Abstract
Over 1,500 missense variants of sodium channel hNav1.5, which are reported in the ClinVar database, are associated with cardiac diseases. For most of the variants, the clinical significance is uncertain (VUS), not provided (NP), or has conflicting interpretations of pathogenicity (CIP). Reclassifying these variants as pathogenic/likely pathogenic (P/LP) variants is important for diagnosing genotyped patients. In our earlier work, several bioinformatics tools and paralogue annotation method consensually predicted that 74 VUS/NP/CIP variants of 54 wild type residues (set w54) are potentially damaging variants (PDVs). Atomic mechanisms underlying dysfunction of the PDVs are unknown. Here we employed a recent cryo-EM structure of the hNav1.5 channel with likely inactivated pore domain (PD) and activated voltage-sensing domains (VSDs), and ad hoc models of the closed and open PD and resting VSDs to explore intersegment contacts of w54 residues. We found that 44 residues from set w54 contact 84 residues with 118 disease missense variants. These include 104 VUS/NP/CIP variants, most of which are associated with the loss-of-function Brugada syndrome (BrS1) or gain-of-function long QT syndrome (LQT3). Matrix representation of the PDVs and their contact variants facilitated recognition of coupled mutations associated with the same disease. In particular, BrS1-associated coupled mutations, which disturb the P-loops region with the selectivity filter slow inactivation gate, would cause the channel dysfunction. Other likely causes of the channel dysfunction include coupled BrS1-associated variants within VSDs that would destabilize their activated states and coupled LQT3-associated variants, which would stabilize the open PD or activated VSDs. Our study proposes mechanisms of channel dysfunction for scores of BrS1- and LQT3-associated variants, confirms status for 82% of PDVs, and suggests damaging status for their contact variants, which are currently categorized as VUS/NP/CIP variants.
Keywords: Brugada syndrome, cardiac arrhythmias, cryo-EM structure, homology modeling, intersegment contacts, long QT syndrome, Monte Carlo minimizations
Introduction
Sodium channels Nav1.5 are responsible for initiation and propagation of the action potential in cardiomyocytes. The pore-forming α-subunit of the Nav1.5 channel, which is encoded by gene SCN5A, folds from a single polypeptide chain of four homologous repeat domains (I-IV). Each repeat has six transmembrane (TM) helices (S1-S6) connected by intra- and extracellular loops, including a large extracellular membrane reentering P-loop. The latter contains membrane-descending helix P1 and membrane-ascending helix P2. In each repeat, helices S1 - S4 form a voltage-sensing domain (VSD), while helices S5, S6 and the P-loop contribute a quarter to the pore domain (PD). The selectivity-filter ring DEKA (Asp, Glu, Lys and Ala) divides the ion permeation pathway into two parts: extracellularly exposed outer pore and the inner pore, which in the open channel is exposed to the cytoplasm. Sodium channels adopt various conformations, which in electrophysiological studies are categorized as resting, open, fast inactivated and slow inactivated states.
Sodium channels allow permeation of inward depolarizing current (INa) during phase 0 of the action potential (Amin et al., 2010; Veerman et al., 2015). Nav1.5 dysfunctions cause various hereditary and acquired arrhythmogenic syndromes. Many genetic gain-of-function mutations increase INa or delay cardiac repolarization, thus prolonging the action potential (Savio-Galimberti et al., 2018). Such mutations are associated with the long QT syndrome (LQT3), a congenital cardiac disorder, which is characterized by prolonged QT interval and higher risk of life-threatening arrhythmia. Clinical manifestations of LQT3 vary from life-long asymptomatic disease course to syncope and cardiac arrest due to torsades de pointes. Mutations, which diminish INa, may decrease cardiac excitability and reduce velocity of electrical conduction, causing various clinical phenotypes such as Brugada syndrome type 1 (BrS1), sick sinus syndrome, sudden infant death syndrome, dilated cardiomyopathy, atrial fibrillation, and arrhythmogenic cardiomyopathy. BrS1 is diagnosed via typical ECG-phenomenon with ST-segment elevation in ventricular leads without any structural alterations of the heart (Coppola et al., 2019; Skinner et al., 2019).
Thousands genetic variants of hNav1.5 and other hNav1.x channels are described in public databases, including ClinVar (Landrum et al., 2016), Swiss-Var (Mottaz et al., 2010) and Humsavar (http://www.uniprot.org/docs/humsavar). In April 2021, the ClinVar database reported 1,513 missense variants of SCN5A. Among these, only 17% are classified as benign/likely benign or pathogenic/likely pathogenic (P/LP) variants according to the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) guideline (Richards et al., 2015). Data on pathogenicity is not provided (NP) for 200 variants, conflicting interpretations of pathogenicity (CIP) are reported for 139 variants, and 911 variants are of uncertain clinical significance (VUS).
The ACMG/AMP guideline recommends using computational predictions as a line of evidence for variant interpretation (Richards et al., 2015). Bioinformatics analysis of variants associated with BrS1 suggests that some mutations, which are currently classified as damaging, should be considered benign due to lack of experimental data on channel dysfunction (Kaltman et al., 2018). Incorrect classification of variants is an important problem in medical genetics. Indeed, in many cases a familial history or the pedigree data are lacking. Low frequency of disease-associated genetic variants in population further complicates correct data interpretation and delays diagnostics.
Earlier we employed several bioinformatics tools and the paralogue annotation method to reclassify 74 ClinVar-reported VUS/NP/CIP variants of 54 WT residues (set w54) as potentially damaging variants (PDVs) of hNav1.5 (Tarnovskaya et al., 2020). Here we used a recent cryo-EM structure of the hNav1.5 channel with likely inactivated PD and activated VSDs (Li et al., 2021), and ad-hoc models of the open and closed PD and resting VSDs to show that 44 residues from set w54 are involved in 84 intersegment contacts with residues for which 118 missense variants are reported in ClinVar, mostly as VUS/NP/CIP variants. Matrix representation of the PDVs and their contact variants facilitated recognition of coupled variants where substitution of either contact partner is associated with the same disease. We suggest that BrS1-associated variants in P-loops and VSDs would cause loss of channel function, respectively, by disturbing the selectivity-filter slow inactivation gate or destabilizing activated state of VSDs. LQT3-associated variants would cause the channel gain-of-function by increasing population of channels with the open pore. Our analysis confirms the status for most of earlier proposed PDVs, suggests that many of the 118 contact variants of the PDVs may be reclassified as P/LP variants, and proposes atomic mechanisms of loss-of-functions and gain-of function for scores of BrS1-associated and LQT3-associated mutations.
Methods
Energy Calculations and Optimization
Our methodology of molecular modeling with the ZMM program is described elsewhere, e.g., (Bruhova and Zhorov, 2010; Korkosh et al., 2014; Tikhonov and Zhorov, 2017). Briefly, we use the AMBER force field (Weiner et al., 1986) and calculate nonbonded interactions with the distance cutoff of 9 Å and a shifting function (Brooks et al., 1985). Electrostatic interactions were calculated with the distance- and environment-dependent dielectric function (Garden and Zhorov, 2010) without the distance cutoff for interactions involving ionized groups. The models were optimized by Monte Carlo energy minimizations (MCM) (Li and Scheraga, 1987) in the space of torsional angles and bond angles of prolines (Zhorov, 1981). MCM trajectories were terminated when 2,000 minimizations did not decrease energy of the apparent global minimum.
Modeling PD in the Open and Closed States
The hNav1.5 structure with quinidine (PDB ID: 6lqa) captured PD in likely inactivated state and VSDs in the activated states (Li et al., 2021). To model PD in the activated and closed states, we used as templates the open- and closed-pore structures of the NavAb channel (PDB codes 5vb8 and 5vb2, respectively) (Lenaeus et al., 2017). The structures of Nav1.5 and NavAb were 3D aligned by minimizing root mean square deviations of Cα atoms in the P1 helices, which are the most 3D-conserved elements of P-loop channels (Tikhonov and Zhorov, 2012), from Cα atoms of sequentially matching residues in the Kv1.2/Kv2.1 channel, the first eukaryotic P-loop channel whose crystal structure was obtained at a relatively high resolution below 2.5 Å (PDB ID: 2R9R) (Long et al., 2007). The NavAb and hNav1.5 channels have rather similar folding of the transmembrane (TM) and P1 helices, but different folding of extracellular loops, especially in the PD. Therefore, we imposed distance constraints between Cα atoms in the TM and P1 helices of hNav1.5 and matching Cα atoms of the template, whereas no distance constraints were imposed to the intra- and extracellular loops. A distance constraint is a flat-bottom parabolic function that imposes the energy penalty with the force constraint of 10 kcal·mol−1 Å−2 if a Cα atom in the model deviates from the template matching atom by more than 2 Å. The 3D alignment was performed by step-wise reducing distances between the Cα atoms in the model and template. At each step, the energy was MC-minimized with all degrees of freedom allowed to vary. More methodological details on large-scale transformations of PD can be found elsewhere (Korkosh et al., 2019).
Modeling VSDs in the Resting States
To model VSDs in the resting states, we employed as a template the crystal structure of the prokaryotic sodium channel NavAb with deactivated VSDs (PDB ID: 6p6x) (Wisedchaisri et al., 2019). Methodology is similar to that used for modeling the channel with the open and closed PD.
Designations and Visualization of Residues
We designate a residue by one-letter code with superscripted residue number and superscripted prefix indicating the channel segment where the residue is located. Experimental structures and models were visualized with the PyMol Molecular Graphics System, version 0.99rc (Schrödinger, New York, NY).
Results and Discussions
Intersegment Contacts in the Cryo-EM Structure
Earlier, we employed several bioinformatics tools and paralogue annotation method to reclassify 74 missense VUS/NP/CIP variants of w54 residues in the hNav1.5 channel as PDVs (Tarnovskaya et al., 2020). Following this publication, eight additional VUS/NP/CIP variants of the w54 residues were reported in ClinVar (version April, 2021). Therefore, currently the PDV set includes 82 variants.
We used a recent cryo-EM structure of the hNav1.5 channel obtained in nanodiscs (Li et al., 2021) to find intersegment contacts of the w54 residues. We instructed PyMol to display 3D neighbor residues within 5 Å from each query residue of set w54. Since the cryo-EM structure lacks hydrogen atoms, the distances were measured between heavy atoms. We have selected those 3D neighbors, which in the amino acid sequence are at least 5 positions away from the query residue and have sidechains facing the query residue sidechain (Cα atoms of glycines were considered as sidechains). We further ensured that each 3D neighbor of the query residue has at least one ClinVar-reported missense variant. We found 64 contact residues, which satisfied the above criteria and 85 missense variants of these residues. If two WT residues form a contact, their variants are also considered to contact each other even though they may be farther than 5 Å from each other.
Table 1 shows PDVs and their contact variants in the PD. For each PDV we show location (channel segment), all contact variants, associated syndromes, and their clinical significance. Table 2 shows analogous data for VSDs. Among 85 contact variants, 74 are VUS/NP/CIP variants and clinical significance of 11 variants is P/LP. Four pairs of residues from set w54 are contacting each other (columns “e” in Table 1). Contacts involving channel segments that undergo significant transition upon channel gating are marked “S” in column “f”.
TABLE 1.
PDVs in the pore domain and their contacts a .
| PDVs | Contacts | ||||||
|---|---|---|---|---|---|---|---|
| Variant b | Syndrome c , d | Variant | Syndrome d | Significance | e | f | |
| IS5A242T | A | IS6Q419P | BrS1, NP | VUS | S | ||
| V | HCM | ||||||
| IS5L250V | BrS1 | IS6V411M | LQT3, BrS1, CVP | P | S | ||
| IS5S262G | A, BrS1 | IP1F358S | NS | VUS | |||
| IS5V263I | Multi | IVS4I1633V | BrS1 | VUS | S | ||
| IS5-P1G274S | BrS1 | IS5-P1D356Y | BrS1 | LP | |||
| N | NP/BrS1 | P/LP | |||||
| IS6A413E | LQT3 | IIS6L935P | BrS1 | NP | S | ||
| S | NS | ||||||
| T | A, LQT3,BrS1 | ||||||
| IIS5G857D | A, BrS1 | IIS5-P1M881I | LQT3 | VUS | 1 | ||
| IIS5-P1M881I | LQT3 | IIS6F919S | NS | LP | 1 | ||
| IIS5G857D | A, BrS1 | VUS | |||||
| IIP1F892L | CVP | IVS6V1451D | BrS1 | VUS | 2 | ||
| I | BrS1 | L | NS, A | VUS | |||
| IIP1R893H | BrS11,3,4,8–10 | IIP2E901K | BrS1 | VUS | 2 | S | |
| C | BrS1 | ||||||
| IIIS5V1324I | A, BrS1, NP | IIS3W1271C | BrS1 | NP | |||
| IIIS5L1342F | BrS1 | IIIP1Y1409C | BrS1 | NP | S | ||
| IIIP1L1410P | NP | IIIS5-P2V1400G | CVP | VUS | |||
| IIIP2I1424F | NP | IIIS5-P1F136°C | BrS1 | NP | |||
| IIIS5-P1V1400G | CVP | VUS | |||||
| IIIS6V1451D | BrS11,3 | IIP1F892L | CVP | VUS | |||
| L | A, NS | I | BrS1 | NP | |||
| IIIS6N1463K | NP | IIS6F934S | BrS1 | VUS | |||
| IIIS6F1465L | LQT3 | IIIS5V1337I | A | VUS | S | ||
| IIIS5I1334V | LQT3, BrS1, NP | CIP | S | ||||
| IVS6F176°C | BrS1 | P | S | ||||
| IVS6L1761F | LQT3 | NP | S | ||||
| H | LQT3 | NP | |||||
| IIIS6I1466T | NP, A | IIS6F934S | BrS1 | VUS | S | ||
| V | NP | IVS6V1764F | BrS1 | NP | S | ||
| IVS6F176°C | BrS1 | P | S | ||||
| IIIS6V1468A | A, BrS1,NP | IIIS5S1333Y | LQT3 | NP | S | ||
| IIIS5V1337I | A | VUS | S | ||||
| IIIS6I1469F | NP | IIIS5I1334V | LQT3, BrS1, NP | CIP | S | ||
| IVS6L1761F | LQT3 | NP | S | ||||
| H | LQT3 | NP | S | ||||
| IVS6V1764F | BrS1 | NP | |||||
| IVS5V1667L | NP | IVS6I1758V | A, BrS1 | VUS | |||
| IVS5-P1D1690N | A, BrS1, NS14,15,16 | IP2R383K | BrS1 | VUS | |||
| IVP1M1701I | BrS1, DCM17 | IVS5M1668T | A | VUS | |||
| IS6M390I | A | VUS | |||||
| V | BrS1, A | VUS | |||||
| IS6M394I | A | VUS | |||||
| L | A | VUS | |||||
| IVS5I1660V | NP, CVP, BrS1, LQT31,3,13,14 | IVS6V1764F | BrS1 | NP | 3 | S | |
| IVS6L1761F | LQT3 | NP | S | ||||
| H | LQT3 | NP | S | ||||
| IIIS4-S5V1323G | BrS1 | NP | S | ||||
| I | A, BrS1 | VUS | S | ||||
| IVS6M1766I | CVP, BrS1 | VUS | S | ||||
| K | NP, BrS1, SSS | VUS | |||||
| V | BrS1 | VUS | |||||
| T | HCM | VUS | |||||
| L | LQT3 | P | |||||
| IIIS6−IVS1F1486L | SIDS | NP | |||||
| IVP1L1704H | BrS1 | IVS5M1668T | A | VUS | |||
| IVP1T1709M | A, BrS1,CVP,NP1,3,18 | IIIP1-P2K1419E | BrS1 | NP | |||
| IP1-P2W374G | BrS1 | NP | |||||
| IP1-P2Q371R | BrS1 | VUS | |||||
| IS6I397T | BrS1, LQT3, NP | CIP | |||||
| IVS6M1766I | BrS1, CVP | IS6N406K | LQT3 | P, LP | 3 | S | |
| K | NP, BrS1, SSS | IS6N406S | BrS1 | NP | |||
| V | BrS1 | IVS5I1660V | A, BrS1, CVP, LQT3 | CIP | |||
| T | HCM | ||||||
| L | LQT3 | ||||||
See Energy Calculations and Optimization Section for definition of contacts.
All PDVs were listed as VUS in ClinVar-2019. In ClinVar-2021, conflicting interpretations of pathogenicity is reported for three variants (R894H, G1262D, I1660V) and two variants (D197G, L225V) are classified as likely pathogenic.
Superscripted numbers refer to clinical and experimental studies of the variants. 1. Huang et al. (2017), 2. Ortiz-Bonnin et al. (2016), 3. Kapplinger et al. (2010), 4. Walsh et al. (2014), 5. Calloe et al. (2013), 6. Liang et al. (2016), 7. Son et al. (2018), 8. Zakliaz’minskaia et al. (2013), 9. Kapplinger et al. (2015), 10. Rudic et al. (2016), 11. Shin et al. (2004), 12. Priganc et al. (2017), 13. Cordeiro et al. (2006), 14. Selga et al. (2015), 15. Núñez et al. (2013), 16. Zeng et al. (2016), 17. Bodian et al. (2017), 18. Li et al. (2018), 19. Brewer et al. (2020).
Abbreviations: A, Arrhythmia; AF, atrial fibrillation; BrS1, Brugada syndrome; CA, Cardiac Arrest; CIP, Conflicting interpretations of pathogenicity; CVP, Cardiovascular phenotype; DCM, Dilated cardiomyopathy; HCM, Hypertrophic cardiomyopathy; LP, Likely Pathogenic, LQT3, Long QT syndrome Type 3; M, Multiple diseases; NP, not provided; NR, Variants are not reported in ClinVar; NS, Not Specified; P, Pathogenic; PFHB-1, Progressive Familial Heart Block type 1A; PFVF1, Paroxysmal Familial Ventricular Fibrillation 1; SIDS, Sudden Infant Death Syndrome; VUS, Variant of Uncertain Significance.
Matching numbers show WT residues that are in contact in the cryo-EM structure.
state-dependent contacts involving residues that move upon activation gating (located C-terminal to gating-hinge in KcsA).
TABLE 2.
PDVs and their contact variants in the open-PD model a .
| PDV | Contacts | |||
|---|---|---|---|---|
| Variant | Syndrome | Variant | Syndrome | Significance |
| IIIS6N1463K | NP | IIS6S94°C | BrS1 | VUS |
| IIS6S941C | CP | VUS | ||
| N | LQT3 | P | ||
| IIIS6F1465L | LQT3 | IIIS5A1330P | LQT3 | NP |
| T | BrS1, LQT3, More | P/LP | ||
| IIIS6I1466T | NP, A | IIS6S941C | CP | VUS |
| V | NP | N | LQT3 | P |
| IIIS6V1468A | A, BrS1,NP | IIIS5A1330P | LQT3 | NP |
| T | BrS1, LQT3, More | P/LP | ||
Contacts in the cryo-EM structure (Table 1) are not shown.
Matrices of Contact Variants
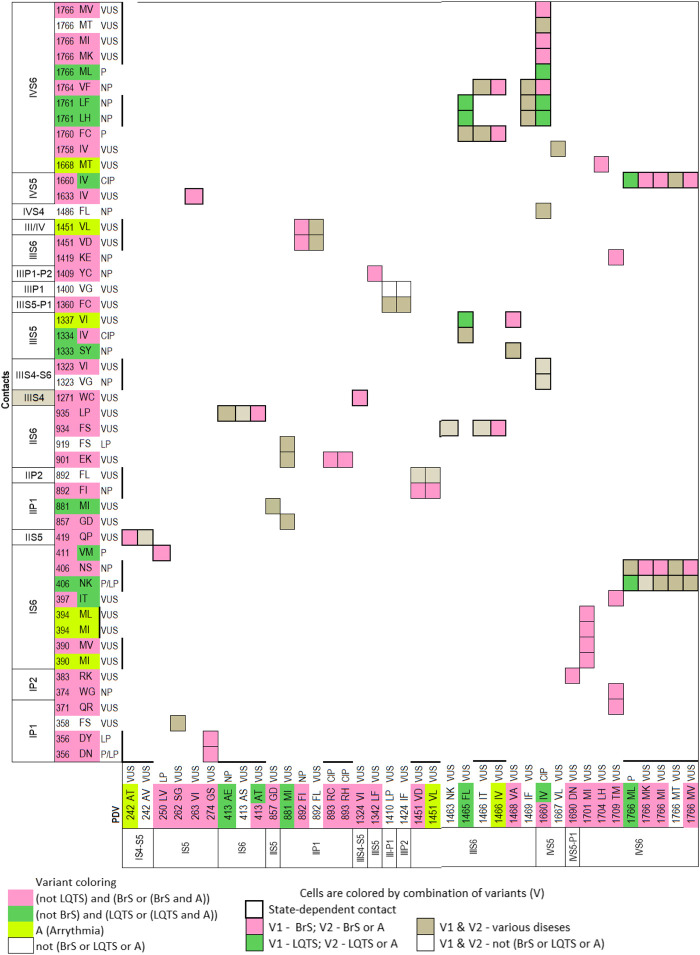
To facilitate recognition of disease-related contacts, we schematically represented them in colored matrices. Each matrix represents an individual domain because inter-domain contacts of PDVs are rare. A matrix of contact variants for the inactivated PD (Figure 1) has four bottom rows, which represent PDVs, their location, and clinical significance; four left columns represent analogous characteristics of contact variants of PDVs. Variants associated with BrS1, LQT3 or non-specified arrhythmia are colored pink, green or yellow, respectively. Variants associated with other diseases are not colored. Variants, which are associated with more than one disease, are shown with two colors. Each cell in the matrix body links a PDV and its contact variant.
FIGURE 1.
Matrix of contact variants in the PD cryo-EM. Bottom rows (X1-X4) represent w54 residues and their PDVs. X1 and X2 show PD segments and UniProt residue numbers. X3 shows one-letter codes of WT residues and their variants. X4 indicates clinical significance of variants. Variants associated with BrS1, LQT3, non-specified Arrhythmias or other diseases are colored, respectively, pink, green, yellow, or not colored. If besides BrS1 or LQT3, a variant is associated with a non-specified arrhythmia, it is colored pink or green, respectively. If various diseases are reported for a variant, two colors are used. Left columns (Y1-Y4) show analogous features of contact variants of w54 residues. A colored cell in the matrix body links a PDV and its contact variant. If a WT residue has m PDVs and n contacts variants, a cluster of m x n colored cells shows all variant combinations. If PDV and its contact variant have the same color, it is applied to the cell. If a cell links pink and yellow variants, it is colored pink. If a cell links green and yellow variants, it is colored green. If a variant has color A and its contact variants colors A and B, color A is applied to the cell. Otherwise the cell is grey. If a residue is located in cytoplasmic halves of S6 helices, which move upon the channel gating, respective cell is bold-boxed.
The matrix highlighted multiple contacts where mutation of either WT residue is associated with the same syndrome. A likely cause is that a disease-related contact contributes to stabilization (or destabilization) of a particular channel state; a weakened or strengthened contact would similarly affect the channel state stability regardless of the mutated contact partner. Structural analysis of all contacts involving PDVs is beyond goals of this study. Below we propose mechanisms of dysfunction for some PDVs and their contact variants.
Contacts in P-Loops
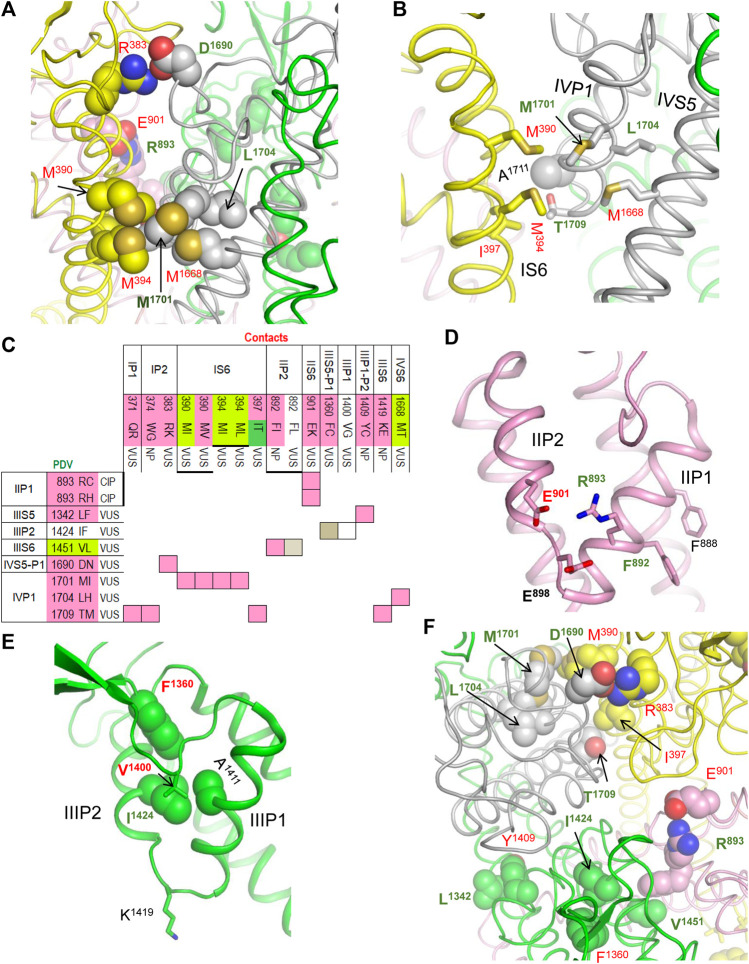
In our models with the closed and open PD, which are described in a later section, conformations of P-loops are rather similar to those in the cryo-EM structure. And although P-loops do undergo conformational changes, e.g., upon slow inactivation, here we consider their folding as relatively state-independent. Figure 2 shows P-loops with some contacts, which apparently stabilize their folding and a matrix of respective contacts variants. Five hydrophobic residues (IS6M390, IS6M397, IVS5M1668, IVP1M1701, and L1704) form a tight cluster (Figures 2A,B). In variants M390I/V, M394I, M1668T, and M1701I, methionine substitutions with smaller residues (Figure 2C) would relax the cluster and increase flexibility of the smallest involved helix, IVP1. Substitutions IVP1L1704H and IS6I397T would also increase flexibility of this helix. The C-end of helix IVP1 approaches the DEKA-ring alanine in the SF region (Figure 2B), which harbors the SF gate. The outer-pore of a prokaryotic sodium channel exhibits flexibility during slow inactivation (Chatterjee et al., 2018). In the Nav1.4 channel, the P-loop of domain IV is more flexible than in other domains (Tsushima et al., 1997a; Tsushima et al., 1997b). The above mutations would increase flexibility of helix IVP1, disturb the SF gate, and facilitate the channel transition to the slow-inactivated state. This proposition is supported by the fact that most substitutions in P-loops are associated with the loss-of-function syndromes, especially with BrS1 (Figure 2C).
FIGURE 2.
Residue contacts that stabilize P-loops in the cryo-EM structure. Backbones and sidechain carbon atoms in repeats I, II, III and IV are yellow, pink, green and gray, respectively. Residues w54 and their contacts are indicated with green and red labels, respectively. (A,B) Intra-membrane views of P-loops in the cryo-EM structure. Residues M390, M394, M1668, M1701 and L1704 form a tight hydrophobic cluster, which apparently stabilizes mutual disposition of helices IS6, IVS5 and IVP1. (C) Matrix of contact variants in P-loops. Majority of coupled variants are associated with BrS1 and arrhythmia, which also may be a loss-of-function disease. (D) Intra-membrane view of helices IIP1 and IIP2 whose mutual disposition is stabilized by salt bridge R893:E901. Side-chain conformation of the selectivity-filter DEKA glutamate E901 is stabilized by its salt bridge with R898. Helical structure of helix IIP1 is stabilized by π-stacking F888 and F892. (E) Intra-membrane view of helices IIIP1 and IIIP2 whose mutual disposition is stabilized by a cluster of four hydrophobic residues. The selectivity-filter DEKA lysine K1419 is shown by sticks. (F) Extracellular view of salt bridges R383:D1690, R893:E901 and hydrophobic contacts I397:T1709, L1342:Y1409 and I1424:F1360.
Several other mutations involving P-loops would also increase their flexibility. For example, in the cryo-EM structure, IIP1F892 and IIP1F888 form a stacking pair (Figure 2D) that can stabilize the α-helix (Butterfield et al., 2002). Mutation IIP1F892L affects the inter-segment contact with IIIS6V1451 (Table 1) and destroys the stacking pair with likely destabilization of helix IIP1. A sequential neighbor of F892, IIP1R893 forms several strong contacts, including a salt bridge with IIP2E901, which stabilizes mutual disposition of helices IIP1 and IIP2 (Figure 2D). Substitution of any partner in the salt bridge is associated with BrS1 (Figure 2C) likely due to destabilization of mutual disposition of helices IIP1 and IIP2. Another strong contact of IIP1R893 involves the SF glutamate IIP1-P2E898, which is proposed to adopt multiple conformations in a cycle of sodium ion permeation (Zhorov, 2021). The cryo-EM structure of Nav1.5 is obtained in complex with quinidine that favors inactivated channels (Li et al., 2021). However, it is unknown whether the SF gate is captured in the open or closed state. A strong electrostatic attraction to IIP1R893 may stabilize the E898 conformation in the slow-inactivated state.
Several other contacts also stabilize folding of P-loops. An example is a knob-into-hole contact IIIP2I1424 : IIIP1A1411 that stabilizes the mutual disposition of helices IIIP1 and IIIP2 with the selectivity-filter lysine IIIP1-P2K1419 (Figure 2E). Mutation I1424F affects contact with A1411, as well as contacts with IIIS5-P1F1360 and IIIS5-P1V1400 whose variants are associated with diseases (Table 1). Another example is a salt bridge IVS5-P1D1690 : IP2R383 (Figures 2A,F), which is absent in BrS1-associated variants IVS5-P1D1690N and R383K. In a functional study, variant D1690N reduced INa and shifted the steady-state activation by +7 mV (Zeng et al., 2016).
Thus, majority of mutations affecting the P1 and P2 helices are associated with the loss-of-function BrS1. Apparently these mutations distort the SF gate and increase population of slow-inactivated channels. For comparison, two residues beyond P1 and P2 helices, IIS5-P1M881 and IIS5G857, form a knob-into-hole contact that would not directly affect the SF region. Both IIS5-P1M881I and IIS5G857D are PDVs (Table 1). However, unlike the above-described BrS1-associated variants that affect folding of P-helices, M881I is associated with the gain-of-function LQT3, while G857D is associated with BrS1.
Contacts in the Channel Models with the Open and Closed PD
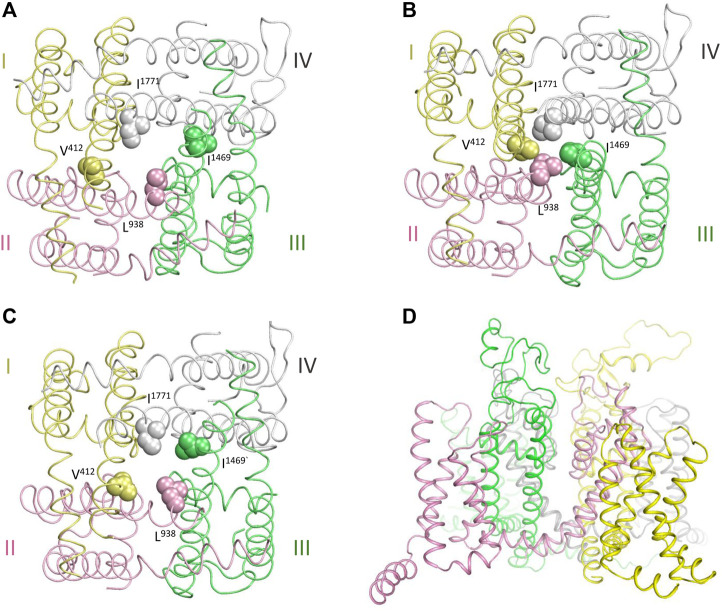
Figures 3A–C show cytoplasmic views of the PD, respectively, in the cryo-EM structure, closed state, and open state. Space-filled hydrophobic residues at the cytoplasmic ends of S6 helices highlight different dimensions of the activation gate in these states. Figure 3D shows intra-membrane view of the full-fledged channel model with the open PD and activated VSDs. PDVs and respective contact variants, which are specific for the models with the closed PD and open PD, are presented in Tables 2, 3.
FIGURE 3.
Cryo-EM structure and homology model of the hNav1.5 channel. Repeats I,II, III and IV are yellow, pink, green and gray, respectively. (A, C) Cytoplasmic views at the pore domain in the cryo-EM structure (A), and NavAb-based models of the channel states with the closed (B) and open (C) pore domain. Space-filled residues at the cytoplasmic ends of S6 helices highlight different dimensions of the activated gate. (D) Intra-membrane view of the channel model with the open PD.
TABLE 3.
PDVs and their contact variants in the closed-PD model a .
| PDV | Contacts | |||
|---|---|---|---|---|
| Variant | Syndrome | Variant | Syndrome | Signif |
| IS6A413E | LQT3 | IVS6N1774D | LQT3 | NP |
| S | NS | S | BrS1 | NP |
| T | A, LQT3,BrS1 | Y | NS | CIP |
| IIIS6N1463K | NP | IIS5S835L | BrS1 | NP |
| A | BrS1 | VUS | ||
| IIIS6F1465L | LQT3 | IVS6I1768V | BrS1, LQT3 | P |
| IIIS6I1469F | NP | IVS6I1768V | BrS1, LQT3 | P |
| IVS6L1772P | BrS1 | VUS | ||
| V | LQT3 | NP | ||
| IVS6M1766I | CVP, BrS1 | IVS6G1661R | BrS1 | NP, CIP |
| K | NP, BrS1, SSS | |||
| V | BrS1 | |||
| T | HCM | |||
| L | LQT3 | |||
Contacts in the cryo-EM structure (Table 1) are not shown.
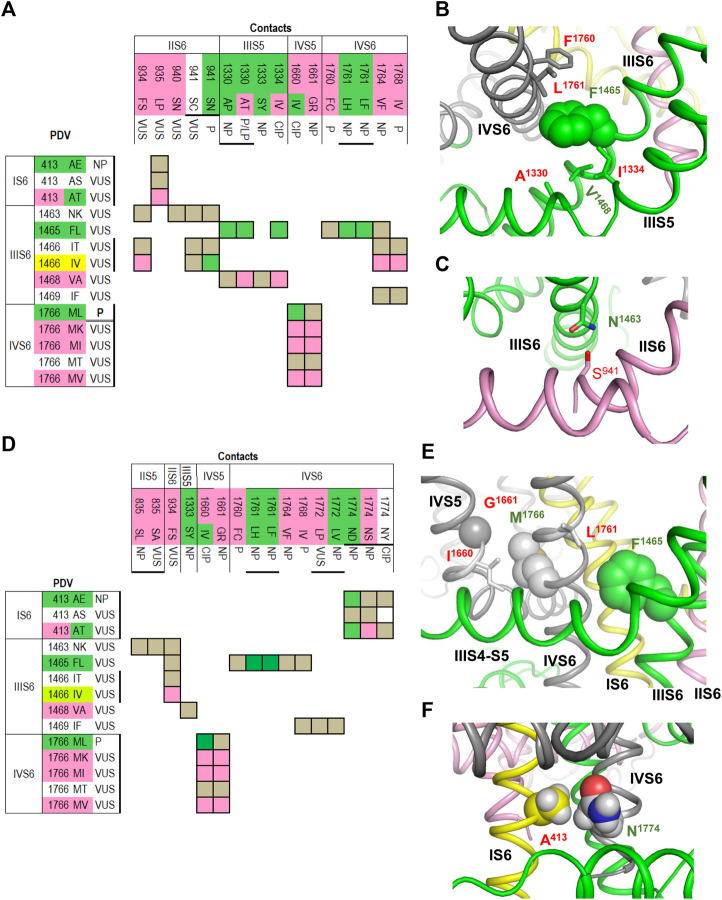
Figure 4A shows matrix of contact variants in the cytoplasmic halves of TM helices of the open PD. A hydrophobic cluster involving IIIS5A1330, IIIS5I1334, IIIS6F1465, IIIS6V1468 and IVS6L1761 (Figure 4B) apparently stabilizes the open PD. Substitutions of A1330 with bigger proline or threonine, or substitution of F1465 with more flexible leucine are associated with the gain-of-function LQT3 likely because respective variants over-stabilize the open PD. In LQT3-associated variants L1761H/F, the open state of PD would be stabilized due to π-stacking of the aromatic substitutions with F1465.
FIGURE 4.
Open and closed conformations of PD. (A–C) Open PD model. (A) Matrix of contact variants shows only those PDVs and their contacts, which are unique in the open PD model. Most of coupled variants are associated with either BrS1 or LQT3. (B) Hydrophobic contacts L1761:F1465:F1760 and A1330:V1468:I1334 stabilize, respectively, mutual disposition of helices IIIS6/IVS6 and IIIS5/IIIS6. (C) H-bond S941:N1463 stabilizes mutual disposition of helices IIS6/IIIS6. (D–F) Closed PD model. (D) Matrix of contact variants shows only those PDV and their contacts, which are unique in the closed-PD model. (E) Mutual disposition of helices IIIS6 and IVS6 in the closed PD is stabilized by hydrophobic contacts F1465:L1761 with side chain conformations different from those in the open-PD model. In coupled LQT3-associated variants, substitutions F1465L or L1761F/H would destabilize the closed PD. Mutual disposition of helices IVS5/IVS6 is stabilized by a hydrophobic contact M1761:I1660 and a knob-into-hole contact M1761:G1661. (F) Mutual disposition of helices IS6/IVS6 is stabilized by a tight hydrophobic contact between A413 and methylene group of N1774. Replacement of any contact partner with a large residue would destabilize the closed PD, a likely cause of LQT3 associated with these mutations.
Variant IVS6F1760C is associated with BrS1. In functional studies, F1760C impaired fast inactivation, accelerated recovery from inactivation (Carboni et al., 2005; O'Leary and Chahine, 2015), and enhanced slow inactivation (Carboni et al., 2005). Mutant channel F1760A, which is used as a model for drug binding assays, causes a ∼ 20 mV depolarizing shift of steady-state inactivation (Carboni et al., 2005). LQT3-associated mutation A1330P increased the window current, whereas mutation A1330T, which is associated with LQT3, BrS1 and other diseases, increased the window current, impaired inactivation and accelerated recovery from inactivation (Smits et al., 2005b).
Figure 4C shows the H-bond between IIIS6N1463 and IIS6S941 that is specific for the open PD. This is an example of inter-repeat H-bonds involving asparagine residues, which are exceptionally conserved in sodium and calcium channels. Earlier such H-bonds were predicted (Tikhonov et al., 2015) and demonstrated (Du et al., 2018) to stabilize the open conformations of these channels. LQT3-associated variant S941N would over-stabilize the H-bond and hence the open state of PD, whereas variant S941C, which is associated with a non-specific cardiovascular phenotype, may destabilize the H-bond and thus the channel open state. Variant S941N was found in an infant died from the sudden infant death syndrome, the disease associated with both Nav1.5 gain-of-function and loss-of-function (Amin et al., 2010). The mutant channel S941N expressed in Xenopus oocytes demonstrated the increased late current (Schwartz et al., 2000).
Contacts between cytoplasmic halves of TM helices in the closed PD are summarized in matrix shown in Figure 4D. Some open-PD contacts, e.g., IVS6L1761 : IIIS6F1465, are also seen in the closed PD, but the residue orientations in the two models are substantially different (cf. Figures 4B, E). A knob-into-hole contact IVS6M1766 : IVS5G1661, which is seen only in the closed-PD model (Table 3), and hydrophobic contact IVS6M1766 : IVS5I1660 (Figure 4E) apparently stabilize the closed PD conformation. It would be destabilized in LQT3-associated variants M1766L and I1660V where large hydrophobic residues are substituted with smaller ones. In functional studies, mutation I1660V decreased the current, which was rescued by 48 h incubation at room temperature or by mexiletine; the variant was suggested to affect the channel folding, while mexiletine or room temperature stabilized the folding (Cordeiro et al., 2006). Mutation M1766L significantly decreased the sodium channel expression, which was partially rescued by mexiletine; however, this mutation also demonstrated a 10-fold increase of the persistent late sodium current (Valdivia et al., 2002).
A tight contact between IS6A413 and the methylene group of IVS6N1774 is seen only in the closed-PD model (Table 3 and Figure 4F). Substitution of either contact partner with a larger residue would destabilize the closed conformation, resulting in LQT3 (Figure 4D). Mutant channel N1774D expressed in the CHO-K1 cells demonstrated increased peak sodium current density, late current and enhanced activation (Kato et al., 2014). Similar electrophysiological changes, as well as markedly increased duration of the action potential, were found in the cardiac myocytes derived from induced pluripotent stem cells from a boy with LQT3-associated mutation N1774D (Hirose et al., 2020). Variant N1774S was found in a Chinese family with BrS1 (Du et al., 2005).
In the cryo-EM structure, IIIS6F1465 forms hydrophobic contacts with four residues whose three variants are associated with LQT3 (Table 1). In our closed-PD model, IVS6I1768 forms a tight contact with F1465 whose substitution F1465L is also associated with LQT3 (Table 3). Substitutions F1465L or I1768V would destabilize the hydrophobic contact and thus the closed conformation of PD. LQT3-associated substitution I1768V would weaken contacts that stabilize both the inactivated and closed-state conformation of the PD. In functional studies, mutant channel I1768V accelerated recovery from inactivation and increased the channel availability (Rivolta et al., 2002). In another study, mutant channel I1768V accelerated recovery from inactivation, impaired slow inactivation, but did not affect steady-state activation and inactivation (Groenewegen et al., 2003).
Contacts in VSDs
PDVs and their contacts in the cryo-EM structure with the activated VSDs and models with resting PDVs are shown in Tables 4, 5, respectively. Many contacts switch upon VSD transitions between the activated-state conformations, which are captured in the cryo-EM structure, and resting-state models. We summarized these contacts in four matrices (Figures 5–7) where cells are labeled “C”, “M” or “B” depending on whether respective contacts are seen only in the cryo-EM structure, only in the resting-state model, or both in the cryo-EM structure and the model. It should be noted that ionized residues in VSDs switch contacts during channel gating. Impact of mutations in such contacts on relative stability of different channel states is difficult to predict. On the other hand, respective contacts are promising targets for further studies.
TABLE 4.
PDVs and their contacts in activated VSDsa.
| PDVs | Contacts | |||||
|---|---|---|---|---|---|---|
| Variantb | Syndromec | Mutation | Syndromed | Significance | e | f |
| IS1M138I | AF | IS4T229S | BrS1 | VUS | S | |
| T | DCM1 | IS4-S5V232I | A, BrS1, CVP | CIP | S | |
| IS1I141N | BrS1 | IS4T229S | BrS1 | VUS | S | |
| F | NS | IS4R225Q | BrS1, LQT3, More | CIP | S | |
| W | BrS1, LQT3, More | P/LP | S | |||
| IS4A226D | BrS1 | VUS | S | |||
| V | BrS1, LQT3, More | CIP | ||||
| IS4V232I | A, BrS1, CVP | CIP | ||||
| IS2R179P | BrS1 | IS1L128P | BrS1 | VUS | 4 | |
| Q | BrS1 | IS2-S3T187S | BrS1 | VUS | ||
| I | BrS1 | NP | ||||
| A | BrS1, CA | CIP | ||||
| IS2-S3T187S | BrS1 | IS2R179P | BrS1 | VUS | 4 | |
| I | BrS1, NP | Q | BrS1 | VUS | ||
| A | BrS1 | |||||
| IS3D197G | BrS1 | IS2K175D | BrS1 | NP | ||
| H | BrS1 | IS2E171G | BrS1 | VUS | ||
| Y | DCM | |||||
| IIS3E795K | BrS1 | IIS4L807P | BrS1, NP | VUS | S | |
| IIS4-S5L825P | BrS1 | IIS4F816L | A | VUS | S | |
| Y | LQT3 | NP | S | |||
| IIIS5V1340L | CVP | VUS | ||||
| I | NP, BrS1, A | VUS | ||||
| IIS5I848F | A,BrS1, LQT31 | IS4A226V | BrS1, LQT3, More | CIP | S | |
| D | BrS1 | VUS | S | |||
| IS4I230T | ||||||
| IIIS1A1221V | A, BrS1 | IIIS2L1239P | BrS1 | NP | ||
| IIIS4R1306L | CVP | IIIS2D1243E | BrS1 | VUS | S | |
| H | NP, BrS1 | NG | BrS1, More | VUS | S | |
| C | A | IIIS2E1225K | BrS1 | VUS | S | |
| S | BrS1 | LQT3, BrS1, A | VUS | S | ||
| IIIS4R1316L | BrS1 | IIIS1E1208K | A, NP | VUS | S | |
| Q | BrS1 | |||||
| IVS2E1574K | BrS11,3 | IVS1I1534S | LQT3 | VUS | S | |
| IVS2C1575Y | A, BrS1 | IVS3F1596I | LQT3, BrS1, More | VUS | ||
| S | A | |||||
| IVS3N1592Y | BrS1 | IVS4R1638P | NP | LP | ||
| IVS4-S5I1643L | NP | IVS3I1637T | NS | VUS | ||
a-e footnotes to Table 1.
TABLE 5.
PDVs and their contacts in resting VSDs a .
| PDV | Contacts | |||
|---|---|---|---|---|
| Variant | Syndrome | Variant | Syndrome | Signif. |
| IS1I141N | BrS1 | IS4V223A | A | LB |
| F | NP | L | BrS1 | NP |
| IS2R179P | BrS1 | IS2-S3H184Q | BrS1, CVP | CIP |
| Q | BrS1 | IS2-S3A185T | BrS1, LQT3, A | CIP |
| V | BrS1, A | VUS | ||
| IS4R219C | A, BrS1 | IS2E161Q | BrS1 | NP |
| H | BrS1, DCM | K | BrS1 | CIP |
| P | LQT3, BrS1, More | |||
| IIS3E795K | BrS1 | IIS4R808P | LQT3 | NP |
| C | A | VUS | ||
| H | LQT3, A | VUS | ||
| IIS4-S5L825P | BrS1 | IIS4A819P | BrS1 | VUS |
| IIS5I848F | A, BrS1, LQT3 | IS4V223A | A | LB |
| L | BrS1 | NP | ||
| IS4L227P | BrS1 | CIP | ||
| IIIS4R1306L | CVP | IIIS3I1278N | LQT3 | NP |
| H | NP, BrS1 | |||
| C | A | |||
| S | BrS1 | |||
| IVS2C1575Y | A, BrS1 | IVS3D1595H | BrS1, LQT3, More | LP |
| S | A | N | BrS1, More | P |
| IVS4-S5I1643L | NP | IS5D252V | BrS1 | VUS |
| IVS3I1637T | NS | VUS | ||
Contacts in the cryo-EM structure are shown in Table 4.
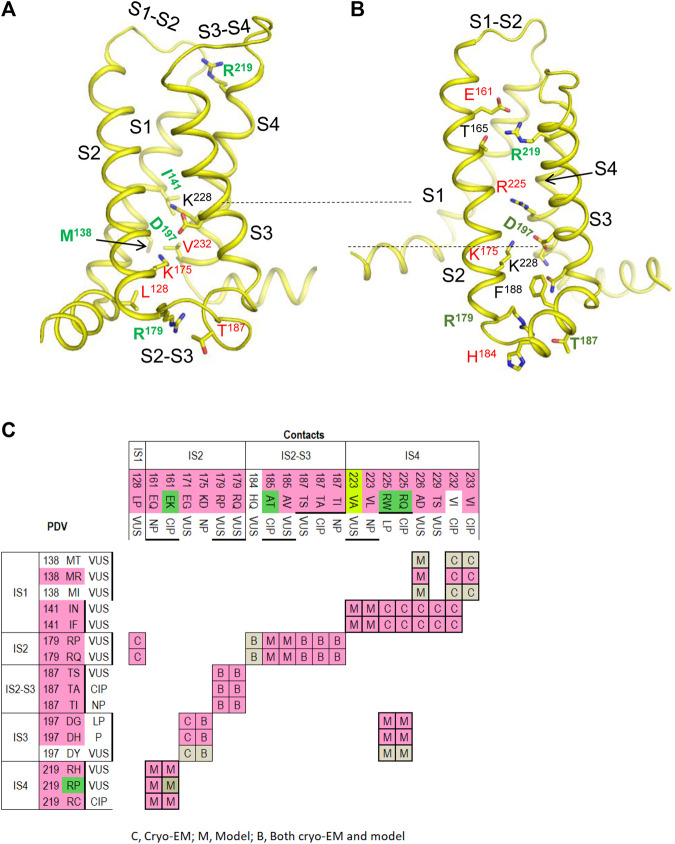
FIGURE 5.
VSD-I in the activated-state structure and resting-state model. (A,B) Intra-membrane views at VSD-I in the activated (A) and resting (B) states. Side chains of some w54 residues and their contacts are indicated with green and red labels, respectively. Dashed lines indicate displacement of K228 upon activation of VSD-I. (C) Matrix of contacts variants. To indicate contacts, which may switch upon activation of VSD-I, each cell is labeled “C”, “M” or “B” depending on whether the contact is found only in the cryo-EM structure, only in the resting-state model, or both in the cryo-EM structure and the model. Overwhelming majority of PDVs and their contact variants are associated with BrS1. See section VSD-I for more details.
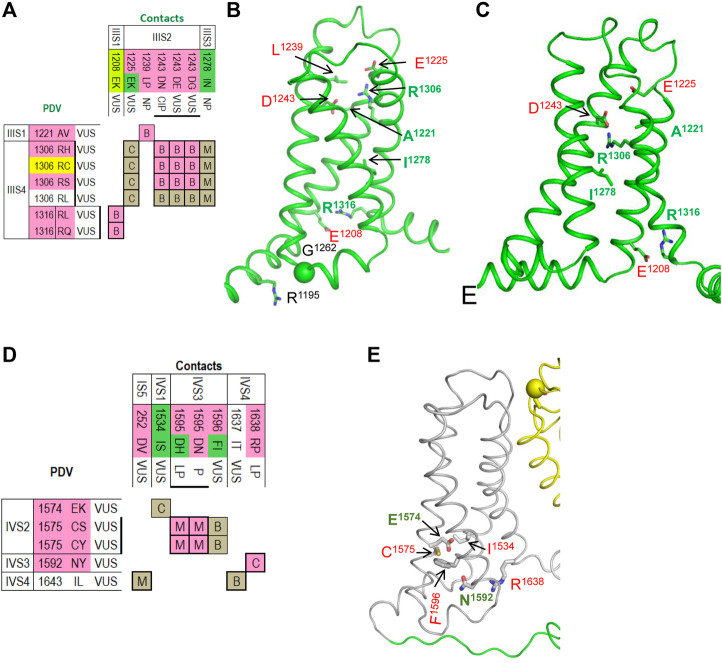
FIGURE 7.
Matrices of contact variants and structures of VSD-III and VSD-VI. (A) Matrix of contact variants of VSD-III. (B,C) VSD-III in the activated-state cryo-EM structure (B) and resting-state model (C). Residues from set w54 and their contacts are indicated with green and red labels, respectively. See section VSD-III for more detail. (D) Matrix of contact variants of VSD-IV. (E) Activated-state structure of VSD-IV. Residues from set w54 and their contacts are indicated with green and red labels, respectively. See section VSD-IV for more details.
VSD-I
Figure 5 shows VSD-I in the activated (A) and resting (B) states with w54 residues and their contacts. Dashed lines indicate displacement of IS4K228 upon transition of the voltage-sensing helix IS4 between the resting and activated states. In the activated VSD-I, R219 lacks specific inter-segment contacts, while in the resting VSD-I, R219 forms a salt bridge with IS2E161 (Figure 5B). BrS1-associated substitutions R19C/H/P or E161Q/K would eliminate the salt bridge, likely destabilizing the resting VSD-I. Mutation E161K, which was found in patients with the sick sinus syndrome, BrS1, or cardiac conduction disease, reduced current density and markedly impaired activation (Smits et al., 2005a).
Variant R219H is associated with mixed arrhythmia and dilated cardiomyopathy, likely due to the proton leak current, which was observed in biophysical studies (Gosselin-Badaroudine et al., 2012; Moreau et al., 2018). Mutations R219P and E161K would destroy the salt ridge and destabilize the resting state, the effect apparently inconsistent with BrS1. However, the same mutations are also associated with LQT3 (Figure 5C).
In the activated VSD-I, IS2R179 is H-bonded with IS2-S3T187 and forms a hydrophobic contact with IS1L128. Variants of these residues are associated with BrS1, indicating that respective mutations destabilize the activated VSD-I. In the resting VSD-I, R179 retains the H-bond with T187 and interacts with IS2-S3H184 and IS2-S3F188. Substitutions R179P/Q and T187S/A/I would eliminate the contacts, thus affecting flexibility of loop IS2-S3. Contact between R179 and T187 is seen in both activated and resting states of VSD-I, whereas variants of these residues are associated with BrS1, implying that flexibility of loop IS2-S3 is important for activation of VSD-I.
In the activated VSD-I, IS3D197 is salt-bridged with IS2K175 and IS4K228 (Figure 5A). In the resting VSD-I, D197 retains the salt bridge with IS2K175 and approaches IS4K228 and IS4R225 (Figure 5B). Substitutions D197G/H/T, K175N, and R225Q/W would eliminate or weaken these contacts in both states. The fact that D197G/H/T and K175N are associated with BrS1 (Figure 5C) suggests that salt bridge D197G : K175 contributes to stabilization of the activated VSD-I. Notably, most of variants in VSD-I are associated with BrS1, but four variants are associated with both BrS1 and LQT3 (Figure 5C). The structural cause of the later ambiguity is unclear.
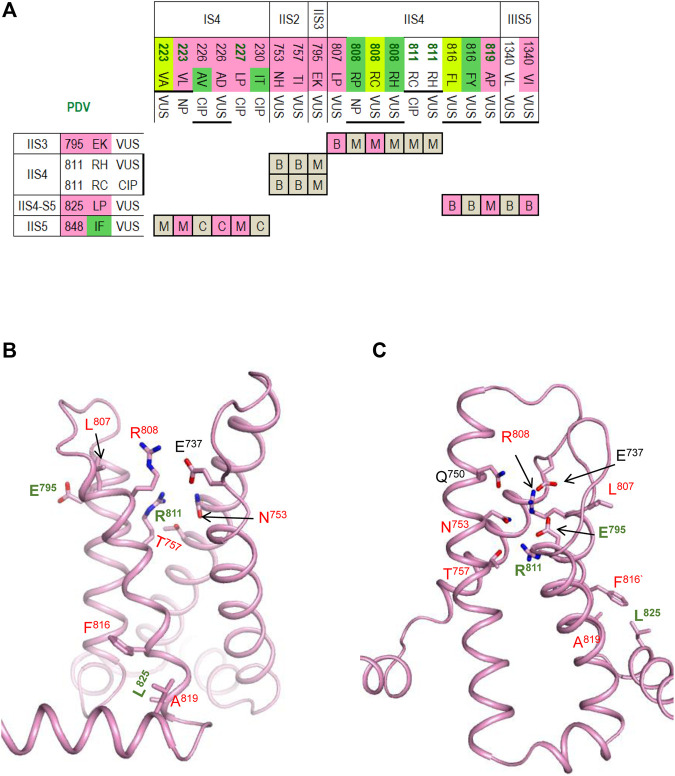
VSD-II
The matrix of contact variants in VSD-II (Figure 6A) shows four w54 residues, including IIS5I848, which forms state-dependent interdomain contacts with four residues in helix IS4, and IIS4-S5L825 that forms interdomain contact with IIS5V1340. Most residue substitutions in the interdomain contacts are associated with BrS1, but three variants are also associated with LQT3. Homozygous mutation IIIS5V1340L was found in two female patients with atrial standstill, while in three unaffected relatives the mutations was heterozygous. In a functional study, V1340L reduced the current density and impaired channel activation (Tan et al., 2018). Another mutation, V1340I, did not alter biophysical characteristics at room temperature, but at 40°C it decreased the current density, caused hyperpolarizing shift of steady-state inactivation, and accelerated recovery from the fast inactivation (Samani et al., 2009).
FIGURE 6.
VSD-II in the activated-state structure and resting-state model. (A) Matrix of contact variants, in addition to contacts within VSD-II, slows contacts IIS4-5/IIIS5 and IIS5/IS4 (B,C) VSD-II in the activated-state cryo-EM structure (B) and resting-state model (C). Residues from set w54 and their contacts are indicated with green and red labels, respectively. VSD-II Section for more details.
The activated VSD-II is stabilized by a salt bridge IIS4R808 : IIS1E737, an H-bond IIS4R811 : IIS2N753, a hydrophobic contact IIS3E795 : IIS4L807, and hydrophobic contacts involving IIS4F816, IIS4A819 and IIS4-S5L825 (Figure 6B). The latter contacts are also seen in the resting VSD-II, but the downshift of helix IIS4 significantly rearranged contacts involving R808 and R811 (Figure 6C). Salt bridges R808 : E737, R811 : E795 and an H-bond R808 : E795 apparently stabilize the resting VSD-II. Mutations R808P/H would eliminate the salt bridges and destabilize the resting state of VSD-II stronger than its activate state in which R808 forms only one salt bridge. This model-based prediction agrees with the fact that variants R808 P/H are associated with LQT3 (Figure 6A).
VSD-III
The matrix of contact variants in VSD-III shows seven PDVs and seven their contact variants (Figure 7A). Activated VSD-III is stabilized by a salt bridge IIIS4R1306 : IIIS1E1225, which is seen only in the cryo-EM structure, two salt bridges seen in both the cryo-EM structure and resting-state model (R1306 : IIIS2D1243 and IIIS4R1316 : IIIS1E1208) and the hydrophobic contact IIIS2L1239 : IIIS1A1221 (Figure 7B). Elimination of the salt bridges in these variants is usually associated with BrS1 (Figure 7A). In the cryo-EM structure, IIS1R1195 and IIS2-S3G1262 are farther than 5 Å from each other (Figure 7B), but PDV G1262S (Table 6) may form an H-bond with R1195 and decrease flexibility of linker IIS2-S3.
TABLE 6.
PDVs whose missense contact variants in the cryo-EM structure or models are not listed in ClinVar.
| # | PDV | Exposure | Comment | |
|---|---|---|---|---|
| Varianta | Syndromeb,c | |||
| 1 | IS6V396L | BrS11,3,4 | Lipids | Tight contacts withIS5F264 IP1L365 and IP1L368 |
| 2 | IIS1L732P | BrS1 | ||
| 3 | IIS1-S2L736P | BrS1 | Extracellular | Loop Flexibility |
| 4 | IIS1-S2H738P | BrS1 | Extracellular | Loop Flexibility |
| 5 | IIS3M794T | NP | Lipids | |
| 6 | IIS4-S5G833R | LQT-3,NP,A, BrS17 | Lipids | |
| 7 | IIS5V924F | NP | Lipids | |
| 8 | IIIS1R1195C | BrS1 | Cytoplasm | R1195 may reach G1262
“ |
| 9 | IIIS1R1195H | “ | Cytoplasm | |
| 10 | IIIS2-S3G1262S | BrS1, LQT-3, PFHB1,3,11,12 | Cytoplasm | Loop flexibility |
| 11 | IIIS6I1448L | A, BrS1, CVP1,3,4 | Lipids | |
a-e See footnotes to Table 1.
The resting state of VSD-III is stabilized by a hydrophobic contact of I1278 with methylene groups of R1306 (Figure 7C). Variant I1278V, which was found in a Thai patient with congenital complete heart block, is considered disease-causative because this variant was not found in unaffected members of the patient’s family (Thongnak et al., 2016). Substitution I1278N would destabilize the resting state in agreement with the fact that this variant is associated with LQT3 (Figure 7A).
VSD-IV
The matrix of contact variants in VSD-IV shows five PDVs and seven their contacts variants (Figure 7D). Several contacts at the cytoplasmic half of VSD-IV stabilize the activated state (Figure 7E); weakening or elimination of these contacts in BrS1-associated PDVs would destabilize the activated VSD-IV. In contrast, LQT3-associated mutation IVS1I1534S would stabilize the activated state due to H-bond between IVS2E1574 and IVS1I1534S. In both activated and resting states of VSD-IV, IVS3F1596 forms a hydrophobic contact with IVS2C1575 (Figure 7D). Variants C1575S/Y are associated with BrS1, whereas variant F1596I is associated with both BrS1 and LQT3. In functional studies, F1596I did not change peak current, but enhanced steady-state inactivation and accelerated recovery from inactivation (Olesen et al., 2012; Hoshi et al., 2015).
Domain Distribution of PDVs and their Contact Variants
Table 7 shows domain distribution of PDVs, their contact variants, and clinical significance of the latter variants. In the PD, 27 WT residues with a total of 38 PDVs are engaged in contacts with 45 WT residues, which have a total of 63 missense disease variants. In the four VSDs, 17 WT residues with a total of 33 PDVs contact 39 WT residues with a total of 55 missense disease variants. While majority of PDVs and their contact variants are seen in the cryo-EM structures, models of the open/closed PD and resting VSDs revealed an additional PDV and 34 contact variants. Thus, in the considered channel states, w54 residues with 82 PDVs form contacts with 84 WT residues that have a total of 118 disease variants. Current number of PDVs exceeds the published number (Tarnovskaya et al., 2020) because new variants of w54 residues were recently reported in ClinVar. For majority of the published PDVs clinical significance is still VUS/NP/CIP, but two variants, IS5L250V and IVS6M1766L, were recently reclassified as P/LP. As many as 104 contact disease variants are categorized as VUS/NP/CIP variants. Further bioinformatics analysis is required to predict potentially damaging effect of these variants, but the fact that they are coupled with earlier predicted PDVs suggests that a large portion of the 104 variants would be reclassified as a P/LP variants.
TABLE 7.
Domain distribution of PDVs and their contact variants.
| PDVs | Contacts | Clinical significance of contact variants | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Domain | State | WT | Variants | WT | Variants | VUS | P/LP | NP | CIP |
| PD | Inactivated a | 27 | 38 | 37 | 49 | 29 | 7 | 11 | 2 |
| Open b | 3 | 5 | 2 | 2 | 1 | ||||
| Closed b | 5 | 9 | 2 | 1 | 5 | 1 | |||
| VSD-I | Activated a | 5 | 13 | 11 | 15 | 8 | 1 | 2 | 4 |
| Resting c | 1 | 3 | 3 | 6 | 2 | 2 | 2 | ||
| VSD-II | Activated a | 4 | 5 | 8 | 10 | 8 | 2 | ||
| Resting c | 6 | 10 | 6 | 2 | 2 | ||||
| VSD-III | Activated a | 3 | 7 | 4 | 6 | 4 | 1 | 1 | |
| Resting c | 1 | 1 | 1 | ||||||
| VSD-IV d | Activated a | 4 | 5 | 4 | 4 | 3 | 1 | ||
| Resting c | 2 | 3 | 1 | 2 | |||||
| Total | 10 | 11 | |||||||
| 54 | 82 | 84 | 118 | 65 | 14 | 25 | 14 | ||
| Set e | w54 | PDV | c84 | cv118 | |||||
Cryo-EM structure.
WT residues that have contacts variants in the inactivated PD (cryo-EM structure) are not counted.
WT residues that have contacts variants in the activated VSD (cryo-EM structure) are not counted.
Location is not shown for WT residues that do not form contacts with disease-associated residues.
Names of the sets are shown in bold.
Table 8 shows domain distribution of disease variants. Most of these are associated with BrS1, followed by LQT3 and unspecified arrhythmia. A total of 32 variants are associated with other diseases, which are specified in Tables 1–5. In the contact variant matrices respective variants are not colored. It should be noted that most of “other” diseases are associated with the Nav1.5 loss-of-function. The cases when variants associated with “other” loss-of function diseases are coupled with BrS1-associated variants provide additional examples of contacts in which mutation of either partner destabilizes the open channel.
TABLE 8.
Domain distribution of disease variants.
| Location | State | BrS1 | LQT3 | Arrythmias | Other diseases | ||||
|---|---|---|---|---|---|---|---|---|---|
| PDV | Contacts | PDV | Contacts | PDV | Contacts | PDV | Contacts | ||
| PD | Inactivated a | 21 | 30 | 6 | 10 | 3 | 6 | 10 | 7 |
| Open b | 2 | 3 | 1 | ||||||
| Closed b | 6 | 3 | 1 | ||||||
| VSD1 | Activated a | 10 | 14 | 2 | 3 | 2 | |||
| Resting c | 3 | 5 | 1 | 2 | 1 | ||||
| VSD2 | Activated a | 3 | 7 | 1 | 3 | 1 | 2 | 1 | |
| Resting c | 4 | 2 | 2 | 2 | |||||
| VSD3 | Activated a | 5 | 5 | 1 | 1 | 1 | 1 | ||
| Resting c | 1 | ||||||||
| VSD4 | Activated a | 4 | 2 | 2 | 1 | 1 | |||
| Resting c | 3 | 1 | |||||||
| Total | 46 | 78 | 8 | 30 | 4 | 11 | 17 | 15 | |
a-c See footnotes to Table 7.
Only eleven of 82 PDVs (13%) are not coupled with other disease variants (Table 6). One PDV (IS6V396L) is involved in tight intra-PD contacts, while 10 PDVs are exposed to lipids, cytoplasm or extracellular space. Three variants (IIS1-S2L736P, IIS1-S2H738P, and IIIS2-S3G1262S) affect flexibility of VSD linkers. The fact that 87% of PDVs are coupled with disease variants supports bioinformatics-based predictions of PDVs (Tarnovskaya et al., 2020) and suggests that many contact variants of PDVs, which are currently annotated as VUS/NP/CIP, may be reclassified as P/LP variants.
Limitations of the Approach
In the absence of cryo-EM structures of the hNav1.5 channel with the open and closed pore domain and resting VSDs, we modeled these states using as templates crystal structures of prokaryotic sodium channel NavAb (see Methods). Some of the proposed intersegment contacts in these states require future validation in functional studies. Ionizable residues in VSDs are involved in multiple state-dependent electrostatic interactions. Structural interpretation of respective mutations may be ambiguous.
Concluding Remarks
Only a small fraction of known missense variants of the hNav1.5 channel are classified as P/LP variants. Prediction of damaging effects of VUS/NP/CIP variants would improve diagnostics of increasingly large number of genotyped cardiac patients. Here we employed a recent cryo-EM structure of the Nav1.5 channels and ad-hoc homology models of the channel with the open or closed PD and resting VSDs to explore intersegment contacts of 54 WT residues whose VUS/NP/CIP variants are reclassified as PDVs (Tarnovskaya et al., 2020). We found that 44 of the 54 WT residues form intersegment contacts with 84 WT residues for which 118 disease variants are reported. Overwhelming majority of the contact variants is categorized as VUS/NP/CIP. We propose that if two WT residues, which are engaged in an intersegment contact, have VUS/NP/CIP variants, these are P/LP variants. Validation of this proposition requires bioinformatics analysis of a large set of contacts involving disease variants and electrophysiological testing of some contacts (Smith and Goldin, 1997; Bénitah et al., 1999). In many contacts, variants of either residue are associated with the same syndrome likely because they similarly affect stability of a particular channel state. To facilitate recognition of such coupled variants, we presented them in colored matrices. Structural analysis suggested atomic mechanism of the channel dysfunction in scores of coupled variants. These include BrS1-associated mutations in P-loops that would destabilize the selectivity-filter slow inactivation gate, BrS1-associated variants in VSDs that would destabilize their activated states, and LQT3-associated mutations in PD that would stabilize the channel states with the open pore (or destabilize states with the closed pore). Understanding these mechanisms may assist structure-based predictions of pathogenic variants and contribute to general physiology of ion channels.
Acknowledgments
Computations were performed using facilities of Compute Canada (www.computeca).
Author Contributions
VK, Molecular modeling, manuscript writing; AZ, Collecting literature on channelopathies, manuscript writing; AK, Project administration, manuscript editing; BZ, Conceptualization, supervising, manuscript writing and editing, funding acquisition.
Funding
This study was supported by grants to BSZ from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2020-07100) and Russian Science Foundation (17-15-01292-Π).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, orclaim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.756415/full#supplementary-material
Abbreviations
BrS1, brugada syndrome type 1; CIP, conflicting interpretations of pathogenicity; LQT3, long QT syndrome type 3; MC, monte carlo; MCM, MC-minimizations; NP, data on pathogenicity are not provided; P1 and P2, membrane-descending and membrane-ascending pore helices in P-loops; PD, pore domain; P/LP, pathogenic/likely pathogenic; PDV, potentially damaging variant (P/LP and PDV are used, respectively, for database-listed variants and those predicted computationally); SF, selectivity filter; TM, transmembrane; VSD, voltage-sensing domain; VUS, variant of uncertain clinical significance; w54, 54 wild-type residues for which PDVs are predicted; WT, wild type.
Reference
- Amin A. S., Asghari-Roodsari A., Tan H. L. (2010). Cardiac Sodium Channelopathies. Pflugers Arch. 460 (2), 223–237. 10.1007/s00424-009-0761-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénitah J. P., Chen Z., Balser J. R., Tomaselli G. F., Marbán E. (1999). Molecular Dynamics of the Sodium Channel Pore Vary with Gating: Interactions between P-Segment Motions and Inactivation. J. Neurosci. 19 (5), 1577–1585. 10.1523/jneurosci.19-05-01577.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodian D. L., Vilboux T., Hourigan S. K., Jenevein C. L., Mani H., Kent K. C., et al. (2017). Genomic Analysis of an Infant with Intractable Diarrhea and Dilated Cardiomyopathy. Cold Spring Harb Mol. Case Stud. 3 (6), a002055. 10.1101/mcs.a002055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer K. R., Kuenze G., Vanoye C. G., George A. L., Jr., Meiler J., Sanders C. R. (2020). Structures Illuminate Cardiac Ion Channel Functions in Health and in Long QT Syndrome. Front. Pharmacol. 11, 550. 10.3389/fphar.2020.00550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C. L., Pettitt B. M., Karplus M. (1985). Structural and Energetic Effects of Truncating Long Ranged Interactions in Ionic and Polar Fluids. J. Chem. Phys. 83, 5897–5908. 10.1063/1.449621 [DOI] [Google Scholar]
- Bruhova I., Zhorov B. S. (2010). A Homology Model of the Pore Domain of a Voltage-Gated Calcium Channel Is Consistent with Available SCAM Data. J. Gen. Physiol. 135 (3), 261–274. 10.1085/jgp.200910288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield S. M., Patel P. R., Waters M. L. (2002). Contribution of Aromatic Interactions to Alpha-helix Stability. J. Am. Chem. Soc. 124 (33), 9751–9755. 10.1021/ja026668q [DOI] [PubMed] [Google Scholar]
- Calloe K., Refaat M. M., Grubb S., Wojciak J., Campagna J., Thomsen N. M., et al. (2013). Characterization and Mechanisms of Action of Novel NaV1.5 Channel Mutations Associated with Brugada Syndrome. Circ. Arrhythm Electrophysiol. 6 (1), 177–184. 10.1161/CIRCEP.112.974220 [DOI] [PubMed] [Google Scholar]
- Carboni M., Zhang Z. S., Neplioueva V., Starmer C. F., Grant A. O. (2005). Slow Sodium Channel Inactivation and Use-dependent Block Modulated by the Same Domain IV S6 Residue. J. Membr. Biol. 207 (2), 107–117. 10.1007/s00232-005-0805-0 [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Vyas R., Chalamalasetti S. V., Sahu I. D., Clatot J., Wan X., et al. (2018). The Voltage-Gated Sodium Channel Pore Exhibits Conformational Flexibility during Slow Inactivation. J. Gen. Physiol. 150 (9), 1333–1347. 10.1085/jgp.201812118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola G., Corrado E., Curnis A., Maglia G., Oriente D., Mignano A., et al. (2019). Update on Brugada Syndrome 2019. Curr. Probl. Cardiol. 46, 100454. 10.1016/j.cpcardiol.2019.100454 [DOI] [PubMed] [Google Scholar]
- Cordeiro J. M., Barajas-Martinez H., Hong K., Burashnikov E., Pfeiffer R., Orsino A. M., et al. (2006). Compound Heterozygous Mutations P336L and I1660V in the Human Cardiac Sodium Channel Associated with the Brugada Syndrome. Circulation 114 (19), 2026–2033. 10.1161/CIRCULATIONAHA.106.627489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du R., Ren F. X., Yang J. G., Yuan G. H., Zhang S. Y., Kang C. L., et al. (2005). Relationship between Congenital Long QT Syndrome and Brugada Syndrome Gene Mutation. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 27 (3), 289–294. [PubMed] [Google Scholar]
- Du Y., Tikhonov D. B., Nomura Y., Dong K., Zhorov B. S. (2018). Mutational Analysis of State-dependent Contacts in the Pore Module of Eukaryotic Sodium Channels. Arch. Biochem. Biophys. 652, 59–70. 10.1016/j.abb.2018.06.008 [DOI] [PubMed] [Google Scholar]
- Garden D. P., Zhorov B. S. (2010). Docking Flexible Ligands in Proteins with a Solvent Exposure- and Distance-dependent Dielectric Function. J. Comput. Aided Mol. Des. 24 (2), 91–105. 10.1007/s10822-009-9317-9 [DOI] [PubMed] [Google Scholar]
- Gosselin-Badaroudine P., Keller D. I., Huang H., Pouliot V., Chatelier A., Osswald S., et al. (2012). A Proton Leak Current through the Cardiac Sodium Channel Is Linked to Mixed Arrhythmia and the Dilated Cardiomyopathy Phenotype. PLoS One 7 (5), e38331. 10.1371/journal.pone.0038331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewegen W. A., Bezzina C. R., van Tintelen J. P., Hoorntje T. M., Mannens M. M., Wilde A. A., et al. (2003). A Novel LQT3 Mutation Implicates the Human Cardiac Sodium Channel Domain IVS6 in Inactivation Kinetics. Cardiovasc. Res. 57 (4), 1072–1078. 10.1016/s0008-6363(02)00838-6 [DOI] [PubMed] [Google Scholar]
- Hirose S., Makiyama T., Melgari D., Yamamoto Y., Wuriyanghai Y., Yokoi F., et al. (2020). Propranolol Attenuates Late Sodium Current in a Long QT Syndrome Type 3-Human Induced Pluripotent Stem Cell Model. Front Cel Dev Biol. 8, 761. 10.3389/fcell.2020.00761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi M., Liu H., Kaufman E. S., Deschênes I. (2015). Polygenic Case of Long QT Syndrome Confirmed through Functional Characterization Informs the Interpretation of Genetic Screening Results. Heartrhythm Case Rep. 1 (4), 201–205. 10.1016/j.hrcr.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Liu M., Yan S. F., Yan N. (2017). Structure-based Assessment of Disease-Related Mutations in Human Voltage-Gated Sodium Channels. Protein Cell 8 (6), 401–438. 10.1007/s13238-017-0372-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltman J. R., Evans F., Fu Y. P. (2018). Re-evaluating Pathogenicity of Variants Associated with the Long QT Syndrome. J. Cardiovasc. Electrophysiol. 29 (1), 98–104. 10.1111/jce.13355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapplinger J. D., Giudicessi J. R., Ye D., Tester D. J., Callis T. E., Valdivia C. R., et al. (2015). Enhanced Classification of Brugada Syndrome-Associated and Long-QT Syndrome-Associated Genetic Variants in the SCN5A-Encoded Na(v)1.5 Cardiac Sodium Channel. Circ. Cardiovasc. Genet. 8 (4), 582–595. 10.1161/CIRCGENETICS.114.000831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapplinger J. D., Tester D. J., Alders M., Benito B., Berthet M., Brugada J., et al. (2010). An International Compendium of Mutations in the SCN5A-Encoded Cardiac Sodium Channel in Patients Referred for Brugada Syndrome Genetic Testing. Heart Rhythm 7 (1), 33–46. 10.1016/j.hrthm.2009.09.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K., Makiyama T., Wu J., Ding W. G., Kimura H., Naiki N., et al. (2014). Cardiac Channelopathies Associated with Infantile Fatal Ventricular Arrhythmias: from the Cradle to the Bench. J. Cardiovasc. Electrophysiol. 25 (1), 66–73. 10.1111/jce.12270 [DOI] [PubMed] [Google Scholar]
- Korkosh V. S., Kiselev A. M., Mikhaylov E. N., Kostareva A. A., Zhorov B. S. (2019). Atomic Mechanisms of Timothy Syndrome-Associated Mutations in Calcium Channel Cav1.2. Front. Physiol. 10, 335. 10.3389/fphys.2019.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkosh V. S., Zhorov B. S., Tikhonov D. B. (2014). Folding Similarity of the Outer Pore Region in Prokaryotic and Eukaryotic Sodium Channels Revealed by Docking of Conotoxins GIIIA, PIIIA, and KIIIA in a NavAb-Based Model of Nav1.4. J. Gen. Physiol. 144 (3), 231–244. 10.1085/jgp.201411226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum M. J., Lee J. M., Benson M., Brown G., Chao C., Chitipiralla S., et al. (2016). ClinVar: Public Archive of Interpretations of Clinically Relevant Variants. Nucleic Acids Res. 44 (D1), D862–D868. 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaeus M. J., Gamal El-Din T. M., Ing C., Ramanadane K., Pomès R., Zheng N., et al. (2017). Structures of Closed and Open States of a Voltage-Gated Sodium Channel. Proc. Natl. Acad. Sci. U S A. 114 (15), E3051–E3060. 10.1073/pnas.1700761114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Yin L., Shen C., Hu K., Ge J., Sun A. (2018). SCN5A Variants: Association with Cardiac Disorders. Front. Physiol. 9, 1372. 10.3389/fphys.2018.01372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Jin X., Wu T., Huang G., Wu K., Lei J., et al. (2021). Structural Basis for Pore Blockade of the Human Cardiac Sodium Channel Nav 1.5 by the Antiarrhythmic Drug Quinidine*. Angew. Chem. Int. Ed. Engl. 60 (20), 11474–11480. 10.1002/anie.202102196 [DOI] [PubMed] [Google Scholar]
- Li Z., Scheraga H. A. (1987). Monte Carlo-Minimization Approach to the Multiple-Minima Problem in Protein Folding. Proc. Natl. Acad. Sci. U S A. 84 (19), 6611–6615. 10.1073/pnas.84.19.6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Sallam K., Wu H., Li Y., Itzhaki I., Garg P., et al. (2016). Patient-Specific and Genome-Edited Induced Pluripotent Stem Cell-Derived Cardiomyocytes Elucidate Single-Cell Phenotype of Brugada Syndrome. J. Am. Coll. Cardiol. 68 (19), 2086–2096. 10.1016/j.jacc.2016.07.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S. B., Tao X., Campbell E. B., MacKinnon R. (2007). Atomic Structure of a Voltage-dependent K+ Channel in a Lipid Membrane-like Environment. Nature 450 (7168), 376–382. 10.1038/nature06265 [DOI] [PubMed] [Google Scholar]
- Moreau A., Gosselin-Badaroudine P., Mercier A., Burger B., Keller D. I., Chahine M. (2018). A Leaky Voltage Sensor Domain of Cardiac Sodium Channels Causes Arrhythmias Associated with Dilated Cardiomyopathy. Sci. Rep. 8 (1), 13804. 10.1038/s41598-018-31772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottaz A., David F. P., Veuthey A. L., Yip Y. L. (2010). Easy Retrieval of Single Amino-Acid Polymorphisms and Phenotype Information Using SwissVar. Bioinformatics 26 (6), 851–852. 10.1093/bioinformatics/btq028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez L., Barana A., Amorós I., de la Fuente M. G., Dolz-Gaitón P., Gómez R., et al. (2013). p.D1690N Nav1.5 Rescues p.G1748D Mutation Gating Defects in a Compound Heterozygous Brugada Syndrome Patient. Heart Rhythm 10 (2), 264–272. 10.1016/j.hrthm.2012.10.025 [DOI] [PubMed] [Google Scholar]
- O'Leary M. E., Chahine M. (2015). MTSET Modification of D4S6 Cysteines Stabilize the Fast Inactivated State of Nav1.5 Sodium Channels. Front. Pharmacol. 6, 118. 10.3389/fphar.2015.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen M. S., Yuan L., Liang B., Holst A. G., Nielsen N., Nielsen J. B., et al. (2012). High Prevalence of Long QT Syndrome-Associated SCN5A Variants in Patients with Early-Onset Lone Atrial Fibrillation. Circ. Cardiovasc. Genet. 5 (4), 450–459. 10.1161/CIRCGENETICS.111.962597 [DOI] [PubMed] [Google Scholar]
- Ortiz-Bonnin B., Rinné S., Moss R., Streit A. K., Scharf M., Richter K., et al. (2016). Electrophysiological Characterization of a Large Set of Novel Variants in the SCN5A-Gene: Identification of Novel LQTS3 and BrS Mutations. Pflugers Arch. 468 (8), 1375–1387. 10.1007/s00424-016-1844-3 [DOI] [PubMed] [Google Scholar]
- Priganc M., Zigová M., Boroňová I., Bernasovská J., Dojčáková D., Szabadosová V., et al. (2017). Analysis of SCN5A Gene Variants in East Slovak Patients with Cardiomyopathy. J. Clin. Lab. Anal. 31 (2), e22037. 10.1002/jcla.22037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., et al. (2015). Standards and Guidelines for the Interpretation of Sequence Variants: a Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17 (5), 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta I., Clancy C. E., Tateyama M., Liu H., Priori S. G., Kass R. S. (2002). A Novel SCN5A Mutation Associated with Long QT-3: Altered Inactivation Kinetics and Channel Dysfunction. Physiol. Genomics 10 (3), 191–197. 10.1152/physiolgenomics.00039.2002 [DOI] [PubMed] [Google Scholar]
- Rudic B., Chaykovskaya M., Tsyganov A., Kalinin V., Tülümen E., Papavassiliu T., et al. (2016). Simultaneous Non-invasive Epicardial and Endocardial Mapping in Patients with Brugada Syndrome: New Insights into Arrhythmia Mechanisms. J. Am. Heart Assoc. 5 (11), e004095. 10.1161/JAHA.116.004095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani K., Wu G., Ai T., Shuraih M., Mathuria N. S., Li Z., et al. (2009). A Novel SCN5A Mutation V1340I in Brugada Syndrome Augmenting Arrhythmias during Febrile Illness. Heart Rhythm 6 (9), 1318–1326. 10.1016/j.hrthm.2009.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savio-Galimberti E., Argenziano M., Antzelevitch C. (2018). Cardiac Arrhythmias Related to Sodium Channel Dysfunction. Handb Exp. Pharmacol. 246, 331–354. 10.1007/164_2017_43 [DOI] [PubMed] [Google Scholar]
- Schwartz P. J., Priori S. G., Dumaine R., Napolitano C., Antzelevitch C., Stramba-Badiale M., et al. (2000). A Molecular Link between the Sudden Infant Death Syndrome and the Long-QT Syndrome. N. Engl. J. Med. 343 (4), 262–267. 10.1056/NEJM200007273430405 [DOI] [PubMed] [Google Scholar]
- Selga E., Campuzano O., Pinsach-Abuin M. L., Pérez-Serra A., Mademont-Soler I., Riuró H., et al. (2015). Comprehensive Genetic Characterization of a Spanish Brugada Syndrome Cohort. PLoS One 10 (7), e0132888. 10.1371/journal.pone.0132888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D. J., Jang Y., Park H. Y., Lee J. E., Yang K., Kim E., et al. (2004). Genetic Analysis of the Cardiac Sodium Channel Gene SCN5A in Koreans with Brugada Syndrome. J. Hum. Genet. 49 (10), 573–578. 10.1007/s10038-004-0182-z [DOI] [PubMed] [Google Scholar]
- Skinner J. R., Winbo A., Abrams D., Vohra J., Wilde A. A. (2019). Channelopathies that Lead to Sudden Cardiac Death: Clinical and Genetic Aspects. Heart Lung Circ. 28 (1), 22–30. 10.1016/j.hlc.2018.09.007 [DOI] [PubMed] [Google Scholar]
- Smith M. R., Goldin A. L. (1997). Interaction between the Sodium Channel Inactivation Linker and Domain III S4-S5. Biophys. J. 73 (4), 1885–1895. 10.1016/S0006-3495(97)78219-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits J. P., Koopmann T. T., Wilders R., Veldkamp M. W., Opthof T., Bhuiyan Z. A., et al. (2005a). A Mutation in the Human Cardiac Sodium Channel (E161K) Contributes to Sick Sinus Syndrome, Conduction Disease and Brugada Syndrome in Two Families. J. Mol. Cel Cardiol. 38 (6), 969–981. 10.1016/j.yjmcc.2005.02.024 [DOI] [PubMed] [Google Scholar]
- Smits J. P., Veldkamp M. W., Bezzina C. R., Bhuiyan Z. A., Wedekind H., Schulze-Bahr E., et al. (2005b). Substitution of a Conserved Alanine in the Domain IIIS4-S5 Linker of the Cardiac Sodium Channel Causes Long QT Syndrome. Cardiovasc. Res. 67 (3), 459–466. 10.1016/j.cardiores.2005.01.017 [DOI] [PubMed] [Google Scholar]
- Son M. J., Kim M. K., Yang K. M., Choi B. H., Lee B. W., Yoo S. H. (2018). Retrospective Genetic Analysis of 200 Cases of Sudden Infant Death Syndrome and its Relationship with Long QT Syndrome in Korea. J. Korean Med. Sci. 33 (32), e200. 10.3346/jkms.2018.33.e200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R. B., Gando I., Bu L., Cecchin F., Coetzee W. (2018). A Homozygous SCN5A Mutation Associated with Atrial Standstill and Sudden Death. Pacing Clin. Electrophysiol. 10.1111/pace.13386 [DOI] [PubMed] [Google Scholar]
- Tarnovskaya S. I., Korkosh V. S., Zhorov B. S., Frishman D. (2020). Predicting Novel Disease Mutations in the Cardiac Sodium Channel. Biochem. Biophys. Res. Commun. 521 (3), 603–611. 10.1016/j.bbrc.2019.10.142 [DOI] [PubMed] [Google Scholar]
- Thongnak C., Limprasert P., Tangviriyapaiboon D., Silvilairat S., Puangpetch A., Pasomsub E., et al. (20162016). Exome Sequencing Identifies Compound Heterozygous Mutations in SCN5A Associated with Congenital Complete Heart Block in the Thai Population. Dis. Markers 2016, 3684965. 10.1155/2016/3684965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov D. B., Bruhova I., Garden D. P., Zhorov B. S. (2015). State-dependent Inter-repeat Contacts of Exceptionally Conserved Asparagines in the Inner Helices of Sodium and Calcium Channels. Pflugers Arch. 467 (2), 253–266. 10.1007/s00424-014-1508-0 [DOI] [PubMed] [Google Scholar]
- Tikhonov D. B., Zhorov B. S. (2012). Architecture and Pore Block of Eukaryotic Voltage-Gated Sodium Channels in View of NavAb Bacterial Sodium Channel Structure. Mol. Pharmacol. 82 (1), 97–104. 10.1124/mol.112.078212 [DOI] [PubMed] [Google Scholar]
- Tikhonov D. B., Zhorov B. S. (2017). Mechanism of Sodium Channel Block by Local Anesthetics, Antiarrhythmics, and Anticonvulsants. J. Gen. Physiol. 149 (4), 465–481. 10.1085/jgp.201611668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsushima R. G., Li R. A., Backx P. H. (1997a). Altered Ionic Selectivity of the Sodium Channel Revealed by Cysteine Mutations within the Pore. J. Gen. Physiol. 109 (4), 463–475. 10.1085/jgp.109.4.463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsushima R. G., Li R. A., Backx P. H. (1997b). P-loop Flexibility in Na+ Channel Pores Revealed by Single- and Double-Cysteine Replacements. J. Gen. Physiol. 110 (1), 59–72. 10.1085/jgp.110.1.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia C. R., Ackerman M. J., Tester D. J., Wada T., McCormack J., Ye B., et al. (2002). A Novel SCN5A Arrhythmia Mutation, M1766L, with Expression Defect Rescued by Mexiletine. Cardiovasc. Res. 55 (2), 279–289. 10.1016/s0008-6363(02)00445-5 [DOI] [PubMed] [Google Scholar]
- Veerman C. C., Wilde A. A., Lodder E. M. (2015). The Cardiac Sodium Channel Gene SCN5A and its Gene Product NaV1.5: Role in Physiology and Pathophysiology. Gene 573 (2), 177–187. 10.1016/j.gene.2015.08.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh R., Peters N. S., Cook S. A., Ware J. S. (2014). Paralogue Annotation Identifies Novel Pathogenic Variants in Patients with Brugada Syndrome and Catecholaminergic Polymorphic Ventricular Tachycardia. J. Med. Genet. 51 (1), 35–44. 10.1136/jmedgenet-2013-101917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner S. J., Kollman P. A., Nguyen D. T., Case D. A. (1986). An All Atom Force Field for Simulations of Proteins and Nucleic Acids. J. Comput. Chem. 7 (2), 230–252. 10.1002/jcc.540070216 [DOI] [PubMed] [Google Scholar]
- Wisedchaisri G., Tonggu L., McCord E., Gamal El-Din T. M., Wang L., Zheng N., et al. (2019). Resting-State Structure and Gating Mechanism of a Voltage-Gated Sodium Channel. Cell 178 (4), 993–e12. e1012. 10.1016/j.cell.2019.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakliaz'minskaia E. V., Shestak A. G., Revishvili A. Sh., Pronicheva I. V., Podoliak D. G., Nechaenko M. A., et al. (2013). Clinic and Genetic Polymorphism of Brugada Syndrome in Russian Patients, Caused by Mutation in SCN5A Gene. Khirurgiia (Mosk) (2), 49–53. [PubMed] [Google Scholar]
- Zeng Z., Xie Q., Huang Y., Zhao Y., Li W., Huang Z. (2016). p.D1690N Sodium Voltage-Gated Channel α Subunit 5 Mutation Reduced Sodium Current Density and Is Associated with Brugada Syndrome. Mol. Med. Rep. 13 (6), 5216–5222. 10.3892/mmr.2016.5162 [DOI] [PubMed] [Google Scholar]
- Zhorov B. S. (2021). Possible Mechanism of Ion Selectivity in Eukaryotic Voltage-Gated Sodium Channels. J. Phys. Chem. B 125 (8), 2074–2088. 10.1021/acs.jpcb.0c11181 [DOI] [PubMed] [Google Scholar]
- Zhorov B. S. (1981). A Vector Method for Calculating the Derivatives of the Energy of the Atom-Atom Interactions of Complex Molecules with Respect to Generalized Coordinates. J. Struct. Chem. 22, 4–8. 10.1007/bf00745970 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.