Abstract
Signaling pathway-driven target gene transcription is critical for fate determination of embryonic stem cells (ESCs), but enhancer-dependent transcriptional regulation in these processes remains poorly understood. Here, we report enhancer architecture-dependent multilayered transcriptional regulation at the Halr1–Hoxa1 locus that orchestrates retinoic acid (RA) signaling-induced early lineage differentiation of ESCs. We show that both homeobox A1 (Hoxa1) and Hoxa adjacent long non-coding RNA 1 (Halr1) are identified as direct downstream targets of RA signaling and regulated by RARA/RXRA via RA response elements (RAREs). Chromosome conformation capture-based screens indicate that RA signaling promotes enhancer interactions essential for Hoxa1 and Halr1 expression and mesendoderm differentiation of ESCs. Furthermore, the results also show that HOXA1 promotes expression of Halr1 through binding to enhancer; conversely, loss of Halr1 enhances interaction between Hoxa1 chromatin and four distal enhancers but weakens interaction with chromatin inside the HoxA cluster, leading to RA signaling-induced Hoxa1 overactivation and enhanced endoderm differentiation. These findings reveal complex transcriptional regulation involving synergistic regulation by enhancers, transcription factors and lncRNA. This work provides new insight into intrinsic molecular mechanisms underlying ESC fate determination during RA signaling-induced early differentiation.
Graphical Abstract
Graphical Abstract.
Enhancer architecture-dependent multilayered transcriptional regulation at the Halr1–Hoxa1 locus that orchestrates RA signaling-induced early lineage differentiation of ESCs.
INTRODUCTION
ESCs can undergo unlimited self-renewal and proliferation, while maintaining the potential to differentiate into different germ layers, activities requiring precise control of gene expression (1,2). Recent studies indicate that in addition to promoters, there are many other types of functional DNA elements such as enhancers, insulators, silencers and transposable elements that control the precise expression of gene in ESCs (3–6). Enhancers also reportedly show significant cell-type specificity during ESC lineage differentiation stimulated by different signaling factors, while results obtained by chromosome conformation capture also reveal significant changes in high-order chromatin structure during these processes (7,8). Although enhancers regulate expression of master transcription factors (TFs) through long-range chromatin interactions to coordinate embryonic development (9–11), the precise mechanisms underlying the regulation of enhancer in ESC lineage differentiation remain largely unclear.
RA signaling is essential for mammalian development and functions in embryogenesis, immunogenesis, hematopoiesis and skeletogenesis (12,13). Abnormal RA signaling underlies numerous disease states, including tumorigenesis (12,14,15). ESCs rapidly differentiate in response to RA signaling, a process requiring transcriptional regulation by the RA receptor RXR/RAR (16,17). Recent studies show that RA signaling induces neural differentiation of ESCs, with HOXA1 playing a central transcriptional role in this process (18–21) and CTCF organizing chromatin interactions (22–24). However, early stages of ESC differentiation induced by RA signaling are reportedly characterized by endodermal differentiation marked by significantly high expression of endodermal master control genes Gata4, Sox17 and Gata6 (25,26). These findings suggest that ESCs may undergo multi-lineage differentiation in response to RA signaling, and that the underlying molecular regulatory mechanisms are unknown.
We and others have found that several enhancers and CTCF binding elements (CBEs) at the Halr1–Hoxa1 locus play important regulatory roles during RA signaling-induced ESC differentiation (23,26–29). In addition, the long non-coding RNA (lncRNA) Halr1 also reportedly regulates HoxA cluster gene expression by orchestrating enhancer interactions under RA-induced early ESC differentiation (27,29). However, regulatory relationships between enhancers, CBEs, lncRNAs and HoxA following RA induction of early ESC differentiation remain unknown. Considering that all these elements are present at the Halr1–Hoxa1 locus, analyses of their activity in this context may provide an excellent model to define mechanisms involved in RA signaling-induced early ESC differentiation.
Here, using the Halr1-Hoxa1 locus as a model, we conducted various analyses of ESC differentiation and showed that RA signaling significantly induced differentiation towards ectoderm and endoderm lineages and that Halr1 and Hoxa1 were significantly activated in this process. We also identified Halr1 and Hoxa1 as direct downstream targets of RA signaling and found that proper Halr1 and Hoxa1 expression is necessary for RA-dependent induction of endodermal ESC differentiation. Moreover, multiple enhancers at the Halr1–Hoxa1 locus were required for RA signaling to induce Halr1 and Hoxa1 expression and mesendoderm differentiation. We also observed reciprocal regulation between Hoxa1 and Halr1: HOXA1 promoted Halr1 expression by binding to enhancer regions, while Halr1 repressed Hoxa1 expression by coordinating interactions of multiple enhancers with Hoxa1 chromatin. Finally, we identified a subset of genes regulated by HOXA1 associated with endoderm development. Overall, these findings reveal a new mechanism for RA signaling-induced early lineage differentiation of ESCs and provide a new perspective on ESC fate decisions.
MATERIALS AND METHODS
ESCs culture
Mouse ESCs E14 were cultured in undifferentiated condition as previously described (23). Briefly, ESCs were grown in culture dishes coated with 0.1% gelatin (Sigma) in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 15% fetal bovine serum (FBS, AusGeneX), 1× nonessential amino acids (100×, Gibco), 1× l-glutamate (100×, Gibco), 1× penicillin–streptomycin (100×, Gibco), 50 μM β-mercaptoethanol (Sigma), 10 ng/ml LIF (ESGRO), 1 μM PD0325901 (MedChemExpress) and 3 μM CHIR99021 (MedChemExpress). The medium was replaced every 1–2 days. ESCs were maintained at 37°C in a 5% CO2 incubator.
Adherent differentiation of ESCs
ESC differentiation was carried out as previously reported (23,26). ESCs were gently washed with 1× phosphate buffer saline solution (PBS, Solarbio), dissociated, and plated at an appropriate density on gelatin-coated plates in LIF/2i withdrawal medium or medium supplemented with 2 μM RA (Solarbio). The differentiated culture mediums were replaced every day.
Embryoid bodies (EBs) formation
For EB formation, 106 ESCs were plated on non-adherent 10 cm plates in LIF/2i withdrawal medium. EBs were harvested using TRIzol reagent (Invitrogen) every day. RNA was prepared, and qRT-PCR was performed as described previously (26,30).
Alkaline phosphatase (AP) staining analysis
ESCs were plated at low density in 12-well plates coated with gelatin for 3 days, washed twice with PBS, and then incubated with reagents from the AP Staining kit (System Biosciences) following the manufacturer's instructions. Digital images were taken using an Olympus Inverted Fluorescence Microscope (26).
RNA extraction, cDNA synthesis and quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from differentiated or undifferentiated ESCs using TRIzol Reagent (Life Technologies). cDNA synthesis was performed using a PrimerScript™ RT reagent Kit with gDNA Eraser (TaKaRa) according to the manufacturer's instructions. PCR reactions were performed using HieffTMqPCR SYBR Green Master Mix (YEASEN) and a BioRad CFX Connect Real-Time system. PCR cycling conditions were as previously reported: 95°C for 5 min, 40 cycles of 95°C for 15 s, 60°C for 15 s and 72°C for 30 s. We then constructed a melting curve of amplified DNA (23,26). Target gene values were normalized to Gapdh expression and the experimental control using ΔΔCt methods (31). Primer sequences used in this study are shown in Supplementary Table S1.
Construction of stably transduced ESC lines
Full-length coding sequences (CDSs) of Rara, Rxra and Hoxa1 were synthesized at GENEWIZ and inserted into the FLAG-tag vector using Nhe1 and Not1 sites. ESCs were transfected with the overexpression vectors using Lipofectamine 3000 (Life Technologies) and then 24 h later were treated with medium containing 5 μM puromycin (MCE) until stably-transduced cells were harvested. qRT-PCR and western blot (WB) were used to identify overexpression cell lines (26). Full-length CDSs of Rara, Rxra and Hoxa1 are shown in Supplementary Table S2.
Western blot analysis in ESCs
Protein was extracted using RIPA lysis buffer (Strong, YEASEN) and electrophoresis was performed using a PAGE Gel Quick Preparation Kit (10%, YEASEN) following the manufacturer's instructions. WB was carried out with the following antibodies: (primary) RARA (Invitrogen, #MA1-810A, 1:1000), RXRA (Invitrogen, #433900, 1:1000), HOXA1 (Invitrogen, #PA5-68809, 1:1000), FLAG (Beytime, #AF-0036, 1:1000) and GAPDH (Santa Cruz, #SC-365062, 1:2000), and then HRP-linked secondary antibodies (Sungene Biotech). HRP activity was detected using Luminol HRP Substrate (Millipore). Digital images were taken using an automatic chemiluminescence imaging analysis system (Tanon) (6,26,32,33).
CRISPR/Cas9-mediated genome editing in ESCs
CRISPR/Cas9-mediated knockout was performed using published protocols (23,26,34). Briefly, target-specific guide RNAs (sgRNAs) were designed using an online tool (http://chopchop.cbu.uib.no/). sgRNAs of the appropriate site and score were selected and sgRNAs were cloned into a Cas9-puro vector using the Bsmb1 site. sgRNA sequences are shown in Supplementary Table S3.
For enhancer and TF binding site knockout cells, ESCs were transfected with two sgRNA plasmids using Lipofectamine 3000 (Life Technologies), and 24 h later, cells were treated with 5 μM puromycin (MCE) for 24 h and then cultured in a medium without puromycin for another 5–7 days. Individual colonies were picked and validated by genomic DNA PCR or subsequent Sanger DNA sequencing. Enhancer knockout lines En1-KO, En2-KO, En3-KO and En1/2/3-KO were reported in our previous study (23). For coding gene knockouts, sgRNAs were designed at both ends of the gene CDS based on a previous report (21). Successful knockout lines were validated by genomic DNA PCR, qRT-PCR and WB. For lncRNA knockouts, sgRNAs were designed at the promoter region based on a previous report (27). Successful knockout lines were validated by genomic DNA PCR and qRT-PCR. PCR primers used for genotyping are listed in Supplementary Table S4.
Chromosome conformation capture (Capture-C) assay and data analysis
Probes used for Capture-C assay were designed using an online tool (http://apps.molbiol.ox.ac.uk/CaptureC/cgi-bin/CapSequm.cgi). Probe sequences are listed in Supplementary Table S5. Capture-C libraries were prepared as described (23,26,35,36). Briefly, undifferentiated or RA-induced differentiated ESCs were fixed with 1% (vol/vol) formaldehyde for 10 min at room temperature, quenched with 125 mM glycine in PBS, and then lysed in cold lysis buffer [10 mM Tris–HCl, pH 7.5, 10 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA, 0.2% NP-40, 1× complete protease inhibitor cocktail (Roche)]. Chromatin was digested with DpnII (New England Biolabs) at 37°C overnight. Fragments were then diluted and ligated with T4 DNA ligase (Takara) at 16°C overnight. Cross-linking was reversed by overnight incubation at 60°C with proteinase K (Bioline). Then 3C libraries were purified by phenol–chloroform followed by chloroform extraction and ethanol-precipitated at −80°C overnight. Sequencing libraries were prepared from 10 μg of the 3C library by sonication to an average size of 200–300 bp and indexed using NEBnext reagents (New England Biolabs), according to the manufacturer's protocol. Enrichment of 2 μg of an indexed library incubated with 3 μM of a pool of biotinylated oligonucleotides was performed using the SeqCap EZ Hybridization reagent kit (Roche/NimbleGen), following the manufacturer's instructions. Two rounds of capture employing 48–72 h and 24 h hybridizations, respectively, were used. Correct library sizes were confirmed by agarose gel electrophoresis, and DNA concentration was determined using a Qubit 2.0 Fluorometer (Thermo Fisher Scientific). All sequencing was performed on Hi-Seq 2500 platforms using paired 150 bp protocols (Illumina).
Capture-C data were analyzed as previously described methods (23,26,35,36). Briefly, clean paired-end reads were reconstructed into single reads using FLASH (37). After in silico DpnII digestion using the DpnII2E.pl script, reads were mapped back to the mm10 mouse genome using Bowtie1. Finally, chimeric reads containing captured reads and Capture-Reporter reads were analyzed using CCanalyser3.pl. Results were visualized using the Integrated Genome Browser (IGV) (38) and an online tool (https://epgg-test.wustl.edu/browser/) (39).
Eukaryotic cognate transcriptome sequencing (RNA-seq) and data analysis
RNA-seq was performed as reported (23,26). Briefly, ESCs were lysed with Trizol reagent (Life Technologies), and RNA was extracted based on the manufacturer's instructions. RNA was sequenced by the Novogene. Clean reads were mapped to the Ensemble mm10 mouse genome using Hisat2 with default parameters. Gene reads were counted by Htseq (40). Fold changes (FC) were computed as a log2 ratio of normalized reads per gene using the DEseq2 R package (41). Gene expression with ∣log2 FC∣ ≥ 1 (FDR < 0.05) was considered significantly altered. Heatmaps were drawn using the heatmap.2 function or Microsoft Excel 97-2003. At least two biological replicates were analyzed for each experimental condition.
Gene ontology (GO) analysis
GO analyses were performed using the following online tools: Metascape (http://metascape.org/gp/index.html#/main/step1) (42) and DAVID Functional Annotation Bioinformatics Microarray Analysis tool (http://david.abcc.ncifcrf.gov/) (43). Briefly, differential genes (FC ≥ 2, FDR < 0.5) were loaded into the analysis module as instructed by the tools, then the mouse was selected as the species for GO analysis to obtain the final output.
Gene set enrichment analysis (GSEA)
GSEA analysis was carried out using an online tool (https://www.gsea-msigdb.org/gsea/index.jsp) as reported (44,45). Gene sets of embryonic skeletal system morphogenesis and stem cell population maintenance were obtained from the KEGG database (https://www.genome.jp/kegg/) (46).
Data set availability
DNA-seq and RNA-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE169058. Previously published datasets used in this study are shown in Supplementary Table S6. These published data sets were downloaded and analyzed using online tools (http://cistrome.org/db/#/, http://promoter.bx.psu.edu/hi-c/ and https://www.ncbi.nlm.nih.gov/geo/) (47–49).
Statistical analysis
Data were analyzed by the unpaired Student's t-test (two-tailed) unless otherwise specified. Statistically significant P-values are indicated in Figure legends as follows: * P < 0.05, ** P < 0.01.
RESULTS
Characterization of RA signaling-induced early lineage differentiation of ESCs
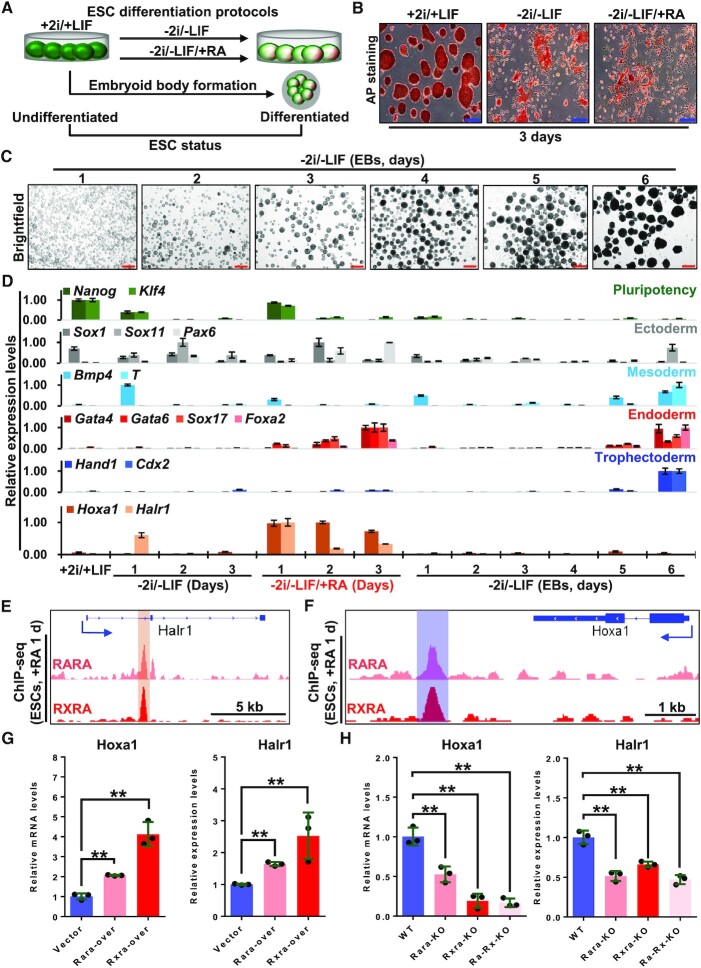
To analyze characteristics of RA-induced differentiation of ESCs, we employed three commonly used ESC differentiation protocols: spontaneous differentiation following LIF/2i removal, differentiation induced by RA after LIF/2i removal, and embryoid body (EB) differentiation (Figure 1A). As differentiation proceeded, differentiated cells showed greater cell morphological differences compared to undifferentiated ESCs (Figure 1B and C). Pluripotency marker genes and germ layer marker genes were used to identify ESC differentiation status (23,50), qRT-PCR results showed that Nanog and Klf4 levels decreased with time in all three protocols, and RA-induced ESC differentiation resulted in significantly higher expression of ectodermal (Sox1 and Pax6) and endodermal (Gata4, Gata6, Foxa2 and Sox17) genes, compared with the other differentiation schemes (Figure 1D), suggesting that RA-induced ESCs’ early differentiation has the significant ectodermal and endodermal profile.
Figure 1.
Hoxa1 and Halr1 are direct downstream target genes of RA signaling. (A) Schematic showing ESC differentiation protocols. (B) Images showing AP staining of undifferentiated and differentiated ESCs. Scale bars: 100 μm. (C) Bright-field images of EBs on days 1–6. Scale bars: 200 μm. (D) qRT-PCR data showing transcript levels of pluripotency and germ layer genes as well as Halr1 and Hoxa1 during ESC differentiation (n = 3 replicates). (E and F) ChIP-seq data showing RARA and RXRA binding peaks at Halr1 and Hoxa1 loci on day 1 of RA-induced ESC differentiation. (G) qRT-PCR data showing Halr1 and Hoxa1 transcript levels in ESCs overexpressing RARA and RXRA on day 1 of RA treatment. Data represent the mean ± SD (n = 3 replicates). Statistical significance was determined by a two-tailed Student's t-test (unpaired), * P< 0.05, ** P < 0.01. (H) qRT-PCR data showing Halr1 and Hoxa1 transcript levels in WT or KO on day 1 of RA treatment. Data represent the mean ± SD (n = 3 replicates). Statistical significance was determined by a two-tailed Student's t-test (unpaired), * P< 0.05, ** P < 0.01.
To further assess the global view of gene expression changes, we performed transcriptome sequencing (RNA-seq) at day one after RA treatment. At that time point, we observed 3070 up-regulated and 3303 down-regulated genes compared to undifferentiated ESCs (Supplementary Figure S1A). Further analysis of germ layer gene expression revealed significant up-regulation of genes associated with the ectoderm, trophectoderm and endoderm, with the greatest up-regulation in endodermal genes (Supplementary Figure S1B and C). GO analysis also showed enrichment of biological processes associated with multicellular organism development, cell differentiation, endoderm formation and cellular responses to RA (Supplementary Figure S1D; Supplementary Table S7). Taken together, these data suggest that RA signaling significantly induces early ESC differentiation into endoderm and ectoderm.
Hoxa1 and Halr1 are direct downstream target genes of RA signaling
To explore the expression patterns of Hoxa1 and Halr1, we next evaluated their transcript levels in various ESC differentiation models. qRT-PCR results showed that Hoxa1 and Halr1 were more robustly expressed following RA-induced ESC differentiation relative to other differentiation systems (Figure 1D). Upon analysis of the RNA-seq data, we found that Hoxa1 and Halr1 were significantly enriched in the biological processes associated with cellular response to RA (Supplementary Figure S1D and E). In addition, we also found that Halr1 transcript levels peaked at day one of differentiation (Supplementary Figure S2A and B). These results suggest that RA signaling can specifically and significantly activate the expression of Hoxa1 and Halr1 during early ESC differentiation.
The transcriptional activation ability of RA signaling is dependent on RA receptors such as RARA and RXRA (16). Thus, we asked whether Hoxa1 and Halr1 expression depended on RARA and RXRA. To do so, we first analyzed ChIP-seq data for RARA/RXRA and found that RA induced a significant co-binding peak for RARA and RXRA in the first intron of Halr1 in ESCs (at day 1 of differentiation) and a significant peak at the Hoxa1 3′-end (Figure 1E and F) (17,51), suggesting possible regulation of Hoxa1 and Halr1 by RARA and RXRA. Next, we overexpressed RXRA and RARA in ESCs and performed qRT-PCR analysis for Hoxa1 and Halr1 transcripts (Supplementary Figure S3). At day 1 of RA differentiation, expression levels of both were significantly up-regulated in cells overexpressing RARA and RXRA compared to the control cells (Figure 1G). We also established RARA and RXRA single or double knockout lines using the CRISPR-Cas9 system and analyzed Hoxa1 and Halr1 transcript levels by qRT-PCR (Supplementary Figure S4). Relative to wild-type (WT) controls, RARA and RXRA deletion, either singly or double KOs, significantly suppressed Hoxa1 and Halr1 transcript levels at day one differentiation (Figure 1H). Taken together, these findings overall suggest that Hoxa1 and Halr1 may be direct target genes of RA signaling.
RAREs are required for RA signaling-induced Hoxa1 and Halr1 expression
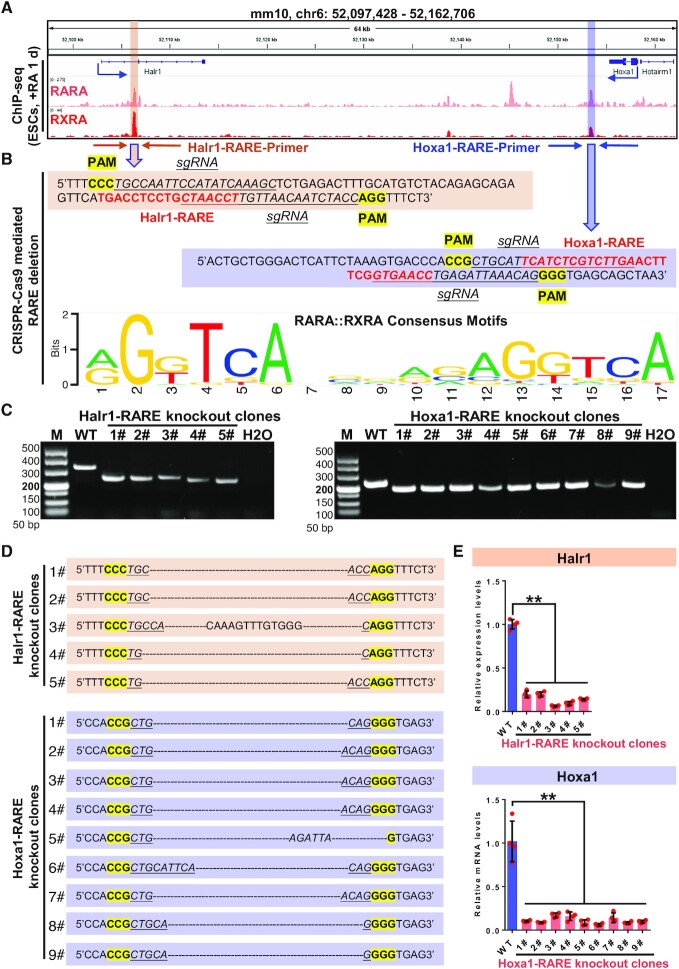
Previous studies have shown that the regulation of target gene expression by RA is dependent on the cis-functional RARE at target gene loci (16,52). Several functional target genes exhibiting RARE elements have been identified, such as Cyp26 (53), Hoxb1 (54–58) and Hoxa7 (59). We analyzed ChIP-seq data showing significant co-binding of RARA/RXRA at the Hoxa1 and Halr1 loci (Figure 2A). However, whether these RAREs at the Halr1-Hoxa1 locus are functional is unclear. To assess their functionality, we examined the region of RARA/RXRA co-binding at these loci using the JASPAR database (60) and identified multiple putative RAREs (Figure 2B). We deleted these RAREs using the CRISPR–Cas9 system (Figure 2B) and then, using genomic-DNA PCR with specific primers and Sanger sequencing, we obtained multiple RARE knockout clones (Figure 2C and D; Supplementary Figure S5). RA treatment of RARE knockout cells resulted in significantly lower Halr1 and Hoxa1 transcript levels relative to WT cells (Figure 2E), strongly suggesting that these RAREs act as cis-functional elements necessary for RA signaling-dependent activation of Hoxa1 and Halr1 expression. Taken together, these results indicated that Halr1 and Hoxa1 are direct downstream targets of RA signaling and regulated by RARA/RXRA via RAREs.
Figure 2.
RAREs are required for RA signaling-induced Hoxa1 and Halr1 expression. (A) ChIP-seq data showing RARA and RXRA binding peaks at Halr1 and Hoxa1 loci in ESCs on day 1 of RA treatment. (B) JASPAR prediction of RAREs at the Halr1–Hoxa1 locus, the RARA/RXRA binding structural domain, and the CRISPR-Cas9-mediated RARE knockout scheme. (C) Agarose gel electrophoresis showing RARE knockout clones using primers shown in (B). (D) Sanger sequencing of PCR products in (C). (E) qRT-PCR showing Halr1 and Hoxa1 transcript levels in RARE knockout cells. Data represent the mean ± SD (n = 4 replicates). Statistical significance was determined by a two-tailed Student's t-test (unpaired), * P< 0.05, ** P < 0.01. M: DNA ladder marker.
Increased enhancer interactions promote RA signaling-induced Hoxa1 and Halr1 activation
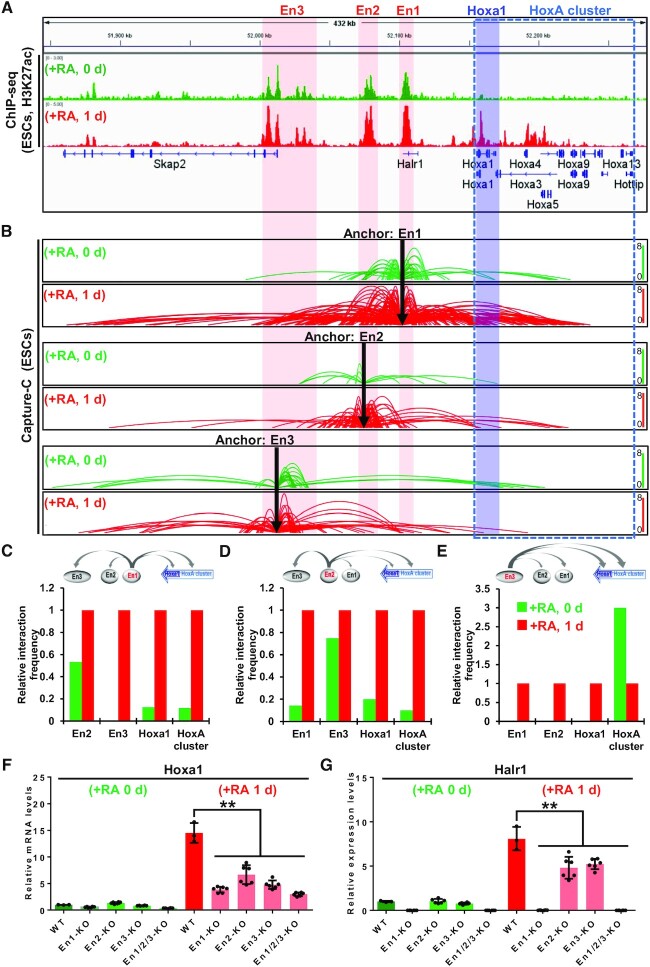
Our previous study reported that there are three enhancers at the Halr1–Hoxa1 locus, and in the RA-induced ESC differentiation state, the three enhancers significantly interact with each other to form an enhancer-enhancer interaction complex (EEIC) that regulates RA signaling-induced HoxA cluster gene expression (23). However, the pattern of interactions between these enhancers in the RA signaling-induced differentiated and undifferentiated states of ESCs and the regulation of Hoxa1 and Halr1 are unclear.
To investigate the chromatin interaction patterns of these enhancers during RA-induced early ESC differentiation, we used the Capture-C method (23,35,36,61), a 3C-based chromosome conformation capture technique (62). The three enhancers at the Halr1–Hoxa1 locus served as anchor regions to perform enhancer capture using the capture-C method in undifferentiated and RA-induced differentiated ESCs (at day one) (Figure 3A and B) (28,63). Chromatin interactions between En1 and En2 under RA-induced ESC differentiation significantly increased, while En3 also showed significantly enhanced interactions with En1, En2 and Hoxa1 chromatin, but reduced interactions with HoxA cluster chromatin (Figure 3C–E). These data suggest that a tight EEIC forms between enhancers during RA-induced early ESC differentiation and that these interactions with the HoxA cluster chromatin are also significantly enhanced. Considering previous results showing that RA signaling induces significant activation of Hoxa1 and Halr1 (Figure 1D), we hypothesize that there may be a positive correlation between increased enhancer interactions and high expression of Hoxa1 and Halr1. To test the hypothesis, we treated both WT and enhancer KO cells with RA for one day and monitored Hoxa1 and Halr1 transcript levels by qRT-PCR (23). WT cells showed significant upregulation of Hoxa1 and Halr1 expression after RA treatment, relative to undifferentiated ESCs; however, RA-treated enhancer KO cells showed significantly reduced Hoxa1 and Halr1 expression (Figure 3F and G). In addition, Enhancer KO cells also showed significant suppression of other genes in the HoxA cluster (Hoxa2-13) (Supplementary Figure S6). Taken together, these data confirm the increase in enhancer interactions following stimulation of RA signaling at the Halr1-Hoxa1 locus, further suggesting that these enhancers are required for the activation of Hoxa1 and Halr1.
Figure 3.
Increased enhancer interactions promote RA signaling-induced Hoxa1 and Halr1 activation. (A) H3K27ac ChIP-seq data in ESCs either untreated or treated 1 day with RA demonstrating enhancers at the Halr1-Hoxa1 locus. (B) Chromatin loops showing enhancer Capture-C data over the course of RA-induced ESC differentiation. (C–E) Relative quantitative results showing enhancer interactions during RA-induced ESC differentiation. (F, G) qRT-PCR results showing Halr1 and Hoxa1 transcript levels after enhancer KO during RA-induced ESC differentiation. Data represent the mean ± SD, n = 3 or 6 replicates for WT or enhancer knockout cells. Statistical significance was determined by a two-tailed Student's t-test (unpaired), * P< 0.05, ** P < 0.01.
Enhancers are required for RA signaling-induced early proper lineage differentiation of ESCs
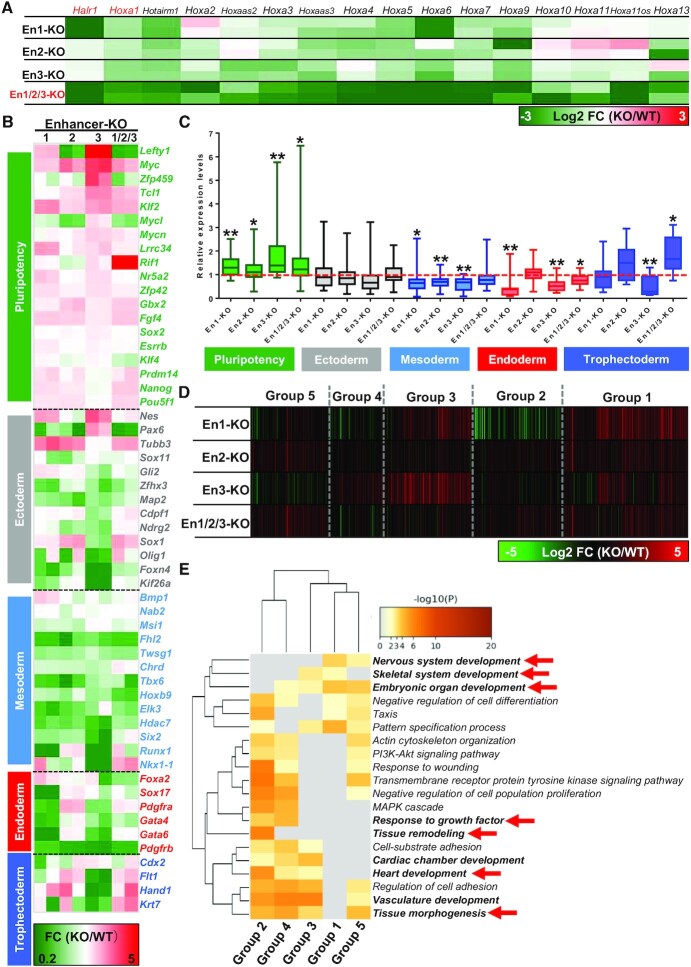
To further investigate whether these enhancers regulate RA signaling-induced early lineage differentiation of ESCs, we next performed RNA-seq analysis in WT and enhancer knockout cells after one day of RA treatment. Enhancer deletion resulted in significant changes in gene expression relative to WT cells (Supplementary Figure S7A). GO analysis of significantly altered gene groups (FD ≥ 2, FDR < 0.05) revealed that tissue morphogenesis, heart development, embryonic organ development and other developmentally-related biological processes were significantly enriched in enhancer KO cells (Supplementary Figure S7B). Moreover, among specific genes related to ESC differentiation, Bmp4, Gata3, Gata4, Wnt3a and Notch1 were enriched (Supplementary Figure S7C; Supplementary Table S8). These results suggest that enhancer deletion regulates RA signaling induced-early differentiation of ESCs.
Next, we also analyzed the changes in Halr1 and HoxA cluster gene expression in RNA-seq data and found that enhancer deletions significantly suppressed Halr1 and HoxA cluster gene expression (Figure 4A), a finding consistent with the qRT-PCR results. Furthermore, analysis of the expression of pluripotency and germ layer genes showed that enhancers’ deletion significantly promotes the expression of pluripotency genes compared to WT cells, whereas the mesendoderm genes were significantly repressed, indicating that enhancers deletion inhibited RA-induced early mesendoderm differentiation of ESCs (Figure 4B and C). Cluster analysis of differentially-expressed genes and subsequent GO analysis revealed that multiple biological processes associated with differentiation were significantly enriched in enhancer deletion cells (Figure 4D and E; Supplementary Table S9), such as embryonic organ development, nervous system development, heart development and tissue morphogenesis. Taken together, these data suggest that in ESCs, these enhancers are required for induction of proper lineage differentiation by RA signaling.
Figure 4.
Enhancers are required for RA signaling-induced early proper lineage differentiation of ESCs. (A) Heatmap showing relative transcript levels of Halr1 and HoxA cluster genes in enhancer KO ESCs at day 1 of RA treatment. (B) Heatmap showing relative transcript levels of pluripotency and germ layer genes in enhancer KO cells at day 1 of RA treatment. (C) Results showing changes in expression of pluripotency or germ layer genes in enhancer KO relative to WT ESCs (RNA-seq data). Expression levels in WT ESCs were set to 1 are and shown as the red dotted line. (D) Cluster analysis of differentially-expressed genes in enhancer KO cells. (E) GO-BP analysis of gene clusters obtained in (D).
HOXA1 promotes expression of Halr1 through binding to enhancer
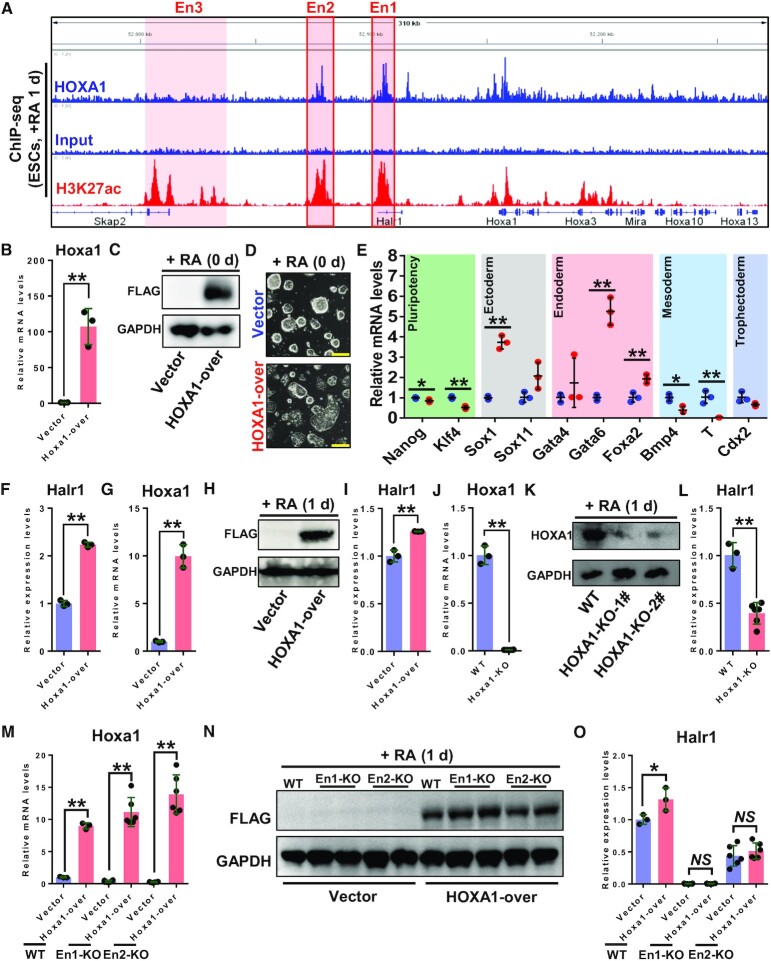
Previous studies have shown that HOXA1 plays an essential regulatory role as TF during RA signaling-induced early ESC differentiation (18,19,21). However, whether HOXA1 regulates lncRNA is unknown. Interestingly, by analyzing HOXA1 ChIP-seq data, we found significant binding of HOXA1 at the Halr1 locus, and further analysis in conjunction with H3K27ac ChIP-seq data revealed significant co-localization of HOXA1 binding with enhancers at the Halr1–Hoxa1 locus (Figure 5A) (21,28). These findings suggest that HOXA1 may be involved in regulating Halr1 expression.
Figure 5.
HOXA1 promotes Halr1 expression through binding to enhancers. (A) Enrichment of HOXA1 and H3K27ac ChIP-seq signals at the Halr1-Hoxa1 locus. (B) qRT-PCR analysis of Hoxa1 transcript levels in ESCs transfected with vector control or HOXA1 overexpression plasmids. (C) WB analysis of HOXA1 protein levels in ESCs transfected as in (B). (D) Bright-field images showing ESC morphology after HOXA1 overexpression. Scale bars: 100 μm. (E) qRT-PCR analysis of pluripotency and germ layer transcripts in ESCs transfected with control vector or HOXA1 overexpression plasmids. (F) qRT-PCR analysis of Halr1 transcript levels in ESCs transfected with vector control or HOXA1 overexpression plasmids. (G) qRT-PCR analysis of Hoxa1 mRNA levels in ESCs transfected with control vector or HOXA1 overexpression plasmids on day 1 of RA treatment. (H) WB analysis of HOXA1 protein levels in cells transfected as in (G). (I) qRT-PCR analysis of Halr1 RNA expression levels in ESCs transfected with control vector or HOXA1 overexpression plasmids on day 1 of RA treatment. (J) qRT-PCR analysis of Hoxa1 transcript levels in WT and Hoxa1 KO cells on day 1 of RA treatment. (K) WB analysis of HOXA1 protein levels in cells transfected as in (J). (L) qRT-PCR analysis of Halr1 transcript levels in WT and Hoxa1 KO cells on day 1 of RA treatment. (M) qRT-PCR analysis of Hoxa1 transcript levels in WT and enhancer KO cells transfected with control vector or HOXA1 overexpression plasmids on day 1 of RA treatment. (N) WB analysis of HOXA1 protein levels in cells transfected as in (M). (O) qRT-PCR analysis of Halr1 transcript levels in WT and enhancer KO cells transfected with control vector or HOXA1 overexpression plasmids on day 1 of RA treatment. NS: not significant. In (B), (E–G), (I), (J), (L), (M) and (O), data represent the mean ± SD (n = 3 or 6 replicates). Statistical significance was determined by a two-tailed Student's t-test (unpaired), * P< 0.05, ** P < 0.01.
To determine whether HOXA1 regulates Halr1 expression, we examined potential direct regulatory effects of HOXA1 on the Halr1 gene using overexpression and loss-of-function studies. First, we overexpressed HOXA1 in undifferentiated ESCs and found significant differences in ESCs’ cell morphology (Figure 5B–D). Further pluripotency gene assays revealed significant downregulation of Nanog and Klf4, while differentiation genes such as Sox1 (ectoderm), Gata6 and Foxa2 (endoderm) were significantly upregulated and mesoderm genes (Bmp4 and T) were significantly downregulated in HOXA1-over ESC, suggesting that HOXA1 overexpression perturbs ESC pluripotency (Figure 5E). qRT-PCR analysis of ESCs overexpressing HOXA1 showed significantly higher Halr1 expression compared with vector control, suggesting that HOXA1 promotes Halr1 expression in ESCs (Figure 5F). Following RA-induced ESC differentiation, HOXA1 overexpression still upregulated Halr1 expression (Figure 5G–I). Further, we used CRISPR-Cas9 technology to knockout HOXA1 and assess effects on Halr1 expression (Supplementary Figure S8). qRT-PCR analysis showed significantly decreased Halr1 transcript levels in HOXA1 KO ESCs after RA treatment compared to comparably treated WT cells (Figure 5J–L). This combined with the HOXA1 ChIP-seq data suggests that HOXA1 directly regulates Halr1 expression.
Considering the significant binding of HOXA1 to enhancer En1 and En2 (Figure 5A), we asked whether the promoting effect of HOXA1 on Halr1 expression was enhancer-dependent. To do so, we overexpressed HOXA1 in WT and in En1 or En2 enhancer KO cells (Figure 5M and N). After 1 day of RA treatment, WT cells overexpressing HOXA1 showed increased Halr1 expression, while enhancers (En1 and En2) knockout cells overexpressing HOXA1 showed no change in Halr1 expression (Figure 5O). These results suggest that enhancers are required for HOXA1 to promote Halr1 expression during RA-induced early differentiation of ESCs. Further, to assess direct regulatory effects, we also knockout the HOXA1 binding sites (H1BSs) at these enhancer regions (En1 and En2) by using CRISPR-Cas9 technology (Supplementary Figure S9A–D). qRT-PCR results showed that deletion of these H1BSs significantly suppressed RA-induced Halr1 expression, indicating the direct regulatory effect of HOXA1 on Halr1 (Supplementary Figure S9E). Taken together, these data suggest that HOXA1 promotes Halr1 expression through these enhancers.
Halr1 orchestrates the Hoxa1 chromatin structure by acting as the ‘brake’ and the ‘binder’
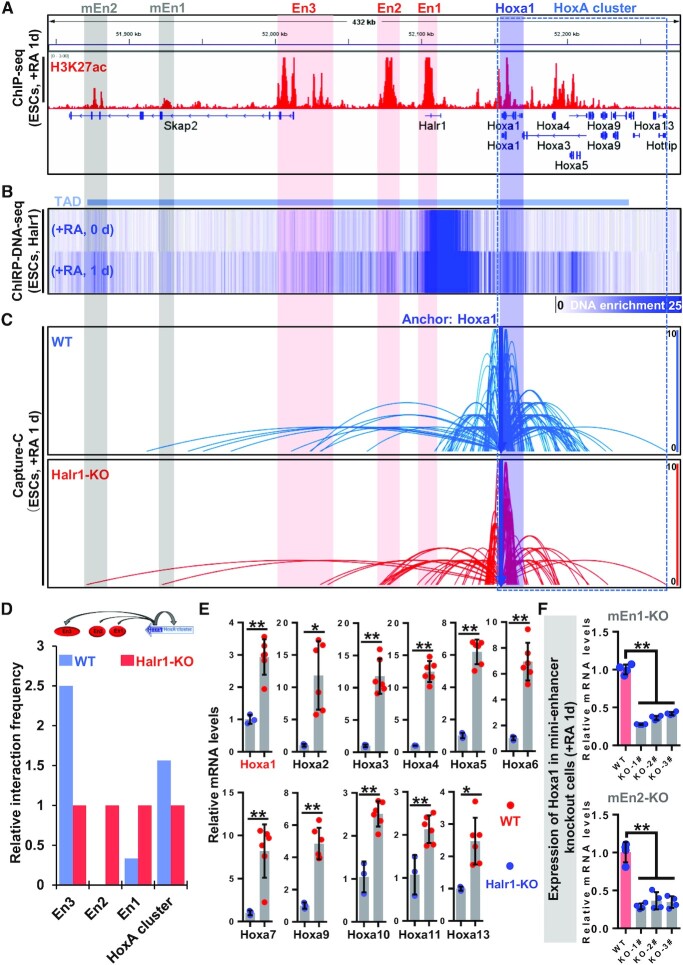
Previous studies have shown that long noncoding RNAs regulate gene expression through multiple modes, such as acting as sponges for microRNAs or as scaffolds that interact with proteins to regulate target gene expression (64–66). Halr1 transcription site has significant H3K27ac labeling and knockout of enhancer En1 abolishes Halr1 expression, suggesting that Halr1 may be an enhancer-associated RNA (eRNA) (Figure 6A).
Figure 6.
Halr1 RNA orchestrates Hoxa1 chromatin interactions during RA signaling-induced early differentiation of ESCs. (A) ChIP-seq data demonstrating H3K27ac enrichment at the Halr1–Hoxa1 locus. (B) ChIRP-seq data demonstrating Halr1 RNA enrichment at the Halr1–Hoxa1 locus. (C) Chromatin loops showing Hoxa1 Capture-C data demonstrating Hoxa1 chromatin interactions after Halr1 deletion. (D) Relative quantitative results demonstrating changes in Hoxa1 chromatin interactions after Halr1 deletion. (E) Heatmap showing transcript levels of HoxA cluster genes in WT and Halr1-KO ESCs on day 1 of RA treatment. (F) Expression levels of Hoxa1 mRNAs in mini-enhancer KO cells. Data represent the mean ± SD (n = 4 replicates). Statistical significance was determined by a two-tailed Student's t-test (unpaired), * P< 0.05, ** P < 0.01.
A recent study showed that Halr1 RNA regulates HoxA cluster expression mainly by binding HoxA cluster chromatin (27). Our analysis of Halr1 ChIRP-DNA-seq data (27), combined with Hi-C data (8,47), revealed that Halr1 is mainly enriched at the Halr1-Hoxa1 locus and is located within the same topologically associating domain (TAD) (Figure 6B; Supplementary Figure S10). To determine if Halr1 regulates Hoxa1 chromatin structure, we again used Capture-C technology to compare chromatin interactions in WT and Halr1-KO cells using the Hoxa1 locus as the anchor region (Figure 6C). Halr1 deletion significantly altered Hoxa1 chromatin interactions, and in Halr1-KO cells, Hoxa1 chromatin interactions with enhancers En1 and En2 significantly increased, while interactions with enhancer En3 and HoxA cluster chromatin decreased (Figure 6D). In addition, we also unexpectedly observed increased interactions of two distal mini-enhancers (mEn1 and mEn2) with Hoxa1 chromatin in Halr1-KO cells (Figure 6C). These results suggest that Halr1 RNA bound to chromatin acts as a scaffold, on one hand, as a ‘binder’ to facilitate interaction of Hoxa1 chromatin with enhancer En3 and maintain the HoxA cluster chromatin structure, and on the other hand, as a ‘brake’ to inhibit interaction of Hoxa1 chromatin with enhancers En1 and En2 and two distal mini-enhancers (mEn1 and mEn2).
Halr1 deletion promotes RA signaling-induced Hoxa1 overactivation and early endoderm differentiation of ESCs
To investigate the regulation of Hoxa1 expression by Halr1, we examined the mRNA expression levels of Hoxa1 by qRT-PCR and showed that the mRNA expression levels of Hoxa1 were significantly increased in Halr1-KO cells when compared with WT cells in the presence of RA treatment (Figure 6E). In addition, expression of other HoxA cluster genes (Hoxa2–13) was also significantly increased in Halr1-KO cells, consistent with previous reports (27,29). Considering that deletion of Halr1 also increases the interaction of Hoxa1 with two distal mini-enhancers (mEn1 and mEn2), it is unclear whether these enhancers regulate RA signaling-induced Hoxa1 expression. Next, we also generated the two distal mini-enhancers knockout cells by using CRISPR–Cas9 technology (Supplementary Figure S11). At day one after RA treatment, qRT-PCR analysis indicated significant decreased Hoxa1 transcript levels relative to comparably treated WT cells (Figure 6F), indicating that the two distal mini-enhancers are also required for RA-induced Hoxa1 expression and are newly identified Hoxa1 enhancers. Taken together, these data suggest that Halr1 plays a critical role in suppressing Hoxa1 expression during RA-induced early differentiation of ESCs.
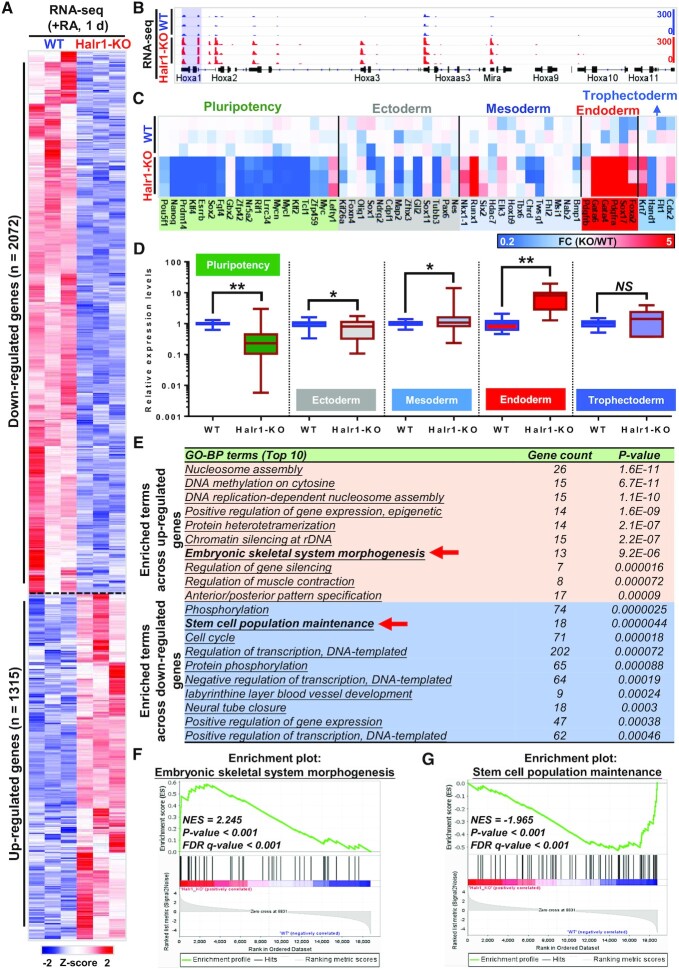
To further investigate the regulatory effect of Halr1 in the early stages of RA-induced ESC differentiation, we performed RNA-seq in WT and Halr1-KO cells at day one after RA treatment. Halr1 deletion significantly altered gene expression (Figure 7A; Supplementary Figure S12A), and HoxA cluster genes were significantly up-regulated (Figure 7B), consistent with qRT-PCR results (Figure 6E). Further analysis of pluripotency and germ layer genes revealed that pluripotency and ectodermal genes were significantly down-regulated, while meso-endoderm was significantly up-regulated, especially endoderm genes were more significantly up-regulated (Figure 7C and D). Further GO-BP analysis revealed that up-regulated genes were significantly enriched in biological processes related to embryonic skeletal system morphogenesis, endoderm formation and endodermal cell differentiation, while down-regulated genes were significantly enriched in processes associated with stem cell population maintenance (Figure 7E; Supplementary Table S10). GSEA of these biological processes also showed significant differences (Figure 7F and G). In summary, these results suggest that Halr1 is required for RA signaling-induced early proper lineage differentiation of ESCs, particularly of the endoderm.
Figure 7.
Halr1 deletion promotes RA signaling-induced Hoxa1 overactivation and enhanced early endodermal differentiation of ESCs. (A) Heatmap showing genes differentially-expressed in WT and Halr1-KO cells. (B) IGV screenshots showing HoxA cluster gene expression levels in WT and Halr1-KO cells. (C) Heatmap showing expression levels of pluripotency and germ layer genes in WT and Halr1-KO cells. (D) Results showing changes in pluripotency and germ layer gene expression in Halr1-KO relative to WT cells (RNA-seq data). (E) GO analysis showing biological processes of differential gene enrichment (Top 10). (F, G) Gene Set Enrichment Analysis (GSEA) of genes differentially expressed in Halr1-KO versus WT ESCs among genes associated with embryonic skeletal system morphogenesis (F) and stem cell population maintenance (G).
Hoxa1 cooperates with Halr1 to regulate RA signaling-induced early lineage differentiation of ESCs
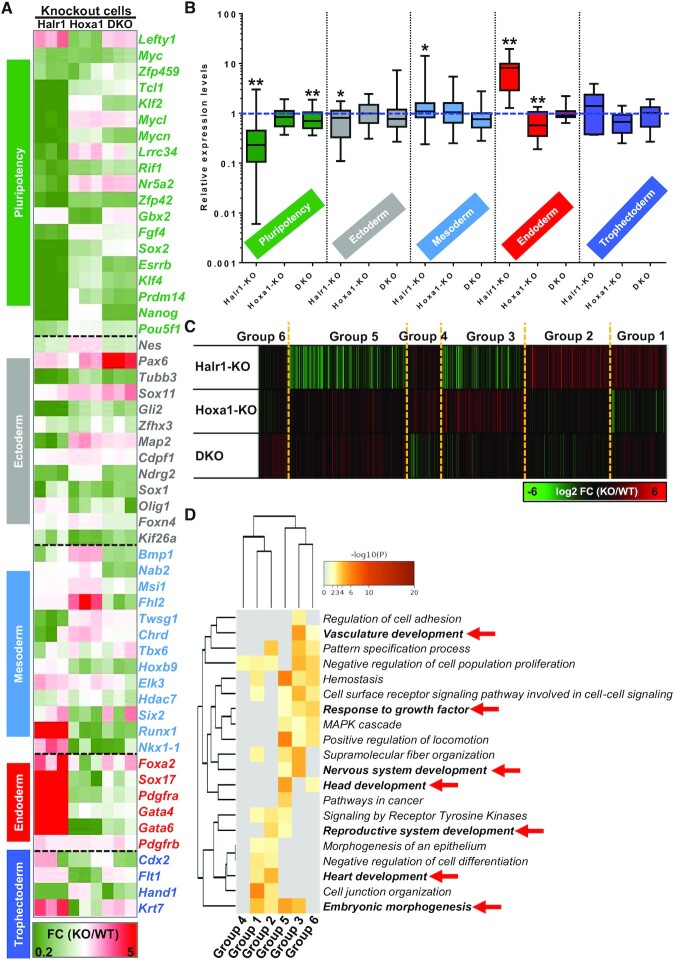
Our findings suggest that RA signaling activates Hoxa1 and Halr1 expression, and concomitantly, HOXA1 promotes Halr1 expression, which in turn represses Hoxa1 expression. Given these complex relationships, we established Hoxa1/Halr1 double-knockout (DKO) lines to investigate effects on RA signaling-induced early ESC differentiation (Supplementary Figure S8). RNA-seq data revealed that Hoxa1/Halr1 DKO significantly altered gene expression (Supplementary Figure S12A), and GO analysis of genes differentially expressed in single or double knockout cells revealed significant enrichment for biological processes associated with ESC development, such as heart development, tissue morphogenesis, gliogenesis, embryonic morphogenesis and cell fate commitment (Supplementary Figure S12B). Further analysis of biological processes associated with endoderm revealed changes in expression of several TFs significantly associated with endoderm development, such as Sox9, Gata3, Gata4 and Gata6 (Supplementary Figure S12C; Supplementary Table S11).
To reveal the co-regulatory effects of Hoxa1 and Halr1 in RA signaling-induced early lineage differentiation of ESCs, we also analyzed pluripotency and germ layer genes expression in these knockout cells (Figure 8A). Among them, Halr1 KO promoted endoderm gene expression relative to WT cells, while Hoxa1 KO suppressed endoderm gene expression, whereas endoderm gene expression in DKO cells did not differ significantly (Figure 8B). These results reveal that RA signaling-induced early proper lineage differentiation of ESC requires expressions of Hoxa1 and Halr1. Further, GO analysis after combining these differential genes in knockout cells revealed significant enrichment in biological processes related to heart development, vasculature development, nervous system development and embryonic morphogenesis (Figure 8C and D; Supplementary Table S12). Taken together, these results suggest that Hoxa1 and Halr1 co-expression maintains RA signaling-induced early ESC proper lineage differentiation.
Figure 8.
Hoxa1 cooperates with Halr1 to regulate RA signaling-induced early lineage differentiation of ESCs. (A) Heatmap showing expression of pluripotency and germ layer genes (RNA-seq data). (B) Changes in expression of pluripotency and germ layer genes in indicated single or double KO relative to WT cells (RNA-seq data). Expression levels in WT ESCs were set to 1 and shown as the blue dotted line. (C) Cluster analysis of genes differentially expressed in Halr1-KO, Hoxa1-KO and DKO cells. (D) GO-BP analysis of gene clusters obtained in (C).
HOXA1 acts as a core transcriptional regulator and directly regulates the expression of a subset of genes associated with endoderm development
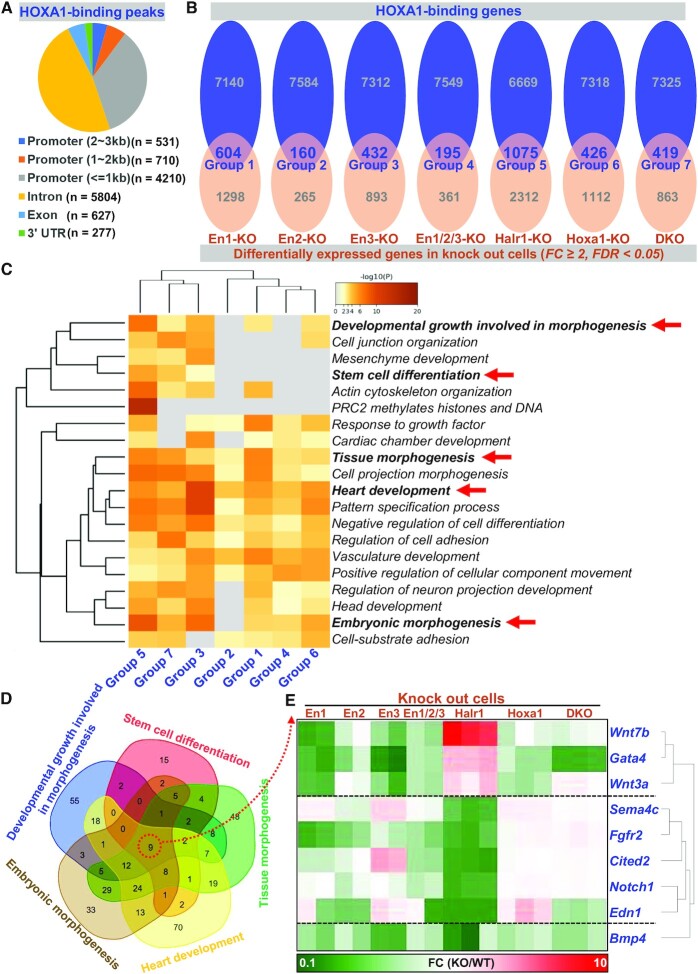
Considering that both enhancers and Halr1 can regulate Hoxa1 expression, and HOXA1, as a transcription factor, has extensive downstream regulation within the genome (18,21). To further characterize the regulatory effects of enhancers and Halr1 through HOXA1 during RA-induced early ESC differentiation. We also analyzed the distribution of HOXA1 binding within the genome (Figure 9A), and HOXA1-bound genes were analyzed together with differential genes in various knockout cells using RNA-seq data. That analysis resulted in seven gene sets (Figure 9B). Subsequent GO analysis revealed significant enrichment in biological processes such as stem cell differentiation, developmental growth involved in morphogenesis, and embryonic morphogenesis (Figure 9C), suggesting that HOXA1 may be directly involved in RA-induced ESC differentiation as an executor in these regulatory processes. Furthermore, to identify gene sets related to endoderm development, we selected five gene sets enriched in developmental processes and after overlap identified a cluster of genes directly regulated by HOXA1, including Gata4, Wnt7b and Wnt3a (Figure 9D and E; Supplementary Table S13).
Figure 9.
The core transcriptional regulator HOXA1 directly regulates expression of a subset of endodermal development-related genes. (A) Data showing the proportion of HOXA1 binding peaks in various genomic loci. (B) Comprehensive analysis of differential genes enriched for regulation by HOXA1. (C) GO-BP analysis of gene clusters obtained in (B). (D) Demonstration of common genes in selected biological process. (E) Heatmap showing expression levels of genes overlapped in (D).
DISCUSSION
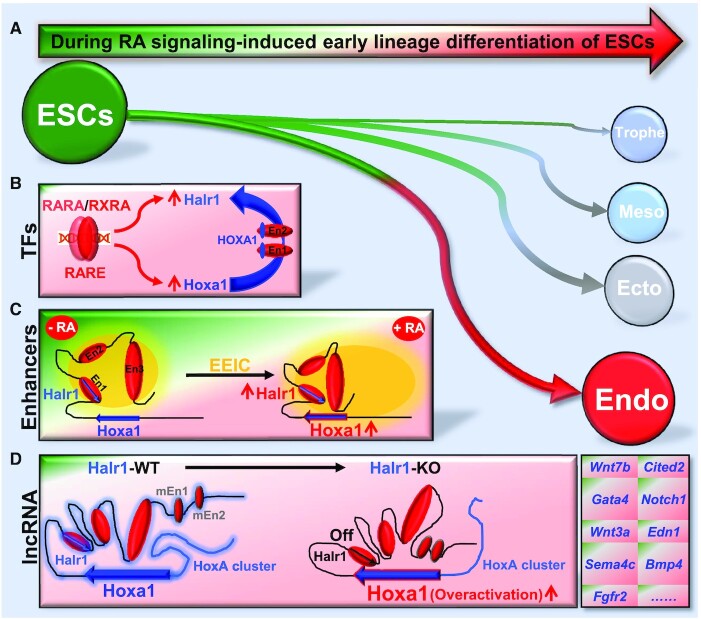
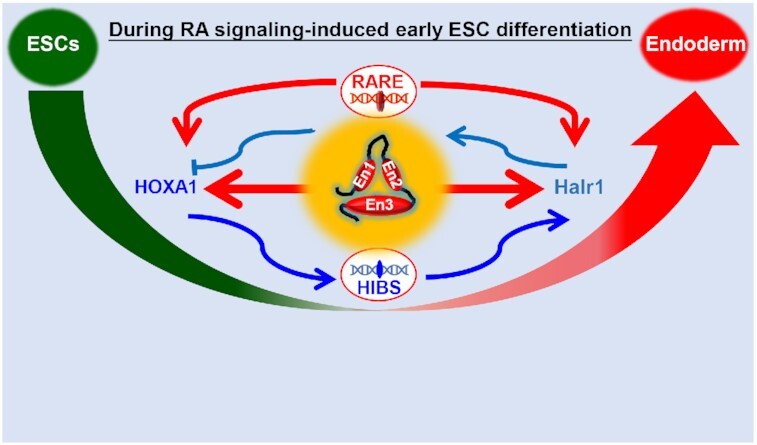
Temporally and spatially precise transcription of genes is critical for ESC fate decisions. However, mechanisms underlying this process remain largely unclear. Previous studies have revealed that many TFs play a central role in controlling gene expression during ESC differentiation (2). However, recent studies of high-order chromatin structure and long non-coding RNAs have provided new insights into the precise intrinsic mechanisms underlying the regulation of gene expression (67–69). In this study, we report enhancer architecture-dependent multilayered transcriptional regulation at the Halr1–Hoxa1 locus orchestrating RA signaling-induced early lineage differentiation of ESCs. Our results reveal a novel mechanism by which RA signaling facilitates correct ESC fate decisions during early differentiation via synergistic activity of enhancer elements, TFs and lncRNA (Figure 10).
Figure 10.
Schematic summary of this study. Schematic showing enhancer architecture-dependent multilayered transcriptional regulation at the Halr1–Hoxa1 locus during RA signaling-induced early lineage differentiation of ESCs (A). (B) TF-driven gene expression: RARA/RXRA activates Halr1 and Hoxa1 expression via RAREs, and HOXA1 promotes Halr1 expression through binding to enhancer regions. (C) Enhancer-dependent transcriptional regulation: RA promotes increased interactions between enhancers, resulting in a tighter EEIC and expression of Halr1 and Hoxa1. (D) lncRNA Halr1-mediated chromatin organization orchestrates Hoxa1 expression: Halr1 RNA bound to DNA acts as a scaffold for chromatin structure, first as a binder to facilitate Hoxa1 chromatin interaction with enhancer En3 and to maintain HoxA cluster chromatin structure, and second as a braking element to inhibit Hoxa1 chromatin interaction with four distal enhancers (En1, En2 and mini-enhancers). Endo, endoderm; Ecto, ectoderm; Meso, mesoderm; Trophe, trophectoderm.
Maintenance of ESC pluripotency requires activation of LIF signaling and inhibition of MEK and GSK3 pathways (70,71). When stimulation of these signaling pathways is withdrawn, the ESCs differentiate. In our random differentiation conditions lacking RA, ESCs show marked ectodermal differentiation with high expression of ectodermal master control TFs Sox1 and Sox11. By contrast, RA-induced ESC differentiation also activated endodermal master control TFs such as Gata4, Gata6, Sox17 and Foxa2, as well as ectodermal differentiation genes (Figure 1D), indicative of mixed lineage differentiation at this early point (Figure 10A). Indeed, it has also been previously reported that RA signaling significantly activates endoderm master control TFs during ESC differentiation, already suggesting RA-induced ESC endoderm differentiation (25,26). In addition, recent reports reveal that in ESCs, RA signaling significantly activates 2cell-like (2C-like) marker expression, such as Zscan4, suggesting that RA also facilitates ESC conversion to a 2C-like cell state (2CLC) (72–75). Interestingly, in our data (Supplementary Figure S13), we also found that the expression of Zscan4 was significantly increased during RA-induced early ESC differentiation, while enhancers and Hoxa1 both modulate Zscan4 expressions. These findings suggest that RA signaling-induced early ESC lineage differentiation may be more complex, and molecular mechanisms underlying these complex activities warrant further analysis
TF-driven transcription plays a central role in ESC differentiation (76,77). In our present study, the results reveal that early ESC differentiation after RA treatment is regulated on multiple levels (Figure 10B). First, we conclude that Hoxa1 and Halr1 are the direct downstream target genes of RA signaling regulated by RARA/RXRA via RAREs. The RARE at the 3′-end of Hoxa1 was previously reported as required for RA-induced Hoxa1 expression (78,79), and our results support this conclusion in our differentiation model. However, a RARE within the Halr1 locus has not until now been reported, and mechanisms underlying high expression of Halr1 induced by RA signaling were not clear. Here, we identify a RARE within the Halr1 locus that is functionally comparable to that of Hoxa1 and required for RA signaling to promote high Halr1 expression. Secondly, our results also further reveal that HOXA1 can directly promote Halr1 expression. HOXA1 is a core TF in RA signaling-induced early ESC differentiation, and previous studies have primarily characterized its regulation of coding genes (18,21), but whether it can regulate the expression of long non-coding RNAs has not been reported. Here, we show that HOXA1 can regulate expression of the lncRNA Halr1, expanding its TF function. Thus, our study reveals a novel model of multilayered regulation driven by TFs during RA signaling-induced early ESC lineage differentiation, whereby RARA/RXRA activates the expression of Hoxa1 and Halr1, while HOXA1 further promotes Halr1 expression. Furthermore, considering that both RA receptors (RARA/RXRA) and HOXA1 bind and exert regulatory effects at the Halr1–Hoxa1 locus during RA signaling-induced early ESC differentiation, the combined data above suggest that the regulation of RA signaling at the Halr1–Hoxa1 locus is more likely to be the result of synergistic regulation by multi-factor, while the more refined underlying mechanisms may require further validation.
Enhancer is essential as the functional DNA element in ESCs maintenance and individual development (3). We previously showed that enhancer interactions during RA-induced ESC early differentiation result in formation of an EEIC that regulates HoxA cluster gene expression (23). In this study, we compared enhancer interactions in ESCs in both undifferentiated and RA-induced differentiation states and showed that RA signaling promoted enhancer interactions, with some results similar to those previously reported (28), revealing dynamic chromatin interactions at the Halr1–Hoxa1 locus (Figure 10C). Subsequently, our further analysis indicated that these enhancers are required for RA-induced Hoxa1 and Halr1 expression and early ESC lineage differentiation, particularly regulating mesendoderm differentiation, which has not been previously reported. We recently reported that two novel enhancers at the HoxB cluster locus are also required for RA-induced early ESC differentiation (80). Overall, these evidences reveal a direct regulatory relationship between enhancer and ESC lineage differentiation and highlight the important role of functional elements such as enhancer in fate determination of ESCs. However, we currently do not have a thorough understanding of the underlying mechanisms by which RA signaling promotes increased enhancer interactions. Recent studies employing using HiChIP technology reveal that mechanistically TFs function to regulate high-order chromatin structure and promote DNA-loop formation (81–83). Considering our previous results showing HOXA1 binding at the enhancer regions (En1 and En2), we therefore speculate that HOXA1 may be involved in regulating enhancer interactions, and these need further study.
Although lncRNAs reportedly regulate ESC multipotency, only a few studies report their orchestration of high-order chromatin structure in this context (27,33). Halr1 (also known as linc-HOXA1, Haunt, Gm15055 and linc1547) reportedly regulates Hoxa1 expression in at least three ways: by binding PURB to transcriptionally repress Hoxa1 (84), by binding to the PRC2 complex to mediate epigenetic repression (29), and by acting as a chromatin RNA to inhibit interaction of enhancers (here, En1 and En2) with Hoxa1 chromatin (27). In this study, we focus on the regulatory effects of Halr1 on chromatin structure (Figure 10D). By analyzing Hi-C and Halr1-ChIRP-DNA-seq data (8,27), we found that RA-induced high expression of Halr1 binds mainly to the Halr1–Hoxa1 locus and acts as a cis-acting element to regulate gene expression within a TAD (Supplementary Figure S10). Further by Capture-C screening, we found that deletion of Halr1 not only facilitated the interactions of Hoxa1 chromatin with enhancer En1 and En2 but also weakened the interactions with enhancer En3 and HoxA cluster (Figure 6). These results suggest that Halr1 not only acts as a ‘brake’ to precisely control the interactions of Hoxa1 chromatin with distal enhancers (27) but also acts as a ‘binder’ to maintain the interactions with enhancer and HoxA cluster. These results also suggest that Halr1 RNA binds to the HoxA cluster as a chromatin RNA to maintain a relatively closed chromatin state and that in its absence, HoxA cluster chromatin becomes opened, allowing interaction with enhancers outside the cluster. Thus, these findings not only expand the role of Halr1′s function as a chromatin RNA but also further highlight the flexible nature of lncRNAs to regulate gene expression by orchestrating high-order chromatin structures.
Previous studies have shown that deletion of Halr1 RNA significantly promotes the expression of HoxA cluster genes, while deletion of enhancer En1 significantly represses the expression of HoxA cluster genes, revealing the opposite regulation of HoxA cluster genes’ expression by Halr1 RNA and its genomic locus, and our findings are consistent with that (27) (Supplementary Figure S14). Considering that we also found that HOXA1 promotes Halr1 expression, together these results suggest a significant positive feedback regulation between Hoxa1 and Halr1 that may resemble a circular regulation. However, it is not clear who is the initiator of the regulation and this needs to be explored further (85). Furthermore, our transcriptome analysis reveals that Halr1 regulates RA signaling-induced early ESC lineage differentiation particularly by promoting ESC endoderm differentiation, a novel role for Halr1 not previously reported. Interestingly, we observed that Halr1 was highly expressed in the early stage of spontaneous differentiation of ESCs (without LIF/2i). This pattern of expression of Halr1 in early ESC differentiation is very similar to that of the previously reported lncRNA Ephemeron (86), a lncRNA expressed only in mouse ESCs. Unexpectedly, we found that Halr1 is not present in human genome and is only expressed in the mouse. We also found significant differences in enhancer distribution and chromatin interactions at the Skap2-Hoxa1 locus between mouse and human (Supplementary Figure S15) (8,63,87,88). These new findings suggest that Halr1 is a species-specific lncRNA and may play other regulatory roles during early mouse ESC differentiation, which requires further investigation.
In conclusion, our studies reveal enhancer architecture-dependent multilayered transcriptional regulation at the Halr1–Hoxa1 locus orchestrating RA signaling-induced early lineage differentiation of ESCs. These findings provide new insight into molecular mechanisms underlying ESC fate decisions.
DATA AVAILABILITY
The raw data produced in this study for the DNA-seq and RNA-seq data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE169058.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of our laboratory for many helpful discussions. We also thank Dr Lingyi Chen (Professor, Nankai University), Dr Jun Zhou (Professor, Nankai University), Dr Xinyi Lu (Professor, Nankai University), Dr Jiaxi Zhou (Professor, Chinese Academy of Medical Sciences) and Dr Weiping Yuan (Professor, Chinese Academy of Medical Sciences) for helpful comments. We also thank Dr Shaohui Chen for his comments during the early stage of this study. We also thank Dr Lamar for editing this article.
Author contributions: W.L., G.S. and L.Z. conceived of and designed the study. W.L., G.S., L.Z., Z.Z. and J.S. supervised the study. G.S., X.Z., J.C., J.Z., M.L. and J.B. performed experiments and analyzed the data. W.W., S.S. and D.G. analyzed sequencing data. G.S., W.W. and B.C. performed bioinformatic analysis. G.S. and W.L. wrote the manuscript. All authors have read and approved the final manuscript.
Contributor Information
Guangsong Su, College of Life Sciences, Nankai University, 94 Weijin Road, 300071 Tianjin City, China.
Wenbin Wang, College of Life Sciences, Nankai University, 94 Weijin Road, 300071 Tianjin City, China.
Xueyuan Zhao, College of Life Sciences, Nankai University, 94 Weijin Road, 300071 Tianjin City, China.
Jun Chen, College of Life Sciences, Nankai University, 94 Weijin Road, 300071 Tianjin City, China.
Jian Zheng, College of Life Sciences, Nankai University, 94 Weijin Road, 300071 Tianjin City, China.
Man Liu, College of Life Sciences, Nankai University, 94 Weijin Road, 300071 Tianjin City, China.
Jinfang Bi, College of Life Sciences, Nankai University, 94 Weijin Road, 300071 Tianjin City, China.
Dianhao Guo, College of Life Sciences, Nankai University, 94 Weijin Road, 300071 Tianjin City, China.
Bohan Chen, College of Life Sciences, Nankai University, 94 Weijin Road, 300071 Tianjin City, China.
Zhongfang Zhao, College of Life Sciences, Nankai University, 94 Weijin Road, 300071 Tianjin City, China.
Jiandang Shi, College of Life Sciences, Nankai University, 94 Weijin Road, 300071 Tianjin City, China.
Lei Zhang, College of Life Sciences, Nankai University, 94 Weijin Road, 300071 Tianjin City, China.
Wange Lu, College of Life Sciences, Nankai University, 94 Weijin Road, 300071 Tianjin City, China; State Key Laboratory of Medicinal Chemical Biology, Nankai University, 94 Weijin Road, 300071 Tianjin City, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2017YFA0102600]; Fundamental Research Funds for the Central Universities (Nankai University) [63201087]. Funding for open access charge: National Key R&D Program of China [2017YFA0102600]; Fundamental Research Funds for the Central Universities (Nankai University) [63201087].
Conflict of interest statement. None declared.
REFERENCES
- 1. Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G.et al.. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005; 122:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Young R.A. Control of the embryonic stem cell state. Cell. 2011; 144:940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zheng H., Xie W.. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 2019; 20:535–550. [DOI] [PubMed] [Google Scholar]
- 4. Plasschaert R.N., Vigneau S., Tempera I., Gupta R., Maksimoska J., Everett L., Davuluri R., Mamorstein R., Lieberman P.M., Schultz D.et al.. CTCF binding site sequence differences are associated with unique regulatory and functional trends during embryonic stem cell differentiation. Nucleic Acids Res. 2014; 42:774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang J., Huang J., Shi G.. Retrotransposons in pluripotent stem cells. Cell Regener. 2020; 9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ji Z., Mohammed H., Webber A., Ridsdale J., Han N., Carroll J.S., Sharrocks A.D.. The forkhead transcription factor FOXK2 acts as a chromatin targeting factor for the BAP1-containing histone deubiquitinase complex. Nucleic Acids Res. 2014; 42:6232–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W.et al.. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015; 518:331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y., Xu X., Lv X., Hugnot J.P., Tanay A.et al.. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017; 171:557–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Osterwalder M., Barozzi I., Tissières V., Fukuda-Yuzawa Y., Mannion B.J., Afzal S.Y., Lee E.A., Zhu Y., Plajzer-Frick I., Pickle C.S.et al.. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018; 554:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mead T.J., Wang Q., Bhattaram P., Dy P., Afelik S., Jensen J., Lefebvre V.. A far-upstream (-70 kb) enhancer mediates Sox9 auto-regulation in somatic tissues during development and adult regeneration. Nucleic Acids Res. 2013; 41:4459–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wei Z., Gao F., Kim S., Yang H., Lyu J., An W., Wang K., Lu W.. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell. 2013; 13:36–47. [DOI] [PubMed] [Google Scholar]
- 12. Das B.C., Thapa P., Karki R., Das S., Mahapatra S., Liu T.C., Torregroza I., Wallace D.P., Kambhampati S., Van Veldhuizen P.et al.. Retinoic acid signaling pathways in development and diseases. Development. 2014; 22:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Erkelens M.N., Mebius R.E.. Retinoic acid and immune homeostasis: a balancing act. Trends Immunol. 2017; 38:168–180. [DOI] [PubMed] [Google Scholar]
- 14. Das B.C., Thapa P., Karki R., Das S., Mahapatra S., Liu T.C., Torregroza I., Wallace D.P., Kambhampati S., Van Veldhuizen P.et al.. Retinoic acid signaling pathways in development and diseases. Bioorg. Med. Chem. 2014; 22:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ablain J., de Thé H.. Retinoic acid signaling in cancer: the parable of acute promyelocytic leukemia. Int. J. Cancer. 2014; 135:2262–2272. [DOI] [PubMed] [Google Scholar]
- 16. Chatagnon A., Veber P., Morin V., Bedo J., Triqueneaux G., Semon M., Laudet V., Buc F., Benoit G.. RAR/RXR binding dynamics distinguish pluripotency from differentiation associated cis-regulatory elements. Nucleic Acids Res. 2015; 43:4833–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Simandi Z., Horvath A., Wright L.C., Cuaranta-Monroy I., De Luca I., Karolyi K., Sauer S., Deleuze J.F., Gudas L.J., Cowley S.M.et al.. OCT4 Acts as an integrator of pluripotency and signal-induced differentiation. Mol. Cell. 2016; 63:647–661. [DOI] [PubMed] [Google Scholar]
- 18. De Kumar B., Parker H.J., Parrish M.E., Lange J.J., Slaughter B.D., Unruh J.R., Paulson A., Krumlauf R.. Dynamic regulation of Nanog and stem cell-signaling pathways by Hoxa1 during early neuro-ectodermal differentiation of ES cells. PNAS. 2017; 114:5838–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Kumar B., Parker H.J., Paulson A., Parrish M.E., Zeitlinger J., Krumlauf R.. Hoxa1 targets signaling pathways during neural differentiation of ES cells and mouse embryogenesis. Dev. Biol. 2017; 432:151–164. [DOI] [PubMed] [Google Scholar]
- 20. Martinez-Ceballos E., Gudas L.J.. Hoxa1 is required for the retinoic acid-induced differentiation of embryonic stem cells into neurons. J. Neurosci. Res. 2008; 86:2809–2819. [DOI] [PubMed] [Google Scholar]
- 21. De Kumar B., Parker H.J., Paulson A., Parrish M.E., Pushel I., Singh N.P., Zhang Y., Slaughter B.D., Unruh J.R., Florens L.et al.. HOXA1 and TALE proteins display cross-regulatory interactions and form a combinatorial binding code on HOXA1 targets. Genome Res. 2017; 27:1501–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kubo N., Ishii H., Xiong X., Bianco S., Meitinger F., Hu R., Hocker J.D., Conte M., Gorkin D., Yu M.et al.. Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat. Struct. Mol. Biol. 2021; 28:152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Su G., Wang W., Chen J., Liu M., Zheng J., Guo D., Bi J., Zhao Z., Shi J., Zhang L.et al.. CTCF-binding element regulates ESC differentiation via orchestrating long-range chromatin interaction between enhancers and HoxA. J. Biol. Chem. 2021; 296:100413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O., Reinberg D.. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015; 347:1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Capo-Chichi C.D., Rula M.E., Smedberg J.L., Vanderveer L., Parmacek M.S., Morrisey E.E., Godwin A.K., Xu X.X.. Perception of differentiation cues by GATA factors in primitive endoderm lineage determination of mouse embryonic stem cells. Dev. Biol. 2005; 286:574–586. [DOI] [PubMed] [Google Scholar]
- 26. Su G., Guo D., Chen J., Liu M., Zheng J., Wang W., Zhao X., Yin Q., Zhang L., Zhao Z.et al.. A distal enhancer maintaining Hoxa1 expression orchestrates retinoic acid-induced early ESCs differentiation. Nucleic Acids Res. 2019; 47:6737–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yin Y., Yan P., Lu J., Song G., Zhu Y., Li Z., Zhao Y., Shen B., Huang X., Zhu H.et al.. Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell. 2015; 16:504–516. [DOI] [PubMed] [Google Scholar]
- 28. Cao K., Collings C.K., Marshall S.A., Morgan M.A., Rendleman E.J., Wang L., Sze C.C., Sun T., Bartom E.T., Shilatifard A.. SET1A/COMPASS and shadow enhancers in the regulation of homeotic gene expression. Genes Dev. 2017; 31:787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu G.Y., Zhao G.N., Chen X.F., Hao D.L., Zhao X., Lv X., Liu D.P.. The long noncoding RNA Gm15055 represses Hoxa gene expression by recruiting PRC2 to the gene cluster. Nucleic Acids Res. 2016; 44:2613–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Acharya D., Hainer S.J., Yoon Y., Wang F., Bach I., Rivera-Pérez J.A., Fazzio T.G.. KAT-independent gene regulation by Tip60 promotes ESC self-renewal but not pluripotency. Cell Rep. 2017; 19:671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmittgen T.D., Livak K.J.. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008; 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 32. Li P., Ding N., Zhang W., Chen L.. COPS2 antagonizes OCT4 to accelerate the G2/M transition of mouse embryonic stem cells. Stem cell reports. 2018; 11:317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao T., Cai M., Liu M., Su G., An D., Moon B., Lyu G., Si Y., Chen L., Lu W.. lncRNA 5430416N02Rik promotes the proliferation of mouse embryonic stem cells by activating Mid1 expression through 3D chromatin architecture. Stem cell reports. 2020; 14:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A.et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davies J.O., Telenius J.M., McGowan S.J., Roberts N.A., Taylor S., Higgs D.R., Hughes J.R.. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat. Methods. 2016; 13:74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hughes J.R., Roberts N., McGowan S., Hay D., Giannoulatou E., Lynch M., De Gobbi M., Taylor S., Gibbons R., Higgs D.R.. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 2014; 46:205–212. [DOI] [PubMed] [Google Scholar]
- 37. Angiuoli S.V., Salzberg S.L.. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics. 2011; 27:334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Freese N.H., Norris D.C., Loraine A.E.. Integrated genome browser: visual analytics platform for genomics. Bioinformatics. 2016; 32:2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou X., Lowdon R.F., Li D., Lawson H.A., Madden P.A., Costello J.F., Wang T.. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat. Methods. 2013; 10:375–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anders S., Pyl P.T., Huber W.. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K.. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019; 10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang D.W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 44. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S.et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E.et al.. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003; 34:267–273. [DOI] [PubMed] [Google Scholar]
- 46. Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M.. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016; 44:D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., Li D., Choudhary M.N.K., Li Y., Hu M.et al.. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018; 19:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mei S., Qin Q., Wu Q., Sun H., Zheng R., Zang C., Zhu M., Wu J., Shi X., Taing L.et al.. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017; 45:D658–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng R., Wan C., Mei S., Qin Q., Wu Q., Sun H., Chen C.H., Brown M., Zhang X., Meyer C.A.et al.. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019; 47:D729–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang P., Andrianakos R., Yang Y., Liu C., Lu W.. Kruppel-like factor 4 (Klf4) prevents embryonic stem (ES) cell differentiation by regulating Nanog gene expression. J. Biol. Chem. 2010; 285:9180–9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Moutier E., Ye T., Choukrallah M.A., Urban S., Osz J., Chatagnon A., Delacroix L., Langer D., Rochel N., Moras D.et al.. Retinoic acid receptors recognize the mouse genome through binding elements with diverse spacing and topology. J. Biol. Chem. 2012; 287:26328–26341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Costa-Giomi M.P., Gaub M.P., Chambon P., Abarzúa P.. Characterization of a retinoic acid responsive element isolated by whole genome PCR. Nucleic Acids Res. 1992; 20:3223–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Loudig O., Babichuk C., White J., Abu-Abed S., Mueller C., Petkovich M.. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol. Endocrinol. 2000; 14:1483–1497. [DOI] [PubMed] [Google Scholar]
- 54. Huang D., Chen S.W., Gudas L.J.. Analysis of two distinct retinoic acid response elements in the homeobox gene Hoxb1 in transgenic mice. Dev. Dyn. 2002; 223:353–370. [DOI] [PubMed] [Google Scholar]
- 55. Huang D., Chen S.W., Langston A.W., Gudas L.J.. A conserved retinoic acid responsive element in the murine Hoxb-1 gene is required for expression in the developing gut. Development. 1998; 125:3235–3246. [DOI] [PubMed] [Google Scholar]
- 56. Marshall H., Studer M., Pöpperl H., Aparicio S., Kuroiwa A., Brenner S., Krumlauf R.. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994; 370:567–571. [DOI] [PubMed] [Google Scholar]
- 57. Nolte C., Jinks T., Wang X., Martinez Pastor M.T., Krumlauf R.. Shadow enhancers flanking the HoxB cluster direct dynamic Hox expression in early heart and endoderm development. Dev. Biol. 2013; 383:158–173. [DOI] [PubMed] [Google Scholar]
- 58. Studer M., Pöpperl H., Marshall H., Kuroiwa A., Krumlauf R.. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science. 1994; 265:1728–1732. [DOI] [PubMed] [Google Scholar]
- 59. Kim M.H., Shin J.S., Park S., Hur M.W., Lee M.O., Park H., Lee C.S.. Retinoic acid response element in HOXA-7 regulatory region affects the rate, not the formation of anterior boundary expression. Int. J. Dev. Biol. 2002; 46:325–328. [PubMed] [Google Scholar]
- 60. Fornes O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A., Modi B.P., Correard S., Gheorghe M., Baranašić D.et al.. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020; 48:D87–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Qian Y., Zhang L., Cai M., Li H., Xu H., Yang H., Zhao Z., Rhie S.K., Farnham P.J., Shi J.et al.. The prostate cancer risk variant rs55958994 regulates multiple gene expression through extreme long-range chromatin interaction to control tumor progression. Sci. Adv. 2019; 5:eaaw6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kempfer R., Pombo A.. Methods for mapping 3D chromosome architecture. Nat. Rev. Genet. 2020; 21:207–226. [DOI] [PubMed] [Google Scholar]
- 63. Xiao S., Xie D., Cao X., Yu P., Xing X., Chen C.C., Musselman M., Xie M., West F.D., Lewin H.A.et al.. Comparative epigenomic annotation of regulatory DNA. Cell. 2012; 149:1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang K.C., Chang H.Y.. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011; 43:904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mercer T.R., Mattick J.S.. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013; 20:300–307. [DOI] [PubMed] [Google Scholar]
- 66. Yang Y.W., Flynn R.A., Chen Y., Qu K., Wan B., Wang K.C., Lei M., Chang H.Y.. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife. 2014; 3:e02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Burton A., Torres-Padilla M.E.. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat. Rev. Mol. Cell Biol. 2014; 15:723–734. [DOI] [PubMed] [Google Scholar]
- 68. Hsieh T.S., Cattoglio C., Slobodyanyuk E., Hansen A.S., Rando O.J., Tjian R., Darzacq X.. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol. Cell. 2020; 78:539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nair L., Chung H., Basu U.. Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nature reviews. Mol. Cell Biol. 2020; 21:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A.. The ground state of embryonic stem cell self-renewal. Nature. 2008; 453:519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Huang G., Ye S., Zhou X., Liu D., Ying Q.L.. Molecular basis of embryonic stem cell self-renewal: from signaling pathways to pluripotency network. Cell. Mol. Life Sci.: CMLS. 2015; 72:1741–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. De Felice M., Falco G., Wang Y., Na Q., Li X., Tee W.W., Wu B.. Retinoic acid induces NELFA-mediated 2C-like state of mouse embryonic stem cells associates with epigenetic modifications and metabolic processes in chemically defined media. Cell Death Differ. 2021; 54:e13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tagliaferri D., Mazzone P., Noviello T.M.R., Addeo M., Angrisano T., Del Vecchio L., Visconte F., Ruggieri V., Russi S., Caivano A.et al.. Retinoic acid induces embryonic stem cells (ESCs) Transition to 2 Cell-Like state through a coordinated expression of dux and duxbl1. Front. Cell Dev. Biol. 2019; 7:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Iturbide A., Ruiz Tejeda Segura M.L., Noll C., Schorpp K., Rothenaigner I., Ruiz-Morales E.R., Lubatti G., Agami A., Hadian K., Scialdone A.et al.. Retinoic acid signaling is critical during the totipotency window in early mammalian development. Nat. Struct. Mol. Biol. 2021; 28:521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang Y., Na Q., Li X., Tee W.-W., Wu B., Bao S.. Retinoic acid induces NELFA-mediated 2C-like state of mouse embryonic stem cells associates with epigenetic modifications and metabolic processes in chemically defined media. Cell Prolif. 2021; 54:e13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tsankov A.M., Gu H., Akopian V., Ziller M.J., Donaghey J., Amit I., Gnirke A., Meissner A.. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015; 518:344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ji Z., Li Y., Liu S.X., Sharrocks A.D.. The forkhead transcription factor FOXK2 premarks lineage-specific genes in human embryonic stem cells for activation during differentiation. Nucleic Acids Res. 2021; 49:1345–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dupé V., Davenne M., Brocard J., Dollé P., Mark M., Dierich A., Chambon P., Rijli F.M.. In vivo functional analysis of the Hoxa-1 3′ retinoic acid response element (3′RARE). Development. 1997; 124:399–410. [DOI] [PubMed] [Google Scholar]
- 79. Langston A.W., Thompson J.R., Gudas L.J.. Retinoic acid-responsive enhancers located 3′ of the Hox A and Hox B homeobox gene clusters. Functional analysis. J. Biol. Chem. 1997; 272:2167–2175. [DOI] [PubMed] [Google Scholar]
- 80. Zheng J., Su G., Wang W., Zhao X., Liu M., Bi J., Zhao Z., Shi J., Lu W., Zhang L.. Two enhancers regulate HoxB genes expression during retinoic acid-induced early embryonic stem cells differentiation through long-range chromatin interactions. Stem Cells Dev. 2021; 30:683–695. [DOI] [PubMed] [Google Scholar]
- 81. Chen L., Cao W., Aita R., Aldea D., Flores J., Gao N., Bonder E.M., Ellison C.E., Verzi M.P.. Three-dimensional interactions between enhancers and promoters during intestinal differentiation depend upon HNF4. Cell Rep. 2021; 34:108679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rowley M.J., Nichols M.H., Lyu X., Ando-Kuri M., Rivera I.S.M., Hermetz K., Wang P., Ruan Y., Corces V.G.. Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell. 2017; 67:837–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cai W., Huang J., Zhu Q., Li B.E., Seruggia D., Zhou P., Nguyen M., Fujiwara Y., Xie H., Yang Z.et al.. Enhancer dependence of cell-type–specific gene expression increases with developmental age. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:21450–21458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Maamar H., Cabili M.N., Rinn J., Raj A.. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 2013; 27:1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lin N., Dang J., Rana T.M.. Haunting the HOXA locus: two faces of lncRNA regulation. Cell stem cell. 2015; 16:449–450. [DOI] [PubMed] [Google Scholar]
- 86. Li M.A., Amaral P.P., Cheung P., Bergmann J.H., Kinoshita M., Kalkan T., Ralser M., Robson S., von Meyenn F., Paramor M.et al.. A lncRNA fine tunes the dynamics of a cell state transition involving Lin28, let-7 and de novo DNA methylation. eLife. 2017; 6:e23468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang Y., Li T., Preissl S., Amaral M.L., Grinstein J.D., Farah E.N., Destici E., Qiu Y., Hu R., Lee A.Y.et al.. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat. Genet. 2019; 51:1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J.. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011; 470:279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data produced in this study for the DNA-seq and RNA-seq data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE169058.