Abstract
We have identified and characterized a novel mouse protein, Bop1, which contains WD40 repeats and is highly conserved through evolution. bop1 is ubiquitously expressed in all mouse tissues examined and is upregulated during mid-G1 in serum-stimulated fibroblasts. Immunofluorescence analysis shows that Bop1 is localized predominantly to the nucleolus. In sucrose density gradients, Bop1 from nuclear extracts cosediments with the 50S-80S ribonucleoprotein particles that contain the 32S rRNA precursor. RNase A treatment disrupts these particles and releases Bop1 into a low-molecular-weight fraction. A mutant form of Bop1, Bop1Δ, which lacks 231 amino acids in the N- terminus, is colocalized with wild-type Bop1 in the nucleolus and in ribonucleoprotein complexes. Expression of Bop1Δ leads to cell growth arrest in the G1 phase and results in a specific inhibition of the synthesis of the 28S and 5.8S rRNAs without affecting 18S rRNA formation. Pulse-chase analyses show that Bop1Δ expression results in a partial inhibition in the conversion of the 36S to the 32S pre-rRNA and a complete inhibition of the processing of the 32S pre-rRNA to form the mature 28S and 5.8S rRNAs. Concomitant with these defects in rRNA processing, expression of Bop1Δ in mouse cells leads to a deficit in the cytosolic 60S ribosomal subunits. These studies thus identify Bop1 as a novel, nonribosomal mammalian protein that plays a key role in the formation of the mature 28S and 5.8S rRNAs and in the biogenesis of the 60S ribosomal subunit.
Biogenesis of the eukaryotic ribosomes occurs in the nucleolus, a complex nuclear organelle that forms around the nucleolar organizer regions located in heterochromatic chromosomal sites containing multiple rRNA genes (69, 81). The organization of the nucleolus and the assembly of ribosomes are coupled to transcription of rDNA by RNA polymerase I, which synthesizes a large primary precursor transcript. This precursor transcript is then processed into the mature 18S, 5.8S, and 28S/25S rRNAs. These rRNAs are assembled into preribosomes with some 80 ribosomal proteins that are transported into the nucleus, and with the 5S rRNA, transcribed by RNA polymerase III outside of the nucleolus (28). A large number of small nucleolar RNAs (snoRNAs) and nonribosomal proteins are also recruited to the nucleolus to participate in the modification, processing, and assembly of the rRNAs and proteins into ribonucleoprotein (RNP) particles. These preribosomal RNP (pre-rRNP) particles mature into nearly complete ribosomal subunits prior to their export out of the nucleus.
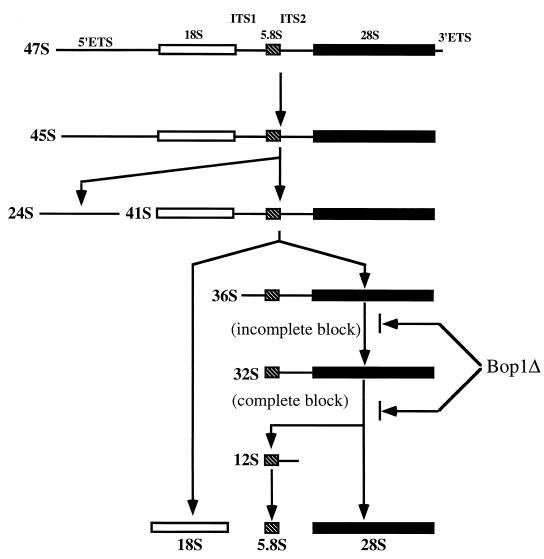
Upon synthesis, the primary precursor rRNA transcript is modified by ribose methylation and pseudouridine conversion and processed through a series of nucleolytic cleavages into the matured rRNAs (21) (see Fig. 6). In vertebrates, the arrangement of the 47S primary precursor transcript begins with a 5′ external transcribed spacer (5′-ETS), followed by the 18S rRNA, internal transcribed spacer 1 (ITS1), 5.8S rRNA, internal transcribed spacer 2 (ITS2), 28S rRNA, and the 3′ external transcribed spacer (3′-ETS). Although in general, processing events occur in a polar fashion from the 5′ to the 3′ end of the nascent transcript, differences in the order of processing events and intermediates generated have been reported for different cell types (8, 31, 56). A similar arrangement of the primary transcript is also found in yeasts (79), and it is generally thought that while specific processing sites might differ between yeasts and vertebrates, parallel processing pathways appear to exist for eukaryotic organisms from yeasts to mammals (75).
FIG. 6.
Schematic representation of the mammalian rRNA-processing pathway. Mammalian rRNA is transcribed as a single precursor which is further processed by successive nucleolytic cleavages that lead to elimination of the external transcribed spacers, 5′ETS and 3′ ETS, and the internal transcribed spacers, ITS1 and ITS2. The sedimentation coefficients (S) of various intermediates and mature products of the processing pathway are indicated. The mammalian 47S precursor is rapidly cleaved at the 5′ETS and at the 3′ETS to give rise to the 45S pre-rRNA. Further processing at the 5′ETS takes place, giving rise to a 41S rRNA precursor, which is rapidly processed to the 18S rRNA and a 36S precursor RNA that contains the sequences of the 5.8S and 28S rRNAs with an intervening sequence (ITS2). The 36S precursor then undergoes cleavage at the 5′ end to give rise to a 32S precursor, which is processed to the 28S rRNA and a 12S RNA; the 12S RNA is then further processed to form the 5.8S rRNA.
A large number of snoRNAs and nonribosomal nucleolar proteins play critical roles in ribosome biogenesis (21, 75). More than 150 snoRNAs have been found in the nucleolus, and some of them play key roles in various pre-rRNA processing events including nucleotide modification and cleavage reactions (45, 76, 83). Although a large number of snoRNAs have been identified and characterized, less is known about the functions of the nonribosomal proteins in the RNP particles, especially in mammalian systems. Many of the recent advances made in the identification of protein factors involved in rRNA processing have come from genetic and biochemical analyses in yeast. Among the yeast proteins known to participate in rRNA processing are endoribonucleases (Rnt1p and RNase MRP) (22, 44), 5′→3′ exoribonucleases (Xrn1p and Rat1p) (35, 39), nearly a dozen 3′→5′ exoribonucleases that comprise the exosome (1, 48), putative ATP-dependent RNA helicases (Drs1p, Sbp4p, Rrp3p, and Rok1p) (18, 54, 78), and a number of noncatalytic nucleolar proteins (Nop1p, Nop2p, Nopp3p, etc.) (34, 36, 66). The total number of nonribosomal proteins involved in rRNA processing is unknown but is likely to be large, as underscored by the complexity of eukaryotic rRNA modification, processing, and ribosome assembly.
In mammalian systems, relatively few nonribosomal nucleolar proteins have been characterized; the best-studied examples include fibrillarin, a common component of the snoRNPs (43, 53); nucleolin (C23), a pre-rRNA binding protein with multiple functions in processing and ribosome assembly (29, 55, 77); B23, associated with ribosome assembly at later stages of maturation (11); and p120, a nucleolar RNA binding protein that cofractionates with 60S-80S pre-rRNA particles (30, 33). In comparison with yeast, the majority of the molecular players in the mammalian rRNA processing machinery remain unidentified, and their characterization will be necessary to understand fully the mechanism of rRNA processing and ribosomal assembly.
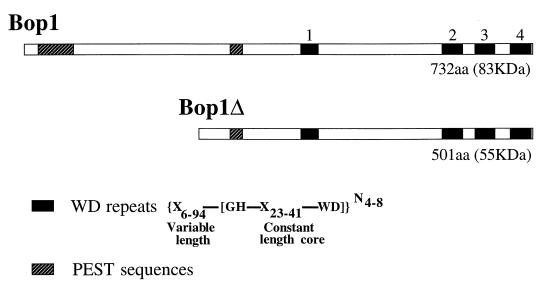
In a previous study, a genetic selection for growth-inhibitory sequences in mouse cells identified a cDNA, bop1Δ, whose inducible expression results in a powerful but reversible block in the G1 phase (58). Further analysis revealed that bop1Δ represents a cDNA fragment that encodes an N-terminally truncated form of a novel protein, Bop1. Sequence analysis showed that bop1 is an evolutionarily conserved gene that encodes an 83-kDa protein with four WD40 repeats (Fig. 1) (58). The WD40 repeat is a sequence motif found in a diverse group of functionally distinct proteins involved in the regulation of myriad cellular processes, including signal transduction, gene transcription, and mRNA modification (51). Moreover, WD40 proteins are often found to form multiprotein complexes, suggesting that the WD40 motifs may be involved in protein-protein interactions (7, 14, 60).
FIG. 1.
Structural features of the Bop1 protein. Schematic representation of the full-length murine Bop1 protein (732 aa) and the amino-terminally truncated Bop1Δ (501 aa). The four WD repeats, whose consensus structure is as indicated, are shown as solid boxes. Repeats 1 and 4 are close to the consensus structure, while repeats 2 and 3 are more divergent. PEST sequences, often associated with short-lived regulatory proteins, are shown by hatched boxes.
In this study, we demonstrated that Bop1 is a novel component of the 28S branch of the rRNA processing machinery. Bop1 is localized to the nucleolus and forms part of the large RNP particles that probably represent preribosomes. Expression of its N-terminally truncated mutant form, Bop1Δ, specifically inhibits the processing of the 32S precursor to form the mature 5.8S and 28S rRNAs and results in a deficiency of the 60S ribosomal subunits. These results identify Bop1 as a previously unknown player in the processing of the 32S pre-rRNA and in the biogenesis of the 60S ribosomal subunits and suggest that Bop1 may interact with and coordinate the activities of proteins that comprise the pre-RNA processing machinery.
MATERIALS AND METHODS
Expression plasmids.
An IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible expression vector, pX11, drives inducible expression with low background in LAP3 cells (59). Expression constructs with cDNAs encoding either Bop1 or Bop1Δ cloned into pX11 have been described previously (58). To express Bop1 or Bop1Δ N-terminally tagged with a hemagglutinin antigen (HA) sequence, a polynucleotide linker encoding the HA tag (23) was inserted into these vectors. To express Bop1 as a glutathione S-transferase (GST) fusion protein in Escherichia coli, an EcoRI restriction fragment of Bop1 (GenBank accession no. U77415) corresponding to nucleotides (nt) 442 to 2461 was cloned into pGEX-4T-1 (Pharmacia). To express Bop1 in insect cells via a baculovirus expression vector, the full-length 2.4-kb bop1 cDNA fragment was cloned in pBlueBacHis2A (Invitrogen), thereby driving the expression of a full-length Bop1 protein with an N-terminal 6-histidine tag.
Cell culture.
BALB/c 3T3 cells were grown in MEM-10 (minimal essential medium with 10% fetal bovine serum). The cells were brought to quiescence by growth to confluence and serum starved in MEM-0.5 for 2 days; serum stimulation was accomplished by changing the medium to MEM-10. LAP3 is a clonal cell line derived from NIH 3T3 cells that constitutively expresses the IPTG-inducible transactivator protein LAP267 (5). To create various clonal cell lines, LAP3 cells were cotransfected with a hygromycin marker plasmid pHyg (73) and either the empty pX11 vector (line 1-1), pX11-Bop1 (line 45), pX11-Bop1Δ (lines 6 and 8), pX11-HA-Bop1 (line 10), or pX11-HA-Bop1Δ (line 13) using the calcium phosphate coprecipitation method (12). Clonal cell lines were selected following transfection by growth in 130 to 150 μg of hygromycin (Boehringer Mannheim) per ml. LAP3-derived cell lines were maintained in Dulbecco's modified Eagle's medium containing 10% calf serum (HyClone) and penicillin-streptomycin (Gibco-BRL). IPTG (dioxane free; Sigma) was added to 1 mM where indicated.
RNA blot analysis.
Where indicated (except for Fig. 11), RNA was isolated using Trizol reagent (Gibco-BRL) as specified by the manufacturer. The samples in the experiment shown in Fig. 11 were withdrawn from sucrose density gradients, treated with proteinase K, and subjected to phenol-chloroform extraction and isopropanol precipitation. Northern blot hybridization was performed by standard methods (68). The bop1 cDNA probe was synthesized by random priming in the presence of [32P]dCTP (Decaprime II kit; Ambion). A 40-mer oligonucleotide (5′-GCGTTCGAAGTGTCGATGATCAATGTGTCCTGCAATTCAC-3′) complementary to nt 68 to 108 of the 5.8S rRNA and a 33-mer oligonucleotide (5′-ACTGGTGAGGCAGCGGTCCGGGAGGCGCCGACG-3′) complementary to nt 1480 to 1512 of the ITS2 region of the pre-rRNA were 5′-end labeled using [γ-32P]ATP and T4 polynucleotide kinase.
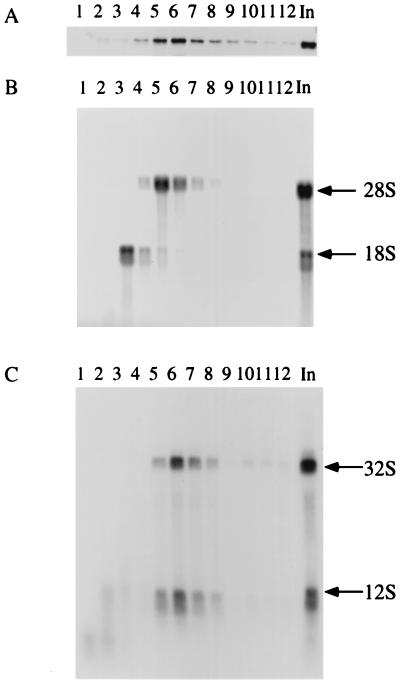
FIG. 11.
Bop1 cofractionates with the 32S precursor and 28S rRNA in the nuclear extract. Nuclear extracts from LAP3 cells were isolated and separated on a 10 to 30% sucrose density gradient. The fractions collected were subjected to parallel analysis of protein and RNA. (A) Proteins from various fractions were electrophoresed on an SDS–10% polyacrylamide gel and immunoblotted with affinity-purified anti-Bop1 antibodies. (B) RNAs from each fraction analyzed in panel A were resolved on a 1% agarose gel and transferred to a nylon filter, which was stained with methylene blue. The staining pattern reveals the fractions containing the 18S and 28S rRNAs. (C) The nylon filter shown in panel B was subjected to hybridization using radioactively labeled sequences from ITS2 as a probe, revealing the 32S precursor RNA. In, unfractionated soluble nuclear extract.
Anti-Bop1 antibodies and affinity purification.
To raise polyclonal anti-Bop1 antibodies, a Bop1 polypeptide corresponding to amino acids (aa) 131 to 732 was expressed as a GST fusion protein in E. coli (see above). The fusion protein formed insoluble inclusion bodies, which were solubilized in 5 M urea and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (6% polyacrylamide). The GST-Bop1 protein band was excised from the gel, and the protein was used for immunization of rabbits at the Immunological Resource Center at University of Illinois at Urbana. Antisera raised against this fusion protein were affinity purified by a standard method (32) by passage through a Bop1-Sepharose column. To prepare the affinity column, full-length Bop1 with an N-terminal histidine tag was expressed in SF9 cells using a baculovirus expression system (Invitrogen) and purified using Pro-Bond resin (Invitrogen) under denaturing conditions as specified by the manufacturer. The protein was further dialyzed against 125 mM sodium phosphate (pH 8)–500 mM NaCl and coupled to cyanogen bromide-activated Sepharose (5 mg of protein per ml of Sepharose slurry) as specified by the manufacturer (Pharmacia). The anti-Bop1 antiserum was diluted 1:10 in phosphate-buffered saline (PBS), passed through the Bop1-Sepharose column, and washed with 10 mM Tris (pH 7.5)–500 mM NaCl. Bound antibodies were first eluted with 100 mM glycine (pH 2.5) and then eluted with 100 mM ethanolamine (pH 11.5). Eluates were combined, dialyzed against PBS, and concentrated with Centricon-30 columns (Amicon) or on a protein A-Sepharose column (68).
Western blot analysis.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (68), and equal amounts of protein, determined by the DC protein assay (Bio-Rad), were resolved by SDS-PAGE. For immunoblot analysis of the sucrose gradient fractions, the proteins from each fraction were precipitated with cold 10% trichloroacetic acid and dissolved in 80 μl of Laemmli buffer, half of which was resolved by SDS-PAGE. Western blot analysis was done by standard methods (2) using affinity-purified anti-Bop1 antibodies.
To analyze proteins associated with ribosomal particles in the nucleus and the cytoplasm, cells were lysed as described below for the preparation of nuclear extracts. The cytoplasmic fraction and nuclear sonicate were cleared at 15,000 × g for 15 min and then centrifuged through 10% sucrose in 10 mM Tris-HCl (pH 7.2)–60 mM KCl–10 mM MgSO4–1 mM dithiothreitol (DTT) at 160,000 × g for 4 h at 5°C. The pellets were dissolved in 1% SDS and used for the isolation of RNA with Trizol and protein analysis by SDS-PAGE. The volumes loaded on a protein gel were normalized for the RNA content of the samples as determined by absorbance at 260 nm and electrophoresis on a formaldehyde-containing agarose gel followed by staining with SYBR Gold (Molecular Probes, Inc.).
Indirect immunofluorescence.
Cells were grown on coverslips, incubated with IPTG for 12 h, fixed with paraformaldehyde, permeabilized with 0.5% Triton X-100, and incubated for 1 h at room temperature with either monoclonal anti-HA antibody (Babco), polyclonal anti-HA antibodies (Babco), affinity-purified anti-Bop1 antibodies, or the anti-fibrillarin monoclonal antibody (72B9) diluted in PBS containing 0.5% bovine serum albumin. After being washed, the cells were incubated with either fluorescein isothiocyanate (FITC)-conjugated or Texas Red-conjugated anti-mouse antibodies (Vector Laboratories) or FITC-conjugated anti-rabbit antibodies (Zymed) and analyzed using a MicroMAX digital camera mounted on a Axioplan II Zeiss microscope with differential interference contrast (DIC) optics.
Metabolic labeling and analysis of RNA transcripts.
Various cell lines were plated in six-well plates at 105 cells per well. One day after being plated, the cells were either left untreated or treated with IPTG for 16 h to induce expression. To measure RNA synthesis, the cells were incubated in medium with [3H]uridine (2.5 μCi/ml) for 30 min and then in nonradioactive medium for 2 h. RNA was isolated using Trizol, and label incorporation was measured by scintillation counting. RNA was subsequently separated on a 1% agarose gel and transferred to a nylon membrane, which was treated with En3Hance (New England Nuclear) and exposed to film. Pulse-chase experiments were carried out using l-[methyl-3H]methionine due to the rapid turnover of the cellular methionine pool. Cells were preincubated for 15 min in methionine-free medium and then incubated for 30 min in medium containing l-[methyl-3H]methionine (50 μCi/ml). The cells were then chased in nonradioactive medium containing 15 μg of methionine per ml for various times, after which the RNA was isolated using Trizol. RNA from the same number of cells was analyzed as described above. Where indicated, cells were treated with 5 μM 5-fluorouridine (FUrd) for 15 min prior to labeling. For analysis of the synthesis of 5.8S RNA, 32P labeling was used to increase the sensitivity of detection. Cells to be labeled with radioactive phosphate were pretreated in phosphate-free medium for 1 h, labeled in medium with [32P]orthophosphate (32Pi) (20 μCi/ml) for 1 h, and chased in nonradioactive medium for 1.5 h. RNA from the same number of cells was separated on a 10% polyacrylamide–7 M urea gel.
Sucrose density gradient fractionation.
To fractionate cytoplasmic ribosomes, cells were harvested by trypsinization 5 min after the addition of 50 μg of cycloheximide per ml to the medium. Equal numbers of cells (determined with a Coulter counter) from each sample were pelleted by low-speed centrifugation and lysed in 20 mM Tris-HCl (pH 7.2)–130 mM KCl–10 mM MgCl2–2.5 mM DTT–0.5% NP-40–0.5% sodium deoxycholate–10 μg of cycloheximide per ml–0.2 mg of heparin per ml–200 U of RNasin per ml (Promega) for 15 min at 4°C. The lysates were centrifuged at 8,000 × g for 10 min, and the supernatants were layered on 10 to 45% (wt/wt) sucrose density gradients in 10 mM Tris-HCl (pH 7.2)–60 mM KCl–10 mM MgCl–1 mM DTT–0.1 mg of heparin per ml. The gradients were centrifuged at 36,000 rpm for 3 h at 5°C in a Beckman SW41Ti rotor and fractionated by upward displacement through a Bio-Rad EM-1 UV monitor for continuous measurement of the absorbance at 254 nm. Sedimentation constants were calculated as described previously (46).
To fractionate nuclear extracts, cells were washed twice in PBS and once in ice-cold hypotonic wash buffer (10 mM Tris [pH 7.4], 10 mM KCl, 2 mM MgCl) and then left to swell for 20 min in hypotonic lysis buffer (10 mM Tris [pH 7.4], 10 mM KCl, 2 mM MgCl, 0.05% Triton X-100, 1 mM EGTA, 1 mM DTT, 40 μg of phenylmethylsulfonyl fluoride per ml, 10 μl of protease inhibitor cocktail [Sigma] per ml) (40). The cells were forced through a 25-gauge needle and centrifuged for 5 min at 700 × g to obtain a pellet of nuclei, which was resuspended in 25 mM Tris (pH 7.5)–100 mM KCl–1 mM DTT–2 mM EDTA–0.05% NP-40–1 mM NaF–40 μg of phenylmethylsulfonyl fluoride per ml–10 μl of protease inhibitor cocktail per ml–0.1 U of RNasin (Promega) per ml and sonicated four times for 15 s each with a microtip (Heat Systems Ultrasonics, Inc.). The nuclear lysate was centrifuged at 15,000 × g for 15 min, and the resulting supernatant was overlaid on 10 to 30% (wt/wt) sucrose gradients in 25 mM Tris (pH 7.5)–100 mM KCl–1 mM DTT–2 mM EDTA and centrifuged at 36,000 rpm for 3 h at 5°C in a Beckman SW41Ti rotor. Where indicated, the nuclear extract was treated with RNase A (100 μg/ml) for 10 min at 30°C before being loaded on the gradient. The gradients were analyzed as above.
RESULTS
Expression of the bop1 gene.
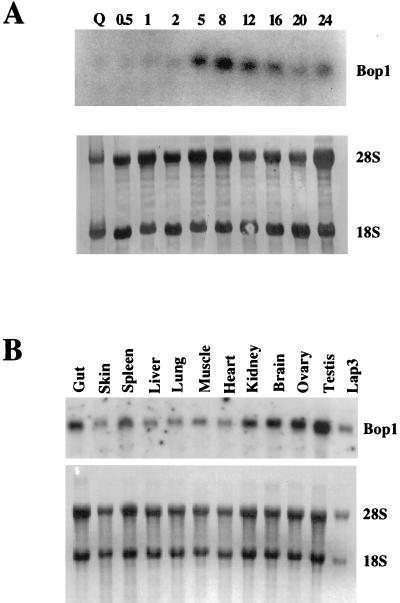
Expression of an N-terminally truncated form of Bop1, Bop1Δ, results in a strong but reversible G1 growth arrest (58). This observation raised the possibility that the activity of Bop1 might affect cell cycle progression and that expression of the bop1 gene might be linked to the proliferative state. To determine if bop1 expression is growth regulated, we analyzed RNA isolated from BALB/c 3T3 cells stimulated to reenter the cell cycle from quiescence (Fig. 2A). bop1 expression was low in quiescent cells and was induced after serum stimulation, starting at 5 h, reaching maximal level at 8 h, and slowly declining thereafter. Thus, bop1 expression increases in cells stimulated to proliferate, with maximal expression in mid-G1. A survey of RNA isolated from various mouse tissues showed the presence of the 2.6-kb bop1 mRNA in all tissues tested, with the highest level in the testis (Fig. 2B).
FIG. 2.
Expression of bop1. (A) BALB/c 3T3 cells were brought to quiescence (Q) by serum starvation and restimulated with 10% fetal bovine serum. RNA was isolated at the indicated times (in hours) after serum stimulation and analyzed by Northern blotting with 32P-labeled bop1 cDNA as probe. The RNA blot was stained with methylene blue (lower panel) to show the relative amounts of rRNAs, indicating equal loading of the samples. (B) RNA was isolated from different adult mouse tissues and analyzed by Northern blotting with labeled bop1 cDNA as probe. The lower panel shows methylene blue staining of the same blot to control for loading of the RNA.
Bop1 and Bop1Δ are localized in the nucleolus.
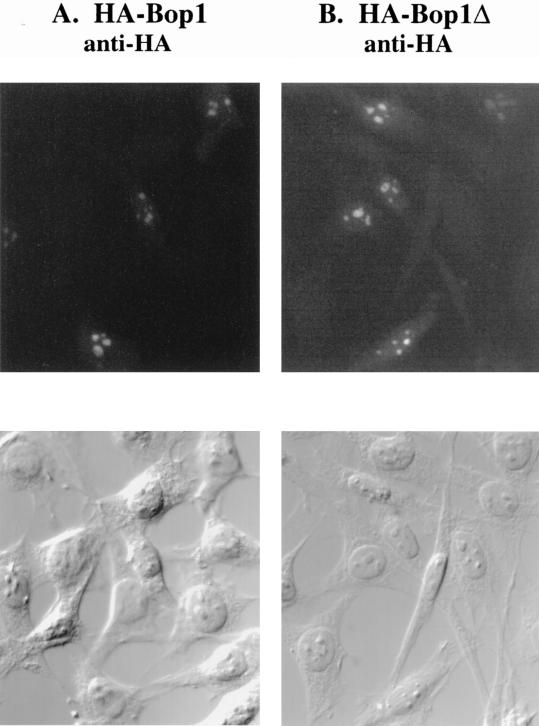
In an effort to determine the function of Bop1, we ascertained its subcellular localization. First, we prepared IPTG-inducible expression constructs that drive the expression of either HA-tagged Bop1 or Bop1Δ in LAP3 cells, an NIH 3T3-derived cell line that supports IPTG-regulated gene expression (see Materials and Methods). Stable pools of transfected LAP3 cells were induced with IPTG and subjected to immunofluorescence analysis using monoclonal anti-HA antibody (Fig. 3A and B). Cells that express either HA-tagged Bop1 or HA-tagged Bop1Δ showed strong antibody staining in the nucleoli, although a weak and diffuse nucleoplasmic staining was also observed. Examination of the same fields using DIC optics confirmed the nucleolar localization. Double staining with polyclonal anti-HA antibodies and a monoclonal antifibrillarin antibody (72B9) (62) also confirmed the nucleolar localization of the HA-tagged Bop1 and Bop1Δ (data not shown).
FIG. 3.
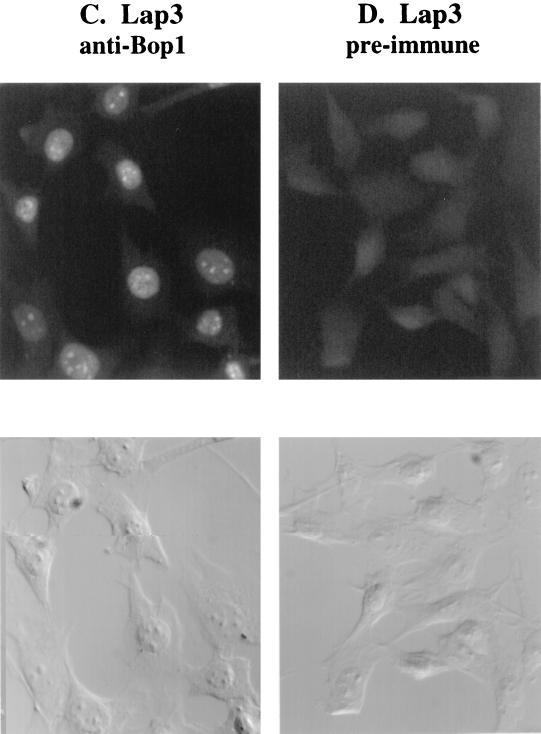
Subcellular localization of Bop1 by indirect immunofluorescence. (A and B) Pools of LAP3 cells stably transfected with vectors that express HA-tagged Bop1 (A) or Bop1Δ (B) were grown on coverslips, induced with IPTG for 12 h, fixed, permeabilized, and stained with monoclonal anti-HA antibody. Antibody-antigen complexes were detected with FITC-conjugated anti-mouse antibody and visualized by fluorescence microscopy. An image of the same field visualized with DIC optics is shown in the lower panels. (C and D) To localize the endogenous Bop1 protein, LAP3 cells were grown on coverslips, fixed, permeabilized, and stained with affinity-purified, polyclonal anti-Bop1 antibodies (C) or the preimmune serum as a control (D).
To reinforce and substantiate the above findings, we determined the subcellular localization of the endogenous Bop1 protein by using affinity-purified anti-Bop1 antibodies (see Materials and Methods). These antibodies reacted specifically with Bop1 in cell lysates on a denaturing Western blot, where Bop1 exhibited an apparent molecular mass of ∼100 kDa (larger than the calculated size of 83 kDa) (Fig. 4). These antibodies were used to demonstrate that various IPTG-inducible cell lines express the appropriate gene products upon induction (Fig. 4). Using these antibodies, endogenous Bop1 was also localized to the nucleolus by immunofluorescence analysis whereas preimmune serum from the same rabbit did not show any staining (Fig. 3C and D). No anti-Bop1 staining was detected in the cytoplasm, although a low level of diffuse staining was also detected in the nucleoplasm. Together, these results show that both endogenous Bop1 and exogenously expressed Bop1 or Bop1Δ are localized predominantly in the nucleolus.
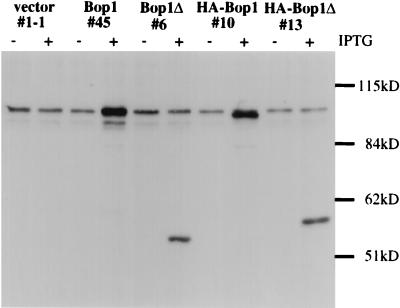
FIG. 4.
Characterization of the affinity-purified anti-Bop1 antibodies. Clonal lines of LAP3 cells stably transfected with either the empty vector pX11 (line 1-1), pX11-Bop1 (line 45), pX11-Bop1Δ (line 6), pX11-HA-Bop1 (line 10), or pX11-HA-Bop1Δ (line 13) were either treated with 1 mM IPTG (+) for 12 h or left untreated (−). Cells were lysed in RIPA buffer, and equal amounts of protein from each sample were resolved by SDS-PAGE (8% polyacrylamide) and transferred to a nitrocellulose filter. The blot was incubated with affinity-purified anti-Bop1 antibodies, and chemiluminescent reagents were used for detection. The affinity-purified antibodies recognized a high-molecular-mass band of ∼100 kDa (larger than the expected size of 83 kDa), which was increased significantly in a Bop1-overexpressing cell line (line 45). The predicted 55-kDa Bop1Δ fragment was detected in line 6, which inducibly expresses Bop1Δ. Addition of the HA tag caused a minor shift in the mobility of both Bop1 and Bop1Δ in clonal lines 10 and 13, respectively.
Involvement of Bop1 in rRNA processing.
The localization of Bop1 in the nucleolus—the site of rRNA synthesis, modification, processing, and ribosome biogenesis—suggests that it may play a role in one or more of these processes. That Bop1Δ is colocalized with Bop1 and its expression inhibits cell growth suggests that it might interfere with the normal function of Bop1, most probably acting as a dominant negative mutant.
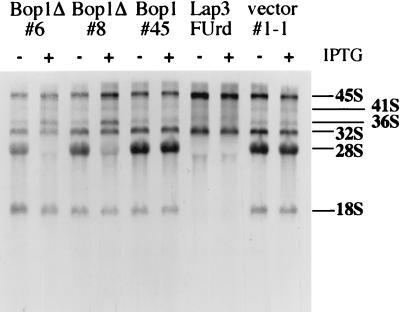
To investigate the function of Bop1, we first examined whether expression of Bop1 or Bop1Δ affects rRNA synthesis. For this purpose, we created a set of cell lines that express Bop1 or Bop1Δ in an IPTG-inducible manner (see Materials and Methods). These inducible cell lines were either untreated or treated with IPTG, metabolically labeled with [3H]uridine for 30 min, and subjected to a 2-h chase. Inducible overexpression of Bop1 (line 45) showed no effect on the synthesis of the large rRNA precursors (45S and 32S) or the mature 18S and 28S rRNAs, resulting in an rRNA pattern indistinguishable from that of the untreated cells or the control cell line (line 1-1; the parental LAP3 cells transfected with the empty cloning vector) (Fig. 5). By contrast, expression of Bop1Δ (lines 6 and 8) strongly inhibited production of the 28S rRNA whereas the levels of 18S rRNA and the 45S precursor were unchanged (Fig. 5). Bop1Δ expression also resulted in accumulation of the 36S rRNA, normally present in small amounts. In control experiments where LAP3 cells were treated with FUrd, a known inhibitor of rRNA processing (84), a complete blockade of production of both 28S and 18S rRNA was observed without affecting the formation of the 45S and 32S precursors (Fig. 5). Together, these results suggested that Bop1Δ did not significantly affect the synthesis and initial processing of the primary rRNA transcript but specifically compromised later processing steps that lead to maturation of the 28S rRNA.
FIG. 5.
Expression of Bop1Δ inhibits the generation of 28S rRNA. Clonal cell lines derived from transfection of LAP3 cells with pX11-Bop1Δ (lines 6 and 8), pX11-Bop1 (line 45), or the pX11 vector (line 1-1) were either treated with IPTG (+) for 16 h or left untreated (−). Thereafter, cells were metabolically labeled with [3H]uridine (2.5 μCi/ml) for 30 min and chased in nonradioactive medium for 2 h. In other samples, LAP3 cells were treated with FUrd for 15 min prior to chase. RNA was then isolated, and equal counts per sample were electrophoresed on 1% agarose gel, transferred to a nylon membrane, and visualized by fluorography.
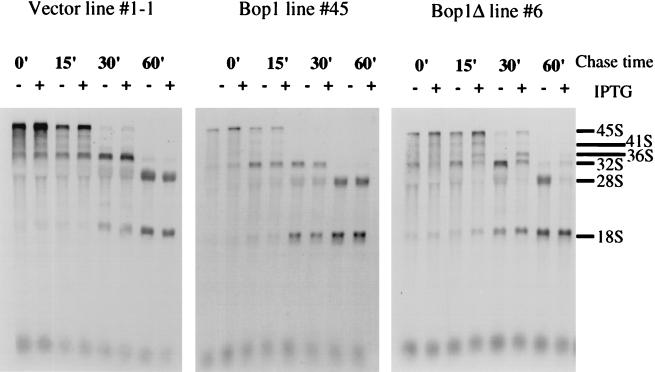
In mammalian cells, the 36S precursor is processed to form the 32S pre-rRNA, which is further processed to produce the mature 28S rRNA and a 12S precursor from which the 5.8S rRNA is generated (Fig. 6). The concomitant block of the 28S rRNA and accumulation of the 36S pre-rRNA described above suggested that Bop1Δ may inhibit the processing steps that convert the 36S pre-rRNA into the mature 28S rRNA. To evaluate this interpretation and to identify the specific rRNA processing blocks due to Bop1Δ expression, pulse-chase experiments were performed by metabolic labeling with 3H-labeled methyl methionine. LAP3-derived cell lines that either inducibly express Bop1 (line 45) or Bop1Δ (line 6) and the control cell line transfected with empty vector (line 1-1) were either left untreated or treated with IPTG, pulse-labeled for 30 min, and chased with excess nonradioactive methionine for various durations (Fig. 7). In the absence of IPTG, each cell line showed the synthesis of the 45S precursor, which was rapidly converted to the 32S pre-rRNA and the mature 18S rRNA within 30 min after pulse-labeling. The 36S pre-rRNA is relatively short-lived and does not accumulate significantly under these conditions. Processing of the 32S pre-rRNA to the mature 28S RNA appeared to be complete by 60 min after pulse-labeling. The addition of IPTG to the control cell line (line 1-1) or to the Bop1-expressing cell line (line 45) did not show any effect on rRNA synthesis or processing. By contrast, expression of Bop1Δ (line 6) resulted in accumulation of the 36S pre-rRNA and diminution of the amount of 32S pre-rRNA. Whereas production of the 18S rRNA appeared normal, maturation of the 28S rRNA was completely blocked. By 60 min after pulse-labeling, both the 36S and 32S pre-rRNAs appeared degraded rather than processed, resulting in the presence of only the 18S rRNA. These results show that expression of Bop1Δ does not affect production of the 18S rRNA but completely inhibits formation of the 28S rRNA. Moreover, Bop1Δ expression elicits an incomplete block in the conversion of the 36S to the 32S pre-rRNA and a complete block in the processing of the 32S to the 28S rRNA (Fig. 6 and 7).
FIG. 7.
Bop1Δ inhibits processing of the 36S and 32S precursors to form the 28S rRNA. Clonal LAP3 cell lines transfected with either the empty pX11 vector (line 1-1), pX11-Bop1 (line 45), or pX11-Bop1Δ (line 6) were either left untreated (−) or treated with 1 mM IPTG for 16 h (+) and pulse-labeled with 3H-labeled methyl methionine for 30 min. After a chase in nonradioactive medium plus excess methionine for the indicated times, RNA was isolated, resolved on a 1% agarose gel, transferred to a membrane, and visualized by fluorography.
Bop1Δ expression inhibits de novo generation of the 12S and 5.8S rRNAs.
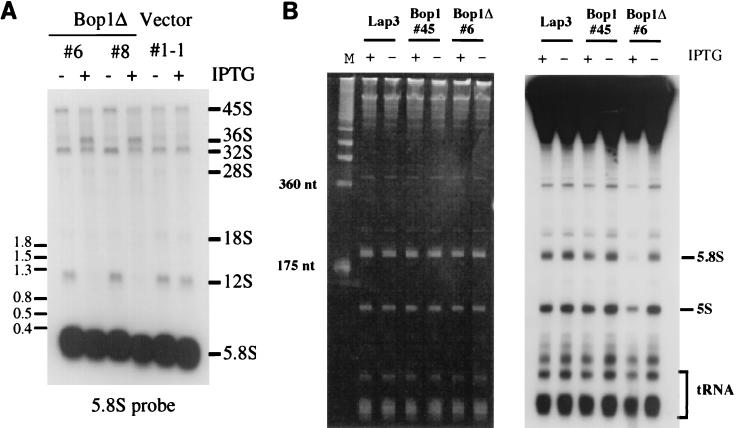
The 32S pre-rRNA is normally processed to form the mature 28S rRNA and the 12S precursor, from which the 5.8S rRNA is generated (Fig. 6) (21). The complete inhibition of 28S rRNA generation by Bop1Δ suggested that formation of the 12S and 5.8S rRNAs might also be inhibited. To test this possibility, we examined the levels of the 12S pre-rRNA and the 5.8S rRNA in Bop1Δ-expressing cells. Total RNA was isolated from various cell lines grown in the presence or absence of IPTG and subjected to RNA blot analysis using the coding region of the 5.8S rRNA as probe (Fig. 8A). This probe should hybridize to and reveal the steady-state levels of the 45S, 41S, 36S, 32S, and 12S precursors, as well as the mature 5.8S rRNA (Fig. 6). These experiments showed that expression of Bop1Δ (lines 6 and 8) resulted in the diminution of the amount of 12S pre-rRNA to an undetectable level, with a concomitant accumulation of the 36S precursor. The steady-state level of the mature 5.8S rRNA appeared unchanged during the course of this experiment, given the high levels of preexisting rRNAs.
FIG. 8.
Bop1Δ expression inhibits generation of the 12S precursor and the 5.8S rRNA. (A) Clonal LAP3 cell lines transfected with either the empty pX11 vector or pX11-Bop1Δ (lines 6 and 8) were either left untreated or treated with IPTG for 16 h. RNA isolated from the same number of cells was separated on a 1% agarose gel, transferred to a nylon membrane, and hybridized with an oligonucleotide probe complementary to a region in 5.8S rRNA. (B) The parental LAP3 cells or clonal cell lines transfected with either pX11-Bop1 (line 45) or pX11-Bop1Δ (line 6) were either left untreated or treated with IPTG for 16 h and metabolically labeled with 32Pi (20 μCi/ml) for 1 h. Following a chase in nonradioactive medium for 1.5 h, RNA was isolated, and equal amounts of RNA from each sample were resolved on a 10% denaturing polyacrylamide gel, which was stained with ethidium bromide for photography (left panel) and dried for autoradiography (right panel).
Inhibition of 12S pre-rRNA formation predicts an inability to generate mature 5.8S rRNA from the newly synthesized rRNA transcript. To test this possibility, we carried out a pulse-labeling experiment to examine the formation of the 5.8S rRNA. Various cell lines grown with or without IPTG were pulse-labeled with 32Pi for 1 h and then chased in nonradioactive medium for 1.5 h, and total RNA isolated from these cells was resolved by denaturing acrylamide gel electrophoresis (Fig. 8B). Whereas overexpression of Bop1 had no effect (line 45), expression of Bop1Δ (line 6) resulted in nearly complete inhibition in 5.8S rRNA synthesis (Fig. 8B). These results show that expression of Bop1Δ leads to a block in the processing of the 32S pre-rRNA into the 28S rRNA as well as into the 12S and 5.8S rRNAs. Interestingly, a small decrease in the amount of 5S RNA was also observed, possibly reflecting a coordinate regulation between pre-rRNA processing and 5S RNA transcription (37). In addition, the recruitment of 5S rRNA to the pre-60S particle was shown to be necessary for efficient processing of the 27S rRNA precursor in yeast (16). It is possible to speculate that the recruited 5S rRNA might be degraded together with the improperly processed 32S rRNA.
Expression of Bop1Δ causes a deficit of the 60S ribosomal subunit.
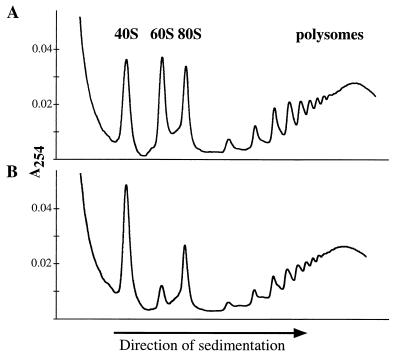
Since both 28S and 5.8S rRNAs are incorporated into the 60S ribosome, a specific block in their generation may result in a deficit in ribosome biogenesis. To examine this possibility, we used sucrose density gradient centrifugation to fractionate cytoplasmic extracts of a cell line that inducibly expressed Bop1Δ (line 6) (Fig. 9). Upon Bop1Δ expression by IPTG treatment, significantly fewer free 60S ribosomal subunits were observed in the cytoplasm whereas the 40S ribosomal subunits were accumulated to a higher level. Compared to untreated cells, the amount of 80S ribosomes was also decreased. The simplest interpretation of these results is that the 60S ribosomal subunits cannot form due to the block in 28S and 5.8S rRNAs synthesis and that the 40S ribosomal subunits accumulate due to a stoichiometric imbalance with the 60S subunit. Although the peaks that corresponded to polysomes appeared somewhat diminished by Bop1Δ expression, no global polysome disassembly was detected. This is consistent with the observation that the overall rate of translation was unaffected by Bop1Δ expression within the duration of these experiments as judged by metabolic labeling with [35S]methionine (data not shown).
FIG. 9.
Expression of Bop1Δ causes a deficit in 60S ribosomal subunits. The inducible Bop1Δ expression cell line, line 6, was grown in the absence (A) or presence (B) of IPTG as indicated for 38 h. Cytoplasmic extracts were isolated and separated on a 10 to 45% sucrose density gradient. Profiles of absorbance at 254 nm (A254) profiles revealed the positions and relative amounts of the ribosomal subunits in the gradient.
Bop1 and Bop1Δ are cosedimented with the 50S-80S pre-rRNP particles.
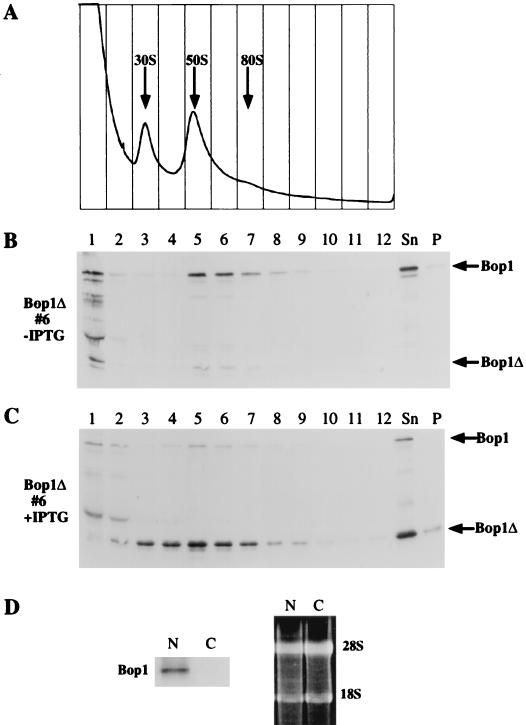
The processing of eukaryotic rRNA occurs coordinately with the assembly of ribosomal particles in the nucleolus (81, 85). Pre-rRNP particles have been identified as several complexes by sucrose density gradients. In the presence of EDTA, these complexes, which contain the 45S, 36S, and 32S pre-rRNAs, sediment in the range of 50S to 80S (42, 47, 82). The apparent involvement of Bop1 in pre-rRNA processing suggested that Bop1 might be associated with the pre-rRNP particles. To investigate this possibility, nuclear extracts prepared from the inducible Bop1Δ-expressing cell line (line 6) were analyzed by sucrose gradient centrifugation (Fig. 10A). Proteins from each gradient fraction were assayed for endogenous Bop1 and ectopically expressed Bop1Δ by Western blot analysis. When cells were grown in the absence of IPTG, endogenous Bop1 protein sedimented primarily in fractions 5 to 7, which corresponded to the 50S-80S particles (Fig. 10B and C). When expressed, Bop1Δ was distributed more broadly from fractions 3 through 7 with a peak at fraction 5, where the peak of the endogenous Bop1 was also found. Thus, both Bop1 and Bop1Δ were found in large nuclear particles with sizes similar to pre-rRNP particles. However, Bop1 is not a component of matured ribosomes, since Western blotting of a cytoplasmic ribosomal preparation did not detect any Bop1 protein (Fig. 10D).
FIG. 10.
Bop1 and Bop1Δ cofractionate with the 50S-80S pre-RNP particles in nuclear extracts. (A to C) The inducible Bop1Δ expression cell line (line 6) was grown in the presence (C) or absence (A and B) of IPTG for 24 h as indicated. Nuclear extracts isolated from these cells were analyzed on 10 to 30% sucrose density gradients, which were fractionated with continuous monitoring of absorbance at 254 nm (A). Individual fractions were electrophoresed on an SDS–10% polyacrylamide gel and subjected to immunoblotting analysis using affinity-purified anti-Bop1 antibodies. Sn, unfractionated soluble nuclear extracts; P, unsoluble pellet. (D) (Left) Immunoblot analysis with anti-Bop1 antibodies detects Bop1 in nuclear RNPs (N) but not cytoplasmic ribosomes (C). (Right) Electrophoretic analysis of RNA in the fractions used for immunoblotting. RNA was extracted from the nucleoprotein complexes and separated by electrophoresis on a formaldehyde-containing agarose gel to demonstrate the presence equivalent amounts of rRNA in both samples.
To substantiate the observation that Bop1 cosediments with the 50S-80S preribosomes, nuclear extracts were fractionated by sucrose density gradients and proteins and RNA from each gradient fractions were analyzed in parallel (Fig. 11). Proteins were resolved by electrophoresis, blotted, and probed with anti-Bop1 antibodies, while RNA from the same samples was electrophoresed, blotted, stained with methylene blue, and probed with the 32P-labeled DNA fragment from the ITS2 region, which can recognize the 47S, 45S, 41S, 36S, and 32S pre-rRNAs. This analysis showed that the Bop1 protein cosedimented with rRNP particles that contained the 32S precursor RNA, found in fractions 5 to 8, and that the peaks for both Bop1 and the 32S rRNA both occurred in fraction 6. The 18S rRNA was found mainly in fractions 3 and 4, whereas the 28S rRNA was found in fractions 4 to 7, with the peak in fraction 5. Thus, these results show that Bop1 cosediments with the rRNP particles containing the 32S pre-rRNA, consistent with a role for Bop1 in the processing of the 32S pre-rRNA.
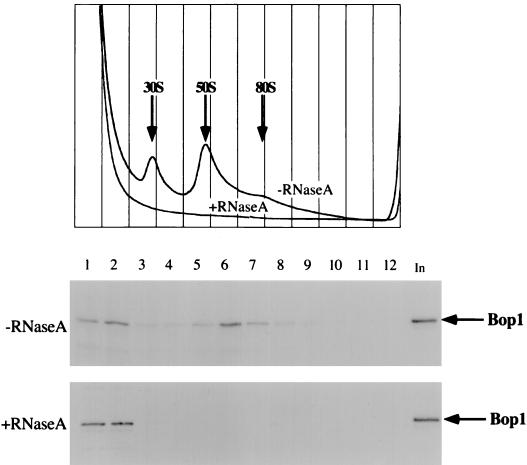
To confirm that Bop1 is a component of rRNP particles, nuclear extracts were either untreated or treated with RNase A before being subjected to fractionation on a sucrose gradient (Fig. 12). As expected, Bop1 cofractionated with rRNP particles in the absence of RNase A. Treatment with RNase completely destroyed the rRNP particles, as shown by the absorbance of the gradient at 254 nm. Bop1 protein was completely shifted to the top of the gradient on nuclease digestion, consistent with its release from rRNP particles. In separate experiments, Bop1Δ was also released from the 50S-80S fractions to the top of the gradient on RNase A treatment (data not shown). Together, these results show that as a nucleolar protein, Bop1 is a component of the RNP particles that cosediment with the pre-rRNP particles containing the 32S pre-rRNA.
FIG. 12.
Bop1 is part of an RNP complex. Nuclear extracts from LAP3 cells were either treated with RNaseA or left untreated as indicated and analyzed on 10 to 30% sucrose density gradients. The gradients were fractionated and monitored for absorbance at 254 nm (top panel). Various fractions were subjected to SDS-PAGE followed by immunoblotting using affinity-purified anti-Bop1 antibodies. In, unfractionated soluble nuclear extract.
DISCUSSION
In this study we identified Bop1 as a novel participant in the mammalian nucleolar rRNA processing and ribosome biogenesis machinery. This conclusion is drawn based on data accumulated through two approaches: (i) biochemical studies show that Bop1 is a nucleolar protein that forms part of a large, RNA-containing protein complex that cosediments with pre-rRNP particles containing the 32S pre-rRNA; and (ii) functional analyses indicate that Bop1 plays a role in the maturation of the 28S and 5.8S rRNAs and the biogenesis of the 60S ribosomal subunit.
Several lines of evidence provide support for the conclusion that Bop1 is a nonribosomal nucleolar protein that constitutes a component of pre-rRNP particles. Immunofluorescence analysis demonstrates that both endogenous and ectopically expressed Bop1 are localized to the nucleolus (Fig. 3). Sucrose density gradient fractionation of nuclear extracts shows that Bop1 forms part of large ribonucleoprotein complexes that sediment at 50S-80S (Fig. 10). Treatment of the nuclear preparations with RNase A, which destroys pre-rRNP particles, releases Bop1 into low-molecular-weight fractions at the top of the gradient (Fig. 12). Thus, while Bop1 is not part of the mature cytosolic ribosomes, these data strongly indicate that it is a component of the pre-rRNP particles.
To analyze the function of Bop1, we took advantage of an N-terminally truncated derivative, Bop1Δ, as a means of interfering with the activity of the wild-type protein in a dominant manner. Bop1Δ displays the same nucleolar localization as Bop1 and a similar sedimentation profile in a sucrose density gradient, indicating that it resides in the same RNP complexes as the wild-type protein and thus retains a subset of its functions. Since Bop1Δ retains the WD40 motifs present in the full-length Bop1 (Fig. 1), it is plausible to speculate that Bop1Δ is able to interact with many of the same proteins with which Bop1 interacts. However, Bop1Δ apparently interferes with the normal function of Bop1. Indeed, expression of Bop1Δ results in a serious defect in the processing of the 28S and 5.8S rRNAs and a deficit of mature 60S ribosome subunits. This conclusion is supported by a preponderance of evidence. (i) Metabolic labeling shows that expression of Bop1Δ leads to a specific inhibition of 28S and 5.8S rRNA maturation while having no effect on the 18S rRNA (Fig. 5 and 8). (ii) Pulse-chase analysis reveals a partial processing block in the conversion of the 36S to the 32S pre-rRNA and a complete block in the processing of the 32S pre-rRNA to the mature 28S rRNA and 12S pre-rRNA and consequently to the 5.8S rRNA (Fig. 7 and 8). (iii) Examination of the steady-state RNA levels shows an accumulation of the 36S rRNA and an inhibition of 12S pre-rRNA (Fig. 8A), confirming the results of pulse-chase labeling. (iv) Sedimentation gradients show a marked decrease in the number of cytosolic 60S ribosomal subunits (Fig. 9). These findings strongly implicate Bop1 as an important player in rRNA maturation, specifically in the conversion of the 32S pre-rRNA into the 28S rRNA and the 12S precursor. Moreover, both Bop1 and Bop1Δ cosediment with pre-rRNPs that contain the 32S rRNA precursors (Fig. 10 and 11), further corroborating the conclusion that Bop1 is involved in the maturation of the 5.8S and 28S rRNAs, a process with which Bop1Δ interferes.
Expression of Bop1Δ under the control of an inducible promoter was previously shown to cause a powerful but reversible G1 growth arrest in mouse fibroblasts (58). The precise mechanism by which Bop1Δ causes this growth inhibition is unclear. Nevertheless, it is possible to envisage at least two distinct mechanisms by which Bop1Δ may affect cell cycle progression. First, growth arrest due to Bop1Δ may be a secondary effect resulting from perturbations in the translation machinery caused by deficiency of 60S ribosomal subunits. For example, deficiencies in the translation initiation factor eIF4E/CDC33 lead to a G1 growth arrest in yeast (9). Alternatively, Bop1 might play a dual role in both pre-rRNA processing and cell cycle progression, thereby mediating a cross talk between these two distinct cellular pathways. The nucleolus has been implicated in functions other than rRNA processing and ribosome biogenesis (57), including the regulation of cellular exit from mitosis (4). Exosomes and Xrn1p exonuclease function in mRNA in addition to rRNA processing (10, 17, 50) and may thus affect the synthesis of cell cycle regulators. The possibility that Bop1 might exert an effect on the cell cycle machinery hints at an as yet poorly understood link between the cellular capacity for coordinating protein synthesis and cell cycle progression. Understanding the precise role of Bop1 in cell cycle progression will require further investigation.
Consistent with a role for Bop1 in rRNA processing and ribosome maturation, Bop1 appears to be ubiquituously expressed irrespective of the tissue type (Fig. 2). Moreover, bop1 mRNA levels begin to rise in mid to late G1, coincident with the timing of rRNA synthesis (24, 65, 70). As expected of a protein that plays a role in ribosome maturation, Bop1 appears to be highly conserved throughout evolution. The mouse Bop1 displays >90% amino acid identity to its human ortholog (KIAA0124) and ∼45% identity to a sequence in Saccharomyces cerevisiae (YMR049c). A BLAST search in the currently available databases also reveals protein coding sequences highly homologous to Bop1 in such diverse organisms as Drosophila melanogaster, Caenorhabditis elegans, and Arabidopsis thaliana.
In this report, we show for the first time the effect of a nucleolar protein on rRNA processing in a mammalian system. Only a few other mammalian proteins have been implicated in rRNA processing to date, based on the function of their yeast homologs. For example, p120 was originally identified as a tumor proliferation antigen (33) and was later implicated in rRNA processing, since deletion of its yeast homolog, Nop2p, causes a block in the processing of the 27S pre-rRNA to the mature 25S rRNA (36). Consistent with a role in rRNA processing, p120 was shown to cofractionate with the 60-80S preribosomal particles in HeLa cell extracts (30).
In yeast, several proteins whose depletion specifically affects processing of the 25S branch of rRNA processing have been identified. For example, depletion of the yeast Nop4 protein results in the inhibition of the 25S rRNA maturation as well as 60S ribosomal-subunit accumulation, while production of the 18S rRNA is unaffected (74). Other examples of similar phenotypes result from depletion of members of the DEAD box family of putative ATP-dependent RNA helicases including DRS1 (63), Dbp6p (41), Dbp7p (15), and Spb4p (18), as well as nucleolar proteins Nip7p (87) and Nop8p (86). These observations underscore the notion that processing of the 25S/28S branch of rRNA requires complexes of numerous proteins, whose nature is only beginning to be understood. In Xenopus, depletion of the U8 snoRNA (56) also has effects similar to Bop1Δ expression, namely, an incomplete inhibition of 36S pre-rRNA processing but a complete block of 32S pre-rRNA processing, leading to inhibition of 28S and 5.8S rRNA production. Immunoprecipitation of Bop1 followed by RNA blotting with an anti-U8 RNA probe failed to detect the U8 RNA (data not shown). In addition, immunoprecipitation of Bop1 from cells metabolically labeled with radioactive phosphate followed by PAGE also did not reveal the presence of small RNAs (data not shown). These results suggest that Bop1 is unlikely to be mediating the interaction of U8 or other snoRNAs with the rRNA precursor.
Processing of pre-rRNA and ribosome biogenesis appear to be highly coordinated (19, 49, 64, 80, 81). Consistent with this notion, there is a dramatic decrease in the amount of mature 60S ribosomal subunits when maturation of the 28S and 5.8S rRNAs is blocked as a result of Bop1Δ expression (Fig. 9). The deficit in the 60S ribosomal subunit may be a direct result of the inability to produce its cognate mature rRNAs. However, we cannot rule out the possibility that Bop1 may also have the distinct function of coordinating 60S ribosomal assembly and that this function is compromised by Bop1Δ.
Analysis of the primary sequence of Bop1 does not suggest an apparent enzymatic activity, although it does show features of a short-lived, regulatory protein that may participate in critical protein-protein interactions. The Bop1 sequence contains clusters of charged amino acid residues, known as PEST sequences, often associated with regulatory and short-lived proteins (13, 61, 67). Similar clusters are also observed in a number of nucleolar proteins (71). A putative nuclear localization signal in Bop1 is located at aa 360 to 366 (PRQRKMR; underlining indicates positively charged residues) (20, 25). Data from the present study with Bop1Δ show that the N-terminal 231 aa are not needed for nucleolar location or complex formation with RNP particles but are necessary for critical functions illustrated by the consequences of Bop1Δ expression.
Bop1 contains four WD40 repeats (Fig. 1), a sequence motif found in a large variety of regulatory proteins and known to mediate protein-protein interactions (26, 27, 51, 52, 72). WD40 proteins have been implicated in a number of cellular processes, including various stages of RNA metabolism (3, 6). At present, at least two other nucleolar proteins that contain WD40 repeats have been identified: the yeast SOF1 (38) and the related (but not orthologous) human protein hU3-55k (60). Inasmuch as a number of WD40 repeat proteins are known to form large multiprotein complexes, Bop1 may mediate protein-protein interactions that are important for the formation and activities of nucleolar RNPs. Future studies aimed at the identification and characterization of the proteins with which Bop1 interacts may yield new insights into the structural organization and mechanism of action of the nucleolar pre-rRNA-processing machinery.
ACKNOWLEDGMENTS
We thank members of our laboratory for helpful discussions, Michael Pollard for providing the antifibrillarin antibody (72B9), Alisa Katzen and Gary Ramsey for helping with microscopy, and Robert Costa, Nissim Hay, Susan Liebman, and Jeff Greenspan for critical reading of the manuscript.
This work was supported by a grant from the National Institutes of Health (CA52220).
REFERENCES
- 1.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 3.Ayadi L, Miller M, Banroques J. Mutations within the yeast U4/U6 snRNP protein Prp4 affect a late stage of spliceosome assembly. RNA. 1997;3:197–209. [PMC free article] [PubMed] [Google Scholar]
- 4.Bachant J B, Elledge S J. Mitotic treasures in the nucleolus. Nature. 1999;398:757–758. doi: 10.1038/19641. [DOI] [PubMed] [Google Scholar]
- 5.Baim S B, Labow M A, Levine A J, Shenk T. A chimeric mammalian transactivator based on the lac repressor that is regulated by temperature and isopropyl β-d-thiogalactopyranoside. Proc Natl Acad Sci USA. 1991;88:5072–5076. doi: 10.1073/pnas.88.12.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben Yehuda S, Dix I, Russell C S, Levy S, Beggs J D, Kupiec M. Identification and functional analysis of hPRP17, the human homologue of the PRP17/CDC40 yeast gene involved in splicing and cell cycle control. RNA. 1998;4:1304–1312. doi: 10.1017/s1355838298980712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorn S P, Soltyk A, Beggs J D, Friesen J D. PRP4 (RNA4) from Saccharomyces cerevisiae: its gene product is associated with the U4/U6 small nuclear ribonucleoprotein particle. Mol Biol Cell. 1989;9:3698–3709. doi: 10.1128/mcb.9.9.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman L H, Rabin B, Schlessinger D. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 1981;9:4951–4966. doi: 10.1093/nar/9.19.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner C, Nakayama N, Goebl M, Tanaka K, Toh-e A, Matsumoto K. CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:3556–3559. doi: 10.1128/mcb.8.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpousis A J, Vanzo N F, Raynal L C. mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet. 1999;15:24–28. doi: 10.1016/s0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
- 11.Chang J H, Olson M O. Structure of the gene for rat nucleolar protein B23. J Biol Chem. 1990;265:18227–18233. [PubMed] [Google Scholar]
- 12.Chen C A, Okayama H. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. BioTechniques. 1988;6:632–638. [PubMed] [Google Scholar]
- 13.Chevaillier P. Pest sequences in nuclear proteins. Int J Biochem. 1993;25:479–482. doi: 10.1016/0020-711x(93)90653-v. [DOI] [PubMed] [Google Scholar]
- 14.Dalrymple M A, Petersen-Bjorn S, Friesen J D, Beggs J D. The product of the PRP4 gene of S. cerevisiae shows homology to beta subunits of G proteins. Cell. 1989;58:811–812. doi: 10.1016/0092-8674(89)90930-6. [DOI] [PubMed] [Google Scholar]
- 15.Daugeron M C, Linder P. Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA. 1998;4:566–581. doi: 10.1017/s1355838298980190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dechampesme A M, Koroleva O, Leger-Silvestre I, Gas N, Camier S. Assembly of 5S ribosomal RNA is required at a specific step of the pre-rRNA processing pathway. J Cell Biol. 1999;145:1369–1380. doi: 10.1083/jcb.145.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decker C J. The exosome: a versatile RNA processing machine. Curr Biol. 1998;8:R238–R240. doi: 10.1016/s0960-9822(98)70149-6. [DOI] [PubMed] [Google Scholar]
- 18.de la Cruz J, Kressler D, Rojo M, Tollervey D, Linder P. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA. 1998;4:1268–1281. doi: 10.1017/s1355838298981158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deshmukh M, Stark J, Yeh L C, Lee J C, Woolford J L., Jr Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5 S rRNA and assembly into ribosomes. J Biol Chem. 1995;270:30148–30156. doi: 10.1074/jbc.270.50.30148. [DOI] [PubMed] [Google Scholar]
- 20.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 21.Eichler D C, Craig N. Processing of eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1994;49:197–239. doi: 10.1016/s0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- 22.Elela S A, Igel H, Ares M J. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 23.Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujikawa-Yamamoto K. RNA dependence in the cell cycle of V79 cells. J Cell Physiol. 1982;112:60–66. doi: 10.1002/jcp.1041120110. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Bustos J, Heitman J, Hall M N. Nuclear protein localization. Biochim Biophys Acta. 1991;1071:83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Higuera I, Fenoglio J, Li Y, Lewis C, Panchenko M P, Reiner O, Smith T F, Neer E J. Folding of proteins with WD-repeats: comparison of six members of the WD-repeat superfamily to the G protein beta subunit. Biochemistry. 1996;35:13985–13994. doi: 10.1021/bi9612879. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Higuera I, Gaitatzes C, Smith T F, Neer E J. Folding a WD repeat propeller. Role of highly conserved aspartic acid residues in the G protein beta subunit and Sec13. J Biol Chem. 1998;273:9041–9049. doi: 10.1074/jbc.273.15.9041. [DOI] [PubMed] [Google Scholar]
- 28.Geiduschek E P, Tocchini-Valentini G P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- 29.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 30.Gustafson W C, Taylor C W, Valdez B C, Henning D, Phippard A, Ren Y, Busch H, Durban E. Nucleolar protein p120 contains an arginine-rich domain that binds to ribosomal RNA. Biochem J. 1998;331:387–393. doi: 10.1042/bj3310387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hadjiolova K V, Nicoloso M, Mazan S, Hadjiolov A A, Bachellerie J P. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur J Biochem. 1993;212:211–215. doi: 10.1111/j.1432-1033.1993.tb17652.x. [DOI] [PubMed] [Google Scholar]
- 32.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 33.Hazlewood J, Fonagy A, Henning D, Freeman J W, Busch R K, Busch H. mRNA levels for human nucleolar protein P120 in tumor and nontumor cells. Cancer Commun. 1989;1:29–34. doi: 10.3727/095535489820875426. [DOI] [PubMed] [Google Scholar]
- 34.Henriquez R, Blobel G, Aris J P. Isolation and sequencing of NOP1. A yeast gene encoding a nucleolar protein homologous to a human autoimmune antigen. J Biol Chem. 1990;265:2209–2215. [PubMed] [Google Scholar]
- 35.Henry Y, Wood H, Morrissey J P, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong B, Brockenbrough J S, Wu P, Aris J P. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol Biol Cell. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson A J, Ittmann M, Pugh B F. The BN51 protein is a polymerase (Pol)-specific subunit of RNA Pol III which reveals a link between Pol III transcription and pre-rRNA processing. Mol Cell Biol. 1995;15:94–101. doi: 10.1128/mcb.15.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jansen R, Tollervey D, Hurt E C. A U3 snoRNP protein with homology to splicing factor PRP4 and G beta domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson A W. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol Cell Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jordan P, Mannervik M, Tora L, Carmo-Fonseca M. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kressler D, de la Cruz J, Rojo M, Linder P. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1855–1865. doi: 10.1128/mcb.18.4.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liau M C, Perry R P. Ribosome precursor particles in nucleoli. J Cell Biol. 1969;42:272–283. doi: 10.1083/jcb.42.1.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lischwe M A, Ochs R L, Reddy R, Cook R G, Yeoman L C, Tan E M, Reichlin M, Busch H. Purification and partial characterization of a nucleolar scleroderma antigen (Mr = 34,000; pI, 8.5) rich in NG,NG-dimethylarginine. J Biol Chem. 1985;260:14304–14310. [PubMed] [Google Scholar]
- 44.Lygerou Z, Allmang C, Tollervey D, Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 45.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 46.McEwen C R. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal Biochem. 1967;20:114–149. doi: 10.1016/0003-2697(67)90271-0. [DOI] [PubMed] [Google Scholar]
- 47.Mirault M E, Scherrer K. Isolation of preribosomes from HeLa cells and their characterization by electrophoresis on uniform and exponential-gradient-polyacrylamide gels. Eur J Biochem. 1971;23:372–386. doi: 10.1111/j.1432-1033.1971.tb01631.x. [DOI] [PubMed] [Google Scholar]
- 48.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 49.Moritz M, Pulaski B A, Woolford J L., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991;11:5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muhlrad D, Decker C J, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neer E J, Schmidt C J, Nambudripad R, Smith T F. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 52.Neer E J, Smith T F. G protein heterodimers: new structures propel new questions. Cell. 1996;84:175–178. doi: 10.1016/s0092-8674(00)80969-1. [DOI] [PubMed] [Google Scholar]
- 53.Ochs R L, Lischwe M A, Spohn W H, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell. 1985;54:123–133. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 54.O'Day C L, Chavanikamannil F, Abelson J. 18S rRNA processing requires the RNA helicase-like protein Rrp3. Nucleic Acids Res. 1996;24:3201–3207. doi: 10.1093/nar/24.16.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olson M O, Guetzow K, Busch H. Localization of phosphoprotein C23 in nucleoli by immunological methods. Exp Cell Res. 1981;135:259–265. doi: 10.1016/0014-4827(81)90161-0. [DOI] [PubMed] [Google Scholar]
- 56.Peculis B A, Steitz J A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- 57.Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pestov D G, Grzeszkiewicz T M, Lau L F. Isolation of growth suppressors from a cDNA expression library. Oncogene. 1998;17:3187–3197. doi: 10.1038/sj.onc.1202260. [DOI] [PubMed] [Google Scholar]
- 59.Pestov D G, Lau L F. Genetic selection of growth-inhibitory sequences in mammalian cells. Proc Natl Acad Sci USA. 1994;91:12549–12553. doi: 10.1073/pnas.91.26.12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pluk H, Soffner J, Luhrmann R, van Venrooij W J. cDNA cloning and characterization of the human U3 small nucleolar ribonucleoprotein complex-associated 55-kilodalton protein. Mol Cell Biol. 1998;18:488–498. doi: 10.1128/mcb.18.1.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 62.Reimer G, Pollard K M, Penning C A, Ochs R L, Lischwe M A, Busch H, Tan E M. Monoclonal autoantibody from a (New Zealand black × New Zealand white) F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum. 1987;30:793–800. doi: 10.1002/art.1780300709. [DOI] [PubMed] [Google Scholar]
- 63.Ripmaster T L, Vaughn G P, Woolford J L., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc Natl Acad Sci USA. 1992;89:11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rotenberg M O, Moritz M, Woolford J L., Jr Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988;2:160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- 65.Rudland P S, Weil S, Hunter A R. Changes in RNA metabolism and accumulation of presumptive messenger RNA during transition from the growing to the quiescent state of cultured mouse fibroblasts. J Mol Biol. 1975;96:745–766. doi: 10.1016/0022-2836(75)90150-3. [DOI] [PubMed] [Google Scholar]
- 66.Russell I D, Tollervey D. NOP3 is an essential yeast protein which is required for pre-rRNA processing. J Cell Biol. 1992;119:737–747. doi: 10.1083/jcb.119.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salama S R, Hendricks K B, Thorner J. G1 cyclin degradation: the PEST motif of yeast Cln2 is necessary, but not sufficient, for rapid protein turnover. Mol Cell Biol. 1994;14:7953–7966. doi: 10.1128/mcb.14.12.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 69.Scheer U, Weisenberger D. The nucleolus. Curr Opin Cell Biol. 1994;6:354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 70.Seuwen K, Steiner U, Adam G. Cellular content of ribosomal RNA in relation to the progression and competence signals governing proliferation of 3T3 and SV40-3T3 cells. Exp Cell Res. 1984;154:10–24. doi: 10.1016/0014-4827(84)90664-5. [DOI] [PubMed] [Google Scholar]
- 71.Shaw P J, Jordan E G. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- 72.Smith T F, Gaitatzes C, Saxena K, Neer E J. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 73.Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun C, Woolford J L., Jr The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 1994;13:3127–3135. doi: 10.1002/j.1460-2075.1994.tb06611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tollervey D. Trans-acting factors in ribosome synthesis. Exp Cell Res. 1996;229:226–232. doi: 10.1006/excr.1996.0364. [DOI] [PubMed] [Google Scholar]
- 76.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 77.Tuteja R, Tuteja N. Nucleolin: a multifunctional major nucleolar phosphoprotein. Crit Rev Biochem Mol Biol. 1998;33:407–436. doi: 10.1080/10409239891204260. [DOI] [PubMed] [Google Scholar]
- 78.Venema J, Bousquet-Antonelli C, Gelugne J P, Caizergues-Ferrer M, Tollervey D. Rok1p is a putative RNA helicase required for rRNA processing. Mol Cell Biol. 1997;17:3398–3407. doi: 10.1128/mcb.17.6.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 80.Warner J R. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol Rev. 1989;53:256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Warner J R. The nucleolus and ribosome formation. Curr Opin Cell Biol. 1990;2:521–527. doi: 10.1016/0955-0674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- 82.Warner J R, Soeiro R. Nascent ribosomes from HeLa cells. Proc Natl Acad Sci USA. 1967;58:1984–1990. doi: 10.1073/pnas.58.5.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weinstein L B, Steitz J A. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 84.Wilkinson D S, Tlsty T D, Hanas R J. The inhibition of ribosomal RNA synthesis and maturation in Novikoff hepatoma cells by 5-fluorouridine. Cancer Res. 1999;35:3014–3020. [PubMed] [Google Scholar]
- 85.Woolford J L J. The structure and biogenesis of yeast ribosomes. Adv Genet. 1991;29:63–118. doi: 10.1016/s0065-2660(08)60107-8. [DOI] [PubMed] [Google Scholar]
- 86.Zanchin N I, Goldfarb D S. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol Cell Biol. 1999;19:1518–1525. doi: 10.1128/mcb.19.2.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zanchin N I, Roberts P, DeSilva A, Sherman F, Goldfarb D S. Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol Cell Biol. 1997;17:5001–5015. doi: 10.1128/mcb.17.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]