Abstract
The HMG-I/Y gene encodes the HMG-I and HMG-Y proteins, which function as architectural chromatin binding proteins important in the transcriptional regulation of several genes. Although increased expression of the HMG-I/Y proteins is associated with cellular proliferation, neoplastic transformation, and several human cancers, the role of these proteins in the pathogenesis of malignancy remains unclear. To better understand the role of these proteins in cell growth and transformation, we have been studying the regulation and function of HMG-I/Y. The HMG-I/Y promoter was cloned, sequenced, and subjected to mutagenesis analysis. A c-Myc–Max consensus DNA binding site was identified as an element important in the serum stimulation of HMG-I/Y. The oncoprotein c-Myc and its protein partner Max bind to this site in vitro and activate transcription in transfection experiments. HMG-I/Y expression is stimulated by c-Myc in a Myc-estradiol receptor cell line in the presence of the protein synthesis inhibitor cycloheximide, indicating that HMG-I/Y is a direct c-Myc target gene. HMG-I/Y induction is decreased in Myc-deficient fibroblasts. HMG-I/Y protein expression is also increased in Burkitt's lymphoma cell lines, which are known to have increased c-Myc protein. Like Myc, increased expression of HMG-I protein leads to the neoplastic transformation of both Rat 1a fibroblasts and CB33 cells. In addition, Rat 1a cells overexpressing HMG-I protein form tumors in nude mice. Decreasing HMG-I/Y proteins using an antisense construct abrogates transformation in Burkitt's lymphoma cells. These findings indicate that HMG-I/Y is a c-Myc target gene involved in neoplastic transformation and a member of a new class of potential oncogenes.
The myc family of oncogenes include c-myc, N-myc, and l-myc (17, 18, 20, 22, 23, 29, 65, 72, 83). The first identified member of the family, v-myc, has been shown to be sufficient for the induction of avian myelocytomatosis, a syndrome that includes leukemias and sarcomas (96). c-myc is the best characterized of the myc genes and has been implicated in the control of normal cell growth, neoplastic transformation, and apoptosis (17, 18, 20, 22, 23, 29, 65, 72, 83). Aberrant expression of c-myc appears to play an important role in the pathogenesis of several human malignancies, most notably Burkitt's lymphoma, in which a translocation event causes deregulated, constitutive c-myc expression (17, 18, 22, 23, 72, 81). Increased c-myc expression has also been identified in numerous other malignancies, including renal cell, colon, ovarian, lung, and breast carcinoma (20, 22, 72). In addition, Rat 1a fibroblasts (56, 84, 86) and CB33 cells (46, 63) are transformed by stable transfection with a plasmid expressing c-myc alone. Because of its prominent role in neoplasia, the c-Myc oncoprotein has been extensively studied, although the precise molecular basis for c-Myc activity remains unclear.
The c-Myc protein functions as a transcription factor that acts in conjunction with its protein partner, Max (2, 11, 12, 21, 54, 55). After dimerization with Max, Myc-Max heterodimers bind with high affinity to the E-box motif CACGTG, presumably in cis-acting elements of genes involved in regulating cell growth (48, 57, 76). To date, few transcriptional targets of c-Myc have been identified (18, 20, 29, 45). Among the putative c-Myc target genes are those encoding ornithine decarboxylase (ODC) (7, 74, 75), carbamoyl-phosphate synthase–aspartate carbamoyltransferase-dihydroorotase (CAD) (13, 69), prothymosin α (26, 27, 35), cdc25A (32), eukaryotic translation initiation factor 4E (eIF-4E) (53, 78), eIF-2α (78), ECA39 (9, 80), LDH-A (82), Rcl (60), MrDb (44), telomerase (97), H-ferritin (100), IRP2 (100), and tumor-associated membrane protein (Tmp) (8). Some of these genes appear to be required for cellular proliferation, such as ODC (4, 5, 74), which encodes an essential enzyme involved in polyamine biosynthesis. ODC also appears to be essential for Myc-mediated apoptosis and displays oncogenic properties (4, 5, 7, 74, 75). The telomerase gene (97), rcl (60), LDH-A (82), cdc25A (32, 33), and Tmp (8) appear to participate in transformation. The cad product is required for DNA synthesis, although no oncogenic properties have been described (13, 69). cad gene expression also decreases in myc-null cells (68). Downregulation of the H-ferritin gene appears to be necessary for Myc-mediated transformation (100). The precise role of these and other putative Myc target genes in mediating Myc function is only beginning to emerge. Thus, the identification and characterization of c-Myc targets should provide insight into c-Myc function in both normal and neoplastic cell growth.
The HMG-I/Y proteins were originally identified as basic, nonhistone, chromosome binding proteins that are encoded by alternately spliced products of the HMG-I/Y gene (31, 50, 51). Recent studies indicate an important role for HMG-I/Y proteins in regulating gene expression (25, 30, 66, 87, 91, 92, 93, 101). HMG-I/Y relieves histone H1-mediated repression of transcription (87, 101). Moreover, HMG-I/Y has been found to be essential for the viral induction of the beta interferon gene (25, 91, 92, 93). Although the HMG-I/Y proteins do not have transcriptional activity alone, through protein-protein and protein-DNA interactions, they organize the framework of a nuclear protein-DNA transcriptional complex. Because these proteins alter the conformation of DNA, they have been termed architectural transcription factors.
Like c-myc, expression of HMG-I/Y also correlates with rapidly proliferating mammalian tissues as well as neoplastic transformation (15, 16, 38, 39, 40, 41, 42, 59, 64, 77, 89, 90, 95). In fibroblasts stimulated by serum or growth factors, HMG-I/Y is a delayed-early gene whose expression follows that of c-myc, with peak expression at 7.5 to 20 h (59, 99). Elevated expression of HMG-I/Y proteins has been observed in several mammalian cancers, including high-grade human prostatic cancer (14, 89, 90) and malignant thyroid cancer in rats and humans (10, 15, 38, 39, 40). Elevated HMG-I/Y expression is also associated with the ability of rat prostatic cell lines to metastasize and has been proposed as a possible diagnostic marker for the metastatic potential of prostatic cancer cells in humans (14). A correlation between expression of HMG-I/Y and progressive transformation in mouse mammary epithelial cells has also been reported (77). Interestingly, HMG-I/Y has been localized to the short arm of chromosome 6 in a region known to be involved in rearrangements, translocations, and other abnormalities correlated with human cancer (31, 50, 51). Although previous studies have shown that HMG-I/Y expression is correlated with neoplastic transformation, the basis for the elevated expression and the biologic consequences of the enhanced expression has been unknown.
To better understand the potential role of the HMG-I/Y gene products in cell growth and neoplasia, we have been studying the transcriptional regulation of HMG-I/Y. In this paper, we show that HMG-I/Y is a direct c-Myc target gene. Like c-Myc, HMG-I/Y proteins are also increased in Burkitt's lymphoma. Serum induction of HMG-I/Y mRNA is blunted in myc null fibroblasts. Ectopic expression of HMG-I leads to the neoplastic transformation of Rat 1a fibroblasts and CB33 cell lines in a manner indistinguishable from that of c-Myc. In addition, fibroblasts with increased HMG-I form tumors in nude mice. Decreasing HMG-I/Y proteins using an antisense approach abrogates transformation in Burkitt's lymphoma cells. Our findings suggest that HMG-I/Y is an important c-Myc target gene involved in neoplastic transformation and a potential human oncogene.
MATERIALS AND METHODS
Cell culture and transfection.
NIH 3T3 cells were maintained as described before (69). NIH 3T3 cells were used for transfection experiments because HMG-I/Y was cloned from BALB/c3T3 fibroblasts as a delayed-early gene; NIH 3T3 cells are similar to BALB/c3T3 cells, with the advantage that they can be transfected with higher efficiency. In addition, previous studies evaluating Myc-responsive genes have used these cells (44, 69). For initial transfection experiments evaluating the stimulation of the HMG-I/Y reporter constructs by basic fibroblast growth factor (bFGF), platelet-derived growth factor (PDGF), and serum, 10 μg of reporter plasmid and 10 μg of YNLacZ, a plasmid expressing β-galactosidase to control for transfection efficiency, were transfected as previously described (3, 79, 94). Calcium phosphate-DNA precipitates were incubated overnight, after which cells were washed and placed in 0.5% fetal bovine serum (FBS) for 48 h. Cells were then washed and stimulated with 20% FBS, 100 ng of BB-PDGF (human recombinant; Collaborative Research) per ml, or 80 ng of recombinant, basic human FGF (Collaborative Research) per ml in Dulbecco's modified Eagle's medium (DMEM). Duplicate 100-μl aliquots of medium were then assayed for growth hormone (GH) at the indicated times according to the manufacturer's instructions (Nichols Institute). At 25 to 30 h, cells were harvested for β-galactosidase assays as described before (3, 79); β-galactosidase activity was used to normalize all GH results for transfection efficiency. Relative induction was determined by dividing the quantity of GH at 25 to 30 h by the quantity of GH at the time of stimulation (0 h).
For transfections comparing the relative activities of each HMG-I/Y reporter plasmid, 5 μg of the HMG-I/Y reporter plasmid and 5 μg of control plasmid expressing β-galactosidase were transfected into NIH 3T3 cells (5 × 105) in 5-cm dishes in quadruplicate as described above. After transfection, cells were placed in Eagle's minimal essential medium (MEM) containing 0.1% FBS for 25 to 30 h. Cells were subsequently washed, and duplicate dishes were stimulated with 20% FBS in DMEM; the remaining duplicate dishes were maintained in 0.1% FBS in DMEM. At 25 to 30 h, the medium was assayed for GH, and cells were harvested for β-galactosidase activity as described before (3, 79). The GH activity in cells maintained in 0.1% FBS was subtracted from the GH activity of cells stimulated with 20% FBS.
For experiments determining the transactivation potential of c-Myc and Max, 3 μg of the reporter constructs, 5 μg of control plasmid expressing β-galactosidase, and various amounts of plasmids expressing Myc, Max, or vector alone (Rous Sarcoma virus [RSV]) were cotransfected, with the total quantity of DNA kept constant at 18 μg. After transfection, cells were placed in 10% FBS in DMEM for 24 to 26 h. The medium was then assayed for GH, and cells were harvested for β-galactosidase activity.
To determine if the HMG-I/Y gene is transcriptionally activated by c-Myc, we used a previously described Rat 1a cell line expressing a Myc-estradiol receptor fusion protein (Myc-ER) that is activated by the addition of hydroxytamoxifen to the growth medium (26, 27, 43, 62). Rat 1a-Myc-ER cells lines were grown to confluency in DMEM with 10% FBS. Cells were made quiescent by growing them to confluency and maintaining them at approximately 100% confluency for 48 h in 0.1% FBS. Cells were then stimulated with hydroxytamoxifen at 200 nM for the indicated time periods. To determine if HMG-I/Y is a direct c-Myc target gene, these cells were also treated with the protein synthesis inhibitor cycloheximide at 10 μM added 30 min before activation of c-Myc by hydroxytamoxifen. For Western analysis of the Myc-ER cells, the cells were made quiescent by starvation in DMEM supplemented with 0.1% FBS for 5 days. The extended starvation was used because the half-life of the HMG-I/Y mRNA and protein is estimated to be greater than 30 h (49; L. M. S. Resar, unpublished data). Cells were then stimulated with hydroxytamoxifen as described above. We also used two Rat-1-ER cell lines (a generous gift from L. Penn) expressing either wild-type Myc or a mutated c-Myc protein that lacks transcriptional, oncogenic, and apoptotic activity (MycΔ105-143-ER) (19). The wild-type and mutated Myc proteins are activated by the addition of β-estradiol or hydroxytamoxifen. The Rat-1 cells were made quiescent by growing cells to confluency and subsequent incubation in MEM without phenol red in 2% charcoal-treated FBS for 48 h. Cells were induced as described above. The Myc-deficient fibroblasts were maintained and serum stimulated as previously described (68).
The Rat 1a cells used for stable cell lines were maintained as previously described (47, 84). Cells were transfected with a plasmid expressing HMG-I (pSG5-HMG-I; 5 μg) and pBABE-puro (1 μg) for puromycin resistance (60) using lipofectin as described by the manufacturer (Gibco-BRL). Pooled resistant cell lines were selected in medium containing puromycin (0.75 μg/ml).
Burkitt's lymphoma and CB33 cells were grown and transfected as previously described (46, 63, 82).
Screening of murine genomic library.
A genomic library made from BALB/c3T3 cells was screened with probes derived from the murine HMG-I/Y cDNA as well as intronic sequences identified from the 5′ untranslated region. One positive clone that contained sequences corresponding to the coding region of HMG-I/Y as well as the published 5′ untranslated cDNA region was isolated. Additional sequences were identified and found to represent intronic sequences in the 5′ untranslated region. Given the existence of HMG-I/Y pseudogenes in human genomic DNA (31, 50, 51), the intronic sequences were also used as probes in order to distinguish the authentic murine HMG-I/Y promoter region from the intronless pseudogenes. A genomic Southern blot was performed using the genomic isolate to confirm the presence of a 7.8-kb BamHI fragment in the authentic murine clone as previously described (50). Phagemid DNA was prepared from genomic phage isolates and subjected to Southern blot analysis using either a carboxyl-terminal HMG-I/Y cDNA probe or a probe derived from an intron in the HMG-I/Y promoter region in order to establish a restriction map.
Cloning the HMG-I/Y promoter region.
A bacteriophage containing the HMG-I/Y cDNA sequences and genomic flanking sequences on the 5′ side was isolated from a BALB/c embryonic genomic DNA library using a cDNA probe and Southern blot analysis (59). A HindIII fragment of approximately 3.5 kb containing the 5′ end of the cDNA approximately 2 kb upstream from the transcription start and 1.5 kb downstream from the transcription start site was isolated and cloned into pBluescript II KS(−) phagemid (Stratagene). An additional 400-bp HindIII-Xho fragment, downstream of the transcription start, was cloned into the HMG-I/Y-pBluescript II KS(−) construct. The remaining 1-kb fragment from the Xho site up to but not including the translation start was obtained by PCR from a murine genomic library and cloned into pBluescript II KS(−). The following primers were used: sense primer, TCTGACCGAGTACTCGAGTTTGAAATCTCGTAA; antisense primer, AGTACGGTACCGTCGACTCTCCTTCTCTATGTGGGG. This 1-kb fragment was joined to the remaining HMG-I/Y promoter region fragment in pBluescript II KS(−) from the 4.6-kb HMG-I/Y-Bluescript II KS(−).
Plasmids.
The 4.6-kb HindIII-Kpn fragment from 4.6-kb HMG-I/Y-Bluescript II KS(−) containing approximately 2 kb upstream from the transcription start and 2.6 kb downstream from the transcription start site was cloned into the pOGH reporter plasmid (Nichols Institute) at the HindIII and Kpn sites and designated 4.6-kb HMG-I/Y-GH. The 4.6-kb HMG-I/Y-GH plasmid was modified by deleting a 2.6-kb BamHI fragment 76 bp downstream from the major transcription start. 5′ deletion mutations were generated using the following restriction sites: SacI (−1635), Pflm (−1476), BsaA1 or Pml (−1377). Additional 5′ deletions were made using exonuclease III (Erase-A-Base; Promega) as described in the kit directions.
The plasmid containing a mutated E-box (−1895 MT Myc-GH) was made using PCR and recombination site-specific mutagenesis as described before (52). The primers specific to pBluescript II KS(−) have been described (88). The primers used to mutate the c-Myc–Max site from CACGTG to CAGCTG (shown in boldface) were as follows: sense, ACCCCCACCGAGCAGCTGCTGCCCTGCGCCCA, and antisense, TGGGCGCAGGGCACCAGCTGCTCGGTGGGGGT. The mutated site was confirmed by sequencing, and the HMG-I/Y promoter fragment was shuttled into pBluescript II KS(−) plasmids and ultimately cloned into pOGH to yield −1895 MT Myc-GH. The −1895 MT Myc-GH construct was also sequenced in the region of the E-box to confirm the presence of the mutated site.
pMax, which carries human Max cDNA, and pMyc, which carries human c-Myc cDNA, have been described (6). The RSV vector was made by deleting the c-Myc coding sequences from pMyc by restriction digestion with SacI and HindIII. The remaining vector was treated with Klenow, and the resulting blunt ends were ligated.
pSG5-HMG-I was made by excision from pBS-HMG-I (66) using HincII and BamHI restriction and ligation to pSG5. Prior to ligation, pSG5 (Stratagene) underwent restriction digestion with EcoRI, Klenow treatment, and subsequent restriction with BamHI.
The HMG-I/Y antisense construct was made using the vector pU1/RIBOZYME (70), which incorporates an autocatalytic hammerhead ribozyme structure within the complementary sequence. The regions of complementarity are predicted to align the autocatalytic ribozyme structure with the consensus sequence (GUC) for ribozyme cleavage within the targeted HMG-I message. The HMG-I sense oligonucleotide sequence 5′ to the ribozyme structure was TGCTCCTCCTCCGAG; the HMG-I sense sequence 3′ to the ribozyme structure was TCCTGCGAGATGCCC. The parent vector pU1/RIBOZYME was used as a control vector.
For transfection of the CB33 cells, the pHEBoCMV vector was used as a control vector as previously described (82). pHEBoCMV-HMG-I was made by cleaving pBS-HMG-I (66) with HindIII and Not1 to release the HMG-I cDNA, which was then cloned into pHEBoCMV at the same restriction sites.
Primer extension and S1 nuclease assay.
The extension and assay were carried out as previously described (3, 81). The primer, a [γ-32P]ATP-labeled oligonucleotide complementary to the 24 nucleotides of the HMG-I/Y RNA in the 5′ untranslated region, 156 to 133 bp from the translation start had the sequence CGGTCGCAAATGCGGATCTGAAAC.
Protein preparation.
Plasmids expressing a polyhistidine-containing, truncated c-Myc expression vector, c-Myc340–439 (tMyc) and Max were expressed in Escherichia coli and purified as previously described (1, 55, 98). All proteins were stored in 10 mM Tris (pH 7.5)–100 mM NaCl–1 nM EDTA–1 mM dithiothreitol–20% glycerol. Protein purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described before (55, 98). Protein quantities were estimated using the Bio-Rad protein assay (Bio-Rad Laboratories) according to the manufacturer's directions.
DNA probes.
To determine if c-Myc and Max bind to the HMG-I/Y promoter E-box, a probe containing the HMG-I/Y E-box and flanking sequences was generated by annealing equimolar amounts of two 21-nucleotide complementary oligonucleotides, ACCGAGCACGTGCTGCCCTGC and GCAGGGCAGCACGTGCTCGGT, and designated the wild type (WT). Sequences shown in boldface are the consensus sequences for the E-box. The probe containing the mutated site (MT) has the sequence ACCGAGCAGCTGCTGCCCTGC; the mutated site is in boldface. A control probe containing the ornithine decarboxylase promoter E-box (ODC), which has previously been shown to bind c-Myc and Max, was also used (7). A 120-ng aliquot of double-stranded probe was 5′-end labeled with T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P]ATP. Labeled probes were separated from unincorporated nucleotides using NICK columns (Pharmacia Biotech) according to the manufacturer's instructions.
For Northern analysis, blots were probed with a carboxyl-terminal HMG-I/Y cDNA probe. cDNA from the human acidic ribosomal phosphoprotein PO (PO) (58, 67) was used as a control probe for sample loading.
Electrophoretic mobility shift assay.
Heterodimers of truncated Myc (tMyc) and Max were formed by incubating 3 μg of tMyc and 1 ng of Max at 43°C for 15 min (55, 98). The reaction conditions for complex formation, in a final volume of 30 μl, were 10 mM Tris-HCl (pH 7.4), 50 mM NaCl, 1 nM dithiothreitol, 1 mM EDTA, 12.5% glycerol, and 1 μg of sheared salmon sperm DNA, and 0.4 to 2 ng of labeled probe was added and allowed to incubate at room temperature for 20 min (61, 98). For competition experiments, unlabeled, double-stranded competitors and 1 μg of sheared salmon sperm DNA were added to give 2-fold, 10-fold, 50-fold, and 100-fold molar excesses of unlabeled competitor over labeled probe. Incubation with the sheared salmon sperm DNA and competitors was allowed to proceed at room temperature for 20 min. Labeled probe (0.4 to 2 ng) was then added and allowed to incubate for an additional 15 min at room temperature. Samples were analyzed as described before (98).
Northern analysis.
For Northern blot analysis, total RNA from the Myc-ER (19, 26, 27, 43, 62) or MycΔ105-143-ER (19) cell line was isolated using the guanidinium-phenol-chloroform extraction method (Trizol; Gibco-BRL). RNA samples (10 to 20 μg) were analyzed by electrophoresis through a 0.8% agarose gel. RNA was transferred and probed as previously described (3, 79). A PhosphorImager (Molecular Dynamics) was used to compare the radiolabeled signals. HMG-I/Y mRNA was normalized to expression of the control ribosomal protein PO mRNA (58, 67).
Western analysis.
For Western blot analysis of HMG-I/Y, total cell lysates collected from plates of exponentially growing cells were boiled in 2× Laemmli buffer, analyzed by SDS–15% PAGE, and subjected to Western analysis (3, 79) using a chicken polyclonal antibody raised against the amino terminus of HMG-I/Y (described below) diluted 1:200. For analysis of HMG-C, a rabbit polyclonal antibody raised against the amino terminus of HMG-C (described below) was diluted 1:500. The actin monoclonal antibody AC15 (Sigma Immunochemicals) was diluted 1:2,500 and used to control sample loading. An antibody to the Ki-67 antigen (DAKO Corporation), which labels proliferating cells, was diluted 1:50 and used as a control for cellular proliferation in the Western analysis of the Burkitt's cells and normal lymphocytes (36, 37, 85). Reactive proteins were detected by enhanced chemiluminescence (Amersham).
HMG-I/Y and HMG-C antibody preparation.
The amino-terminal 22-amino-acid coding sequence of HMG-I/Y was generated by PCR from BALB/c3T3 mouse cDNA libraries. The primers for HMG-I/Y were 5′-GCTCGGGATCCCCATGAGCGAGTCGGGCTCAAAGTCCA and 3′-GAGCCGGATCCTCAAGTCCCATCCTTTTCCTGTTTGGA. The amino-terminal 24-amino-acid coding sequence of HMG-C was also generated by PCR from BALB/c3T3 mouse cDNA libraries. The primers for HMG-C were 5′-GCTCGGGATCCCCATGAGCGCACGCGGTGAGGGCGCCG and 3′-GAGCCGGATCCTCATGGCACCGGGGCGGCAGGTTGTCC. The PCR products were cut with BamHI and cloned into the carboxyl-terminal region of glutathione-S-transferase (GST) via the BamHI site of vector pGEX-3X (Pharmacia). The orientation and sequence were confirmed by sequencing. The resultant fusion proteins were expressed in the bacterial strain BL-21(DE3) and purified by glutathione-Sepharose 4B chromatography (Pharmacia). The purified GST-HMG-I/Y protein was used to immunize chickens (HRP, Inc.). Chicken immunoglobulin Y (IgY) antibody was purified from egg yolk with the EGGstract kit (Promega). The purified GST-HMG-C(N) protein was used to immunize rabbits (HRP, Inc.).
DNA sequencing.
HMG-I/Y genomic isolates were cloned into pBluescript II KS(−), and both strands were sequenced by the dideoxynucleotide chain termination method (3, 79) and by automated sequencing. Sequence of GC-rich regions was confirmed using the chemical sequencing method of Maxam and Gilbert (3, 79).
Soft agar assay.
The soft agar assay was performed as previously described (82) except that 5 × 104 Rat 1a cells were suspended in 8 ml of 0.3% agarose and poured onto a 10-ml 0.7% agarose bed in 100-mm tissue culture dishes. Rat 1a colonies greater than 100 μm were counted after 3 to 4 weeks. For the Burkitt's and CB33 cells, the soft agar assay was performed as described (82) except that 105 cells were suspended in 8 ml of 0.3% agarose. Colonies greater than 1 mm were counted after 3 to 4 weeks. Burkitt's cells transfected with antisense constructs were incubated for 2 to 4 months and counted at 3 to 4 weeks as well as after 2 to 4 months.
Cellular growth rates.
The growth rates of the Rat 1a cells were determined as previously described (82). Cells were seeded at 104 into six separate 10-cm tissue culture dishes. Duplicate dishes were harvested every 24 h for 3 days, and the cells were counted. The growth rates of the Burkitt's cells were determined as described above except that cells were seeded at 105 in 1-cm tissue culture dishes and counted daily for 3 days. Growth rates of the CB33 cells were determined as described above except that cells were seeded at 5 × 105 in 2.5-cm tissue culture dishes and counted daily for 3 days.
Cell cycle analysis.
The cell cycle profiles of the various Rat 1a cell lines growing on top of soft agar were analyzed using bromodeoxyuridine-propidium iodine staining as previously described (60).
Tumorigenicity assays.
Tumorigenicity assays were performed as previously described (56) but with the following modifications. Rat 1a cells (107) were suspended in 200 μl of serum-free DMEM and injected subcutaneously into 6- to 8-week-old athymic nude mice (Ncr-nu mice; National Cancer Institute). Animals were monitored at periodic intervals for the appearance of tumors up to 50 to 55 days following injection.
Tumor pathologic examination.
Pathologic examination of the tumors was performed after tumors were fixed by immersion in Bouin's fixative. Tissues were routinely processed for paraffin embedment, sectioned at 5.0 μm, and stained with hematoxylin and eosin (H & E).
RESULTS
Isolation, mapping, and sequence analysis of the HMG-I/Y promoter region.
Since previous work had documented the existence of several HMG-I/Y pseudogenes in the human genome (31, 50, 51), our efforts to clone the authentic murine HMG-I/Y promoter region included screening with a genomic isolate that contained introns in the 5′ untranslated region as well as the 3′ region. A single isolate that contained over 10 kb of sequence, including the coding region, the 5′ region upstream of the first exon, and the 3′ untranslated regions, was identified from a murine embryonic library (59). The murine HMG-I/Y promoter region was mapped using Southern analysis and ultimately sequenced. By primer extension and S1 nuclease analysis (data not shown), a major transcription start site and three minor transcription start sites were identified (Fig. 1A). Of note, multiple transcription start sites were also identified in the human HMG-I/Y promoter region in K562 cells (73). The murine start sites that we identified, however, differ slightly from the start sites identified in the human promoter sequences (73). These differences may be related to species-specific or cell type-specific differences.
FIG. 1.
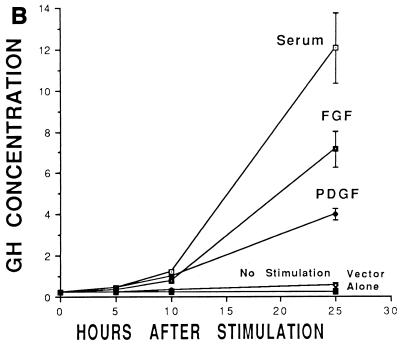
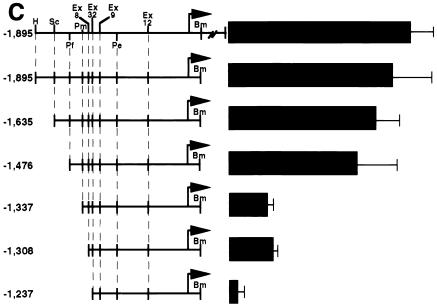
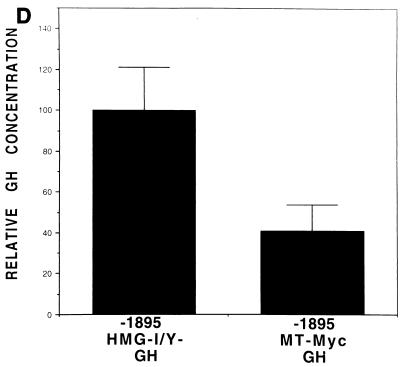
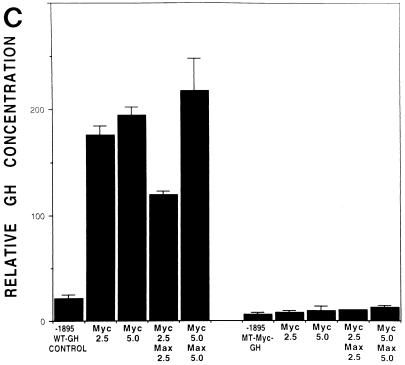
c-Myc–Max consensus DNA binding site is an important serum- or growth factor-responsive element in the HMG-I/Y promoter. (A) Restriction map of the 5′ noncoding region of the murine HMG-I/Y gene. The transcription start sites, E-box, and AP2 site are indicated. (B) Stimulation of HMG-I/Y promoter sequences by serum, FGF, and PDGF. NIH 3T3 cells were cotransfected with HMG-I/Y-GH- and β-galactosidase-expressing plasmids. Each time point represents the average for four samples; dishes were transfected in duplicate, and two aliquots of medium were taken from each dish at each time point. Error bars indicate standard deviations. Experiments were repeated three to five times with similar results. The GH concentration is expressed in micrograms per milliliter. (C) HMG-I/Y promoter mutational analysis. Progressive 5′ deletion mutations of the HMG-I/Y promoter construct are depicted on the left, and the relative GH activities are shown on the right. The full-length, wild-type promoter construct was assigned an activity of 100%; the activities of the deletion constructs are expressed as a percentage of the wild-type promoter construct activity. Transfections were performed in quadruplicate and repeated three to five times. The solid bar represents the average for three to five experiments; error bars indicate standard deviations. Restriction enzyme site abbreviations: H, HindIII; Sc, SacI; Pf, Pflm; Pm, Pml; Ps, PstI; Bm, BamHI. Ex, site of exonuclease digestion; the numbers are the original clone numbers. Note the significant drop (78%) in relative GH activity between constructs −1476 and −1337. A second decrease in activity is observed between −1308 and −1237. (D) Decreased serum stimulation (60%) of the HMG-I/Y promoter with the E-box mutation (−1896 MT Myc-GH) relative to the wild-type HMG-I/Y promoter (−1896 HMG-I/Y–GH). A control plasmid expressing β-galactosidase and plasmids expressing either −1895 HMG-I/Y–GH or −1896 MT Myc-GH were cotransfected into NIH 3T3 cells. Cells were made quiescent by starvation in 0.1% MEM for 25 to 30 h and subsequently stimulated with 20% FBS for 25 to 30 h. Transfections were performed in quadruplicate, and experiments were repeated five times. The solid bars show the mean values from five different experiments; error bars show the standard deviations. As before, the wild-type promoter construct was assigned an activity of 100%. Note that the E-box mutation decreases the serum responsiveness of the HMG-I/Y promoter by 60%.
Serum- and growth factor-dependent responsiveness of the HMG-I/Y promoter sequences.
Because HMG-I/Y is a delayed-early gene whose expression peaks about 15 h after serum or growth factor stimulation, we sought to identify the cis-acting elements that are required for this stimulation. To determine whether nucleotide sequences upstream of the HMG-I/Y gene confer serum and growth factor responsiveness, a cloned fragment of the murine HMG-I/Y genomic DNA containing start site 1 (−1,895 bp from the major transcription start site) up to the translation start site (approximately +2,600 bp) was ligated upstream of a reporter gene (HMG-I/Y–GH) expressing human GH and transfected into NIH 3T3 cells. The time course of reporter gene activation was measured after stimulation of serum-deprived cells with 20% FBS, PDGF, or bFGF. We found that the 4.6-kb HMG-I/Y promoter region was significantly induced by serum (Fig. 1B). There was 2-fold-higher induction at 5 h, 5-fold-higher induction at 10 h, and about 50-fold-higher induction at 24 h. Of note, induction of HMG-I/Y mRNA begins at 2.5 to 5 h following serum or growth factor stimulation, and HMG-I/Y mRNA levels remain elevated for at least 15 to 36 h (59, 99). In addition, the half-life of HMG-I/Y mRNA is believed to be greater than 30 h (49). Thus, the increase in secreted GH protein would be expected to follow the HMG-I/Y mRNA induction as observed here. The HMG-I/Y promoter construct was also induced by PDGF and bFGF. Neither the GH vector alone after stimulation with serum nor the 4.6-kb HMG-I/Y-GH construct in the absence of serum showed significantly enhanced GH secretion. We conclude that the region between −1895 and +2600 from the predominant transcription start site (start site 1) contains sequences responsible for the serum or growth factor induction of the HMG-I/Y promoter.
Identification of a serum growth factor-responsive element in the HMG-I/Y promoter.
To identify the elements mediating the serum- or growth factor-dependent stimulation of the HMG-I/Y promoter, deletional analysis was undertaken (Fig. 1C). First, most of the 5′ untranslated region 76 bp downstream from the transcription start at the BamHI site up to the translation start was deleted, creating −1896 HMG-I/Y–GH, and found to have no impact on the serum responsiveness of the remaining sequences. We therefore used this plasmid to generate subsequent HMG-I/Y promoter constructs with deletion mutations. Progressive 5′ deletions showed that there is a significant decrease (78%) in the serum stimulation of the HMG-I/Y–GH construct after deletion of the region between bp −1475 and −1337. This region contains putative sites for known transcription factors, including an E-box at bp −1337 that represents a consensus DNA binding site for the c-Myc transcription factor and its protein partner Max (Fig. 1A) (11, 12, 48, 57).
Since c-Myc is a transcription factor whose gene expression is known to precede that of HMG-I/Y (59, 71, 99), the contribution of this E-box to the serum induction of the HMG-I/Y promoter was further investigated. First, this site was mutated from CACGTG to CAGCTG, a mutation that has been shown to abrogate c-Myc and Max binding (55). A construct analogous to −1895 HMG-I/Y–GH with the mutated c-Myc site (−1895 MT Myc-GH) was tested for serum responsiveness in transfection experiments. Serum responsiveness decreased 60% in the −1895 MT Myc-GH construct compared to the wild-type −1895 HMG-I/Y–GH (Fig. 1D), suggesting that this E-box is involved in the serum-dependent stimulation of the HMG-I/Y promoter.
c-Myc and Max recombinant proteins bind to the E-box in the HMG-I/Y promoter.
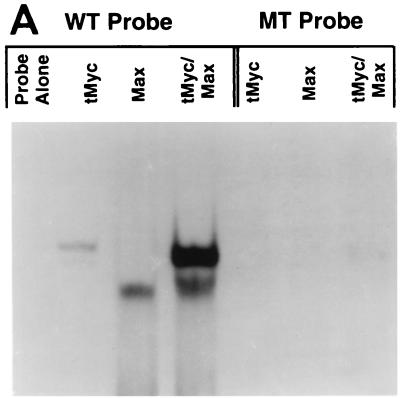
To determine if c-Myc and Max can bind to the E-box at position −1337 in the HMG-I/Y promoter region, electrophoretic mobility shift assay reactions were performed with an oligonucleotide containing the HMG-I/Y E-box (WT) and flanking sequences in the presence of recombinant Myc and Max proteins. Truncated c-Myc (c-Myc340–439 or tMyc) and Max were prepared as previously described (55, 98). These proteins have been shown to bind oligonucleotides containing the consensus Myc-Max site as tMyc and Max homodimers or tMyc-Max heterodimers. tMyc alone, Max alone, and tMyc-Max heterodimers bind to the WT HMG-I/Y E-box but not to the analogous nucleotide with the mutated E-box (MT oligonucleotide) (Fig. 2A). To prove that binding by tMyc and Max to the HMG-I/Y E-box was specific, competition electrophoretic mobility shift experiments were performed using unlabeled WT HMG-I/Y E-box probe or unlabeled MT E-box probe as competitors. The WT probe competes for binding of tMyc and Max to the HMG-I/Y E-box at a 2- and 10-fold molar excess over the probe, although the MT probe shows no competitive binding except when present at a 50- to 100-fold molar excess over the WT probe, indicating that tMyc and Max bind to the HMG-I/Y E-box specifically (Fig. 2B).
FIG. 2.
c-Myc and Max proteins bind specifically to the E-box in the HMG-I/Y promoter and activate transcription. (A) Binding of tMyc, Max, and heterodimers of tMyc and Max to the wild-type probe (WT) containing the core binding sequence CACGTG but not to the mutated E-box probe (MT) containing a double point mutation in the core sequence (CAGCTG). The first four lanes contain WT probe as follows: probe alone or with tMyc, Max, or tMyc and Max, respectively. The next four lanes contain the MT probe as follows: probe alone or with tMyc, Max, or tMyc and Max, respectively. (B) Specificity of binding of tMyc-Max to the WT probe. DNA sequence specificity was tested by comparative competitions with the indicated molar excess of unlabeled WT probe versus MT probe. Lanes contain labeled WT probe and heterodimers of tMyc and Max with no competitor or an increasing molar excess of the unlabeled WT or MT probe. Note that the WT but not the MT probe competes effectively at 2- and 10-fold molar excesses. (C) Plasmids expressing the control vector alone, c-Myc alone, or c-Myc and Max were cotransfected with the HMG-I/Y–GH promoter constructs containing the wild-type but not the mutated E-box HMG-I/Y promoter sequences by over 10-fold. Each bar shows the mean relative GH activity from four samples; error bars indicate the standard deviation. Transfections were performed in growing cells in duplicate, and two aliquots of medium were taken from each dish. Transfections were repeated three times with similar results. Note that c-Myc and Max transactivate the wild-type but not the mutated E-box HMG-I/Y promoter sequences by over 10-fold.
Activation of the HMG-I/Y promoter by c-Myc and Max.
To test whether c-Myc and Max can stimulate the expression of the HMG-I/Y promoter, cotransfection experiments with plasmids expressing c-Myc and Max were performed. Cotransfection of plasmids expressing c-Myc and Max resulted in transactivation of the wild-type HMG-I/Y promoter expression by over 10-fold (Fig. 2C). In contrast, cotransfection of plasmids expressing c-Myc and Max had no effect on the activity of HMG-I/Y promoter constructs with a mutated E-box, including −1895 MT Myc-GH (Fig. 2C) and −1337 HMG-I/Y–GH (data not shown), suggesting that transactivation of the HMG-I/Y promoter by c-Myc and Max is dependent upon c-Myc and Max binding to the E-box at position −1337. Of note, transfection of c-Myc alone was also able to transactivate expression of the HMG-I/Y promoter, presumably due to dimerization with endogenous Max in NIH 3T3 cells.
HMG-I/Y expression is stimulated by c-Myc.
To explore further whether the HMG-I/Y gene is transcriptionally activated by c-Myc, we used a previously described cell line expressing a Myc-estradiol receptor fusion protein (Myc-ER) (26, 27, 43, 62) that is activated by the addition of the estrogen analogue hydroxytamoxifen to the growth medium. Activation of Myc-ER in growing cells causes induction of HMG-I/Y expression (Fig. 3A and B). An increase in HMG-I/Y mRNA was detected 4 h after the addition of hydroxytamoxifen, peaking at 10 to 12 h at a level about 30 times higher than that found in uninduced cells (Fig. 3A and B). The same blot was probed with ribosomal protein PO (58, 67) to control for RNA loading in each lane (Fig. 3A and B). HMG-I/Y mRNA was also induced over 30-fold following exposure to hydroxytamoxifen in the presence of the protein synthesis inhibitor cycloheximide, indicating that HMG-I/Y is directly stimulated by c-Myc in these cells (Fig. 3A and B). HMG-I/Y mRNA increased minimally (1.6- to 2.8-fold) after incubation with cycloheximide alone (Fig. 3A and B). This pattern of induction following stimulation by hydroxytamoxifen and cycloheximide is similar to that observed for other putative direct c-Myc target genes, including ODC (7, 74, 75), the eIF-2α gene (78), the eIF-4E gene (78), the prothymosin α gene (26, 27), cdc25A (32), rcl (60), MrDb (44), IRP2 (100), and Tmp (8). The HMG-I/Y proteins also increased in the Myc-ER cells after activation of c-Myc (Fig. 3C).
FIG. 3.
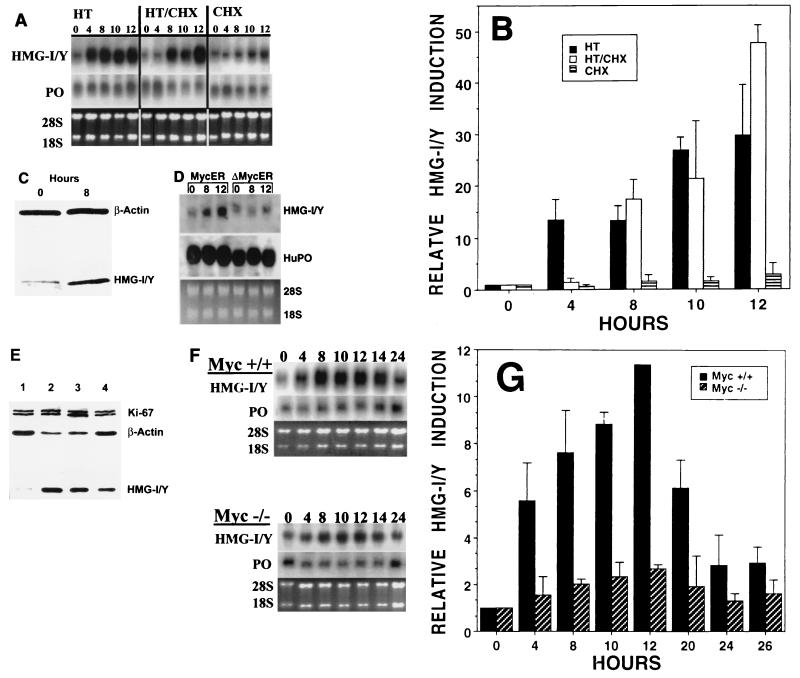
Direct stimulation of HMG-I/Y expression by c-Myc. (A) Northern blot analysis showing HMG-I/Y expression in a cell line expressing Myc-estradiol receptor fusion protein (Myc-ER) following incubation with the estrogen analogue hydroxytamoxifen (HT), HT and the protein synthesis inhibitor cycloheximide (CHX), or CHX alone. The ribosomal protein PO mRNA was used to control for sample loading. PO and the ethidium bromide-stained gel with the 28S and 18S rRNA bands are shown. Note the 30-fold induction of HMG-I/Y expression by 10 to 12 h after incubation in hydroxytamoxifen and activation of c-Myc. HMG-I/Y expression is also stimulated over 30-fold by c-Myc after incubation with HT and CHX for 10 to 12 h, indicating that HMG-I/Y is directly activated by c-Myc in these cells. HMG-I/Y expression changes minimally (1.6- to 2.8-fold) after incubation with cycloheximide alone. (B) Graphic representation of HMG-I/Y induction in Myc-ER cells following incubation with hydroxytamoxifen and cycloheximide. Experiments were repeated three to four times with similar results. The solid bars show the mean values from these experiments; error bars denote the standard deviations. Note that HMG-I/Y increases by over 30-fold after c-Myc activation in the presence of CHX, indicating that HMG-I/Y is a direct c-Myc target gene. (C) Western analysis showing induction of the HMG-I/Y protein after activation of Myc by incubation in hydroxytamoxifen for 8 h in the Myc-ER cells. β-Actin was used as a control for sample loading. (D) Northern analysis showing that HMG-I/Y expression is stimulated threefold following activation of wild-type c-Myc (Myc-ER), but not by a mutated c-Myc (ΔMyc-ER) that lacks transcriptional, transforming, and apoptotic activity in Rat-1 cells expressing the wild-type or mutated Myc-ER proteins. Cells were incubated with hydroxytamoxifen for the indicated time periods (in hours). PO was used to control for sample loading; the positions of human (Hu) PO, 28S, and 18S RNA are shown. (E) HMG-I/Y proteins are increased in Burkitt's lymphoma cells compared to EBV-transformed lymphocytes from healthy individuals. Western blot analysis of HMG-I/Y protein and controls Ki-67 and β-actin in Burkitt's lymphoma cell lines and EBV-transformed B lymphocytes from a healthy individual. Lanes: 1, healthy; 2 to 4, Burkitt's lymphoma cell lines: ST486, Ramos, and DW6, respectively. Note the marked increase in HMG-I/Y proteins in all Burkitt's lymphoma cell lines. (F) HMG-I/Y induction by serum is reduced in myc-null (Myc−/−) fibroblasts (68). Northern analysis showing that HMG-I/Y is stimulated 8.9- to 11.4-fold in wild-type (Myc+/+) fibroblasts, but only 1.6- to 2.7-fold in the Myc-deficient (Myc−/−) cells. (G) Graphic representation of the decreased HMG-I/Y induction in Myc-deficient cells. This experiment was repeated four times with similar results. The solid bars show the mean values from these experiments; error bars denote the standard deviations. These results are consistent with our findings that HMG-I/Y is a direct c-Myc target gene.
To further investigate the specificity of stimulation of HMG-I/Y expression by c-Myc, we determined whether HMG-I/Y expression could be induced by a mutated c-Myc protein that behaves in a dominant-negative fashion (MycΔ105–143) (19). This Myc mutant is nononcogenic (86) and lacks both transcriptional (54) and apoptotic (28) activity. We compared HMG-I/Y expression in Rat-1 cells expressing Myc-ER to that in Rat-1 cells expressing MycΔ105–143-ER after stimulation with hydroxytamoxifen. Our results show that HMG-I/Y expression increased only in the cells with wild-type c-Myc (Fig. 3D).
HMG-I/Y proteins are increased in Burkitt's lymphoma cells compared to normal lymphocytes.
Since HMG-I/Y is a direct c-Myc target gene, we examined Burkitt's lymphoma cell lines, which are known to have increased c-Myc protein (81, 82), to determine if HMG-I/Y proteins are also increased. As predicted, HMG-I/Y proteins are increased in all three Burkitt's lymphoma cell lines examined compared to Epstein-Barr virus (EBV)-transformed B lymphocytes from a normal individual, consistent with our finding that HMG-I/Y expression is stimulated by c-Myc (Fig. 3E). HMG-I/Y proteins are increased 5- to 10-fold in the Burkitt's cells compared to normal B lymphocytes using the proliferation-responsive antigen Ki-67 (36, 37, 85) to control for loading and 5- to 20-fold using β-actin to control for loading. Thus, HMG-I/Y proteins are significantly elevated in Burkitt's lymphoma cells, and this increase is not a result of increased cellular proliferation in these cells.
Serum induction of HMG-I/Y is decreased in Myc-deficient fibroblasts.
We also explored HMG-I/Y expression in Myc-deficient fibroblasts stimulated with serum compared to wild-type fibroblasts stimulated with serum (68). We observed that HMG-I/Y is induced 8.9- to 11.4-fold in wild-type (Myc+/+) fibroblasts 10 to 12 h after serum stimulation (Fig. 3F and G). In contrast, HMG-I/Y induction is significantly reduced to only 1.6- to 2.7-fold in Myc-deficient (Myc−/−) fibroblasts (Fig. 3F and G), providing additional evidence that HMG-I/Y is a direct c-Myc target gene.
Cells with increased HMG-I expression form transformed colonies in soft agar and tumors in athymic nude mice.
Because expression of both c-myc and HMG-I/Y is correlated with cell growth and neoplastic transformation, we hypothesized that HMG-I/Y may participate in c-Myc-mediated neoplastic transformation. To explore the potential role of HMG-I in neoplastic transformation, we constructed three different polyclonal Rat 1a cell lines overexpressing HMG-I to determine if ectopic expression of the HMG-I protein leads to transformation in Rat 1a cells. All three polyclonal Rat 1a cell lines overexpressing HMG-I formed colonies in soft agar in a manner analogous to Rat 1a-myc cells, a previously described polyclonal Rat 1a cell line overexpressing c-Myc (Fig. 4A and C) (47, 84, 86). Rat 1a cells that overexpress a mutated HMG-I protein that no longer binds DNA are not transforming (L. M. S. Resar, unpublished data). Cell cycle distribution and DNA-synthetic capability (bromodeoxyuridine incorporation) of Rat 1a-HMG-I, Rat 1a-myc, and control Rat 1a-pSG5 cells cultured adherently or nonadherently on top of soft agar were determined. We observed that all Rat 1a cell lines grew similarly when cultured adherently (data not shown). When cells were cultured nonadherently on top of soft agar for 48 h, the cell cycle profiles of the Rat 1a-myc and Rat 1a-HMG-I cells were similar (Fig. 4B). Both Rat 1a-myc and Rat 1a-HMG-I cells exhibited an increased percentage of cells in the S phase of the cell cycle (35 and 28%, respectively) compared to the control Rat 1a-pSG5 cells (5%), indicating that both Rat 1a-myc and Rat 1a-HMG-I cells grow in a transformed fashion on top of soft agar (Fig. 4B).
FIG. 4.
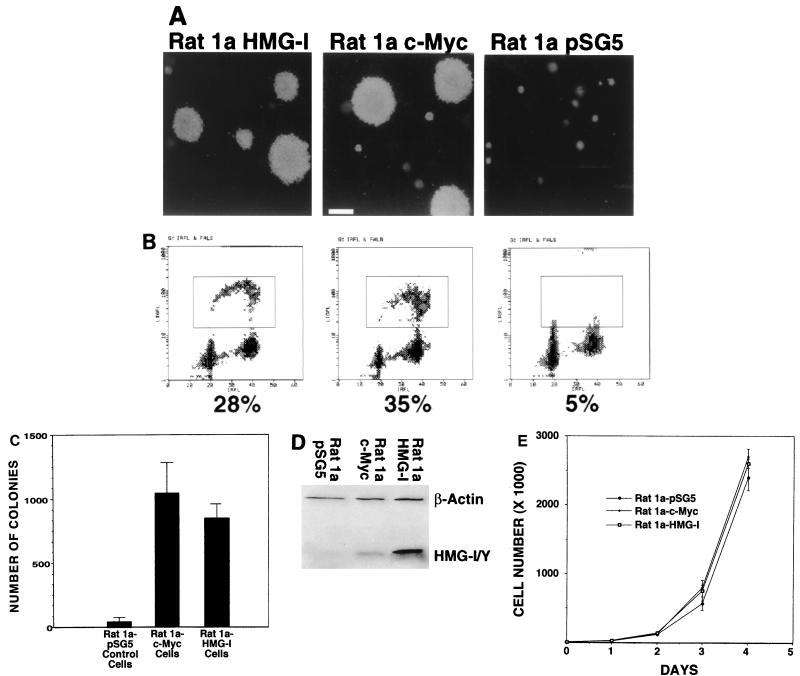
Rat 1a cells overexpressing HMG-I form colonies in the soft agar assay. (A) Rat 1a cells overexpressing HMG-I (Rat 1a-HMG-I) or c-Myc (Rat 1a-myc) or control Rat 1a cells transfected with the vector alone (Rat 1a pSG5) were subjected to analysis in the soft agar assay. Both Rat 1a-HMG-I and Rat 1a-myc cells formed colonies capable of anchorage-independent growth in the soft agar assay. Bar, 100 μm. (B) Rat 1a-HMG-I and Rat 1a-myc cells exhibit similar cell cycle profiles when grown on top of soft agar. This experiment was repeated three times with similar results. (C) The number of colonies formed by Rat 1a-HMG-I and Rat 1a-myc cells was similar. Assays were performed in duplicate, and the results are taken from two separate experiments. The solid bar represents the mean from two different experiments; the error bars indicate the standard deviation. (D) Rat 1a-HMG-I and Rat 1a-myc cells overexpress HMG-I protein. Western analysis shows that both Rat 1a-HMG-I and Rat 1a-myc cells overexpress HMG-I compared to control Rat 1a cells transfected with pSG5 vector alone. The lanes were blotted with the HMG-I/Y antibody as well as a β-actin antibody to control for sample loading. (E) Cell growth rates of Rat 1a cell lines. This experiment was performed with duplicate plates and repeated twice. The data points represent the average counts from duplicate plates; the error bars depict the standard deviations from a representative experiment. Note that all Rat 1a cell lines grow at similar rates.
Western analysis of the Rat 1a cell lines transfected with a plasmid expressing HMG-I compared to the Rat 1a cells transfected with pSG5 control vector alone show that the Rat 1a-HMG-I cells overexpress the HMG-I protein (Fig. 4D). In addition, the Rat 1a-myc cells also exhibit increased HMG-I/Y protein expression, further substantiating our finding that HMG-I/Y expression is stimulated by c-Myc (Fig. 4D). To determine if all the Rat 1a cell lines grow similarly in tissue culture, we performed growth curves for all the stable cell lines. All cell lines grew at a similar rate, indicating that the transformed phenotype of the Rat 1a cells overexpressing HMG-I was not a result of an increased growth rate (Fig. 4E).
To further explore the oncogenic properties of HMG-I, CB33 cells were also transfected with a plasmid expressing HMG-I. CB33-HMG-I cells formed transformed foci like CB33-Myc cells in the soft agar assay (46, 63). CB33-Myc cells are a previously described c-Myc-transformed human lymphoblastoid cell line (46, 63) (Fig. 5A and B). Western analysis shows that HMG-I is overexpressed in the CB33-HMG-I cells as well as in the C33-Myc cells; the latter result again supports our finding that HMG-I/Y is stimulated by c-Myc (Fig. 5C). All CB33 cell lines also grow at a similar rate, indicating that the transformed phenotype observed in the CB33-HMG-I cells was not a result of an increased growth rate (Fig. 5D). Thus, our results show that HMG-I has transforming activity similar to c-Myc in two different experimental cell lines, including Rat 1a and CB33 cells.
FIG. 5.
CB33 cells overexpressing HMG-I form colonies in the soft agar assay. (A) CB33 cells overexpressing HMG-I (CB33-HMG-I) or c-Myc (CB33-Myc) or control CB33 cells transfected with the vector alone (CB33-Control) were subjected to analysis in the soft agar assay. Both CB33-HMG-I and CB33-Myc cells formed colonies capable of anchorage-independent growth in the soft agar assay. (B) The number of colonies formed by the CB33-HMG-I and CB33-Myc cells was similar. Assays were performed in duplicate, and the results are taken from two or three separate experiments. The solid bar represents the mean from two or three different experiments; the error bars indicate the standard deviation. (C) CB33-HMG-I and CB33-Myc cells overexpress HMG-I protein. Western analysis shows that both CB33-HMG-I and CB33-Myc cells overexpress HMG-I compared to control CB33 cells transfected with vector alone. The lanes were blotted with the HMG-I/Y antibody as well as a β-actin antibody to control for sample loading. (D) Growth rates of the CB33 cell lines. This experiment was performed with quadruplicate cell counts taken per time period. The data points represent the average counts from two separate experiments; the error bars depict the standard deviations. Note that all CB33 cell lines grow at similar rates.
To verify the tumorigenic potential of the Rat 1a cells overexpressing HMG-I, we introduced these cells into athymic nude mice. Tumors were visible in four of five mice injected with Rat 1a-HMG-I cells (Fig. 6A). No tumors formed in the mice injected with Rat 1a cells transfected with vector alone. The tumors were visible by day 30, and the average size of the tumors was 10 mm by day 50 (Fig. 6D). The pathology results show that the tumors are fibrosarcomas (Fig. 6B and C), which is identical to tumors formed by Rat 1a-myc cells.
FIG. 6.
Rat 1a-HMG-I cells form tumors in nude mice. (A) Rat 1a-HMG-I, Rat 1a-myc, and control Rat 1a-pSG5 cells were injected into nude mice. Only mice injected with Rat 1a cells overexpressing HMG-I or c-Myc formed tumors. This photograph shows a representative mouse injected with Rat 1a-HMG-I cells. (B) Pathologic evaluation of the tumors showed that all tumors formed from Rat 1a-HMG-I or Rat 1a-myc cells were fibrosarcomas. This photograph shows a 12× magnification of the large subcutaneous tumor in a mouse injected with Rat 1a-HMG-I cells (H & E). (C) Tumor at 300× magnification. Note the bundles of spindle-shaped cells (H & E). (D) Characteristics of tumors from nude mice injected with Rat 1a-HMG-I and Rat 1a-myc cells.
Decreasing HMG-I/Y inhibits Myc-mediated transformation in Burkitt's lymphoma cells.
To determine if HMG-I/Y is involved in Myc-mediated transformation, an HMG-I antisense construct was made using a ribozyme expression vector (70). The antisense construct was transfected into Burkitt's lymphoma cells and resulted in a significant, specific decrease in HMG-I/Y protein expression (Fig. 7A). HMG-C protein, an HMG-I/Y family member encoded by a separate gene, was unaffected. Transformation was almost completely abrogated in these cells, suggesting that HMG-I/Y is important for transformation in these cells (Fig. 7B). The Burkitt's lymphoma cells with decreased HMG-I/Y also grow at a slower rate, which suggests an important role for HMG-I/Y in regulating cell growth (Fig. 7C). A similar diminution in growth rate has been reported for Myc-deficient fibroblasts (68). Because the Burkitt's lymphoma cells with decreased HMG-I/Y grow more slowly than control Burkitt's cells, it is possible that transformation was inhibited by a decrease in the growth rate. Of note, these cells failed to exhibit colony formation after incubation for several months. Transformation could also be inhibited through another transformation-specific mechanism independent of the decreased growth rate, although this experimental approach does not distinguish between these two possible mechanisms.
FIG. 7.
Decreasing HMG-I/Y protein level inhibits Myc-mediated transformation in Burkitt's lymphoma cells. (A) Western blot of Burkitt's lymphoma cells (Ramos) transfected with the antisense ribozyme construct or the vector control. Note the marked, specific decrease in HMG-I/Y proteins in cells transfected with the antisense HMG-I construct. HMG-C protein was unaffected. (B) Transformation in soft agar in Burkitt's cells is abrogated by decreasing HMG-I/Y protein levels. The soft agar assay was performed in quadruplicate. The number of foci and the standard deviations were taken from three different experiments. (C) Growth rates of Burkitt's lymphoma cells transfected with control vector or the HMG-I antisense vector. Note the decreased growth rate of the Burkitt's lymphoma cells with decreased HMG-I/Y protein levels.
DISCUSSION
Although the c-Myc oncoprotein appears to be an important regulator of cell growth, the identification of relevant target genes is limited. The putative c-Myc target gene encoding CAD is required for DNA synthesis (13, 69). ODC (7, 74, 75), cdc25A (32, 33), rcl (60), LDH-A (82), Tmp (8), the H-ferritin gene (100), and the telomerase gene (97) are additional putative effector genes that appear to be involved in neoplastic transformation, although their precise roles in Myc function are only beginning to emerge. We observed that HMG-I/Y is a c-Myc target gene that is sufficient for transformation in both Rat 1a and CB33 cells. In addition, HMG-I is tumorigenic in nude mice. In fact, the oncogenic properties of HMG-I/Y and c-myc in these two cell lines are highly similar. Moreover, decreasing the level of HMG-I/Y proteins in Burkitt's lymphoma cells inhibits transformation in the soft agar assay, suggesting that HMG-I/Y may be important for Myc-mediated neoplastic transformation in these cells.
Several observations led us to suggest that HMG-I/Y is a relevant c-Myc target gene. First, previously published Northern analyses of murine fibroblasts stimulated by serum or growth factors have shown that HMG-I/Y expression follows that of c-myc (59, 99). HMG-I/Y proteins are also known to be increased in PC-13 cells cotransformed by c-Myc and polyoma leukemia virus carrying the polyoma leukemia virus middle T gene (42). Moreover, expression of both c-myc and HMG-I/Y has been shown to correlate with cell growth and neoplastic transformation. Our studies identified a c-Myc/Max consensus DNA binding site as an important serum- or growth factor-dependent regulatory element in the HMG-I/Y promoter. We also show that c-Myc and Max proteins bind to the HMG-I/Y E-box and transactivate the HMG-I/Y promoter. An E-box has also been identified in the human HMG-I/Y promoter sequences at a similar site (R. Reeves, personal communication), and the conservation of this site in the human and murine genes suggests that this element is important in regulating HMG-I/Y expression. In addition, HMG-I/Y mRNA increases following activation of Myc in a pattern similar to that reported for other putative c-Myc target genes. Increased HMG-I/Y expression occurred in the presence of the protein synthesis inhibitor cycloheximide, indicating that HMG-I/Y is a direct c-Myc target gene. HMG-I/Y induction is blunted in Myc-deficient fibroblasts. In addition, HMG-I/Y proteins are increased in Burkitt's lymphoma cell lines, and decreasing HMG-I/Y protein levels in these cells inhibits transformation. Finally, like c-Myc, HMG-I transforms both Rat 1a fibroblasts and CB33 cells. Moreover, the Rat 1a cells with increased HMG-I protein are tumorigenic in nude mice.
We initially observed that the region of the HMG-I/Y promoter between bp −1895 and +75 from the major transcription start site was required for the stimulation of HMG-I/Y after exposure to serum or individual growth factors. Our studies do not exclude the possibility that additional upstream sequences, intronic sequences, or downstream sequences play a role in regulating HMG-I/Y expression. Previous investigators have reported induction of the human HMG-I/Y promoter sequence between −22 and +222 from the major transcription start site following stimulation with a phorbol ester (tetradecanoyl phorbol acetate [TPA]) (73). The analogous region in the murine promoter did not mediate significant induction following exposure to serum or TPA (Resar, unpublished data) in NIH 3T3 cells. Since our studies examined additional 5′ sequences in the promoter and stimulation by serum and growth factors, the results obtained using the human promoter are difficult to compare with our own (73). The E-box within the HMG-I/Y promoter sequence is located in a far upstream position from the transcription start site, which is distinct from the arrangement of other putative Myc target genes. Although some putative Myc target genes have binding sites within intronic sequences, recent studies suggest that distal enhancer positions may favor activation by Myc and Max over USF (24). Thus, the HMG-I/Y E-box, which is located over 1,000 bp from the transcription start site, would be predicted to favor regulation by Myc and Max over USF.
Our results also show that the HMG-I/Y gene is regulated by additional factors. Mutation of the E-box abolished 60% but not 100% of the serum stimulation of the HMG-I/Y gene. Thus, additional proteins which may cooperate or act independently of c-Myc and Max are likely to contribute to HMG-I/Y regulation. Further deletional analysis, however, has not revealed additional cis-acting elements that contribute more than the E-box to the serum induction of HMG-I/Y (Resar, unpublished data). Of note, an AP2 consensus site is located downstream of the E-box in the ODC and α-prothymosin promoters, and evidence suggests that AP2 may serve to downregulate ODC and α-prothymosin expression (34). An AP2 consensus site is likewise located downstream of the HMG-I/Y E-box and may contribute to the downregulation of HMG-I/Y (Fig. 1A).
In summary, we have identified a new c-Myc target gene that is sufficient for transformation in Rat 1a and CB33 cells. Moreover, Rat 1a cells with increased expression of HMG-I protein are tumorigenic in nude mice. HMG-I/Y proteins are also increased in several human cancers. These findings suggest that HMG-I/Y is a relevant c-Myc target gene and represents a new class of potential oncogenes.
ACKNOWLEDGMENTS
This work is dedicated to the memory of Daniel Nathans. We are indebted to him for invaluable guidance, support, and inspiration. This work was initiated while L.M.S.R. was on sabbatical leave in his laboratory. We also thank Chi V. Dang for advice, insightful discussions, and reagents and Jonathon Simons for guidance and reagents used in the nude mice experiments.
This work was supported in part by grants 5K11CA59793 and R29CA76130, by the Concern Foundation (L.M.S.R. and Y.X.), and by grant 1T32CA604441 (L.J.W., C.E.D., and M.M.).
REFERENCES
- 1.Abate C, Luk D, Gentz R, Rauscher III F J, Curran T. Expression and purification of the leucine zipper and DNA-binding domains of Fos and Jun: both Fos and Jun contact DNA directly. Proc Natl Acad Sci USA. 1990;87:1032–1036. doi: 10.1073/pnas.87.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amin C A, Wagner J, Hay N. Sequence-specific transcriptional activation by Myc and repression by Max. Mol Cell Biol. 1993;13:383–390. doi: 10.1128/mcb.13.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1990. [Google Scholar]
- 4.Auvinen M, Laine A, Paasinen-Sohns A, Kangas A, Kangas L, Saksela O, Andersson L C, Holtta E. Human ornithine decarboxylase-overproducing NIH3T3 cells induce rapidly growing, highly vascularized tumors in nude mice. Cancer Res. 1997;57:3016–3025. [PubMed] [Google Scholar]
- 5.Auvinen M, Paasinen A, Andersson L C, Holtta E. Ornithine decarboxylase activity is critical for cell transformation. Nature. 1992;360:355–358. doi: 10.1038/360355a0. [DOI] [PubMed] [Google Scholar]
- 6.Barrett J, Birrer M J, Kato G J, Dosaka-Akita H, Dang C V. Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol Cell Biol. 1992;12:3130–3137. doi: 10.1128/mcb.12.7.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bello-Fernandez C, Packham G, Cleveland J L. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci USA. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Porath I, Yanuka O, Benvisty N. The Tmp gene, encoding a membrane protein, is a c-Myc target with a tumorigenic activity. Mol Cell Biol. 1999;19:3529–3539. doi: 10.1128/mcb.19.5.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benvenisty N, Leder A, Kuo A, Leder P. An embryonically expressed gene is a target for c-Myc regulation via the c-Myc-binding sequence. Genes Dev. 1992;6:2513–2523. doi: 10.1101/gad.6.12b.2513. [DOI] [PubMed] [Google Scholar]
- 10.Berlingieri M T, Manfioletti G, Santoro M, Bandiera A, Visconti R, Giancotti V, Fusco A. Inhibition of HMG-C protein synthesis suppresses retrovirally induced neoplastic transformation. Mol Cell Biol. 1995;15:1545–1553. doi: 10.1128/mcb.15.3.1545. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 12.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Max. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 13.Boyd K E, Farnham P J. Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol Cell Biol. 1997;17:2529–2537. doi: 10.1128/mcb.17.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bussemakers M J G, van de Ven W J M, Debruyne F M J, Schalken J. Identification of high mobility group I(Y) as potential progression marker for prostate cancer by differential hybridization analysis. Cancer Res. 1991;51:606–611. [PubMed] [Google Scholar]
- 15.Chiapetta G, Bandiera A, Berlingiera M T, Visconti R, Manfioletti G, Battista S, Martinez-Tello F J, Santoro M, Giancotti V, Fusco A. The expression of the high mobility group HMGI(Y) protein correlates with the malignant phenotype of human thyroid neoplasias. Oncogene. 1995;10:1307–1314. [PubMed] [Google Scholar]
- 16.Cochran B H, Reffel A C, Stiles C D. Molecular cloning of gene sequences regulated by platelet-derived growth factor. Cell. 1983;33:939–947. doi: 10.1016/0092-8674(83)90037-5. [DOI] [PubMed] [Google Scholar]
- 17.Cole M D. Myc meets its Max. Cell. 1991;65:715–716. doi: 10.1016/0092-8674(91)90377-b. [DOI] [PubMed] [Google Scholar]
- 18.Cole M D, McMahon S B. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 19.Daksis J I, Lu R Y, Facchini L M, Marhin W W, Penn L J Z. Myc induces cyclin D1 expression in the absence of de novo protein synthesis and links mitogen-stimulated signal transduction to the cell cycle. Oncogene. 1994;9:3635–3645. [PubMed] [Google Scholar]
- 20.Dang C V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dang C V, Barrett J, Villa-Garcia M, Resar L M S, Kato G J, Fearon E R. Intracellular leucine zipper interactions suggest c-Myc heterodimerization. Mol Cell Biol. 1990;11:954–962. doi: 10.1128/mcb.11.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dang C V, Lee L A. c-myc function in neoplasia. R. G. Austin, Tex: Landes; 1995. [Google Scholar]
- 23.DePino R A, Schreiber-Agus N, Alt F W. Myc family oncogenes in the development of normal and neoplastic cells. Adv Cancer Res. 1991;57:1–46. doi: 10.1016/s0065-230x(08)60994-x. [DOI] [PubMed] [Google Scholar]
- 24.Desbarats L, Gaubatz S, Eilers M. Discrimination between different E-box-binding proteins at an endogenous target gene of c-myc. Genes Dev. 1996;10:447–460. doi: 10.1101/gad.10.4.447. [DOI] [PubMed] [Google Scholar]
- 25.Du W, Thanos D, Maniatis T. Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell. 1993;74:887–898. doi: 10.1016/0092-8674(93)90468-6. [DOI] [PubMed] [Google Scholar]
- 26.Eilers M, Picard D, Yamamoto K R, Bishop J M. Chimeras between the MYC oncoprotein and steroid receptors cause hormone-dependent transformation in cells. Nature. 1989;340:66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- 27.Eilers M S, Schirm S, Bishop J M. The MYC protein activates transcription of the α-prothymosin gene. EMBO J. 1991;10:133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evan G I, Littlewood T D. The role of c-myc in cell growth. Curr Opin Gene Dev. 1993;3:44–49. doi: 10.1016/s0959-437x(05)80339-9. [DOI] [PubMed] [Google Scholar]
- 29.Facchini L M, Penn L Z. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- 30.Falvo J V, Thanos D, Maniatis T. Reversal of intrinsic DNA bends in the IFNβ gene enhancer by transcription factors and the architectural protein HMG I(Y) Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 31.Friedmann M, Holth L T, Zoghbi H Y, Reeves R. Organization, inducible expression and chromosomal localization of the human HMG-I(Y) nonhistone protein gene. Nucleic Acids Res. 1993;21:4259–4267. doi: 10.1093/nar/21.18.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galaktionov K, Chen X, Beach D. cdc25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 33.Galaktionov K, Lee A K, Eckstein J, Draetta G, Meckler J, Loda M, Beach D. CDC25 phosphatases as potential human oncogenes. Science. 1995;269:1575–1577. doi: 10.1126/science.7667636. [DOI] [PubMed] [Google Scholar]
- 34.Gaubatz S, Imhof A, Dosch R, Weiner O, Mitchell P, Buettner R, Eilers M. Transcriptional activation of Myc is under negative control by transcription factor AP-2. EMBO J. 1995;14:1508–1519. doi: 10.1002/j.1460-2075.1995.tb07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaubatz S, Meichle A, Eilers M. An E-box element localized in the first intron mediates regulation of the prothymosin α gene by c-myc. Mol Cell Biol. 1994;14:3853–3862. doi: 10.1128/mcb.14.6.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerdes J, Schwab U, Lemke H, Stein H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;13:13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- 37.Gerdes J, Carsten Schlueter L L, Duchrow M, Wohlenberg C, Gerlach C, Stahmer I, Kloth S, Brandt E, Flad H-D. Immunobiochemical and molecular biologic characterization of the cell proliferation-associated nuclear antigen that is defined by monoclonal antibody Ki-67. Am J Pathol. 1991;138:867–873. [PMC free article] [PubMed] [Google Scholar]
- 38.Giancotti V, Bandiera A, Buratti E, Fusco A, Marzari R, Coles B, Goodwin G H. Comparison of multiple forms of the high mobility group I proteins in rodent and human cells. Identification of the human high mobility group I-C protein. Eur J Biochem. 1991;198:211–216. doi: 10.1111/j.1432-1033.1991.tb16003.x. [DOI] [PubMed] [Google Scholar]
- 39.Giancotti V, Bandiera A, Ciani L, Santro D, Crane-Robinson C, Goodwin G H, Boiocchi M, Dolcetti R, Casetta B. High-mobility-group (HMG) proteins and histone H1 subtypes expression in normal and tumor tissues of mouse. Eur J Biochem. 1993;213:825–832. doi: 10.1111/j.1432-1033.1993.tb17825.x. [DOI] [PubMed] [Google Scholar]
- 40.Giancotti V, Berlingieri M T, DiFiore P P, Fusco A, Veccio G, Crane-Robinson C. Changes in nuclear proteins following transformation of rat thyroid epithelial cells by a murine sarcoma retrovirus. Cancer Res. 1985;45:6051–6057. [PubMed] [Google Scholar]
- 41.Giancotti V, Buratti E, Perissin L, Zorzet S, Balman A, Portella G, Fusco A, Goodwin G H. Analysis of the HMGI nuclear proteins in mouse neoplastic cells induced by different procedures. Exp Cell Res. 1989;184:538–545. doi: 10.1016/0014-4827(89)90352-2. [DOI] [PubMed] [Google Scholar]
- 42.Giancotti V, Pani B, D'Andrea P, Berlingieri M T, DiFiore P P, Fusco A, Veccio G, Philip R, Crane-Robinson C, Nicolas R H, Wright C A, Goodwin G H. Elevated levels of a specific class of nuclear phosphoproteins in cells transformed with v-ras and v-mos oncogenes and by co-transfection with c-myc and polyoma middle T genes. EMBO J. 1987;6:1981–1987. doi: 10.1002/j.1460-2075.1987.tb02461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graeber T G, Osmanian C, Jacks T, Housman D E, Koch C J, Lowe S W, Giaccia A. Hypoxia-mediated selection of cells with diminished apoptotic potential on solid tumors. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 44.Grandori C, Mac J, Siebelt F, Ayer D E, Eisenman R N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 45.Grandori C, Eisenman R N. Myc target genes. Trends Biochem Sci. 1997;22:177–181. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 46.Gu W, Cechova K, Tassi V, Dalla-Favara R. Opposite regulation of gene transcription and cell proliferation by c-Myc and Max. Proc Natl Acad Sci USA. 1993;90:2935–2939. doi: 10.1073/pnas.90.7.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoang A T, Cohen K J, Barrett J F, Bergstrom D A, Dang C V. Participation of cyclin A in Myc-induced apoptosis. Proc Natl Acad Sci USA. 1994;91:6875–6877. doi: 10.1073/pnas.91.15.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holozoneti T D, Kandil A N. Determination of the c-Myc DNA-binding site. Proc Natl Acad Sci USA. 1991;88:6162–6166. doi: 10.1073/pnas.88.14.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holth L T, Thorlacius A E, Reeves R. Effects of epidermal growth factor and estrogen on the regulation of the HMG-I/Y gene in human mammary epithelial cell lines. DNA Cell Biol. 1997;16:1299–1309. doi: 10.1089/dna.1997.16.1299. [DOI] [PubMed] [Google Scholar]
- 50.Johnson K R, Cook S A, Davisson M T. Chromosomal localization of the murine gene and 2 related sequences encoding high-mobility group I and Y proteins. Genomics. 1992;12:503–509. doi: 10.1016/0888-7543(92)90441-t. [DOI] [PubMed] [Google Scholar]
- 51.Johnson K R, Lehn D A, Elton T S, Barr P J, Reeves R. The chromosomal high mobility group protein HMG-I(Y): complete murine cDNA sequence, genomic structure, and tissue expression. J Biol Chem. 1988;18:18338–18342. [PubMed] [Google Scholar]
- 52.Jones D H, Winistorfer S C. Recombinant circle PCR for site-specific mutagenesis without PCR product purification. BioTechniques. 1992;12:528–534. [PubMed] [Google Scholar]
- 53.Jones R M, Branda J, Johnston K A, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt E V. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-Myc. Mol Cell Biol. 1996;16:4754–4764. doi: 10.1128/mcb.16.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kato G, Barrett J, Villa-Garcia M, Dang C V. An amino-terminal c-Myc domain required for neoplastic transformation. Mol Cell Biol. 1990;10:5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kato G J, Lee W M F, Chen L, Dang C V. Max: functional domains and interaction with c-Myc. Genes Dev. 1992;6:81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- 56.Keath E J, Caimi P G, Cole M D. Fibroblast lines expressing activated c-myc oncogenes are tumorigenic in nude mice and syngeneic animals. Cell. 1984;39:339–348. doi: 10.1016/0092-8674(84)90012-6. [DOI] [PubMed] [Google Scholar]
- 57.Kerkhoff E, Bister K, Klempnauer K H. Sequence-specific DNA binding by Myc proteins. Proc Natl Acad Sci USA. 1991;88:4323–4327. doi: 10.1073/pnas.88.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laborda J. 36B4 cDNA used as an estradiol-independent mRNA control is the cDNA for human acidic ribosomal phosphoprotein PO. Nucleic Acids Res. 1991;19:3998. doi: 10.1093/nar/19.14.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lanahan A, Williams J B, Sanders L K, Nathans D. Growth factor-induced delayed early-response genes. Mol Cell Biol. 1992;12:3919–3929. doi: 10.1128/mcb.12.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lewis B C, Shim H, Li Q, Wu C S, Lee L A, Maity A, Dang C V. Identification of putative c-Myc-responsive genes: characterization of rcl, a novel growth-related gene. Mol Cell Biol. 1997;17:4967–4978. doi: 10.1128/mcb.17.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Littlewood T D, Amati B, Land H, Evan G I. Max and cMyc/Max DNA-binding activities in cell extracts. Oncogene. 1992;7:1783–1792. [PubMed] [Google Scholar]
- 62.Littlewood T D, Hancock D C, Danielian P S, Parker M G, Evan G I. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lombardi L, Newcomb E W, Dalla-Favara R. Pathogenesis of Burkitt lymphoma: expression of an activated c-myc oncogene causes the tumorigenic conversion of EBV-infected human B lymphocytes. Cell. 1987;49:161–170. doi: 10.1016/0092-8674(87)90556-3. [DOI] [PubMed] [Google Scholar]
- 64.Lund T, Holtman J, Frederiksen M, Laland S G. On the presence of two new high mobility group-like proteins in HeLa S3 cells. FEBS Lett. 1983;152:163–167. doi: 10.1016/0014-5793(83)80370-6. [DOI] [PubMed] [Google Scholar]
- 65.Lüscher B, Eisenman R N. New light on myc and myb. 1. myc. Genes Dev. 1990;4:2025–2035. doi: 10.1101/gad.4.12a.2025. [DOI] [PubMed] [Google Scholar]
- 66.Maher J F, Nathans D. Multivalent DNA-binding properties of the HMG-I proteins. Proc Natl Acad Sci USA. 1995;93:6716–6720. doi: 10.1073/pnas.93.13.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Masiakowski P, Breathnath R, Bloch J, Gannon F, Krust A, Chambon P. Cloning of cDNA sequences of hormone-regulated genes from the MCF-7 human breast cancer cell line. Nucleic Acids Res. 1982;10:7895–7903. doi: 10.1093/nar/10.24.7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mateyak M K, Obaya A J, Adachi S, Sedivy J M. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 69.Miltenberger R J, Sukow K A, Farnham P J. An E box-mediated increase in cad transcription at the G1/S-phase boundary is suppressed by inhibitory c-Myc mutants. Mol Cell Biol. 1995;15:2527–2535. doi: 10.1128/mcb.15.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Montgomery R A, Dietz H C. Inhibition of fibrillin 1 expression using U1 snRNA as a vehicle for the presentation of antisense targeting sequence. Hum Mol Genet. 1997;6:519–525. doi: 10.1093/hmg/6.4.519. [DOI] [PubMed] [Google Scholar]
- 71.Nathans D, Christy B A, DuBois R, Lanahan A, Sanders L K, Nakabeppu Y. Transcription factors induced by growth-signaling agents. In: Brugge J, editor. Origins of human cancer: a comprehensive review. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1991. pp. 353–364. [Google Scholar]
- 72.Nesbit C E, Tersak J M, Prochownik E V. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3076. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 73.Ogram S A, Reeves R. Differential regulation of a multipromoter gene. J Biol Chem. 1995;270:14235–14242. doi: 10.1074/jbc.270.23.14235. [DOI] [PubMed] [Google Scholar]
- 74.Packham G, Cleveland J L. Ornithine decarboxylase is a mediator of c-Myc-induced apoptosis. Mol Cell Biol. 1994;14:5741–5747. doi: 10.1128/mcb.14.9.5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pena A, Reddy C D, Wu S, Hickok N J, Premkumar Reddy E, Yumet G, Soprano D R, Soprano K J. Regulation of human ornithine decarboxylase expression by the c-Myc-Max protein complex. J Biol Chem. 1993;268:27277–27285. [PubMed] [Google Scholar]
- 76.Prendergast G C, Ziff E B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1990;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 77.Ram T J, Reeves R, Hosick H L. Elevated high mobility group I(Y) gene expression is associated with progressive transformation of mouse mammary epithelial cells. Cancer Res. 1993;53:2655–2660. [PubMed] [Google Scholar]
- 78.Rosenwald I B, Rhoads D B, Callanan L D, Isselbacher K J, Schmidt E V. Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2 alpha in response to growth induction by c-Myc. Proc Natl Acad Sci USA. 1993;90:6175–6178. doi: 10.1073/pnas.90.13.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 80.Schuldiner O, Eden A, Ben-Yosef T, Yanuka O, Simchen G, Benvenisty N. ECA39, a conserved gene regulated by c-Myc in mice, is involved in G1/S cell cycle regulation of yeast. Proc Natl Acad Sci USA. 1996;93:7143–7148. doi: 10.1073/pnas.93.14.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen-Ong G L C, Keath E J, Picolli S P, Cole M D. Novel myc oncogene RNA from abortive immunoglobin-gene recombination in mouse plasmacytomas. Cell. 1982;31:443–453. doi: 10.1016/0092-8674(82)90137-4. [DOI] [PubMed] [Google Scholar]
- 82.Shim H, Dolde C, Lewis B C, Wu C-S, Dang G, Jungmann R A, Dalla-Favera R, Dang C V. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Slamon D J, deKernion J B, Verma I M, Cline M T. Cellular oncogenes in human malignancies. Science. 1984;224:256–263. doi: 10.1126/science.6538699. [DOI] [PubMed] [Google Scholar]
- 84.Small M B, Hay N, Schwab M, Bishop J M. Neoplastic transformation by the human gene N-myc. Mol Cell Biol. 1987;7:1638–1645. doi: 10.1128/mcb.7.5.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Starborg M, Gell K, Brundell E, Hoog C. The murine Ki-67 cell proliferation antigen accumulates in the nucleolar and heterochromatic regions of interphase cells and at the periphery of the mitotic chromosomes in a process essential for cell cycle progression. J Cell Sci. 1996;109:143–153. doi: 10.1242/jcs.109.1.143. [DOI] [PubMed] [Google Scholar]
- 86.Stone J, deLange T, Ramsey G, Jakabovitz E, Bishop J M, Varmus H E, Lee W. Definition of regions in human c-Myc involved in transformation and nuclear localization. Mol Cell Biol. 1987;7:1691–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Strick R, Laemmli U K. SARs are cis DNA elements of chromosome dynamics: synthesis of a SAR repressor protein. Cell. 1995;83:1137–1148. doi: 10.1016/0092-8674(95)90140-x. [DOI] [PubMed] [Google Scholar]
- 88.Sun B, Hirsh D A, Jackson D, Beachy P A. Ultrabithorax protein is necessary, but not sufficient for full activation of decapentaplegic expression of visceral mesoderm. EMBO J. 1995;14:520–535. doi: 10.1002/j.1460-2075.1995.tb07028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tamimi Y, van der Poel H G, Denyn M M, Umbus R, Karthaus H F, Dubruyne F M, Shalken J A. Increased expression of high mobility group protein I(Y) in high grade prostatic cancer determined by in situ hybridization. Cancer Res. 1993;53:5512–5516. [PubMed] [Google Scholar]
- 90.Tamimi Y, van der Poel H G, Karthaus H F M, Dubruyne F M J, Schalken J A. A retrospective study of high mobility group I(Y) as progressive marker for prostate cancer determined by in situ hybridization. Br J Cancer. 1996;74:573–578. doi: 10.1038/bjc.1996.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thanos D, Du W, Maniatis T. The high mobility group protein HMG I(Y) is an essential structural component of viral-inducible enhancer complex. Cold Spring Harbor Symp Quant Biol. 1993;58:73–81. doi: 10.1101/sqb.1993.058.01.011. [DOI] [PubMed] [Google Scholar]
- 92.Thanos D, Maniatis T. Virus induction of human IFN-β gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 93.Thanos D, Maniatis T. The high mobility group protein HMGI(Y) is required for NF-kappaB-dependent virus induction of the human IFN-β gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 94.Van der Eb A J, Graham F L. Assay of transforming activity of tumor virus DNA. Methods Enzymol. 1980;65:826–839. doi: 10.1016/s0076-6879(80)65077-0. [DOI] [PubMed] [Google Scholar]
- 95.Vartiane E, Palvimo J, Mahonen A, Linnala-Kankkuren A, Maenpaa P H. Selective decrease in low-M, HMG proteins HMGI and HMGY during differentiation of mouse teratocarcinoma cells. FEBS Lett. 1988;228:45–48. doi: 10.1016/0014-5793(88)80581-7. [DOI] [PubMed] [Google Scholar]
- 96.Vennstrom B, Sheiness D, Zabielski J, Bishop J M. Isolation and characterization of c-myc, a cellular homolog of the oncogene (v-myc) of avian myelocytomatosis virus strain 29. J Virol. 1982;31:514–524. doi: 10.1128/jvi.42.3.773-779.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang J, Xie L Y, Allan S, Beach D, Hannon G J. Myc activates telomerase. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wechsler D S, Dang C V. Opposite orientations of DNA bending by c-Myc and Max. Proc Natl Acad Sci USA. 1992;89:7635–7339. doi: 10.1073/pnas.89.16.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Williams J B, Lanahan A A. A mammalian delayed-early response gene encodes HNP36, a novel, conserved nucleolar protein. Biochem Biophys Res Commun. 1995;206:916–926. doi: 10.1006/bbrc.1995.2133. [DOI] [PubMed] [Google Scholar]
- 100.Wu K-J, Polack A, Dalla-Favara R. Coordinated regulation of iron-controlling genes, H-Ferritin and IRP2, by c-Myc. Science. 1999;283:676–679. doi: 10.1126/science.283.5402.676. [DOI] [PubMed] [Google Scholar]
- 101.Zhao K, Kas E, Gonzales E, Laemmli U K. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J. 1993;12:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]