Abstract
Dysregulation of the gut microbiome has been implicated in the progression of many diseases. This study explored the role of microbial and metabolic signatures, and their interaction between the Human inflammatory bowel disease (IBD) and healthy controls (HCs) based on the combination of machine learning and traditional statistical analysis, using data collected from the Human Microbiome Project (HMP) and the Integrative Human Microbiome Project (iHMP). It was showed that the microbial and metabolic signatures of IBD patients were significantly different from those of HCs. Compared to HCs, IBD subjects were characterized by 25 enriched species and 6 depleted species. Furthermore, a total of 17 discriminative pathways were identified between the IBD and HC groups. Those differential pathways were mainly involved in amino acid, nucleotide biosynthesis, and carbohydrate degradation. Notably, co-occurrence network analysis revealed that non-predominant bacteria Ruminococcus_obeum and predominant bacteria Faecalibacterium_prausnitzii formed the same broad and strong co-occurring relationships with pathways. Moreover, the essay identified a combinatorial marker panel that could distinguish IBD from HCs. Receiver Operating Characteristic (ROC) and Decision Curve Analysis (DCA) confirmed the high accuracy (AUC = 0.966) and effectiveness of the model. Meanwhile, an independent cohort used for external validation also showed the identical high efficacy (AUC = 0.835). These findings showed that the gut microbes may be relevant to the pathogenesis and pathophysiology, and offer universal utility as a non-invasive diagnostic test in IBD.
Keywords: Inflammatory bowel disease, Gut microbiome, Metabolic pathways, Machine learning, Diagnosis
1. Introduction
IBD, the most common complication in adults, encompassing both Crohn's disease (CD) and ulcerative colitis (UC), is a weakening and multifactorial complex disease characterized by abnormalities in the immune system, intestinal homeostasis disorders, and changes in gut microbiota [1]. Periods of clinical remission and disease flare-ups is commonly observed in IBD patients and seriously affects the survival and quality of life [2].
Currently, the underlying molecular basis of IBD remains mostly obscure, although several hypotheses have attempted to explain its pathophysiological mechanisms. However, accumulating evidence suggests that the gut microbiota in the pathogenesis and therapeutics as crucial factors in the Pathogenesis of IBD: The germfree state reduces severity of symptoms in animal models of chronic intestinal inflammation [3,4]; the reactivity to intestinal microbes have been changed in humans with IBD [5], and the antibodies in the blood are widely known biomarker in CD and UC, but the antibodies are only present as the intestine microbiota altered [6]. Although the cause of IBD remain largely unclear, strategies to reduction of microbiota exposure (such as elemental diets or antibiotic therapy) can have beneficial effects against particular cases of IBD [[7], [8], [9]].
In the study, metagenomic datasets generated by the HMP [10,11] and iHMP [12] were used to investigate role of microbial signatures and metabolic signatures and their interaction between the IBD and HCs. Furthermore, the data was sought to reveal the relationship between specific microbial composition, abundance and metabolic pathways, as well as how regulated severity of IBD, either directly or indirectly by adjusting the gut microbiota, the co-occurrence network analysis were performed. Moreover, to apply the expected differences to clinical practice, a risk classification method involving machine-learning algorithms to was proposed to assess IBD risk based on the biological significance and accuracy contribution of bacterial genus in the cohort, and the accuracy of model was further verified using an external independent cohort, which showed the identical high accuracy.
2. Methods
2.1. Metagenomic sequence analysis
Metagenomic cohort datasets with a total 28 patients with IBD and 35 healthy individuals and another an independent cohort including 357 patients with IBD and 81 healthy individuals for external validation were obtained through the NIH Human Microbiome Project database (https://portal.hmpdacc.org/). Taxonomic profiles of the microbiome was analyzed using MetaPhlan2 pipeline [13] phylogenetic clade identification (default parameters). Functional and pathway composition was performed with HUMAnN2 pipeline [14] using the UniRef90 database and MetaCyc database. Alpha diversity analysis, nonparametric multi-dimensional scaling (NMDS) and principal co-ordinates analysis (PCoA) were conducted and visualized using the vegan and ggplot2 packages in R. In addition, the permutational multivariate analysis of variance (PERMANOVA) and were used to test group differences. The linear discriminant analysis effect size (LEfSe) [15] was used to identify the differential taxonomic classification of microbial between the two groups with a linear discriminant analysis (LDA) score >2.
2.2. Statistical analysis
Statistical analyses were carried out using R v4.0.2 and STAMP [16]. Random forest classifier (R package) was utilized to identify biomarkers, and construct model for identifying potential microorganisms-disease associations between IBD and HCs [17]. Internal validation was performed to tune and validate the model by 5 times repeat, 10-fold cross-validation. The microbial multivariate associations analysis was performed via logistic regression. The receiver operating characteristic (ROC) curve was used to obtain accuracy of the constructed score model, then the result used the area under the ROC curve (AUC) and Decision Curve Analysis (DCA) [18] as the metrics for evaluation. The statistical analysis were carried out using R with level of statistical significance: p-value < 0.05. The co-occurrence network was constructed based on the relative abundance of bacterial species and MetaCyc pathways using the Spearman's correlation coefficient (r > 0.6 or < −0.6; P < 0.05). The generated co-occurrence network was visualized in Cytoscape [19].
3. Results
3.1. Gut microbiome differences between IBD and HC subjects
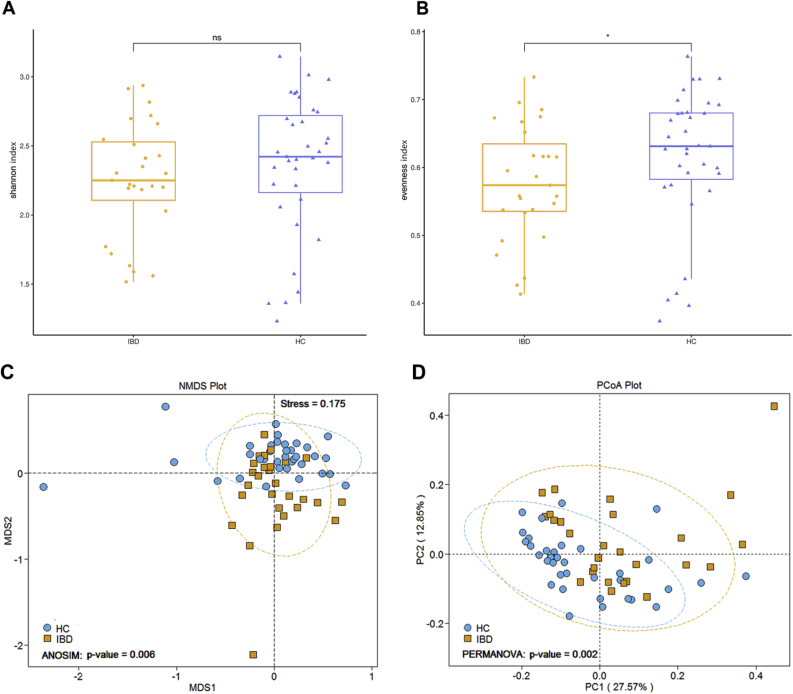
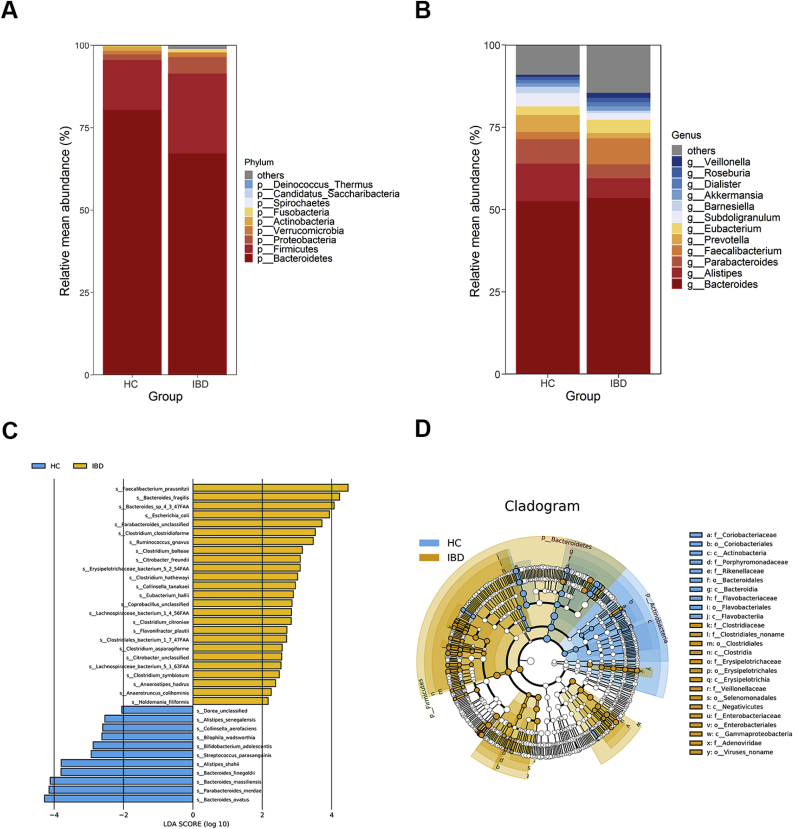
Here, metagenomic sequencing was used to compare the microbial compositions of patients with IBD and HCs. Firstly, alpha diversity analysis showed that there was no significant difference in shannon index, but species evenness in IBD patients is significantly lower relative to HCs (Fig. 1A and B). To explore whether the overall gut bacterial phenotypes of two groups were dissimilar. Beta diversity was evaluated using Principal coordinates analysis (PCoA) and Non-Metric Multidimensional Scaling (NMDS) according to the Bray–Curtis distance, which showed that bacterial signatures in the IBD and HCs were extremely significantly distinct (NMDS: ANOSIM, P = 0.006 < 0.01, Fig. 1C; PCoA: PERMANOVA, P = 0.002 < 0.01, Fig. 1D). Next, a total of 36 discriminative bacterial species between the MDD and HC groups using Linear Discriminant Analysis Effect Size (LEfSe) were identified. Compared with HCs, IBD subjects were characterized by 25 enriched species mainly belonging to the family Clostridiaceae (6 species: Clostridium_asparagiforme, Clostridium_bolteae, Clostridium_citroniae, Clostridium_clostridioforme, Clostridium_hathewayi, and Clostridium_symbiosum), Lachnospiraceae (4 species: Anaerostipes_hadrus, Lachnospiraceae_bacterium_1_4_56FAA, Lachnospiraceae_bacterium_5_1_63FAA, and Ruminococcus_gnavus), Erysipelotrichaceae (3 species: Coprobacillus_unclassified, Erysipelotrichaceae_bacterium_5_2_54FAA, and Holdemania_filiformis), and Enterobacteriaceae (3 species: Citrobacter_freundii, Citrobacter_unclassified, Escherichia_coli), and by 6 depleted species mainly belonging to the family Bacteroidaceae (3 species: Bacteroides_finegoldii, Bacteroides_massiliensis, and Bacteroides_ovatus) and Rikenellaceae (2 species: Alistipes_senegalensis and Alistipes_shahii) (Fig. 2C). The majority of the higher and lower species in IBD be-longed to the phylum Firmicutes (72%, 18 of 25) and Bacteroidetes (55%, 6 of 11) relative to HCs, respectively, as well as, taxonomic cladogram showed the dominant phyla identified in IBD and HCs were Firmicutes and Bacteroidetes, respectively (Fig. 2D). Meanwhile, compared to HCs in the overall gut microbial communities, the abundance of the phylum Firmicutes, Proteobacteria, and Verrucomicrobia and the genus Faecalibacterium, Eubacterium markedly increase, whereas the phylum Bacteroidetes and the genus Alistipes, Parabacteroides, Prevotella, Subdoligranulum, Barnesiella and Akkermansia exhibited a significant decrease (Fig. 2A and B).
Fig. 1.
Altered gut microbiome signatures in IBD versus HCs. (A) There were no significance shannon index. differences between the two groups (Wilcoxon, ns: P > 0.05). (B) Species evenness between the two groups were significantly different (Wilcoxon, ∗: P < 0.05). (C) NMDS analysis (ANOSIM, P = 0.006 < 0.01) and (D) PCoA analysis (PERMANOVA, P = 0.002 < 0.01) showed bacterial communities between the two groups were extremely significantly distinct based on Bray-Curtis distance.
Fig. 2.
Microbial composition of intestinal microbiota between IBD and HCs. Relative mean abundance of the dominant phyla (A) and genera (>1%) (B) in these two groups. (C) Linear Discriminant Analysis Effect Size (LEfSe) analysis for differentially abundant species. (D) Taxonomic cladogram of Least Discriminant Analysis (LDA) coupled with effective size measurement showing differentially microbiota hierarchy.
3.2. Alterations of gut microbial functional signatures in IBD
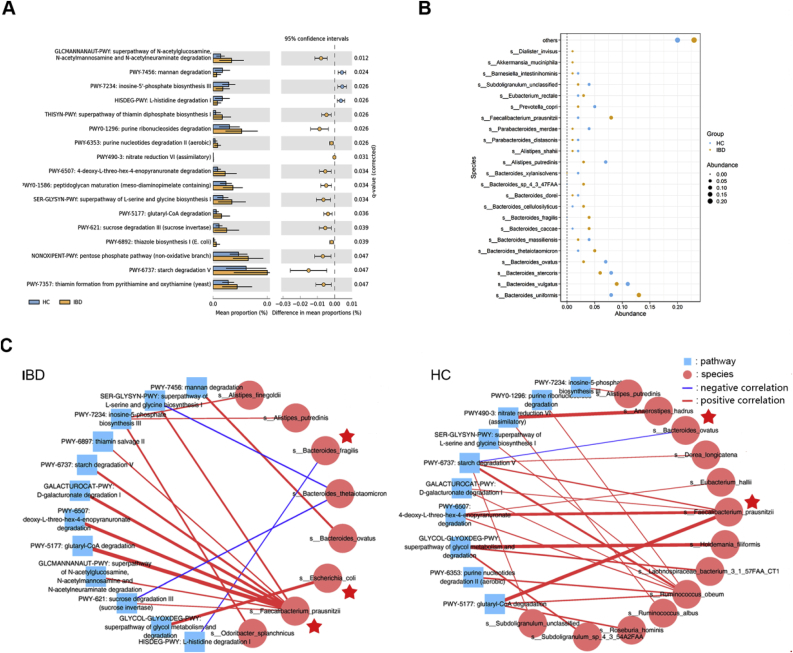
Then, a total of 17 discriminative pathways based on MetaCyc databases between the MDD and HC groups using Humann2 pipeline and STAMP software were identified (Fig. 3A). Compared with HCs, IBD patients were enriched in 14 pathways and depleted in 3 pathways (Corrected P < 0.05). The down-regulated pathways, including polysaccharide degradation, proteinogenic amino acid degradation, and purine nucleotide biosynthesis, would probably play a vital role in IBD. Those differential pathways were mainly involved in four biological processes: amino acid biosynthesis, carbohydrate degradation, cofactor, carrier, and vitamin Biosynthesis, nucleoside and nucleotide biosynthesis.
Fig. 3.
MetaCyc pathway based on humann2 pipeline that discriminate IBD from HCs, the highly abundance bacterial species (>1%), and co-occurrence network constructed from the relative abundances of differential microbial taxa and distinct pathway in IBD subjects versus HCs. (A) STAMP analysis identified that relative abundances of 17 MetaCyc pathway differentiating between the two groups. Compared with HC, the IBD group was characterized by 14 up-regulated metabolites and 3 down-regulated pathway. Those pathways were mainly involved in amino acid biosynthesis, carbohydrate degradation, cofactor, carrier, and vitamin Biosynthesis, nucleoside and nucleotide biosynthesis. (Corrected P < 0.05). (B) Bubble chart of 23 dominant bacterial species with minimun 1% mean relative abundance. These species mainly belonging to the genus Bacteroides (12 species) Alistipes (2 species), and Parabacteroides (2 species). (C) Co-occurrence coefficients among microbiome components at the species level, MetaCyc pathways and the cohort were calculated by spearman correlation analysis, and networks (P < 0.05; Spearman's correlation >0.6 or < −0.6, the line thickness indicates the level of the correlation coefficient) are depicted using Cytoscape. Overall, the species Faecalibacterium_prausnitzii, Ruminococcus_obeum formed broad and strong co-occurring relationships with pathways in two groups. Blue square and red circle indicated pathway and species-level, respectively. Red star represent the dominant species displayed in the (B).
3.3. Co-occurrence network analysis of functional pathways and gut bacteria components
Afterwards, the study explored the potential correlations of abundances of these differential gut bacterial species and functional pathways. Overall, co-occurrence analysis showed that bacterial species widely formed strong co-occurring relationships with pathways (Fig. 3C). The species dominent Faecalibacterium_prausnitzii in the IBD, and Ruminococcus_obeum and Faecalibacterium_prausnitzii in the HCs formed broad and strong positive relationships with many pathways in the two groups, meanwhile Bacteroides_fragilis and Bacteroides_thetaiotaomicron, and Bacteroides_ovatus displayed a negative correlations in IBD and HCs, respectively, whereas compare to HCs, Bacteroides_ovatus of the IBD group showed a strong positive correlations with another pathway. In addition, the dominant species Faecalibacterium_prausnitzii, Bacteroides_fragilis, Escherichia_coli, Bacteroides_ovatus displyed in Fig. 3B were found reside in co-occurring network, in contrast, the lowly abundance species, such as Ruminococcus_obeum (mean abundance 0.04% in HCs), remain strongly significant and comparable across all groups. These findings suggest that different intestinal microbiota and signaling pathways or molecules may form synergistic and niche-related co-occurring network in patients with IBD.
3.4. Combinatorial biomarkers model with high accuracy for discriminating IBD from HC
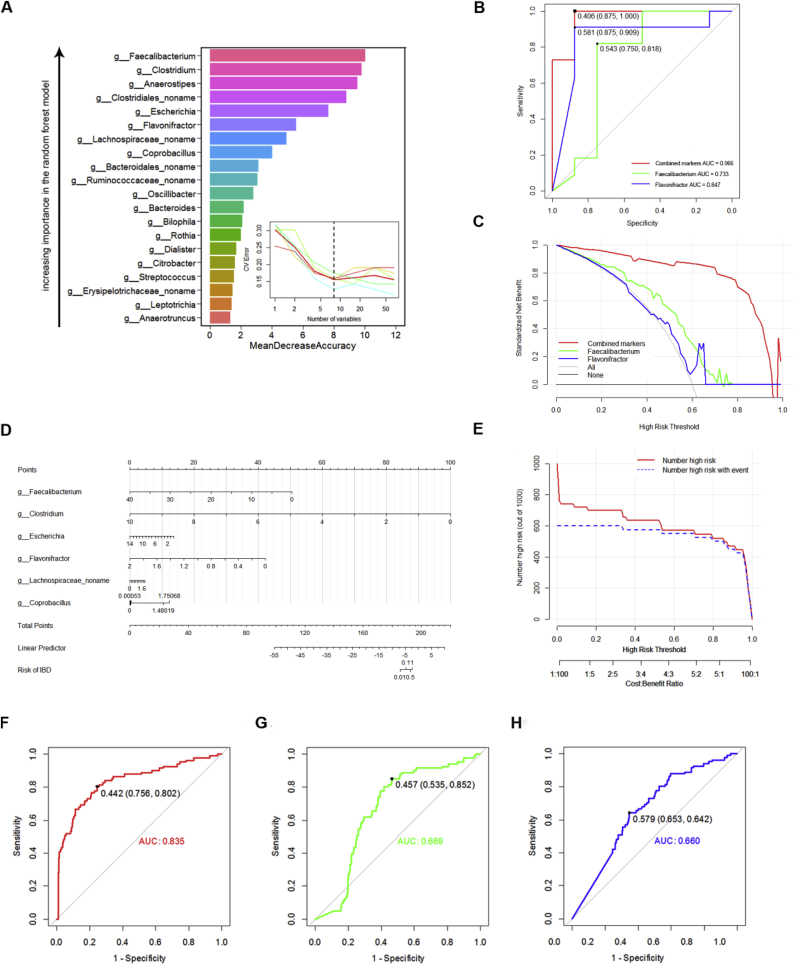
Random forest classifier was used to identify potential biomarker discriminate subjects with IBD from HCs. A five times repeated, 10-fold cross validation was used to identify the more representative bacterial genus, which can showed the most significant deviations in IBD and HCs. The top 8 genera selected as disease markers were used to construct the model via logistic regression (Fig. 4A). The datasets were divided into training and validation sets using a 70–30% split, the samples from the validation set were used to confirm the diagnostic performance independently. The model of combined 6 markers (the genus Faecalibacterium, Clostridium, Anaerostipes, Clostridiales_noname, Escherichia, Flavonifractor, Lachnospiraceae_noname, Coprobacillus) yielded more robust diagnostic performance (AUC = 0.966) over that of separate microbial markers (Faecalibacterium and Flavonifractor) in the final models with a c-statistic of 0.965 (Fig. 4B). The decision curve was used to evaluate the benefit of the combined markers and single one method, which displayed combined markers model provided superior net benefit and reduction than that of treating everyone or treating no one, with a probability threshold of 0.6 or greater (Fig. 4C). A nomogram for assessment of IBD risk based on combinatorial biomarkers was finally shown in Fig. 4D, which would be more feasible and economical in clinical practice and could be convenient to predict the risk of IBD by assigning points for variable by drawing a line upward from the corresponding variable to the line of Points and summing the points associated with the line of Total Points. The performance of model was further conformed by Clinical Impact Curve (CIC) with 1,000 people Risk stratification based on self SIMPLE model (Fig. 4E). Here, the above mentioned methods was defined as Host-Microbiome And Disease Risk Classification Method (HADRCM).
Fig. 4.
A universal model for IBD based on Host-Microbiome And Disease Risk Classification Method (HADRCM). (A) The top 20 bacterial genera ranked in descending order of importance to the accuracy in the diagnosis model were identified by applying Random forest classifier. The insert showed that the number of genera was selected as disease markers using five times repeated, 10-fold cross validation. (B) Multivariate logistic regression was conducted to accurately classify IBD and HCs using 6 filtered variables in the final models. The models for single marker and combined markers were evaluated by the Receiver-Operator Characteristic (ROC) curve and the area under the ROC curve. This combinatorial marker including 6 markers yielded more robust diagnostic performance over that of separate microbial markers (AUC = 0.966). (C) The Decision Curve Analysis (DCA) showed the standardized net benefit against different decision thresholds for the diagnosis models. (D) Nomogram for predicting IBD risk based on combinatorial biomarkers. (E) Clinical Impact Curve exhibited 1,000 people Risk stratification based on SIMPLE model. The x-axis showed the risk threshold and ratio of cost and benefit. The number of high risk is almost consistent with true positive number when risk threshold close to 0.6. Receiver-operating characteristic (ROC) curves for performance of models in an external independent cohort. ROC curves using the combination of 6 biomarkers (F) and 2 individual markers (G, H) were plotted for the datasets, and the areas under the ROC curves (AUCs) were calculated.
To further validate our models above, an external cohort was used to confirm the diagnostic performance independently. Consequently, compared to 2 individual marker, the marker panel of these six biomarkers could still effectively discriminate between the two groups with AUC of 0.835, however, a relatively poor diagnostic performance with AUC of 0.669 and 0.660 were achieved using the 2 individual marker, respectively (Fig. 4F–H). These above mentioned showed using HADRCM could yield a robust model with high accuracy for cohort study and would be more feasible, economical, and convenient in clinical practice.
4. Discussion
This study outlined landscapes and interaction networks of differential microbe and pathways in the IBD and HCs group, Moreover, it provided a novel method named as HADRCM based on machine-learning and statistical analysis. Meanwhile, using HADRCM, a combinatorial marker panel that could distinguish IBD from HC subjects with high accuracy was identified and independently validated, and nomogram of the model could be conveniently applied in clinical practice. Furthermore, because the two subtypes (CD and UC) of IBD have different microbial communities, this approach based on theoretical basis can be also applicable to distinguish IBD subtypes. Our findings suggest that gut microbiota disturbances could potentially contribute to IBD pathogenesis by modulating the host's amino acid, nucleotide, cofactor, vitamin and carbohydrate metabolism, which provides a new avenue by which to understand the basis of IBD.
Compared with the 16s rRNA amplicon sequencing, metagenomic sequencing can provide species level and more valuable information, especially potential functional mechanisms in the microbiota. Our findings were consistent with that of previous research, in which a decrease of alpha diversity in IBD patients was observed [20], however, the species level was not reported. Here, 25 differential bacterial species responsible for this discrimination were identified, which were primarily from the phyla Firmicutes (72.2%), however depleted species mainly belonging to the phyla Bacteroidetes (55%). The data presented in this study confirm previously reported changes of phyla Firmicutes and Bacteroidetes in IBD patients versus HCs [21,22]. Bacteroidetes play significant roles in gut microbiota–host interactions, especially on metabolic pathways [23]. Consistent with previous reports, many Bacteroides species were significantly correlated amino acid, nucleotide, cofactor, vitamin and carbohydrate metabolism. Therefore, the down-regulation Bacteroides species may account for lower amino acid levels in IBD.
In the co-expression network analysis, Faecalibacterium_prausnitzii was found to form strong co-occurring relationships with lots of pathways assigned to carbohydrate and amino acid metabolism and was the dominant species in both groups. Furthermore, previous gut microbiome studies suggested Faecalibacterium have special effects in IBD [[24], [25], [26]].
Currently, machine learning is increasingly used in the disease diagnostics field [27,28]. However, many studies only apply a single algorithm or simple statistics, and only obtain low accuracy. And it is rare and difficult to be applied in actual clinical medical diagnosis. In this study, random forest, logistic regression and statistical analysis were combined to construct diagnostics model and confirmed the excellent performance using ROC and DCA. The union of the methods was defined as HADRCM. Using HADRCM, some microbial markers could effectively discriminate the IBD individuals from HCs. Moreover, a combinatorial marker panel could distinguish IBD individuals from HCs with AUC of 0.966, which still generate a vigorous performance with AUC of 0.835 in an independent external cohort. Ultimately, this output nomogram can provide the non-invasive diagnosis proof for clinicians. Hence, the HADRCM would be a promising alternative in the cohort study.
There are several limitations of this study: (i) The specific microbial species mechanisms need to be further clarified in animal studies. (ii) All data were collected from HMP, which has a relatively wide geographical representation and there is no information similar to BMI of the samples. (iii) Strain-level have no been explored and compared in the cohort.
Despite these limitations, the results from this cohort emphasize the importance of the gut microbiota as a risk factor accounting for the pathogenesis of IBD and the diagnostic implication and accuracy of combination of biomarkers as a non-invasive clinical evaluation indicator for IBD, meanwhile, the union of various machine learning and statistical analysis would be a very promising approach for translational medicine, precision medicine and next generation medicine.
Data availability
All data could be obtained from HMP and iHMP databases and the accession numbers are made available in the Supplementary Table 1.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
CRediT authorship contribution statement
Guangcai Liang: Conceptualization, Methodology, Software, Data curation, Writing – original draft, preparation, Visualization, Validation, Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interestsor personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2021.10.003.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Xavier R.J., Podolsky D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 2.Ordas I., Eckmann L., Talamini M., Baumgart D.C., Sandborn W. J. Ulcerative colitis. Lancet. 2012;380:1606–1619. doi: 10.1016/S0140-6736(12)60150-0. [DOI] [PubMed] [Google Scholar]
- 3.Blumberg R., Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012;4:137rv7. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strober W., Fuss I.J., Blumberg R.S. The immunology of mucosal models of inflammation. Annu Rev Immunol. 2002;20:495–549. doi: 10.1146/annurev.immunol.20.100301.064816. [DOI] [PubMed] [Google Scholar]
- 5.Hegazy A.N., West N.R., Stubbington M.J.T., Wendt E., Suijker K.I.M., Datsi A., et al. Circulating and tissue-resident CD4(+) T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology. 2017;153:1320–1337. doi: 10.1053/j.gastro.2017.07.047. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd-Price J., Arze C., Ananthakrishnan A.N., Schirmer M., Avila-Pacheco J., Poon T.W., et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gecse K.B., Bemelman W., Kamm M.A., Stoker J., Khanna R., Ng S.C., et al. A global consensus on the classification, diagnosis and multidisciplinary treatment of perianal fistulising Crohn's disease. Gut. 2014;63:1381–1392. doi: 10.1136/gutjnl-2013-306709. [DOI] [PubMed] [Google Scholar]
- 8.Cummings, J. H. & Kong, S. C. Probiotics, prebiotics and antibiotics in inflammatory bowel disease. Novartis Found Symp 263, 99-111; discussion -4, 211-8 (2004). [PubMed]
- 9.Zachos M., Tondeur M., Griffiths A.M. Enteral nutritional therapy for induction of remission in Crohn's disease. Cochrane Database Syst Rev. 2007 doi: 10.1002/14651858.CD000542.pub2. CD000542. [DOI] [PubMed] [Google Scholar]
- 10.Huttenhower C., Gevers D., Knight R., Abubucker S., White O. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Human Microbiome Project C. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Proctor L.M., Creasy H.H., Fettweis J.M., Lloyd-Price J., Mahurkar A., Zhou W., et al. The integrative human microbiome Project. Nature. 2019;569:641–648. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Truong D.T., Franzosa E.A., Tickle T.L., Scholz M., Weingart G., Pasolli E., et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 14.Franzosa E.A., McIver L.J., Rahnavard G., Thompson L.R., Schirmer M., Weingart G., et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15:962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parks D.H., Tyson G.W., Hugenholtz P., Beiko R.G. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bento A.P., Gaulton A., Hersey A., Bellis L.J., Chambers J., Davies M., et al. Classification and regression by randomForest. R News. 2002;23:18–22. [Google Scholar]
- 18.Vickers A.J., Elkin E.B. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dovrolis N., Michalopoulos G., Theodoropoulos G.E., Arvanitidis K., Gazouli M. The interplay between mucosal microbiota composition and host gene-expression is linked with infliximab response in inflammatory bowel diseases. Microorganisms. 2020;8:438. doi: 10.3390/microorganisms8030438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh P., Teal T.K., Marsh T.L., Tiedje J.M., Mosci R., Jernigan K., et al. Intestinal microbial communities associated with acute enteric infections and disease recovery. Microbiome. 2015;3:45. doi: 10.1186/s40168-015-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santoru M.L., Piras C., Murgia A., Palmas V., Camboni T., Liggi S., et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep. 2017;7:9523. doi: 10.1038/s41598-017-10034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maier E., Anderson R.C., Roy N.C. Understanding how commensal obligate anaerobic bacteria regulate immune functions in the large intestine. Nutrients. 2014;7:45–73. doi: 10.3390/nu7010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinken A., Ravcheev D.A., Baldini F., Heirendt L., Thiele I. Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome. 2019;7 doi: 10.1186/s40168-019-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sokol H., Seksik P., Furet J.P., Firmesse O., Nion-Larmurier I., Beaugerie L., et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 26.Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermudez-Humaran L.G., Gratadoux J.J., et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh T.G., Kim S.M., Caussy C., Fu T., Guo J., Bassirian S., et al. A universal gut-microbiome-derived signature predicts cirrhosis. Cell Metab. 2020;32:878–888 e6. doi: 10.1016/j.cmet.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren Z., Li A., Jiang J., Zhou L., Yu Z., Lu H., et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68:1014–1023. doi: 10.1136/gutjnl-2017-315084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data could be obtained from HMP and iHMP databases and the accession numbers are made available in the Supplementary Table 1.