Abstract
Background & Aims
Microbiota dysbiosis and mucosa-associated bacteria are involved in colorectal cancer progression. We hypothesize that an interaction between virulent pathobionts and epithelial defense promotes tumorigenesis.
Methods
Chemical-induced CRC mouse model was treated with antibiotics at various phases. Colonic tissues and fecal samples were collected in a time-serial mode and analyzed by gene microarray and 16S rRNA sequencing. Intraepithelial bacteria were isolated using a gentamicin resistance assay, and challenged in epithelial cultures.
Results
Our study showed that antibiotic treatment at midphase but not early or late phase reduced mouse tumor burden, suggesting a time-specific host–microbe interplay. A unique antimicrobial transcriptome profile showing an inverse relationship between autophagy and oxidative stress genes was correlated with a transient surge in microbial diversity and virulence emergence in mouse stool during cancer initiation. Gavage with fimA/fimH/htrA-expressing invasive Escherichia coli isolated from colonocytes increased tumor burden in recipient mice, whereas inoculation of bacteria deleted of htrA or triple genes did not. The invasive E.coli suppressed epithelial autophagy activity through reduction of microtubule-associated protein 1 light-chain 3 transcripts and caused dual oxidase 2–dependent free radical overproduction and tumor cell hyperproliferation. A novel alternating spheroid culture model was developed for sequential bacterial challenge to address the long-term changes in host–microbe interaction for chronic tumor growth. Epithelial cells with single bacterial encounter showed a reduction in transcript levels of autophagy genes while those sequentially challenged with invasive E.coli showed heightened autophagy gene expression to eliminate intracellular microbes, implicating that bacteria-dependent cell hyperproliferation could be terminated at late phases. Finally, the presence of bacterial htrA and altered antimicrobial gene expression were observed in human colorectal cancer specimens.
Conclusions
Invasive pathobionts contribute to cancer initiation during a key time frame by counterbalancing autophagy and oxidative stress in the colonic epithelium. Monitoring gut microbiota and antimicrobial patterns may help identify the window of opportunity for intervention with bacterium-targeted precision medicine.
Keywords: Colon Carcinoma, Gut Microbiome, Intracellular Microbes, Xenophagy, Reactive Oxygen Species, Organoids and Spheroids, Intestinal Epithelial Cells
Abbreviations used in this paper: ABX, antibiotic; afa, afimbrial adhesin; AOM, azoxymethane; ATPase, adenosine triphosphatase; cdt, cytolethal distending toxin; CFU, colony forming unit; cnf, cytotoxic necrotizing factor; CRC, colorectal cancer; Ct, cycle threshold; DEG, differentially expressed genes; DUOX, dual oxidase; dsb, oxidoreductase disulfide bond; DSS, dextran sodium sulfate; EM, epithelial monolayer; ES, epithelial spheroid; fim, fimbrial adhesin; fli, flagellin; htrA, high temperature requirement A; IRGM, immunity related GTPase M; LB, Luria-Bertani; lpf, long polar fimbriae; MAP1LC3B, microtubule associated protein 1 light chain 3 beta; MOI, multiplicity-of-infection; NADPH, nicotinamide adenine dinucleotide phosphate; NTUCM, National Taiwan University College of Medicine; NTUH, National Taiwan University Hospital; omp, outer membrane protein; P, passage; pap, P-fimbriae; PBS, phosphate-buffered saline; PCA, principal component analysis; PCoA, Principal coordinate analysis; PCR, polymerase chain reaction; pksR, polyketide synthase R; ROS, reactive oxygen species; rRNA, ribosomal RNA; SQSTM1, Sequestosome-1; stx, Shiga toxin; WT, wild-type
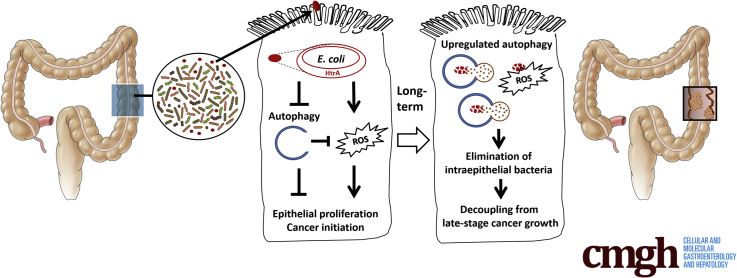
Graphical abstract
Summary.
Invasive pathobionts contribute to cancer initiation during a key time frame by counterbalancing autophagy and oxidative stress in the colonic epithelium. Monitoring gut microbiota and antimicrobial patterns may help identify the window of opportunity for intervention with bacterium-targeted precision medicine.
An altered intestinal microbiota and enrichment of mucosal bacteria have been reported in human colorectal cancers (CRCs).1,2 Despite a strong link between microbiota dysbiosis and colon carcinogenesis, the timing of dysfunctional host–microbe interactions that contribute to tumor progression has not been elucidated. Previous studies using germ-free models and long-term antibiotic treatment in conventionalized animals have shown a causal role for gut bacteria in CRC3, 4, 5; however, whether there is a critical phase in which gut microbiota modify tumor progression and the associated mechanisms remain unclear.
Gut commensals are mostly lumen-dwelling, mixing in the stool and outer mucus layers. Nevertheless, high densities of mucosa-associated bacteria have been identified in CRC specimens.6,7 The abundance of Escherichia coli, Bacteroides fragilis, and Fusobacterium nucleatum in familial adenomatous polyps and colorectal tumor biopsy specimens have been reported.8,9 The presence of intraepithelial E coli also has been documented in experimental models of intestinal diseases.10, 11, 12 Previous studies have shown that mucosa-associated bacteria promoted tumor growth through indirect mechanisms such as stimulation of macrophage cyclooxygenase-2 production and modulation of the immune microenvironment.8,13 Other studies have shown that monoassociation of genotoxin-producing polyketide synthase (pks)-positive E. coli causes DNA damage and cell-cycle arrest in epithelial cells, and increases the colon tumor load in gnotobiotic mice.14, 15, 16 Whether intraepithelial microbes play direct roles in cell hyperproliferation, a major hallmark of cancer, still is unknown.
To date, the timing and detailed mechanisms of the dysregulated host–microbe interplay that contributes to carcinogenesis remain poorly understood. We hypothesized that an interaction between virulent pathobionts and epithelial defense lead to cancer initiation. In the present study, a time-series approach was used to identify crucial players from the host and microbial sides. A unique antimicrobial transcriptome profile was observed in the colonic tissues, which correlated with temporal alterations in the microbiota composition and virulence emergence in the stool during cancer initiation. Moreover, invasive E. coli was associated with hyperproliferative and tumorigenic abilities through manipulation of epithelial antimicrobial responses.
Results
A Critical Window for Antibiotic Treatment During Cancer Initiation Reduced Tumor Burden in Mice
Mice were administered carcinogen to induce CRCs and macroscopic inspection was performed on day 90. Additional groups included mice with antibiotic (ABX) treatment for 1 week: early ABX (days 46–53), mid-ABX (days 56–63), and late ABX (days 66–73), which were compared with control mice not treated with ABX (Figure 1A). Mid-ABX treatment significantly reduced tumor multiplicity and area, whereas treatment at early or late phases had no effect on tumor burden (Figure 1Ba and Bb). In contrast to the presence of high-grade dysplasia and carcinoma in the control mice, low-grade dysplasia was observed in the tumor tissues of mid-ABX mice (Figure 1Bc and C). The data suggest a critical window (ie, midphase) for ABX intervention to modulate tumor growth.
Figure 1.
A critical window for antibiotic treatment during cancer initiation reduced tumor burden in mice. (A) Mice were administered 3 cycles of AOM/DSS and divided into 4 groups, including those without ABX treatment as the control (Ctrl) group and those with ABX treatment during early, middle, or late phases. All mice were killed on day 90. (B) Only ABX treatment at midphase, but not early or late phases, reduced tumor burden. (a) Tumor multiplicity, (b) tumor area, and (c) tumor grade are shown. Each dot represents the data of 1 mouse. ∗P < .05 vs Ctrl. N = 12–14/group. Percentages of tumors in low- and high-grade dysplasia and carcinoma are shown for each group. (C) Histopathologic images of colon tumors on day 90. Scale bar: 100 μm. (D) The natural course of tumor formation in control mice. (a) Tumor area and (b) tumor grade were determined before (day 0) and after carcinogen administration (days 46, 56, and 66). Low- to high-grade dysplasia was observed on day 46, but carcinoma was found only after day 56 (midphase). ∗P < .05 vs day 0. N = 10/group. (E) Histopathologic images of colon tumors at the indicated time points. Scale bar: 100 μm. (F) Principal component analysis (PCA) of transcriptome profiles in colonic tissues by gene array. Each dot represents a sample collected on days 0, 46, 56, or 66. N = 3/group. (G) Heat map and Venn diagram of DEGs from colonic tissues. The gene profiles on days 46, 56, and 66 were compared with day 0, and labeled as D46/0, D56/0, and D66/0, respectively. Among the 536 DEGs after carcinogen administration, 62 genes were unique to D56/0. (H) The messenger RNA levels of the 18 most significant genes were selected from the 62 DEGs unique to D56/0 for quantitative PCR analysis, and showed up-regulation (open bars) and down-regulation (closed bars) of specific gene transcription. Changes in autophagy and oxidative stress gene expression were observed on day 56 (midphase). ∗P < .05. N = 3/group.
To decipher the natural course of tumorigenesis, colon morphology and transcriptome profiles were examined in the control mice. Tumors were observed in the colon on days 46, 56, and 66 after carcinogen administration (Figure 1Da). Low- to high-grade dysplasia was observed on day 46, while carcinoma was found on days 56 and 66 (Figure 1Db and E). Dysplasia refers to a precancerous change in epithelial cells with pleomorphism and a hyperchromatic nucleus, which may regress spontaneously or may progress to advanced carcinoma.17,18 Therefore, our results indicate the onset of the cancerous transition at approximately day 56 (ie, midphase) in the mouse CRC model.
Unique Host Gene Profiles Related to Antimicrobial Responses During Cancer Initiation
The microarray data showed distinct colon transcriptome profiles on days 46, 56, and 66 compared with that on day 0 (Figure 1F and G). The up-regulated and down-regulated gene patterns on day 56 vs day 0 (D56/0) were different from those for D46/0 and D66/0. Among the 536 differentially expressed genes (DEGs) with altered expression at the 3 time points compared with those on day 0, only 62 DEGs were unique to D56/0, as shown in the Venn diagram (Figure 1G). Based on ingenuity pathway analysis of the top networks, the 18 most significant genes that were unique to D56/0 were chosen for quantification using real-time polymerase chain reaction (PCR) (Figure 1H). The top networks associated with the unique transcriptome profile for D56/0 included cancer, antimicrobial response, cell morphology, and cell-mediated immune response. Some of the DEGs (eg, Tsku, Cyp2C44, Ugdh, Kcnk5, Fgf7, and Pck1) were reported oncogenes, and other DEGs (eg, Rad51ap1, Hells, Arl5c, Rrm2, and Tnfsf9) were known tumor-suppressor genes (Figure 1H). Among the identified genes, DEGs related to autophagy pathways (eg, immunity-related guanosine triphosphatase M 2 [Irgm2]; P < .05) and free radical responses such as nicotinamide adenine dinucleotide phosphate (NADPH) oxidase family members dual oxidase (Duox) and glutathione peroxidases (eg, Duox2, Duoxa2, and Gpx2; P < .05) were observed for D56/0 (Figure 1H). Although autophagy activity and free radical production are both host defense mechanisms for eliminating microbes,19, 20, 21 an inverse relationship was noted between the 2 pathways for D56/0.
Alterations in Fecal Microbiota During the Course of Tumorigenesis
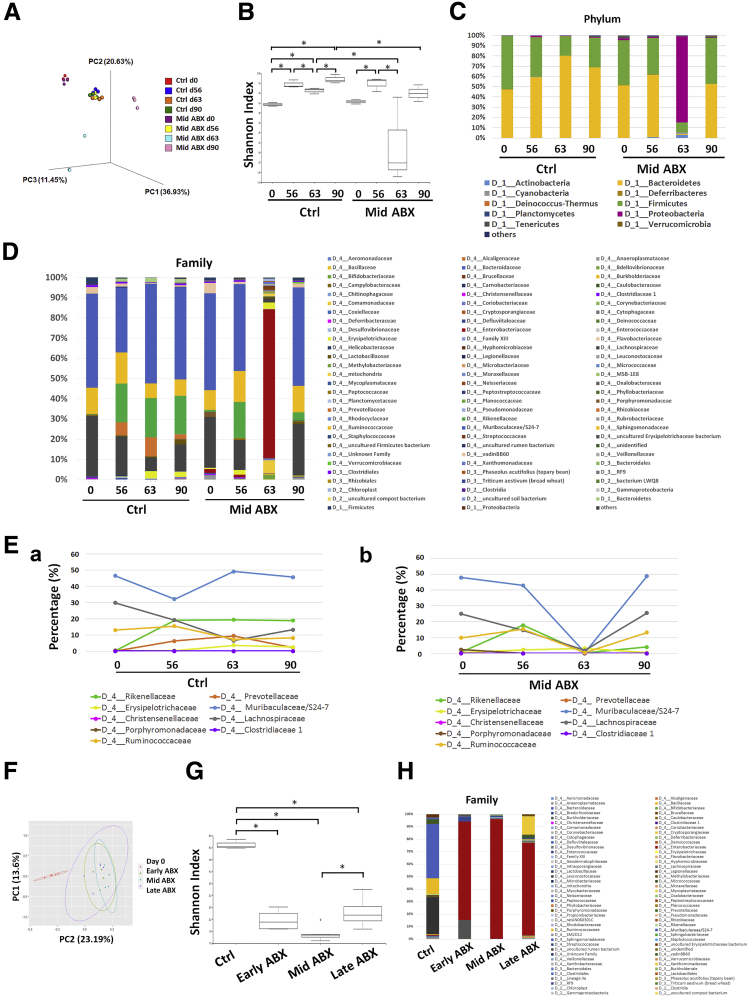
Fecal samples were collected from control and mid-ABX mice at various time points for comparison of microbiota by next-generation sequencing. The principal coordinate analysis plot showed that the fecal microbiota composition in the control and mid-ABX mice on days 0 and 56 (before ABX treatment) were similar at each time point, showing no difference in the baseline populations between the mouse groups before ABX treatment (Figure 2A). In the control mice, the microbial composition on days 56, 63, and 90 after carcinogen administration was different from that on day 0 (Figure 2A). Disturbance of the gut microbiota was identified in the mid-ABX mice on day 63, and this phenomenon still was apparent until 90 days (Figure 2A and B). Moreover, an increase in Shannon’s diversity was found on day 56 compared with that on day 0 in both the control and mid-ABX groups (Figure 2B).
Figure 2.
Alterations in the fecal microbiota during tumorigenesis. Fecal samples were collected from control (Ctrl) and mid-ABX mice at various time points (ie, days 0, 56, 63, and 90) for taxonomy-based comparison of intestinal microbiota. (A) Principal coordinate analysis (PCoA) score plot based on the relative abundance of operational taxonomic units (OTUs) (97% similarity level). Bray Curtis dissimilarity matrices were generated to show the differences in the overall bacterial diversity across the samples. Each symbol represents a sample. (B) Shannon index showed different fecal microbial diversity in Ctrl and mid-ABX mice at various times. ∗P < .05. N = 3/group. (C and D) Relative abundance of bacterial phyla and families in the stool of Ctrl and mid-ABX mice at various times. Mid-ABX treatment resulted in relative abundance of the Enterobacteriaceae family (mainly Escherichia and unidentified genera). The label “D” with numbers indicated the taxonomic rank in the hierarchy. (E) Time-series plot of particular fecal bacterial taxa in the (a) Ctrl and (b) mid-ABX mice. Alterations in the relative bacterial abundance were indicated by the linear discriminant analysis score. Mid-ABX treatment depleted all of these bacteria on day 63. (F) PCoA plot of the relative abundance of OTUs in mouse stool after early, middle, and late ABX treatment. The 3 ABX regimens significantly disturbed the gut microbiota. (G) Shannon Index showed reductions in fecal microbial diversity by 3 ABX regimens. ∗P < .05. N = 4–6/group. (H) Percentages of bacterial family in the fecal microbiota of mice treated with early, middle, or late ABX. The Enterobacteriaceae family became predominant in stool samples in all 3 ABX regimens. N = 4–6/group.
The bacterial phyla Bacteroidetes and Firmicutes constituted more than 95% of the fecal commensal population in control mice on day 0, along with Proteobacteria, Actinobacteria, Deferribacteres, and Tenericutes. After carcinogen administration, an increased ratio of Bacteroidetes:Firmicutes was found on days 56, 63, and 90 (Figure 2C). Based on the bacterial family percentages and the linear discriminant analysis scores, decreases in the relative abundance of Muribaculaceae/S24-7, Lachnospiraceae, Porphyromonadaceae, and Clostridiaceae associated with increases in Prevotellaceae, Rikenellaceae, and Christensenellaceae were observed in the fecal microbiota of the control mice on day 56 in comparison with that on day 0 (Figure 2D and E). An additional decrease in Ruminococcaceae and increase in Erysipelotrichaceae were observed from days 56 to 63 (Figure 2D and E). Families that were absent at earlier times, such as Moraxellaceae and Xanthomonadaceae, increased on day 90 in the control mice bearing tumors.
Analysis of the fecal bacterial composition of mid-ABX mice on day 63 was unfeasible for some of the stool samples owing to microbial depletion after ABX treatment, as evidenced by the absence of the 16S ribosomal RNA (rRNA) gene. For the fecal samples containing sufficient bacterial DNA for sequencing, the Enterobacteriaceae family (Escherichia and an unidentified genus) was the predominant microbial population in the stool of mid-ABX mice on day 63 (Figure 2D). Further analysis across the early-, mid-, and lateABX groups was performed to evaluate the status of microbial disturbance by ABX at various time points. The fecal microbial diversity was decreased by the 3 ABX regimens (Figure 2F and G). Similar to the observations for the mid-ABX group, Enterobacteriaceae became the predominant bacterial family remaining in the stool after the early and late ABX treatment (Figure 2H). Because all 3 ABX regimens reduced fecal bacterial diversity but only microbial disturbance during the midphase window halted tumor progression, we next investigated the host–microbe interaction during this critical phase.
Positive Correlation Between Virulence-Expressing Enterobacteriaceae and Tumor Burden in Mice
Because of the complex nature of gut microbiota, we took a reductionist approach to identify bacteria with tumorigenic characteristics. Bacterial virulence factors related to adherence, mobility, genotoxins, and stress survival first were assessed in the mouse stool. Fimbriae subunits (fimA, fimC, and fimH), and stress survival proteins (high temperature requirement A (htrA) and outer membrane protein C (ompC) were detected in the stool samples of control mice on day 63 (Figure 3A), but were undetectable at earlier time points, such as days 0 and 56. The levels of fimA, fimC, fimH, htrA, and ompC in the stool of mid-ABX mice were higher than in those of the control mice on day 63 (Figure 3A), which correlated with the relative abundance of the Enterobacteriaceae family in the stool. No signs of polyketide synthase R (pksR), flagellin C (fliC), long polar fimbriae A (lpfA), oxidoreductase disulfide bond A (dsbA), and P-fimbriae (papC) were found in the fecal samples of either the control or mid-ABX mice (Figure 3A).
Figure 3.
Characterization of virulence-expressing bacteria in the stool and colonocytes of tumor-developing mice. Bacterial virulent factors in mouse stool and the presence of intraepithelial bacteria were assessed. (A) Virulent factors were observed in the stool of control (Ctrl) and mid-ABX (ABX) mice on day 63, but absent at earlier time points (ie, days 0 and 56). ∗P < .05, ∗∗∗P < .001 vs Ctrl. N = 5/group. (B) Among the mice in the mid-ABX group, a positive correlation was found between the absolute abundance of fecal bacteria on day 63 and the tumor burden on day 90 by regression analysis. Each dot represents the data of 1 mouse in the mid-ABX group. The Enterobacteriaceae family was the predominant fecal bacteria population after mid-ABX treatment. (C) Intracellular bacterial counts in colonocytes of untreated (UT), Ctrl, and mid-ABX mice, and of mice given a single dose of AOM or DSS. Negligible counts were observed in UT mice, and higher intraepithelial bacterial counts were observed in Ctrl mice than in mid-ABX mice. ∗P < .05 vs UT, #P < .05 vs Ctrl. N = 10/group. (D) Representative electromicroscopic images showing intracellular bacteria (bac) in colonocytes. The brush border (BB) and tight junction (TJ) were labeled to show orientation of epithelial cells. Scale bar: 1 μm. (E) The intraepithelial bacteria included E.coli, Enterococcus (Enc), Enterobacter (Enb), and Staphylococcus (Sta). Three strains of each were tested for virulence expression. (F) The (a) intracellular and (b) apical counts of E.coli LI60C3, Enc LI91C2, Enb HM763C3, and Sta IX60C1 after incubation with human Caco-2 cells for 4 hours. N = 8/group. (G) Comparison of intracellular bacterial counts among 5 strains of mouse E.coli and human-derived LF82 in Caco-2 cells. ∗P < .05 vs without (w/o); #P < .05 vs LI60C3. N = 6–8/group. (H) Longer duration of intracellular survival for E.coli LI60C3 than Enb HM763C3 and Enc LI91C2. N = 8/group. (I) Mouse-isolated E.coli LI60C3 strain did not harbor genotoxin-producing genes such as pks, cnf, and cdt, or Shiga toxin (stx) genes. Positive controls for the virulence factors included bacterial strains known for virulence expression or gene-constructed plasmids. (J) Increasing invasive ability of E.coli LI60C3 after sequential passaging in Caco-2 cells. P0 indicates the first isolate of bacteria from mouse colonocytes, and P1 and P2 are bacteria internalized into Caco-2 cells once or twice. ∗P < .05, ∗∗P < .01 vs P0. N = 8/group. (K) Virulence factors of LI60C3 after sequential passaging. fimA/C/H and ompC, N = 3/group; htrA, fliC, and lpfA, N = 6/group. Experiments were repeated twice. Adj, adjusted; rDNA, ribosomal DNA; Rsqr, R2.
Among the mice in the mid-ABX group, a positive correlation was found between the absolute abundance of fecal bacteria on day 63 and the tumor burden on day 90 (Figure 3B). Mid-ABX mice with a low to negligible tumor area (bottom quartile, 0.13 ± 0.08 mm2) had undetectable levels of the bacterial 16S rRNA gene in stool samples, whereas those with a higher tumor area (top quartile, 25.86 ± 4.56 mm2) showed the presence of bacterial 16S rRNA (Figure 3B). The data suggested that the fimA+/fimC+/fimH+/htrA+/ompC+ microbes remaining in the stool after mid-ABX treatment could be associated with tumor growth. Therefore, we isolated the virulence-expressing microbes for evaluation of tumorigenic potentials.
Invasive Ability and Virulence Expression of Internalized Bacteria Harvested From Mouse Colonocytes
Because the microarray data pointed to intracellular antimicrobial responses, we suspected that the virulence-expressing bacteria were microbes residing in the gut mucosa. The presence of intraepithelial bacteria was assessed in the tumor-developing mice by using a gentamicin-resistance assay. Gentamicin is a broad-spectrum antibiotic that kills extracellular bacteria while leaving the intracellular microbes intact.10, 11, 12 High intracellular bacterial counts were observed in the colonocytes of carcinogen-treated mice whereas no microbes were detected in the colonocytes of untreated mice (Figure 3C). To elucidate the individual effects of the mutagen and colitogenic agent on bacterial internalization, separate groups of mice were given each chemical by a single dose. The presence of intracellular bacteria was observed in the mice given dextran sodium sulfate (DSS), but not in those injected with azoxymethane (AOM) (Figure 3C). The presence of intracellular bacteria in mouse colonocytes during the midphase of tumor induction also was visualized by transmission electron microscopy (Figure 3D).
The intraepithelial bacterial colonies were isolated for 16S rRNA gene sequencing and mass spectrometry, revealing 4 bacterial species, E. coli, Enterococcus faecalis, Enterobacter cloacae, and Staphylococcus lentus. These bacteria showed susceptibility to gentamicin, ruling out the possibility that extracellular microbes were not killed during the procedure of gentamicin-resistance assay (Table 1). The virulence factors subsequently were examined in the 4 bacterial species, with 3 strains examined for each species. The presence of fimbrial adhesin (fimA, fimC, and fimH), htrA, ompC, fliC, and lpfA was found in the 3 strains of intraepithelial E coli (designated LI60C3, HM784C1, and HM926C2), whereas dsbA, papC, afaC, and pksR were absent (Figure 3E). Only htrA and afaC were found in the intraepithelial E. cloacae strains, and none of the virulence factors were detected in the E. faecalis or S. lentus strains (Figure 3E).
Table 1.
Antibiogram of Mouse-Isolated Bacteria
| Antimicrobial agent |
E.coli LI60C3 |
E.coli HM784C1 |
E.cloacae HM763C3 |
E.faecalis LI91C2 |
S.lentus IX60C1 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC, ug/mL | Result | MIC, ug/mL | Result | MIC, ug/mL | Result | MIC, ug/mL | Result | MIC, ug/mL | Result | |
| Gentamicin | ≤1 | S | ≤1 | S | ≤2 | S | 4 | S | ≤2 | S |
| Ampicillin | ≥32 | R | ≤2 | S | >16 | R | 1 | S | ||
| Piperacillin | 8 | S | ≤4 | S | >64 | R | ||||
| Cefazolin | ≥64 | R | ≤4 | S | >16 | R | ≤2 | S | ||
| Cefmetazole | 32 | I | ≤1 | S | >32 | R | ||||
| Cefotaxime | ≤1 | S | ≤1 | S | >16 | R | ||||
| Ceftazidime | ≤1 | S | ≤1 | S | >16 | R | ||||
| Cefepime | ≤1 | S | ≤1 | S | 8 | S | ||||
| Ertapenem | ≤0.5 | S | ≤0.5 | S | 1 | I | ||||
| Imipenem | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ||||
| Meropenem | ≤0.25 | S | ≤0.25 | S | ≤1 | S | ||||
| Amikacin | ≤2 | S | ≤2 | S | ≤8 | S | ||||
| Ciprofloxacin | ≤0.25 | S | ≤0.25 | S | ≤1 | S | 1 | S | ≤0.5 | S |
| Levofloxacin | ≤0.12 | S | ≤0.12 | S | ≤1 | S | ||||
| Tigecycline | ≤0.5 | S | ≤0.5 | S | ||||||
| Trimethoprim | ≤20 | S | ≤20 | S | ≤0.5 | S | ≤0.5 | S | ≤0.5 | S |
| Penicillin | 4 | S | ||||||||
| Oxacillin | >4 | 1 | R | |||||||
| Erythromycin | ≤0.25 | S | 0.5 | S | ||||||
| Clindamycin | >2 | >2 | S | |||||||
| Vancomycin | ≤0.1 | S | ≤1 | S | ||||||
| Tetracycline | 8 | R | ≤0.5 | S | ||||||
| Nitrofurantoin | >8 | R | ≤16 | S | ||||||
| Daptomycin | ≤16 | S | ≤1 | S | ||||||
| Linezolid | 2 | S | 2 | S | ||||||
| Teicoplanin | ≤1 | S | ≤1 | S | ||||||
The invasive ability of the 4 intraepithelial bacterial species was compared by exposing microbes to human cell lines in antibiotic-free medium for 4 hours. The group with the highest intracellular bacterial counts was E. coli (LI60C3), followed by E. faecalis (LI91C2) and E. cloacae (HM763C3), and the lowest was the S. lentus (IX60C1) (Figure 3Fa). The apical bacterial counts were comparable for all species, except the S lentus numbers were significantly lower (Figure 3Fb). Given that E. coli exposure resulted in the highest intraepithelial counts among the different species, additional strains of mouse-isolated E. coli (ie, LI60I2, HM784C1, HM926C2, and HM936C4) were assessed for their invasive abilities. The intracellular bacterial counts for all the mouse E. coli isolates, except HM784C1, were comparable with those of human-derived E. coli LF82 (Figure 3G). The duration of bacterial survival inside epithelial cells also was determined, showing constant intracellular counts of E. coli LI60C3 in Caco-2 cells after 5 days (Figure 3H). No sign of genotoxin-encoding genes such as pksR, cytotoxic necrotizing factor 1 (cnf1), and cytolethal distending toxin (cdtB), or Shiga toxin genes such as Shiga toxin (stx)1/2 was found in E. coli LI60C3 (Figure 3I).
To mimic in vivo dynamics, the invasive ability of E. coli LI60C3 was evaluated further after sequential passaging in epithelial cells. An initial isolate of mouse bacteria (denoted passage 0 [P0]) was used for apical challenge in Caco-2 cells, and then the internalized bacteria were harvested after cell lysis as the first passage (P1). P1 bacteria were used for subsequent challenge in newly plated epithelial cells, and the internalized bacteria were harvested again for a second passage (P2); this process then was repeated. An increase in the intracellular bacterial counts was observed for sequential passages of LI60C3 (P1 and P2) compared with the initial isolate from mouse colonocytes (P0) (Figure 3J). Gradual increases in bacterial levels of fimA/H, htrA, and ompC, but not of fliC or lpfA, were observed after sequential passaging (Figure 3K).
Invasion of Virulence-Expressing E coli Stimulated Epithelial Hyperproliferation and Mouse Tumor Growth
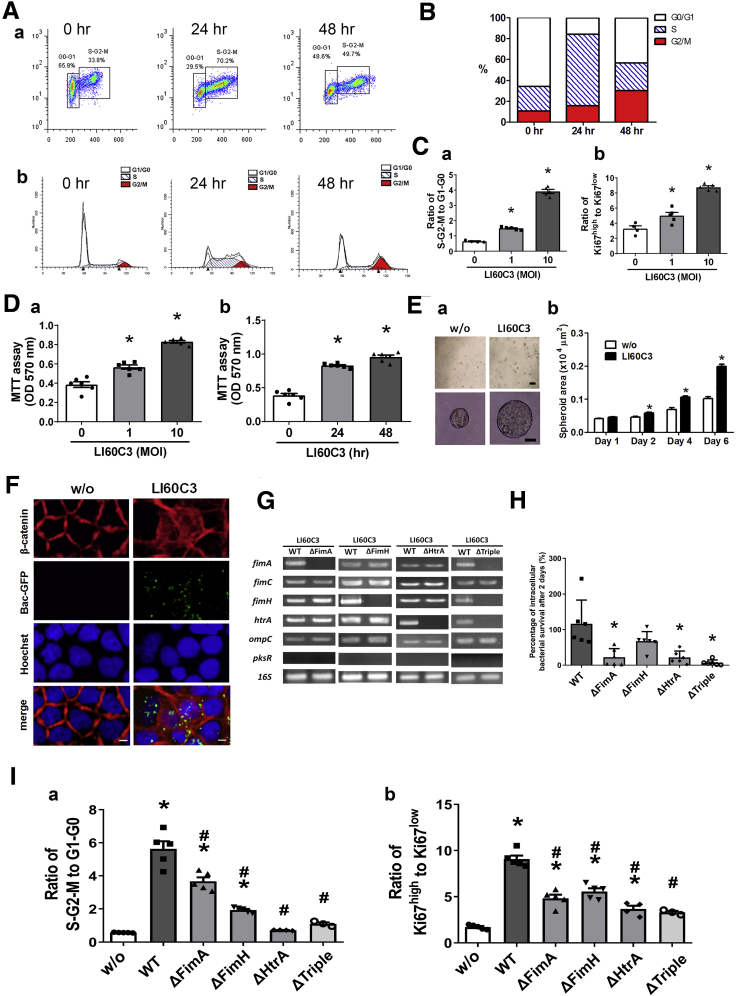
Based on the in vitro data, the invasive E. coli LI60C3 strain was chosen for verification of epithelial proliferative and tumorigenic ability. By using flow cytometry, a significant increase in the ratio of cells in the S/G2/M phases and high Ki67 intensity were observed in Caco-2 cells after exposure to LI60C3 (Figure 4A–C). Increased cellular viability and a larger spheroid size were observed for Caco-2 cells after bacterial infection, with the effects occurring in a dose- and time-dependent manner (Figure 4D and E). Nuclear translocation of β-catenin also was noted in the bacteria-infected cells (Figure 4F).
Figure 4.
Invasive htrA-positive E.coli induced epithelial cell hyperproliferation. Caco-2 cells were exposed to E.coli LI60C3 for 4 hours, and epithelial proliferation was assessed at later time points. (A) Cell cycles of bacteria-exposed Caco-2 cells (MOI, 10) at 0, 24, and 48 hours by flow cytometry. (a) Dot plots of Ki67 (Y-axis) and propidium iodide (PI) (X-axis) staining. (b) Histograms of PI staining. (B) Percentages of cells in G1/G0, S, and G2/M phases showing accelerated cell cycles after bacterial exposure. (C) Faster cell cycles after exposure to increasing MOIs of LI60C3. The cell ratios of (a) S/G2/M phase to G1/G0 phase or (b) high to low Ki67 intensity are shown. ∗P < .05 vs 0. N = 5/group. Experiments were repeated twice. (D) Increased Caco-2 cell viability after bacterial exposure in a (a) dose- and (b) time-dependent manner. ∗P < .05 vs 0. N = 8/group (E) Larger spheroid area of Caco-2 cells after bacterial exposure. (a) Spheroid images. Scale bar: 200 μm (upper), 20 μm (lower). (b) Spheroid area. ∗P < .05 vs w/o. (F) Nuclear translocation of β-catenin (red) after exposure to LI60C3 tagged with green fluorescent protein (GFP) (green). Scale bar: 10 μm. (G) Specific gene deletion for WT E.coli LI60C3 was confirmed by PCR analysis. Bacteria were engineered with single- or triple-gene deletion of fimA, fimH, and htrA (ie, ΔFimA, ΔFimH, ΔHtrA, or ΔTriple). (H) Caco-2 cells were challenged with WT and gene-deleted bacteria at a MOI of 1, and the intracellular bacterial counts were determined at various time points after exposure. The percentage of intracellular bacterial survival after 2 days was shown. N = 6/group. ∗P < .05 vs WT. (I) Faster cell cycles after exposure to WT compared with gene-deleted bacteria. The cell ratios of (a) S/G2/M phase to G1/G0 phase or (b) high to low Ki67 intensity. ∗P < .05 vs w/o, #P < .05 vs WT. N = 5/group. Experiments were repeated twice. MTT,3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide.
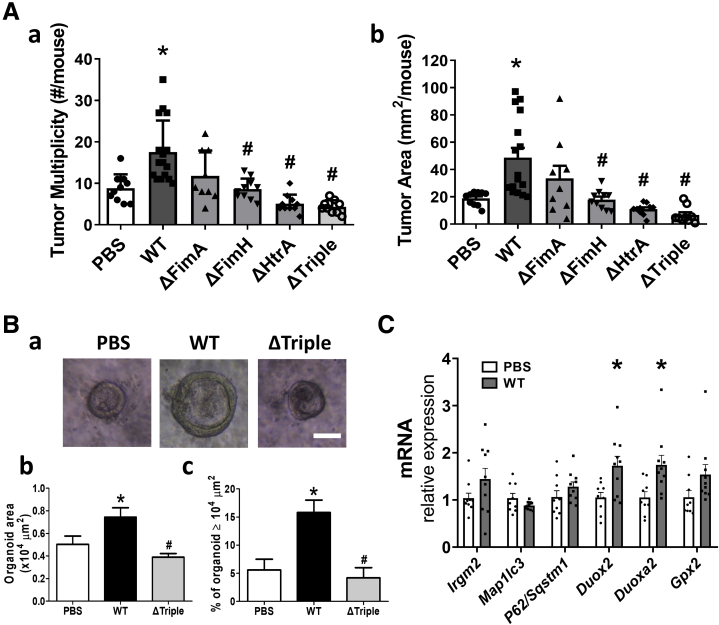
The wild-type (WT) LI60C3 was engineered with single- or triple-gene deletion of fimA, fimH, and htrA (ie, ΔFimA, ΔFimH, ΔHtrA, or ΔTriple) (Figure 4G) to assess whether bacterial internalization was essential for epithelial proliferative effects. Reduction of intracellular counts of gene-deleted bacteria in epithelial cells was confirmed (Figure 4H). The cell-cycle rates in Caco-2 cells exposed to the gene-deleted bacterial strains were lower than those exposed to the isogenic WT bacteria (Figure 4Ia and Ib). Next, mice were inoculated with bacteria at midphase for tumor inspection on day 90. Significant increases in tumor burden were observed in the mice inoculated with the WT LI60C3 strain compared with those given saline (Figure 5Aa and Ab), whereas tumor burden was lower in the mice inoculated with ΔFimH, ΔHtrA, or ΔTriple (Figure 5Aa and Ab). Moreover, primary intestinal organoid cultures were developed from mouse colonic crypts 2 days after bacterial inoculation to evaluate epithelial proliferation in vivo. Larger colonic organoid sizes were seen in the WT group compared with those given saline or ΔTriple (Figure 5B).
Figure 5.
Gavage with wild-type E.coli LI60C3 but not gene-deleted bacteria increased tumor burden in recipient mice. (A) Higher (a) tumor multiplicity and (b) tumor area in mice inoculated with WT LI60C3 compared with those with gene-deleted bacteria. ∗P < .05 vs PBS, #P < .05 vs WT. N = 10–15/group. (B) Colonic organoid cultures derived from mice 2 days after bacterial inoculation. (a) Organoid images. Scale bar: 50 μm. (b) Organoid area. (c) Percentage of organoids larger than 10,000 μm2. Increased growth rate of organoids derived from mice inoculated with WT compared with those with PBS and gene-deleted bacteria. (C) Altered antimicrobial gene expression in mouse colonocytes 2 days after bacterial inoculation. ∗P < .05 vs PBS. N = 10/group. mRNA, messenger RNA.
To investigate host–microbe interaction, antimicrobial gene expression was analyzed in mouse colonocytes 2 days after inoculation with saline or WT LI60C3 (Figure 5C). The antimicrobial machinery against intracellular bacteria is composed of free radical–producing NADPH oxidases and autophagy effectors, such as immunity-related guanosine triphosphatase M (IRGM), microtubule-associated protein 1 light chain 3β (MAP1LC3B), and P62/sequestosome-1 (SQSTM1).19,20 The messenger RNA levels of autophagy effectors (ie, Irgm2, Map1lc3b, and P62/Sqstm1) showed no difference between the 2 mouse groups, while increased transcripts of oxidative stress genes (ie, Duox2 and Duoxa2) were observed in the mouse colonocytes inoculated with WT LI60C3 (Figure 5C). Overall, the data showed that invasive E coli stimulated epithelial hyperproliferation and promoted tumor growth.
Invasive E coli Suppressed Autophagy and Caused Free Radical Overproduction That Led to Epithelial Hyperproliferation
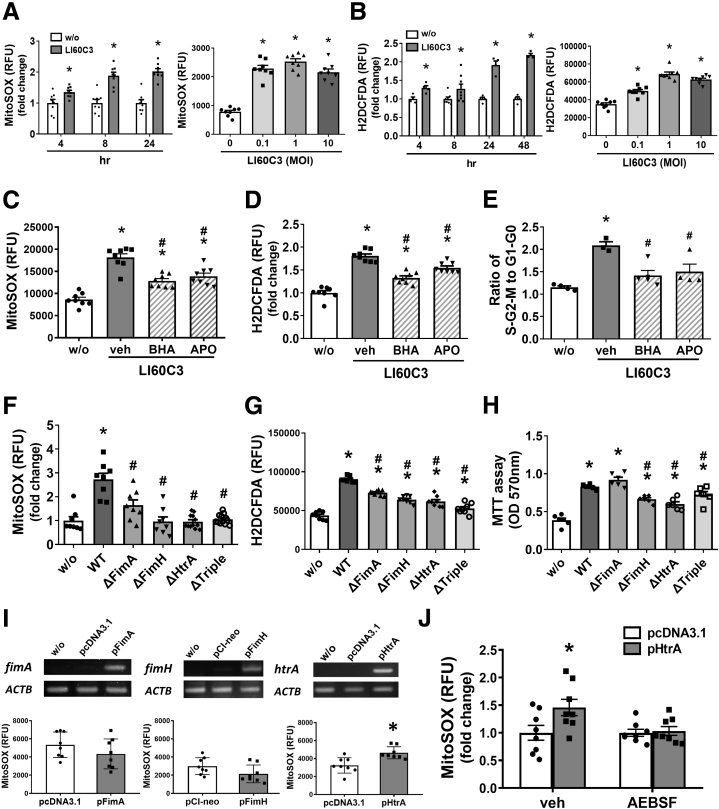
The mechanisms of bacteria-induced epithelial hyperproliferation were next elucidated by focusing on the involvement of reactive oxygen species (ROS) and autophagy. Paradoxic roles for free radicals and autophagy were documented in colon tumorigenesis.22, 23, 24, 25 Here, bacterial infection induced mitochondrial superoxide and cellular ROS synthesis in Caco-2 cells in a time-dependent manner (Figure 6A and B). Pretreatment with butylated hydroxyanisole (an antioxidant) or apocynin (an inhibitor to NADPH oxidase) attenuated the bacteria-induced cell-cycle acceleration (Figure 6C–E), suggesting that bacteria-induced ROS production promoted epithelial hyperproliferation. In contrast to WT LI60C3, the ΔFimA, ΔFimH, ΔHtrA, or ΔTriple failed to increase the ROS levels (Figure 6F–H). We sought to determine whether E. coli virulence factors per se may induce epithelial ROS production (uncoupled to virulence factor–dependent bacterial adhesion or survival) and transfected the individual microbial products into Caco-2 cells. Expression of HtrA increased the cellular free radical levels, whereas expression of FimA or FimH did not (Figure 6I). Bacterial HtrA is a heat shock protein that functions as both a chaperone and a serine protease. Pretreatment with 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF) (a serine protease inhibitor) reduced the ROS levels triggered by bacterial HtrA (Figure 6J).
Figure 6.
Invasive E.coli–induced epithelial hyperproliferation was dependent on free radicals. Synthesis of ROS after exposure to E.coli LI60C3 was determined in Caco-2 cells. (A) Increased levels of mitochondrial superoxide after bacterial infection for the indicated times (left) and doses (right). (B) Increased intracellular ROS levels after bacterial infection for the indicated times (left) and doses (right). ∗P < .05 vs w/o or 0. N = 6–8/group. (C and D) Pretreatment with antioxidants, such as butylated hydroxyanisole (BHA, a free radical scavenger) or apocynin (APO, a nicotinamide adenine dinucleotide phosphate oxidase inhibitor), decreased the bacteria-induced ROS synthesis in Caco-2 cells. ∗P < .05 vs w/o, #P < .05 vs vehicle (veh). N = 8/group. (E) Antioxidants prevented the bacteria-induced epithelial cell-cycle acceleration. ∗P < .05 vs w/o, #P < .05 vs vehicle. N = 4/group. (F and G) Higher free radical levels in Caco-2 cells exposed to WT LI60C3 compared with gene-deleted bacteria. ∗P < .05 vs w/o, #P < .05 vs WT. N = 8/group. (H) Caco-2 cell viability was higher after exposure to WT LI60C3 than gene-deleted bacteria. ∗P < .05 vs w/o, #P < .05 vs WT. N = 6/group. (I) Increased free radical levels in Caco-2 cells after transfection with plasmids encoding bacterial HtrA (pHtrA) but not FimA (pFimA) or FimH (pFimH). The expression of virulence factors (upper panel) and the synthesis of free radicals in the transfected cells (lower panel). ∗P < .05 vs mock plasmids. N = 8/group. (J) Pretreatment with AEBSF (a serine protease inhibitor) prevented the HtrA-induced free radical synthesis in Caco-2 cells. ∗P < .05 vs mock plasmids. N = 8/group. Experiments were repeated at least twice. MTT,3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide; RFU,relative fluorescence units.
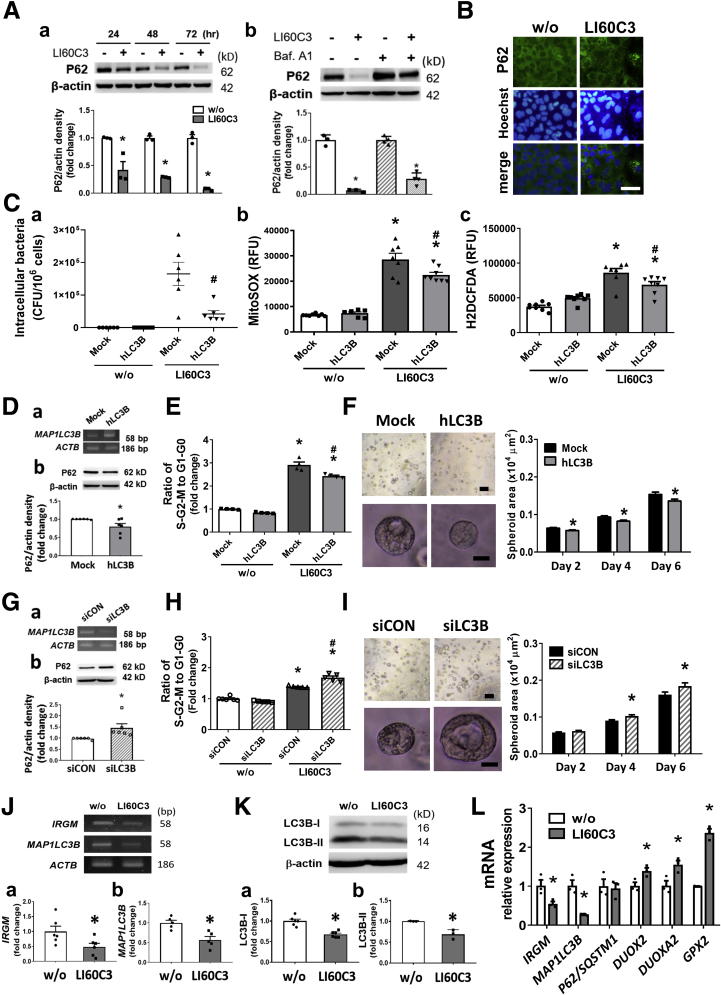
The autophagy flux activity next was investigated in bacteria-exposed Caco-2 cells. Degradation of P62/SQSTM-1 (a ubiquitin-binding scaffold protein) signifies autophagosome fusion with lysosomes. Exposure to LI60C3 induced P62 degradation in cells, which was blocked by pretreatment with bafilomycin A1 (an inhibitor of vacuolar H+-adenosine triphosphatase [ATPase]) (Figure 7A and B). To verify whether autophagy flux activity may eliminate intraepithelial bacteria and modulate oxidative stress responses, we overexpressed the human MAP1LC3B gene by plasmid transfection in Caco-2 cells before microbial exposure. Decreased intracellular bacterial counts and ROS levels were observed in the cells overexpressing MAP1LC3B (Figure 7C). Overexpression of MAP1LC3B increased autophagy flux activity as evidenced by the reduction in P62 levels (Figure 7D), and diminished epithelial proliferation and spheroid growth in the bacteria-infected cells (Figure 7E and F). In contrast, inhibition of autophagic flux by gene silencing of MAP1LC3B intensified bacteria-induced cell-cycle acceleration and spheroid growth (Figure 7G–I). Our results suggested that free radical synthesis and autophagy flux activity both were stimulated by bacterial invasion. However, antagonistic roles of the 2 antimicrobial pathways were observed, of which free radicals promoted epithelial proliferation whereas autophagy inhibited epithelial proliferation.
Figure 7.
Invasive E coli suppressed autophagy and caused free radical overproduction, which led to epithelial hyperproliferation. (A) Autophagy flux activity was examined in bacteria-infected Caco-2 cells. (a) Time-dependent autophagosomal degradation of P62 was observed after bacterial exposure. (b) P62 degradation was reduced by pretreatment with bafilomycin A1 (a lysosomal inhibitor). ∗P < .05 vs w/o. N = 4/group. (B) Immunofluorescent images showing homogenous distribution of P62 staining (green) in the cytoplasm of uninfected cells and punctate staining in bacteria-infected cells. Scale bar: 50 μm. (C) Overexpression of human MAP1LC3B (hLC3B) by plasmid transfection reduced (a) intracellular bacterial counts, (b) mitochondrial superoxide, and (c) intracellular ROS levels in microbe-infected cells. ∗P < .05 vs w/o, #P < .05 vs mock plasmid. N = 6–8/group. (D) Overexpression of hLC3B was confirmed by PCR (panel a) and caused reduction of P62 (panel b). ∗P < .05 vs mock. (E) Overexpressing hLC3B decreased bacteria-induced cell-cycle acceleration. ∗P < .05 vs w/o, #P < .05 vs mock. N = 6/group. (F) Overexpressing hLC3B decreased spheroid growth. Scale bar: 200 μm (upper), 20 μm (lower). ∗P < .05 vs mock. N = 6/group. (G) Knockdown of MAP1LC3B by small interfering (si)RNA (siLC3B) in Caco-2 cells was confirmed by (a) PCR and caused (b) accumulation of P62. ∗P < .05 vs siCON. (H) siLC3B potentiated bacteria-induced epithelial cell-cycle acceleration. ∗P < .05 vs w/o, #P < .05 vs siCON. N = 6/group. (I) siLC3B increased bacteria-induced spheroid growth. Scale bar: 200 μm (upper), 20 μm (lower). ∗P < .05 vs siCON. N = 6/group. (J) Bacterial exposure suppressed autophagy gene expression. Gel blots and densitometric analysis of (a) IRGM and (b) MAP1LC3B transcripts by semiquantitative PCR. ∗P < .05 vs w/o. N = 6/group. (K) Western blots showing reduced protein levels of (a) LC3B-I (cytoplasmic form) and (b) LC3B-II (autophagosome-associated form) after bacterial exposure. Epithelial cells were pretreated with bafilomycin A1 to prevent autolysosome degradation of proteins. (L) Quantitative PCR results showing decreased autophagy genes and increased oxidative stress gene expression in bacteria-infected cells. ∗P < .05 vs w/o. N = 3/group. Experiments were repeated at least twice. mRNA, messenger RNA; RFU, relative fluorescence units.
Previous studies have documented that invasive pathogens impaired cellular autophagy as a means of microbial evasion.26, 27, 28, 29 Here, we hypothesized that pathobionts could manipulate autophagy activity and cause imbalances between the 2 defense machineries to alter epithelial proliferation. Decreased messenger RNA levels of IRGM and MAP1LC3B, and reductions in protein levels of both light chain 3 (LC3)-I (cytosolic form) and LC3-II (autophagosome-associated form), were observed in Caco-2 cells after bacterial exposure (Figure 7J and K). The reductions in IRGM and MAP1LC3B gene expression were associated with increases in the DUOX2, DUOXA2, and GPX2 levels in bacteria-infected Caco-2 cells (Figure 7L). Taken together, the data indicated that invasive E coli suppressed cellular autophagy and caused free radical overproduction, which led to epithelial hyperproliferation.
Heightened Epithelial Autophagy Eliminated Intracellular Microbes in a Long-Term Alternating Spheroid Culture Model
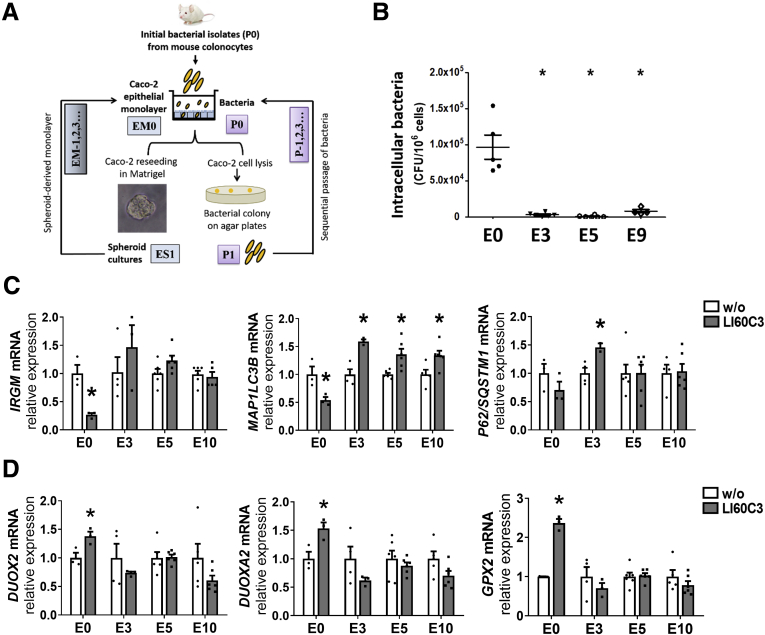
Considering that epithelial autophagy eliminates intracellular bacteria while invasive E. coli actively suppress autophagy to benefit their own survival, the counterbalance between the host and microbes may determine the critical window for bacteria-driven epithelial hyperproliferation. To address the long-term changes in host–microbe interplay for chronic tumor growth, a new model of alternating spheroid cultures was developed for sequential challenge with bacteria (Figure 8A). In this novel platform, epithelial monolayers (EM0) were apically challenged with the initial isolate of E. coli LI60C3 (P0), and then reseeded as spheroids (designated as [epithelial spheroid] ES1 with 1 bacterial encounter). The spheroid cultures replated as monolayers (denoted as EM1) were apically challenged with bacteria isolated from the EM0 cell lysates (denoted P1), followed by reseeding as spheroid cultures (ES2) and isolation of intracellular bacteria (P2). A total of 10 cycles were conducted to acquire sequentially passaged bacteria up to P9 and epithelial cultures up to E10 (Figure 8A).
Figure 8.
A novel alternating spheroid model showing that repeated invasion of E.coli up-regulated epithelial autophagy, which eliminated intracellular microbes in the long term. (A) An alternating spheroid culturing platform was developed for sequential bacterial challenge to address the long-term counterbalance between invasive pathobionts and host defense in chronic tumor growth. Our pilot study showed that epithelial cells cultured as spheroids were able to sustain replating after repeated bacterial invasion whereas those cultured as monolayers did not survive. Caco-2 EMs apically challenged with an initial isolate of mouse E.coli (P0) were either reseeded into ES cultures or underwent cell lysis for enumeration of intracellular bacteria. The spheroids were harvested and replated into monolayers for the next encounter of passaged bacteria for up to 10 cycles. The same MOI value was used for each challenge. (B) Decreased intracellular bacterial counts were observed in the third, fifth, and ninth cycles of epithelial culture after exposure to the corresponding bacterial passages. The 10th cycle of spheroid-derived monolayer was not challenged with bacteria. Each dot represents 1 sample. ∗P < .05 vs E0. N = 6/group. (C and D) Quantitative PCR results showing changes in autophagy and oxidative stress genes in the 3rd, 5th, and 10th cycles of epithelial culture. ∗P < .05 vs w/o. N = 4–6/group. Experiments were repeated twice. mRNA, messenger RNA.
We observed a more than 96% decrease in the intracellular bacterial counts after the third round of bacterial invasion, despite using the same multiplicity-of-infection (MOI) value for each challenge (Figure 8B). In contrast to the decreased IRGM and MAP1LC3B levels after a single bacterial exposure in epithelial cells (E0), up-regulated expression of autophagy genes was observed after repeated bacterial invasion (Figure 8C). Increased MAP1LC3B transcripts were noted in the E3, E5, and E10 cultures, and up-regulated P62/SQSTM1 levels were found in the E3 culture (Figure 8C). No change in DUOX2, DUOXA2, or GPX2 expression was observed after repeated bacterial invasion in the alternating spheroid cultures (Figure 8D). The data indicated that sequential challenge of invasive E coli led to heightened autophagy gene expression in the host epithelia that eliminated intracellular microbes in the long term.
Bacterial Virulence Factors and Altered Antimicrobial Gene Expression Were Found in Human CRCs
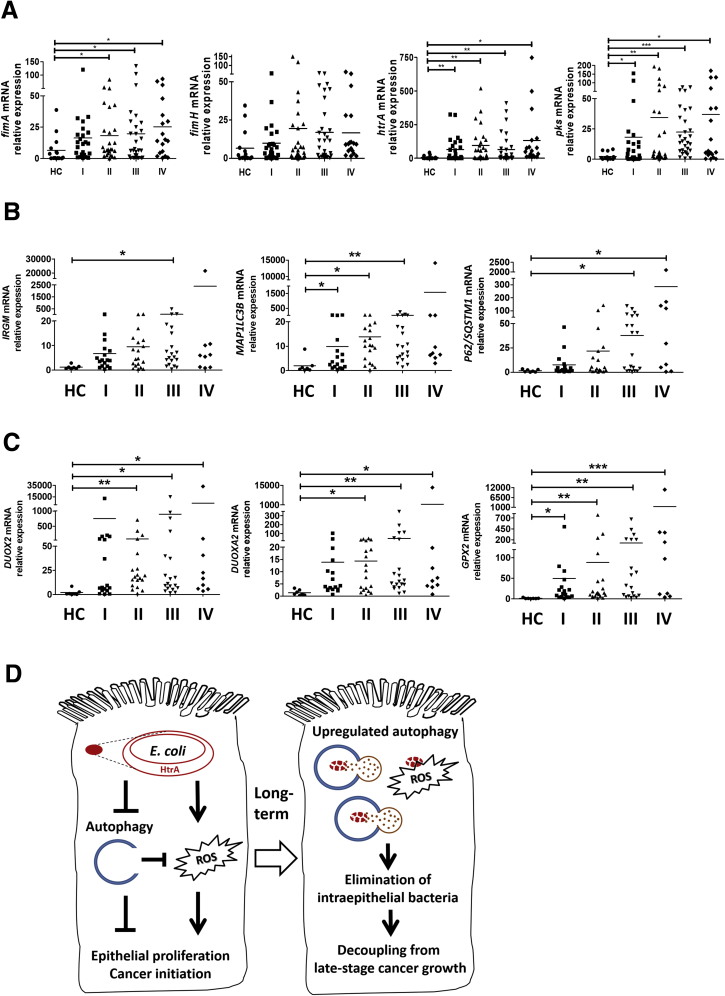
Lastly, the mucosal presence of bacterial virulence factors and changes in antimicrobial gene expression were assessed in human CRC specimens (Tables 2 and 3). An increase in the fimA level was observed in the stages II to IV tumor specimens compared with healthy mucosal tissues (Figure 9A). The htrA level was 10 times higher in stages I to IV tumors than in the healthy mucosa (Figure 9A). Although pks was absent in the mouse stool and invasive E coli, we observed relatively high pks levels in the human CRCs (Figure 9A). Moreover, increased expression of autophagy genes (ie, IRGM, MAP1LC3B, and P62/SQSTM1) and oxidative stress genes (ie, DUOX2, DUOXA2, and GPX2) was found in human CRC specimens compared with healthy mucosal tissues (Figure 9B and C). Taken together, the findings suggest that invasive HtrA-positive E coli suppressed cellular autophagy and caused ROS overproduction, which led to epithelial hyperproliferation and cancer initiation. In the long term, host epithelia up-regulated autophagy activity after repeated bacterial invasion to eliminate the intracellular microbes, which may terminate the bacteria-dependent epithelial hyperproliferation at late cancer stages (Figure 9D).
Table 2.
Characteristics of Healthy Controls and Patients With Colorectal Cancers for Analysis of Bacterial Virulence Factors
| Tumor stages | Control | Stage I | Stage II | Stage III | Stage IV |
|---|---|---|---|---|---|
| Patients, n | 15 | 24 | 26 | 30 | 18 |
| Age, y (mean) | 22–66 (40.5) | 41–81 (60.1) | 40–88 (63.96) | 31–87 (64.7) | 31–83 (67.27) |
| Sex, males; females | 7; 8 | 16; 8 | 20; 6 | 15; 15 | 12; 6 |
| Tumor location (%) | |||||
| Cecum | – | 3 (12.5) | 2 (7.7) | 1 (3.3) | 1 (5.6) |
| Ascending colon | – | 5 (20.8) | 3 (11.5) | 6 (20.0) | 2 (11.1) |
| Transverse colon | – | 1 (4.2) | 1 (3.8) | 2 (6.7) | 1 (5.6) |
| Descending colon | – | 2 (8.3) | 4 (15.4) | 1 (3.3) | 2 (11.1) |
| Sigmoid–rectum | – | 7 (29.2) | 10 (38.5) | 12 (40.0) | 6 (33.3) |
| Rectum | – | 6 (25.0) | 6 (23.1) | 8 (26.7) | 6 (33.3) |
Table 3.
Characteristics of Healthy Controls and Patients With Colorectal Cancers for Analysis of Autophagy and Oxidative Stress Genes
| Tumor stages | Control | Stage I | Stage II | Stage III | Stage IV |
|---|---|---|---|---|---|
| Patients, n | 7 | 17 | 19 | 20 | 9 |
| Age, y (mean) | 31–76 (54.6) | 38–81 (61.4) | 26–88 (62.2) | 31–87 (63.5) | 31–83 (63.8) |
| Sex, males; females | 5; 2 | 10; 7 | 15; 4 | 10; 10 | 5; 4 |
| Tumor location (%) | |||||
| Cecum | – | 1 (5.8) | 1 (5.3) | 1 (5.0) | 0 (0.0) |
| Ascending colon | – | 2 (11.8) | 1 (5.3) | 2 (10.0) | 2 (22.2) |
| Transverse colon | – | 2 (11.8) | 1 (5.3) | 1 (5.0) | 1 (11.1) |
| Descending colon | – | 2 (11.8) | 3 (15.8) | 1 (5.0) | 1 (11.1) |
| Sigmoid–rectum | – | 6 (35.3) | 7 (36.8) | 8 (40.0) | 2 (22.2) |
| Rectum | – | 4 (23.5) | 4 (21.0) | 7 (35.0) | 3 (33.4) |
| Colon, unspecified | – | 0 (0.0) | 2 (10.5) | 0 (0.0) | 0 (0.0) |
Figure 9.
Presence of bacterial virulence factors and altered antimicrobial gene expression in human CRC specimens. Mucosal samples of healthy control (HC) subjects and CRC specimens from patients at the 4 stages (I, II, III, and IV) were analyzed by quantitative PCR. (A) Relative expression of microbial virulence factors fimA, fimH, htrA, and pks. (B) Relative expression of human autophagy genes IRGM, MAP1LC3B, and P62/SQSTM1. (C) Relative expression of human oxidative stress genes DUOX2, DUOXA2, and GPX2. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 vs HC. (D) Schematic illustration of aberrant host–microbe interplay at a critical phase of tumorigenesis. The invasive HtrA-positive E coli suppressed cellular autophagy, resulting in free radical overproduction, which led to epithelial hyperproliferation and cancer initiation. After long-term repeated bacterial invasion, the host epithelia showed up-regulated autophagy to eliminate the intracellular microbes, which terminated the phenomenon of bacteria-dependent epithelial hyperproliferation. In sum, invasive E coli contributed to cancer initiation but was decoupled from late-stage cancer growth. mRNA, messenger RNA.
Discussion
Our study showed a key time frame for dysfunctional host–microbe interactions that contributed to cancer initiation through bacterial counterbalancing of antimicrobial responses in the colonic epithelium. The temporal action of epithelial hyperproliferation caused by microbiota dysbiosis and E. coli invasion was linked to consequences of carcinoma transformation. The findings confirmed a direct role for invasive bacteria in promoting epithelial proliferation through manipulation of host defense. Moreover, a novel alternating spheroid model was developed for sequential bacterial challenge to address the long-standing counterbalance between host and microbes in chronic tumor growth. We showed that host epithelia displayed up-regulated autophagy activity after long-term repeated bacterial infection to eliminate intracellular microbes, which supported a critical window for bacteria-dependent tumor hyperproliferation and its decoupling from late-stage cancer growth.
A driver–passenger role of bacteria for CRC development has been described,30, 31, 32 but the crucial timing of aberrant host–microbe interplay is unknown. Here, we observed a surge in fecal microbial diversity and virulence emergence during a key phase of cancer initiation. The findings are compatible with clinical reports, which show biphasic modulation of the fecal microbiota—high diversity and increased numbers of bacteria in adenoma33,34 and reduced bacterial diversity in carcinoma.35,36 The critical timing of cancer initiation (malignant transformation from dysplasia to carcinoma) correlated with the observation of an altered antimicrobial transcriptome profile in colon tissue. Because dysplasia is a precancerous lesion that may regress or progress to carcinoma,17,18 an aberrant host–microbe interaction during the midphase may promote the onset of cancer malignancy. On the other hand, an intervention (eg, ABX) during the key phase may abort the tumorigenesis. Overall, the data suggest that longitudinally monitoring the gut microbiota and colon transcriptome profiles, as well as the presence of invasive E coli and their virulence factors, could be valuable in clinical practice to signify the proper time point for bacterium-targeted intervention in high-risk patients.
Virulence-expressing E. coli isolated from colonic epithelial cells increased the tumor burden in the recipient mice. Invasive E. coli dose-dependently accelerated the cell-cycle transition and spheroid growth of epithelial cell lines in vitro, supporting the conclusion that the bacteria-induced epithelial hyperproliferation was independent of immune cell activation and inflammation. Gradual increases in invasive ability and virulence factor expression were observed in the tumorigenic E. coli after sequential passaging in epithelial cells, pointing to the possibility that intestinal pathobionts were derived from intraepithelial commensals striving to survive against antimicrobial pressures. The virulence factors including adhesin fimbriae fimA/H and stress survival htrA and ompC levels were increased after sequential passaging in epithelial cells, whereas a decrease in the fliC gene implicated reduced motility of the internalized bacteria. HtrA is a heat shock protein that functions as both a chaperone and a serine protease, and OmpC acts as porins to allow water and solute to diffuse into bacterial cells, which are likely essential for intracellular survival of microbes. The virulence factor HtrA, beyond its role in enhancing bacterial survival under stress conditions, was responsible for the induction of free radicals via its serine protease domain for promoting epithelial hyperproliferation. We also observed bacterial fimA, htrA, and pksR in human CRC specimens. Although previous studies showed that genotoxicity conferred by pks-positive E. coli promoted tumor growth,14, 15, 16 a knowledge gap still exists between DNA damage-dependent cell–cycle arrest and tumor cell hyperproliferation. Our study shows that HtrA-positive E. coli triggers epithelial cell-cycle acceleration for tumor growth, independent of genotoxin production or immune cell activation.
In addition to virulence factors, the counterbalance between invasive pathobionts and epithelial defense fuels tumor progression. Free radicals rapidly kill microbes while antibacterial autophagy (referred to as xenophagy) requires hours or days to remove intracellular microbes. With respect to tumor growth, the roles of autophagy and free radicals are controversial.22, 23, 24 Cellular autophagy and mitochondrial ROS promoted CRC growth in some studies,37,38 whereas other studies documented a tumor-suppressive role for autophagy through reduction of inflammation and oxidative stress.39,40 We identified that invasive E. coli suppressed epithelial autophagy, which although beneficial to its own survival in the short term, inversely boosted the intracellular ROS levels and triggered epithelial hyperproliferation. A recent article showed that Apc-mutant mice with epithelium-specific disruption of autophagy showed a lower tumor burden than Apc-mutant mice. However, those autophagy-disrupted Apc-mutant mice showed a higher tumor burden than Apc-mutant mice if inoculated with colibactin-producing E. coli.41 Thus, epithelial autophagy exerts opposing roles in colon tumorigenesis depending on the presence or absence of mucosa-associated bacteria, which emphasizes a crucial microbial factor in neoplasia transformation. Invasive pathogens were known to evade autophagic clearance to achieve persistent infection in the host.26, 27, 28, 29 Unlike pathogen eradication, which is mostly dependent on adaptive immunity, epithelial autophagy up-regulation was shown to be a form of heightened innate defense sufficient to eliminate intracellular pathobionts in the long term. This phenomenon was observed in the novel alternating spheroid platform developed in our laboratory to address the long-standing counterbalance between host and microbes, of which the experimental design was to ensure epithelial viability after repeated bacterial infection to mimic chronic tumor growth. Moreover, human CRC specimens showed increased MAP1LC3B expression as early as stage I while P62 levels were up-regulated at later cancer stages. The human data are consistent with the in vitro finding that invasive E coli manipulated epithelial autophagy activity through modulating gene expression of upstream cascading effectors (ie, IRGM and MAP1LC3B). Overall, the results suggested a chronic wax-and-wane pattern between host defense and pathobiont invasion, and provided an explanation for the decoupling of E coli–driven innate dysregulation and cancer growth at late stages.
In conclusion, a unique host oncogenic profile related to antimicrobial responses was associated with microbiota dysbiosis during cancer initiation. An invasive pathobiont manipulated the epithelial autophagy and oxidative stress responses, which were linked to consequences for carcinoma transformation. Our findings emphasized a critical time point in bacteria-driven cancer initiation, which could be a window of opportunity to intervene with microbe-targeted precision medicine.
Materials and Methods
Mouse Models of CRC
Mice were administered AOM and DSS42, 43, 44 and were given ABX mixtures (vancomycin, neomycin, metronidazole, and ampicillin) by daily oral gavage and drinking water supplementation at various phases, including early (days 46–53), mid (days 56–63), and late (days 66–73) regimens. The experiment was conducted on 3 separate batches of mice, and each batch was divided into 4 groups including control, early, mid-, and late ABX (N = 5/group). All animals were killed on day 90 for macroscopic inspection and histologic grading of tumors. In another experiment, mice were killed before (day 0) or after carcinogen administration (days 46, 56, and 66) to determine the colon transcriptome profiles and the timing of the dysplasia transition to carcinoma.
Inoculation of WT and Gene-Deleted Bacteria in Mice
Mice subjected to AOM/DSS and administered ABX mixtures at midphase were orogastrically inoculated with either phosphate-buffered saline (PBS), WT bacteria, or gene-deleted bacteria (see later) at 109 colony forming units (CFU) per mouse on day 64. The animals were killed on day 90 to determine tumor burden.
Gene Microarray
Mouse colonic tissues were collected on days 0, 46, 56, and 66 after administration of 3 cycles of AOM/DSS. Tissue RNA were extracted by using the RNeasy Mini Kit (Qiagen, Germantown, MD) and sent for microarray using the Clariom S Assay (ThermoFisher, Waltham, MA) to analyze more than 22,100 mouse genes in the National Health Research Institute (Miao-Li, Taiwan). The gene array data were submitted to the GEO repository (GSE144719). The transcript expression levels were measured by quantitative PCR using primer pairs based on National Center for Biotechnology Information sequences, and calculated according to the cycle threshold (CT) value and normalized using glyceraldehyde 3-phosphate dehydrogenase as the internal control.
Microbiota Analysis
Mouse fecal samples (wet weight, 0.1–0.5 g) were collected and extracted for bacterial DNA using a QIAamp DNA stool mini kit (Qiagen). Fecal bacterial DNA was analyzed by high-throughput bacterial 16S rRNA gene sequencing with the Illumina Genome Analyzer IIx platform (Illumina Inc., San Diego, CA) in the Medical Microbiota Core at the National Taiwan University College of Medicine (NTUCM).45 The sequencing data were submitted to the GEO database (GSE166030 and GSE166038).
Bacterial Virulence Factor Analysis in Mouse Fecal Samples and Isolated Bacteria
Fecal samples were extracted using a QIAamp DNA stool mini kit. Isolated bacterial colonies from mouse colonic epithelial cells were grown in Luria–Bertani broth and used directly as DNA templates. The levels of virulence factors in bacterial DNA were measured by PCR, and calculated according to the Ct value and normalized using 16S rRNA as the internal control. The virulence factors tested were related to adherence ability, such as afimbrial adhesin C (afaC), fimbrial adhesin (fimA/C/H), long polar fimbriae A (lpfA), and P-fimbriae (papC); mobility such as flagellin C (fliC); stress responses such as oxidoreductase dsbA, high temperature requirement A (htrA), and outer membrane protein C (ompC); genotoxins such as polyketide synthase R (pksR), cytolethal distending toxin (cdt), and cytotoxic necrotizing factor (cnf); and protein synthesis inhibition, such as Shiga toxins.7,14,46, 47, 48, 49
Human Sample Collection for Host Gene and Bacterial Virulence Analysis
Colonic specimens from healthy individuals and CRC patients at 4 tumor stages collected in a BioBank at the National Taiwan University Hospital (NTUH) were analyzed. Written informed consent was obtained from all study subjects, and approval for this study was granted by the Research Ethics Committee of NTUH (9361701157). The tissue RNA was reverse-transcribed and analyzed by quantitative PCR. The host genes were calculated according to the Ct value and normalized using β-actin as the internal control. Moreover, bacterial DNA extracted from the human colonic tissues was analyzed by quantitative PCR, and the virulence levels were calculated according to the Ct value and normalized using 16S rRNA as the internal control.
Approval for the animal study was granted by the Institutional Animal Care and Use Committee of the NTUCM (20180030).
Quantification of Intraepithelial Bacteria by a Gentamicin-Resistance Assay
Intestinal epithelial cells were isolated for a gentamicin-resistance assay as described.10, 11, 12 Briefly, purified epithelial cells were incubated with 300 μg/mL gentamicin for 1 hour to kill extracellular bacteria. Cell lysates were plated onto agar plates overnight, and the number of bacterial colonies was normalized to those of Trypan blue–negative epithelial cells, and are presented as log10 CFU/106 cells. Moreover, individual colonies were extracted for bacterial DNA using a standard kit (Qiagen) and amplified by PCR with universal 16S rRNA gene primers for sequencing and classification. Further analysis to identify the bacterial species was conducted by using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. The antibiogram of bacterial species was generated by the Department of Medical Biotechnology at the NTUH, and the bacteria showed susceptibility to gentamicin with a minimum inhibitory concentration of 2 μg/mL or less. The 16S rRNA gene sequences of Escherichia coli LI60C3, Enterobacter cloacae HM763C3, Enterococcus faecalis LI91C2, and Staphylococcus lentus IX60C1 were submitted to the DNA databank of Japan (accession numbers: LC369044, LC375378, LC375379, and LC375380). These bacterial strains were stored in glycerol at -80°C until further use.
Construction of Gene-Deleted Bacteria
The fimA, fimH, and htrA genes of the E. coli LI60C3 strain were sequenced and submitted to the DNA databank of Japan (accession numbers: LC374589, LC374590, and LC374591). The bacteria were gene-deleted using the phage λ-red and Flp recombinase method.14 PCR products were generated by using primers with 36- to 50-nt extensions that are homologous to regions adjacent to the virulence genes to be inactivated, and template plasmids carrying antibiotic-resistance genes that are flanked by Flp recognition sites. The bacteria were transformed with Red/ET (Gene Bridges, Heidelberg, Germany) plasmids and the linear PCR products, after induction of recombination to allow the PCR product to be inserted into the chromosome. Gene-deleted bacteria were confirmed by PCR and sequencing.
Human Cell Lines
Human Caco-2 cell lines with monolayered features were used for bacteria-epithelial cocultures, and gene knockdown and overexpression experiments.50,51 These cells were purchased from and authenticated by the American Type Culture Collection/Bioresource Collection and Research Center, and short tandem repeat (STR)–PCR profile was performed by the Bioresource Collection and Research Center. In some experiments, human cell lines were pretreated with butylated hydroxyanisole (a free radical scavenger; 200 μmol/L; Sigma, St. Louis, MO), or apocynin (a NADPH oxidase inhibitor; 1 mmol/L; Calbiochem, San Diego, CA), 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF, a serine protease inhibitor; 1 mmol/L; Sigma), or bafilomycin A1 (an inhibitor to lysosomal H+-ATPase; 0.1–1 μmol/L; Cayman, Ann Arbor, MI) before bacterial exposure.
Gene Silencing by Small Interfering RNA
Cells were transfected with 50 nmol/L of small interfering RNA oligonucleotides using Lipofectamine RNAiMAX reagent (Life Technologies, Carlsbad, CA) in serum-free basal medium. After confirmation of knockdown efficiency by reverse-transcription PCR, the transfected cells were exposed to bacteria for measurement of intracellular bacterial counts.
Plasmid Constructs and Cell Transfection
The expression vectors pcDNA3.1 (Addgene_79663) and pCI-neo (Addgene_25099) were used for gene construction. The coding sequences of all plasmid constructs were confirmed by automated sequencing analysis. Cells were transfected with pcDNA3.1-MAP1LC3B, pcDNA3.1-FimA, pcDNA3.1-HtrA, and pCI-neo-FimH plasmids (2.5 μg/well each), or with mock plasmids using Lipofectamine 3000 Reagent (Life Technologies) for 24 hours. Transfection efficiency was confirmed by reverse-transcription PCR.
Bacteria-Epithelial Cell Co-cultures
Human cell lines were apically exposed to bacteria in antibiotic-free medium at various MOIs for 4 hours at 37°C. To obtain intracellular bacterial counts, the monolayers were treated with 1 mmol/L EDTA to obtain single cells for incubation in culture medium containing gentamicin (300 μg/mL) to remove extracellular bacteria for 1 hour. After lysis with 1% Triton X-100 (Sigma) in PBS for 10 minutes on ice, cell lysates were plated on agar plates at 37°C overnight. The numbers of viable intracellular bacteria are expressed as log10 CFU/106 cells.10
In other settings, cells exposed to bacteria for 4 hours were washed with PBS twice, incubated with gentamicin-containing medium for 24–48 hours, and evaluated for cell viability and cell-cycle progression by using a tetrazolium dye MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) assay and flow cytometry.42,43,52 Briefly, the cells were fixed with ice-cold 70% ethanol, and incubated with anti-human Ki67 (1:400; Cell Signaling, Beverly, MA) followed by secondary antibody conjugated to Alexa-Fluor 488, and then stained with propidium iodide for 30 minutes at room temperature. A minimum of 10,000 propidium iodide–stained nuclei were analyzed by flow cytometry, and the percentages of cells in the G0–G1, S, and G2–M phases of the cell cycle and the ratio of cells with high to low intensity of Ki67 were determined using Mod Fit LT cell-cycle analysis software (Verity Software, Topsham, ME). Moreover, bacteria-exposed cells were incubated in gentamicin-containing medium for 24–72 hours, and transcript levels of autophagy and oxidative stress genes were examined by PCR.
Sequential Passaging of Bacteria in Epithelial Cells
An initial isolate of mouse bacteria (denoted P0) was used for apical challenge in Caco-2 cells, and then the internalized bacteria were harvested after cell lysis with 1% Triton X-100 in PBS and grown on agar plates as the first passage (P1). P1 bacteria were used for subsequent apical challenge of newly plated epithelial cells, and the internalized bacteria were harvested again for a second passage (P2); this process then was repeated. The bacterial internalization counts and virulence factor levels were determined after each passage in Caco-2 cells.
Colonic Spheroid and Organoid Cultures
Human cell lines were plated as 3-dimensional spheroid cultures based on previous protocols.42,43,52 Briefly, Caco-2 cells cocultured with bacteria for 4 hours were trypsinized and resuspended in culture medium containing gentamicin (300 μg/mL) to eliminate extracellular bacteria. The cells were immediately mixed with ice-cold Matrigel (354234; Corning, New York, NY) at a 3:1 ratio with cell culture medium, and a volume of 300 μL was seeded per well in 24-well plates. After gel polymerization, 300 μL culture medium containing gentamicin (300 μg/mL) was overlaid and was changed every 2 days. Spheroids were cultured for up to 6 days, and the images of the spheroids were captured using a camera.
Primary mouse colonic crypt cells in the nontumor area were collected 2 days after bacterial inoculation, and cultured as organoids.42, 43, 44 Briefly, cell clusters of the crypt fractions were isolated by using an EDTA-detachment method. After counting, approximately 1500 cell clusters were plated in 300 μL ice-cold Matrigel (356235; BD Biosciences, Franklin Lakes, NJ) (3:1 ratio with crypt culture medium) per well of 24-well plates. After the polymerization of Matrigel, 300 μL crypt culture medium was overlaid. The crypt culture medium is composed of advanced Dulbecco’s modified Eagle medium/F12 (Life Technologies, CA) supplemented with 100 ng/mL mouse recombinant Wnt3A (R&D Systems, Minneapolis, MN), 50 ng/mL epidermal growth factor (Life Technologies), 100 ng/mL Noggin (PeproTech, Cranbury, NJ), 500 ng/mL R-spondin-1 (Peprotech), 1 μmol/L Jag-1 (AnaSpec, Fremont, CA), 10 μmol/L Y-27632 (Sigma), 2 mmol/L L-glutamine, 100 U/mL penicillin, 100 μg/mL Streptomycin (Life Technologies), and 2% heat-inactivated fetal bovine serum (Biological Industries, Kibbutz Beit-Haemek, Israel). The overlaying medium was refreshed every 2–3 days. The organoids were grown for up to 6–8 days, and images were captured under a light microscope and analyzed using imaging software (AxioVision release 4.8, Zeiss, Oberkochen, Germany). The total area of 180–200 spheroids and organoids was quantified per treatment or mouse group.
Alternating Spheroid Culture for Sequential Challenge With Bacteria
A new model of 3-dimensional epithelial cultures for sequential challenge with bacteria was developed to investigate the long-term counterbalance between host and microbes by mimicking chronic bacterial invasion in the intestine in vivo. Our pilot study showed that epithelial cells cultured as spheroids were able to sustain replating after sequential bacterial invasion whereas those cultured as monolayers showed excessive cell death. Caco-2 epithelial monolayers (denoted as EM0) were grown for 3 days until confluency, and then apically challenged with an initial isolate of bacteria (P0) at an MOI of 1 in antibiotic-free medium for 4 hours. The monolayers were trypsinized into single cells. One fifth of the cells were subjected to a gentamicin-resistance assay, and cell lysates were plated to determine the intracellular bacterial counts and isolate the first passage of bacteria (P1). Four fifths of the cells were resuspended in gentamicin-containing culture medium mixed with Matrigel for reseeding into spheroid cultures for 4 days (denoted as ES1, the number indicated 1 bacterial encounter). The ES1 spheroids were replated as monolayers (denoted as EM1) for apical challenge with P1 bacteria, and then reseeded into spheroid cultures (ES2) and for isolation of intracellular bacteria (P2). In total, alternating cycles of spheroid cultures were challenged with the sequentially passaged bacteria 10 times. Quantitative PCR analysis and a gentamicin-resistance assay were performed during the 3rd, 5th, and 10th cycles of epithelial culture.
Cellular Free Radical Assays
Cells grown on a black 96-well culture plate (Greiner Bio-One, Kremsmunster, Austria) were exposed to bacteria for 4 hours. After washing off extracellular bacteria, cells were placed in gentamicin-containing medium for 4–24 hours. The cells then were incubated with cell-permeable MitoSOX Red (Invitrogen, Waltham, MA) or 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA; Sigma) for reading of relative fluorescent units according to the manufacturer’s instruction. MitoSOX, which selectively targets functional mitochondria, shows red fluorescence after oxidization by superoxide. The MitoSOX assay was detected at Excitation/Emission 510/580 on the Paradigm Detection Platform (Beckman Coulter, Brea, CA). The H2DCFDA assay was detected at Ex/Em 485/535 on SpectraMax (Molecular Devices, San Jose, CA).
Measurement of Autophagy Flux Activity
Cells were pretreated with or without bafilomycin A1 (an inhibitor to lysosomal H+-ATPase) to prevent degradation of autophagy proteins before bacterial exposure. The bacteria-exposed cells were switched to gentamicin-containing medium for 24–72 hours and harvested to detect autophagic flux activity by Western blot. Autophagic flux was characterized by conversion of microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B) from cytosolic LC3-I to the autophagosomal membrane-associated LC3 (LC3-II, a lower migrating form), and by lysosomal degradation of P62/sequestosome 1 (SQSTM1). The primary antibodies used for Western blot included rabbit anti-human MAP1LC3B (1:2000; Cell Signaling), rabbit anti-human P62/SQSTM1 (1:5000; Novus Biologicals, Minneapolis, MN), and mouse anti-human β-actin (1:10,000; Sigma). The secondary antibodies used were goat anti-rabbit IgG or horse anti-mouse IgG (1:2000; Cell Signaling).
Statistical Analysis
All data are expressed as the means ± SEM, except that dot plots were used for human specimens and bacterial counts. The data were compared by analysis of variance with the Tukey multiple comparison test or the Newman–Keuls post hoc test. When comparing the difference among groups or between 2 groups of bacterial CFUs, the nonparametric Kruskal–Wallis test or the Mann–Whitney U test was used. A probability value less than .05 was considered significant.
Acknowledgments
The authors thank the staff of the imaging and sequencing facility at the First Research Core, Animal Center, Medical Microbiota Core, and Genomic Core at the NTUCM for their technical assistance.
CRediT Authorship Contributions
Linda Chia-Hui Yu, PhD (Conceptualization: Lead; Funding acquisition: Lead; Investigation: Lead; Resources: Lead; Supervision: Lead; Writing – original draft: Lead)
Shu-Chen Wei, MD, PhD (Formal analysis: Equal; Funding acquisition: Equal; Methodology: Equal; Resources: Lead; Validation: Equal)
Yi-Hsuan Li, MSc (Formal analysis: Lead; Investigation: Equal; Validation: Lead; Visualization: Equal)
Po-Yu Lin, MSc (Formal analysis: Equal; Investigation: Supporting; Methodology: Equal; Validation: Equal)
Xin-Yu Chang, MSc (Investigation: Equal; Methodology: Supporting; Validation: Equal; Visualization: Supporting)
Jui-Ping Weng, MSc (Formal analysis: Supporting; Investigation: Equal; Methodology: Equal; Validation: Equal)
Yin-Wen Shue, MSc (Formal analysis: Equal; Investigation: Supporting; Validation: Supporting)
Liang-Chuan Lai, PhD (Data curation: Equal; Formal analysis: Equal; Software: Lead)
Jin-Town Wang, MD, PhD (Data curation: Equal; Formal analysis: Supporting; Validation: Equal)
Yung-Ming Jeng, MD, PhD (Investigation: Equal; Methodology: Equal; Visualization: Supporting)
Yen-Hsuan Ni, MD, PhD (Conceptualization: Lead; Data curation: Lead; Funding acquisition: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the National Health Research Institute, Taiwan, grants NHRI-EX105-10520BI, NHRI-EX106-10520BI, NHRI-EX107-10520BI, NHRI-EX108-10823BI, NHRI-EX109-10823BI, and NHRI-EX110-10823BI; Ministry of Science and Technology, Taiwan, grants MoST 107-2320-B-002-041-MY3 and MoST 110-2320-B-002-011-MY3; and National Taiwan University Core Research Projects grants NTU-CCP-106R890504 and NTU-CC-109L893102.
Contributor Information
Linda Chia-Hui Yu, Email: lchyu@ntu.edu.tw.
Yen-Hsuan Ni, Email: yhni@ntu.edu.tw.
References
- 1.Flemer B., Lynch D.B., Brown J.M., Jeffery I.B., Ryan F.J., Claesson M.J., O'Riordain M., Shanahan F., O'Toole P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66:633–643. doi: 10.1136/gutjnl-2015-309595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao R., Kong C., Huang L., Li H., Qu X., Liu Z., Lan P., Wang J., Qin H. Mucosa-associated microbiota signature in colorectal cancer. Eur J Clin Microbiol Infect Dis. 2017;36:2073–2083. doi: 10.1007/s10096-017-3026-4. [DOI] [PubMed] [Google Scholar]
- 3.Uronis J.M., Muhlbauer M., Herfarth H.H., Rubinas T.C., Jones G.S., Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zackular J.P., Baxter N.T., Chen G.Y., Schloss P.D. Manipulation of the gut microbiota reveals role in colon tumorigenesis. mSphere. 2016;1 doi: 10.1128/mSphere.00001-15. e00001–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman O.I., Lobner E.M., Bierwirth S., Sorbie A., Waldschmitt N., Rath E., Berger E., Lagkouvardos I., Clavel T., McCoy K.D., Weber A., Heikenwalder M., Janssen K.P., Haller D. Activated ATF6 induces intestinal dysbiosis and innate immune response to promote colorectal tumorigenesis. Gastroenterology. 2018;155:1539–1552 e12. doi: 10.1053/j.gastro.2018.07.028. [DOI] [PubMed] [Google Scholar]
- 6.Bonnet M., Buc E., Sauvanet P., Darcha C., Dubois D., Pereira B., Dechelotte P., Bonnet R., Pezet D., Darfeuille-Michaud A. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res. 2014;20:859–867. doi: 10.1158/1078-0432.CCR-13-1343. [DOI] [PubMed] [Google Scholar]
- 7.Prorok-Hamon M., Friswell M.K., Alswied A., Roberts C.L., Song F., Flanagan P.K., Knight P., Codling C., Marchesi J.R., Winstanley C., Hall N., Rhodes J.M., Campbell B.J. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut. 2014;63:761–770. doi: 10.1136/gutjnl-2013-304739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kostic A.D., Chun E., Robertson L., Glickman J.N., Gallini C.A., Michaud M., Clancy T.E., Chung D.C., Lochhead P., Hold G.L., El-Omar E.M., Brenner D., Fuchs C.S., Meyerson M., Garrett W.S. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dejea C.M., Fathi P., Craig J.M., Boleij A., Taddese R., Geis A.L., Wu X., DeStefano Shields C.E., Hechenbleikner E.M., Huso D.L., Anders R.A., Giardiello F.M., Wick E.C., Wang H., Wu S., Pardoll D.M., Housseau F., Sears C.L. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592–597. doi: 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pai Y.C., Weng L.T., Wei S.C., Wu L.L., Shih D.Q., Targan S.R., Turner J.R., Yu L.C. Gut microbial transcytosis induced by tumor necrosis factor-like 1A-dependent activation of a myosin light chain kinase splice variant contributes to IBD. J Crohns Colitis. 2021;15:258–272. doi: 10.1093/ecco-jcc/jjaa165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L.L., Peng W.H., Kuo W.T., Huang C.Y., Ni Y.H., Lu K.S., Turner J.R., Yu L.C. Commensal bacterial endocytosis in epithelial cells is dependent on myosin light chain kinase-activated brush border fanning by interferon-gamma. Am J Pathol. 2014;184:2260–2274. doi: 10.1016/j.ajpath.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu L.C., Shih Y.A., Wu L.L., Lin Y.D., Kuo W.T., Peng W.H., Lu K.S., Wei S.C., Turner J.R., Ni Y.H. Enteric dysbiosis promotes antibiotic-resistant bacterial infection: systemic dissemination of resistant and commensal bacteria through epithelial transcytosis. Am J Physiol Gastrointest Liver Physiol. 2014;307:G824–G835. doi: 10.1152/ajpgi.00070.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raisch J., Rolhion N., Dubois A., Darfeuille-Michaud A., Bringer M.A. Intracellular colon cancer-associated Escherichia coli promote protumoral activities of human macrophages by inducing sustained COX-2 expression. Lab Invest. 2015;95:296–307. doi: 10.1038/labinvest.2014.161. [DOI] [PubMed] [Google Scholar]
- 14.Arthur J.C., Perez-Chanona E., Muhlbauer M., Tomkovich S., Uronis J.M., Fan T.J., Campbell B.J., Abujamel T., Dogan B., Rogers A.B., Rhodes J.M., Stintzi A., Simpson K.W., Hansen J.J., Keku T.O., Fodor A.A., Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cougnoux A., Dalmasso G., Martinez R., Buc E., Delmas J., Gibold L., Sauvanet P., Darcha C., Dechelotte P., Bonnet M., Pezet D., Wodrich H., Darfeuille-Michaud A., Bonnet R. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014;63:1932–1942. doi: 10.1136/gutjnl-2013-305257. [DOI] [PubMed] [Google Scholar]
- 16.Tomkovich S., Yang Y., Winglee K., Gauthier J., Muhlbauer M., Sun X., Mohamadzadeh M., Liu X., Martin P., Wang G.P., Oswald E., Fodor A.A., Jobin C. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res. 2017;77:2620–2632. doi: 10.1158/0008-5472.CAN-16-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdelrazeq A.S. Spontaneous regression of colorectal cancer: a review of cases from 1900 to 2005. Int J Colorectal Dis. 2007;22:727–736. doi: 10.1007/s00384-006-0245-z. [DOI] [PubMed] [Google Scholar]
- 18.Choi J.W., Kim P., Kim J.K., Kim Y.R., Fukumura D., Yun S.H. Longitudinal tracing of spontaneous regression and anti-angiogenic response of individual microadenomas during colon tumorigenesis. Theranostics. 2015;5:724–732. doi: 10.7150/thno.10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogawa M., Mimuro H., Yoshikawa Y., Ashida H., Sasakawa C. Manipulation of autophagy by bacteria for their own benefit. Microbiol Immunol. 2011;55:459–471. doi: 10.1111/j.1348-0421.2011.00343.x. [DOI] [PubMed] [Google Scholar]
- 20.Abuaita B.H., Schultz T.L., O'Riordan M.X. Mitochondria-derived vesicles deliver antimicrobial reactive oxygen species to control phagosome-localized Staphylococcus aureus. Cell Host Microbe. 2018;24:625–636 e5. doi: 10.1016/j.chom.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu C. Autophagy: a potential target for rescuing sepsis-induced hepatic failure. Chin J Physiol. 2019;62:53–62. doi: 10.4103/CJP.CJP_25_19. [DOI] [PubMed] [Google Scholar]
- 22.Cicchini M., Karantza V., Xia B. Molecular pathways: autophagy in cancer--a matter of timing and context. Clin Cancer Res. 2015;21:498–504. doi: 10.1158/1078-0432.CCR-13-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moloney J.N., Cotter T.G. ROS signalling in the biology of cancer. Semin Cell Dev Biol. 2018;80:50–64. doi: 10.1016/j.semcdb.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 24.Galadari S., Rahman A., Pallichankandy S., Thayyullathil F. Reactive oxygen species and cancer paradox: to promote or to suppress? Free Radic Biol Med. 2017;104:144–164. doi: 10.1016/j.freeradbiomed.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Gorrini C., Harris I.S., Mak T.W. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 26.Raju D., Hussey S., Ang M., Terebiznik M.R., Sibony M., Galindo-Mata E., Gupta V., Blanke S.R., Delgado A., Romero-Gallo J., Ramjeet M.S., Mascarenhas H., Peek R.M., Correa P., Streutker C., Hold G., Kunstmann E., Yoshimori T., Silverberg M.S., Girardin S.E., Philpott D.J., El Omar E., Jones N.L. Vacuolating cytotoxin and variants in Atg16L1 that disrupt autophagy promote Helicobacter pylori infection in humans. Gastroenterology. 2012;142:1160–1171. doi: 10.1053/j.gastro.2012.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baxt L.A., Goldberg M.B. Host and bacterial proteins that repress recruitment of LC3 to Shigella early during infection. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ganesan R., Hos N.J., Gutierrez S., Fischer J., Stepek J.M., Daglidu E., Kronke M., Robinson N. Salmonella Typhimurium disrupts Sirt1/AMPK checkpoint control of mTOR to impair autophagy. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vdovikova S., Luhr M., Szalai P., Nygard Skalman L., Francis M.K., Lundmark R., Engedal N., Johansson J., Wai S.N. A novel role of Listeria monocytogenes membrane vesicles in inhibition of autophagy and cell death. Front Cell Infect Microbiol. 2017;7:154. doi: 10.3389/fcimb.2017.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tjalsma H., Boleij A., Marchesi J.R., Dutilh B.E. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 31.Weng M.T., Chiu Y.T., Wei P.Y., Chiang C.W., Fang H.L., Wei S.C. Microbiota and gastrointestinal cancer. J Formos Med Assoc. 2019;118(Suppl 1):S32–S41. doi: 10.1016/j.jfma.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 32.Yu L.C., Wei S.C., Ni Y.H. Impact of microbiota in colorectal carcinogenesis: lessons from experimental models. Intest Res. 2018;16:346–357. doi: 10.5217/ir.2018.16.3.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen X.J., Rawls J.F., Randall T., Burcal L., Mpande C.N., Jenkins N., Jovov B., Abdo Z., Sandler R.S., Keku T.O. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1:138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanapareddy N., Legge R.M., Jovov B., McCoy A., Burcal L., Araujo-Perez F., Randall T.A., Galanko J., Benson A., Sandler R.S., Rawls J.F., Abdo Z., Fodor A.A., Keku T.O. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 2012;6:1858–1868. doi: 10.1038/ismej.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahn J., Sinha R., Pei Z., Dominianni C., Wu J., Shi J., Goedert J.J., Hayes R.B., Yang L. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105:1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huipeng W., Lifeng G., Chuang G., Jiaying Z., Yuankun C. The differences in colonic mucosal microbiota between normal individual and colon cancer patients by polymerase chain reaction-denaturing gradient gel electrophoresis. J Clin Gastroenterol. 2014;48:138–144. doi: 10.1097/MCG.0b013e3182a26719. [DOI] [PubMed] [Google Scholar]
- 37.Weinberg F., Hamanaka R., Wheaton W.W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G.M., Budinger G.R., Chandel N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levy J., Cacheux W., Bara M.A., L'Hermitte A., Lepage P., Fraudeau M., Trentesaux C., Lemarchand J., Durand A., Crain A.M., Marchiol C., Renault G., Dumont F., Letourneur F., Delacre M., Schmitt A., Terris B., Perret C., Chamaillard M., Couty J.P., Romagnolo B. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol. 2015;17:1062–1073. doi: 10.1038/ncb3206. [DOI] [PubMed] [Google Scholar]
- 39.Liu E.Y., Xu N., O'Prey J., Lao L.Y., Joshi S., Long J.S., O'Prey M., Croft D.R., Beaumatin F., Baudot A.D., Mrschtik M., Rosenfeldt M., Zhang Y., Gillespie D.A., Ryan K.M. Loss of autophagy causes a synthetic lethal deficiency in DNA repair. Proc Natl Acad Sci U S A. 2015;112:773–778. doi: 10.1073/pnas.1409563112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quach C., Song Y., Guo H., Li S., Maazi H., Fung M., Sands N., O'Connell D., Restrepo-Vassalli S., Chai B., Nemecio D., Punj V., Akbari O., Idos G.E., Mumenthaler S.M., Wu N., Martin S.E., Hagiya A., Hicks J., Cui H., Liang C. A truncating mutation in the autophagy gene UVRAG drives inflammation and tumorigenesis in mice. Nat Commun. 2019;10:5681. doi: 10.1038/s41467-019-13475-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas C., Salesse L., Hoang M.H.T., Bonnet M., Sauvanet P., Larabi A., Godfraind C., Gagniere J., Pezet D., Rosenstiel P., Barnich N., Bonnet R., Dalmasso G., Nguyen H.T.T. Autophagy of intestinal epithelial cells inhibits colorectal carcinogenesis induced by colibactin-producing Escherichia coli in Apc(Min/+) mice. Gastroenterology. 2020;158:1373–1388. doi: 10.1053/j.gastro.2019.12.026. [DOI] [PubMed] [Google Scholar]
- 42.Kuo W.T., Lee T.C., Yang H.Y., Chen C.Y., Au Y.C., Lu Y.Z., Wu L.L., Wei S.C., Ni Y.H., Lin B.R., Chen Y., Tsai Y.H., Kung J.T., Sheu F., Lin L.W., Yu L.C. LPS receptor subunits have antagonistic roles in epithelial apoptosis and colonic carcinogenesis. Cell Death Differ. 2015;22:1590–1604. doi: 10.1038/cdd.2014.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo W.T., Lee T.C., Yu L.C. Eritoran suppresses colon cancer by altering a functional balance in Toll-like receptors that bind lipopolysaccharide. Cancer Res. 2016;76:4684–4695. doi: 10.1158/0008-5472.CAN-16-0172. [DOI] [PubMed] [Google Scholar]
- 44.Huang Y.J., Lee T.C., Pai Y.C., Lin B.R., Turner J.R., Yu L.C. A novel tumor suppressor role of myosin light chain kinase splice variants through downregulation of the TEAD4/CD44 axis. Carcinogenesis. 2021;42:961–974. doi: 10.1093/carcin/bgab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chua H.H., Chou H.C., Tung Y.L., Chiang B.L., Liao C.C., Liu H.H., Ni Y.H. Intestinal dysbiosis featuring abundance of Ruminococcus gnavus associates with allergic diseases in infants. Gastroenterology. 2018;154:154–167. doi: 10.1053/j.gastro.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Carvalho F.A., Barnich N., Sivignon A., Darcha C., Chan C.H., Stanners C.P., Darfeuille-Michaud A. Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J Exp Med. 2009;206:2179–2189. doi: 10.1084/jem.20090741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bilge S.S., Clausen C.R., Lau W., Moseley S.L. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J Bacteriol. 1989;171:4281–4289. doi: 10.1128/jb.171.8.4281-4289.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balaji B., O'Connor K., Lucas J.R., Anderson J.M., Csonka L.N. Timing of induction of osmotically controlled genes in Salmonella enterica Serovar Typhimurium, determined with quantitative real-time reverse transcription-PCR. Appl Environ Microbiol. 2005;71:8273–8283. doi: 10.1128/AEM.71.12.8273-8283.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monday S.R., Bohach G.A. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J Clin Microbiol. 1999;37:3411–3414. doi: 10.1128/jcm.37.10.3411-3414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang C.Y., Yu L.C. Distinct patterns of interleukin-12/23 and tumor necrosis factor alpha synthesis by activated macrophages are modulated by glucose and colon cancer metabolites. Chin J Physiol. 2020;63:7–14. doi: 10.4103/CJP.CJP_75_19. [DOI] [PubMed] [Google Scholar]
- 51.Lee T.C., Huang Y.C., Lu Y.Z., Yeh Y.C., Yu L.C. Hypoxia-induced intestinal barrier changes in balloon-assisted enteroscopy. J Physiol. 2018;596:3411–3424. doi: 10.1113/JP275277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang C.Y., Huang C.Y., Pai Y.C., Lin B.R., Lee T.C., Yu L.C. Glucose metabolites exert opposing roles in tumor chemoresistance. Front Oncol. 2019;9:1282. doi: 10.3389/fonc.2019.01282. [DOI] [PMC free article] [PubMed] [Google Scholar]