ABSTRACT
In natural fertilisation, the female reproductive tract allows only a strictly selected sperm subpopulation to proceed in the vicinity of an unfertilised oocyte. Female-mediated sperm selection (also known as cryptic female choice (CFC)) is far from a random process, which frequently biases paternity towards particular males over others. Earlier studies have shown that CFC is a ubiquitous phenomenon in the animal kingdom and often promotes assortative fertilisation between genetically compatible mates. Here, I demonstrate that CFC for genetic compatibility likely also occurs in humans and is mediated by a complex network of interacting male and female genes. I also show that the relative contribution of genetic compatibility (i.e. the male–female interaction effect) to reproductive success is generally high and frequently outweighs the effects of individual males and females. Together, these facts indicate that, along with male- and female-dependent pathological factors, reproductive failure can also result from gamete-level incompatibility of the reproductive partners. Therefore, I argue that a deeper understanding of these evolutionary mechanisms of sperm selection can pave the way towards a more inclusive view of infertility and open novel possibilities for the development of more personalised infertility diagnostics and treatments.
Keywords: cryptic female choice, evolution, fertilisation, genetic incompatibility, infertility, mate choice, personalised reproductive medicine, sexual selection, sperm function
Introduction
Modern ARTs have helped millions of infertile couples to bypass their reproductive challenges. Thus, development of ART is indisputably one of the greatest achievements of medicine. However, despite the demonstrated efficiency of these treatments, the success rate of ART is still far from perfect, and many couples either fail to achieve pregnancy or need several treatment cycles to attain parenthood (Sakkas et al., 2015; De Geyter et al., 2018). Furthermore, diagnosis of infertility is extremely challenging (e.g. Gelbaya et al., 2014; Oehninger and Ombelet, 2019), and in a significant proportion of couples, the reason for infertility remains unexplained (Ray et al., 2012).
According to the current diagnostic practice, infertility is expected to arise from male- and female-dependent pathological factors or a combination of male and female factors (Gardner et al., 2018). However, in addition to male and female pathologies, natural fertilisation success is also heavily dependent on the ability of sperm to traverse the female reproductive tract in the vicinity of an unfertilised oocyte (Fitzpatrick and Lüpold, 2014; Sakkas et al., 2015). Importantly, it has been estimated that in humans, only about 1 out of 1 000 000 sperm are able to enter female oviducts, and only a few of these cells eventually manage to enter the fertilisation site, the ampulla (Eisenbach and Giojalas, 2006). Therefore, natural fertilisation is a highly selective process, in which only very few sperm cells are able to reach the unfertilised oocyte (Holt and Fazeli, 2015; Hanevik et al., 2016). Consequently, the fertilisation success of sperm is dependent on not only the intrinsic quality of the ejaculate (or the pathology of the female reproductive system) but also on the ability of sperm to successfully interact with the female reproductive tract and the oocyte (Fitzpatrick and Lüpold, 2014). In this sense, functionally relevant phenotypic evaluation of ejaculates may be practically impossible in the absence of the selective factors of the female reproductive tract.
Sperm are incapable of fertilising an oocyte immediately after ejaculation and fertilisation competence is achieved only in the female reproductive tract via a series of physiological changes known as sperm capacitation. Capacitated sperm show intense flagella beating (hyperactivation) and directional motility (chemotaxis) towards the chemical factors secreted by unfertilised oocytes. Only capacitated sperm can undergo the acrosome reaction, penetrate the zona pellucida and eventually bind and fertilise an oocyte. All these processes are strongly dependent on the secretions of the female reproductive tract, the oocyte and its surrounding cumulus and granulosa cells. Together, these female-induced biochemical factors allow highly specific sperm selection, possibly even at the level of individual spermatozoa (Holt and Fazeli, 2015). Traditionally, it has been thought that the female-induced sperm selection mechanisms have evolved primarily to eliminate fertilisation-incompetent sperm or to reduce the risk of polyspermy (Fitzpatrick and Lüpold, 2014; Kekäläinen and Evans, 2018). However, in this article, I show that female-mediated sperm selection can also facilitate assortative fusion between genetically compatible gametes. Based on this evidence, I argue that reproductive failure does not necessarily exclusively represent a pathological condition, but can also result from sexual selection (‘mate choice’) at the level of the gametes. Thus, better integration of this evolutionary concept into current infertility diagnostics may provide novel insights into the development of more accurate and personalised infertility diagnostics and treatments.
Cryptic female choice and gamete-mediated mate choice
Mate choice has traditionally been assumed to occur only at the level of the individuals (i.e. between males and females). However, in many species, it has been demonstrated to continue after mating in the form of cryptic female choice (CFC) (Firman et al., 2017). CFC refers to various female-driven mechanisms that act primarily prior to (or during) fertilisation and bias fertilisation towards the sperm of specific males. In many animal species, CFC is mediated by various female-derived reproductive secretions or via gamete surface molecules, both of which can have major impact on the fertilisation dynamics (Fig. 1). Together, these chemical factors mediate CFC at the level of the gametes (gamete-mediated mate choice (GMMC)) (reviewed by Kekäläinen and Evans, 2018). GMMC has previously been demonstrated to occur primarily in externally fertilising species, such as marine mussels, in which egg-derived sperm chemoattractants selectively change sperm swimming behaviour and thereby promote assortative fertilisation between genetically compatible gametes (e.g. Evans et al., 2012; Oliver and Evans, 2014). Furthermore, several fish studies have demonstrated that a similar fertilisation bias towards particular males can also be mediated by ovarian fluid (Urbach et al., 2005; Dietrich et al., 2008; Rosengrave et al., 2008; Gasparini and Pilastro, 2011; Rosengrave et al., 2016; Geßner et al., 2017).
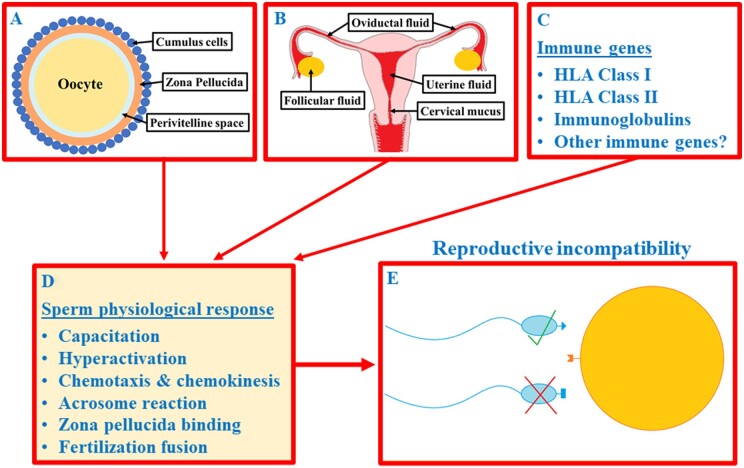
Figure 1.
Schematic illustration of the potential mechanisms of gamete-mediated mate choice in humans. (A) Surface proteins and glycans of the oocyte–cumulus complex and sperm chemoattractants released by these cells (e.g. chemokines, peptides and odourants); (B) other female-derived reproductive secretions; (C) various genes of the immune system. Together, these female-derived factors cause a number of physiological changes in sperm (D) that can selectively bias fertilisation towards the sperm of genetically compatible males (E).
In internally fertilising species, GMMC occurs within the female reproductive tract, which has hampered experimental attempts to demonstrate GMMC in such species, including humans. These technical difficulties most likely largely explain why experimental evidence of GMMC in humans has been lacking. However, Fitzpatrick et al. (2020) recently demonstrated that in humans, follicular fluid highly selectively attracts the sperm of specific males over others, and in this way, facilitates mate choice at the level of the gametes (Fig. 1). Additionally, Jokiniemi et al. (2020a,b) demonstrated (also in humans) that sperm performance in different female reproductive secretions (follicular fluid and cervical mucus) is strongly dependent on the male–female combination. In other words, female reproductive secretions were found to selectively increase sperm performance of some males but decrease it in for others. In both studies, sperm performance was also found to be higher in human leucocyte antigen (HLA) dissimilar male–female combinations, which suggests that the female reproductive tract may non-randomly promote gamete fusion between HLA compatible partners. Magris et al. (2021) also demonstrated that in addition to HLA, sperm performance in the female reproductive tract is dependent on the structural similarity of male and female immunoglobulins (antibodies). Together with the earlier findings in different animal species, these results indicate that one of the primary functions of GMMC may be to ‘evaluate’ the immunogenetic compatibility of the reproductive partners prior to gamete fusion.
Genetic interactions, genetic (in)compatibility and reproductive success
The effect of each individual gene on a phenotype is often assumed to be additive, when the combined effect of alleles at two or more gene loci should equal the sum of their separate effects (additive genetic effect). In other words, the phenotypic effect of genes is expected to be independent of all the other genes. However, in all sexually reproducing organisms, the genotypes of the individuals consist of complex networks of genetic interactions (non-additive genetic effects). These interactions can occur among alleles at the same locus, when one allele of a gene masks or overrides the effect of another allele of the same gene (dominance). Additionally, genetic interactions occur between different loci in a phenomenon known as epistasis, in which the phenotypic effect of one locus is enhanced or suppressed by the genotypes at the other locus (or there is a change in the direction of phenotypic effects) (Mackay, 2014). In both dominance and epistasis, the final effect of a gene (or allele) on the phenotype depends on the genotype of the associated genes (or alleles).
Non-additive genetic effects, and especially epistasis, have traditionally been assumed to act as important mechanisms maintaining the reproductive isolation between species (Hart et al., 2018). However, recent studies have demonstrated that epistatic interactions are also common within single species, including humans (Rohlfs et al., 2010; Corbett-Detig et al., 2013; Mackay, 2014; Wang et al., 2017). Importantly, non-additive genetic effects (of the male–female combination) are often much more important determinants of oocyte fertilisation success, embryo survival and fertility traits in general, compared with additive genetic effects of males or females (Palucci et al., 2007; Dziminski et al., 2008; Rodríguez-Muñoz and Tregenza, 2009; Agbali et al., 2010). In other words, reproductive success is frequently more strongly dependent on the male–female compatibility than on individual males and females. This indicates that the same genetic mechanisms responsible for preventing crossbreeding between individuals of different species can also lead to variation in reproductive compatibility between individual males and females within each species. Accordingly, certain male (sperm) genotypes that have high reproductive success with certain female (oocyte) genotypes can have much lower reproductive success with other female genotypes.
Molecular mechanisms of gamete-mediated mate choice
Widespread evidence for female-mediated fertilisation bias towards the sperm of genetically compatible males suggests that the female reproductive tract and oocytes can identify compatible sperm genotypes based on specific gamete surface molecular markers (Holt and Fazeli, 2015). Consequently, some earlier studies have demonstrated that gamete compatibility genes are expressed on the surface of the sperm and oocytes, and particularly male and female gamete surface proteins have been widely believed to play an important role as molecular targets in compatibility recognition (e.g. Stapper et al., 2015; Springate and Frasier, 2017) (Table I). Genes coding gamete surface proteins are among the fastest-evolving genes known (Swanson and Vacquier, 2002; Springate and Frasier, 2017), and GMMC is expected to act as an important driver of this evolutionary process, facilitating continual coevolution (reciprocal evolutionary change) between sperm and oocyte proteins (Springate and Frasier, 2017, see below). Due to this coevolutionary process, different variants (alleles) of the coevolving gene pairs often have differential compatibility, which can ultimately lead to complete incompatibility (i.e. reproductive failure) between certain allele pairs (Ziegler et al., 2005).
Table I.
Oocyte and sperm genes that are known to mediate physical interactions between gametes and are essential for fertilisation in mammals.
| Oocyte | Function | Location | Effect | Ref. |
|---|---|---|---|---|
| CD9 | Sperm–oocyte fusion | Oocyte surface | Deletion: fertility −40% | 1 |
| CD81 | Sperm–oocyte fusion | Oocyte surface | Deletion: fertility −38% | 1 |
| Juno | Sperm–oocyte membrane adhesion | Oocyte surface | Deletion: 100% infertility | 2 |
| ZP1-ZP3 | Sperm–oocyte binding/coevolution | Zona pellucida | Sperm–egg compatibility | 3,4 |
|
| ||||
| Sperm | ||||
|
| ||||
| Izumo1 | Sperm–oocyte membrane adhesion | Sperm surface after AR | Deletion: 100% infertility | 5 |
| FIMP | Sperm–oocyte fusion | Sperm equatorial segment | Deletion: severe subfertility | 6 |
| THEM95 | Sperm–oocyte fusion | Sperm plasma membrane | Deletion: 100% infertility | 7 |
| SOF1 | Sperm–oocyte fusion | Sperm plasma membrane | Deletion: 100% infertility | 7 |
| SPACA6 | Sperm–oocyte fusion | Sperm plasma membrane | Deletion: 100% infertility | 7 |
| DCST1/DCST2 | Sperm–oocyte fusion | Sperm plasma membrane | Deletion: 100% infertility | 8 |
| C4BPA (ZP3R) | Sperm–oocyte binding/coevolution | Sperm plasma membrane | Sperm–egg compatibility | 3,4 |
| PKDREJ | Zona pellucida (ZP) binding | Sperm acrosome | Mutation: lower fertility | 9 |
| CRISP1/CRISP2 | Sperm–oocyte interaction | Sperm plasma membrane | Blocking: lower fertility | 9 |
| PH-20 | Cumulus penetration + ZP binding | Sperm plasma membrane | Deletion: delayed fertilisation | 9,10 |
| Zonadhesin | ZP binding | Sperm acrosome | Blocking: lower fertility | 9 |
See also Gahlay and Rajput (2020) for a comprehensive list of sperm genes involved in the interaction of the sperm with the female reproductive tract interaction.
AR, Acrosome reaction.
References: 1. Rubinstein et al., 2006; 2. Bianchi et al., 2014; 3. Rohlfs et al., 2010; 4. Hart et al., 2018; 5. Inoue et al., 2005; 6. Fujihara et al., 2020; 7. Noda et al., 2020; 8. Inoue et al., 2021; 9. Springate and Frasier, 2017; 10. Baba et al., 2002.
In addition to proteins, it has been shown that gamete compatibility is also dependent on their surface carbohydrates (glycans). Ghaderi et al. (2011) demonstrated in mice that reproductive incompatibility between males and females is caused by a female immune response against certain (‘mismatched’) sperm surface glycans. Similarly, Kekäläinen and Evans (2017) showed in a marine mussel that egg-derived chemical factors trigger structural changes in sperm surface glycans and sperm fertilisation capability, and that the strength of these physiological changes is strongly dependent on the male-female combination. Kekäläinen and Evans (2017) also demonstrated that the compatibility verification process of the gametes likely commences before the physical contact of the sperm and oocytes, via chemical signals secreted by the female reproductive tract and unfertilised oocytes (reviewed by Kekäläinen and Evans, 2018). This is important, because during the fertilisation process, sperm are exposed to multiple female-derived reproductive secretions, including follicular fluid, oviductal fluid, uterine fluid and cervical mucus, indicating that the selection of genetically compatible sperm can occur in different parts of the female reproductive tract.
Potential (in)compatibility genes in humans and other mammals
The identities of interacting male and female genes responsible for gamete-level incompatibilities are still largely unclear, and only a few potentially interacting candidate genes have been found. This is largely due to the fact that only one directly interacting sperm–oocyte ‘binding protein’ pair has so far been identified (Izumo1–Juno: Bianchi et al., 2014; Bianchi and Wright, 2020). However, it is likely that many other gamete surface proteins play important roles as mediators of sperm and oocyte interactions. Supporting this view, gamete surface ‘reproductive’ proteins have been demonstrated to diverge (evolve) rapidly, and continual coevolution between interacting sperm and oocyte proteins is likely a key driver of this divergence (Clark et al., 2009). In this coevolutionary process, one or both ‘members’ of the interacting protein pairs adaptively compensates for changes in the other, which can eventually lead to variation in reproductive compatibility between certain allele pairs of the male and female genes encoding these proteins (Hart et al., 2018).
Accumulating numbers of studies have highlighted that coevolution between sperm and oocyte genes is likely common in mammals, including humans (Vicens and Roldan, 2014; Hart et al., 2018). For example, Grayson (2015) showed that Izumo1 and Juno are coevolving under similar selection pressures, which are at least partly driven by sexual selection. This indicates that some Izumo1-Juno allele pairs have higher compatibility (gamete fusion success) than others, causing variation in the compatibility between reproductive partners. Rohlfs et al. (2010) also demonstrated in humans that the zona pellucida (glycoprotein layer surrounding the oocyte) gene ZP3 coevolves with its putative binding partner, ZP3R, in sperm (Table I). Furthermore, recent genome editing studies have revealed several other sperm- and oocyte-specific genes that play an important role in gamete interaction (Abbasi et al., 2020; Fujihara et al., 2020; Lamas-Toranzo et al., 2020; Noda et al., 2020). Although the binding partners of these genes remain to be demonstrated in future studies, all of them have a potential to increase our understanding of the molecular mechanisms of gamete incompatibility.
Many of the key molecules responsible in gamete recognition and binding are not directly situated on gamete surfaces but are dispersed in various female reproductive secretions (Bernabò et al., 2014). Accordingly, sperm behaviour and function in the female reproductive tract are strongly dependent on a large array of female-derived soluble factors, such as chemokines, small peptides and odourant molecules (Brenker et al., 2012). Furthermore, it has been demonstrated that many female-derived factors are transferred from the female reproductive fluids onto the sperm plasma membrane prior to fertilisation (Al-Dossary et al., 2013). For example, two key oocyte surface proteins (CD9 and CD81) known to be involved in sperm–oocyte fusion are also released from oocytes via exosomes (oocyte-derived extracellular vesicles) and interact with sperm before the physical contact of the gametes (Ohnami et al., 2012). Interestingly, many sperm plasma membrane protein genes, such as SPAM1, PMCA4a, CRISP1 and CATSPER, are also expressed in the female reproductive tract (Griffiths et al., 2008; Al-Dossary et al., 2013; Ernesto et al., 2015; Martinez et al., 2020) and have an important role in regulating sperm function (reviewed by Hernández-Silva and Chirinos, 2019). For example, female-derived CRISP1 proteins were found to regulate sperm Ca2+ channels critical for sperm motility (Ernesto et al., 2015). Crucially, fertilisation of the oocytes of CRISP1 knockout female mice was severely impaired, indicating that female-expressed CRISP1 proteins have a key function in determining the fertilisation capability of sperm.
Together this evidence indicates that genetic compatibility of the reproductive partners may be dependent on the complex network of interacting male and female genes. These genes may not be expressed exclusively on the sperm or oocyte surfaces, but are likely already acting before the physical contact of the gametes via female reproductive tract secretions. This, in turn, indicates that the reproductive compatibility of the partners may be a result of a large number of functionally redundant and possibly relatively weak receptor–ligand interactions (Wright and Bianchi, 2016), which collectively determine the overall compatibility of the partners.
Clinical significance and future challenges
The primary reason for fertilisation failure in conventional IVF is an unsuccessful sperm–oocyte interaction (Sabetian et al., 2014). It has commonly been assumed that this is primarily caused by some defects in sperm or oocyte membrane proteins mediating the interaction (Sabetian and Shamsir, 2017) or other abnormalities in the ability of sperm to bind and penetrate the zona pellucida (Hamada et al., 2011). However, Firman and Simmon (2015) demonstrated in mice that the success rate of IVF is also dependent on oocyte-driven mechanisms of sperm selection that bias fertilisation towards the sperm of genetically compatible (non-sibling) males. Similarly, Stapper et al. (2015) found in sea urchins that eggs non-randomly fused with the sperm that had cell surface protein (bindin) genotypes similar to their own. Finally, Lenz et al. (2018) showed in sticklebacks that after controlled IVF, eggs can distinguish sperm genotypes even at the level of individual alleles (haplotypes) and assortatively fuse with complementary sperm haplotypes. Together with the above-mentioned facts, these findings indicate that fertilisation failure does not necessarily represent a pathological condition, but can also result from genetic incompatibility avoidance at the level of the gametes.
Besides affecting the probability of the fertilisation, the compatibility of the gametes at fertilisation has also been demonstrated to be positively associated with embryo survival (Dziminski et al., 2008; Rodríguez-Muñoz and Tregenza, 2009; Agbali et al., 2010; Aguirre et al., 2016; Byrne et al., 2021). Therefore, it is likely that the genetic compatibility of the reproductive partners has a major impact on both fertilisation success and the probability of achieving successful pregnancy and, in this way, influences the overall success rate of infertility treatments. However, according to the definition currently used by the World Health Organization (WHO), infertility is seen as a disease of the reproductive system and is thus assumed to be caused by male- or female-derived pathological factors. In light of previous findings, this may be an overly simplistic view, since it misses the important fact that some male-female (gamete) combinations often ‘match’ better than the others. Therefore, I argue that we need a more inclusive definition of infertility, one which takes into account the possibility that the probability of conception is also affected by the evolutionary mechanisms that strive to ensure the compatibility of the parental genes prior to gamete fusion. This broader definition of infertility can open novel possibilities to better understand the current reliability challenges of infertility diagnostics and to understand why the current diagnostic tests frequently fail to find any clear reason for reproductive failure (Ray et al., 2012) (Table II).
Table II.
Potential clinical relevance of investigating genetic incompatibility of the reproductive partners and key challenges for future researchers and clinicians working in the fields of human reproduction and ARTs.
| Clinical relevance: |
|---|
| Improved accuracy of infertility diagnostics |
| Improved predictive value of the semen analyses |
| More personalised infertility treatments, tailored to each individual couple |
| Reduced overall costs of ART procedures |
|
|
| Future research challenges: |
|
|
| Identify functionally important male and female genes responsible for gamete-level incompatibility |
| Understand detailed mechanisms of sperm selection in the female reproductive tract |
| Clarify how the female reproductive tract and oocytes ‘identify’ compatible sperm genotypes |
| Develop analytical methods for genome-wide characterisation of genetic interactions and genetic interaction networks responsible for gamete-level incompatibility |
| Investigate the effect of genetic interactions and genetic incompatibility on the health of the offspring |
|
|
| Future clinical challenges: |
|
|
| Develop clinical tests for parental genetic compatibility |
| Develop more realistic functional tests for sperm fertilisation capability and male fertility |
| Clarify how to prevent the negative impact of genetic incompatibility on reproductive success and offspring health |
Fertility traits and reproduction success in general are often dominated by non-additive genetic effects (e.g. Alves et al., 2020). Therefore, in order to predict the probability of conception in individual couples, it is critically important to gain an understanding of how specific male and female genes interact during the fertilisation process. Consequently, deeper understanding of epistatic and dominance interactions between reproductive partners has a great potential to improve the accuracy of infertility diagnostics and facilitate development of more personalised diagnostic tools (Table II). Personalised reproductive medicine is still in its infancy, and routine clinical tests for parental genetic compatibility are lacking (Beim et al., 2017). However, the rapidly decreasing costs of modern whole-genome sequencing techniques raise an important possibility of including genome-wide characterisation of incompatibility genes in future diagnostics routines. Importantly, recent advances in analytical methods now enable robust identification of genetic interactions and genetic interaction networks from the genome-wide data (e.g. Fang et al., 2019; Sun et al., 2020).
As highlighted above, accumulating evidence indicates that the definitive reproductive incompatibility of the partners is affected by large number of male and female genes, many of which are expressed in female reproductive tract secretions. Consequently, these female-derived secretions could potentially enable diagnosis of the reproductive incompatibility of the partners without the need to fertilise the oocytes (cf. Jokiniemi et al., 2020a,b; Magris et al., 2021). Furthermore, female reproductive secretions could also open novel possibilities for the development of biologically more realistic functional tests for sperm fertilisation capability and male fertility. Thus, besides allowing the evaluation of the reproductive compatibility of the couples, such functional tests could also increase the overall predictive value of semen analyses (Table II). In practice, sperm functional tests could involve, for example, measuring sperm physiological response to follicular fluid or cervical mucus, both of which can be relatively easily collected during routine ART procedures. Additionally, it has been demonstrated that sperm functional response in ‘non-reproductive’ biological fluids, such as serum, could potentially be used as a reliable indicator of sperm motility and function in female-derived reproductive fluids (Lee et al., 1994; Mandal et al., 2006; Dungdung et al., 2016). This raises an intriguing possibility that the reproductive compatibility of the partners could be screened as a part of the initial infertility testing, which could provide novel opportunities to tailor the following infertility treatments to each couple. However, more studies are needed to experimentally investigate the diagnostic potential of proposed reproductive incompatibility tests and to identify the most suitable candidate genes and other biomarkers to be utilised in such tests.
Conclusion
According to the current definition, infertility is a disease of the male or female reproductive system. However, an infertility diagnosis can be extremely challenging, and the exact reason for infertility often remains unknown. Recent evolutionary studies have demonstrated that, in addition to being dependent on individual males and females, fertilisation success is also strongly dependent on the reproductive compatibility of the partners (non-additive genetic effects) and that the definitive ‘test’ for male–female compatibility occurs in the female reproductive tract prior to the fertilisation. Therefore, it seems likely that reproductive failure is not exclusively a pathological condition but is also affected by mate choice at the level of the gametes, which reduces the probability of conception between genetically incompatible partners. GMMC is likely based on complex network of interacting male and female genes, which are expressed both on the surface of the gametes and in the female reproductive tract secretions. Besides mediating sperm selection towards those of compatible partners prior to the physical contact of the gametes, female-derived reproductive secretions may also offer novel tools to diagnose the reproductive incompatibility of the partners and thus facilitate development of biologically more realistic fertility tests. Overall, a deeper understanding of molecular basis of reproductive incompatibility may open novel possibilities to overcome the barriers to truly personalised infertility diagnostics and treatments.
Data availability
No new data were generated or analysed in support of this research.
Author’s roles
The manuscript was written by the author only.
Acknowledgements
I would like to thank Annalaura Jokiniemi for comments on the earlier version of the manuscript.
Funding
Funding was received from the Academy of Finland (308485).
Conflict of interest
The author declares that no conflicts of interest exist.
References
- Abbasi F, Kodani M, Emori C, Kiyozumi D, Mori M, Fujihara Y, Ikawa M.. CRISPR/Cas9-mediated genome editing reveals Oosp family genes are dispensable for female fertility in mice. Cells 2020;9:821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbali M, Reichard M, Bryjová A, Bryja J, Smith C.. Mate choice for nonadditive genetic benefits correlate with MHC dissimilarity in the rose bitterling (Rhodus ocellatus). Evolution 2010;64:1683–1696. [DOI] [PubMed] [Google Scholar]
- Aguirre JD, Blows MW, Marshall DJ.. Genetic compatibility underlies benefits of mate choice in an external fertilizer. Am Nat 2016;187:647–657. [DOI] [PubMed] [Google Scholar]
- Al-Dossary AA, Strehler EE, Martin-DeLeon PA.. Expression and secretion of plasma membrane Ca2+-ATPase 4a (PMCA4a) during murine estrus: association with oviductal exosomes and uptake in sperm. PLoS One 2013;8:e80181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves K, Brito LF, Baes CF, Sargolzaei M, Robinson JAB, Schenkel FS.. Estimation of additive and non-additive genetic effects for fertility and reproduction traits in North American Holstein cattle using genomic information. J Anim Breed Genet 2020;137:316–330. [DOI] [PubMed] [Google Scholar]
- Baba D, Kashiwabara S, Honda A, Yamagata K, Wu Q, Ikawa M, Okabe M, Baba T.. Mouse sperm lacking cell surface hyaluronidase PH-20 can pass through the layer of cumulus cells and fertilize the egg. J Biol Chem 2002;277:30310–30314. [DOI] [PubMed] [Google Scholar]
- Beim PY, Parfitt D-E, Tan L, Sugarman EA, Hu-Seliger T, Clementi C, Levy B.. At the dawn of personalized reproductive medicine: opportunities and challenges with incorporating multigene panel testing into fertility care. J Assist Reprod Genet 2017;34:1573–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabò N, Ordinelli A, Di Agostino R, Mattioli M, Barboni B.. Network analyses of sperm-egg recognition and binding: ready to rethink fertility mechanisms? OMICS 2014;18:740–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E, Doe B, Goulding D, Wright GJ.. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 2014;508:483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E, Wright GJ.. Find and fuse: unsolved mysteries in sperm–egg recognition. PLoS Biol 2020;18:e3000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenker C, Goodwin N, Weyand I, Kashikar ND, Naruse M, Krähling M, Müller A, Kaupp UB, Strünker T.. The CatSper channel: a polymodal chemosensor in human sperm. EMBO J 2012;31:1654–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne PG, Keogh JS, O'Brien DM, Gaitan-Espitia JD, Silla AJ.. Evidence that genetic compatibility underpins female mate choice in a monandrous amphibian. Evolution 2021;75:529–541. [DOI] [PubMed] [Google Scholar]
- Clark NL, Gasper J, Sekino M, Springer SA, Aquadro CF, Swanson WJ.. Coevolution of interacting fertilization proteins. PLoS Genet 2009;5:e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Zhou J, Clark AG, Hartl DL, Ayroles JF.. Genetic incompatibilities are widespread within species. Nature 2013;504:135–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Geyter C, Calhaz-Jorge C, Kupka MS, Wyns C, Mocanu E, Motrenko T, Scaravelli G, Smeen J, Vidakovic S, Goossens V; European IVF-monitoring Consortium (EIM) for the European Society of Human Reproduction and Embryology (ESHRE). ART in Europe, 2014: results generated from European registries by ESHRE: The European IVF-monitoring Consortium (EIM) for the European Society of Human Reproduction and Embryology (ESHRE). Hum Reprod 2018;33:1586–1601. [DOI] [PubMed] [Google Scholar]
- Dietrich GJ, Wojtczak M, Słowińska M, Dobosz S, Kuźmiński H, Ciereszko A.. Effects of ovarian fluid on motility characteristics of rainbow trout (Oncorhynchus mykiss Walbaum) spermatozoa. J Appl Ichthyol 2008;24:503–507. [Google Scholar]
- Dungdung S, Bhoumik A, Saha S, Ghosh P, Das K, Mukherjee S, Nath D, Chakrabarty J, Kundu C, Jaiswal B.. Sperm motility regulatory proteins: a tool to enhance sperm quality. IntechOpen 2016;doi:10.5772/62470. [Google Scholar]
- Dziminski MA, Roberts D, Simmons LW.. Fitness consequences of parental compatibility in the frog Crinia georgiana. Evolution 2008;62:879–886. [DOI] [PubMed] [Google Scholar]
- Eisenbach M, Giojalas LC.. Sperm guidance in mammals—an unpaved road to the egg. Nat Rev Mol Cell Biol 2006;7:276–285. [DOI] [PubMed] [Google Scholar]
- Ernesto JI, Weigel Muñoz M, Battistone MA, Vasen G, Martínez-López P, Orta G, Figueiras-Fierro D, De la Vega-Beltran JL, Moreno IA, Guidobaldi HA. et al. CRISP1 as a novel CatSper regulator that modulates sperm motility and orientation during fertilization. J Cell Biol 2015;210:1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JP, Garcia-Gonzalez F, Almbro M, Robinson O, Fitzpatrick JL.. Assessing the potential for egg chemoattractants to mediate sexual selection in a broadcast spawning marine invertebrate. Proc R Soc B 2012;279:2855–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Wang W, Paunic V, Heydari H, Costanzo M, Liu X, Liu X, VanderSluis B, Oately B, Steinbach M. et al. Discovering genetic interactions bridging pathways in genome-wide association studies. Nat Commun 2019;10:4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firman R, Gasparini C, Manier MK, Pizzari T.. Postmating female control: 20 years of cryptic female choice. Trends Ecol Evol 2017;32:368–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firman RC, Simmon LW.. Gametic interactions promote inbreeding avoidance in house mice. Ecol Lett 2015;18:937–943. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JL, Lüpold S.. Sexual selection and the evolution of sperm quality. Mol Hum Reprod 2014;20:1180–1189. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JL, Willis C, Devigili A, Young A, Carroll M, Hunter HR, Brison DR.. Chemical signals from eggs facilitate cryptic female choice in humans. Proc R Soc B 2020;287:20200805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara Y, Lu Y, Noda T, Oji A, Larasati T, Kojima-Kita K, Yu Z, Matzuk RM, Matzuk MM, Ikawa M.. Spermatozoa lacking Fertilization Influencing Membrane Protein (FIMP) fail to fuse with oocytes in mice. Proc Natl Acad Sci USA 2020;117:9393–9400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlay GK, Rajput N.. The enigmatic sperm proteins in mammalian fertilization: an overview. Biol Reprod 2020;103:1171–1185. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Weissman A, Howles CM, Shoham Z.. Textbook of Assisted Reproductive Techniques, 5th edn.London and New York: Taylor & Francis, 2018. [Google Scholar]
- Gasparini C, Pilastro A.. Cryptic female preference for genetically unrelated males is mediated by ovarian fluid in the guppy. Proc R Soc B 2011;278:2495–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbaya TA, Potdar N, Jeve YB, Nardo LG.. Definition and epidemiology of unexplained infertility. Obstet Gynecol Surv 2014;69:109–115. [DOI] [PubMed] [Google Scholar]
- Geßner C, Nakagawa S, Zavodna M, Gemmell NJ.. Sexual selection for genetic compatibility: the role of the major histocompatibility complex on cryptic female choice in Chinook salmon (Oncorhynchus tshawytscha). Heredity (Edinb) 2017;118:442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaderi D, Springer SA, Ma F, Cohen M, Secrest P, Taylor RE, Varki A, Gagneux P.. Sexual selection by female immunity against paternal antigens can fix loss of function alleles. Proc Natl Acad Sci USA 2011;108:17743–17748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson P. Izumo1 and Juno: the evolutionary origins and coevolution of essential sperm–egg binding partners. R Soc Open Sci 2015;2:150296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths GS, Miller KA, Galileo DS, Martin-DeLeon PA.. Murine SPAM1 is secreted by the estrous uterus and oviduct in a form that can bind to sperm during capacitation: acquisition enhances hyaluronic acid-binding ability and cumulus dispersal efficiency. Reproduction 2008;135:293–301. [DOI] [PubMed] [Google Scholar]
- Hamada A, Esteves SC, Agarwal A.. Unexplained male infertility: potential causes and management. Hum Androl 2011;1:2–16. [Google Scholar]
- Hanevik HI, Hessen DO, Sunde A, Breivik J.. Can IVF influence human evolution? Hum Reprod 2016;31:1397–1402. [DOI] [PubMed] [Google Scholar]
- Hart MW, Stover DA, Guerra V, Mozaffari SV, Ober C, Mugal CF, Kaj I.. Positive selection on human gamete-recognition genes. PeerJ 2018;6:e4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Silva G, Chirinos M.. Proteins from male and female reproductive tracts involved in sperm function regulation. Zygote 2019;27:5–16. [DOI] [PubMed] [Google Scholar]
- Holt WV, Fazeli A.. Do sperm possess a molecular passport? Mechanistic insights into sperm selection in the female reproductive tract. Mol Hum Reprod 2015;21:491–501. [DOI] [PubMed] [Google Scholar]
- Inoue N, Hagihara Y, Wada I.. Evolutionarily conserved sperm factors, DCST1 and DCST2, are required for gamete fusion. eLife 2021;10:e66313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue N, Ikawa M, Isotani A, Okabe M.. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 2005;434:234–238. [DOI] [PubMed] [Google Scholar]
- Jokiniemi A, Kuusipalo L, Ritari J, Koskela S, Partanen J, Kekäläinen J.. Gamete-level immunogenetic incompatibility in humans–towards deeper understanding of fertilization and infertility? Heredity (Edinb) 2020a;125:281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokiniemi A, Magris M, Ritari J, Kuusipalo L, Lundgren T, Partanen J, Kekäläinen J.. Post-copulatory genetic matchmaking: HLA-dependent effects of cervical mucus on human sperm function. Proc R Soc B 2020b;287:20201682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kekäläinen J, Evans JP.. Female-induced remote regulation of sperm physiology may provide opportunities for gamete-level mate choice. Evolution 2017;71:238–248. [DOI] [PubMed] [Google Scholar]
- Kekäläinen J, Evans JP.. Gamete-mediated mate choice: towards a more inclusive view of sexual selection. Proc R Soc B 2018;285:20180836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas-Toranzo I, Hamze JG, Bianchi E, Fernández-Fuertes B, Pérez-Cerezales S, Laguna-Barraza R, Fernández-González R, Lonergan P, Gutiérrez-Adán A, Wright GJ. et al. TMEM95 is a sperm membrane protein essential for mammalian fertilization. eLife 2020;9:e53913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-L, Kao C-C, Wei Y-H.. Antithrombin III enhances the motility and chemotaxis of boar sperm. Comp Biochem Physiol Comp Physiol 1994;107:277–282. [PubMed] [Google Scholar]
- Lenz TL, Hafer N, Samonte IE, Yeates SE, Milinski M.. Cryptic haplotype-specific gamete selection yields offspring with optimal MHC immune genes. Evolution 2018;72:2478–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat Rev Genet 2014;15:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magris M, Jokiniemi A, Kuusipalo L, Ritari J, Koskela S, Partanen J, Kekäläinen J.. Structural dissimilarity of partners’ immune genes increases sperm viability in women’s reproductive tract. J Evol Biol 2021;34:1125–1132. [DOI] [PubMed] [Google Scholar]
- Mandal M, Saha S, Ghosh AK, Majumder GC.. Identification and characterization of a sperm motility promoting glycoprotein from buffalo blood serum. J Cell Physiol 2006;209:353–362. [DOI] [PubMed] [Google Scholar]
- Martinez CA, Alvarez-Rodriguez M, Wright D, Rodriguez-Martinez H.. Does the pre-ovulatory pig oviduct rule sperm capacitation in vivo mediating transcriptomics of Catsper channels? Int J Mol Sci 2020;21:1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Lu Y, Fujihara Y, Oura S, Koyano T, Kobayashi S, Matzuk MM, Ikawa M.. Sperm proteins SOF1, TMEM95, and SPACA6 are required for sperm-oocyte fusion in mice. Proc Natl Acad Sci USA 2020;117:11493–11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehninger S, Ombelet W.. Limits of current male fertility testing. Fertil Steril 2019;111:835–841. [DOI] [PubMed] [Google Scholar]
- Ohnami N, Nakamura A, Miyado M, Sato M, Kawano N, Yoshida K, Harada Y, Takezawa Y, Kanai S, Ono C. et al. CD81 and CD9 work independently as extracellular components upon fusion of sperm and oocyte. Biol Open 2012;1:640–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver M, Evans JP.. Chemically moderated gamete preferences predict offspring fitness in a broadcast spawning invertebrate. Proc R Soc B 2014;281:20140148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palucci V, Schaeffer LRS, Miglior FM, Osborne V.. Non-additive genetic effects for fertility traits in Canadian Holstein cattle. Genet Sel Evol 2007;39:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Shah A, Gudi A, Homburg R.. Unexplained infertility: an update and review of practice. Reprod Biomed Online 2012;24:591–602. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Muñoz R, Tregenza T.. Genetic compatibility and hatching success in the sea lamprey (Petromyzon marinus). Biol Lett 2009;5:286–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfs RV, Swanson WJ, Weir BS.. Detecting coevolution through allelic association between physically unlinked loci. Am J Hum Genet 2010;86:674–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengrave P, Gemmell NJ, Metcalf V, McBride K, Montgomerie R.. A mechanism for cryptic female choice in Chinook salmon. Behav Ecol 2008;19:1179–1185. [Google Scholar]
- Rosengrave P, Montgomerie R, Gemmell N.. Cryptic female choice enhances fertilization success and embryo survival in chinook salmon. Proc R Soc B 2016;283:20160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein E, Ziyyat A, Prenant M, Wrobel E, Wolf J-P, Levy S, Le Naour F, Boucheix C.. Reduced fertility of female mice lacking CD81. Dev Biol 2006;290:351–358. [DOI] [PubMed] [Google Scholar]
- Sabetian S, Shamsir MS, Naser MA.. Functional features and protein network of human sperm-egg interaction. Syst Biol Reprod Med 2014;60:329–337. [DOI] [PubMed] [Google Scholar]
- Sabetian S, Shamsir MS.. Deficiency in sperm–egg protein interaction as a major cause of fertilization failure. J Membr Biol 2017;250:133–144. [DOI] [PubMed] [Google Scholar]
- Sakkas D, Ramalingam M, Garrido N, Barratt CLR.. Sperm selection in natural conception: what can we learn from Mother Nature to improve assisted reproduction outcomes? Hum Reprod Update 2015;21:711–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springate L, Frasier TR.. Gamete compatibility genes in mammals: candidates, applications and a potential path forward. R Soc Open Sci 2017;4:170577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapper AP, Beerli P, Levitan DR.. Assortative mating drives linkage disequilibrium between sperm and egg recognition protein loci in the sea urchin Strongylocentrotus purpuratus. Mol Biol Evol 2015;32:859–870. [DOI] [PubMed] [Google Scholar]
- Sun S, Dong B, Zou Q.. Revisiting genome-wide association studies from statistical modelling to machine learning. Brief Bioinform 2020;22:bbaa263. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD.. The rapid evolution of reproductive proteins. Nat Rev Genet 2002;3:137–144. [DOI] [PubMed] [Google Scholar]
- Urbach D, Folstad I, Rudolfsen G.. Effects of ovarian fluid on sperm velocity in Arctic charr (Salvelinus alpinus). Behav Ecol Sociobiol 2005;57:438–444. [Google Scholar]
- Vicens A, Roldan ERS.. Coevolution of positively selected IZUMO1 and CD9 in rodents: evidence of interaction between gamete fusion proteins? Biol Reprod 2014;90:113–119. [DOI] [PubMed] [Google Scholar]
- Wang H, Choi Y, Tayo B, Wang X, Morris N, Zhang X, Broeckel U, Hanis C, Kardia S, Redline S. et al. Genome-wide survey in African Americans demonstrates potential epistasis of fitness in the human genome. Genet Epidemiol 2017;41:122–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GJ, Bianchi E.. The challenges involved in elucidating the molecular basis of sperm–egg recognition in mammals and approaches to overcome them. Cell Tissue Res 2016;363:227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, Kentenich H, Uchanska-Ziegler B.. Female choice and the MHC. Trends Immunol 2005;26:496–502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analysed in support of this research.