Abstract
Studies of transcription regulation are often focused on binding of transcription factors (TFs) to a small number of promoters of interest. It is often assumed that TFs are in great excess to their binding sites (TFBSs) and competition for TFs between DNA sites is seldom considered. With increasing evidence that TFBSs are exceedingly abundant for many TFs and significant variations in TF and TFBS numbers occur during growth, the interplay between a TF and all TFBSs should not be ignored. Here, we use additional decoy DNA sites to quantitatively analyze how the relative abundance of a TF to its TFBSs impacts the steady-state level and onset time of gene expression for the auto-activated Escherichia coli PhoB response regulator. We show that increasing numbers of decoy sites progressively delayed transcription activation and lowered promoter activities. Perturbation of transcription regulation by additional TFBSs did not require extreme numbers of decoys, suggesting that PhoB is approximately at capacity for its DNA sites. Addition of decoys also converted a graded response to a bi-modal response. We developed a binding competition model that captures the major features of experimental observations, providing a quantitative framework to assess how variations in TFs and TFBSs influence transcriptional responses.
INTRODUCTION
Cells respond to environmental perturbations by allocating cellular resources to produce the right amount of response proteins at the right time. Gene expression levels usually correlate with cell fitness, and concentrations of many proteins are optimized to meet cellular demands and provide balanced cost and benefit (1–4). Biochemical characteristics of proteins, such as enzyme activities and the binding stoichiometry within multi-protein complexes, impact the cost and benefit, thus placing constraints on cellular protein levels (2–5). Protein expression levels are primarily modulated via transcription regulation by transcription factors (TFs). The fundamental biochemical reaction in TF regulation is the binding of TFs to regulatory DNA. TF affinity, TF abundance and the total number of regulatory DNA sites can all affect the binding reaction and the regulatory output. Extensive progress has been made in characterizing TF binding activities and understanding how TF abundance and affinities impact gene regulation. For example, binding affinities of TFs to different target promoter sites are found to determine the temporal order of gene expression and have been evolved for hierarchical expression of genes following their functional orders (6–8). However, how the total number of transcription factor binding sites (TFBSs) affects transcription regulation has been less explored. The quantitative consequences on gene expression levels and response dynamics when the relative abundance of TFs to TFBSs is altered are incompletely understood.
The total number of TFBSs creates a cellular demand for the TF and may place constraints on optimal TF abundance. Except for a small fraction of global TFs, prokaryotic TFs usually regulate a limited set of genes. In Escherichia coli, >70% of TFs regulate five or less target operons according to RegulonDB (9). Thus, it is often assumed that the number of TF protein molecules is in great excess to the number of TFBSs. Recent genome-wide binding studies of many prokaryotic TFs are starting to change our view about the relative abundance of TFs relative to TFBSs. A large number of TFBSs have been discovered in gene coding regions distant to regulatory promoters (10–14). For example, a total of >10 000 (66%) binding sites for 154 TFs from Mycobacterium tuberculosis are located outside the promoter window (11). Most of the intragenic TFBSs do not appear to be associated with direct transcription regulation (10,11) and are believed to arise from random occurrence due to lack of functional selection (15). Although non-functional individually, these intragenic sites, often referred to as natural decoys (16), collectively impact the relative abundance of TFs to TFBSs and may compete with functional sites for the availability of TF proteins. This interplay between TFs and all TFBSs causes the output of a transcription unit to be dependent not only on transcriptional components but also on downstream targets and genomic environments, which is termed as ‘retroactivity’ (17). TF competition has also been exploited with synthetic decoy binding sites to modulate TF activity for purposes of cancer therapy or metabolic engineering (18–21).
Understanding the regulatory potential of natural and synthetic decoys requires precise knowledge of how the relative abundance of TFs to TFBSs impacts transcription output. Despite many theoretical studies on the decoy effects (22,23), quantitative characterization of the interplay between TFs and TFBSs has been limited to only a few systems, such as the E. coli LacI repressor (24) and a synthetic auto-repressed system based on LacI (25). In comparison to repressors, transcription activators in E. coli are generally expressed in lower numbers (5), likely reflecting a TF demand different from that of repressors. Nearly 20% of E. coli TFs are positively autoregulated according to RegulonDB (9). Positive autoregulation is a common regulatory motif important for response speed, sensitivity, expression output and noise. We chose to focus our study on a natural auto-activated system, the E. coli PhoR/PhoB system, which has been extensively studied in quantitative detail.
The PhoR/PhoB system belongs to the family of two-component systems, a versatile signaling scheme prevalent in prokaryotes (26,27). The sensor histidine kinase PhoR responds to limitation of environmental phosphate (Pi) and modulates its kinase and phosphatase activities to control the phosphorylation level of the response regulator (RR) PhoB. Phosphorylated PhoB (PhoBp) is the active form of the TF that activates expression of itself and other genes responsible for phosphorus assimilation. The timing of expression of these genes, such as phoB, phoA and phnC, is determined by the affinities of binding sites for PhoBp in their promoters and matches their functional order of need during the Pi starvation response (7). PhoB has been well characterized in the context that its abundance, phosphorylation, and DNA-binding properties can be adjusted by experimental means and the regulatory impact of these variations have been assessed both experimentally and theoretically (4,7,28–30). PhoB autoregulation enables an ∼20-fold increase of PhoB concentrations upon stimulation by Pi depletion (30). A global binding study revealed a total of 54 genomic binding sites with 20 in gene coding regions (31). DNA binding sites of PhoBp have been well characterized (7,32), allowing us to design synthetic decoys to alter the relative abundance of PhoB to its TFBSs. In this work, we show that decoys reduce transcription output of PhoB regulated genes in an engineered non-autoregulatory system. This decoy effect depends on TF abundance and promoter affinities and can be well predicted by a quantitative binding competition model. The non-stimulated expression level of PhoB is near capacity to the demand and the autoregulated system is susceptible to decoy titration. In the wild-type autoregulated system, decoys reduce and delay the response, as well as convert a graded response to a bimodal response.
MATERIALS AND METHODS
Strains and plasmids
Strains and plasmids used in this study are listed in Supplementary Table S1. E. coli strain BW25113 (33) is the parent strain from which all strains were derived. Strain RU1616, also known as LAC with an IPTG-inducible promoter Plac promoter replacing the autoregulated phoB promoter, has been described previously (30). To create YFP reporters, fragments containing the PphoB* promoter, the PphoB promoter with the auto-repression site disrupted (28), and the PphnC promoter were excised from pRG368 and pRG400 by PstI/XbaI digestion, and ligated to pRG261 to replace the PphoA promoter in the integration plasmid. The resulting plasmids pLH1 and pLH2, together with pRG261, were respectively integrated into the HK022 phage attachment site in the chromosome of either BW25113 or RU1616 using recombination strategies (34). It was confirmed that only a single copy of DNA was integrated.
A vector plasmid for cloning decoy sites, pRG475, was generated by Golden Gate assembly of multiple PCR fragments containing the ColE1 replication origin, a Ptet-cfp, an ampicillin resistance gene and two BsaI sites. DNA fragments with different numbers of decoy PhoB-binding sites (each a 22-bp consensus sequence for PhoB dimer binding) were cloned between the two BsaI sites of pRG475 by Golden Gate cloning. An ∼170 bp non-specific DNA with no decoy sites was ordered from Integrated DNA Technologies (IDT) and used to create pLH10. For cloning one, two and three decoy sites, the desired number of annealed oligos with complementary ends were assembled into pRG475 to generate pRG491, pRG492 and pLH6. DNA with multiple decoy sites was then amplified by PCR to generate compatible ends for further assembly into higher numbers of decoy sites, yielding pLH8 (five consensus sites) and pLH9 (7 consensus sites). Detailed DNA sequences and cloning procedures are provided in Supplementary materials. Plasmid DNA stability was evaluated by DNA sequencing before and after Pi starvation and reporter activation assays; decoy repeats were stable and recombination was not observed after the reporter assay.
Bacterial growth conditions and phosphate starvation
Bacteria were grown in LB broth or MOPS minimal media (35) with appropriate antibiotics, 0.4% (w/v) glucose and amino acids (40 μg/ml each) as described previously (36). For reporter strains that carry the spectinomycin resistance gene on the chromosome, 12.5 μg/ml instead of 50 μg/ml of spectinomycin was included in the MOPS.
For phosphate starvation, cells from overnight MOPS cultures (Pi-replete, 2 mM KH2PO4) were diluted 20 fold into fresh Pi-replete MOPS media for inoculation. Once the optical density (OD) measured at 600 nm reached 0.4–0.6, bacteria were washed with non-activating MOPS medium (20–50 μM Pi) twice and resuspended in activating MOPS medium (4 μM Pi) for Pi starvation. The starting OD was 0.15 and the inoculated cultures were transferred to either 96-well plates for fluorescence reporter assays, or flasks for flow cytometry and immunoblots. Aliquots corresponding to approximately 0.3 OD*ml cells were taken from bulk cultures for the subsequent SDS-PAGE and quantitative western analyses as described (30).
Determination of plasmid copy number by qPCR
Cell cultures under exponential growth in Pi-replete MOPS medium were harvested for total DNA extraction using the QIAamp DNA mini kit (Qiagen). Extracted DNA was used as template in qPCR with four sets of primers targeting three chromosomal genes, dxs, secD and tdk, and one plasmid-borne gene bla. Reactions were performed with GoTaq qPCR mix (Promega) using a QuantStudio 3 cycler (Thermofisher). Details of primers and qPCR conditions are provided in Supplementary materials. For target genes, differences in threshold cycle values, ΔCt, were calculated using dxs as the reference. Amplification efficiency was determined for dxs and bla to be ∼1, indicating one doubling of DNA amount per PCR cycle. Thus, the plasmid copy number is based on ΔCt of bla and calculated as 2^ΔCt, representing the relative copies of plasmid to chromosome. To evaluate whether the number of decoy sites or strain background impacts plasmid copy number, DNA samples from RU2079/pLH10 were used as a reference to measure differences of ΔCt, ΔΔCt, for samples with different decoy plasmids or strain backgrounds, leading to calculation of relative ratios of plasmid copy numbers.
Fluorescence reporter assays
Inoculated cultures in 96-well plates were monitored for YFP fluorescence (excitation 488 nm, emission 530 nm) and OD 600 nm every 5–7 min using a Varioskan plate reader (Thermo Scientific) with constant shaking. The time when bacteria were inoculated in activating MOPS medium (4 μM Pi) was considered as time 0, and there is usually an ∼10-min interval between time 0 and the first measurement because of the time required for loading the 96-well plates and temperature equilibration. CFP expressed from Ptet-cfp on decoy plasmids did not interfere with YFP fluorescence detection. YFP fluorescence and growth OD data were processed as described previously (7,28). Briefly, background YFP fluorescence was derived from a control strain, BW25113 containing pLH10 and pCL1920 (37), and subtracted from reporter fluorescence. First derivatives of smoothed fluorescence (dFluo./dt) were calculated numerically and normalized to OD to represent the promoter activity [(dFluo./dt)/OD] (28). Because the stress response elicited by Pi starvation can greatly reduce the promoter activity during the late stage of starvation (36), a time window of ∼40 min after the promoter activity reached a plateau, but before the transcription repression by stress, was identified as the plateaued region, and the average promoter activity within this time window was calculated to represent the reporter transcription output. A reference strain with 0 decoy sites was included in every assay and the plateaued promoter activity of samples was compared to the reference strain to calculate the relative promoter activity. The fluorescence onset time was identified as the time point when fluorescence switches on and the second derivative of fluorescence peaks (7).
Flow cytometry analyses
Inoculated cultures were grown under Pi starvation for 2.5 h and diluted 300 fold for flow cytometry analyses using a Gallios Flow Cytometer (Beckman Coulter). YFP (FL1, excitation 488 nm) and CFP (FL10, excitation 408 nm) were measured for ∼60 000 cells per sample. Cell counts were normalized to total cell counts to represent relative cell population. Data with outlier values of front scattering or side scattering (<1%) were excluded for fluorescence analyses. Spearman correlation between CFP and YFP was calculated from the measured log10 values of CFP and YFP. For LAC strains with constant PhoB expression, the entire population was used for the correlation calculation while only the YFP-activated fluorescent population was used for the autoregulated strains with wild-type (WT) phoB. The YFP-activated fluorescent population was determined from gating based on the histogram of bimodal YFP distribution.
Modeling of decoy competition
A deterministic model was used to describe the TF titration effects. The model includes two major regulatory modules: (i) the phosphorylation cycle that determines the total PhoBp concentration [PhoBp]T based on the input of total PhoB concentration [PhoB]T and (ii) the transcription regulation module that describes the competitive binding of PhoBp to various DNA sites and outputs the promoter occupancy.
The phosphorylation module was modeled similarly to that described previously (30,38,39). Steady-state levels of [PhoBp]T, are determined by [PhoB]T, and two composite enzyme activity parameters Cp and Ct as follows:
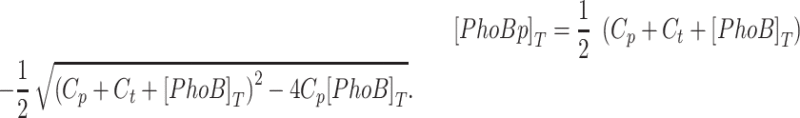 |
(1) |
To model transcription regulation, promoter activity is presumed to be proportional to the binding probability of PhoBp to the promoter binding site, termed as promoter occupancy for simplicity. Decoys are believed not to affect other promoter elements, such as binding of RNA polymerase and DNA access due to nucleoid association, thus these effects are not considered. DNA binding reactions are fast, so a quasi-equilibrium approximation was used for the competitive DNA binding events. PhoBp binds DNA as a dimer with a Hill coefficient of 2 for two half-sites (40), thus, occupancy of any given full DNA site, p, can be described by the following equation:
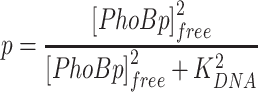 |
(2) |
in which KDNA is the dissociation constant for the particular DNA site and [PhoBp]free is the concentration of unbound PhoBp. For simplicity, cooperativity between different full sites is not considered. The total concentration of PhoBp is the sum of bound and unbound PhoBp:
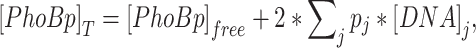 |
(3) |
in which [DNA]j is the concentration of DNA binding sites that have the affinity of KDNA, j and the binding occupancy pj. [DNA]j is derived by multiplying the concentration of a single site with the total number of binding sites and the growth-dependent copy number of genome equivalent (41). Equivalent genome copy number is based on the copy number of the qPCR reference gene dxs, and the value is 1.5 (see details in Supplementary materials). In addition to the reporter site and decoy sites, all the endogenous DNA sites within the genome are treated as an ensemble of sites with a single affinity for simplicity. For non-autoregulatory systems with constant [PhoB]T, Equations (1–3) are numerically solved to derive [PhoBp]free and the reporter promoter occupancy. Simulated promoter occupancy data are further divided by the reference promoter occupancy at 8.2 μM [PhoB]T to compare with the experimentally measured relative promoter activities.
Modeling of autoregulation
For the autoregulated PhoB, [PhoB]T is determined by the PhoB production rate as well as the growth dilution and degradation rates. When autoregulation is via a sole positive feedback, PhoB production rate is dependent on promoter occupancy. At assay conditions, PhoB is very stable and degradation of PhoB is minimal (36), thus only growth-dilution (rate constant kdil) is considered:
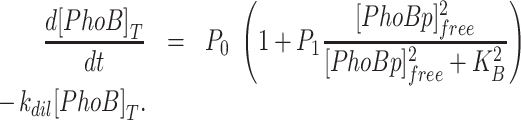 |
(4) |
P 0 is the basal production rate and P1 is the maximal induction fold. Both were parameterized relative to kdil from PhoB concentrations as described previously (28). At steady state, d[PhoB]T/dt equals 0 and solutions for Equations (1–4) can be numerically obtained to assess the steady-state solutions. For more complex autoregulatory schemes such as the coupled positive and negative feedbacks, additional regulation factors are used to modify the RR production rate. Details for these regulatory schemes and simulation of [PhoB]T kinetics are included in the Supplementary materials. Parameter values for all modeling are listed in Supplementary Table S2 and details of parameter estimation are described in Supplementary materials.
RESULTS
Design of the decoy system for quantitation of TF titration
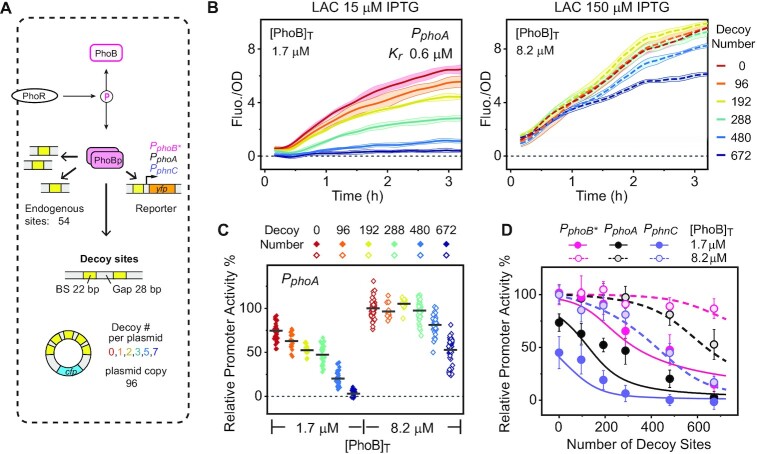
For the autoregulated PhoR/PhoB system, our previous analyses estimated concentrations of the response regulator PhoB at 0.45 μM (∼270 molecules) in the absence of stimulus and 9.3 μM (∼5500 molecules) when stimulated (7,30), corresponding to ∼5- and 100-fold the total number of PhoB binding sites. To alter the relative abundance of PhoB to its DNA binding sites and investigate its impact on transcriptional responses, DNA decoy binding sites were introduced via a multi-copy plasmid (Figure 1A) into the autoregulated strains with wild-type (WT) phoB or the engineered non-autoregulatory strains (LAC) in which PhoB levels can be modulated by IPTG (30). The decoy site consists of a 22-bp consensus sequence to compete for binding of a PhoB dimer, and multiple decoy repeats (between 0 and 7 per plasmid) were inserted to create a range of total decoy numbers. There is a 28-bp gap between decoy sites to minimize the complexity of binding cooperativity between immediately adjacent PhoB-binding sites.
Figure 1.
Titration effects of decoy sites on response output in engineered non-autoregulatory strains. (A) Schematic diagram of the experimental design of TF titration by decoys. The active TF, phosphorylated PhoB (PhoBp), can bind to both endogenous chromosomal sites and the plasmid-borne decoy sites. (B and C) Reduction of phoA reporter fluorescence by decoys. Reporter outputs (B) were tracked in strain RU1988 (LAC, PphoA-yfp) with plasmids harboring different numbers of decoy sites. Time 0 refers to the time when bacteria were resupended in Pi-limited media. Lines with corresponding shaded ranges indicate mean ± SD from 8 individual wells from one representative experiment. Relative promoter activities (C) were derived using the reference promoter activity at 8.2 μM PhoB with no decoy sites. Diamond symbols illustrate data from individual cultures and horizontal bars indicate the averages. (D) Dependence of decoy effects on DNA affinity and total PhoB concentrations. Three reporter strains with decreasing promoter affinities for PhoBp, RU1989 (PphoB*), RU1988 (PphoA) and RU1990 (PphnC), were assayed. Data are shown as mean ± SD. All data were from at least two independent experiments with eight individual cultures per experiment. Solid and dashed lines represent modeled data at corresponding total PhoB concentrations.
In order to quantitate the decoy competition effects, binding affinity and the total number of decoy sites were measured. Binding affinity of phosphorylated PhoB (PhoBp) to the decoy site was determined similarly as other PhoB-binding sites using electrophoretic mobility shift assays (28), yielding a dissociation constant Kdecoy of 0.2 μM (Supplementary Figure S1). The plasmid copy number was measured in relation to the chromosomal reference gene dxs by qPCR to derive the total number of decoy sites (Supplementary Figure S2 and Supplementary materials). The average plasmid copy number is 96 ± 31 and different numbers of decoy repeats or reporter strain backgrounds do not significantly alter the copy number (Supplementary Figure S2). For decoy plasmids with 0, 1, 2, 3, 5 and 7 decoy sites per plasmid, this yields a series of decoy numbers of 0, 96, 192, 288, 480 and 672.
Effects of decoys in the non-autoregulatory strains are consistent with binding competition
Effects of DNA-binding decoys on transcription output were evaluated with chromosomal YFP transcription reporters at two constant PhoB concentrations, one at 8.2 μM (∼4900 molecules), which enables full promoter activation, and one at 1.7 μM (∼1000 molecules), which yields partial but still considerable promoter activation (7). Upon stimulation with Pi depletion, YFP fluorescence gradually increased (Figure 1B and Supplementary Figure S3A). Increasing numbers of decoys gradually reduced transcription of the PphoApromoter at both PhoB levels (Figure 1B). To quantify transcription levels, the rate of fluorescence increase is normalized to cell density (OD600) to represent transcription rate, i.e., promoter activity, and promoter activities within a plateaued region (Supplementary Figure S3B) are used to represent reporter transcription output. To compare transcription output across strains with different number of decoys or different TF abundance, plateaued promoter activities are compared against the corresponding promoter activity of a reference strain to derive the relative promoter activities (Supplementary Figure S3C). Strains carrying the plasmid with 0 decoys are used as reference strains because this plasmid does not alter the reporter promoter activity (Supplementary Figure S3C), suggesting that the vector backbone of decoy plasmids does not impact transcription regulation.
Transcription reduction by decoys depends on PhoB concentrations. At 8.2 μM PhoB, apparent reduction of promoter activity was not observed until addition of 480 decoys, ∼1/10 the number of PhoB molecules, while 96 decoy sites clearly decreased the promoter activity at 1.7 μM PhoB (Figure 1C and D). Similar patterns of decoy effects have also been observed with two other promoters (Supplementary Figure S4), with each containing a single operator site with different affinities for PhoBp. Three promoters, PphnC, PphoA and PphoB*, have increasing affinities to PhoBp with KD values of 1.5, 0.6 and 0.25 μM, respectively. Decoy effects correlate with PhoBp affinities, and the promoter with a higher affinity is less susceptible to transcription reduction by decoys (Figure 1D).
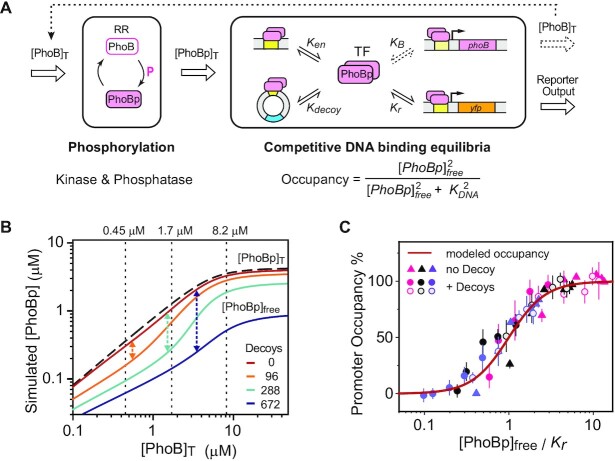
Qualitatively, the observed transcription reduction by decoys can be easily explained by decoys titrating away PhoBp molecules. We sought to quantitatively evaluate this decoy titration effect with a deterministic model and examine how well model prediction matches experimental data. Two modules, including phosphorylation and competitive DNA binding, are considered (Figure 2A). The phosphorylation module describes the phosphorylation, or activation, of the TF PhoB. Unlike many other TFs whose active form is difficult to track in cellular conditions, cellular phosphorylation of the RR PhoB has been well described in previous modeling and measurements (30,39). Steady-state phosphorylation output of the PhoR/PhoB two-component system can be approximated with a function dependent on the total PhoB concentration [PhoB]T (Equation (1) in Materials and Methods). For every input of [PhoB]T, the module outputs [PhoBp]T (dashed line in Figure 2B). For example, at [PhoB]T levels of 1.7 and 8.2 μM, [PhoBp]T levels are 1.3 and 3.5 μM, corresponding to ∼75% and 40% of total PhoB concentrations. [PhoBp]T is the input for the binding competition module. The numbers and affinities of all competing DNA sites either have been measured, e.g., decoy sites and the binding site in the reporter promoter, or can be estimated, e.g., endogenous chromosomal sites (see details in Supplementary materials). A quasi-equilibrium approximation is used to determine the concentration of free unbound PhoBp ([PhoBp]free, solid lines in Figure 2B). Competition by decoys decreases [PhoBp]free, and our simulation indicates that the largest relative changes of [PhoBp]free (arrowed lines in Figure 2B) at 96, 288 and 672 decoys occurs at [PhoB]T levels of 0.5, 1.4 and 3.3 μM, with PhoBp molecules at ∼240, 600 and 1300, ∼two-fold the number of decoys. This suggests that the largest relative titration effect on PhoBp occurs when the PhoBp number is about the same as the total protein-binding capacity of decoy DNA sites.
Figure 2.
Modeling of the decoy effects on transcription output. (A) Phosphorylation and competitive DNA-binding modules of the decoy titration model. (B) Simulated titration effects of decoys on concentrations of free unbound PhoBp. Dashed line indicates the total concentration of PhoBp derived from the phosphorylation module. Solid lines represent [PhoBp]free with different number of decoys. Arrowed lines indicate the largest relative change of [PhoBp]free caused by different number of decoys. Vertical dotted lines highlight three [PhoB]T levels measured for the non-stimulated WT, and LAC induced with 15 μM and 150 μM IPTG. The corresponding PhoBp levels represent ∼80%, 75% and 42% of total [PhoB]T levels. (C) Agreement between experimental data and the binding competition model. For the entire collection of data with various promoter affinities (colored similarly as Figure 2), [PhoB]T levels (triangles) and decoy numbers (circles), [PhoBp]free are calculated and further divided by the promoter affinity Kr to get a uniform scale. The maroon line represents the theoretical binding curve modeled with the equation shown in (A).
To model transcription output and compare with experimental data, occupancy of the reporter promoter can be derived from [PhoBp]free and the promoter affinity. Because transcription output was experimentally measured as relative promoter activities in relation to the reference strain, the modeled occupancy was compared to the reference occupancy of a specific promoter with 0 decoy sites at 8.2 μM [PhoB]T, thus the resulting values are considered as modeled promoter activities that can be compared with measured data. With all parameters known, the model predicts [PhoBp]free and the relative promoter activities for every experimentally tested condition. Predicted activities (dashed and solid lines in Figure 1D) agree well with the experimental data. Aside from decoy effects, the relative abundance of PhoB can also be altered by changing PhoB protein concentrations. PhoB abundance impacts relative promoter activities differently depending on PhoBp affinities to reporter promoters, and these effects are well captured by model predictions (Supplementary Figure S4E). Relative promoter activities can be converted to promoter occupancy by multiplying by the reference occupancy (see details in Supplementary Figure S4). An alternative way to examine the agreement between experimental and predicted data is to plot promoter occupancy as a function of scaled [PhoBp]free calculated from [PhoB]T and decoy numbers (Figure 2C). All of the theoretically predicted curves conform to a single binding curve (maroon line) and the experimental data agree well with this simple binding competition model. This provides a quantitative framework to assess the decoy effects on the autoregulatory WT system.
Decoys reduce and delay transcription activation in the autoregulated WT
The WT PhoB expression level increases >20-fold from 0.45 μM in the absence of stimulus to 9.3 μM when stimulated. As shown in Figure 2B, at 0.45 μM [PhoB]T, the predicted [PhoBp]free reduced from 0.3 μM at 0 decoys to 0.15 μM with only 96 decoy sites. This will greatly reduce occupancy of the autoregulated phoB promoter, which has a PhoBp affinity of 0.25 μM, affecting both PhoB expression rate and steady-state levels. Therefore, decoys are expected to alter the autoregulation kinetics and amplify the transcription reduction effects in autoregulatory strains by lowering PhoB expression as well as by limiting its availability to the reporter promoter via binding competition.
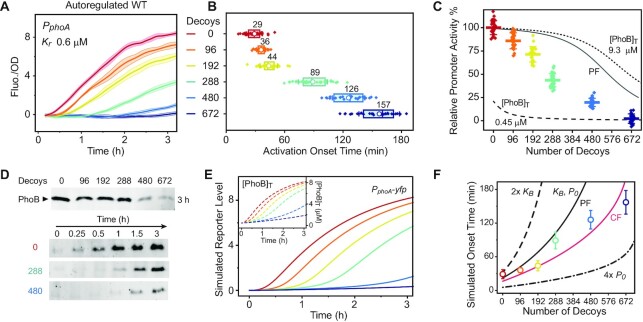
Indeed, all three reporters displayed reduced promoter activities and delayed transcription activation in the presence of decoys (Figure 3A and Supplementary Figure S5). Onset time of transcription activation, defined as the switch point of YFP fluorescence that starts to rise (7), increased from 29 to 36 min with 96 decoys, and 126 min with 480 decoys for the PphoA promoter (Figure 3B). Considering the doubling time of less than 1 h, a 7-min delay with merely 96 decoys is non-trivial. Relative promoter activities also decreased greatly with increasing number of decoys and were almost completely titrated away by 672 decoys (Figure 3C and Supplementary Figure S5). Not surprisingly, promoter activities of the autoregulatory strain fall between the theoretical curves predicted from two constitutive PhoB levels at 0.45 and 9.3 μM. Promoter activities at higher numbers of decoys are closer to the curve with low [PhoB]T. Immunoblot analyses indicate that a high number of decoys results in a significantly reduced level of PhoB (Figure 3D), which also contributes to reduction of reporter output. Slow-down of PhoB protein accumulation is apparent with addition of decoys, accounting for the observed delay in reporter activation.
Figure 3.
Effects of decoy sites on the output level and timing of transcription in the autoregulated WT. (A–C) Effects of decoys on PphoA-yfp activities in the WT RU2079 strain. Decoys decreased reporter fluorescence (A), delayed response as shown in activation onset times (B) and reduced the relative promoter activities (C). Data from individual wells are shown by diamond symbols and the averages are shown in circles in (B) and horizontal bars in (C). Lines are modeled curves: solid, positively autoregulated; dashed and dotted, constitutive PhoB expression at indicated concentrations. (D) Reduced and delayed PhoB expression in the presence of decoys. Immunoblots were used to show total PhoB levels 3 h after activation (top) and PhoB expression kinetics (bottom). (E) Simulated decoy effects in the autoregulatory system. Both PphoA transcription levels and total PhoB concentrations (inset) are simulated for a system with a sole positive feedback. (F) Dependence of onset time on autoregulation parameters. Colored circles indicate the mean ± SD of experimentally measured onset times from (B). Simulated onset time for PphoA is defined as the time when [PhoBp]free reaches the binding affinity of PphoA, 0.6 μM. Black lines are simulated with a sole positive feedback (PF) with indicated autoregulation parameters and the red line is modeled with a coupled feedback (CF).
Built upon the binding competition model, autoregulation is modeled by feeding back transcription of the phoB promoter and the production rate of [PhoB]T to the phosphorylation module. First, we consider the simplest form of positive autoregulation, a sole positive feedback with TF binding to an activation site at the autoregulated promoter. Decoys lead to a decrease in both PhoB expression and reporter output (Figure 3E). Simulated reporter dynamics qualitatively recapitulate the decoy effects observed experimentally, such as output reduction and response delay, but the predicted steady-state promoter activities (solid line, Figure 3C) are much higher than measured data. Our simulation indicates that the response delay caused by decoys greatly lengthens the time required for reaching the steady state well beyond the experimental timescale of 3 h (Supplementary Figure S6A–C). Discrepancies between experimental and modeled data may result from different timescales and/or additional transcriptional regulation, such as a starvation-induced general stress response (42,43) that is not considered in the simple positive feedback model. Moreover, instead of a sole positive feedback, a coupled feedback involving an additional weak repression site has been discovered in the phoB promoter (28). The effect of decoy titration on the repression site is expected to increase rather than decrease the steady-state [PhoB]T (Supplementary Figure S6D–F), inconsistent with experimental observations.
It has been documented that the starvation-induced stress response can cause significant transcription repression at the late stage of Pi starvation (36). Thus, we sought to include the stress response in the coupled feedback model to reconcile the apparent discrepancy between modeled and experimental data. An extremely simplified model of stress response can account for the observed decrease of PhoB expression at high numbers of decoys (Supplementary Figure S6G-S6J). The response delay causes cells to enter the late stationary phase without enough expression of PhoB, and the stress-related inhibition overshadows the transcription-increasing effect from TF titration on the repression site. Accurate prediction of the WT response requires a better quantitative understanding of the starvation stress response, which is beyond the scope of this study. However, the major features of decoy effects on transcription output and temporal response can be captured by our autoregulation model.
Response speed, or the timing of regulated gene expression, is important for appropriate response output and feedback regulation greatly impacts response dynamics (44). Positive autoregulation is known to slow the response (45,46), and decoy titration further amplifies the delay. Our simulation indicates how the autoregulation parameters, such as promoter strength and TF binding affinity, impact the transcription activation onset time in the presence of decoys (Figure 3F). A high basal transcription rate, P0, results in a high basal concentration of PhoB, which provides fast response with short onset time across a wide range of decoy numbers (dash dotted line in Figure 3F), but also carries fitness cost under non-stimulating Pi-replete conditions (4). The autoregulated phoB promoter has one of the strongest affinities among PhoB-binding sites. Low affinity to the autoregulated promoter (dashed line in Figure 3F), i.e., a high value of KB, causes a modest increase in the onset time in the absence of decoys, while addition of decoys increases the onset time, more dramatically than occurs with a strong affinity promoter. It appears that the autoregulated promoter with a stronger affinity is relatively better buffered, or less susceptible to the change of onset time caused by alteration of relative abundance of a TF to TFBSs.
Decoys promote bimodal responses in the autoregulated WT
All decoy titration effects described above were observed in bulk bacterial cultures. We also used flow cytometry to examine decoy effects in single cells. For individual cells, the plasmid copy number, and thus the total number of decoys, is not a single value, but rather a distribution owing to cell-to-cell variability. As all decoy plasmids harbor a constitutively expressed cfp gene, the population distribution of CFP fluorescence will reflect the distribution of plasmid DNA amount. All decoy plasmids showed similar uniform distributions of CFP fluorescence (Supplementary Figure S7A). When there is no decoy, cells display a single distribution of PphoA-yfp reporter output (Figure 4) and have a positive correlation (Spearman correlation coefficient r = 0.61) between CFP and YFP due to the shared cellular machinery for expressing fluorescent proteins. Subpopulations of cells with higher plasmid copy numbers, or higher CFP, have more decoys and thus are expected to have less YFP reporter output, decreasing the CFP-YFP correlation. The higher the number of decoys, the stronger the titration effect and the less the CFP-YFP correlation. Such decreased correlation by decoys was observed in fluorescent cell populations with the correlation coefficient reduced from 0.61 with 0 decoys to 0.04 with 672 decoys (Figure 4A and Supplementary Figure S7E).
Figure 4.
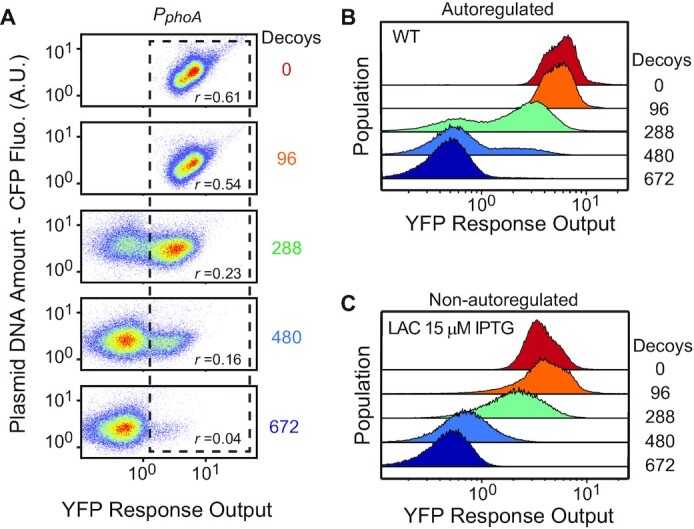
Bimodal distribution of reporter fluorescence in the presence of decoy sites. (A) Flow cytometry analyses of RU2079 (PphoA-yfp) carrying different decoy plasmids. Every decoy plasmid contains a constitutively expressed cfp gene. The vertical axis shows CFP fluorescence that reflects plasmid copies while the horizontal axis shows the reporter YFP fluorescence. Dashed rectangles highlight the cell populations with activated YFP fluorescence, and correlations between CFP and YFP fluorescence for these activated populations are indicated by the Spearman correlation coefficient r. (B and C) Histograms of PphoA-yfp fluorescence for the autoregulated WT (RU2079) and non-autoregulated LAC (RU1988) strain.
More remarkably, addition of decoys results in bimodal expression of YFP reporters. Two discrete populations of cells, corresponding to activated cells with high YFP fluorescence and non-activated cells with background fluorescence, are apparent with intermediate numbers (288 and 480) of decoys (Figure 4B). The PhoR/PhoB system has been shown to have a graded response to varying stimulus strength (4). In the absence of decoys, cells display uniform distributions of YFP reporter output with a gradual increase of mean fluorescence at successive time points after Pi starvation (Supplementary Figure S7B). Addition of decoys converts the graded response to a bimodal response. Bimodality is not limited to the PphoA reporter but also observed for PphoB* and PphnC reporters (Supplementary Figure S7C, D). Positive autoregulation is necessary for bimodal responses because the non-autoregulatory strain displays graded responses with decoys at different constitutive PhoB expression levels (Figure 4C and Supplementary Figure S7).
Bimodality is often associated with bistability. We investigated how bistability can originate from decoy titration with a positive autoregulation model. The phosphorylation module defines the relation between output [PhoBp]T and input [PhoB]T, while the binding competition and autoregulation module determines the occupancy of the autoregulated phoB promoter and the corresponding [PhoB]T level for every input concentration of [PhoBp]T (Figure 5A). Intersections of the two curves give steady-state solutions satisfying both modules. Decoy titration lowers promoter occupancy and the autoregulated [PhoB]T at low concentrations of PhoBp, making the curve derived from autoregulation module more sigmoidal and ultrasensitive. This allows existence of two stable steady states at intermediate numbers of decoys and the system becomes bistable (Figure 5B). It should be noted that the bistability simulation is not intended for quantitative recapitulation of experimental data and a weaker phoB promoter affinity is used to show bistability. The actual cellular behavior is subject to complex regulation by the coupled feedback and stress response. Nevertheless, the simple autoregulation model reveals a mechanism by which decoys promote ultrasensitivity, converting a graded response to bimodal.
Figure 5.
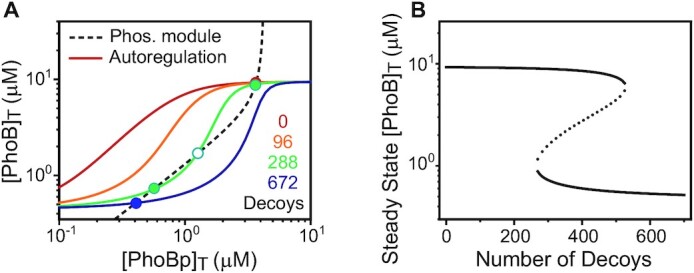
Modeling of bistability caused by decoy competition. (A) Steady-state output in the autoregulated system. The relationship between [PhoB]T and [PhoBp]T can be described by both the phosphorylation module (dashed line) and autoregulation module (solid lines). Intersections of the two curves satisfy both modules, representing the steady-state solutions. Decoy sites lead to ultrasensitivity in autoregulation, causing multiple steady states. Filled circles represent stable steady states while the empty circle indicates the unstable steady state. (B) Effects of decoy sites on steady-state [PhoB]T. Two solid lines illustrate stable steady states with unstable states (dashed line) in the middle. All curves are simulated using a sole feedback model with parameter values in Supplementary Table S2 except for: Kdecoy = 0.125 μM, and KB = 0.5 μM.
Correlation of TF abundance with the number of TFBSs in E. coli
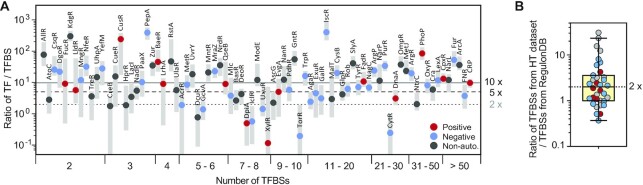
Our studies suggest that PhoB, with a TF/TFBS ratio of ∼5 under non-stimulated conditions, is affected by the titration effect of extra TFBSs. This provides a benchmark to estimate how likely it is that TF regulation will be influenced by binding site titration based on the relative abundance of TF to TFBS. We assessed the relative abundance of TF protein molecules in relation to the numbers of their binding sites for each E. coli TF (Figure 6A) with available TFBS data from RegulonDB (9) and TF abundance from mass spectrometry measurements (47). The median of TF protein levels across a wide range of growth conditions is used to represent TF abundance. The numbers of TFBSs have a moderate correlation with TF abundance (Pearson correlation coefficient r = 0.51, P value, 3.8 × 10–8), suggesting that TFs with higher demand, i.e., more binding sites, are expressed at higher levels. The ratio of median TF abundance to TFBS varies from one TF to another (circles in Figure 6A), with the median (N = 119) at ∼10. About 33% of TFs (39/119) have a TF/TFBS ratio smaller than 5, and 17% of TFs (20/119) with a ratio smaller than 2, the mathematical minimum required for occupying every TFBS because most E. coli TFs function as dimers or higher-order oligomers. These TFs are potentially susceptible to titration effects of TFBSs.
Figure 6.
Relationship between TF protein abundance and the number of TFBSs. (A) Ratio of TF abundance to the number of TFBSs for selected TFs in E. coli. TF abundance is derived from mass spectrum measurements (47). Grey ranges represent the ranges of TF/TFBS ratios across 19 growth conditions (excluding stationary phase growth) and circles indicate the medians for each TF. Horizontal lines indicate the TF/TFBS ratios of 2, 5 and 10. For graphic clarity, only selected TFs are shown for TFs with less than 5 binding sites. The full set of TF data is provided in Supplementary Table S3. (B) Underestimation of TFBSs. TFBS numbers from the high throughput (HT) dataset analyzed by RegulonDB (9) are compared to the TFBS numbers curated from the experimental dataset of regulonDB (see Supplementary materials for details). Each circle represents one TF among the 36 with HT data. Colors of circles correspond to autoregulation modes.
TF protein levels can vary across different growth conditions or signaling states, and certain growth conditions can lead to even smaller TF/TFBS ratios than the median calculated above. Even though protein abundance measurements did not cover all signaling states of every TF, the reported protein levels include measurements from a wide range of growth conditions, resulting in a range of TF/TFBS ratios (grey ranges in Figure 6A). There are ∼37% of TFs that have a TF/TFBS ratio ranging below 2 under one or a few growth conditions, suggesting that protein levels of a large fraction of TFs are at the binding capacity of TFBSs under these conditions. Further, binding site data from RegulonDB are biased toward functional sites, thus the TFBS numbers are likely underestimated. We evaluated the extent of TFBS underestimation by comparing TFBSs identified from high throughput studies with those curated at RegulonDB. TFBSs characterized by global binding studies have been documented for a small fraction (36/221) of TFs on RegulonDB (9) and a median of 2-fold more binding sites have been discovered for these 36 TFs (Figure 6B). Considering such underestimation of TFBSs, limited availability of TFs does not appear to be uncommon and a large fraction of E. coli TFs may be expressed at capacity to the TF demand, i.e., the number of TFBSs, thus may be susceptible to influences from natural or synthetic decoy TFBSs.
DISCUSSION
There is increasing evidence that many TFs are not in great excess to their binding sites, but rather, are expressed in limited numbers and shared among regulatory and non-functional TFBSs (47–50). As a result, transcription output is not only determined by the cis-regulatory elements of the particular promoter but also subject to titration by other TFBSs via competition for TF availability. In this study, we investigated the quantitative consequences of how the shared demand, or the relative abundance of TFs to TFBSs, alters the regulatory behaviors of the auto-activated TF PhoB.
The relative abundance of PhoB to its binding sites has a major impact on the timing of expression of PhoB-regulated genes by affecting the auto-activated expression rate of PhoB. Although PhoB levels under fully stimulated conditions appear to be in great excess to the number of binding sites with a TF/TFBS ratio of ∼100, response speed is largely dependent on production of PhoB protein starting from the unstimulated level at which PhoB is not in great excess. It appears that under non-stimulated conditions the PhoB level is near capacity in the sense that addition of only 96 decoys in excess of the endogenous 54 sites reduces the non-stimulated TF/TFBS ratio from ∼5 to <2. Correspondingly, the expression onset times are delayed by 5–7 min. The magnitude of this change is physiologically relevant and similar to the 5–10 min response time range in the temporal hierarchy of gene expression that has evolved to match the order of functional need of phosphorus utilization gene products (7). Further increasing the number of decoy sites can cause substantial response delay on the order of hours. Under such circumstances, cells may never reach the steady state or accumulate sufficient response proteins before transcription is diminished by the stress response. Precise timing of gene expression is important for many adaptive responses such as virulence, quorum sensing, nutrient uptake and metabolism, and is often achieved by different TF affinities for target genes (6,51–53). The tuning potential of TF affinities on response speed is dependent on the relative abundance of TFs to TFBSs. Additional TFBSs outside the target genes, either naturally present or synthetically introduced, play a significant role in setting the response timescale.
Competition for TF molecules by additional TFBSs provides a simple means to tune the population behavior from unimodal to bimodal. The mechanism is based on sequestration of active regulatory molecules by non-functional binding decoys to generate ultrasensitivity and promote bistability. Such mechanisms have been demonstrated in synthetic gene circuits with either protein (54,55) or DNA (56) as sequestrating decoys. Our studies establish the presence of the sequestration-based bistability in the native PhoR/PhoB two-component system and our model provides a quantitative explanation of how the relative TF abundance changes single-cell behaviors. A similar sequestration scheme based on protein-protein interactions between sigma factor and anti-sigma factor has been proposed to give bimodal output for the MprA/MprB system in Mycobacterium (57). Positive autoregulation is common in two-component systems, but bimodal responses are not usually observed because auto-activated expression of bifunctional histidine kinases can also increase the phosphatase activity to reduce the level of activated response regulator TFs (57–59). Phenotypic heterogeneity enables bet-hedging by bacterial cells for maximal survival (60,61), thus bimodal responses can be advantageous and desired under certain conditions. TF sequestration provides an alternative mechanism for bistability. Enhancement of TF sequestration can be achieved by increasing the number of TFBSs as well as decreasing TF abundance. Additional control of TF abundance can potentially impact the extent of TF sequestration and regulate the heterogeneity of responses for a single signaling system under different environmental conditions. This mechanism allows more flexible tuning of responses for evolutional design of different two-component systems to satisfy diverse signaling needs.
Here, we show that the quantitative consequences of altering the relative abundance of TF to TFBSs can be evaluated with the basic kinetic binding competition model. One advantage of using the response regulator PhoB for our study is that phosphorylation of this RR has been studied extensively (30) and the active form of the TF, PhoBp, can be readily modeled with no free parameters. Similar to the theoretical prediction regarding titration of transcription repressors (24), our simulation indicates that the largest relative titration effect on transcription activators occurs when the active TF copy number, instead of the total TF copy number, is similar to the binding capacity of all TFBSs. The modeled transcription output agrees well with experimental data for the engineered non-autoregulatory strains that have constitutive expression levels of PhoB near the mid- and high-end of the range achieved by autoregulation in a WT strain. For the autoregulated WT with low TF level under non-stimulated conditions, more accurate modeling may require a more detailed phosphorylation model (28) and/or the statistical thermodynamic binding model for TF competition (24,49). Moreover, the current model treats all binding sites, decoys or endogenous genomic sites, as respective single values while the decoy plasmid number or affinities of different endogenous sites may be best approximated with a distribution of different values (24). TF competition is also subject to other influences, such as cooperative binding from multiple binding sites commonly seen in bacterial promoters, and differences in DNA access due to binding of nucleoid proteins. All these require more specific analyses of individual TFs and TFBSs. Nevertheless, our model captures the major features of decoy titration and provides a quantitative framework for understanding the interplay between TFs and TFBSs.
The interplay between TFs and TFBSs, although investigated with synthetic decoys, has physiological relevance in general understanding of gene regulation. Cells experience changes of either TF abundance or the TFBS number regularly. E. coli cells can have multiple rounds of chromosome replication before division and the copies of replication origins per cell can range from 2 to above 6 depending on the growth rate (41). The total number of TFBSs will vary accordingly, dependent on the replication states and genomic distances of TFBSs to the replication origins. Additional TFBSs could also be introduced when extrageneous DNA fragments are horizontally acquired. These changes in TF abundance and TFBS number may lead to alteration of response due to TFBS titration effects. This may place constraints on the design of TF regulatory pathways. A high TF/TFBS ratio can potentially allow the pathway to buffer such changes and maintain proper responses while low TF abundance can be advantageous in minimizing resource costs. A large fraction of E. coli TFs appear to have low TF/TFBS ratios under certain growth conditions, suggesting that TFBS titration effects could be common among these TFs. Certainly, the interplay between TFs and TFBSs depends on the access of DNA to TF proteins because binding of nucleoid associated proteins can restrict the access of chromosomal DNA. Recent protein occupancy data (62) indicate that such transcriptionally silent, extended protein-protected chromosomal regions constitute ∼10% of E. coli genome, representing a non-trivial effect requiring further investigation.
In experimental studies, plasmids or viral DNA constructs that usually have different copy numbers than their chromosomally encoded counterparts are commonly used for investigating gene function or engineering synthetic circuits. Small variations in gene copies or extra binding sites can potentially disrupt the inherent balance between TFs and TFBSs, leading to alterations in system responses (63,64). Experimental results with unnatural DNA copies, overexpression of TFs or mutations that alter the stability of TF proteins need to be interpreted with great caution because even a modest change in the relative abundance of TF can be potentially amplified for auto-activated TFs with low capacity for TFBSs. Furthermore, the substantial perturbation of response output caused by TF insufficiency suggests that TFs that regulate key steps in virulence pathways might be sensitive targets for antimicrobial therapeutics (65,66). In another applied context, understanding the interplay between TF and TFBS becomes increasingly important in designing synthetic gene circuits. While different numbers of TFBSs in different genetic backgrounds may result in inconsistent output and hinder the standardization of gene circuits, synthetic decoys can be designed to modulate the circuit output (67). Our analyses provide a quantitative framework for better predicting and designing the desired outputs.
Supplementary Material
Contributor Information
Rong Gao, Center for Advanced Biotechnology and Medicine, Department of Biochemistry and Molecular Biology, Rutgers University - Robert Wood Johnson Medical School, Piscataway, NJ 08854, USA.
Libby J Helfant, Center for Advanced Biotechnology and Medicine, Department of Biochemistry and Molecular Biology, Rutgers University - Robert Wood Johnson Medical School, Piscataway, NJ 08854, USA.
Ti Wu, Center for Advanced Biotechnology and Medicine, Department of Biochemistry and Molecular Biology, Rutgers University - Robert Wood Johnson Medical School, Piscataway, NJ 08854, USA.
Zeyue Li, Center for Advanced Biotechnology and Medicine, Department of Biochemistry and Molecular Biology, Rutgers University - Robert Wood Johnson Medical School, Piscataway, NJ 08854, USA.
Samantha E Brokaw, Center for Advanced Biotechnology and Medicine, Department of Biochemistry and Molecular Biology, Rutgers University - Robert Wood Johnson Medical School, Piscataway, NJ 08854, USA.
Ann M Stock, Center for Advanced Biotechnology and Medicine, Department of Biochemistry and Molecular Biology, Rutgers University - Robert Wood Johnson Medical School, Piscataway, NJ 08854, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R35GM131727 to A.M.S.]. Funding for open access charge: NIGMS [R35GM131727].
Conflict of interest statement. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1. Keren L., Hausser J., Lotan-Pompan M., Vainberg Slutskin I., Alisar H., Kaminski S., Weinberger A., Alon U., Milo R., Segal E.. Massively parallel interrogation of the effects of gene expression levels on fitness. Cell. 2016; 166:1282–1294. [DOI] [PubMed] [Google Scholar]
- 2. Dekel E., Alon U.. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005; 436:588–592. [DOI] [PubMed] [Google Scholar]
- 3. Eames M., Kortemme T.. Cost-benefit tradeoffs in engineered lac operons. Science. 2012; 336:911–915. [DOI] [PubMed] [Google Scholar]
- 4. Gao R., Stock A.M.. Evolutionary tuning of protein expression levels of a positively autoregulated two-component system. PLoS Genet. 2013; 9:e1003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li G.W., Burkhardt D., Gross C., Weissman J.S.. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014; 157:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zaslaver A., Mayo A.E., Rosenberg R., Bashkin P., Sberro H., Tsalyuk M., Surette M.G., Alon U.. Just-in-time transcription program in metabolic pathways. Nat. Genet. 2004; 36:486–491. [DOI] [PubMed] [Google Scholar]
- 7. Gao R., Stock A.M.. Temporal hierarchy of gene expression mediated by transcription factor binding affinity and activation dynamics. mBio. 2015; 6:e00686-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lam F.H., Steger D.J., O'Shea E.K. Chromatin decouples promoter threshold from dynamic range. Nature. 2008; 453:246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Santos-Zavaleta A., Sanchez-Perez M., Salgado H., Velazquez-Ramirez D.A., Gama-Castro S., Tierrafria V.H., Busby S.J.W., Aquino P., Fang X., Palsson B.O.et al.. A unified resource for transcriptional regulation in Escherichia coli K-12 incorporating high-throughput-generated binding data into RegulonDB version 10.0. BMC Biol. 2018; 16:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shimada T., Ishihama A., Busby S.J., Grainger D.C.. The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res. 2008; 36:3950–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Minch K.J., Rustad T.R., Peterson E.J., Winkler J., Reiss D.J., Ma S., Hickey M., Brabant W., Morrison B., Turkarslan S.et al.. The DNA-binding network of Mycobacterium tuberculosis. Nat. Commun. 2015; 6:5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Visweswariah S.S., Busby S.J.. Evolution of bacterial transcription factors: how proteins take on new tasks, but do not always stop doing the old ones. Trends Microbiol. 2015; 23:463–467. [DOI] [PubMed] [Google Scholar]
- 13. Ishihama A., Shimada T., Yamazaki Y.. Transcription profile of Escherichia coli: genomic SELEX search for regulatory targets of transcription factors. Nucleic Acids Res. 2016; 44:2058–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coutte L., Antoine R., Slupek S., Solans L., Derop J., Bonnefond A., Hot D., Locht C.. Combined RNAseq and ChIPseq analyses of the BvgA virulence regulator of Bordetella pertussis. mSystems. 2020; 5:e00208-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mrazek J., Karls A.C.. In silico simulations of occurrence of transcription factor binding sites in bacterial genomes. BMC Evol. Biol. 2019; 19:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kemme C.A., Nguyen D., Chattopadhyay A., Iwahara J.. Regulation of transcription factors via natural decoys in genomic DNA. Transcription. 2016; 7:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Del Vecchio D., Ninfa A.J., Sontag E.D.. Modular cell biology: retroactivity and insulation. Mol. Syst. Biol. 2008; 4:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang T., Tague N., Whelan S.A., Dunlop M.J.. Programmable gene regulation for metabolic engineering using decoy transcription factor binding sites. Nucleic Acids Res. 2021; 49:1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wan X., Pinto F., Yu L., Wang B.. Synthetic protein-binding DNA sponge as a tool to tune gene expression and mitigate protein toxicity. Nat. Commun. 2020; 11:5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang B., Guo F., Dong S.H., Zhao H.. Activation of silent biosynthetic gene clusters using transcription factor decoys. Nat. Chem. Biol. 2019; 15:111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leong P.L., Andrews G.A., Johnson D.E., Dyer K.F., Xi S., Mai J.C., Robbins P.D., Gadiparthi S., Burke N.A., Watkins S.F.et al.. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:4138–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Das S., Choubey S.. Tunability enhancement of gene regulatory motifs through competition for regulatory protein resources. Phys. Rev. E. 2020; 102:052410. [DOI] [PubMed] [Google Scholar]
- 23. Burger A., Walczak A.M., Wolynes P.G.. Abduction and asylum in the lives of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4016–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brewster R.C., Weinert F.M., Garcia H.G., Song D., Rydenfelt M., Phillips R.. The transcription factor titration effect dictates level of gene expression. Cell. 2014; 156:1312–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ali M.Z., Parisutham V., Choubey S., Brewster R.C.. Inherent regulatory asymmetry emanating from network architecture in a prevalent autoregulatory motif. Elife. 2020; 9:e56517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Buschiazzo A., Trajtenberg F.. Two-Component sensing and regulation: How do histidine kinases talk with response regulators at the molecular level?. Annu. Rev. Microbiol. 2019; 73:507–528. [DOI] [PubMed] [Google Scholar]
- 27. Gao R., Bouillet S., Stock A.M.. Structural basis of response regulator function. Annu. Rev. Microbiol. 2019; 73:175–197. [DOI] [PubMed] [Google Scholar]
- 28. Gao R., Stock A.M.. Overcoming the cost of positive autoregulation by accelerating the response with a coupled negative feedback. Cell Rep. 2018; 24:3061–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao R., Stock A.M.. Quantitative kinetic analyses of shutting off a two-component system. mBio. 2017; 8:e00412-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao R., Stock A.M.. Probing kinase and phosphatase activities of two-component systems in vivo with concentration-dependent phosphorylation profiling. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang C., Huang T.W., Wen S.Y., Chang C.Y., Tsai S.F., Wu W.F., Chang C.H.. Genome-wide PhoB binding and gene expression profiles reveal the hierarchical gene regulatory network of phosphate starvation in Escherichia coli. PLoS One. 2012; 7:e47314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wanner B.L. Neidhardt F.C., Curtiss R. III, Ingraham J.L., Lin E.C.C., Low K.B. Jr, Magasanik B., Reznikoff W.S., Riley M., Schaechter M., Umbarger H.E.. Escherichia coli and Salmonella: Cellular and Molecular Biology. 1996; 1:Washington, D.C.American Society for Microbiology Press; 1357–1381. [Google Scholar]
- 33. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haldimann A., Wanner B.L.. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 2001; 183:6384–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Neidhardt F.C., Bloch P.L., Smith D.F.. Culture medium for enterobacteria. J. Bacteriol. 1974; 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gao R., Godfrey K.A., Sufian M.A., Stock A.M.. Counterbalancing regulation in response memory of a positively autoregulated two-component system. J. Bacteriol. 2017; 199:e00390-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lerner C.G., Inouye M.. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990; 18:4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Siryaporn A., Perchuk B.S., Laub M.T., Goulian M.. Evolving a robust signal transduction pathway from weak cross-talk. Mol. Syst. Biol. 2010; 6:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Batchelor E., Goulian M.. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ritzefeld M., Walhorn V., Kleineberg C., Bieker A., Kock K., Herrmann C., Anselmetti D., Sewald N.. Cooperative binding of PhoB(DBD) to its cognate DNA sequence-a combined application of single-molecule and ensemble methods. Biochemistry. 2013; 52:8177–8186. [DOI] [PubMed] [Google Scholar]
- 41. Bremer H., Dennis P.P. Neidhardt F.C., Curtiss R. III, Ingraham J.L., Lin E.C.C., Low K.B., Magasanik B., Reznikoff W.S., Riley M., Schaechter M., Umbarger H.E.. Escherichia coli and Salmonella: Cellular and Molecular Biology. 1996; 2:Washington, D.C.American Society for Microbiology Press; 1553–1569. [Google Scholar]
- 42. Battesti A., Majdalani N., Gottesman S.. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011; 65:189–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Spira B., Yagil E.. The relation between ppGpp and the PHO regulon in Escherichia coli. Mol. Gen. Genet. 1998; 257:469–477. [DOI] [PubMed] [Google Scholar]
- 44. Alon U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 2007; 8:450–461. [DOI] [PubMed] [Google Scholar]
- 45. Maeda Y.T., Sano M.. Regulatory dynamics of synthetic gene networks with positive feedback. J. Mol. Biol. 2006; 359:1107–1124. [DOI] [PubMed] [Google Scholar]
- 46. Mitrophanov A.Y., Hadley T.J., Groisman E.A.. Positive autoregulation shapes response timing and intensity in two-component signal transduction systems. J. Mol. Biol. 2010; 401:671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schmidt A., Kochanowski K., Vedelaar S., Ahrne E., Volkmer B., Callipo L., Knoops K., Bauer M., Aebersold R., Heinemann M.. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 2016; 34:104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Teng S.W., Wang Y., Tu K.C., Long T., Mehta P., Wingreen N.S., Bassler B.L., Ong N.P.. Measurement of the copy number of the master quorum-sensing regulator of a bacterial cell. Biophys. J. 2010; 98:2024–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rydenfelt M., Cox R.S. 3rd, Garcia H., Phillips R.. Statistical mechanical model of coupled transcription from multiple promoters due to transcription factor titration. Phys. Rev. E. 2014; 89:012702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Das D., Dey S., Brewster R.C., Choubey S.. Effect of transcription factor resource sharing on gene expression noise. PLoS Comput. Biol. 2017; 13:e1005491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Williams C.L., Cotter P.A.. Autoregulation is essential for precise temporal and steady-state regulation by the Bordetella BvgAS phosphorelay. J. Bacteriol. 2007; 189:1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Waters C.M., Bassler B.L.. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 2006; 20:2754–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang B., Muir T.W.. Regulation of virulence in Staphylococcus aureus: molecular mechanisms and remaining puzzles. Cell Chem. Biol. 2016; 23:214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen D., Arkin A.P.. Sequestration-based bistability enables tuning of the switching boundaries and design of a latch. Mol. Syst. Biol. 2012; 8:620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Buchler N.E., Cross F.R.. Protein sequestration generates a flexible ultrasensitive response in a genetic network. Mol. Syst. Biol. 2009; 5:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee T.H., Maheshri N.. A regulatory role for repeated decoy transcription factor binding sites in target gene expression. Mol. Syst. Biol. 2012; 8:576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tiwari A., Balazsi G., Gennaro M.L., Igoshin O.A.. The interplay of multiple feedback loops with post-translational kinetics results in bistability of mycobacterial stress response. Phys. Biol. 2010; 7:036005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Groisman E.A. Feedback control of two-component regulatory systems. Annu. Rev. Microbiol. 2016; 70:103–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Goulian M. Two-component signaling circuit structure and properties. Curr. Opin. Microbiol. 2010; 13:184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Veening J.W., Smits W.K., Kuipers O.P.. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 2008; 62:193–210. [DOI] [PubMed] [Google Scholar]
- 61. Carey J.N., Mettert E.L., Roggiani M., Myers K.S., Kiley P.J., Goulian M.. Regulated stochasticity in a bacterial signaling network permits tolerance to a rapid environmental change. Cell. 2018; 173:196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Freddolino P.L., Amemiya H.M., Goss T.J., Tavazoie S.. Dynamic landscape of protein occupancy across the Escherichia coli chromosome. PLoS Biol. 2021; 19:e3001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. van Dijk D., Sharon E., Lotan-Pompan M., Weinberger A., Segal E., Carey L.B.. Large-scale mapping of gene regulatory logic reveals context-dependent repression by transcriptional activators. Genome Res. 2017; 27:87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mileyko Y., Joh R.I., Weitz J.S.. Small-scale copy number variation and large-scale changes in gene expression. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:16659–16664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Daly S.M., Elmore B.O., Kavanaugh J.S., Triplett K.D., Figueroa M., Raja H.A., El-Elimat T., Crosby H.A., Femling J.K., Cech N.B.et al.. ω-Hydroxyemodin limits Staphylococcus aureus quorum sensing-mediated pathogenesis and inflammation. Antimicrob. Agents Chemother. 2015; 59:2223–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sully E.K., Malachowa N., Elmore B.O., Alexander S.M., Femling J.K., Gray B.M., DeLeo F.R., Otto M., Cheung A.L., Edwards B.S.et al.. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 2014; 10:e1004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brophy J.A., Voigt C.A.. Principles of genetic circuit design. Nat. Methods. 2014; 11:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.