Abstract
Objectives
We aimed to determine susceptibilities of Elizabethkingia spp. to 25 commonly tested and 8 novel antibiotics, and to compare the performance of different susceptibility testing methods.
Methods
Clinical isolates of Elizabethkingia spp., Chryseobacterium spp. and Flavobacterium spp. collected during 2002–18 (n = 210) in a nationwide surveillance programme in Taiwan were speciated by 16S rRNA sequencing. MICs were determined by broth microdilution. The broth microdilution results of 18 common antibiotics were compared with those obtained by the VITEK 2 automated system.
Results
Among the Elizabethkingia spp. identified (n = 108), Elizabethkingia anophelis was the most prevalent (n = 90), followed by Elizabethkingia meningoseptica (n = 7) and Elizabethkingia miricola cluster [E. miricola (n = 6), Elizabethkingia bruuniana (n = 3) and Elizabethkingia ursingii (n = 2)]. Most isolates were recovered from respiratory or blood specimens from hospitalized, elderly patients. PFGE showed two major and several minor E. anophelis clones. All isolates were resistant to nearly all the tested β-lactams. Doxycycline, minocycline and trimethoprim/sulfamethoxazole inhibited >90% of Elizabethkingia spp. Rifampin inhibited E. meningoseptica (100%) and E. anophelis (81.1%). Fluoroquinolones and tigecycline were active against E. meningoseptica and E. miricola cluster isolates. Novel antibiotics, including imipenem/relebactam, meropenem/vaborbactam, ceftazidime/avibactam, cefepime/zidebactam, delafloxacin, eravacycline and omadacycline were ineffective but lascufloxacin inhibited half of Elizabethkingia spp. The very major discrepancy rates of VITEK 2 were >1.5% for ciprofloxacin, moxifloxacin and vancomycin. Major discrepancy rates were >3% for amikacin, tigecycline, piperacillin/tazobactam and trimethoprim/sulfamethoxazole.
Conclusions
MDR, absence of standard interpretation criteria and poor intermethod concordance necessitate working guidelines to facilitate future research of emerging Elizabethkingia spp.
Introduction
Elizabethkingia spp. are aerobic, non-motile, non-spore-forming Gram-negative bacilli that do not ferment glucose.1 In addition to their natural reservoirs such as soil and water, Elizabethkingia spp. have also been recovered from hospital environments. At least six species have been classified in the genus since its designation as a novel taxon in 2005.2 Three of the species are emerging opportunistic pathogens that cause serious infections, particularly in immunocompromised patients. Elizabethkingia meningoseptica, previously known as Chryseobacterium meningosepticum, is a well-known aetiological agent of nosocomial pneumonia, meningitis and sepsis.3Elizabethkingia miricola and Elizabethkingia anophelis were recently proposed in 2003 and 2011, respectively.4,5
While E. miricola causes sporadic cases, E. anophelis has caused moderate to large-scale nosocomial outbreaks.6–14 The reported incidence of Elizabethkingia spp. infections is increasing in Asian countries, including Taiwan.7–11 However, the unreliability of phenotypic methods to differentiate Chryseobacterium from Elizabethkingia and to speciate Elizabethkingia spp. isolates may confound epidemiological studies.15 In addition, the numbers of longitudinal and nationwide surveillance studies are limited.
Elizabethkingia spp. exhibit high-level MDR.6–14 A variety of antibiotics have been tested in vitro. However, antibiotic susceptibilities obtained in different studies may not be comparable due to the use of different strains, testing methods and interpretative criteria. Broth microdilution methods were used in many studies as the gold standard to determine MICs of Elizabethkingia spp.7,10,11 However, automated susceptibility testing systems are much more commonly used in clinical laboratories and discrepancies between broth microdilution and automated systems have been reported.16
The present study was conducted to investigate the epidemiology, clinical characteristics and antibiotic susceptibility profiles of Elizabethkingia isolates in Taiwan using 16S rRNA sequencing of isolates collected during 2002–18 by the Taiwan Surveillance of Antimicrobial Resistance (TSAR) programme. We assessed the antimicrobial susceptibilities of Elizabethkingia spp. to 25 antibiotics evaluated in previous studies7–11 and used in clinical practice, and to 8 novel antibiotics that are either in clinical development or have been recently approved by the USA or EU.17 Because of reported discrepancies between broth microdilution and automated systems,16 we used both methods and compared results for 18 of the 25 commonly tested antibiotics.
Materials and methods
Bacterial isolates
The TSAR programme is a nationwide surveillance system that collects clinical isolates biennially from 11 medical centres and 15 regional hospitals in all four regions of Taiwan. The collection protocol has been described previously.18 All isolates from participating hospitals are stored at −80°C and subcultured to ensure purity prior to subsequent testing. The study period was from 2002 to 2018 (corresponding to TSAR periods III to XI). Isolates identified as Chryseobacterium spp., Elizabethkingia spp. or Flavobacterium spp. by participating hospitals were selected and subjected to speciation.
16s rRNA gene sequencing
The species of all isolates in the present study were identified by 16S rRNA gene sequencing using Oxford Nanopore Technologies (ONT) MinION sequencing, modified from that published by Liou et al.19 The 16S rRNA gene was amplified by PCR with 12 sets of barcodes attached to the universal primers (8F: AGAGTTTGATCCTGGCTCAG; 1492R: GGTTACCTTGTTACGACTT). The 12 unique barcoded DNAs were purified and pooled for the second barcode ligation. The pooled DNAs were repaired and dA-tailed using NEBNext FFPE Repair Mix (New England BioLabs, NEB; M6630) and NEBNext Ultra II End Repair/dA-Tailing Module (NEB; E7546). The ONT Native Barcoding Kit (EXP-NBD103) was used to multiplex the eight pools of PCR-barcoded DNAs. A total of 96 (12 × 8) dual-barcoded DNAs were purified and pooled in equimolar amounts for downstream library construction using a 1D Ligation Sequencing Kit (ONT, SQK-LSK109). The sequencing adapter was ligated to the DNAs using NEBNext Quick Ligation Module (NEB; E6056). The library was loaded into a SpotON flowcell R9.4.1 (FLO-MIN106D) and sequencing was executed via MinKNOW (release v 19.05.0). Data processing was performed following the workflow published by Liou et al.19 with basecalling and read-polishing applications changed to Guppy (v 3.0.3) and Medaka (v 0.7.0), respectively. These data were compared with those deposited in NCBI for species identification. We compared the results of ONT MinION sequencing of 16S rRNA areas with Sanger sequencing for five E. anophelis, five E. meningoseptica and five E. miricola cluster isolates. The overall accuracy was 99.87%. The number of discordant nucleotides ranged from 0 to 4 among the 1478 bp compared and the discordant nucleotides were not located in the area for speciation.
Rapid identification by PCR
Two-step PCR reactions were performed for speciation (See Figure S1 and Table S1, available as Supplementary data at JAC Online). The first multiple-PCR step used two sets of primers targeting 23S rRNA to identify E. anophelis and E. meningoseptica. The amplification procedure consisted of initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 30 s and elongation at 72°C for 1 min 10 s, then a final cycle of elongation at 72°C for 5 min. The second set of PCR primers was designed based on the findings of the previous study showing that pheT could be used for speciation.20 For isolates other than E. anophelis and E. meningoseptica by the first PCR reaction, the second PCR was performed to identify E. miricola cluster isolates. Amplification conditions were the same as those used in the first PCR step except the annealing step was done at 50°C. A previously reported PCR assay by Chew et al.7 was also tested in parallel to test the accuracy of our PCR typing scheme.
Determination of clonality by PFGE
PFGE was performed on all Elizabethkingia spp. isolates after digestion with ApaI.21 All electrophoresis runs were performed on 1% agarose gels in 0.5× Tris-borate-EDTA buffer at 14°C, in a Bio-Rad CHEF Mapper XA system operating with initial and final switch times of 5 and 35 s, respectively, for 22 h. Stained gels were photographed and analysed using BioNumerics software (v 5.1; Applied Maths, Saint-Martens-Latem, Belgium). ATCC® BAA-664™ was used as a standard for DNA pattern normalization. Dendrograms were generated to determine the relatedness of isolates. Isolates having >80% similarity were assigned a PFGE cluster name (pulsotype) if there were three or more isolates within the cluster.
Antimicrobial susceptibility to commonly tested antibiotics
MICs of 25 common antimicrobial agents were determined by reference broth microdilution testing following the guidelines of CLSI.22 In addition, the MICs of 18 of the 25 antimicrobial agents were determined by an automated system (VITEK 2). Broth microdilution testing was performed using the Sensititre GNX3F (Trek Diagnostics, West Sussex, England), except moxifloxacin, tetracycline, rifampin and vancomycin, for which in-house-prepared 96-well microtitre plates were used. The following quality control strains were included: Escherichia coli ATCC 25922 (rifampin, tetracycline and GNX3F), Pseudomonas aeruginosa ATCC 27853 (moxifloxacin, tetracycline and GNX3F), Staphylococcus aureus ATCC 29213 (vancomycin and GNX3F) and Enterococcus faecalis ATCC 29212 (moxifloxacin, rifampin and vancomycin).
For automated testing, VITEK 2 was used with AST-P605 and AST-N322 cards (bioMérieux, France). The quality control strains were S. aureus ATCC 29213 and E. faecalis ATCC 29212, as well as E. coli ATCC 25922 and P. aeruginosa ATCC 27853.
Antimicrobial susceptibility to novel antibiotics
Novel antibiotics included imipenem/relebactam, meropenem/vaborbactam, ceftazidime/avibactam, cefepime/zidebactam, lascufloxacin, delafloxacin, eravacycline and omadacycline. Relebactam, vaborbactam, avibactam, eravacycline and omadacycline were obtained from MedChemExpress (USA), zidebactam and lascufloxacin were from MedKoo Biosciences (USA) and delafloxacin was from Sigma–Aldrich (USA). Broth microdilution testing was performed using in-house-prepared 96-well microtitre plates. E. coli ATCC 25922 was used as the quality control strain for eravacycline, lascufloxacin and omadacycline; P. aeruginosa ATCC 27853 was used for imipenem/relebactam, meropenem/vaborbactam, ceftazidime/avibactam, cefepime/zidebactam and delafloxacin.
Data analysis
Susceptibilities were calculated using Whonet software (Stelling and O’Brien).18 Due to the lack of specific interpretive criteria for Elizabethkingia spp., especially for novel antibiotics, CLSI or US FDA breakpoints for other species were adapted and specified according to previous protocols (Tables S2 and S3).8,11 VITEK 2 results were compared with those of reference broth microdilution. Very major discrepancy (VMD) was defined as resistance in broth microdilution but susceptibility in VITEK 2; major discrepancy (MD) was defined as susceptibility in broth microdilution but resistance in VITEK 2. MD rates ≤3.0% and VMD rates ≤1.5% are considered as the minimum performance standard.
Ethics
The TSAR bacterial isolates were recovered from clinical samples taken as part of standard care and the study was approved by the Research Ethics Committee of the National Health Research Institutes (EC1010602-E, EC1030406-E, EC1050606-E).
Results and discussion
Epidemiology
Among 210 isolates previously identified as Elizabethkingia spp., Chryseobacterium spp. or Flavobacterium spp. by hospital clinical laboratories during 2002–18, 16S rRNA sequencing showed that 108 were Elizabethkingia spp. and 90 were Chryseobacterium spp. The other 12 isolates were Candidatus spp., Klebsiella aerogenes, Microbacterium arborescens, Pedobacter spp., Pseudomonas plecoglossicida, Pseudomonas putida, Rheinheimera spp., Sphingomonas spp. and Stenotrophomonas maltophilia. The number of Elizabethkingia spp. isolates increased each year (Figure S2). Among the 108 Elizabethkingia spp. isolates, E. anophelis (n = 90) was the most common, followed by E. meningoseptica (n = 7). E. miricola (n = 6), Elizabethkingia bruuniana (n = 3) and Elizabethkingia ursingii (n = 2) isolates were grouped as ‘E. miricola cluster’, according to previous studies.11,20,23Elizabethkingia occulta, the other member of the cluster, was not identified in this study.
Patient characteristics are listed in Table 1. Most isolates were recovered from respiratory or blood specimens in elderly (≥65-year-old) patients. The specimens were from patients not only in ICUs but also in wards and outpatient departments of regional hospitals and medical centres in different regions of Taiwan. Previous studies implied that clinical manifestations of E. anophelis infections may differ from those caused by other Elizabethkingia spp. For example, Elizabethkingia spp. other than E. anophelis are common colonizers of cystic fibrosis patients and are less pathogenic.20,23 However, E. meningoseptica and E. miricola cluster isolates from blood cultures and from ICU patients were not uncommon in our study. Since the clinical outcomes were not collected in our study, we are not able to provide further clinical implications of different species.
Table 1.
Patient characteristics in cases of Elizabethkingia spp. infection
| E. anophelis | E. meningoseptica | E. miricola clusterb | |
|---|---|---|---|
| Total (n) | 90 | 7 | 11 |
| Age, years, n (%)a | |||
| <18 | 1 (1.1) | 0 (0) | 0 (0) |
| 18–64 | 20 (22.2) | 1 (14.3) | 2 (18.2) |
| ≥65 | 65 (72.2) | 6 (85.7) | 9 (81.8) |
| Hospital type, n (%) | |||
| Medical centres | 63 (70) | 4 (57.1) | 5 (45.5) |
| Regional hospitals | 27 (30) | 3 (42.9) | 6 (54.5) |
| Region of hospitals, n (%) | |||
| North | 42 (46.7) | 3 (42.9) | 7 (63.6) |
| Central | 22 (24.4) | 2 (28.6) | 1 (9.1) |
| South | 20 (22.2) | 2 (28.6) | 3 (27.3) |
| East | 6 (6.7) | 0 (0) | 0 (0) |
| Patient location, n (%)a | |||
| ICU | 44 (48.9) | 4 (57.1) | 8 (72.7) |
| Non-ICU ward | 39 (43.3) | 1 (14.3) | 1 (9.1) |
| OPD/ER | 4 (4.4) | 0 (0) | 2 (18.2) |
| Specimen type, n (%) | |||
| Respiratory | 46 (51.1) | 3 (42.9) | 3 (27.3) |
| Blood | 41 (45.6) | 3 (42.9) | 8 (72.7) |
| Pus/discharge | 1 (1.1) | 0 (0) | 0 (0) |
| Urine | 1 (1.1) | 0 (0) | 0 (0) |
| Other | 1 (1.1) | 1 (14.3) | 0 (0) |
OPD, outpatient department; ER, emergency room.
Not all age and hospital location data were available.
E. miricola cluster comprised E. miricola (6), E. bruuniana (3) and E. ursingii (2); no E. occulta isolates were identified.
PFGE revealed one major E. anophelis cluster (>80% in similarity) in multiple hospitals and several minor clusters (Figure S3). The major cluster grouped into two subclusters, A1 and A2; the isolates of A1 and A2 were from multiple hospitals, indicating possible outbreaks. Other Elizabethkingia spp. were identified in sporadic cases unrelated to the E. anophelis cluster. Previous reports showed the ability of E. anophelis to cause outbreaks, in contrast to other Elizabethkingia spp.12,13 Outbreaks of E. meningoseptica reported in previous studies may be attributable to E. anophelis due to the predominance of this species in the clinical settings and the lack of knowledge of this new species at the time of study.24
Rapid identification by PCR
The 210 isolates described above were used to test the accuracies of our PCR scheme and that of Chew et al.7 Using 16S rRNA sequencing as the reference gold standard, our first PCR reaction and the PCR assay by Chew et al.7 both accurately differentiated all E. anophelis from E. meningoseptica. Additionally, our second PCR step was 100% accurate in identifying E. miricola cluster isolates. Ten isolates of E. coli, K. pneumoniae, P. aeruginosa and Acinetobacter baumannii randomly selected from the TSAR collection all tested negative in our PCR scheme (data not shown).
A recent study found that MALDI-TOF MS accurately identified all E. anopheles, E. meningoseptica and E. miricola when an amended database was used.11 Since MALDI-TOF and 16S rRNA sequencing are not readily available in all clinical laboratories, the PCR scheme could be a rapid and simple alternative method to accurately differentiate these species. Our PCR scheme demonstrated good accuracy in the identification of E. anophelis, E. meningoseptica and E. miricola cluster isolates. In addition, one-tube testing for E. anophelis and E. meningoseptica and additional identification of E. miricola cluster isolates may be more practical than other PCR schemes.
Susceptibility to 25 commonly tested and 8 novel antibiotics
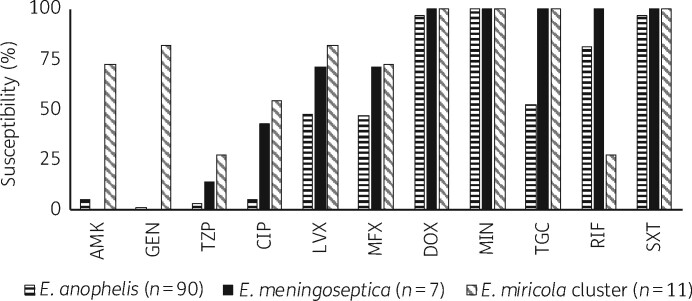
Drug susceptibilities of the Elizabethkingia spp. isolates and MIC ranges for all tested antibiotics are shown in Figures 1 and 2 and Table S4. The commonly tested antibiotics for which <5% of all Elizabethkingia spp. were susceptible are shown in Table S4. All 108 isolates were resistant to all β-lactams, including carbapenems, due to intrinsic MBL genes blaB and blaGOB.10 Vancomycin, an agent with discordant in vitro testing results and varied treatment outcomes for E. meningoseptica reported in the literature,25 was inactive against our Elizabethkingia spp. isolates (MIC ≥ 16 mg/L). Doxycycline, minocycline and trimethoprim/sulfamethoxazole inhibited >90% of all isolates. Previous studies showed consistently high rates of susceptibility to doxycycline and minocycline.3,7–9,11 However, susceptibility to trimethoprim/sulfamethoxazole varied among these studies,3,7–9,11 which may be attributed to differences in strains, methodology, definition of 80% reduction in growth22 and interpretive criteria. In contrast, ciprofloxacin, levofloxacin, moxifloxacin and tigecycline were more active against E. meningoseptica/E. miricola cluster isolates, with respective susceptibilities of 42.9%/54.5%, 71.4%/81.8%, 71.4%/72.7% and 100%/100%, compared with 5.6%, 47.8%, 46.7% and 52.2% for E. anophelis. E. miricola cluster isolates were more susceptible to aminoglycosides, including amikacin and gentamicin, but not tobramycin. Rifampin inhibited 81.1% of E. anophelis and 100% of E. meningoseptica.
Figure 1.
Susceptibility of Elizabethkingia spp. to commonly tested antibiotics. In the absence of CLSI breakpoints, susceptibility criteria were adapted from previous studies and are listed in Table S2. The commonly tested antibiotics for which <5% of all Elizabethkingia spp. were susceptible are not shown in Figure 1; refer to Table S4 for their susceptibility. They included cefepime, cefotaxime, ceftazidime, doripenem, imipenem, meropenem, ampicillin/sulbactam, ticarcillin/clavulanic acid, vancomycin, colistin, polymyxin B, tobramycin, tetracycline and aztreonam. AMK, amikacin; GEN, gentamicin; TZP, piperacillin/tazobactam; CIP, ciprofloxacin; LVX, levofloxacin; MFX, moxifloxacin; DOX, doxycycline; MIN, minocycline; TGC, tigecycline; RIF, rifampin; SXT, trimethoprim/sulfamethoxazole.
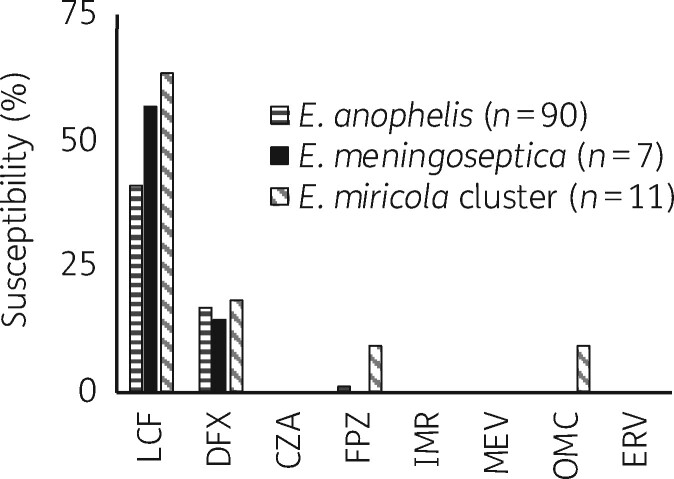
Figure 2.
Susceptibility of Elizabethkingia spp. to novel antibiotics. Due to the lack of CLSI breakpoints, susceptibility criteria were adapted from previous studies and are listed in Table S3. LCF, lascufloxacin; DFX, delafloxacin; CZA, ceftazidime/avibactam; FPZ, cefepime/zidebactam; IMR, imipenem/relebactam; MEV, meropenem/vaborbactam; OMC, omadacycline; ERV, eravacycline.
Our Elizabethkingia spp. isolates were also highly resistant to seven of the eight novel antibiotics tested, which further underscores the clinical challenges posed by Elizabethkingia spp. The inability of new β-lactamase inhibitors to enhance the activity of β-lactams was not unexpected because these inhibitors are known to have low activity against MBLs,24 which are intrinsically present in Elizabethkingia spp. Lascufloxacin inhibited 41.1%, 57.1% and 63.6% of E. anophelis, E. meningoseptica and E. miricola, respectively. However, its activity was not better than that of levofloxacin, to which E. anophelis, E. meningoseptica and E. miricola exhibited susceptibility rates of 47.8%, 71.4% and 81.8%, respectively.
Performance of VITEK 2 for 18 commonly tested antibiotics
VMD rates were >1.5% for ciprofloxacin, moxifloxacin and vancomycin; the MD rates were >3% for amikacin, piperacillin/tazobactam, tigecycline and trimethoprim/sulfamethoxazole (Table 2). Similar discrepancy rates were also observed within each species with some variations. Vancomycin was inactive against all Elizabethkingia spp., but VITEK 2 gave false-positive results of vancomycin susceptibility in 3.7% of isolates. Figure S4 illustrates the poor correlation between the two testing methods for many antibiotics. Whether results obtained by the automated susceptibility testing system could help determine the treatment of Elizabethkingia spp. warrants further investigation. Notably, modest susceptibilities to fluoroquinolones, relatively high discrepancy rates and emerging resistance to other fluoroquinolones in Elizabethkingia spp.9,10 indicate that this drug class should be used cautiously.
Table 2.
| Agent | Total (n = 108) |
E. anophelis (n = 90) |
E. meningoseptica (n = 7) |
E. miricola cluster (n = 11)c |
||||
|---|---|---|---|---|---|---|---|---|
| MD | VMD | MD | VMD | MD | VMD | MD | VMD | |
| Amikacin | 12.1 | 0 | 5.6 | 0 | 0 | 0 | 72.7 | 0 |
| Ampicillin/sulbactam | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cefepime | 0.9 | 0 | 1.1 | 0 | 0 | 0 | 0 | 0 |
| Cefotaxime | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ceftazidime | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ciprofloxacin | 0 | 8.3 | 0 | 8.9 | 0 | 0 | 0 | 9.1 |
| Colistin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Gentamicin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Imipenem | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Levofloxacin | 0 | 0.9 | 0 | 1.1 | 0 | 0 | 0 | 0 |
| Meropenem | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Minocycline | 0.9 | 0 | 0 | 0 | 0 | 0 | 9.1 | 0 |
| Moxifloxacin | 0.9 | 5.6 | 0 | 6.7 | 0 | 0 | 9.1 | 0 |
| Piperacillin/tazobactam | 6.5 | 0 | 3.3 | 0 | 14.3 | 0 | 27.3 | 0 |
| Rifampin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tigecycline | 11.1 | 0 | 7.8 | 0 | 42.9 | 0 | 18.2 | 0 |
| Trimethoprim/sulfamethoxazole | 45.4 | 0 | 50 | 0 | 57.1 | 0 | 0 | 0 |
| Vancomycin | 0 | 3.7 | 0 | 1.1 | 0 | 14.3 | 0 | 18.2 |
Data are presented as percentages (%).
MD rates of >3.0% or VMD rates of >1.5% are underlined.
E. miricola cluster comprised E. miricola (6), E. bruuniana (3) and E. ursingii (2); no E. occulta isolates were identified.
In conclusion, Elizabethkingia spp., especially E. anophelis, emerged in multiple healthcare settings in Taiwan, causing both sporadic cases and outbreaks. Isolates were resistant to commonly tested and newly developed antibiotics. The absence of interpretive criteria specific to these organisms and low concordance between testing methods further confound therapeutic decision-making. In view of the increasing threat of Elizabethkingia spp. as emerging opportunistic pathogens, working guidelines and consensus statements on MIC testing and interpretation will be essential to guide clinical practice and to facilitate future clinical and basic research.
Supplementary Material
Acknowledgements
We express our sincere appreciation to the following hospitals for their participation in the TSAR: Buddhist Tzu Chi General Hospital; Cathay General Hospital; Changhua Christian Hospital; Cheng-Ching Hospital; Chung Shan Medical University Hospital; Da Chien General Hospital; Ditmanson Medical Foundation Chia-Yi Christian Hospital; Far Eastern Memorial Hospital; Hua-Lien Hospital; Jen-Ai Hospital; Kaohsiung Armed Forces General Hospital; Kaohsiung Chang Gung Memorial Hospital of the C.G.M.F.; Kaohsiung Medical University Chung-Ho Memorial Hospital; Kaohsiung Veterans General Hospital; Kuang Tien General Hospital; Lo-Hsu Foundation, Inc. Lotung Poh-Ai Hospital; Mennonite Christian Hospital; Min-Sheng Healthcare; National Cheng Kung University Hospital; Saint Mary’s Hospital Luodong; Show Chwan Memorial Hospital; Tungs’ Taichung MetroHarbor Hospital; Taichung Veterans General Hospital; Tainan Sin-Lau Hospital, the Presbyterian Church in Taiwan; Taipei City Hospital Heping Fuyou Branch; Taipei City Hospital Zhongxiao Branch; Taipei Veterans General Hospital; and Tri-Service General Hospital.
We also thank Shu-Man Shih, Institute of Population Health Sciences, National Health Research Institutes for drawing Figure S4.
Funding
This project was supported by an intramural grant from the National Health Research Institutes (IV-108-PP-09, IV-107-PP-09, IV-107-SP-01) and Ministry of Science and Technology (107-2320-B-400-010-MY3 and 109-2321-B-415-004-).
Transparency declarations
None to declare.
Supplementary data
Tables S1 and S2 are available as Supplementary data at JAC Online.
References
- 1. Nicholson AC, Gulvik CA, Whitney AM. et al. Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Antonie van Leeuwenhoek 2018; 111: 55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim KK, Kim MK, Lim JH. et al. Transfer of Chryseobacterium meningosepticum and Chryseobacterium miricola to Elizabethkingia gen. nov. as Elizabethkingia meningoseptica comb. nov. and Elizabethkingia miricola comb. nov. Int J Syst Evol Microbiol 2005; 55: 1287–93. [DOI] [PubMed] [Google Scholar]
- 3. Hsu MS, Liao CH, Huang YT. et al. Clinical features, antimicrobial susceptibilities, and outcomes of Elizabethkingia meningoseptica (Chryseobacterium meningosepticum) bacteremia at a medical center in Taiwan, 1999-2006. Eur J Clin Microbiol Infect Dis 2011; 30: 1271–8. [DOI] [PubMed] [Google Scholar]
- 4. Li Y, Kawamura Y, Fujiwara N. et al. Chryseobacterium miricola sp. nov., a novel species isolated from condensation water of space station Mir. Syst Appl Microbiol 2003; 26: 523–8. [DOI] [PubMed] [Google Scholar]
- 5. Kampfer P, Matthews H, Glaeser SP. et al. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int J Syst Evol Microbiol 2011; 61: 2670–5. [DOI] [PubMed] [Google Scholar]
- 6. Green O, Murray P, Gea-Banacloche JC.. Sepsis caused by Elizabethkingia miricola successfully treated with tigecycline and levofloxacin. Diagn Microbiol Infect Dis 2008; 62: 430–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chew KL, Cheng B, Lin RTP. et al. Elizabethkingia anophelis is the dominant Elizabethkingia species found in blood cultures in Singapore. J Clin Microbiol 2018; 56: e01445-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Han MS, Kim H, Lee Y. et al. Relative prevalence and antimicrobial susceptibility of clinical isolates of Elizabethkingia species based on 16S rRNA gene sequencing. J Clin Microbiol 2017; 55: 274–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin JN, Lai CH, Yang CH. et al. Clinical manifestations, molecular characteristics, antimicrobial susceptibility patterns and contributions of target gene mutation to fluoroquinolone resistance in Elizabethkingia anophelis. J Antimicrob Chemother 2018; 73: 2497–502. [DOI] [PubMed] [Google Scholar]
- 10. Jian MJ, Cheng YH, Chung HY. et al. Fluoroquinolone resistance in carbapenem-resistant Elizabethkingia anophelis: phenotypic and genotypic characteristics of clinical isolates with topoisomerase mutations and comparative genomic analysis. J Antimicrob Chemother 2019; 74: 1503–10. [DOI] [PubMed] [Google Scholar]
- 11. Cheng YH, Perng CL, Jian MJ. et al. Multicentre study evaluating matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically isolated Elizabethkingia species and analysis of antimicrobial susceptibility. Clin Microbiol Infect 2019; 25: 340–5. [DOI] [PubMed] [Google Scholar]
- 12. Perrin A, Larsonneur E, Nicholson AC. et al. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat Commun 2017; 8: 15483.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Teo J, Tan SY, Tay M. et al. First case of E. anophelis outbreak in an intensive-care unit. Lancet 2013; 382: 855–6. [DOI] [PubMed] [Google Scholar]
- 14. Figueroa Castro CE, Johnson C, Williams M. et al. Elizabethkingia anophelis: clinical experience of an academic health system in southeastern Wisconsin. Open Forum Infect Dis 2017; 4: ofx251.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin JN, Lai CH, Yang CH. et al. Comparison of four automated microbiology systems with 16S rRNA gene sequencing for identification of Chryseobacterium and Elizabethkingia species. Sci Rep 2017; 7: 13824.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sader HS, Fritsche TR, Jones RN.. Accuracy of three automated systems (MicroScan WalkAway, VITEK, and VITEK 2) for susceptibility testing of Pseudomonas aeruginosa against five broad-spectrum β-lactam agents. J Clin Microbiol 2006; 44: 1101–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gould IM, Gunasekera C, Khan A.. Antibacterials in the pipeline and perspectives for the near future. Curr Opin Pharmacol 2019; 48: 69–75. [DOI] [PubMed] [Google Scholar]
- 18. Kuo SC, Chang SC, Wang HY. et al. Emergence of extensively drug-resistant Acinetobacter baumannii complex over 10 years: nationwide data from the Taiwan Surveillance of Antimicrobial Resistance (TSAR) program. BMC Infect Dis 2012; 12: 200.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liou CH, Wu HC, Liao YC. et al. nanoMLST: accurate multilocus sequence typing using Oxford Nanopore Technologies MinION with a dual-barcode approach to multiplex large numbers of samples. Microb Genom 2020; 6: e000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kelly AJ, Karpathy SE, Gulvik CA. et al. A real-time multiplex PCR assay for detection of Elizabethkingia species and differentiation between Elizabethkingia anophelis and E. meningoseptica. J Clin Microbiol 2019; 57: e01619-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang X, Wang D, Wang Y. et al. Occurrence of antimicrobial resistance genes sul and dfrA12 in hospital environmental isolates of Elizabethkingia meningoseptica. World J Microbiol Biotechnol 2012; 28: 3097–102. [DOI] [PubMed] [Google Scholar]
- 22.CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically—Eleventh Edition: M07. 2018. [Google Scholar]
- 23. Kenna DTD, Fuller A, Martin K. et al. rpoB gene sequencing highlights the prevalence of an E. miricola cluster over other Elizabethkingia species among UK cystic fibrosis patients. Diagn Microbiol Infect Dis 2018; 90: 109–14. [DOI] [PubMed] [Google Scholar]
- 24. Lin JN, Lai CH, Yang CH. et al. Elizabethkingia infections in humans: from genomics to clinics. Microorganisms 2019; 7: 295.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jean SS, Hsieh TC, Ning YZ. et al. Role of vancomycin in the treatment of bacteraemia and meningitis caused by Elizabethkingia meningoseptica. Int J Antimicrob Agents 2017; 50: 507–11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.