Abstract

Monoclonal antibody (mAb) pharmaceuticals consist of a plethora of different proteoforms with different functional characteristics, including pharmacokinetics and pharmacodynamics, requiring their individual assessment. Current binding techniques do not distinguish between coexisting proteoforms requiring tedious production of enriched proteoforms. Here, we have developed an approach based on mobility shift-affinity capillary electrophoresis–mass spectrometry (ACE–MS), which permitted us to determine the binding of coexisting mAb proteoforms to Fc receptors (FcRs). For high-sensitivity MS analysis, we used a sheathless interface providing adequate mAb sensitivity allowing functional characterization of mAbs with a high sensitivity and dynamic range. As a model system, we focused on the interaction with the neonatal FcR (FcRn), which determines the half-life of mAbs. Depending on the oxidation status, proteoforms exhibited different electrophoretic mobility shifts in the presence of FcRn, which could be used to determine their affinity. We confirmed the decrease of the FcRn affinity with antibody oxidation and observed a minor glycosylation effect, with higher affinities for galactosylated glycoforms. Next to relative binding, the approach permits the determination of individual KD values in solution resulting in values of 422 and 139 nM for double-oxidized and non-oxidized variants. Hyphenation with native MS provides unique capabilities for simultaneous heterogeneity assessment for mAbs, FcRn, and complexes formed. The latter provides information on binding stoichiometry revealing 1:1 and 1:2 for antibody/FcRn complexes. The use of differently engineered Fc-only constructs allowed distinguishing between symmetric and asymmetric binding. The approach opens up unique possibilities for proteoform-resolved antibody binding studies to FcRn and can be extended to other FcRs and protein interactions.
Monoclonal antibodies (mAbs) have been demonstrated to be beneficial for treating various diseases ranging from cancer to cardiovascular to infectious diseases.1 Most of the therapeutic strategies using mAbs require binding to Fc receptors (FcRs) to trigger an immune response and for efficient recycling.2,3 The neonatal Fc receptor (FcRn), in particular, is involved in antibody recycling and transport through polarized membranes.3 Recycling of mAbs from circulation determines the half-life of antibodies in serum (i.e., pharmacokinetics), whereas transcytosis is crucial for neonates as they are not able to produce enough IgGs during their first months.4 These cellular transport and recycling mechanism are based on a pH-dependent binding of the antibody to the FcRn.
Antibody therapeutics contain not only one defined molecule but also several different proteoforms of the same antibody. These proteoforms range from different glycoforms attached to the highly conserved N-glycosylation site in the Fc region5,6 to additional post-translational modifications (PTMs) on the protein backbone such as oxidation or deamidation. The latter ones can occur either during manufacturing or during sample storage. Antibody binding to FcRn is proteoform-dependent, and proteoforms that are not able to bind to the receptor (e.g., due to a modification close to the binding site) are directed to the lysosome for degradation.7 Binding to FcRn occurs via the CH2–CH3 domain of human IgG.8,9 In this region, two Met are present in human IgG1 (Met252 and Met428), which are crucial for the binding. Oxidation of Met252 (and to a minor extent Met428) drastically affects the binding to FcRn reducing its serum half-life.10−12 The role of Fc glycosylation in FcRn binding has been more controversial. While some studies showed distinct FcRn binding between different glycoforms (e.g., deglycosylated, galactosylated, or sialylated),13−15 others reported no binding difference.16−18 Recently, glycosylation of the Fab region, which occurs in about 15–25% of mAbs, has also been shown to influence the binding with the FcRn due to a decreased complex association.19 However, most of these reports focus on specific features and do not consider potential confounding by other proteoforms (e.g., the influence of oxidation during glycosylation studies). This highlights the importance of proteoform-selective binding assessment to draw reliable conclusions.
For studying the interaction between mAbs and FcRn, the enzyme-linked immunosorbent assay (ELISA)20 and surface plasmon resonance (SPR)10,21 are the most widely employed techniques. In SPR, quantitative information on the antibody and Fc receptor affinity can be obtained (KD, kon, and koff).22 With ELISA, an affinity value (EC50) can be obtained, which describes the concentration necessary to obtain half of the maximum binding effect.23 However, the major drawback of these techniques is that no distinction between different proteoforms can be made, and an overall KD or EC50 value for all the proteoforms of an antibody sample is obtained. Therefore, binding information on single proteoforms requires tedious proteoform isolation or engineering procedures,23,24 and yet faces the problem of potential confounding by modifications that go unnoticed. In an effort to overcome this challenge, the use of affinity liquid chromatography hyphenated with mass spectrometry (affinity LC–MS) using FcRn columns has been proposed.25 In this approach, the antibody is injected at the binding pH and eluted by gradually increasing the pH to induce dissociation. Proteoforms with a lower affinity will elute earlier than those with higher affinity ones, permitting to study differences in binding in a straightforward way. However, no KD or EC50 values can be determined and, therefore, only relative differences in the retention time under stress conditions can be studied.26 Furthermore, immobilization of the FcRn receptor to a stationary phase does not permit further structural characterization of the interaction regarding, for example, stoichiometry and limit their application to FcRn, as columns with other immobilized FcRs are not commercially available. In short, currently there is no optimal solution for the proteoform-resolved study of antibody binding to FcRn and other FcRs.
Capillary electrophoresis (CE) separates analytes in an open-tube capillary without a stationary phase. As separations occur in solution, non-covalent (protein) interactions can be maintained during the analysis under appropriate conditions, opening great possibilities to study protein binding under physiologically relevant conditions.27,28 One approach that has shown immense potential to study the affinity of individual proteoforms is the mobility-shift approach.29 In this approach, the capillary is filled with one (free) interaction partner (e.g., receptor), whereas the second one is injected into the system (e.g., a mixture of proteoforms). During separation, proteoforms interact with the receptor influencing their effective electrophoretic mobility, which can be exploited to determine KDs.30−32 ACE has been applied for monitoring protein interactions with low- and high-molecular-mass ligands.27−29 Traditionally, affinity CE is performed with UV detection, bringing significant limitations as many proteoforms often co-migrate in one peak. Hyphenation with MS overcomes this issue as structural heterogeneity is assessed and resolved. Mobility shift affinity capillary electrophoresis–mass spectrometry (ACE–MS) has so far only been employed to study the interaction of very small peptides with antibiotics33−35 or stromal cell-derived factor-1 with different sulfated oligosaccharides.36 Recently, we have for the first time shown the capabilities of mobility-shift ACE–MS for monitoring protein–protein interactions using two small model proteins, that is, trypsinogen (24 kDa) and aprotinin (6.5 kDa).31 Hyphenation with MS was done using a sheath–liquid interface resulting in a limited sensitivity and hampering the possibility to apply the approach to antibodies and FcRs (approx. 150 and 50 kDa, respectively).
Sheathless interfaces for CE–MS operate with nano-electrospray ionization (nano-ESI) and have been demonstrated to provide a better ionization efficiency and suffer less from ionization suppression opening possibilities to application of larger proteins such as antibodies.37 In this work, we developed a new approach based on mobility-shift ACE–MS using sheathless interfacing for the proteoform-specific assessment of the interaction between mAb and the FcRn receptor. Different engineered antibodies and oxidized samples were employed to develop and demonstrate the capabilities of the approach. We show that we can simultaneously monitor the heterogeneity of the antibody and the FcRn and specifically assess their binding in solution. This allowed us to determine specific KD values for the different proteoforms in a mixture without the need of isolation or proteoform engineering. Furthermore, we determined an antibody/FcRn complex stoichiometry of 1:2 with symmetrical FcRn binding.
Experimental Section
Chemicals
All reagents employed were of analytical grade. Ammonium hydroxide, dithiothreitol (DTT), 7.5 M ammonium acetate (AmAc) solution, hydrogen chloride, guanidinium hydrochloride (Gua-HCl), tris-hydroxymethyl aminomethane (Tris), calcium chloride, and H2O2 were obtained from Sigma-Aldrich (Steinheim, Germany). Formic acid (FA), acetonitrile (ACN), and iodoacetic acid (IAA) were purchased from Thermo Scientific (Pittsburgh, PA). NAP-5 columns were obtained from GE Healthcare (Chicago, IL). Deionized water was obtained using a Milli-Q purification system (EMD Millipore, Burlington, MA). Bio-Spin 6 columns were purchased from Bio-Rad (Hercules, CA). 10 kDa Vivaspin MWCO filters were provided by Sartorius (Göttingen, Germany). Trypsin proteomic-grade was obtained from Roche Diagnostics (Mannheim, Germany).
Samples
Standard and engineered mAbs, Fc-only constructs, and FcRn were provided by Roche Diagnostics (Penzberg, Germany). The FcRn consisted of the human beta-2-microglobulin with a (G4S)4 linker,38 followed by the extracellular domain of the human IgG receptor FcRn large subunit p51, a His 10-tag, and an Avi-tag. Oxidation of the mAb samples (1 μg/μL) was achieved by adding different amounts of 1% v/v H2O2 solution resulting in a final H2O2 concentration of 0.009% v/v for NGmAb or 0.2% v/v for mAb1 and AAA-mAb and incubation for 24 h at 25 °C. The mAbs and Fc-only samples were buffer exchanged to 50 mM AmAc (adjusted with 50 mM AmAc to pH 6.0) using NAP-5 columns. The columns were equilibrated with a complete buffer fill four times. Next, 300 μL (1 mg) of mAb samples was loaded onto the column and allowed to drip through. After adding an additional 350 μL of 50 mM AmAc pH 6.0, the sample was eluted with 500 μL 50 mM AmAc pH 6.0. The concentration of the samples was determined using a nanophotometer (Implen, Munich, Germany) and diluted to obtain a concentration of 1 μg/μL for mAb samples or mixtures of oxidized and non-oxidized mAbs, and 0.75 μg/μL for Fc-only samples prior injection. Lysozyme from chicken egg (Sigma-Aldrich) was desalted and buffer exchanged to 50 mM AmAc pH 6.0 using 10 kDa Vivaspin columns. Lysozyme was diluted to a final concentration of 2 μg/μL. Similarly, the FcRn receptor (4.9 μg/μL) was desalted and buffer exchanged to 50 mM AmAc pH 6.0 using 10 kDa Vivaspin columns and diluted to the desired concentration (0.5–12 μM).
Affinity CE–MS
Affinity CE experiments were carried out on a Sciex (Framingham, MA) CESI 8000 instrument using OptiMS neutrally coated capillaries (Sciex) with a porous tip (length, 91 cm; 30 μm i.d.; 150 μm o.d.). The neutrally coated capillaries were conditioned according to the supplier specifications. Prior to the first use, the capillary tip was inserted in a falcon tube containing 50 mL of water, and the capillary was flushed for 5 min(100 psi, forward) with 0.1 M HCl, 10 min (100 psi, forward) with 50 mM acetic acid (adjusted with 50 mM AmAc to pH 3.0), and 30 min (100 psi, forward) with water (1 psi equals 6895 Pa). After the last rinse, the capillary was equilibrated for 16–18 h to allow rehydration of the neutral coating. This step was performed only before the first injection. For the preparation of the BGE, 50 mL of a 50 mM AmAc solution was adjusted to pH 6.0 by the addition of 2.2 mL of 50 mM HAc. Before each run, the capillary was flushed for 2 min with 0.1 M HCl (100 psi, forward pressure), 2 min with Milli-Q (100 psi, forward pressure), 2 min with 50 mM AmAc pH 6.0 (100 psi, reverse pressure), and 2 min with 50 mM AmAc pH 6.0 (100 psi, forward pressure). Following that, the capillary was filled for 2 min (100 psi, forward pressure) with 50 mM AmAc pH 6.0 containing different concentrations of the FcRn receptor or without receptor. Afterward, a marker protein (lysozyme) was injected (1.5 psi, 15 s), followed by the antibody sample (2.5 psi, 15 s) and a plug of BGE with or without receptor (1 psi, 25 s). The separation was carried out for 45 min with 20 kV (normal polarity) at 25 °C. After the separation was complete, the voltage was ramped down to 1 kV (normal polarity) in 5 min.
The outlet of the separation capillary was placed in a nanoelectrospray ionization source at the entrance of an Orbitrap exactive plus extended mass range mass spectrometer (Thermo, Waltham, MA). The mass spectrometer was operated in positive ionization mode using the following parameters: a capillary voltage of 1.8–2.2 kV; an ion injection time of 200 ms; a HCD energy of 100 eV; a skimmer voltage of 15 V; a S-Lens voltage of 15 V; and an ultrahigh vacuum of 3.11 × 10–10 mbar. The mass spectra were recorded in the mass range of 1000–15,000 m/z with a resolution of 17,500 at m/z 200. MS control and data acquisition were performed using the Xcalibur (Thermo Fisher Scientific) software. For data analysis, the Intact mass software from Protein Metrics (Cupertino, CA) was used.
Electrophoretic mobilities were calculated using the migration time of the beginning of the peak after alignment of the electropherograms using the marker protein (lysozyme). For calculation of KDs, the electrophoretic mobility shift compared to the measurement without FcRn was calculated for each proteoform. Following that, the data obtained were plotted using software GraphPad Prism using the one side-specific binding model.
Assessment of mAb Oxidation Levels by LC–MS/MS
To determine the oxidation levels of the mAbs and Fc-only samples, 50 μg (50 μL) of sample was denatured with an equal amount of denaturation buffer (8 M Gua-HCl, 0.4 M Tris, adjusted with HCl to pH 8.5). Afterward, the samples were reduced by adding DTT to a final concentration of 20 mM and incubated for 60 min at 50 °C. Following that, the samples were alkylated with IAA at a final concentration of 50 mM and kept for 30 min in the dark. For buffer exchange, prior to trypsin digestion, the samples were loaded on Bio-Spin 6 columns. Beforehand, the columns were centrifuged for 2 min at 1000×g to remove the storage liquid and three times equilibrated with 500 μL digestion buffer (50 mM Tris, 2 mM CaCl2, adjusted with HCl to pH 7.5), and centrifuged for 2 min at 1000×g in between. The reduced and alkylated samples were loaded onto the column and centrifuged for 4 min at 1000×g collecting thereby the eluate. 25 μg trypsin was dissolved in 100 μL 10 mM HCl solution. 3 μL of trypsin solution was added to each sample and incubated for 18 h at 37 °C. To stop the digestion, 17 μL of 10% v/v TFA solution was added, and the samples were diluted 1:1 (v/v) with MilliQ prior to LC–MS measurement. 10 μL sample was injected into an Acquity UPLC (Waters, Milford, MA, USA) equipped with a CSH C18, 1.7 μm, 130 Å, 2.1 × 150 mm column (waters). The separation was performed using 0.1% v/v FA in H2O as mobile phase A and 0.1% v/v FA in ACN as mobile phase B at a column temperature of 65 °C. A linear gradient from 1% B to 35% B in 45 min was used for the separation of peptides, followed by a cleaning for 3 min at 8% B and a re-equilibration for 4 min at 1% B. The liquid chromatograph was coupled to an orbitrap velos mass spectrometer operated in positive ionization mode. The isolation width for collision-induced dissociation (CID) fragmentation was 1 m/z, the fragmentation energy was set to 35 eV, the activation q was 0.25, and the activation time was 10 ms. The resolution was set to 30,000, and an m/z range of 200–2000 in full scan mode was monitored. The amount of oxidation for the sites Met252 and Met428 was calculated by determining the area under the curve of the non-oxidized and oxidized peptide extracted ion chromatograms and determining the relative ratio of the oxidized peptide in each sample.
Results and Discussion
Development of a Mobility-Shift ACE–MS Approach for Proteoform-Resolved Antibody–FcRn Binding
The binding of antibodies to FcRn takes place in the endosome at a pH between 5.5 and 6.0. Mobility shift affinity approaches rely on different mobilities of the protein and receptor, and in consequence the protein–receptor complex. To study which conditions show the largest difference in mobility between FcRn and mAb1, a mixture of both proteins was injected (1 μg/μL each) and analyzed using a background electrolyte consisting of 50 mM AmAc at pH 5.5 or 6.0. At these pHs, both the FcRn receptor (calculated pI: 6.24) and mAb1 (calculated pI: 8.47) are positively charged. Therefore, separations were performed using a neutrally coated capillary to avoid adsorption onto the capillary wall. 50 mM AmAc at pH 5.5 resulted only in a minor difference in mobility between FcRn and mAb1 (Figure S1). Using AmAc at 6.0, FcRn exhibited significantly lower mobilities than mAb1 (Figure S1), indicating that a shift in the mobility could be observed for the FcRn–antibody complex.
For affinity CE experiments, FcRn was added to the BGE, and after filling the capillary with the receptor, a mixture of antibody proteoforms was injected. Antibody proteoforms with high affinity toward FcRn would interact with the receptor during the separation and exhibit a lower effective mobility, while proteoforms with no interaction should not change their effective mobility compared to their analysis in the absence of a receptor. To correct for changes in the ionic strength and viscosity of the BGE with addition of different amounts of FcRn, a marker protein showing no interaction with FcRn (lysozyme; molecular mass: 14.3 kDa; calculated pI: 10.36) was employed.
Initial experiments were performed with an engineered antibody, which bears no glycans in the Fc domain (NGmAb). Oxidation of Met252 is known to influence the binding between the antibody and the FcRn receptor. Therefore, to fully explore the capabilities of the approach, analyses were performed with the NGmAb material, which was intentionally oxidized as confirmed by bottom-up proteomics. NGmAb oxidation levels for Met252 were 41%, comprising a mixture of antibodies containing 0, 1, or 2 oxidations at this site. First, the mixture of mono- and double-oxidized as well as non-oxidized species were analyzed in the absence of a FcRn receptor using 50 mM AmAc at pH 6.0.
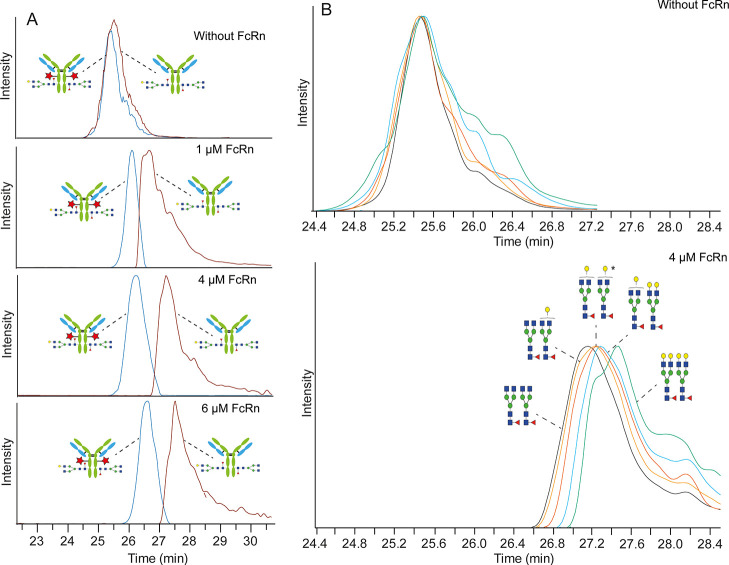
Upon oxidation, NGmAb showed unchanged electrophoretic mobility under standard CE conditions (Figure 1A), which was in line with expectations as oxidation comes with only a relatively small increase in intact mass while not affecting the mAb charge.
Figure 1.
Sheathless CE–MS separation obtained for a H2O2-stressed (A) NGmAb or (B) AAA-mAb sample using a BGE without FcRn (upper panel) and containing FcRn (lower panel). The blue trace corresponds to the extracted ion electropherograms (EIEs) of the double-oxidized (two red stars), the green trace corresponds to the EIEs of mono-oxidized (one red star), and the brown trace shows the EIEs of the non-oxidized mAbs present in the mixture. Signal intensities are normalized.
Reported KDs for antibodies and FcRn are between 3.2 and 284.7 nM.39 Therefore, for affinity experiments, FcRn was added to the BGE at concentrations in the range of nanomoles to a few micromoles. Analysis of the oxidized NGmAb sample using 4 μM FcRn in the BGE resulted in a clear shift of the effective electrophoretic mobility for all the species indicating that all of them interact with FcRn (Figure 1A). More interestingly, the different species start to separate indicating different binding affinities. The double-oxidized form of the NGmAb migrated in the first place indicating that it has the lowest binding affinity, while the unmodified NGmAb, which migrates at the last, has the highest binding affinity. The mono-oxidized NGmAb showed an intermediate shift between the double and non-oxidized antibody indicating a decrease in the binding affinity. However, it is important to note that the observed decrease corresponds to the inaccessibility of one of the Fc chains for binding to FcRn in solution, but does not imply a decrease of the in vivo half-life as still the antibody can interact with the FcRn via the non-modified Fc chain. In addition, even if a decrease in the binding affinity was observed for the oxidized forms, still binding to FcRn was observed for all the species.
To confirm that these observations correspond to the specific binding of the antibody to the FcRn, and are not the results of non-specific interactions or any other artifacts, we analyzed an antibody, which has been silenced for binding with FcRn (AAA-mAb or triple-A mutant). The antibody was intentionally oxidized prior to analysis resulting in an oxidation level of 99.6% for Met252 (i.e., double-oxidized antibodies). Analysis of a 1:1 mixture between double-oxidized and non-oxidized AAA-mAb in the absence of FcRn resulted in two overlapping signals at 27.5 min resulting from the double-oxidized and the unmodified triple-A antibody (Figure 1B). Of note, different antibodies (e.g., triple-A and NGmAb) exhibit different electrophoretic mobilities, and therefore the obtained migration times cannot be taken as absolute values of affinity but relative to their analysis in the absence of a receptor (i.e., mobility shift). Addition of up to a 12 μM FcRn receptor to the BGE did not induce any shift in the mobility or additional separation indicating that the AAA-mAb is not binding to FcRn (Figure 1B). These results confirmed that the shift in the mobility observed for the NGmAb antibody, indeed, is induced by the specific binding of the antibody with the FcRn receptor.
To demonstrate the applicability of the method for simultaneous binding assessment of multiple proteoforms, a standard glycosylated mAb was employed. As in previous cases, a 1:1 mixture of double-oxidized (97.7% for Met252) and non-oxidized mAb1 was analyzed, resulting in comigration in the absence of FcRn (Figure 2A). The addition of a receptor in the BGE at concentrations between 0.25 and 4 μM induced a shift in the mobility for both species with a partial separation between the double-oxidized and the non-oxidized mAb for the latest (Figure 2A). A further increase in the concentration to 6 μM did not result in a higher mobility shift and/or further separation suggesting that a plateau is reached above 4 μM (Figure 2A). One characteristic of mobility shift affinity CE approaches is their capability to provide quantitative information about binding. Next, to monitor the differences in relative binding for different proteoforms, we also wanted to evaluate the capabilities of the method to calculate their absolute binding—that is, the determination of affinity constants in a proteoform-selective manner. After determining and plotting the shift in the effective electrophoretic mobility for the measured concentrations of FcRn, a binding curve was obtained. By fitting the curve using non-linear regression, KD values of 139 (±67) nM for the non-oxidized and 422 (±157) nM for the double-oxidized mAb were obtained (Figure S2). The reported KD values are in the range of 3.2–284.7 nM39 depending on the specific structural features of the antibody (Fc and Fab portion), which are in line with the obtained values. However, the errors associated with our KD determination were significantly elevated indicating that more points (especially in the increasing part of the binding hyperbola) and further correction of electrophoretic mobilities would be needed for accurate KD determination. Yet, this initial exploration shows that the current approach could be used to determine KDs of different antibody proteoforms.
Figure 2.
Sheathless CE–MS separation obtained for a 1:1 mixture of H2O2-stressed and reference mAb1 using a BGE without or with different amounts of FcRn. (A) Illustration of the effect of oxidation. The blue trace corresponds to the EIEs of the double-oxidized (two red stars) and the brown trace shows the EIEs of the non-oxidized mAbs. In both cases, the antibody with G0F and G1F glycoforms was extracted. (B) Illustration of the effect of glycosylation. Different colors show EIEs of different glycoforms of the non-oxidized antibody. Signal intensities are normalized. *Either G1F/G1F or G2F/G0F.
In addition to oxidation, different glycoforms can potentially affect the binding with FcRn. mAb1 comprises different glycoforms dominated by complex-type glycans containing different core fucosylation and galactosylation levels. The influence of these glycans on FcRn binding was investigated considering their shift in their electrophoretic mobility (Figure 2B). While different glycoforms do not influence the electrophoretic mobility (Figure 2B, upper panel), shifted profiles were observed in the presence of FcRn (Figure 2B, lower panel). The shift difference between glycoforms was significantly lower than that observed for oxidoforms, which indicates some degree of influence on FcRn binding. In line with recent publications, we did not observe any effect on fucosylation in binding, while higher galactosylation slightly increased the binding affinity.13−15
Structural Assessment by Online Native MS
Affinity separations were performed with online native MS detection permitting, next to functional assessment, structural characterization. The use of 50 mM AmAc as BGE provided good signal intensities for the antibody proteoforms. For the NGmAb, which is non-glycosylated, next to the main form, two additional signals corresponding to the antibody with one (+162 Da) and two times glycation (+324 Da) were observed. After addition of the receptor to the BGE, additional signals appeared in the mass spectra corresponding to the free FcRn and in complex with the antibody, which were more perceptible with increasing amounts of FcRn in the BGE. The constant presence of FcRn in the MS also caused a slight decrease of the antibody signal intensity. A plateau was reached at around 4 μM of FcRn depending on the antibody. However, at higher concentrations of FcRn (up to 12 μM tested), the ionization suppression was moderate (intensity of the signal around six times lower compared to that observed in the analysis without the FcRn receptor), and good sensitivity was achieved for the detection of the antibody proteoforms and their complexes. The constant FcRn signal, on the other hand, permitted the simultaneous characterization of the receptor heterogeneity during the affinity experiments. FcRn was detected as a complex pattern of signals at around 49 kDa, corresponding to different glycoforms at the N-glycosylation site at position 220 on our FcRn construct (Figures 3 and S3). The glycoforms were assigned based on their average mass and on the previously reported glycopeptide data40 (Figure S3). Next to the antibody and the receptor, NGmAb in complex with FcRn was detected in a stoichiometry of 1:1 or 1:2 (Figure 3). At a concentration of FcRn of 2 μM, only NGmAb in complex with the 1 FcRn molecule (Figure S4A) was observed, while at 4 μM NGmAb in complex with 2 FcRn molecules was also noticeable (Figure S4B). A further increase of the FcRn concentration up to 12 μM higher resulted in an increase of the signals in particular for the corresponding NGmAb–FcRn 1:2 complex (Figure S4C,D). Figure 3 shows the zoomed spectra of the complex of the NGmAb with one FcRn molecule. Several signals were observed corresponding to the antibody in complex with different FcRn glycoforms (Figure 3), which can be clearly represented after simulation of the glycoprofile of the NGmAb-FcRn for the most abundant FcRn glycoforms (Figure 3, red traces). The NGmAb in complex with 2 FcRn was also observed at around 243 kDa, as a complex pattern of signals, where the main glycoforms were detected (Figure 3).
Figure 3.
Deconvoluted mass spectrum of the non-stressed NGmAb using a BGE containing 12 μM FcRn. Blue, zoom of FcRn containing several different glycoforms. Green, zoom of NGmAb containing 0, 1, or 2 glycations. Vermillion, zoom of NGmAb in complex with 1 FcRn. Yellow, zoom of NGmAb with 2 FcRn molecules. Heterogeneity of complexes comes from different glycoforms of the FcRn (most abundant ones are simulated in red).
Using the glycosylated standard mAb1 resulted in a more complex pattern of signals for the complex formed between the mAb1 and FcRn. However, we were able to detect the antibody in complex with one FcRn (Figure S5). The deconvoluted mass spectrum of this complex is dominated by the glycosylation of the antibody (G0/G0 to G2F/G2F) (Figure S5), which still contains different glycoforms of the receptor resulting in broader signals. The complex (Figure S5 red lines) was simulated using the two major glycoforms of the FcRn receptor resulting in a good match between the theoretical and the observed spectra. The complex of mAb1 with two FcRn molecules could not be detected, most probably due to the high complexity arising from the glycosylation of the antibody and the two FcRn molecules resulting in a higher spread of the signals and, therefore, a reduced signal intensity.
Monitoring the heterogeneity of all the species including the antibody, receptor, and the complex results in multiple benefits. Next to establishing the stoichiometry, it permits studying if the variability of the FcRn receptor (e.g., different glycoforms) has an influence on binding with the antibody, that is, it permits studying the interaction in a receptor proteoform-specific manner. For our particular example of FcRn, we could not observe any clear preference of certain FcRn glycoform in binding with NGmAb based on the obtained data, suggesting no influence of the FcRn glycosylation on the binding with the antibody. The observed glycation on the NGmAb (Figure 3) did also not show any shift in mobility as expected.
Evaluation of Antibody/FcRn Binding Stoichiometry and Symmetry Using Fc-Only Constructs
Unlike other Fc receptors, the binding of antibodies to FcRn occurs in a 1:2 stoichiometry. This has been demonstrated by crystal structures of rat FcRn and rat IgG2a.41 However, whereas some reports suggest a dimerization of the rat FcRn and binding only to one side of the Fc portion,41,42 other studies using column binding assays43 or using mutated Fcs44 suggested binding of one FcRn per Fc chain. For human FcRn, similar opposite observations have been made. While no FcRn dimers could be observed in crystallization experiments,45 recent publications show strong evidence for the self FcRn interaction in a pH-dependent manner.46 Recently, cryo-EM analysis of mAbs/FcRn showed 1:1 and 1:2 stoichiometries with one FcRn molecule binding to each side of the mAb-Fc.47
Our MS results show 1:2 binding for NGmAb but still this does not answer the question on binding symmetry. To test whether we can also study if we have an asymmetric or symmetric binding of the FcRn with our mobility-shift CE–MS approach, we analyzed two different engineered Fc constructs.48 These constructs consist of the Fc portion (two Fc/2 chains) connected by disulfide bridges. One of the constructs comprised of two unmodified Fc/2 chains (wt/wt) and should allow for FcRn binding on both sides. The second construct carried a triple-A mutation in one of the Fc/2 chains (wt/AAA) permitting only binding of the FcRn to one of the two Fc/2 chains. Injection of a mixture of both constructs without FcRn led to a separation of both Fc constructs due to their different pI (and therefore, different electrophoretic mobilities) with the wt/wt migrating earlier than the wt/AAA (Figure 4A). After filling of the capillary with a 6 μM FcRn receptor, a clear shift in the mobility for both constructs was observed (Figure 4B). For wt/AAA, the mobility shift was minor, while for the wt/wt constructs a larger shift was observed.
Figure 4.

Sheathless CE–MS separation obtained for 1:1 mixture Fc constructs [wt/wt (green) or wt/AAA (green and red)] using a BGE (A) without FcRn or (B) containing 6 μM FcRn. The orange trace represents the EIE of the wt/wt Fc-construct, and the brown trace shows the EIE of the wt/AAA Fc-construct. Signal intensities are normalized.
These results indicate that the construct that allowed a two side (symmetrical) binding showed a larger shift than the one to which only one FcRn molecule can bind. These results suggest that under these conditions, the observed 1:2 complex corresponds most likely to the one FcRn molecule bound to each side of the mAb-Fc as reported recently.47 Looking at the mass spectra, no dimers of the FcRn were observed supporting this observation. However, dissociation during ionization cannot be excluded. Regarding the complexes, we could only observe the complex between the Fc constructs (wt/wt and wt/AAA) and one FcRn (Figure S6). As for mAb1, the complex with two FcRn receptors for the wt/wt construct could not be detected due to the large heterogeneity coming from the glycosylation of the Fc construct and the two glycosylated FcRn molecules. Furthermore, from this experiment, it can be concluded that the missing Fab portion does not abrogate binding of the Fc to FcRn, and still a mobility shift can be observed. Although the absolute affinity of this molecules would be different from their complete counterparts containing the Fab domain, the proposed approach can be applied to monitor the effect of Fc modifications in FcRn binding opening possibilities to the application of polyclonal samples.
Conclusions
We developed a unique approach based on mobility shift ACE–MS to study the affinity of individual antibody proteoforms in a sample to FcRn. This approach represents the first affinity ACE–MS for functional studies on mAbs and overcomes most of the limitations faced with current binding techniques. We have focused on the FcRn antibody interactions, which are of special interest for antibody recycling and therefore are important for antibody therapeutic pharmacokinetics. We demonstrated that proteoforms with different binding affinities (i.e., oxidoforms) exhibit different shifts in the electrophoretic mobility in the presence of FcRn. This permits determining their individual binding affinity without necessity of enrichment or purification of different forms. Differences in relative binding between species can be studied in a straightforward and easy-to-interpret way by analyzing the mixture in the absence and the presence of a receptor. In addition, we also showed that KD values could be individually determined for the double-oxidized and non-oxidized mAb1 in mixtures, providing lower KD values for the non-oxidized mAb1. However, absolute KD determinations require larger sets of analyses in order to obtain accurate values. Including a second electrophoretic marker would allow more accurate electrophoretic mobility determinations, and therefore decrease KD errors. Future studies are ensured in this direction. The MS detection allows characterization of not only the antibody and its proteoforms but also the heterogeneity of the receptor and the complexes formed. Another advantage of MS detection is the possibility to also characterize and study the binding affinity of overlapping species. Analyzing a standard glycosylated mAb showed a slight difference in binding of Fc-glycoforms to FcRn with galactosylated variants showing higher binding as recently suggested in a few reports. Similar observations were made through MS monitoring of the complex between mAb and FcRn receptors, where no preferred binding of FcRn or antibody glycoforms could be monitored. In the particular case of the FcRn receptor heterogeneity, our data did not show specific FcRn glycoforms in higher abundance in the complex compared to the background FcRn signal, but more works would be needed to confirm this due to the complexity of the spectra. Reversing the approach (i.e., adding the mAb in the BGE and injecting the receptor) could provide information on the proteoform receptor binding in a more accurate way, and will be further studied. Furthermore, we investigated the binding stoichiometry between mAbs and the FcRn. Our MS spectra revealed 1:1 and 1:2 mAb/FcRn complexes, which, under the applied conditions, seem to be symmetrically bound. We used two different Fc-only constructs, which allowed one or two side binding of the FcRn. We found binding of both Fc-only constructs to FcRn with the one allowing both side binding provides a larger mobility shift compared to the construct allowing only one side binding.
The flexibility and simplicity of switching receptors (just changing the BGE) next to the low amount of receptor necessary compared to classical affinity LC make this approach very attractive for effector function antibody monitoring in a proteoform-selective manner. Due to the binding similarity of FcRn to other FcRs, we believe that this approach could be extended to study their binding. More studies are warranted on the interaction of mAbs with other Fc receptors.
Acknowledgments
We thank Protein Metrics for the temporal software license of their Intact mass software for data analysis. This work was supported by the Analytics for Biologics project (grant agreement ID 765502) of the European Commission and the LUMC Fellowship 2020 to E.D.-V.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.1c03560.
Details of the mobility of FcRn and mAb1 under different pH conditions; binding curves of double-oxidized and non-oxidized mAb1; glycosylation of FcRn; deconvoluted mass spectra for NGmAb with different amounts of FcRn; deconvoluted mass spectrum for mAb1 in complex with one FcRn; and deconvoluted mass spectrum of Fc constructs and in complex with one FcRn molecule (PDF)
Author Contributions
C.G. and E.D.-V. performed the measurements and processed the data. E.D.-V. and M.H. conceived the idea and designed the experiments. T.C. and T.S. provided the Fc-constructs. A.K. and T.S. provided the FcRn construct. C.G. and E.D.-V. drafted the manuscript. C.G., M.H., T.C., A.K., T.S., D.R., M.H., M.W., and E.D.-V. reviewed this manuscript equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Lu R.-M.; Hwang Y.-C.; Liu I.-J.; Lee C.-C.; Tsai H.-Z.; Li H.-J.; Wu H.-C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020, 27, 1. 10.1186/s12929-019-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenderman L. Inside-Out Control of Fc-Receptors. Front. Immunol. 2019, 10, 544. 10.3389/fimmu.2019.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyzik M.; Sand K. M. K.; Hubbard J. J.; Andersen J. T.; Sandlie I.; Blumberg R. S. The Neonatal Fc Receptor (FcRn): A Misnomer?. Front. Immunol. 2019, 10, 1540. 10.3389/fimmu.2019.01540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi S.; Bournazos S.; Thulin N. K.; Li C.; Gajewski A.; Sherwood R. W.; Zhang S.; Harris E.; Jagannathan P.; Wang L.-X.; Ravetch J. V.; Wang T. T. FcRn, but not Fc gamma Rs, drives maternal-fetal transplacental transport of human IgG antibodies. Proc. Natl. Acad. Sci. 2020, 117, 12943–12951. 10.1073/pnas.2004325117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-H.; Feng H.; Guo R.; Li P.; Laserna A. K. C.; Ji Y.; Ng B. H.; Li S. F. Y.; Khan S. H.; Paulus A.; Chen S.-M.; Karger A. E.; Wenz M.; Ferrer D. L.; Huhmer A. F.; Krupke A. Intact NIST monoclonal antibody characterization—Proteoforms, glycoforms—Using CE-MS and CE-LIF. Cogent Chem. 2018, 4, 1480455. 10.1080/23312009.2018.1480455. [DOI] [Google Scholar]
- Kabat E. A.; Wu T. T.; Reid-Miller M.; Perry H. M.; Gottesman K. S.. Sequences of Proteins of Immunological Interest; U.S. Department of Health and Human Services, Public Health Service, National Institutes of Health: Bethesda, MD, 1991. [Google Scholar]
- Ghetie V.; Ward E. S. Multiple Roles for the Major Histocompatibility Complex Class I– Related Receptor FcRn. Annu. Rev. Immunol. 2000, 18, 739–766. 10.1146/annurev.immunol.18.1.739. [DOI] [PubMed] [Google Scholar]
- Martin W. L.; West A. P.; Gan L.; Bjorkman P. J. Crystal Structure at 2.8 Å of an FcRn/Heterodimeric Fc Complex: Mechanism of pH-Dependent Binding. Mol. Cell 2001, 7, 867–877. 10.1016/s1097-2765(01)00230-1. [DOI] [PubMed] [Google Scholar]
- Kim J.-K.; Firan M.; Radu C. G.; Kim C.-H.; Ghetie V.; Ward E. S. Mapping the site on human IgG for binding of the MHC class I-related receptor FcRn. Eur. J. Immunol. 1999, 29, 2819–2825. . [DOI] [PubMed] [Google Scholar]
- Bertolotti-Ciarlet A.; Wang W.; Lownes R.; Pristatsky P.; Fang Y.; McKelvey T.; Li Y.; Li Y.; Drummond J.; Prueksaritanont T.; Vlasak J. Impact of methionine oxidation on the binding of human IgG1 to Fc Rn and Fc gamma receptors. Mol. Immunol. 2009, 46, 1878–1882. 10.1016/j.molimm.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Mo J.; Yan Q.; So C. K.; Soden T.; Lewis M. J.; Hu P. Understanding the Impact of Methionine Oxidation on the Biological Functions of IgG1 Antibodies Using Hydrogen/Deuterium Exchange Mass Spectrometry. Anal. Chem. 2016, 88, 9495–9502. 10.1021/acs.analchem.6b01958. [DOI] [PubMed] [Google Scholar]
- Wang W.; Vlasak J.; Li Y.; Pristatsky P.; Fang Y.; Pittman T.; Roman J.; Wang Y.; Prueksaritanont T.; Ionescu R. Impact of methionine oxidation in human IgG1 Fc on serum half-life of monoclonal antibodies. Mol. Immunol. 2011, 48, 860–866. 10.1016/j.molimm.2010.12.009. [DOI] [PubMed] [Google Scholar]
- Wada R.; Matsui M.; Kawasaki N. Influence of N-glycosylation on effector functions and thermal stability of glycoengineered IgG1 monoclonal antibody with homogeneous glycoforms. mAbs 2019, 11, 350–372. 10.1080/19420862.2018.1551044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashivets T.; Thomann M.; Rueger P.; Knaupp A.; Buchner J.; Schlothauer T. Multi-Angle Effector Function Analysis of Human Monoclonal IgG Glycovariants. PLoS One 2015, 10, e0143520 10.1371/journal.pone.0143520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennewein M. F.; Goldfarb I.; Dolatshahi S.; Cosgrove C.; Noelette F. J.; Krykbaeva M.; Das J.; Sarkar A.; Gorman M. J.; Fischinger S.; Boudreau C. M.; Brown J.; Cooperrider J. H.; Aneja J.; Suscovich T. J.; Graham B. S.; Lauer G. M.; Goetghebuer T.; Marchant A.; Lauffenburger D.; Kim A. Y.; Riley L. E.; Alter G. Fc Glycan-Mediated Regulation of Placental Antibody Transfer. Cell 2019, 178, 202–215.e14. 10.1016/j.cell.2019.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons L. C.; Reilly D.; Klimowski L.; Shantha Raju T.; Meng G.; Sims P.; Hong K.; Shields R. L.; Damico L. A.; Rancatore P.; Yansura D. G. Expression of full-length immunoglobulins in Escherichia coli: rapid and efficient production of aglycosylated antibodies. J. Immunol. Methods 2002, 263, 133–147. 10.1016/s0022-1759(02)00036-4. [DOI] [PubMed] [Google Scholar]
- Pawlowski J. W.; Bajardi-Taccioli A.; Houde D.; Feschenko M.; Carlage T.; Kaltashov I. A. Influence of glycan modification on IgG1 biochemical and biophysical properties. J. Pharm. Biomed. Anal. 2018, 151, 133–144. 10.1016/j.jpba.2017.12.061. [DOI] [PubMed] [Google Scholar]
- Jefferis R. Isotype and glycoform selection for antibody therapeutics. Arch. Biochem. Biophys. 2012, 526, 159–166. 10.1016/j.abb.2012.03.021. [DOI] [PubMed] [Google Scholar]
- Volkov M.; Van Schie K.; Bondt A.; Kissel T.; Brinkhaus M.; Bentlage A.; Koeleman C.; De Taeye S.; Dolhain R.; Wuhrer M.; Toes R.; Vidarsson G.; Van der Woude D. ACPA Illustrating the impact of IgG Fab-glycosylation on transplacental transfer of. Ann. Rheum. Dis. 2021, 80, 1044. 10.1136/annrheumdis-2021-eular.1727. [DOI] [Google Scholar]
- Lu Y.; Vernes J.-M.; Chiang N.; Ou Q.; Ding J.; Adams C.; Hong K.; Truong B.-T.; Ng D.; Shen A.; Nakamura G.; Gong Q.; Presta L. G.; Beresini M.; Kelley B.; Lowman H.; Wong W. L.; Meng Y. G. Identification of IgG1 variants with increased affinity to FcγRIIIa and unaltered affinity to FcγRI and FcRn: Comparison of soluble receptor-based and cell-based binding assays. J. Immunol. Methods 2011, 365, 132–141. 10.1016/j.jim.2010.12.014. [DOI] [PubMed] [Google Scholar]
- Magistrelli G.; Malinge P.; Anceriz N.; Desmurs M.; Venet S.; Calloud S.; Daubeuf B.; Kosco-Vilbois M.; Fischer N. Robust recombinant FcRn production in mammalian cells enabling oriented immobilization for IgG binding studies. J. Immunol. Methods 2012, 375, 20–29. 10.1016/j.jim.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Neuber T.; Frese K.; Jaehrling J.; Jäger S.; Daubert D.; Felderer K.; Linnemann M.; Höhne A.; Kaden S.; Kölln J.; Tiller T.; Brocks B.; Ostendorp R.; Pabst S. Characterization and screening of IgG binding to the neonatal Fc receptor. mAbs 2014, 6, 928–942. 10.4161/mabs.28744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; DiLillo D. J.; Bournazos S.; Giddens J. P.; Ravetch J. V.; Wang L.-X. Modulating IgG effector function by Fc glycan engineering. Proc. Natl. Acad. Sci. 2017, 114, 3485. 10.1073/pnas.1702173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke J.; Emrich T.; Rueger P.; Schlothauer T.; Kling L.; Knaupp A.; Hertenberger H.; Wolfert A.; Spick C.; Lau W.; Drabner G.; Reiff U.; Koll H.; Papadimitriou A. A novel approach to investigate the effect of methionine oxidation on pharmacokinetic properties of therapeutic antibodies. mAbs 2014, 6, 1229–1242. 10.4161/mabs.29601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahoual R.; Heidenreich A.-K.; Somsen G. W.; Bulau P.; Reusch D.; Wuhrer M.; Haberger M. Detailed Characterization of Monoclonal Antibody Receptor Interaction Using Affinity Liquid Chromatography Hyphenated to Native Mass Spectrometry. Anal. Chem. 2017, 89, 5404–5412. 10.1021/acs.analchem.7b00211. [DOI] [PubMed] [Google Scholar]
- Cymer F.; Schlothauer T.; Knaupp A.; Beck H. Evaluation of an FcRn affinity chromatographic method for IgG1-type antibodies and evaluation of IgG variants. Bioanalysis 2017, 9, 1305–1317. 10.4155/bio-2017-0109. [DOI] [PubMed] [Google Scholar]
- Dawod M.; Arvin N. E.; Kennedy R. T. Recent advances in protein analysis by capillary and microchip electrophoresis. Analyst 2017, 142, 1847–1866. 10.1039/c7an00198c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Štěpánová S.; Kašička V. Analysis of proteins and peptides by electromigration methods in microchips. J. Sep. Sci. 2017, 40, 228–250. 10.1002/jssc.201600962. [DOI] [PubMed] [Google Scholar]
- Yu F.; Zhao Q.; Zhang D.; Yuan Z.; Wang H. Affinity Interactions by Capillary Electrophoresis: Binding, Separation, and Detection. Anal. Chem. 2019, 91, 372–387. 10.1021/acs.analchem.8b04741. [DOI] [PubMed] [Google Scholar]
- Hutanu A.; Hauser P. C.; Moritz B.; Kiessig S.; Noël A.; Stracke J. O.; Wild M.; Schwarz M. A. Methionine oxidation of proteins analyzed by affinity capillary electrophoresis in presence of silver(I) and gold(III) ions. Electrophoresis 2021, 42, 1209–1216. 10.1002/elps.202000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Vega E.; Haselberg R.; Somsen G. W.; de Jong G. J. Simultaneous Assessment of Protein Heterogeneity and Affinity by Capillary Electrophoresis–Mass Spectrometry. Anal. Chem. 2015, 87, 8781–8788. 10.1021/acs.analchem.5b01701. [DOI] [PubMed] [Google Scholar]
- Stein M.; Haselberg R.; Mozafari-Torshizi M.; Wätzig H. Experimental design and measurement uncertainty in ligand binding studies by affinity capillary electrophoresis. Electrophoresis 2019, 40, 1041–1054. 10.1002/elps.201800450. [DOI] [PubMed] [Google Scholar]
- Lynen F.; Zhao Y.; Becu C.; Borremans F.; Sandra P. Considerations concerning interaction characterization of oligopeptide mixtures with vancomycin using affinity capillary electrophoresis-electrospray mass spectrometry. Electrophoresis 1999, 20, 2462–2474. . [DOI] [PubMed] [Google Scholar]
- Dunayevskiy Y. M.; Lyubarskaya Y. V.; Chu Y.-H.; Vouros P.; Karger B. L. Simultaneous Measurement of Nineteen Binding Constants of Peptides to Vancomycin Using Affinity Capillary Electrophoresis–Mass Spectrometry. J. Med. Chem. 1998, 41, 1201–1204. 10.1021/jm970578s. [DOI] [PubMed] [Google Scholar]
- Machour N.; Place J.; Tron F.; Charlionet R.; Mouchard L.; Morin C.; Desbène A.; Desbène P.-L. Analysis of virtual two-dimensional gels based upon affinity capillary electrophoresis hyphenated to ion trap-mass spectrometry. Electrophoresis 2005, 26, 1466–1475. 10.1002/elps.200410213. [DOI] [PubMed] [Google Scholar]
- Fermas S.; Gonnet F.; Sutton A.; Charnaux N.; Mulloy B.; Du Y.; Baleux F.; Daniel R. Sulfated oligosaccharides (heparin and fucoidan) binding and dimerization of stromal cell-derived factor-1 (SDF-1/CXCL 12) are coupled as evidenced by affinity CE-MS analysis. Glycobiology 2008, 18, 1054–1064. 10.1093/glycob/cwn088. [DOI] [PubMed] [Google Scholar]
- Haselberg R.; Ratnayake C. K.; de Jong G. J.; Somsen G. W. Performance of a sheathless porous tip sprayer for capillary electrophoresis-electrospray ionization-mass spectrometry of intact proteins. J. Chromatogr. A 2010, 1217, 7605–7611. 10.1016/j.chroma.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Chen X.; Zaro J. L.; Shen W.-C. Fusion protein linkers: property, design and functionality. Adv. Drug Delivery Rev. 2013, 65, 1357–1369. 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piche-Nicholas N. M.; Avery L. B.; King A. C.; Kavosi M.; Wang M.; O’Hara D. M.; Tchistiakova L.; Katragadda M. Changes in complementarity-determining regions significantly alter IgG binding to the neonatal Fc receptor (FcRn) and pharmacokinetics. MAbs 2017, 10, 81–94. 10.1080/19420862.2017.1389355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P. F.; Larraillet V.; Schlothauer T.; Kettenberger H.; Hilger M.; Rand K. D. Investigating the interaction between the neonatal Fc receptor and monoclonal antibody variants by hydrogen/deuterium exchange mass spectrometry. Mol. Cell. Proteom. 2015, 14, 148–161. 10.1074/mcp.m114.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister W. P.; Gastinel L. N.; Simister N. E.; Blum M. L.; Bjorkman P. J. Crystal structure at 2.2 Å resolution of the MHC-related neonatal Fc receptor. Nature 1994, 372, 336–343. 10.1038/372336a0. [DOI] [PubMed] [Google Scholar]
- Vaughn D. E.; Milburn C. M.; Penny D. M.; Martin W. L.; Johnson J. L.; Bjorkman P. J. Identification of critical IgG binding epitopes on the neonatal Fc receptor11Edited by I. A. Wilson. J. Mol. Biol. 1997, 274, 597–607. 10.1006/jmbi.1997.1388. [DOI] [PubMed] [Google Scholar]
- Huber A. H.; Kelley R. F.; Gastinel L. N.; Bjorkman P. J. Crystallization and Stoichiometry of Binding of a Complex between a Rat Intestinal Fc Receptor and Fc. J. Mol. Biol. 1993, 230, 1077–1083. 10.1006/jmbi.1993.1220. [DOI] [PubMed] [Google Scholar]
- Martin W. L.; Bjorkman P. J. Characterization of the 2:1 Complex between the Class I MHC-Related Fc Receptor and Its Fc Ligand in Solution. Biochemistry 1999, 38, 12639–12647. 10.1021/bi9913505. [DOI] [PubMed] [Google Scholar]
- West A. P.; Bjorkman P. J. Crystal Structure and Immunoglobulin G Binding Properties of the Human Major Histocompatibility Complex-Related Fc Receptor. Biochemistry 2000, 39, 9698–9708. 10.1021/bi000749m. [DOI] [PubMed] [Google Scholar]
- Soltermann F.; Foley E. D. B.; Pagnoni V.; Galpin M.; Benesch J. L. P.; Kukura P.; Struwe W. B. Quantifying Protein-Protein Interactions by Molecular Counting with Mass Photometry. Angew. Chem. 2020, 59, 10774–10779. 10.1002/anie.202001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.; Estevez A.; Schlothauer T.; Wecksler A. T. Antigen physiochemical properties allosterically effect the IgG Fc-region and Fc neonatal receptor affinity. mAbs 2020, 12, 1802135. 10.1080/19420862.2020.1802135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolaut A.; Schlothauer T.. Fc-Region Variants with Modified FcRn- and Maintained Protein A-Binding Properties. U.S. Patent 0,037,153A1, 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.