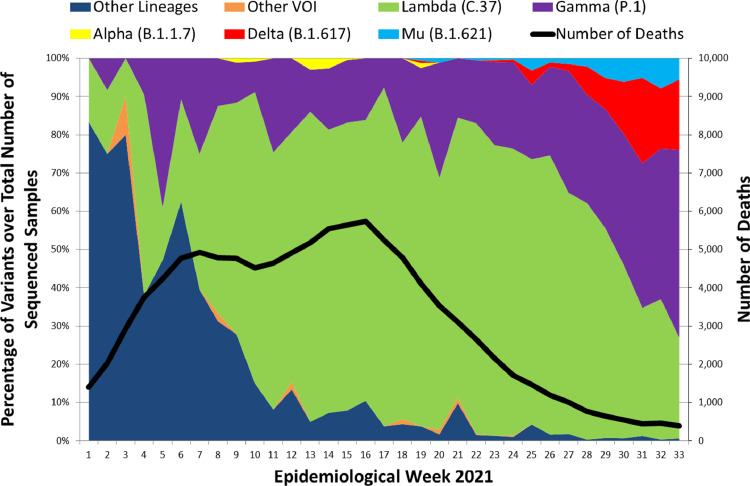
Persistent ongoing transmission of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) at a global level has led to mutations in its genetic code that confer differentiated behaviors from the original Wuhan virus, giving rise to novel variants.1 One of them is Lambda (C.37), designated as a variant of interest (VOI) on June 15, 2021 by the World Health Organization (WHO). First identified in Peru in December 2020, Lambda became the predominant variant in the Coastal and Andean regions of Peru during the first half of 2021, surpassing for several months other circulating variants of concern (VOC) such as Gamma (P.1), Alfa (B.1.1.7) and currently Delta (B.1.617.2).2 Only the north Amazon area of Peru was dominated by Gamma throughout its second wave. Taking on account the country as a whole, Lambda showed a steep increase during March, reaching a peak between April and June, then decreasing by epidemiological week 31 when it was surpassed in proportion of cases by the Gamma variant (Fig. 1). These trends were recognized via genomic surveillance. In Peru, 382 samples are analysed per week, mostly randomly selected samples from hospitalized and ambulatory patients across all Peruvian Regions, using the Illumina COVIDSEQ platform.
Fig. 1.
Percentages of SARS-CoV-2 variants over total number of samples sequenced by Epidemiological Week in Peru, January–August 2021.
(Source: Instituto Nacional de Salud – Perú. Secuenciación Genómica del virus SARS-CoV-2 en el Perú. https://web.ins.gob.pe/es/covid19/secuenciamiento-sars-cov2 / Ministerio de Salud - Perú. Sala de Situación COVID-19. http://www.dge.gob.pe/covid19.html).
Given the rapid spread of Gamma in Brazil´s Amazonas State,3 it was remarkable that this VOC did not become dominant throughout the whole Peru during the first six months of the year. One possibility is that the Lambda variant held off its expansion; initially due to “founder effect” (having been detected one month earlier) and later due to some selection advantages.4 Lambda mutations, as T76I and L452Q located in the spike protein, could be responsible for higher infectivity rates; while F490S, RSYLTPGD246-253N, and particularly L452Q, very similar to the Delta variant mutation L452R, would give Lambda the property to evade the immune system.5 Acevedo et al. also found in its Lambda pseudovirus trial a decrease in the antibodies produced by the CoronaVac vaccine by a factor or 3.05, compared to Gamma and Alfa variants.6 These features could explain the expansion and predominance of Lambda in Peru during the coronavirus disease (COVID-19) second wave, despite the presence of the other highly transmissible variants.
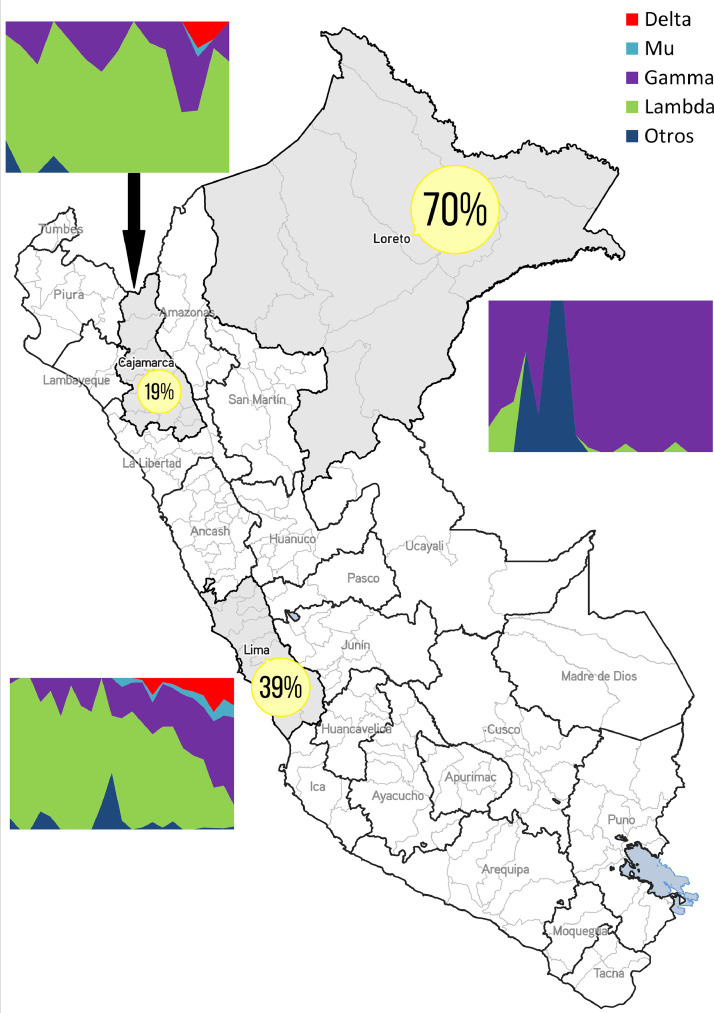
In contrast, Gamma was always dominant in the north Amazon basin adjacent to the Brazilian border (Loreto Region). Beside its geographic location, this region had the highest SARS-CoV-2 seroprevalence in Peru right after the devastating first COVID-19 epidemic wave, with a staggering 70%.7 Likewise, Gamma has recently become predominant in Peru largest conurbation (Lima and Callao), a region that experienced a seroprevalence increase from 39% in December 2020 to around 61% in July 2021 (personal communication, P. Pachas, Ministerio de Salud–Peru, in July 2021). Thus, it is possible that any Lambda advantage over Gamma variant is reduced in settings where a very high proportion of people have developed antibodies. Noteworthy, the emergence of the Gamma variant in Manaus also occurred in the context of a very high seroprevalence, suggesting that this VOC could evade immunity caused by non-Gamma previous infections, even in highly exposed populations.8 To illustrate these differences, we are showing three regions with contrasting seroprevalences at the end of 2020, and how the variants distribution has unfolded divergently over time (Fig. 2). In addition, according to the Global Initiative on Sharing All Influenza Data (GISAID) database (https://www.gisaid.org), Lambda has shown less variability with only one sub-lineage (C.37.1 reported in Europe), while Gamma presents 10 recent sub-lineages carrying spike protein mutations, 3 of which are circulating in Peru (P.1.1, P.1.4 and P.1.7).
Fig. 2.
Map of Peru showing COVID-19 seroprevalence during the last months of 2020, and their variants distributions March-August 2021 for three Regions (Lima, Loreto, Cajamarca)
(Source: Ministerio de Salud - Perú. Estudio Incidencia, prevalencia y factores de riesgo para la infección por virus SARS-CoV-2, estudio poblacional en el Perú, 2020–2021 - EpiCovid – Perú. https://www.dge.gob.pe/portalnuevo/wp-content/uploads/2020/12/Prevalencia-Lima-y-Callao.pdf / Instituto Nacional de Salud – Perú. Secuenciación Genómica del virus SARS-CoV-2 en el Perú. https://web.ins.gob.pe/es/covid19/secuenciamiento-sars-cov2).
The simultaneous coexistence of highly transmissible variants could lead to an evolutionary competition phenomenon, displaying complex epidemiological dynamics. Some variants have mutations that enhance its infectious capability; while the advantage of others resides not on their fitness but in their immune evasion competency like Gamma.9 The latter could be more advantageous than sheer infectivity in settings where a high proportion of people have been previously infected or vaccinated. Therefore, the dominance of one variant over others would be not only based on their inherent genomic characteristics, but also on epidemiological features that could produce a more “advantageous” environment for a given strain. However, with the recent entry of Delta and Mu (B.1.621) variants in Peru, most probably these will displace the still predominant Gamma and Lambda variants in the coming months.
In Peru, the introduction of the Delta variant was faced employing epidemiological fences, contact tracing, and greatly enhancing vaccination in the Region where it was first detected. In fact, full vaccination coverage was only 6.0% when the Delta variant was identified in June, and four months later is 48.4%.10 The importance of genomic surveillance for detecting and characterizing SARS-CoV-2 variants, should always be highlighted, and the need to complement it with epidemiological, clinical and serological data in order to respond adequately.
Declaration of Competing Interest
None.
Role of the funding source
Authors declare there is no funding source for this study.
Footnotes
Editor note: The Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
References
- 1.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Padilla-Rojas C., Jimenez-Vasquez V., Hurtado V., Mestanza O., Molina I.S., Barcena L., et al. Genomic analysis reveals a rapid spread and predominance of Lambda (C.37) SARS-COV-2 lineage in Peru despite circulation of variants of concern. J Med Virol. 2021:1–5. doi: 10.1002/jmv.27261. Aug 9Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D da S., Mishra S., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372(6544):815–821. doi: 10.1126/science.abh2644. May 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2—what do they mean? JAMA. 2021;325(6):529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- 5.Kimura I., Kosugi Y., Wu J., Yamasoba D., Butlertanaka E.P., Tanaka Y.L., et al. SARS-CoV-2 Lambda variant exhibits higher infectivity and immune resistance bioRxiv 2021.07.28.454085; doi: 10.1101/2021.07.28.454085. [DOI] [PMC free article] [PubMed]
- 6.Acevedo M.L., Alonso-Palomares L., Bustamante A., Gaggero A., Paredes F., Cortés C.P., et al. Infectivity and immune escape of the new SARS-CoV-2 variant of interest Lambda. medRxiv 2021.06.28.21259673; doi: 10.1101/2021.06.28.21259673. [DOI]
- 7.Álvarez-Antonio C., Meza-Sánchez G., Calampa C., Casanova W., Carey C., Alava F., et al. Seroprevalence of anti-SARS-CoV-2 antibodies in Iquitos, Peru in July and August 2020: a population-based study. Lancet Glob Health. 2021;9(7):e925–e931. doi: 10.1016/S2214-109X(21)00173-X. July. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romano C.M., Felix A.C., Paula de A.V., Jesus de J.G., Andrade P.S., Cândido D., et al. SARS-CoV-2 reinfection caused by the P.1 lineage in Araraquara city, Sao Paulo State, Brazil. Rev Inst Med Trop São Paulo. 2021;63:e36. doi: 10.1590/S1678-9946202163036. Apr 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plante J.A., Mitchell B.M., Plante K.S., Debbink K., Weaver S.C., Menachery V.D. The variant gambit: cOVID-19′s next move. Cell Host Microbe. 2021;29(4):508–515. doi: 10.1016/j.chom.2021.02.020. Apr 14Epub 2021 Mar 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Repositorio Único Nacional de Informacion en Salud – Ministerio de Salud – Peru. Cobertura de vacunación contra la COVID-19. Available from: https://www.minsa.gob.pe/reunis/data/vacunas-covid19.asp.