Abstract
Objective:
We sought to determine histologic and gene expression features of clinical improvement in early diffuse cutaneous systemic sclerosis (dcSSc; scleroderma).
Methods:
Fifty-eight forearm biopsies were evaluated from 26 individuals with dcSSc in two clinical trials. Histologic/immunophenotypic assessments of global severity, alpha-smooth muscle actin (aSMA), CD34, collagen, inflammatory infiltrate, follicles, and thickness were compared to gene expression and clinical data. Support vector machine learning was performed using scleroderma gene expression subset (normal-like, fibroproliferative, inflammatory) as classifiers and histology scores as inputs. Comparison of w-vector mean absolute weights was used to identify histologic features most predictive of gene expression subset. We then tested for differential gene expression according to histologic severity and compared those to clinical improvement (according to the Combined Response Index in Systemic Sclerosis).
Results:
aSMA was highest and CD34 lowest in samples with highest local modified Rodnan skin score. CD34 and aSMA changed significantly from baseline to 52-weeks in clinical improvers. CD34 and aSMA were the strongest predictors of gene expression subset, with highest CD34 staining in the normal-like subset (p<0.001) and highest aSMA staining in the inflammatory subset (p=0.016). Analysis of gene expression according to CD34 and aSMA binarized scores identified a 47-gene fibroblast polarization signature that decreases over time only in improvers (vs. non-improvers). Pathway analysis of these genes identified gene expression signatures of inflammatory fibroblasts.
Conclusion:
CD34 and aSMA stains describe distinct fibroblast polarization states, are associated with gene expression subsets and clinical assessments, and are useful biomarkers of clinical severity and improvement in dcSSc.
Keywords: Systemic sclerosis, fibroblasts, inflammation, autoimmune diseases
Introduction:
Systemic sclerosis (SSc; scleroderma) is an autoimmune disorder characterized by vasculopathy, inflammation, and fibrosis of the skin and internal organs [1]. Among rheumatic diseases, SSc carries the highest mortality rate, in part due to limited treatment options that do not address both the fibrotic and inflammatory disease features [2]. Progress in the field is limited by patient heterogeneity and imperfect outcome measurements [3], and there is a growing need to discover novel treatment targets.
Most SSc treatment trials have used the modified Rodnan skin score (MRSS) as the primary outcome measurement tool. This validated measure of skin thickness has limitations including inter-observer variability [4]. The Combined Response Index in Systemic Sclerosis (CRISS) is a composite outcome measure that incorporates new scleroderma renal crisis, decline in forced vital capacity (FVC) >15 %-predicted, new heart failure, and pulmonary hypertension requiring treatment, as well as change in MRSS, patient and physician global assessments, Health Assessment Questionnaire Disability Index (HAQ-DI), and FVC. The CRISS output is a probability of clinical improvement (0—1), and a threshold of ≥0.6 has been proposed [5].
Many SSc trials use skin histology and/or gene expression as exploratory outcomes. Skin biopsies have good face, content, criterion, and construct validity [6]; however, no standardized approach exists for interpreting histology in clinical trials. Previous studies have described correlations of skin biopsy weight [7], collagen, and alpha-smooth muscle actin (aSMA) with MRSS [8]. CD34, a dermal fibroblast marker, is decreased in SSc versus normal skin [9]. Skin gene expression may also describe SSc clinical severity and heterogeneity. Previous studies identified three gene expression subsets in diffuse cutaneous (dc)SSc skin: normal-like, fibroproliferative, and inflammatory [10, 11]. These subsets have been incorporated into stratified clinical trial analyses to better understand patient heterogeneity and treatment response [12–18].
The purpose of this study was to determine which histologic features of SSc lesional skin are most informative of gene expression subset and to use those histologic features to focus subsequent gene expression analyses to gain insights relevant to clinical improvement. We aimed (1) to define SSc skin histologic correlates of clinical improvement, (2) to assess the power of histologic features to predict gene expression subsets using an unbiased machine learning approach, and (3) to integrate histology-based gene expression analyses with 52-week clinical improvement.
Methods:
Patient data.
Fifty-eight forearm skin biopsies from 26 individuals with early, dcSSc were assessed by physical exam, DNA microarray, and histology in the context of two clinical trials at Hospital for Special Surgery (New York): the nilotinib in SSc trial (N=8) [15] and the belimumab in SSc trial (N=18) [12]. The nilotinib trial was an open-label, single-arm pilot trial where background immunosuppressive treatment was not permitted and all participants received nilotinib, a tyrosine kinase inhibitor. The belimumab trial was a randomized, controlled pilot trial where all participants received background mycophenolate mofetil and were randomized to receive either intravenous belimumab or placebo. Clinical data were collected including disease duration, autoantibodies, clinical assessments of lung and renal SSc involvement, c-reactive protein (CRP), erythrocyte sedimentation rate (ESR), FVC, total MRSS, local (biopsy-site) MRSS (scored by a single assessor using “averaging” approach [19]), physician and patient global assessments, and HAQ-DI. 52-week CRISS was calculated.
Sample processing.
Two 3-mm punch biopsies of extensor-surface, forearm lesional skin were obtained (nilotinib: baseline, 26-, 52-weeks; belimumab: baseline, 52-weeks) [12, 15]. Subsequent biopsies were performed 1-cm from baseline procedure. One biopsy was formalin-fixed, paraffin-embedded, and stained for hematoxylin and eosin (H&E), aSMA (Leica PA0943, RTU), and CD34 (Leica PA0212, RTU). The other biopsy was analyzed by DNA microarray as described previously and in supplemental methods [12, 15]. Microarray data were log2-lowess normalized and filtered for probes with intensity ≥1.5-fold over local background. Probes with >20% missing data were excluded. Missing expression values were imputed using GenePattern module (ImputeMissingValues.KNN) with default parameters, and probe expression set was collapsed to gene expression set using respective GenePattern modules [20]. Nilotinib samples were processed in a single batch. Belimumab samples were processed in two batches. These three batches did not exhibit a significant batch bias (as determined by gPCA, p=0.434, Supplemental Figure 1); therefore, no batch adjustment was performed. Expression data are accessible at NCBI GEO (accession no. GSE65405 and GSE97248, respectively).
Histologic evaluation.
Each sample was assessed using a histology scoring system that includes seven histologic/immunophenotypic features: thickness (epidermis to subcutis, measured by micrometer), follicle count, and a semi-quantitative (0—3) assessment of global histologic severity, infiltrate intensity, and collagen density detectable by H&E stain, and two fibroblast markers: CD34 and aSMA (Supplemental Figure 2). Similar to a patient or physician global assessment, the pathologist global assessment of histologic severity is a summary assessment of the histologic features assessed by H&E stain. Blinded to prior scores, a dermatopathogist (CM) and second pathologist (YZ), analyzed a sub-sample of biopsies (N=12) for reliability.
Statistical analysis.
Intraclass correlation coefficients (ICC) were calculated for inter-rater and intra-rater reliability for each histology domain. Median [interquartile range] score of each histology feature was calculated by local MRSS. In a paired analysis (baseline and 52-week), Wilcoxon signed-rank test was used to evaluate histologic change, stratified by CRISS 0.6 probability threshold to differentiate clinical improvers and non-improvers [5]. Spearman correlation was used to correlate histologic change with 52-week CRISS and change in each clinical outcome included in CRISS. Kruskal-Wallis and Mann-Whitney U-tests, Bonferroni-adjusted for multiple comparisons, were used to determine differences in clinical characteristics and histologic features by gene expression subset assignment.
Predicting gene expression subset assignment using histologic features.
Supplemental Figure 3 outlines the data processing pipeline. Samples were assigned to gene expression subsets using multinomial elastic net supervised classifier (GLMnet), as previously developed [21]. For each sample, the classifier assigns a probability for belonging to each gene expression subset (sum of probabilities equals 100%), and samples are assigned to the subset with the highest probability. Then, using histologic features as inputs and gene expression subsets as classifiers, support vector machine learning was performed to determine histologic features most predictive of gene expression subset. To binarize continuous variables (i.e., thickness), quantiles were generated. The area under the curve (AUC) of the receiver operating characteristic (ROC) curves generated for each gene expression subset were calculated to evaluate algorithm performance. Mean absolute weight for each histology score was determined using „w-vector‟ to identify histologic features and associated scores most predictive of subset assignment.
Differential gene expression by histologic features.
Using binarized scores for the histologic/immunophenotypic features with the highest weight for classifying samples (CD34 and aSMA), differentially expressed genes (DEG) were identified using an unequal sample size and unequal variance t-test between groups. Significant genes were defined as those with Bonferroni adjusted p-value <0.05. Hierarchical clustering was performed for DEG identified according to binarized histology scores. DEG identified by histology scores were similarly analyzed in paired samples, stratified by improvement status. Average DEG expression was plotted by clinical improvement status. Hierarchical clustering, supervised by baseline or 52-weeks, was performed for significant genes in improvers and non-improvers.
Pathway analysis.
Pathway analysis was performed for 47 identified genes differentially expressed according to aSMA and CD34 scores (goseq R package) [22]. The background gene set was the 16,645 genes expressed in the skin samples (genes with intensity >1.5 fold over background Cy3 and Cy5 in the microarray). All gene ontology (GO) pathways were considered. Benjamini-Hochberg adjusted false discovery rate <5% was considered significant.
Results:
Histologic features underlying biopsy-site assessments of SSc severity.
Fifty-six biopsies were analyzed from 26 individuals with dcSSc (median disease duration 0.8 years). Patient characteristics are presented in Table 1. Reliability of histology scores are presented in Supplemental Table 1. To describe histologic features underlying biopsy-site clinical assessments of SSc severity, median histology scores were calculated for each local MRSS score (Table 2). Higher global severity, aSMA, and collagen density and lower CD34 scores were observed for samples with highest (worse) local MRSS. The reverse was true for lowest local MRSS samples where global severity, aSMA, and collagen density was low and CD34 was high.
Table 1.
Baseline characteristics of study cohort
| Patient Characteristics | Total Cohort (N=26) |
|---|---|
| Age, mean (SD) | 50.7 (13.6) |
| Disease duration* (years), (median, IQR) | 0.8 [0.6, 1.2] |
| Sex, female, n (%) | 20 (77%) |
| Race, n (%) | |
| White | 19 (73%) |
| Black | 5 (19%) |
| Asian | 2 (8%) |
| SSc-specific autoantibodies, positive, n (%) | |
| Anti-topoisomerase I (Scl-70) | 6 (23%) |
| Anti-centromere | 2 (8%) |
| Anti-RNA polymerase III | 15 (58%) |
| ILD present, n (%) | 8 (31%) |
| FVC %-predicted, mean (SD) | 86 (16) |
| DLCO %-predicted, mean (SD) | 79 (17) |
| History of renal crisis, n (%) | 2 (8%) |
| MRSS, total, median (IQR) | 25 [22, 31] |
| MRSS, forearm (local), mean (SD) | 2 (0.75) |
| Physician Global Assessment (0–10), median (IQR) | 5.8 [5.0, 6.8] |
| Patient Global Assessment (0–10), median (IQR) | 2.9 [1.1, 4.7] |
| HAQ-DI, mean (SD) | 0.85 (0.59) |
Time since first non-Raynaud’s disease symptom. SSc=systemic sclerosis; ILD=interstitial lung disease; FVC=forced vital capacity; DLCO=diffusing capacity for carbon monoxide in the lungs; MRSS=modified Rodnan skin score; HAQ-DI=Health Assessment Questionnaire Disability Index; CRISS=Combined Response Index in Systemic Sclerosis; IQR=interquartile range.
Table 2.
Histologic features underlying local (biopsy-site) modified Rodnan skin scores for 26 individuals with diffuse cutaneous systemic sclerosis contributing 58 skin biopsy samples
| Histologic Feature | Local MRSS = 0 (N=3) |
Local MRSS = 1 (N=23) |
Local MRSS = 2 (N=17) |
Local MRSS = 3 (N=15) |
|---|---|---|---|---|
| Global | 0.5 [0, 2.5] | 1.5 [0.5, 2.5] | 2 [2, 2.5] | 2 [2, 3] |
| aSMA | 0 [0, 1] | 0.5 [0, 1.5] | 1 [0, 2] | 2 [1, 2] |
| CD34 | 3 [1.5, 3] | 2 [1, 3] | 1 [1, 2] | 1 [0, 1] |
| Collagen | 0 [0, 2.5] | 1 [0.5, 2] | 2 [1.5, 2.5] | 2.5 [2, 3] |
| Infiltrate | 0.5 [0.5, 1] | 0.5 [0, 1] | 0.5 [0.5, 1] | 0.5 [0.5, 1] |
| Follicles | 2 [2, 3] | 1 [0, 2] | 0 [0, 1] | 0 [0, 1] |
| Thickness | 1.9 [1.3, 2] | 2.6 [2.1, 3.0] | 2.4 [2.1, 2.7] | 2.8 [2.2, 3.0] |
MRSS=modified Rodnan skin score; aSMA=alpha-smooth muscle actin. Thickness is measured in micrometers from epidermis to subcutis. Median [interquartile range] is reported.
Histologic correlates of clinical improvement.
Paired baseline and 52-week skin biopsies were available for 24 individuals. Samples were stratified according to clinical improvers (CRISS≥0.6) versus non-improvers (Supplemental Table 2). As expected, clinical improvers had significant improvements in total and local MRSS, physician global assessment, patient global assessment, and HAQ-DI (Figure 1A). Among improvers, there were significant changes in baseline versus 52-week global histologic severity (2.5 to 1.5, p<0.01), aSMA (2 to 0.5, p=0.04), CD34 (1 to 2, p<0.001), and collagen density (2 to 1, p<0.01) (Figure 1B). Among six non-improvers, there were no significant changes in any baseline versus 52-week histology scores. Because the CRISS threshold for improvement is still provisional, we also compared histology changes to CRISS as a continuous measure. Consistent with the previous analysis using a CRISS threshold of ≥0.6, increasing (more favorable) CRISS correlated with decreasing global histologic severity (r=−0.52, p=0.01), aSMA (r=−0.44, p=0.03), and collagen density (r=−0.44, p=0.03), and increasing CD34 (r=0.53, p=0.01) (Table 3).
Figure 1. Clinical and histologic correlates of the Combined Response Index in Systemic Sclerosis (CRISS) ≥ 0.6 (N=18).
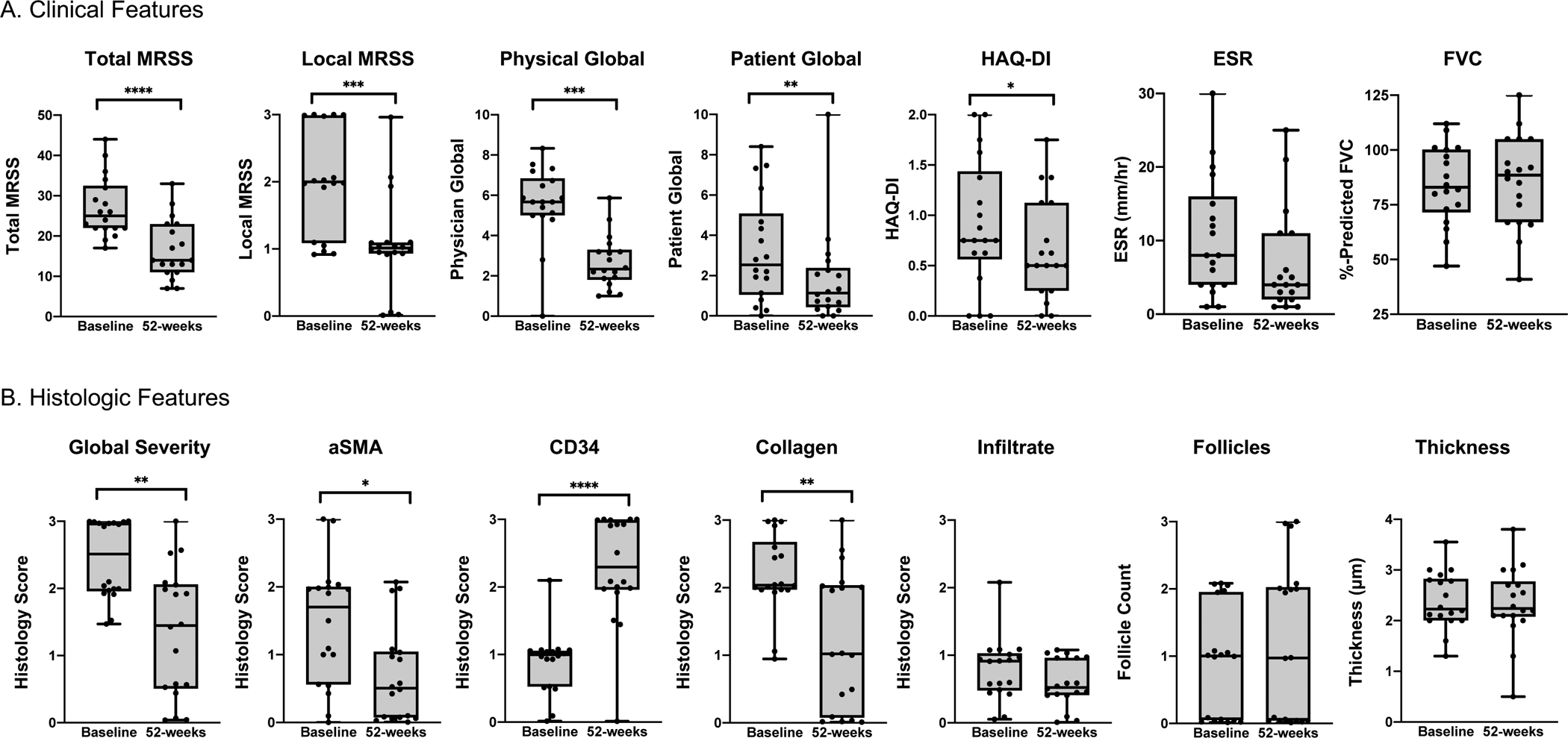
Among 18 individuals with diffuse cutaneous systemic sclerosis with 52-week CRISS ≥ 0.6, (A) clinical and (B) histologic changes in paired baseline and 52-week samples are demonstrated. Boxplots have whiskers from minimum to maximum values, a horizontal line at median value, and box edges at lower (Q1) and upper quartiles (Q3). P-values represent results of Wilcoxon signed-rank test. ****indicates p≤0.0001, ***p≤0.001, **p≤0.01, *p≤0.05. Adjusting for multiple comparisons, local MRSS, total MRSS, physician global assessment, patient global assessment, global histologic severity and CD34 remain statistically significant.
Table 3.
Correlation of 52-week histology change‡ with CRISS and change in clinical findings among 24 individuals with diffuse cutaneous systemic sclerosis
| Histology | CRISS | Total MRSS | Physician Global | Patient Global | HAQ-DI | FVC |
|---|---|---|---|---|---|---|
| Global | −0.52* | 0.43 * | 0.11 | 0.07 | 0.27 | −0.31 |
| aSMA | −0.44* | 0.46 * | 0.20 | 0.30 | 0.14 | −0.02 |
| CD34 | 0.53 * | −0.30 | −0.42* | −0.46* | −0.51* | 0.35 |
| Collagen | −0.44* | 0.32 | 0.09 | 0.22 | 0.12 | −0.25 |
| Infiltrate | −0.09 | 0.11 | 0.04 | 0.14 | 0.08 | −0.16 |
| Follicles | −0.09 | 0.23 | 0.26 | 0.10 | −0.21 | 0.39 |
| Thickness | −0.20 | 0.25 | 0.33 | 0.41 * | 0.13 | −0.10 |
Denotes significant p-value <0.05. Spearman correlation coefficients reported.
Histology change categorized as decreased, unchanged, or increased score from baseline to 52-weeks. CRISS=Combined Response Index in Systemic Sclerosis; MRSS=modified Rodnan skin score; HAQ-DI=Health Assessment Questionnaire Disability Index; FVC=%-predicted force vital capacity; aSMA=alpha-smooth muscle actin.
Histologic features to predict gene expression subset.
24, 16 and 18 samples were assigned to normal-like, fibroproliferative, or inflammatory gene expression subsets, respectively. Skin biopsy sites from normal-like, fibroproliferative or inflammatory samples were associated with increasing clinical severity with median local MRSS of 1, 2, and 3; respectively, (p<0.01) (Figure 2A). There was also a significant trend of increasing total MRSS and ESR and decreasing disease duration across the three gene expression subsets (Figure 2A). Histologic/immunophenotypic features also associated with gene expression subset. CD34 staining was highest in normal-like compared to fibroproliferative and inflammatory samples (median score 2, 1, 0.5, respectively; p<0.001) (Figure 2B). Conversely, aSMA staining was lowest in normal-like versus fibroproliferative and inflammatory samples (0.5, 0.75, 2, respectively; p=0.02). There were also significant differences in global severity (1.5, 2.25, 2, respectively; p=0.01) and collagen density (1, 2, 2.5, respectively; p=0.01).
Figure 2. Clinical and histologic correlates of three gene expression subsets among 26 individuals with diffuse cutaneous systemic sclerosis.
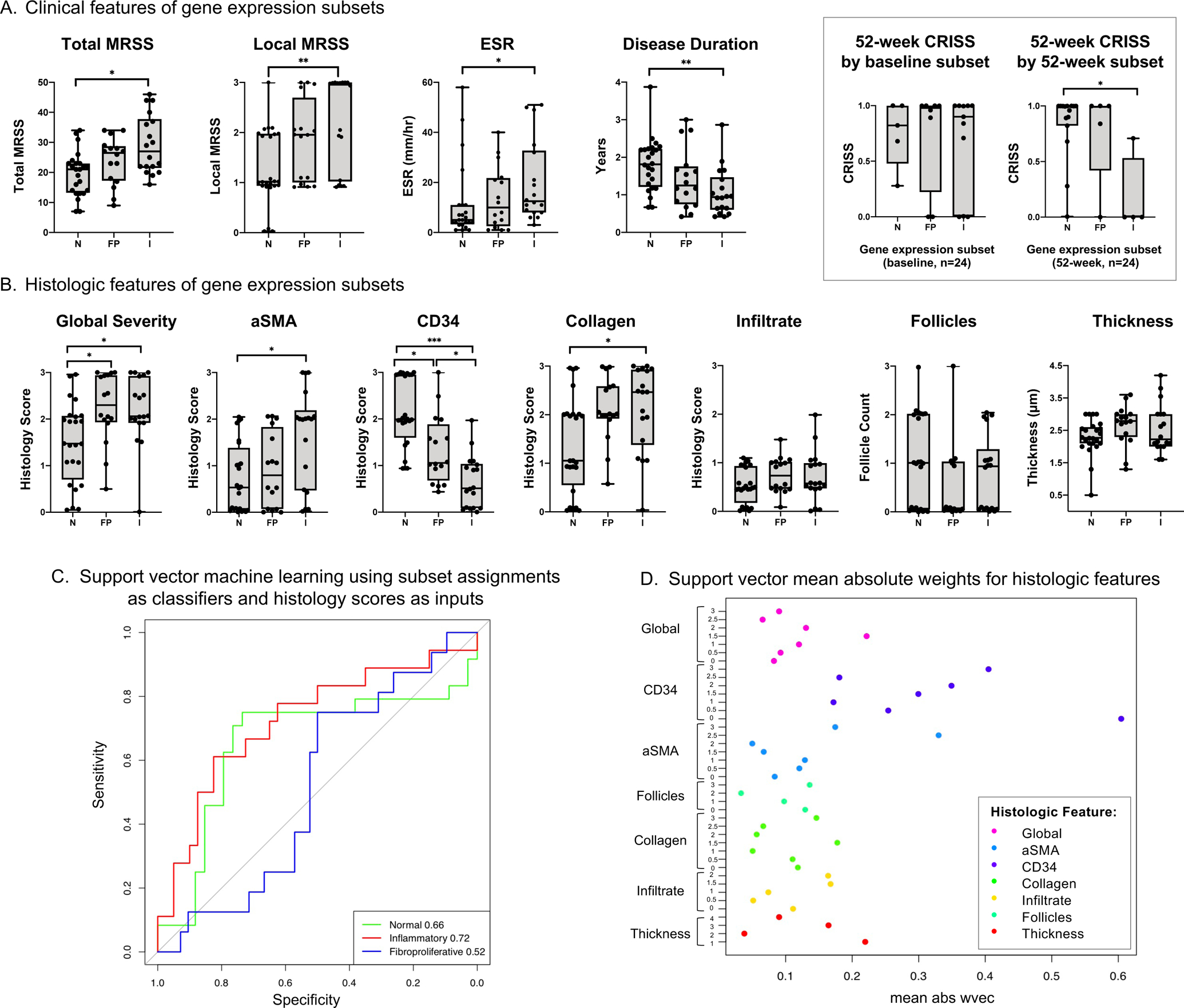
A. Clinical features of samples assigned to each gene expression subset (N = normal-like, FP = fibroproliferative, I = inflammatory). Box displays 52-week Combined Response Index in Systemic Sclerosis (CRISS), stratified by baseline and 52-week gene expression subset for 24 individuals with paired biopsy samples. Boxplots have whiskers from minimum to maximum values, a horizontal line at median value, and box edges at lower (Q1) and upper quartiles (Q3). B. Histologic features of samples assigned to each gene expression subset. C. Support vector machine learning was performed using gene expression subset as classifiers and the seven histology feature scores (global severity, aSMA, CD34, collagen density, infiltrate, follicle count, and thickness) as inputs. For continuous variables (i.e., thickness), quantiles were generated. Area under the curve (AUC) of the receiver operating characteristic curves assessed algorithm performance and are shown in the lower right legend. The p-values for the AUC for normal-like, inflammatory, and fibroproliferative subsets are 0.02, 0.003, and 0.58, respectively. D. Support vector mean absolute weights for each binarized histology score from „w-vector‟ model identified histologic features most predictive of subset assignment. P-values represent results of Mann-Whitney U-test, adjusted for multiple comparisons using Bonferroni correction. ***indicates p≤0.001, **p≤0.01, *p≤0.05.
We next tested the performance of a machine learning algorithm using histologic features to predict gene expression subset. The AUC of the ROC curve of models predicting inflammatory, normal-like, and fibroproliferative gene expression subsets were 0.72, 0.66, and 0.52, respectively (Figure 2C). The histology features with strongest predictive values (highest mean weight) were CD34 and aSMA (Figure 2D). In subsequent machine learning models, using only either CD34 or aSMA as inputs, CD34 predicted fibroproliferative subsets (AUC 0.77) and normal-like subsets (AUC 0.76), while aSMA predicted inflammatory subsets (AUC 0.73).
Gene expression signature of high aSMA vs. high CD34 scleroderma skin.
aSMA was highest and CD34 was lowest in samples with high MRSS and high inflammatory gene expression, and reversed with clinical improvement (Table 2, Figures 1B, 2B, 5). Taken together, these data support a strong and clinically relevant inverse relationship between the two dermal fibroblast markers. To visualize this, we plotted aSMA and CD34 scores for all samples (Figure 3A). Samples with highest aSMA and lowest CD34 scores were often assigned to the inflammatory gene expression subset, while the inverse was true of the normal-like subset. We next sought to uncover gene expression signatures that characterize fibroblast polarization. We identified one DEG when constraining the analysis to aSMA high versus low and 32 DEGs when constraining the analysis to CD34 high versus low. When we further focused the analysis to samples with either extreme of the immunophenotype (aSMAlow/CD34high vs. aSMAhigh/CD34low), we identified 36 DEGs. The union of these results yielded a total of 47 genes which we refer to as aSMA/CD34 polarization genes (Figure 3B).
Figure 5. Model integrating analysis of clinical, histologic and gene expression features of clinical improvement.
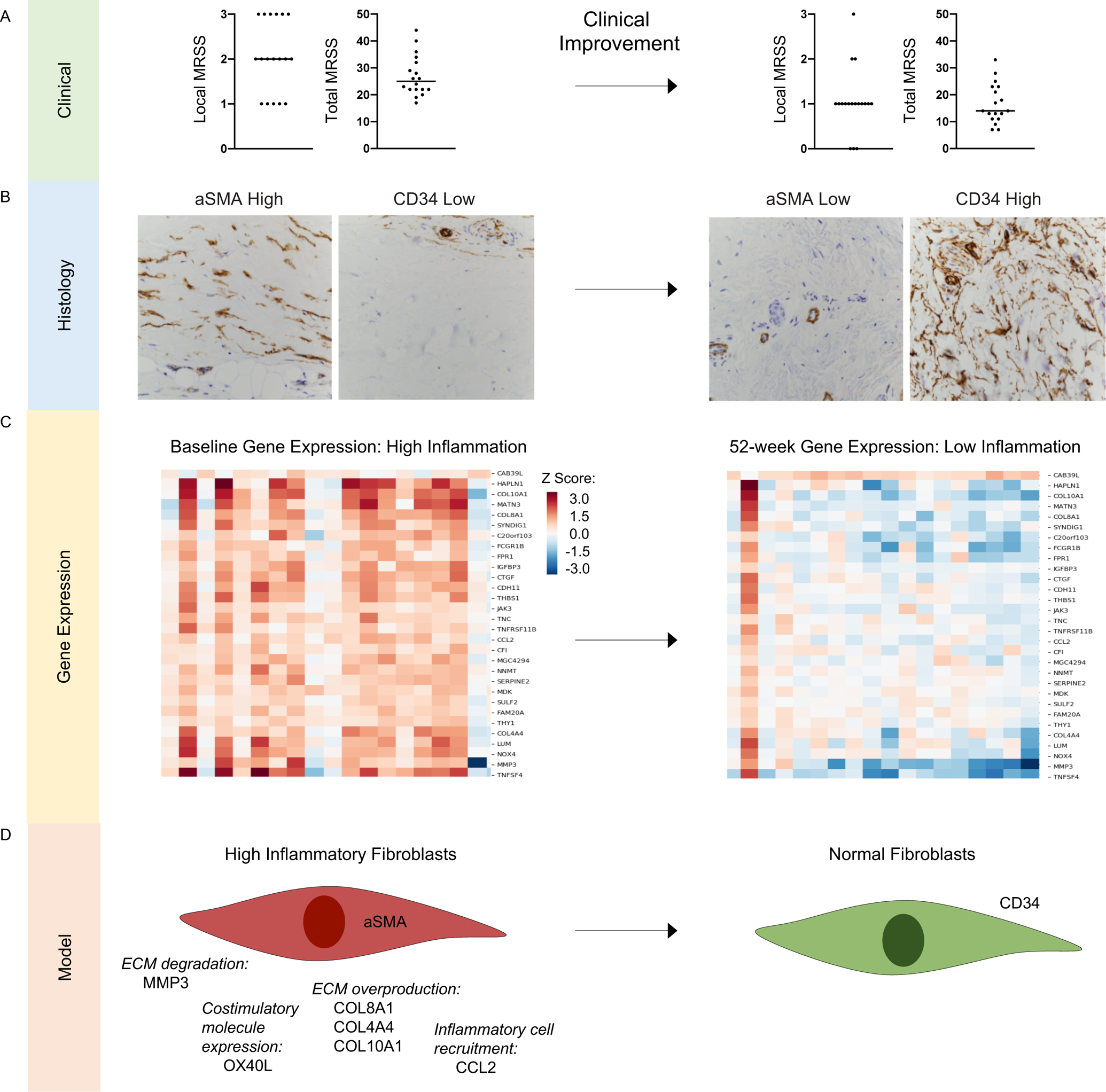
A. Baseline (left) and 52-week (right) total and local modified Rodnan skin score (MRSS) among 18 individuals with diffuse cutaneous systemic sclerosis (SSc) who experienced 52-week clinical improvement, defined by the Combined Response Index in SSc (CRISS) ≥0.6. Baseline median total MRSS is 25 and local MRSS is 2; 52-week median total MRSS is 14 and local MRSS is 1. B. Representative fibroblast stains (magnification 40X) at baseline (aSMA high and CD34 low) and 52-weeks (aSMA low and CD34 high) in an individual who experienced clinical improvement. C. Gene expression heatmap of 30 significant aSMA/CD34 polarization genes differentially expressed from baseline to 52 weeks (N=18 skin samples) from individuals who experienced clinical improvement from baseline to 52 weeks. D. Model proposing that fibroblast polarization is a hallmark of clinical severity in SSc. ECM=extracellular matrix.
Figure 3. Gene expression according to fibroblast polarization.
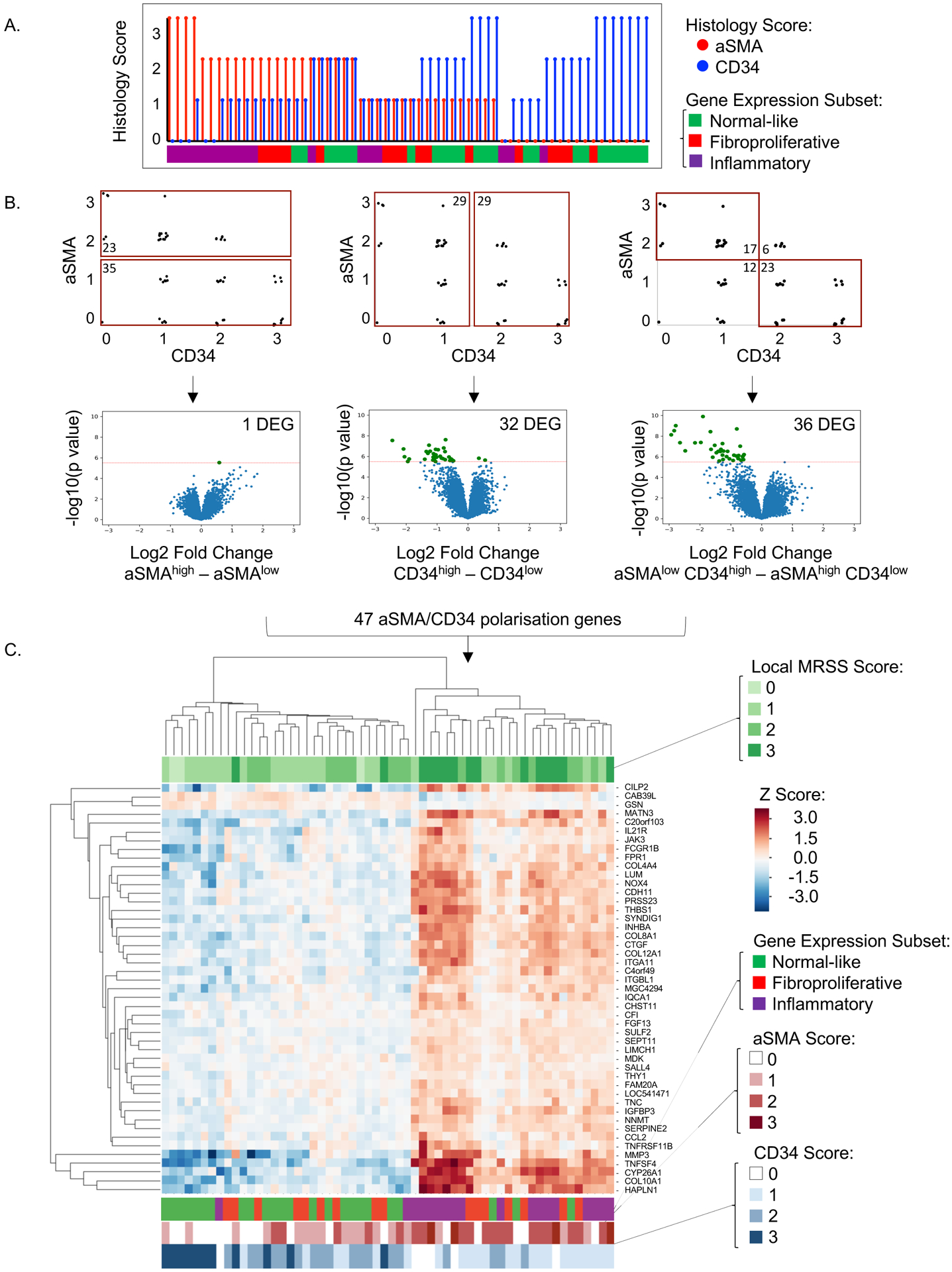
A. aSMA and CD34 scores with corresponding gene expression subset assignments for N=58 samples. aSMA scores (red vertical lines) were sorted in descending order and associated CD34 scores (blue vertical lines) were visualized for each patient sample. The horizonal bar below depicts each sample‟s gene expression subset assignment. B. Upper panel: Three sample gating strategies according to aSMA and CD34 staining. aSMAhigh vs. aSMAlow (N=58 samples), CD34high vs. CD34low (N=58 samples), and aSMAlow/CD34high vs. aSMAhigh/CD34low (N=40 samples). Lower panel: Volcano plots of significantly differentially expressed genes according to aSMA and CD34 scores. Significant differentially expressed genes are highlighted in green and quantified in top right corner. The threshold for significance (horizonal red line) was 0.000003, determined using Bonferroni correction for multiple comparisons of 16645 genes evaluated. C. Unsupervised hierarchical clustering using single linkage method and Euclidean distance metric of 47 aSMA/CD34 polarization genes identified in analysis outlined in B. Top horizontal bar indicates local MRSS for each skin biopsy sample. Bottom horizontal bars indicate gene expression subset, aSMA score (red), and CD34 score (blue) for each sample.
We compared unsupervised hierarchical clustering of the aSMA/CD34 polarization genes (Figure 3C) to local MRSS (Figure 3C top color bar), gene expression subset, and immunophenotypic assessments of CD34 and aSMA (Figure 3C bottom color bar). Pathway analysis confirmed these genes relate to fibroblast activation state, including FGF13, COL4A4, MMP3, TNFSF4 (OX40L), THY1 (CD90), and JAK3. The top ten most highly differentially expressed genes includes COL8A1, COL10A1, SERPINE2, SYNDIG1, TNFSF4, MATN3, and HAPLN1. Supplemental Table 3 summarizes the functional enrichment analysis for 47 aSMA/CD34 polarization genes. Significant GO biologic pathways include “extracellular matrix organization” (adjusted p-value<0.0001), “cell adhesion” (adjusted p-value<0.001), “regulation of leukocyte activation” (adjusted p-value=0.017), “interleukin-12 production” (adjusted p-value=0.011), and “skeletal system development” (adjusted p-value<0.001).
Expression of aSMA/CD34 polarization genes and clinical improvement.
We compared gene expression of 47 aSMA/CD34 polarization genes at baseline versus 52-weeks in 18 improvers and six non-improvers (Figure 4A). Among improvers, 30 of the 47 aSMA/CD34 polarization genes were significantly differentially expressed between baseline and 52-weeks. There were no significant differences in baseline versus 52-week expression of any gene in non-improvers. Average expression of the 47 aSMA/CD34 polarization genes was plotted for improvers vs. non-improvers for baseline and 52-week samples. Average gene expression in improvers (vs. non-improvers) was higher at baseline and lower at 52-weeks (Figure 4B). A heat map of the 30 significant genes differentially expressed from baseline to 52-weeks further demonstrates the increased expression at baseline relative to 52-weeks in improvers but not non-improvers (Figure 4C). Of these 30 genes, MMP3, TNFRSF11B, and THBS1 were most strongly correlated with CRISS, that is, the more these decreased from baseline to 52-weeks, the more likely a patient was to have clinically improved (Supplemental Figure 4). Taken together, this work indicates clinically severe scleroderma skin harbors aSMA high and CD34 low fibroblasts with associated high inflammatory gene expression, and these histologic and gene expression signatures of fibroblast polarization can reverse with clinical improvement (Figure 5).
Figure 4. Change in 47 aSMA/CD34 polarization genes from baseline to 52 weeks among individuals with and without 52-week clinical improvement.
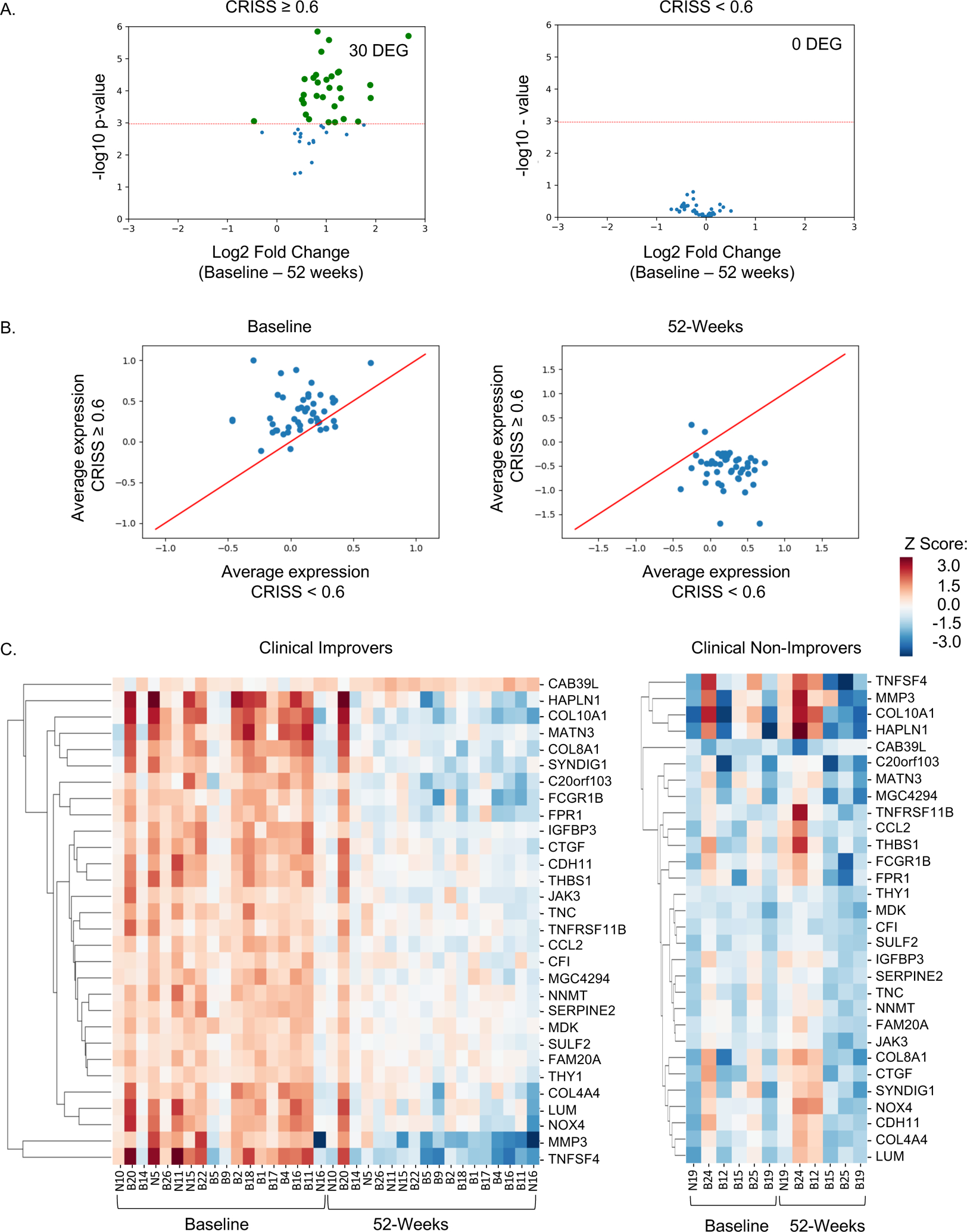
A. Volcano plot of 47 aSMA/CD34 polarization genes (log2 fold change of baseline vs. 52-weeks) in 18 individuals with diffuse cutaneous systemic sclerosis who were classified as clinical improvers (CRISS ≥0.6; N=36 samples), upper panel, or 6 individuals classified as non-improvers (CRISS <0.6; N=12 samples), lower panel. The threshold for significance (horizontal red line) was 0.001, determined using Bonferroni correction for multiple comparisons of 47 aSMA/CD34 polarization genes evaluated. Significant differently expressed genes are highlighted in green and quantified in top corner. B. Average expression of 47 aSMA/CD34 polarization genes at baseline and 52-weeks by clinical improvement status. C. Hierarchical clustering using single linkage method and Euclidean distance metric, supervised by either baseline or 52-weeks, of 30 significant differentially expressed genes identified in A in improvers and non-improvers.
Discussion:
There has been an unmet need to standardize the approach to skin histology assessment in SSc research. We report that of seven tested histologic/immunophenotypic features, global severity, aSMA, and collagen negatively correlated with clinical improvement, measured by CRISS, while CD34 positively correlated. In a parallel, unbiased machine learning analysis, two fibroblast markers (aSMA and CD34) also emerged as most strongly predictive of gene expression subset. These findings are consistent with prior investigations that describe positive correlations between aSMA and collagen with local MRSS [8] and decreased CD34 in SSc and morphea [23, 24]. Further, in a cross-sectional study of individuals with morphea (N=50) and healthy controls (N=50), Lee et al. found that individuals with morphea (vs. healthy controls) had higher aSMA and lower CD34 scores, and these were associated with assessments of fibrosis severity (mild vs. severe [25]) [26]. We add to the literature by demonstrating that baseline aSMA/CD34 immunophenotype among improvers changes to resemble at 52-weeks the aSMAlow/CD34high immunophenotype of normal skin.
The aSMA/CD34 polarization transcripts included genes under investigation as possible SSc treatment targets: TNFSF4 [27], JAK3 [28], CDH11 [29], and TGF-β regulated genes [30]. TNFSF4/OX40L, a costimulatory molecule expressed on antigen presenting cells [31] and SSc fibroblasts, is a genetic risk factor for dcSSc and SSc-associated autoantibodies [32], and its blockade leads to fibrosis regression in mice [27]. Additionally, several TGF-β regulated genes (e.g., THBS1, SERPINE2, and CTGF) were increased in samples with aSMAhigh/CD34low immunophenotype. Expression of THBS1 (thrombospondin-1) has been shown to correlate with MRSS [33]. SERPINE2/PN-1 is induced by TGF-β in models of cardiac fibrosis [34] and induces collagen promoter activity in 3T3 fibroblasts [35]. In an open-label study, 15 individuals with dcSSc received fresolimumab, a neutralizing antibody against TGF-β, and post-treatment (vs. baseline) dermal SERPINE2 and CTGF expression decreased and MRSS improved [36]. We also identified DEGs involved in MEK/ERK signaling: Integrin Subunit Alpha 1 (ITGA1) and Hyaluronan And Proteoglycan Link Protein 1 (HAPLN1). Inhibiting MEK/ERK pathway in vitro, reduces fibroblast contractility, suggesting that the MEK/ERK pathway is dysregulated in SSc fibroblasts [37]. Nazari et al. described an inverse staining pattern between CD34 and two other fibroblast markers: podoplanin and CD90/Thy1 [9]. We similarly observed significantly decreased expression of CD90/Thy1 in CD34 high vs. low samples. Together these studies suggest fibroblasts can be polarized towards an inflammatory fibroblast/myofibroblast state in the context of scleroderma-related inflammation. Indeed, this transition can be induced in vitro in response to tumor necrosis factor, IL-1β, or acute skin injury [9]. Our data add to the growing literature implicating inflammatory fibroblasts in SSc by showing that inflammatory fibroblast polarization can be reversed as scleroderma improves clinically.
Our data also suggest that gene expression profiles might be useful for identifying patients more likely to improve, as demonstrated by higher baseline expression of aSMA/CD34 polarization genes in improvers vs. non-improvers. Lofgren et al. developed an SSc-specific 415-gene expression signature and defined a SSc skin severity score (4S) based upon these genes [38]. The study results showed that the 4S score correlated with MRSS, and the 4S score at 12-months predicted 24-month MRSS. The aSMA/CD34 polarization genes identified herein includes eight overlapping genes with the 4S gene expression signature: CHST11, FPR1, GSN, HAPLN1, LUM, PRSS23, THY1, and TNFSF4. Our results may help to refine the 415-gene signature and improve the ability for gene expression to function as an outcome measure and predictive tool. By synchronizing histology with gene expression and clinical data, we also suggest that fibroblast polarization is the likely foundation of this gene expression signature.
Study strengths include dermatopathologist collaboration and use of a histology-centered approach to gene expression analysis to better understand disease heterogeneity and clinical improvement. We also acknowledge study limitations. Data was retrospectively analyzed from single-center trials for early, dcSSc with a high proportion of RNA polymerase III autoantibody positivity. This limits generalizability. Also, while there was no statistically significant batch bias, minor batch effects could still exist and potentially influence results of downstream analyses. The majority (18 of 26) of individuals were classified as 52-week improvers. As a result, our analysis of non-improvers was likely underpowered and findings regarding these individuals can only be considered descriptive. We used the provisional classification of CRISS ≥0.6 to distinguish clinical improvers vs. non-improvers; however, CRISS does not allow us to describe histologic and gene expression features of clinical stability versus worsening, and this cutoff may evolve over time as clinical trials aggregate data. Additionally, it is not possible to know the precise cell types that express each inflammatory gene identified. Future approaches using single-cell RNA sequencing are needed to better understand the cellular sources of these genes.
In conclusion, histologic features reflect disease severity, while dually enhancing our understanding of fibroblasts as contributors to SSc disease heterogeneity and behavior over time.
Supplementary Material
Supplemental Figure 1. Principal component analysis of Belimumab and Nilotinib samples. Belimumab samples were processed in two batches (red circles, green triangles) and nilotinib samples were processed in a single batch (blue crosses). Principal component analysis of these three batches did not exhibit a significant batch bias (as determined by gPCA, p=0.434).
Supplemental Figure 2. Systemic sclerosis histologic features analyzed. These images demonstrate examples of high scores for each of the seven histologic/immunophenotypic features analyzed. The scores shown, from left to right, include: global histologic severity (score: 3; low power view), alpha-smooth muscle actin (aSMA) (score: 2; magnification 40X), CD34 (score: 3; magnification 40X), collagen density (score: 3; magnification 4X), infiltrate intensity (score: 2; magnification 10X), follicle count (score: 2; low power view), thickness (score: 2.3μm; low power view).
Supplemental Figure 3. Diagram of data processing pipeline. A. Skin biopsy samples were analyzed by microarray and assigned to gene expression subset (normal-like, fibroproliferative, inflammatory) using a multinomial elastic net (GLMnet) classifier [21]. B. Skin biopsy samples were assessed for seven histologic/immunophenotypic features: thickness, follicle count, global histologic severity, infiltrate intensity, collagen density, and two fibroblast markers: CD34 and aSMA. Support vector machine learning (leave-one-out cross validation) was performed using each histologic feature score as inputs and gene expression subsets as classifiers.
Supplemental Figure 4. Change in gene expression compared to change in clinical severity. Heatmap demonstrates Pearson correlation coefficients for Log2(fold-change baseline versus 52 weeks) and Combined Response Index in Systemic Sclerosis (CRISS) for 30 aSMA/CD34 polarization genes. These 30 genes were identified as differentially expressed from baseline to 52 weeks among 18 clinical improvers.
Key Messages:
What is known about this subject?
Systemic sclerosis (SSc) histologic features (collagen score, alpha-smooth muscle actin (aSMA), and biopsy weight) have been shown to correlate with the modified Rodnan skin score (MRSS).
What does this study add?
CD34 staining decreases with worsening clinical severity and subsequently increases with clinical improvement. Conversely, alpha-smooth muscle actin (aSMA) staining increases with worsening clinical severity and subsequently decreases with clinical improvement.
Fibroblast polarization, according to aSMA and CD34 staining intensity, can be used to distinguish between scleroderma gene expression subsets.
We identify a robust fibroblast polarization gene expression signature that decreases over time in those with clinical improvement, but not in those who do not improve.
How might this impact clinical practice or future developments?
Dermal fibroblast polarization between aSMA and CD34 may be used to measure clinical improvement among individuals with diffuse cutaneous SSc.
Acknowledgements:
Part of this work was presented at the International Workshop on Scleroderma Research (2019), Systemic Sclerosis World Congress (2020), and the 2019 American College of Rheumatology meeting (Arthritis Rheumatol. 2019; 71 (suppl 10)).
Competing Interests:
Dr. Spiera reports receiving funds for the following activities: Research support: GlaxoSmithKline, Genentech/Roche, Novartis, Corbus Pharmaceuticals, Cytori Therapeutics, Evidera, Actelion Pharmaceuticals, ChemoCentryx, Boehringer Ingelheim Pharmaceuticals, Forbius, InfaRx, Sanofi, Kadmon Corporation; Consulting: GlaxoSmithKline, Janssen Pharmaceuticals, Sanofi Aventis, ChemoCentryx, Forbius, CSL Behring; Dr. Whitfield reports grants and personal fees from Celdara Medical, grants and personal fees from Bristol Myers Squib, personal fees from Acceleron, personal fees from Abbvie, grants and personal fees from Corbus and other fees from Boehringer Ingelheim, outside the submitted work; Dr. Orange reports receiving funds from Pfizer and personal fees from Astra Zeneca, outside the submitted work; Dr. Gordon reports receiving funds for the following activities: Consulting: Eicos Sciences; Research Support: Corbus Pharmaceuticals, Cumberland Pharmaceuticals, and Eicos Sciences, outside the submitted work.
Funding:
This study was supported by the Hospital for Special Surgery Rheumatology Council (JKG) and the Scleroderma Clinical Trials Consortium (SCTC) Working Group Grant (KS). This study utilizes data from the nilotinib and belimumab trials in systemic sclerosis that were investigator-initiated studies supported by research grants from Novartis and GlaxoSmithKline, respectively.
Footnotes
Ethical Approval: Hospital for Special Surgery Institutional Review Board approved this study (approval numbers: 2014-268 and 2019-0089), and patient informed consent was obtained.
Patient and Public Involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans for this study.
Data Availability Statement:
Data are available in a public, open access repository. All gene expression data has been deposited in Gene Expression Omnibus (GEO) (accession no. GSE65405 and GSE97248).
References:
- 1.Varga J, Abraham D. Systemic sclerosis: a prototypic multisystem fibrotic disorder. J Clin Invest. 2007. March; 117(3):557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferri C, Valentini G, Cozzi F, Sebastiani M, Michelassi C, La Montagna G, et al. Systemic sclerosis: demographic, clinical, and serologic features and survival in 1,012 Italian patients. Medicine (Baltimore). 2002. March; 81(2):139–153. [DOI] [PubMed] [Google Scholar]
- 3.Gordon JK, Domsic RT. Clinical Trial Design Issues in Systemic Sclerosis: an Update. Curr Rheumatol Rep. 2016. June; 18(6):38. [DOI] [PubMed] [Google Scholar]
- 4.Clements P, Lachenbruch P, Siebold J, White B, Weiner S, Martin R, et al. Inter and intraobserver variability of total skin thickness score (modified Rodnan TSS) in systemic sclerosis. J Rheumatol. 1995. July; 22(7):1281–1285. [PubMed] [Google Scholar]
- 5.Khanna D, Berrocal VJ, Giannini EH, Seibold JR, Merkel PA, Mayes MD, et al. The American College of Rheumatology Provisional Composite Response Index for Clinical Trials in Early Diffuse Cutaneous Systemic Sclerosis. Arthritis Rheumatol. 2016. February; 68(2):299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumanovics G, Pentek M, Bae S, Opris D, Khanna D, Furst DE, et al. Assessment of skin involvement in systemic sclerosis. Rheumatology (Oxford). 2017. September 1; 56(suppl_5):v53–v66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furst DE, Clements PJ, Steen VD, Medsger TA Jr., Masi AT, D’Angelo WA, et al. The modified Rodnan skin score is an accurate reflection of skin biopsy thickness in systemic sclerosis. J Rheumatol. 1998. January; 25(1):84–88. [PubMed] [Google Scholar]
- 8.Kissin EY, Merkel PA, Lafyatis R. Myofibroblasts and hyalinized collagen as markers of skin disease in systemic sclerosis. Arthritis Rheum. 2006. November; 54(11):3655–3660. [DOI] [PubMed] [Google Scholar]
- 9.Nazari B, Rice LM, Stifano G, Barron AM, Wang YM, Korndorf T, et al. Altered Dermal Fibroblasts in Systemic Sclerosis Display Podoplanin and CD90. Am J Pathol. 2016. October; 186(10):2650–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pendergrass SA, Lemaire R, Francis IP, Mahoney JM, Lafyatis R, Whitfield ML. Intrinsic gene expression subsets of diffuse cutaneous systemic sclerosis are stable in serial skin biopsies. J Invest Dermatol. 2012. May; 132(5):1363–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martyanov V, Whitfield ML. Molecular stratification and precision medicine in systemic sclerosis from genomic and proteomic data. Curr Opin Rheumatol. 2016. January; 28(1):83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon JK, Martyanov V, Franks JM, Bernstein EJ, Szymonifka J, Magro C, et al. Belimumab for the Treatment of Early Diffuse Systemic Sclerosis: Results of a Randomized, Double-Blind, Placebo-Controlled, Pilot Trial. Arthritis Rheumatol. 2018. February; 70(2):308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khanna D, Spino C, Johnson S, Chung L, Whitfield ML, Denton CP, et al. Abatacept in Early Diffuse Cutaneous Systemic Sclerosis: Results of a Phase II Investigator-Initiated, Multicenter, Double-Blind, Randomized, Placebo-Controlled Trial. Arthritis Rheumatol. 2020. January; 72(1):125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martyanov V, Kim GJ, Hayes W, Du S, Ganguly BJ, Sy O, et al. Novel lung imaging biomarkers and skin gene expression subsetting in dasatinib treatment of systemic sclerosis-associated interstitial lung disease. PLoS One. 2017; 12(11):e0187580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon JK, Martyanov V, Magro C, Wildman HF, Wood TA, Huang WT, et al. Nilotinib (Tasigna) in the treatment of early diffuse systemic sclerosis: an open-label, pilot clinical trial. Arthritis Res Ther. 2015. August 18; 17:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinchcliff M, Huang CC, Wood TA, Matthew Mahoney J, Martyanov V, Bhattacharyya S, et al. Molecular signatures in skin associated with clinical improvement during mycophenolate treatment in systemic sclerosis. J Invest Dermatol. 2013. August; 133(8):1979–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martyanov V, Nesbeth Y, Cai G, Wood TA, Reder J, Constantine S, et al. Effect of Anabasum (JBT-101) on Gene Expression in Skin Biopsies from Subjects with Diffuse Cutaneous Systemic Sclerosis (dcSSc) and the Relationship of Baseline Molecular Subsets to Clinical Benefit in the Phase 2 Trial [abstract]. Arthritis Rheum. 2017; 69. [Google Scholar]
- 18.Franks J, Martyanov V, Wood TA, Crofford L, Keyes-Elstein L, Furst DE, et al. Machine Learning Classification of Peripheral Blood Gene Expression Identifies a Subset of Patients with Systemic Sclerosis Most Likely to Show Clinical Improvement in Response to Hematopoietic Stem Cell Transplant [abstract]. Arthritis Rheum. 2018; 70. [Google Scholar]
- 19.Khanna D, Furst DE, Clements PJ, Allanore Y, Baron M, Czirjak L, et al. Standardization of the Modified Rodnan Skin Score for Use in Clinical Trials of Systemic Sclerosis. Journal of Scleroderma and Related Disorders. 2017; 2(1):11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006. May; 38(5):500–501. [DOI] [PubMed] [Google Scholar]
- 21.Franks JM, Martyanov V, Cai G, Wang Y, Li Z, Wood TA, et al. A Machine Learning Classifier for Assigning Individual Patients with Systemic Sclerosis to Intrinsic Molecular Subsets. Arthritis Rheumatol. 2019. March 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010; 11(2):R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aiba S, Tabata N, Ohtani H, Tagami H. CD34+ spindle-shaped cells selectively disappear from the skin lesion of scleroderma. Arch Dermatol. 1994. May; 130(5):593–597. [PubMed] [Google Scholar]
- 24.Skobieranda K, Helm KF. Decreased expression of the human progenitor cell antigen (CD34) in morphea. Am J Dermatopathol. 1995. October; 17(5):471–475. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto N, Nishioka S, Sasai Y. Polarization microscopic investigation of collagen and acid glycosaminoglycans in the skin of progressive systemic sclerosis (PSS). Acta Histochem. 1995. April; 97(2):195–202. [DOI] [PubMed] [Google Scholar]
- 26.Lee JS, Park HS, Yoon HS, Chung JH, Cho S. CD34 stromal expression is inversely proportional to smooth muscle actin expression and extent of morphea. J Eur Acad Dermatol Venereol. 2018. December; 32(12):2208–2216. [DOI] [PubMed] [Google Scholar]
- 27.Elhai M, Avouac J, Hoffmann-Vold AM, Ruzehaji N, Amiar O, Ruiz B, et al. OX40L blockade protects against inflammation-driven fibrosis. Proc Natl Acad Sci U S A. 2016. July 5; 113(27):E3901–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W, Bhattacharyya S, Marangoni RG, Carns M, Dennis-Aren K, Yeldandi A, et al. The JAK/STAT pathway is activated in systemic sclerosis and is effectively targeted by tofacitinib. Journal of Scleroderma and Related Disorders. 2020; 5(1):40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu M, Pedroza M, Lafyatis R, George AT, Mayes MD, Assassi S, et al. Identification of cadherin 11 as a mediator of dermal fibrosis and possible role in systemic sclerosis. Arthritis Rheumatol. 2014. April; 66(4):1010–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varga J, Pasche B. Transforming growth factor beta as a therapeutic target in systemic sclerosis. Nat Rev Rheumatol. 2009. April; 5(4):200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delgado-Vega AM, Abelson AK, Sanchez E, Witte T, D’Alfonso S, Galeazzi M, et al. Replication of the TNFSF4 (OX40L) promoter region association with systemic lupus erythematosus. Genes Immun. 2009. April; 10(3):248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gourh P, Arnett FC, Tan FK, Assassi S, Divecha D, Paz G, et al. Association of TNFSF4 (OX40L) polymorphisms with susceptibility to systemic sclerosis. Ann Rheum Dis. 2010. March; 69(3):550–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice LM, Ziemek J, Stratton EA, McLaughlin SR, Padilla CM, Mathes AL, et al. A longitudinal biomarker for the extent of skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheumatol. 2015. November; 67(11):3004–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Zhao D, Guo Z, Li T, Qili M, Xu B, et al. Overexpression of SerpinE2/protease nexin-1 Contribute to Pathological Cardiac Fibrosis via increasing Collagen Deposition. Sci Rep. 2016. November 23; 6:37635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strehlow D, Jelaska A, Strehlow K, Korn JH. A potential role for protease nexin 1 overexpression in the pathogenesis of scleroderma. J Clin Invest. 1999. April; 103(8):1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice LM, Padilla CM, McLaughlin SR, Mathes A, Ziemek J, Goummih S, et al. Fresolimumab treatment decreases biomarkers and improves clinical symptoms in systemic sclerosis patients. J Clin Invest. 2015. July 1; 125(7):2795–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, Leask A, Abraham DJ, Pala D, Shiwen X, Khan K, et al. Heparan sulfate-dependent ERK activation contributes to the overexpression of fibrotic proteins and enhanced contraction by scleroderma fibroblasts. Arthritis Rheum. 2008. February; 58(2):577–585. [DOI] [PubMed] [Google Scholar]
- 38.Lofgren S, Hinchcliff M, Carns M, Wood T, Aren K, Arroyo E, et al. Integrated, multicohort analysis of systemic sclerosis identifies robust transcriptional signature of disease severity. JCI Insight. 2016. December 22; 1(21):e89073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Principal component analysis of Belimumab and Nilotinib samples. Belimumab samples were processed in two batches (red circles, green triangles) and nilotinib samples were processed in a single batch (blue crosses). Principal component analysis of these three batches did not exhibit a significant batch bias (as determined by gPCA, p=0.434).
Supplemental Figure 2. Systemic sclerosis histologic features analyzed. These images demonstrate examples of high scores for each of the seven histologic/immunophenotypic features analyzed. The scores shown, from left to right, include: global histologic severity (score: 3; low power view), alpha-smooth muscle actin (aSMA) (score: 2; magnification 40X), CD34 (score: 3; magnification 40X), collagen density (score: 3; magnification 4X), infiltrate intensity (score: 2; magnification 10X), follicle count (score: 2; low power view), thickness (score: 2.3μm; low power view).
Supplemental Figure 3. Diagram of data processing pipeline. A. Skin biopsy samples were analyzed by microarray and assigned to gene expression subset (normal-like, fibroproliferative, inflammatory) using a multinomial elastic net (GLMnet) classifier [21]. B. Skin biopsy samples were assessed for seven histologic/immunophenotypic features: thickness, follicle count, global histologic severity, infiltrate intensity, collagen density, and two fibroblast markers: CD34 and aSMA. Support vector machine learning (leave-one-out cross validation) was performed using each histologic feature score as inputs and gene expression subsets as classifiers.
Supplemental Figure 4. Change in gene expression compared to change in clinical severity. Heatmap demonstrates Pearson correlation coefficients for Log2(fold-change baseline versus 52 weeks) and Combined Response Index in Systemic Sclerosis (CRISS) for 30 aSMA/CD34 polarization genes. These 30 genes were identified as differentially expressed from baseline to 52 weeks among 18 clinical improvers.
Data Availability Statement
Data are available in a public, open access repository. All gene expression data has been deposited in Gene Expression Omnibus (GEO) (accession no. GSE65405 and GSE97248).


