Abstract
Cytomegalovirus (CMV) is the most common cause of congenital infection in the developed world. We have designed and evaluated an assay that includes an internal control for amplification and detection of CMV DNA in amniotic fluid and neonatal urine samples. We present data on the use of this assay in the diagnosis of congenital CMV infection. A total of 145 amniotic and fetal fluid samples were examined by this assay; 83 were from healthy pregnant women and 62 were from women who were being investigated because of concerns over the pregnancy (diagnostic group). CMV DNA was detected in three amniotic fluid samples from the diagnostic group but was not detected in any samples taken from healthy pregnant women. Thirty-nine urine samples were obtained from 19 neonates with suspected congenital infection; CMV DNA was detected in urine from 6 of these patients. The assay provides useful information about CMV infection in the fetus and the neonate; when used in conjunction with other diagnostic tools it will enable mothers and obstetricians to make informed decisions about the management of pregnancies complicated by CMV infection.
Cytomegalovirus (CMV) is the most common cause of congenital infection in the developed world. In the United States approximately 1% of all neonates excrete CMV (4); of these 10% will be severely affected with a range of symptoms, the most common of which are intrauterine growth retardation, thrombocytopenia, microcephaly, and neurological deficit such as developmental delay and intracranial calcification (4). In the United Kingdom 400 CMV-affected neonates are born annually (9). Of the 90% of these babies who are asymptomatic at birth, approximately 15% will develop a significant handicap such as deafness or neurological problems (9, 26). The diagnosis and management of cytomegalovirus infection in pregnancy pose a number of problems. Primary infection is more likely to result in fetal damage than reactivation of latent infection or reinfection (1, 4, 7), but affected babies born to immune mothers have been described (10). During pregnancy transmission to the fetus occurs in 40% of cases of primary maternal infection (4). Among women in whom the virus reactivates during pregnancy, the transmission rate to the fetus is less than 2% (26). Diagnosis is complicated, as serological evidence of active infection in the mother provides no information about whether the baby has been infected and, if the baby is infected, whether that child will develop severe disease or problems in later life. Consequently, appropriate management of active CMV infection during pregnancy is unclear, and often, insufficient information is available about the risk to the fetus to enable the mother to make an informed decision.
We have developed an assay that detects the presence of CMV DNA in amniotic fluid and urine. Amniotic fluid is largely derived from the fetus and has been shown to be a reliable sample for use in the diagnosis of viral infections in utero (5, 12, 13, 16). The results of investigation of amniotic fluid, when used in conjunction with other investigations such as serological confirmation of active maternal CMV infection and detailed fetal ultrasonography, will enable specific and accurate diagnosis of fetal infection and help inform clinicians and parents of the prognosis for the baby.
MATERIALS AND METHODS
Patients and samples.
Amniotic fluid was obtained from two separate groups of women: a control cohort of healthy pregnant women who had elected to have an amniocentesis because of concern about maternal age and a group of women who were under investigation during their pregnancy for reasons such as detection of an abnormality on ultrasound scan, a history of maternal illness, or abnormal results for a sample subjected to biochemical screening (referred to here as the diagnostic group). Ethical approval for this study was granted by the Central Public Health Laboratory Service Ethics Committee and South Glamorgan Health Authority Local Ethical Committee. Healthy control patients were counseled and signed a consent form agreeing to the use of surplus amniotic fluid for study by molecular methods for detection of CMV. Culture of amniotic fluid was not included in the consent form for the healthy women. The diagnostic group of patients was counseled and gave consent for an amniocentesis so that investigations could be undertaken to enable a diagnosis to be made. All samples that were received directly in the laboratory were cultured. Samples received from medical genetics laboratories had previously been spun to remove cells for cytogenetic studies, and these were not suitable for culture for virus. Samples received from medical genetics laboratories were handled in an aseptic manner. Samples that came from other hospitals for which only PCR was requested were also unsuitable for culture.
A total of 145 amniotic or fetal fluid samples were examined; 83 of these were from healthy pregnant women and 62 were from the diagnostic group. Of the 62 samples from women who were being investigated because of concerns about the pregnancy, 60 were amniotic fluid, 1 was fetal pleural fluid, and 1 was fetal ascitic fluid.
A total of 39 urine samples were obtained from 19 neonates with suspected congenital CMV infection.
Preparation and storage of samples.
All samples were stored at 4°C until they were processed. Amniotic fluid and urine samples that were cultured were inoculated into MRC-5 cells within 24 h of arrival in the laboratory. Samples for molecular analysis were processed within 7 days. When this was not possible, the samples were spun and the supernatant and the cell fraction were stored at −70°C until they were processed. Prior to analysis the samples were spun at 10,000 × g for 5 min and the supernatant was removed. The DNA was extracted by boiling the supernatant for 5 min; the boiled supernatant was then transferred to ice. Four blood-stained samples were diluted 1 in 2 with molecular-grade water and were spun after boiling in order to obtain a cell-free sample suitable for PCR.
For amplification by nested PCR, cell-free boiled fractions were used without further preparation. For samples to be analyzed in a single-round PCR with analysis by plate hybridization, a preoptimized concentration of internal control (IC), approximately 50 copies, was added at this stage.
Preparation of bacteriophage lambda IC.
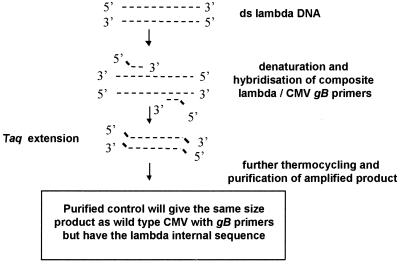
An IC was prepared. When the IC was added to clinical samples it was amplified with the CMV-specific first-round primers but with an internal sequence different from that of the wild-type product. This IC could be distinguished from wild-type CMV at the detection stage. The IC was prepared as outlined in Fig. 1 by using composite primers with CMV- and bacteriophage lambda-specific regions. The primers were designed so that the amplified product from the bacteriophage lambda target sequence would be the same size as the wild-type CMV-specific product (150 bp) and have a similar melting temperature. The percent G+C contents for the designed IC and wild-type CMV were 56 and 55%, respectively. The sequences of the composite primers are as follows (5′ to 3′): primer λgB1, GAGGACAACGAAATCCTGTTGGGCAAGGTGAACGATGCGTAATGT, andprimer λgB2, GTCGACGGTGGAGATACTGCTGAGGTAAACGTACCATGTCCACCT. The CMV-specific regions were as described previously for amplification of the glycoprotein B (gB) gene (gB) and are given in italics (3, 8).
FIG. 1.
Schematic representation of preparation of CMV PCR IC. ——, CMV sequence; –––, bacteriophage lambda sequence.
Primers λgB1 and λgB2 were used in a single-round reaction with approximately 10 copies of purified bacteriophage lambda (Sigma Chemical Company Ltd., Poole, Dorset, United Kingdom) as a target. The conditions for the first-round PCR were as described previously (8): denaturation at 94°C for 4 min, followed by 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min. A final incubation at 72°C for 7 min ensured that all amplified products were complete. The amplified products from four separate reactions were pooled, and the IC was purified by a column-based procedure (Nucleon QC for PCR; Biosciences, Glasgow, Scotland). The amount of IC was estimated by measurement of the optical density (OD) at 260 nm, and the appropriate amount to be used in a single-round PCR was optimized by using a chessboard approach. A dilution of purified IC equivalent to approximately 50 copies was found to give reproducible positive results without inhibition of wild-type CMV detection. Amplified IC and wild-type products were distinguished by using CMV- and bacteriophage lambda-specific probes in the plate hybridization assay as detailed below.
First-round PCR.
The laboratory facilities where the amplification methods were performed adhere to strict guidelines in order to minimize the risk of contamination (14).
Primers for the first-round PCR were from the gB region of CMV, and the PCR conditions were as described previously (8), with 10 μl of prepared cell-free sample added to the reaction mixture. Three dilutions of purified human CMV DNA (Sigma Chemical Company Ltd.) at known concentrations (equivalent to 500, 50, and 5 copies) and four negative controls (in which water replaced the clinical material) as a check for contamination were incorporated into every run. The primers used for the first-round PCR have been well validated, and their sequences are homologous to a conserved region of the viral genome (3). These primers have been used extensively for analysis of CMV in clinical studies by the authors and other groups of investigators (8, 18, 19, 25). As expected from available sequences (GenBank), the primers do not detectably amplify genomic DNA or high-copy-number targets prepared from the other seven human herpesviruses (data not shown). The validated sensitivity for this assay is ≤10 copies of the CMV target sequence (8, 18–20, 25). The first-round PCR products were either subjected to a second round of PCR (nested PCR) or analyzed by the plate hybridization method.
Plate hybridization for detection of first-round PCR products.
For each sample two wells were required: one for detection of amplified wild-type CMV and one for detection of amplified IC. Plate hybridization was undertaken according to the manufacturer's instructions (Gen-ETI-K DEIA; Sorin Diagnostics, Wokingham, United Kingdom). This DNA immunoassay is based on the hybridization of amplified DNA with a biotin-labelled specific probe. Hybridized product is “captured” onto a solid phase (microtiter plate), and after incubation and washing of the sample, the addition of anti-double-stranded DNA identifies the wells in which hybridization has taken place. Protein A conjugated with horseradish peroxidase is used to detect the double-stranded DNA–antibody complex. The addition of tetramethylbenzidine substrate produces a color change that is read at an OD of 450 nm.
The optimal concentration of probe for this assay was determined to be 500 pg per well. The hybridization for the CMV wild type and IC was optimal at 50°C. The probe sequences used are as follows (5′-3′; biotin labeled at the 5′ end): CMV probe, ACCACCGCACTGAGGAATGTCAG, and IC probe, AGGTGAACGATGCGTAATGT. The CMV probe sequence selected for use in this study is the same as that of one of the nested PCR primers (see below).
Determination of positive and negative results was made according to the manufacturer's instructions. Briefly, the cutoff was taken as the mean for the negative control + 0.15 OD unit. All positive results exceeded the cutoff value by 0.5 OD unit, the background OD range varied between 0.01 and 0.10 OD units.
The process of extraction, amplification, and plate detection could easily be completed within a working day; however, the plates need to be coated with the probe and incubated for 18 to 24 h at 4°C.
Nested PCR.
Primers were from the gB region of CMV, and the PCR conditions were as described previously (8, 18–20, 25), with 1 μl of first-round product added to the second-round reaction mixture. Primers used for the first-round and second-round PCRs are as follows (5′ to 3′): first-round primer 1, GAGGACAACGAAATCCTGTTGGGCA; first-round primer 2, GTCGACGGTGGAGATACTGCTGAGG; second-round primer 3, ACCACCGCACTGAGGAATGTCAG; and second-round primer 4, TCAATCATGCGTTTGAAGAGGTA. The nested PCR products were analyzed by agarose gel electrophoresis with ethidium bromide staining. A positive result was defined as one in which a clear 100-bp product was visible on the gel, an example of which that shows detectable second-round products is given in Fig. 2.
FIG. 2.
Ethidium bromide-stained agarose gel demonstrating detectable second-round CMV products. Lanes M, molecular size marker.
Tissue culture.
Amniotic fluid and urine samples were inoculated into two tubes of confluent MRC-5 cells (Centre for Applied Microbiology and Research, Porton Down, United Kingdom) and were incubated at 37°C for up to 21 days by standard tissue culture methods (24). The presence of CMV was confirmed by indirect immunofluorescence. Washed cells suspended in phosphate-buffered saline were spotted onto a glass microscope slide and were immunostained (indirect monoclonal antibody for CMV; Syva Microtrak; Behring Diagnostic, Milton Keynes, United Kingdom) according to the manufacturer's instructions.
Serological investigations.
The presence of CMV-specific immunoglobulin M (IgM) in serum was determined with the IMX system (Abbott Diagnostics, Maidenhead, United Kingdom). Samples were processed according to the manufacturer's instructions. The complement fixation test (CFT) was performed by a standard protocol (complement was supplied by Serion Immunodiagnostica, Würzburg, Germany). CMV antigen for CFT was obtained from Central Public Health Laboratory, (London, United Kingdom).
RESULTS
Urine samples.
Urine samples taken from 19 neonates were examined for the presence of CMV DNA by nested PCR and single-round PCR with plate hybridization detection. Urine from 16 of these neonates was also cultured for the presence of CMV. For samples from five of the neonates, the presence of CMV was detected by all three methods, and for the sample from one neonate, CMV was detected by both molecular methods but the virus failed to grow in tissue culture. Urine from the remaining 13 neonates did not contain detectable CMV (Table 1).
TABLE 1.
Results of tissue culture and molecular assay with neonatal urine samples
| Patient no. | Detection of CMV DNA in urine by:
|
Virus isolation | Outcome | ||
|---|---|---|---|---|---|
| Nested PCR | Plate hybridization assay
|
||||
| CMV DNA | IC | ||||
| 1 | Not detected | Not detected | Detected | NDa | No CCMVIb |
| 2 | Not detected | Not detected | Detected | ND | No CCMVI |
| 3 | Not detected | Not detected | Detected | Negative | No CCMVI |
| 4 | Detected | Detected | Detected | Positive | Symptomatic CMV |
| 5 | Not detected | Not detected | Detected | ND | No CCMVI |
| 6 | Not detected | Not detected | Detected | Negative | No CCMVI |
| 7 | Not detected | Not detected | Detected | Negative | No CCMVI |
| 8 | Detected | Detected | Detected | Positive | Asymptomatic CCMVI |
| 9 | Detected | Detected | Detected | Negative | Symptomatic CCMVI |
| 10 | Not detected | Not detected | Detected | Negative | No CCMVI |
| 11 | Detected | Detected | Detected | Positive | Symptomatic CCMVI |
| 12 | Detected | Detected | Detected | Positive | Symptomatic CCMVI |
| 13 | Not detected | Not detected | Detected | Negative | No CCMVI |
| 14 | Detected | Detected | Not detected | Positive | Symptomatic CCMVI |
| 15 | Not detected | Not detected | Detected | Negative | No CCMVI |
| 16 | Not detected | Not detected | Detected | Negative | No CCMVI |
| 17 | Not detected | Not detected | Detected | Negative | No CCMVI |
| 18 | Not detected | Not detected | Detected | Negative | No CCMVI |
| 19 | Not detected | Not detected | Detected | Negative | No CCMVI |
ND, not done.
CCMVI, congenital CMV infection.
Amniotic fluid and fetal fluid samples.
In total, 145 amniotic and other fetal fluid samples were examined for the presence of CMV DNA after PCR amplification. For 132 samples (74 from the control group and 58 from the diagnostic group), this was by nested PCR, and for 144 samples (82 from the control group and 62 from the diagnostic group), this was by single-round PCR with plate hybridization detection. A total of 131 fluid samples were examined by both of the molecular methods and the results were compared. Of the 62 samples from the diagnostic group, 47 were inoculated into cell culture (Table 2). None of the amniotic fluid samples from the control group were cultured. CMV DNA was detected by both of the molecular assays in three amniotic fluid samples from the diagnostic group. For only one of these samples were culture results available, and virus failed to grow from that sample. However, CMV was grown from the neonate's urine, taken within the first week of life, and the baby was severely affected at birth. CMV DNA was not detected in any of the control amniotic fluid samples.
TABLE 2.
Comparison of both molecular methods and tissue culture results with amniotic fluid samples
| Sample | No. of samples
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Nested PCR
|
Plate hybridization with IC
|
Tissue culture
|
||||||
| CMV DNA detected | CMV DNA not detected | CMV DNA detected | CMV DNA not detected | IC detected | IC not detected | Positive | Negative | |
| Control group AFa | 0 | 74 | 0 | 82 | 82 | 0 | NTb | NT |
| Diagnostic group AF | 3 | 55 | 3 | 59 | 62 | 0 | 0 | 47 |
AF, amniotic fluid.
NT, not tested.
Comparison of single-round and nested PCRs.
The end point detection sensitivities were ≤100 copies of wild-type CMV DNA for single-round PCR coupled with plate hybridization and ≤10 copies of wild-type CMV DNA for nested PCR. These were determined by amplification of a known concentration of control material. There was a 100% correlation between the two amplification methods used. The IC was positive in all tests with samples in which CMV DNA was not detected, thus ensuring that intrinsic inhibitory factors were not affecting the PCR. In one urine sample in which CMV DNA was present, no signal was detected for the IC. This was probably due to competitive inhibition of the primers between wild-type CMV and the IC and is as expected. The IC for the assay was set up in such a way as to ensure that a negative result was not due to inhibitors of the PCR. Thus, a valid positive result does not depend on significant amplification of the very low copy number of the IC. In tests with all other samples in which CMV DNA was detected, the IC also produced a signal. Four samples with detectable CMV DNA (three amniotic fluid samples and one urine sample) were stored for more than 8 weeks at 4°C. Throughout this period these samples remained strongly positive by both amplification methods.
Follow-up of patients with positive amniotic fluid results.
CMV DNA was detected in samples from nine patients: three amniotic fluid samples from the diagnostic group and urine samples from six neonates. For all three patients in whose amniotic fluid CMV DNA was detected, the fetus had clinically apparent disease. The first mother had no clinical signs or symptoms during pregnancy but was further investigated at 16 weeks of gestation after routine biochemical screening tests revealed a high-risk index for trisomy 21, although chromosomal analysis was normal. The baby was born with severe disease, and CMV was isolated from the urine taken within the first week of life. The second mother was investigated after gross fetal abnormalities were detected on an ultrasound scan at 30 weeks of gestation. Intrauterine death occurred at 34 weeks of gestation, and a postmortem examination of the fetus confirmed disseminated CMV disease. In the last mother alpha-fetoprotein levels were elevated, suggesting a high risk for trisomy 21 at 16 weeks of gestation, but again, chromosomal studies were unremarkable. Fetal central nervous system irregularities were detected on an ultrasound scan and later by magnetic resonance imaging. Due to concern about maternal age (13 years) and the fetal central nervous system abnormalities, a late-gestation termination was performed. A request for a postmortem examination was refused. All three mothers had detectable CMV-specific IgM (IMX), consistent with active CMV infection.
Follow-up of patients with CMV-positive urine.
In five of six neonates in whose urine CMV DNA was detected there was clinical evidence of CMV disease (Table 1, patients 4, 9, 11, 12, and 14). Two of the mothers of these babies had CMV-specific IgM antibody (IMX) present in their serum, and in two neonates fetal IgM was detected (Table 3). Patient 8 (Table 1) was excreting CMV in urine but was clinically asymptomatic at birth. This neonate was investigated because maternal CMV hepatitis was diagnosed at 11 weeks of gestation. The diagnosis was made on the basis of clinical jaundice and the presence of maternal IgM antibody specific to CMV and the absence of markers for acute hepatitis A, B, and C and Epstein-Barr virus infection. This mother also had a history of contact with a neonate with congenital CMV.
TABLE 3.
Results of serological investigations of neonatesa
| Patient no. | Clinical details | Serological investigations | Additional information |
|---|---|---|---|
| 1 | Petechial rash | CFT titer, <8 | |
| 2 | Increased tone, poor feeding | Neonatal IgM negative; CFT titer, 64 | |
| 3 | Hydrocephalus | ND | |
| 4 | Petechial rash | Maternal IgM positive; neonatal IgM negative; CFT titer, 128 | Confirmed CCMV infection |
| 5 | Premature, 29 weeks of gestation | ND | |
| 6 | Maternal illness, hydropic baby | Maternal IgM negative | |
| 7 | Microcephaly | Neonatal IgM negative | |
| 8 | Maternal CMV hepatitis | Maternal IgM positive; CFT titer, 8 | Well baby excreting CMV at birth |
| 9 | Jaundice, hepatosplenomegaly | Serological evidence of maternal infectionb | Confirmed CCMV infection |
| 10 | Maternal illness | Maternal IgM positive; neonatal IgM negative; CFT titer, 64 | Well baby, no evidence of CCMVI |
| 11 | IUGR | Neonatal IgM positive; CFT titer, >512 | Confirmed CCMVI |
| 12 | Petechial rash, hepatosplenomegaly | Maternal IgM positive; CFT titer, 64 | Confirmed CCMVI |
| 13 | Investigation of maternal illness | Maternal IgM positive | Well baby, no evidence of CCMVI |
| 14 | IUGR, petechial rash, hepatosplenomegaly | Neonatal IgM positive; CFT titer, 256 | Confirmed CCMVI |
| 15 | Thrombocytopenia, anemia | CFT titer, <8 | |
| 16 | Admitted to neonatal intensive care | Fetal IgM negative; CFT titer, 64 | |
| 17 | Admitted to neonatal intensive care | CFT titer, 32 | |
| 18 | Hydropic baby | CFT titer, <8 | |
| 19 | Fetal pleural effusion | CFT titer, <8 | CMV DNA not detected in AF |
Abbreviations: IUGR, intrauterine growth retardation; AF, amniotic fluid; CCMVI, congenital CMV infection; ND, not done.
Serological investigations were carried out at another center.
A total of 39 urine samples from 16 of the neonates were cultured for the virus. CMV was isolated from urine obtained from five of the six babies in whom CMV DNA was detected by PCR. When multiple samples were available from the same baby, virus isolation results were found to be variable, whereas both molecular methods gave consistent results for individual patient samples.
Results of serological investigations.
As shown in Table 3, serological investigations are often inconclusive and unsatisfactory in the investigation of congenital CMV infection in neonates, and these data clearly demonstrate the need for a more sensitive and specific method of diagnosis of fetal and neonatal infection.
DISCUSSION
Single-round PCR with detection of amplified products by plate hybridization was sensitive and reliable for detection of CMV DNA in amniotic fluid and urine samples. Follow-up of patients in the study revealed no cases of congenital CMV infection that were missed by the assay. The assay was found to be 10-fold less sensitive than the nested PCR (see Results section); this does not affect its use as a diagnostic tool, as in all positive samples an abundance of viral nucleic acid was present. The use of the IC provides essential information about the presence of inhibitory factors and allows interpretation of a negative result. The molecular assay described here can be performed within the normal working day and provides reliable results in a much shorter time frame than that required for either traditional tissue culture or the shell vial assay and can be used for diagnosis in both the prenatal and postnatal periods. Standard tissue culture methods can take up to 21 days before a positive culture result is seen, while shell vial assays are able to provide a positive result within 72 h. The assay described here can be completed within a working day. The expense of molecular reagents and royalties are offset by the high labor and overhead costs of tissue culture. The timeliness of reliable results provides added value, which means that the benefit of molecular assays justifies the cost.
There is a small risk of fetal loss (an approximately 0.3% risk in a tertiary-care referral center) as a result of the amniocentesis; this is significantly lower than the risk from more invasive techniques such as cordocentesis. Fetal blood sampling is less reliable in the diagnosis of congenital CMV infection, as it is dependent on the fetus being able to produce an IgM antibody response; the ability to do this is determined by gestational age and the existence of a functioning immune system.
The management of women who have active CMV infection during their pregnancies is fraught with difficulty. Although the risks to the fetus from a primary infection are much greater than the risks from a secondary infection or reactivation, it is often impossible to determine the type of infection, as stored sera predating the pregnancy are rarely available. It is our experience that the presence of specific IgM antibody is not seen just in patients with primary infection, although others advocate the use of radioimmunoassay for CMV-specific IgM to distinguish primary infection from reactivation or reinfection (11). Even for patients in whom a diagnosis of a primary infection is made, determination of whether the baby will be clinically affected is impossible from maternal serological investigations alone. For 90% of women who have a primary CMV infection during pregnancy, the babies born will be healthy, although they may excrete virus in their urine. For only 10% of women with primary infection will the baby be severely affected; maternal factors which determine clinical outcome have yet to be established. Maternal immunity has been shown to provide some protection, but sequelae associated with secondary maternal infection or reactivation of latent infection have been reported (7, 10). Amniotic fluid is derived largely from fetal renal excretion and secretion from the amniotic membranes and has been shown to be a useful specimen that provides direct information about the baby in utero (5, 12, 13, 16).
In the present study, for patients who had CMV DNA in their amniotic fluid, the pregnancy resulted in a clinically affected fetus or neonate. The absence of CMV DNA from control amniotic fluid samples confirms the results of other investigators that the presence of viral DNA in this sample is a significant finding (17).
For two of the women for whom the diagnosis of congenital CMV infection was made prenatally, both women had been offered and accepted an amniocentesis because biochemical screening tests indicated a high risk for Down syndrome. For both of these women chromosome studies revealed a normal karyotype. An amniocentesis was performed for a third woman because of the observance of gross abnormalities on an ultrasound scan. In all three women active CMV infection was confirmed by the presence of CMV-specific IgM. Severe congenital CMV infection in the neonate or fetus was confirmed for two of the patients by isolation of the virus from urine in the first week of life and postmortem investigations. In the case of one fetus there was no confirmation of fetal infection, as permission for postmortem examination was refused by the family.
It has yet to be determined if the presence of virus in amniotic fluid is indicative of CMV disease or whether it may also be found in a woman whose fetus remains healthy and unaffected. There have been reports of the detection of CMV DNA in amniotic fluid samples but with the result of the pregnancy being a clinically infected but apparently unaffected neonate (5, 15, 23). Others have been unable to detect CMV in amniotic fluid samples when the pregnancy resulted in the birth of an infected asymptomatic neonate (2, 6, 21–23). When amplification methods were used, only one group included an IC (22). Use of such a control increases the diagnostic certainty of a negative CMV DNA result. The time of sampling in relation to the mother's infection is probably crucial, as an amniocentesis performed too early after infection may result in a false-negative result.
When congenital CMV infection is strongly suspected in a pregnancy, use of amniocentesis for detection of DNA in amniotic fluid has distinct advantages over the use of other, more invasive techniques such as cordocentesis and antibody assay of fetal blood. The latter techniques carry much higher fetal loss rates than that from amniocentesis, and the validity of the results is dependent on the fetus being able to produce IgM antibody. Gestational age and an adequately functioning immune system determine this; 50% of neonates born with congenital CMV infection do not produce IgM antibody (5, 20).
The assay described here provides a useful, rapid method for determination of the presence of virus in samples relevant to fetal infection. In the three examples in which CMV DNA was detected in amniotic fluid, fetal abnormalities were apparent. It has yet to be determined whether the presence of viral DNA is itself a marker of systemic fetal disease or whether the amount of virus present in amniotic fluid correlates with the presence of a fetal abnormality. Until this issue is clarified and as 90% of babies infected in utero will be normal at birth, the assay should be used to complement other screening and diagnostic tools to help determine the likely long-term outcome of infection. With careful interpretation of all relevant test results, the obstetrician can be more confident in advising women with CMV infection during pregnancy on the potential risks of having an affected baby.
ACKNOWLEDGMENT
This work was supported by The Welsh Scheme for the Development of Health and Social Research.
REFERENCES
- 1.Alford C A, Hayes K, Britt W. Primary cytomegalovirus infection in pregnancy: comparison of antibody responses to virus-encoded proteins between women with and without intrauterine infection. J Infect Dis. 1988;158:917–925. doi: 10.1093/infdis/158.5.917. [DOI] [PubMed] [Google Scholar]
- 2.Catanzarite V, Danker W M. Prenatal diagnosis of congenital cytomegalovirus infection: false amniocentesis at 20 weeks gestation. Prenatal Diagn. 1993;13:1021–1025. doi: 10.1002/pd.1970131103. [DOI] [PubMed] [Google Scholar]
- 3.Darlington J, Super M, Patel K, Grundy J E, Griffiths J D, Emery V C. Use of the polymerase chain reaction to analyse sequence variation within a major neutralising epitope of glycoprotein B (gp58) in clinical isolates of human cytomegalovirus. J Gen Virol. 1991;72:1985–1989. doi: 10.1099/0022-1317-72-8-1985. [DOI] [PubMed] [Google Scholar]
- 4.Demmler G J. Infectious Disease Society for America and Centers for Disease Control. Summary of workshop on surveillance for congenital cytomegalovirus disease. Rev Infect Dis. 1991;13:315–329. doi: 10.1093/clinids/13.2.315. [DOI] [PubMed] [Google Scholar]
- 5.Donner C, Leisnard C, Content J, Busine A, Aderca J, Rodesch F. Prenatal diagnosis of 52 pregnancies at risk for congenital cytomegalovirus infection. Obstet Gynecol. 1993;82:481–486. [PubMed] [Google Scholar]
- 6.Donner C, Leisnard C, Brancart F, Rodesch F. Accuracy of amniotic fluid testing before 21 weeks gestation in prenatal diagnosis of congenital cytomegalovirus infection. Prenat Diagn. 1994;14:1055–1059. doi: 10.1002/pd.1970141108. [DOI] [PubMed] [Google Scholar]
- 7.Fowler K B, Stagno S, Pass R F, Britt W J, Boll T J, Alford C A., Jr The outcome of congenital cytomegalovirus infection in relation to maternal antibody status. N Engl J Med. 1992;326:663–667. doi: 10.1056/NEJM199203053261003. [DOI] [PubMed] [Google Scholar]
- 8.Fox J D, Zuckermann M A, Brink N S, Neild P, Gazzard B G, Tedder R S, Miller R F. Detection of herpesviral DNA by nested polymerase chain reaction in CSF of HIV-infected individuals with neurological disease: a prospective evaluation. J Infect Dis. 1995;172:1087–1090. doi: 10.1093/infdis/172.4.1087. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths P D. Current management of cytomegalovirus disease. J Med Virol. 1993;41(Suppl. 1):106–111. doi: 10.1002/jmv.1890410521. [DOI] [PubMed] [Google Scholar]
- 10.Griffiths P D, Baboonian C, Rutter D, Peckham C. Congenital and maternal cytomegalovirus infection in a London population. Br J Obstet Gynaecol. 1991;98:135–140. doi: 10.1111/j.1471-0528.1991.tb13358.x. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths P D, Stagno S, Pass R F, Smith R J, Alford C A., Jr Infection with cytomegalovirus during pregnancy: specific IgM antibody as a marker of recent primary infection. J Infect Dis. 1982;145:647–653. doi: 10.1093/infdis/145.2.647. [DOI] [PubMed] [Google Scholar]
- 12.Grose C, Meehan T, Weiner C P. Prenatal diagnosis of congenital cytomegalovirus infection by virus isolation after amniocentesis. Pediatr Infect Dis J. 1992;11:605–607. [PubMed] [Google Scholar]
- 13.Hagay Z J, Biran G, Ornoy A, Reece E A. Congenital cytomegalovirus infection: a long standing problem still seeking a solution. Am J Obstet Gynecol. 1996;174:241–245. doi: 10.1016/s0002-9378(96)70401-5. [DOI] [PubMed] [Google Scholar]
- 14.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;239:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 15.Kyriazopoulou V, Bondis J, Frantzidou F, Athanasiadis A, Diza E, Simitsopoulou M, Souliou E. Prenatal diagnosis of fetal cytomegalovirus infection in seropositive pregnant women. Eur J Obstet Gynecol Reprod Biol. 1996;69:91–95. doi: 10.1016/0301-2115(95)02541-3. [DOI] [PubMed] [Google Scholar]
- 16.Lynch L, Daffos F, Emanuel D, Giovangrandi Y, Meisel R, Forestier F, Cathomas G, Berkowitz R L. Prenatal diagnosis of fetal cytomegalovirus infection. Am J Obstet Gynecol. 1991;165:714–718. doi: 10.1016/0002-9378(91)90315-i. [DOI] [PubMed] [Google Scholar]
- 17.McLean L K, Chehab F F, Goldberg J D. Detection of viral nucleic acid in amniotic fluid of low-risk pregnancies by polymerase chain reaction. Am J Obstet Gynecol. 1995;173:1283–1286. doi: 10.1016/0002-9378(95)91371-8. [DOI] [PubMed] [Google Scholar]
- 18.Miller R F, Fox J D, Thomas P, Waite J C, Sharvell Y C, Gazzard B G, Harrison M J G, Brink N S. Acute lumbosacral polyradiculopathy due to cytomegalovirus in advanced HIV disease: CSF findings in 17 patients. J Neurol Neurosurg Psychiatry. 1996;61:456–460. doi: 10.1136/jnnp.61.5.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell S M, Fox J D, Tedder R S, Gazzard B, Lightman S. Vitreous sampling and viral genome detection for the diagnosis of viral retinitis. J Med Virol. 1994;43:336–340. doi: 10.1002/jmv.1890430404. [DOI] [PubMed] [Google Scholar]
- 20.Nelson C T, Demmler G J. Cytomegalovirus infection in the pregnant mother, fetus and newborn infant. Clin Perinatol. 1997;24:151–160. [PubMed] [Google Scholar]
- 21.Nicolini U, Kustermann A, Tassis B, Fogliani R, Galimberti A, Percivalle E, Revello M G, Gerna G. Prenatal diagnosis of congenital human cytomegalovirus infection. Prenat Diagn. 1994;14:903–906. doi: 10.1002/pd.1970141002. [DOI] [PubMed] [Google Scholar]
- 22.Revello M G, Baldanti F, Furione M, Sarasini A, Percivalle E, Zavattoni M, Gerna G. Polymerase chain reaction for prenatal diagnosis of congenital human cytomegalovirus infection. J Med Virol. 1995;47:462–466. doi: 10.1002/jmv.1890470428. [DOI] [PubMed] [Google Scholar]
- 23.Ruellan-Eugene G, Barjot P, Campet M, Vabret A, Herlicoviez M, Muller G, Levy G, Guillois B, Freymuth F. Evaluation of virological procedure to detect fetal human cytomegalovirus infection: avidity of IgG antibodies, virus detection in amniotic fluid and maternal serum. J Med Virol. 1996;50:9–15. doi: 10.1002/(SICI)1096-9071(199609)50:1<9::AID-JMV3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt N J. Cell culture procedures for diagnostic virology. In: Schmidt N J, Emmons R W, editors. Diagnostic procedures for viral, rickettsial and chlamydial infections. 6th ed. Washington, D.C.: American Public Health Association; 1989. pp. 51–100. [Google Scholar]
- 25.Wakefield A J, Fox J D, Sawyerr A M, Taylor J E, Sweenie C H, Smith M, Emery V, Hudson M, Tedder R S, Pounder R. Detection of herpesvirus DNA in the large intestine of patients with ulcerative colitis and Crohn's disease using the nested polymerase chain reaction. J Med Virol. 1992;38:183–190. doi: 10.1002/jmv.1890380306. [DOI] [PubMed] [Google Scholar]
- 26.Yow M D, Williamson D W, Leeds L J, Thompson P, Woodward R M, Walmus B F, Lester J W, Six H R, Griffiths P D. Epidemiological characteristics of cytomegalovirus infection in mothers and their infants. Am J Obstet Gynecol. 1988;158:1189–1195. doi: 10.1016/0002-9378(88)90252-9. [DOI] [PubMed] [Google Scholar]