timing program which was temporally and spatially established at G1 phase (90). In the contrast, because DNA damages might occur anywhere in the genome, translocation, redistribution and/or local concentration of repair proteins are required, which, in case of ATAD5, can lead to more tight chromatin association at the damage sites. However, how detergent-insoluble signals of ATAD5 increases upon DNA damage while associating with protein-dense chromatin or nuclear matrix requires further investigation.
We observed MMS sensitivity of ATAD5-depleted cells using a clonogenic survival assay but not with an acute survival assay (Supplementary Figure S4C and Figure 4H). Using the clonogenic survival assay, higher MMS sensitivity was also reported in ATAD5-depleted cancer cells (11,29). In addition, a recent genome-scale CRISPR-CAS9 screen against various DNA-damaging agents in a non-cancer cell line reported sensitivity of ATAD5-knockout cells to alkylating agents (30). In the same report, genes that promote survival against alkylating agents were more enriched in the HR and Fanconi anemia groups than the BER/SSBR group. When the replication fork collides with an unrepaired SSB or nick on DNA, a one-ended DSB is generated, which could be repaired by the HR pathway (91). Therefore, the MMS sensitization phenotype of ATAD5-depleted cells observed in the clonogenic survival assay may reflect HR defects. Several previous reports strongly suggest a role of ATAD5 (Elg1) in HR: (i) ATAD5-depleted cells are sensitive to bleomycin and CPT, both of which can generate DSBs (29); (ii) DSB-induced HR is reduced by depletion of ATAD5 (Elg1) in both budding yeast and human cells (11,27) and (iii) ATAD5-depleted cells are highly sensitive to PARP inhibitors (29,92), which is consistent with the requirement of PARP-mediated repair for survival of HR-deficient cells (93,94). Collectively, the positive role of ATAD5 (Elg1) in HR is evident but its molecular mechanism remains to be elucidated.
Altogether, we have described a new role of the replisome protein ATAD5 in maintaining genomic stability beyond its role in PCNA regulation at replication forks, which can contribute to the tumor-suppressive function of ATAD5. In addition, although the effect of cell-cycle-specific ATAD5 depletion on genomic stability, which was studied in budding yeast (16,28), was not investigated here, our data provide another example of PCNA accumulation on chromatin causing defects in DNA metabolism and genomic integrity.
DATA AVAILABILITY
All the reagents are available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members in the Center for Genomic Integrity, IBS for helpful discussions and comments on the manuscript.
Contributor Information
Su Hyung Park, Center for Genomic Integrity, Institute for Basic Science (IBS), Ulsan 44919, Republic of Korea.
Youyoung Kim, Center for Genomic Integrity, Institute for Basic Science (IBS), Ulsan 44919, Republic of Korea; Department of Biological Sciences, Ulsan National Institute of Science and Technology (UNIST), Ulsan 44919, Republic of Korea.
Jae Sun Ra, Center for Genomic Integrity, Institute for Basic Science (IBS), Ulsan 44919, Republic of Korea.
Min woo Wie, Center for Genomic Integrity, Institute for Basic Science (IBS), Ulsan 44919, Republic of Korea; Department of Biological Sciences, Ulsan National Institute of Science and Technology (UNIST), Ulsan 44919, Republic of Korea.
Mi-Sun Kang, Center for Genomic Integrity, Institute for Basic Science (IBS), Ulsan 44919, Republic of Korea.
Sukhyun Kang, Center for Genomic Integrity, Institute for Basic Science (IBS), Ulsan 44919, Republic of Korea.
Kyungjae Myung, Center for Genomic Integrity, Institute for Basic Science (IBS), Ulsan 44919, Republic of Korea; Department of Biomedical Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan 44919, Republic of Korea.
Kyoo-young Lee, Center for Genomic Integrity, Institute for Basic Science (IBS), Ulsan 44919, Republic of Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institute for Basic Science [IBS-R022-D1]. Funding for open access charge: Institute for Basic Science [IBS-R022-D1].
Conflict of interest statement. None declared.
REFERENCES
- 1. Moldovan G.L., Pfander B., Jentsch S.. PCNA, the maestro of the replication fork. Cell. 2007; 129:665–679. [DOI] [PubMed] [Google Scholar]
- 2. Yao N.Y., O’Donnell M.. The RFC clamp loader: structure and function. Subcell. Biochem. 2012; 62:259–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shiomi Y., Nishitani H.. Control of genome integrity by RFC complexes; conductors of PCNA loading onto and unloading from chromatin during DNA replication. Genes (Basel). 2017; 8:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Majka J., Burgers P.M.. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acid Res. Mol. Biol. 2004; 78:227–260. [DOI] [PubMed] [Google Scholar]
- 5. Kang M.S., Ryu E., Lee S.W., Park J., Ha N.Y., Ra J.S., Kim Y.J., Kim J., Abdel-Rahman M., Park S.H.et al.. Regulation of PCNA cycling on replicating DNA by RFC and RFC-like complexes. Nat. Commun. 2019; 10:2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee K.Y., Fu H., Aladjem M.I., Myung K.. ATAD5 regulates the lifespan of DNA replication factories by modulating PCNA level on the chromatin. J. Cell Biol. 2013; 200:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kubota T., Nishimura K., Kanemaki M.T., Donaldson A.D.. The Elg1 replication factor C-like complex functions in PCNA unloading during DNA replication. Mol. Cell. 2013; 50:273–280. [DOI] [PubMed] [Google Scholar]
- 8. Bell D.W., Sikdar N., Lee K.Y., Price J.C., Chatterjee R., Park H.D., Fox J., Ishiai M., Rudd M.L., Pollock L.M.et al.. Predisposition to cancer caused by genetic and functional defects of mammalian Atad5. PLoS Genet. 2011; 7:e1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maleva Kostovska I., Wang J., Bogdanova N., Schurmann P., Bhuju S., Geffers R., Durst M., Liebrich C., Klapdor R., Christiansen H.et al.. Rare ATAD5 missense variants in breast and ovarian cancer patients. Cancer Lett. 2016; 376:173–177. [DOI] [PubMed] [Google Scholar]
- 10. Kuchenbaecker K.B., Ramus S.J., Tyrer J., Lee A., Shen H.C., Beesley J., Lawrenson K., McGuffog L., Healey S., Lee J.M.et al.. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat. Genet. 2015; 47:164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sikdar N., Banerjee S., Lee K.Y., Wincovitch S., Pak E., Nakanishi K., Jasin M., Dutra A., Myung K.. DNA damage responses by human ELG1 in S phase are important to maintain genomic integrity. Cell Cycle. 2009; 8:3199–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shiomi Y., Nishitani H.. Alternative replication factor C protein, Elg1, maintains chromosome stability by regulating PCNA levels on chromatin. Genes Cell. 2013; 18:946–959. [DOI] [PubMed] [Google Scholar]
- 13. Lee K.Y., Park S.H.. Eukaryotic clamp loaders and unloaders in the maintenance of genome stability. Exp. Mol. Med. 2020; 52:1948–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Park S.H., Kang N., Song E., Wie M., Lee E.A., Hwang S., Lee D., Ra J.S., Park I.B., Park J.et al.. ATAD5 promotes replication restart by regulating RAD51 and PCNA in response to replication stress. Nat. Commun. 2019; 10:5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim S., Kang N., Park S.H., Wells J., Hwang T., Ryu E., Kim B.G., Hwang S., Kim S.J., Kang S.et al.. ATAD5 restricts R-loop formation through PCNA unloading and RNA helicase maintenance at the replication fork. Nucleic Acids Res. 2020; 48:7218–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson C., Gali V.K., Takahashi T.S., Kubota T.. PCNA retention on DNA into G2/M phase causes genome instability in cells lacking Elg1. Cell Rep. 2016; 16:684–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shemesh K., Sebesta M., Pacesa M., Sau S., Bronstein A., Parnas O., Liefshitz B., Venclovas C., Krejci L., Kupiec M.. A structure-function analysis of the yeast Elg1 protein reveals the importance of PCNA unloading in genome stability maintenance. Nucleic Acids Res. 2017; 45:3189–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abbotts R., Wilson D.M. 3rd. Coordination of DNA single strand break repair. Free Radic. Biol. Med. 2017; 107:228–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wallace S.S. Base excision repair: a critical player in many games. DNA Repair (Amst). 2014; 19:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krokan H.E., Bjoras M.. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013; 5:a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Whitaker A.M., Schaich M.A., Smith M.R., Flynn T.S., Freudenthal B.D.. Base excision repair of oxidative DNA damage: from mechanism to disease. Front Biosci (Landmark Ed). 2017; 22:1493–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beard W.A., Horton J.K., Prasad R., Wilson S.H.. Eukaryotic base excision repair: new approaches shine light on mechanism. Annu. Rev. Biochem. 2019; 88:137–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsumoto Y., Kim K., Hurwitz J., Gary R., Levin D.S., Tomkinson A.E., Park M.S.. Reconstitution of proliferating cell nuclear antigen-dependent repair of apurinic/apyrimidinic sites with purified human proteins. J. Biol. Chem. 1999; 274:33703–33708. [DOI] [PubMed] [Google Scholar]
- 24. Klungland A., Lindahl T.. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 1997; 16:3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woodrick J., Gupta S., Camacho S., Parvathaneni S., Choudhury S., Cheema A., Bai Y., Khatkar P., Erkizan H.V., Sami F.et al.. A new sub-pathway of long-patch base excision repair involving 5' gap formation. EMBO J. 2017; 36:1605–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. El-Khamisy S.F., Masutani M., Suzuki H., Caldecott K.W.. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003; 31:5526–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ogiwara H., Ui A., Enomoto T., Seki M.. Role of Elg1 protein in double strand break repair. Nucleic Acids Res. 2007; 35:353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paul Solomon Devakumar L.J., Gaubitz C., Lundblad V., Kelch B.A., Kubota T.. Effective mismatch repair depends on timely control of PCNA retention on DNA by the Elg1 complex. Nucleic Acids Res. 2019; 47:6826–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Giovannini S., Weller M.C., Hanzlikova H., Shiota T., Takeda S., Jiricny J.. ATAD5 deficiency alters DNA damage metabolism and sensitizes cells to PARP inhibition. Nucleic Acids Res. 2020; 48:4928–4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Olivieri M., Cho T., Alvarez-Quilon A., Li K., Schellenberg M.J., Zimmermann M., Hustedt N., Rossi S.E., Adam S., Melo H.et al.. A Genetic Map of the Response to DNA Damage in Human Cells. Cell. 2020; 182:481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ben-Aroya S., Koren A., Liefshitz B., Steinlauf R., Kupiec M.. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:9906–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilson D.M. 3rd, Thompson L.H.. Molecular mechanisms of sister-chromatid exchange. Mutat Res. 2007; 616:11–23. [DOI] [PubMed] [Google Scholar]
- 33. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee K.Y., Yang K., Cohn M.A., Sikdar N., D’Andrea A.D., Myung K.. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J. Biol. Chem. 2010; 285:10362–10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y.et al.. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013; 154:1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bradley M.O., Kohn K.W.. X-ray induced DNA double strand break production and repair in mammalian cells as measured by neutral filter elution. Nucleic Acids Res. 1979; 7:793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gautier A., Juillerat A., Heinis C., Correa I.R., Kindermann M., Beaufils F., Johnsson K.. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 2008; 15:128–136. [DOI] [PubMed] [Google Scholar]
- 38. Kong X., Mohanty S.K., Stephens J., Heale J.T., Gomez-Godinez V., Shi L.Z., Kim J.S., Yokomori K., Berns M.W.. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009; 37:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S.. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002; 419:135–141. [DOI] [PubMed] [Google Scholar]
- 40. Lee K.Y., Myung K.. PCNA modifications for regulation of post-replication repair pathways. Mol. Cells. 2008; 26:5–11. [PMC free article] [PubMed] [Google Scholar]
- 41. Kashiwaba S., Kanao R., Masuda Y., Kusumoto-Matsuo R., Hanaoka F., Masutani C.. USP7 is a suppressor of PCNA ubiquitination and oxidative-stress-induced mutagenesis in human cells. Cell. Rep. 2015; 13:2072–2080. [DOI] [PubMed] [Google Scholar]
- 42. Zlatanou A., Despras E., Braz-Petta T., Boubakour-Azzouz I., Pouvelle C., Stewart G.S., Nakajima S., Yasui A., Ishchenko A.A., Kannouche P.L.. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol eta in response to oxidative DNA damage in human cells. Mol. Cell. 2011; 43:649–662. [DOI] [PubMed] [Google Scholar]
- 43. Yang Y., Durando M., Smith-Roe S.L., Sproul C., Greenwalt A.M., Kaufmann W., Oh S., Hendrickson E.A., Vaziri C.. Cell cycle stage-specific roles of Rad18 in tolerance and repair of oxidative DNA damage. Nucleic Acids Res. 2013; 41:2296–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Caldecott K.W. XRCC1 and DNA strand break repair. DNA Repair (Amst). 2003; 2:955–969. [DOI] [PubMed] [Google Scholar]
- 45. Karimi-Busheri F., Daly G., Robins P., Canas B., Pappin D.J., Sgouros J., Miller G.G., Fakhrai H., Davis E.M., Le Beau M.M.et al.. Molecular characterization of a human DNA kinase. J. Biol. Chem. 1999; 274:24187–24194. [DOI] [PubMed] [Google Scholar]
- 46. Jilani A., Ramotar D., Slack C., Ong C., Yang X.M., Scherer S.W., Lasko D.D.. Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3'-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J. Biol. Chem. 1999; 274:24176–24186. [DOI] [PubMed] [Google Scholar]
- 47. Winters T.A., Henner W.D., Russell P.S., McCullough A., Jorgensen T.J.. Removal of 3'-phosphoglycolate from DNA strand-break damage in an oligonucleotide substrate by recombinant human apurinic/apyrimidinic endonuclease 1. Nucleic Acids Res. 1994; 22:1866–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Parsons J.L., Dianova II, Dianov G.L.. APE1 is the major 3'-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 2004; 32:3531–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Izumi T., Hazra T.K., Boldogh I., Tomkinson A.E., Park M.S., Ikeda S., Mitra S.. Requirement for human AP endonuclease 1 for repair of 3'-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis. 2000; 21:1329–1334. [PubMed] [Google Scholar]
- 50. Chen D.S., Herman T., Demple B.. Two distinct human DNA diesterases that hydrolyze 3'-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 1991; 19:5907–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Whitehouse C.J., Taylor R.M., Thistlethwaite A., Zhang H., Karimi-Busheri F., Lasko D.D., Weinfeld M., Caldecott K.W.. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001; 104:107–117. [DOI] [PubMed] [Google Scholar]
- 52. Sattler U., Frit P., Salles B., Calsou P.. Long-patch DNA repair synthesis during base excision repair in mammalian cells. EMBO Rep. 2003; 4:363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Benitez-Bribiesca L., Sanchez-Suarez P.. Oxidative damage, bleomycin, and gamma radiation induce different types of DNA strand breaks in normal lymphocytes and thymocytes. A comet assay study. Ann. N Y Acad. Sci. 1999; 887:133–149. [DOI] [PubMed] [Google Scholar]
- 54. Baker M.A., He S.Q.. Elaboration of cellular DNA breaks by hydroperoxides. Free Radic. Biol. Med. 1991; 11:563–572. [DOI] [PubMed] [Google Scholar]
- 55. Waisertreiger I., Popovich K., Block M., Anderson K.R., Barlow J.H.. Visualizing locus-specific sister chromatid exchange reveals differential patterns of replication stress-induced fragile site breakage. Oncogene. 2020; 39:1260–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ishii Y., Bender M.A.. Effects of inhibitors of DNA synthesis on spontaneous and ultraviolet light-induced sister-chromatid exchanges in Chinese hamster cells. Mutat. Res. 1980; 79:19–32. [DOI] [PubMed] [Google Scholar]
- 57. Rice P.A. Holding damaged DNA together. Nat. Struct. Biol. 1999; 6:805–806. [DOI] [PubMed] [Google Scholar]
- 58. Mol C.D., Izumi T., Mitra S., Tainer J.A.. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature. 2000; 403:451–456. [DOI] [PubMed] [Google Scholar]
- 59. Wilson S.H., Kunkel T.A.. Passing the baton in base excision repair. Nat. Struct. Biol. 2000; 7:176–178. [DOI] [PubMed] [Google Scholar]
- 60. Kim S.J., Wie M., Park S.H., Kim T.M., Park J.H., Kim S., Myung K., Lee K.Y.. ATAD5 suppresses centrosome over-duplication by regulating UAF1 and ID1. Cell Cycle. 2020; 19:1952–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wienholz F., Vermeulen W., Marteijn J.A.. Amplification of unscheduled DNA synthesis signal enables fluorescence-based single cell quantification of transcription-coupled nucleotide excision repair. Nucleic Acids Res. 2017; 45:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hanzlikova H., Kalasova I., Demin A.A., Pennicott L.E., Cihlarova Z., Caldecott K.W.. The Importance of Poly(ADP-Ribose) Polymerase as a Sensor of Unligated Okazaki Fragments during DNA Replication. Mol. Cell. 2018; 71:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mjelle R., Hegre S.A., Aas P.A., Slupphaug G., Drablos F., Saetrom P., Krokan H.E.. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair (Amst). 2015; 30:53–67. [DOI] [PubMed] [Google Scholar]
- 64. Bauer N.C., Corbett A.H., Doetsch P.W.. The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 2015; 43:10083–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Meas R., Smerdon M.J.. Nucleosomes determine their own patch size in base excision repair. Sci. Rep. 2016; 6:27122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bondarenko V.A., Steele L.M., Ujvari A., Gaykalova D.A., Kulaeva O.I., Polikanov Y.S., Luse D.S., Studitsky V.M.. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell. 2006; 24:469–479. [DOI] [PubMed] [Google Scholar]
- 67. Churchman L.S., Weissman J.S.. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011; 469:368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smith D.J., Whitehouse I.. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012; 483:434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gali V.K., Dickerson D., Katou Y., Fujiki K., Shirahige K., Owen-Hughes T., Kubota T., Donaldson A.D.. Identification of Elg1 interaction partners and effects on post-replication chromatin re-formation. PLoS Genet. 2018; 14:e1007783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Janke R., King G.A., Kupiec M., Rine J.. Pivotal roles of PCNA loading and unloading in heterochromatin function. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E2030–E2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Thakar T., Leung W., Nicolae C.M., Clements K.E., Shen B.H., Bielinsky A.K., Moldovan G.L.. Ubiquitinated-PCNA protects replication forks from DNA2-mediated degradation by regulating Okazaki fragment maturation and chromatin assembly. Nat. Commun. 2020; 11:2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Okano S., Lan L., Caldecott K.W., Mori T., Yasui A.. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 2003; 23:3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Green C.M., Almouzni G.. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 2003; 22:5163–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gaillard P.H.L., Martini E.M., Kaufman P.D., Stillman B., Moustacchi E., Almouzni G.. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996; 86:887–896. [DOI] [PubMed] [Google Scholar]
- 75. Ogi T., Limsirichaikul S., Overmeer R.M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N.G.et al.. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 2010; 37:714–727. [DOI] [PubMed] [Google Scholar]
- 76. Scharer O.D. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect. Biol. 2013; 5:a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kang M.S., Kim J., Ryu E., Ha N.Y., Hwang S., Kim B.G., Ra J.S., Kim Y.J., Hwang J.M., Myung K.et al.. PCNA unloading is negatively regulated by BET proteins. Cell Rep. 2019; 29:4632–4645. [DOI] [PubMed] [Google Scholar]
- 78. Wessel S.R., Mohni K.N., Luzwick J.W., Dungrawala H., Cortez D.. Functional analysis of the replication fork proteome identifies BET proteins as PCNA regulators. Cell Rep. 2019; 28:3497–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Niu Y., DesMarais T.L., Tong Z., Yao Y., Costa M.. Oxidative stress alters global histone modification and DNA methylation. Free Radic Biol. Med. 2015; 82:22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Masutani C., Kusumoto R., Iwai S., Hanaoka F.. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J. 2000; 19:3100–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Maga G., Villani G., Crespan E., Wimmer U., Ferrari E., Bertocci B., Hubscher U.. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007; 447:606–608. [DOI] [PubMed] [Google Scholar]
- 82. Hedglin M., Pandey B., Benkovic S.J.. Stability of the human polymerase delta holoenzyme and its implications in lagging strand DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E1777–E1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pena-Diaz J., Bregenhorn S., Ghodgaonkar M., Follonier C., Artola-Boran M., Castor D., Lopes M., Sartori A.A., Jiricny J.. Noncanonical mismatch repair as a source of genomic instability in human cells. Mol. Cell. 2012; 47:669–680. [DOI] [PubMed] [Google Scholar]
- 84. Park J.M., Yang S.W., Yu K.R., Ka S.H., Lee S.W., Seol J.H., Jeon Y.J., Chung C.H.. Modification of PCNA by ISG15 plays a crucial role in termination of error-prone translesion DNA synthesis. Mol. Cell. 2014; 54:626–638. [DOI] [PubMed] [Google Scholar]
- 85. Huang T.T., Nijman S.M., Mirchandani K.D., Galardy P.J., Cohn M.A., Haas W., Gygi S.P., Ploegh H.L., Bernards R., D’Andrea A.D.. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 2006; 8:339–347. [DOI] [PubMed] [Google Scholar]
- 86. Cohn M.A., Kowal P., Yang K., Haas W., Huang T.T., Gygi S.P., D’Andrea A.D.. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell. 2007; 28:786–797. [DOI] [PubMed] [Google Scholar]
- 87. Leonhardt H., Rahn H.P., Weinzierl P., Sporbert A., Cremer T., Zink D., Cardoso M.C.. Dynamics of DNA replication factories in living cells. J. Cell Biol. 2000; 149:271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Pardoll D.M., Vogelstein B., Coffey D.S.. A fixed site of DNA replication in eucaryotic cells. Cell. 1980; 19:527–536. [DOI] [PubMed] [Google Scholar]
- 89. Kitamura E., Blow J.J., Tanaka T.U.. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 2006; 125:1297–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rivera-Mulia J.C., Gilbert D.M.. Replicating large genomes: divide and conquer. Mol. Cell. 2016; 62:756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ensminger M., Iloff L., Ebel C., Nikolova T., Kaina B., Lbrich M.. DNA breaks and chromosomal aberrations arise when replication meets base excision repair. J. Cell Biol. 2014; 206:29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bajrami I., Frankum J.R., Konde A., Miller R.E., Rehman F.L., Brough R., Campbell J., Sims D., Rafiq R., Hooper S.et al.. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 2014; 74:287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C.et al.. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005; 434:917–921. [DOI] [PubMed] [Google Scholar]
- 94. Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T.. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005; 434:913–917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the reagents are available upon request.