ABSTRACT
The iconic Australian marsupial, the koala (Phascolarctos cinereus), has suffered dramatic population declines as a result of habitat loss and fragmentation, disease, vehicle collision mortality, dog attacks, bushfires and climate change. In 2012, koalas were officially declared vulnerable by the Australian government and listed as a threatened species. In response, research into diseases affecting koalas has expanded rapidly. The two major pathogens affecting koalas are Chlamydia pecorum, leading to chlamydial disease and koala retrovirus (KoRV). In the last eight years, these pathogens and their diseases have received focused study regarding their sources, genetics, prevalence, disease presentation and transmission. This has led to vast improvements in pathogen detection and treatment, including the ongoing development of vaccines for each as a management and control strategy. This review will summarize and highlight the important advances made in understanding and combating C. pecorum and KoRV in koalas, since they were declared a threatened species. With complementary advances having also been made from the koala genome sequence and in our understanding of the koala immune system, we are primed to make a significant positive impact on koala health into the future.
Keywords: Chlamydia, Chlamydia pecorum, koala retrovirus, KoRV, koala, Phascolarctos cinereus
Recent advances in understanding the two major pathogens of koalas, Chlamydia pecorum and koala retrovirus (KoRV), have benefited both koala conservation and general chlamydial and retroviral research fields.
INTRODUCTION
Disease is a major conservation challenge for the well-loved Australian marsupial, the koala (Phascolarctos cinereus) (Fig. 1). Once believed to number in the millions (Phillips and Service 1990), experts now estimate the koala population in Australia to be approximately 330 000 animals, with major and continued population decline anticipated in the northern half of their range (Melzer et al. 2000; McAlpine et al. 2015; Adams-Hosking et al. 2016; Beyer et al. 2018). This has resulted in koalas from the northern half of Australia being added to the Australian Environmental Protection Biodiversity Conservation Act in 2012 (Australia 2011) (Fig. 2) and all koalas being listed as “vulnerable” on the Red List of Threatened Species worldwide (Woinarski and Burbidge 2016). While multiple threats have been identified as impacting koala populations, including habitat degradation/loss, disease, dog attacks, motor vehicle strikes, bushfires and climate change, the significant impact of disease, particularly from Chlamydia and koala retrovirus (KoRV), is routinely highlighted (Australia 2011; Hemming et al. 2018). In response, targeted research into koala diseases has expanded rapidly since 2012. Major advances in understanding the source, genetics, prevalence, disease presentation and transmission of the two major koala pathogens, Chlamydia pecorum and KoRV, has led to complementary advances in pathogen detection and treatment. The overall progress that has been made in koala disease research since 2012 has dramatically improved our knowledge of the koala disease landscape and equipped us with the tools to make a significant improvement in koala conservation moving forward.
Figure 1.

Koalas (Phascolarctos cinereus). Photo credit to Endeavour Veterinary Ecology.
Figure 2.
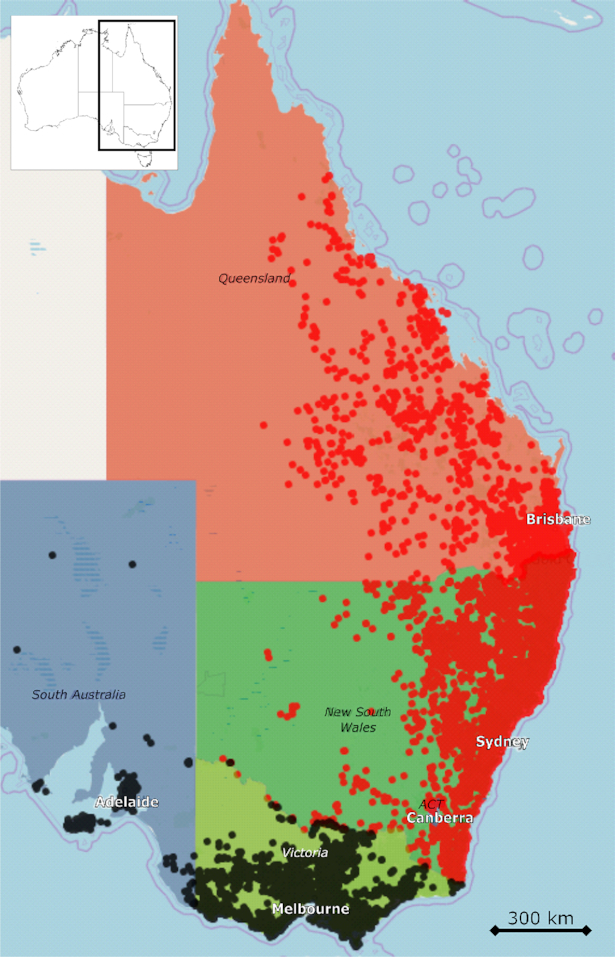
Current range of koalas in Australia. Locations where koalas are listed as vulnerable (from Queensland, New South Wales and Australian Capital Territory (ACT)) are indicated by red dots while locations where koalas do not have a conservation listing (Victoria and South Australia) are indicated by black dots. Map and koala location data are from Atlas of Living Australia website at http://www.ala.org.au. Accessed 10 February 2020.
CHLAMYDIA PECORUM
Chlamydia pecorum is the bacterium that causes chlamydial disease in koalas. While Chlamydia pneumoniae has also been reported in koalas, recent molecular surveys have detected virtually no C. pneumoniae in koalas (Burach et al. 2014; Johnston et al. 2015; Patterson et al. 2015; Legione et al. 2016b; Santamaria and Schlagloth 2016; Hulse et al. 2018; Palmieri et al. 2019), indicating that this species is currently not a major contributor to disease. Like all Chlamydia, C. pecorum has a unique biphasic life cycle. When extracellular, the chlamydiae exist as metabolically inactive infectious particles known as elementary bodies (EBs). EBs are taken up by susceptible host cells into inclusions, where they prevent phagosome-lysozyme fusion and replicate in this intracellular compartment as reticulate bodies (RBs). Once enough RBs have formed, cells convert back into EBs to lyse the cell and infect other cells.
C. pecorum is classically associated with ocular and urogenital disease in koalas (Fig. 3). When the infection establishes in the conjunctiva of the eye, chronic conjunctivitis and keratoconjunctivitis can lead to corneal scarring and eventual blindness (Fig. 3A–D) (Blanshard and Bodley 2008; Jelocnik et al. 2019). When the infection establishes in the urinary tract (including the urethra, bladder, ureters and kidneys), the accompanying urethritis, cystitis, ureteritis and/or pyelonephritis can cause severe pain, polyuria and/or urinary incontinence leading to “wet-bottom” (urine staining the rump fur, Fig. 3E and F), as well as skin ulceration and secondary infection (Polkinghorne, Hanger and Timms 2013; Jelocnik et al. 2019). Finally, when infection establishes in the reproductive tract, inflammation in both females (eg. salpingitis, endometritis, vaginitis) and males (eg. epididymitis, orchitis, urethritis) can lead to scarring and infertility (Blanshard and Bodley 2008; Johnston et al. 2015; Palmieri et al. 2019). C. pecorum infection is, however, not only limited to the classic body sites within the koala, with recent reports implicating this pathogen in fatal pneumonia (Mackie et al. 2016), polyarthritis (Burnard, Gillett and Polkinghorne 2018) and colonization of the gastrointestinal tract (Burach et al. 2014; Wedrowicz et al. 2016; Phillips et al. 2018).
Figure 3.
Chlamydia pecorum disease in koalas. (A), Healthy koala eye, (B), Mild conjunctivitis, (C), Moderate conjunctivitis/keratoconjunctivitis with chemosis, purulent periocular discharge and a mildly proliferative prolapsed nictitating membrane, (D), Severe conjunctivitis with purulent discharge, marked proliferation of conjunctival tissues, completing obscuring the eye, (E), Healthy koala rump, (F), Koala with cystitis, rump fur stained by urine due to incontinence (“wet bottom”). Photo credit to Australia Zoo Wildlife Hospital.
Origin of C. pecorum in koalas
Although records of koalas before and during early European settlement in Australia are sparse, there is a general belief that chlamydiosis has been a component of koalas’ natural history (Phillips 2000). However, recent molecular epidemiology investigations of C. pecorum across several animal hosts have introduced the hypothesis that at least some koala Chlamydia infections may be spill-over events from introduced domestic livestock. Both multilocus sequence typing (MLST) and whole genome comparisons of C. pecorum have detected genetically similar strains between koalas and sheep (Jelocnik et al. 2013, 2014; Bachmann et al. 2015). Additionally, two koalas from French Island, Victoria (VIC) (a closed island koala population believed to be Chlamydia-free) have now been found to be infected with C. pecorum strains more closely related to known cattle (which are farmed on the island) and pig livestock strains compared to known koala strains from the mainland (Legione et al.2016a). More epidemiological tracking is needed to clarify whether these apparent livestock/koala spill-over events are common and if these events are having an appreciable impact on koala disease. However, these findings do highlight that C. pecorum has a broad host range and the introduction of domestic livestock to Australia cannot be ruled out as a source of chlamydial infection for native animals like the koala.
Advances in genetic understanding of C. pecorum
In the last decade, advances in whole genome sequencing and bioinformatics have allowed for new analyses of C. pecorum strains infecting koalas. From the individual gene target perspective, the Major Outer Membrane Protein (MOMP), coded by the ompA gene, has remained a favorite target for strain typing chlamydial diversity in koala populations (Kollipara et al. 2013c; Legione et al.2016b; Nyari et al. 2017; Wedrowicz et al. 2018). Surveys from across Australia have identified 15 unique ompA genotypes in koalas (Fig. 4). Based on the phylogeny of the ompA gene, there appears to be a general separation of ompA genotypes into two major groups; one group consisting of two clades detected mostly in the northern half of the country (genotypes A, E’, F, F’, H, J and K) and another group consisting of two clades detected mostly in the southern half of the country (genotypes B, C, G, I, L, M, N, O) (Fig. 4). However, the geographical separation of the genotypes is not absolute, with genotypes F, F’ and G being detected in koalas from both northern (Queensland (QLD) and New South Wales (NSW)) and southern (VIC and South Australia (SA)) geographic ranges.
Figure 4.
Maximum likelihood tree diagramming the relatedness of Chlamydia pecorum ompA genotypes. The Jones, Taylor, Thornton matrix model was used to construct the tree with 100 bootstrap confidence. Bootstrap values above 60 are shown on the nodes. A C. pecorum isolate from cattle in Japan was included as an outlier. Australian states with koalas positive for each genotype are indicated in the genotype label. QLD – Queensland, NSW – New South Wales, VIC – Victoria, SA – South Australia.
Concern that the ompA gene did not reliably reflect the genetic diversity and relationship of C. pecorum strains alone led to the development of a C. pecorum MLST scheme (Jelocnik et al. 2013). Targeting seven C. pecorum house-keeping genes, enoA, oppA_3, gidA, hemN, hflX, fumC and gatA, the MLST scheme has been used as an alternative system for C. pecorum strain typing (Jelocnik et al. 2014; Fernandez et al. 2019). Although not as widely utilized as the traditional ompA genotyping system, MLST typing of koala C. pecorum strains has allowed for greater comparison of koala isolates to other hosts around the world, underpinning new investigations in strain tracking and transmission between hosts. Alterative C. pecorum gene targets with variable tandem repeat regions, such as incA and ORF663, have also been investigated to characterize koala C. pecorum strains (Higgins et al. 2012; Mohamad et al. 2014). For both genes, initial analysis has revealed that koala C. pecorum strains do possess variable numbers of tandem repeats and that there may be a link between a higher number of repeats and less virulent strains (Higgins et al. 2012; Mohamad et al. 2014). However, with the continued advancement of whole genome analysis over single gene studies, non-ompA typing and targeting studies have trailed off in recent years.
Whole genome analysis of koala C. pecorum strains has advanced our understanding of several important genetic and biological properties of this koala pathogen. Sequencing has revealed that the polymorphic membrane protein (pmp) gene cluster and the plasticity zone of the C. pecorum genome are hot spots for single nucleotide polymorphisms (SNPs) between strains (Bachmann et al. 2015; Jelocnik et al. 2015). A plasmid with potential virulence genes has been detected in the majority of C. pecorum strains, with an association between plasmid carriage and disease outcomes starting to emerge (Jelocnik et al. 2015, 2016; Phillips et al. 2018). In addition, shotgun sequencing directly from clinical samples has revealed the presence of multiple distinct C. pecorum strains, often at the urogenital site, within an individual koala (Bachmann et al. 2015). This finding reminds us that infections are often complex within an animal and most simple typing schemes likely underrepresent the actual C. pecorum complexity present.
Finally, whole genome sequencing of C. pecorum strains initially illuminated an important biological difference between this species and its related human pathogen, Chlamydia trachomatis. C. trachomatis is a tryptophan auxotroph, having only the tryptophan synthase (trpBA) gene for the final conversion of indole to tryptophan (Ziklo et al. 2016b). This results in C. trachomatis being very sensitive to interferon-gamma (IFN-ɣ) mediated depletion of host tryptophan (Islam et al. 2018). Alternatively, genome sequencing revealed that C. pecorum possesses a nearly complete tryptophan biosynthesis operon (Mojica et al. 2011; Bachmann et al. 2014). This finding led to in vitro testing where C. pecorum was found to survive in tryptophan-free media supplemented with a more diverse pool of tryptophan precursors than C. trachomatis could utilize (Islam et al. 2018). C. pecorum was also found to be completely resistant to IFN-ɣ in human epithelial cells (Islam et al. 2018). Translating this genetic difference back to the koala, recent modelling of koala immune parameter data and gene expression analysis has suggested that IFN-ɣ may not play a major role in C. pecorum control in koalas (Phillips et al.2019). This situation is an excellent example of how genome analysis can be the basis of advancing important biological understandings in chlamydial research.
Prevalence of C. pecorum infection and chlamydial disease
An important area of koala chlamydial research that has received focused attention since 2012 has been the survey and characterization of C. pecorum infections and/or chlamydial disease in koalas across Australia (Figs 5 and 6, Tables 1 and 2). A review of C. pecorum positivity in koala populations before 2012 found rates from 0% C. pecorum detected (from isolated island koala populations) up to 87%–90% positivity in koala populations from QLD and VIC (Polkinghorne, Hanger and Timms 2013). Since that review, there have been 18 additional studies that have investigated some aspect of C. pecorum presence and/or chlamydial disease rates in koalas across the range (Table 1A) (Griffith and Higgins 2012; Funnell et al. 2013; Griffith et al. 2013; Kollipara et al. 2013c; Patterson et al. 2015; Legione et al. 2016a,b; Speight et al. 2016; Gonzalez-Astudillo et al. 2017; Nyari et al. 2017; Speight et al. 2018; Wedrowicz et al. 2018; Fabijan et al. 2019b; Fernandez et al. 2019; Gonzalez-Astudillo et al. 2019; Hulse et al. 2019b; Maher et al. 2019; Palmieri et al. 2019). These studies have used a range of both established and novel C. pecorum detection and genotyping PCR assays (Table 1B) (Devereaux et al. 2003; Robertson et al. 2009; Griffith 2010; Pantchev et al. 2010; Marsh et al. 2011; Wan et al. 2011; Jelocnik et al. 2013; Kollipara et al. 2013c; Jelocnik et al. 2017; Hulse et al. 2018).
Figure 5.

Prevalence of Chlamydia pecorum infection reported in studies from 2012 to 2019. Grey ellipticals indicate the mapped area investigated, pie charts and percentages represent the C. pecorum positivity reported in ‘n’ number of koalas tested. C. pecorum ompA genotype (A-O) or multilocus sequence type (MLST) ST type detected in the study area are given when reported. Details for each study can be found in Table 1.
Figure 6.

Chlamydial disease prevalence reported in studies from 2012 to 2019. Grey ellipticals indicate the mapped area investigated, pie charts and percentages represent the chlamydial disease reported in ‘n’ number of koalas examined. Details for each study can be found in Table 2. *See clinical disease note in Table 2.
Table 1A.
Chlamydia pecorum infection prevalence reported between 2012 and 2019 in Australian koalas.
| State | Region | No. Koalas | No. Chlamydia Positive (%) | Chlamydia genotypes detected | C. pneumoniae positive | Reference |
|---|---|---|---|---|---|---|
| QLD | Brisbane | 62 | 509* (71%) | ND | 0/677 tissues | Palmieri et al. 2019 |
| Moreton Bay | 160 | 49 (31%) | E’, F, G | ND | Nyari et al. 2017 | |
| SEQLD | 250 | 167 (67%) | ND | ND | Hulse et al. 2019b | |
| SEQLD | 244 | 96 (39%) | A, E’, F, G, H | ND | Kollipara et al.2013c | |
| SEQLD | 13 | 5 (38%) | A’, F | ND | Wedrowicz et al. 2018 | |
| St Bees | 36 | 10 (28%) | F | ND | Kollipara et al.2013c | |
| NSW | Byron Bay | 5 | 1 (20%) | F | ND | Kollipara et al.2013c |
| Gunnedah | 140 | 93 (66%) | ST69, ST73, ST198 | ND | Fernandez et al. 2019 | |
| Port Macquarie | 73 | 46 (63%) | F, F’, J | ND | Kollipara et al.2013c | |
| SENSW | 11 | 3 (27%) | F | ND | Wedrowicz et al. 2018 | |
| Tanilba Bay | 41 | 24 (59%) | A, F, G, I, J, K | ND | Kollipara et al. 2013c | |
| VIC | Cape Otway | 41 | 2 (5%) | L | ND | Wedrowicz et al. 2018 |
| Far West | 168 | 36 (21%) | B | 0 | Legione et al.2016b | |
| French Island | 64 | 0 (0%) | ND | ND | Patterson et al. 2015 | |
| French Island | 237 | 2 (1%) | N | 0 | Legione et al.2016b | |
| Greater Gippsland | 30 | 11 (37%) | B, C, F, M | 0 | Legione et al.2016b | |
| Mallacoota | 5 | 0 (0%) | – | ND | Wedrowicz et al. 2018 | |
| Mornington Peninsula | 13 | 6 (46%) | B, C | 0 | Legione et al.2016b | |
| Mt Eccles national park | 120 | 30 (25%) | ND | ND | Patterson et al. 2015 | |
| Raymond Island | 104 | 43 (41%) | ND | ND | Patterson et al. 2015 | |
| Raymond Island | 153 | 50 (33%) | B | 0 | Legione et al.2016b | |
| Raymond Island | 26 | 21 (81%) | B | ND | Wedrowicz et al. 2018 | |
| South Gippsland | 198 | 117 (59%) | B, C, F, I, M, O | ND | Wedrowicz et al. 2018 | |
| South West Coast | 210 | 15 (7%) | B, L | 1 | Legione et al.2016b | |
| SA | Adelaide Hills | 4 | 4 (100%) | B, G | ND | Kollipara et al.2013c |
| Kangaroo Island | 170 | 0 (0%) | – | ND | Fabijan et al.2019b | |
| Mount Lofty | 75 | 35 (47%) | ND | ND | Fabijan et al.2019b | |
| Mount Lofty (+ Eyre Peninsula) | 65 | 57 (88%) | ND | ND | Speight et al. 2016 |
*677 tissues tested.
Table 2.
Chlamydial disease prevalence reported between 2012 and 2019 in Australian koalas.
| State | Region | No. Koalas | No. chlamydial disease (%) | Reference | Notes |
|---|---|---|---|---|---|
| QLD | Brisbane | 62 | 44 (71%) | Palmieri et al. 2019 | Males only, Moggill Koala Hospital |
| Moreton Bay | 160 | 44 (28%) | Nyari et al. 2017 | ||
| SEQLD | 250 | 65 (29%) | Hulse et al. 2019 | Males only | |
| SEQLD | 20250** | 21619 (52%) | Gonzalez-Astudillo et al. 2017 | Moggill Koala Hospital records from 1997–2013 | |
| SEQLD | 519 | 304 (59%) | Gonzalez-Astudillo et al. 2019 | Moggill Koala Hospital, Currumbin Wildlife Sanctuary Hospital, and Australia Zoo Wildlife Hospital, records from 2013–2016 | |
| NSW | Port Macquarie | 3781*** | 771 (20%) | Griffith et al. 2013 | Port Macquarie koala hospital records 1975–2004 |
| VIC | French Island | 64 | 28 (44%) | Patterson et al. 2015 | Disease was observed wet bottom |
| Mt Eccles national park | 120 | 46 (38%) | Patterson et al. 2015 | Disease was observed wet bottom | |
| Raymond Island | 104 | 45 (44%) | Patterson et al. 2015 | Disease was observed wet bottom | |
| SA | Adelaide | 85 | 10 (12%) | Speight et al. 2018 | Survey of deceased koalas at Veterinary School |
| Kangaroo Island | 170 | 0 (0%) | Fabijan et al. 2019a | 12 koalas (7%) had ocular clinical scores of 1 (very mild) | |
| Mount Lofty | 75 | 3 (4%) | Fabijan et al. 2019a | All three cases were severe disease | |
| Mount Lofty (+ Eyre Peninsula) | 65 | 41 (63%) | Speight et al. 2016 |
**41 606 aetiologies determined from 20 250 koalas from Moggill koala hospital records from 1997 to 2013, with Chlamydia infection being the most common; ***Port Macquarie koala hospital records 1975–2004, with chlamydiosis being the second most common aetiology.
Table 1B.
Tests used to determine C. pecorum infections prevalence.
| Study reference | Gene target and primers for C. pecorum test | Reference for C. pecorum test | Gene target and primers for genotyping | Reference for genotyping |
|---|---|---|---|---|
| Kollipara et al. 2013c | 16S gene - RT-Pec.spF/R | Marsh et al. 2011 | ompA - ompA-F/R | Kollipara et al. 2013c |
| Patterson et al. 2015 | 16S gene - 16SG F/R | Robertson et al. 2009 | ND | ND |
| Legione et al.2016b | 16S gene - 16SG F/R | Robertson et al. 2009 | ompA - ompA-F/R | Kollipara et al. 2013c |
| Speight et al. 2016 | 16S gene - 16Sf/16Sr | Wan et al. 2011 | ND | ND |
| Nyari et al. 2017 | 16S gene - RT-Pec.spF/R | Marsh et al. 2011 | ompA - CpeNTVD3/4 | Devereaux et al. 2003 |
| Wedrowicz et al. 2018 | ompA - CppecOMP1-F/R/S | Pantchev et al. 2010 | ompA - ompA-F/R | Kollipara et al. 2013c |
| Hulse et al. 2019 | ompB - CpecOmpB-F/R | Hulse et al. 2018 | ND | ND |
| Palmieri et al. 2019 | ompB - CpecOmpB-F/R | Hulse et al. 2018 | ND | ND |
| Fabijan et al. 2019b | HP gene - B3/F3 | Jelocnik et al. 2017 | ND | ND |
| Fernandez et al. 2019 | ompB - F/R | Griffith 2010 | MLST - gatA, oppA_3, hflX, gidA, enoA, hemN, fumC | Jelocnik et al. 2013 |
On one end of the spectrum, when more than five koalas have been tested in an area, only Kangaroo Island, South Australia (SA) has remained apparently C. pecorum-free (Figs 5 and 6) (Fabijan et al. 2019b). In this population, while no overt cases of chlamydial disease were observed, 7% of examined koalas presented with very mild ocular clinical scores (Table 2) (Fabijan et al. 2019b). It is well established that koalas can have chlamydial disease signs with no detectable C. pecorum organisms present (Wan et al. 2011; Nyari et al. 2017; Legione et al. 2018; Quigley et al. 2018a), so whether this disparity on Kangaroo Island represents a situation where C. pecorum organisms were present at low levels and were naturally cleared before testing or whether clinical signs were due to another aetiological agent remains an open question. French Island, VIC is another island koala population that had historically tested C. pecorum-free, but with signs of chlamydial disease (44% of koalas observed with “wet bottom”) (Tables 1 and 2) (Patterson et al. 2015). Subsequent extended survey and analysis has since found these koalas to be infected with a C. pecorum strain more genetically similar to known livestock strains compared to koalas strains, now designated ompA genotype N (Legione et al.2016a,b). As island koala populations continue to be examined in greater depth and detail, hope fades that any wild koala population will continue to test C. pecorum-free.
Across mainland Australia, where more than five koalas from an area have been tested, C. pecorum infection rates have ranged from 21%–88% (Fig. 5, Table 1). This range is seen not only across the country, but also within regions where multiple studies have been conducted over different years (southeast QLD: 31%–71%; Raymond Island, VIC: 33%–81%; Gippsland, VIC: 25%–59%; Mount Lofty, SA: 47%–88%) (Fig. 5). This suggests that C. pecorum infection rates are dynamic within a koala population, most likely influenced by a range of host and environmental pressures. As noted in the C. pecorum ompA genotyping discussion, there is a trend for some C. pecorum ompA genotypes, such as genotypes B, C, L and M, to be found only in southern (VIC and SA) koalas while other strains, like genotypes A, E’ and F’ are present only in northern (QLD and NSW) koalas (Fig. 5, Table 1). However, there does not appear to be any pattern between ompA genetic differences and C. pecorum infection rates in koala populations at the locations surveyed to date.
Studies have also focused on documenting clinical chlamydial disease in koala populations across Australia. These studies generally reflect C. pecorum infection prevalence, with chlamydial disease prevalence ranging from 4%–71% across the country (Fig. 6 and Table 2). Included in these data are three surveys of koala hospital records: (i) Moggill Koala Hospital (in Brisbane, QLD) reporting a chlamydial disease prevalence of 52% (n = 20 250) of admitted koalas between 1997 and 2013 (Gonzalez-Astudillo et al. 2017), (ii) Moggill Koala Hospital, Currumbin Wildlife Sanctuary Hospital and Australia Zoo Wildlife Hospital (all southeast QLD) reporting a combined chlamydial disease prevalence of 59% (n = 519) in necropsied koalas between 2013 and 2016 (Gonzalez-Astudillo et al. 2019) and (iii) Port Macquarie Koala Hospital (Port Macquarie, NSW) reporting a chlamydial disease prevalence of 20% (n = 3781) of admitted koalas between 1975 and 2004 (Griffith et al. 2013). While koala hospital surveys offer useful insights into disease prevalence in an area over time, it should be remembered these types of surveys are inherently biased towards animals that show overt signs of distress and are brought into care. Less externally apparent signs of chlamydial disease may not be recognized in the community, leading to fewer of these koalas being brought into hospital care and recorded in prevalence measures. Attempting to address this bias, one study that focused on characterizing chlamydial disease signs not readily observable found 75% (n = 62) of koalas examined from Port Macquarie Koala Hospital without “wet bottom” or conjunctivitis had lesions attributable to chlamydiosis only observable by ultrasonography and/or at necropsy (Marschner et al. 2014). Another study found that 51% (n = 65) of koalas from the Mount Lofty Ranges and Eyre Peninsula, SA had chlamydial lesions that were only detectable microscopically or by histopathology during necropsy (Speight et al. 2016). These studies, as well as others (Patterson et al. 2015; Nyari et al. 2017), highlight that only a limited spectrum of chlamydial disease clinical signs can be easily visualized without specialist equipment or internal examination and suggest that the full impact and burden of chlamydial disease may be underrepresented when only classical visual assessment methods are used.
Finally, the situation where koalas present with the classic signs of chlamydial disease, but with no detectable C. pecorum, should also be acknowledged. Cross-sectional studies that have evaluated both chlamydial infection and disease in the same animal routinely report individuals with disease signs clinically attributable to chlamydial disease but without C. pecorum being detected by molecular testing of samples (Patterson et al. 2015; Nyari et al. 2017; Quigley et al. 2018a). The definitive reason for these discrepancies is currently unknown, but theories to account for these differences include an alternative pathogen(s) in koalas causing the same clinical signs (Patterson et al. 2015), variation in organism shedding during the course of disease or the resolution of the C. pecorum infection before/without the resolution of clinical signs (Nyari et al. 2017).
Chlamydial disease and host response
Chlamydiosis is a well-characterized disease in koalas. In the last 10 years, two areas of research have made notable advancements in our understanding of chlamydial disease in koalas: disease progression over time and the koala immune response to infection. Based on the recognition that koala chlamydiosis research was lacking population-level, long-term disease studies (Grogan et al. 2017; McCallum et al. 2018), researchers followed koala populations over several years to investigate more complex questions related to disease progression. In addition, the release of the first complete koala genome (Johnson et al. 2018), as well as specialized koala transcriptome datasets (Hobbs et al. 2014; Morris et al. 2014; Abts, Ivy and DeWoody 2015; Morris et al. 2016), have allowed for koala-specific immune targets to be specifically investigated. Together, these advances have provided the foundation to delve more deeply into chlamydial disease progression in koalas.
Unlike C. trachomatis infections in people, which are most often asymptomatic (Ziklo et al. 2016a), longitudinal monitoring of a wild koala population in southeast QLD found that 66% (n = 38) of koalas with C. pecorum infections progressed to clinical disease (Robbins et al. 2019). This observation addressed a long-standing question from cross-sectional studies where koalas are routinely found to be C. pecorum positive by PCR but with no apparent clinical signs (Nyari et al. 2017; Quigley et al. 2018a) – will these koalas resolve the asymptomatic infection without intervention or will they progress to disease? This longitudinal study suggests that 2/3 of koalas with progress to disease while 1/3 with recover without intervention (Robbins et al. 2019). Disease progression at the urogenital site has been found to be associated with urogenital C. pecorum load (Robbins et al. 2019). Urogenital C. pecorum load was also found to be an important factor in disease progression in several modeling studies (Quigley et al. 2018b). Additionally, antibiotic treatment of chlamydial disease was shown to provide only short-term protection, with treated koalas acquiring new C. pecorum infections and disease signs within six months of previous disease resolution (Robbins et al. 2019). Taken together, these longitudinal data indicate that C. pecorum infection appears to progress to disease regularly and recovery from disease after treatment seems to offer the koala little protection against future infection and disease.
Cytokines play a critical role in the immune system, regulating and directing the immune response to invading pathogens. Assays targeting koala cytokines, including the pro-inflammatory cytokine Tumour Necrosis Factor alpha (TNF-α), pro-inflammatory Th1 response cytokine IFN-ɣ, pro-inflammatory Th17 response cytokine Interleukin 17A (IL-17A), Th2 response cytokine Interleukin 4 (IL-4) and the anti-inflammatory Th2 response cytokine Interleukin 10 (IL-10), have all been developed (Mathew et al. 2013a,b; Maher et al. 2014; Mathew et al. 2014). In addition, assays for koala Interleukin 6 (IL-6), known to be expressed by epithelial cells during chlamydial infection (Cunningham et al. 2013), as well as koala-specific CD4 and CD8β markers, have also been developed (Maher et al. 2014). Comparing peripheral blood mononuclear cells (PBMCs) from koalas with current chlamydial disease, asymptomatic chlamydial infection and no chlamydial infection/disease determined that koalas with active disease had significantly higher expression of TNF-α, INF-ɣ and IL-10 (Mathew et al. 2013a,b). These results were further supported by a subsequent study that detected significantly higher expression of IL-17A and modest increases in expression of TNF-α and IFN-ɣ in currently chlamydial diseased koalas (Mathew et al. 2014). However, cytokine involvement in the immune response to C. pecorum, particularly in relation to the IFN-γ, is still an unclear area in koala chlamydial disease research. Investigation of IFN-γ during chlamydial disease has expanded to include both targeted gene studies (Mathew et al. 2013b) and total RNA expression within the cell (Phillips et al. 2019), with the importance of IFN-γ appearing different between the different study methods. Given the complexity of intracellular pathways and networks, it is not surprising that discrepancies will arise between studies. Future research in this area will be necessary to clarify the roles of all important cytokines, not just IFN-γ, for the safe implementation of treatment options, like vaccination, to koalas.
Chlamydia pecorum transmission
Chlamydia are well-established as sexually transmitted pathogens and sexual contact appears to be the primary transmission route of C. pecorum between koalas (Polkinghorne, Hanger and Timms 2013). However, investigations have expanded our awareness of non-sexual C. pecorum transmission, primarily between mothers (dams) and offspring (joeys). Two recent studies focused on joeys that were either still dependent (with their dams, less than one year old) or still sexually immature (between 9 and 13 months old) and both studies found a 27% C. pecorum positivity in the joeys ((n = 15) (Nyari et al. 2017); (n = 11) (Russell et al. 2018)). The dependent joey study (Russell et al. 2018) was based on koalas in care, so while dam-joey transmission was suspected as the primary route of C. pecorum transmission to the joeys, handling of dams and joeys by the same animal handler could not be completely ruled out. Alternatively, the sexually immature joey study (Nyari et al. 2017) was conducted from a monitored wild koala population where routes of C. pecorum transmission to joeys other than dam-to-joey were unlikely. These studies reveal that koalas less than one year old are acquiring chlamydial infections through non-sexual transmission routes and future management strategies will need to take this into account.
Another C. pecorum transmission concern is when koalas are translocated between different geographical sites with different chlamydial characteristics (Waugh et al.2016a). A translocation study was carried out in VIC where 30 C. pecorum negative (also Chlamydia-antibody negative) koalas were moved from French Island, VIC (where C. pecorum had not been detected up to this point) to forests near Ballarat, VIC (on the mainland) (Santamaria and Schlagloth 2016). After the first breeding season on the mainland (six months post-translocation), 25 of the translocated koalas were examined to find 48% (12/25) were now Chlamydia-antibody positive, with no detectable C. pecorum infections and six live joeys in the group. After the second breeding season (19 months post-translocation), 17 of the translocated koalas were examined to find 94% (16/17) were now Chlamydia-antibody positive, with 56% (9/16 (one koala was not swabbed for culture)) now having a chlamydial infection (seven with C. pecorum, one with both C. pecorum and C. pneumoniae and one with C. pneumoniae) and only one live joey in the group (Santamaria and Schlagloth 2016). The study concluded that moving koalas without carefully determining the chlamydial disease status of both the translocating koalas and the translocation site should be regarded as detrimental to the animals and translocation should only be undertaken with the greatest possible care and monitoring (Santamaria and Schlagloth 2016).
Detection of C. pecorum
As our understanding of both chlamydial infection and disease has progressed since 2012, so has the development of tools to detect C. pecorum and chlamydial disease in koalas. Progress has been made in understanding which samples should be tested and how both molecular and non-molecular techniques can contribute to pathogen and disease detection. Recognizing that non-invasive koala sampling, such as scat detection and testing, can be a low cost, non-disruptive survey method, effort has been put into testing whether C. pecorum detection in scat is comparable to direct urogenital swab sampling by quantitative PCR (qPCR) (Wedrowicz et al. 2016). In a small study, testing found a high level of concordance (83%; 5/6) between paired scat and urogenital swab samples, although the same C. pecorum ompA genotype was not always found between samples from the same koala (Wedrowicz et al. 2016). Perhaps unsurprisingly, C. pecorum copy numbers were consistently higher from urogenital swab samples compared to paired scat samples (Wedrowicz et al. 2016). Overall, this data suggests that non-invasive koala sampling has lower C. pecorum sensitivity compared to samples taken directly from a koala. This difference should be kept in mind when comparing prevalence estimates generated from different sampling methods.
Molecular tests for C. pecorum are the gold standard for detection, with established assays targeting the C. pecorum 16S rRNA gene (Marsh et al. 2011; Wan et al. 2011), ompA gene (Marsh et al. 2011; Higgins et al. 2012) and the CpecG_0573 (hypothetical protein (HP)) gene (Fabijan et al. 2019b; Robbins et al. 2019). Advances to these molecular tests have come in the forms of multiplexing and rapid point-of-care (POC) adaptations. Increasing the throughput of testing with multiplexing, real-time PCR assays have been developed to detect either the combination of the koala beta-actin gene and genus level Chlamydia, Mycoplasma and Ureaplasma DNA or species-specific C. pecorum, C. pneumoniae and Bordetella bronchiseptica (Hulse et al. 2018). Further accelerating detection from qPCR to POC diagnostics, two loop‐mediated isothermal amplification (LAMP) assays have recently been developed for C. pecorum detection. The first LAMP assay, released in 2017, targets a hypothetical protein gene, CpecG_0573, determined to be unique to C. pecorum based on genomic analysis (Jelocnik et al. 2017), while the second assay, released in 2019, targets the mreC gene (coding for a cell shape determining protein) from C. pecorum (Hulse et al. 2019a). Both assays report no cross-reactivity with a range of non-target organisms and rapid sample preparation protocols for minimal sample preparation, making them both candidates for POC diagnostics in the field or veterinary clinic (Jelocnik et al. 2017; Hulse et al.2019a).
Non-molecular tests have also been evaluated in recent years for C. pecorum infection or disease detection. For a time, an enzyme immunoassay, Clearview, was available for C. pecorum POC testing. Comparison to qPCR found that Clearview was 93% specific for C. pecorum, but only 43% sensitive (needing +400 copies of C. pecorum genomic DNA per test for detection) (Hanger et al. 2013). However, since this evaluation, the Clearview test is no longer available commercially. Detecting chlamydial disease, either in cases where overt clinical signs are absent or from koala samples like scat, has been another area that has received attention in recent years. A review was conducted at the Port Macquarie Koala Hospital to determine if ultrasonography was an accurate method to detect urogenital tract structural disease in koalas (Marschner et al. 2014). By comparing ultrasonography observations to paired necropsy results, the study found strong positive agreement in results from 86% of kidneys, 90% of bladders and 93% of ovarian bursal cysts, indicating ultrasonography was an effective diagnostic tool for assessing structural damage caused by chlamydial disease in koalas (Marschner et al. 2014). Another study sought to determine if koala detection dogs could recognize koala scats from actively diseased koalas compared to healthy koalas (Cristescu et al. 2019). While sample sizes were limited, the detection dogs could distinguish scat from clinically diseased koalas (n = 5) verses koalas that showed no clinical signs (n = 13), most likely recognizing a volatile organic compound difference in the scats (Cristescu et al. 2019). These alternative infection and disease determining techniques continue to broaden the toolkit available to koala disease researchers for better diagnosis and monitoring of chlamydial infection and disease in koala populations.
Treatments for C. pecorum – antibiotics and vaccines
Because the koala gastrointestinal tract is full of specialized bacteria needed for the digestion of eucalyptus leaves, koalas are especially sensitive to caeco-colic dysbiosis/typhlocolitis syndrome from antibiotic-induced microbial dysbiosis (Gillett and Hanger 2019). This disruption to the koala gut microbiome can be fatal, making antibiotic use to treat chlamydial disease in koalas a delicate balancing act. Traditional C. pecorum antibiotic treatments include the use of chloramphenicol and enrofloxacin, with enrofloxacin treatment failure a known issue (Polkinghorne, Hanger and Timms 2013). More recent research into the minimum inhibitory concentration (MIC) needed to control C. pecorum isolates from koalas revealed that the dosage of enrofloxacin needed to kill C. pecorum was above the conventional dose rate, possibly explaining these previous treatment failures (Black, Higgins and Govendir 2015). This has made chloramphenicol the treatment of choice for chlamydial disease in koalas. Chloramphenicol has traditionally been recommended at a dosage of 60 mg/kg for 45 days and even at this dosage, severe urogenital disease did not respond well (Govendir et al. 2012). Recent re-evaluation of this treatment regimen found that after koalas with a poor prognosis were removed from treatment on animal welfare grounds, a 60 mg/kg dosage of chloramphenicol for only 14 to 28 days had a successful treatment rate of 95% (Robbins et al. 2018). However, despite the success of chloramphenicol for koala chlamydial disease treatment, significant side effects, including bone marrow depression and fatal caeco-colic dysbiosis, are still seen with prolonged use (Govendir et al. 2012). Additionally, the supply of chloramphenicol base (the most effective form of the antibiotic (Black et al. 2013)) is not secure, with a commercial suspension of this antibiotic's production in 2013–2014. This shortage lead to investigations into alternative antibiotics suitable for koala use.
Alternative antibiotics such as doxycycline, florfenicol and penicillin G have all been considered for chlamydial disease treatment in koalas. Doxycycline given at 5 mg/kg diluted 50:50 in sterile saline once a week for four weeks has been found to reverse the signs of clinical cystitis, eliminate “wet bottom” and clear C. pecorum infection in a small group of koalas (n = 3) (Phillips et al. 2019). Florfenicol treatment at dosages tolerable in the field had limited success, with only 26% (n = 5) of treated koalas resolving their clinical signs and being released without further treatment, with another 32% (n = 6) requiring additional treatment with chloramphenicol to resolve their disease signs and the remaining animals (36%, n = 7) failing to clinically improve (Budd et al. 2017). Finally, penicillin G did not make it out of in vitro testing, where cell culture testing found this antibiotic induced a chlamydial stress response (into persistence) in C. pecorum and was not bactericidal (Leonard, Dewez and Borel 2016). Given these results, there is still a great need for alternatives to antibiotic treatment for chlamydial disease management in koalas.
The quest for a koala chlamydial vaccine has recently been reviewed (Phillips, Quigley and Timms 2019). Modelling has been done to predict the effect a chlamydial vaccine could have on declining koala populations in southeast QLD (an area endemic with Chlamydia) (Craig et al. 2014). With other koala threats remaining constant, modeling predicted that a vaccine with 75% protective efficacy, covering around 10% of the koala population each year and targeting young female koalas could reverse current population declines in five to six years (Craig et al. 2014). Several chlamydial vaccine formulations have been tested, including different adjuvants and C. pecorum target proteins, to determine which combinations produced the strongest humoral and cellular immune responses and offered the best disease protection (Kollipara et al. 2012; Kollipara et al.2013a,b; Khan et al. 2014; Waugh et al. 2015; Khan et al. 2016a,b; Waugh et al. 2016c,d; Desclozeaux et al. 2017; Nyari et al. 2018, 2019). Presently, a recombinant MOMP C. pecorum vaccine has shown the most promising protection overall, as well as having the therapeutic potential to replace antibiotics for mild ocular disease (Desclozeaux et al. 2017; Lizárraga, Carver and Timms 2019; Nyari et al. 2019). Further efforts to refine the recombinant MOMP vaccine into a peptide-based vaccine have shown promising results (Nyari et al. 2018), but more work on this formulation is still needed. In addition, long-term monitoring of vaccinated and control koalas (n = 106) has revealed that chlamydial vaccination also had positive effects on koala lifespan, adding a median of 3.5 years to vaccinated koala's lives (lifespan of 12.25 years for vaccinated koalas verses 8.8 years for unvaccinated koalas) (Hernandez-Sanchez et al. 2015). It was believed that, in additional to avoiding chronic illness, this generalized longer life may have been due to cross-reactive adaptive immune responses (heterologous immunity) together with epigenetic reprogramming of the innate immune system (trained innate immunity) (Hernandez-Sanchez et al. 2015). Finally, the benefits of chlamydial vaccination have not only been realized in the individual animals vaccinated, but also potentially in their future offspring. Preliminary evidence from five dam-joey pairs found that the joeys of vaccinated dams (n = 3) remained chlamydial infection and disease free as they became independent from their mothers while joeys from unvaccinated dams (n = 2) developed either chlamydial infection or both infection and disease as they became independent (Russell et al. 2018). Although passive immunity from mother to offspring is a well-recognized immunological process, further research into its place in koala chlamydial vaccination is needed before conclusive statements can be made. However, collectively, the evidence that chlamydial vaccination has a positive impact on koalas is continuing to grow with each study.
KOALA RETROVIRUS (KoRV)
As early as 1988, researchers noted by electron microscopy that koalas with leukemia had detectable Gammaretrovirus-like (type C retrovirus) particles budding from their cancerous cells (Canfield, Sabine and Love 1988). However, it took until 2000 for a retrovirus in koalas to be fully recognized, sequenced and named koala retrovirus (KoRV) (Fig. 7) (Hanger et al. 2000). At this early stage, it was recognized that KoRV shared remarkable homology with gibbon ape leukemia virus (GALV) and had a proviral integration pattern consistent with an endogenous retrovirus (a virus that has incorporated into germline cells and is transmitted from parent to offspring in the chromosomal DNA) (Hanger et al. 2000). Continued investigation confirmed that KoRV was indeed endogenously incorporated into the koala genome in northern koalas (from QLD and NSW), while still believed to be exogenous (transmitted horizontally between koalas through infection) in the south (VIC and SA) (Tarlinton et al. 2005; Tarlinton, Meers and Young 2006). In addition, KoRV was found to produce intact viral particles in infected animals, indicating that the virus appeared to be both endogenous and transmissible (Tarlinton et al. 2005; Tarlinton, Meers and Young 2006). Importantly for koala health, it was also determined that increased plasma levels of KoRV RNA could be associated with koalas who developed leukemia and lymphoma, as well as clinical chlamydiosis (Tarlinton et al. 2005). By 2012, KoRV was clearly recognized as a pathogen of koalas. However, focused study of this virus since 2012 has revealed a complex diversity and epidemiology. As our understanding of KoRV has evolved over the last eight years, several reviews have tried to capture the current state of knowledge about this virus (Tarlinton 2012; Denner and Young 2013; Xu and Eiden 2015; Denner 2016; Kinney and Pye 2016; Greenwood et al. 2018; Higgins and Maher 2019). However, research into KoRV has progressed as fast as reviews could be written, with new discoveries routinely being made in understanding where KoRV came from, what it is doing and how we may be able to intervene to help the koala.
Figure 7.
Schematic of koala retrovirus (KoRV) with discussed features highlighted. LTR - Long terminal repeats, gag - the group-specific antigen gene, pol - protease-polymerase gene, env - envelope gene, VRA – variable region A (receptor binding domain), FPPR – fusion peptide-proximal region, IS/CKS-17 – immunosuppressive domain, MPER – membrane-proximal external region. Arrows indicate PCR primer binding sites for common assays.
Origin of KoRV in koalas
KoRV was first recognized as a close relative of GALV. Given that GALV appeared as a pathogenic exogenous virus in a captive gibbon research facility in Thailand and has never been detected in gibbons other than within this one spill-over event (Brown and Tarlinton 2017) and koalas are native to Australia, there has been an active interest in identifying the reservoir host of the precursor Gammaretrovirus that became KoRV. The first reasonable candidate emerged from the native Australian rodent, the grassland melomys, Melomys burtoni. A screen of 42 vertebrate species in Australia (including rodents and bats) identified a novel Gammaretrovirus with 83% identity to KoRV and 93% identity to GALV in M. burtoni, leading to this virus being designated Melomys burtoni retrovirus (MbRV) (Simmons et al. 2014). Despite the overlap in geographical ranges between melomys and koalas, MbRV appeared to be a defective endogenous retrovirus, not producing any detectable viral RNA or virus particles, reducing its likelihood of being the source of KoRV (Simmons et al. 2014). An extended Gammaretrovirus search in Southeast Asia detected another virus in M. burtoni from Indonesia, distinct from MbRV, which was named Melomys woolly monkey virus (MelWMV) (Alfano et al. 2016). However, MelWMV also appeared to be a defective endogenous retrovirus (missing major gene regions), so again, not a likely source for GALV and KoRV (Alfano et al. 2016). Most recently, gammaretroviruses have been identified from black flying-foxes (Pteropus alecto) from QLD and were designated flying-fox retrovirus (FFRV) (McMichael et al. 2019) and Hervey pteropid gammaretrovirus (HPG) (Hayward et al. 2020). Analysis of the full-length, intact FFRV and HPG sequences found that these gammaretroviruses are distinct from other known bat viruses and instead phylogenetically grouped quite closely with KoRV and GALV (McMichael et al. 2019; Hayward et al. 2020). Bats have been hypothesized as sources of novel retroviruses before, with a defective retrovirus of some similarity to KoRV having been detected in the microbat Megaderma lyra (Megaderma lyra retrovirus, MIRV) in China previously (Cui et al. 2012). Bats are well-known for their ability to travel long distances and the black flying-fox has a known geographic range that includes Australia, Papua New Guinea and Indonesia (Roberts et al. 2017). So, as the search for the reservoir host of the Gammaretrovirus that spilled over into koalas to become KoRV continues, this research continues to expand our knowledge of rodent and bat gammaretroviruses and the possible interactions between these animals and koalas.
Advances in the genetic understanding of KoRV
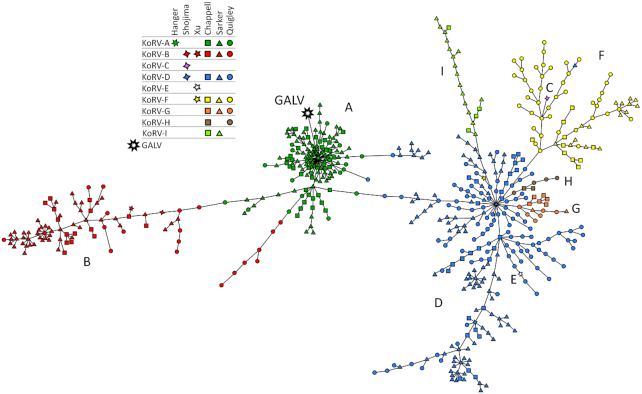
Some of the most exciting and technically advanced work that has been done with KoRV in recent years relates to our understanding of this virus on a genetic level. In 2012, KoRV was considered a single virus while, by 2020, there are now nine subtypes recognized (Fig. 8). Beyond investigating the diversity of this virus, work has also focused on understanding basic viral properties, understanding viral integration into the koala genome, expanding our knowledge of defective KoRV variations, and learning what other endogenous retroviruses (ERVs) in the koala that KoRV may interact with. Collectively, this body of work has not only advanced KoRV understanding but has added important information to our knowledge of retrovirology in general.
Figure 8.
KoRV diversity detected from koalas, visualized by minimum spanning tree (PHYLOViZ, http://online.phyloviz.net/index). Reference strains including Hanger et al. 2000 (original KoRV/KoRV-A), Shojima et al. 2013 (KoRV-B/J = OJ-4, 11–4; KoRV-C = OJ-4, 11–2; KoRV-D = OJ-4, 11-1), Xu et al. 2013 (KoRV-B) and Xu et al. 2015 (KoRV-E and -F), Chappell et al. 2017; Sarker et al. 2019 and Quigley 2020 (accession numbers MN931399-MN931590).
Gammaretroviruses are categorized based on the cellular receptors they use for infection. The original KoRV isolate (now known as KoRV-A) was shown to use the same receptor for cell entry as GALV – the sodium-dependent phosphate transporter membrane protein PiT1 (Oliveira, Farrell and Eiden 2006). Next, within months of each other in 2013, two reports were published on a new KoRV subtype that both used a different cell receptor, the thiamine transporter 1 (THTR1), to enter cells (Shojima et al. 2013b; Xu et al. 2013). The first report came from captive koalas at Los Angeles Zoo suffering from lymphoma and this KoRV strain was designated KoRV-B (Xu et al. 2013). The second report came from QLD koalas reared at the Kobe Municipal Oji Zoo in Hyogo, Japan and the KoRV strain was designated KoRV-J (Shojima et al.2013b). Very quickly, phylogenetic analysis determined that KoRV-B and KoRV-J represented the same subtype and were grouped together with the designation KoRV-B (Fig. 8) (Shimode et al. 2014). Furthermore, phylogenetic analysis indicated that additional KoRV isolates from the Kobe Zoo study contained enough differences in their putative receptor binding domains (within the variable region A [VRA] of their envelope genes) that they should be designated as novel subtypes – KoRV-C and KoRV-D (Fig. 8) (Shimode et al. 2014). In the same year, KoRV-B was also detected for the first time in a wild Australian koala (a female from the Port Macquarie Koala Hospital (NSW)) (Hobbs et al. 2014). Additional studies followed from koalas at the San Diego and Los Angeles Zoos that isolated unique KoRV variants that also used different cell receptors from PiT1 and THTR1 and these KoRV subtypes were designated KoRV-E and KoRV-F (Fig. 8) (Xu et al. 2015). Finally, a survey study of wild southeast QLD koalas targeting the VRA region of the KoRV envelope gene found additional receptor binding domain diversity and established the subtypes KoRV-G, KoRV-H and KoRV-I (Fig. 8) (Chappell et al. 2017).
Based on the phylogenetic diversity of KoRV envelope VRA regions, KoRV currently segregates into 3 major clades representing KoRV-A (PiT1 receptor), KoRV-B (THTR1 receptor) and KoRV-C-I (unknown receptor(s)), with these groupings robustly reproduced in larger survey studies that have encompassed wild koalas from across Australia (QLD and SA – (Sarker et al. 2019); QLD, NSW and VIC – Quigley (accession numbers MN931399-MN931590) ) (Fig. 8). As more KoRV sequences have been characterized, it has become clear that the original KoRV-C sequence (Shojima et al. 2013b) now belongs to the characterized KoRV-D or -F subtype and the original KoRV-E and KoRV-F sequences (Xu et al. 2015) now belong to the large and diverse KoRV-D subtype (Fig. 8). In fact, given the diversity of the VRA region within the KoRV-D subtype and the uncertainty of KoRV-C to KoRV-I cell receptor use, support for the use of all the subtype designations (especially KoRV-G and KoRV-H) is variable (Fig. 8) (Sarker et al. 2019). Regardless, despite nine subtypes of KoRV officially being described, there are generally three (KoRV-A, -B, -D) to seven (KoRV-A, -B, -D, -F, -G, -H, -I) subtypes routinely reported (Chappell et al. 2017; Quigley et al. 2019; Sarker et al. 2019) (Table 3A).
Table 3A.
KoRV infection prevalence reported between 2012 and 2019 in wild and captive koalas.
| KoRV subtype | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Origin | Region | #koalas | Total pol gene | A | B | D | F | G | H | I | Reference |
| QLD | northern (1891-1980s) | 16 | 15 (94%) | Avila-Arcos et al. 2013 | |||||||
| Blair Athol | 27 | 27 (100%) | Simmons et al. 2012 | ||||||||
| SEQLD | 250 | 250 (100%) | Simmons et al. 2012 | ||||||||
| SEQLD | 12 | 12 (100%) | Wedrowicz et al. 2018 | ||||||||
| SEQLD | 290 | 290 (100%) | 83 (29%) | Quigley et al. 2018 | |||||||
| SEQLD | 16 | 16 (100%) | 4 (25%) | 14 (88%) | 4 (25%) | – | – | – | Quigley et al. 2019 | ||
| SEQLD | 33 | 33 (100%) | 33 (100%) | 33 (100%) | – | 11 (33%) | – | 32 (97%) | Sarker et al. 2019 | ||
| SEQLD | 18 | 18 (100%) | 14 (78%) | 17 (94%) | 8 (44%) | 2 (11%) | 1 (6%) | 1 (6%) | Chappell et al. 2017 | ||
| NSW | NENSW | 12 | 12 (100%) | Wedrowicz et al. 2018 | |||||||
| Pilliga | 57 | 57 (100%) | Simmons et al. 2012 | ||||||||
| Port Macquarie | 15 | 15 (100%) | Wedrowicz et al. 2016 | ||||||||
| Port Macquarie | 43 | 43 (100%) | Simmons et al. 2012 | ||||||||
| VIC | Southern (1891-1980) | 3 | 1 (33%) | Avila-Arcos et al. 2013 | |||||||
| Mallacoota | 3 | 0 (0%) | Wedrowicz et al. 2018 | ||||||||
| Raymond Island | 136 | 38 (28%) | Legione et al. 2017 | ||||||||
| Raymond Island | 29 | 10 (35%) | Simmons et al. 2012 | ||||||||
| Raymond Island | 18 | 4 (22%) | Wedrowicz et al. 2018 | ||||||||
| Greater Gippsland | 33 | 6 (18%) | Legione et al. 2017 | ||||||||
| Gippsland | 20 | 11 (55%) | Simmons et al. 2012 | ||||||||
| Strezlecki Ranges | 26 | 18 (69%) | Simmons et al. 2012 | ||||||||
| South Gippsland | 203 | 64 (32%) | Wedrowicz et al. 2018 | ||||||||
| South Gippsland/ Raymond Island | 19 | 9 (47%) | Wedrowicz et al. 2016 | ||||||||
| Central Gippsland | 17 | 13 (76%) | Wedrowicz et al. 2018 | ||||||||
| Snake Island | 12 | 6 (50%) | Simmons et al. 2012 | ||||||||
| French Island | 94 | 23 (24%) | Legione et al. 2017 | ||||||||
| French Island | 28 | 6 (21%) | Simmons et al. 2012 | ||||||||
| Phillip Island | 6 | 0 (0%) | Wedrowicz et al. 2016 | ||||||||
| Phillip Island | 11 | 0 (0%) | Simmons et al. 2012 | ||||||||
| Mornington Peninsula | 15 | 4 (27%) | Legione et al. 2017 | ||||||||
| South West Coast | 178 | 30 (17%) | Legione et al. 2017 | ||||||||
| Cape Otway | 11 | 2 (18%) | Wedrowicz et al. 2018 | ||||||||
| Far West | 167 | 52 (31%) | Legione et al. 2017 | ||||||||
| Far north Vic | 15 | 6 (40%) | Legione et al. 2017 | ||||||||
| General mainland | 43 | 36 (82%) | Simmons et al. 2012 | ||||||||
| SA | Kangaroo Island | 170 | 72 (42%) | 0 (0%) | Fabijan et al. 2019a | ||||||
| Kangaroo Island | 162 | 24 (15%) | Simmons et al. 2012 | ||||||||
| Mt Lofty | 28 | 28 (100%) | 28 (100%) | 28 (100%) | – | 12 (43%) | – | 18 (64%) | Sarker et al. 2019 | ||
| Mt Lofty | 75 | 49 (65%) | 0 (0%) | Fabijan et al. 2019a | |||||||
| USA | Los Angeles Zoo | 13 | 13(100%) | 6(46%) | Xu et al. 2013 | ||||||
| Japan | 9 unnamed zoos: QLD/NSW koalas | 40 | 40 (100%) | 27 (68%) | Shojima et al. 2013b | ||||||
| 9 unnamed zoos: VIC koalas | 11 | 4 (36%) | 0 (0%) | Shojima et al. 2013b | |||||||
| Kobe Oji Zoo, Saitama Children's Zoo, Hirakawa Zoological Park | 20 | 20 (100%) | 12 (60%) | Hashem et al. 2019; Kayesh et al. 2019 | |||||||
| Germany | Duisburg Zoo | 5 | 5 (100%) | 2 (40%) | Fiebig, Keller and Denner 2016 | ||||||
| Belgium | Antwerp Zoo | 1 | 1 (100%) | 1 (100%) | Fiebig, Keller and Denner 2016 | ||||||
Notes: Avila-Arcoset al.2013 data is from museum koala skins; Legione et al.2017total KoRV data was further tested to confirm 88% of cases were positive for KoRV-A while no KoRV-B was detected. Dashes indicate the subtype was not detected, blank spaces indicate the study did not test for the indicated target.
The biology of a typical Gammaretrovirus is well understood by virologists (Maclachlan and Dubovi 2010). Like all gammaretroviruses, KoRV is composed of a simple genome (∼8.5 kb long) with long terminal repeats (LTRs) at each end of a linear single-stranded RNA genome containing three genes; the group-specific antigen (gag) gene, the protease-polymerase (pro-pol or pol) gene and the envelope (env) gene (Fig. 6) (Maclachlan and Dubovi 2010; Denner and Young 2013). To understand if KoRV behaves the same way as other gammaretroviruses during infection, studies have investigated specific properties of KoRV genes and proteins.
The Gag protein, known to be important in virus budding from infected cells, was the first KoRV protein to be investigated. Using a constructed infectious clone of KoRV, researchers were able to show that, like other gammaretroviruses, Gag plays a critical role in KoRV budding by recruiting the endosomal sorting complexes required for transport (ESCRT) machinery through interaction with a specific L-domain region in KoRV Gag to allow virions to be released from infected cells (Shimode et al. 2013; Shojima et al.2013a). In addition, an alternative form of Gag, known as glycosylated Gag or glyco-gag, is known to be important for some gammaretroviruses to combat host restriction factors like APOBEC3 (Fig. 6). Interestingly, although the koala genome encodes for genes that appear to be APOBECs (XP_020850070.1, XP_020855279.1, XP_020849984.1 and XP_020819701.1), targeted investigation found no evidence that KoRV expresses glyco-gag (at least in human cell culture) and KoRV infectivity was restricted by both human APOBEC3G and mouse APOBEC3 (Nitta et al. 2015). In fact, the strong restriction of KoRV by human APOBEC3G, which is highly expressed in hematopoietic cells commonly targeted by gammaretroviruses, supports the assertion that there is a low likelihood of zoonotic transmission of KoRV to humans (Nitta et al. 2015). This assertion has so far held up, with no reports of veterinarians or koala animal carriers contracting a KoRV infection.
The other KoRV protein that has received targeted study has been the Env protein. Like other gammaretroviruses, KoRV is known to make two RNA transcripts from its genome; a full length genome transcript and an Env mRNA (Maclachlan and Dubovi 2010; Hobbs et al. 2014). Recent investigations have quantified the Env mRNA to be 5-fold more abundant than the unspliced genomic transcript (Yu et al. 2019). Within the Env protein, there are two regions; the surface (SU or gp70) protein followed by the transmembrane (TM or p15E) protein (Fig. 6). Within the TM protein, there are several epitopes known to be important for neutralizing antibody responses (the fusion peptide-proximal region (FPPR) and membrane-proximal external region (MPER)) (Fiebig et al. 2003), as well as a major immunosuppressive domain (IS or CKS-17 region) (Fig. 6) (Blinov et al. 2013). Studies of these important Env regions across the different KoRV subtypes and between northern and southern koalas have found striking conservation of these epitopes (Ishida et al.2015b; Olagoke et al. 2019). These data suggest that all KoRV subtypes may have similar immunosuppressive capability (at least based on CKS-17 interactions) and all subtypes may be targetable by the same neutralizing antibody responses. Together, this information provides a critical foundation for treatment options like vaccination (discussed below).
Another important area of KoRV biology that has advanced in recent years is our understanding of KoRV integration into the koala genome. Retroviruses in general must reverse transcribe and insert their genomes into the host genome (to become proviruses) in order to replicate and survive. Studies of koala genomes from the north have estimated KoRV proviral integration in the range of 133–165 copies per cell (Simmons et al. 2012; Hobbs et al. 2017; Johnson et al. 2018) while koala genomes from the south only appear to contain KoRV proviral integration at a rate of ∼1 copy/10 000 cells (Simmons et al. 2012). The vast majority of KoRV proviruses detected in individual koalas from the north appear to be endogenous in nature, with focused study and analysis of a northern koala family finding the proviral pattern detected in a joey could be accounted for by the proviral pattern of their parents (dam and sire) (Ishida et al.2015b). Additional studies, incorporating both modern wild koalas and museum koala samples dating back 130 years, have found that the sites of KoRV proviral integration were largely unique to each unrelated koala examined and were all KoRV-A in subtype (Tsangaras et al. 2014; Ishida et al. 2015a; Cui et al. 2016). This suggests that KoRV is in the early stages of retroviral invasion of the koala germline, with many unique insertion events proliferating at low frequency throughout the koala population (Ishida et al. 2015a). Finally, analysis of KoRV proviral LTR regions date initial KoRV endogenization as occurring no more than 22 200 – 49 900 years ago, although a much more recent time of integration is possible (Ishida et al. 2015a). This has created an exciting opportunity for virologists to study the process of viral endogenization in real time, as many other known mammalian ERVs integrated into their host's genome millions of years ago.
As with many viruses, variations of KoRV that appear to be defective in replication and/or transmission have also been detected (Xu et al. 2015; Hobbs et al. 2017; Sarker et al. 2020) (Quigley et al. 2020 under review). One study worth highlighting was a recent investigation of 97 southern Australian koalas that found KoRV proviral genes could be detected from 99% of koalas tested, but only 79% koalas had a complete detectable provirus (LTR, gag, pol, and env) and only 51% of koalas had detectable KoRV plasma RNA, suggesting that defective KoRV may be common in the south (Sarker et al. 2020). It should be noted that sequence missing from the pol gene of these defect proviruses is the same region targeted in common qPCR assay for KoRV (Tarlinton et al. 2005; Simmons et al. 2012) (Table 3B), raising questions about whether KoRV-negative results from southern koalas can really be taken as indicating KoRV-free koalas. Whether these defective KoRV variants have had any effect on the apparent lack of KoRV endogenization and prevalence in southern koalas remains an area of active investigation.
Table 3B.
Tests used to determine KoRV infection prevalence.
| Study reference | Gene target and KoRV subtype targeted (primers) for test | Reference for KoRV test |
|---|---|---|
| Avila-Arcos et al. 2013 | gag-pol and env genes, KoRV-A (multiple sets) | Avila-Arcos et al. 2013 |
| Simmons et al. 2012 | pol gene, All KoRV (F/R) external set and internal set | Tarlinton et al. 2005; Simmons et al. 2012 |
| Shojima et al.2013b | pol and env genes, KoRV-pol (F/R), KoRV-A (F/R), KoRV-J (F/R) | Shojima et al.2013b |
| Xu et al. 2013 | env gene, KoRV-A (P3/P7), KoRV-B (P1/P5, P2/P4) | Xu et al. 2013 |
| Fiebig, Keller and Denner 2016 | env gene, KoRV-A (F/R), KoRV-B (P2/P4) | Fiebig, Keller and Denner 2016; Xu et al. 2013 |
| Wedrowicz et al. 2016 | env gene, KoRV-A (P3/P7) | Xu et al. 2013 |
| Chappell et al. 2017 | env gene, All KoRV (env22.F/env514.R) | Chappell et al. 2017 |
| Legione et al. 2017 | pol and env genes, All KoRV (F/R), KoRV-A (P3/P7), KoRV-B (P1/P5) | Tarlinton et al. 2005; Xu et al. 2013 |
| Quigley et al. 2018 | env gene, KoRV-A (UF/A_R), KoRV-B (UF/B_R) | Waugh et al. 2017 |
| Wedrowicz et al. 2018 | env gene, KoRV-A (P3/P7) | Xu et al. 2013 |
| Fabijan et al. 2019a | pol gene, All KoRV (F/R) external set and internal set | Tarlinton et al. 2005; Simmons et al. 2012 |
| Hashem et al. 2019; Kayesh et al. 2019 | env gene, KoRV-A (UF/A_R), KoRV-B (UF/B_R) | Waugh et al. 2017 |
| Quigley et al. 2019 | env gene, All KoRV (env22.F/env514.R) | Chappell et al. 2017 |
| Sarker et al. 2019 | env gene, All KoRV (env22.F/env514.R) | Chappell et al. 2017 |
The last aspect of KoRV genetics that has been recently investigated has been its interaction with other ERVs in the koala genome. Studies to date have identified four ERVs in the koala genome, three of which appear to fall into established ERV families (ERV, ERVL and ERVK) and have been designated ERV.1, ERVL.1, ERVK.14 (Yu et al. 2019), and a fourth ERV designated Phascolarctos endogenous retroelement (PhER) (Hobbs et al. 2017). Detailed analysis has uncovered that KoRV and PhER have recombined at least 16 times to generate novel ERVs in the representative genome-sequenced koala, with the most prevalent version designated recKoRV1 (Hobbs et al. 2017; Lober et al. 2018). RecKoRV1 has been detected in koalas from across Australia, with a higher prevalence in northern koalas (Lober et al. 2018). Of special note, a unique case has been reported from two south Australian koalas, where recKoRV1 was detected in animals that had no detectable intact KoRV provirus (Lober et al. 2018). How this finding will fit with our current understanding of KoRV biology and detection is still an open question.
Prevalence of KoRV in koalas
As KoRV gained recognition as an important koala pathogen to investigate, surveys of koala populations from across Australia have included KoRV detection (Fig. 8, Table 3). Between 2012 and 2019, KoRV prevalence has been determined in 10 studies of Australian koalas and four studies of koalas in international zoos (Table 3A) (Simmons et al. 2012; Avila-Arcos et al. 2013; Shojima et al.2013b; Fiebig, Keller and Denner 2016; Wedrowicz et al. 2016; Chappell et al. 2017; Legione et al. 2017; Quigley et al.2018a; Wedrowicz et al. 2018; Fabijan et al. 2019a; Hashem et al. 2019; Kayesh et al. 2019; Quigley et al. 2019; Sarker et al. 2019). As with C. pecorum detection, KoRV detection has relied on a range of PCR assays (Table 3B).
Early studies, before the range of KoRV subtypes were recognized, focused on detecting KoRV using a general pol gene assay (now known to detect all the subtypes together) (Fig. 7). More recent studies have either combined KoRV pol gene pre-screening with specific KoRV-A and KoRV-B assays, have assayed directly for KoRV-A (and occasionally KoRV-B) or have surveyed for all KoRV subtypes simultaneously by deep amplicon sequencing (Figs 7 and 9, Table 3). These studies have found that northern koalas are always found to be infected with KoRV/KoRV-A (100% detectable infection) while southern koala infection rates range from 0% to 100%, depending on the area (Fig. 9, Table 3).
Figure 9.

Prevalence of koala retrovirus (KoRV) provirus infection reported in studies from 2012 to 2019. Grey ellipticals indicate the mapped area investigated, pie charts represent the KoRV positivity by subtype reported in ‘n’ number of koalas tested. Details for each study can be found in Table 3.
In northern koalas, beyond the endogenous 100% KoRV-A rate, believed exogenous subtype prevalence has been examined in southeast QLD. KoRV-B and KoRV-D appear to be the predominant non-KoRV-A subtypes, with KoRV-B detected in 25%-100% of koalas and KoRV-D detected in 88%-100% of koalas surveyed (Fig. 9, Table 3) (Chappell et al. 2017; Quigley et al. 2018a; Quigley et al. 2019; Sarker et al. 2019). Additional subtypes reported were not detected in all studies and included KoRV-F (25%-44% prevalence), KoRV-G (11%–33% prevalence), KoRV-H (6% prevalence) and KoRV-I (6%–97% prevalence) (Fig. 9, Table 3).
In the south, most studies have focused on total KoRV or KoRV-A detection alone. At one extreme, when more than five koalas were tested, Phillip Island was found to have a total KoRV or KoRV-A prevalence of 0% in two separate studies (Simmons et al. 2012; Wedrowicz et al. 2016). Although koala sampling numbers were not large (n = 6 and 11), this is the only island population of koalas where KoRV has not been detected. At the other extreme, a study of Mount Lofty, SA koalas used deep amplicon sequencing (a much more sensitive method) and found KoRV prevalence comparable to northern koalas, with all the koalas (n = 28) being KoRV-A, KoRV-B and KoRV-D positive (100%), KoRV-G with 43% prevalence and KoRV-I with 64% prevalence (Sarker et al. 2019). Through the rest of VIC and SA, the total KoRV or KoRV-A prevalence ranged from 17%–82% (Fig. 9, Table 3).
Finally, there have also been reports of KoRV prevalence from koalas in captive zoo populations internationally. Koalas in zoos appear to reflect the KoRV prevalence of their source region. Most international zoo koalas, including Los Angeles Zoo, USA, several Japanese zoos, Duisburg Zoo, Germany and Antwerp Zoo, Belgium, have KoRV prevalence patterns consistent with northern koalas (their koalas most likely origin), with 100% KoRV-A and 40%-68% KoRV-B (where there were at least five koalas) (Table 3) (Shojima et al. 2013b; Xu et al. 2013; Fiebig, Keller and Denner 2016; Hashem et al. 2019; Kayesh et al. 2019). Similarly, a collection of Japanese zoo koalas known to be from VIC had reported KoRV-A prevalence of only 36% and no KoRV-B detected, similar to current VIC wild koala numbers (Table 3) (Shojima et al. 2013b).
Disease associated with KoRV and host response
Retroviruses, including Gammaretrovirus members, are the causative agents of certain types of cancers and immunosuppressive or immune-mediated diseases (Maclachlan and Dubovi 2010). In koalas, KoRV was originally identified because of its apparent link to leukemia (Canfield, Sabine and Love 1988) and this association has been maintained over time (Tarlinton et al. 2005; Ito et al. 2019). Additional links to immunosuppression, seen in associations between chlamydial disease rates and KoRV rates in koalas, have also been reported (Legione et al. 2017; Waugh et al. 2017; Quigley et al. 2018a). While reports continue to emerge showing KoRV in association with cancers and immunosuppressive outcomes in koalas, the data to date are of a correlative nature, not yet showing clear causation. However, as studies progress, clues into possible disease mechanisms of KoRV in koalas continue to be suggested.
With the division of KoRV into subtypes, KoRV-B has emerged as the major subtype often associated with lymphomas, leukemias and neoplasms in koalas (Xu et al. 2013; Quigley et al. 2018a). Although it is not clear why only KoRV-B has been associated with cancer in koalas, two biological features may offer hypotheses worthy of future investigation. The first feature is the point of distinction between KoRV subtypes found in the LTR region. The U3 region within the upstream LTR in KoRV contains an enhancer region for viral transcription (Fig. 7). Within KoRV-A, a single copy of this enhancer region is typically found, while the original KoRV-B strain was found to contain four copies of this enhancer region and a KoRV-F strain (now recognized as KoRV-D) has been found with five copies of this enhancer region (Xu et al. 2013, 2015). This suggested that non-KoRV-A strains may have higher transcription rates, a finding that was confirmed by comparing the proviral and expression loads of multiple KoRV subtypes in koalas over time (Quigley et al. 2019). Because KoRV randomly inserts into the host genome to replicate, increased viral transcription may lead to increased non-specific host transcription downstream of insertion and increased potential for oncogenic outcomes. However, this hypothesis would suggest that all non-KoRV-A subtypes, not just KoRV-B, should display increased oncogenic potential. A second biological feature that may separate KoRV-B and KoRV-D clade subtypes may be in the frequency of their defective variants. Defective KoRV-B and KoRV-D strains have been characterized (Xu et al. 2015), with the single koala genome available found to contain intact KoRV-B provirus but only detective KoRV-D and KoRV-E proviruses (Hobbs et al. 2017). Transmission studies have found that KoRV-B was consistently transmitted from dam-to-joey in both captive and wild settings (Xu et al. 2013; Quigley et al. 2018a) while the original KoRV-E and KoRV-F strains (now known to both represent KoRV-D/F strains) were not transmitted uniformly from either the dam- or sire-to-joey in a captive setting (Xu et al. 2015). The difference between these transmission rates is currently unknown, but if it is found that defective provirus is more common in the KoRV-D subtype clade, that may limit these subtypes in their ability to transcribe infectious particles and also limit the oncogenic potential of their U3 enhancer regions compared to KoRV-B. However, despite these interesting observations and hypotheses, the definitive reason for the increased oncogenic association of only KoRV-B is still an open research question.
Immunosuppression is the suppression of the immune system and its ability to fight infections. In koalas, the most prevalent infection by which to gauge immunosuppression is chlamydial disease. In the north, where KoRV-A is endogenous, koalas with detectable KoRV-B provirus showed a significant association with having chlamydial disease (Waugh et al. 2017; Quigley et al. 2018a). Cytokine investigation in northern koalas found a significant upregulation of IL-17A in KoRV-B-infected koalas (Maher and Higgins 2016), which corresponds with IL-17A as an immune marker for chlamydial disease severity and pathogenesis in northern koalas (Mathew et al. 2014). In the south, where KoRV-A appears to be exogenous, koalas with detectable KoRV-A provirus showed a significant association with having chlamydial disease (Legione et al. 2017). Interestingly, cytokine investigation in southern koalas found a significant downregulation of IL-17A and IFN-ɣ in KoRV positive koalas (by total KoRV pol gene assay) (Maher et al. 2019). Whether the discrepancy between IL-17A gene expression levels between northern and southern koalas can be explained by KoRV endogenization status, KoRV subtype infection profiles or some other non-KoRV related factor is an area of future investigation. Finally, chlamydial disease is not the only infection that KoRV appears to affect, with southern koalas also more likely to present with periodontitis (inflammation of the gums caused by bacteria in the dental plaque) when KoRV positive (by total KoRV pol gene assay) (Butcher et al. 2020).
Finally, the effect of KoRV expression over time on koala health has been investigated. While it had been previously shown that increases in total KoRV RNA levels in plasma were associated with the development of neoplasia (Tarlinton et al. 2005), details of KoRV subtype expression have only recently been investigated. Following a group of female northern koalas for multiple samplings over several years, koalas that remained healthy for the study (n = 11) were found to have stable KoRV-D/KoRV-A expression ratios while koalas that developed chlamydial cystitis (n = 5) had variable KoRV expression profiles, dominated by KoRV-A or KoRV-B and lacking KoRV-D, prior to disease onset (Quigley et al. 2019). Collectively, research has shown that different subtypes of KoRV have different effects and that both provirus and expressed virus contribute to pathology in koalas.
Transmission of KoRV
Transmission of ERVs in the germline of the host genome, from parents to offspring, is a hallmark feature of endogenous retroviruses (Greenwood et al. 2018). As such, KoRV-A transmission in northern koalas follows the understood endogenous process. Transmission of the proposed exogenous variants (KoRV-B to -I in the north and KoRV-A to -I in the south) is an area of more uncertainty. As discussed in the disease section above, KoRV-B has been shown in both wild and captive koalas to be universally transmitted from dam-to-joey, with no apparent influence of the infection status of the sire (Xu et al. 2013; Quigley et al. 2018a), while KoRV-D strains do not show a predictable dam-to-joey or sire-to-joey transmission pattern (Xu et al. 2015). Dam-to-joey transmission is likely a significant transmission route for exogenous KoRV, as transcriptomic analysis of the koala mammary gland found KoRV transcripts to be 3% of all RNA in these cells (the fourth most abundant transcript detected) and proteomic analysis of koala milk found KoRV proteins (Gag, Pol and Env) to be ∼1% of all peptides in milk (collectively the fourteenth most abundant peptides) (Morris et al. 2016). Beyond mother to offspring transmission, the KoRV-B adult-to-adult contact transmission rate in a northern koala population with a general prevalence of 25% KoRV-B positivity was determined to be 3% new adult KoRV-B infections per year when 49 koalas were followed for three years by conventional proviral PCR (Quigley et al. 2018a).
Related to transmission has been the hypothesis that KoRV is currently spreading across Australia in a north to south direction. This hypothesis is based on the endogenous status of KoRV-A in the north (Tarlinton, Meers and Young 2006), similarity to other retroviruses that suggests KoRV may have entered the koala population from a spill-over event based in northern Australia (Greenwood et al. 2018) and prevalence surveys that show that KoRV has a 100% prevalence in the north, but not in the south (Tarlinton, Meers and Young 2006; Simmons et al. 2012). However, KoRV is clearly in southern Australia (Fig. 9), raising questions about why KoRV detection in the south is not higher. Whether the recent discoveries regarding the potential prevalence of defective KoRV strains, especially in southern koalas, can address this question, remains to be seen. However, with much still to learn about the receptors used by the KoRV-D subtype clade, there is still a lot to investigate regarding the transmission of KoRV in general and of different subtypes specifically.
Detection of KoRV
Before 2012, when KoRV was considered a single entity, detection of a conserved region of the pol gene by PCR assay was the established method of total KoRV detection (Fig. 7) (Tarlinton et al. 2005). Since the definition of KoRV subtypes, several KoRV subtype-specific PCR assays targeting the env gene have been developed. Specific subtype primer sets have utilized a universal forward env gene primer with subtype-specific reverse primers based on the diversity in the VRA region of the env gene. KoRV-A and KoRV-B specific primer sets were originally designed based on published isolates (Xu et al. 2013; Waugh et al. 2017) and have recently been updated to capture more of the known diversity within these subtypes (Fig. 7) (Quigley et al. 2019). New assays to specifically detect KoRV-D and KoRV-F have also been designed and published (Fig. 7) (Quigley et al. 2019). Another very useful KoRV assay uses an env gene primer set (env22.F and env514.R) that spans either side of the VRA and allows for deep amplicon sequencing (similar to microbiome analysis for bacteria) of all KoRV subtypes present within a sample (Fig. 7) (Chappell et al. 2017). Finally, primer sets have also been designed to cover the complete KoRV genome (P8 and P9), allowing for long-range PCR of complete genomes or detection and analysis of defective variants (Fig. 7) (Xu et al. 2013, 2015).
Treatment for KoRV – vaccines
Currently, there are no treatments available for KoRV in koalas. Antiretroviral drug therapies currently available for human retrovirus treatment require daily drug administration, a situation that is limiting for captive koala colonies and unfeasible in wild koala populations. As such, the field has turned to vaccine development as the best hope for a KoRV management strategy.
Before an animal is born, its immune system learns to recognize all the antigens that make up “self” and develops a tolerance to these to avoid auto-immunity. Given the endogenous nature of KoRV-A in northern koalas (meaning that KoRV-A antigens may be considered “self” as part of the genome), the first issue to be addressed was determining whether koalas produced, or could produce antibodies to KoRV. Initially, western blot analysis failed to detect KoRV antibodies from 16 captive and wild koalas (Fiebig et al.2015a). However, more detailed analysis using ELISA-based techniques from 235 wild koalas determined that the majority of koalas do naturally produce anti-KoRV antibodies, with anti-KoRV IgG levels peaking at approximately seven years of age and gradually declining thereafter (Olagoke et al. 2019). Additionally, some of these naturally occurring anti-KoRV antibodies mapped to known Gammaretrovirus neutralizing (and conserved) regions on the Env protein (FPPR, CSK-17 and MPER epitopes; Fig. 7), making it unsurprising that there was an inverse relationship between anti-KoRV IgG levels and circulating KoRV viral load (Olagoke et al. 2019). However, this raised the conundrum of how koalas produced antibodies to endogenous KoRV proteins that could be considered self-proteins. A possible explanation came with the discovery of KoRV antisense Piwi-interacting RNAs (piRNAs) in koalas. piRNAs guide the adaptive genome immune system to silence established transposons (and active ERVs) during germline development (Russell and LaMarre 2018; Ozata et al. 2019). Study within koalas has detected KoRV-A piRNAs apparently derived from isolated proviral insertions in the genome (Yu et al. 2019). This system, which is conserved from flies to mice, has the potential of silencing KoRV expression during fetal development, protecting developing joeys from KoRV and allowing KoRV proteins to be recognized as foreign in mature animals. These exciting findings set the stage for KoRV vaccine development.
The first KoRV vaccine trials tested whether recombinant KoRV (rKoRV) Env protein given to rats and goats could generate neutralizing antibodies (Denner 2013; Fiebig et al.2015b). These studies determined that binding antibodies could be generated to the FPPR and MPER regions of the Env protein, however, only the antibodies to the MPER region were neutralizing (Denner 2013; Fiebig et al. 2015b). This lead to a small safety trial where three northern koalas were given a rKoRV-A Env vaccine (Waugh et al.2016b). All three vaccinated koalas showed no signs of adverse reaction and the one koala that did not have detectable Env antibodies prior to vaccination was found to be generating Env antibodies post-vaccination (Waugh et al. 2016b). Subsequently, vaccination of six southern koalas also found no adverse vaccination effects, significant anti-KoRV antibody responses (including neutralizing antibody production) and decreases in KoRV circulating load post-vaccination (Olagoke et al. 2018). These promising pilot studies have established that vaccination for KoRV is safe in koalas and represents a promising avenue for KoRV management. The successes so far have paved the way for larger vaccine trials that are currently underway.
FUTURE DIRECTIONS
With so many advances made to C. pecorum and KoRV understanding in the past eight years, the future of disease management in koalas has never looked more promising. The major opportunities for continued advancement continue along the same lines as the most significant recent advances – genetic understanding and improved treatments. Genome sequencing technologies, both at the metagenomic level for C. pecorum sequencing directly from clinical samples and additional koala genome sequencing for advanced KoRV study, are continuing to gain greater bioinformatics support and reduced costs. Advances in koala genome sequencing will also greatly improve our understanding of koala genetics, the genetic differences between northern and southern populations (where bottleneck effects are believed to have hindered the genetic robustness of southern koala populations) and how these host genetics affect Chlamydia and KoRV in the different regions. With similar methodologies, more detailed transcriptomic studies of both pathogens, in unique disease situations and over time, will further illuminate key markers of disease and key targets for therapeutic interventions. Greater pathogen understanding will be needed to develop new and improved treatment options. Alternatives to antibiotics for C. pecorum, such as a novel HtrA serine protease inhibitor (Lawrence et al. 2016), are in early development. Finally, effective vaccines for both C. pecorum and KoRV are in active development, with enormous potential to shift the disease landscape in the koala population. Australians, as well as animal-lovers from around the world, care about koalas and want to see these unique animals thrive. Continued progress in understanding and controlling the major diseases koalas suffer from is an actionable, obtainable goal that will hopefully allow many future generations to enjoy these marsupials in the wild.
ACKNOWLEDGEMENTS
The authors would like to gratefully acknowledge Martina Jelocnik, Samuel Phillips, Amy Robbins, David Lizarraga and Sharon Nyari for critical reading and comments on this review. The authors also wish to thank the many groups that have supported our overall koala disease work, including the Queensland Government (Department of Transport and Main Roads), the Moreton Bay Rail project team, the Queensland Department of Environment and Heritage Protection, Moreton Bay Regional Council, Gold Coast City Council, Gympie Regional Council, NSW Koala Strategy, Friends of the Koala - Lismore, Koala Action Inc, Royal Society of Queensland, Endeavour Veterinary Ecology, Australia Zoo Wildlife Hospital, Lone Pine Koala Sanctuary, Currumbin Wildlife Sanctuary, Wildlife HQ Zoo, VIDO – Canada and the NSW Office of Environment and Heritage. The authors declare no conflicts of interest.
Contributor Information
Bonnie L Quigley, Genecology Research Centre, University of the Sunshine Coast, 90 Sippy Downs Drive, Sippy Downs, Queensland, 4556, Australia.
Peter Timms, Genecology Research Centre, University of the Sunshine Coast, 90 Sippy Downs Drive, Sippy Downs, Queensland, 4556, Australia.
FUNDING
This work was supported by the Wildlife Disease Association small grants program (to BLQ) and Australian Research Council Linkage scheme (to PT).
Conflict of interest
None declared.
REFERENCES
- Abts KC, Ivy JA, DeWoody JA. Immunomics of the koala (Phascolarctos cinereus). Immunogenetics. 2015;67:305–21. [DOI] [PubMed] [Google Scholar]
- Adams-Hosking C, McBride MF, Baxter Get al. Use of expert knowledge to elicit population trends for the koala (Phascolarctos cinereus). Diversity Distrib. 2016;22:249–62. [Google Scholar]
- Alfano N, Michaux J, Morand Set al. Endogenous Gibbon Ape Leukemia Virus identified in a rodent (Melomys burtoni subsp.) from Wallacea (Indonesia). J Virol. 2016;90:8169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Australia CO. ‘The koala - saving our national icon’. Report of the Senate Standing Committees on Environment and Communications on the inquiry into the status, health and sustainability of Australia's koala population. https://www.aph.gov.au/Parliamentary_Business/Committees/Senate/Environment_and_Communications/Completed_inquiries/2010-13/koalas/report/index, 2011. [Google Scholar]
- Avila-Arcos MC, Ho SY, Ishida Yet al. One hundred twenty years of koala retrovirus evolution determined from museum skins. Mol Biol Evol. 2013;30:299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann NL, Fraser TA, Bertelli Cet al. Comparative genomics of koala, cattle and sheep strains of Chlamydia pecorum. BMC Genomics. 2014;15:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann NL, Sullivan MJ, Jelocnik Met al. Culture-independent genome sequencing of clinical samples reveals an unexpected heterogeneity of infections by Chlamydia pecorum. J Clin Microbiol. 2015;53:1573–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer HL, de Villiers D, Loader Jet al. Management of multiple threats achieves meaningful koala conservation outcomes. J Appl Ecol. 2018;55:1966–75. [Google Scholar]
- Black LA, Higgins DP, Govendir M. In vitro activity of chloramphenicol, florfenicol and enrofloxacin against Chlamydia pecorum isolated from koalas (Phascolarctos cinereus). Aust Vet J. 2015;93:420–3. [DOI] [PubMed] [Google Scholar]
- Black LA, McLachlan AJ, Griffith JEet al. Pharmacokinetics of chloramphenicol following administration of intravenous and subcutaneous chloramphenicol sodium succinate, and subcutaneous chloramphenicol, to koalas (Phascolarctos cinereus). J Vet Pharmacol Ther. 2013;36:478–85. [DOI] [PubMed] [Google Scholar]
- Blanshard W, Bodley K. Koalas. In: Vogelnest L, Portas T. (eds.). Current Therapy in Medicine of Australian Mammals. Clayton, Victoria, Australia: CSIRO Publishing, 2008, 227–328. [Google Scholar]
- Blinov VM, Krasnov GS, Shargunov AVet al. Immunosuppressive domains of retroviruses: cell mechanisms of the effect on the human immune system. Mol Biol. 2013;47:613–21. [Google Scholar]
- Brown K, Tarlinton RE. Is gibbon ape leukaemia virus still a threat? Mammal Review. 2017;47:53–61. [Google Scholar]
- Budd C, Flanagan C, Gillett Aet al. Assessment of florfenicol as a possible treatment for chlamydiosis in koalas (Phascolarctos cinereus). Aust Vet J. 2017;95:343–9. [DOI] [PubMed] [Google Scholar]
- Burach F, Pospischil A, Hanger Jet al. Chlamydiaceae and Chlamydia-like organisms in the koala (Phascolarctos cinereus)–organ distribution and histopathological findings. Vet Microbiol. 2014;172:230–40. [DOI] [PubMed] [Google Scholar]
- Burnard D, Gillett A, Polkinghorne A. Chlamydia pecorum in joint tissue and synovial fluid of a koala (Phascolarctos cinereus) with arthritis. J Wildl Dis. 2018;54:646–9. [DOI] [PubMed] [Google Scholar]
- Butcher RG, Pettett LM, Fabijan Jet al. Periodontal disease in free-ranging koalas (Phascolarctos cinereus) from the Mount Lofty Ranges, South Australia, and its association with koala retrovirus infection. Aust Vet J. 2020;98:200–6. [DOI] [PubMed] [Google Scholar]
- Canfield PJ, Sabine JM, Love DN. Virus particles associated with leukaemia in a koala. Aust Vet J. 1988;65:327–8. [DOI] [PubMed] [Google Scholar]
- Chappell KJ, Brealey JC, Amarilla AAet al. Phylogenetic diversity of koala retrovirus within a wild koala population. J Virol. 2017;91:e01820–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AP, Hanger J, Loader Jet al. A 5-year Chlamydia vaccination programme could reverse disease-related koala population decline: predictions from a mathematical model using field data. Vaccine. 2014;32:4163–70. [DOI] [PubMed] [Google Scholar]
- Cristescu RH, Miller RL, Schultz AJet al. Developing noninvasive methodologies to assess koala population health through detecting Chlamydia from scats. Mol Ecol Resour. 2019;19:957–69. [DOI] [PubMed] [Google Scholar]
- Cui J, Tachedjian G, Tachedjian Met al. Identification of diverse groups of endogenous gammaretroviruses in mega- and microbats. J Gen Virol. 2012;93:2037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P, Lober U, Alquezar-Planas DEet al. Comprehensive profiling of retroviral integration sites using target enrichment methods from historical koala samples without an assembled reference genome. PeerJ. 2016;4:e1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K, Stansfield SH, Patel Pet al. The IL-6 response to Chlamydia from primary reproductive epithelial cells is highly variable and may be involved in differential susceptibility to the immunopathological consequences of chlamydial infection. BMC Immunol. 2013;14:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denner J, Young PR. Koala retroviruses: characterization and impact on the life of koalas. Retrovirology. 2013;10:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denner J. Immunising with the transmembrane envelope proteins of different retroviruses including HIV-1. Human Vaccines, Immunotherapeutics. 2013;9:462–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denner J. Transspecies transmission of gammaretroviruses and the origin of the Gibbon Ape Leukaemia Virus (GaLV) and the Koala Retrovirus (KoRV). Viruses. 2016;8:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desclozeaux M, Robbins A, Jelocnik Met al. Immunization of a wild koala population with a recombinant Chlamydia pecorum Major Outer Membrane Protein (MOMP) or Polymorphic Membrane Protein (PMP) based vaccine: new insights into immune response, protection and clearance. PLoS One. 2017;12:e0178786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereaux LN, Polkinghorne A, Meijer Aet al. Molecular evidence for novel chlamydial infections in the koala (Phascolarctos cinereus). Syst Appl Microbiol. 2003;26:245–53. [DOI] [PubMed] [Google Scholar]
- Fabijan J, Caraguel C, Jelocnik Met al. Chlamydia pecorum prevalence in South Australian koala (Phascolarctos cinereus) populations: Identification and modelling of a population free from infection. Sci Rep. 2019b;9:6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabijan J, Miller D, Olagoke Oet al. Prevalence and clinical significance of koala retrovirus in two South Australian koala (Phascolarctos cinereus) populations. J Med Microbiol. 2019a;68:1072–80. [DOI] [PubMed] [Google Scholar]
- Fernandez CM, Schmertmann LJ, Higgins DPet al. Genetic differences in Chlamydia pecorum between neighbouring sub-populations of koalas (Phascolarctos cinereus). Vet Microbiol. 2019;231:264–70. [DOI] [PubMed] [Google Scholar]
- Fiebig U, Dieckhoff B, Wurzbacher Cet al. Induction of neutralizing antibodies specific for the envelope proteins of the koala retrovirus by immunization with recombinant proteins or with DNA. Virol J. 2015b;12:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig U, Keller M, Denner J. Detection of koala retrovirus subgroup B (KoRV-B) in animals housed at European zoos. Arch Virol. 2016;161:3549–53. [DOI] [PubMed] [Google Scholar]
- Fiebig U, Keller M, Moller Aet al. Lack of antiviral antibody response in koalas infected with koala retroviruses (KoRV). Virus Res. 2015a;198:30–4. [DOI] [PubMed] [Google Scholar]
- Fiebig U, Stephan O, Kurth Ret al. Neutralizing antibodies against conserved domains of p15E of porcine endogenous retroviruses: basis for a vaccine for xenotransplantation? Virology. 2003;307:406–13. [DOI] [PubMed] [Google Scholar]
- Funnell O, Johnson L, Woolford Let al. Conjunctivitis associated with Chlamydia pecorum in three koalas (Phascolarctos cinereus) in the Mount Lofty Ranges, South Australia. J Wildl Dis. 2013;49:1066–9. [DOI] [PubMed] [Google Scholar]
- Gillett A, Hanger J. Koala. In: Vogelnest L, Portas T. (eds.). Current Therapy in Medicine of Australian Mammals. Clayton, Victoria, Australia: CSIRO Publishing, 2019, 463–86. [Google Scholar]
- Gonzalez-Astudillo V, Allavena R, McKinnon Aet al. Decline causes of koalas in South East Queensland, Australia: a 17-year retrospective study of mortality and morbidity. Sci Rep. 2017;7:42587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Astudillo V, Henning J, Valenza Let al. A necropsy study of disease and comorbidity trends in morbidity and mortality in the koala (Phascolarctos cinereus) in South-East Queensland, Australia. Sci Rep. 2019;9:17494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govendir M, Hanger J, Loader JJet al. Plasma concentrations of chloramphenicol after subcutaneous administration to koalas (Phascolarctos cinereus) with chlamydiosis. J Vet Pharmacol Ther. 2012;35:147–54. [DOI] [PubMed] [Google Scholar]
- Greenwood AD, Ishida Y, O'Brien SPet al. Transmission, evolution, and endogenization: Lessons learned from recent retroviral invasions. Microbiol Mol Biol Rev. 2018;82:e00044–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. Studies Into the Diagnosis, Treatment and Management of Chlamydiosis in Koalas. Thesis, Sydney, NSW: The University of Sydney, 2010. [Google Scholar]
- Griffith JE, Dhand NK, Krockenberger MBet al. A retrospective study of admission trends of koalas to a rehabilitation facility over 30 years. J Wildl Dis. 2013;49:18–28. [DOI] [PubMed] [Google Scholar]
- Griffith JE, Higgins DP. Diagnosis, treatment and outcomes for koala chlamydiosis at a rehabilitation facility (1995-2005). Aust Vet J. 2012;90:457–63. [DOI] [PubMed] [Google Scholar]
- Grogan LF, Ellis W, Jones Det al. Current trends and future directions in koala chlamydial disease research. Biol Conserv. 2017;215:179–88. [Google Scholar]
- Hanger J, Loader J, Wan Cet al. Comparison of antigen detection and quantitative PCR in the detection of chlamydial infection in koalas (Phascolarctos cinereus). Vet J. 2013;195:391–3. [DOI] [PubMed] [Google Scholar]
- Hanger JJ, Bromham LD, McKee JJet al. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to Gibbon ape leukemia virus. J Virol. 2000;74:4264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem MA, Kayesh MEH, Yamato Oet al. Coinfection with koala retrovirus subtypes A and B and its impact on captive koalas in Japanese zoos. Arch Virol. 2019;164:2735–45. [DOI] [PubMed] [Google Scholar]
- Hayward JA, Tachedjian M, Kohl Cet al. Infectious KoRV-related retroviruses circulating in Australian bats. Proc Natl Acad Sci USA. 2020;117:9529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming V, Hoffmann M, Jarrad Fet al. NSW Koala research plan: Expert elicitation of knowledge gaps. In: Research CoEaE (ed). Office of Environment and Heritage. Melbourne, Victoria, Austriala: The University of Melbourne, 2018, 122. [Google Scholar]
- Hernandez-Sanchez J, Brumm J, Timms Pet al. Vaccination of koalas with a prototype chlamydial vaccine is safe, does not increase the incidence of lymphoma-related disease and maybe associated with increased lifespan in captive koalas. Vaccine. 2015;33:4459–63. [DOI] [PubMed] [Google Scholar]
- Higgins D, Maher I. Koala retrovirus. In: Vogelnest L, Portas T. (eds.). Current Therapy in Medicine of Australian Mammals. Clayton, Victoria, Australia: CSIRO Publishing, 2019, 487–93. [Google Scholar]
- Higgins DP, Beninati T, Meek Met al. Within-population diversity of koala Chlamydophila pecorum at ompA VD1-VD3 and the ORF663 hypothetical gene. Vet Microbiol. 2012;156:353–8. [DOI] [PubMed] [Google Scholar]
- Hobbs M, King A, Salinas Ret al. Long-read genome sequence assembly provides insight into ongoing retroviral invasion of the koala germline. Sci Rep. 2017;7:15838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs M, Pavasovic A, King AGet al. A transcriptome resource for the koala (Phascolarctos cinereus): insights into koala retrovirus transcription and sequence diversity. BMC Genomics. 2014;15:786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse LS, Beagley K, Ellis Wet al. Epidemiology of Chlamydia-induced reproductive disease in male koalas (Phascolarctos cinereus) from southeast Queensland, Australia as assessed from penile urethral swabs and semen. J Wildl Dis. 2019b;56:82–92. [PubMed] [Google Scholar]
- Hulse LS, Hickey D, Mitchell JMet al. Development and application of two multiplex real-time PCR assays for detection and speciation of bacterial pathogens in the koala. J Vet Diagn Invest. 2018;30:523–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse LS, McDonald S, Johnston SDet al. Rapid point-of-care diagnostics for the detection of Chlamydia pecorum in koalas (Phascolarctos cinereus) using loop-mediated isothermal amplification without nucleic acid purification. MicrobiologyOpen. 2019a;8:e916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, McCallister C, Nikolaidis Net al. Sequence variation of koala retrovirus transmembrane protein p15E among koalas from different geographic regions. Virology. 2015b;475:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Zhao K, Greenwood ADet al. Proliferation of endogenous retroviruses in the early stages of a host germ line invasion. Mol Biol Evol. 2015a;32:109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MM, Jelocnik M, Huston WMet al. Characterization of the in vitro Chlamydia pecorum response to gamma interferon. Infect Immun. 2018;86:e00714–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N, Yoshida T, Ichikawa Ret al. Clinical and pathological characteristics of acute myelogenous leukemia in a female koala with diabetes mellitus. J Vet Med Sci. 2019;81:1229–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelocnik M, Bachmann NL, Kaltenboeck Bet al. Genetic diversity in the plasticity zone and the presence of the chlamydial plasmid differentiates Chlamydia pecorum strains from pigs, sheep, cattle, and koalas. BMC Genomics. 2015;16:893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelocnik M, Bachmann NL, Seth-Smith Het al. Molecular characterisation of the Chlamydia pecorum plasmid from porcine, ovine, bovine, and koala strains indicates plasmid-strain co-evolution. PeerJ. 2016;4:e1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelocnik M, Frentiu FD, Timms Pet al. Multilocus sequence analysis provides insights into molecular epidemiology of Chlamydia pecorum infections in Australian sheep, cattle, and koalas. J Clin Microbiol. 2013;51:2625–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelocnik M, Gillett A, Hanger Jet al. Chlamydiosis in koalas. In: Vogelnest L, Portas T. (eds.). Current Therapy in Medicine of Australian Mammals. Clayton, Victoria, Australia: CSIRO Publishing, 2019, 496–506. [Google Scholar]
- Jelocnik M, Islam MM, Madden Det al. Development and evaluation of rapid novel isothermal amplification assays for important veterinary pathogens: Chlamydia psittaci and Chlamydia pecorum. PeerJ. 2017;5:e3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelocnik M, Walker E, Pannekoek Yet al. Evaluation of the relationship between Chlamydia pecorum sequence types and disease using a species-specific multi-locus sequence typing scheme (MLST). Vet Microbiol. 2014;174:214–22. [DOI] [PubMed] [Google Scholar]
- Johnson RN, O'Meally D, Chen Zet al. Adaptation and conservation insights from the koala genome. Nat Genet. 2018;50:1102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SD, Deif HH, McKinnon Aet al. Orchitis and epididymitis in koalas (Phascolarctos cinereus) infected with Chlamydia pecorum. Vet Pathol. 2015;52:1254–7. [DOI] [PubMed] [Google Scholar]
- Kayesh MEH, Yamato O, Rahman MMet al. Molecular dynamics of koala retrovirus infection in captive koalas in Japan. Arch Virol. 2019;164:757–65. [DOI] [PubMed] [Google Scholar]
- Khan SA, Desclozeaux M, Waugh Cet al. Antibody and cytokine responses of koalas (Phascolarctos cinereus) vaccinated with recombinant chlamydial Major Outer Membrane Protein (MOMP) with two different adjuvants. PLoS One. 2016b;11:e0156094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SA, Polkinghorne A, Waugh Cet al. Humoral immune responses in koalas (Phascolarctos cinereus) either naturally infected with Chlamydia pecorum or following administration of a recombinant chlamydial major outer membrane protein vaccine. Vaccine. 2016a;34:775–82. [DOI] [PubMed] [Google Scholar]
- Khan SA, Waugh C, Rawlinson Get al. Vaccination of koalas (Phascolarctos cinereus) with a recombinant chlamydial major outer membrane protein adjuvanted with poly I:C, a host defense peptide and polyphosphazine, elicits strong and long lasting cellular and humoral immune responses. Vaccine. 2014;32:5781–6. [DOI] [PubMed] [Google Scholar]
- Kinney ME, Pye GW. Koala Retrovirus: A Review. J Zoo Wildl Med. 2016;47:387–96. [DOI] [PubMed] [Google Scholar]
- Kollipara A, George C, Hanger Jet al. Vaccination of healthy and diseased koalas (Phascolarctos cinereus) with a Chlamydia pecorum multi-subunit vaccine: evaluation of immunity and pathology. Vaccine. 2012;30:1875–85. [DOI] [PubMed] [Google Scholar]
- Kollipara A, Polkinghorne A, Beagley KWet al. Vaccination of koalas with a recombinant Chlamydia pecorum major outer membrane protein induces antibodies of different specificity compared to those following a natural live infection. PLoS One. 2013a;8:e74808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollipara A, Polkinghorne A, Wan Cet al. Genetic diversity of Chlamydia pecorum strains in wild koala locations across Australia and the implications for a recombinant C. pecorum major outer membrane protein based vaccine. Vet Microbiol. 2013c;167:513–22. [DOI] [PubMed] [Google Scholar]
- Kollipara A, Wan C, Rawlinson Get al. Antigenic specificity of a monovalent versus polyvalent MOMP based Chlamydia pecorum vaccine in koalas (Phascolarctos cinereus). Vaccine. 2013b;31:1217–23. [DOI] [PubMed] [Google Scholar]
- Lawrence A, Fraser T, Gillett Aet al. Chlamydia serine protease inhibitor, targeting HtrA, as a new treatment for koala Chlamydia infection. Sci Rep. 2016;6:31466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legione AR, Amery-Gale J, Lynch Met al. Chlamydia pecorum infection in free-ranging koalas (Phascolarctos cinereus) on French Island, Victoria, Australia. J Wildl Dis. 2016a;52:426–9. [DOI] [PubMed] [Google Scholar]
- Legione AR, Amery-Gale J, Lynch Met al. Variation in the microbiome of the urogenital tract of Chlamydia-free female koalas (Phascolarctos cinereus) with and without ‘wet bottom’. PLoS One. 2018;13:e0194881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legione AR, Patterson JL, Whiteley Pet al. Koala retrovirus genotyping analyses reveal a low prevalence of KoRV-A in Victorian koalas and an association with clinical disease. J Med Microbiol. 2017;66:236–44. [DOI] [PubMed] [Google Scholar]
- Legione AR, Patterson JL, Whiteley PLet al. Identification of unusual Chlamydia pecorum genotypes in Victorian koalas (Phascolarctos cinereus) and clinical variables associated with infection. J Med Microbiol. 2016b;65:420–8. [DOI] [PubMed] [Google Scholar]
- Leonard CA, Dewez F, Borel N. Penicillin G-induced chlamydial stress response in a porcine strain of Chlamydia pecorum. Int J Microbiol. 2016;2016:3832917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizárraga D, Carver S, Timms P. Navigating to the most promising directions amid complex fields of vaccine development: a chlamydial case study. Expert Rev Vaccines. 2019;18:1323–37. [DOI] [PubMed] [Google Scholar]
- Lober U, Hobbs M, Dayaram Aet al. Degradation and remobilization of endogenous retroviruses by recombination during the earliest stages of a germ-line invasion. Proc Natl Acad Sci USA. 2018;115:8609–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie JT, Gillett AK, Palmieri Cet al. Pneumonia due to Chlamydia pecorum in a koala (Phascolarctos cinereus). J Comp Pathol. 2016;155:356–60. [DOI] [PubMed] [Google Scholar]
- Maclachlan NJ, Dubovi EJ. Fenner's Veterinary Virology, Chapter 14 Retroviridae. Saint Louis, USA: Elsevier Science & Technology, 2010. [Google Scholar]
- Maher IE, Griffith JE, Lau Qet al. Expression profiles of the immune genes CD4, CD8beta, IFNgamma, IL-4, IL-6 and IL-10 in mitogen-stimulated koala lymphocytes (Phascolarctos cinereus) by qRT-PCR. PeerJ. 2014;2:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher IE, Higgins DP. Altered immune cytokine expression associated with KoRV B infection and season in captive koalas. PLoS One. 2016;11:e0163780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher IE, Patterson J, Curnick Met al. Altered immune parameters associated with Koala Retrovirus (KoRV) and Chlamydial infection in free ranging Victorian koalas (Phascolarctos cinereus). Sci Rep. 2019;9:11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner C, Flanagan C, Higgins DPet al. Validation of ultrasonography in detecting structural disease of the urogenital tract of the koala, Phascolarctos cinereus. Aust Vet J. 2014;92:177–8. [DOI] [PubMed] [Google Scholar]
- Marsh J, Kollipara A, Timms Pet al. Novel molecular markers of Chlamydia pecorum genetic diversity in the koala (Phascolarctos cinereus). BMC Microbiol. 2011;11:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew M, Beagley KW, Timms Pet al. Preliminary characterisation of tumor necrosis factor alpha and interleukin-10 responses to Chlamydia pecorum infection in the koala (Phascolarctos cinereus). PLoS One. 2013a;8:e59958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew M, Pavasovic A, Prentis PJet al. Molecular characterisation and expression analysis of interferon gamma in response to natural Chlamydia infection in the koala, Phascolarctos cinereus. Gene. 2013b;527:570–7. [DOI] [PubMed] [Google Scholar]
- Mathew M, Waugh C, Beagley KWet al. Interleukin 17A is an immune marker for chlamydial disease severity and pathogenesis in the koala (Phascolarctos cinereus). Dev Comp Immunol. 2014;46:423–9. [DOI] [PubMed] [Google Scholar]
- McAlpine C, Lunney D, Melzer Aet al. Conserving koalas: A review of the contrasting regional trends, outlooks and policy challenges. Biol Conserv. 2015;192:226–36. [Google Scholar]
- McCallum H, Kerlin DH, Ellis Wet al. Assessing the significance of endemic disease in conservation—koalas, chlamydia, and koala retrovirus as a case study. Conserv Lett. 2018;11:e12425. [Google Scholar]
- McMichael L, Smith C, Gordon Aet al. A novel Australian flying-fox retrovirus shares an evolutionary ancestor with Koala, Gibbon and Melomys gamma-retroviruses. Virus Genes. 2019;55:421–4. [DOI] [PubMed] [Google Scholar]
- Melzer A, Carrick F, Menkhorst Pet al. Overview, critical assessment, and conservation implications of koala distribution and abundance. Conserv Biol. 2000;14:619–28. [Google Scholar]
- Mohamad KY, Kaltenboeck B, Rahman Kh Set al. Host adaptation of Chlamydia pecorum towards low virulence evident in co-evolution of the ompA, incA, and ORF663 Loci. PLoS One. 2014;9:e103615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica S, Huot Creasy H, Daugherty Set al. Gemone sequence of the obligate intracellular animal pathogen Chlamydia pecorum E58. J Bacteriol. 2011;193:3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K, Prentis PJ, O'Meally Det al. The koala immunological toolkit: sequence identification and comparison of key markers of the koala (Phascolarctos cinereus) immune response. Australian J of Zoology. 2014;62:195–9. [Google Scholar]
- Morris KM, O'Meally D, Zaw Tet al. Characterisation of the immune compounds in koala milk using a combined transcriptomic and proteomic approach. Sci Rep. 2016;6:35011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta T, Ha D, Galvez Fet al. Human and murine APOBEC3s restrict replication of koala retrovirus by different mechanisms. Retrovirology. 2015;12:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyari S, Booth R, Quigley BLet al. Therapeutic effect of a Chlamydia pecorum recombinant major outer membrane protein vaccine on ocular disease in koalas (Phascolarctos cinereus). PLoS One. 2019;14:e0210245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyari S, Khan SA, Rawlinson Get al. Vaccination of koalas (Phascolarctos cinereus) against Chlamydia pecorum using synthetic peptides derived from the major outer membrane protein. PLoS One. 2018;13:e0200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyari S, Waugh CA, Dong Jet al. Epidemiology of chlamydial infection and disease in a free-ranging koala (Phascolarctos cinereus) population. PLoS One. 2017;12:e0190114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olagoke O, Miller D, Hemmatzadeh Fet al. Induction of neutralizing antibody response against koala retrovirus (KoRV) and reduction in viral load in koalas following vaccination with recombinant KoRV envelope protein. NPJ Vaccines. 2018;3:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olagoke O, Quigley BL, Eiden MVet al. Antibody response against koala retrovirus (KoRV) in koalas harboring KoRV-A in the presence or absence of KoRV-B. Sci Rep. 2019;9:12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira NM, Farrell KB, Eiden MV. In vitro characterization of a koala retrovirus. J Virol. 2006;80:3104–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch Aet al. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. 2019;20:89–108. [DOI] [PubMed] [Google Scholar]
- Palmieri C, Hulse L, Pagliarani Set al. Chlamydia pecorum infection in the male reproductive system of koalas (Phascolarctos cinereus). Vet Pathol. 2019;56:300–6. [DOI] [PubMed] [Google Scholar]
- Pantchev A, Sting R, Bauerfeind Ret al. Detection of all Chlamydophila and Chlamydia spp. of veterinary interest using species-specific real-time PCR assays. Comp Immunol Microbiol Infect Dis. 2010;33:473–84. [DOI] [PubMed] [Google Scholar]
- Patterson JL, Lynch M, Anderson GAet al. The prevalence and clinical significance of Chlamydia infection in island and mainland populations of Victorian koalas (Phascolarctos cinereus). J Wildl Dis. 2015;51:309–17. [DOI] [PubMed] [Google Scholar]
- Phillips B, Service ANPaW. Koalas: the little Australians we'd all hate to lose. Australian Government Publishing Service Press: Canberra, 1990. [Google Scholar]
- Phillips S, Quigley BL, Aziz Aet al. Antibiotic treatment of Chlamydia-induced cystitis in the koala is linked to expression of key inflammatory genes in reactive oxygen pathways. PLoS One. 2019;14:e0221109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S, Quigley BL, Timms P. Seventy years of Chlamydia vaccine research - Limitations of the past and directions for the future. Front Microbiol. 2019;10:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S, Robbins A, Loader Jet al. Chlamydia pecorum gastrointestinal tract infection associations with urogenital tract infections in the koala (Phascolarctos cinereus). PLoS One. 2018;13:e0206471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips SS. Population trends and the koala conservation debate. Conserv Biol. 2000;14:650–9. [Google Scholar]
- Polkinghorne A, Hanger J, Timms P. Recent advances in understanding the biology, epidemiology and control of chlamydial infections in koalas. Vet Microbiol. 2013;165:214–23. [DOI] [PubMed] [Google Scholar]
- Quigley BL, Carver S, Hanger Jet al. The relative contribution of causal factors in the transition from infection to clinical chlamydial disease. Sci Rep. 2018b;8:8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley BL, Ong VA, Hanger Jet al. Molecular dynamics and mode of transmission of Koala Retrovirus (KoRV) as it invades and spreads through a wild Queensland koala population. J Virol. 2018a;92:e01871–01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley BL, Phillips S, Olagoke Oet al. Changes in endogenous and exogenous Koala Retrovirus subtype expression over time reflect koala health outcomes. J Virol. 2019;93:e00849–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins A, Hanger J, Jelocnik Met al. Longitudinal study of wild koalas (Phascolarctos cinereus) reveals chlamydial disease progression in two thirds of infected animals. Sci Rep. 2019;9:13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins A, Loader J, Timms Pet al. Optimising the short and long-term clinical outcomes for koalas (Phascolarctos cinereus) during treatment for chlamydial infection and disease. PLoS One. 2018;13:e0209679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B, Eby P, Tsang SMet al. Pteropus alecto. The IUCN Red List of Threatened Species e.T18715A22080057. https://doi.org/22080010.22082305/IUCN.UK.22082017-22080052.RLTS.T22018715A22080057.en. Downloaded on 22080024 February 22082020, 2017.
- Robertson T, Bibby S, O'Rourke Det al. Characterization of Chlamydiaceae species using PCR and high resolution melt curve analysis of the 16S rRNA gene. J Appl Microbiol. 2009;107:2017–28. [DOI] [PubMed] [Google Scholar]
- Russell I, Timms P, Hanger Jet al. Prevalence of Chlamydia pecorum in juvenile koalas (Phascolarctos cinereus) and evidence for protection from infection via maternal immunization. J Wildl Dis. 2018;54:863–5. [DOI] [PubMed] [Google Scholar]
- Russell SJ, LaMarre J. Transposons and the PIWI pathway: genome defense in gametes and embryos. Reproduction. 2018;156:R111–24. [DOI] [PubMed] [Google Scholar]
- Santamaria F, Schlagloth R. The effect of Chlamydia on translocated Chlamydia-naïve koalas: a case study. Australian Zoologist. 2016;38:192–202. [Google Scholar]
- Sarker N, Fabijan J, Seddon Jet al. Genetic diversity of Koala retrovirus env gene subtypes: insights into northern and southern koala populations. J Gen Virol. 2019;100:1328–39. [DOI] [PubMed] [Google Scholar]
- Sarker N, Tarlinton R, Owen Het al. Novel insights into viral infection and oncogenesis from Koala Retrovirus (KoRV) infection of HEK293T cells. Gene. 2020;733:144366. [DOI] [PubMed] [Google Scholar]
- Shimode S, Nakagawa S, Yoshikawa Ret al. Heterogeneity of koala retrovirus isolates. FEBS Lett. 2014;588:41–6. [DOI] [PubMed] [Google Scholar]
- Shimode S, Nakaoka R, Hoshino Set al. Identification of cellular factors required for the budding of koala retrovirus. Microbiol Immunol. 2013;57:543–6. [DOI] [PubMed] [Google Scholar]
- Shojima T, Hoshino S, Abe Met al. Construction and characterization of an infectious molecular clone of Koala retrovirus. J Virol. 2013a;87:5081–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojima T, Yoshikawa R, Hoshino Set al. Identification of a novel subgroup of Koala retrovirus from Koalas in Japanese zoos. J Virol. 2013b;87:9943–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G, Clarke D, McKee Jet al. Discovery of a novel retrovirus sequence in an Australian native rodent (Melomys burtoni): a putative link between gibbon ape leukemia virus and koala retrovirus. PLoS One. 2014;9:e106954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons GS, Young PR, Hanger JJet al. Prevalence of koala retrovirus in geographically diverse populations in Australia. Aust Vet J. 2012;90:404–9. [DOI] [PubMed] [Google Scholar]
- Speight KN, Hicks P, Graham Cet al. Necropsy findings of koalas from the Mount Lofty Ranges population in South Australia. Aust Vet J. 2018;96:188–92. [DOI] [PubMed] [Google Scholar]
- Speight KN, Polkinghorne A, Penn Ret al. Prevalence and pathologic features of Chlamydia pecorum infections in South Australian koalas (Phascolarctos cinereus). J Wildl Dis. 2016;52:301–6. [DOI] [PubMed] [Google Scholar]
- Tarlinton R, Meers J, Hanger Jet al. Real-time reverse transcriptase PCR for the endogenous koala retrovirus reveals an association between plasma viral load and neoplastic disease in koalas. J Gen Virol. 2005;86:783–7. [DOI] [PubMed] [Google Scholar]
- Tarlinton R. Koala Retrovirus endogenisation in action. In: Witzany G (ed). Viruses: Essential Agents of Life. Dordrecht, The Netherlands: Springer, 2012. [Google Scholar]
- Tarlinton RE, Meers J, Young PR. Retroviral invasion of the koala genome. Nature. 2006;442:79–81. [DOI] [PubMed] [Google Scholar]
- Tsangaras K, Siracusa MC, Nikolaidis Net al. Hybridization capture reveals evolution and conservation across the entire Koala retrovirus genome. PLoS One. 2014;9:e95633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C, Loader J, Hanger Jet al. Using quantitative polymerase chain reaction to correlate Chlamydia pecorum infectious load with ocular, urinary and reproductive tract disease in the koala (Phascolarctos cinereus). Aust Vet J. 2011;89:409–12. [DOI] [PubMed] [Google Scholar]
- Waugh C, Austin R, Polkinghorne Aet al. Treatment of Chlamydia-associated ocular disease via a recombinant protein based vaccine in the koala (Phascolarctos cinereus). Biologicals. 2016c;44:588–90. [DOI] [PubMed] [Google Scholar]
- Waugh C, Gillett A, Polkinghorne Aet al. Serum antibody response to koala retrovirus antigens varies in free-ranging koalas (Phascolarctos cinereus) in Australia: implications for vaccine design. J Wildl Dis. 2016b;52:422–5. [DOI] [PubMed] [Google Scholar]
- Waugh C, Hanger J, Timms Pet al. Koala translocations and Chlamydia: Managing risk in the effort to conserve native species. Biol Conserv. 2016a;197:247–53. [Google Scholar]
- Waugh C, Khan SA, Carver Set al. A prototype recombinant-protein based Chlamydia pecorum vaccine results in reduced chlamydial burden and less clinical disease in free-ranging koalas (Phascolarctos cinereus). PLoS One. 2016d;11:e0146934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh CA, Hanger J, Loader Jet al. Infection with koala retrovirus subgroup B (KoRV-B), but not KoRV-A, is associated with chlamydial disease in free-ranging koalas (Phascolarctos cinereus). Sci Rep. 2017;7:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh CA, Timms P, Andrew Det al. Comparison of subcutaneous versus intranasal immunization of male koalas (Phascolarctos cinereus) for induction of mucosal and systemic immunity against Chlamydia pecorum. Vaccine. 2015;33:855–60. [DOI] [PubMed] [Google Scholar]
- Wedrowicz F, Mosse J, Wright Wet al. Using non-invasive sampling methods to determine the prevalence and distribution of Chlamydia pecorum and koala retrovirus in a remnant koala population with conservation importance. Wildlife Research. 2018;45:366–80. [Google Scholar]
- Wedrowicz F, Saxton T, Mosse Jet al. A non-invasive tool for assessing pathogen prevalence in koala (Phascolarctos cinereus) populations: detection of Chlamydia pecorum and koala retrovirus (KoRV) DNA in genetic material sourced from scats. Conservation Genetics Resources. 2016;8:511–21. [Google Scholar]
- Woinarski J, Burbidge AA. Phascolarctos cinereus. The IUCN Red List of Threatened Species e.T16892A21960344. https://doi.org/21960310.21962305/IUCN.UK.21962016-21960341.RLTS.T21916892A21960344.en. Downloaded on 21960311 February 21962020, 2016.
- Xu W, Eiden MV. Koala retroviruses: evolution and disease dynamics. Annu Rev Virol. 2015;2:119–34. [DOI] [PubMed] [Google Scholar]
- Xu W, Gorman K, Santiago JCet al. Genetic diversity of koala retroviral envelopes. Viruses. 2015;7:1258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Stadler CK, Gorman Ket al. An exogenous retrovirus isolated from koalas with malignant neoplasias in a US zoo. Proc Natl Acad Sci U S A. 2013;110:11547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Koppetsch BS, Pagliarani Set al. The piRNA response to retroviral invasion of the koala genome. Cell. 2019;179:632–43.e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziklo N, Huston WM, Hocking JSet al. Chlamydia trachomatis genital tract infections: when host immune response and the microbiome collide. Trends Microbiol. 2016a;24:750–65. [DOI] [PubMed] [Google Scholar]
- Ziklo N, Huston WM, Taing Ket al. In vitro rescue of genital strains of Chlamydia trachomatis from interferon-gamma and tryptophan depletion with indole-positive, but not indole-negative Prevotella spp. BMC Microbiol. 2016b;16:286. [DOI] [PMC free article] [PubMed] [Google Scholar]