Abstract
The adenovirus E1B 55-kDa protein binds to cellular tumor suppressor p53 and inactivates its transcriptional transactivation function. p53 transactivation activity is dependent upon its ability to bind to specific DNA sequences near the promoters of its target genes. It was shown recently that p53 is acetylated by transcriptional coactivators p300, CREB bidning protein (CBP), and PCAF and that acetylation of p53 by these proteins enhances p53 sequence-specific DNA binding. Here we show that the E1B 55-kDa protein specifically inhibits p53 acetylation by PCAF in vivo and in vitro, while acetylation of histones and PCAF autoacetylation is not affected. Furthermore, the DNA-binding activity of p53 is diminished in cells expressing the E1B 55-kDa protein. PCAF binds to the E1B 55-kDa protein and to a region near the C terminus of p53 encompassing Lys-320, the specific PCAF acetylation site. We further show that the E1B 55-kDa protein interferes with the physical interaction between PCAF and p53, suggesting that the E1B 55-kDa protein inhibits PCAF acetylase function on p53 by preventing enzyme-substrate interaction. These results underscore the importance of p53 acetylation for its function and suggest that inhibition of p53 acetylation by viral oncoproteins prevent its activation, thereby contributing to viral transformation.
The cellular tumor suppressor p53 exerts its tumor suppression functions largely by acting as a transcriptional transactivator. In response to a variety of stimuli, such as DNA damage and expression of cellular or viral oncoproteins, p53 is stabilized and binds to specific DNA sequences in the vicinity of the promoter of its target genes and activates their transcription. The genes activated by p53 include p21 ( also called WAF1 or Cip1) (cyclin-dependent kinase inhibitor), cyclin G, GADD45, Mdm2, and Bax1 (apoptosis inducer). The products of these genes are implicated in regulation of cell cycle progression, DNA replication, and apoptosis (13, 25, 31).
Growth arrest or apoptosis imposed by p53 could severely hinder the replication of small DNA tumor viruses, as such replication requires host cells to enter the S phase. Thus, it is not surprising that a number of viral oncoproteins, such as the adenovirus (Ad) E1B 55-kDa protein, human papillomavirus (HPV) E6, and simian virus 40 large T antigen, bind to and repress the biological functions of p53 (30, 34, 49, 66). The E6 proteins of highly oncogenic HPV types 16 and 18 (HPV16 and HPV18) associate with p53 and target it for ubiquitination and subsequent degradation (51). Simian virus 40 large T antigen binds to the sequence-specific DNA binding domain of p53 (52, 56). This interaction interferes with sequence-specific DNA binding of p53 and therefore inhibits p53-mediated transcriptional transactivation (2, 10, 41).
Inhibition of p53 transactivation function is thought to be the key step in cell transformation induced by Ad (45, 69). The transforming function of Ad maps to the early region 1 (E1) of the 36-kb Ad genome (45). The E1 region encompasses two independent transcription units, E1A and E1B. E1A encodes two major polypeptides, 289R and 243R, whereas E1B transcript specifies two overlapping open reading frames which encode E1B 19-kDa and 55-kDa proteins. The E1A proteins bind to retinoblastoma protein pRb and inhibit its function in regulating cell cycle progression (11) and also appear to affect p53 functions (54). The E1B 19-kDa protein functions as an inhibitor of apoptosis (7, 36; reviewed in reference 46). The E1B 55-kDa protein suppresses p53 transactivation activity and also p53-mediated apoptosis (38, 55, 58, 69, 71). Both E1B proteins are required to fully transform cells in cooperation with E1A (3, 14, 60).
In Ad-transformed cells as well as in vitro, the E1B 55-kDa protein tightly associates with p53 (24, 48, 70, 72). Linker insertion mutagenesis of Ad type 2 (Ad2) E1B 55-kDa protein indicated that two regions around position H180 and between positions A262 and H326 are important for p53-E1B 55-kDa protein interaction (70). In a reciprocal study using in vitro immunoprecipitation (IP) assays, the amino-terminal 123 residues of murine p53 were shown to be responsible for binding to E1B 55-kDa protein (24). Several hydrophobic amino acid residues including Trp-23 and Pro-27 of human p53 are important for binding to the E1B 55-kDa protein, and these hydrophobic residues are also critical for p53 transactivation activity (33). These studies thus suggest that the interaction between E1B 55-kDa protein and p53 is important to inactivate p53 transactivation function. Further studies demonstrated that transcriptional repression function of E1B 55-kDa protein correlates with its ability to transform cells and that binding to p53 is necessary but not sufficient for transcriptional repression and transformation activities of the Ad2 E1B 55-kDa oncoprotein (71). Moreover, phosphorylation of three residues (Ser-490 and -491 and Thr-495) near the carboxyl terminus of Ad5 E1B 55-kDa protein is also required for transcriptional repression and transformation (57). The E1B 55-kDa protein from highly oncogenic Ad12 also represses p53 transactivation activity (55). It shares a high level of sequence identity with its Ad2 or Ad5 counterpart after position Lys-136 of the Ad12 protein, whereas sequence identity in the amino-terminal region up to residue Tyr-135 between Ad12 and Ad2 or Ad5 ranges from very weak to nonrecognizable (32). Since it is the conserved sequence within the E1B 55-kDa protein that is required for binding to p53, it is expected that Ad12 E1B 55-kDa protein might bind to p53 as well as does its Ad2 or Ad5 counterpart. However, previous studies using immunofluorescence microscopy and IP failed to detect interaction between Ad12 E1B 55-kDa protein and p53 (37, 61, 73). Nonetheless, these results do not prove that there is no interaction between p53 and Ad12 E1B 55-kDa protein; one possibility is that specific antibodies used in those studies could block the binding sites between p53 and E1B 55-kDa protein. Indeed, interaction between p53 and Ad12 E1B 55-kDa protein was detected in IP assays as well as in immunofluorescence microscopy using different antibodies (12, 16, 67).
How does E1B 55-kDa oncoprotein inhibit the p53-mediated transcriptional transactivation function? There are several possible scenarios. First, the E1B 55-kDa protein repression domain may be tethered to the transcriptional machinery through its interaction with DNA-bound p53. In agreement with this, it was shown previously that Ad2 E1B 55-kDa protein has a generalized transcriptional repression activity, and electrophoretic mobility shift assays indicated that E1B 55-kDa protein can supershift a p53-DNA complex (71). Furthermore, purified Ad2 E1B 55-kDa protein can specifically suppress p53 transactivation function in an in vitro transcription assay (39, 40). Second, E1B 55-kDa protein might interact with histone deacetylases and bring them to chromatin through interaction between E1B and DNA-bound p53 in a manner similar to that of a number of known transcription repressors (6, 27, 59). Third, the E1B 55-kDa oncoprotein might inhibit posttranslational modifications of p53, as covalent modifications of p53 such as phosphorylation and acetylation play important roles in activating p53 (42).
It has been shown recently that the acetylases p300, CREB binding protein (CBP), and PCAF acetylate p53 and enhance its sequence-specific DNA-binding activity (17, 35, 47). Furthermore, such acetylation is induced in response to DNA damage (35, 47), suggesting that this modification may represent a physiological response to activate p53; consequently, suppression of p53 acetylation may inhibit its sequence-specific DNA-binding activity and render it unable to transactivate transcription.
In this study, we show that E1B 55-kDa protein from both Ad2 and Ad12 inhibits acetylation of p53 by PCAF in vitro and in vivo, whereas it does not affect acetylation of histones by PCAF or its autoacetylation. Moreover, the DNA-binding activity of p53 in cells expressing E1B 55-kDa protein is greatly reduced. PCAF interacts with a region near the C terminus of p53 as well as with both Ad2 and Ad12 E1B 55-kDa proteins. In addition, E1B 55-kDa protein appears to interfere with the physical interaction between PCAF and p53. These results suggest that abrogation of p53 acetylation by viral oncoproteins such as the E1B 55-kDa protein inactivates p53 and thereby contributes to viral infection and cell transformation.
MATERIALS AND METHODS
Cell culture.
Sf9 cells were maintained at 27°C in Grace's insect medium (Gibco-BRL), supplemented with 10% fetal bovine serum (FBS), 1× lactalbumin hydrolysate, 1× yeastolate, and 0.1% pluronic F-48 (Gibco-BRL). A hybridoma cell line producing monoclonal antibody 2A6 against Ad2 or Ad5 E1B 55-kDa protein (a kind gift of Arnold Levine) was cultured in RPMI medium supplemented with 10% FBS. Monolayer cell lines, G401, G401-CC3, and 293, were cultured in Dulbecco's modified essential medium supplemented with 10% FBS, 100 U of penicillin, and 0.1 mg of streptomycin per ml. For culturing G401 and G401-CC3, hypoxanthine (15 μg/ml) and thymidine (10 μg/ml) were added to the medium. In addition, 250 μg of G418 per ml was added to the medium for growing G401-CC3. G401 is a rhabdoid kidney tumor cell line (65); G401-CC3 is a derivative of G401, which was stably transfected with a vector expressing Ad12 E1B 55-kDa protein (55). 293 was derived from human embryonic kidney cells by transformation with Ad5 DNA fragments, and this cell line expresses both E1A and E1B proteins (15).
Protein expression.
The Ad E1B 55-kDa proteins were expressed in insect cells and in Escherichia coli. To construct recombinant baculovirus, a BamHI-SalI fragment containing the entire coding region of Ad2 or Ad12 E1B 55-kDa protein was excised from plasmid pGEM-T/Ad2 HA-E1B or pGEM-T/Ad12 HA-E1B, and this fragment was cloned into pFastBacHTb (Gibco-BRL) donor vector. To tag proteins with both the FLAG peptide and six His residues in the donor vector, an oligonucleotide containing codons for the 9 amino acid (aa) residues of the FLAG peptide was first inserted into BamHI and EcoRI sites of pFastBac1, and then the BamHI-HindIII fragment encompassing the FLAG coding sequence and multiple cloning sites of pFastBac1 was cloned into BamHI and HindIII sites of pFastBacHTa to make the plasmid FLAG-pFastBacHTa. The human p53 coding region was cloned into the EcoRI site of FLAG-pFastBacHTa. Recombinant baculoviruses (bacmids) were generated by transforming DH10Bac competent cells (Gibco-BRL) with various donor plasmids. The DNA of these bacmids was isolated from E. coli and was transfected into Sf9 insect cells. Viruses were produced typically at 72 to 96 h posttransfection. To amplify the recombinant baculovirus, the supernatant of initial culture was used to further infect Sf9 cells. After one round of amplification, the viral titer was determined using plaque assay. We typically got a titer of ∼1010 PFU/ml. To determine optimal expression, Sf9 cells were infected with varying amounts of virus and harvested at different time points postinfection. The Ad2 and Ad12 E1B 55-kDa proteins with amino-terminal six-His tag and p53 with both six-His and FLAG tags at the amino terminus were expressed in the insect cells (see Fig. 1A).
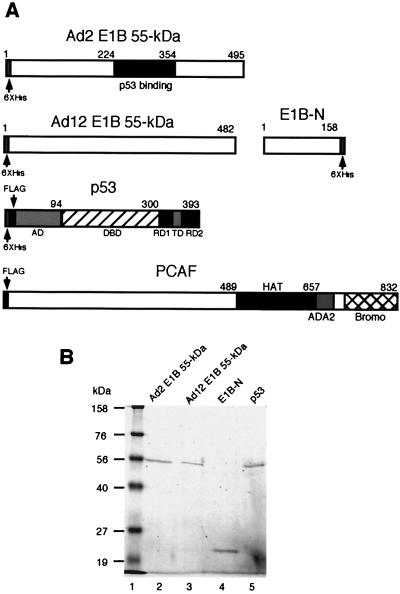
FIG. 1.
Purification of E1B 55-kDa protein, E1B-N, and p53. (A) Schematic drawings of proteins used in this study. His or FLAG tag was attached either at the N terminus or at the C terminus of a protein as indicated. Known functional domains of these proteins are denoted: DBD, sequence-specific BD; RD, regulatory domain; TD, tetramerization domain; HAT, HAT domain; ADA2, alteration and/or deficiency in activation (4, 63); Bromo, bromodomain (8). (B) SDS-PAGE analysis of purified proteins. Full-length Ad2, Ad12 E1B 55-kDa, and p53 proteins were expressed in insect Sf9 cells, and the N terminus of Ad12 E1B (E1B-N) was expressed in E. coli. The proteins were purified using Ni-NTA agarose, an aliquot of each protein was analyzed by SDS–10% PAGE, and the gel was stained with Coomassie brilliant blue. The protein identity is indicated on top of each lane.
The wild-type (WT) Ad12 E1B 55-kDa protein and its N-terminal portion were also expressed in E. coli. The entire coding region for Ad12 E1B 55-kDa protein was carboxyl terminally tagged with six-His residues by PCR with primers Ad12E1B-Nde (5′-ACATATGGAGCGAGAAATCCCACCT-3′, NdeI site in boldface) and Ad12E1B-His (5′-AGAATTCTCAGTGGTGGTGGTGGTGGTGggatccGTTGTCGTCTTCATCACTTGA-3′, EcoRI site in boldface, BamHI site in lowercase, and six-His region underlined). The PCR fragment was cloned into NdeI and EcoRI sites of pET-22b(+) (Novagen). The coding sequence for the amino-terminal portion up to residue K-158 of Ad12 E1B 55-kDa protein was generated by PCR with primers Ad12E1B-Nde and E1BH3 (5′-AAAAGCTTCTTAATAGCACACTCCATATCCTC-3′, HindIII site in boldface and Ad12 sequence underlined), which was cloned into the NdeI and HindIII sites of pET-22b(+). These constructs were verified by DNA sequencing. These plasmids were introduced into E. coli strain BL21 (DE3), and the recombinant proteins were expressed at 30°C upon induction with isopropyl-β-d-thiogalactopyranoside (IPTG; 0.5 mM) for 3 h. The N-terminal fragment of the Ad2 E1B 55-kDa protein (aa 1 to 161) was cloned into pQE30 (Qiagen) and expressed in E. coli strain XL1-Blue with IPTG induction.
Protein purification.
All proteins were affinity purified with Ni-NTA (nickel-nitrilotriacetic acid) agarose (Qiagen). All purification steps were carried out at 4°C. To purify proteins expressed in Sf9 cells, extracts were prepared from infected cells by one cycle of freeze and thaw in buffer A (20 mM Tris-HCl [pH 7.9], 0.5 M NaCl, 20 mM imidazole, 0.5% Nonidet P-40, 10% glycerol, 10 mM β-mercaptoethanol) supplemented with 1× protease inhibitor cocktail (16 μg of benzamidin HCl per ml, 10 μg of phenanthroline per ml, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 10 μg of pepstatin A per ml, 1 mM phenylmethylsulfonyl fluoride [PMSF]). The extracts were incubated for 30 min with slow rotation. The extracts were then centrifuged and filtered through an 0.22-μm-pore-size filter. The cleared extracts were mixed with Ni-NTA agarose and incubated for 4 h with slow rotation. The resulting agarose was washed three times with buffer A containing 60 mM imidazole, and the proteins were eluted using buffer A containing 200 mM imidazole. Eluted polypeptides were dialyzed in storage buffer (20 mM Tris-HCl [pH 7.9], 0.5 M NaCl, 50% glycerol, 0.1 mM 1,4-dithiothreitol), 0.1 mM EDTA, 0.5 mM PMSF, 1 mM benzamidine) overnight.
To purify proteins expressed in E. coli, the bacterial pellets were resuspended in buffer A supplemented with 10 μg of leupeptin per ml, 10 μg of aprotinin per ml, and 0.5 mM PMSF. After sonication and centrifugation, the extracts were filtered through an 0.22-μm-pore-size filter and were incubated with Ni-NTA agarose for 4 h with slow rotation. The other steps in the purification protocol are the same as described above.
Acetyltransferase assay.
The human PCAF was purified as described previously (68) and used as acetylase. Purified p53 was subject to acetylation by PCAF in the presence or absence of E1B 55-kDa protein as specified in Fig. 2. Protein samples were incubated at 30°C for 30 to 60 min in a total volume of 20 μl containing 50 mM Tris-HCl (pH 8.0), 10% glycerol, 1 mM 1,4-dithiothreitol, 1 mM PMSF, 0.1 mM EDTA, 10 mM sodium butyrate, and 90 pmol of 1-14C-acetyl coenzyme A (55 mCi/mmol; Amersham). The reaction mixtures were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the gels were stained with Coomassie brilliant blue, dried, and were subjected to autoradiography.
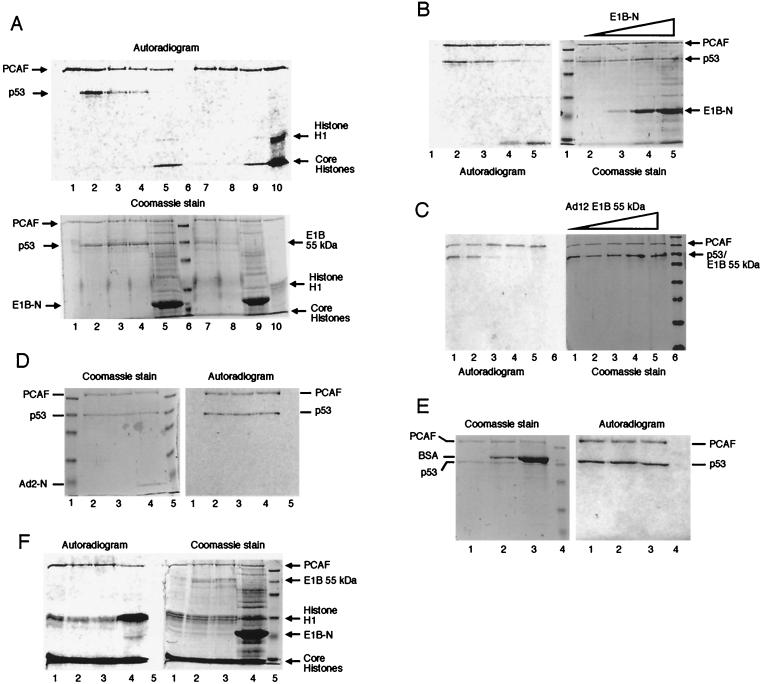
FIG. 2.
E1B 55-kDa protein specifically inhibits acetylation of p53 by PCAF. (A) E1B 55-kDa protein inhibits acetylation of p53 by PCAF but not its autoacetylation. PCAF (4.3 pmol) was incubated alone (lane 1) or with 5.4 pmol of p53 (lanes 2 to 5) in the absence (lanes 1 and 2) or presence (lanes 3 to 5) of E1B (Ad2 E1B 55-kDa protein [3.5 pmol, lane 3], Ad12 E1B 55-kDa protein [3.5 pmol, lane 4], and E1B-N [60 pmol, lane 5]). Lane 6 is molecular weight markers. The same amount of PCAF (4.3 pmol) was incubated with full-length E1B 55-kDa protein (Ad2 [3.5 pmol, lane 7]) and Ad12 [3.5 pmol, lane 8]) and E1B-N (60 pmol, lane 9) in the absence of p53. Lane 10 shows acetylation of histones (2.0 μg; Sigma) by PCAF. The top portion is an autoradiogram of the Coomassie blue-stained gel (10%, bottom portion). (B) Inhibition of p53 acetylation by PCAF with increasing concentrations of E1B-N. PCAF (4.3 pmol) was incubated with p53 (5.4 pmol, lane 2) or with p53 plus increasing amounts of E1B-N at 6.0 pmol (lane 3), 30 pmol (lane 4), and 60 pmol (lane 5). The left part is an autoradiogram of the Coomassie blue-stained gel (10%) shown on the right. (C) Inhibition of p53 acetylation by PCAF with increasing concentrations of Ad12 E1B. PCAF (4.3 pmol) was incubated with p53 (5.4 pmol, lane 1) or with p53 plus increasing amounts of Ad12 E1B 55-kDa protein at 1.0, 4.0, 6.0, and 8.0 pmol (lanes 2 to 5, respectively). The left part is an autoradiogram of the Coomassie blue-stained gel (10%) shown on the right. (D) The N-terminal domain of Ad2 E1B 55-kDa protein does not inhibit acetylation of p53 by PCAF. PCAF (4.3 pmol) was incubated with p53 (5.4 pmol, lanes 2 to 4) in the absence (lane 2) or presence (lanes 3 and 4) of Ad2 E1B N-terminal domain (aa 1 to 161) (8 pmol [lane 3] and 60 pmol [lane 4]). Lanes 1 and 5 are molecular weight markers. The Coomassie blue-stained gel (10%) and its autoradiogram are shown on the left and right, respectively. (E) BSA does not affect acetylation of p53 by PCAF. PCAF (4.3 pmol) was incubated with p53 (5.4 pmol, lanes 1 to 3) in the absence (lane 1) or presence (lanes 2 and 3) of BSA (8 pmol [lane 2] and 60 pmol [lane 3]). Lane 4 is molecular weight markers. The Coomassie blue-stained gel (10%) and its autoradiogram are shown on the left and right, respectively. (F) E1B does not affect acetylation of histones by PCAF. Histones (2.0 μg) were subjected to acetylation by PCAF (4.3 pmol) in the absence of E1B (lane 1) or the presence of 3.5 pmol of Ad2 E1B 55-kDa protein (lane 2), 3.5 pmol of Ad12 E1B 55-kDa protein (lane 3), or 300 pmol of E1B-N (lane 4). Lane 5 is molecular weight markers. At left is an autoradiogram, and at right is the Coomassie blue stain of the same gel (10%).
Detection of in vivo p53 acetylation by PCAF.
G401, G401-CC3, and 293 cells were grown for 24 h to 60 to 70% confluence. Cells were washed twice with phosphate-buffered saline (PBS) and then grown in complete medium supplemented with the deacetylase inhibitor trichostatin A (TSA; Sigma) at a final concentration of 5 μM. At different time points following the addition of TSA, cells were washed twice with ice-cold PBS and lysed on ice in ice-cold lysis buffer (50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 50 mM NaCl, 1% Triton X-100, 50 mM NaF, 10 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 10 μg of aprotin per ml, 10 μg of leupeptin per ml, 5 μg of pepstatin per ml, 0.5 mM PMSF, and 5 μM TSA). Lysates were clarified at 20,000 × g for 30 min at 4°C. Before IP, each sample (containing 2.5 mg of total cellular proteins) was precleared by protein G-agarose (Roche Molecular Biochemicals) for 1 h. Subsequently, 2.5 μg of affinity-purified mouse monoclonal anti-p53 antibody DO-1 (Santa Cruz Biotechnology) was added to the precleared lysates and incubated on ice for 1 h. Then protein G-agarose was added, and the IP mixture was further incubated at 4°C for 1 h with rotation. The beads were collected by centrifugation and then washed five times with ice-cold lysis buffer. The amount of p53 protein in the immunoprecipitates was estimated by Western blotting using goat polyclonal anti-p53 antibody p53 FL-393-G (Santa Cruz Biotechnology) as primary antibody. To determine the acetylation status of p53 in these cells, the same immunoprecipitates with an equal amount of p53 protein were subjected to SDS-PAGE and acetylated p53 was detected with rabbit anti-acetylated p53 (lysine-320) antiserum (Upstate Biotechnology) in Western blot analysis using the enhanced chemiluminescence method (ECL; Amersham-Pharmacia) with an appropriate peroxidase-conjugated secondary antibody.
Electrophoretic mobility shift assay.
The DNA-binding activity of p53 in the nuclear extracts of several cell lines was assayed using the NUSHIFT kit (Geneka Biotechnology, Inc.) according to the protocol provided by the manufacturer. The nuclear extracts of G401, G401-CC3, and 293 cells containing approximately equal amounts of p53 were incubated with the oligonucleotide containing consensus p53-binding sites, 5′-AGCTGGACATGCCCGGGCATGTCC-3′ (the consensus binding sequence is in bold-face), which was labeled with [γ-32P]ATP. An excessive amount (200-fold over that of labeled probe) of unlabeled WT oligonucleotide or a mutant oligonucleotide, 5′-AGCTGGATCGCCCCGGGC ATGTCC-3′ (mutated nucleotides are underlined), was used in the competition assay. The Raji nuclear extract was supplied in the kit and used as positive control. Two microliters of PAb421 anti-p53 monoclonal antibody (100 μg/ml; Calbiochem) was added per assay in some reactions to supershift the p53-DNA complex. The reactions were resolved on a 5% polyacrylamide gel and run at 4°C. The gel was dried and subjected to autoradiography.
IP.
In each IP experiment, approximately 0.5 to 1 μg of each purified protein was incubated with an appropriate antibody in NET-gel buffer (20 mM Tris-HCl [pH 7.4], 0.1 M NaCl, 0.5% Nonidet P-40, 10% glycerol, and 5 mM EDTA) for at least 1 h with rotation at 4°C. The amount of antibody used per IP assay is different depending on specific antibodies; the typical amount is 100 μl of hybridoma supernatant (2A6; anti-Ad2 E1B 55-kDa protein), 3 μl of purified monoclonal antibody against p53 (DO-1 [Santa Cruz Biotechnology] or PAb421 [Calbiochem]), 6 μl of anti-FLAG M2 antibody (Sigma), or 3 μl of rabbit antiserum against the N terminus of Ad12 E1B 55-kDa protein. Protein A-agarose beads (15 μl; Roche Molecular Biochemicals) were added into the protein-antibody mixture and incubated at 4°C for 1 h with rotation. The beads were collected by centrifugation and washed three times. The first wash was done with NET-gel buffer supplemented with NaCl to a final concentration of 0.5 M; the second wash was with NET-gel buffer supplemented with 0.1% SDS, and the third wash was carried out with 10 mM Tris-HCl (pH 7.4) with 0.1% Nonidet P-40. The beads were pelleted by centrifugation and mixed with 30 μl of 1× SDS loading buffer. The precipitated proteins were separated by SDS-PAGE and transferred onto a nitrocellulose membrane in 25 mM Tris base–190 mM glycine at 50 V for 3 h at 4°C. The coprecipitated proteins were detected using an appropriate antibody with the enhanced chemiluminescence (ECL) kit.
To detect E1B-PCAF interaction in vivo, 293 cells were washed twice with PBS and lysed on ice with ice-cold RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, pH 7.5). Lysates were clarified by centrifugation and then precleared with protein G-agarose. The IP antibody was incubated with the cell lysates at 4°C for 1 h. Protein G-agarose was added and incubated at 4°C for 1 h. The beads were collected by centrifugation and then washed five times with ice-cold RIPA buffer. For detecting Ad12 E1B-PCAF interaction, G401-CC3 cell lysates were subjected to IP using anti-E1B, and the immunoprecipitates were analyzed by Western blotting using anti-PCAF. In addition, G401-CC3 cells were transfected with pCX-Flag-PCAF by lipofection. Forty-eight hours after transfection, cell lysates were made and then subjected to IP as described above for 293 cells.
Yeast two-hybrid assay.
Various DNA fragments spanning different regions of the Ad2 or Ad12 E1B 55-kDa protein open reading frame were fused either to yeast GAL4 activation domain (AD) in plasmids pGAD-C(x) or to GAL4 DNA-binding domain (BD) in plasmids pGBDU-C(x) (22). Similarly, a DNA fragment encoding the full-length human PCAF was cloned into pGAD-C3 and pGBDU-C3 (22). A series of plasmids containing varying N-terminal deletions of human p53 cDNA that were fused to GAL4 AD were described previously (21) and were kindly provided by Stanley Fields. The p53 AD (aa 1 to 145) and sequence-specific BD (aa 76 to 315) were separately cloned into pGAD-C1 and pGBDU-C1.
The yeast strain PJ69-4A (MATa trp1-190 leu2-3,112 ufa3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) was used for all two-hybrid assays as described previously (22). Plasmids are introduced into PJ69-4A by the standard lithium acetate transformation method. To test potential protein-protein interaction, transformants were screened for growth in medium lacking histidine but in the presence of 5 mM 3-aminotriazol (3-AT) (His+ phenotype) or lacking adenine (Ade+ phenotype) or assayed for β-galactosidase activity (blue phenotype) in the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) or by quantitative measurement using Galacto-Light reagents (Tropix) and a luminometer.
Reverse two-hybrid assay.
The BamHI-BglII fragment encoding Ad2 E1B 55-kDa protein aa 1 to 437 (missing 58 aa residues in the C terminus) from pGEM-T/Ad2 HA-E1B and the EcoRI fragment encoding the full-length Ad12 E1B from pGEM-T/Ad12 HA-E1B were cloned into plasmid pCu424, which has a copper-inducible promoter and the Trp1-selectable marker (28). These plasmids were separately introduced into yeast strain PJ69-4A together with plasmids pGBDU-PCAF and pGAD-p53. Yeast minimal medium lacking copper was supplemented with ascorbic acid (1 mM) and bathocuproinedisulfonate (33 μM). Different amounts of CuSO4 were added for induction of gene expression from pCu424 constructs.
Preparation of yeast cell extracts.
Fifty milliliters of overnight culture in selective dropout medium with an optical density at 600 nm (OD600) of between 0.4 and 0.5 was centrifuged, and the pellet was resuspended in 100 μl of cold trichloroacetic acid (TCA) buffer (20 mM Tris HCl [pH 8.0], 50 mM ammonium acetate, 2 mM EDTA, 1 mM PMSF, 1× protease inhibitor solution containing 0.1 μg of pepstatin A per ml, 0.03 μM leupeptin, 145 μM benzamidine, and 0.37 μM aprotinin) per 7.5 OD600 units (OD600 units = volume [ml] × OD600 [1 ml]). An equal volume of glass beads and 20% TCA were added; the mixture was vortexed for 1 min with a 30-s pause on ice, and the vortexing was repeated five times. The suspension of broken cells was transferred into another tube, and the glass beads were washed with 250 μl each of TCA and 20% TCA. The combined broken-cell suspension was centrifuged, and the pellet was resuspended in 10 μl of TCA-Laemmli loading buffer (3.5% [wt/vol] SDS, 14% [vol/vol] glycerol, 120 mM Tris base, 8 mM EDTA, 0.01% bromophenol blue, 0.7 M β-mercaptoethanol, 2 mM PMSF, 1× protease inhibitor solution) per OD600 unit. The samples were heated at 100°C for 10 min and centrifuged. The supernatant was subjected to SDS-PAGE and Western blot analysis to detect proteins under study.
RESULTS
Expression and purification of E1B 55-kDa protein and p53.
Recombinant baculoviruses expressing Ad2, Ad12 E1B 55-kDa protein, or p53 were constructed using the Bac-to-Bac baculovirus expression system (Gibco-BRL). We found that the levels of protein expression were highest at 72 h postinfection for Ad2 and Ad12 E1B 55-kDa protein as well as for p53. We also found that the level of p53 expression was significantly higher than that of either Ad2 or Ad12 E1B 55-kDa protein. The reason for this is unknown but could stem from hindrance of Sf9 cell growth by the 55-kDa oncoprotein, as it also slows the growth of human cells (data not shown).
The Ad12 E1B 55-kDa protein and its amino-terminal portion (Ad12 E1B-N, aa 1 to 158) were also expressed in E. coli. The expression level of WT Ad12 E1B 55-kDa protein was very low at 37°C, due to severe degradation, whereas Ad12 E1B-N was expressed at high levels but remained largely insoluble in the inclusion body at 37°C. Thus, we shifted bacterial growth temperature to 30°C for their expression.
Since all the recombinant proteins were tagged with six His residues, they were affinity purified using Ni-NTA agarose. Figure 1 shows Coomassie brilliant blue-stained SDS-polyacrylamide gels of purified proteins. Both Ad2 and Ad12 E1B 55-kDa proteins migrate in parallel with glutamic dehydrogenase (molecular mass of 55.6 kDa) in the protein marker lane, whereas the p53 carrying both the six-His and FLAG tags migrates slightly faster than this marker protein. The identity of these proteins was confirmed by Western blotting using antibodies 2A6 (48), rabbit polyclonal antiserum against Ad12 E1B 55-kDa protein (32), and anti-p53 antibody DO-1 (data not shown).
E1B 55-kDa protein specifically inhibits p53 acetylation by PCAF in vitro.
p53 is covalently modified by phosphorylation and acetylation, and such modifications have profound effects on p53 functions (42). p53 is acetylated by transcriptional coactivator p300 at Lys-373 and Lys-382 and by PCAF at Lys-320. Importantly, acetylation of p53 at these sites enhances sequence-specific DNA-binding activity of p53 (17, 35, 47), thereby activating p53. As Ad E1B 55-kDa oncoprotein represses p53-mediated transcriptional transactivation (55, 69), we reasoned that E1B might do so by inhibiting covalent modifications of p53. To test this possibility, we carried out acetylation assays using PCAF as the acetylase and p53 as the substrate in the absence or presence of E1B 55-kDa protein. The effects of E1B 55-kDa proteins on PCAF acetylation are presented in Fig. 2. Consistent with previous reports, PCAF acetylates itself and p53 (Fig. 2A, lanes 1 and 2) (47, 68). The presence of either purified Ad2 E1B 55-kDa protein (lane 3), Ad12 E1B 55-kDa protein (lane 4), or Ad12 E1B N-terminal domain (E1B-N) (lane 5) resulted in striking (70 to 95%) reduction of p53 acetylation, whereas PCAF autoacetylation remained largely unaffected by E1B (lanes 3 to 5 and 7 to 9). PCAF does not acetylate WT E1B 55-kDa protein (lanes 7 and 8) or E1B-N (lane 9) (the acetylated species that ran at the dye front in lanes 5 and 9 containing E1B-N are unknown). As a positive control, histones were used as substrate for PCAF acetylation (lane 10). The inhibition of p53 acetylation by E1B was concentration dependent. As shown in Fig. 2B, the addition of increasing amounts of E1B-N gradually inhibited p53 acetylation by PCAF. At high E1B-N concentrations, p53 acetylation was hardly detectable (Fig. 2B, lane 5). Similar dosage-dependent inhibition was also observed with WT E1B 55-kDa protein (Fig. 2C). The observed inhibition of p53 acetylation by PCAF is unlikely to be due to a contaminant in the E1B preparation since full-length E1B protein was purified from Sf9 cells and E1B-N was purified from E. coli. Furthermore, the purified N-terminal fragment of Ad2 E1B 55-kDa protein (aa 1 to 161) did not inhibit acetylation catalyzed by PCAF (Fig. 2D, lanes 3 and 4). (Note that this protein fragment did not stain well with Coomassie blue. We loaded 0.17 μg [∼8 pmol] in Fig. 2D, lane 3, and 1.2 μg in lane 4 [∼60 pmol]). This Ad2 E1B fragment shares little sequence identity with the Ad12 E1B N-terminal fragment (E1B-N) and does not bind to either p53 or PCAF (see below). In additional control experiments, high concentrations of bovine serum albumin (BSA) did not inhibit p53 acetylation by PCAF or its autoacetylation (Fig. 2E, lanes 2 and 3).
To investigate whether E1B 55-kDa protein inhibits histone acetylation by PCAF, purified Ad2 and Ad12 E1B 55-kDa proteins as well as E1B-N (Fig. 2F, lanes 2 to 4) were incubated with histones and PCAF. Acetylation of neither core histones nor histone H1 was inhibited by WT E1B (lanes 2 and 3) or E1B-N (lane 4), although p53 acetylation was severely inhibited or completely abolished at the E1B concentrations used in the assays (Fig. 2A). We conclude that E1B 55-kDa protein does not affect PCAF histone acetylase (HAT) activity. Curiously, acetylation of histone H1 was dramatically increased in the presence of a high concentration of E1B-N (Fig. 2F, lane 4). The reason for this is unknown.
Acetylation of p53 is suppressed in vivo.
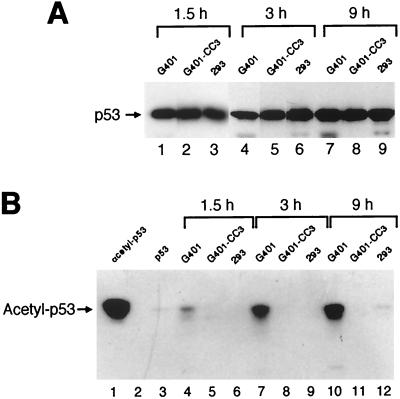
We then tested if E1B 55-kDa oncoprotein would affect acetylation of p53 by PCAF in vivo. We first quantified the total amount of p53 protein in cell lysates with an IP-Western blot protocol using mouse monoclonal anti-p53 antibody DO-1 for IP and goat polyclonal antibody anti-full-length p53 for Western blot analysis. The results are shown in Fig. 3A. The same immunoprecipitates containing approximately the same amounts of p53 were then subjected to SDS-PAGE and Western blot analysis, and acetylated p53 was detected with rabbit polyclonal antiserum against p53 acetylated at Lys-320, the specific acetylation site of PCAF (35, 47). As shown in Fig. 3B, acetylated p53 was detectable in G401 cells at 1.5 h and accumulated gradually at 3 and 9 h after addition of deacetylase inhibitor TSA (lanes 4, 7, and 10). In contrast, acetylated p53 was not detectable or was dramatically reduced in G401-CC3 cells that express Ad12 E1B 55-kDa protein and 293 cells that express Ad5 E1B 55-kDa protein (Fig. 3B, lanes 5, 6, 8, 9, 11, and 12), although the same amount of total p53 was present in each sample (Fig. 3A). The specificity of anti-acetyl-p53 antibody was confirmed, as it was reactive only to purified p53 that was acetylated by PCAF in vitro (lane 1), but not to the same amount of p53 that was not treated with PCAF (lane 3). Thus, p53 acetylation was severely suppressed by the E1B 55-kDa protein in vivo.
FIG. 3.
E1B 55-kDa proteins inhibit acetylation of p53 in vivo. (A) Quantification of total p53 protein. G401 cells do not express E1B 55-kDa protein, while G401-CC3 expresses Ad12 E1B 55-kDa protein and 293 produces both Ad5 E1A and E1B proteins. Cells were harvested at 1.5 h (lanes 1 to 3), 3 h (lanes 4 to 6), or 9 h (lanes 7 to 9) after addition of deacetylase inhibitor TSA (5 μM), and cell extracts were subjected to IP with mouse monoclonal antibody DO-1 and Western blot analysis with goat anti-p53 polyclonal antibody. (B) Detection of acetylated p53. The same immunoprecipitates containing approximately equal amounts of total p53 protein as shown in panel A were resolved by SDS-PAGE, and the acetylated fraction of p53 was detected by Western blot analysis with an anti-acetyl-p53 (Lys-320) antibody (Upstate Biotechnology). Lanes 1 and 3 show p53 (9 pmol) acetylated by PCAF in vitro and nontreated, respectively. Lane 2 is an empty lane.
The sequence-specific DNA binding of p53 is inhibited in G401-CC3 and 293 cells.
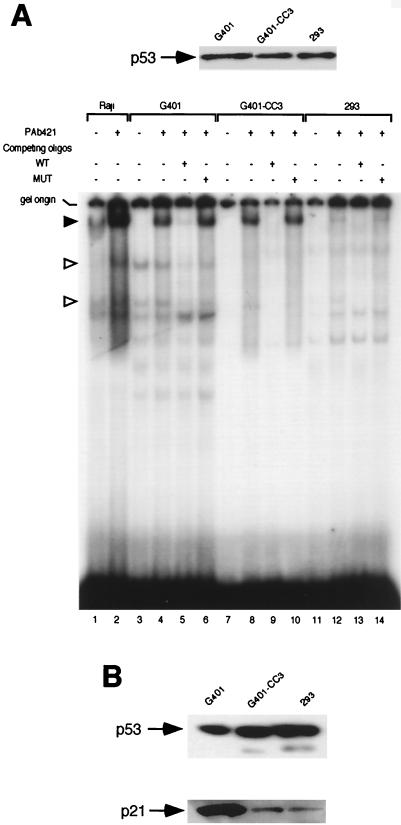
Since acetylation of p53 by p300 and PCAF stimulates sequence-specific DNA-binding activity of p53 (17, 35, 47), and we showed above that E1B 55-kDa protein inhibits acetylation of p53, one would expect that p53 from cells expressing E1B may be less competent to bind to its cognate DNA sequence. To test this possibility, we monitored the sequence-specific DNA-binding activity of p53 in nuclear extracts of G401, G401-CC3, and 293 cells in an electrophoretic mobility shift assay using a radiolabeled oligonucleotide bearing consensus p53-binding sites (oligonucleotide WT in Fig. 4). As shown in Fig. 4, two specific bands denoted with a hollow arrowhead, presumably p53-DNA complexes, were readily detectable in G401 nuclear extracts (lanes 3 and 4). These two bands disappeared almost completely in the presence of competing unlabeled oligonucleotide WT (lane 5). However, these two bands were not detectable in the nuclear extracts of G401-CC3 (lane 7) and 293 (lane 11) cells, although these nuclear extracts contained about the same amount of p53 protein as that of G401 (Fig. 4A, top). As expected, monoclonal antibody PAb421, which recognizes an epitope in p53 RD2 (aa 371 to 381), enhances DNA binding by p53 (lanes 2 and 4; the supershifted complex is denoted with a solid arrowhead). PAb421 was able to stimulate p53's DNA-binding activity in G401-CC3 nuclear extracts (lanes 8 and 10). Surprisingly, it had virtually no effect on p53's DNA-binding activity in 293 nuclear extracts (lanes 12 and 14). As both G401-CC3 and 293 express E1B 55-kDa protein, these results indicate that reduced acetylation of p53 in the presence of E1B impairs p53's sequence-specific DNA-binding activity. As binding of p53 to its cognate DNA sequence is required for its transactivation function, inhibition of p53 acetylation by E1B may result in diminished transactivation by p53. Indeed, the level of p21 protein, whose expression is stimulated by p53, was significantly lower in G401-CC3 and 293 cells than in G401 cells in the presence of approximately equal amounts of p53 (Fig. 4B).
FIG. 4.
p53 sequence-specific DNA-binding activity in cells expressing E1B 55-kDa protein. (A) Electrophoretic mobility shift assay of p53 DNA-binding activity. The nuclear extracts of cell lines G401, G401-CC3, and 293, which contained about equal amounts of p53 as shown on the top of panel A, were incubated with a radioactive oligonucleotide bearing consensus p53-binding sites (WT). Competing unlabeled WT oligonucleotide or mutant oligonucleotide (MUT) as well as monoclonal anti-p53 antibody PAb421 was added in some reactions as indicated. Raji nuclear extract was used as positive control. Specific p53-DNA complexes are denoted with arrowheads. (B) p21 levels in G401, G401-CC3, and 293 cells. The p53 levels in the total cell extracts of these three cell lines were estimated by Western blotting (top), and the p21 protein concentration in total cell extracts containing about equal amounts of p53 was determined by Western blotting using an anti-p21 polyclonal antibody (C-19; Santa Cruz Biotechnology).
PCAF binds directly to E1B 55-kDa protein and p53.
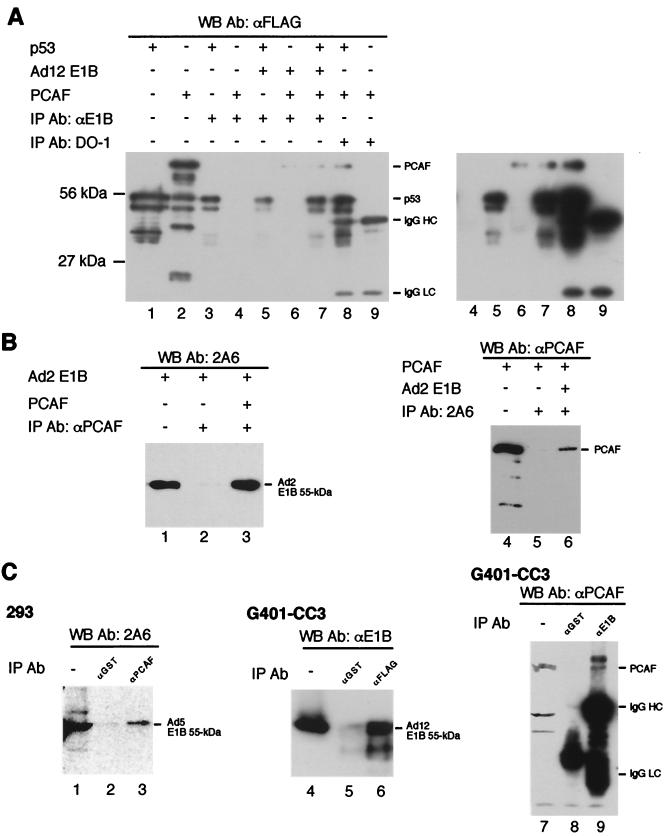
To understand the mechanisms by which E1B inhibits p53 acetylation by PCAF, we studied possible interactions among PCAF, p53, and E1B by IP and yeast two-hybrid assays. Figure 5A shows IP results with the rabbit polyclonal antibody anti-E1B against Ad12 E1B 55-kDa protein and mouse monoclonal antibody DO-1 against p53. The precipitates were subjected to Western blot analysis using mouse monoclonal antibody M2 against the FLAG tag. Purified p53 and PCAF were both tagged with FLAG at the amino terminus and loaded directly on the gel without IP (lanes 1 and 2; about 10% of the amount used for binding reactions). The small species in these two lanes are likely due to partial degradation of p53 (lane 1) and PCAF (lane 2). The polyclonal antibody anti-E1B precipitates PCAF in the presence of Ad12 E1B (lane 6), but this antibody does not directly precipitate PCAF (lane 4). M2 (anti-FLAG) did not recognize any protein species in the preparation of Ad12 E1B 55-kDa protein (data not shown), thereby excluding the possibility that the band seen in lanes 6 and 7 was due to cross-reaction of M2 with Ad12 E1B preparation. Thus, Ad12 E1B 55-kDa protein binds directly to PCAF. The simultaneous presence of PCAF, p53, and Ad12 E1B did not affect PCAF precipitation by anti-E1B (lane 7). Similarly, DO-1 precipitates PCAF along with p53 (lane 8) but does not precipitate PCAF directly (lane 9). A longer exposure of the blot allowed clear visualization of PCAF (see lanes 6 to 8 in the right panel). Therefore, PCAF also binds directly to p53. We also tested if p53 binds to Ad12 E1B 55-kDa protein. p53 was clearly precipitated by anti-E1B (lane 5), but this antibody can recognize p53 directly (lane 3). Thus, whether Ad12 E1B 55-kDa protein binds directly to p53 remains inconclusive based on these IP results. However, we showed that the two proteins colocalize in the cell (see below).
FIG. 5.
Interactions among p53, E1B, and PCAF. For in vitro assay, purified p53 (5.4 pmol), E1B 55-kDa protein (7 pmol), and PCAF (6 pmol) were incubated and subjected to IP with different antibodies indicated on the top of each panel. The precipitates were subjected to Western blot analysis with indicated antibodies. The abbreviations IgG HC and IgG LC denote IgG heavy and light chains, respectively. (A) Ad12 E1B 55-kDa protein binds directly to PCAF. About 10% of the amount of both p53 (lane 1) and PCAF (lane 2) used for the binding reactions was loaded directly on the gel without IP. p53 (lane 3) or PCAF (lane 4) was incubated with rabbit antiserum raised against Ad12 E1B 55-kDa protein (anti-E1B) and subjected to IP. IP of p53 and Ad12 E1B 55-kDa protein (lane 5); PCAF and Ad12 E1B 55-kDa protein (lane 6); and PCAF, p53, and Ad12 E1B 55-kDa protein (lane 7) was done using anti-E1B. Monoclonal antibody DO-1 against p53 was incubated with PCAF and p53 (lane 8) or PCAF alone (lane 9) and subjected to IP. The immunoprecipitates were detected with mouse monoclonal antibody M2 against FLAG tag (anti-FLAG). For a clear visualization of the PCAF band, a longer exposure of the same blot is shown for the right portion (from lanes 4 to 9) of the image. (B) Ad2 E1B binds to PCAF. About 10% of the amount of both Ad2 E1B (lane 1) and PCAF (lane 4) used for the binding reactions was loaded directly on the gel without IP. Rabbit polyclonal antibody against PCAF (anti-PCAF) was incubated with Ad2 E1B (lane 2) or Ad2 E1B plus PCAF (lane 3) and subjected to IP. The immunoprecipitates were probed with mouse monoclonal anti-E1B antibody 2A6 in a Western blot analysis. Similarly, 2A6 was incubated with PCAF (lane 5) or Ad2 E1B plus PCAF (lane 6) and subjected to IP. PCAF in the immunoprecipitates was detected with anti-PCAF. (C) E1B 55-kDa proteins bind to PCAF in vivo. Cell lysate of 293 cells was subjected to IP with anti-PCAF (lane 3) or anti-GST (lane 2). The precipitated E1B 55-kDa protein was detected by 2A6. Lane 1 shows a direct load of 293 cell lysate (10% of that used for IP). To detect interaction between PCAF and Ad12 E1B 55-kDa protein, plasmid pCX-Flag-PCAF was transfected into G401-CC3 cells which constitutively express Ad12 E1B 55-kDa protein. The lysate of transfected cells was subjected to IP with anti-FLAG (lane 6) or anti-GST (lane 5), and the precipitates were analyzed in a Western blot assay using anti-E1B. Lane 4 is a direct load of G401-CC3 cell lysate (10% of that used for IP). In reciprocal experiments, G401-CC3 cell lysate was subjected to IP with anti-E1B (lane 9) or anti-GST (lane 8), and the precipitates were subjected to a Western blot analysis using anti-PCAF. Lane 7 is a direct load of G401-CC3 cell lysate (10% of that used for IP). WB, Western blot; Ab, antibody.
The interactions between Ad2 55-kDa protein and PCAF were studied by IP using rabbit polyclonal antibody against PCAF (anti-PCAF) as IP antibody and monoclonal antibody 2A6 against Ad2 55-kDa protein for Western blot analysis (Fig. 5B). The purified Ad2 E1B 55-kDa protein was loaded directly on the gel (lane 1, 10% of the amount used for binding reactions) as a positive control. Anti-PCAF did not precipitate Ad2 E1B directly (lane 2), but it can recover Ad2 E1B in the presence of PCAF (lane 3). In the reciprocal IP experiments using antibody 2A6, PCAF coprecipitated with Ad2 E1B (lane 6), but not directly by 2A6 (lane 5).
To determine PCAF-E1B interaction in vivo, anti-PCAF was again used as IP antibody for human 293 cell lysates. As shown in Fig. 5C, Ad5 E1B 55-kDa protein was precipitated by anti-PCAF (lane 3), but not by irrelevant antibody anti-glutathione S-transferase (anti-GST) (lane 2). Lane 1 is a direct load of 293 cell lysates. To detect interactions between PCAF and Ad12 E1B 55-kDa protein we chose to transfect G401-CC3 cells with plasmid pCX-Flag-PCAF (68) just to avoid the visualization of immunoglobulin heavy and light chains because both anti-PCAF and anti-E1B were raised in rabbits. The transfected cell lysates were subjected to IP with mouse monoclonal antibody M2 against the FLAG tag and anti-GST, and the immunoprecipitates were detected with anti-E1B (rabbit polyclonal antiserum). As shown in Fig. 5C, Ad12 E1B 55-kDa protein was precipitated by M2 (anti-FLAG, lane 6), but not by anti-GST (lane 5). Lane 4 is a direct load of G401-CC3 cell lysates. Furthermore, PCAF was present in the immunoprecipitates using anti-E1B (lane 9), but not in that using anti-GST (lane 8). Lane 7 is a direct load of G401-CC3 cell lysates. Therefore, both Ad5 and Ad12 E1B 55-kDa proteins interact with PCAF in vivo.
Ad12 E1B 55-kDa protein and p53 colocalize in the cell.
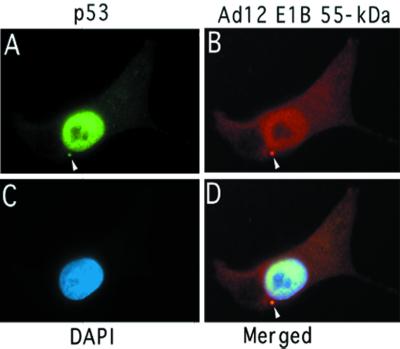
Whether Ad12 E1B 55-kDa protein and p53 interact with each other is less clear, as inconsistent results were reported in the literature (12, 16, 55, 61, 73). To clarify this issue, we tested whether p53 and Ad12 E1B 55-kDa protein colocalize in cells. G401-CC3 cells, which constitutively express Ad12 E1B 55-kDa protein, were grown on a glass coverslip, fixed, and then stained with anti-p53 antibody DO-1 and anti-Ad12 E1B antibody anti-E1B. p53 was visualized with fluorescein-conjugated anti-mouse immunoglobulin G (IgG) antibody (Fig. 6A), and Ad12 E1B was visualized with Texas red-conjugated anti-rabbit IgG antibody (Fig. 6B). Both p53 and Ad12 E1B 55-kDa protein localize primarily to the nucleus, and the colocalization of the two proteins in the nucleus is quite apparent in the merged image (Fig. 6D). Furthermore, the two proteins colocalize in a dense cytoplasmic body (denoted with arrowheads in Fig. 6A, B, and D). The staining pattern of anti-E1B shown in Fig. 6B was seen only in cells expressing Ad12 E1B 55-kDa protein, as we showed previously (32), excluding the possibility that the pattern was due to cross-reaction of cellular proteins with anti-E1B. Collectively, these results indicate that Ad12 E1B 55-kDa protein also interacts with p53, consistent with recent studies (12, 16, 67).
FIG. 6.
Subcellular colocalization of Ad12 E1B 55-kDa protein and p53. G401-CC3 cells that constitutively express Ad12 E1B 55-kDa protein were grown on glass coverslip and fixed. Antibody staining of the cells was done as described previously (32). (A) p53 was visualized with anti-p53 antibody DO-1 and fluorescein-conjugated anti-mouse IgG antibody. (B) The Ad12 E1B 55-kDa protein was stained with anti-Ad12 E1B antibody anti-E1B and visualized with Texas red-conjugated anti-rabbit IgG antibody. (C) Nuclear staining with 4′,6′-diamidino-2-phenylindole (DAPI). (D) Merged image of panels A, B, and C. Colocalization of p53 and Ad12 E1B 55-kDa protein in a cytoplasmic body is indicated by an arrowhead.
PCAF binds to the C-terminal domain of p53.
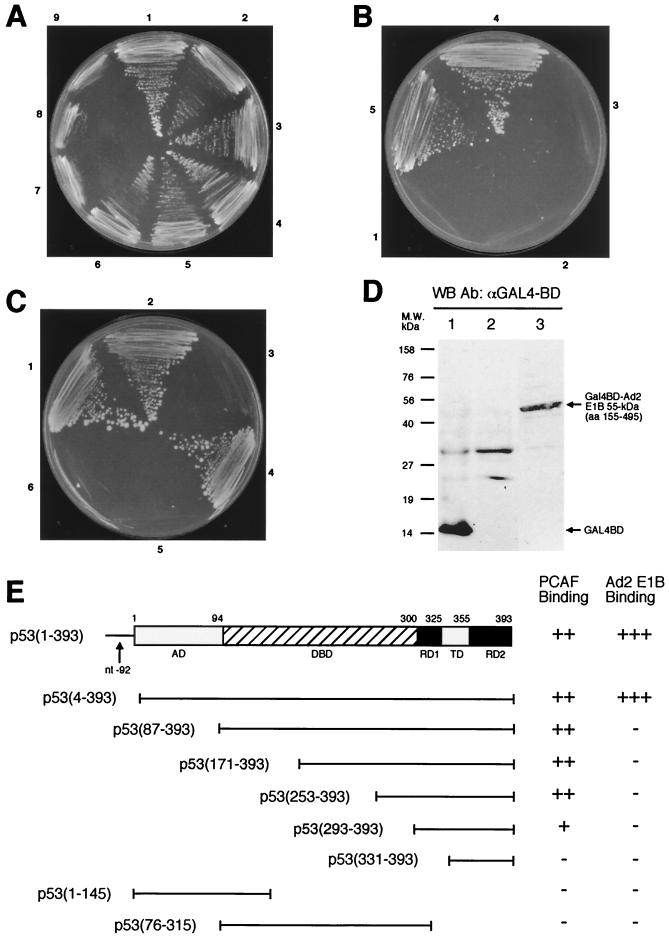
We employed the yeast two-hybrid assay to assess which domain of p53 binds to PCAF. The PCAF-GAL4 BD hybrid interacts with p53-GAL4 AD hybrid, resulting in a His+ phenotype (Fig. 7A), as well as expression of β-galactosidase (data not shown). Progressive deletion of the N terminus of p53 until aa 253 did not attenuate PCAF-p53 interaction (Fig. 7A, sectors 1 to 5). The p53 construct spanning residues 293 and 393 can still bind to PCAF (sector 6), although this construct appears to bind to PCAF with reduced affinity based on β-galactosidase activity (data not shown). The deletion mutant carrying only residues 331 to 393 no longer bound to PCAF (sector 7), and neither did p53 mutants having only the N-terminal 145 aa (sector 8) or the DBD (aa 76 to 315, sector 9). All the p53 constructs were expressed in yeast as judged by Western blot analysis (data not shown), as was the PCAF-GAL4 BD hybrid (see Fig. 9C). Figure 7E summarizes the results of assays for PCAF-p53 interaction. Since PCAF acetylates Lys-320 of p53, these results suggest that a direct physical interaction may be necessary for acetylation. The primary sequence and/or structure of the substrate may determine the specificity of PCAF acetylation. Our results are in agreement with biochemical data reported recently (35).
FIG. 7.
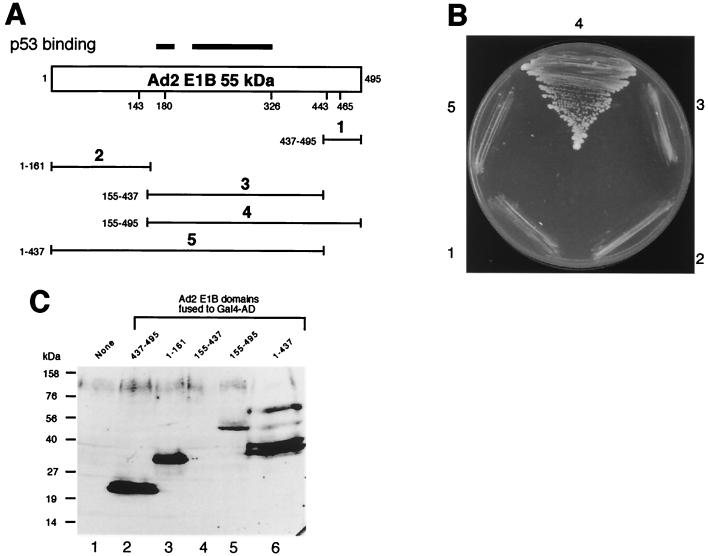
Yeast two-hybrid assays of p53, PCAF, and E1B interactions. (A) PCAF binds to the C-terminal domain of p53. Yeast cells were transformed with a hybrid between full-length PCAF and GAL4 BD and various GAL AD hybrids containing p53 (1–393) (sector 1), p53 (4–393) (sector 2), p53 (87–393) (sector 3), p53 (171–393) (sector 4), p53 (253–393) (sector 5), p53 (293–393) (sector 6), p53 (331–393) (sector 7), p53 (1–145) (sector 8), and p53 (76–315) (sector 9), and the transformed cells were grown under His− selection in the presence of 5 mM 3-AT. (B) The central region of Ad2 E1B 55-kDa protein interacts with p53. Yeast cells were transformed with p53 (1–393)-GAL4 AD hybrid and various hybrids between GAL4 BD and different domains of Ad2 E1B 55-kDa protein: aa 437 to 495 (sector 1), aa 1 to 161 (sector 2), aa 155 to 437 (sector 3), aa 155 to 495 (sector 4), and aa 1 to 437 (sector 5). The transformed cells were grown under His− selection as for panel A. (C) The N-terminal domain of p53 is necessary for binding to Ad2 E1B 55-kDa protein. A fusion between GAL4 BD and Ad2 E1B (aa 155 to 495) (sectors 1 to 3) or full-length Ad12 E1B (sectors 4 to 6) was introduced into yeast along with GAL4 AD hybrids containing p53 (1–393) (sectors 1 and 6), p53 (4–393) (sectors 2 and 5), p53 (87–393) (sector 3), and Ad12 E1B full-length protein (sector 4). (D) Expression of GAL4 BD–E1B 55-kDa protein hybrids. The E1B-GAL4 BD hybrids were detected using anti-GAL4 BD antibody (Santa Cruz Biotechnology). Lane 1 shows lysates of yeast harboring only GAL4 BD plasmid and serves as control. Lanes 2 and 3 were yeast cell extracts prepared from yeast cells that contained GAL4 AD-p53 (1–393) hybrid and one of the GAL4 BD–E1B hybrids: full-length Ad12 E1B (lane 2) and Ad2 E1B (aa 155 to 495) (lane 3). (E) Summary of p53-PCAF and p53-E1B interaction. Relative activities of reporter gene expression as measured by growth under His− selection in the presence of 5 mM 3-AT (A to C) or β-galactosidase expression (data not shown) are illustrated at the right. β-Galactosidase activities were assessed with in situ X-Gal staining on plates or quantitative measurement using Galacto-Light reagents (Tropix) and a luminometer (data not shown). The p53 domains are denoted as in Fig. 1. WB, Western blot; Ab, antibody; nt, nucleotide.
FIG. 9.
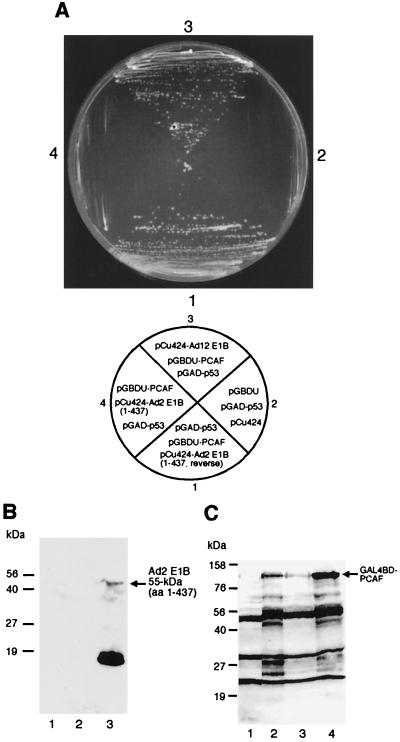
E1B 55-kDa protein interferes with the PCAF-p53 interaction. (A) For reverse two-hybrid assay, full-length PCAF-GAL4 BD hybrid and p53(1–393)-GAL4 AD hybrid were introduced into yeast cells together with pCu424 containing the gene for Ad2 E1B 55-kDa protein (aa 1 to 437) cloned in reverse orientation (sector 1) or in correct orientation (sector 4) or the gene for WT Ad12 E1B 55-kDa protein in correct orientation (sector 3). Sector 2 serves as a negative control, in which the same yeast strain was transformed with pGBDU-C3, pGAD-p53, and pCu424. The plasmids used in each transformation shown on the top are depicted below. (B) Expression of Ad2 E1B (aa 1 to 437). Extracts were prepared from yeast cells that were transformed with pGAD-p53, pGBDU, and pCu424 (lane 1); pGAD-p53, pGBDU-PCAF, and pCu424-Ad2 E1B (1 to 437, reverse) (lane 2); and pGAD-p53, pGBDU-PCAF, and pCu424-Ad2 E1B (1 to 437) (lane 3). The extracts were then subjected to Western blot analysis with antibody 2A6 against Ad2 E1B 55-kDa protein. Note that Ad2 E1B was expressed only when the gene was cloned in the correct orientation (lane 3). The dark band in lane 3 may be partially degraded Ad2 E1B, as this band was absent in lanes 1 and 2. (C) Expression of PCAF-GAL4 BD hybrid. The hybrid was detected with anti-GAL4 BD antibody. The plasmids used for transformation were pGAD-p53, pGBDU, and pCu424 (lane 1); pGAD-p53, pGBDU-PCAF, and pCu424-Ad2 E1B (1 to 437) (lane 2); pGAD-p53, pGBDU-PCAF, and pCu424-Ad12 E1B (lane 3); and pGAD-p53, pGBDU-PCAF, and pCu424-Ad2 E1B (1 to 437, reverse) (lane 4).
We also tested interaction between E1B 55-kDa protein and p53 as well as between E1B and PCAF using the two-hybrid assay. We were unable to make the GAL4 BD or AD fusions with the full-length Ad2 E1B gene; these constructs were unstable, as reported previously (9, 32). Nonetheless, several constructs containing a partial Ad2 55-kDa protein sequence were fused with GAL4 BD and AD, and they were used for two-hybrid assays. As shown in Fig. 7B, C, and E, two overlapping constructs spanning residues 1 to 437 and 155 to 495 exhibited strong interactions with p53 (Fig. 7B, sectors 4 and 5). However, Ad2 E1B 55-kDa protein carrying residues 1 to 161 (Fig. 7B, sector 2) and 437 to 495 (sector 1) did not bind to p53. Thus, the central portion of Ad2 E1B 55-kDa protein binds to the N terminus of p53 (inability of the Ad2 construct containing aa 155 to 437 to bind to p53 was due to the failure of this construct to express itself in yeast [see Fig. 8C, lane 4]). Both p53(1–393) and p53(4–393) constructs exhibited interaction with the Ad2 E1B (aa 155 to 495) construct (Fig. 7C, sectors 1 and 2). However, p53 constructs lacking the N-terminal domain, such as p53(87–393) (Fig. 7C, sector 3), and other p53 constructs shown in Fig. 7E (data not shown) did not show interaction with Ad2 E1B. Curiously, p53(1–145) fused to GAL4 AD did not bind to Ad2 E1B, although in biochemical assays, aa 1 to 123 of murine p53 were shown to be sufficient for binding to Ad2 E1B (24). This could stem from altered conformation of this p53-GAL4 AD hybrid. Collectively, these results indicate that the amino-terminal domain of p53 is required for binding to the central portion of Ad2 E1B, in full agreement with previous reports (33, 70). However, we were unable to detect an interaction between p53 and Ad12 E1B 55-kDa protein (Fig. 7C, sectors 5 and 6), although Ad12 E1B 55-kDa protein self-interaction was detected (sector 4). This may be explained by the extremely low expression level of Ad12 E1B-GAL4 BD fusion, being virtually undetectable in Western blots (Fig. 7D, lane 2), while Ad2 E1B-GAL4 BD fusion can be detected (lane 3).
FIG. 8.
Mapping of the domain of Ad2 E1B 55-kDa protein required for binding to PCAF. (A) Schematic drawings of different Ad2 E1B fragments fused to GAL4 AD. The regions required for interacting with p53 are denoted with heavy lines on the top of the full-length Ad2 E1B 55-kDa protein. The Ad2 E1B fragments are numbered from 1 to 5, and the residues contained in each construct are indicated on the left. (B) The domain near the C terminus of Ad2 E1B is required for PCAF binding. The PCAF-GAL4 BD hybrid was tested for its interaction with Ad2 E1B. The GAL4 AD was fused to different Ad2 E1B fragments as shown in panel A: aa 437 to 495 (sector 1), aa 1 to 161 (sector 2), aa 155 to 437 (sector 3), aa 155 to 495 (sector 4), and aa 1 to 437 (sector 5). (C) Expression of GAL4 AD-Ad2 E1B hybrids. The hybrids in yeast cell extracts were detected using anti-GAL4 AD antibody (Santa Cruz Biotechnology). The Ad2 E1B domain in each hybrid is indicated on the top of each lane. Lane 1 shows yeast lysate without any GAL4 AD hybrid.
A domain near the C terminus of Ad2 E1B 55-kDa protein is necessary for binding to PCAF.
We also tested PCAF-E1B interaction using the two-hybrid assay. Five Ad2 E1B 55-kDa protein constructs containing various regions of the protein were used (Fig. 8A). As shown in Fig. 8B, only the Ad2 55-kDa protein construct carrying residues 155 to 495 exhibited interaction with PCAF (Fig. 8B, sector 4), although all these Ad2 E1B constructs except construct 3 containing aa 155 to 437 were well expressed in yeast (Fig. 8C). As both Ad2 E1B constructs 4 and 5 bind to p53 (Fig. 7), these results appear to suggest that the PCAF-binding site of E1B is distinct from the p53-binding site shown on the top of Fig. 8A. Thus, a unique region near the C terminus of Ad2 E1B 55-kDa protein is likely to be necessary for binding to PCAF.
E1B 55-kDa protein interferes with PCAF-p53 interaction.
One possible mechanism by which E1B 55-kDa protein inhibits acetylation of p53 by PCAF might be that PCAF fails to interact with p53 in the presence of E1B. To investigate this possibility, we employed a reverse two-hybrid assay. PCAF-GAL4 BD hybrid and p53-GAL4 AD fusion were introduced into yeast together with pCu424 expressing residues 1 to 437 of the Ad2 E1B 55-kDa protein (again, the WT Ad2 E1B could not be cloned in this plasmid) or the full-length Ad12 E1B 55-kDa protein. Since p53 and PCAF can interact with each other, yeast growth would be expected in medium lacking histidine in the presence of p53-GAL4 AD and PCAF-GAL4 BD hybrids. If E1B interferes with p53-PCAF interaction, the His+ phenotype might be lost when E1B-expressing pCu424 is present in addition to p53-GAL4 AD and PCAF-GAL4 BD hybrids. Indeed, as shown in Fig. 9A, Ad2 E1B 55-kDa protein (aa 1 to 437) expression led to the loss of the His+ phenotype in the presence of PCAF-GAL4 BD hybrid along with p53(1–393)-GAL4 AD fusion (Fig. 9A, sector 4). In contrast, when the DNA fragment encoding Ad2 E1B (1–437) was cloned in reverse orientation in pCu424, significant yeast growth was seen (Fig. 9A, sector 1). Failure of yeast growth in the presence of Ad2 E1B cannot be ascribed to simple toxicity of E1B to yeast, as yeast containing Ad2 E1B constructs grows very well in nonselective media, and the E1B expression in yeast can be detected (Fig. 9B). Interestingly, the His+ phenotype was retained when pCu424-Ad12 E1B was introduced into yeast along with p53-GAL4 AD and PCAF-GAL4 BD (Fig. 9A, sector 3). As a negative control, the p53 hybrid was cotransformed with empty vectors pGBDU and pCu424. As expected, no yeast growth was detected (Fig. 9A, sector 2). These results indicate that Ad2 E1B 55-kDa protein interferes with p53-PCAF interaction. Ad2 E1B was expressed when cloned in correct orientation (Fig. 9B, lane 3). The GAL4 BD-PCAF fusion was also expressed in all cases (Fig. 9C). However, we were unable to detect Ad12 E1B expression from pCu424-Ad12 E1B plasmid. Thus, the failure of pCu424-Ad12 E1B to prevent p53-PCAF interaction (Fig. 9A, sector 3) might reflect a low level of expression of Ad12 E1B 55-kDa protein in yeast (also Fig. 7D, lane 2).
DISCUSSION
Key to its function as a tumor suppressor, p53 regulates the expression of a specific set of genes involved in cell growth control. p53 activates transcription of genes containing specific DNA-binding sites for p53. Under normal physiological conditions, p53 exists in low abundance, apparently in an inactive, latent state with low sequence-specific DNA-binding activity (29). Multiple types of modifications occur on p53 in response to DNA damage or other genotoxic stresses. Covalent modifications of p53, such as phosphorylation by kinases, dephosphorylation by phosphatases, and acetylation by acetylases, lead to its stabilization (53), specific interaction with other regulatory proteins such as 14-3-3 (64), and enhanced DNA-binding activity (17, 35, 47). Consequently, any interference with covalent modifications of p53 could compromise its functions. The results presented here show that the E1B 55-kDa protein specifically inhibits p53 acetylation by PCAF both in vitro and in vivo (Fig. 2 and 3). Since acetylation of p53 has been shown to enhance its sequence-specific DNA-binding activity and also to be induced in response to DNA damage (17, 35, 47), the E1B oncoprotein represses p53 functions at least in part by inhibiting its acetylation. Indeed, we have found that the DNA-binding activity of p53 in cells expressing E1B 55-kDa protein is greatly reduced (Fig. 4).
How does E1B specifically inhibit acetylation of p53 by PCAF? We found that E1B interacts with both p53 and PCAF (Fig. 5 to 8). In addition, p53 also binds to PCAF (Fig. 7). Interestingly, E1B can efficiently prevent the interaction between p53 and PCAF (Fig. 9), suggesting that E1B may inhibit the acetylation of p53 by PCAF by blocking the enzyme-substrate interaction. Consistent with this, E1B does not affect the acetylation of histones by PCAF (Fig. 2). Our data indicate that Ad2 E1B fragment aa 1 to 437 is able to prevent p53-PCAF interaction (Fig. 9). Since this E1B fragment binds to p53 but not to PCAF (Fig. 7 and 8), binding of E1B to p53 may be sufficient for E1B to interfere with the p53-PCAF interaction. Nonetheless, E1B 55-kDa protein also binds to PCAF in vitro and in vivo (Fig. 5 and 8). Thus, by association with both p53 and PCAF, E1B protein might shift equilibrium of protein-protein complexes from p53-PCAF to p53-E1B and PCAF-E1B, thereby effectively sequestering p53 and PCAF and preventing PCAF from acetylating p53. It appears that PCAF and p53 bind to different sites in E1B (Fig. 7 and 8). This finding might permit an assessment of the relative importance of p53-E1B and PCAF-E1B interactions in inhibiting acetylation of p53 through extensive mutagenesis of the p53- and PCAF-binding sites in the E1B 55-kDa protein.
While the role of E1B-PCAF interaction in inhibiting acetylation of p53 is not clear at present, blocking the catalytic core of PCAF (the HAT domain) by E1B is unlikely to be the cause of inhibition; rather, binding of E1B to PCAF might induce conformational change of PCAF so that its substrate specificity is altered. We have found that a mutant PCAF lacking aa 62 to 464 can acetylate both p53 and histones because it still contains the HAT domain (44). Interestingly, acetylation of p53 but not that of histones by this mutant PCAF is also inhibited by E1B (data not shown). Thus, binding of E1B to a domain in the C-terminal portion of PCAF may play a role in the observed inhibition of p53 acetylation, as this mutant PCAF lacks most of the sequence in its N-terminal region. Future study will address which domain of PCAF is responsible for binding to E1B 55-kDa protein.
The acetylation of p53 by PCAF is severely impaired in cells expressing E1B 55-kDa protein (Fig. 3). Accumulation of acetylated p53 occurs normally in G401 cells upon treatment with the specific deacetylase inhibitor TSA (Fig. 3B). By contrast, acetylated p53 is virtually undetectable under the same conditions in cell line G401-CC3, derived from G401 cells by stable transfection with an Ad12 E1B 55-kDa protein-expressing vector (55). The acetylation of p53 in 293 cells is also suppressed. While 293 cells express both E1A and E1B proteins, G401-CC3 cells express only E1B 55-kDa protein. Therefore, E1B 55-kDa protein is most likely to be responsible for the observed inhibition of p53 acetylation by PCAF, consistent with the in vitro studies (Fig. 2). Effects of E1A oncoproteins on the activities of PCAF, p300, and CBP have been reported previously (1, 5, 19, 44). While E1A was found to bind to PCAF (44), it does not appear to affect the HAT activity of PCAF. Instead, E1A protein was found to stimulate HAT activities of p300 and CBP under certain circumstances (1). Conversely, recent studies demonstrated that the E1A oncoproteins may repress HAT activity of both PCAF and p300 in vitro (5, 19). Regardless of the potential role of E1A proteins in regulation acetylation, our results clearly indicate that E1B 55-kDa protein inhibits acetylation of p53 by PCAF both in vivo and in vitro.
Whereas acetylation of p53 enhances its sequence-specific DNA-binding activity (17, 35, 47), a number of other modifications of the p53 C-terminal domain can also activate its DNA-binding function (20). We found that anti-p53 antibody PAb421 can effectively enhance p53 DNA binding in G401-CC3 nuclear extracts (Fig. 4), despite reduced p53 acetylation in this cell line (Fig. 3). Thus, it is conceivable that binding of PAb421 to the C-terminal domain of p53 may have a similar effect on activating p53 DNA-binding activity as acetylation of the lysine residues within the p53 C-terminal domain. Interestingly, we found that PAb421 cannot activate p53 DNA binding in 293 nuclear extracts (Fig. 4), suggesting that E1A may have additional inhibitory effects on p53 DNA-binding activity. Indeed, p53 appears to form high-molecular-weight oligomers when E1A proteins are expressed in G401 cells (54). Such modified p53 protein may be less competent for DNA binding, as its transcriptional transactivation function was greatly repressed by E1A (54).
The E1B 55-kDa protein represses transcriptional transactivation by p53 by binding directly to DNA-bound p53 without destabilizing p53-DNA complexes, thereby tethering the E1B transcriptional repression domain to promoters containing p53 binding sites (71). Furthermore, E1B 55-kDa protein appears to enhance p53-DNA interaction and could repress transactivation mediated by p53 in an in vitro assay using purified RNA polymerase II components (39, 40). We show here that E1B 55-kDa protein inhibits effectively p53 acetylation. Inhibition of p53 acetylation (thus reducing its affinity to its binding sites [Fig. 4]) and direct targeting of DNA-bound p53 might reflect two levels of repression of p53 functions by E1B. At the first level, E1B represses acetylation of p53 by PCAF, and possibly also by p300 and CBP. At the second level, E1B could still target any p53 that binds to specific sites within target promoters due to incomplete inhibition of acetylation or other means, thereby resulting in direct transcriptional repression. The two-level, fail-safe repression mechanisms on p53 suggest that E1B 55-kDa oncoprotein is a particularly powerful repressor of p53. Whether E1B also inhibits other types of covalent modifications occurring on p53 remains to be established. Our unpublished data indicated that phosphorylation of p53 at serine 392 was not affected by E1B 55-kDa protein in a Western blot analysis using a specific antibody against p53 phosphorylated at serine 392.
The Ad E1A and E1B proteins play important roles in cell transformation (3, 45). The E1A oncoproteins stimulate cell proliferation by binding to pRB, p300, and CBP (reviewed in reference 11). The E1B 55-kDa protein acts in cell transformation by inactivating the p53 pathway. Although inactivation of pRB and p53 pathways is essential to the transformation of a normal cell into a tumor cell (18), deregulation of other cellular regulatory circuitry may also be involved in cell transformation and development of cancer. As PCAF is implicated in regulation of cell differentiation, cell cycle progression, and transcriptional regulation (43, 50, 62), deregulation of the PCAF pathway might also play a role in cell transformation. Intriguingly, just as E1A and E1B 55-kDa proteins inhibit p53 transactivation function, the same viral oncoproteins bind to PCAF, although it is likely that E1A and E1B affect different aspects of the PCAF functions. Furthermore, E1A appears to compete with PCAF for access to p300 (68), which could affect critical functions of PCAF in regulating cellular pathways. Additionally, the Tax oncoprotein of the human T-cell leukemia virus type 1 recruits PCAF for transactivating viral promoters (23). Such exploitation of PCAF by viral oncoproteins might perturb cellular physiology, thus contributing to cell transformation.
Increasing evidence supports critical roles for protein acetylation in cellular physiology, including regulation of protein-DNA and protein-protein interaction, as well as protein stability. Like phosphorylation, acetylation can regulate key cellular processes in response to extracellular signals. Thus, it has been proposed previously that acetylation as a biologically relevant modification may be as important as phosphorylation (26). Consistent with this view, our data demonstrate that the Ad E1B 55-kDa oncoproteins specifically inhibit p53 acetylation by PCAF, while its histone acetylation and autoacetylation activities were not affected. These data suggest that the inhibition of p53 acetylation by viral proteins may represent an important mechanism of p53 inactivation. Future investigation on how E1B 55-kDa oncoprotein affects PCAF functions will provide insight into the biological functions mediated by PCAF, as well as how modulation of cellular acetylase activities by viral oncoproteins contributes to cell transformation and oncogenesis.
ACKNOWLEDGMENTS
We thank Arnold Berk for Ad2 and Ad12 55-kDa protein expression constructs and a recombinant baculovirus for expressing Ad2 E1B 55-kDa protein, Bert Vogelstein for human p53 expression vectors, Stanley Fields for pGAD plasmids containing various p53 segments, Arnold Levine for hybridoma cell line 2A6, and Sherif Abou Elela for expert help with the yeast two-hybrid technique and for plasmids. We acknowledge Chong Jiang's contributions in the initial phase of this study. We are also grateful to Pierre Bourgaux, Benoit Chabot, Raymund Wellinger, and Alan Weiner for reading the manuscript and for insightful suggestions.
This work was supported by the Fonds de la Recherche en Santé du Québec (FRSQ) and by Medical Research Council of Canada (MRC) grants (MOP-14109 to D.L. and MT-14608 to X.-J.Y.). D.L. is a Chercheur-Boursier Junior I of FRSQ, and X.-J.Y. is an MRC Scholar.
REFERENCES
- 1.Ait-Si-Ali S, Ramirez S, Barre F X, Dkhissi F, Magnaghi-Jaulin L, Girault J A, Robin P, Knibiehler M, Pritchard L L, Ducommun B, Trouche D, Harel-Bellan A. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 2.Bargonetti J, Friedman P N, Kern S E, Vogelstein B, Prives C. Wild-type but not mutant p53 immunopurified proteins bind to sequences adjacent to the SV40 origin of replication. Cell. 1991;65:1083–1091. doi: 10.1016/0092-8674(91)90560-l. [DOI] [PubMed] [Google Scholar]
- 3.Barker D D, Berk A J. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology. 1987;156:107–121. doi: 10.1016/0042-6822(87)90441-7. [DOI] [PubMed] [Google Scholar]
- 4.Candau R, Zhou J X, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 6.Davie J R. Covalent modifications of histones: expression from chromatin templates. Curr Opin Genet Dev. 1998;8:173–178. doi: 10.1016/s0959-437x(98)80138-x. [DOI] [PubMed] [Google Scholar]
- 7.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 8.Dhalluin C, Carlson J E, Zeng L, He C, Aggarwal A K, Zhou M M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 9.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farmer G, Bargonetti J, Zhu H, Friedman P, Prywes R, Prives C. Wild-type p53 activates transcription in vitro. Nature. 1992;358:83–86. doi: 10.1038/358083a0. [DOI] [PubMed] [Google Scholar]
- 11.Flint J, Shenk T. Viral transactivating proteins. Annu Rev Genet. 1997;31:177–212. doi: 10.1146/annurev.genet.31.1.177. [DOI] [PubMed] [Google Scholar]
- 12.Gallimore P H, Lecane P S, Roberts S, Rookes S M, Grand R J, Parkhill J. Adenovirus type 12 early region 1B 54K protein significantly extends the life span of normal mammalian cells in culture. J Virol. 1997;71:6629–6640. doi: 10.1128/jvi.71.9.6629-6640.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottlieb T M, Oren M. p53 in growth control and neoplasia. Biochim Biophys Acta. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- 14.Graham F L, Abrahams P J, Mulder C, Heijneker H L, Warnaar S O, De Vries F A, Fiers W, Van Der Eb A J. Studies on in vitro transformation by DNA and DNA fragments of human adenoviruses and simian virus 40. Cold Spring Harbor Symp Quant Biol. 1975;39:637–650. doi: 10.1101/sqb.1974.039.01.077. [DOI] [PubMed] [Google Scholar]
- 15.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 16.Grand R J, Grant M L, Gallimore P H. Enhanced expression of p53 in human cells infected with mutant adenoviruses. Virology. 1994;203:229–240. doi: 10.1006/viro.1994.1480. [DOI] [PubMed] [Google Scholar]
- 17.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 18.Hahn W C, Counter C M, Lundberg A S, Beijersbergen R L, Brooks M W, Weinberg R A. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 19.Hamamori Y, Sartorelli V, Ogryzko V, Puri P L, Wu H Y, Wang J Y, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 20.Hupp T R, Meek D W, Midgley C A, Lane D P. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 21.Iwabuchi K, Li B, Bartel P, Fields S. Use of the two-hybrid system to identify the domain of p53 involved in oligomerization. Oncogene. 1993;8:1693–1696. [PubMed] [Google Scholar]
- 22.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang H, Lu H, Schlitz R L, Pise-Masison C A, Ogryzko V V, Nakatani Y, Brady J N. PCAF interacts with tax and stimulates tax transactivation in a histone acetyltransferase-independent manner. Mol Cell Biol. 1999;19:8136–8145. doi: 10.1128/mcb.19.12.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao C C, Yew P R, Berk A J. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology. 1990;179:806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 25.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 26.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 28.Labbe S, Zhu Z, Thiele D J. Copper-specific transcriptional repression of yeast genes encoding critical components in the copper transport pathway. J Biol Chem. 1997;272:15951–15958. doi: 10.1074/jbc.272.25.15951. [DOI] [PubMed] [Google Scholar]
- 29.Lane D. Awakening angels. Nature. 1998;394:616–617. doi: 10.1038/29166. [DOI] [PubMed] [Google Scholar]
- 30.Lane D P, Crawford L V. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 31.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 32.Liao D, Yu A, Weiner A M. Coexpression of the adenovirus 12 E1B 55 kDa oncoprotein and cellular tumor suppressor p53 is sufficient to induce metaphase fragility of the human RNU2 locus. Virology. 1999;254:11–23. doi: 10.1006/viro.1998.9512. [DOI] [PubMed] [Google Scholar]
- 33.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 34.Linzer D I, Levine A J. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, Scolnick D M, Trievel R C, Zhang H B, Marmorstein R, Halazonetis T D, Berger S L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe S W, Ruley H E. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 37.Mak I, Mak S, Benchimol S. Expression of the cellular p53 protein in cells transformed by adenovirus 12 and viral DNA fragments. Virology. 1988;163:201–204. doi: 10.1016/0042-6822(88)90248-6. [DOI] [PubMed] [Google Scholar]
- 38.Marcellus R C, Teodoro J G, Charbonneau R, Shore G C, Branton P E. Expression of p53 in Saos-2 osteosarcoma cells induces apoptosis which can be inhibited by Bcl-2 or the adenovirus E1B-55 kDa protein. Cell Growth Differ. 1996;7:1643–1650. [PubMed] [Google Scholar]
- 39.Martin M E, Berk A J. Adenovirus E1B 55K represses p53 activation in vitro. J Virol. 1998;72:3146–3154. doi: 10.1128/jvi.72.4.3146-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin M E, Berk A J. Corepressor required for adenovirus E1B 55,000-molecular-weight protein repression of basal transcription. Mol Cell Biol. 1999;19:3403–3414. doi: 10.1128/mcb.19.5.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mietz J A, Unger T, Huibregtse J M, Howley P M. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. EMBO J. 1992;11:5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prives C. Signaling to p53: breaking the MDM2-p53 circuit. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 43.Puri P L, Sartorelli V, Yang X J, Hamamori Y, Ogryzko V V, Howard B H, Kedes L, Wang J Y, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 44.Reid J L, Bannister A J, Zegerman P, Martinez-Balbas M A, Kouzarides T. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 1998;17:4469–4477. doi: 10.1093/emboj/17.15.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ricciardi R P. Transformation and tumorigenesis mediated by the adenovirus E1A and E1B oncogenes. In: Barbanti-Brodano G, et al., editors. DNA tumor viruses: oncogenic mechanisms. New York, N.Y: Plenum Press; 1995. pp. 195–225. [Google Scholar]
- 46.Roulston A, Marcellus R C, Branton P E. Viruses and apoptosis. Annu Rev Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- 47.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1B-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 49.Sarnow P, Sullivan C A, Levine A J. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology. 1982;120:510–517. doi: 10.1016/0042-6822(82)90054-x. [DOI] [PubMed] [Google Scholar]
- 50.Sartorelli V, Puri P L, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang J Y, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 51.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 52.Schmieg F I, Simmons D T. Characterization of the in vitro interaction between SV40 T antigen and p53: mapping the p53 binding site. Virology. 1988;164:132–140. doi: 10.1016/0042-6822(88)90628-9. [DOI] [PubMed] [Google Scholar]
- 53.Shieh S Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 54.Steegenga W T, van Laar T, Riteco N, Mandarino A, Shvarts A, van der Eb A J, Jochemsen A G. Adenovirus E1A proteins inhibit activation of transcription by p53. Mol Cell Biol. 1996;16:2101–2109. doi: 10.1128/mcb.16.5.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steegenga W T, Van Laar T, Shvarts A, Terleth C, Van der Eb A J, Jochemsen A G. Distinct modulation of p53 activity in transcription and cell-cycle regulation by the large (54 kDa) and small (21 kDa) adenovirus E1B proteins. Virology. 1995;212:543–554. doi: 10.1006/viro.1995.1512. [DOI] [PubMed] [Google Scholar]
- 56.Tan T H, Wallis J, Levine A J. Identification of the p53 protein domain involved in formation of the simian virus 40 large T-antigen–p53 protein complex. J Virol. 1986;59:574–583. doi: 10.1128/jvi.59.3.574-583.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teodoro J G, Branton P E. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J Virol. 1997;71:3620–3627. doi: 10.1128/jvi.71.5.3620-3627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teodoro J G, Shore G C, Branton P E. Adenovirus E1A proteins induce apoptosis by both p53-dependent and p53-independent mechanisms. Oncogene. 1995;11:467–474. [PubMed] [Google Scholar]
- 59.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 60.van den Elsen P J, Houweling A, van der Eb A J. Morphological transformation of human adenoviruses is determined to a large extent by gene products of region E1a. Virology. 1983;131:242–246. doi: 10.1016/0042-6822(83)90549-4. [DOI] [PubMed] [Google Scholar]
- 61.van den Heuvel S J, van Laar T, The I, van der Eb A J. Large E1B proteins of adenovirus types 5 and 12 have different effects on p53 and distinct roles in cell transformation. J Virol. 1993;67:5226–5234. doi: 10.1128/jvi.67.9.5226-5234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vassilev A, Yamauchi J, Kotani T, Prives C, Avantaggiati M L, Qin J, Nakatani Y. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol Cell. 1998;2:869–875. doi: 10.1016/s1097-2765(00)80301-9. [DOI] [PubMed] [Google Scholar]
- 63.Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis C D, Berger S L. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waterman M J, Stavridi E S, Waterman J L, Halazonetis T D. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat Genet. 1998;19:175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- 65.Weissman B E, Saxon P J, Pasquale S R, Jones G R, Geiser A G, Stanbridge E J. Introduction of a normal human chromosome 11 into a Wilms' tumor cell line controls its tumorigenic expression. Science. 1987;236:175–180. doi: 10.1126/science.3031816. [DOI] [PubMed] [Google Scholar]
- 66.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 67.Wienzek S, Roth J, Dobbelstein M. E1B 55-kilodalton oncoproteins of adenovirus types 5 and 12 inactivate and relocalize p53, but not p51 or p73, and cooperate with E4orf6 proteins to destabilize p53. J Virol. 2000;74:193–202. doi: 10.1128/jvi.74.1.193-202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 69.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 70.Yew P R, Kao C C, Berk A J. Dissection of functional domains in the adenovirus 2 early 1B 55K polypeptide by suppressor-linker insertional mutagenesis. Virology. 1990;179:795–805. doi: 10.1016/0042-6822(90)90147-j. [DOI] [PubMed] [Google Scholar]
- 71.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 72.Zantema A, Fransen J A, Davis-Olivier A, Ramaekers F C, Vooijs G P, DeLeys B, Van der Eb A J. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology. 1985;142:44–58. doi: 10.1016/0042-6822(85)90421-0. [DOI] [PubMed] [Google Scholar]
- 73.Zantema A, Schrier P I, Davis-Olivier A, van Laar T, Vaessen R T M J, van der Eb A J. Adenovirus serotype determines association and localization of the large E1B tumor antigen with cellular tumor antigen p53 in transformed cells. Mol Cell Biol. 1985;5:3084–3091. doi: 10.1128/mcb.5.11.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]