Abstract
The resistance that Triple-Negative Breast Cancer (TNBC), the most aggressive breast cancer subtype, develops against radiotherapy is a complex phenomenon involving several regulators of cell metabolism and gene expression; understanding it is the only way to overcome it. We focused this review on the contribution of the two leading classes of regulatory non-coding RNAs, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), against ionizing radiation-based therapies. We found that these regulatory RNAs are mainly associated with DNA damage response, cell death, and cell cycle regulation, although they regulate other processes like cell signaling and metabolism. Several regulatory RNAs regulate multiple pathways simultaneously, such as miR-139-5p, the miR-15 family, and the lncRNA HOTAIR. On the other hand, proteins such as CHK1 and WEE1 are targeted by several regulatory RNAs simultaneously. Interestingly, the study of miRNA/lncRNA/mRNA regulation axes increases, opening new avenues for understanding radioresistance. Many of the miRNAs and lncRNAs that we reviewed here can be used as molecular markers or targeted by upcoming therapeutic options, undoubtedly contributing to a better prognosis for TNBC patients.
Keywords: breast cancer, triple negative breast-cancer, radioresistance, non-coding RNAs, long non-coding RNAs, microRNAs
Introduction
Breast cancer (BC) is the malignant tumor with the highest number of cases diagnosed worldwide and the most common cause of death in women (1). Although it is a heterogeneous disease, breast tumors can be classified based on the expression level of hormonal receptors for estrogen (ER), progesterone (PR), and human epidermal growth factor receptor 2 (HER2) in subtypes depending on the presence (+) or absence (–) of hormonal receptors, namely Luminal A (ER+, PR+/-, HER2-), Luminal B (ER+, PR+/-, HER2+), and HER2-enriched (ER -, PR -, HER2+). A fourth subtype that lacks the expression of all the mentioned hormonal receptors is named Triple Negative (ER -, PR -, HER2-) or Basal-like (2).
Triple-Negative Breast Cancer (TNBC) is further classified into four subtypes; Basal-like 1 (BL1), Basal-like 2 (BL2), Mesenchymal (M), and Luminal Androgen Receptor positive (LAR), where each subtype considers cancerous stage, gene pattern expression, propagation, metastasis, histologic differences and response to common chemotherapeutic neoadjuvants (3). Among the breast cancer subtypes, TNBC is the most aggressive, has a poor prognosis and a high risk of recurrence and metastasis (4–6), and complicates targeted therapies in patients due to the absence of hormonal receptors (ER, PR, HER2) (6).
TNBC patients commonly receive systemic treatments such as chemotherapy or local therapies, including conventional surgery and radiotherapy either in isolation or in combination with other types of treatments for increased effectiveness and prognosis after surgery (6–9).
Radiotherapy has proved efficient for breast cancer patients after mastectomy, at least in levels I and II, reducing recurrence and mortality (10). This type of therapy employs ionizing radiation (IR), e.g., X-rays, gamma rays, α, and β particles, ion carbon or electron, neutron, and proton beams (11, 12) to improve the diagnosis.
IR affects cells directly and indirectly. The direct effect is promoting DNA damage like single-strand breaks (SSBs), double-strand breaks (DSBs), also called clustered DNA lesions, genomic instability, and inducing apoptosis. On the other hand, the indirect effect is caused by reactive oxygen species (ROS) generated from the interconnection between IR and water, promoting complex DNA lesions that alter cell homeostasis, modifying proteins and lipids, eventually lead to cell death (13–15). Nevertheless, the implementation of radiotherapy is still controversial (16), and its efficacy may be limited by the presence of tumor cells resistant to ionizing radiation (17) due to alterations in the pathways and genes involved in the DNA damage response system (DDR).
The alteration of these elements that generally play an important role in preserving cell viability through the repairing genetic material modifies the response of tumor cells to radiotherapy (18).
Recently, it has been observed that not only the irradiated cells themselves react by modifying their metabolism, but that they communicate with neighboring, unirradiated cells through gap junctions and secreted small molecules in a mechanism known as ‘radiation-induced bystander effect’ (RIBE). Through RIBE, bystander cells can rescue irradiated cells, increasing their survival (19). It has been shown that angiogenesis, invasion, metastasis, and proliferative signaling maintenance can also be induced through RIBE, affecting the outcome of IR therapies and enhancing radioresistance (20).
Several groups have reported mechanisms that lead cells to resistance to TNBC therapies, such as hypoxia, cell cycle regulation (21), signaling pathways linked to radiosensitivity like mTOR (22) and EGFR/PI3K/Akt (23), among others. Here we want to highlight the role of two classes of non-coding RNAs (ncRNAs), microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), in the development of radioresistance.
MicroRNAs are small, 21-25 nucleotide-long, single-stranded RNA molecules (24) that negatively regulate mRNA through binding their 3’ UTR and blocking translation (25, 26). They are involved in virtually every cellular process: cell cycle control, differentiation, proliferation, apoptosis, autophagy, and DNA repair, among others, and thus have a role in cancer, either as oncogenes –dubbed oncomirs– or tumor suppressors (27).
Several studies show that miRNAs promote resistance to treatments in other cancer types (28); notably, they can promote radioresistance or radiosensitivity. For instance, miR-214 is upregulated in ovarian cancer, leading to PTEN mRNA degradation and PI3K/Akt activation, thus promoting radioresistance (29). miR-183-5p promotes radioresistance by decreasing ATG5 mRNA expression, interacting with downstream signaling genes from PI3K and Wnt signaling pathways, and upregulating them in colorectal cancer (30, 31). Likewise, miR-365 enhances radiosensitivity by inhibiting the CDC25A expression in non-small cell lung cancer cells, consequently improving the prognosis after IR treatment (32).
Long non-coding (lncRNAs) RNAs are 200+ nucleotide-long molecules (33), transcribed mainly by RNA pol II (34). There are recent reports of their involvement in the regulation of gene expression, metastasis, and invasion of cancer cells (33), miRNA silencing (35), apoptosis, autophagy, cell cycle regulation, and DNA repair (17, 36, 37). As the number of described lncRNAs increases (38), so does the number that regulates the biological processes mentioned above.
lncRNAs have been described in various cancer types. For example, NEAT1 is implicated in the DNA repair process by homologous recombination pathway regulating CHK1, CHK2, BRCA1, and RPA2 expression in multiple myeloma (39). FAM83H-AS1 promotes metastasis and proliferation by interacting and regulating HuR protein stability in ovarian cancer (40). ANRIL promotes proliferation, cell metastasis and inhibits apoptosis by suppressing miR-125a expression in nasopharyngeal carcinoma cells (41). POU3F3 inhibits autophagy signaling by decreasing SMAD4 in colorectal cancer and is involved in cell proliferation and migration (42). Finally, upregulated WTAPP1 promotes invasion and migration in non-small cell lung cancer by interacting with HAND2-S1 and decreasing its expression (43).
Most interestingly, these two classes of ncRNAs can interact with each other, adding to the complexity and importance of their regulation on mRNAs. Several lncRNAs have regions complementary to miRNA sequences that compete for their binding with the target mRNA. This binding sequesters miRNAs to complementary lncRNAs and prevents them from binding to their mRNA targets, turning lncRNAs into miRNA sponges, effective positive mRNA regulators (44, 45).
Interactions of this kind have been reported in diverse biological processes. lncRNA PCAT1 downregulates miR-128 in cervical cancer, promoting proliferation, migration, invasion and thus decreasing radiosensitivity (46). LncRNA lnc-RI competitively binds with miR-4727 regulating Non-Homologous End Joining (NHEJ) through LIG4 mRNA stabilization, affecting cell cycle and radiosensitivity in colorectal cancer (47). LncRNA TRPM2-AS in gastric cancer serves as a sponge for miR-612, promoting radioresistance by upregulation of the DNA double-strand break repair protein FOXM1 (48). Several interactions like these have been reported in TNBC. For instance, lncRNA WEE-AS1promotes proliferation by downregulating miR-32-5p (49), while LINC00173 downregulates miR-490-3p and promotes a more aggressive phenotype (50). Recently, Yuan and colleagues identified MAL2 and NEAT1 as key miRNA regulators in TNBC through an in silico approach (51).
Both miRNAs (52) and lncRNAs (53) have been employed as radiotherapy response biomarkers; however, more research is needed to understand their role in radioresistance fully. A complete grasp of this process and its elements will provide a knowledge base for increasing radiotherapy’s effectiveness in breast and other cancer types. This review describes the different miRNAs, lncRNAs, and their associations that regulate resistance against ionizing radiation-based therapies in breast cancer. We found that these ncRNAs are mainly involved in DNA damage response, but they are also involved in cell death, cell cycle regulation, and other functional aspects. In the following sections, we summarize the currently described ncRNAs involved in the alteration of these processes.
Methods
We searched the Medline database for journal articles in English, published from 2001 to 2021, using combinations of the following keywords: lncRNA, miRNA, breast cancer, radiotherapy, radioresistance, and radiosensitivity.
We obtained 45 articles reporting the diverse roles of ncRNAs in radioresistance. We thoroughly read each paper and extracted data about the type of ncRNAs, targets, and pathways involved in cell radiosensitivity or radioresistance mechanisms, the type of cell line used in both in vivo or in vitro assays; subsequently, we constructed three ncRNA interaction networks using Cytoscape, available at NDEx. (https://www.ndexbio.org/#/). These networks correspond to those processes most regulated by ncRNAs: DNA damage, apoptosis and autophagy, and cell cycle.
ncRNAs Involved in DNA Damage
DNA damage response system (DDR) is a complex network comprising several processes to locate and correct DNA damage to maintain genomic integrity. This extensive network includes mechanisms for damage detection, signal transduction, DNA repair tolerance processes, and cell cycle control. For detailed descriptions of the proteins that participate in these processes, please refer to Giglia-Mari et al. (54).
DNA is an intrinsically reactive molecule and is highly susceptible to damage or chemical alterations due to endogenous processes and factors, such as replication errors, spontaneous deamination of bases, oxidative damage by ROS and formation of abasic sites; or by exogenous agents, for example, DNA breaks by IR, alkylation of bases by chemical agents, modification of bases by ultraviolet (UV) radiation, among others (55, 56). The main repair mechanisms for these damages are nucleotide excision repair (NER), base excision repair (BER), homologous recombination (HR), non-homologous end junction (NHEJ), and mismatch repair (MMR). These processes are extensively explained by Christmann et al. (57).
DNA double-strand breaks (DSB) are the most predominant and damaging lesions caused by IR (58). The most common DSB repair mechanisms are the Homologous Recombination (HR) and the Non-Homologous End Joining (NHEJ) pathways (59). The cell cycle phase determines the triggering of one or the other, but in both cases, they require the intervention of other DDR proteins (54). In addition to the proteins involved in DDR, many ncRNAs are essential to the damage response mechanisms (60–62). Furthermore, these ncRNAs modulate the DDR elements’ activity after irradiation, promoting radioresistant or radiosensitive phenotypes (9).
H2AX as an Indicator of Radiosensitivity
Phosphorylation of the histone variant H2AX is an early event in DDR and, thus, a reliable marker of ongoing DNA repair. However, H2AX foci decrease upon completion of the DNA repair process, so extended detection indicates radiosensitivity (63). The effect of multiple ncRNAs that target DDR proteins can be assessed through H2AX detection.
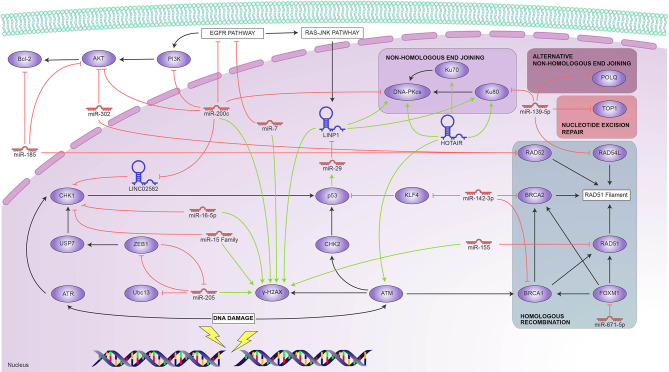
P. Zhang and collaborators (64) found BC cells that overexpress miR-205 exhibit persistent H2AX foci, indicating their low capacity to repair damage after IR. The authors suggest that ZEB1 mediates the effect of miR-205 by partially restoring repair. They demonstrated that miR-205 inhibition increases the expression levels of ZEB1 and Ubc13[u1] [u2]. Therefore, miR-205 radiosensitizes BC cells by inhibiting HR by targeting ZEB1 and Ubc13.
Similarly, Mei and colleagues (65) reported that BC cells transfected with the miR-15 family of mimics showed persistent higher levels of gamma-H2AX after irradiation, indicating unrepaired DNA damage. It is well-known that gamma-H2AX foci decrease shortly after radiation; these authors suggest that the miR-15 family be involved in inhibiting DNA repair, thus acting as radiosensitizers.
Masoudi-Khoram et al. (66) used gamma-H2AX and RAD51 as markers to evaluate DNA damage by IR in two BC-derived cell lines. They found that RAD51 expression increased post-radiation while gamma-H2AX expression reached a peak 4 hours after irradiation and then rapidly decreased. They identified miR-16-5p as a possible important mediator of radiation response and suggested that miR-16-5p could promote radiosensitive breast cancer cells to IR.
In a study with diverse cancer-derived cell lines, Koo and colleagues demonstrated that miR-200c overexpression in the breast cancer cell line, MDA-MB-468 provoked an increase of gamma-H2AX foci and prolonged focus formations after irradiation. This effect was associated with a discernible downregulation of p-DNA-PKcs involved in NHEJ repair (67).
Lin et al. (68) found that overexpression of miR-200c enhanced IR-induced DNA strand breaks in BC cell culture. They found a correlation between increased miR-200c expression and the presence of H2AX foci. Years later, Wang et al. (69) discovered that lncRNA LINC02582 is a downstream target of miR-200c. LINC02582 interacts with USP7 to deubiquitinate and stabilize CHK1, a critical effector in response to DNA damage that facilitates DNA repair, promoting radioresistance (70). However, their results demonstrated that miR-200c expression reduced the CHK1 protein level since it targets LINC02582. They suggest the miR-200c/LINC02582/USP7/CHK1 signaling axis as a potential target to improve breast cancer response to radiation therapy.
In another study with diverse cancer cell types, including BC cells, Lee et al. (71) described miR-7 as a radiosensitizer. Its overexpression causes downregulation of EGFR, AKT, ERK, and STAT3. They inhibited miR-7, which led to positive regulation of EGFR and its downstream effectors to validate these results. Besides, they reported that ectopic overexpression of the miR-7 caused marked prolongation of radiation-induced gamma-H2AX foci formation. The authors associated this phenomenon with a decrease in DNA-PKcs phosphorylation with an activated EGFR-associated signaling pathway.
Zhang et al. (72) found a positive correlation between the expression of LINP1, Ku80, and DNA-PKcs after IR and identified that the lncRNA LINP1 binds Ku80 and DNA-PKcs, promoting radioresistance. They hinted that DSB repair is enhanced by LINP1 across the NHEJ pathway due LINP1 to providing a scaffold for Ku80 and DNA-Pkcs. The authors confirmed this by measuring DNA damage through gamma-H2AX. When LINP1 was removed, gamma-H2AX foci were more persistent. Besides, they discovered that activation of EGFR upregulates LINP1 transcription through activation of the RAS-MEK-ERK pathway; in this manner, cells with EGFR activation improve DNA repair through the LINP1/Ku80/DNA PKcs axis. Also, they identified a negative feedback mechanism where p53 and miR-29 are involved. P53 regulates the expression of miR-29 directly, and, in turn, this negatively regulates LIPN1; this is an uncommon miRNA-lncRNA interaction since lncRNAs sponge miRNAs in most of the currently described instances.
ncRNAs That Target HR Proteins
RAD51, catalyzes the strand transfer between a broken sequence and its homolog to re-synthesize the damaged region (73). Gasparini et al. (74), demonstrated that miR-155 effectively reduces HR repair by targeting RAD51 directly; thus, miR-155 contributes to increased sensitivity to IR. These findings were established both in vivo and in vitro. They found that miR-155 overexpression is associated with lower RAD51 expression; besides, they found a higher survival rate in a TNBC patient cohort due to the anti-correlation between miR-155 overexpression and its target RAD51.
Another study (75) demonstrated that miR-302a downregulation confers radioresistance and that restoration of its expression sensitizes breast cancer cells to radiotherapy since miR-302a targets RAD52, an essential participant in HR repair, and AKT1 (76). Chai et al. (77) show that miR-185 was downregulated in radioresistant BC cells and that there is an inverse correlation with the expression of AKT1 and RAD52. Besides, induced overexpression of miR-185 decreases the expression of AKT1, RAD52, and Bcl-2.
In another work that involved alteration of HR participants, Troschel and collaborators (78) reported that miR-142-3p can sensitize breast cancer cells to radiotherapy by downregulating BRCA1 and BRCA2, two proteins that mediate DSB repair by HR. BRCA1 and BRCA2 play a role as mediators of recombination, promote ssDNA resection, and are believed to be required for subnuclear assembly of RAD51 (79).
Another workgroup found that miR-671-5p was inversely correlated with FOXM1. Through HCR assay, the authors measured the DNA repair capability in breast cancer cell lines. In cells with miR-671-5p inhibited after IR, the HCR activity was significant compared to the control, and FOXM1 expression also increased. Their western blot results showed that miR-671-5p suppressed the expression of genes downstream from FOXM1 involved in the DNA repair pathway; these are RAD51 and BRIP1, the latter contributes to the DNA repair function of BRCA1 (80). Thus, their results hint that miR-671-5p radiosensitizes breast cancer cells by targeting the FOXM1 target, affecting downstream genes involved in DNA repair (81).
ncRNAs That Target Other DDR Proteins
The lncRNA HOTAIR has recently emerged as a multifunctional regulator. Quian et al. (82) demonstrated that HOTAIR could induce resistance to radiotherapy in breast cancer cells. They found that the Ku70 and Ku80 proteins, DNA-PKs, and ATM were upregulated due to HOTAIR overexpression, thus promoting repair and reducing IR sensitivity. In response to DSB, Ku70 and Ku80 associate with broken end chains and then recruit DNA-PKcs to the damage sites, i.e., Ku proteins act as a scaffold for other proteíns that participate in the NHEJ pathway (83, 84).
Surprisingly, it was reported that miR-139-5p modulated resistance to radiation in breast cancer by affecting multiple genes involved in DDR. Five of its six confirmatory targets have roles in diverse DDR pathways essential for post-radiation damage repair. These pathways include microhomology-mediated end-junction (MMJE) with POLQ and XRCC5, BER in which miR-139-5 targets POLQ, NHEJ with XRCC5, HR for RAD54L. Additionally, it regulates DNA topology during repair targeting TOP2A and TOP1 and seems to have a ROS defense role by targeting MAT2AT (85).
The above findings are summarized in Figure 1 , which shows the main elements involved in DDR and the ncRNAs that reportedly regulate them. We found it interesting that only one report involved ATM, one of the primary DNA-damage sensors; ATM responded to HOTAIR overexpression, as did the Ku proteins. There are, however, reports of several ncRNAs that target virtually every downstream pathway—notably, miR139-5p targets proteins that participate in HR, NER, and NHEJ. miR-200c is also multifunctional in DDR; it targets CHK1 through LINC02582 and the DNA PKcs involved in NHEJ. This repair pathway is also targeted by the LINP1 lncRNA, itself regulated by P53 through miR-29; conversely, our search only yielded reports of HR being regulated by miRNAs, such as miR-155 and miR-142. All these ncRNAs are potential radioresistance markers and attractive targets towards induced radiosensitivity.
Figure 1.
Reported ncRNAs that regulate DNA damage response and their targets. Green arrowheads represent positive regulation and red bars, negative regulation.
ncRNAs Involved in Apoptosis and Autophagy
Dysregulation of cell death plays a key role during carcinogenesis. Multiple alterations occur within apoptotic pathways leading to an overall reduction of apoptosis in tumor cells and the rise of apoptosis-resistant phenotypes (86, 87).
Apoptosis
Apoptosis is the most common form of controlled cell death, in which the cell gradually collapses and ultimately dies. It can be triggered by the intrinsic pathway, initiated by either the absence or excess of growth factors, hormones, and cytokines, or by the extrinsic pathway, set off by interaction between death ligands and death receptors such as those from the Tumour Necrosis Factor (TNF) family. For further detail on apoptosis and its effectors, please refer to Cao & Tait (88).
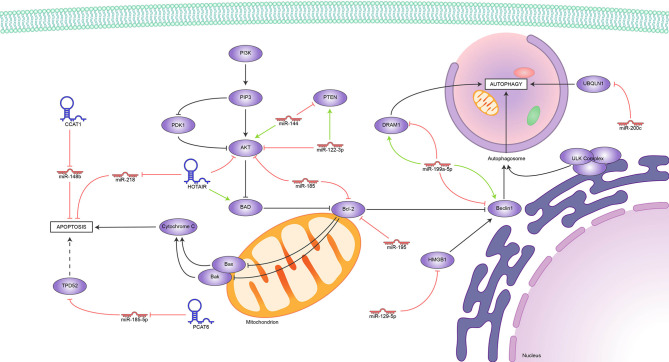
Several ncRNAs regulate the apoptotic response to IR in BC. Yu and colleagues (89) observed an association between miR-144 overexpression and cell survival after irradiation. Subsequent experiments revealed that miR-144 overexpression increased Bcl2 levels and inhibited the pro-apoptotic protein Bax and caspase activity; meanwhile, PTEN and pAkt showed aberrant expression levels, suggesting that miR-144 regulate the radiation-induced apoptotic response by targeting the PTEN/Akt signaling pathway.
Overexpression of the multifunctional lncRNA HOTAIR was also observed in BC cells following irradiation; high expression of this lncRNA has been associated with radioresistance acquisition, even though the exact role of HOTAIR in this process remains unclear. In-vitro experiments showed alterations in the proliferative and apoptotic cells ratio, altered Akt expression, and downregulation of the pro-apoptotic Bad protein. These findings suggest that HOTAIR induces radioresistance by inhibiting apoptosis via the PI3K/Akt-Bad signaling pathway (90). A more recent study suggests another possible mechanism for HOTAIR-induced radioresistance. Knockdown of HOTAIR resulted in an increase of radiation-induced apoptosis, DNA damage, cell cycle arrest, and an upregulation of miR-218. Since miR-218 upregulation promoted cell apoptosis, this data suggests that the HOTAIR-miR-218 axis plays a critical role in radiation-induced apoptosis (36).
Other authors found an upregulation of the lncRNA CCAT1 in radioresistant BC tissues where miR-148b was found to be downregulated. The interaction between CCAT1 and miR-148b was confirmed through luciferase reporter assay. Downregulation of CCAT1 increased radiosensitivity through inhibiting proliferation and promoting apoptosis, implying that the CCAT1-miR148b interaction regulates the acquisition of radioresistance in BC cells (91).
On the other hand, ncRNAs have also been found to sensitize BC cells to radiotherapy by inducing apoptosis. Zhu and colleagues (92) observed that the upregulation of miR-195 enhanced radiosensitivity in BC cells via increasing radiation-induced apoptosis by downregulation of Bcl2. More recently, Chai and colleagues (77) reported downregulation of miR-185 in radioresistant BC cells; conversely, overexpressed miR-185 radiosensitized BC cells. miR-185 overexpression led to Bcl2 downregulation, thus identifying Bcl2 as a downstream target of miR-185. Further experiments showed that Bcl2 silencing radiosensitized BC cells, confirming the role of the miR-185-Bcl2 axis in radioresistance.
In another study, miR-122-3p overexpression was found to sensitize BC cells to ionizing radiation. It was also found that miR-122-3p overexpression induced apoptosis after irradiation while suppressing migration and invasion. Additionally, the aberrant expression levels of PTEN/PI3K/AKT and EMT pathways proteins suggest that miR-122-3p might control radiation-induced apoptosis by regulating the PTEN/PI3K/AKT pathway (93).
Autophagy
Autophagy is a set of adaptations usually aimed at avoiding cell death by sequestering and recycling a portion of the cytoplasm and organelles. Still, it can be triggered to remove damaged or senescent organelles to maintain energy balance or as a result of nutrient deprivation, ultimately leading to cell death. Descriptions of the involved proteins and their functions can be found in reviews such as those by Doherty & Baehrecke (94), Kim & Lee (95), and Maiuri et al. (96). Autophagy plays a dual role during carcinogenesis, leading to cell death or promoting cell survival via inhibiting apoptosis (97).
Several workgroups have demonstrated that ncRNAs play a role in the regulation of autophagy in BC after irradiation. Yi and colleagues (98) observed that the overexpression of miR-199a-5p in MCF7 cells inhibited radiation-induced autophagy. Inhibition of Beclin1 and DRAM1 due to miR-199a-5p was also observed, identifying them as downstream targets and suggesting a potential mechanism for radiation-induced autophagy. However, experiments in the MDA-MB-231 cell line showed that miR-199a-5p overexpression upregulated Beclin1 and DRAM1, promoting radiation-induced autophagy. Further experiments showed that miR-199a-5p regulates cell cycle arrest after IR; additionally, it altered the radiation response of BC after IR. This evidence confirms a role for miR-199a-5p in radiation-induced autophagy through a still undetermined underlying molecular mechanism.
In the same way, miR-200c sensitized BC cells to IR. miR-200c overexpression inhibited radiation-induced autophagy in BC cells; moreover, UBQLN1, a protein associated with promoting autophagosome formation, was identified as a downstream target of miR-200c. This finding suggested that miR-200c enhances radiosensitivity in BC cells by suppressing radiation-induced autophagy through the regulation of UBQLN1 (99).
Luo and colleagues (100) found that the overexpression of miR-129-5p sensitized BC cells to IR, while autophagy acted as a protective response. Subsequently, miR-129-5p was found to inhibit autophagy during the early stages of autophagosome formation, promoting apoptosis. HMGB1 was identified as a potential downstream target for miR-129-5p using online databases. HMGB1 knockdown reduced cell survival and radiation-induced autophagy, suggesting that miR-129-5p may radiosensitize BC cells by inhibiting radiation-induced autophagy via directly targeting HMGB1.
Unsurprisingly, we found reports of several lncRNAs that induce radioresistance by blocking apoptosis and others that perform the opposite function, all of them represented in Figure 2 . So far, the evidence appoints Bcl2 as the hub of this regulation; it is upregulated indirectly by miR144 and downregulated by miR-185 and miR-195. Meanwhile, the HOTAIR-miR-218, CCAT1-miR148b, and PCAT6-miR-185-5p axes block apoptosis through mechanisms still under study. Conflicting reports on the role of miR-199-5p show how much more there is to know about the role of ncRNAs in the delicate balance between apoptosis and autophagy in tumor development.
Figure 2.
Reported ncRNAs that regulate apoptosis, autophagy, and their targets. Green arrowheads represent positive regulation and red bars, negative regulation.
ncRNAs Involved in Cell Cycle
The equilibrium between cell proliferation and death is tightly controlled by the cell cycle, a complex regulatory network that progresses through alternating cell growth, subcellular component synthesis, and cell division phases. Cell cycle progression is regulated primarily by a family of proteins called cyclins that bind and activate their effector counterparts, cyclin-dependent kinases, or CDKs. In an undisturbed cell, timely cyclin expression activates the necessary CDKs, which, in turn, phosphorylate multiple targets that control phase-specific processes. Cyclin D is expressed from early G1 and activates CDK6, both cyclin E in late G1-S and cyclin A in early S-G2 phase activate CDK2, and cyclin B activates CDK1 in G2-M. For additional details on these and other cell cycle regulators and their alterations in cancer, please refer to Foster (101).
Cell cycle checkpoints are, essentially, fail-safe mechanisms that prevent cell cycle progression in response to stimuli such as cell overgrowth, suboptimal chromosome segregation during mitosis, and, notably, DNA damage (102). The ATM/ATR–p53 signaling pathway, part of the DDR, induces G1 or G2 arrest, allowing for DNA repair prior to replication or preventing the cell from undergoing mitosis with a set of altered chromosomes, respectively. However, these mechanisms are dysregulated in cancer cells and let cells with accumulating mutations proliferate (103). In this way, several ncRNAs are upregulated in BC and BC-derived cell lines, associated with a radioresistant phenotype both in patients and cell cultures, suggesting active participation of ncRNAs in the modulation of the response to radiotherapy.
G1/S Checkpoint
Zhang and collaborators (104) found that LINC00963 expression led to the upregulation of the cell cycle regulatory proteins cyclin D1 and CDK6, leading to higher p27 levels and cell cycle progression. Furthermore, elevated LINC00963 expression was significantly associated with tumor size and metastasis. These authors searched for potential miRNA targets and found that LINC00963 sponged miR-324-3p and upregulated ACK1, which belongs to a family of non-receptor-tyrosine-kinases and functions as a driver of tumor progression.
Liu and colleagues (17) found a strong association between cell survival in vitro and increased LINC00511 expression, besides its significant over-expression in BC patients. Subsequent in-vitro experiments correlated its expression with radioresistance and a higher cell proliferation rate. These authors performed a bioinformatic search for miRNA targets and found that LINC00511 sponges miR-185 upregulating STXBP4. This protein has been proven to promote cell cycle progression through TP63 activation (105).
On the other hand, some ncRNAs were recently shown to increase radiosensitivity. For instance, the multifunctional lncRNA HOTAIR increased its expression in BC cells upon radiation exposure. Experimental HOTAIR knockdown increased DNA damage and led to cell cycle arrest. It was also observed that HOTAIR exerted its radiosensitizing effect through the downregulation of miR-218, although the corresponding upregulated target is still to be elucidated (36).
G2/M and Spindle Checkpoints
Mei and colleagues found that miR-15a, 15b, and 16 influence radiosensitivity of MCF7 and MDA-MB-231 breast cancer cells, observable through the enhanced duration of H2AX foci and release of the G2 arrest induced by radiation. They demonstrated the interaction between these miRNAs and the cell cycle regulator WEE1 and CHK1 mRNAs through luciferase assays, but they did not find the dramatic reduction they expected at the protein level, hinting at a more complex mechanism (65). In a differential miRNA expression study, miR-16-5p was upregulated in correlation with radiosensitivity in the radiosensitive T47D and the radioresistant MDA-MB-231cell lines. Through bioinformatic analyses, these authors predicted its interaction with targets such as WEE1, Chk1, and CDC27 (66). miR-16-5p had been previously observed to inhibit proliferation in prostate (106) and breast (107) cancers by targeting AKT.
Low CDC27, a component of the anaphase-promoting complex, is a radioresistance marker in TBNC (108). According to a study in MDA-MB-231 cells, this affects the corresponding miR-27a overexpression, which targets CDC27 and increases cell proliferation even under ionizing radiation (109).
According to these reports, ncRNAs mainly regulate the spindle checkpoint, as it is strongly controlled by the miR-15 family and the closely related miR-16; meanwhile, LINC00963 regulates the G1/S checkpoint, as seen in Figure 3 . Interestingly, we found no reports of ncRNAs that influence the S or G2 phases, which leaves ample room for research in this area, given the importance of cell cycle control in cancer. In this regard, the mechanisms employed by HOTAIR and LINC5011 to control cell proliferation are still to be determined.
Figure 3.
Reported ncRNAs that regulate cell cycle and their targets. Green arrowheads represent positive regulation and red bars, negative regulation.
Other ncRNA Targets
Besides those reviewed in the preceding sections, our search yielded reports on ncRNAs that induce either radioresistance or radiosensitivity by regulating processes such as cell signaling, metabolism, and inflammation, although these were not as abundant. We briefly summarize them in this section and Table 1 , hoping to encourage further research in these aspects.
Table 1.
Reported ncRNAs that regulate cell signaling, cell metabolism, and inflammation.
| ncRNA | Target | Pathway | Reference |
|---|---|---|---|
| ncRNA promoting radiosensitivity | |||
| miR-634 | STAT3 | JAK-STAT signaling pathway | Yang et al. (110) |
| miR-223 | EGFR | EGFR signaling pathway | Fabris et al. (111) |
| miR-7 | EGFR, Akt | EGFR signaling pathway, PI3K-AKT signaling pathway | Lee et al. (71) |
| miR-125b | c-JUN | JNK signaling pathway | Metheerairut et al. (112) |
| miR-671-5p | FOXM1 | Cellular senescence | Tan et al. (81) |
| Let-7 | – | Embryonic development | Wang et al. (113) |
| miR-22 | Sirt1 | AMPK signaling pathway | Zhang et al. (114) |
| miR-770-5p | PBK (PDZ-binding kinase) | – | Lee et al. (115) |
| ncRNA promoting radiorensistance | |||
| miR-124 | STAT3 | JAK-STAT signaling pathway | Fu et al. (116) |
| miR-122 | ZNF611, ZNF304, RIPK1, HRAS, Dusp8, TNFRSF21 | TNF signaling pathway, RAS-MAPK signaling pathway | Pérez-Añorve et al. (117) |
| miR-34 | p53 | p53 signaling pathway | Kato et al. (118) |
| miR-33a | ABCA1/ABCG1 | Lipid metabolism | Wolfe et al. (119) |
| HOTAIR | miR-449b-5p, HSPA1A | JNK signaling pathway | Zhang et al. (120) |
| miR-210, miR-10b, miR-182, miR-142, miR-221, miR-21, miR-93, miR-15b | – | – | Grinan-Lison et al. (52) |
| miR-620 | HPGD/PGE2 | Metabolism of prostaglandins | Huang et al. (121) |
| PCAT6 | miR-185-5p, TPD52 | Membrane traffic | Shi et al. (122) |
| NEAT1 | NOQ1 | Oxidative stress | Lin et al. (123) |
| miR-668 | IkB-alfa | NF-Kappa B signaling pathway | Luo et al. (124) |
Cell Signaling
STAT3 is a transcription factor that regulates gene expression in response to several stimuli such as growth factors and interleukins. In breast cancer, it regulates several target oncogenes and participates in cancer progression, metastasis, apoptosis, and resistance to therapies (125), and it is targeted by ncRNAs modifying the response to radiotherapy in breast cancer.
miR-124 was negatively regulated in HER2-positive breast cancer cells; this miRNA directly targets STAT3, which regulates HER2 expression. So, miR-124 overexpression caused STAT3 downregulation and enhanced radiotherapy response by increasing cell death. The weak miR-124 expression could enhance STAT3 expression and promote radioresistance in HER2-positive breast cancer (116). Similarly, Yang and coworkers (110) observed that miR-634 was significantly decreased in breast cancer cell lines upon radiation. A miR-634 transfection assay showed an increase in apoptosis and a drastic decrease in cell survival capacity. They demonstrated that miR-634 suppresses breast cancer cells by targeting STAT3, increasing radiotherapy sensitivity.
The EGFR pathway is also associated with breast cancer progression since it regulates multiple tumorigenic processes (126). Fabris et al. (111) observed IR-induced miR-223 expression following BC mass removal. Further experiments revealed that miR-223 directly targets EGF, suggesting it may affect the activation of the EGFR pathway. Additionally, miR-223 overexpression was found to antagonize the pro-tumorigenic signals induced by wound fluids via negative regulation of EGF.
Overexpression of miR-122 was observed in therapy-induced radioresistant BC cells; additionally, it sensitized BC cells to IR. Contrastingly, miR-122 knockdown resulted in the acquisition of radioresistance in BC cells. Several proteins involved in diverse pathways such as the transcription factor ZNF611, the TNF pathway elements TNFRS21 and RIPK1, and the Ras-MAPK pathway mediators DUSP8 and HRAS were identified as miR-122 potential targets. These findings suggest that miR-122 may play a multifunctional role in acquiring radioresistance (117).
The JNK signaling pathway promoted cell survival in cancer by interacting with multiple pathways (127). Metheetrairut and colleagues (112) found that miR-125b sensitized BC cells to IR. miR-125b was also found to promote radiation-induced senescence in BC cells. Furthermore, c-JUN regulation by miR-125b was found to be involved in radiosensitivity in BC cells; additionally, members of the MAPK signaling pathway were targeted by miR-125b, suggesting that regulation of the MAPK-c-JUN axis by miR-125b might modulate radiosensitivity in BC cells.
Alterations in the p53 pathway play a key role during carcinogenesis (128). Kato and colleagues (118) observed radiation-induced expression of miR-34 mediated by p53 in BC cells. Furthermore, various BC cell lines showed differential miR-34 expression, and cell lines with low miR-34 levels were radiosensitive. Further experimentation revealed that miR-34 might prevent cells from radiation-induced cell death.
FOXM1 is a transcription factor necessary for many biological processes as cell proliferation, cell cycle progression, and cell differentiation. It is a master regulator of DNA damage response, and it is also associated with EMT phenotype in cancer; likewise, it promotes metastasis and tumor progression (129, 130). Tan et al. (81) demonstrated that miR-671-5p radio- and chemosensitize breast cancer cells by targeting FOXM1. They worked with 21T cells and found that miR-671-5p was decreased during breast cancer progression, contrary to FOXM1. In addition, they found that miR-671-5poverexpression reduces FOXM1 expression and affects the downstream genes involved in EMT (TGF-β and VEGF) and DNA repair during BC progression. This way, miR-671-5p inhibits cell proliferation and invasion and sensitizes breast cancer cells to IR.
Cell Metabolism
Cholesterol regulation has proven to be involved in cancer progression (131). Wolfe and colleagues (119) found that miR-33a expression regulates HDL-induced radioresistance through targeting ABCA1. miR-33a expression was found to be lower in irradiated BC cells than in non-irradiated BC cells. Additionally, the expression of the ABCA1 protein was inversely correlated with that of miR-33a. Furthermore, knockdown of miR-33a in BC cell lines with higher miR-33a expression levels resulted in radiosensitization, whereas miR-33a mimic transfection in BC cell lines with low miR-33a expression led to the inhibition of HDL-induced radiosensitization via regulation of ABCA1. miR-33a was also associated with an adverse outcome in BC patients.
The Lin28/Let-7 axis, primarily active during embryonic development, regulates multiple genes involved in several tumorigenic processes (132). It may also be involved in the regulation of radioresistance in BC. Cell lines expressing higher levels of the Lin28 protein showed increased survival compared to those expressing lower levels of Lin28. Meanwhile, Lin28 knockdown showed an increase in radiosensitivity. Lin28 was also associated with the regulation of apoptosis; on the other hand, Let7 was confirmed to be directly regulated by Lin28, thus suggesting possible mechanisms for acquiring radioresistance via Lin28 (113).
Sirt1 is a histone deacetylase that acts as a regulator in multiple physiological processes such as cell growth, apoptosis, DNA damage and, tumor development; in addition, it promotes tumorigenesis and is upregulated in breast cancer (133–135). Zhang and collaborators (114) reported that Sirt1 is a direct target of miR-22, and their expression is antagonistic, so miR-22 improves radiosensitivity to breast cancer cells by targeting Sirt1. While miR-22 expression was downregulated in breast cancer cells after IR, Sirt1 was upregulated. However, they found that overexpression of miR-22 regulated Sirt1 expression negatively, blocking its function, such as suppressing tumorigenesis and enhancing the radiosensitivity of breast cancer.
Lee and colleagues (115) discovered that miR-770-5p radiosensitizes breast cancer cells by targeting PDZ-binding kinase (PBK). PBK is a serine-threonine kinase that has been reported to be upregulated in rapidly proliferating cells, as well as in a variety of tumors, furthermore, it was shown to promote transformation and has metastatic properties (136–138). In this study, miR-770-5p was shown to be upregulated by IR response and to be inversely correlated with PBK expression both in vitro and in vivo. Despite the oncogenic potential of PBK, the authors report that miR-770-5p can directly target PBK in radiation response, confers radiosensitivity to breast cancer.
In addition to the HOTAIR roles described in the previous sections, Zhang et al. (120) identified that it confers radioresistance to breast cancer cells through the HOTAIR/miR-449-5p/HSPA1A axis. HSPA1A is a chaperone overexpressed in a large variety of tumor lines, including breast cancer (139), and its expression exhibited a positive correlation with that of HOTAIR in irradiated breast cancer cells. Also, HOTAIR acts as a sponge for miR-449-5p, preventing it from exerting its role as a negative HSPA1 regulator, allowing the development of a radioresistant phenotype.
Cancer stem cells (CSCs) play a key role during tumor development (140). Griñán‐Lisón and colleagues (52) identified several miRNAs that may modulate some CSCs properties, such as proliferation, metastasis, and response to IR. miR-142, miR-15b, miR-210, miR-21, miR-221, miR-10b, miR-182, and miR-93, involved in multiple pathways, showed aberrant expression in various BC cell lines and patients. Their results showed that IR affected BC cell lines differentially, decreasing stemness properties in MCF7 and SKBR3 cells and increasing them in the TNBC cell line MDA-MB-231, along with miR-10b, miR-210, and miR-221 expression. Similarly, miR-10b was overexpressed in patients positive for Ki67 that received IR, while miR-210 and miR-221 were detected in the only TNBC patient with recurrence in the study.
In the same way, the lncRNA PCAT6 was found to be upregulated in TNBC tissues. Subsequent experiments showed that PCAT6 knockdown promoted radiosensitivity in BC cells by inhibiting cell survival and promoting apoptosis. miR-185-5p was later identified as a potential target for PCAT6 and shown to be negatively regulated by it. Also, miR-185-5p was found to target TPD52 directly. Knockdown of both PCAT6 and TPD52 resulted in an increased radiosensitivity in TNBC cells, indicating PCAT6 plays a role in radioresistance via regulating the miR-185-5p-TPD52 axis (122).
Lin and colleagues (123) found that NQO1 expression and activity were higher in radioresistant BC-derived cells, modulated by the cancer stem cell-derived NEAT1 lncRNA instead of the more traditional JNK signaling. This finding suggested that the regulation of NEAT1 in NQO1 expression was potentially mediated by suppressing NQO1-targeting miRNAs because the mRNA level was not changed in the radioresistant MDA-MB-231 cells. Still, their results suggested that NEAT might regulate the protein stability of NOQ1 in 231-RR cells through a yet undescribed mechanism. At the time of writing, this is the only report of a lncRNA associated with radioresistance exerting its function through a pathway other than gene up-regulation through miRNA sponging.
Inflammation
Inflammation has also been related to cancer progression (141), and Huang and collaborators (121) found that ncRNAs can also regulate it. Mainly, miR-620 regulates 15-hydroxyprostaglandin dehydrogenase (15-PGDH/HPGD) negatively, which induces radioresistance driven by prostaglandin E2 (PGE2) accumulation, as 15-PGDH normally antagonizes COX-2 by degrading it.
Multiple inflammatory effects during carcinogenesis are mediated by the activation of the NF-κB pathway (142). M. Luo and colleagues (124) observed that increased expression levels of miR-668 in BC cells led to the acquisition of radioresistance while its knockdown sensitized resistant BC cells to IR. miR-668 inhibited lκBα, activating the NF-κB pathway and increasing intranuclear p65, which, in turn, enhances NF-κB binding activity. Thus, miR-668 might regulate radioresistance in BC cells by activating the NF-κB pathway.
Concluding Remarks
Breast cancer is the most common malignancy in women and one of the leading causes of cancer death worldwide. Fortunately, radiotherapy is an effective treatment that provides local tumor control, increases survival, and reduces mortality. However, the acquisition of radioresistant phenotypes can compromise the success of therapy. In this review, we summarize the ncRNAs that participate in conferring radioresistance to breast cancer.
Recently, ncRNAs have emerged as important regulators of multiple cellular processes, and resistance to cancer treatment is no exception; we found reports of ncRNAs involved mainly in the regulation of the DDR mechanisms, followed by cell death, cell cycle, and other processes where the role of ncRNAs is studied in the same depth. A significant number of these works concerned miRNAs, although the proportion of reports on lncRNAs is likely to grow in the upcoming years since lncRNAs have a lower research age. In addition, we observed a growing trend in the number of reports of the response to IR through lncRNA-miRNA-target axes. While it is probable that most of the ncRNAs that regulate radioresistance follow this model, there are other mechanisms of action to explore.
We found of particular interest that several of the reported ncRNAs exhibit a multi-modulator capacity, targeting genes involved in various pathways. Some of them even perform dual roles inducing either radiosensitivity or radioresistance in different contexts. For example, miR-139-5p modulates five different targets involved in four different DDR pathways and, additionally, can regulate DNA topology during the repair process. miR-185 regulates AKT and BCL-2, involved in the regulation of apoptosis, and RAD52, involved in HR. The miR-15 family, comprising the closely related miR-15a, miR-15b, and miR-16, is also multifunctional; it targets CHK1, promoting the formation of the γH2AX foci, while also regulating the cell cycle by targeting WEE1. miR-200c targets multiple proteins involved in the DDR and is also involved in autophagy by regulating the UBQLN1 protein; additionally, it may participate in other major pathways such as the PI3K-AKT and the EGFR. As for lncRNAs, HOTAIR regulates proteins involved in NHEJ, and its overexpression promotes radiation-induced apoptosis, possibly by targeting the PTEN-AKT pathway and miR-218, suggesting it may be a hub where the regulation of DDR, apoptosis, and cell cycle converge.
Accumulating evidence highlights the importance of the interaction between miRNAs and lncRNA. We found reports of miRNA/lncRNA/mRNA axes with a role in BC radioresistance, such as miR-200c/LINC02582/CHK1 and HOTAIR/miR-449-5p/HSPA1A. Our findings point to an increase in this kind of report since mRNA targets are yet to be identified. For instance, miR-185 was identified as a downstream target of the lncRNA LINC00511, promoting cell cycle progression and modulating DDR by regulating unidentified mRNAs. We also found reports with solid association data between a given ncRNA and radioresistance, such as HOTAIR, NEAT1, and miR-199a-5p, whose targets and interactions are still to be determined.
On the other hand, the regulation exerted by multiple ncRNAs converges in some protein targets, evincing the importance of their roles in the acquisition of radioresistance. CHK1 was directly regulated by lncRNA LINC02582, miR-16-5p, and the miR-15 family, and WEE1 was found to be controlled by the miR-15 family and miR-16-5p. Similarly, γH2AX foci formation was induced by several ncRNAs, including miR-155, LINP1, miR-200c, miR-7, miR-16-5p, the miR-15 family, and miR-205.
Many of the ncRNAs mentioned in this review are molecular marker candidates and promising therapeutic targets. Strategies aimed at downregulating ncRNAs that confer radioresistance or re-establishing the expression of those that elicit radiosensitivity are evident possibilities for adjuvant therapies that improve the outcome of radiotherapy alone. However, to get to that point, we need to fully characterize the mechanisms these ncRNAs employ. We anticipate more profound studies on ncRNA function for the upcoming years, as more research groups aim to validate the soaring results that bioinformatic analyses yield through the use of in vivo and in vitro models. Overall, the study of ncRNAs has great potential in the development of adjuvant and targeted therapies in the quest for higher survival rates and better prognosis not only for BC patients but for all cancer patients.
Author Contributions
EL-U conceived the review. AA-M, JS-P, CL-C, and LB-E searched and organized the information. AL-A, JS-P, LB-E, and EL-U wrote the manuscript. AL-A, JS-P, DL, and LB-E prepared the figures. CL-C contributed substantially over the discussion and revision of the manuscript. EL-U and CP-P edited the final version. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by DGAPA-PAPIIT, Universidad Nacional Autónoma de México with Grant Number IA204620, awarded to EL-U.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
LB-E is a student from Posgrado en Ciencias Biológicas, Universidad Nacional Autónoma de México (UNAM) and received a fellowship from CONACYT.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Llombart-Cussac A, Cortés J, Paré L, Galván P, Bermejo B, Martínez N, et al. HER2-Enriched Subtype as a Predictor of Pathological Complete Response Following Trastuzumab and Lapatinib Without Chemotherapy in Early-Stage HER2-Positive Breast Cancer (PAMELA): An Open-Label, Single-Group, Multicentre, Phase 2 Trial. Lancet Oncol (2017) 18:545–54. doi: 10.1016/S1470-2045(17)30021-9 [DOI] [PubMed] [Google Scholar]
- 3. Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PloS One (2016) 11:1–22. doi: 10.1371/journal.pone.0157368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ménard S, Fortis S, Castiglioni F, Agresti R, Balsari A. HER2 as a Prognostic Factor in Breast Cancer. Oncology (2001) 61:67–72. doi: 10.1159/000055404 [DOI] [PubMed] [Google Scholar]
- 5. Abulkhair O, Moghraby JS, Badri M, Alkushi A. Clinicopathologic Features and Prognosis of Triple-Negative Breast Cancer in Patients 40 Years of Age and Younger in Saudi Arabia. Hematol Oncol Stem Cell Ther (2012) 5:101–6. doi: 10.5144/1658-3876.2012.101 [DOI] [PubMed] [Google Scholar]
- 6. He MY, Rancoule C, Rehailia-Blanchard A, Espenel S, Trone JC, Bernichon E, et al. Radiotherapy in Triple-Negative Breast Cancer: Current Situation and Upcoming Strategies. Crit Rev Oncol Hematol (2018) 131:96–101. doi: 10.1016/j.critrevonc.2018.09.004 [DOI] [PubMed] [Google Scholar]
- 7. Lu L, Dong J, Wang L, Xia Q, Zhang D, Kim H, et al. Activation of STAT3 and Bcl-2 and Reduction of Reactive Oxygen Species (ROS) Promote Radioresistance in Breast Cancer and Overcome of Radioresistance With Niclosamide. Oncogene (2018) 37:5292–304. doi: 10.1038/s41388-018-0340-y [DOI] [PubMed] [Google Scholar]
- 8. Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA - J Am Med Assoc (2019) 321:288–300. doi: 10.1001/jama.2018.19323 [DOI] [PubMed] [Google Scholar]
- 9. Podralska M, Ciesielska S, Kluiver J, van den Berg A, Dzikiewicz-Krawczyk A, Slezak-Prochazka I. Non-Coding RNAs in Cancer Radiosensitivity: MicroRNAs and Lncrnas as Regulators of Radiation-Induced Signaling Pathways. Cancers (Basel) (2020) 12:1–27. doi: 10.3390/cancers12061662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McGale P, Taylor C, Correa C, Cutter D, Duane F, Ewertz M, et al. Effect of Radiotherapy After Mastectomy and Axillary Surgery on 10-Year Recurrence and 20-Year Breast Cancer Mortality: Meta-Analysis of Individual Patient Data for 8135 Women in 22 Randomised Trials. Lancet (2014) 383:2127–35. doi: 10.1016/S0140-6736(14)60488-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Giulietti A. Laser-Driven Particle Acceleration for Radiobiology and Radiotherapy: Where We Are and Where We Are Going. Med Appl Laser-Generated Beams Part IV Rev Prog Strateg Futur (2017) 10239:1023904. doi: 10.1117/12.2270945 [DOI] [Google Scholar]
- 12. Malouff TD, Mahajan A, Mutter RW, Krishnan S, Hoppe BS, Beltran C, et al. Carbon Ion Radiation Therapy in Breast Cancer: A New Frontier. Breast Cancer Res Treat (2020) 181:291–6. doi: 10.1007/s10549-020-05641-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lomax ME, Folkes LK, O’Neill P. Biological Consequences of Radiation-Induced DNA Damage: Relevance to Radiotherapy. Clin Oncol (2013) 25:578–85. doi: 10.1016/j.clon.2013.06.007 [DOI] [PubMed] [Google Scholar]
- 14. Hatzi VI, Laskaratou DA, Mavragani IV, Nikitaki Z, Mangelis A, Panayiotidis MI, et al. Non-Targeted Radiation Effects In Vivo: A Critical Glance of the Future in Radiobiology. Cancer Lett (2015) 356:34–42. doi: 10.1016/j.canlet.2013.11.018 [DOI] [PubMed] [Google Scholar]
- 15. Sridharan DM, Asaithamby A, Bailey SM, Costes SV, Doetsch PW, Dynan WS, et al. Understanding Cancer Development Processes After HZE-Particle Exposure: Roles of ROS, DNA Damage Repair and Inflammation. Radiat Res (2015) 183:1–26. doi: 10.1667/RR13804.1 [DOI] [PubMed] [Google Scholar]
- 16. Yao Y, Chu Y, Xu B, Hu Q, Song Q. Radiotherapy After Surgery has Significant Survival Benefits for Patients With Triple-Negative Breast Cancer. Cancer Med (2019) 8:554–63. doi: 10.1002/cam4.1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu L, Zhu Y, Liu AM, Feng Y, Chen Y. Long Noncoding RNA LINC00511 Involves in Breast Cancer Recurrence and Radioresistance by Regulating STXBP4 Expression via miR-185. Eur Rev Med Pharmacol Sci (2019) 23:7457–68. doi: 10.26355/eurrev_201909_18855 [DOI] [PubMed] [Google Scholar]
- 18. Nickoloff JA, Boss MK, Allen CP, LaRue SM. Translational Research in Radiation-Induced DNA Damage Signaling and Repair. Transl Cancer Res (2017) 6:S875–91. doi: 10.21037/tcr.2017.06.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen S, Zhao Y, Han W, Chiu SK, Zhu L, Wu L, et al. Rescue Effects in Radiobiology: Unirradiated Bystander Cells Assist Irradiated Cells Through Intercellular Signal Feedback. Mutat Res Fundam Mol Mech Mutagen (2011) 706:59–64. doi: 10.1016/j.mrfmmm.2010.10.011 [DOI] [PubMed] [Google Scholar]
- 20. Heeran AB, Berrigan HP, O’Sullivan J. The Radiation-Induced Bystander Effect (RIBE) and its Connections With the Hallmarks of Cancer. Radiat Res (2019) 192:668–79. doi: 10.1667/RR15489.1 [DOI] [PubMed] [Google Scholar]
- 21. Wang H, Mu X, He H, Zhang XD. Cancer Radiosensitizers. Trends Pharmacol Sci (2018) 39:24–48. doi: 10.1016/j.tips.2017.11.003 [DOI] [PubMed] [Google Scholar]
- 22. Choi J, Yoon YN, Kim N, Park CS, Seol H, Park IC, et al. Predicting Radiation Resistance in Breast Cancer With Expression Status of Phosphorylated S6K1. Sci Rep (2020) 10:1–8. doi: 10.1038/s41598-020-57496-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaidar-Person O, Lai C, Kuten A, Belkacemi Y. “The Infinite Maze” of Breast Cancer, Signaling Pathways and Radioresistance. Breast (2013) 22:411–8. doi: 10.1016/j.breast.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 24. He L, Hannon GJ. MicroRNAs: Small RNAs With a Big Role in Gene Regulation. Nat Rev Genet (2004) 5:522–31. doi: 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 25. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of Post-Transcriptional Regulation by microRNAs: Are the Answers in Sight? Nat Rev Genet (2008) 9:102–14. doi: 10.1038/nrg2290 [DOI] [PubMed] [Google Scholar]
- 26. Asiaf A, Ahmad ST, Arjumand W, Zargar MA. MicroRNAs in Breast Cancer: Diagnostic and Therapeutic Potential. Methods Mol Biol (2018) 1699:23–43. doi: 10.1007/978-1-4939-7435-1_2 [DOI] [PubMed] [Google Scholar]
- 27. Svoronos AA, Engelman DM, Slack FJ. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res (2016) 76:3666–70. doi: 10.1158/0008-5472.CAN-16-0359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Si W, Shen J, Zheng H, Fan W. The Role and Mechanisms of Action of microRNAs in Cancer Drug Resistance. Clin Epigenet (2019) 11:1–24. doi: 10.1186/s13148-018-0587-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Q, Zhang S. MIR-214 Promotes Radioresistance in Human Ovarian Cancer Cells by Targeting PETN. Biosci Rep (2017) 37:1–8. doi: 10.1042/BSR20170327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Falzone L, Scola L, Zanghì A, Biondi A, Di Cataldo A, Libra M, et al. Integrated Analysis of Colorectal Cancer microRNA Datasets: Identification of microRNAs Associated With Tumor Development. Aging (Albany NY) (2018) 10:1000–14. doi: 10.18632/aging.101444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng S, Zhong Y-F, Tan D-M, Xu Y, Chen H-X, Wang D. miR-183-5p Enhances the Radioresistance of Colorectal Cancer by Directly Targeting ATG5. J Biosci (2019) 44:92. doi: 10.1007/s12038-019-9918-y [DOI] [PubMed] [Google Scholar]
- 32. Li H, Jiang M, Cui M, Feng G, Dong J, Li Y, et al. MiR-365 Enhances the Radiosensitivity of non-Small Cell Lung Cancer Cells Through Targeting CDC25A. Biochem Biophys Res Commun (2019) 512:392–8. doi: 10.1016/j.bbrc.2019.03.082 [DOI] [PubMed] [Google Scholar]
- 33. Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res (2017) 77:3965–81. doi: 10.1158/0008-5472.CAN-16-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prensner JR, Chinnaiyan AM. The Emergence of lncRNAs in Cancer Biology. Cancer Discov (2011) 1:391–407. doi: 10.1158/2159-8290.CD-11-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sahu A, Singhal U, Chinnaiyan AM. Long Noncoding RNAs in Cancer: From Function to Translation. Trends Cancer (2015) 1:93–109. doi: 10.1016/j.trecan.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu X, Ding D, Zhang J, Cui J. Knockdown of lncRNA HOTAIR Sensitizes Breast Cancer Cells to Ionizing Radiation Through Activating miR-218. Biosci Rep (2019) 29:1–9. doi: 10.1042/BSR20181038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Su M, Wang H, Wang W, Wang Y, Ouyang L, Pan C, et al. LncRNAs in DNA Damage Response and Repair in Cancer Cells. Acta Biochim Biophys Sin (Shanghai) (2018) 50:433–9. doi: 10.1093/ABBS/GMY022 [DOI] [PubMed] [Google Scholar]
- 38. Chakraborty S, Deb A, Maji RK, Saha S, Ghosh Z. LncRBase: An Enriched Resource for lncRNA Information. PloS One (2014) 9:1–11. doi: 10.1371/journal.pone.0108010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taiana E, Favasuli V, Ronchetti D, Todoerti K, Pelizzoni F, Manzoni M, et al. Long non-Coding RNA NEAT1 Targeting Impairs the DNA Repair Machinery and Triggers Anti-Tumor Activity in Multiple Myeloma. Leukemia (2020) 34:234–44. doi: 10.1038/s41375-019-0542-5 [DOI] [PubMed] [Google Scholar]
- 40. Dou Q, Xu Y, Zhu Y, Hu Y, Yan Y, Yan H. LncRNA FAM83H-AS1 Contributes to the Radioresistance, Proliferation, and Metastasis in Ovarian Cancer Through Stabilizing HuR Protein. Eur J Pharmacol (2019) 852:134–41. doi: 10.1016/j.ejphar.2019.03.002 [DOI] [PubMed] [Google Scholar]
- 41. Hu X, Jiang H, Jiang X. Downregulation of lncRNA ANRIL Inhibits Proliferation, Induces Apoptosis, and Enhances Radiosensitivity in Nasopharyngeal Carcinoma Cells Through Regulating miR-125a. Cancer Biol Ther (2017) 18:331–8. doi: 10.1080/15384047.2017.1310348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shan TD, Xu JH, Yu T, Li JY, Zhao LN, Ouyang H, et al. Knockdown of Linc-POU3F3 Suppresses the Proliferation, Apoptosis, and Migration Resistance of Colorectal Cancer. Oncotarget (2016) 7:961–75. doi: 10.18632/ONCOTARGET.5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang L, Jin C, Yang G, Wang B, Hua P, Zhang Y. LncRNA WTAPP1 Promotes Cancer Cell Invasion and Migration in NSCLC by Downregulating lncRNA HAND2-As1. BMC Pulm Med (2020) 20:1–6. doi: 10.1186/s12890-020-01180-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen X, Liang H, Zhang CY, Zen K. MiRNA Regulates Noncoding RNA: A Noncanonical Function Model. Trends Biochem Sci (2012) 37:457–9. doi: 10.1016/j.tibs.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 45. Yang C, Wu D, Gao L, Liu X, Jin Y, Wang D, et al. Competing Endogenous RNA Networks in Human Cancer: Hypothesis, Validation, and Perspectives. Oncotarget (2016) 7:13479–90. doi: 10.18632/oncotarget.7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ge X, Gu Y, Li D, Jiang M, Zhao S, Li Z, et al. Knockdown of lncRNA PCAT1 Enhances Radiosensitivity of Cervical Cancer by Regulating miR-128/GOLM1 Axis. Onco Targets Ther (2020) 13:10373–85. doi: 10.2147/OTT.S263728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu R, Zhang Q, Shen L, Chen S, He J, Wang D, et al. Long Noncoding RNA lnc-RI Regulates DNA Damage Repair and Radiation Sensitivity of CRC Cells Through NHEJ Pathway. Cell Biol Toxicol (2020) 36:493–507. doi: 10.1007/s10565-020-09524-6 [DOI] [PubMed] [Google Scholar]
- 48. Xiao J, Lin L, Luo D, Shi L, Chen W, Fan H, et al. Long Noncoding RNA TRPM2-AS Acts as a microRNA Sponge of miR-612 to Promote Gastric Cancer Progression and Radioresistance. Oncogenesis (2020) 9:1–15. doi: 10.1038/s41389-020-0215-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang R, Huang Z, Qian C, Wang M, Zheng Y, Jiang R, et al. LncRNA WEE2-AS1 Promotes Proliferation and Inhibits Apoptosis in Triple Negative Breast Cancer Cells via Regulating miR-32-5p/TOB1 Axis. Biochem Biophys Res Commun (2020) 526:1005–12. doi: 10.1016/j.bbrc.2020.01.170 [DOI] [PubMed] [Google Scholar]
- 50. Fan H, Yuan J, Li X, Ma Y, Wang X, Xu B, et al. LncRNA LINC00173 Enhances Triple-Negative Breast Cancer Progression by Suppressing miR-490-3p Expression. BioMed Pharmacother (2020) 125:109987. doi: 10.1016/j.biopha.2020.109987 [DOI] [PubMed] [Google Scholar]
- 51. Huang Y, Wang X, Zheng Y, Chen W, Zheng Y, Li G, et al. Construction of an mRNA-miRNA-IncRNA Network Prognostic for Triple-Negative Breast Cancer. Aging (Albany NY) (2021) 13:1153–75. doi: 10.18632/aging.202254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Griñán-Lisón C, Olivares-Urbano MA, Jiménez G, López-Ruiz E, Val C, Morata-Tarifa C, et al. miRNAs as Radio-Response Biomarkers for Breast Cancer Stem Cells. Mol Oncol (2020) 14:556–70. doi: 10.1002/1878-0261.12635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yao Z, Zhang Y, Xu D, Zhou X, Peng P, Pan Z, et al. Research Progress on Long non-Coding RNA and Radiotherapy. Med Sci Monit (2019) 25:5757–70. doi: 10.12659/MSM.915647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Giglia-Mari G, Zotter A, Vermeulen W. DNA Damage Response. Cold Spring Harb Perspect Biol (2011) 3:1–19. doi: 10.1101/cshperspect.a000745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chatterjee N, Walker GC. Mechanisms of DNA Damage, Repair, and Mutagenesis. Environ Mol Mutagen (2017) 58:235–63. doi: 10.1002/em.22087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sancar A, Lindsey-Boltz LA, Ünsal-Kaçmaz K, Linn S. Molecular Mechanisms of Mammalian DNA Repair and the DNA Damage Checkpoints. Annu Rev Biochem (2004) 73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723 [DOI] [PubMed] [Google Scholar]
- 57. Christmann M, Tomicic MT, Roos WP, Kaina B. Mechanisms of Human DNA Repair: An Update. Toxicology (2003) 193:3–34. doi: 10.1016/S0300-483X(03)00287-7 [DOI] [PubMed] [Google Scholar]
- 58. Mladenov E, Magin S, Soni A, Iliakis G. DNA Double-Strand Break Repair as Determinant of Cellular Radiosensitivity to Killing and Target in Radiation Therapy. Front Oncol (2013) 3:113. doi: 10.3389/fonc.2013.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Scott SP, Pandita TK. The Cellular Control of DNA Double-Strand Breaks. J Cell Biochem (2006) 99:1463–75. doi: 10.1002/jcb.21067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Durut N, Mittelsten Scheid O. The Role of Noncoding RNAs in Double-Strand Break Repair. Front Plant Sci (2019) 10:1155. doi: 10.3389/fpls.2019.01155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Storici F, Tichon AE. RNA Takes Over Control of DNA Break Repair. Nat Cell Biol (2017) 19:1382–4. doi: 10.1038/ncb3645 [DOI] [PubMed] [Google Scholar]
- 62. Zhang C, Peng G. Non-Coding RNAs: An Emerging Player in DNA Damage Response. Mutat Res Rev Mutat Res (2015) 763:202–11. doi: 10.1016/j.mrrev.2014.11.003 [DOI] [PubMed] [Google Scholar]
- 63. Mah LJ, El-Osta A, Karagiannis TC. γh2AX: A Sensitive Molecular Marker of DNA Damage and Repair. Leukemia (2010) 24:679–86. doi: 10.1038/leu.2010.6 [DOI] [PubMed] [Google Scholar]
- 64. Zhang P, Wang L, Rodriguez-Aguayo C, Yuan Y, Debeb BG, Chen D, et al. MiR-205 Acts as a Tumour Radiosensitizer by Targeting ZEB1 and Ubc13. Nat Commun (2014) 5:1–10. doi: 10.1038/ncomms6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mei Z, Su T, Ye J, Yang C, Zhang S, Xie C. The miR-15 Family Enhances the Radiosensitivity of Breast Cancer Cells by Targeting G2 Checkpoints. Radiat Res (2015) 183:196–207. doi: 10.1667/RR13784.1 [DOI] [PubMed] [Google Scholar]
- 66. Masoudi-Khoram N, Abdolmaleki P, Hosseinkhan N, Nikoofar A, Mowla SJ, Monfared H, et al. Differential miRNAs Expression Pattern of Irradiated Breast Cancer Cell Lines Is Correlated With Radiation Sensitivity. Sci Rep (2020) 10:1–12. doi: 10.1038/s41598-020-65680-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Koo T, Cho BJ, Kim DH, Park JM, Choi EJ, Kim HH, et al. MicroRNA-200c Increases Radiosensitivity of Human Cancer Cells With Activated EGFR-Associated Signaling. Oncotarget (2017) 8:65457–68. doi: 10.18632/oncotarget.18924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lin J, Liu C, Gao F, Mitchel REJ, Zhao L, Yang Y, et al. MiR-200c Enhances Radiosensitivity of Human Breast Cancer Cells. J Cell Biochem (2013) 114:606–15. doi: 10.1002/jcb.24398 [DOI] [PubMed] [Google Scholar]
- 69. Wang B, Zheng J, Li R, Tian Y, Lin J, Liang Y, et al. Long Noncoding RNA LINC02582 Acts Downstream of miR-200c to Promote Radioresistance Through CHK1 in Breast Cancer Cells. Cell Death Dis (2019) 10:1–15. doi: 10.1038/s41419-019-1996-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen Y, Sanchez Y. Chk1 in the DNA Damage Response: Conserved Roles From Yeasts to Mammals. DNA Repair (Amst) (2004) 3:1025–32. doi: 10.1016/j.dnarep.2004.03.003 [DOI] [PubMed] [Google Scholar]
- 71. Lee KM, Choi EJ, Kim IA. MicroRNA-7 Increases Radiosensitivity of Human Cancer Cells With Activated EGFR-Associated Signaling. Radiother Oncol (2011) 101:171–6. doi: 10.1016/j.radonc.2011.05.050 [DOI] [PubMed] [Google Scholar]
- 72. Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang Z, et al. Long Noncoding RNA LINP1 Regulates Repair of DNA Double-Strand Breaks in Triple-Negative Breast Cancer. Nat Struct Mol Biol (2016) 23:522–30. doi: 10.1038/nsmb.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Daboussi F, Dumay A, Delacôte F, Lopez BS. DNA Double-Strand Break Repair Signalling: The Case of RAD51 Post-Translational Regulation. Cell Signal (2002) 14:969–75. doi: 10.1016/S0898-6568(02)00052-9 [DOI] [PubMed] [Google Scholar]
- 74. Gasparini P, Lovat F, Fassan M, Casadei L, Cascione L, Jacob NK, et al. Protective Role of miR-155 in Breast Cancer Through RAD51 Targeting Impairs Homologous Recombination After Irradiation. Proc Natl Acad Sci USA (2014) 111:4536–41. doi: 10.1073/pnas.1402604111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liang Z, Ahn J, Guo D, Votaw JR, Shim H. MicroRNA-302 Replacement Therapy Sensitizes Breast Cancer Cells to Ionizing Radiation. Pharm Res (2013) 30:1008–16. doi: 10.1007/s11095-012-0936-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Symington LS. Role of RAD52 Epistasis Group Genes in Homologous Recombination and Double-Strand Break Repair. Microbiol Mol Biol Rev (2002) 66:630–70. doi: 10.1128/mmbr.66.4.630-670.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chai Y, Yang X, Li Y, Qu Q. MicroRNA-185 Overexpression Sensitizes Breast Cancer Cells to Ionizing Radiation: A Potential Therapeutic Role in Breast Cancer. Int J Clin Exp Pathol (2017) 10:274–81. [Google Scholar]
- 78. Troschel FM, Böhly N, Borrmann K, Braun T, Schwickert A, Kiesel L, et al. miR-142-3p Attenuates Breast Cancer Stem Cell Characteristics and Decreases Radioresistance In Vitro . Tumor Biol (2018) 40:1–10. doi: 10.1177/1010428318791887 [DOI] [PubMed] [Google Scholar]
- 79. Zhang J. The Role of BRCA1 in Homologous Recombination Repair in Response to Replication Stress: Significance in Tumorigenesis and Cancer Therapy. Cell Biosci (2013) 3:1–14. doi: 10.1186/2045-3701-3-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, et al. BACH1, a Novel Helicase-Like Protein, Interacts Directly With BRCA1 and Contributes to its DNA Repair Function. Cell (2001) 105:149–60. doi: 10.1016/S0092-8674(01)00304-X [DOI] [PubMed] [Google Scholar]
- 81. Tan X, Li Z, Ren S, Rezaei K, Pan Q, Goldstein AT, et al. Dynamically Decreased miR-671-5p Expression Is Associated With Oncogenic Transformation and Radiochemoresistance in Breast Cancer. Breast Cancer Res (2019) 21:1–14. doi: 10.1186/s13058-019-1173-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Qian L, Fei Q, Zhang H, Qiu M, Zhang B, Wang Q, et al. lncRNA HOTAIR Promotes DNA Repair and Radioresistance of Breast Cancer via EZH2. DNA Cell Biol (2020) 39:2166–73. doi: 10.1089/dna.2020.5771 [DOI] [PubMed] [Google Scholar]
- 83. Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA Double-Strand Break Repair Pathway Choice. Cell Res (2008) 18:134–47. doi: 10.1038/cr.2007.111 [DOI] [PubMed] [Google Scholar]
- 84. Tomimatsu N, Tahimic CGT, Otsuki A, Burma S, Fukuhara A, Sato K, et al. Ku70/80 Modulates ATM and ATR Signaling Pathways in Response to DNA Double Strand Breaks. J Biol Chem (2007) 282:10138–45. doi: 10.1074/jbc.M611880200 [DOI] [PubMed] [Google Scholar]
- 85. Pajic M, Froio D, Daly S, Doculara L, Millar E, Graham PH, et al. miR-139-5p Modulates Radiotherapy Resistance in Breast Cancer by Repressing Multiple Gene Networks of DNA Repair and ROS Defense. Cancer Res (2018) 78:501–15. doi: 10.1158/0008-5472.CAN-16-3105 [DOI] [PubMed] [Google Scholar]
- 86. Kaczanowski S. Apoptosis: Its Origin, History, Maintenance and the Medical Implications for Cancer and Aging. Phys Biol (2016) 13:31001. doi: 10.1088/1478-3975/13/3/031001 [DOI] [PubMed] [Google Scholar]
- 87. Pistritto G, Trisciuoglio D, Ceci C, Garufi A, D’Orazi G. Apoptosis as Anticancer Mechanism: Function and Dysfunction of its Modulators and Targeted Therapeutic Strategies. Aging (Albany NY) (2016) 8:603–19. doi: 10.18632/aging.100934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cao K, Tait SWG. Apoptosis and Cancer: Force Awakens, Phantom Menace, or Both? Int Rev Cell Mol Biol (2018) 337:135–52. doi: 10.1016/bs.ircmb.2017.12.003 [DOI] [PubMed] [Google Scholar]
- 89. Yu L, Yang Y, Hou J, Zhai C, Song Y, Zhang Z, et al. MicroRNA-144 Affects Radiotherapy Sensitivity by Promoting Proliferation, Migration and Invasion of Breast Cancer Cells. Oncol Rep (2015) 34:1845–52. doi: 10.3892/or.2015.4173 [DOI] [PubMed] [Google Scholar]
- 90. Zhou Y, Wang C, Liu X, Wu C, Yin H. Long non-Coding RNA HOTAIR Enhances Radioresistance in MDA-MB231 Breast Cancer Cells. Oncol Lett (2017) 13:1143–8. doi: 10.3892/ol.2017.5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lai Y, Chen Y, Lin Y, Ye L. Down-Regulation of LncRNA CCAT1 Enhances Radiosensitivity via Regulating miR-148b in Breast Cancer. Cell Biol Int (2018) 42:227–36. doi: 10.1002/cbin.10890 [DOI] [PubMed] [Google Scholar]
- 92. Zhu J, Ye Q, Chang L, Xiong W, He Q, Li W. Upregulation of miR-195 Enhances the Radiosensitivity of Breast Cancer Cells Through the Inhibition of BCL-2. Int J Clin Exp Med (2015) 8:9142–8. doi: 10.1007/s12038-019-9918-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhang J, Cui Y, Lin X, Zhang G, Li Z. MiR-122-3p Sensitizes Breast Cancer Cells to Ionizing Radiation via Controlling of Cell Apoptosis, Migration and Invasion. Int J Clin Exp Pathol (2017) 10:215–23. [Google Scholar]
- 94. Doherty J, Baehrecke EH. Life, Death and Autophagy. Nat Cell Biol (2018) 20:1110–7. doi: 10.1038/s41556-018-0201-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kim KH, Lee M-S. Autophagy—a Key Player in Cellular and Body Metabolism. Nat Rev Endocrinol (2014) 10:322–37. doi: 10.1038/nrendo.2014.35 [DOI] [PubMed] [Google Scholar]
- 96. Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-Eating and Self-Killing: Crosstalk Between Autophagy and Apoptosis. Nat Rev Mol Cell Biol (2007) 8:741–52. doi: 10.1038/nrm2239 [DOI] [PubMed] [Google Scholar]
- 97. D’Arcy MS. Cell Death: A Review of the Major Forms of Apoptosis, Necrosis and Autophagy. Cell Biol Int (2019) 43:582–92. doi: 10.1002/cbin.11137 [DOI] [PubMed] [Google Scholar]
- 98. Yi H, Liang B, Jia J, Liang N, Xu H, Ju G, et al. Differential Roles of miR-199a-5p in Radiation-Induced Autophagy in Breast Cancer Cells. FEBS Lett (2013) 587:436–43. doi: 10.1016/j.febslet.2012.12.027 [DOI] [PubMed] [Google Scholar]
- 99. Sun Q, Liu T, Yuan Y, Guo Z, Xie G, Du S, et al. MiR-200c Inhibits Autophagy and Enhances Radiosensitivity in Breast Cancer Cells by Targeting UBQLN1. Int J Cancer (2015) 136:1003–12. doi: 10.1002/ijc.29065 [DOI] [PubMed] [Google Scholar]
- 100. Luo J, Chen J, He L. Mir-129-5p Attenuates Irradiation-Induced Autophagy and Decreases Radioresistance of Breast Cancer Cells by Targeting HMGB1. Med Sci Monit (2015) 21:4122–9. doi: 10.12659/MSM.896661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Foster I. Cancer: A Cell Cycle Defect. Radiography (2008) 14:144–9. doi: 10.1016/j.radi.2006.12.001 [DOI] [Google Scholar]
- 102. Barnum KJ, O’Connell MJ. Cell Cycle Regulation by Checkpoints. Methods Mol Biol (2014) 1170:29–40. doi: 10.1007/978-1-4939-0888-2_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kastan MB, Bartek J. Cell-Cycle Checkpoints and Cancer. Nature (2004) 432:316–23. doi: 10.1038/nature03097 [DOI] [PubMed] [Google Scholar]
- 104. Zhang N, Zeng X, Sun C, Guo H, Wang T, Wei L, et al. LncRNA LINC00963 Promotes Tumorigenesis and Radioresistance in Breast Cancer by Sponging miR-324-3p and Inducing ACK1 Expression. Mol Ther Nucleic Acids (2019) 18:871–81. doi: 10.1016/j.omtn.2019.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Rokudai S, Li Y, Otaka Y, Fujieda M, Owens DM, Christiano AM, et al. STXBP4 Regulates APC/C-Mediated P63 Turnover and Drives Squamous Cell Carcinogenesis. Proc Natl Acad Sci USA (2018) 115:E4806–14. doi: 10.1073/pnas.1718546115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang F, Wang W, Lu L, Xie Y, Yan J, Chen Y, et al. MicroRNA-16-5p Regulates Cell Survival, Cell Cycle and Apoptosis by Targeting AKT3 in Prostate Cancer Cells. Oncol Rep (2020) 44:1282–92. doi: 10.3892/or.2020.7682 [DOI] [PubMed] [Google Scholar]
- 107. Ruan L, Qian X. MiR-16-5p Inhibits Breast Cancer by Reducing AKT3 to Restrain NF-κB Pathway. Biosci Rep (2019) 39(8):20191611. doi: 10.1042/BSR20191611 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 108. Talvinen K, Karra H, Pitkänen R, Ahonen I, Nykänen M, Lintunen M, et al. Low Cdc27 and High Securin Expression Predict Short Survival for Breast Cancer Patients. APMIS (2013) 121:945–53. doi: 10.1111/apm.12110 [DOI] [PubMed] [Google Scholar]
- 109. Ren YQ, Fu F, Han J. MiR-27a Modulates Radiosensitivity of Triple- Negative Breast Cancer (TNBC) Cells by Targeting CDC27. Med Sci Monit (2015) 21:1297–303. doi: 10.12659/MSM.893974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yang B, Kuai F, Chen Z, Fu D, Liu J, Wu Y, et al. MiR-634 Decreases the Radioresistance of Human Breast Cancer Cells by Targeting STAT3. Cancer Biother Radiopharm (2020) 35:241–8. doi: 10.1089/cbr.2019.3220 [DOI] [PubMed] [Google Scholar]
- 111. Fabris L, Berton S, Citron F, D’Andrea S, Segatto I, Nicoloso MS, et al. Radiotherapy-Induced miR-223 Prevents Relapse of Breast Cancer by Targeting the EGF Pathway. Oncogene (2016) 35:4914–26. doi: 10.1038/onc.2016.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Metheetrairut C, Adams BD, Nallur S, Weidhaas JB, Slack FJ. Cel-Mir-237 and its Homologue, hsa-miR-125b, Modulate the Cellular Response to Ionizing Radiation. Oncogene (2017) 36:512–24. doi: 10.1038/onc.2016.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang L, Yuan C, Lv K, Xie S, Fu P, Liu X, et al. Lin28 Mediates Radiation Resistance of Breast Cancer Cells via Regulation of Caspase, H2A.X and Let-7 Signaling. PloS One (2013) 8:e67373. doi: 10.1371/journal.pone.0067373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhang X, Li Y, Wang D, Wei X. miR-22 Suppresses Tumorigenesis and Improves Radiosensitivity of Breast Cancer Cells by Targeting Sirt1. Biol Res (2017) 50:27. doi: 10.1186/s40659-017-0133-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lee HC, Her N-G, Kang D, Jung SH, Shin J, Lee M, et al. Radiation-Inducible miR-770-5p Sensitizes Tumors to Radiation Through Direct Targeting of PDZ-Binding Kinase. Cell Death Dis (2017) 8:e2693–3. doi: 10.1038/cddis.2017.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Fu Y, Xiong J. MicroRNA-124 Enhances Response to Radiotherapy in Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer Cells by Targeting Signal Transducer and Activator of Transcription 3. Croat Med J (2016) 57:457–64. doi: 10.3325/cmj.2016.57.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Perez-Añorve IX, Gonzalez-De la Rosa CH, Soto-Reyes E, Beltran-Anaya FO, Del Moral-Hernandez O, Salgado-Albarran M, et al. New Insights Into Radioresistance in Breast Cancer Identify a Dual Function of miR-122 as a Tumor Suppressor and oncomiR. Mol Oncol (2019) 13:1249–67. doi: 10.1002/1878-0261.12483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kato M, Paranjape T, Ullrich R, Nallur S, Gillespie E, Keane K, et al. The Mir-34 microRNA Is Required for the DNA Damage Response In Vivo in C. Elegans and In Vitro in Human Breast Cancer Cells. Oncogene (2009) 28:2419–24. doi: 10.1038/onc.2009.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wolfe AR, Bambhroliya A, Reddy JP, Debeb BG, Huo L, Larson R, et al. MiR-33a Decreases High-Density Lipoprotein-Induced Radiation Sensitivity in Breast Cancer. Int J Radiat Oncol Biol Phys (2016) 95:791–9. doi: 10.1016/j.ijrobp.2016.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zhang S, Wang B, Xiao H, Dong J, Li Y, Zhu C, et al. LncRNA HOTAIR Enhances Breast Cancer Radioresistance Through Facilitating HSPA1A Expression via Sequestering miR-449b-5p. Thorac Cancer (2020) 11:1801–16. doi: 10.1111/1759-7714.13450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Huang X, Taeb S, Jahangiri S, Korpela E, Cadonic I, Yu N, et al. miR-620 Promotes Tumor Radioresistance by Targeting 15-Hydroxyprostaglandin Dehydrogenase (HPGD). Oncotarget (2015) 6:22439–51. doi: 10.18632/oncotarget.4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Shi R, Wu P, Liu M, Chen B, Cong L. Knockdown of lncRNA PCAT6 Enhances Radiosensitivity in Triple-Negative Breast Cancer Cells by Regulating miR-185-5p/TPD52 Axis. Onco Targets Ther (2020) 13:3025–37. doi: 10.2147/OTT.S237559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lin LC, Lee H, Chien PJ, Huang YH, Chang MY, Lee YC, et al. Nad(P)h:Quinone Oxidoreductase 1 Determines Radiosensitivity of Triple Negative Breast Cancer Cells and Is Controlled by Long non-Coding Rna Neat1. Int J Med Sci (2020) 17:2214–24. doi: 10.7150/ijms.45706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Luo M, Ding L, Li Q, Yao H. miR-668 Enhances the Radioresistance of Human Breast Cancer Cell by Targeting IκBα. Breast Cancer (2017) 24:673–82. doi: 10.1007/s12282-017-0756-1 [DOI] [PubMed] [Google Scholar]
- 125. Ma JH, Qin L, Li X. Role of STAT3 Signaling Pathway in Breast Cancer. Cell Commun Signal (2020) 18:1–13. doi: 10.1186/s12964-020-0527-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of Epidermal Growth Factor Receptor in Breast Cancer. Breast Cancer Res Treat (2012) 136:331–45. doi: 10.1007/s10549-012-2289-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wu Q, Wu W, Fu B, Shi L, Wang X, Kuca K. JNK Signaling in Cancer Cell Survival. Med Res Rev (2019) 39:2082–104. doi: 10.1002/med.21574 [DOI] [PubMed] [Google Scholar]
- 128. Duffy MJ, Synnott NC, Crown J. Mutant P53 as a Target for Cancer Treatment. Eur J Cancer (2017) 83:258–65. doi: 10.1016/j.ejca.2017.06.023 [DOI] [PubMed] [Google Scholar]
- 129. Raychaudhuri P, Park HJ. FoxM1: A Master Regulator of Tumor Metastasis. Cancer Res (2011) 71:4329–33. doi: 10.1158/0008-5472.CAN-11-0640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zona S, Bella L, Burton MJ, Nestal de Moraes G, Lam EWF. FOXM1: An Emerging Master Regulator of DNA Damage Response and Genotoxic Agent Resistance. Biochim Biophys Acta - Gene Regul Mech (2014) 1839:1316–22. doi: 10.1016/j.bbagrm.2014.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Sharma B, Agnihotri N. Role of Cholesterol Homeostasis and its Efflux Pathways in Cancer Progression. J Steroid Biochem Mol Biol (2019) 191:105377. doi: 10.1016/j.jsbmb.2019.105377 [DOI] [PubMed] [Google Scholar]
- 132. Balzeau J, Menezes MR, Cao S, Hagan JP. The LIN28/let-7 Pathway in Cancer. Front Genet (2017) 8:31. doi: 10.3389/fgene.2017.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Lee H, Kim KR, Noh SJ, Park HS, Kwon KS, Park BH, et al. Expression of DBC1 and SIRT1 Is Associated With Poor Prognosis for Breast Carcinoma. Hum Pathol (2011) 42:204–13. doi: 10.1016/j.humpath.2010.05.023 [DOI] [PubMed] [Google Scholar]
- 134. Liu T, Liu PY, Marshall GM. The Critical Role of the Class III Histone Deacetylase SIRT1 in Cancer. Cancer Res (2009) 69:1702–5. doi: 10.1158/0008-5472.CAN-08-3365 [DOI] [PubMed] [Google Scholar]
- 135. Santolla MF, Avino S, Pellegrino M, De Francesco EM, De Marco P, Lappano R, et al. SIRT1 Is Involved in Oncogenic Signaling Mediated by GPER in Breast Cancer. Cell Death Dis (2015) 6:1–12. doi: 10.1038/cddis.2015.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Ayllón V, O’Connor R. PBK/TOPK Promotes Tumour Cell Proliferation Through P38 MAPK Activity and Regulation of the DNA Damage Response. Oncogene (2007) 26:3451–61. doi: 10.1038/sj.onc.1210142 [DOI] [PubMed] [Google Scholar]
- 137. Dougherty JD, Garcia ADR, Nakano I, Livingstone M, Norris B, Polakiewicz R, et al. PBK/TOPK, a Proliferating Neural Progenitor-Specific Mitogen-Activated Protein Kinase Kinase. J Neurosci (2005) 25:10763–85. doi: 10.1523/JNEUROSCI.3207-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Joel M, Mughal AA, Grieg Z, Murrell W, Palmero S, Mikkelsen B, et al. Targeting PBK/TOPK Decreases Growth and Survival of Glioma Initiating Cells In Vitro and Attenuates Tumor Growth In Vivo . Mol Cancer (2015) 14:1–15. doi: 10.1186/s12943-015-0398-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Vostakolaei MA, Hatami-Baroogh L, Babaei G, Molavi O, Kordi S, Abdolalizadeh J. Hsp70 in Cancer: A Double Agent in the Battle Between Survival and Death. J Cell Physiol (2021) 236:3420–44. doi: 10.1002/jcp.30132 [DOI] [PubMed] [Google Scholar]
- 140. Clara JA, Monge C, Yang Y, Takebe N. Targeting Signalling Pathways and the Immune Microenvironment of Cancer Stem Cells — a Clinical Update. Nat Rev Clin Oncol (2020) 17:204–32. doi: 10.1038/s41571-019-0293-2 [DOI] [PubMed] [Google Scholar]
- 141. Gupta SC, Kunnumakkara AB, Aggarwal S, Aggarwal BB. Inflammation, a Double-Edge Sword for Cancer and Other Age-Related Diseases. Front Immunol (2018) 9:2160. doi: 10.3389/fimmu.2018.02160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Aggarwal BB. Nuclear Factor-κB. Cancer Cell (2004) 6:203–8. doi: 10.1016/j.ccr.2004.09.003 [DOI] [PubMed] [Google Scholar]