Abstract
Background
Cumulative epidemiologic evidence has shown that early-life adiposity is strongly inversely associated with breast cancer risk throughout life, independent of adult obesity. However, the molecular mechanisms remain poorly understood.
Methods
We assessed the association of early-life adiposity, defined as self-reported body size during ages 10-20 years from a validated 9-level pictogram, with the transcriptome of breast tumor (N = 835) and tumor-adjacent histologically normal tissue (N = 663) in the Nurses’ Health Study. We conducted multivariable linear regression analysis to identify differentially expressed genes in tumor and tumor-adjacent tissue, respectively. Molecular pathway analysis using Hallmark gene sets (N = 50) was further performed to gain biological insights. Analysis was stratified by tumor estrogen receptor (ER) protein expression status (n = 673 for ER+ and 162 for ER− tumors).
Results
No gene was statistically significantly differentially expressed by early-life body size after multiple comparison adjustment. However, pathway analysis revealed several statistically significantly (false discovery rate < 0.05) upregulated or downregulated gene sets. In stratified analyses by tumor ER status, larger body size during ages 10-20 years was associated with decreased cellular proliferation pathways, including MYC target genes, in both ER+ and ER− tumors. In ER+ tumors, larger body size was also associated with upregulation in genes involved in TNFα/NFkB signaling. In ER− tumors, larger body size was additionally associated with downregulation in genes involved in interferon α and interferon γ immune response and Phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling; the INFγ response pathway was also downregulated in ER− tumor-adjacent tissue, though at borderline statistical significance (false discovery rate = 0.1).
Conclusions
These findings provide new insights into the biological and pathological underpinnings of the early-life adiposity and breast cancer association.
Adult obesity is an established risk factor for postmenopausal breast cancer (1). In contrast, early-life (childhood and adolescent) obesity, assessed by perceived body size using a validated 9-level pictogram (2,3) or body mass index (BMI), is inversely associated with breast cancer risk in both pre- and postmenopausal women (4-12), independent of adult obesity (5-10,12). Large body size or high BMI at adolescence was associated with an approximately 15%-30% reduced overall breast cancer risk compared with a small body size or low BMI at adolescence (5-9,12). A similar association has been reported for tumor estrogen receptor positive (ER+) or luminal-like breast cancer (5-8,10); however, the inverse association for ER negative (ER−) breast cancer has been found in some studies (5,8,10) but not others (6,7). A recent Mendelian randomization analysis found that a genetic risk score of childhood BMI, derived from childhood BMI-related variants identified in genome-wide association studies, was also inversely associated with breast cancer risk (13), suggesting a possible etiologic role of early-life adiposity in breast cancer.
Despite the consistent strong inverse association observed in epidemiologic studies, the underlying molecular mechanisms remain unclear. Several hypotheses have been proposed, including higher frequency of anovulatory cycles (14), earlier breast tissue differentiation and maturation (15), or influences on insulin-like growth factor 1 (16-19) or on adult mammographic density (20,21). Nevertheless, no definitive conclusion has yet been reached. Thus, to improve our understanding in the epidemiologic, molecular, and pathological underpinnings of early-life obesity in breast cancer risk, we assessed the association of early-life adiposity with the transcriptome of breast tumor and tumor-adjacent histologically normal tissue collected as part of the Nurses’ Health Study (NHS).
Methods
Study Population and Assessment of Early-Life Adiposity
Participants were identified from the ongoing prospective cohort studies NHS and NHSII. The NHS was established in 1976 when 121 700 US female registered nurses, aged 30-55 years, completed an initial mailed questionnaire. The NHSII was established in 1989 when 116 429 US female registered nurses, aged 25 to 42 years, completed an initial questionnaire. Both cohorts have been followed biennially by mailed questionnaire to update information on exposure status and ascertain newly diagnosed diseases, including cancers. Selection of breast cancer patients and tumor tissue block collection have been described previously (22-25) (Supplementary Methods, available online). The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health as well as those of participating registries as required.
Information on early-life adiposity and covariates was obtained from questionnaires (Supplementary Methods, available online). Adiposity was assessed using a validated 9-level pictogram (Supplementary Figure 1, available online) that best corresponded to their body size at ages 5, 10, and 20 years, respectively (2,3). In a previous validation study, women’s recalled body size at ages 5, 10, and 20 years correlated well with measured BMI at the same ages, respectively (Pearson r = 0.60-0.66) (26).
RNA Expression Microarray and Quality Control Analysis
RNA extraction and transcriptomic profiling have been described previously (22,23,25) (Supplementary Methods, available online). Briefly, RNA was extracted from multiple 1-mm or 1.5-mm cores taken from tumor or tumor-adjacent tissues from formalin-fixed paraffin-embedded (FFPE) blocks. Transcriptomic profiling was done in 2 batches using 2 types of microarray chips: Glue Grant Human Transcriptome Array 3.0 prerelease version (Affymetrix, Santa Clara) (23,25) and Glue Grant Human Transcriptome Array 2.0 (22). After conducting stringent quality control analysis (Supplementary Methods, available online), 835 tumor and 663 tumor-adjacent tissue samples were included in this analysis. Gene expression data were deposited into the Gene Expression Omnibus (accession number: GSE115577).
Statistical Analysis
We performed both differential gene expression (DGE) analysis and gene set enrichment analyses (GSEA). All analyses were conducted in ER+ tumors, ER+ tumor-adjacent, ER− tumors, and ER− tumor-adjacent separately. For DGE, we conducted multivariable linear regression using the Bioconductor package LIMMA (27). To maximize power, we combined the 2 batches of the microarrays and included only common genes (N = 17 791) and adjusted batch effects using the ComBat procedure (28). We further removed the bottom 25th percentile of low expressed genes, leaving 13 343 in downstream analyses. The primary exposure variable is the average body size during ages 10-20 years because it has shown the strongest inverse association with breast cancer risk (5); it was treated as a continuous variable in regression models. Age at diagnosis, year of diagnosis, age at menarche, first-degree family history of breast cancer, recent BMI, menopausal status before diagnosis, average alcohol consumption, and physical activity from baseline to diagnosis were adjusted in the regression models. Furthermore, to capture gene expression heterogeneity due to variations in tumor tissue components (eg, epithelium, stroma), we conducted surrogate variable analysis, using the Bioconductor SVA package, to estimate surrogate variables that may capture the unwanted gene expression heterogeneity and further adjusted these surrogate variables, if identified, in regression models (29). Finally, in a secondary analysis, among a subset of tumor samples with existing protein expression data (measured using immunohistochemistry) for several breast epithelium-specific markers, including cytokeratin (CK) 5/6, CK5/14 and CK7/18, we assessed whether protein expression of these markers may vary across early-life body size categories, considering that variations in epithelium-specific components (eg, luminal, basal) may confound the analysis.
Considering that postmenopausal BMI is associated with increased risk of breast cancer (1) but premenopausal adult BMI is inversely associated with breast cancer, we performed a sensitivity analysis by including an interaction term of menopausal status and recent BMI at diagnosis in regression models (instead of recent BMI alone), thus potentially better controlling for the confounding effect of recent BMI. However, results remained essentially the same (data not shown).
We conducted GSEA using a competitive gene set test procedure that accounts for intergene correlation (30). The 50 Hallmark gene sets (version 6.1; Broad Institute) were used in the analysis (31). These gene sets were generated using information from more than 4000 founder gene sets collected by Molecular Signature Database through a combination of an automated approach and expert curation to provide a collection of well-delineated biological gene sets for enrichment analysis. Thus, each hallmark gene set conveys a specific biological process and displays coherent expression while minimizing noise and redundancy.
For DGE, we considered q-value less than 0.05 as genome-wide significance (32); for GSEA, gene sets were considered statistically significant if the false discovery rate (FDR), estimated using the Benjamini-Hochberg procedure (33), was less than 0.05. All P values were based on 2-sided tests. Analyses were conducted using R software, version 3.5.3.
Results
The distributions of participants’ characteristics by the average body size during ages 10-20 years were similar for most characteristics except that the leanest group had higher proportions of older and postmenopausal women and included more women from the NHS than NHSII (Table 1). The average body size during ages 10-20 years was positively correlated with BMI at breast cancer diagnosis. Most (81.9%) of the participants had a medium body shape (body size level = 1.5-4) during ages 10-20 years, and 10.9% had the leanest body shape and 7.2% had a large body shape (body size level > 4). More than 90.0% of the tumors were stage I or II; neither tumor stage nor grade varied substantially across body size categories (Supplementary Table 1, available online). In a subset of the tumor samples, protein expression levels of several breast epithelium-specific markers, such as CK5/6, CK5/14, and CK7/18, were similar across early-life body size categories (Supplementary Table 4, available online). Ki67 protein expression did not vary statistically significantly across body size categories (Supplementary Table 4, available online).
Table 1.
Participant characteristics in the NHS according to average body size during ages 10-20 years
| Participant characteristics | Overall (N = 835) | Shape 1 (n = 91) | Shape 1.5-2 (n = 261) | Shape 2.5-3 (n = 259) | Shape 3.5-4 (n = 164) | Shape ≥4.5 (n = 60) |
|---|---|---|---|---|---|---|
| NHS cohort, No. (%) | ||||||
| NHSI | 501 (60.0) | 72 (79.1) | 164 (62.8) | 145 (56.0) | 84 (51.2) | 36 (60.0) |
| NHSII | 334 (40.0) | 19 (20.9) | 97 (37.2) | 114 (44.0) | 80 (48.8) | 24 (40.0) |
| Age at diagnosis, mean (SD), y | 59.1 (11.3) | 64.1 (10.7) | 59.0 (11.4) | 58.4 (11.1) | 57.4 (11.1) | 59.4 (11.3) |
| Age at menarche, mean (SD), y | 12.4 (1.4) | 12.9 (1.3) | 12.5 (1.3) | 12.4 (1.4) | 12.2 (1.5) | 12.1 (1.3) |
| Median year of diagnosis | 1999 | 2000 | 1999 | 2000 | 2000 | 2000 |
| ER, No. (%) | ||||||
| Positive | 673 (80.6) | 73 (80.2) | 204 (78.2) | 205 (79.2) | 140 (85.4) | 51 (85.0) |
| Negative | 162 (19.4) | 18 (19.8) | 57 (21.8) | 54 (20.8) | 24 (14.6) | 9 (15.0) |
| Race, No. (%) | ||||||
| White | 797 (95.4) | 87 (95.6) | 247 (94.6) | 244 (94.2) | 161 (98.2) | 58 (96.7) |
| Others | 38 (4.6) | 4 (4.4) | 14 (5.4) | 15 (5.8) | 3 (1.8) | 2 (3.3) |
| Family history of breast cancer, No. (%) | ||||||
| No | 679 (83.1) | 78 (85.7) | 219 (83.9) | 217 (83.8) | 129 (78.7) | 54 (90.0) |
| Yes | 138 (16.9) | 13 (14.3) | 42 (16.1) | 42 (16.2) | 35 (21.3) | 6 (10.0) |
| Menopausal statusa, No. (%) | ||||||
| Premenopausal | 229 (27.4) | 14 (15.4) | 67 (25.7) | 75 (29.0) | 56 (34.1) | 17 (28.3) |
| Postmenopausal | 571 (68.4) | 75 (82.4) | 179 (68.6) | 175 (67.5) | 100 (61.0) | 42 (70.0) |
| Unknown | 35 (4.2) | 2 (2.2) | 15 (5.7) | 9 (3.5) | 8 (4.9) | 1 (1.7) |
| Recent BMIa, mean (SD), kg/m2 | 26.1 (5.1) | 24.6 (4.0) | 24.8 (4.1) | 26.1 (4.8) | 27.8 (6.1) | 28.8 (6.4) |
| Cumulative average physical activity, mean (SD), MET-h/wk | 18.6 (17.3) | 18.9 (15.0) | 19.3 (17.7) | 17.4 (14.4) | 19.2 (21.3) | 18.3 (19.0) |
| Cumulative average alcohol consumption, mean (SD), g/d | 5.8 (8.8) | 5.8 (8.6) | 5.6 (8.2) | 5.8 (9.2) | 6.6 (9.3) | 5.2 (8.5) |
Defined as 1 cycle before breast cancer diagnosis. BMI = body mass index; ER = estrogen receptor; MET = metabolic equivalents; NHS = Nurses’ Health Study; NHSI = Nurses’ Health Study cohort I; NHSII = Nurses’ Health Study cohort II.
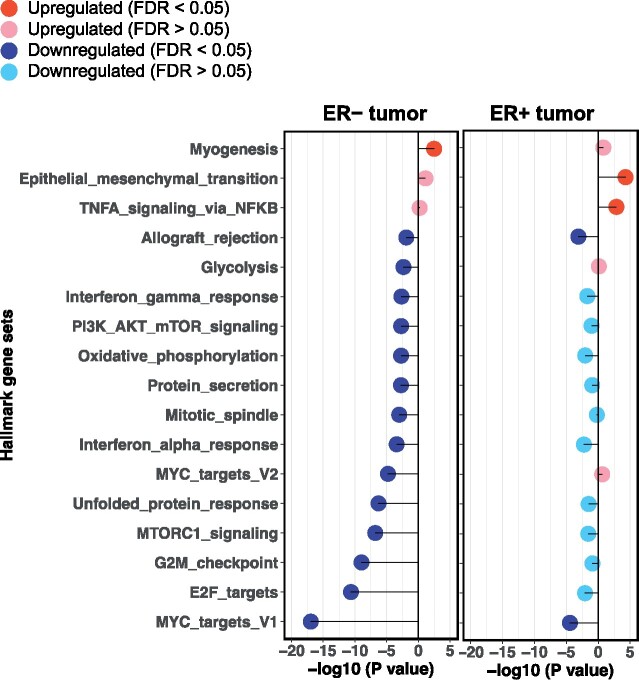
No individual genes were statistically significantly differentially expressed by body size during ages 10-20 years in ER+ or ER− tumors or tumor-adjacent tissue to ER+ or ER− tumors (all q-values > 0.05; Supplementary Table 2, available online). However, in GSEA, we identified several Hallmark gene sets that were statistically significantly upregulated or downregulated by early-life body size (n = 4 and 15 gene sets at FDR < 0.05 in ER+ and ER− tumors, respectively; Table 2). In ER+ tumors, per 1-unit increase in body size pictogram was associated with upregulation in pathways of epithelial to mesenchymal transition (EMT) (FDR = 0.001) and TNFα/NFkB signaling (FDR = 0.02) and downregulation in MYC targets variant 1 (FDR = 0.001) and allograft rejection (FDR = 0.01). In contrast, the majority of the identified gene sets in ER− tumors, including pathways related to proliferation, immune response and inflammation, and PI3K signaling, were downregulated in relation to larger body size. Per 1-unit increase in body size pictogram was linked to downregulation in cellular proliferation pathways, including MYC targets variant 1 (FDR = 5.41 × 10−16) and cell cycle progression E2F targets (FDR = 5.89 × 10−10) and G2/M checkpoint (FDR = 1.76 × 10−8). There were 6 common genes (HMGA1, STMN1, MKI67, MCM3, KPNA2, RAD21) at a nominal P less than .05 between E2F targets and G2/M checkpoint gene sets, whereas KPNA2 was the only common gene among the 3 proliferation-related gene sets. Furthermore, per 1-unit increase in body size pictogram was associated with lower gene expression in genes involved in interferon (IFN)α (FDR = 0.003) and IFNγ response (FDR = 0.01). Four genes (MX1, TRIM25, LY6E, and EIF2AK2) were common at a nominal P less than .05 in both IFNα and IFNγ response. Genes involved in PI3K signaling via AKT to mTORC1 were also down-expressed in ER− tumors (FDR = 0.01). We also performed GSEA by combining ER+ with ER− tumors and combining ER+ tumor-adjacent with ER− tumor-adjacent, respectively; results generally reflected a weighted average of the 2 hormone-receptor specific strata (Supplementary Table 3, available online).
Table 2.
Statistically significantlya upregulated or downregulated gene sets by early-life body size
| Pathway name | No. of genesb | Direction | P | FDR |
|---|---|---|---|---|
| ER+ tumors (n = 673) | ||||
| HALLMARK_MYC_TARGETS_V1 | 187 | Down | 3.60 × 10−5 | 0.001 |
| HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 174 | Up | 5.30 × 10−5 | 0.001 |
| HALLMARK_ALLOGRAFT_REJECTION | 149 | Down | 7.65 × 10−4 | 0.01 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 168 | Up | 1.43 × 10−3 | 0.02 |
| ER− tumors (n = 162) | ||||
| HALLMARK_MYC_TARGETS_V1 | 187 | Down | 1.08 × 10−17 | 5.41 × 10−16 |
| HALLMARK_E2F_TARGETS | 147 | Down | 2.35 × 10−11 | 5.89 × 10−10 |
| HALLMARK_G2M_CHECKPOINT | 149 | Down | 1.05 × 10−9 | 1.76 × 10−8 |
| HALLMARK_MTORC1_SIGNALING | 165 | Down | 1.57 × 10−7 | 1.96 × 10−6 |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 105 | Down | 5.00 × 10−7 | 5.00 × 10−6 |
| HALLMARK_MYC_TARGETS_V2 | 49 | Down | 1.57 × 10−5 | 1.31 × 10−4 |
| HALLMARK_INTERFERON_ALPHA_RESPONSE | 76 | Down | 3.73 × 10−4 | 0.003 |
| HALLMARK_MITOTIC_SPINDLE | 166 | Down | 9.34 × 10−4 | 0.01 |
| HALLMARK_PROTEIN_SECRETION | 83 | Down | 1.79 × 10−3 | 0.01 |
| HALLMARK_OXIDATIVE_PHOSPHORYLATION | 185 | Down | 1.84 × 10−3 | 0.01 |
| HALLMARK_PI3K_AKT_MTOR_SIGNALING | 92 | Down | 1.90 × 10−3 | 0.01 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 153 | Down | 2.11 × 10−3 | 0.01 |
| HALLMARK_MYOGENESIS | 178 | Up | 3.32 × 10−3 | 0.01 |
| HALLMARK_GLYCOLYSIS | 166 | Down | 4.42 × 10−3 | 0.02 |
| HALLMARK_ALLOGRAFT_REJECTION | 149 | Down | 1.22 × 10−2 | 0.04 |
Only gene sets with FDR less than 0.05 were presented. No gene set with FDR less than 0.05 was identified in either ER+ tumor-adjacent histologically normal or ER− tumor-adjacent histologically normal samples. ER = estrogen receptor; ER− = estrogen receptor-negative; ER+ = estrogen receptor positive; FDR = false discovery rate.
Number of genes that contributed to the enrichment of the gene set in this dataset.
When comparing gene sets identified in ER+ vs ER− tumors (Figure 1), multiple associations were in the same gene expression direction, but only 2 common (ie, MYC targets variant 1 and allograft rejection) gene sets demonstrated statistically significant associations (FDR < 0.05). For instance, per 1-unit increase in body size pictogram was associated with lower gene expression in genes involved in MYC targets variant 1 in both ER+ and ER− tumors, though the association was stronger in ER− tumors. Nine and 18 individual genes in this MYC-mediated gene set were at a nominal P less than .05 in ER+ and ER− tumors, respectively; however, no common genes were identified.
Figure 1.
Comparison of statistically significantly upregulated or downregulated Hallmark gene sets by early-life body size in estrogen receptor-positive (ER+) tumors vs estrogen receptor-negative (ER−) tumors. Only gene sets that were statistically significantly (false discovery rate [FDR] < 0.05) upregulated or downregulated in ER+ tumors and/or ER− tumors are presented (ie, each gene set in the figure was found statistically significantly upregulated or downregulated in either ER+ tumors or ER− tumors, or in both). Upregulated gene sets are denoted by −log10 (P value) greater than 0, and downregulated gene sets are denoted with −log10 (P value) less than 0.
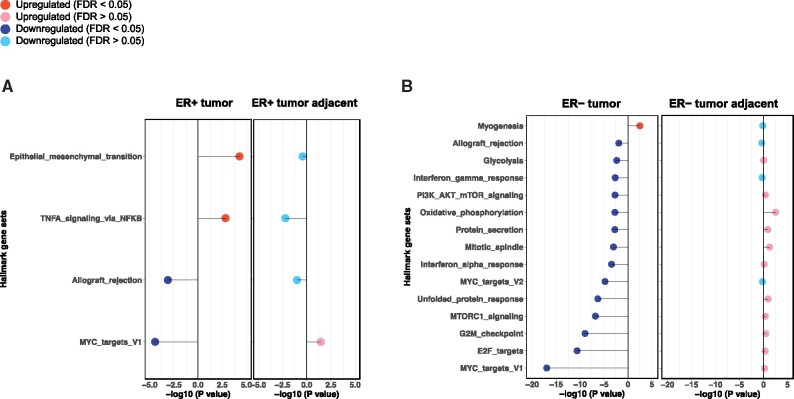
No gene set with FDR less than 0.05 was identified in either ER+ tumor-adjacent or ER− tumor-adjacent tissue. For those statistically significantly dysregulated gene sets identified in ER+ or ER− tumors, the majority of them demonstrated opposite gene expression directions (Figure 2).
Figure 2.
Comparison of statistically significantly upregulated or downregulated Hallmark gene sets by early-life somatotype. A) Estrogen receptor-positive (ER+) tumors vs ER+ tumor adjacent and (B) estrogen receptor-negative (ER−) tumors vs ER− tumor adjacent. Only gene sets that were statistically significantly (false discovery rate [FDR] < 0.05) upregulated or downregulated in tumor and/or tumor-adjacent are presented. Upregulated gene sets are denoted by −log10 (P value) greater than 0, and downregulated gene sets are denoted with −log10 (P value) less than 0.
Discussion
In this large cohort of women with breast cancer, we assessed the association of early-life adiposity with breast tumor tissue (both tumor and tumor-adjacent histologically normal) genome-wide gene expression. Although no individual gene was statistically significantly differentially expressed by early-life body size, several Hallmark gene sets or pathways were identified in gene set enrichment analysis. Specifically, a statistically significant association observed in both ER+ and ER− tumors was that large body size during ages 10-20 years was linked to lower tumor cellular proliferation, with a stronger association in ER− tumors. Additionally, a number of associations varied by tumor ER status. For example, in ER+ tumors, upregulation of gene sets involved in EMT and tumor necrosis factor-α (TNFα)/mediated nuclear factor kappa B (NFkB) signaling was seen, whereas in ER− tumor, downregulation in pathways related to IFNα and IFNγ response and Phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling was observed.
The influence of early-life exposure on adult health outcomes has been of increasing interest in recent years (34). There have been multiple hypotheses, from a life-course perspective, regarding how early-life exposure might affect later life health outcomes through increasing cumulative life-time exposure and/or acting during critical development periods (35). It is likely that being overweight or obese during adolescence when breast tissue undergoes critical development may cause substantial and long-lasting effects and thus influence later breast cancer risk, with or without interaction with adult obesity. For instance, we observed a statistically significant association of larger early-life adiposity and lower gene expression of cellular proliferation pathways in ER+ and ER− tumors.
By contrast, we conducted a similar analysis of BMI at diagnosis and postmenopausal breast tumor transcriptomic analysis from the same NHS and NHSII dataset and reported that higher BMI at diagnosis was linked to increased gene expression in cellular proliferation pathways in both ER+ and ER− breast tumors (25), particularly in ER+ tumors (Supplementary Figure 2, available online). Similar patterns were observed for IFN response in ER+ and/or ER− tumors such that in ER− tumors, genes involved in IFNα and IFNγ response were downregulated in early-life adiposity but upregulated in postmenopausal adiposity (25). Furthermore, genes involved in PI3K/AKT/mTOR signaling were downregulated in relation to early-life obesity (particularly among ER− tumors) but upregulated in relation to higher postmenopausal BMI (statistically significant in both ER+ and ER− tumors) (25). Collectively, the opposite gene expression directions in gene set analysis appear to be consistent with the observed epidemiologic associations of early-life (inverse) or postmenopausal (positive) obesity with breast cancer risk. In analysis of early-life adiposity, we adjusted BMI at diagnosis in statistical models (ie, findings are likely independent of adult obesity). Thus, it is biologically plausible that during the “sensitive” period of breast tissue development (ie, puberty), being overweight or obese may induce permanent changes in normal breast epithelial cells (that serve as cell-of-origin for most breast cancers) that may directly or indirectly affect breast cancer risk in later life and also affect the phenotype of the resulting tumors. Our results suggest that the long-lasting effect may act through both similar (although seemingly in opposite directions) as well as additional pathways as adult obesity’s impact on breast cancer risk. External validation of our findings in other cohorts is warranted.
In addition to the lower cellular proliferation observed in ER+ tumors, we found that higher early-life adiposity was associated with upregulation in genes involved in TNFα/NFkB signaling. Chronic expression of TNFα, a potent NFkB activator, can drive breast cancer metastasis by inducing EMT and tumor cell migration (36). NFkB is a key transcription factor involved in mediating inflammation, cellular stress, and tumor progression. Transcriptional positive crosstalk between ER and NFkB has been shown to promote breast cancer cell survival and reduce response to therapeutic agents such as tamoxifen (37). TNFα/NFkB signaling may be expected to be downregulated in ER+ tumors given the inverse association of early-life adiposity and breast cancer risk as well as its important role in tumor progression and invasion; however, we observed that TNFα/NFkB signaling was upregulated in ER+ tumors though non-statistically significantly downregulated in ER+ tumor-adjacent histologically normal tissue(P = .006, FDR = 0.16), which might suggest the accumulation of inflammatory cytokines in the tumor microenvironment during tumor progression. However, the gene expression levels of several inflammatory genes, such as TNFα, did not demonstrate statistically significant differences by tumor stage or grade (data not shown).
In ER− tumors, we found several gene sets that were not observed in ER+ tumors, including downregulation in IFNα and IFNγ response and PI3K/AKT/mTOR signaling pathways. Recent molecular analyses indicate that aberrations in the PI3K/AKT/mTOR signaling pathway play a key role in ER− breast cancer (38). The PI3K/AKT/mTOR signaling pathway also has been linked to obesity: obesity-related hormones (eg, insulin), cytokines, and growth factors (eg, insulin-like growth factor-1) can interact with mTOR by activating the PI3K/AKT pathway (39,40). The other strong association observed in ER− cancer was downregulation in pathways of IFNα and IFNγ immune response in relation to higher early-life adiposity. IFNγ has both anti- and protumorigenic roles (41); suppression of IFNγ or IFNγ knock-out in experimental studies demonstrated improved insulin sensitivity and decreased adipocyte size or suppressed adipocyte differentiation (42-44). Further, IFNγ response was also downregulated in ER− tumor-adjacent tissue samples though at borderline statistical significance (FDR = 0.1). Taken together, IFNγ-mediated glucose metabolism may play a role in the early-life adiposity and breast cancer association, independent of ER expression (though possibly even stronger in ER− cancer). Thus, the additional pathways identified in ER− tumors (ie, PI3K/AKT/mTOR and IFNγ) that were linked to early-life adiposity, if replicated, may highlight pathways independent of ER expression, such as energy balance or insulin-mediated glucose metabolism in the association of early-life obesity and ER− breast cancer.
To our knowledge, our study is the first to investigate the association of early-life adiposity and breast tumor tissue transcriptome. The strengths of this study include a large sample size, detailed and updated information on early-life body size, important breast cancer risk factors (thus allowing us to control for confounders, including BMI at diagnosis), and detailed tumor characteristics, including the centrally reviewed ER protein expression status. Furthermore, although FFPE tumor tissue blocks were collected and processed at multiple institutions over a 20-year period, we carefully conducted the quality control process (Supplementary Methods) and confirmed the high correlation between ESR1, PGR, and ERBB2 gene expression and protein expression of ER, PR, and HER2 immunohistochemistry staining (23), demonstrating reasonable RNA quality from FFPE tumor blocks in this study.
A major limitation is the lack of validation of the identified gene sets in an independent dataset. However, to our knowledge, there are currently no other studies that have investigated this association, and our large sample size decreases the likelihood of false positives. Furthermore, tumor tissue microarrays were created from whole tissue sections, thus causing potential confounding because of heterogeneity in tissue components across samples. We addressed this by controlling for transcriptome surrogate variables that capture unwanted variations in gene expression (29) in statistical models. Further, in secondary analyses, we evaluated protein expression of several breast epithelium-specific markers and observed that epithelial component was similar across tissue samples. The other limitation is that “early-life” was defined as a wide range of age (10-20 years); however, the reason for using this time of life is mainly because the inverse association of early-life adiposity with breast cancer risk appears to be strongest for the average of body size during ages 10-20 years compared with that at 1 time point (eg, body size at age 5, 10, or 20 years, respectively) (5). Finally, the study population was predominantly White women, whereas the inverse association of early-life adiposity with breast cancer risk has been observed in minority women (8,45-47); thus, future studies in ethnically diverse populations are warranted.
To summarize, we identified several Hallmark pathways whose gene expression in tumors were statistically significantly associated with early-life adiposity. Such gene sets were either common or specific to ER+ or ER− breast cancer. Early-life adiposity was linked to lower cellular proliferation in both ER+ and ER− tumors. We also observed that pathways of IFNα and IFNγ response were downregulated in tumors, particularly ER− tumors, among women with higher early-life adiposity. In ER− tumors, the PI3K/AKT/mTOR signaling pathway, one of the most important signaling pathways in breast cancer etiology and prognosis, was downregulated in women with higher early-life adiposity. Future studies that include racially and ethnically diverse populations and incorporate additional omics data (eg, DNA methylation) are warranted to verify these results and further investigate the underlying biology.
Funding
This work has been funded by the following grants: Komen Foundation Grant SAC110014 and the NIH/NCI U19/GAME-ON DRIVE (CA148065) initiative, UM1 CA186107, P01 CA87969, UM1 CA176726, U01 CA176726, and R01 CA166666.
Notes
Role of the funders: The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Disclosures: Kornelia Polyak serves on the scientific advisory boards of Farcast Biosciences and Acrivon Therapeutics. The rest of the authors have no conflict of interest to disclose.
Author contributions: Cheng Peng, Formal analysis; Writing - review and editing; Catherine Guranich, Data curation; Formal analysis; Writing – review and editing; Yujing Heng, Data curation; Formal analysis; Writing – review and editing; Gabrielle Baker, Data curation; Writing – review and editing; Christopher Rubadue, Data curation; Writing – review and editing; Kimberly Glass, Formal analysis; Writing – review and editing; A. Heather Eliassen, Resources; Writing – review and editing; Rulla Tamimi, Resources; Writing – review and editing; Kornelia Polyak, Formal analysis; Writing – review and editing; Susan Hankinson, Conceptualization; Formal analysis; Funding acquisition; Resources; Writing – review and editing.
Acknowledgments: We would like to thank the participants and staff of the Nurses’ Health Study and the Nurses’ Health Study II for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CG, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Data Availability
The gene expression and patients’ characteristics data underlying this article are available in Gene Expression Omnibus and can be accessed with accession number GSE115577.
Supplementary Material
References
- 1. Neuhouser ML, Aragaki AK, Prentice RL, et al. Overweight, obesity, and postmenopausal invasive breast cancer risk: a secondary analysis of the Women's Health Initiative Randomized Clinical Trials. JAMA Oncol. 2015;1(5):611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stunkard AJ, Sorensen T, Schulsinger F.. Use of the Danish Adoption Register for the study of obesity and thinness. Res Publ Assoc Res Nerv Ment Dis. 1983;60:115–120. [PubMed] [Google Scholar]
- 3. Sorensen TI, Stunkard AJ.. Does obesity run in families because of genes? An adoption study using silhouettes as a measure of obesity. Acta Psychiatr Scand Suppl. 1993;370:67–72. [DOI] [PubMed] [Google Scholar]
- 4. Ahlgren M, Melbye M, Wohlfahrt J, Sorensen TI.. Growth patterns and the risk of breast cancer in women. N Engl J Med. 2004;351(16):1619–1626. [DOI] [PubMed] [Google Scholar]
- 5. Baer HJ, Tworoger SS, Hankinson SE, Willett WC.. Body fatness at young ages and risk of breast cancer throughout life. AmJ Epidemiol. 2010;171(11):1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bardia A, Vachon CM, Olson JE, et al. Relative weight at age 12 and risk of postmenopausal breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(2):374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fagherazzi G, Guillas G, Boutron-Ruault MC, Clavel-Chapelon F, Mesrine S.. Body shape throughout life and the risk for breast cancer at adulthood in the French E3N cohort. Eur J Cancer Prev. 2013;22(1):29–37. [DOI] [PubMed] [Google Scholar]
- 8. Ma H, Ursin G, Xu X, et al. Body mass index at age 18 years and recent body mass index in relation to risk of breast cancer overall and ER/PR/HER2-defined subtypes in White women and African-American women: a pooled analysis. Breast Cancer Res. 2018;20(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shawon MSR, Eriksson M, Li J.. Body size in early life and risk of breast cancer. Breast Cancer Res. 2017;19(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Warner ET, Hu R, Collins LC, et al. Height and body size in childhood, adolescence, and young adulthood and breast cancer risk according to molecular subtype in the Nurses' Health Studies. Cancer Prev Res. 2016;9(9):732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weiderpass E, Braaten T, Magnusson C, et al. A prospective study of body size in different periods of life and risk of premenopausal breast cancer. Cancer Epidemiol. 2004;13(7):1121–1127. [PubMed] [Google Scholar]
- 12. Xue F, Rosner B, Eliassen H, Michels KB.. Body fatness throughout the life course and the incidence of premenopausal breast cancer. Int J Epidemiol. 2016;45(4):1103–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao C, Patel CJ, Michailidou K, et al. Mendelian randomization study of adiposity-related traits and risk of breast, ovarian, prostate, lung and colorectal cancer. Int J Epidemiol. 2016;45(3):896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stoll BA. Teenage obesity in relation to breast cancer risk. Int J Obes. 1998;22(11):1035–1040. [DOI] [PubMed] [Google Scholar]
- 15. Cabanes A, Wang M, Olivo S, et al. Prepubertal estradiol and genistein exposures up-regulate BRCA1 mRNA and reduce mammary tumorigenesis. Carcinogenesis. 2004;25(5):741–748. [DOI] [PubMed] [Google Scholar]
- 16. Kajantie E, Fall CH, Seppala M, et al. Serum insulin-like growth factor (IGF)-I and IGF-binding protein-1 in elderly people: relationships with cardiovascular risk factors, body composition, size at birth, and childhood growth. J Clin Endocrinol Metab. 2003;88(3):1059–1065. [DOI] [PubMed] [Google Scholar]
- 17. Key TJ, Appleby PN, Reeves GK, Roddam AW.. Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol. 2010;11(6):530–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martin RM, Holly JM, Davey Smith G, Gunnell D.. Associations of adiposity from childhood into adulthood with insulin resistance and the insulin-like growth factor system: 65-year follow-up of the Boyd Orr Cohort. J Clin Endocrinol Metab. 2006;91(9):3287–3295. [DOI] [PubMed] [Google Scholar]
- 19. Poole EM, Tworoger SS, Hankinson SE, Schernhammer ES, Pollak MN, Baer HJ.. Body size in early life and adult levels of insulin-like growth factor 1 and insulin-like growth factor binding protein 3. Am J Epidemiol. 2011;174(6):642–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rice MS, Bertrand KA, VanderWeele TJ, et al. Mammographic density and breast cancer risk: a mediation analysis. Breast Cancer Res. 2016;18(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yochum L, Tamimi RM, Hankinson SE.. Birthweight, early life body size and adult mammographic density: a review of epidemiologic studies. Cancer Causes Control. 2014;25(10):1247–1259. [DOI] [PubMed] [Google Scholar]
- 22. Kensler KH, Sankar VN, Wang J, et al. PAM50 molecular intrinsic subtypes in the Nurses' Health Study Cohorts. Cancer Epidemiol Biomarkers Prev. 2019;28(4):798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J, Heng YJ, Eliassen AH, et al. Alcohol consumption and breast tumor gene expression. Breast Cancer Res. 2017;19(1):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tamimi RM, Baer HJ, Marotti J, et al. Comparison of molecular phenotypes of ductal carcinoma in situ and invasive breast cancer. Breast Cancer Res. 2008;10(4):R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heng YJ, Wang J, Ahearn TU, et al. Molecular mechanisms linking high body mass index to breast cancer etiology in post-menopausal breast tumor and tumor-adjacent tissues. Breast Cancer Res Treat. 2019;173(3):667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Must A, Willett WC, Dietz WH.. Remote recall of childhood height, weight, and body build by elderly subjects. Am J Epidemiol. 1993;138(1):56–64. [DOI] [PubMed] [Google Scholar]
- 27. Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1):1–25. [DOI] [PubMed] [Google Scholar]
- 28. Johnson WE, Li C, Rabinovic A.. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. [DOI] [PubMed] [Google Scholar]
- 29. Leek JT, Storey JD.. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3(9):e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu D, Smyth GK.. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40(17):e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P.. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Storey JD, Tibshirani R.. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benjamini Y, Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 34. Ben-Shlomo Y, Kuh D.. A life course approach to chronic disease epidemiology: conceptual models, empirical challenges and interdisciplinary perspectives. Int J Epidemiol. 2002;31(2):285–293. [PubMed] [Google Scholar]
- 35. Clarke MA, Joshu CE.. Early life exposures and adult cancer risk. Epidemiol Rev. 2017;39(1):11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu Y, Zhou BP.. TNF-alpha/NF-kappaB/Snail pathway in cancer cell migration and invasion. Br J Cancer. 2010;102(4):639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frasor J, Weaver A, Pradhan M, et al. Positive cross-talk between estrogen receptor and NF-kappaB in breast cancer. Cancer Res. 2009;69(23):8918–8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Costa RLB, Han HS, Gradishar WJ.. Targeting the PI3K/AKT/mTOR pathway in triple-negative breast cancer: a review. Breast Cancer Res Treat. 2018;169(3):397–406. [DOI] [PubMed] [Google Scholar]
- 39. Bjornsti MA, Houghton PJ.. The TOR pathway: a target for cancer therapy. Nat Rev Cancer. 2004;4(5):335–348. doi: 10.1038/nrc1362 [DOI] [PubMed] [Google Scholar]
- 40. Dutcher JP. Mammalian target of rapamycin inhibition. Clin Cancer Res. 2004;10(18):6382S–6387S. [DOI] [PubMed] [Google Scholar]
- 41. Castro F, Cardoso AP, Goncalves RM, Serre K, Oliveira MJ.. Interferon-Gamma at the crossroads of tumor immune surveillance or evasion. Front Immunol. 2018;9:847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McGillicuddy FC, Chiquoine EH, Hinkle CC, et al. Interferon gamma attenuates insulin signaling, lipid storage, and differentiation in human adipocytes via activation of the JAK/STAT pathway. J Biol Chem. 2009;284(46):31936–31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wong N, Fam BC, Cempako GR, et al. Deficiency in interferon-gamma results in reduced body weight and better glucose tolerance in mice. Endocrinology. 2011;152(10):3690–3699. [DOI] [PubMed] [Google Scholar]
- 44. O'Rourke RW, White AE, Metcalf MD, et al. Systemic inflammation and insulin sensitivity in obese IFN-gamma knockout mice. Metabolism. 2012;61(8):1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bandera EV, Chandran U, Zirpoli G, et al. Body size in early life and breast cancer risk in African American and European American women. Cancer Causes Control. 2013;24(12):2231–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sangaramoorthy M, Phipps AI, Horn-Ross PL, Koo J, John EM.. Early-life factors and breast cancer risk in Hispanic women: the role of adolescent body size. Cancer Epidemiol Biomarkers Prev. 2011;20(12):2572–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Slattery ML, Sweeney C, Edwards S, et al. Body size, weight change, fat distribution and breast cancer risk in Hispanic and non-Hispanic White women. Breast Cancer Res Treat. 2007;102(1):85–101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene expression and patients’ characteristics data underlying this article are available in Gene Expression Omnibus and can be accessed with accession number GSE115577.