Abstract
Kinase suppressor of Ras (KSR) is an evolutionarily conserved component of Ras-dependent signaling pathways. Here, we report the identification of B-KSR1, a novel splice variant of murine KSR1 that is highly expressed in brain-derived tissues. B-KSR1 protein is detectable in mouse brain throughout embryogenesis, is most abundant in adult forebrain neurons, and is complexed with activated mitogen-activated protein kinase (MAPK) and MEK in brain tissues. Expression of B-KSR1 in PC12 cells resulted in accelerated nerve growth factor (NGF)-induced neuronal differentiation and detectable epidermal growth factor (EGF)-induced neurite outgrowth. Sustained MAPK activity was observed in cells stimulated with either NGF or EGF, and all effects on neurite outgrowth could be blocked by the MEK inhibitor PD98059. In B-KSR1-expressing cells, the MAPK–B-KSR1 interaction was inducible and correlated with MAPK activation, while the MEK–B-KSR1 interaction was constitutive. Further examination of the MEK–B-KSR1 interaction revealed that all genetically identified loss-of-function mutations in the catalytic domain severely diminished MEK binding. Moreover, B-KSR1 mutants defective in MEK binding were unable to augment neurite outgrowth. Together, these findings demonstrate the functional importance of MEK binding and indicate that B-KSR1 may function to transduce Ras-dependent signals that are required for neuronal differentiation or that are involved in the normal functioning of the mature central nervous system.
Cellular proliferation and differentiation must be precisely controlled for the proper development, growth, and homeostasis of a multicellular organism. One protein that plays a pivotal role in regulating these processes is the Ras GTPase. In response to a diverse array of extracellular signals, Ras is converted from its inactive GDP-bound form to its active GTP-bound form. Activated Ras then interacts directly with a specific set of effector molecules to achieve transmission, amplification, and integration of these signals (for reviews see references 18, 19, and 30). Through genetic and biochemical studies, numerous proteins functioning downstream of Ras have been identified. These proteins include Ral-specific guanine nucleotide exchange factors, phosphatidylinositol-3 phosphate kinase, Akt kinase, Raf kinases, MEK, mitogen-activated protein kinase (MAPK), and kinase suppressor of Ras (KSR) (for reviews see references 7, 13, and 29). While much is known regarding the function of many of these molecules, the role that KSR plays in the transmission of Ras-dependent signals is poorly understood.
KSR constitutes a novel protein family that is related to, but distinct from, the Raf kinase family (16, 25, 26). KSR proteins are found in Drosophila, Caenorhabditis elegans, and mammals but not yeast. Members of the KSR family contain five conserved protein domains (CA1 to 5 [26]). CA1 is a 40-amino-acid domain unique to the KSR proteins, CA2 is a proline-rich domain, CA3 is a cysteine-rich domain, CA4 is a serine-threonine-rich domain, and CA5 constitutes the catalytic domain. CA1 to 4 are located in the amino-terminal region of the protein, whereas CA5 is found in the carboxy-terminal region. Surprisingly, while the KSR proteins are predicted to be protein kinases, no physiological substrates of KSR have been identified, nor has KSR been conclusively demonstrated to possess intrinsic kinase activity (6, 20, 24, 31, 33, 34).
KSR was first identified to be a positive effector of Ras signaling by genetic studies performed in Drosophila and C. elegans (16, 25, 26). Evaluating the contribution of mammalian KSR to Ras signaling, however, has been more difficult since experiments addressing KSR function in mammalian cells have yielded conflicting results. In some reports, expression of murine KSR1 enhanced the biological activity of activated Ras by accelerating the activation of MEK and MAPK (20, 27, 31). In contrast, other studies found that KSR1 expression inhibited Ras signaling by either blocking MEK and MAPK activation (6, 14, 33) or inhibiting Elk-1 phosphorylation (24). The discrepancy in these findings appears to be due to the level of KSR protein expressed. For example, in Xenopus oocytes, KSR1 functioned as a positive regulator of Ras signaling when expressed at low levels, whereas at high levels of expression, KSR1 blocked Ras-mediated signal transduction (3). Likewise, even though KSR is required for Ras-dependent R7 photoreceptor formation in Drosophila (26), overexpression of Drosophila melanogaster KSR1 (DmKSR1) in the fly eye can block R7 formation (3). Thus, the biological function of KSR as a positive effector of Ras signaling appears to be dependent on maintaining KSR protein expression at low or near physiological levels.
A model for how KSR might influence Ras signaling has emerged from the findings that in mammalian cells murine KSR1 interacts with numerous cellular proteins and translocates from the cytosol to the plasma membrane in response to Ras activation (20, 23, 31). Therefore, it has been proposed that KSR may function as a scaffolding protein to coordinate the assembly of a signaling complex. Proteins reported to associate with KSR1 include 14-3-3 (3, 23, 31), p50cdc37 (23), hsp90 (23), G-protein βγ (2), Raf-1 (27), MEK (3, 6, 23, 33), and MAPK (3, 33). The interactions between KSR1 and 14-3-3, p50cdc37, hsp90, and MEK appear to be constitutive, while the associations with G-protein βγ, MAPK, and Raf-1 are induced upon Ras activation. In addition, the binding of 14-3-3, p50cdc37, hsp90, G-protein βγ, MEK, and MAPK is direct, while the interaction with Raf-1 appears to be indirect, mediated perhaps through MEK or 14-3-3. The binding sites on KSR1 for these associated molecules have been localized to two phosphorylated serine residues (Ser297 and Ser392) for 14-3-3 (3), the CA3 domain for G-protein βγ (2), an FXFP motif in the CA4 domain for MAPK (12), and the CA5 catalytic domain for p50cdc37, hsp90, and MEK (23, 33).
To date, much of our knowledge regarding mammalian KSR has been obtained from the analysis of proliferating cells that overexpress exogenous KSR1. Therefore, to address whether these studies accurately reflect the true biological role of mammalian KSR, we initiated experiments to examine the properties of endogenous KSR1. From these studies, we have identified a novel splice variant of murine KSR1 that is highly expressed in brain-derived tissues, B-KSR1. Experiments characterizing the B-KSR1 isoform reveal that B-KSR1 is in a complex with MEK and MAPK under physiological conditions and that the interaction with MEK is a critical aspect of B-KSR1 function. Our findings further indicate that KSR proteins may function in Ras signaling pathways that are distinct from those involved in cellular proliferation. In particular, B-KSR1 may play a critical role in transducing Ras-dependent signals that are required for promoting or maintaining neuronal differentiation or that are involved in the normal functioning of the mature central nervous system.
MATERIALS AND METHODS
Northern blot analysis.
Northern blot analysis was performed by standard procedures using poly(A)-selected RNA from adult mouse tissues and a 32P-labeled probe corresponding to the amino-terminal domain of murine KSR1 (amino acid residues 1 to 539 [27]). RNA transcripts hybridizing to the probe were visualized by autoradiography.
Isolation of B-KSR1 cDNA.
A DNA fragment corresponding to the amino-terminal domain of murine KSR1 (amino acid residues 1 to 539 [27]) was used as a probe to screen a mouse brain cDNA library (Stratagene). Three full-length and four partial cDNA clones were isolated from 106 PFU screened. By sequence analysis, all seven clones encoded an alternatively spliced variant of murine KSR1 named B-KSR1.
Generation of DNA constructs and recombinant adenoviruses.
B-KSR1 deletion and point mutant constructs were generated by standard procedures (22) using the appropriate oligonucleotides and the murine B-KSR1 cDNA clone. The full-length wild-type (WT) construct (B-KSR1/WT) encodes amino acid residues 1 to 863, the N-terminal domain construct encodes residues 1 to 553, and the C-terminal catalytic domain construct encodes residues 541 to 863. All B-KSR1 clones were constructed to contain two copies of a polyomavirus-derived epitope tag (Pyo; amino acids MEYMPME) at the amino terminus. DNA fragments containing the WT and mutant KSR1 sequences were isolated and inserted into pcDNA3 (Invitrogen) for expression in mammalian cells. Sequences encoding the catalytic domain of B-KSR1 were inserted into the pAdTrack-CMV vector for the generation of recombinant adenoviruses according to the procedures of He et al. (11).
Antibodies and immunohistochemical staining.
KSR1 antibodies used in this study include a goat polyclonal antibody generated against the carboxy terminus (residues 855 to 871) of murine KSR1 (Santa Cruz Biotechnology), a rabbit polyclonal antibody directed against the carboxy terminus of B-KSR1, and a rat monoclonal antibody and a rabbit polyclonal antibody generated against a glutathione S-transferase fragment of murine KSR1 encoding amino acid residues 118 to 248 (3). 14-3-3 and MAPK antibodies were from Santa Cruz Biotechnology, MEK antibody was from Transduction Laboratories, and antibodies specific for phosphorylated MAPK (P-MAPK) were from New England Biolabs. For immunohistochemical analysis of adult mouse brain, 30-μm-thick sagittal sections were cut from paraformaldehyde-fixed tissues and processed free floating in antibody directed against B-KSR1. Bound immunoglobulin was visualized with the avidin-biotin-peroxidase method using 3,3-diaminobenzidine tetrahydrochloride as the chromogen.
Cell transfection and isolation of B-KSR1-expressing cell lines.
Plasmid DNAs (5 μg) were transfected into 293 and PC12 cells by using Lipofectamine. 293 cells were analyzed 40 to 48 h following transfection. Transfected PC12 cells were selected in medium containing G418, and drug-resistant lines were established. All cell lines were screened for the presence of Pyo-tagged B-KSR1 by immunoblot analysis.
Neurite outgrowth assays.
PC12 cells were plated on collagen-coated plates and cultured overnight in Dulbecco's modified Eagle's medium containing 10% horse serum and 5% calf serum. Culture media was then removed and medium containing 1% horse serum, 0.5% calf serum, and either nerve growth factor (NGF; 50 ng/ml) or epidermal growth factor (EGF; 100 ng/ml) was added. Cells were examined for neurite outgrowth as previously described (8).
ATP binding assays.
Transfected 293 cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride, 20 μM leupeptin, 0.15 U of aprotinin per ml, 5 mM sodium vanadate), and the B-KSR1 catalytic domain proteins were immunoprecipitated as previously described (27). The complexes were washed once with RIPA buffer, three times with NP-40 buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1% NP-40, 1 mM PMSF, 20 μM leupeptin, 0.15 U of aprotinin per ml, 5 mM sodium vanadate), and once with phosphate-buffered saline (PBS). Modification of proteins with 5′-p-fluorosulfonylbenzoyl adenosine (FSBA) and detection of reaction products were accomplished by using an ATP binding protein detection kit from Boehringer Mannheim. Briefly, proteins were reacted with 1.3 mM FSBA in PBS–10% dimethyl sulfoxide for 20 min at 30°C. In some experiments, MgATP (10 mM) was included during the incubation to competitively block the interaction of FSBA with ATP binding sites. The reaction was terminated by addition of 4× gel loading buffer (200 mM Tris [pH 6.8], 8% SDS, 400 mM dithiothreitol, 40% glycerol, 0.1% bromophenol blue). The samples were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and the modified proteins detected by immunoblotting using the FSBA antibody.
In vitro protein kinase assays.
B-KSR1 and Raf-1 proteins were immunoprecipitated from lysates of transfected 293 cells. The immunoprecipitated proteins were washed once with RIPA buffer followed by three washes with NP-40 buffer and one wash with kinase buffer (30 mM HEPES [pH 7.4], 7 mM MgCl2, 7 mM MnCl2, 1 mM dithiothreitol, 1 μM ATP). The complexes were then incubated in 40 μl of kinase buffer containing 20 μCi of [γ-32P]ATP and 1 μg of the indicated substrate. After incubation for 20 min at 25°C, the assays were terminated by the addition of 4× gel loading buffer. The samples were then resolved by SDS-PAGE, and the phosphoproteins were visualized by autoradiography.
RESULTS
Identification of B-KSR1, a novel splice variant of murine KSR1.
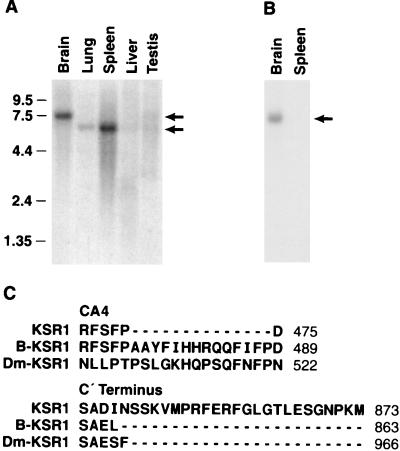
To further characterize mammalian KSR, we examined the distribution of KSR1 mRNA in various murine tissues by Northern blot analysis. When RNA samples were hybridized with a probe corresponding to the amino-terminal domain of murine KSR1 (27), a 6.4-kb band that correlated with the predicted size of the KSR1 transcript was detected in lung, spleen, and testis (Fig. 1A). In addition, a strongly reactive 7.4-kb band was observed in brain and a similar-sized weakly reactive band was found in testis. Alternatively spliced transcripts of numerous genes, such as c-src and B-raf (1, 17), have been found in neuronal tissues. Therefore, to determine whether the 7.4-kb band in brain represented a novel KSR transcript, an adult mouse brain cDNA library was screened for the presence of KSR1-reactive clones. Seven full-length or partial cDNA clones were isolated, all of which encoded a novel splice variant of the previously reported murine KSR1 (26). Sequence analysis revealed that the B-KSR1 cDNA contained two additional exons not found in KSR1 (Fig. 1C). The first exon is located in the CA4 domain and encodes a 14-amino-acid insert. The second exon is located at the protein's carboxy terminus and encodes two amino acids, a stop codon (resulting in the removal of the last 24 amino acids of KSR1) and an additional 1 kb of 3′ untranslated sequences. When RNA samples were hybridized with a probe corresponding to the 3′ untranslated sequences unique to B-KSR1, only a 7.4-kb band was detected in brain (Fig. 1B), confirming that the larger KSR1-reactive transcript identified in brain is B-KSR1. Interestingly, the features of the B-KSR1 isoform, i.e., extension of the CA4 domain and truncation of the carboxy terminus, are also observed in the Drosophila and C. elegans KSR1 proteins.
FIG. 1.
Identification of the B-KSR1 isoform. (A) A Northern blot containing RNA isolated from various mouse tissues was hybridized with a probe corresponding to the amino-terminal region of murine KSR1. Size markers are indicated in kilobases. (B) As for panel A except that the blot was hybridized with a probe corresponding to a 1-kb fragment of 3′ untranslated sequence unique to B-KSR1. (C) The CA4 domain and C terminus are two regions of B-KSR1 affected by alternative splicing. The amino acid sequence of these two regions in B-KSR1 is aligned to the corresponding regions of KSR1 and DmKSR1.
B-KSR1 is highly expressed in brain-derived tissue.
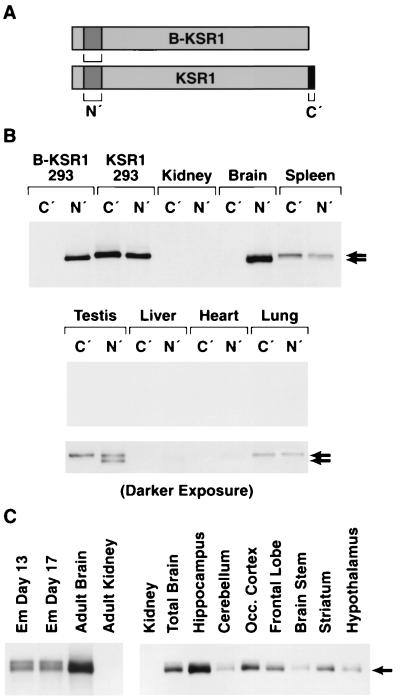
To examine the in vivo distribution of the KSR1 and B-KSR1 proteins, lysates were prepared from various mouse tissues, equalized for protein content, and then incubated with either of two KSR antibodies. The first antibody (αC′) recognizes the carboxy terminus of KSR1 and should interact only with KSR1, whereas the second antibody (αN′) recognizes an amino-terminal KSR1 epitope and should interact with both KSR1 and B-KSR1 (Fig. 2A). As predicted, in control experiments using lysates from 293 cells expressing either KSR1 or B-KSR1, the αN′ reacted with both KSR1 and B-KSR1, while αC′ interacted only with KSR1 (Fig. 2B). When immunoprecipitates from mouse tissues were examined by immunoblot analysis, KSR proteins were readily detectable in brain and spleen (Fig. 2B). A longer exposure of the blot revealed KSR proteins in testis and lung; however, little to no protein was detected in kidney, liver, and heart (Fig. 2B). The highest overall expression was observed in brain, with B-KSR1 being the sole isoform expressed. Spleen and lung expressed the KSR1 isoform, while testis had equivalent expression of both isoforms (note that B-KSR1 migrates slightly faster than KSR1). Examination of various tissue culture lines revealed that NIH 3T3 and BALB/c 3T3 fibroblast cells expressed low levels of KSR1, whereas PC12 pheochromocytoma cells contained low levels of B-KSR1 (data not shown).
FIG. 2.
Expression of B-KSR1 and KSR1 proteins in various mouse tissues. (A) Immunoprecipitation assays were performed using two antibodies, αN′ (N′) and αC′ (C′). The presence of these epitopes on B-KSR1 and KSR1 is depicted. (B) Lysates prepared from 293 cells expressing either B-KSR1 or KSR1 and lysates from various adult mouse tissues were equalized for protein content and immunoprecipitated with either αN′ or αC′. The immunoprecipitates were then examined by immunoblot analysis using αN′. Short and long exposures are shown. (C) Kidney and brain tissue from adult mice, brain tissue from mice at embryonic (Em) days 13 and 17, and tissues from specialized brain regions were isolated, and lysates were prepared. The lysates were equalized for protein content and immunoprecipitated with αN′. The immunoprecipitates were then examined by immunoblot analysis using an antibody recognizing KSR.
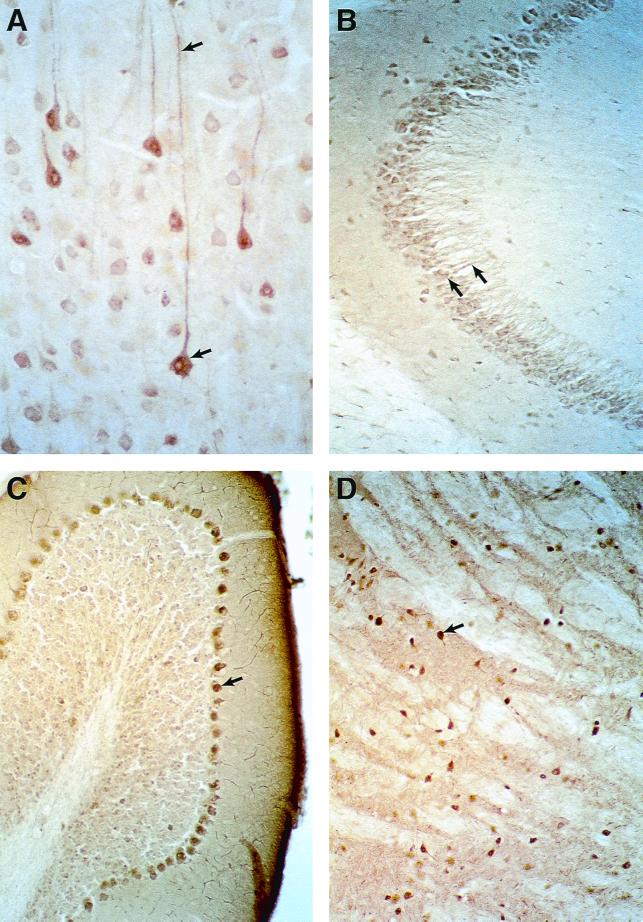
To further characterize the expression of B-KSR1 in neuronal tissues, lysates from mouse brains collected at various developmental time points were examined. As shown in Fig. 2C, B-KSR1 was detected at embryonic day 13, one of the earliest times when brain-specific tissue can be isolated, and at embryonic day 17. The protein expression level was equivalent at both embryonic times but increased in the adult brain. To determine whether B-KSR1 expression is restricted to a specific brain structure, lysates from isolated brain regions were examined. Although B-KSR1 was present in all samples, the highest level of expression was detected in hippocampus (Fig. 2C). Significant expression was also observed in striatum, occular cortex, and frontal lobe. To confirm these findings and to further localize the expression of B-KSR1 within these tissues, immunohistochemical staining was performed. Examination of sagittal brain sections revealed that B-KSR1 was widely distributed in adult forebrain neurons. Strong staining was observed in layer V pyramidal neurons of the neocortex (Fig. 3A), CA3 pyramidal neurons of the hippocampus (Fig. 3B), and neurons of the caudate putamen and globus pallidus (Fig. 3C). Intense staining of Purkinje cells was detected in the cerebellum; however, the granule cell and molecular layers were relatively devoid of B-KSR1 immunoreactivity (Fig. 3D). Neuronal staining was predominantly cytoplasmic and could be detected throughout the dendritic processes (Fig. 3A and B).
FIG. 3.
Expression of B-KSR1 in mouse brain. (A) Bright-field photomicrograph of B-KSR1 immunoreactive neurons in layer V of the somatosensory cortex. Note the presence of staining throughout the long dendritic processes of these neurons. (B) Distinct cellular and dendritic staining is also observed in pyramidal neurons of the hippocampus, shown here in the mossy fiber termination zone arising from CA3 neurons. (C) B-KSR1-positive neurons in the globus pallidus. Abundant staining of neurons was also observed in the caudate putamen (not shown). (D) Sections through the cerebellum reveal intense staining of Purkinje cells.
B-KSR1 is associated with 14-3-3, MEK, and activated MAPK in brain tissues.
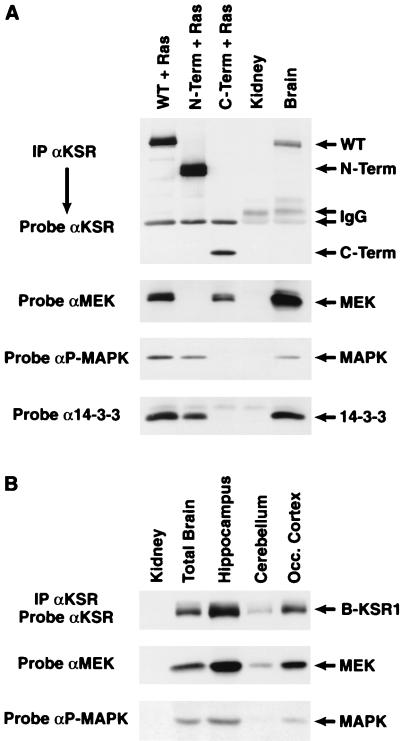
Since KSR1 has been reported to interact with numerous signaling molecules, we next examined whether B-KSR1 also exists as part of a multiprotein complex. B-KSR1 proteins were immunoprecipitated from lysates of 293 cells expressing activated Ras and either full-length B-KSR1, the isolated B-KSR1 amino-terminal domain, or the isolated B-KSR1 carboxy-terminal catalytic domain. The B-KSR1 proteins were expressed in the presence of activated Ras in order to detect both constitutive and Ras-dependent interactions. The immunoprecipitates were then examined for the presence of associated, endogenous 14-3-3, MEK, and activated MAPK. Consistent with the findings reported for KSR1 (3, 33), 14-3-3 and activated MAPK interacted with the amino-terminal domain of B-KSR1, while MEK associated with the catalytic domain (Fig. 4A). To determine whether these interactions occur under physiological conditions, B-KSR1 immunoprecipitates prepared from mouse brain lysates were examined. As shown in Fig. 4A, we found that endogenous 14-3-3, MEK, and activated MAPK all associated with endogenous B-KSR1. Moreover, in comparison to transfected 293 cells, an even greater interaction with MEK was observed in the brain sample. The complex formation between B-KSR1, MEK, and activated MAPK was also detected in immunoprecipitates prepared from lysates of embryonic brain (data not shown) and specific brain regions (Fig. 4B), further indicating the biological relevance of these interactions.
FIG. 4.
B-KSR1 associates with 14-3-3, MEK, and activated MAPK in vivo. (A) B-KSR1 proteins were immunoprecipitated (IP) from lysates of 293 cells coexpressing activated RasV12 and either full-length B-KSR1 (WT) or the isolated amino-terminal domain (N-Term) or isolated catalytic domain (C-Term) of B-KSR1, using a Pyo antibody. In addition, B-KSR1 was immunoprecipitated from lysates of adult mouse brain and kidney tissues using a KSR1 antibody (αKSR). The immunoprecipitates were then examined by immunoblot analysis using KSR, MEK, P-MAPK, and 14-3-3 antibodies to detect endogenous proteins interacting with B-KSR1. (B) Lysates from specialized brain regions were analyzed as for panel A and examined by immunoblot analysis with KSR, MEK, and P-MAPK antibodies.
Expression of B-KSR1 in PC12 cells augments neuronal differentiation.
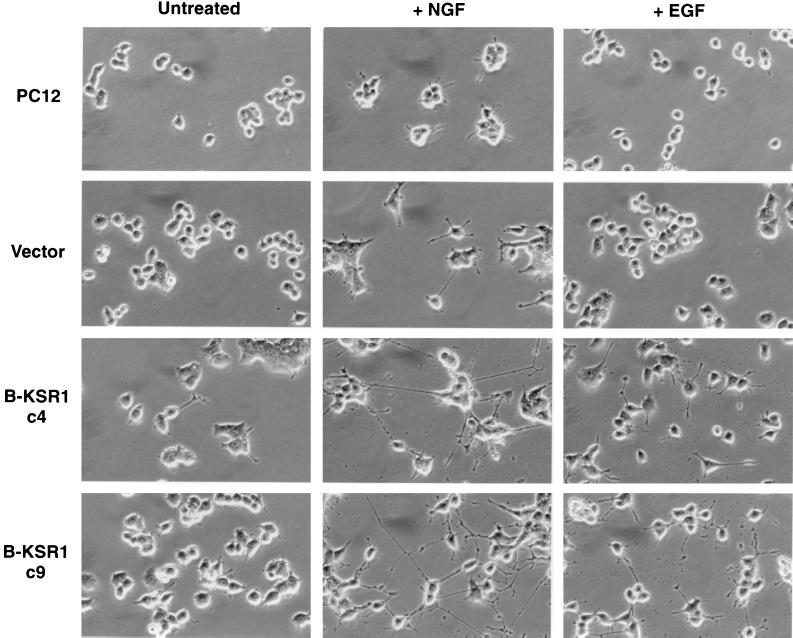
The high expression of B-KSR1 in nonproliferating neural tissues suggests that B-KSR1 may function in promoting or maintaining a differentiated phenotype. To examine this possibility, we generated PC12 cell lines that stably express epitope-tagged B-KSR1. PC12 cells can be induced to differentiate into sympathetic neuron-like cells when treated with NGF and thereby provide a useful model system for studying the signaling pathways involved in differentiation. Five cell lines that stably expressed B-KSR1 three- to sevenfold over endogenous levels and three vector-transfected lines were isolated and used for this analysis. Interestingly, we have been unable to establish cell lines that express B-KSR1 more than sevenfold over endogenous levels and, despite several attempts, have failed to generate stable lines that express either KSR1 or the isolated catalytic domain of B-KSR1. When cellular growth rates were examined, we found that all of the isolated B-KSR1 lines had a doubling time (average of 37.5 h) that was similar to that of the parental (38.5 h) and vector-transfected (37 h) cells. However, in comparison to parental or vector-transfected cells, the B-KSR1 lines displayed a more flattened phenotype in the unstimulated state and showed an accelerated induction of neurite outgrowth in response to NGF (Fig. 5). By 2 days of NGF treatment, the neurite projections extending from the B-KSR1 cells had well-defined growth cones and were three to four times the diameter of the cell body. In contrast, the neurite projections in vector-transfected cells were approximately one cell body length. Surprisingly, when the B-KSR1 cell lines were treated with EGF, a mitogenic factor that does not normally promote PC12 differentiation, detectable neurite outgrowth was also observed (Fig. 5).
FIG. 5.
Neurite outgrowth in B-KSR1 PC12 cell lines. Parental, vector-transfected, and B-KSR1-expressing (c4 and c9) PC12 cell lines were left untreated or stimulated with NGF (50 ng/ml) or EGF (100 ng/ml). Cells were photographed 2 days after treatment. The experiment was repeated twice with similar results.
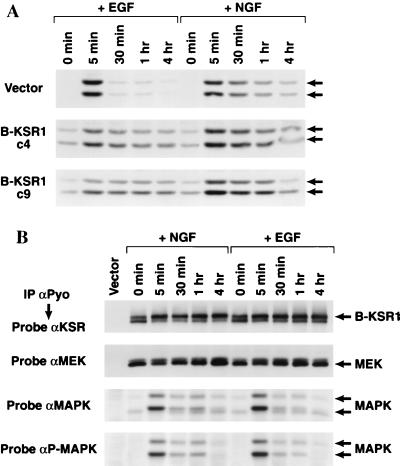
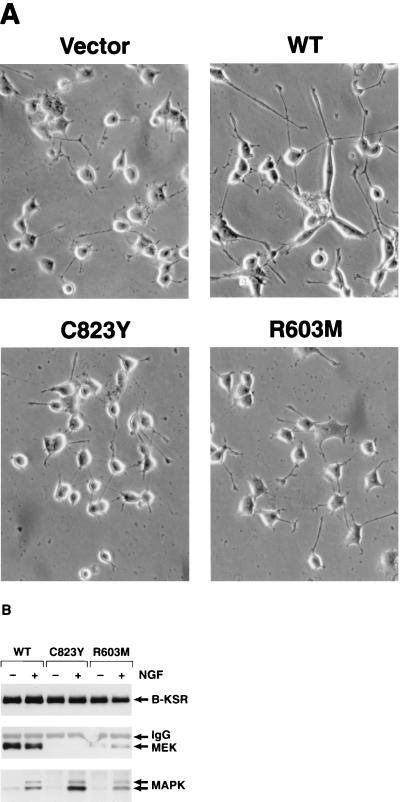
Since PC12 differentiation often correlates with sustained MAPK activity (5, 28), we next examined whether MAPK activation was altered in the B-KSR1 cell lines. Lysates were prepared from serum-starved cells that had been treated with NGF or EGF for various time periods. The activation state of MAPK was then determined by probing the lysates with an antibody that specifically recognizes activated (phosphorylated) MAPK. In comparison to vector-transfected cells, the B-KSR1 cell lines contained a higher basal level of activated MAPK (Erk1 and Erk2) and exhibited sustained MAPK activation in response to both NGF and EGF stimulation (Fig. 6A). In vector-transfected cells, sustained MAPK activation was observed only in response to NGF treatment.
FIG. 6.
(A) Activation of MAPK in B-KSR1 PC12 cell lines. Vector-transfected and B-KSR1 PC12 cell lines (c4 and c9) were serum starved for 24 h and then stimulated for the indicated times with either EGF or NGF. Cell lysates were prepared and examined by immunoblot analysis using P-MAPK antibodies. (B) Association of B-KSR1 with MEK and MAPK in B-KSR1 PC12 cells. B-KSR1-expressing PC12 cells (c4) were serum starved for 24 h and then stimulated for the indicated times with either NGF (50 ng/ml) or EGF (100 ng/ml). Lysates were prepared and immunoprecipitated (IP) using a Pyo antibody (αPyo). The immunoprecipitates were then examined by immunoblot analysis using KSR, MEK, MAPK, and P-MAPK antibodies.
We next sought to determine the effect of NGF and EGF treatment on the kinetics of complex formation between B-KSR1, MEK, and MAPK. Serum-starved B-KSR1 cells were left untreated or treated with NGF or EGF for various time periods. B-KSR1 immunoprecipitates prepared from the lysed cells were then examined for the presence of MEK, MAPK, and P-MAPK. As shown in Fig. 6B, both NGF and EGF treatments induce a shift in the electrophoretic mobility of B-KSR1 that correlates with the hyperphosphorylation of B-KSR1 (data not shown). When the B-KSR1 immunoprecipitates were examined for the presence of MAPK, we found that the association of MAPK was significantly enhanced by treatment with either NGF or EGF. The interaction between B-KSR1 and MAPK was observed in untreated cells, peaked at 5 min in response to both factors, and was still detected by 4 h after treatment. In contrast, equivalent amounts of MEK were detected in all B-KSR1 immunoprecipitates. Therefore, while the association between B-KSR1 and MAPK is inducible, the B-KSR1–MEK interaction is constitutive.
MEK activity is required for the augmenting effect of B-KSR1 on neuronal differentiation.
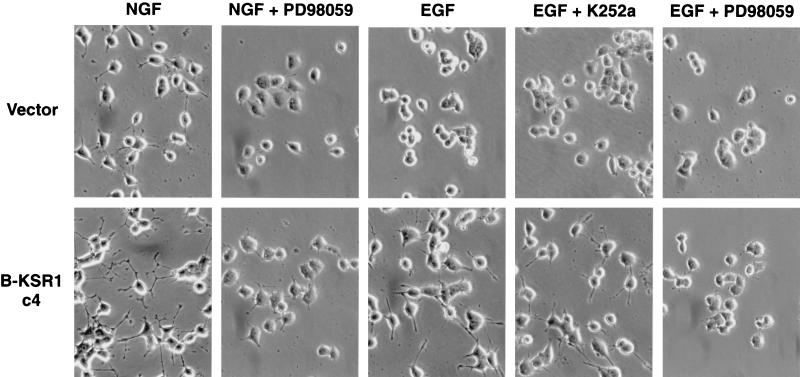
To further characterize the B-KSR1 PC12 cell lines, we used two pharmacological inhibitors, the Trk-NGF receptor inhibitor K252a and the MEK inhibitor PD98059. K252a was used to examine whether neurite outgrowth induced by EGF treatment was due to the autocrine activation of the NGF receptor. While pretreatment with K252a did inhibit NGF-induced differentiation of vector-transfected and B-KSR1-expressing cells (data not shown), it did not block EGF-induced differentiation of B-KSR1 cells (Fig. 7), indicating that the observed effect was not mediated by the NGF receptor. Next, the MEK inhibitor PD98059 was used to evaluate the importance of MEK activity for the accelerated neurite outgrowth observed in the B-KSR1 cells. When cells were pretreated with PD98059, NGF- and EGF-induced neurite outgrowth was completely blocked. These findings demonstrate that the stimulatory effect of B-KSR1 requires MEK activity and further indicate that B-KSR1 is not activating an alternative pathway that results in augmented neuronal differentiation and sustained MAPK activation.
FIG. 7.
Neuronal differentiation of B-KSR1 PC12 cells requires MEK-MAPK activity. Vector-transfected and B-KSR1-expressing PC12 cells were left untreated or treated for 3 h with the MEK inhibitor PD98059 or with the Trk-NGF receptor inhibitor K252a. Cells were then stimulated with NGF (50 ng/ml) or EGF (100 ng/ml) and photographed 2 days after treatment. The experiment was repeated twice with similar results.
LOF mutations in the B-KSR1 catalytic domain reduce MEK binding.
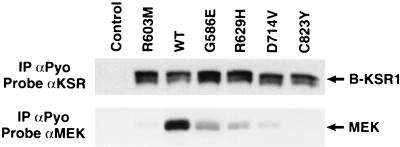
Because the MEK–B-KSR1 interaction is constitutive and MEK activity is required for the stimulatory effect of B-KSR1 in PC12 cells, we initiated experiments to evaluate the importance of MEK binding to B-KSR1 function. Previously, it has been shown that many of the mutations that inhibit Drosophila, C. elegans, and murine KSR1 function (loss-of-function [LOF] mutations) occur in the KSR catalytic domain (16, 25–27), and as shown in Fig. 4, MEK interacts with the catalytic domain of B-KSR1. Therefore, we examined the effect of various catalytic domain LOF mutations on MEK binding. The LOF mutations that were incorporated into B-KSR1 include G586E, R629H, C823Y (three mutations originally identified in C. elegans), R603M (a mutation in the kinase subdomain II that would be expected to abolish catalytic activity), and D714V (a mutation in the conserved kinase domain DFG motif). The B-KSR1 mutant proteins were then transiently transfected into 293 cells and examined for the ability to interact with MEK. Compared to wild-type B-KSR1/WT, immunoprecipitates of B-KSR1 proteins containing the R603M, G586E, R629H, and D714V mutations all exhibited reduced levels of MEK (Fig. 8). In addition, no MEK binding was detected in B-KSR1/C823Y immunoprecipitates. Thus, all of the conserved LOF mutations inhibit MEK binding. Interestingly, while these experiments were ongoing, Stewart et al. (23) reported that MEK binding was reduced when the KSR1 protein was mutated at sites analogous to the R603M, R629H, and C823Y mutations examined here. Together these findings indicate the importance of MEK binding and suggest that the biochemical basis of the LOF phenotype may be due to the disruption of the MEK-KSR complex.
FIG. 8.
LOF mutations in the B-KSR1 catalytic domain severely impair MEK binding. Lysates were prepared from control 293 cells and 293 cells expressing B-KSR1/WT or B-KSR1 proteins containing the R603M, G586E, R629H, D714V, or C823Y mutation. B-KSR1 proteins were immunoprecipitated (IP) with a Pyo-specific antibody (αPyo) and examined by immunoblot analysis using KSR and MEK antibodies.
MEK binding is required for B-KSR1 function in PC12 cells.
To more directly evaluate the contribution of MEK binding to B-KSR1 function, we examined the effect of two B-KSR1 LOF mutants on PC12 differentiation. Pooled populations of G418-resistant cells were established that had been transfected with constructs encoding B-KSR1/WT, B-KSR1/R603M, or B-KSR1/C823Y. As had been observed with the clonal cell lines, the pooled cell lines expressing B-KSR1/WT exhibited accelerated neurite outgrowth in response to NGF treatment (Fig. 9A). However, enhanced neuronal differentiation was not seen in cell populations expressing either the R603M or C823Y LOF mutants. In addition, these mutants failed to induce neurite outgrowth in response to EGF (data not shown). When complex formation between B-KSR1, MEK, and MAPK was examined, we found that the MEK–B-KSR1 interaction was greatly reduced in cells expressing B-KSR1/K603M and was completely abolished in cells expressing B-KSR1/C823Y (Fig. 9B). In contrast, the NGF-induced association with MAPK was detected in all cell lines, and an enhanced interaction was observed between MAPK and B-KSR1/C823Y. Therefore, while the ligand-induced interaction between MAPK and B-KSR1 is not dependent on MEK binding, the interaction with MEK is required for the augmenting effect of B-KSR1 on PC12 differentiation.
FIG. 9.
The augmenting effect of B-KSR1 on PC12 differentiation correlates with MEK binding. (A) G418-resistant pooled populations of vector-transfected and WT, R603M, or C823Y B-KSR1-expressing cells were left untreated or stimulated with NGF (50 ng/ml). Cells were photographed 2 days after treatment. The experiment was repeated twice with similar results. (B) WT, R603M, or C823Y B-KSR1-expressing cells were serum starved for 24 h and then stimulated for 5 min with NGF (50 ng/ml). Lysates were prepared and immunoprecipitated with a Pyo antibody. The immunoprecipitates were then examined by immunoblot analysis using KSR, MEK, and MAPK antibodies.
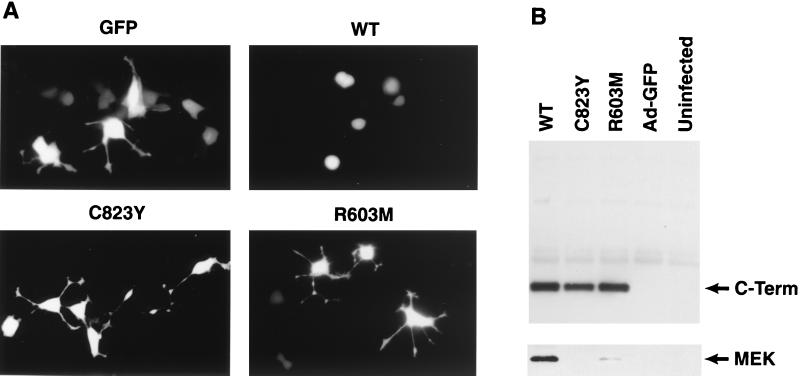
To further analyze the functional significance of the MEK–B-KSR1 interaction, we performed experiments utilizing the isolated catalytic domain of B-KSR1. Previously, it was shown that expression of the isolated DmKSR1 catalytic domain could block R7 photoreceptor formation in the Drosophila eye (27). In addition, the KSR1 catalytic domain can inhibit Ras signaling when expressed in NIH 3T3 cells or in Xenopus oocytes (3, 27, 33). To determine whether the B-KSR1 catalytic domain also functions as a dominant inhibitory protein and to examine whether MEK binding is required for this effect, we generated recombinant adenoviruses expressing the B-KSR1/WT catalytic domain and the B-KSR1 catalytic domain containing either the R603M or C823Y LOF mutation. PC12 cells were infected with the recombinant adenoviruses, stimulated with NGF, and then monitored for neuronal differentiation. In comparison to control adenovirus-infected cells, neurite outgrowth was blocked in cells expressing the WT catalytic domain but was not affected in cells expressing R603M or C823Y catalytic domain proteins (Fig. 10A). All of the B-KSR1 catalytic domain proteins were expressed to equivalent levels in the infected PC12 cells; however, only the WT catalytic domain showed a significant interaction with MEK (Fig. 10B). Thus, the dominant inhibitory activity of the B-KSR1 catalytic domain correlates with MEK binding.
FIG. 10.
Expression of the isolated B-KSR1 catalytic domain blocks neuronal differentiation in PC12 cells. PC12 cells were infected with recombinant adenoviruses expressing green fluorescent protein (Ad-GFP) or coexpressing GFP and the WT, R603M, or C823Y B-KSR1 catalytic domain proteins (C-Term). (A) Cells were treated with NGF (50 ng/ml) 18 h after infection and were photographed 2 days after NGF treatment. (B) The B-KSR1 catalytic domain proteins were immunoprecipitated from lysates of uninfected and adenovirus-infected PC12 cells. The immunoprecipitates were then examined by immunoblot analysis using Pyo and MEK antibodies. The experiment was repeated twice with similar results.
Examination of the kinase properties of B-KSR1.
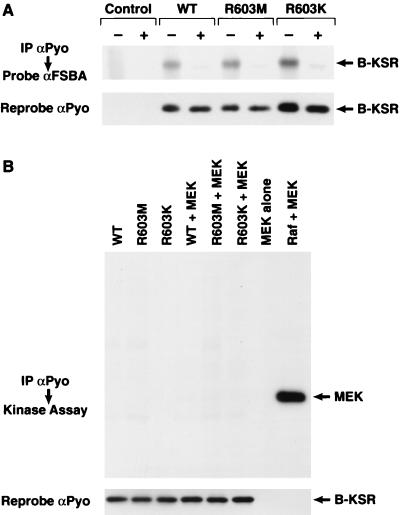
In view of the finding that the biological activity of B-KSR1 correlates with MEK binding, the question arises as to whether B-KSR1 functions as a protein kinase. In addition, sequence analysis reveals that B-KSR1 contains an arginine residue at a position in the kinase subdomain II that is invariantly occupied by a lysine residue in all other functionally active kinases. Therefore, to further evaluate the kinase properties of B-KSR1, we first examined whether B-KSR1 could bind ATP. For this analysis, B-KSR1 catalytic domain proteins that had been coexpressed with activated Ras in 293 cells were immunoprecipitated and incubated with FSBA. The proteins covalently modified by FSBA were then detected by immunoblot analysis using an FSBA antibody. As shown in Fig. 11A, the WT protein was recognized by the FSBA antibody, indicating that it was capable of binding ATP. A mutant protein that contained a methionine residue at amino acid position 603 (a mutation that would be expected to inactivate catalytic activity) also bound ATP, as did a mutant containing a lysine at position 603 (a mutation that would be expected to restore kinase activity). Since B-KSR1 was able to bind ATP, we next examined whether B-KSR1 could autophosphorylate or phosphorylate any of the proteins with which it is known to interact. For this analysis, in vitro kinase assays were performed with wild-type B-KSR1, as well as the kinase-inactive R603M and kinase-restored R603K mutants. As shown in Fig. 11B, none of the B-KSR1 proteins were able to autophosphorylate or to phosphorylate MEK. In comparison, MEK was efficiently phosphorylated by purified Raf-1. By similar analysis, we found that B-KSR1 did not phosphorylate inactive Raf-1 and partially activated Raf-1, MAPK, Hsp90, and 14-3-3 (data not shown), indicating that these proteins are not substrates for B-KSR1.
FIG. 11.
Examination of the kinase properties of B-KSR1. (A) ATP binding assay. Lysates were prepared from 293 cells coexpressing activated Ras and either the WT, R603M, or R603K B-KSR1 catalytic domain protein. The B-KSR1 proteins were immunoprecipitated (IP) from cell lysates using a Pyo antibody (αPyo) and reacted with FSBA in the absence (−) or presence (+) of MgATP. The modified proteins were then analyzed by immunoblotting with antibodies specific to FSBA and Pyotag (αFSBA and αPyo). (B) Immune complex kinase assays. 293 cells were infected with the recombinant adenoviruses, and B-KSR1 was immunoprecipitated from cell lysates with a Pyo antibody. Kinase assays were then performed using kinase-inactive MEK as a substrate or without substrate to analyze the autophosphorylation of B-KSR1. Control reactions include incubating MEK alone or in the presence of purified, activated Raf-1. The immune complexes were subsequently examined by immunoblot analysis using the Pyo antibody to verify uniform expression of the B-KSR1 proteins.
DISCUSSION
To further elucidate the biological function of KSR, we have examined the endogenous expression pattern and physiological properties of mammalian KSR. From these experiments we identified B-KSR1, a novel splice variant of murine KSR1. The existence of the B-KSR1 isoform was first indicated by Northern blot experiments and subsequently confirmed by the cloning of B-KSR1 from a mouse brain cDNA library. Sequence analysis of the B-KSR1 clone revealed that it contains two additional exons not present in KSR1, one located in the serine-threonine-rich CA4 domain and one found at the protein's carboxy terminus. The B-KSR1 isoform appears to also be present in human brain tissue, since a partial KSR clone isolated from a human brain-derived cDNA library contains the characteristic features of the B-KSR1 splice variant (26). Interestingly, the changes resulting from the alternative splicing of the B-KSR1 isoform, i.e., extension of the CA4 domain and truncation of the carboxy terminus, are also observed in the Drosophila and C. elegans KSR1 proteins. Thus, B-KSR1 is the isoform with the highest degree of homology to Drosophila and C. elegans KSR1.
Analysis of the tissue distribution of the KSR proteins revealed that the expression pattern of KSR1 and B-KSR1 is restricted. The most abundant expression was observed in brain, with B-KSR1 being the sole isoform expressed. This observation is supported by the fact that no KSR1-encoding cDNAs were isolated from the mouse brain library. Significant levels of KSR protein were also detected in spleen; however, KSR1 was the sole isoform expressed in this tissue. Interestingly, both isoforms were expressed at low levels in testis, consistent with the presence of both the 6.4- and 7.4-kb KSR transcripts in testicular RNA samples. Little or no protein expression was observed in other tissue samples. Likewise, we have found that KSR protein expression is low in most tissue culture lines, including NIH 3T3, 293, and PC12 cells. In brain-derived tissue, B-KSR1 protein was detectable at embryonic day 13, one of the earliest times when brain tissue can be isolated. However, the most abundant expression of B-KSR1 was found in the adult hippocampus. Immunohistochemical staining revealed that B-KSR1 was widely distributed in forebrain neurons of the adult mouse a finding that further emphasizes the tissue-specific expression of the B-KSR1 isoform.
Previous studies in mammalian cells have suggested that KSR may function, in part, as a scaffolding protein. Indeed, KSR1 has been found to associate with numerous cellular proteins; however, most of these interactions have been detected in cells overexpressing exogenous KSR1. In this study, we provide the first conclusive evidence that a KSR protein does exist as part of a multiprotein complex under physiological conditions. Furthermore, our findings indicate that complex formation with MEK and MAPK is a critical aspect of B-KSR1 function. MEK and activated MAPK were associated with B-KSR1 at all developmental time points and in all brain structures expressing B-KSR1. Consistent with the findings reported for KSR1, MAPK interacted with the amino terminus of B-KSR1, while MEK associated with the catalytic domain. For KSR1, an FXFP motif in the CA4 domain has been identified as the binding site for MAPK (12). Interestingly, as a result of the CA4 domain insert, B-KSR1 contains an additional MAPK binding site not present in KSR1. Therefore, B-KSR1 provides two docking sites for MAPK, which, in turn, would increase the efficiency by which MAPK is localized to the B-KSR1 signaling complex.
In experiments designed to further address the functional importance of the MEK–B-KSR1 interaction, we found that all LOF mutations in the B-KSR1 catalytic domain severely reduce MEK binding. Moreover, B-KSR1 mutants defective in MEK binding were unable to augment neurite outgrowth in PC12 cells. Thus, complex formation with MEK appears to be required for KSR function, raising the question of whether KSR is indeed a protein kinase. To date, a substrate for KSR1 has not been identified, nor has KSR1 been demonstrated to have intrinsic kinase activity. Likewise, we have been unable to detect any enzymatic activity intrinsic to B-KSR1. In immune complex kinase assays, B-KSR1 did not autophosphorylate, nor did it phosphorylate Raf-1, MEK, MAPK, hsp90, or 14-3-3, all of which interact or would be expected to interact with B-KSR1. The KSR family does possess many of the structural characteristics of protein kinases (9); however, a peculiarity of the mammalian KSR proteins is that they contain an arginine residue at a position in the ATP binding domain that is normally occupied by a lysine residue (26). This conserved lysine residue is thought to be involved in the transfer of phosphate from ATP to the substrate (15), and for all kinases analyzed to date, mutation of this residue inactivates the enzyme (9). Even the conservative change of lysine to arginine has been found to inactivate the protein-tyrosine kinase p60src (15), as well as the serine-threonine kinase v-mos (10). To possibly restore kinase activity, we mutated the arginine residue of B-KSR1 to a lysine and still were unable to detect any catalytic activity intrinsic to B-KSR1. In light of these observations, it is interesting to speculate that even the function of the Drosophila and C. elegans KSR proteins, which contain a lysine in the ATP binding domain, may be accomplished by mechanisms other than phosphorylation. Alternatively, it is possible that the mammalian KSR proteins have accumulated mutations in addition to the lysine exchange that abolish catalytic activity. Finally, however, we cannot exclude the possibility that mammalian KSR proteins do function as kinases and that phosphorylation of a yet unidentified KSR substrate may be required for KSR function and/or MEK binding.
Previous studies have shown that KSR1 translocates from the cytoplasm to the plasma membrane upon Ras activation (20, 23, 31). Thus, we predict, as others have for KSR1 (23, 33), that an important function of the full-length B-KSR1 protein may be to bind MEK and shuttle it from the cytosol to the plasma membrane, where it can be phosphorylated by activated, membrane-associated Raf-1. In this case, mutations in the KSR catalytic domain disrupting the MEK-KSR interaction would lead to the formation of nonfunctional complexes lacking MEK. The studies presented here also reveal that the interaction with MEK is required for the dominant inhibitory activity of the isolated catalytic domain of B-KSR1. As has been observed for the isolated catalytic domains of KSR1 and DmKSR1, we found that the isolated catalytic domain of B-KSR1 blocked the transmission of Ras-dependent signals. Since the B-KSR1–MEK interaction is constitutive, deletion of the B-KSR1 amino terminus might result in the sequestering of MEK in the cytosol, thereby disrupting signal propagation from Ras and Raf-1 to MEK and MAPK. Therefore, as has been observed for Ste5 in yeast (4, 21), B-KSR1 appears to play an important role in coordinating the interaction between components of the MAPK module.
To date, most studies evaluating mammalian KSR function have been performed under mitogenic conditions and in proliferating cells. However, KSR was originally identified in genetic screens examining the perturbation of differentiative pathways such as R7 formation in Drosophila and vulva development in C. elegans. Moreover, we find that the highest level of KSR protein expression is observed in nonproliferating neural tissues. Thus, the biological function of B-KSR1 may be required for promoting or maintaining a differentiated phenotype. Indeed, we find that NGF-induced neurite outgrowth was significantly more rapid in PC12 cells stably expressing B-KSR1 than in vector-transfected or parental PC12 cells. Surprisingly, treatment with EGF, a proliferative factor that does not normally induce neuronal differentiation, also resulted in detectable neurite outgrowth in the B-KSR1 lines. The EGF-induced neurite outgrowth was not due to the autocrine activation of the NGF receptor since the Trk-NGF receptor inhibitor K252a did not block neurite formation.
Neuronal differentiation of PC12 cells often requires the sustained activation of MAPK. Under normal conditions, sustained MAPK activity is induced by NGF treatment but not by EGF stimulation. However, EGF has been shown to promote sustained MAPK activation and neuronal differentiation in PC12 cells overexpressing the EGF receptor (28). In this case, it is thought that differentiation occurs because MAPK activity has been elevated above a critical level for a sufficient time period, thus allowing MAPK to enter the nucleus and regulate transcription factors required for differentiation. Based on the following observations, we predict that the effect of B-KSR1 on PC12 differentiation is also mediated by the MAPK pathway. First, both NGF- and EGF-induced differentiation of the B-KSR1 cell lines is blocked by the MEK inhibitor PD98059. Second, B-KSR1 is complexed with MEK and activated MAPK in these cells. Third, the ability of B-KSR1 to augment neurite outgrowth correlates with MEK binding. Fourth, in B-KSR1-expressing cells, the activity of MAPK is elevated in unstimulated cells and is sustained in response to both NGF and EGF treatment. Thus, overexpression of B-KSR1 appears to influence the kinetics of MAPK activation in PC12 cells. If B-KSR1 is limiting in PC12 cells, additional B-KSR1–MEK complexes might accelerate and extend the activation of MAPK such that the critical threshold of MAPK activity is reached in EGF-treated cells and is achieved more rapidly in cells stimulated with NGF. B-KSR1 may also modulate the activity of another protein involved in differentiation and/or MAPK activation. Rap1 is a protein that has been reported to promote the sustained activation of MAPK activity in PC12 cells (32). The effect of Rap1 is thought to be mediated by the B-Raf kinase, which is an activator of the MAPK pathway. In our studies, however, we have not detected a significant interaction between B-KSR1 and B-Raf in either PC-12 cells or brain tissues, nor have we been able to demonstrate a conclusive effect of B-KSR1 overexpression on Rap1 activity (data not shown). Therefore, further experiments are required to determine the precise mechanism by which B-KSR1 mediates its effect on MAPK activation and PC12 differentiation.
In conclusion, the high levels of B-KSR1 protein in fully differentiated neurons, together with the complex formation of B-KSR1 with MEK and MAPK in these cells indicates that B-KSR1 may function in Ras signaling pathways that are distinct from those involved in cellular proliferation. In particular, B-KSR1 may play a critical role in transducing Ras-dependent signals that are required for promoting or maintaining neuronal differentiation or that may be involved in the normal functioning of the mature central nervous system. Thus, the intriguing possibility exists that B-KSR1 may participate in the regulation of critical cellular events occurring in the adult brain, including synaptic neurotransmission and neuronal plasticity required for learning and memory.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We gratefully acknowledge Margaret Ashcroft for advice and helpful comments throughout this project; Marc Therrien for guidance in cloning B-KSR1; Dan Court for excellent technical assistance; and Monica Murakami, Peter Johnson, and Vaughn Cleghon for critical reading of the manuscript.
This work was supported in part by the National Cancer Institute, DHHS, under contract with ABL.
REFERENCES
- 1.Barnier J V, Papin C, Eychene A, Lecoq O, Calothy G. The mouse B-raf gene encodes multiple protein isoforms with tissue specific expression. J Biol Chem. 1995;270:23381–23389. doi: 10.1074/jbc.270.40.23381. [DOI] [PubMed] [Google Scholar]
- 2.Bell B, Xing H, Yan H, Gautam N, Muslin A J. KSR1 binds to G-protein βγ subunits and inhibits βγ-induced mitogen-activated protein kinase activation. J Biol Chem. 1999;274:7982–7986. doi: 10.1074/jbc.274.12.7982. [DOI] [PubMed] [Google Scholar]
- 3.Cacace A M, Michaud N R, Therrien M, Mathes K, Copeland T, Rubin G M, Morrison D K. Identification of constitutive and Ras-inducible phosphorylation sites of KSR: implication for 14-3-3 bindings, mitogen-activated protein kinase binding, and KSR overexpression. Mol Cell Biol. 1999;19:229–240. doi: 10.1128/mcb.19.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi K Y, Satterberg B, Lyons D M, Elion E A. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 5.Cowley S, Paterson H, Kemp P, Marshall C J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 6.Denouel-Galy A, Douville E M, Warne P H, Papin C, Laugier D, Calothy G, Downward J, Eychene A. Murine Ksr interacts with MEK and inhibits Ras-induced transformation. Curr Biol. 1997;8:46–55. doi: 10.1016/s0960-9822(98)70019-3. [DOI] [PubMed] [Google Scholar]
- 7.Downward J. Ras signalling and apoptosis. Curr Opin Genet Dev. 1998;8:49–54. doi: 10.1016/s0959-437x(98)80061-0. [DOI] [PubMed] [Google Scholar]
- 8.Greene L A, Sobeih M M, Teng K K. Methodologies for the culture and experimental use of the PC12 rat pheochromocytoma cell line. Cambridge, Mass: MIT Press; 1991. [Google Scholar]
- 9.Hanks S, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 10.Hannink M, Donoghue D. Lysine residue 121 in the proposed ATP-binding site of the v-mos protein is required for transformation. Proc Natl Acad Sci USA. 1985;82:7894–7898. doi: 10.1073/pnas.82.23.7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He T-C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs D, Glossip D, Xing H, Muslin A J, Kornfeld K. Multiple docking sites on substrate proteins forms a modular system that mediates the recognition by ERK MAP kinase. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- 13.Joneson T, Bar-Sargi D. Ras effectors and their role in mitogenesis and oncogenesis. J Mol Med. 1997;75:587–583. doi: 10.1007/s001090050143. [DOI] [PubMed] [Google Scholar]
- 14.Joneson T, Fulton J A, Volle D J, Chaika O V, Bar-Sagi D, Lewis R E. Kinase suppressor of Ras inhibits the activation of extracellular ligand-regulated (ERK) mitogen-activated protein (MAP) kinase by growth factors, activated Ras, and Ras effectors. J Biol Chem. 1998;273:7743–7748. doi: 10.1074/jbc.273.13.7743. [DOI] [PubMed] [Google Scholar]
- 15.Kamps M, Sefton B. Neither arginine nor histidine can carry out the function of lysine-295 in the ATP-binding site of p60src. Mol Cell Biol. 1986;6:751–757. doi: 10.1128/mcb.6.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kornfeld K, Hom D B, Horvitz H R. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell. 1995;83:903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- 17.Levy J B, Dorai T, Wang L H, Brugge J S. The structurally distinct form of pp60csrc detected in neuronal cells is encoded by a unique c-src mRNA. Mol Cell Biol. 1987;7:4142–4245. doi: 10.1128/mcb.7.11.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall C J. Ras effectors. Curr Opin Cell Biol. 1996;2:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- 19.McCormick F. Activators and effectors of ras p21 proteins. Curr Opin Genet Dev. 1994;4:71–76. doi: 10.1016/0959-437x(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 20.Michaud N R, Therrien M, Cacace A, Edsall L C, Spiegel S, Rubin G M, Morrison D K. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc Natl Acad Sci USA. 1997;94:12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Stewart S, Sundaram M, Zhang Y, Lee J, Han M, Guan K-L. Kinase suppressor of Ras forms a multiprotein signaling complex and modulates MEK localization. Mol Cell Biol. 1999;19:5523–5534. doi: 10.1128/mcb.19.8.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugimoto T, Stewart S, Han M, Guan K-L. The kinase suppressor of Ras (KSR) modulates growth factor and Ras signaling by uncoupling elk-1 phosphorylation from MAP kinase activation. EMBO J. 1997;17:1717–1727. doi: 10.1093/emboj/17.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundaram M, Han M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995;83:889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- 26.Therrien M, Chang H C, Solomon N M, Karim F D, Wassarman D A, Rubin G M. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 27.Therrien M, Michaud N R, Rubin G M, Morrison D K. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- 28.Traverse S, Seedorf K, Paterson H, Marshall C J, Cohen P, Ullrich A. EGF triggers neuronal differentiation of PC12 cells that overexpress the EGF receptor. Curr Biol. 1994;4:694–701. doi: 10.1016/s0960-9822(00)00154-8. [DOI] [PubMed] [Google Scholar]
- 29.Wassarman D, Therrien M, Rubin G M. The Ras signaling pathway in Drosophila. Curr Opin Gen Dev. 1995;5:44–50. doi: 10.1016/s0959-437x(95)90052-7. [DOI] [PubMed] [Google Scholar]
- 30.Wittinghofer A. Signal transduction via Ras. Biol Chem. 1998;379:933–937. [PubMed] [Google Scholar]
- 31.Xing H, Kornfeld K, Muslin A J. The protein kinase KSR interacts with 14-3-3 protein and Raf. Curr Biol. 1997;7:294–300. doi: 10.1016/s0960-9822(06)00152-7. [DOI] [PubMed] [Google Scholar]
- 32.York R D, Yao H, Dillon T, Ellig C L, Eckert S P, McCleskey E W, Stork P J S. Rap1 mediates sustained MAP kinase activation induced by nerve growth factor. Nature. 1998;392:622–626. doi: 10.1038/33451. [DOI] [PubMed] [Google Scholar]
- 33.Yu W, Fantl W J, Harrowe G, Williams L T. Regulation of the MAP kinase pathway by mammalian Ksr through direct interaction with MEK and ERK. Curr Biol. 1997;8:56–64. doi: 10.1016/s0960-9822(98)70020-x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Yao B, Delikat S, Bayoumy S, Lin X-H, Basu S, McGinley M, Chan-Hui P-Y, Lichenstein H, Kolesnick R. Kinase suppressor of Ras is ceramide-activated protein kinase. Cell. 1997;89:63–72. doi: 10.1016/s0092-8674(00)80183-x. [DOI] [PubMed] [Google Scholar]