Abstract
The Repeat Expansion Diseases, a large group of human diseases that includes the fragile X-related disorders (FXDs) and Huntington's disease (HD), all result from expansion of a disease-specific microsatellite via a mechanism that is not fully understood. We have previously shown that mismatch repair (MMR) proteins are required for expansion in a mouse model of the FXDs, but that the FANCD2 and FANCI associated nuclease 1 (FAN1), a component of the Fanconi anemia (FA) DNA repair pathway, is protective. FAN1’s nuclease activity has been reported to be dispensable for protection against expansion in an HD cell model. However, we show here that in a FXD mouse model a point mutation in the nuclease domain of FAN1 has the same effect on expansion as a null mutation. Furthermore, we show that FAN1 and another nuclease, EXO1, have an additive effect in protecting against MSH3-dependent expansions. Lastly, we show that the loss of FANCD2, a vital component of the Fanconi anemia DNA repair pathway, has no effect on expansions. Thus, FAN1 protects against MSH3-dependent expansions without diverting the expansion intermediates into the canonical FA pathway and this protection depends on FAN1 having an intact nuclease domain.
INTRODUCTION
More than 40 human genetic disorders are caused by an expansion of a disease-specific tandem array or microsatellite (1). This includes the FMR1 disorders, aka the fragile X-related disorders (FXDs), that result from expansion of a CGG-repeat tract in the FMR1 gene (2). Emerging evidence from genome-wide association studies (GWAS) (3–6) and mouse models of these diseases (reviewed in (7,8)), suggests that expansion in these diseases may share many common features. We have previously shown using a knockin mouse model of the FXDs that the mismatch repair (MMR) proteins MutSβ (9) and all three mammalian MutL complexes, MutLα, MutLβ and MutLγ (10), are required for expansion in this model. Evidence from GWAS/transcriptome-wide association studies (TWAS) in cohorts of patients with other Repeat Expansion Diseases supports a role for some of these proteins in modulating somatic expansion (3–6). This suggests that the FXD mouse model may be useful for understanding the mutational mechanism responsible for these diseases.
In contrast to the role of the MMR proteins in promoting expansion, we showed previously that the FANCD2 and FANCI associated nuclease 1 (FAN1) protects against expansion in the FXD mouse model (11). FAN1 is also protective in mouse and human cell models of Huntington's disease (HD) (12,13). Although no comparable study of the FXDs has been reported to date, a protective role for FAN1 is consistent with data from GWAS/TWAS of multiple polyglutamine expansion diseases, including HD (3,4,6) and see (14) for a recent review. A similar role for FAN1 was not seen in a small study of another Repeat Expansion Disease, X-linked dystonia-parkinsonism (XDP). However, given the small sample size and the homogeneous nature of the XDP patient population who all share the same founder mutation and originate from a single island in the Philippines (15), this leaves open the possibility that FAN1 is a modifier of expansion risk in many, if not all, of the Repeat Expansion Diseases.
FAN1 was initially identified as a 5′ to 3′ exonuclease and 5′ flap endonuclease that is an important component of the Fanconi anemia (FA) pathway of DNA repair (16–22). Given the importance of the MMR pathway in promoting expansion, how FAN1 acts to protect against repeat expansions is unclear, particularly since its nuclease activity was reported to be dispensable for this effect in a U2OS osteosarcoma cell model of HD expansions (13) and we have previously demonstrated both a nuclease-dependent and nuclease-independent role for EXO1 nuclease in its protection against repeat expansion (23). To help address the role of FAN1 in protecting against repeat expansion we generated FXD mice with a point mutation in the nuclease domain of FAN1, along with mice that have both this mutation in FAN1 and null mutations in other MMR proteins. We also examined the effect of a null mutation in Fancd2, which encodes a protein essential for the recruitment of downstream participants in the FA pathway, including FAN1, in response to DNA damage (24). FANCD2 is of particular interest in the context of the FXDs since, in addition to its role in repair of interstrand DNA crosslinks (ICLs), it is also important for the repair of stalled replication forks (16,18) which have been suggested to play a role in repeat expansion (25). However, we show here that FAN1 acts to prevent MSH3-dependent expansions, that it requires an intact nuclease domain to do so and that this protection is independent of the critical FA protein, FANCD2.
MATERIALS AND METHODS
Reagents and services
All reagents were from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. Primers were from Life Technologies (Grand Island, NY). Single-stranded donor DNA, crRNA, tracrRNA and HiFi Cas9 nuclease V3 enzyme were from Integrated DNA Technologies (Coralville, IA). Capillary electrophoresis of fluorescently labeled PCR products was carried out by the Roy J Carver Biotechnology Center, University of Illinois (Urbana, IL). DNA sequencing was carried out by Psomagen (Rockville, MD).
Mouse generation, breeding and maintenance
Fan1 D963A mice were generated using CRISPR-Cas9 ribonucleoprotein (RNP)-mediated homology-directed recombination (HDR) in C57BL/6J zygotes. Briefly, a single guide RNA (gRNA) (5′-GAATTCCACACCACCAAGTC-3′) was designed to target the Fan1 codon D963 in exon 13. The off-target effects of this gRNA were evaluated by three different web based software tools: Alt-R Custom Cas9 crRNA Design Tool (https://www.idtdna.com), CHOPCHOP (version 3) (http://chopchop.cbu.uib.no/) (26), and Cas-OFFinder (http://www.rgenome.net/cas-offinder/) (27). No off-targets with fewer than 3 mismatches were identified by any of the algorithms. We designed a single-stranded donor DNA (5′- GTCCTCAAGGTGGCTAACTCTGCTCTTGTATCGTGCTTAGGATCTTGTCTCCTGCCTCGGGGGTCCTGTCCTCAGTGGTGTGTGCAGGCGCCTGGCTGCTGACTTTCGGCACTGCCGAGGGGGCCTCCCAGCATTAGTCGTGTGGAACTCTCAGAGCCACCATTGCAAGGTCAGTAGAGAACACCATTTGTAATTTCAGC-3′) carrying the desired mutations shown underlined. As illustrated in Supplementary Figure S1A, successful HDR results in the replacement of the adjacent A and C residues in the codon for D963 with C and A, thus converting the aspartate codon (GAC) into one that specifies alanine (GCA). Three additional single base changes would also be introduced downstream. These changes are silent and were introduced to avoid CRISPR recutting of the modified allele. C57BL/6J zygotes were collected from the oviducts of hormonally stimulated females at embryonic day 0.5 (E0.5). Equal amounts of crRNA (200 μM) and tracrRNA were heated at 95°C for 5 minutes and slowly cooled to room temperature in Duplex Buffer (30 mM HEPES, pH 7.5; 100 mM Potassium Acetate). The crRNA:tracrRNA (200 ng/μL), Cas9 protein (250 ng/μL), and DNA oligo donor (250 ng/μl) were mixed in Opti-MEM medium (Thermo Fisher Scientific, Waltham, MA) and electroporated into zygotes using a NEPA electroporator (NepaGene, Chiba, Japan) as per the manufacturer's recommendations. Injected zygotes were cultured in KSOM medium at 37°C in 5% CO2 until the two-cell embryo stage and transferred into the oviducts of pseudo-pregnant female mice. The resulting offspring were genotyped by PCR as described below and sequenced to identify founders carrying the Fan1 D963A mutation (Supplementary Figure S1). We also sequenced across the exon 12/13 and exon 13/14 junctions of the cDNA generated from this allele to ensure that the mutations introduced did not affect splicing (Supplementary Figure S2). While there were no significant predicted off-target effects of the gRNAs used, we still took the precaution of using 3 different embryos of these mice to establish 3 independent lineages. These mice were then crossed to FXD mice we have described previously (28) to generate animals heterozygous for the point mutation. These mice were then crossed again with new FXD mice to generate mice homozygous for the FXD allele. Littermates were then crossed twice more to obtain enough FXD mice homozygous for the mutant Fan1 allele. We used Fan1D/D littermates to further control for any potential off-target effects of the gene editing procedure. However, they showed expansions that were indistinguishable from unedited FXD mice that had been backcrossed to C57BL/6J mice for more than 20 generations.
The generation of the FXD mice that were null for Fan1, Exo1 or Msh3 were described previously (9,11,23,28). Fancd2 mice (29) were provided by Andre Nussenzweig (NIH, Bethesda, MD). All mice were on a C57BL/6J background. Mice were maintained in a manner consistent with the Guide for the Care and Use of Laboratory Animals (NIH publication no. 85–23, revised 1996) and in accordance with the guidelines of the NIDDK Animal Care and Use Committee, who approved this research (ASP-K021-LMCB-15).
DNA isolation
DNA from mouse tails at 3-week-old was extracted for genotyping using the KAPA Mouse Genotyping Kit (KAPA Biosystems, Wilmington, MA). DNA from different organs of 6-month-old mice was isolated using a Maxwell®16 Mouse Tail DNA Purification kit (Promega, Madison, WI) according to the manufacturer's instructions. A 5 cm region of the jejunum starting 10 cm downstream of stomach was used as the small intestine sample and the DNA was isolated from this segment as described above for intact organs.
Genotyping and analysis of repeat number
Fan1 D963A genotyping was carried out with KAPA mouse genotyping kit (KAPA Biosystems) according to the manufacturer’s instructions with primers Fan1DA-AF (5′- CCAGCATTAGTCGTGTGGAAC-3′)/Fan1DA-AR (5′-CTCACACCAAAGGTATGATCCA-3′) primer pair to detect the mutated Fan1 A (GCA) allele and the primers Fan1DA-DF (5′-TTGTACTGGTGTCCTCTGATGG -3′)/ Fan1DA-DR (5′-TGAGAATTCCACACCACCAAGT -3′) to detect the WT Fan1 D (GAC) allele. The underlined primer bases are allele specific. Fan1, Exo1, Msh3 genotyping was carried out as previously described (9,11,23). Fancd2 genotyping was carried out as described in Parmar et al. (29). Fmr1 PM allele genotyping and repeat size analysis was carried out as described previously (23). The PCR products were resolved by capillary electrophoresis on an ABI Genetic Analyzer. The resultant fsa file was then displayed using a previously described custom R script (23) that is available on request. We quantified somatic expansions using the average number of repeats added to the original inherited allele as assessed from the tail DNA taken at weaning (23). For expansions in different brain regions we also used the ‘Expansion Index’ as a metric. This metric is better suited to comparisons of regions where at least one region shows expansions limited to a small number of cells in the population. Statistical analyses of the differences in repeat numbers in different tissues of mice of different genotypes were performed using GraphPad Prism 8.4. Statistical significance was assessed using the two-tailed unpaired t-test for comparisons involving just two genotypes with the Holm-Sidak correction for multiple comparisons. For analysis of more than two groups we used either a mixed-effects model when not all organs were available for all animals, or a repeated measures (RM) two-way ANOVA, both with the Geisser-Greenhouse correction and Tukey ’s post hoc correction for multiple comparisons.
RESULTS
FAN1 with a D963A point mutation in its nuclease domain is unable to protect against expansion
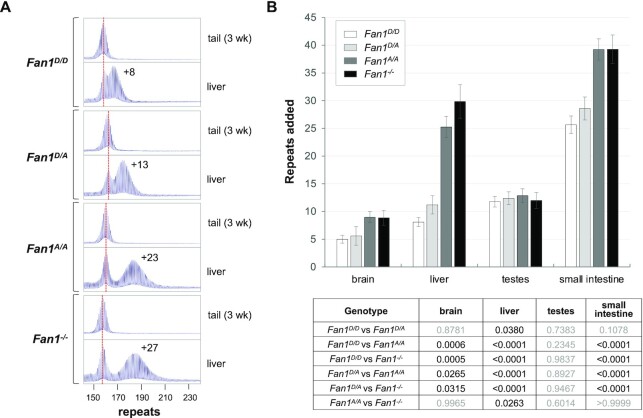
The C-terminal region of FAN1 contains a PDXn(D/E)XK nuclease motif that is highly conserved in organisms from bacteria to man (Supplementary Figure S3). The first aspartate residue in that motif corresponds to D960 in the human protein and D963 in the mouse. A D-to-A mutation eliminates FAN1’s nuclease activity, without affecting protein stability, folding or DNA binding (17,19,21,30–32). To test the role of the nuclease domain in FAN1’s protection against repeat expansion, we made a D963A mutation in the mouse FAN1 using a CRISPR-Cas9 HDR approach in embryos. The mutation was verified by PCR and sequencing of both the DNA and the mature mRNA as shown in Supplementary Figures S1 and S2. Mice carrying the D963A mutation were then backcrossed for 2–3 generations to generate size matched FXD mice carrying either the WT (D963) or mutant (A963) Fan1 alleles. We then compared the number of repeats added to the Fmr1 allele in different tissues of 6-month-old male Fan1D/D, Fan1D/A, Fan1A/A and Fan1-/- mice, all with inherited Fmr1 alleles ranging from 157–163 repeats. As can be seen in Figure 1 and Supplementary Figure S4, Fan1-/- mice showed significantly more expansions than the Fan1D/D (WT) mice in brain, liver and small intestine. As we previously reported, there was no significant effect of the loss of FAN1 on expansion in testes (11). The D963A mutant mice also showed significant differences with WT mice in brain, liver and small intestine, whereas there are no significant differences between the null and D963A mutant mice in the extent of expansion in most tissues. Fan1 mice heterozygous for the point mutation also showed significantly more expansions relative to WT animals in liver and small intestine but not in brain (Figure 1). Thus, even the loss of a single allele with an intact nuclease domain has a significant effect in some tissues.
Figure 1.
A point mutation in the nuclease domain of FAN1 eliminates its protective effect against expansion in many organs. (A) Typical repeat PCR profiles from tail taken at weaning (3-week tail) and liver of 6-month old Fan1D/D, Fan1D/A, Fan1A/A and Fan1-/-mice. The dotted line represents the size of the original inherited allele as ascertained from the tail DNA taken at weaning (3-week tail). The numbers associated with some of the traces indicates the number of repeats added during the lifetime of the mouse. (B) Comparison of the number of repeats added to the original allele in different organs of 6-month old Fan1D/D, Fan1D/A, Fan1A/A and Fan1-/- mice with ∼161 repeats in the original allele. The brain data represents the average of five Fan1D/D, five Fan1D/A, five Fan1A/A and seven Fan1-/- male mice with 157–163 repeats, the data from other organs represents the average of nine Fan1D/D, five Fan1D/A, eight Fan1A/A and seven Fan1-/- male mice in the same repeat range. The error bars indicate the standard deviations of the mean. The significance of the effects were assessed using a mixed-effects model with correction for multiple testing as described in the Materials and Methods. The adjusted P values of genotype effect are listed in the table below.
While the extent of expansion in the brain and small intestine of homozygous D963A and null mice were indistinguishable, ∼21% fewer repeats were added in the liver of the D963A mutant mice when compared to null mice (P = 0.026). Moreover, when five different pairs of mice matched exactly for repeat size were examined, in each case the null mice showed more expansion than the D963A mice (Supplementary Figure S5). It is possible that the decrease in the number of repeats added reflects a small nuclease-independent protective effect in this organ. Since FAN1 interacts with many MMR proteins (21,33), it is possible that nuclease-deficient FAN1 might, if sufficiently abundant, indirectly protect against expansions by reducing the availability of these proteins to participate in the expansion process. Such a process may explain the fact that when overexpressed, the human D960A mutant protein is able to reduce expansions apparently as effectively as the wildtype protein (13).
The D963A mutation did not affect the steady state levels of Fan1 mRNA (Supplementary Figure S6), but the low levels of the endogenous protein and the lack of good mouse antibodies did not allow us to evaluate the effect of the mutation on protein stability directly ourselves. However, alanine substitutions are known to eliminate the side chain beyond the β carbon without altering main-chain conformation or introducing steric or electrostatic effects (34). Furthermore, the last 645 amino acids of murine FAN1 that includes the nuclease motif has 84% amino acid identity and 92% amino acid similarity with human FAN1 (Supplementary Figure S3C) and despite the evolutionary distance, even the structures of the human and P. aeruginosa proteins are remarkably similar with the root mean square deviation between the two structures being 3.1 Å for 456 Cα atoms (35,36). The similarities in the X-ray crystal structures of human FAN1 and FAN1 with the D960A mutation (32,35,37), together with the high sequence identity of the C-terminal end of the human and mouse proteins, suggest that the mouse and human FAN1 and the D963A/D960A mutated proteins are likely to have similar conformations and stability. The fact that a mutation in the murine Fan1 gene that results in the replacement of the 41 terminal amino acids of the protein with 7 intron encoded amino acids does not affect protein stability (18), lends weight to that idea. Thus it is reasonable to think that the effect of the D963A mutation on repeat expansion reflects the importance of the FAN1 nuclease domain for its role in protecting against expansion.
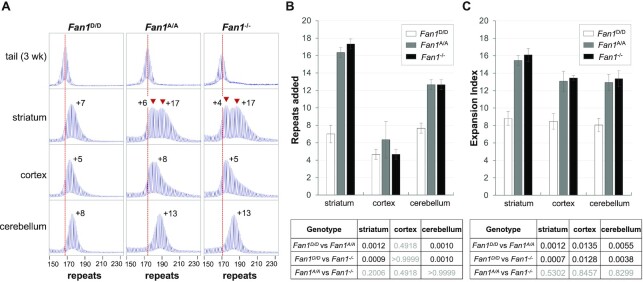
To further understand repeat expansion dynamics in the brain where the consequences of repeat expansion are most deleterious, we compared the number of repeats added in different brain regions of Fan1D/D, Fan1A/A and Fan1-/- mice. In many Repeat Expansion Diseases and mouse models of these diseases, some of the largest expansions in the central nervous system (CNS) are found in the striatum (38–40). It is also a major source of pathology in some of these diseases (41–45). Notably then, it is also the striatum that shows the largest effect of both the point mutation in FAN1 and the null mutation, with a >2-fold increase in the extent of expansion seen in mutant mice relative to Fan1D/D animals (Figure 2). Notably, the PCR profile of both the homozygous mutant mice shows a biphasic distribution of allele sizes with the largest fraction of alleles showing a gain of an average of 16 repeats. A smaller fraction of alleles shows a smaller increase in the average allele size, similar to that seen in Fan1D/D mice. This would be consistent with the majority of striatal cells showing a protective effect of FAN1. Single cell transcriptomics studies suggest that neurons, astrocytes and oligodendrocytes, that together represent 70% of cells in the striatum, express much higher levels of Fan1 mRNA than other striatal cells including the immune and vascular cells (46). It may be that the cells that are insensitive to the loss of FAN1 represent the latter cell types. A smaller, but nonetheless still significant effect of the loss of FAN1 was seen in the cerebellum in both sets of Fan1 mutant mice. It should be noted that in mice of all three genotypes most, if not all, alleles in this brain region have expanded (Figure 2A). Since post-mitotic neurons make up ∼80% of cells in the cerebellum (47), this would be consistent with the idea that expansions can occur independently of chromosomal replication. In the cortex no significant change in the extent of expansion was seen using the ‘repeats added’ metric (Figure 2B). However, there were significant changes seen using the Expansion Index metric (Figure 2C), a metric that is useful in regions like the cortex where only a small subset of cells in the population are affected by the loss of FAN1. This does not reflect reduced levels of FAN1 in the cortex as a whole since RT-PCR (Supplementary Figure S7) and transcriptomics data (48) suggest that, at least at the transcriptional level, the average amount of FAN1 expression in these regions is similar. Should the transcript levels accurately reflect protein levels, the small effect of FAN1 in the cortex may indicate the importance of another nuclease in protecting against expansion in many cortical cell types. This may have implications for efforts to reduce somatic expansion in this part of the brain in diseases with significant cortical dysfunction. In mice heterozygous for the point mutation only the striatum had significantly more expansions relative to WT animals (Supplementary Figure S8).
Figure 2.
Comparison of the effect of the D963A and null mutations on the extent of repeat expansion in different brain regions of FXD mice. (A) Typical repeat PCR profiles from the tail taken at 3 weeks of age and different brain regions of 6-month old Fan1D/D, Fan1A/A and Fan1-/- mice matched for initial repeat number. The dotted line represents the size of the original inherited allele as ascertained from the tail DNA taken at weaning. The peaks associated with the two different expansion classes are indicated by the arrowheads. The numbers associated with the tissue samples indicates the number of repeats added during the lifetime of the mouse. (B) Comparison of the number of repeats added in different brain regions of 6-month old Fan1D/D, Fan1A/A and Fan1-/- mice with 168–174 repeats in the original allele. (C) Comparison of the Expansion Index of different brain regions of the mice shown in panel B. In both panels the data represent the average of data from 3 animals of each genotype. The error bars indicate the standard deviations of the mean. Significance was assessed using a RM two-way ANOVA (genotype and tissue as variables) with correction for multiple testing as described in the Materials and Methods. The adjusted P values of genotype effect are listed in the table below.
FAN1 protects against MutSβ-dependent expansions
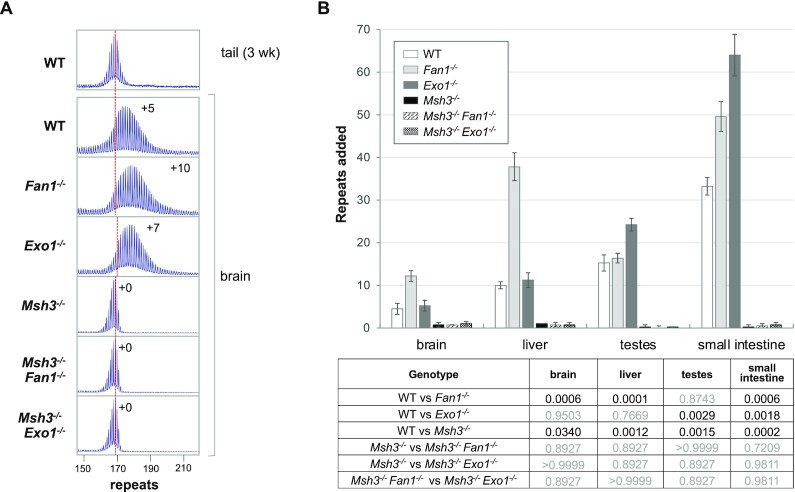
To assess whether FAN1 protects against MutSβ dependent expansions or acts to protect against expansions generated by a mechanism normally masked by the action of FAN1, we crossed our FXD Fan1-/- mice to mice lacking MSH3, the binding partner of MSH2 in the MutSβ heterodimer, a MMR complex essential for expansion. As can be seen from Figure 3 and Supplementary Figure S9, no expansions were seen for the either the Msh3-/- mice or the Msh3-/-Fan1-/- double mutant mice in any of the organs examined, including brain. Thus, FAN1 is acting to prevent MMR-dependent repeat expansion in the FXD mouse model. This is consistent with a recent report from an HD mouse model which showed that FAN1 had no effect in a genetic background deficient in another MMR factor required for expansion, MLH1 (12). This extends the parallels between FXD mouse model and mouse models of other Repeat Expansion Diseases.
Figure 3.
FAN1 and EXO1 protect against MSH3-dependent expansions. (A) Typical repeat PCR profiles from the brains of 6-month old WT, Fan1-/-, Exo1-/-, Msh3-/- as well as Msh3-/-Fan1-/-and Msh3-/-Exo1-/- double knockout mice matched for initial repeat number. The dotted line represents the size of the original inherited allele as ascertained from the tail DNA taken at weaning. The numbers alongside some of the traces indicates the number of repeats added during the lifetime of the mouse. (B) Comparison of the number of repeats added in different organs of 6-month old WT, Fan1-/-, Exo1-/-, Msh3-/- and Msh3-/-Fan1-/-and Msh3-/-Exo1-/- double knockout mice with with 168–174 repeats in the original allele. The data represent the average of five Fan1-/- males and four males of each of the other genotypes. The error bars indicate the standard deviations of the mean. Significance was assessed using a RM two-way ANOVA (genotype and tissue as variables) with correction for multiple testing as described in the Materials and Methods. The adjusted P values of genotype effect are listed in the table.
We had previously shown that EXO1, another enzyme with 5′-3′ exonucleolytic activity, also protected against repeat expansion (23). To evaluate whether it was doing so by acting in the same pathway, we generated Msh3-/-Exo1-/- FXD mice and examined instability in these animals over the same time period. As can be seen in Figure 3 and Supplementary Figure S9, the Msh3-/-Exo1-/- double mutant animals also showed no expansions. Thus both nucleases act to prevent MMR-dependent expansions.
FAN1 and EXO1 double mutant mice have more expansions in small intestine than either single mutant
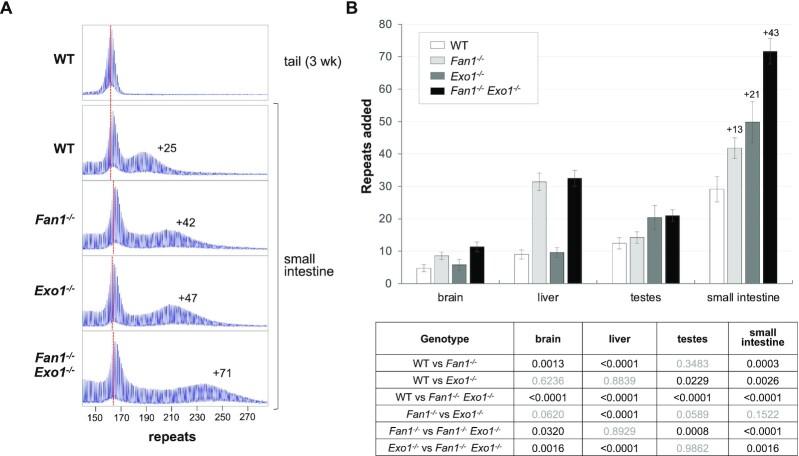
We then generated FXD mice with null mutations in both Fan1 and Exo1 and compared them to age and size matched WT mice and to mice nullizygous for either one of these genes. Note that in very expansion prone organs like the liver of Fan1-/- mice, the pool of expanded alleles becomes more heterogenous over time. This results in a decrease in the peak height of the expanding alleles seen in the PCR profile, although the total number of expanded alleles does not decrease. In addition, Exo1-/- mice lack mature gametes due to apoptosis of spermatocytes during meiosis I (49). Since expansion in the testes is limited to the spermatogonial stem cell pool (50), the effect of the loss of EXO1 can be seen in the larger number of repeats added to the expanding allele even though the population of cells containing such alleles is smaller than in WT or Fan1-/- testes (Supplementary Figures S9 and S10).
As can be seen in Figure 4 and Supplementary Figure S10, loss of EXO1 only has a significant effect in testes and small intestine, as we showed previously (23), while an effect of the loss of FAN1 is only seen in brain, liver and small intestine (11). In the small intestine, where both enzymes are protective, mice nullizygous for Exo1 added an average of 21 more repeats than WT mice by 6 months of age, while Fan1 null mice added 13 more. The double mutant mice added 43 more repeats, slightly more than would be expected if the effect of EXO1 and FAN1 were strictly additive. This may reflect the tendency of larger repeat tracts to expand more than smaller ones rather than any synergistic interaction of these enzymes. Whatever the explanation, it suggests that EXO1 and FAN1 act redundantly to protect against repeat expansion. It also illustrates just how frequent the underlying trigger for expansion can be in this tissue, with ∼72 repeats being added to the original inherited allele in 6 months when the protective effect of EXO1 and FAN1 is absent. This corresponds to an expansion of ∼3 repeats a week.
Figure 4.
Loss of FAN1 and EXO1 has an additive effect on expansion. (A) Typical repeat PCR profiles from small intestine of 6-month old WT, Fan1-/-, Exo1-/- and Fan1-/-Exo1-/- double mutant mice matched for initial repeat number. The dotted line represents the size of the original inherited allele as ascertained from the tail DNA taken at weaning (3-week tail). The numbers associated with the samples from the small intestine indicates the number of repeats added to alleles in that organ during the lifetime of the mouse. (B) Comparison of the number of repeats added in different organs of age and size matched WT, Fan1-/-, Exo1-/- and double mutant Fan1-/-Exo1-/- mice. The data represent the average of nine WT, five Exo1-/-, five Fan1-/- and six double mutant Fan1-/-Exo1-/-male mice with 161–168 repeats. The numbers above the bars in the small intestine indicate the average number of repeats added relative to wildtype for all three of the mutant genotypes. The error bars indicate the standard deviations of the mean. Significance was assessed using a RM two-way ANOVA (genotype and tissue as variables) with correction for multiple testing as described in the Materials and Methods. The adjusted P values of genotype effect are listed in the table below.
Exo1 mRNA levels are very high in testes, ∼36 times higher than in small intestine, ∼72 times higher than in brain and ∼135 times higher than in liver (Supplementary Figure S11). In contrast, Fan1 mRNA is most abundant in brain, with ∼19 times higher levels than Exo1 (Supplementary Figure S11). This may explain EXO1’s importance in protecting against expansion in testes and FAN1’s importance in brain. However, mRNA levels do not always correlate well with steady state levels of proteins, and thus a definitive explanation of the organ-specificity will require improved techniques for accurately quantifying FAN1 and EXO1 proteins.
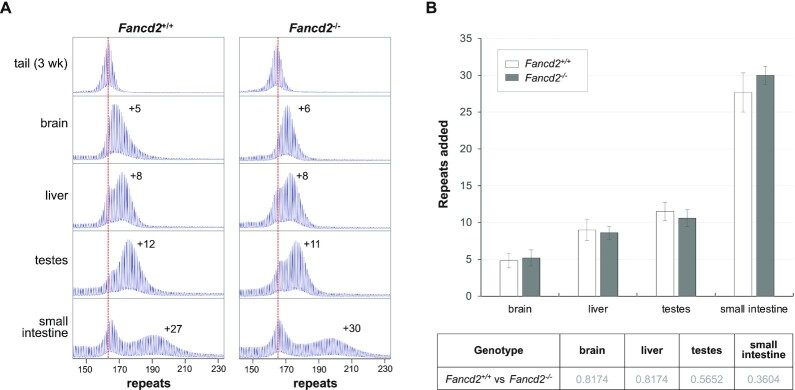
A null mutation in Fancd2 has no effect on expansions
FANCD2 is a central key player in the FA pathway (17,20,51), a major pathway in which FAN1 is active (22). As can be seen in Figure 5, loss of FANCD2 has no effect on the extent of expansion in any of the tissues tested including brain, liver and the small intestine, organs in which the protective effect of FAN1 is most apparent. Thus, FAN1 protects against repeat expansion independently of FANCD2.
Figure 5.
Loss of FANCD2 has no effect on repeat expansion. (A) Typical repeat PCR profiles from tail DNA taken at weaning (3-week tail) and different organs of 6-month old WT and Fancd2-/- mice with ∼165 repeats. The dotted line represents the size of the original inherited allele. The numbers associated with the samples from the animals at 6-months of age indicates the number of repeats added in the indicated organ during the lifetime of the mouse. (B) Comparison of the number of repeats added in different organs of 6-month old WT and Fancd2-/- mice with ∼165 repeats. The data represent the average of six WT and five Fancd2-/- male mice with 162–167 repeats. The error bars indicate the standard deviations of the mean. Significance was assessed using two-tailed unpaired t tests with correction for multiple testing as described in the Materials and Methods. No significant difference was found between the two genotypes.
DISCUSSION
FAN1 is an important modifier of expansion risk in many Repeat Expansion Diseases, where deleterious SNPs have shown to be associated with significantly earlier age of onset and SNPs associated with increased FAN1 expression are associated with a later age at onset (3,4,6). As such, there is intense interest in understanding how this protein protects against expansion and whether this knowledge can be leveraged in some way to delay disease onset particularly in those severe neurodegenerative disorders like HD. A previous study of the effect of the nuclease activity of this protein, suggested that FAN1’s protective effect did not depend on its nuclease activity, since expression of a D960A mutant protein showed the same level of protection against expansions in a cell model of HD expansion as did the WT protein (13).
However, we show here that in an FXD mouse model, the D963A mutation is indistinguishable from the Fan1 null mutation with respect to repeat expansions (Figure 1). This observation is consistent with a requirement for an intact nuclease domain for most of FAN1’s protective effect against expansions. In liver there may be a small, nuclease-independent protective role of the protein, perhaps related to FAN1’s ability to interact with MMR proteins required for expansion (21,33). However, since a nuclease-independent effect is not seen in other organs, including brain where the effects of repeat expansion are most apparent, it is unlikely to be disease-relevant. While our manuscript was in revision a role for the FAN1 nuclease was reported in a human U2OS model system of CAG repeat expansions (52) and in HD induced pluripotent stem cells (Preprint, bioRxiv 439716). This further extends the similarities of the mechanisms responsible for instability of CGG and CAG repeats. FAN1 has both exonucleolytic and endonucleolytic activity (20) and is active on hairpins formed by the HD and Fragile X repeats in vitro (Preprint, bioRxiv 439995). However, since the D963A mutation affects both exo and endonuclease activities and given that FAN1 acts on a wide variety of substrates (Preprint, bioRxiv 439995), further work is required to determine which activity is important for FAN1’s protective effect.
The absence of expansions in Msh3-/-Fan1-/- and Msh3-/-Exo1-/- double mutant mice demonstrate that FAN1 and EXO1 both act to block MSH3-dependent expansions (Figure 3). The lack of an effect of a null mutation in Fancd2 (Figure 5) shows that FAN1 is not acting to channel the expansion intermediates into a FANCD2-dependent FA pathway, but rather is acting independently of FANCD2 to prevent expansions. FAN1 has recently been implicated in MMR (53) and binding to MLH1 is required for the MMR activity of both FAN1 and EXO1 (53,54). In the case of FAN1, this binding is mediated by a SPYF motif located 126 amino acids from the N-terminus of the protein (52,55). Recent experiments in U2OS cells (52) and in vitro (55) show that FAN1’s interaction with MLH1 is also essential for its protective effect against CAG repeat expansions. It has been suggested that FAN1’s competition with other MMR proteins for binding to MLH1 plays an important role in its protection against expansion (52,53). However, given that the D963A mutation has the same effect on expansion as the null allele despite the presence of an intact MLH1-binding domain, the primary way that FAN1 protects against repeat expansion in the FXD mouse model may be the same way that EXO1 protects against mismatches during normal MMR i.e., via recruitment to a MutL-generated nick with the subsequent nucleolytic removal of the nicked strand (56).
Supplementary Material
ACKNOWLEDGEMENTS
The authors want to thank Yangu Zhao from Mammalian Developmental Biology Section, NIDDK for help with CRISPR design and members of the Usdin laboratory for the careful reading of the manuscript and helpful comments. We would like to acknowledge all the hard work done by the staff who take care of our mice and without whom this work would not have been possible.
Contributor Information
Xiaonan Zhao, Laboratory of Cell and Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA.
Huiyan Lu, Laboratory Animal Sciences section, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA.
Karen Usdin, Laboratory of Cell and Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases [DK057808 to K.U.]. Funding for open access charge: Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases [DK057808 to K.U.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Paulson H. Repeat expansion diseases. Handb. Clin. Neurol. 2018; 147:105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lozano R., Rosero C.A., Hagerman R.J.. Fragile X spectrum disorders. Intractable Rare Dis. Res. 2014; 3:134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Genetic Modifiers of Huntington's Disease Consortium CAG repeat not polyglutamine length determines timing of Huntington's disease odnset. Cell. 2019; 178:887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bettencourt C., Hensman-Moss D., Flower M., Wiethoff S., Brice A., Goizet C., Stevanin G., Koutsis G., Karadima G., Panas M.et al.. DNA repair pathways underlie a common genetic mechanism modulating onset in polyglutamine diseases. Ann. Neurol. 2016; 79:983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Flower M., Lomeikaite V., Ciosi M., Cumming S., Morales F., Lo K., Hensman Moss D., Jones L., Holmans P., Investigators T.-H.et al.. MSH3 modifies somatic instability and disease severity in Huntington's and myotonic dystrophy type 1. Brain. 2019; 142:1876–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim K.H., Hong E.P., Shin J.W., Chao M.J., Loupe J., Gillis T., Mysore J.S., Holmans P., Jones L., Orth M.et al.. Genetic and functional analyses point to FAN1 as the source of disease modifier effects. Am. J. Hum. Genet. 2020; 107:96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao X., Kumari D., Miller C.J., Kim G.Y., Hayward B., Vitalo A.G., Pinto R.M., Usdin K.. Modifiers of somatic repeat instability in mouse models of Friedreich ataxia and the fragile X-related disorders: implications for the mechanism of somatic expansion in Huntington's disease. J. Huntingtons Dis. 2021; 10:149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wheeler V.C., Dion V.. Modifiers of CAG/CTG repeat Iistability: insights from mammalian models. J. Huntingtons Dis. 2021; 10:123–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao X.N., Kumari D., Gupta S., Wu D., Evanitsky M., Yang W., Usdin K.. Mutsbeta generates both expansions and contractions in a mouse model of the Fragile X-associated disorders. Hum. Mol. Genet. 2015; 24:7087–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller C.J., Kim G.Y., Zhao X., Usdin K.. All three mammalian MutL complexes are required for repeat expansion in a mouse cell model of the Fragile X-related disorders. PLos Genet. 2020; 16:e1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao X.N., Usdin K.. FAN1 protects against repeat expansions in a Fragile X mouse model. DNA Repair (Amst.). 2018; 69:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loupe J.M., Pinto R.M., Kim K.H., Gillis T., Mysore J.S., Andrew M.A., Kovalenko M., Murtha R., Seong I., Gusella J.F.et al.. Promotion of somatic CAG repeat expansion by Fan1 knock-out in Huntington's disease knock-in mice is blocked by Mlh1 knock-out. Hum. Mol. Genet. 2020; 29:3044–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goold R., Flower M., Moss D.H., Medway C., Wood-Kaczmar A., Andre R., Farshim P., Bates G.P., Holmans P., Jones L.et al.. FAN1 modifies Huntington's disease progression by stabilizing the expanded HTT CAG repeat. Hum. Mol. Genet. 2019; 28:650–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deshmukh A.L., Porro A., Mohiuddin M., Lanni S., Panigrahi G.B., Caron M.C., Masson J.Y., Sartori A.A., Pearson C.E.. FAN1, a DNA repair nuclease, as a modifier of repeat expansion disorders. J. Huntingtons Dis. 2021; 10:95–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Laabs B.H., Klein C., Pozojevic J., Domingo A., Bruggemann N., Grutz K., Rosales R.L., Jamora R.D., Saranza G., Diesta C.C.E.et al.. Identifying genetic modifiers of age-associated penetrance in X-linked dystonia-parkinsonism. Nat. Commun. 2021; 12:3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chaudhury I., Stroik D.R., Sobeck A.. FANCD2-controlled chromatin access of the Fanconi-associated nuclease FAN1 is crucial for the recovery of stalled replication forks. Mol. Cell. Biol. 2014; 34:3939–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kratz K., Schopf B., Kaden S., Sendoel A., Eberhard R., Lademann C., Cannavo E., Sartori A.A., Hengartner M.O., Jiricny J.. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010; 142:77–88. [DOI] [PubMed] [Google Scholar]
- 18. Lachaud C., Moreno A., Marchesi F., Toth R., Blow J.J., Rouse J.. Ubiquitinated Fancd2 recruits Fan1 to stalled replication forks to prevent genome instability. Science. 2016; 351:846–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu T., Ghosal G., Yuan J., Chen J., Huang J.. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010; 329:693–696. [DOI] [PubMed] [Google Scholar]
- 20. MacKay C., Declais A.C., Lundin C., Agostinho A., Deans A.J., MacArtney T.J., Hofmann K., Gartner A., West S.C., Helleday T.et al.. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010; 142:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smogorzewska A., Desetty R., Saito T.T., Schlabach M., Lach F.P., Sowa M.E., Clark A.B., Kunkel T.A., Harper J.W., Colaiacovo M.P.et al.. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell. 2010; 39:36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou W., Otto E.A., Cluckey A., Airik R., Hurd T.W., Chaki M., Diaz K., Lach F.P., Bennett G.R., Gee H.Y.et al.. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat. Genet. 2012; 44:910–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao X., Zhang Y., Wilkins K., Edelmann W., Usdin K.. MutLgamma promotes repeat expansion in a Fragile X mouse model while EXO1 is protective. PLoS Genet. 2018; 14:e1007719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tan W., Deans A.J.. The ubiquitination machinery of the Fanconi Anemia DNA repair pathway. Prog. Biophys. Mol. Biol. 2021; 163:5–13. [DOI] [PubMed] [Google Scholar]
- 25. Polleys E.J., House N.C.M., Freudenreich C.H.. Role of recombination and replication fork restart in repeat instability. DNA Repair (Amst.). 2017; 56:156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E.. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019; 47:W171–W174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bae S., Park J., Kim J.S.. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014; 30:1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Entezam A., Biacsi R., Orrison B., Saha T., Hoffman G.E., Grabczyk E., Nussbaum R.L., Usdin K.. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007; 395:125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parmar K., Kim J., Sykes S.M., Shimamura A., Stuckert P., Zhu K., Hamilton A., Deloach M.K., Kutok J.L., Akashi K.et al.. Hematopoietic stem cell defects in mice with deficiency of Fancd2 or Usp1. Stem Cells. 2010; 28:1186–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thongthip S., Bellani M., Gregg S.Q., Sridhar S., Conti B.A., Chen Y., Seidman M.M., Smogorzewska A.. Fan1 deficiency results in DNA interstrand cross-link repair defects, enhanced tissue karyomegaly, and organ dysfunction. Genes Dev. 2016; 30:645–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Porro A., Berti M., Pizzolato J., Bologna S., Kaden S., Saxer A., Ma Y., Nagasawa K., Sartori A.A., Jiricny J.. FAN1 interaction with ubiquitylated PCNA alleviates replication stress and preserves genomic integrity independently of BRCA2. Nat. Commun. 2017; 8:1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao Q., Xue X., Longerich S., Sung P., Xiong Y.. Structural insights into 5′ flap DNA unwinding and incision by the human FAN1 dimer. Nat. Commun. 2014; 5:5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cannavo E., Gerrits B., Marra G., Schlapbach R., Jiricny J.. Characterization of the interactome of the human MutL homologues MLH1, PMS1, and PMS2. J. Biol. Chem. 2007; 282:2976–2986. [DOI] [PubMed] [Google Scholar]
- 34. Cunningham B.C., Wells J.A.. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989; 244:1081–1085. [DOI] [PubMed] [Google Scholar]
- 35. Yan P.X., Huo Y.G., Jiang T.. Crystal structure of human Fanconi-associated nuclease 1. Protein Cell. 2015; 6:225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jin H., Roy U., Lee G., Scharer O.D., Cho Y.. Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease. J. Biol. Chem. 2018; 293:6482–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang R., Persky N.S., Yoo B., Ouerfelli O., Smogorzewska A., Elledge S.J., Pavletich N.P.. DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1. Science. 2014; 346:1127–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mouro Pinto R., Arning L., Giordano J.V., Razghandi P., Andrew M.A., Gillis T., Correia K., Mysore J.S., Grote Urtubey D.M., Parwez C.R.et al.. Patterns of CAG repeat instability in the central nervous system and periphery in Huntington's disease and in spinocerebellar ataxia type 1. Hum. Mol. Genet. 2020; 29:2551–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kennedy L., Evans E., Chen C.M., Craven L., Detloff P.J., Ennis M., Shelbourne P.F.. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum. Mol. Genet. 2003; 12:3359–3367. [DOI] [PubMed] [Google Scholar]
- 40. Shelbourne P.F., Keller-McGandy C., Bi W.L., Yoon S.R., Dubeau L., Veitch N.J., Vonsattel J.P., Wexler N.S.Group, U.S.-V.C.R. Group, U.S.-V.C.R. Arnheim N.et al.. Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum. Mol. Genet. 2007; 16:1133–1142. [DOI] [PubMed] [Google Scholar]
- 41. Graveland G.A., Williams R.S., DiFiglia M.. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington's disease. Science. 1985; 227:770–773. [DOI] [PubMed] [Google Scholar]
- 42. Ferrante R.J., Kowall N.W., Beal M.F., Richardson E.P. Jr, Bird E.D., Martin J.B.. Selective sparing of a class of striatal neurons in Huntington's disease. Science. 1985; 230:561–563. [DOI] [PubMed] [Google Scholar]
- 43. Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., Richardson E.P. Jr. Neuropathological classification of Huntington's disease. J. Neuropathol. Exp. Neurol. 1985; 44:559–577. [DOI] [PubMed] [Google Scholar]
- 44. Waldvogel H.J., Kim E.H., Thu D.C., Tippett L.J., Faull R.L.. New perspectives on the neuropathology in Huntington's disease in the human brain and its relation to symptom variation. J. Huntingtons Dis. 2012; 1:143–153. [DOI] [PubMed] [Google Scholar]
- 45. Morigaki R., Goto S.. Striatal vulnerability in Huntington's disease: neuroprotection versus Neurotoxicity. Brain Sci. 2017; 7:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gokce O., Stanley G.M., Treutlein B., Neff N.F., Camp J.G., Malenka R.C., Rothwell P.E., Fuccillo M.V., Sudhof T.C., Quake S.R.. Cellular Taxonomy of the Mouse Striatum as Revealed by Single-Cell RNA-Seq. Cell Rep. 2016; 16:1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Valerio-Gomes B., Guimaraes D.M., Szczupak D., Lent R.. The Absolute Number of Oligodendrocytes in the Adult Mouse Brain. Front Neuroanat. 2018; 12:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaplan E., Zubedat S., Radzishevsky I., Valenta A.C., Rechnitz O., Sason H., Sajrawi C., Bodner O., Konno K., Esaki K.et al.. ASCT1 (Slc1a4) transporter is a physiologic regulator of brain d-serine and neurodevelopment. Proc. Natl. Acad. Sci. U. S. A. 2018; 115:9628–9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wei K., Clark A.B., Wong E., Kane M.F., Mazur D.J., Parris T., Kolas N.K., Russell R., Hou H. Jr, Kneitz B.et al.. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003; 17:603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao X., Lu H., Dagur P.K., Usdin K.. Isolation and analysis of the CGG-repeat size in male and female gametes from a Fragile X mouse model. Methods Mol. Biol. 2020; 2056:173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Knipscheer P., Raschle M., Smogorzewska A., Enoiu M., Ho T.V., Scharer O.D., Elledge S.J., Walter J.C.. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009; 326:1698–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goold R., Hamilton J., Menneteau T., Flower M., Bunting E.L., Aldous S.G., Porro A., Vicente J.R., Allen N.D., Wilkinson H.et al.. FAN1 controls mismatch repair complex assembly via MLH1 retention to stabilize CAG repeat expansion in Huntington's disease. Cell Rep. 2021; 36:109649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kratz K., Artola-Boran M., Kobayashi-Era S., Koh G., Oliveira G., Kobayashi S., Oliveira A., Zou X., Richter J., Tsuda M.et al.. FANCD2-associated nuclease 1 partially compensates for the lack of Exonuclease 1 in mismatch repair. Mol. Cell. Biol. 2021; 41:e0030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dherin C., Gueneau E., Francin M., Nunez M., Miron S., Liberti S.E., Rasmussen L.J., Zinn-Justin S., Gilquin B., Charbonnier J.B.et al.. Characterization of a highly conserved binding site of Mlh1 required for exonuclease I-dependent mismatch repair. Mol. Cell. Biol. 2009; 29:907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Porro A., Mohiuddin M., Zurfluh C., Spegg V., Dai J., Iehl F., Ropars V., Collotta G., Fishwick K.M., Mozaffari N.L.et al.. FAN1-MLH1 interaction affects repair of DNA interstrand cross-links and slipped-CAG/CTG repeats. Sci. Adv. 2021; 7:eabf7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Goellner E.M., Putnam C.D., Kolodner R.D.. Exonuclease 1-dependent and independent mismatch repair. DNA Repair (Amst.). 2015; 32:24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.