Abstract
While glycans are crucial for biological processes, existing analysis modalities make it difficult for researchers with limited computational background to include these diverse carbohydrates into workflows. Here, we present glycowork, an open-source Python package designed for glycan-related data science and machine learning by end users. Glycowork includes functions to, for instance, automatically annotate glycan motifs and analyze their distributions via heatmaps and statistical enrichment. We also provide visualization methods, routines to interact with stored databases, trained machine learning models and learned glycan representations. We envision that glycowork can extract further insights from glycan datasets and demonstrate this with workflows that analyze glycan motifs in various biological contexts. Glycowork can be freely accessed at https://github.com/BojarLab/glycowork/.
Keywords: data science, glycobioinformatics, glycobiologymachine learning, Python
Introduction
Discovering patterns in biological data requires (i) large datasets and (ii) data science, bioinformatics or machine learning. This combination has led to great advances in systems biology (Chuang et al. 2010; Zou and Laubichler 2018). Usually, there is limited overlap between groups engaging in data collection and those developing algorithms. In mature systems biology fields, this gap is bridged by user-friendly software that facilitates experimental users to analyze their datasets for routine applications, such as with the Bioconductor (Huber et al. 2015) or Biopython (Cock et al. 2009) platforms.
Glycobiology—the analysis of glycans in biological contexts (Varki 2017)—has recently seen a surge in data gathering and algorithmic development. Moderately large datasets from glycomics (Cummings and Pierce 2014), glycan arrays (Oyelaran and Gildersleeve 2009) or lectin arrays (Ribeiro and Mahal 2013) can by now be gathered on a rather routine basis, depending on the application. Many algorithms for the analysis of glycan-related data, such as subtree mining for the analysis of glycan array data (Coff et al. 2020; Haab and Klamer 2020) or glycan-focused machine learning (Bojar et al. 2021; Burkholz et al. 2021), have been recently developed. Furthermore, resources in glycobioinformatics have been centralized in the context of the GlySpace Alliance (Aoki-Kinoshita et al. 2020) for increased synergy. This includes a large-scale glycan repository in the form of GlyTouCan and portals, such as GlyCosmos or GlyGen, to various glycobioinformatics resources.
Yet while both factors necessary for effective analysis—data and algorithms—are present in glycobiology, most algorithm development efforts are inaccessible to the typical user, who might not be well-versed in computational workflows. While accessible graphical user interfaces for some applications have been developed (Grant et al. 2016; Huang et al. 2021), these approaches often lack the flexibility and throughput that is required for many analyses. Thus, with the exception of platforms such as glypy (Klein and Zaia 2019), geared more toward analyzing glycan-focused mass spectrometry, glycobioinformatics methods cannot be used with the same accessibility that bioinformatics procedures exhibit in other systems biology disciplines.
Therefore, we have developed glycowork, a computational framework designed to be accessible to end users with minimal computational background. While background functions that work with glycans as graphs are available to experts, we provide high-level wrapper functions for analyses that only require the input of glycans in a human-readable format, such as the IUPAC-condensed format. Glycowork is open-source (https://github.com/BojarLab/glycowork/) and we have prepared an extensive documentation with example workflows (https://bojarlab.github.io/glycowork/). We envision glycowork to advance glycobioinformatics, distilling insights from the increasing number of available glycan datasets.
Glycowork—Principles and applications
Glycowork is written in the Python programming language (version 3.6+) and uses pandas dataframes, lists of glycans or single glycans as inputs for its functions. We have structured glycowork into four modules: data loading and handling (glycan_data), sequence alignment (alignment), sequence processing and motif analysis (motif), and glycan-focused machine learning (ml) (Figure 1A).
Fig. 1.
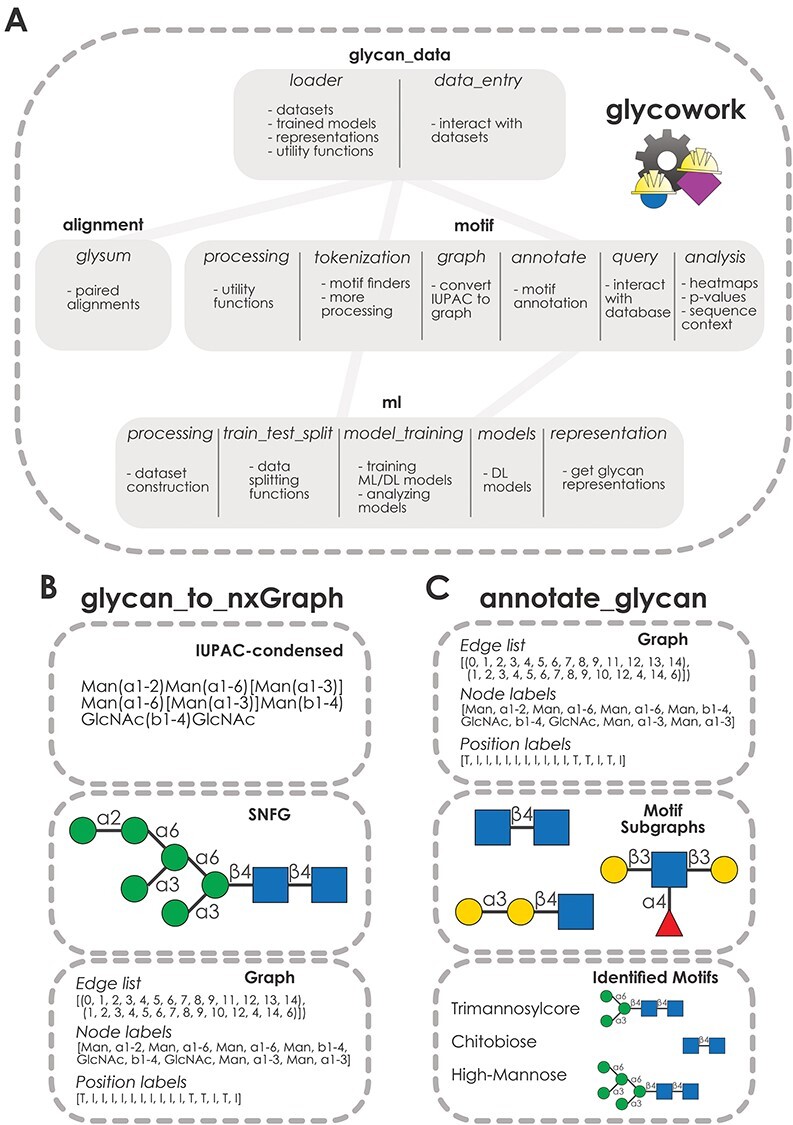
Structure of the glycowork package. (A) Modular structure of glycowork. Modules are depicted as boxes containing submodules. Dependencies between modules are indicated by connecting lines. (B) Workflow of the glycan_to_nxGraph function from the glycowork.motif.graph submodule. An example glycan in IUPAC-condensed notation is converted into a graph. The resulting edge, node and position lists are shown, with “T” indicating a terminal position and “I” indicating an internal position. (C) Workflow of the annotate_glycan function from the glycowork.motif.annotate submodule. Glycan graphs and graphs for known motifs are used to identify occurring motifs via subgraph isomorphism tests.
Functions in glycowork use glycans in the IUPAC-condensed nomenclature as input and convert these to graph objects (Burkholz et al. 2021) for further processing and analysis (Figure 1B). Crucially, this step removes the ambiguity of the IUPAC nomenclature, as the uniqueness of graphs can be tested. We decided against using other nomenclatures, such as GlycoCT (Herget et al. 2008) or WURCS (Tanaka et al. 2014), as we argue that combining a human-readable nomenclature (IUPAC-condensed) with a machine-readable nomenclature (glycan graphs) is necessary and sufficient for all relevant tasks in glycobioinformatics. Furthermore, working with glycans as graphs allowed us to leverage advances in graph theory that have accrued over decades of research, such as the NetworkX package (Hagberg et al. 2008) that can be applied without modifications to our glycan graphs.
Glycan graphs consist of the connectivities in a glycan (edge list), the contained monosaccharides/linkages (node labels) and the information whether these are internal or terminal (position labels; Figure 1B). Glycowork is designed so that users with limited bioinformatics experience can exclusively work with glycans in IUPAC-condensed nomenclature, while all graph operations proceed in the background. We are confident that this will facilitate the accessibility of our open-source package.
Many functions in glycowork leverage the power of graph theory, for instance by unambiguously annotating glycan motifs via subgraph isomorphism using the annotate_glycan function (Figure 1C). The concept of graph isomorphism tests whether two graph objects describe the same graph (similar to how two IUPAC-condensed descriptions of a glycan can describe the same glycan), in the sense that monosaccharides have the same neighbors in both graphs, which can be used to detect the presence of a subgraph, a motif, in a glycan graph. In glycowork, this is done using the position labels, to ensure that motifs such as the O-glycan core motifs are only recognized at the reducing end. Glycowork comes equipped with 150 named motifs from the academic literature and can also analyze relevant disaccharide motifs, as further described below.
Glycowork contains continuously updated glycan datasets, such as glycan array data of influenza viruses or information about species-specific glycans. These could be used for developing algorithms or uncovering new insights into glycan properties. Additionally, learned representations of glycans from a deep learning model (Burkholz et al. 2021) are provided, allowing us to visualize clusters of similar glycans (Figure 2A). Furthermore, glycowork contains functions to cluster groups via heatmaps according to the presence and abundance of glycan motifs.
Fig. 2.
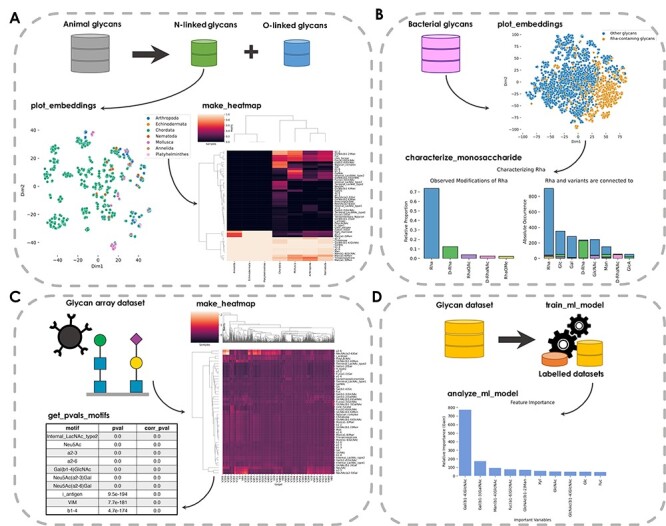
Example workflows from glycowork. (A) Investigating N-linked glycans in animals. The plot_embeddings function displays N-linked glycans, with colors corresponding to taxonomic phyla. The make_heatmap function displays glycan motif distributions for each phylum. (B) Analysis of rhamnose sequence neighborhood in bacteria. Bacterial glycans are colored based on the presence of rhamnose (Rha). Proportions of rhamnose and its variants (left) and their observed neighboring monosaccharides (right), as stacked bar graphs, are visualized via the characterize_monosaccharide function. (C) Glycan-binding specificities of influenza viruses. Measured glycan-binding of various influenza strains is represented as a heatmap. The get_pvals_motifs function displays, for each motif, a P-value and a corrected P-value. Shown are the top 10 motifs, with the full table available in Table SI. (D) Glycan classification using machine learning. The train_ml_model function constructs a model to discriminate between “animal” and “non-animal” glycans. The analyze_ml_model function displays important criteria for glycan classification. Full-scale heatmaps shown in A and C are found in Figures S1 and S2.
Beside these analyses, users can analyze the neighboring sequence of a monosaccharide in a specific taxonomic group such as bacteria (Figure 2B). Comparing the sequence context of different groups of interest might shed light onto evolutionary or functional differences in their glycans. Here, we show this type of analysis with the sequence neighborhood of the monosaccharide rhamnose (Rha) in bacteria. This yields the observation that Rha, D-Rha and D-RhaNAc are all typically found in homogeneous sequence environments (i.e. connected to more Rha/D-Rha/D-RhaNAc, respectively).
Glycan arrays are a common method to determine viral glycan-binding specificity (Smith and Cummings 2014). Glycowork can analyze this data type by generating motif-based heatmaps, in which motifs are colored by their associated Z-score—in this case illustrating the split between Neu5Ac(α2–3)-binding avian influenza viruses and Neu5Ac(α2–6)-preferring mammalian influenza viruses (Figure 2C). This can be extended by identifying statistically significant binding motifs (Figure 2C), which points to the importance of sialic acid-containing motifs as reported previously (Viswanathan et al. 2010).
Another example of the functionalities of glycowork is glycan-focused machine learning. By providing a list of glycans and corresponding labels, glycowork trains machine learning models with a single line of code. As an example, we trained a model to predict whether a glycan stems from an animal or a different organism and then analyzed the model as to which motifs were most predictive for this classification (Figure 2D). In this case, the presence of type-2 LacNAc (Gal(β1–4)GlcNAc) was most predictive for an animal glycan. Analogously, state-of-the-art deep learning models can be trained with only a few lines of codes, for which we direct the user to the full documentation of glycowork.
Conclusion
As the most diverse biological sequence, glycans require dedicated analyses. Until now, technical limitations have directed the focus of researchers to rather short and/or uniform glycans that are amenable to manual analysis, such as N-glycans or short O-glycans. Yet with the addition of more (complex) sequences (Malaker et al. 2021), and the combination of glycans with systems biology data (Kearney et al. 2021), manual analysis is becoming increasingly unrealistic. We envision that accessible analysis platforms such as glycowork will allow researchers to connect knowledge from different glycobiology areas to fuel discoveries and extend the scope of already known phenomena.
We are planning to improve glycowork in future work by expanding its functionalities. This includes implementations of more existing glycobioinformatics techniques, such as the Multiple Carbohydrate Alignment with Weights tool (Hosoda et al. 2017). We will also update the stored datasets in glycowork as new glycans become available, to maximize the utility of sequence context analysis, database queries and others.
We encourage interested readers to find more details and examples in the documentation of glycowork (https://bojarlab.github.io/glycowork/). We also would like to invite the community to suggest—or even implement—changes, improvements or additions, to maximize the utility of glycowork for glycobioinformatics and allow researchers to include glycan data analysis into their routine workflows.
Authors’ contributions
Conceptualization: D.B., Data Curation: L.T., R.B., D.B., Funding Acquisition: D.B., Investigation: L.T., D.B., Resources: D.B., Software: L.T., R.B., D.B., Supervision: D.B., Visualization: L.T., D.B., Writing—Original Draft Preparation: L.T., D.B., Writing—Review & Editing: L.T., R.B., D.B.
Supplementary Material
Acknowledgements
The authors would like to thank Frédérique Lisacek and Benjamin P. Kellman for helpful discussions.
Contributor Information
Luc Thomès, Department of Chemistry and Molecular Biology and Wallenberg Centre for Molecular and Translational Medicine, University of Gothenburg, 41390 Gothenburg, Sweden.
Rebekka Burkholz, Department of Biostatistics, Harvard School of Public Health, Boston, 02115 MA, USA.
Daniel Bojar, Department of Chemistry and Molecular Biology and Wallenberg Centre for Molecular and Translational Medicine, University of Gothenburg, 41390 Gothenburg, Sweden.
Funding
Branco Weiss Fellowship – Society in Science awarded to D.B.; Knut and Alice Wallenberg Foundation; University of Gothenburg, Sweden.
Conflict of interest statement
The authors declare no competing interests.
Data availability statement
All used code and data can be found at https://github.com/BojarLab/glycowork/
References
- Aoki-Kinoshita KF, Lisacek F, Mazumder R, York WS, Packer NH. 2020. The GlySpace alliance: Toward a collaborative global glycoinformatics community. Glycobiology. 30:70–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojar D, Powers RK, Camacho DM, Collins JJ. 2021. Deep-learning resources for studying glycan-mediated host-microbe interactions. Cell Host Microbe. 29:132–144.e3. [DOI] [PubMed] [Google Scholar]
- Burkholz R, Quackenbush J, Bojar D. 2021. Using graph convolutional neural networks to learn a representation for glycans. Cell Rep. 35:109251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang H-Y, Hofree M, Ideker T. 2010. A decade of systems biology. Annu Rev Cell Dev Biol. 26:721–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. 2009. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coff L, Chan J, Ramsland PA, Guy AJ. 2020. Identifying glycan motifs using a novel subtree mining approach. BMC Bioinformatics. 21:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings RD, Pierce JM. 2014. The challenge and promise of glycomics. Chem Biol. 21:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant OC, Xue X, Ra D, Khatamian A, Foley BL, Woods RJ. 2016. Gly-Spec: a webtool for predicting glycan specificity by integrating glycan array screening data and 3D structure. Glycobiology. 26:1027–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haab BB, Klamer Z. 2020. Advances in tools to determine the glycan-binding specificities of lectins and antibodies. Mol Cell Proteomics. 19:224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg AA, Schult DA, Swart PJ. 2008. Exploring network structure, dynamics, and function using NetworkX. Proceedings of the 7th Python in Science Conference (SciPy2008). Pasadena, CA, USA. 11–5.
- Herget S, Ranzinger R, Maass K, Lieth CW. 2008. GlycoCT—a unifying sequence format for carbohydrates. Carbohydr Res. 343:2162–2171. [DOI] [PubMed] [Google Scholar]
- Hosoda M, Akune Y, Aoki-Kinoshita KF. 2017. Development and application of an algorithm to compute weighted multiple glycan alignments. Bioinformatics. 33:1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-F, Aoki K, Akase S, Ishihara M, Liu YS, Yang G, Kizuka Y, Mizumoto S, Tiemeyer M, Gao XD, et al. 2021. Global mapping of glycosylation pathways in human-derived cells. Dev Cell. 56:1195–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 12:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney CJ, Vervoort SJ, Ramsbottom KM, Todorovski I, Lelliott EJ, Zethoven M, Pijpers L, Martin BP, Semple T, Martelotto L, et al. 2021. SUGAR-seq enables simultaneous detection of glycans, epitopes, and the transcriptome in single cells. Sci Adv. 7:eabe3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Zaia J. 2019. glypy: An open source glycoinformatics library. J Proteome Res. 18:3532–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaker SA, Riley NM, Shon DJ, Pedram K, Krishnan V, Dorigo O, Bertozzi CR. 2021. Revealing the human mucinome. bioRxiv. doi: 10.1101/2021.01.27.428510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyelaran O, Gildersleeve JC. 2009. Glycan arrays: recent advances and future challenges. Curr Opin Chem Biol. 13:406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JP, Mahal LK. 2013. Dot by dot: analyzing the glycome using lectin microarrays. Curr Opin Chem Biol. 17:827–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF, Cummings RD. 2014. Investigating virus–glycan interactions using glycan microarrays. Curr Opin Virol. 7:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Aoki-Kinoshita KF, Kotera M, Sawaki H, Tsuchiya S, Fujita N, Shikanai T, Kato M, Kawano S, Yamada I, et al. 2014. WURCS: The Web3 Unique Representation of Carbohydrate Structures. J Chem Inf Model. 54:1558–1566. [DOI] [PubMed] [Google Scholar]
- Varki A. 2017. Biological roles of glycans. Glycobiology. 27:3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan K, Chandrasekaran A, Srinivasan A, Raman R, Sasisekharan S, Sasisekharan R. 2010. Glycans as receptors for influenza pathogenesis. Glycoconj J. 27:561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Laubichler MD. 2018. From systems to biology: A computational analysis of the research articles on systems biology from 1992 to 2013. PLoS One. 13:e0200929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All used code and data can be found at https://github.com/BojarLab/glycowork/


